Abstract
Background:
The aim of this study was to evaluate the effect of preimplantation genetic testing for aneuploidy (PGT-A) on patient-important reproductive outcomes after in vitro fertilization (IVF).
Methods:
Randomized and non-randomized studies have been sought in Ovid, MEDLINE, EMBASE, Web of Science, Scopus, and Cochrane Central Register of Controlled Trials since each database’s inception through May 2021. Main keywords used for the search strategy included “Embryo transfer”, “In vitro fertilization”, “DNA sequencing”, and “Comparative genome hybridization”. Studies were screened independently and in duplicate.
Results:
Ten studies were finally analyzed, representing a total of 2630 embryo transfers. The pooled OR for live birth rates were 1.45 (95%CI 0.24–8.78, I2 96%) and 1.66 (95%PI 0.15–18.01, 95%CI 0.98–2.83, I2 81%) derived from the NRSIs and the RCTs, respectively, in which the miscarriage rate were 1.25 (95%CI 0.19–8.33, I2 70%) and 0.57 (95%PI 0.06–5.34, 95%CI 0.27–1.21, I2 53%), and clinical pregnancy rates were 3.08 (95%CI 2.22–4.29, I2 0%) and 1.43 (95%PI 0.38–5.42, 95%CI 0.96–2.13, I2 68%). Influence analyses showed a greater treatment effect when excluding studies without patients at advanced maternal age.
Conclusion:
There seems to be no significant difference in reproductive outcomes when using PGT-A in the general population; however, the procedure seems advantageous for patients at advanced maternal age. Nevertheless, this warrants caution when recommending the procedure to all couples seeking ART, as the current possible benefits may not justify the additional costs for all groups of patients.
Keywords: Assisted reproductive techniques, Comparative genomic hybridization, Embryo transfer, In vitro fertilization, Preimplantation genetic diagnosis
Introduction
Assisted reproductive technology (ART) is an alternative for couples who are unable to conceive naturally. Despite the evolution of applied techniques for the success of in vitro fertilization (IVF), the improvement in live birth rate (LBR) has not been substantial. The success rate is currently less than 50% with evidence showing decreasing rates as low as 11% in women over 40 years old compared to approximately 28% in the general US population (1–3).
This drastic reduction in success rates with advanced maternal age (AMA) is closely related to the incidence of aneuploidy, which is significantly more common in these women, along with implantation failure and recurrent pregnancy loss, as other common factors related to failure of ART (4–6).
Preimplantation genetic testing for aneuploidy (PGT-A) was proposed as a method to improve pregnancy rates in patients undergoing in vitro fertilization by eliminating the influence of aneuploidy on LBR (7). Using PGT-A, researchers sought to improve reproductive outcomes in patients with AMA, implantation failure, recurrent pregnancy loss, severe male factor infertility, and infertility of unknown origin. Proposed additional benefits included the reduction of time to achieve pregnancy as well as improving reproductive outcomes in IVF cycles with donated eggs (8, 9).
Multiple techniques have been proposed for PGT-A; scientists first began performing biopsies on cleavage stage embryos and studying blastomeres using Fluorescence in situ hybridization (FISH), but results were unsatisfactory (10). Additionally, the technique used to obtain cells for analysis has generated controversy as well; for example, a negative effect on reproductive success was reported regarding biopsies performed on day three embryos (11, 12).
Given the limitation of the initial PGT-A technologies, techniques were developed to evaluate all 24 chromosomes. These new techniques are now known as comprehensive chromosome screening (CCS) and include metaphase comparative genomic hybridization (mCGH), array comparative genomic hybridization (aCGH), next generation sequencing (NGS), single nucleotide polymorphism (SNP) genotyping method, and real-time quantitative polymerase chain reaction (q-PCR) (13–16). Following the introduction of CCS techniques, implantation rates increased, and abortion rates decreased. Additionally, the use of PGT-A increased the rates of single embryo transfers and reduced the risk of multiple pregnancy and premature birth associated with multiple embryo transfers (17).
Despite the promising results seen at the start of the chromosomal analysis era, inconsistent literature reports, increasing costs, systematic errors in outcome evaluation, and the ethical dilemma when dealing with mosaic embryos have raised questions about the utility and generalized applicability of PGT-A for embryonic selection (18–21). A more in-depth analysis of the safety, risks, and benefits of these techniques in the real world is necessary; thus, this meta-analysis was performed to explore the impact of PGT-A before frozen-thawed (FET) or fresh embryo transfer (ET) on the most important reproductive outcomes of patients considering live birth rates and miscarriage rates as our main concern.
Methods
This meta-analysis was done in line with the guidelines of Preferred Reporting Items for Meta-Analyses (PRISMA), (22) and the PRISMA checklist can be found in supplementary file 1. Our protocol was registered in PROSPERO under CRD42020198866.
Eligibility criteria: In this study, randomized controlled trials (RCTs) and prospective non-randomized studies of interventions (NRSIs) were included which evaluated the effect of PGT-A with CCS on reproductive outcomes of couples undergoing FET or ET, compared to only morphological assessment of blastocysts before transfer. NRSIs were included in the current study as very few RCTs are available due to the fact that possible detrimental effects of PGT-A on birth rates is a relatively novel issue and the ethical concerns related to the procedure hinder the researchers to conduct RCTs.
The included studies comprised the research on couples, irrespective of the cause of infertility, who underwent in vitro fertilization (IVF) or intracytoplasmic sperm injection (ICSI) with transfers in the blastocyst stage and research reporting live birth rate (LBR), miscarriage rate, and/or clinical pregnancy (CP) per embryo transfer as their primary or secondary outcomes; moreover, in studies evaluating multiple IVF cycles, merely the first cycle results were considered eligible. Also, studies summarizing multiple cycles without reporting the first cycle results, using low-yield PGT-A techniques (e.g., FISH) and day three biopsy, assessing the effect of solely detecting mosaicisms, or those without adequate reporting of outcomes of interest were excluded.
Information sources and search strategy: An experienced librarian, provided with input from the lead researcher, has designed and conducted the search in Ovid, MEDLINE, EMBASE, Web of Science, Scopus, and Cochrane Central Register of Controlled Trials since each database’s inception through May 2021; references from studies and reviews were also screened for eligibility. A general search strategy combining MeSH terms and free text was built, with the first group of terms referring to the possible techniques used for PGT-A (e.g., single nucleotide polymorphism, genomic hybridization, DNA sequencing, etc.), the second to IVF and ICSI techniques, and the third to embryo transfer. No language restriction or design-specific filters were applied in the databases. The full search strategy used in Scopus is presented in supplementary file 2 as an example.
Study selection process: Four reviewers working independently and in duplicate assessed each manuscript’s title, abstract, and full text for eligibility. Prior to each screening phase, pilots were conducted until achieving an acceptable level of inter-rater reliability (Fleiss’ kappa >0.70) (23). Aiming for a high sensitivity in the title and abstract screening, disagreements between reviewers were solved by including the study in the next phase. Afterwards, the full text of included abstracts were obtained and the same procedure was followed to select the final studies for inclusion; disagreements at this level were solved by consensus or intervention of a third reviewer.
Data collection and outcomes of interest: A web-based extraction form was created and evaluated by all reviewers before data extraction. General information of the included studies (author, year, country, funding, and design) was extracted alongside information on the PGT-A technique used, studied subgroups (e.g. advanced maternal age, recurrent pregnancy loss), treatment cycles, embryo transfer techniques (fresh ET or FET), and number of embryos transferred.
Seeking to combine studies performing multiple and single embryo transfers, reproductive outcomes per embryo transfer, comprising live birth rates, miscarriage rates, and clinical pregnancy were considered as the main outcomes of interest. The definition of the outcomes given by the authors of each study was extracted to evaluate the possible heterogeneity derived from different classifications. For the studies not stating definitions, outcomes were identified according to the guide for the standardization of definitions and reports in infertility (24). All reviewers extracted the data independently and in duplicate and disagreements were solved by consensus or intervention of a third reviewer.
Risk of bias in individual studies and certainty of evidence: Four reviewers, (LS, AFR, FDGC, PC) working independently and in duplicate, evaluated the risk of bias in individual RCTs using the Cochrane risk of bias tool 2.0 (RoB2.0). This tool evaluates six domains including bias arising from the randomization process, deviations from the intended intervention, missing outcome data, mis-measurement of the outcomes, and selection of the reported results. The overall risk of bias was classified as low, moderate (labeled “with some concerns” by the tool), or high, according to the tool’s criteria (25).
For non-randomized studies of interventions (NRSIs), the risk of bias was evaluated using the Cochrane ROBINS-I scale, considering bias arising due to confounding variables, selection of participants, misclassification of interventions, deviations from the intended intervention, missing data, mismeasurement of outcomes, and selection of the reported outcomes. The overall risk of bias was classified as low, moderate, or serious according to the tool criteria (26). All decisions were based on the reproductive outcomes of the studies; if two outcomes had different risk of bias assessments, the outcome with the highest risk was considered for the final decision. Any disagreement between the reviewers was solved either by consensus or intervention of a third reviewer.
Certainty in the evidence for the primary outcomes was evaluated using the GRADE approach, considering the risk of bias of included studies, the inconsistency of effect estimates, the indirectness associated with the study design, the possibility of publication bias, and residual confounders (27). The results were divided according to the study designs (RCTs and NRSIs) and are summarized in a table of findings, generated using the GRADEpro GDT software (28).
Summary measures and synthesis of results: LBR, miscarriage, and CP rates were extracted alongside the embryo transfers and the treatment cycles. For the primary analysis, odds ratios (ORs) were calculated using the pregnancy events per embryo transfer; these ORs were pooled by Der-Simonian and Laird approach (DL) for random effects meta-analyses. As expected, a considerable degree of clinical and methodological heterogeneity was derived from the different subgroups of patients seeking ART (i.e., patients at an advanced maternal age, with recurrent pregnancy loss, etc.) and different stimulation and embryo transfer protocols.
Statistical heterogeneity of the pooled estimates was explored using the X2 statistic with p-value threshold of <0.10 and the I2 statistic, considering the thresholds described in the Cochrane handbook (29). Sensitivity analyses assessing the robustness of findings were planned, first by pooling LBR and CP events per cycle instead of per embryo transfer, and second, by pooling events per clinical pregnancies for the miscarriage rate. The use of fixed-effects meta-analyses depended on the changes of statistical heterogeneity when performing the sensitivity analyses. Subgroup analyses considering populations with advanced maternal age (AMA) and recurrent pregnancy loss (RPL) were planned; however, no formal subgroup effect assessment was possible due to lack of sufficient independent report for each subgroup among the included studies. Nonetheless, further exploration of heterogeneity was performed using influence analyses as described by Viechtbauer and Cheung (30) included in the “dmetar” package (31).
In order to consider how new studies could affect the obtained effect estimates, 95% prediction intervals (95% PI) for the pooled random-effects estimates were calculated through the method described by Higgins et al. (32) if four or more studies were synthetized in each meta-analysis. Statistical analyses were performed in R statistical software vs. 4.0.3 with R studio vs. 1.3.1056 using the packages “metaforest”, “meta”, and “dmetar”.
Results
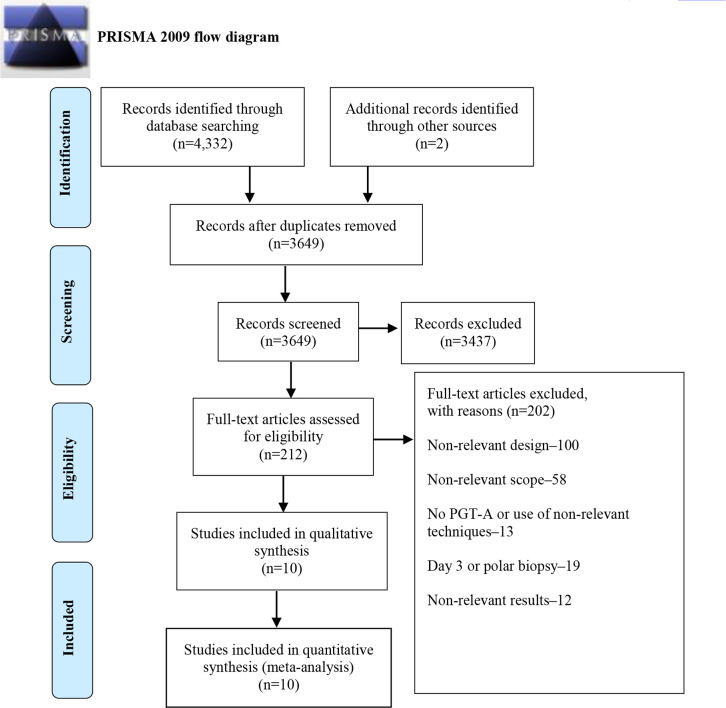
Our search strategy yielded 3649 studies after deduplication. From these, 212 full texts were selected for screening. Six RCTs and four NRSIs were included and analyzed, comprising a total of 2123 couples and 2630 embryo transfers. The study selection process and reasons for exclusion are summarized in figure 1. Most studies included patients with good prognosis with overall ages ranging from 25 to 42 years (8, 16, 33–40).
Figure 1.
PRISMA flow diagram
The technologies used for PGT-A included array-based comparative genomic hybridization (aCGH) in four studies (33, 36, 38, 39), next generation sequencing (NGS) in three studies, (34, 35, 37) real-time quantitative polymerase chain reaction (qPCR) in two studies, (8, 16) and single nucleotide polymorphism (SNP)-based microarray in one study (40). Most of the authors reported performing frozen/thawed embryo transfer (FET) for both groups, except two studies where they reported a combination of fresh embryo transfer (ET) and FET (8, 38). One study reported only fresh embryo transfers (39). Regarding the number of embryos transferred per patient, five of the included studies performed a single transfer for all patients of both groups (35–39); the other studies varied in allowing up to two transfers (34, 38), not limiting the number of embryo transfers (33), and performing double transfers in the control group and single transfers in the PGT-A group (8, 40). The rest of the general characteristics of the included studies are summarized in table 1.
Table 1.
General characteristics of included studies
| Study | Design | Country * | Funding | PGT-A technology | Mean age (SD) | Type of embryo transfer |
|---|---|---|---|---|---|---|
| Schoolcraft et al. 2010 | NRSI | UK | Government | aCGH | 37.7 (0.5) | Frozen/thawed |
| Yang et al. 2012 | RCT | China, USA | NR | aCGH | 31.4 (2.7) | Fresh |
| Scott Jr. et al. 2013 | RCT | USA | Industry | qPCR | 32.2 (0.5) | Frozen/thawed |
| Forman et al. 2013 | RCT | USA | Industry | qPCR | 34.8 (4.3) | Frozen/thawed Fresh |
| Greco et al. 2014 | NRSI | Spain | NR | aCGH | 32 (0.9) | Frozen/thawed Fresh |
| Liss et al. 2018 | NRSI | Poland | Academic | NGS | 35.8 (3.7) | Frozen/thawed |
| Ozgur et al. 2019 | RCT | Turkey | Private | NGS | 28.5 (3.71) | Frozen/thawed |
| Munné et al. 2019 | RCT | USA | Industry | NGS | 33.7 (3.5) | Frozen/thawed |
| Sato et al. 2019 | NRSI | Japan | Government | aCGH | 39.2 (2.1) | Frozen/thawed |
| Sui et al. 2020 | RCT | China | Private | SNP microarray | 35.8 (5.1) | Frozen/thawed |
Country was based on the setting of the study, or the reported country of the corresponding author if the setting was not reported.
NRSI: Non-randomized study of intervention; NR: Non-reported; RCT: Randomized clinical trial; PGT-A: Preimplantation genetic testing for aneuploidy; aCGH: Array comparative genomic hybridization; qPCR: Real-time polymerase chain reaction; NGS: Next generation sequencing
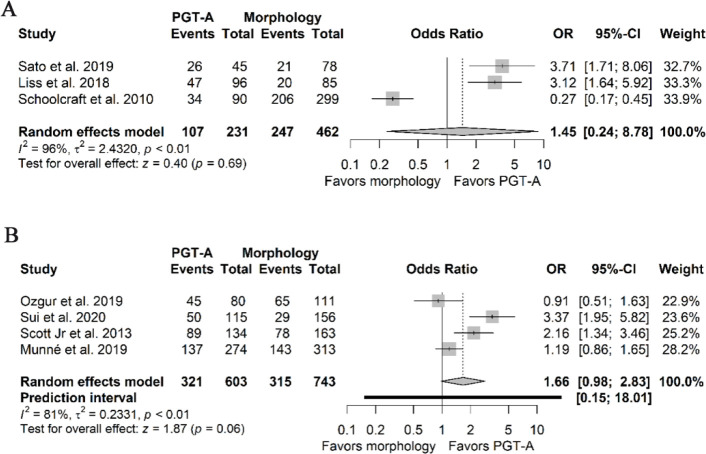
Live-birth rate per embryo transfer: Three out of four of the included NRSIs and four out of six RCTs reported that the LBR was associated with the interventions (16, 33–37, 40). Only two studies reported their definitions for LBR (34, 35). One defined the outcome as having at least one infant born alive which survived for at least one month, and the other explained the outcome as all infants born alive after 20 weeks. The pooled ORs were 1.45 (95%CI 0.24–8.78, I2 96%) and 1.66 (95%PI 0.15–18.01, 95%CI 0.98–2.83, I2 81%), derived from the estimates of the NRSIs (Figure 2A) and the RCTs (Figure 2B), respectively. The pooled estimates derived from RCTs and NRSIs had very low certainty, mainly due to considerable risk of bias, inconsistency, and imprecision.
Figure 2.
Forest plots of live-birth rate per embryo transfer. A) Pooled effect derived from NRSIs. B) Pooled effect derived from RCTs.
NRSIs: Non-randomized studies of interventions; RCTs: Randomized clinical trials; PGT-A: Preimplantation genetic testing for aneuploidy; OR: Odds ratio; 95%CI: 95% confidence interval
Narrative assessment of the included studies showed that three studies indicated improvement in LBR by using PGT-A (16, 34, 40), while the rest of the studies showed no significant difference in LBR when using PGT-A or morphology alone (33, 35–37). In general, the population in the studies with no significant differences was older; moreover, greater proportion of female factors were found as causes of infertility among them and lower number of previous miscarriages were recorded.
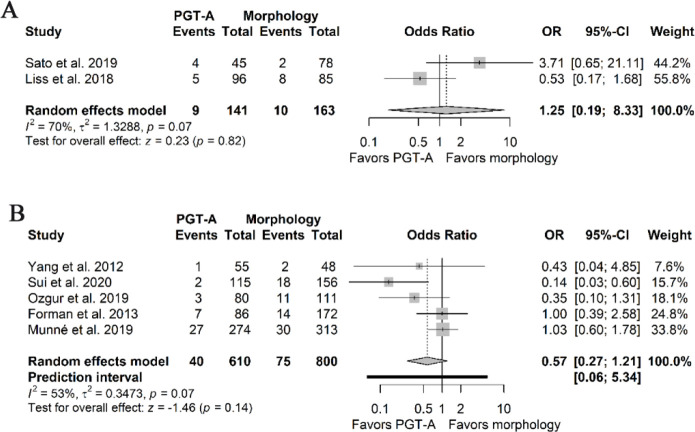
Miscarriage rate per embryo transfer: Five of the RCTs (8, 16, 35, 39) and two of the NRSIs (34, 36) reported the miscarriage rate per ET of their patients and most studies considered miscarriage as fetal mortality after a gestational sac with fetal heartbeat was detected, or an infant born dead before 20 weeks of gestation. The pooled ORs were 1.25 (95%CI 0.19–8.33, I2 70%) and 0.57 (95%PI 0.06–5.34, 95%CI 0.27–1.21, I2 53%) derived from the NRSIs and the RCTs, respectively (Figures 3A and B). The certainty of pooled estimates was rated as very low, due to extremely small sample sizes, considerable risk of bias, and inconsistency in both cases. Most studies considered miscarriage rates as secondary outcomes.
Figure 3.
Forest plots of miscarriage rate per embryo transfer. A) Pooled effect derived from NRSIs. B) Pooled effect derived from RCTs.
NRSIs: Non-randomized studies of interventions; RCTs: Randomized clinical trials; PGT-A: Preimplantation genetic testing for aneuploidy; OR: Odds ratio; 95%CI: 95% confidence interval
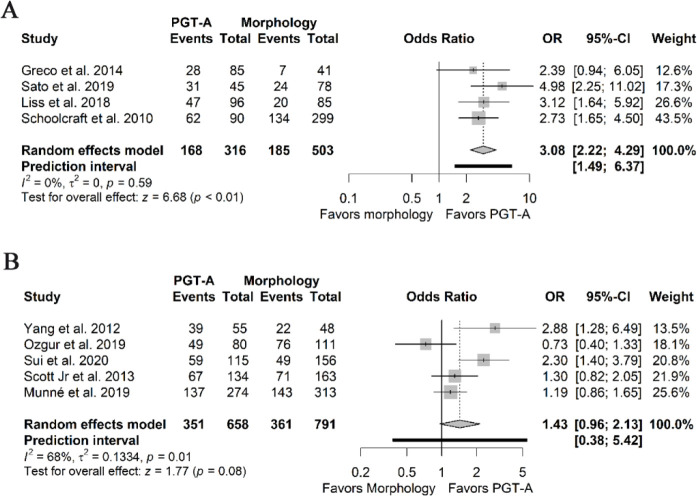
Clinical pregnancy per embryo transfer: Five RCTs and all NRSIs reported the CP rate. Most studies considered this outcome as ultrasonographic evidence of a fetal heartbeat at 4–6 weeks of gestation. The pooled ORs were 3.08 (95%CI 2.22–4.29, I2 0%) and 1.43 (95%PI 0.38–5.42, 95%CI 0.96–2.13, I2 68%), derived from the estimates of the NRSIs (Figure 4A) and RCTs (Figure 4B), respectively. The certainty of pooled estimates was rated as very low, due to serious risk of bias and inconsistency for the RCTs, and due to a very serious risk of bias and a plausible residual confounding which could have spuriously favored the use of PGT-A for the NRSIs. The main difference between the RCTs and NRSIs stemmed from the effect of maternal age on the PGT-A effectiveness estimation, as most RCTs controlled for this factor, either by excluding patients with AMA (35, 39) or by stratifying the randomization based on maternal age blocks (8, 16, 37).
Figure 4.
Forest plots of clinical pregnancy per embryo transfer. A) Pooled effect derived from NRSIs. B) Pooled effect derived from RCTs.
NRSIs: Non-randomized studies of interventions; RCTs: Randomized clinical trials; PGT-A: Preimplantation genetic testing for aneuploidy; OR: Odds ratio; 95%CI: 95% confidence interval
Subgroup and sensitivity analyses
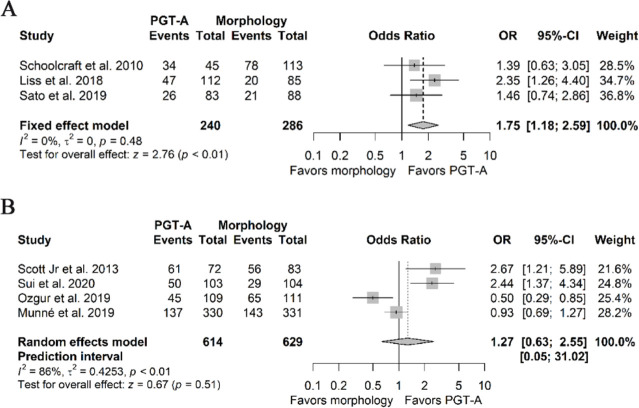
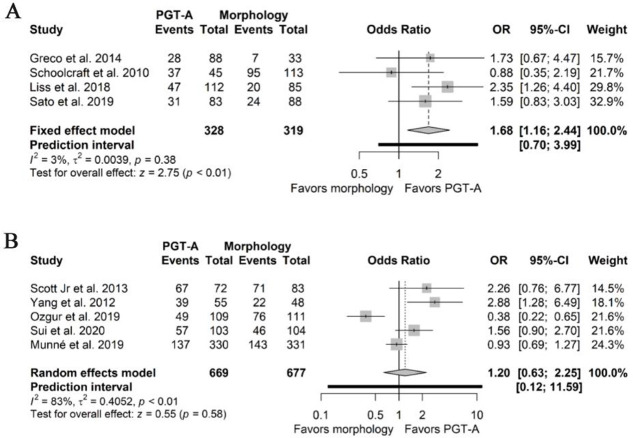
Change of outcome measure: Changing the outcome measure from embryo transfer to cycles had a significant impact on the estimates for LBR and CP of the NRSIs (Supplementary Files 4A and 5A). The pooled odds ratios for LBR changed significantly favoring the use of PGT-A (1.78 95%CI 1.18–2.59, I2 0%) and the effect estimates for CP remained significant, favoring PGT-A (1.68 95%CI 1.16–2.44, I2 3%). In both cases, the statistical heterogeneity was reduced, possibly because the difference between the number of embryos transferred between the groups across the studies was not considered in this calculation. Regarding the effect estimates of LBR and CP from the RCTs (Supplementary Files 4B and 5B), both remained non-significant (1.27 95%PI 0.05–31.02, 95%CI 0.63–2.25, I2 86% and 1.20 95%PI 0.12–11.59, 95%CI 0.63–2.25, I2 83%, respectively). Additionally, statistical heterogeneity increased in both estimates.
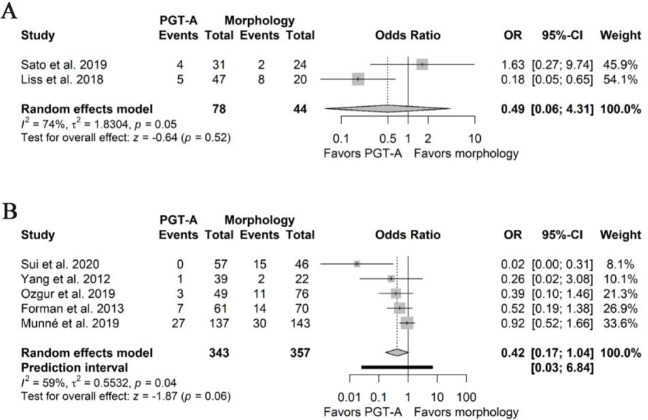
Pooled effects for miscarriage rates remained non-significant when changing the outcome measure from embryo transfer to clinical pregnancies (Supplementary File 6A and 6B), in both NRSIs and RCTs (0.49 95%CI 0.06–4.31, I2 74% and 0.42 95%PI 0.03–6.84, 95%CI 0.17–1.04, I2 59%, respectively). Statistical heterogeneity was relatively unaffected for the estimate derived from the NRSIs and was slightly increased for the one derived from the RCTs.
Advanced maternal age: Subgroup analyses for assessing the intervention effect on patients with AMA (defined as >35 years) were not possible as most studies included patients with a wide age range varying from 25 to 42 years. Only four studies were fit for indirect assessment of the effect of maternal age on reproductive outcomes, two which excluded patients with AMA (35, 39), and two with stratification of the population by maternal age (8, 37); however, only one study reported reproductive outcomes for each age group (37). Individual OR derived from the single study reporting the LBR and miscarriage rates per ET for patients with and without AMA showed a clear difference between subgroups, 1.74 (1.07–2.84) vs. 0.86 (0.56–1.34) for LBR in patients 35 years or older, and younger than 35 years, respectively and 0.71 (0.31–1.65) vs. 1.38 (0.66–2.92) for miscarriage rate in the same populations.
Through influence analyses for RCTs in pooling the LBR and CP outcomes, a significant contribution to heterogeneity and a large treatment effect were demonstrated in studies which excluded patients with AMA (8, 35). On the other hand, the influence of these studies was not as strong for the miscarriage rate estimate.
Excluding both influential studies from the LBR and CP meta-analyses resulted in estimates trending towards a superiority of PGT-A, though the estimates remained non-significant.
Recurrent pregnancy loss: Only one NRSI (36) and one RCT (40) reported reproductive outcomes in patients with recurrent pregnancy loss; thus, no clear conclusions on the possible modifying effect of this factor can be reached. Both studies found significant advantages of PGT-A in LBR and CP when applied in patients with RPL; having adjusted OR, the obtained values from NRSI for LBR and CP were 3.89 (1.16–13.1) and 5.14 (1.52–17.3), respectively. Additionally, adjusted risk ratios (RRs) reported by the RCT were 2.34 (1.59–3.45) and 1.63 (1.22–2.19) for LBR and CP, respectively. However, only the RCT found a significant effect of PGT-A on miscarriage rate, reporting an adjusted RR of 0.15 (0.04–0.64).
Finally, influence analysis of the pooled estimates from the RCTs demonstrated that the study by Sui et al., which only included patients with RPL, was the main influence on the overall heterogeneity of the models for LBR, CP, and miscarriage rate; therefore, excluding this study from the meta-analysis resulted in overall effect estimates trending towards no difference between the use of PGT-A and morphology alone.
Risk of bias and GRADE assessment: The results of the risk of bias assessment are summarized in figures 5A and 5B; one of the included RCTs was at a low risk of bias, (8) two had some concerns, (16, 35) and two were at high risk of bias (37, 39); the main areas of concerns were about randomization process, deviation from the intended interventions, and possible selection of reported results. Regarding the NRSI, two of the studies demonstrated a critical risk of bias, (33, 38) one showed the moderate risk, (36) and the other serious risk (34). The main affected domains across the studies were the lack of statistical or methodological control for critical sources of bias and the evidence of selection of reported results derived from multiple analyses. The table of findings summarizing the conclusions and rationale behind the GRADE assessment for each outcome can be found in the supplementary file 3.
Figure 5.
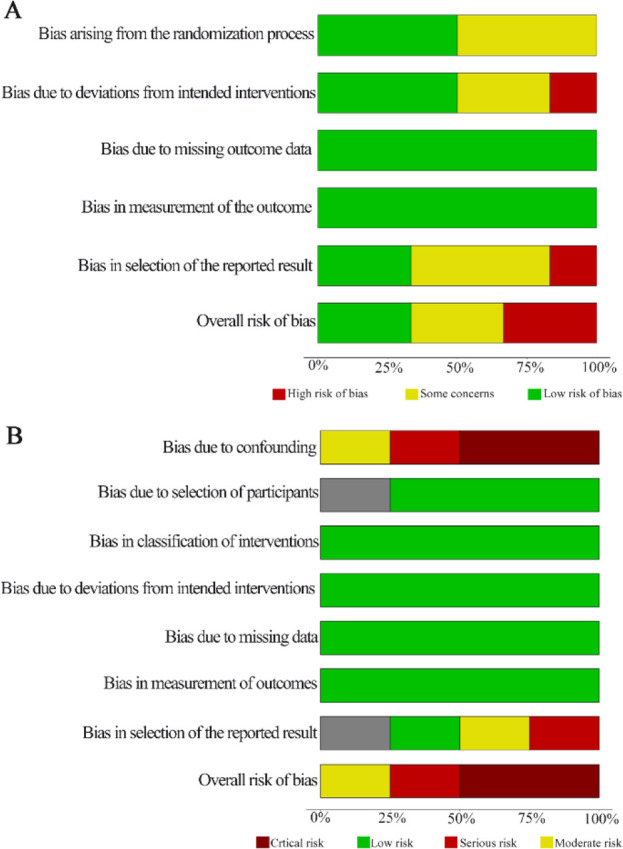
Graph summaries of risk of bias. A) ROB 2.0 results for RCTs. B) ROBINS-I results for NRSIs.
RCTs: Randomized controlled trials; NRSIs: Non-randomized studies of interventions
Discussion
Findings: In our study, no significant difference was found in reproductive outcomes after ART between women undergoing PGT-A for the selection of embryos versus morphology alone. This is a significant finding considering that the present study focused on patient-important reproductive outcomes. Interestingly, when analyzing the possible influence of AMA and a history of RPL on the pooled estimates through sensitivity and influence analyses, it was found that the possible reason behind the heterogeneity of our estimates could have been related to the inclusion of studies where couples of AMA were excluded. The tendency towards PGT-A superiority when excluding these studies indicates a greater benefit of chromosomal screening in patients with AMA, which relates to the increased incidence of aneuploidy in these patients (41, 42). This finding calls for further exploration of the effectiveness of PGT-A as it is related to maternal age.
Comparison with current evidence: The initial studies that evaluated the usefulness of selecting embryos according to their level of euploidy generated great expectations regarding the improvement of reproductive results (38). However, many of these studies are now being questioned due to methodological flaws in their outcome assessment (35, 43).
In 2015, a systematic review and meta-analysis concluded that embryo selection through CCS improves implantation rates in patients with normal ovarian reserve (44). That same year, another systematic review found a possible beneficial effect of embryo selection, using PGT-A, on implantation and clinical pregnancy rates in young patients (45). Five years later, a review argued the need for limiting the use of PGT-A in day-to-day clinical practice and leaving these techniques only for research purposes until their safety is well established (20). Our study reached a similar conclusion, which further supports the idea that the techniques used for PGT-A should only be used in certain situations where there is a clear beneficial potential. Additionally, the selection of our eligibility criteria compared with the criteria used in past reviews may be a stronger indicator of the lack of a true beneficial effect of the use of PGTA in the general population.
A systematic review and network meta-analysis was published in 2021 with the purpose of identifying which groups of patients could benefit the most from PGT-A; however, only two of their eleven references were about live-birth rate and most of them used implantation rates as a measure of effectiveness of ART, which is a practice that has been currently questioned (46, 47). In addition, the clinical pregnancy rate does not represent the final result that every couple who is treated with ART seeks to take a healthy newborn home. This, coupled with the small number and low quality of the RCTs pooled by previous systematic reviews, warrants caution in accepting PGT-A as a beneficial procedure.
Several reviews have been published since 2020, where authors called for taking a step back in the use of these tools in day-to-day clinical practice and instead leaving these techniques only for research purposes until their safety is well stablished (20, 48, 49). As shown by our findings, when pooled estimates are focused on patient-important outcomes, the effect of using PGT-A is less impactful, either in women with a good prognosis, (35, 37) patients of AMA (33) with a history of implantation failure, or women with RPL (36, 38).
One of the RCTs included in our synthesis used more modern technologies as compared with previous studies. In addition, their authors considered LBR as the measure for effectiveness. (35). However, they did not find improvement in LBR or a lower abortion rate when PGT-A was used. On the other hand, the results of another RCT published in the same year showed no difference in LBR or abortion rates with the use of PGT-A in patients with a history of implantation failure or those with a history of RPL. Notably, an improvement in LBR was found when they reported the results obtained per patient (36).
The results of other included RCTs showed no difference in the ongoing pregnancy and abortion rate, when selecting an embryo to transfer via NGS (37). Nonetheless, the authors reported better ongoing pregnancy rates in a group of women between 35 and 40 years when evaluating the results per embryo transfer. This further supports the plausible spurious advantage of PGT-A derived from previous inadequate analyses.
Strengths: Strengths of this meta-analysis include a strict inclusion criterion, and the inclusion of both experimental and observational designs which allowed a broad exploration of current evidence. The most important outcomes for couples were analyzed, which truly represent the objectives of ART and are in accordance with the expectations of our patients. The decisions behind our exclusions were based on recent findings which have identified the most useful techniques and procedures, as well as the harms caused by other techniques. Moreover, the selected studies were confined to those in which embryo biopsy was performed in the blastocyst stage as this is the most efficient and least harmful way to obtain enough cells for the correct chromosomal analysis (50, 51). In addition, the incidence of aneuploidy is lower in embryos in the blastocyst stage than in earlier stage embryos (52).
Limitations: Limitations of this review include the considerable risk of bias of included studies and small number of studies, which limited our capability to strongly recommend the use of PGTA for embryo selection. As shown by our wide prediction intervals, future estimates are expected to differ significantly as more high-quality studies become available. Furthermore, the use of PGT-A has been related to the possibility of reducing the number of multiple pregnancies. However, it could not be possible to determine the effect of PGT-A on the rate of multiple pregnancy due to lack of reporting.
Finally, the actual efficiency of PGT-A may be affected by the wide variety of available CCS technologies. Combining the results derived from different available techniques may have contributed significantly to the heterogeneity, reducing the true effect of PGT-A. Nonetheless, a formal analysis regarding the influence of CCS on our outcomes of interest was not possible due to the lack of well-reported studies exploring this possibility.
Conclusion
The lack of evidence pointing to a clear advantage of selecting embryos for transfer through PGT-A and its effect on LBR, CP, and miscarriage rates, compared to morphological assessment alone warrants caution when recommending this procedure for all couples seeking ART, as the current possible benefits may not justify the additional costs and risk linked to the procedures. Identifying the characteristics of patients who benefit from the use of PGT-A is paramount for a cost-effective application of new technologies. The findings of this meta-analysis suggest a selective positive effect of PGT-A on reproductive outcomes of patients with AMA and RPL. Further studies exploring the association of these maternal factors and PGT-A efficacy are needed to confirm the findings.
Acknowledgement
We would like to thank all members from the Knowledge and Evaluation Research (KER) Unit from the School of Medicine of the Universidad Autónoma de Nuevo León, Monterrey, México, for their guidance and technical support in conducting this meta-analysis.
Footnotes
Conflict of Interest
Authors declare no conflict of interest.
Supplementary
Supplementary File 4.
Forest plots of live birth rate per cycle. A) Pooled effect derived from NRSIs. B) Pooled effect derived from RCTs.
NRSIs: Non-randomized studies of interventions; RCTs: Randomized clinical trials; PGT-A: Preimplantation genetic testing for aneuploidy; OR: Odds ratio; 95%CI: 95% confidence interval
Supplementary File 5.
Forest plots of clinical pregnancy per cycle. A) Pooled effect derived from NRSIs. B) Pooled effect derived from RCTs.
NRSIs: Non-randomized studies of interventions; RCTs: Randomized clinical trials; PGT-A: Preimplantation genetic testing for aneuploidy; OR: Odds ratio; 95%CI: 95% confidence interval
Supplementary File 6.
Forest plots of miscarriage rate per clinical pregnancy. A) Pooled effect derived from NRSIs. B) Pooled effect derived from RCTs.
NRSIs: Non-randomized studies of interventions; RCTs: Randomized clinical trials; PGT-A: Preimplantation genetic testing for aneuploidy; OR: Odds ratio; 95%CI: 95% confidence interval
Supplementary File 1.
PRISMA checklist
| Section and topic | Item # | Checklist item | Location where item is reported |
|---|---|---|---|
| Title | |||
| Title | 1 | Identify the report as a systematic review. | 1 |
| Abstract | |||
| Abstract | 2 | See the PRISMA 2020 for abstracts checklist. | 3 |
| Introduction | |||
| Rationale | 3 | Describe the rationale for the review in the context of existing knowledge. | 4 |
| Objectives | 4 | Provide an explicit statement of the objective(s) or question(s) the review addresses. | 5 |
| Methods | |||
| Eligibility criteria | 5 | Specify the inclusion and exclusion criteria for the review and how studies were grouped for the syntheses. | 5–6 |
| Information sources | 6 | Specify all databases, registers, websites, organisations, reference lists and other sources searched or consulted to identify studies. Specify the date when each source was last searched or consulted. | 6 |
| Search strategy | 7 | Present the full search strategies for all databases, registers and websites, including any filters and limits used. | 6 |
| Selection process | 8 | Specify the methods used to decide whether a study met the inclusion criteria of the review, including how many reviewers screened each record and each report retrieved, whether they worked independently, and if applicable, details of automation tools used in the process. | 6 |
| Data collection process | 9 | Specify the methods used to collect data from reports, including how many reviewers collected data from each report, whether they worked independently, any processes for obtaining or confirming data from study investigators, and if applicable, details of automation tools used in the process. | 7 |
| Data items | 10 a | List and define all outcomes for which data were sought. Specify whether all results that were compatible with each outcome domain in each study were sought (e.g. for all measures, time points, analyses ), and if not, the methods used to decide which results to collect. | 7 |
| 10 b | List and define all other variables for which data were sought (e.g. participant and intervention characteristics, funding sources ). Describe any assumptions made about any missing or unclear information. | 7 | |
| Study risk of bias assessment | 11 | Specify the methods used to assess risk of bias in the included studies, including details of the tool(s) used, how many reviewers assessed each study and whether they worked independently, and if applicable, details of automation tools used in the process. | 7–8 |
| Effect measures | 12 | Specify for each outcome the effect measure(s) (e.g. risk ratio, mean difference ) used in the synthesis or presentation of results. | 8 |
| 13 a | Describe the processes used to decide which studies were eligible for each synthesis (e.g. tabulating the study intervention characteristics and comparing against the planned groups for each synthesis (item #5 )). | 8 | |
| 13 b | Describe any methods required to prepare the data for presentation or synthesis, such as handling missing summary statistics, or data conversions. | 8 | |
| Synthesis methods | 13 c | Describe any methods used to tabulate or visually display results of individual studies and syntheses. | 8 |
| 13 d | Describe any methods used to synthesize results and provide a rationale for the choice(s). If meta-analysis was performed, describe the model(s), method(s) to identify the presence and extent of statistical heterogeneity, and software package(s) used. | 8 | |
| 13 e | Describe any methods used to explore possible causes of heterogeneity among study results (e.g. subgroup analysis, meta-regression ). | 9 | |
| 13 f | Describe any sensitivity analyses conducted to assess robustness of the synthesized results. | 9 | |
| Reporting bias assessment | 14 | Describe any methods used to assess risk of bias due to missing results in a synthesis (arising from reporting biases). | NR |
| Certainty assessment | 15 | Describe any methods used to assess certainty (or confidence) in the body of evidence for an outcome. | 7–8 |
| Results | |||
| Study selection | 16 a | Describe the results of the search and selection process, from the number of records identified in the search to the number of studies included in the review, ideally using a flow diagram. | 9 |
| 16 b | Cite studies that might appear to meet the inclusion criteria, but which were excluded, and explain why they were excluded. | 9 | |
| Study characteristics | 17 | Cite each included study and present its characteristics. | 9 |
| Risk of bias in studies | 18 | Present assessments of risk of bias for each included study. | 13 |
| Results of Individual studies | 19 | For all outcomes, present: (a) summary statistics for each group (where appropriate) and (b) an effect estimate and its precision (e.g. confidence/credible interval ), ideally using structured tables or plots for each study. | 10–13 |
| 20 a | For each synthesis, briefly summarize the characteristics and risk of bias among contributing studies. | 10–13 | |
| Results of syntheses | 20 b | Present results of all statistical syntheses conducted. If meta-analysis was done, present for each the summary estimate and its precision (e.g. confidence/credible interval ) and measures of statistical heterogeneity. If comparing groups, describe the direction of the effect. | 10–13 |
| 20 c | Present results of all investigations of possible causes of heterogeneity among study results. | 10–13 | |
| 20 d | Present results of all sensitivity analyses conducted to assess the robustness of the synthesized results. | 11–13 | |
| Reporting biases | 21 | Present assessments of risk of bias due to missing results (arising from reporting biases) for each synthesis assessed. | NR |
| Certainty of evidence | 22 | Present assessments of certainty (or confidence) in the body of evidence for each outcome assessed. | 10–13 |
| Discussion | |||
| Discussion | 23 a | Provide a general interpretation of the results in the context of other evidence. | 14 |
| 23 b | Discuss any limitations of the evidence in the review. | 17 | |
| 23 c | Discuss any limitations of the review processes. | 17 | |
| 23 d | Discuss implications of the results for practice, policy, and future research. | 17–18 | |
| Other information | |||
| Registration and protocol | 24 a | Provide registration information for the review, including register name and registration number, or state that the review was not registered. | 5 |
| 24 b | Indicate where the review protocol can be accessed, or state that a protocol was not prepared. | 5 | |
| 24 c | Describe and explain any amendments to information provided at registration or in the protocol. | NR | |
| Support | 25 | Describe sources of financial or non-financial support for the review, and the role of the funders or sponsors in the review. | 19 |
| Competing interests | 26 | Declare any competing interests of review authors. | 19 |
| Availability of data, code, and other materials | 27 | Report which of the following are publicly available and where they can be found: template data collection forms, data extracted from included studies, data used for all analyses, analytic code, and any other materials used in the review. | NR |
From: Page MJ, McKenzie JE, Bossuyt PM, Boutron I, Hoffmann TC, Mulrow CD, et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. BMJ 2021; 372: n71. doi: 10.1136/bmj.n71
For more information, visit: http://www.prisma-statement.org/
Supplementary File 2.
Full search strategy
| # 4 | 207 | #3 AND #2 AND #1 Indexes=SCI-EXPANDED, ESCI Timespan=All years |
| # 3 | 17, 632 | TS = (“single embryo transfer” or “embryo transfer” or “embryo transplantation” or “transfer, embryo” or “blastocyst transfer” or “embryo transfers” or “transfers, embryo” or “tubal embryo stage transfer” or “tubal embryo transfer”) Indexes=SCI-EXPANDED, ESCI Timespan=All years |
| # 2 | 42, 094 | TS = (“sperm injections, intracytoplasmic “ or “injection, intracytoplasmic sperm” or “injections, intracytoplasmic sperm” or “injections, sperm, intracytoplasmic” or “intracytoplasmic sperm injection” or “intracytoplasmic sperm injections” or “sperm injection, intracytoplasmic” or “intracytoplasmic sperm injection” or “ICSI” or “intra cytoplasmic sperm injection “ or “babies, test-tube” or “baby, test-tube” or “fertilization, test-tube” or “fertilizations in vitro” or “fertilization in vitro” or “fertilizations, test-tube” or “extracorporeal fertilization” or “in vitro fertilization” or “in vitro fertilization” or “IVF (in vitro fertilization)” or “in vitro fertilizations” or “test tube babies” or “test tube fertilization” or “test-tube babies” or “test-tube baby” or “test-tube fertilization” or “test-tube fertilizations”) Indexes=SCI-EXPANDED, ESCI Timespan=All years |
| # 1 | 242, 623 | TS = (“polymorphism, single nucleotide” or “nucleotide polymorphism, single” or “nucleotide polymorphisms, single” or “polymorphisms, single nucleotide” or “SNPs” or “single nucleotide polymorphism” or “single nucleotide polymorphisms” or “single nucleotid polimorphism” or “array based comparative genomic hybridization” or “array comparative genomic hybridization” or “array-based comparative genomic hybridization” or “comparative genome hybridization” or “comparative genome hybridizations” or “comparative genomic hybridizations” or “genome hybridization, comparative” or “genome hybridizations, comparative” or “genomic hybridization, comparative” or “genomic hybridizations, comparative” or “hybridization, comparative genome” or “hybridization, comparative genomic” or “hybridizations, comparative genome” or “hybridizations, comparative genomic” or “comparative genomic hybridization” or “metaphase comparative genomic hybridization” or “next generation secuencing” or “DNA sequencing, high-throughput” or “deep sequencing” or “deep sequencings” or “high throughput DNA sequencing” or “high throughput nucleotide sequencing” or “high throughput RNA sequencing” or “high-throughput DNA sequencing” or “high-throughput RNA sequencing” or “Illumina sequencing” or “Ion proton sequencing” or “Ion torrent sequencing” or “massively parallel sequencing” or “massively-parallel sequencing” or “next generation sequencing” or “next-generation sequencing” or “nucleotide sequencing, high-throughput” or “pyrosequencing” or “RNA sequencing, high-throughput” or “sequencing, deep” or “sequencing, high-throughput DNA” or “sequencing, high-throughput nucleotide” or “sequencing, high-throughput RNA” or “sequencing, Illumina” or “sequencing, Ion proton” or “sequencing, Ion torrent” or “sequencing, massively-parallel” or “sequencing, next-generation “ or “high-throughput nucleotide sequencing” or “quantitative PCR” or “quantitative polymerase chain reaction” or “quantitative polymerase chain reaction”) Indexes=SCI-EXPANDED, ESCI Timespan=All years |
|
OVID
Ovid MEDLINE(R) ALL 1946 to July 31, 2020 Embase 1974 to 2020 July 31 | ||
|
| ||
| 1 | (“polymorphism, single nucleotide” or “nucleotide polymorphism, single” or “nucleotide polymorphisms, single” or “polymorphisms, single nucleotide” or “SNPs” or “single nucleotide polymorphism” or “single nucleotide polymorphisms” or “single nucleotide polymorphisms” or “array based comparative genomic hybridization” or “array comparative genomic hybridization” or “array-based comparative genomic hybridization” or “comparative genome hybridization” or “comparative genome hybridizations” or “comparative genomic hybridizations” or “genome hybridization, comparative” or “genome hybridizations, comparative” or “genomic hybridization, comparative” or “genomic hybridizations, comparative” or “hybridization, comparative genome” or “hybridization, comparative genomic” or “hybridizations, comparative genome” or “hybridizations, comparative genomic” or “comparative genomic hybridization” or “metaphase comparative genomic hybridization” or “next generation sequencing” or “DNA sequencing, high-throughput” or “deep sequencing” or “deep sequencings” or “high throughput DNA sequencing” or “high throughput nucleotide sequencing” or “high throughput RNA sequencing” or “high-throughput DNA sequencing” or “high-throughput RNA sequencing” or “Illumina sequencing” or “Ion proton sequencing” or “Ion torrent sequencing” or “massively parallel sequencing” or “massively-parallel sequencing” or “next generation sequencing” or “next-generation sequencing” or “nucleotide sequencing, high-throughput” or “pyrosequencing” or “RNA sequencing, high-throughput” or “sequencing, deep” or “sequencing, high-throughput DNA” or “sequencing, high-throughput nucleotide” or “sequencing, high-throughput RNA” or “sequencing, Illumina” or “sequencing, Ion proton” or “sequencing, Ion torrent” or “sequencing, massively-parallel” or “sequencing, next-generation” or “high-throughput nucleotide sequencing” or “quantitative PCR” or “quantitative polymerase chain reaction” or “quantitative polymerase chain reaction”). mp. [mp=ti, ab, hw, tn, ot, dm, mf, dv, kw, fx, dq, nm, kf, ox, px, rx, an, ui, sy] | 661499 |
| 2 | (“sperm injections, intracytoplasmic” or “injection, intracytoplasmic sperm” or “injections, intracytoplasmic sperm” or “injections, sperm, intracytoplasmic” or “intracytoplasmic sperm injection” or “intracytoplasmic sperm injections” or “sperm injection, intracytoplasmic” or “intracytoplasmic sperm injection” or “ICSI” or “intra cytoplasmic sperm injection” or “babies, test-tube” or “baby, test-tube” or “fertilization, test-tube” or “fertilizations in vitro” or “fertilization in vitro” or “fertilizations, test-tube” or “extracorporeal fertilization” or “in vitro fertilization” or “in vitro fertilization” or “IVF (in vitro fertilization)” or “in vitro fertilizations” or “test tube babies” or “test tube fertilization” or “test-tube babies” or “test-tube baby” or “test-tube fertilization” or “test-tube fertilizations”). mp. [mp=ti, ab, hw, tn, ot, dm, mf, dv, kw, fx, dq, nm, kf, ox, px, rx, an, ui, sy] | 118432 |
| 3 | (“single embryo transfer” or “embryo transfer” or “embryo transplantation” or “transfer, embryo” or “blastocyst transfer” or “embryo transfers” or “transfers, embryo” or “tubal embryo stage transfer” or “tubal embryo transfer”).mp. [mp=ti, ab, hw, tn, ot, dm, mf, dv, kw, fx, dq, nm, kf, ox, px, rx, an, ui, sy] | 55491 |
| 4 | 1 and 2 and 3 Embase <1974 to 2020 July 31> Ovid MEDLINE(R) ALL <1946 to July 31, 2020> |
781 533 248 |
Supplementary File 3.
GRADE table of findings
| Certainty assessment | No of patients | Effect | Certainty | Importance | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| No. of studies | Study design | Risk of bias | Inconsistency | Indirectness | Imprecision | Other considerations | Preimplantation genetic diagnosis | Morphology assessment alone | Relative (95% CI) | Absolute (95% CI) | ||
| Live birth rate per embryo transfer | ||||||||||||
| 5 | Randomized trials | Serious a | Serious b | Not serious | Serious c | None | 321/603 (53.2%) | 315/743 (42.4%) | OR 1.66 (0.98 to 2.83) | 13 more per 100 (from 0 fewer to 25 more) | ⊕◯◯◯ Very low |
Critical |
| Live birth rate per embryo transfer | ||||||||||||
| 3 | Observational studies | Very serious d | Serious b | Not serious | Serious e | All plausible residual confounding would suggest spurious effect, while no effect was observed | 107/231 (46.3%) | 247/462 (53.5%) | OR 1.45 (0.24 to 8.78) | 9 more per 100 (from 32 fewer to 38 more) | ⊕◯◯◯ Very low |
Critical |
| Clinical pregnancy per embryo transfer | ||||||||||||
| 5 | Randomized trials | Serious a | Serious f | Not serious | Not serious | None | 351/658 (53.1%) | 361/791 (45.6%) | OR 1.43 (0.96 to 2.13) | 9 more per 100 (from 1 fewer to 18 more) | ⊕⊕◯◯ Low |
Important |
| Clinical pregnancy per embryo transfer | ||||||||||||
| 4 | Observational studies | Very serious d | Not serious | Not serious | Serious c | All plausible residual confounding would suggest spurious effect, while no effect was observed | 168/316 (53.2%) | 185/503 (36.8%) | OR 3.08 (2.22 to 4.29) | 27 more per 100 (from 20 more to 35 more) | ⊕⊕◯◯ low |
Important |
| Miscarriage rate per embryo transfer | ||||||||||||
| 5 | Randomized trials | Serious a | Serious f | Not serious | Serious e | None | 40/610 (6.6%) | 75/800 (9.4%) | OR 0.57 (0.27 to 1.21) | 4 fewer per 100 (from 7 fewer to 2 more) | ⊕◯◯◯ very low |
Important |
| Miscarriage rate per embryo transfer | ||||||||||||
| 2 | Observational studies | Very serious d | Very serious b | Not serious | Very serious e | None | 9/141 (6.4%) | 10/163 (6.1%) | OR 1.25 (0.19 to 8.33) | 1 more per 100 (from 5 fewer to 29 more) | ⊕◯◯◯ very low |
Important |
CI: Confidence interval; OR: Odds ratio
Most included studies were between some concerns and high risk of bias;
There is a high level of heterogeneity and poor overlap of 95% CI;
There is a small sample of embryo transfer, which makes the estimate imprecise;
Included studies were all between moderate and critical risk of bias;
Imprecise 95% CI;
Poor overlap of 95% CI
References
- 1.Solim ZNN, Stavers KC, Mustafa F, Lim R. Assessing characteristics which influence the success rate of in vitro fertilization. J Basic Clin Reprod Sci. 2021;10(3). [Google Scholar]
- 2.Eskew AM, Jungheim ES. A history of developments to improve in vitro fertilization. Mo Med. 2017;114(3):156–9. [PMC free article] [PubMed] [Google Scholar]
- 3.Theobald R, SenGupta S, Harper J. The status of preimplantation genetic testing in the UK and USA. Hum Reprod. 2020;35(4):986–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harper JC, Boelaert K, Geraedts J, Harton G, Kearns WG, Moutou C, et al. ESHRE PGD consortium data collection V: cycles from january to december 2002 with pregnancy follow-up to october 2003. Hum Reprod. 2006;21(1):3–21. [DOI] [PubMed] [Google Scholar]
- 5.Nagaoka SI, Hassold TJ, Hunt PA. Human aneuploidy: mechanisms and new insights into an age-old problem. Nat Rev Genet. 2012;13(7):493–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hassold T, Hunt P. To err (meiotically) is human: the genesis of human aneuploidy. Nat Rev Genet. 2001;2(4):280–91. [DOI] [PubMed] [Google Scholar]
- 7.Verlinsky Y, Cieslak J, Freidine M, Ivakhnenko V, Wolf G, Kovalinskaya L, et al. Pregnancies following pre-conception diagnosis of common aneuploidies by fluorescent in-situ hybridization. Hum Reprod. 1995;10(7):1923–7. [DOI] [PubMed] [Google Scholar]
- 8.Forman EJ, Hong KH, Ferry KM, Tao X, Taylor D, Levy B, et al. In vitro fertilization with single euploid blastocyst transfer: a randomized controlled trial. Fertil Steril. 2013;100(1):100–7.e1. [DOI] [PubMed] [Google Scholar]
- 9.Sermon K, Capalbo A, Cohen J, Coonen E, De Rycke M, De Vos A, et al. The why, the how and the when of PGS 2.0: current practices and expert opinions of fertility specialists, molecular biologists, and embryologists. Mol Hum Reprod. 2016;22(8): 845–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cornelisse S, Zagers M, Kostova E, Fleischer K, van Wely M, Mastenbroek S. Preimplantation genetic testing for aneuploidies (abnormal number of chromosomes) in in vitro fertilisation. Cochrane Database Syst Rev. 2020;9(9):CD005291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Capalbo A, Rienzi L, Cimadomo D, Maggiulli R, Elliott T, Wright G, et al. Correlation between standard blastocyst morphology, euploidy and implantation: an observational study in two centers involving 956 screened blastocysts. Hum Reprod. 2014;29(6):1173–81. [DOI] [PubMed] [Google Scholar]
- 12.Scott RTJ, Upham KM, Forman EJ, Zhao T, Treff NR. Cleavage-stage biopsy significantly impairs human embryonic implantation potential while blastocyst biopsy does not: a randomized and paired clinical trial. Fertil Steril. 2013;100(3):624–30. [DOI] [PubMed] [Google Scholar]
- 13.Fiorentino F, Spizzichino L, Bono S, Biricik A, Kokkali G, Rienzi L, et al. PGD for reciprocal and Robertsonian translocations using array comparative genomic hybridization. Hum Reprod. 2011;26(7):1925–35. [DOI] [PubMed] [Google Scholar]
- 14.Schoolcraft WB, Treff NR, Stevens JM, Ferry K, Katz-Jaffe M, Scott RTJ. Live birth outcome with trophectoderm biopsy, blastocyst vitrification, and single-nucleotide polymorphism microarray-based comprehensive chromosome screening in infertile patients. Fertil Steril. 2011;96(3):638–40. [DOI] [PubMed] [Google Scholar]
- 15.Wells D. Next-generation sequencing: the dawn of a new era for preimplantation genetic diagnostics. Fertil Steril. 2014;101(5):1250–1. [DOI] [PubMed] [Google Scholar]
- 16.Scott RTJ, Upham KM, Forman EJ, Hong KH, Scott KL, Taylor D, et al. Blastocyst biopsy with comprehensive chromosome screening and fresh embryo transfer significantly increases in vitro fertilization implantation and delivery rates: a randomized controlled trial. Fertil Steril. 2013;100(3): 697–703. [DOI] [PubMed] [Google Scholar]
- 17.Coetsier T, Dhont M. Avoiding multiple pregnancies in in-vitro fertilization: who’s afraid of single embryo transfer? Hum Reprod. 1998;13(10):2663–4. [DOI] [PubMed] [Google Scholar]
- 18.Maxwell SM, Grifo JA. Should every embryo undergo preimplantation genetic testing for aneuploidy? A review of the modern approach to in vitro fertilization. Best Pract Res Clin Obstet Gynaecol. 2018;53:38–47. [DOI] [PubMed] [Google Scholar]
- 19.Mantravadi K, Sr., Debnath S, Sanjay Relekar NR, Rao DG., SrPGT-A doesn’t seem to benefit recurrent implantation failure couples to optimize live births. Fertil Steril. 2020;114(3):e436. [Google Scholar]
- 20.Sciorio R, Dattilo M. PGT-A preimplantation genetic testing for aneuploidies and embryo selection in routine ART cycles: Time to step back? Clin Genet. 2020;98(2):107–15. [DOI] [PubMed] [Google Scholar]
- 21.Neumann K, Sermon K, Bossuyt P, Goossens V, Geraedts J, Traeger-Synodinos J, et al. An economic analysis of preimplantation genetic testing for aneuploidy by polar body biopsy in advanced maternal age. BJOG. 2020;127(6):710–8. [DOI] [PubMed] [Google Scholar]
- 22.Moher D, Liberati A, Tetzlaff J, Altman DG, PRISMA Group . Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. BMJ. 2009;339:b2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McGinn T, Wyer PC, Newman TB, Keitz S, Leipzig R, For GG. Tips for learners of evidencebased medicine: 3. Measures of observer variability (kappa statistic). CMAJ. 2004;171(11):1369–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Duffy JMN, Bhattacharya S, Bhattacharya S, Bofill M, Collura B, Curtis C, et al. Standardizing definitions and reporting guidelines for the infertility core outcome set: an international consensus development study. Fertil Steril. 2021;115(1):201–12. [DOI] [PubMed] [Google Scholar]
- 25.Sterne JA, Savović J, Page MJ, Elbers RG, Blencowe NS, Boutron I, et al. RoB 2: a revised tool for assessing risk of bias in randomised trials. BMJ. 2019;366:l4898. [DOI] [PubMed] [Google Scholar]
- 26.Sterne JAC, Hernán MA, Reeves BC, Savović J, Berkman ND, Viswanathan M, et al. ROBINS-I: a tool for assessing risk of bias in non-randomised studies of interventions. BMJ. 2016;355:i4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schünemann H, Brożek J, Guyatt G, Oxman A. GRADE handbook for grading quality of evidence and strength of recommendations. Updated October 2013. The GRADE Working Group., 2013; 2013. [Google Scholar]
- 28.Mcmaster university . GRADEpro GDT: GRAD-Epro guideline development tool [Software] [Internet]. Evidence Prime, Inc.; 2020. Available from: gradepro.org [Google Scholar]
- 29.Higgins JPT, Green S. (editors).Cochrane Handbook for Systematic Reviews of Interventions Version 5.1.0 [updated March 2011]. The Cochrane Collaboration, 2011. Available from www.handbook.cochrane.org [Google Scholar]
- 30.Viechtbauer W, Cheung MWL. Outlier and influence diagnostics for meta-analysis. Res Synth Methods. 2010;1(2):112–25. [DOI] [PubMed] [Google Scholar]
- 31.Harrer M, Cuijpers P, Furukawa TA, Ebert DD. Doing meta-analysis with R: A hands-on guide. 1st ed. New York: Prot Lab Erlangen; 2019. 500 p. [Google Scholar]
- 32.Higgins JPT, Thompson SG, Spiegelhalter DJ. A re-evaluation of random-effects meta-analysis. J R Stat Soc Ser A Stat Soc. 2009;172(1):137–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schoolcraft WB, Fragouli E, Stevens J, Munne S, Katz-Jaffe MG, Wells D. Clinical application of comprehensive chromosomal screening at the blastocyst stage. Fertil Steril. 2010;94(5):1700–6. [DOI] [PubMed] [Google Scholar]
- 34.Liss J, Pastuszek E, Pukszta S, Hoffmann E, Kuczynski W, Lukaszuk A, et al. Effect of next-generation sequencing in preimplantation genetic testing on live birth ratio. Reprod Fertil Dev. 2018;30(12):1720–7. [DOI] [PubMed] [Google Scholar]
- 35.Ozgur K, Berkkanoglu M, Bulut H, Yoruk GDA, Candurmaz NN, Coetzee K. Single best euploid versus single best unknown-ploidy blastocyst frozen embryo transfers: a randomized controlled trial. J Assist Reprod Genet. 2019;36(4):629–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sato T, Sugiura-Ogasawara M, Ozawa F, Yamamoto T, Kato T, Kurahashi H, et al. Preimplantation genetic testing for aneuploidy: a comparison of live birth rates in patients with recurrent pregnancy loss due to embryonic aneuploidy or recurrent implantation failure. Hum Reprod. 2019;34(12):2340–8. [DOI] [PubMed] [Google Scholar]
- 37.Munné S, Kaplan B, Frattarelli JL, Child T, Nakhuda G, Shamma FN, et al. Preimplantation genetic testing for aneuploidy versus morphology as selection criteria for single frozen-thawed embryo transfer in good-prognosis patients: a multicenter randomized clinical trial. Fertil Steril. 2019;112(6): 1071–9.e7. [DOI] [PubMed] [Google Scholar]
- 38.Greco E, Bono S, Ruberti A, Lobascio AM, Greco P, Biricik A, et al. Comparative genomic hybridization selection of blastocysts for repeated implantation failure treatment: a pilot study. Biomed Res Int. 2014;2014:457913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang Z, Liu J, Collins GS, Salem SA, Liu X, Lyle SS, et al. Selection of single blastocysts for fresh transfer via standard morphology assessment alone and with array CGH for good prognosis IVF patients: results from a randomized pilot study. Mol Cytogenet. 2012;5(1):24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sui YL, Lei CX, Ye JF, Fu J, Zhang S, Li L, et al. In vitro fertilization with single-Nucleotide polymorphism microarray-based preimplantation genetic testing for aneuploidy significantly improves clinical outcomes in infertile women with recurrent pregnancy loss: A randomized controlled trial. Reprod Dev Med. 2020;4(1):32–41. [Google Scholar]
- 41.Munné S, Alikani M, Tomkin G, Grifo J, Cohen J. Embryo morphology, developmental rates, and maternal age are correlated with chromosome abnormalities. Fertil Steril. 1995;64(2):382–91. [PubMed] [Google Scholar]
- 42.Rubio C, Simón C, Vidal F, Rodrigo L, Pehlivan T, Remohí J, et al. Chromosomal abnormalities and embryo development in recurrent miscarriage couples. Hum Reprod. 2003;18(1):182–8. [DOI] [PubMed] [Google Scholar]
- 43.Practice committees of the American society for reproductive medicine and the society for assisted reproductive technology . The use of preimplantation genetic testing for aneuploidy (PGT-A): a committee opinion. Fertil Steril. 2018;109(3):429–36. [DOI] [PubMed] [Google Scholar]
- 44.Dahdouh EM, Balayla J, García-Velasco JA. Comprehensive chromosome screening improves embryo selection: a meta-analysis. Fertil Steril. 2015; 104(6):1503–12. [DOI] [PubMed] [Google Scholar]
- 45.Lee E, Illingworth P, Wilton L, Chambers GM. The clinical effectiveness of preimplantation genetic diagnosis for aneuploidy in all 24 chromosomes (PGD-A): systematic review. Hum Reprod. 2015; 30(2):473–83. [DOI] [PubMed] [Google Scholar]
- 46.Griesinger G. Beware of the ’implantation rate’! Why the outcome parameter “implantation rate” should be abandoned from infertility research. Hum Reprod. 2016;31(2):249–51. [DOI] [PubMed] [Google Scholar]
- 47.Simopoulou M, Sfakianoudis K, Maziotis E, Tsioulou P, Grigoriadis S, Rapani A, et al. PGT-A: who and when? a systematic review and network meta-analysis of RCTs. J Assist Reprod Genet. 2021;38(8):1939–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mochizuki L, Gleicher N. The PGS/PGT-A controversy in IVF addressed as a formal conflict resolution analysis. J Assist Reprod Genet. 2020;37(3):677–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gleicher N, Patrizio P, Brivanlou A. Preimplantation Genetic Testing for Aneuploidy – a Castle Built on Sand. Trends Mol Med. 2021;27(8):731–42. [DOI] [PubMed] [Google Scholar]
- 50.McArthur SJ, Leigh D, Marshall JT, de Boer KA, Jansen RPS. Pregnancies and live births after trophectoderm biopsy and preimplantation genetic testing of human blastocysts. Fertil Steril. 2005; 84(6):1628–36. [DOI] [PubMed] [Google Scholar]
- 51.Kokkali G, Traeger-Synodinos J, Vrettou C, Stavrou D, Jones GM, Cram DS, et al. Blastocyst biopsy versus cleavage stage biopsy and blastocyst transfer for preimplantation genetic diagnosis of beta-thalassaemia: a pilot study. Hum Reprod. 2007;22(5):1443–9. [DOI] [PubMed] [Google Scholar]
- 52.Dekel-Naftali M, Aviram-Goldring A, Litmanovitch T, Shamash J, Yonath H, Hourvitz A, et al. Chromosomal integrity of human preimplantation embryos at different days post fertilization. J Assist Reprod Genet. 2013;30(5):633–48. [DOI] [PMC free article] [PubMed] [Google Scholar]