Abstract
Forest soils provide a multitude of habitats for diverse communities of bacteria. In this study, we selected three tropical forests in Kenya to determine the diversity and community structure of soil bacteria inhabiting these regions. Kakamega and Irangi are rainforests, whereas Gazi Bay harbors mangrove forests. The three natural forests occupy different altitudinal zones and differ in their environmental characteristics. Soil samples were collected from a total of 12 sites and soil physicochemical parameters for each sampling site were analyzed. We used an amplicon-based Illumina high-throughput sequencing approach. Total community DNA was extracted from individual samples using the phenol-chloroform method. The 16S ribosomal RNA gene segment spanning the V4 region was amplified using the Illumina MiSeq platform. Diversity indices, rarefaction curves, hierarchical clustering, principal component analysis (PCA), and non-metric multidimensional scaling (NMDS) analyses were performed in R software. A total of 13,410 OTUs were observed at 97% sequence similarity. Bacterial communities were dominated by Proteobacteria, Bacteroidetes, Firmicutes, Actinobacteria, and Acidobacteria in both rainforest and mangrove sampling sites. Alpha diversity indices and species richness were higher in Kakamega and Irangi rainforests compared to mangroves in Gazi Bay. The composition of bacterial communities within and between the three forests was also significantly differentiated (R = 0.559, p = 0.007). Clustering in both PCA and NMDS plots showed that each sampling site had a distinct bacterial community profile. The NMDS analysis also indicated that soil EC, sodium, sulfur, magnesium, boron, and manganese contributed significantly to the observed variation in the bacterial community structure. Overall, this study demonstrated the presence of diverse taxa and heterogeneous community structures of soil bacteria inhabiting three tropical forests of Kenya. Our results also indicated that variation in soil chemical parameters was the major driver of the observed bacterial diversity and community structure in these forests.
Keywords: Tropical forests, Soil bacteria, 16S rRNA, Diversity, Community structure, Soil chemical parameters
Tropical forests; Soil bacteria; 16S rRNA; Diversity; Community structure; Soil chemical parameters.
1. Introduction
Forest soils are among the most biologically diverse ecosystems on earth (Amoo and Babalola, 2019). They harbor a myriad of microbial communities that perform critical ecological functions such as decomposition of organic matter, nitrogen fixation, and cycling of carbon and other soil nutrients (Lladó et al., 2017). Among the forest soil microbiota, bacterial communities encompass a large taxonomic diversity and form an integral part of these ecosystems (Lladó et al., 2017; Wei et al., 2018). The most abundant phyla are the Proteobacteria, Bacteroidetes, Actinobacteria, Acidobacteria, and Firmicutes (Lauber et al., 2009). Members of these phyla are recognized as the key drivers of ecological systems, hence their level of dominance and composition in a community serve as key biological indicators of the main soil functions (Lladó et al., 2017; Praeg et al., 2019).
Typically, both the diversity and community structure of soil bacteria are greatly influenced by the environmental characteristics of their habitat (Shen et al., 2020; Wei et al., 2018; Z. Xia et al., 2016). Forest soils, moreover, provide a wide range of bacterial habitats associated with the heterogeneity of physicochemical parameters and aboveground vegetation (Chandra et al., 2016; Yavitt et al., 2021). Spatial variations in these environmental factors create specific niches within a forest that are often inhabited by distinct bacterial communities (Bickel and Or, 2020; Lladó et al., 2017). Heterogeneity in soil physicochemical parameters such as pH, salinity, organic matter content, nutrient availability, and texture have been reported as important determinants of the diversity and structure of bacterial communities inhabiting various forest habitats (Wei et al., 2018; Z. Xia et al., 2016).
Soil pH is the key environmental driver of bacterial diversity (Lladó et al., 2017), and has a direct influence on the presence and abundance of the dominant bacterial groups across habitats (Yu et al., 2021). For example, members of the phyla Bacteroidetes and Actinobacteria have been reported to increase at high pH levels, while Acidobacteria increase in soils with low pH levels (Lauber et al., 2009; Lladó et al., 2017). Additionally, soils with a pH of or near 7 are reported to harbor a higher bacterial diversity compared to low-pH soils (Fierer and Jackson, 2006; Hinsinger et al., 2009). Bacterial communities also respond strongly to soil salinity, where increasing electrical conductivity (EC) values are associated with decreasing diversity (Fang et al., 2020; Zhang et al., 2019). Soil nutrients, both macro-and micronutrients, are essential for proper bacterial growth, and hence high abundance and diversity are expected in soils with high nutrient levels. However, considering that different bacteria have specific nutrient requirements, the community structure is often shaped by the availability of essential nutrients (Yu et al., 2021). Shifts in species composition of vegetation communities have also been correlated with shifts in bacterial communities across a range of soil habitats (Chandra et al., 2016; Muwawa et al., 2021; Wei et al., 2018). However, the impact of aboveground vegetation on the diversity and structure of bacterial communities is low when compared with the overriding effects of soil chemical parameters (Cheng et al., 2020; Naether et al., 2012).
In this study, three tropical forests in Kenya were selected to determine the diversity and community structure of forest soil bacteria and investigate how soil physicochemical parameters influenced these bacterial communities. Kakamega and Irangi are tropical rainforests colonized by diverse species of trees, shrubs, and grasslands, while Gazi Bay harbors mangrove species that differ based on intertidal and vegetation zonation. These forests provide important ecosystem services, some of which are mediated by soil bacteria (Lladó et al., 2017; Rodríguez-Veiga et al., 2020). Therefore, a clear understanding of bacterial diversity and community composition could provide deeper insight into the ecological processes associated with these forests (Aislabie et al., 2013). We used Illumina high throughput sequencing of 16S rRNA amplicons to determine the diversity and community structure of soil bacteria inhabiting three forests in Kenya. We also investigated the effects of various soil chemical parameters on the bacterial communities within these forests.
2. Materials and methods
2.1. Research authorization
This research was authorized by the National Commission for Science, Technology & Innovation in Kenya. Sampling in the forest reserves was permitted by both the Kenya Wildlife Service and Kenya Forest Service.
2.2. Description of sampling sites and sample collection
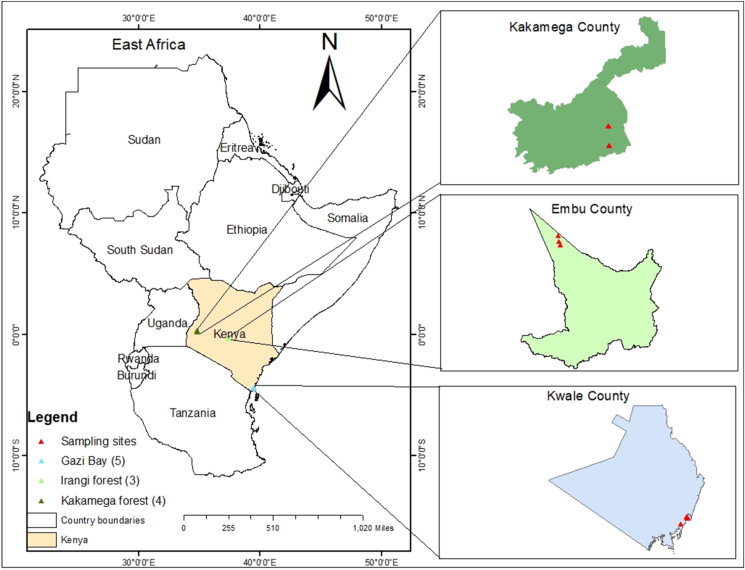
Sampling sites included three natural forests in Kenya (Figure 1), which are characterized by year-round and diverse communities of aboveground vegetation. The forests were selected based on their varying altitudes and environmental characteristics. In Kenya, high altitudes are described as regions above 1800 m above sea level, mid altitudes range from 1000–1800 m, while low altitudes are less than 1000 m (Hassan, 1998). Irangi forest is a high-altitude rainforest located on the eastern side of Mount Kenya in Embu County. It is a montane forest of indigenous species and has distinct vegetation zones. The forest is characterized by a cold and humid climate and temperatures that sometimes fall below 12 °C (Zhou et al., 2018). Kakamega forest is a mid-altitude rainforest situated in Kakamega and Nandi Counties in the western region of Kenya. This tropical rainforest covers a total area of about 26500 ha and has diverse vegetation of indigenous and exotic trees and natural grasslands. The forest has a warm and humid climate with an annual temperature of 25 °C (Mandela et al., 2018). Gazi Bay is a low-altitude region that harbors mangroves. The forest covers a total area of 700 ha along the Indian Ocean coastline in Kwale County and exhibits a typical zonation of mangrove species within the intertidal zone (Lang'at, 2008). The climate in Gazi Bay is hot and humid with an average annual temperature of 28 °C (Muwawa et al., 2021). Soil sampling was done in the year 2018. Within each forest, the selection of the sampling sites was greatly influenced by the type of aboveground vegetation. A stratified random sampling technique was used to collect soil samples from a total of 12 sites. Soil samples in the Kakamega forest were collected from four sites, namely Kalunya grassland, Kalunya canopy, Buyangu canopy, and Buyangu vegetation. Sampling sites in the Irangi forest were Irangi grassland, Irangi vegetation, and Irangi canopy. In Gazi Bay, samples were collected from Gazi grassland, Gazi lowtide1 and 2, Gazi hightide, and Gazi seagrass. Descriptions of the sampling sites are provided in Table 1. Surface litter was removed and approximately 1 kg of soil was collected within the top soil layer at a depth of 0–20 cm. The samples were kept in sterile plastic bags using a sterile hand shovel. One batch comprising 500 g soil from each sampling site was stored at −20 °C for metagenomics analysis, while the other batch of 500 g was stored at 4 °C for physicochemical analysis.
Figure 1.
Map showing the sampling sites of forest soils in Kenya.
Table 1.
Geographical location and descriptions of the sampling sites in Kakamega, Irangi, and Gazi Bay forests.
| Forest | Sampling site | Sample ID | Sample type | GPS coordinates | Altitude (m) | Site characteristics |
|---|---|---|---|---|---|---|
| Kakamega | Kalunya grassland | K1a.Duplicate | Soil | N00°14.611′ E034°51.944′ | 1591 | Open grassland with long grasses |
| Kalunya canopy | K2a.Duplicate | Soil | N00°14.579′ E034°52.126′ | 1603 | Closed canopy of trees, thick undergrowth, and forest floor with decomposing plant materials | |
| Buyangu canopy | K3a.Duplicate | Soil | N00°20.679′ E034°51.769′ | 1608 | Closed canopy of trees, thick undergrowth, and forest floor with decomposing plant materials | |
| Buyangu vegetation | K4a.Duplicate | Soil | N00°20.708′ E034°51.879′ | 1611 | Open canopy of tall trees, thin undergrowth, and forest floor with decomposing plant materials | |
| Irangi | Irangi grassland | K5a.Duplicate | Soil | S00°19.442′ E037°27.306′ | 2119 | Open grassland with short grasses |
| Irangi vegetation | K6a.Duplicate | Soil | S00°16.638′ E037°26.834′ | 2215 | Open canopy of tall trees, thin undergrowth, and forest floor with decomposing plant materials | |
| Irangi canopy | K7a.Duplicate | Soil | S00°18.329′ E037°27.056′ | 2201 | Closed canopy of trees, thick undergrowth, and forest floor with decomposing plant materials | |
| Gazi Bay | Gazi grassland | K8a.Duplicate | Sediment | S04°24.957′ E039°30.514′ | 7 | Open grassland with mangrove propagules |
| Gazi lowtide1 | K9a.Duplicate | Sediment | S04°25.032′ E039°30.666′ | 10 | Thick mangrove growth and decomposing plant materials | |
| Gazi lowtide2 | K10a.Duplicate | Sediment | S04°28.427′ E039°27.802′ | 2 | Thick mangrove growth and decomposing plant materials | |
| Gazi hightide | K11a.Duplicate | Sediment | S04°25.308′ E039°30.751′ | 12 | Thin mangrove undergrowth | |
| Gazi seagrass | K12a.Duplicate | Sediment | S04°25.739′ E039°30.739′ | 2 | Subtidal seagrass vegetation |
2.3. Extraction of total community DNA
From each sample, 0.2 g of soil was weighed and placed into 2 ml Eppendorf tubes. Soil samples were suspended in 1 ml of sterile water. They were centrifuged at 13,200 rpm for 10 min and the supernatant was discarded. Total community DNA was extracted in triplicates using the phenol-chloroform method (Sambrook et al., 1989). The respective triplicates were pooled during the DNA precipitation step. DNA pellets were air-dried and stored at −20 °C.
2.4. Amplicon library preparation and sequencing
Amplicon library preparation and sequencing were performed at Molecular Research (MR) DNA Lab (www.mrdnalab.com, Shallowater, TX, USA). The V4 region of the 16S rRNA gene was amplified using PCR primers 515F (GTGCCAGCMGCCGCGGTAA) and 806R (GGACTACHVGGGTWTCTAAT), with a barcode on the forward primer (Caporaso et al., 2011). PCR was conducted using the HotStarTaq Plus Master Mix Kit (Qiagen, USA). Thermal cycling conditions included an initial denaturation of 94 °C for 3 min, followed by 30 cycles of 94 °C for 30 s, 53 °C for 30 s, and 72 °C for 1 min, and a final elongation step of 72 °C for 5 min. A 2% agarose gel electrophoresis was used to determine the success of PCR amplification and the relative intensity of bands. Multiple samples were pooled together in equal ratios based on their DNA concentrations and molecular weight. The pooled samples were purified using calibrated AMPure XP beads (Beckman Coulter) and were used to prepare the 16S rRNA library by following the TruSeq DNA library preparation protocol (Illumina). Sequencing was performed at MR DNA (www.mrdnalab.com) on Illumina's Miseq 2 × 300 bp Version 3 as per the manufacturer's guidelines.
2.5. Processing of sequence data and taxonomic classification
Sequences generated from the Illumina sequencing platform were processed using the MR DNA analysis pipeline (MR DNA, Shallowater, TX, USA). In brief, the raw FastQ files were depleted of barcodes and primers, while short sequences <200 bp, sequences with homopolymer runs exceeding 6 bp and those with ambiguous base calls were eliminated. Sequences were also denoised, chimeras removed, and the operational taxonomic units (OTUs) were defined by clustering at 97% sequence similarity. The final OTUs were taxonomically classified using BLASTn against a curated database derived from RDPII (rdp.cme.msu.edu) and NCBI (www.ncbi.nlm.nih.gov).
2.6. Soil physicochemical analysis
Soil samples were analyzed for different physicochemical parameters at SGS Kenya Limited Laboratory Services (Mombasa, Kenya). The levels of acidity or alkalinity (pH), EC, organic carbon (OC), total nitrogen (N), and soil texture were analyzed as per ICARDA guidelines (Estefan et al., 2013). Concentrations of other major and minor nutrients were determined using Mehlich 3 method (Mehlich, 1984) These elements included potassium (K), phosphorous (P), magnesium (Mg), sodium (Na), sulfur (S), calcium (Ca), boron (B), iron (Fe), copper (Cu), manganese (Mn) and zinc (Zn).
2.7. Statistical analysis
Statistical analyses were performed using the Vegan package version 2.5.7 (Oksanen et al., 2020). Alpha diversity parameters including species richness, Shannon, and inverse Simpson indices were determined. Rarefaction curves were generated after rarifying our data to 29,682 OTUs per sample. The relative abundance of the dominant bacterial phyla was determined by selecting the phyla that amounted to at least 1% across all sampling sites. A heat map was also generated for hierarchical clustering of the bacterial orders using the Pearson correlation distance metric in gplots version 3.1.1 (Warnes et al., 2020). Selected orders were the predominant taxa that had achieved a total abundance of not less than 1% and data were first subjected to z-score standardization. Differences in community composition between samples were analyzed using the analysis of similarity (ANOSIM) test through 999 permutations. A principal component analysis (PCA) was used to assess the correlations between soil chemical parameters and distributions of bacterial taxa in the three forests. Environmental variables that showed a significant influence on bacterial composition were fitted onto a non-metric multidimensional scaling (NMDS) ordination plot of the Bray-Curtis dissimilarity matrix. The goodness of fit of NMDS ordination was assessed using a squared correlation coefficient (r2) through 999 permutations. All analyses were implemented in R version 4.1.2 (R Core Team, 2021).
3. Results
3.1. Assemblage and taxonomic assignment of bacterial sequences
A total of 909,445 16S rRNA sequence reads were obtained from 12 sampling sites representing three forest soils. These reads were clustered into 13,410 OTUs at 97% genetic distance. Further, the OTUs were assigned to 33 phyla, 69 classes, and 138 orders. Each sampling site had a significant bacterial composition as indicated by the distribution and abundance of the bacterial taxa. Within the Kakamega forest, a higher number of OTUs were assigned to the taxa in Buyangu closed canopy compared to other sampling sites, which recorded 24 phyla, 54 classes, and 101 orders (Table 2). Similarly, OTUs were distributed more to the bacterial taxa in the closed canopy of the Irangi forest and were assigned to 25 phyla, 51 classes, and 97 orders. In Gazi Bay, OTUs were distributed more in the lowtide1 region with thick mangrove growth and were assigned to 28 phyla, 55 classes, and 108 orders. Overall, OTUs were more distributed in the sampling sites with thick vegetation in both rainforests and mangroves.
Table 2.
Distribution of bacterial taxa, OTU richness, and diversity indices in Kakamega, Irangi, and Gazi Bay forests.
| Forest | Sampling site | Sample ID | Sample type | Taxonomic assignment |
Richness and diversity indices |
||||
|---|---|---|---|---|---|---|---|---|---|
| Phyla | Classes | Orders | Richness S | Shannon index H | InvSimpson 1/D | ||||
| Kakamega | Kalunya_grassland | K1a.Duplicate | Soil | 21 | 46 | 91 | 2886 | 5.54 | 74.20 |
| Kalunya_canopy | K2a.Duplicate | Soil | 21 | 48 | 95 | 3667 | 6.45 | 138.60 | |
| Buyangu_canopy | K3a.Duplicate | Soil | 24 | 54 | 101 | 3655 | 6.83 | 239.13 | |
| Buyangu_vegetation | K4a.Duplicate | Soil | 21 | 49 | 96 | 3005 | 6.96 | 378.57 | |
| Irangi | Irangi_grassland | K5a.Duplicate | Soil | 21 | 50 | 95 | 2969 | 6.65 | 229.07 |
| Irangi_vegetation | K6a.Duplicate | Soil | 23 | 51 | 94 | 3186 | 6.70 | 198.00 | |
| Irangi_canopy | K7a.Duplicate | Soil | 25 | 51 | 97 | 3031 | 5.18 | 32.47 | |
| Gazi Bay | Gazi_grassland | K8a.Duplicate | Sediment | 22 | 51 | 100 | 2629 | 5.54 | 31.80 |
| Gazi_lowtide1 | K9a.Duplicate | Sediment | 28 | 55 | 108 | 3179 | 4.43 | 14.82 | |
| Gazi_lowtide2 | K10a.Duplicate | Sediment | 23 | 49 | 85 | 2020 | 3.61 | 9.18 | |
| Gazi_hightide | K11a.Duplicate | Sediment | 26 | 53 | 101 | 3048 | 4.37 | 13.89 | |
| Gazi_seagrass | K12a.Duplicate | Sediment | 21 | 45 | 85 | 2235 | 5.27 | 46.27 | |
3.2. Alpha diversity indices of bacterial communities
Bacterial communities in Kakamega, Irangi, and Gazi Bay forests exhibited high levels of diversity and species richness as shown in Table 2. The closed canopies of Kalunya (3,667) and Buyangu (3,655) in Kakamega forest recorded the highest number of species, followed by an open canopy of Buyangu vegetation (3,005) and Kalunya grassland (2,886). In contrast, the open canopy of Irangi vegetation recorded the highest species richness (3,186), followed by Irangi closed canopy (3,031) and Irangi grassland (2,969). Overall, open and closed canopies within the rainforests had higher species richness compared to the grasslands. Within the Gazi Bay mangrove forest, species richness was highest in the lowtide1 region (3,179), followed by hightide (3,048), grassland (2,629), and seagrass (2,235) regions, while the least number of species were observed in lowtide2 region (2,020).
Shannon and inverse Simpson indices showed that, within the Kakamega forest, bacterial communities in Buyangu vegetation were the most diverse followed by Buyangu and Kalunya canopies and then Kalunya grassland (Table 2). Irangi vegetation and grassland exhibited higher species diversity than the Irangi canopy. Our results also showed that the bacterial taxa in seagrass and grassland regions within the Gazi Bay mangrove forest were the most diverse, whilst lowtide2 had the least diverse taxa.
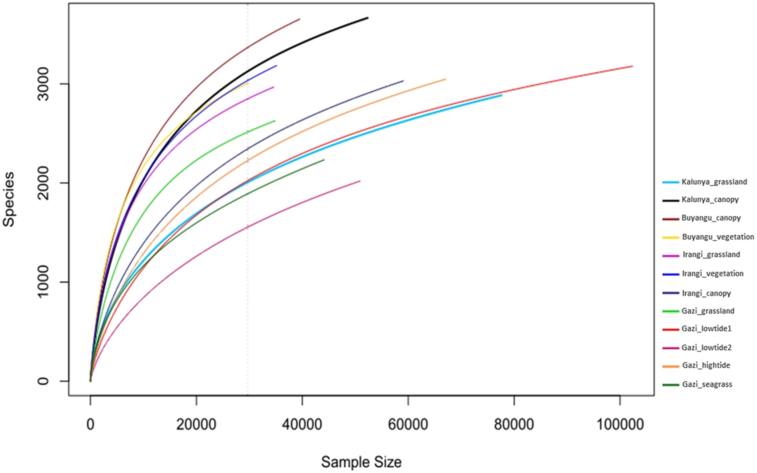
Results from the rarefaction curves showed that, amongst the rainforests, bacterial taxa were richest in the Buyangu canopy and lowest in Kalunya grassland (Figure 2). Within the mangrove forest, Gazi Bay grassland had the highest number of species while the Gazi lowtide2 region recorded the least number of species. The rarefaction curves also tended to show an asymptotic behavior, indicating that the sampling depth was sufficient and our sequence data represented the true diversity of soil bacterial communities in these forests.
Figure 2.
Rarefaction curves showing the rarefied number of species as a function of sample size. The dashed vertical line represents the lowest sample size.
3.3. Bacterial community composition
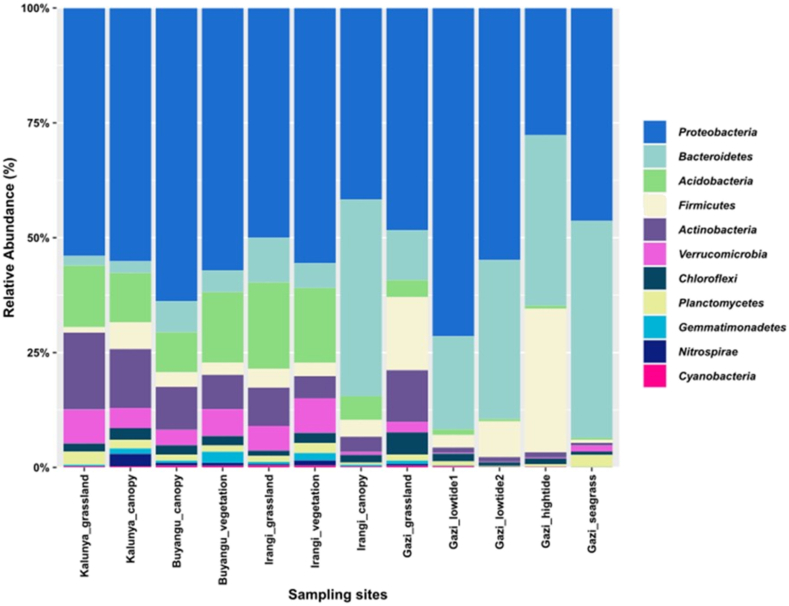
Bacterial community composition was significantly different between the three forest soils (R = 0.559, p = 0.007). At the phylum level, analysis was conducted on 11 taxa that had scored a relative abundance of above 1% across all sampling sites (Figure 3). The top 10 most abundant phyla were Proteobacteria (27.5–71.0%), Bacteroidetes (2.1–46.9%), Firmicutes (0.7–31.0%), Acidobacteria (0.4–18.7%), Actinobacteria (0.6–16.7), Verrucomicrobia (0.1–7.5%), Chloroflexi (0.8–4.8%), Planctomycetes (0.2–2.8%), Gemmatimonadetes (0.0–2.4%), and Nitrospirae (0.0–2.7%). The members of Proteobacteria exhibited high relative abundance in all the sampling sites, indicating their adaptability in both rainforests and mangrove ecosystems. However, other predominant phyla showed unique compositions between the sampling sites. For instance, members of Bacteroidetes were relatively high in the sampling sites within Gazi Bay. The Irangi canopy also displayed high levels of Bacteroidetes in contrast to other sampling sites in the Kakamega and Irangi forests. Further comparison showed that the relative abundances of Acidobacteria, Actinobacteria, and Verrucomicrobia were higher in Kakamega and Irangi forests than in Gazi Bay soils. Within the mangrove region, Gazi grassland differed from other sampling sites due to the relatively high abundance of Actinobacteria. The members of Firmicutes were also comparatively high in Gazi grassland and lowtide2 regions.
Figure 3.
Relative abundance of the most dominant phyla observed in forest soil samples.
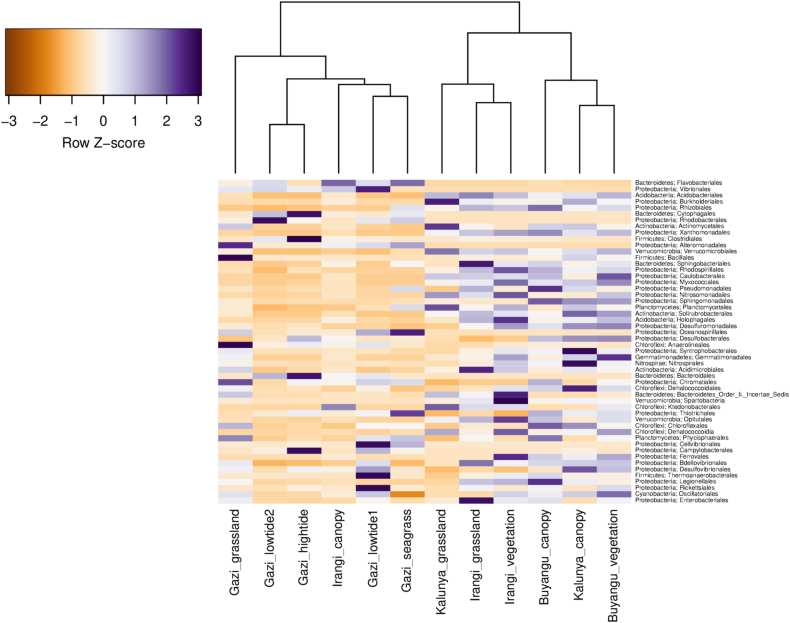
The order level characterization of the bacterial taxa clustered the sampling sites into two main groups (Figure 4). Fifty-two (52) bacterial orders scored a total abundance of above 1% and were used in the heat map visualization of the taxa. The first cluster mainly consisted of the sampling sites in the mangroves of Gazi Bay. The inclusion of Irangi canopy, rainforest taxa, in the Gazi Bay cluster was rather surprising. Despite clustering together, bacterial community composition was highly differentiated by the sampling sites. For instance, Gazi grassland was dominated by Clostridiales, Bacillales, and Anaerolineales, while Rhodobacterales were the most abundant in Gazi lowtide2. Bacterial orders including Cytophagales, Clostridiales, Campylobacterales, and Bacteroidales were abundant in Gazi hightide. In addition, members of Vibrionales, Cellvibrionales, Thermoanaerobacterales, and Rickettsiales were abundant in lowtide1, whereas Gazi seagrass was dominated by Flavobacteriales, Oceanospirillales, and Thiotrichales. Within the Irangi canopy, the most abundant order was Flavobacteriales.
Figure 4.
Heat map showing hierarchical clustering of the dominant bacterial orders observed in forest soil samples.
The second cluster was composed of sampling sites within the rainforests of Kakamega and Irangi. Similar to the first cluster, the dominant orders were differentially abundant between the sampling sites. Within the Kakamega forest, Burkholderiales, Verrucomicrobiales, Actinomycetales, and Planctomycetales exhibited high abundance in Kalunya grassland. Buyangu canopy was dominated by Pseudomonadales, Sphingomonadales, Rhizobiales, and Legionellales, while Syntrophobacterales, Nitrospirales, and Dehalococcoidales were highly abundant in the Kalunya canopy. Members of Gemmatimonadales and Caulobacterales were highly abundant in Buyangu vegetation. Within the Irangi forest, Irangi vegetation recorded several predominant orders including Spartobacteria, Bacteroidetes Order II Incertae sedis, Holophagales, and Ferrovales. In contrast, the notable bacterial orders in Irangi grassland included Sphingobacteriales, Acidimicrobiales, and Enterobacteriales. Overall, the bacterial community structure was significantly differentiated by the forest type and aboveground vegetation.
3.4. Soil physicochemical properties
The physicochemical properties of the sampled soils are presented in Table 3. Irangi canopy had the lowest pH (4.53) among the rainforest sampling sites, while the Kalunya canopy was within the neutral range of 7.17. The soil pH within Gazi Bay ranged from 4.78 in the Gazi grassland to 6.64 in the Gazi lowtide1 region. Soil EC values within Gazi Bay (8.53–18.69) were higher than in Kakamega (0.71–3.66) and Irangi (0.57–2.05) forests. Within-site variation was also evident in Gazi Bay where both soil pH and EC levels were notably low in Gazi grassland. In addition, sampling sites with low EC levels such as Gazi grassland, seagrass, and lowtide1 regions were shown to have higher bacterial alpha diversity indices compared to other mangrove sites. Within the rainforests, the Irangi canopy had comparatively low levels of soil pH and EC that could have influenced its bacterial community composition. Differences in soil nutrients such as K, Ca, P, Fe, Cu, and Zn were minimal across the sampling sites, indicating that the soils from the three forests are nutritionally rich. In addition, the levels of OC and N, the key energy sources for bacteria, were not significantly different between the three forest soils. However, the levels of Na, S, Mg, B, and Mn showed remarkable variation. Different soil textures were also recorded ranging from sand, sandy loam, loamy sand to loam (Table 3).
Table 3.
Physicochemical parameters of soil samples collected from Kakamega, Irangi, and Gazi Bay forests in Kenya.
| Kakamega |
Irangi |
Gazi Bay |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Kalunya grassland | Kalunya canopy | Buyangu canopy | Buyangu vegetation | Irangi grassland | Irangi vegetation | Irangi canopy | Gazi grassland | Gazi lowtide1 | Gazi lowtide2 | Gazi hightide | Gazi seagrass | |
| pH | 5.92 | 7.17 | 6.42 | 5.17 | 5.01 | 6.94 | 4.53 | 4.78 | 6.64 | 5.19 | 6.36 | 6.40 |
| EC (mmhos/cm) | 0.71 | 1.25 | 3.66 | 3.29 | 1.24 | 2.05 | 0.57 | 8.53 | 11.78 | 18.69 | 13.72 | 11.96 |
| OC (%) | 2.74 | 2.25 | 2.83 | 2.71 | 2.62 | 2.57 | 2.61 | 1.70 | 0.93 | 2.50 | 2.59 | 0.56 |
| N (%) | 0.24 | 0.25 | 0.25 | 0.29 | 0.24 | 0.28 | 0.29 | 0.19 | 0.12 | 0.23 | 0.28 | 0.07 |
| P (ppm) | 7.09 | 4.93 | 66.70 | 40.17 | 3.37 | 8.30 | 3.37 | 41.80 | 17.21 | 43.97 | 23.15 | 7.65 |
| K (ppm) | 189.45 | 114.54 | 297.77 | 416.63 | 194.09 | 262.86 | 207.55 | 286.22 | 283.45 | 636.57 | 494.32 | 443.05 |
| Ca (ppm) | 423.85 | 1697.03 | 4157.03 | 4422.94 | 302.79 | 612.34 | 560.32 | 3083.88 | 389.84 | 2325.84 | 2677.96 | 14213.24 |
| Mg (ppm) | 119.04 | 247.95 | 533.72 | 502.86 | 78.12 | 130.35 | 96.25 | 1008.53 | 549.82 | 2058.95 | 1348.84 | 879.94 |
| Na (ppm) | 70.88 | 129.06 | 104.76 | 64.31 | 80.12 | 64.77 | 175.97 | 2486.80 | 3121.09 | 9206.59 | 5714.60 | 4442.84 |
| S (ppm) | 11.70 | 22.29 | 32.06 | 54.56 | 45.29 | 70.74 | 74.67 | 280.63 | 371.05 | 1681.17 | 1070.32 | 867.41 |
| Mn (ppm) | 64.84 | 128.84 | 308.99 | 151.73 | 55.30 | 302.25 | 77.59 | 13.52 | 0.73 | 1.78 | 2.85 | 2.44 |
| Cu (ppm) | 1.51 | 2.51 | 3.18 | 0.93 | 0.36 | 0.91 | 0.58 | 0.64 | 0.10 | 15.27 | 0.05 | 1.14 |
| Fe (ppm) | 97.12 | 108.57 | 163.18 | 293.80 | 86.86 | 165.40 | 72.11 | 131.01 | 170.39 | 298.35 | 328.19 | 142.30 |
| B (ppm) | 14.91 | 14.86 | 14.85 | 15.41 | 14.30 | 14.53 | 14.74 | 20.32 | 20.25 | 35.71 | 28.05 | 21.36 |
| Zn (ppm) | 1.04 | 3.45 | 5.81 | 138.69 | 3.25 | 6.33 | 5.73 | 1.28 | 0.38 | 14.87 | 1.04 | 0.85 |
| Texture | Sandy loam | Loam | Sand | Loamy sand | Sandy loam | Sand | Sandy loam | Sand | Sand | Loamy sand | Sandy loam | Sand |
Means followed by the same letter are not significantly different at P ≤ 0.05. ∗∗∗P ≤ 0.001, ∗∗P ≤ 0.01 and ∗P ≤ 0.05 ns = not significant.
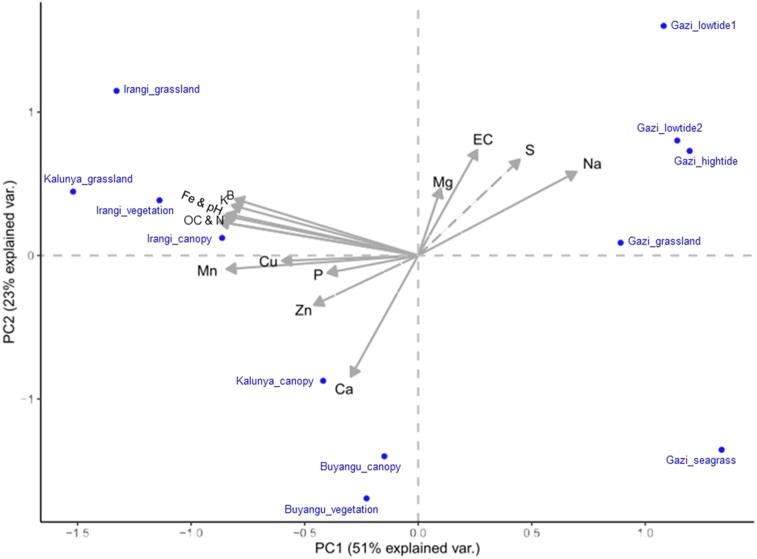
The PCA analysis separated all the sampling sites from each other where the first three principal components (PCs) explained 84.5% of the total variation in bacterial community composition. The first two PC axes of the PCA biplot were 51.4% and 23.0%, respectively (Figure 5). The sampling sites in the mangroves (Gazi Bay) and rainforests (Kakamega and Irangi) contributed to variations in the positive and negative loadings of PC1, respectively. Environmental variables that contributed to the most variability in the negative loading in PC1 were soil pH, OC, N, K, Mn, Fe, and B. Soil EC and S levels mainly contributed to the variation in the positive loading in PC2, while Ca accounted for variation in the negative loading of PC2. The variation resulting from Na was in the positive loading of both PC1 and PC2. The concentrations of both soil P and Mg contributed to the variation in the positive loading of PC3, whose explained variation in the bacterial community composition was 10.1%.
Figure 5.
Correlation between soil chemical parameters and bacterial communities in three forest soils in Kenya.
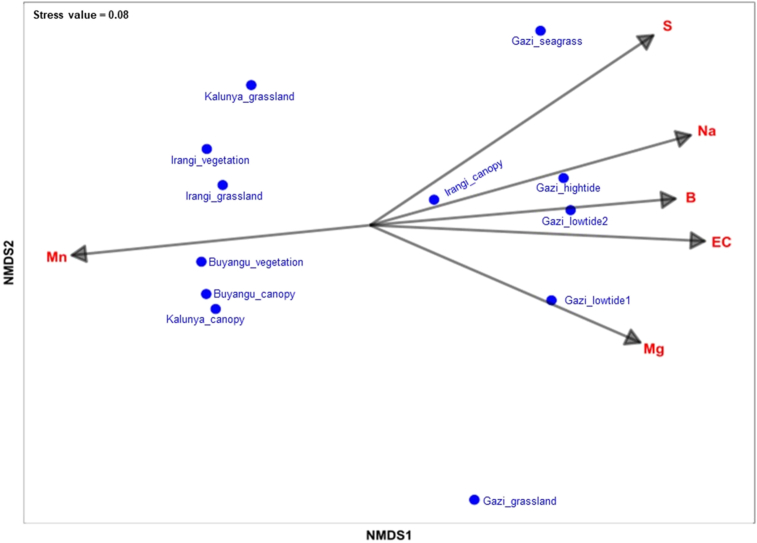
3.5. Key environmental drivers of bacterial community structure
Discrete clustering of the sampling sites in the PCA plot and NMDS ordination revealed significant community variability within and between the studied forests. The NMDS analysis also showed that six environmental variables significantly influenced the bacterial community structure within the studied forests. These environmental vectors scored r2 greater than 0.5 and permutation-based p value less than 0.05 (p < 0.05). Soil EC (r2 = 0.728, p = 0.003), Na (r2 = 0.695, p = 0.005), S (r2 = 0.643, p = 0.007), B (r2 = 0.608, p = 0.009) and Mg (r2 = 0.521, p = 0.031) had a decisive influence on shaping the composition of bacterial taxa within the Gazi Bay (Figure 6). The levels of Mn were significantly higher in Kakamega and Irangi forests and significantly influenced the bacterial composition in these sampling sites (r2 = 0.579, p = 0.012). Overall, the two-dimension NMDS plot showed a stress value of 0.08 indicating a good ordination of the microbial data. Although low pH levels were observed in the Irangi canopy (4.53) and Gazi Bay grasslands (4.78), NMDS analysis showed that this environmental variable did not significantly contribute to shaping the observed bacterial community structure.
Figure 6.
Non-metric multidimensional scaling plot of soil chemical parameters that contribute to variation in bacterial community composition within three forest soils in Kenya.
4. Discussion
Our results showed that forest soils in Kakamega, Irangi, and Gazi Bay harbor diverse taxa of soil bacteria. The presence of heterogeneous communities that were unique to each sampling site confirmed the existence of multiple ecological niches within these forests (Alma'abadi et al., 2015; Valliammai et al., 2021). However, the rainforest soils in Kakamega and Irangi had a higher alpha diversity than the mangrove soils in Gazi Bay. This variation is attributable to varying environmental factors between the two types of forests (Chandra et al., 2016; Z. Xia et al., 2016). Nevertheless, rarefaction curves showed that sampling across the three forests was equally remarkable, and a significant proportion of bacterial communities was assessed.
The dominant phyla reported in this study often represent the major taxa of functional importance in diverse soil ecosystems (Lauber et al., 2009; Liu et al., 2019; Lladó et al., 2017). A high abundance of Proteobacteria in all the sampling sites showed that the members of this phylum are versatile and well adapted to a broad range of habitats. This finding is consistent with previous studies that have reported a high abundance of Proteobacteria in diverse soil environments such as forests, grasslands, garden, and saline soils (Lladó et al., 2017; Mhete et al., 2020). In addition, the dominance of Proteobacteria suggested their importance in forest soil functions such as carbon, nitrogen, iron, and sulfur cycling (Lladó et al., 2017; Tong et al., 2021).
Community compositions of Bacteroidetes, Acidobacteria, Actinobacteria, and Firmicutes were generally variable and were subject to the type of forest. Further visualization of these phyla at the order level also revealed a high degree of inter-sample variability. Based on our results, Bacteroidetes could be the major players in the decomposition of cellulose and other carbon substrates in forest soils within the Gazi Bay and Irangi canopy (Lladó et al., 2017; Mhete et al., 2020). On the other hand, Acidobacteria and Actinobacteria were highly abundant in Kakamega and Irangi forest soils, suggestive of their ecological roles in organic matter turnover and nutrient cycling in the rainforests (Lladó et al., 2017; Tong et al., 2021). Within Gazi Bay, and in particular the grassland and hightide regions, members of Firmicutes were more likely to be the active decomposers of recalcitrant carbon substrates and inorganic nutrients (Lladó et al., 2017; Naghoni et al., 2017).
Soil pH values revealed the presence of acidified forest soils indicative of significant decomposition of organic matter and acidic deposition. However, the Kalunya canopy showed a neutral pH of 7.17 which could be due to the presence of sole loamy soils which are associated with higher pH buffering capacity compared to soils with sand particles (Hazlett et al., 2020). Overall, soil pH seemed optimal for microbial growth across all sampling sites and did not influence the alpha diversity indices and structure of bacterial communities (Deng et al., 2018; Z. Xia et al., 2016). Unlimited availability of most soil nutrients in all the sampling sites could also have contributed to the observed bacterial diversity and community composition (Klimek et al., 2016; Z. Xia et al., 2016). Moreover, results on OC and N, the major determinants of organic matter decomposition, indicated almost equal rates of cycling across the studied sites, and hence lack of significant influence on the observed species richness and bacterial diversity (Bickel and Or, 2020; Lladó et al., 2017; Praeg et al., 2019). All soil textures ranging from loamy to sandy soils also displayed high levels of diversity and species richness. A related study by Chau et al. (2011) reported that soil texture had no significant influence on bacterial diversity, but they observed that bacterial species richness was significantly higher in sandy than silty and clayey soils. In contrast, Q. Xia et al. (2020) reported that the abundance of some bacterial taxa was found to be higher in soils containing higher fractions of silt and clay than in soils with high sand content.
Differences in vegetation cover and specific soil chemical parameters are presented as the contributing factors, either solely or in combination, to the bacterial community differentiation observed within and between the studied forests (Klimek et al., 2016; Praeg et al., 2019; Yang et al., 2018). For instance, observed variations in species richness and bacterial diversity, as well as unique community structures in the grasslands, closed and open canopies demonstrated the significant influence of vegetation in shaping the microbial community composition within the rainforests and this finding is consistent with previous studies conducted in diverse soil ecosystems (Chandra et al., 2016; de Gannes et al., 2016; Klimek et al., 2016). Similarly, samples collected within Gazi Bay exhibited differential composition and community structure of bacterial taxa that could be due to, among other factors, variations in the intertidal zones and associated differences in vegetation cover (Muwawa et al., 2021; Zhu et al., 2018).
In this study, varied levels of six chemical parameters contributed to the variation in bacterial community composition between mangrove soils in Gazi Bay and rainforest soils in Kakamega and Irangi. The levels of soil Mn were significantly associated with the rainforests, indicating that bacterial communities in these forests have a higher rate of forest litter decomposition and Mn cycling compared to mangroves in Gazi Bay (Piazza et al., 2019). High EC levels appeared to negatively influence bacterial diversity and species richness, especially in the Gazi lowtide2 region. These negative effects of salinity on microbial diversity have been previously observed across many soil environments (Bañeras Vives et al., 2012; dC Rubin et al., 2017; Fang et al., 2020). Soil EC values also indicated that bacterial communities within the mangrove ecosystems of Gazi Bay were well adapted to saline environments (Gomes et al., 2011; Liu et al., 2019). Moreover, our results are consistent with previous reports that soil EC levels are positively correlated with the concentrations of Na, S, Mg, and B (dC Rubin et al., 2017; Kord et al., 2010; Tavakkoli et al., 2010). These environmental variables are based on intertidal regions and contribute to variations in microbial composition in the mangroves (Muwawa et al., 2021).
5. Conclusion
This study has provided baseline data on the diversity and heterogeneity of community structure of soil bacteria populating three tropical forests in Kenya. Taxonomic profiling revealed significant bacterial community differentiation associated with environmental heterogeneity (vegetation zonation and edaphic factors) within and between the forests. Moreover, our results indicated that variation in soil chemical parameters contributed significantly to the observed bacterial diversity and community structure. Given that the studied forest ecosystems represent diverse microbial habitats, future studies on environmental factors that contribute to both spatial and temporal variability of soil microbial communities (both prokaryotes and eukaryotes) are recommended.
Declarations
Author contribution statement
Eucharia Kenya, Fathiya Khamis and Mary Mwangi: Conceived and designed the experiments; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Grace Kinyanjui, Alex Kipnyargis: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Franklin Kinyua: Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Romano Mwirichia: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Funding statement
Eucharia Kenya was supported by the National Research Fund, Kenya (NRF/1/MMC/471).
Data availability statement
Data associated with this study has been deposited at the SRA archive under the accession number Bioproject PRJNA856080.
Declaration of interest’s statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
The authors would like to acknowledge the support received from research assistants at Kakamega, Irangi, and Gazi Bay while collecting soil samples from these forest reserves.
References
- Aislabie J., Deslippe J.R., Dymond J. Soil microbes and their contribution to soil services. Ecosyst. Serv. New Zealand–Cond. Trends. 2013;1(12):143–161. Manaaki Whenua Press, Lincoln, New Zealand. [Google Scholar]
- Alma’abadi A.D., Gojobori T., Mineta K. Marine metagenome as a resource for novel enzymes. Dev. Reprod. Biol. 2015;13(5):290–295. doi: 10.1016/j.gpb.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amoo A.E., Babalola O.O. Impact of land use on bacterial diversity and community structure in temperate pine and indigenous forest soils. Diversity. 2019;11(11):217. [Google Scholar]
- Bañeras Vives L., Ruiz Rueda O., López i Flores R., Quintana Pou X., Hallin S. The role of plant type and salinity in the selection for the denitrifying community structure in the rhizosphere of wetland vegetation. Int. Microbiol. 2012;15(2):89–99. doi: 10.2436/20.1501.01.162. [DOI] [PubMed] [Google Scholar]
- Bickel S., Or D. Soil bacterial diversity mediated by microscale aqueous-phase processes across biomes. Nat. Commun. 2020;11(1):1–9. doi: 10.1038/s41467-019-13966-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J.G., Lauber C.L., Walters W.A., Berg-Lyons D., Lozupone C.A., Turnbaugh P.J., Fierer N., Knight R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. USA. 2011;108(Supplement 1):4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra L.R., Gupta S., Pande V., Singh N. Impact of forest vegetation on soil characteristics: a correlation between soil biological and physico-chemical properties. 3 Biotech. 2016;6(2):1–12. doi: 10.1007/s13205-016-0510-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chau J.F., Bagtzoglou A.C., Willig M.R. The effect of soil texture on richness and diversity of bacterial communities. Environ. Forensics. 2011;12(4):333–341. [Google Scholar]
- Cheng J., Zhao M., Cong J., Qi Q., Xiao Y., Cong W., Deng Y., Zhou J., Zhang Y. Soil pH exerts stronger impacts than vegetation type and plant diversity on soil bacterial community composition in subtropical broad-leaved forests. Plant Soil. 2020;450(1):273–286. [Google Scholar]
- dC Rubin S.S., Marín I., Gómez M.J., Morales E.A., Zekker I., San Martín-Uriz P., Rodríguez N., Amils R. Prokaryotic diversity and community composition in the Salar de Uyuni, a large scale, chaotropic salt flat. Environ. Microbiol. 2017;19(9):3745–3754. doi: 10.1111/1462-2920.13876. [DOI] [PubMed] [Google Scholar]
- de Gannes V., Bekele I., Dipchansingh D., Wuddivira M.N., De Cairies S., Boman M., Hickey W.J. Microbial community structure and function of soil following ecosystem conversion from native forests to teak plantation forests. Front. Microbiol. 2016;7:1976. doi: 10.3389/fmicb.2016.01976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng J., Yin Y., Zhu W., Zhou Y. Variations in soil bacterial community diversity and structures among different revegetation types in the Baishilazi Nature Reserve. Front. Microbiol. 2018;9:2874. doi: 10.3389/fmicb.2018.02874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estefan G., Sommer R., Ryan J. third ed. ICARDA; Beirut, Lebanon: 2013. Methods of Soil, Plant, and Water Analysis: A Manual for the West Asia and North Africa Region. [Google Scholar]
- Fang J., Deng Y., Che R., Han C., Zhong W. Bacterial community composition in soils covered by different vegetation types in the Yancheng tidal marsh. Environ. Sci. Pollut. Control Ser. 2020;27(17):21517–21532. doi: 10.1007/s11356-020-08629-z. [DOI] [PubMed] [Google Scholar]
- Fierer N., Jackson R.B. The diversity and biogeography of soil bacterial communities. Proc. Natl. Acad. Sci. USA. 2006;103(3):626–631. doi: 10.1073/pnas.0507535103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes N.C., Cleary D.F., Calado R., Costa R. Mangrove bacterial richness. Commun. Integr. Biol. 2011;4(4):419–423. doi: 10.4161/cib.4.4.15253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan R.M. Cab International in association with the International Maize and Wheat Improvement Centre and the Kenya Agricultural Research Institute; 1998. Maize Technology Development and Transfer: A GIS Application for Research Planning in Kenya; p. 230. [Google Scholar]
- Hazlett P., Emilson C., Lawrence G., Fernandez I., Ouimet R., Bailey S. Reversal of forest soil acidification in the northeastern United States and eastern Canada: site and soil factors contributing to recovery. Soil Syst. 2020;4(3):54. [Google Scholar]
- Hinsinger P., Bengough A.G., Vetterlein D., Young I.M. Rhizosphere: biophysics, biogeochemistry and ecological relevance. Plant Soil. 2009;321(1):117–152. [Google Scholar]
- Klimek B., Chodak M., Jaźwa M., Solak A., Tarasek A., Niklińska M. The relationship between soil bacteria substrate utilisation patterns and the vegetation structure in temperate forests. Eur. J. For. Res. 2016;135(1):179–189. [Google Scholar]
- Kord M., Derakhshan L., Memarian H., Tajabadipour A., Gilkes R.J. 19th World Congress of Soil Science, Soil Solutions for a Changing World. 2010. Effects of high boron concentration on boron uptake and growth of pistachio seedlings; pp. 1–6. [Google Scholar]
- Lang’at J.K.S. Kenya Marine and Fisheries Research Institute; Kenya: 2008. Variability Of Mangrove Forests along the Kenyan Coast. Mangrove Reforestation Report. [Google Scholar]
- Lauber C.L., Hamady M., Knight R., Fierer N. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl. Environ. Microbiol. 2009;75(15):5111–5120. doi: 10.1128/AEM.00335-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M., Huang H., Bao S., Tong Y. Microbial community structure of soils in Bamenwan mangrove wetland. Sci. Rep. 2019;9(1):1–11. doi: 10.1038/s41598-019-44788-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lladó S., López-Mondéjar R., Baldrian P. Forest soil bacteria: diversity, involvement in ecosystem processes, and response to global change. Microbiol. Mol. Biol. Rev. 2017;81(2):e00063. doi: 10.1128/MMBR.00063-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandela H.K., Tsingalia M.H., Gikungu M., Lwande W.M. Distance effects on diversity and abundance of the flower visitors of ocimum kilimandscharicum in the Kakamega forest ecosystem. Int. J. Biodiv. 2018 [Google Scholar]
- Mehlich A. Mehlich 3 soil test extractant: a modification of Mehlich 2 extractant. Commun. Soil Sci. Plant Anal. 1984;15(12):1409–1416. [Google Scholar]
- Mhete M., Eze P.N., Rahube T.O., Akinyemi F.O. Soil properties influence bacterial abundance and diversity under different land-use regimes in semi-arid environments. Scientific African. 2020;7 [Google Scholar]
- Muwawa E.M., Obieze C.C., Makonde H.M., Jefwa J.M., Kahindi J.H., Khasa D.P. 16S rRNA gene amplicon-based metagenomic analysis of bacterial communities in the rhizospheres of selected mangrove species from Mida Creek and Gazi Bay, Kenya. PLoS One. 2021;16(3) doi: 10.1371/journal.pone.0248485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naether A., Foesel B.U., Naegele V., Wüst P.K., Weinert J., Bonkowski M., Alt F., Oelmann Y., Polle A., Lohaus G. Environmental factors affect acidobacterial communities below the subgroup level in grassland and forest soils. Appl. Environ. Microbiol. 2012;78(20):7398–7406. doi: 10.1128/AEM.01325-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naghoni A., Emtiazi G., Amoozegar M.A., Cretoiu M.S., Stal L.J., Etemadifar Z., Fazeli S.A.S., Bolhuis H. Microbial diversity in the hypersaline lake meyghan, Iran. Sci. Rep. 2017;7(1):1–13. doi: 10.1038/s41598-017-11585-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J., Blanchet F.G., Friendly M., Kindt R., Legendre P., McGlinn D., Minchin P.R., O’Hara R.B., Simpson G.L., Solymos P., Stevens M., Henry H., Wagner H. 2020. Vegan: Community Ecology Package. R Package Version 2; pp. 5–7. [Google Scholar]
- Piazza A., Ciancio Casalini L., Pacini V.A., Sanguinetti G., Ottado J., Gottig N. Environmental bacteria involved in manganese (II) oxidation and removal from groundwater. Front. Microbiol. 2019;10:119. doi: 10.3389/fmicb.2019.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praeg N., Pauli H., Illmer P. Microbial diversity in bulk and rhizosphere soil of Ranunculus glacialis along a high-alpine altitudinal gradient. Front. Microbiol. 2019;10:1429. doi: 10.3389/fmicb.2019.01429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . R Foundation for Statistical Computing; Vienna: 2021. R: A Language and Environment for Statistical Computing.https://www.R-project.org/ [Google Scholar]
- Rodríguez-Veiga P., Carreiras J., Smallman T.L., Exbrayat J.-F., Ndambiri J., Mutwiri F., Nyasaka D., Quegan S., Williams M., Balzter H. Carbon stocks and fluxes in Kenyan forests and wooded grasslands derived from earth observation and model-data fusion. Rem. Sens. 2020;12(15):2380. [Google Scholar]
- Sambrook J., Fritsch E.F., Maniatis T. second ed. Cold Spring Harbor Laboratory Press; New York, USA: 1989. Molecular Cloning. A Laboratory Manual. [Google Scholar]
- Shen C., Gunina A., Luo Y., Wang J., He J.-Z., Kuzyakov Y., Hemp A., Classen A.T., Ge Y. Contrasting patterns and drivers of soil bacterial and fungal diversity across a mountain gradient. Environ. Microbiol. 2020;22(8):3287–3301. doi: 10.1111/1462-2920.15090. [DOI] [PubMed] [Google Scholar]
- Tavakkoli E., Rengasamy P., McDonald G.K. High concentrations of Na+ and Cl–ions in soil solution have simultaneous detrimental effects on growth of faba bean under salinity stress. J. Exp. Bot. 2010;61(15):4449–4459. doi: 10.1093/jxb/erq251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong A.-Z., Liu W., Liu Q., Xia G.-Q., Zhu J.-Y. Diversity and composition of the Panax ginseng rhizosphere microbiome in various cultivation modes and ages. BMC Microbiol. 2021;21(1):1–13. doi: 10.1186/s12866-020-02081-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valliammai M.G., Gopal N.O., Anandham R. Elucidation of microbial diversity and lignocellulolytic enzymes for the degradation of lignocellulosic biomass in the forest soils of Eastern and Western Ghats of Tamil Nadu, India. Biofuels, Bioprod. Biorefining. 2021;15(1):47–60. [Google Scholar]
- Warnes G.R., Bolker B., Bonebakker L., Gentleman R., Huber W., Liaw A., Lumley T., Maechler M., Magnusson A., Moeller S. Gplots: Various R Programming Tools for Plotting Data. R package version 3.1.1. 2019. https://CRAN.R-project.org/package=gplots
- Wei H., Peng C., Yang B., Song H., Li Q., Jiang L., Wei G., Wang K., Wang H., Liu S. Contrasting soil bacterial community, diversity, and function in two forests in China. Front. Microbiol. 2018;9:1693. doi: 10.3389/fmicb.2018.01693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Q., Rufty T., Shi W. Soil microbial diversity and composition: links to soil texture and associated properties. Soil Biol. Biochem. 2020;149 [Google Scholar]
- Xia Z., Bai E., Wang Q., Gao D., Zhou J., Jiang P., Wu J. Biogeographic distribution patterns of bacteria in typical Chinese forest soils. Front. Microbiol. 2016;7:1106. doi: 10.3389/fmicb.2016.01106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F., Wu J., Zhang D., Chen Q., Zhang Q., Cheng X. Soil bacterial community composition and diversity in relation to edaphic properties and plant traits in grasslands of southern China. Appl. Soil Ecol. 2018;128:43–53. [Google Scholar]
- Yavitt J.B., Roco C.A., Debenport S.J., Barnett S.E., Shapleigh J.P. Community organization and metagenomics of bacterial assemblages across local scale pH gradients in Northern forest soils. Microb. Ecol. 2021;81(3):758–769. doi: 10.1007/s00248-020-01613-7. [DOI] [PubMed] [Google Scholar]
- Yu Z., Liang K., Huang G., Wang X., Lin M., Chen Y., Zhou Z. Soil bacterial community shifts are driven by soil nutrient availability along a teak plantation chronosequence in tropical forests in China. Biology. 2021;10(12):1329. doi: 10.3390/biology10121329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K., Shi Y., Cui X., Yue P., Li K., Liu X., Tripathi B.M., Chu H. Salinity is a key determinant for soil microbial communities in a desert ecosystem. mSystems. 2019;4(1):e00225. doi: 10.1128/mSystems.00225-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Chen S., Hu G., Mwachala G., Yan X., Wang Q. Species richness and phylogenetic diversity of seed plants across vegetation zones of Mount Kenya, East Africa. Ecol. Evol. 2018;8(17):8930–8939. doi: 10.1002/ece3.4428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu P., Wang Y., Shi T., Zhang X., Huang G., Gong J. Intertidal zonation affects diversity and functional potentials of bacteria in surface sediments: a case study of the Golden Bay mangrove, China. Appl. Soil Ecol. 2018;130:159–168. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data associated with this study has been deposited at the SRA archive under the accession number Bioproject PRJNA856080.