Abstract
Electronic health records (EHRs) have given rise to large and complex databases of medical information that have the potential to become powerful tools for clinical research. However, differences in coding systems and the detail and accuracy of the information within EHRs can vary across institutions. This makes it challenging to identify subpopulations of patients and limits the widespread use of multi-institutional databases. In this study, we leveraged machine learning to identify patterns in medication usage among hospitalized pediatric patients receiving renal replacement therapy and created a predictive model that successfully differentiated between intermittent (iHD) and continuous renal replacement therapy (CRRT) hemodialysis patients. We trained six machine learning algorithms (logistical regression, Naïve Bayes, k-nearest neighbor, support vector machine, random forest, and gradient boosted trees) using patient records from a multi-center database (n = 533) and prescribed medication ingredients (n = 228) as features to discriminate between the two hemodialysis types. Predictive skill was assessed using a 5-fold cross-validation, and the algorithms showed a range of performance from 0.7 balanced accuracy (logistical regression) to 0.86 (random forest). The two best performing models were further tested using an independent single-center dataset and achieved 84–87% balanced accuracy. This model overcomes issues inherent within large databases and will allow us to utilize and combine historical records, significantly increasing population size and diversity within both iHD and CRRT populations for future clinical studies. Our work demonstrates the utility of using medications alone to accurately differentiate subpopulations of patients in large datasets, allowing codes to be transferred between different coding systems. This framework has the potential to be used to distinguish other subpopulations of patients where discriminatory ICD codes are not available, permitting more detailed insights and new lines of research.
Keywords: Machine learning, Electronic health records, Medications, Hemodialysis, Pediatrics
1. Introduction
Electronic health records (EHRs) are digitized forms of health information and documentation that facilitate the systematic search of medical records. EHR systems are increasingly used for clinical research. However, EHRs are notoriously noisy, with detail and accuracy varying between institutions, including lack of secondary diagnoses and comorbidities or mismatches between medical chart records [1-4]. Inconsistencies in coding systems both within and between institutions create additional challenges [1-5]. For example, EHR data use various coding systems, including International Classification of Diseases 9/10 (ICD9/ICD10) systems for diagnosis, procedures, lab results, and medications [1,6]. ICD10 codes are vastly more detailed, with 141,747 procedural and diagnostic codes compared to 15,000 for ICD9 [1-3]. The differences in the level of detail and accuracy between ICD9 and ICD10 codes for the same diagnoses and procedures lead to significant challenges in combining EHR databases. To date, there are two main solutions when combining EHR databases with differing levels of data resolution: 1) removal of lower granularity data; 2) aggregation of higher granularity data to lower granularity. Both approaches result in a loss of critical information with removal of data limiting the number of available records and aggregation of data resulting in a loss of patient information.
Machine learning (ML) offers an alternative approach through the development of models that can robustly disaggregate low granularity data into higher granularity data through learned patterns in patient information [7]. This could limit the loss of information found in existing approaches and facilitate the study of rarer medical conditions with small population sizes. Models developed using ML methods have been used to generate higher resolution from lower resolution data in other fields, in particular image processing and climatology [8,9]. To our knowledge, this approach has not been applied to EHRs to identify patient subpopulations.
Existing ML studies based on EHRs have been used to automate the interpretation of clinical notes (unstructured data) by combining coded (structured) information and natural language processing [10-12]. The models developed in these studies have shown great accuracy in the identification of populations, disease prediction and medication usage based on diagnosis [10-12]. These models are often based on multiple sources of detailed data tailored to specific study questions and are not easily generalizable across EHR databases of varying structure and granularity. Increased generalization can be achieved by restricting models to commonly available information. Disease and medications are correlative and have been used previously in predictive models based on a combination of structed and unstructured EHR data [11,12]. The results of these previous studies suggest that it may be possible to create a simple, predictive and generalizable model to identify subpopulations of patients using only medications and basic demographic information. This approach has the potential to substantially increase the number of records used to study rare conditions with small patient populations. As medication data is a key part of all EHR databases, this facilitates transferability across databases.
In this study, we leverage ML to identify patterns in medication usage among subpopulations of patients within a multi-institutional ICD9/10 coded EHR database. As a test case, we utilized an inpatient pediatric hemodialysis cohort to identify subpopulations based on hemodialysis (HD) modality. This population was chosen for testing as it has a small population size and distinctive subpopulations that cannot be identified in ICD9 coded EHRs. Currently, studies of these subpopulations must be limited to ICD10 coded EHRs, EHRs with both structured and unstructured data, or single institutions where detailed patient chart information can be used to verify modality. Using ML models, we were able to identify two subpopulations of inpatient HD pediatric patients; patients undergoing intermittent hemodialysis (iHD) and continuous renal replacement therapy (CRRT). While CRRT and iHD are both used to replace kidney function, CRRT is a favored modality for critically ill patients, while iHD is used for more stable patients with both chronic as well as acute conditions. The ability to differentiate between these distinctive subpopulations allows for fine-grained analyses. The methods developed in this manuscript will be used in an ongoing study to identify common medications in iHD and CRRT pediatric patients in order to guide future dosing studies. This approach overcomes issues inherent within large databases that combine EHRs from different institutions and offers a general framework that can be used to identify a variety of patient subpopulations for future clinical studies.
2. Methods
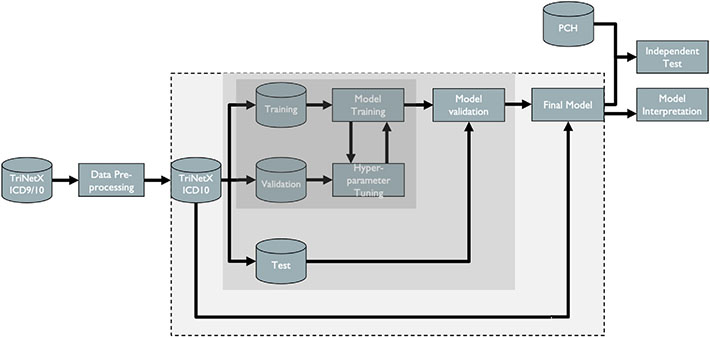
In order to build and test the machine learning algorithms, the following steps were required: 1) acquisition of relevant medical health record data; 2) data pre-processing; 3) model development, 4) model validation; and 5) post-hoc model interpretation (Fig. 1).
Fig. 1.
Flowchart of ML methods. Cylinders represent datasets and rectangles processing steps. The dark grey box includes all steps in the tuning process where model parameters are selected. The light grey box includes steps in the validation process used to prevent overfitting. The box with a dashed outline includes the development of the final model using the entire dataset and the selected model parameters.
2.1. Dataset
2.1.1. Data acquisition
The TriNetX database (subsequently referred to as the TriNetX dataset) was the primary data source used to train the ML algorithms. TriNetX is a global health research network that provides electronic medical records (diagnoses, procedures, medications, laboratory values, genomic information) from 3083 pediatric dialysis patients from 41 healthcare organizations. A request was submitted to TriNetX to obtain records for all patients with one or more of the following dialysis ICD9/10 codes: 5A1D90Z, 5A1D70Z, 5A1D80Z, 90945, 90947, 39.95, 71192002, Z99.2 and 302497006. De-identified EHRs were returned in several distinct files, including patient demographics, encounters (hospitalizations), and medications.
2.1.2. Data pre-processing
Data were merged after acquisition using patient and encounter identification numbers to produce a dataset including only hemodialysis encounters. The merged dataset was then cleaned to provide a subset of data for training the ML algorithms. As the ICD-9 code 39.95 does not discriminate between intermittent and continuous dialysis, we selected encounters corresponding to only ICD-10 codes 5A1D70Z (iHD) and 5A1D90Z (CRRT) for model training. The dataset included information on medications separated into two files: 1) dose, formulation, and brand; 2) ingredients. Therefore, we chose to use medication ingredients to avoid different doses and formulations of the same medication being considered as different medications across patients and/or encounters.
Medications that were used in at least 5% of encounters were selected, resulting in 228 medications, each specified by the corresponding RxNorm code. Medications were binary encoded for each encounter (0 = not prescribed; 1 = prescribed). In addition to medication ingredients, sex and age at the time of encounter were also included as features. As the exact age at the time of encounter was not included in the original data, it was estimated as the difference between the birth year and encounter start date. The outcome was also binary encoded (0 = iHD, 1 = CRRT).
A second retrospective dataset (PCH dataset) was obtained from Intermountain Primary Children’s Hospital in Salt Lake City, UT. As the PCH dataset did not contain RxNorm codes for the prescribed medications, we set up a cross-table linking the medication name in the PCH data to the associated RxNorm code for the medication ingredient(s). The PCH dataset was independent of the TriNetX dataset and was not used to train or tune the ML algorithms. The PCH dataset was used as a second independent test on the model’s predictive skill and was pre-processed using the steps described above.
2.2. Machine learning
2.2.1. Model development
We tested a range of different ML algorithms to help select the best algorithm to classify dialysis patients into iHD or CRRT categories. These included logistic regression, k-nearest neighbors, radial-basis support vector machines, and Naive Bayes classifiers. Two ensemble algorithms were also tested (random forests and gradient boosted trees). Ensemble algorithms use subsets of data to build a collection of decision trees. While each individual tree is considered to be a weak learner with high bias, the bias of predictions made using the collective ensemble of trees is generally low. Both random forest and gradient boosted tree algorithms use subsets of data to build individual trees. The random forest algorithm creates a “forest” of multiple independent trees. In contrast, the gradient boosted algorithm creates an additive sequence of trees with each new tree designed to reduce errors from the preceding tree. One advantage of these algorithms is that interactions between features are inherently incorporated into the decision trees. In order to provide a baseline for model comparison, a featureless model was built. In this featureless model, the encounters were simply predicted as the majority case (iHD rather than CRRT). All models were built with medication ingredients and patient demographics as the input features and hemodialysis modality as the outcome.
2.2.2. Model validation
As ML algorithms do not have traditional statistical goodness of fit tests, we assessed the predictive skill of each algorithm using a 5-fold cross-validation (outer cross-validation) with 10 repeats. In each fold, the TriNetX dataset of known iHD and CRRT pediatric patients was split into two subsets, with 80% of the data in one subset labeled as the training set and the remaining 20% in the testing subset. Models were fit using the training subset and then used to predict iHD and CRRT classification in the testing subset. The discrepancy between the predicted and observed outcomes for the testing subset provided an estimate of the model’s predictive skill. As the dataset includes patients with multiple encounters (Table 1), block sampling was used to generate all subsets of data in the cross-validations. This ensures that encounters for the same patient are not split across training and testing sets.
Table 1.
Training set patient demographics and encounters.
| Patient demographics (training set) | |
| Total encounters | 533 |
| iHD (5A1D70Z) | 365 |
| CRRT (5A1D90Z) | 168 |
| Total patients: | 390 |
| Sex: | |
| Male | 186 |
| Female | 204 |
| Age: | |
| 0–1 | 39 |
| 2–5 | 97 |
| 6–11 | 90 |
| 12–18 | 204 |
| Race: | |
| American Indian or Alaska Native | 4 |
| Asian | 10 |
| Black or African American | 75 |
| Native Hawaiian or Pacific Islander | 1 |
| White | 231 |
| Unknown | 69 |
| Ethnicity: | |
| Hispanic or Latino | 72 |
| Not Hispanic or Latino | 264 |
| Unknown | 54 |
| Medication (ingredients) | 228 |
Prediction skill was estimated using the Receiver Operating Characteristic (ROC) and measured using the area under the ROC curve (AUROC) [13]. We used the following AUROC thresholds to assess models: between 0.7 and 0.8 = ‘Acceptable discrimination’; 0.8 and 0.9 = ‘Excellent discrimination’; above 0.9 = ‘Outstanding discrimination’ [14]. In addition, we calculated the sensitivity, specificity, and accuracy. Finally, we calculated the balanced accuracy as the arithmetic mean of the sensitivity and specificity to account for the imbalance in the dataset.
The two ensemble algorithms tested have an additional set of hyperparameters that control various aspects of the learning processes (e.g., the number of submodels in the random forest or the rate at which weights were updated in the gradient boosted tree). Optimal values for these hyperparameters were chosen by tuning. For tuning, each training dataset was further split into two subsets (training and validation) in a 3-fold inner cross-validation. Models were trained using the inner training set across a range of values for the hyperparameters. These models were then used to predict hemodialysis modality for the validation set. AUROC values were calculated for all predictions. Hyperparameter values that resulted in the highest AUROC values were selected for the final model. Table 2 lists the set of hyperparameters. Although the outcome was binary, predictions were made on a continuous 0–1 scale. In order to compare predicted and observed classes, this scale was converted into a non-continuous, binary classification (iHD vs. CRRT) using a threshold. Thresholds were optimized by selecting the value that resulted in the maximum balanced accuracy for the test dataset as part of the repeated cross-validation.
Table 2.
Hyperparameter values for the random forest and gradient boosted tree algorithms following tuning.
| Algorithm | Hyperparameter | Value |
|---|---|---|
| Random Forest | mtry: number of features to use for each split in individual trees | 20 |
| num.trees: number of individual trees to build | 450 | |
| Gradient Boosted Tree | eta: learning rate | .02 |
| max_depth: complexity of individual trees | 12 | |
| nrounds: number of boosting iterations | 750 | |
| subsample: proportion of observations to use in each boosting iteration | 0.85 | |
| colsample_bytree: proportion of features to use in each boosting iteration | 0.57 | |
| colsample_bylevel: proportion of selected features to be used for each split | 0.67 |
Additional cross-validations were performed for different subpopulations (sex, race and ethnicity, Table SI 1-3). In each cross-validation, patients from one-subpopulation were placed in the test dataset, and all other patients placed in the training set. A random forest model was built using the training set and the set of hyperparameters selected from the tuning process. This model was then used to predict dialysis modality for the sub-population test set. The final, tuned model was further used to predict iHD and CRRT classification in the PCH dataset as a second, fully independent test of predictive skill. Both of these tests were assessed using balanced accuracy.
2.2.3. Post-hoc model interpretation
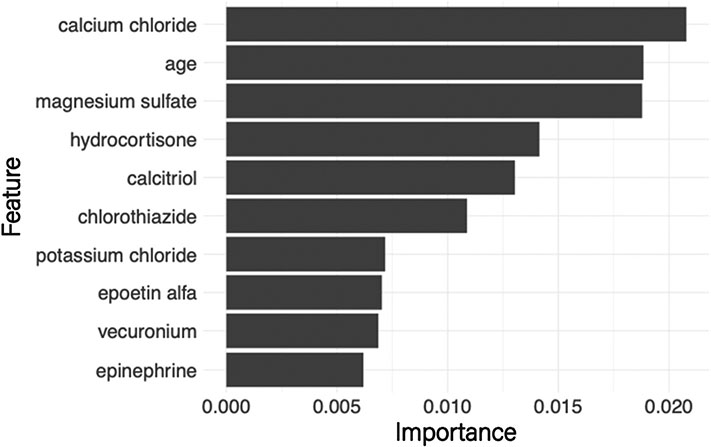
To help interpret model results, permutation-based feature importance was estimated for the random forest and gradient boosted algorithms [15]. Feature importance is calculated as part of model training by randomly permuting the values of one feature in the testing subset and estimating the loss of predictive skill. A considerable reduction in predictive skill would indicate that the permuted feature was essential in fitting the original model. Feature importance was used to help identify the set of patient demographics and medication ingredients that best discriminated between iHD and CRRT (Fig. 2).
Fig. 2.
Feature importance plot based on the random forest model. The width of the bars indicates the reduction in model predictive skill when permuting the values of that feature (medications and age).
A global surrogate model was built based on the best performing algorithm and used to help interpret the model results. Surrogate models are common in engineering, where they are used to approximate a complex model with a more straightforward approach [16]. There is increasing interest in using these models in machine learning to help interpret black-box models where the rules that are ‘learned’ may not be clear, e.g., boosted regression trees or neural networks [17]. While there has been some criticism of overreliance on the interpretation of machine learning models, we included a surrogate model and a partial dependency plot to help illustrate how the rules learned by the random forest model relate to clinical knowledge [18]. The standard practice was followed in which the full black-box model was used to predict hemodialysis modality. Then, a simple decision tree model was trained using the same input features (medications and patient demographics) and predicted hemodialysis modality from the black box model. The resulting decision tree is restricted to the first few splits to show an overview of both the essential features and the effect on the classification of iHD and CRRT.
All machine learning was carried out in R4.1.1 using the mlr3 package to run cross-validation and hyperparameter tuning [19,20]. Random forests and gradient boosted trees were built using the ranger and xgboost packages, respectively [21,22]. All model code can be accessed at the following repository: https://github.com/amcknite/ehr_pediatric_hemodialysis.
2.3. Regulatory
TriNetX is compliant with the Health Insurance Portability and Accountability Act (HIPAA), the US federal law which protects the privacy and security of healthcare data. TriNetX is certified to the ISO 27001:2013 standard and maintains an Information Security Management System (ISMS) to ensure the protection of the healthcare data it has access to and to meet the requirements of the HIPAA Security Rule. Any data displayed on the TriNetX Platform in aggregate form, or any patient-level data provided in a dataset generated by the TriNetX Platform, only contains de-identified data as per the de-identification standard defined in Section §164.514(a) of the HIPAA Privacy Rule. The process by which the data is de-identified is attested to through a formal determination by a qualified expert as defined in Section §164.514(b)(1) of the HIPAA Privacy Rule.
The PCH dataset containing de-identified EHRs from pediatric patients receiving peritoneal dialysis, iHD, or CRRT was obtained from Intermountain Primary Children’s Hospital in Salt Lake City, UT, and approved by the University of Utah Institutional Review Board (IRB protocol number 00074616).
3. Results
A total of 33 encounters included patients with more than one dialysis code (iHD and CRRT). Within these encounters, it was not possible to identify the period of time each code was applicable to the patient. These encounters were subsequently dropped from the dataset. We obtained a final dataset of 533 encounters with associated medication ingredients and patient demographics following data preprocessing. This dataset consisted of 365 iHD and 168 CRRT encounters (Table 1). A total of 228 medication ingredients were included, each specified by a corresponding RxNorm code. The independent PCH dataset contained 174 patients, with 133 iHD and 41 CRRT encounters.
The results of the cross-validation are shown in Table 3. For all models, accuracy values are inflated relative to balanced accuracy due to the imbalance in the dataset. The baseline featureless model had an AUROC score and balanced accuracy of 0.5. All tested ML algorithms had improved AUROC scores between 0.72 and 0.92 and balanced accuracy between 0.69 and 0.86. There was a general increase in model predictive skill as the complexity of the underlying algorithm increased, with the highest performance achieved by the ensemble algorithms (Table 3). The random forest model had a slightly higher score compared to the other ensemble algorithms. The thresholds selected to distinguish between iHD and CRRT were relatively high (0.77 for random forest predictions; 0.75 for gradient boosted trees), reflecting the predominance of iHD encounters in the dataset. Cross validation results for specific sub-populations (SI Tables 1-3) show similar performance values for different sex, race and ethnicity groups. The model did not perform as well for Asian patients (balanced accuracy performance = 0.68). These patients are poorly represented in the data (less than 2.6% of the total patients), but suggests caution when applying this model to this subpopulation. The trained random forest and gradient boosted tree models were then used to predict hemodialysis modality for the independent PCH validation dataset, with a balanced accuracy of 0.85 and 0.84, respectively.
Table 3.
Performance metrics for the tested machine learning algorithms. Values represented as mean (S.D.).
| Algorithm | AUROC | Sensitivity | Specificity | Accuracy | Balanced Accuracy |
|---|---|---|---|---|---|
| Featureless | 0.50 (0.00) |
1.00 (0.00) |
0.00 (0.00) |
0.68 (0.05) |
0.50 (0.00) |
| Logistic Regression | 0.72 (0.06) |
0.75 (0.06) |
0.62 (0.10) |
0.71 (0.05) |
0.69 (0.06) |
| Naïve Bayes | 0.84 (0.04) |
0.85 (0.04) |
0.76 (0.08) |
0.82 (0.04) |
0.80 (0.04) |
| Regularized Regression (ElasticNet) | 0.89 (0.02) |
0.88 (0.04) |
0.73 (0.07) |
0.83 (0.03) |
0.80 (0.03) |
| k-nearest Neighbor | 0.85 (0.03) |
0.88 (0.05) |
0.60 (0.09) |
0.79 (0.04) |
0.74 (0.04) |
| Support Vector Machine | 0.90 (0.03) |
0.85 (0.04) |
0.86 (0.07) |
0.85 (0.03) |
0.85 (0.04) |
| Random Forest | 0.92 (0.03) |
0.90 (0.03) |
0.82 (0.07) |
0.87 (0.03) |
0.86 (0.04) |
| Gradient Boosted Trees | 0.92 (0.02) |
0.89 (0.04) |
0.82 (0.08) |
0.87 (0.03) |
0.85 (0.04) |
The random forest model was used for further investigation, including a) feature importance scores for the top ten features in the dataset (Fig. 2); b) a partial dependency plot of hemodialysis modality on age (Fig. 3); c) a global surrogate model that provides a simple decision tree to predict hemodialysis modality (Fig. 4). The global surrogate model had a cross-validated balanced accuracy of 0.85, suggesting that this successfully approximates the main decisions of the full random forest.
Fig. 3.
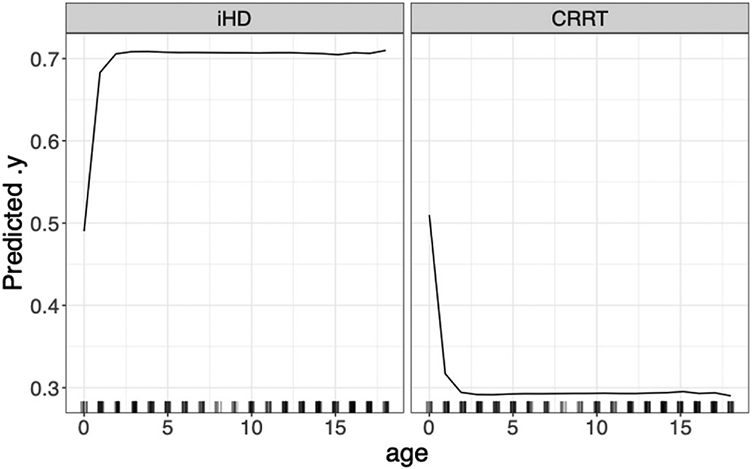
Partial dependency of dialysis type based on patient age. Left panel indicates the probability of iHD with age; right panel the probability of CRRT. Note the marked transition to increased probability of iHD for patients above 1 year old.
Fig. 4.
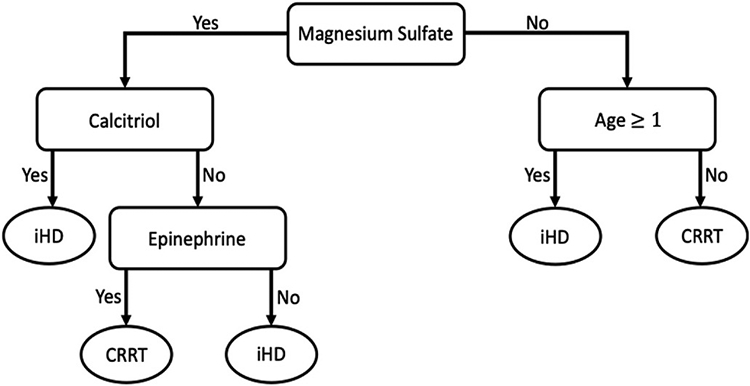
Surrogate decision tree of top 4 deciding features based on the random forest model.
4. Discussion
This study utilized machine learning techniques to develop a predictive model to differentiate between pediatric CRRT and iHD patients using medications from EHRs. The best performing models (random forest and gradient boosted trees) had balanced accuracy scores of 0.85 and 0.84, respectively. These scores indicate that these models were able to predict dialysis modality with an error rate of ~15%. Model tests performed using the entirety of the independent PCH dataset resulted in similar performance scores, and demonstrates that the trained model can be applied to other databases, with negligible loss in predictive skill. Sensitivity values for these models further indicate that the models could correctly predict CRRT patients with an error rate of 10–13%. Prediction skill for the iHD modality is slightly lower, with sensitivity values of 0.85 and 0.84, respectively. Overall, the models were considered to have excellent discrimination and were able to predict hemodialysis modality with minimal error.
All models were built and tested using the medication ingredients rather than individual prescriptions that included brand, formulation, and concentration. This was done to reduce redundancy through multiple medication codes representing variations in formulations and concentrations for each individual medication. Models were tested using RxNorm codes linked to specific prescriptions as well as medication ingredient. Cross-validation of models built with the dataset containing medication prescription had lower predictive skill (e.g., AUROC of 0.78 and 0.8 for the random forest and gradient boosted models respectively), and steep decreases in sensitivity. Aggregating medications by ingredient drastically reduced redundancy resulting in a marked improvement in model performance (Table 3). We acknowledge that there are still some redundancies in combination medications containing more than one ingredient. Despite this, our models show strong predictive skills. Further, by restricting input features to commonly available information (i.e., medication ingredients and demographics) our model should be easily transferable to other databases as evidenced by the high levels of predictive skill when applied to the independent PCH dataset.
Published studies have incorporated additional information from EHRs (e.g., primary diagnoses, procedures, and lab results) to strengthen model identification of specific populations of pediatric patients [23]. In a study of glomerular disease, EHR from a single center was used to develop a model incorporating diagnosis, kidney biopsy, and transplant procedure codes as patient identifiers [23]. Using single-center testing has been shown to increase model accuracy as EHR information can be manually verified with detailed institutional medical charts. However, it is not possible to use a single center dataset for CRRT patient identification due to the small patient population size, which necessitates the use of multiple centers. It is not clear if including additional EHR information such as procedural codes would have improved the classification obtained by our model. Manual EHR or medical chart review in combination with multiple sources of structured data have also been used to identify hematologic malignancy and type 2 diabetic pediatric patients across multiple institutions [24,25]. The patients in these studies had clear and definitive primary diagnoses, in contrast to CRRT patients who are heterogeneous and often have multiple diagnoses. Notably, chronic renal disease represented only 4–9% of pediatric CRRT patients compared to primary diagnoses such as sepsis 11–20% and solid organ transplant 20–22% [26,27]. As a result, if renal-related primary diagnoses were the only search parameters included in the model, there is the potential to miss large proportions of patients and significantly reduce population size. Excluding diagnosis data further reduces the amount of missing data and mismatches in dates corresponding to diagnosis, labs, and procedures, and may improve the transferability of the model between EHR databases [3,4].
Our model training set was restricted to ICD10 codes for dialysis modality identification. ICD10 codes were first implemented in the U.S. starting in 2015, but their predecessors, ICD9, are still used within some U.S. hospital systems [1,28]. The accuracy of ICD9 and ICD10 codes to describe diagnoses are considered similar [29]. ICD10 codes for the primary diagnosis of acute myocardial infarction in EHRs were found to be accurate when compared to patient charts (positive predictive value 82.5–93.8%) [30,31]. The introduction of ICD10 increased the granularity of procedural codes providing 141,747 procedural and diagnostic codes compared to 15,000 for ICD9 [30-32]. As a result, iHD and CRRT can only be distinguished using ICD10 codes and not ICD9 where a general hemodialysis code 39.95 is used for all modalities. Although ICD10 codes were first introduced in 2015, ICD10 hemodialysis codes were not present within the TriNetX dataset until late 2017. This severely reduced the number of patients with identifiable hemodialysis modalities available to train the model. Despite this, the ML algorithms used in this study were able to produce models with high predictive skill. Model testing against gender, race, and ethnic subpopulations (SI Tables 1-3) and the independent PCH dataset suggests that there are no inherent biases.
Previous studies have used rule-based algorithms to identify subpopulations of patients [10-12]. In contrast, this study used ML algorithms as these provide several advantages. The validation of models developed through rule-based algorithms is based on the manual review of a randomly selected subset of EHRs. The ML models in this study were subjected to a more thorough repeated k-fold cross validation that ensured all records were used in validation. Rule-based algorithms use manually developed rules for specific situations, which can be highly time-consuming and subjective. In contrast, rules developed through ML are faster to develop and easily generated for a range of situations. The disadvantage is that ML rules can be difficult to understand due to the ‘black-box’ nature of these models. However, as we demonstrate here, there are several methods that can be used for post-hoc interpretation of these rules, such as partial dependency plots and surrogate models. These methods, when used carefully, can provide additional verification and review of the models by clinical researchers.
We use partial dependency plots and feature importance to understand the factors differentiating between iHD and CRRT patients in the study dataset. These plots were estimated using the random forest model as it had the best overall performance. The permutation-based feature importance scores (Fig. 2) indicate that, although the input features are overwhelmingly medication ingredients, age of the patient remains an important feature in differentiating between the two hemodialysis modalities. Feature importance scores indicate medication ingredients that are most useful in differentiating between hemodialysis modality, but do not indicate the direction of association. For this, the global surrogate model was used to approximate the set of rules learned by the random forest model (Fig. 4). For example, the first decision in the surrogate model tree is whether or not the ingredient magnesium sulfate is given to the patient. This decision alone correctly differentiates approximately 73% of CRRT encounters (left-hand side) and 88% of iHD encounters (right-hand side), with further refinements to this prediction through subsequent nodes of the decision tree.
The medication ingredients identified by the surrogate model decision tree are common in renal failure patients. Magnesium sulfate supplementation is standard in late-stage chronic kidney disease and patients on intermittent hemodialysis where magnesium reabsorption through renal tubules is impaired [33,34]. Late-stage chronic kidney disease patients also have reduced production of active vitamin D and a resultant decrease in calcium absorption [35-37]. Calcitriol is synthetic vitamin D3 and is used to supplement vitamin D loss [35-37]. In addition, CRRT patients have reduced calcium due to the use of citrate anticoagulation [38]. Infusions of calcium chloride or calcium gluconate are used to supplement this calcium loss in these patients [38]. Epinephrine is used to treat severe allergic reactions that may occur during iHD but is used more frequently to treat hypotension in critically ill patients [39-42]. The medications selected by the surrogate model generally agree with those ranked in the variable importance plot (Fig. 1), there are some differences (e.g., calcium chloride is not shown). These discrepancies result from the approximation in the surrogate model, which is based on predicted dialysis modality rather than the observed values.
There are two notable limitations to the methods used in this study. First, the underlying assumption of any predictive model is that the data is stationary over time. Applying these models retrospectively may require additional work to ensure that the prescribing practices have remained relatively constant. Second, while the total TriNetX dataset encompasses billions of records, the subset we obtained for training our models is relatively small (533 encounters). This could potentially be increased in future models by combining larger, multi-institutional datasets provided by TriNetX and the Pediatric Health Information System databases. However, combining these large databases is complicated by redundancy of encounters between the databases as records from individual hospitals may be included in both databases. It is impossible to determine the specific hospital linked to each patient in TriNetX, although this is possible in the Pediatric Health Information System database, and redundant hospitals could be removed if identified.
Overall, the results support the use of ML to differentiate between patients on different modalities of hemodialysis based on medication ingredient profiles from EHRs. Although there is a range of model performance, all algorithms performed better than the baseline featureless model, suggesting that the combination of limited patient demographic information and medications is adequate to successfully predict hemodialysis modality. There is a notable increase in model performance for all non-linear algorithms over the logistic regression model. This highlights the complexity of the dataset and the difficulties of using traditional statistical methods for analysis. This is further illustrated by the non-linearity of the partial dependency plot of age on hemodialysis modality (Fig. 3), with a rapid increase in the probability of iHD for patients above 1 year of age. The two best-performing algorithms achieved balanced accuracy scores of ~0.85 and AUROC scores of ~0.90, and were considered to show outstanding performance [14]. In addition to the cross-validation results, the high levels of accuracy achieved by the two top-performing models for the independent PCH dataset indicates that the trained models can be successfully generalized to other datasets.
The ability to differentiate between two subpopulations of pediatric hemodialysis modalities allows for more targeted research. Hemodialysis patients are often pooled, or assumptions are made regarding dialysis modality based on the location of care (PICU=CRRT) despite distinct differences between these patient populations [26,43-45]. Identifying hemodialysis subpopulations could lead to targeted analysis of patient demographics, medications, and length of stay across multiple institutions. Medication administration offers a rich dataset that could complement patient demographics, allowing the identification of patients undergoing different dialysis modalities while limiting issues that arise with mismatching data in EHRs.
Importantly, this model can be used to back predict CRRT and iHD pediatric patients using ICD9 codes in historic and multi-institutional EHR databases, significantly increasing the sample size for retrospective studies. This will allow the utilization of historical records, significantly increase population size and increase diversity within both iHD and CRRT populations.
5. Conclusion
EHRs are rich in information and can be powerful datasets for clinical research. However, issues inherent in EHRs including variations in coding systems and differing levels of granularity can limit their application. To address these limitations, we used ML methods to identify subpopulations using common information in EHRs (i.e., medications, demographics). Using these methods, we were able to successfully discriminate between pediatric patients undergoing different modalities of hemodialysis. Our models were based on commonly available information increasing the transferability between EHR databases and other patient subpopulations. Our framework can improve the granularity of information in older databases permitting retroactive studies on newly identifiable patient populations, and has the potential to significantly increase the number of EHRs for the analysis of small patient populations.
Supplementary Material
Acknowledgments
The authors would like to thank Dr. Jonathan Constance and Jacob Wilkes for invaluable discussions about the subject of research. Funding: This work was supported by the National Institute of Child Health and Human Development (R01HD097775), the University of Utah College of Pharmacy Donald R. Gehlert fellowship and the National Institute of Diabetes and Digestive and Kidney Diseases (F31DK130542).
Footnotes
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.imu.2022.101104.
References
- [1].Henry J, Pylypchuk Y, Searcy T, Patel V. Adoption of electronic health record systems among U.S. Non-federal acute care hospitals: 2008-2015. In: Office of the National Coordinator for Health Information Technology, editor35. Washington D.C.: ONC Data Brief; 2016. [Google Scholar]
- [2].Roth JA, et al. Introduction to machine learning in digital healthcare epidemiology. Infect Control Hosp Epidemiol 2018;39(12):1457–62. [DOI] [PubMed] [Google Scholar]
- [3].Kim HS, Kim DJ, Yoon KH. Medical big data is not yet available: why we need realism rather than exaggeration. Endocrinol Metab (Seoul) 2019;34(4):349–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Hersh WR, et al. Caveats for the use of operational electronic health record data in comparative effectiveness research. Med Care 2013;51(8 Suppl 3):S30–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Laper SM, Restrepo NA, Crawford DC. The challenges in using electronic health records for pharmacogenomics and precision medicine research. Pac Symp Biocomput 2016;21:369–80. [PMC free article] [PubMed] [Google Scholar]
- [6].Shickel B, et al. Deep EHR: a survey of recent advances in deep learning techniques for electronic health record (EHR) analysis. IEEE J Biomed Health Inform 2018;22(5):1589–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Bzdok D, Altman N, Krzywinski M. Statistics versus machine learning. Nat Methods 2018;15(4):233–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Ledig C, T L, Huszár F. Photo-realistic single image super-resolution using a generative adversarial network. In: IEEE conference on computer vision and pattern recognition; 2017. [Honolulu]. [Google Scholar]
- [9].Serifi A, Günther T, Ban N. Spatio-temporal downscaling of climate data using convolutional and error-predicting neural networks. Front. Clim 2021;3:656479. [Google Scholar]
- [10].Abhyankar S, et al. Combining structured and unstructured data to identify a cohort of ICU patients who received dialysis. J Am Med Inf Assoc 2014;21(5):801–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Liu D, et al. Medi-Care AI: predicting medications from billing codes via robust recurrent neural networks. Neural Network 2020;124:109–16. [DOI] [PubMed] [Google Scholar]
- [12].Backenroth D, et al. Monitoring prescribing patterns using regression and electronic health records. BMC Med Inf Decis Making 2017;17(1):175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Bradley A. The use of the area under the ROC curve in the evaluation of machine learning algorithms. Pattern Recogn 1997;30:1145–59. [Google Scholar]
- [14].Hosmer DW, Lemeshow S. Applied logistic regression. In: Wiley series in probability and statistics. New York Toronto: Wiley; 2000. p. 1. online resource (xii, 373. [Google Scholar]
- [15].Friedman J, Popescu B. Predictive learning via rule-based ensembles. Ann Appl Stat 2008;2(3):916–54. [Google Scholar]
- [16].Craven M, Shavlik J. Extracting tree-structured representations of trained networks. In: Advances in neural information processing systems; 1996. p. 24–30. [Google Scholar]
- [17].Molnar C, C G, Bischl B. iml: an R package for interpretable machine learning. Journal of Open Source Software 2018;3(26). [Google Scholar]
- [18].Rudin C. Stop explaining black box machine learning models for high stakes decisions and use interpretable models instead. Nat Mach Intell 2019;1(5):206–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Lang M, B M, Richter J, Schratz P, Pfisterer F, Coors S, Au Q, Casalicchio G, Kotthoff L, Bischl B, mlr3. A modern object-oriented machine learning framework in R. Journal of Open Source Software 2019. [Google Scholar]
- [20].Team RCR. A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2020. [Google Scholar]
- [21].Wright MN, Ziegler A, ranger. A fast implementation of random forests for high dimensional data in C++ and R. J Stat Software 2017;77(1):1–17. [Google Scholar]
- [22].Chen T, Guestrin C. XGBoost: A Scalable Tree Boosting System [New York, NY, USA]. In: 22nd ACM SIGKDD international conference on knowledge discovery and data mining; 2016. 10.48550/arXiv.1603.02754. [DOI] [Google Scholar]
- [23].Denburg MR, et al. Using electronic health record data to rapidly identify children with glomerular disease for clinical research. J Am Soc Nephrol 2019;30(12):2427–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Zheng T, et al. A machine learning-based framework to identify type 2 diabetes through electronic health records. Int J Med Inf 2017;97:120–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Phillips CA, et al. Development and evaluation of a computable phenotype to identify pediatric patients with leukemia and lymphoma treated with chemotherapy using electronic health record data. Pediatr Blood Cancer 2019;66(9):e27876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Riley AA, et al. Pediatric continuous renal replacement therapy: have practice changes changed outcomes? A large single-center ten-year retrospective evaluation. BMC Nephrol 2018;19(1):268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Aygun F. Evaluation of continuous renal replacement therapy and risk factors in the pediatric intensive care unit. Saudi J Kidney Dis Transpl 2020;31(1):53–61. [DOI] [PubMed] [Google Scholar]
- [28].Adler-Milstein J, et al. Electronic health record adoption in US hospitals: the emergence of a digital "advanced use" divide. J Am Med Inf Assoc 2017;24(6):1142–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Quan H, et al. Assessing validity of ICD-9-CM and ICD-10 administrative data in recording clinical conditions in a unique dually coded database. Health Serv Res 2008;43(4):1424–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Pendergrass SA, Crawford DC. Using electronic health records to generate phenotypes for research. Curr Protoc Hum Genet 2019;100(1):e80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Ando T, et al. Positive predictive value of ICD-10 codes for acute myocardial infarction in Japan: a validation study at a single center. BMC Health Serv Res 2018;18(1):895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Topaz M, Shafran-Topaz L, Bowles KH. ICD-9 to ICD-10: evolution, revolution, and current debates in the United States. Perspect Health Inf Manag 2013;10:1d. [PMC free article] [PubMed] [Google Scholar]
- [33].van de Wal-Visscher ER, Kooman JP, van der Sande FM. Magnesium in chronic kidney disease: should we care? Blood Purif 2018;45(1–3):173–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].William JH, Richards K, Danziger J. Magnesium and drugs commonly used in chronic kidney disease. Adv Chron Kidney Dis 2018;25(3):267–73. [DOI] [PubMed] [Google Scholar]
- [35].Shroff R, et al. Clinical practice recommendations for treatment with active vitamin D analogues in children with chronic kidney disease Stages 2-5 and on dialysis. Nephrol Dial Transplant 2017;32(7):1114–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].McAlister L, et al. The dietary management of calcium and phosphate in children with CKD stages 2-5 and on dialysis-clinical practice recommendation from the Pediatric Renal Nutrition Taskforce. Pediatr Nephrol 2020;35(3):501–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Christodoulou M, Aspray TJ, Schoenmakers I. Vitamin D supplementation for patients with chronic kidney disease: a systematic review and meta-analyses of trials investigating the response to supplementation and an overview of guidelines. Calcif Tissue Int 2021;109(2):157–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Davenport A, Tolwani A. Citrate anticoagulation for continuous renal replacement therapy (CRRT) in patients with acute kidney injury admitted to the intensive care unit. NDT Plus 2009;2(6):439–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Saha M, Allon M. Diagnosis, treatment, and prevention of hemodialysis emergencies. Clin J Am Soc Nephrol 2017;12(2):357–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Ebo DG, et al. Haemodialysis-associated anaphylactic and anaphylactoid reactions. Allergy 2006;61(2):211–20. [DOI] [PubMed] [Google Scholar]
- [41].Myburgh JA, et al. A comparison of epinephrine and norepinephrine in critically ill patients. Intensive Care Med 2008;34(12):2226–34. [DOI] [PubMed] [Google Scholar]
- [42].Dalal R, Grujic D. Epinephrine. In: StatPearls. Treasure Island (FL): StatPearls; 2022. [Google Scholar]
- [43].Rizkalla NA, et al. Patterns of medication exposures in hospitalized pediatric patients with acute renal failure requiring intermittent or continuous hemodialysis. Pediatr Crit Care Med 2013;14(9):e394–403. [DOI] [PubMed] [Google Scholar]
- [44].Lee KH, et al. Continuous renal replacement therapy (CRRT) in children and the specialized CRRT team: a 14-year single-center study. J Clin Med 2019;9(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Hayes LW, et al. Outcomes of critically ill children requiring continuous renal replacement therapy. J Crit Care 2009;24(3):394–400. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.