Abstract
Genes encoding cytochrome P450 enzymes (CYPs) are extremely polymorphic and multiple CYP variants constitute clinically relevant biomarkers for the guidance of drug selection and dosing. We previously reported the distribution of the most relevant CYP alleles using population-scale sequencing data. Here, we update these findings by making use of the increasing wealth of data, incorporating whole exome and whole genome sequencing data from 141,614 unrelated individuals across 12 human populations. We furthermore extend our previous studies by systematically considering also uncharacterized rare alleles and reveal that they contribute between 1.5% and 17.5% to the overall genetically encoded functional variability. By using established guidelines, we aggregate and translate the available sequencing data into population-specific patterns of metabolizer phenotypes. Combined, the presented data refine the worldwide landscape of ethnogeographic variability in CYP genes and aspire to provide a relevant resource for the optimization of population-specific genotyping strategies and precision public health.
Subject terms: Predictive markers, Genetic markers
Introduction
Drug response can vary substantially between individuals with up to 50% of patients undergoing pharmacotherapy suffering from low treatment efficacy or adverse drug reactions [1, 2]. Overall, around 30% of this variability has been attributed to genetic factors and variations in cytochrome P450 (CYP) genes alone have been estimated to be relevant for 10–20% of all drug therapies [3]. Notably, of the 57 CYP enzymes encoded in the human genome, eight (CYP2A6, CYP2B6, CYP2C8, CYP2C9, CYP2C19, CYP2D6, CYP3A4 and CYP3A5) are responsible for the metabolism of most drugs in clinical use [4]. With the exception of CYP3A4, these enzymes lack important endogeneous substrates and, consequently, these CYP genes are extremely polymorphic with a plethora of single nucleotide variations (SNVs) and structural variants [5, 6]. Genetic drift, population admixture and isolation compound the genetic complexity and result in substantial ethnogeographic differences in CYP gene variability across human populations [7].
Besides common polymorphisms, recent advances in Next Generation Sequencing (NGS) have facilitated the identification of tens of thousands of rare variants across the human pharmacogenome [8–10]. However, the vast majority of studies to date only analyzed the frequency and distribution of common genetic candidate polymorphisms in CYP genes. We previously used exome sequencing data from 56,945 individuals to analyze the distribution of clinically relevant CYP star alleles across five major human populations [11]. Here, we update these data for 98 star alleles across the eight clinically most relevant CYP genes. Specifically, we analyzed fully consistent and compatible whole exome and whole genome sequencing data from 141,614 individuals and extend our analyses to twelve well-defined ethnogeographic groups. In addition, we comprehensively map and functionally interpret the rare genetic variability within these CYP genes and integrate both star alleles and uncharacterized variants to infer interethnic differences in phenotype distributions and human drug metabolism.
Materials and methods
Data sources
Star alleles of the analyzed CYP genes were defined based on PharmVar [12]. Variant frequency data from a total of 141,614 individuals were derived from the aggregated publicly available sequencing resource gnomAD [13]. Haplotype frequencies were derived considering population-specific linkage disequilibria between the respective polymorphisms based on data from the 1000 Genomes Project using LDlink [14]. For Ashkenazim and Koreans, linkage information from Europe and East Asia were used, respectively. As suballeles of a given star allele do not differ in allele function, they were aggregated throughout this study. Variant calls from short-read sequencing data can be problematic for CYP2B6 [15] and CYP2D6 [16], resulting in possible underestimations of variant frequency. To ameliorate these issues, we matched the extracted frequency information from gnomAD to data from the National Center for Biotechnology Information (NCBI) Allele Frequency Aggregator (ALFA), which includes sequencing data generated using longer reads. Notably, of all analyzed CYP2B6 and CYP2D6 variants, only rs2279343 was considerably underestimated in gnomAD (10.7% and 24.1% for Europeans and Africans in gnomAD vs 23.1% and 33.1% in ALFA, respectively; Supplementary Table 1). Thus, frequencies of the respective alleles were calculated using data from ALFA instead. Frequencies of the *1 reference allele (fREF) were calculated as fREF = 1 − ∑i fi, with fi being the frequency of each considered variant allele i. Common and rare variants are defined as having minor allele frequencies (MAF) of ≥1% and <1%, respectively.
Evaluation of variant functionality
The functional effects of star allele variants were obtained from the literature. For genetic variants for which effects have not been described, functional consequences were estimated using the ADME-optimized prediction framework (APF) [17]. In brief, APF generates an ensemble score for each variant by integrating five computational algorithms (LRT, MutationAssessor, PROVEAN, VEST3 and CADD) using parameter configurations specifically optimized for pharmacogenomic assessments. Notably, while APF is in principle also applicable to non-coding variations, it has not yet been benchmarked for this purpose and is thus applied here only to variants that affect the amino acid sequence of the respective gene product.
Inferring phenotypes from functional variants
To infer phenotypes, we considered exonic variants with known or putative functional effects, copy number variations in CYP2A6 and CYP2D6, as well as selected intronic (CYP3A4*22 and CYP3A5*3) and regulatory (CYP2A6*9, CYP2B6*22 and CYP2C19*17) variants outside of exons. All variations and haplotypes were assigned a functionality score based on the respective CPIC guidelines where available. For uncharacterized variations we used APF scores as activity score predictions. Both established (CPIC) and predicted (APF) activity scores of variants were then aggregated to infer metabolizer phenotype distributions for each population by calculating diplotype frequencies assuming Hardy-Weinberg equilibrium.
Results
Update on the global distribution of clinically relevant CYP alleles
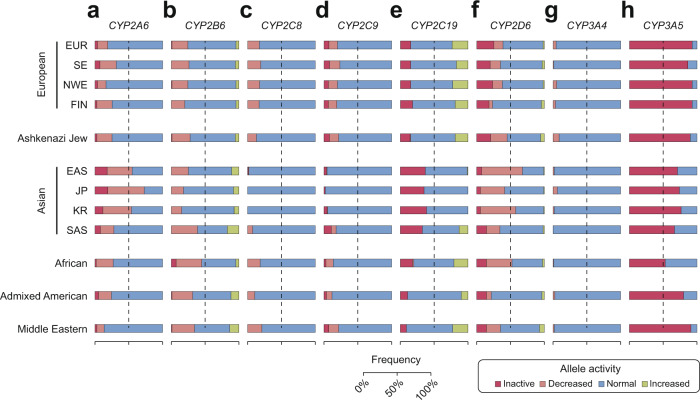
First, we analyzed the ethnogeographic distribution of a total of 98 well-characterized star alleles in eight CYP genes across 12 populations. Of the 16 alleles studied in CYP2A6, *2, *4 (gene deletion) and *5 abolish function, whereas *7, *9, *17, *18, *21, *23 and *35 constitute reduced function alleles (Table 1). Overall, East Asian populations harbor the highest frequency of inactive and decreased function alleles, primarily due to the high frequencies of *4, *7 and *9 (Fig. 1a). Particularly in Japanese, frequencies of these three alleles were highest among all populations with MAFs of 19%, 13.7% and 30%, respectively. In contrast, the loss-of-function allele CYP2A6*2 was not detected in East Asian populations, whereas it was common in European populations (MAF ≥ 2.5%). Within Europe, CYP2A6 gene deletions were common in Southern Europeans (MAF = 4%), whereas frequencies were considerably lower throughout the rest of Europe (MAF ≤ 1%). Among the population-specific CYP2A6 alleles, *23, *25 and *28 were exclusively found in Africans with MAFs pivoting around 1.4%, whereas *8 and *19 were only detected in Asia (MAF = 1.8–2.4% and 0.8–1.3%).
Table 1.
Important variant and allele frequencies of the human CYP2A6 gene.
| Allele | Defining variants | Variant type | Functional consequence | European | AJ | Asian | AFR | AMR | ME | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Overall | SE | NWE | FIN | EAS | JP | KR | SAS | ||||||||
| *1 | None | Normal | 77 | 55 | 79.1 | 73.9 | 72.6 | 41.7 | 23.8 | 42.8 | 68.6 | 69.8 | 74.3 | 84.8 | |
| *2 | rs1801272 | Missense (L160H) | Inactive | 2.6 | 2.8 | 2.8 | 2.5 | 2.2 | 0 | 0 | 0 | 1.2 | 0.4 | 1.6 | 1.3 |
| *4 | CYP2A6 deleted | Inactive | 1 | 4 | 1 | 1 | 0 | 17 | 19 | 10.8 | 7 | 1.5 | 4 | 1 | |
| *5 | rs5031017 | Missense (G479V) | Inactive | <0.1 | 0 | <0.1 | 0.1 | <0.1 | 0.7 | 0 | 1 | <0.1 | <0.1 | <0.1 | 0 |
| *7 | rs5031016 | Missense (I471T) | Decreased | <0.1 | 0.1 | <0.1 | <0.1 | 0.5 | 8.9 | 13.7 | 12.5 | 0.1 | <0.1 | <0.1 | 0.8 |
| *8 | rs28399468 | Missense (R485L) | Normal | <0.1 | 0 | 0 | 0 | 0 | 2.3 | 2.4 | 1.8 | 0 | 0 | 0 | 0 |
| *9 | rs28399433 | TATA box | Decreased | 6.8 | 5.5 | 6.3 | 11.2 | 7.1 | 22.5 | 30 | 24.1 | 14.2 | 8.3 | 14.1 | 6.1 |
| *14 | rs28399435 | Missense (S29N) | 4.1 | 3.1 | 4.5 | 1.3 | 2.1 | <0.1 | 0 | 0 | 3.6 | 0.8 | 1.5 | 1.6 | |
| *17 | rs28399454 | Missense (V365M) | Decreased | <0.1 | <0.1 | <0.1 | 0 | <0.1 | 0 | 0 | 0 | <0.1 | 10.9 | 0.6 | 0.9 |
| *18 | rs1809810 | Missense (Y392F) | Decreased | 1.4 | 1.3 | 1.6 | 1.4 | 0.7 | 0.3 | 0.8 | 0 | 1.2 | 0.5 | 1 | 1.5 |
| *19 | rs5031016, rs1809810 | Missense (I471T, Y392F) | <0.1 | 0.1 | <0.1 | <0.1 | 0.2 | 0.8 | 1.3 | 1.2 | <0.1 | <0.1 | <0.1 | 0.4 | |
| *21 | rs6413474 | Missense (K476R) | Decreased | 1.3 | 1.1 | 1.3 | 2.6 | 2.9 | <0.1 | 0 | 0 | 1.9 | 0.2 | 0.5 | 0.9 |
| *23 | rs56256500 | Missense (R203C) | Decreased | <0.1 | <0.1 | <0.1 | 0 | 0 | 0 | 0 | 0 | <0.1 | 1.4 | <0.1 | 0 |
| *25 | rs28399440 | Missense (F118L) | <0.1 | 0 | <0.1 | 0 | 0 | 0 | 0 | 0 | 0 | 1.4 | <0.1 | 0 | |
| *28 | rs28399463, rs8192730 | Missense (N418D, E419D) | <0.1 | <0.1 | <0.1 | <0.1 | 0 | <0.1 | <0.1 | <0.1 | <0.1 | 1.5 | <0.1 | 0 | |
| *35 | rs143731390 | Missense (N438Y) | Decreased | 5.7 | 7.1 | 3.3 | 7 | 11.7 | 5.7 | 9.1 | 5.7 | 2.1 | 3.3 | 2 | 0.6 |
AFR African, EAS East Asian, SAS South Asian, AMR admixed Americans, FIN Finnish, AJ Ashkenazi Jewish, ME Middle Eastern, SE South European, NWE Northwest European, JP Japanese, KR Korean.
Fig. 1. Ethnogeographic distribution of the inferred functional consequences of well-characterized CYP star alleles.
Frequencies of inactive (dark red), reduced activity (light red), normal (blue) and increased activity (green) alleles shown in Tables 1–5 were aggregated for CYP2A6 (a), CYP2B6 (b), CYP2C8 (c), CYP2C9 (d), CYP2C19 (e), CYP2D6 (f), CYP3A4 (g) and CYP3A5 (h) genes across 12 populations. Star alleles with unclear functional consequences were considered as normal. EUR European, SE South European, NEW North-West European, FIN Finnish, EAS East Asian, JP Japanese, KR South Korean, SAS South Asian.
In contrast to CYP2A6 which harbors multiple common loss-of-function alleles, the only common inactive CYP2B6 allele is CYP2B6*18 in Africa (MAF = 7%; Table 2). The other CYP2B6 loss-of-function alleles *8, *12, *13 and *28 are rare in all populations studied. Notably, the decreased function allele CYP2B6*6 (MAF = 14.5–32%) and the increased function allele CYP2B6*4 (MAF = 1–15.3%) are common across all ethnogeographic groups studied. Furthermore, CYP2B6*9 is common in most populations with MAFs between 2.5% and 11.1% with the exception of Finnish (MAF = 0%) and Koreans (MAF = 0.1%). Overall, inactive and reduced function CYP2B6 alleles are most prevalent in African and South Asian populations, whereas frequencies are lowest in Koreans and Japanese (Fig. 1b).
Table 2.
Important variant and allele frequencies of the human CYP2B6 gene.
| Allele | Defining variants | Variant type | Functional consequence | European | AJ | Asian | AFR | AMR | ME | |||||||
| Overall | SE | NWE | FIN | EAS | JP | KR | SAS | |||||||||
| *1 | None | Normal | 55.7 | 54.4 | 55.8 | 62.1 | 50.8 | 74.4 | 74.4 | 80.7 | 47.8 | 43.6 | 57.3 | 47.1 | ||
| *2 | rs8192709 | Missense (R22C) | Normal | 5.6 | 7.3 | 4.8 | 4.9 | 8 | 4.6 | 4.6 | 2.9 | 3.1 | 3.7 | 3.1 | 10.1 | |
| *4 | rs2279343 | Missense (K262R) | Increased | 3.4 | 3.4 | 3.4 | 3.4 | 3.4 | 10.6 | 7.7 | 7.2 | 15.3 | 1 | 10.2 | 10.4 | |
| *5 | rs3211371 | Missense (R487C) | Normal | 11.8 | 10.3 | 11.9 | 12.1 | 10.4 | 0 | 1.6 | 0.2 | 7.5 | 2.6 | 4.4 | 6.8 | |
| *6 | rs2279343, rs3745274 | Missense (K262R, Q172H) | Decreased | 19.6 | 19.7 | 19.3 | 19.7 | 19.4 | 21.5 | 15.5 | 14.5 | 28.1 | 32 | 20.3 | 22.8 | |
| *7 | rs2279343, rs3745274, rs3211371 | Missense (K262R, Q172H, R487C) | Decreased | <0.1 | 0 | 0.4 | 0 | 0.3 | 0.2 | 0.2 | 0.3 | 0.1 | 0 | 0 | 0.3 | |
| *8 | rs12721655 | Missense (K139E) | Inactive | 0.4 | 0.2 | 0.5 | <0.1 | 0 | 0 | 0 | 0 | 0 | <0.1 | 0.1 | 0 | |
| *9 | rs3745274 | Missense (Q172H) | Decreased | 4.4 | 5.8 | 4.5 | 0 | 7.2 | 3.4 | 2.5 | 0.1 | 10.3 | 5.1 | 11.1 | 10.2 | |
| *12 | rs36060847 | Missense (G99E) | Inactive | <0.1 | <0.1 | <0.1 | 0 | 0.7 | 0 | 0 | 0 | 0 | <0.1 | <0.1 | 0 | |
| *13 | rs2279343, rs3745274, rs12721655 | Missense (K262R, Q172H, K139E) | Inactive | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | <0.1 | 0 | 0 | 0 | |
| *18 | rs28399499 | Missense (I328T) | Inactive | <0.1 | <0.1 | <0.1 | 0 | 0 | 0 | 0 | 0 | <0.1 | 7 | 0.4 | 0.6 | |
| *19 | rs34826503 | Missense (R336C) | Decreased | <0.1 | <0.1 | 0 | 0 | 0 | <0.1 | 0 | <0.1 | 0 | 0.2 | <0.1 | 0 | |
| *20 | rs36056539 | Missense (T168I) | Decreased | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.2 | <0.1 | 0 | |
| *22 | rs34223104 | Regulatory | Increased | 1.3 | 1.9 | 1 | 0.7 | 1.7 | 0.3 | 0 | 0 | 1.8 | 3 | 1.4 | 3.2 | |
| *26 | rs2279343, rs3745274, rs3826711 | Missense (K262R, Q172H, P167A) | Decreased | 0 | 0 | 0 | 0 | 0 | 0.4 | 0 | 0.3 | 0 | 0 | 0 | 0 | |
| *28 | rs34097093 | Stop-gain (R378X) | Inactive | <0.1 | <0.1 | <0.1 | <0.1 | 0 | 0 | 0 | 0 | 0 | <0.1 | <0.1 | 0 | |
AFR African, EAS East Asian, SAS South Asian, AMR admixed Americans, FIN Finnish, AJ Ashkenazi Jewish, ME Middle Eastern, SE South European, NWE Northwest European, JP Japanese, KR Korean.
For CYP2C8, the genetic variability was overall considerably lower than for CYP2A6 and CYP2B6. European, Middle Eastern and African populations feature similar frequency of decreased function CYP2C8 alleles (Fig. 1c). In Africans, the major allele is CYP2C8*2 (MAF = 15.2%), whereas *3 (MAF = 2–15.2%) and *4 (MAF = 1.8–5.8%) are the primary minor alleles in the other populations (Table 3). In contrast, CYP2C8 is extremely conserved in East Asian populations with >99.4% of all alleles corresponding to the reference sequence.
Table 3.
Important variant and allele frequencies of the human CYP2C gene family.
| Allele | Defining variants | Variant type | Functional consequence | European | AJ | Asian | AFR | AMR | ME | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Overall | SE | NWE | FIN | EAS | JP | KR | SAS | ||||||||
| CYP2C8 | |||||||||||||||
| *1 | None | Normal | 82.9 | 81 | 82.6 | 83.1 | 86.9 | 99.3 | 100 | 100 | 92.5 | 81.6 | 89.7 | 78.8 | |
| *2 | rs11572103 | Missense (I269F) | Decreased | 0.3 | 0.6 | 0.2 | <0.1 | 1.4 | <0.1 | 0 | 0 | 1.9 | 15.2 | 0.8 | 3.5 |
| *3 | rs10509681, rs11572080 | Missense (K399R, R139K) | Controversial | 11.3 | 13.2 | 11.8 | 11.1 | 9.9 | <0.1 | 0 | 0 | 4.1 | 2 | 6.8 | 15.2 |
| *4 | rs1058930 | Missense (I264M) | Decreased | 5.4 | 5.2 | 5.3 | 5.8 | 1.8 | <0.1 | 0 | 0 | 1.5 | 1.1 | 2.7 | 2.2 |
| *5 | rs72558196 | Frameshift | Inactive | 0 | 0 | 0 | 0 | 0 | 0.2 | 0 | 0 | 0 | 0 | 0 | 0 |
| *7 | rs72558195 | Stop-gain (R186X) | Inactive | <0.1 | <0.1 | <0.1 | <0.1 | 0 | <0.1 | 0 | 0 | <0.1 | <0.1 | <0.1 | 0.3 |
| *11 | rs78637571 | Stop-gain (E274X) | Inactive | 0 | 0 | 0 | 0 | 0 | 0.4 | 0 | 0 | 0 | 0 | <0.1 | 0 |
| CYP2C9 | |||||||||||||||
| *1 | None | Normal | 79.9 | 76.7 | 79.7 | 81.5 | 78 | 95.9 | 98 | 94.9 | 82.1 | 78.9 | 88.4 | 77.5 | |
| *2 | rs1799853 | Missense (R144C) | Decreased | 12.6 | 14.2 | 13.1 | 11.4 | 13.5 | <0.1 | 0 | <0.1 | 4.7 | 2.2 | 6.8 | 13.6 |
| *3 | rs1057910 | Missense (I359L) | Inactive | 6.8 | 8.5 | 6.5 | 6.3 | 8.4 | 3.3 | 0.7 | 4.4 | 11 | 1.2 | 3.8 | 7.3 |
| *5 | rs28371686 | Missense (D360E) | Decreased | <0.1 | <0.1 | <0.1 | 0 | 0 | 0 | 0 | 0 | <0.1 | 1.1 | <0.1 | 0 |
| *6 | rs9332131 | Frameshift | Inactive | <0.1 | <0.1 | <0.1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | <0.1 | 0 |
| *8 | rs7900194 | Missense (R150H) | Decreased | <0.1 | <0.1 | <0.1 | 0 | 0 | <0.1 | 0 | 0 | <0.1 | 6 | 0.2 | 0 |
| *9 | rs2256871 | Missense (H251R) | Normal | <0.1 | <0.1 | <0.1 | 0 | 0 | <0.1 | 0 | 0 | <0.1 | 7.5 | 0.3 | 0.9 |
| *11 | rs28371685 | Missense (R335W) | Decreased | 0.3 | 0.2 | 0.3 | 0.6 | 0 | <0.1 | 0 | 0 | 0.2 | 1.9 | 0.2 | 0.6 |
| *12 | rs9332239 | Missense (P489S) | Decreased | 0.3 | 0.3 | 0.2 | 0.2 | <0.1 | 0 | 0 | 0 | <0.1 | <0.1 | 0.1 | 0 |
| *13 | rs72558187 | Missense (L90P) | Inactive | 0 | 0 | 0 | 0 | 0 | 0.2 | 0.7 | 0.2 | 0 | 0 | 0 | 0 |
| *14 | rs72558189 | Missense (R125H) | Decreased | <0.1 | <0.1 | <0.1 | 0 | <0.1 | <0.1 | 0 | <0.1 | 1.9 | 0 | <0.1 | 0 |
| *16 | rs72558192 | Missense (T299A) | Decreased | <0.1 | 0 | 0 | 0 | 0 | 0.4 | 0 | 0 | 0 | 0 | 0 | 0 |
| *29 | rs182132442 | Missense (P279T) | Decreased | <0.1 | <0.1 | <0.1 | <0.1 | 0 | 0.1 | 0 | 0.4 | <0.1 | <0.1 | <0.1 | 0 |
| *31 | rs57505750 | Missense (I327T) | Decreased | 0 | 0 | 0 | 0 | 0 | <0.1 | 0.7 | 0 | 0 | <0.1 | <0.1 | 0 |
| CYP2C19 | |||||||||||||||
| *1 | None | Normal | 61.5 | 67.6 | 62.5 | 63.6 | 65.7 | 62.1 | 64.8 | 61.7 | 67.1 | 55.8 | 79.1 | 68.2 | |
| *2 | rs4244285 | Splicing defect | Inactive | 14.7 | 14.7 | 14.8 | 17.5 | 13.2 | 30.8 | 26.7 | 28 | 32.4 | 17.8 | 10.1 | 8.7 |
| *3 | rs4986893 | Stop-gain (W212X) | Inactive | <0.1 | <0.1 | <0.1 | <0.1 | 0 | 6.3 | 8.6 | 10.2 | 0.4 | <0.1 | <0.1 | 0 |
| *4 | rs28399504 | Start lost | Inactive | 0.3 | 0.4 | 0.3 | 0 | 1.6 | <0.1 | 0 | <0.1 | <0.1 | <0.1 | 0.3 | 0 |
| *5 | rs56337013 | Missense (R433W) | Inactive | <0.1 | <0.1 | 0 | 0 | 0 | <0.1 | 0 | <0.1 | <0.1 | 0 | 0 | 0 |
| *6 | rs72552267 | Missense (R132Q) | Inactive | <0.1 | <0.1 | <0.1 | 0 | 0 | <0.1 | 0 | 0 | 0 | <0.1 | <0.1 | 0 |
| *7 | rs72558186 | Splicing defect | Inactive | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | <0.1 | 0 | 0 | NA |
| *8 | rs41291556 | Missense (W120R) | Inactive | 0.3 | 0.2 | 0.3 | <0.1 | <0.1 | 0 | 0 | 0 | <0.1 | <0.1 | <0.1 | 0 |
| *9 | rs17884712 | Missense (R144H) | Decreased | <0.1 | <0.1 | <0.1 | 0 | <0.1 | 0 | 0 | 0 | <0.1 | 1.3 | <0.1 | 0 |
| *10 | rs6413438 | Missense (P227L) | Decreased | <0.1 | 0 | <0.1 | 0 | 0.1 | <0.1 | 0 | 0 | 0 | 0.3 | <0.1 | 0 |
| *13 | rs17879685 | Missense (R410C) | Normal | <0.1 | 0 | <0.1 | 0 | 0 | 0 | 0 | 0 | <0.1 | 1.8 | <0.1 | 0 |
| *15 | rs17882687 | Missense (I19L) | Normal | <0.1 | <0.1 | <0.1 | 0 | 0.4 | 0 | 0 | 0 | <0.1 | 1.9 | <0.1 | 0.3 |
| *17 | rs12248560 | Regulatory | Increased | 23.1 | 17 | 22 | 18.8 | 19 | 0.7 | 0 | 0 | 13.6 | 20.9 | 10.1 | 22.8 |
| *22 | rs140278421 | Missense (R186P) | Inactive | <0.1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.1 | <0.1 | 0 |
AFR African, EAS East Asian, SAS South Asian, AMR admixed Americans, FIN Finnish, AJ Ashkenazi Jewish, ME Middle Eastern, SE South European, NWE Northwest European, JP Japanese, KR Korean.
Similar patterns with overall low variability in East Asian populations are observed for CYP2C9 (Fig. 1d), consistent with previous reports of high linkage between CYP2C8 and CYP2C9 haplotypes [18]. The reduced function allele CYP2C9*2 was globally common (MAF = 2.2–14.2%) with the exception of East Asian populations (MAF < 0.1%; Table 3). CYP2C9 furthermore harbors various population-specific alleles. CYP2C9*6 (MAF = 1%), *8 (MAF = 6%), *9 (MAF = 7.5%) and *11 (MAF = 1.9%) are only common in Africans, while *13 (MAF = 0.7%), *16 (MAF = 0.4%), *29 (MAF = 0.4%) and *31 (MAF = 0.7%) are only found in Asian populations, albeit at lower frequencies.
In CYP2C19, only the splice site loss-of-function allele CYP2C19*2 is common in all populations analyzed (MAF = 8.7–32.4%; Table 3). The regulatory increased function allele CYP2C19*17 is frequent in European, admixed American, Middle Eastern and African populations with MAFs between 10.1% and 23.1%, whereas it is rare across East Asia (MAF = 0–0.7%). Notable among the population-specific variations are the loss-of-function variants CYP2C19*3 and *4 as well as the decreased function variant *9, which are exclusively found in East Asian (MAF ≥ 6.3%), Ashkenazim (MAF = 1.6%) and African populations (MAF = 1.3%), respectively. Overall, unlike for the other CYP2C genes, CYP2C19 loss-of-function alleles are most common and increased function alleles are most rare in East Asians, whereas patterns of allele activity are very similar across all other analyzed populations (Fig. 1e).
CYP2D6 constitutes the most polymorphic pharmacogene and harbors a multitude of common variants with clinical relevance, particularly for the treatment with antidepressants, antipsychotics, opioid analgesics and antihypertensives. Here, we studied 17 CYP2D6 alleles, 13 of which are associated with altered allele function. Decreased function alleles are most prevalent in East Asians due to high frequencies of CYP2D6*10 (MAF ≥ 35%), whereas loss-of-function alleles are most abundant in European populations primarily because of high frequencies of the splicing variant CYP2D6*4 (MAF up to 20.3%) (Table 4 and Fig. 1f). Notably, reduced CYP2D6 allele function are also very common in Africans; however, in these populations the primary drivers are *17 (MAF = 20.5%) and *29 (MAF = 8.9%), which are almost exclusively found in this population. Increased function due to the functional gene duplications CYP2D6*1xN and *2xN is most common in Middle Eastern populations (aggregated MAF = 7%), Ashkenazi Jews (aggregated MAF = 5.6%) and Europeans (aggregated MAF up to 4.7%) but were rare in East Asian populations (aggregated MAF < 1%).
Table 4.
Important variant and allele frequencies of the human CYP2D6 gene.
| Allele | Defining variants | Variant type | Functional consequence | European | AJ | Asian | AFR | AMR | ME | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EUR | SE | NWE | FIN | EAS | JP | KR | SAS | ||||||||
| *1 | None | Normal | 24.1 | 27.3 | 27.7 | 31.8 | 7.9 | 17.5 | 42 | 27.9 | 25.3 | 20.8 | 48.9 | 6.7 | |
| *1xN | Amplification of *1 | Increased | 0.9 | 1.6 | 1 | 2.5 | 3 | 0.3 | 0.5 | <0.1 | 0.6 | 1.5 | 1.1 | 3.1 | |
| *2 | rs16947, rs1135840 | Missense (R296C, S486T) | Normal | 33.6 | 37.2 | 33.5 | 39.1 | 40.4 | 14.6 | 15.8 | 13.6 | 38 | 22.5 | 26 | 50.6 |
| *2xN | Amplification of *2 | Increased | 1.2 | 1.2 | 0.9 | 2.2 | 2.6 | 0.4 | 0.4 | 0.8 | 1.1 | 1.3 | 1.9 | 3.9 | |
| *3 | rs35742686 | Frameshift | Inactive | 1.7 | 1.1 | 1.7 | 3.6 | 0.4 | 0 | 0 | 0 | 0.1 | 0.3 | 0.5 | 0.3 |
| *4 | rs3892097 | Splicing defect | Inactive | 19.6 | 17.1 | 20.3 | 10 | 18.2 | 0.3 | 0 | 0.2 | 10.4 | 8 | 11.1 | 9.8 |
| *5 | CYP2D6 deleted | Inactive | 2.9 | 2.3 | 4.1 | 2.2 | 1.1 | 5.2 | 4.9 | 4.9 | 3.2 | 6.2 | 2.1 | 2.3 | |
| *6 | rs5030655 | Frameshift | Inactive | 1.1 | 1.1 | 1.2 | 2.1 | 0.8 | 0 | 0 | 0 | 0.2 | 0.2 | 0.4 | 0.6 |
| *7 | rs5030867 | Missense (H324P) | Inactive | <0.1 | <0.1 | <0.1 | 0 | <0.1 | 0 | 0 | 0 | 0.8 | <0.1 | <0.1 | 1.3 |
| *9 | rs5030656 | Inframe deletion (K281del) | Decreased | 2.6 | 1.9 | 3 | 1.3 | 0.3 | 0 | 0 | 0 | 0.2 | 0.4 | 1.2 | 0 |
| *10 | rs1065852, rs1135840 | Missense (P34S, S486T) | Decreased | 1.7 | 2.4 | 1.5 | 1.1 | 6.7 | 57.3 | 35 | 50.2 | 5.5 | 4.8 | 1.4 | 6.4 |
| *14 | rs5030865 | Missense (G169R) | Decreased | <0.1 | 0 | 0 | 0 | 0 | 1.3 | 0.8 | 0.5 | <0.1 | 0 | <0.1 | 0 |
| *17 | rs16947, rs28371706 | Missense (R296C, T107I) | Decreased | <0.1 | 0.4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 20.5 | 0.7 | 0 |
| *29 | rs16947, rs1135840, rs61736512, rs59421388 | Missense (R296C, S486T, V136I, V338M) | Decreased | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 8.9 | 0.4 | 0 |
| *41 | rs28371725 | Splicing defect | Decreased | 9.3 | 10.9 | 10 | 3.2 | 17.6 | 3.2 | 0.7 | 1.8 | 13.6 | 2.6 | 4 | 14.3 |
| *33 | rs28371717 | Missense (A237S) | Normal | 1.2 | 0.5 | 1.1 | 1 | 1 | 0 | 0 | 0 | 0.7 | 0.2 | 0.2 | 0.6 |
| *43 | rs28371696 | Missense (R26H) | <0.1 | <0.1 | <0.1 | <0.1 | 0 | <0.1 | 0 | <0.1 | 0.6 | 1.8 | 0.2 | 0 | |
AFR African, EAS East Asian, SAS South Asian, AMR admixed Americans, FIN Finnish, AJ Ashkenazi Jewish, ME Middle Eastern, SE South European, NWE Northwest European, JP Japanese, KR Korean.
Among the major drug metabolizing CYPs, CYP3A4 is the only enzyme with an important endogenous substrate and, as a consequence, CYP3A4 is the most conserved among the studied CYPs (Table 5 and Fig. 1g). In Europeans, admixed Americans, Ashkenazi Jews and Middle Easterners, CYP3A4*22 is the only common allele of functional relevance with frequencies ranging between 0.9% in South Europeans to 9% in Ashkenazim. In contrast, in East Asian populations the population-specific *16 and *18 alleles are common, the former of which only in Japanese (MAF = 3.9%), whereas *22 is absent. Notably, South Asian populations do not harbor common CYP3A4 alleles.
Table 5.
Important variant and allele frequencies of the human CYP3A gene family.
| Allele | Defining variants | Variant type | Functional consequence | European | AJ | Asian | AFR | AMR | ME | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Overall | SE | NWE | FIN | EAS | JP | KR | SAS | ||||||||
| CYP3A4 | |||||||||||||||
| *1 | None | Normal | 94.7 | 98.4 | 93.9 | 93.4 | 91 | 97.5 | 93.4 | 98.2 | 99.4 | 96.2 | 97.1 | 98.1 | |
| *2 | rs55785340 | Missense (S222P) | <0.1 | 0 | <0.1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| *3 | rs4986910 | Missense (M445T) | 0.7 | 0.6 | 0.7 | 1.8 | <0.1 | 0 | 0 | 0 | <0.1 | <0.1 | 0.2 | 0 | |
| *4 | rs55951658 | Missense (I118V) | 0 | 0 | 0 | 0 | 0 | 0.5 | 0 | 0.2 | <0.1 | 0 | <0.1 | 0 | |
| *8 | rs72552799 | Missense (R130Q) | Decreased | 0.1 | <0.1 | <0.1 | 0.2 | 0 | 0 | 0 | 0 | <0.1 | <0.1 | <0.1 | 0 |
| *15 | rs4986907 | Missense (R162Q) | <0.1 | <0.1 | <0.1 | 0 | <0.1 | 0 | 0 | 0 | <0.1 | 2.6 | 0.2 | 0.3 | |
| *16 | rs12721627 | Missense (T185S) | Decreased | 0 | 0 | 0 | 0 | 0 | <0.1 | 3.9 | 0.3 | 0 | 0 | 0 | 0 |
| *18 | rs28371759 | Missense (L293P) | Decreased | 0 | 0 | 0 | 0 | 0 | 1.9 | 2.6 | 1.4 | <0.1 | 0.1 | <0.1 | 0 |
| *22 | rs35599367 | Splicing defect | Decreased | 4.4 | 0.9 | 5.4 | 3.6 | 9 | 0 | 0 | 0 | 0.6 | 0.9 | 2.5 | 1.6 |
| CYP3A5 | |||||||||||||||
| *1 | None | Normal | 6.4 | 12.6 | 6.4 | 6.8 | 9.9 | 28.6 | 26 | 23.5 | 33.2 | 47 | 19.5 | 8.5 | |
| *2 | rs28365083 | Missense (T398N) | 0.6 | 0.4 | 0.6 | 0.2 | <0.1 | 0 | 0 | 0 | 0 | 0.1 | 0.1 | 0 | |
| *3 | rs776746 | Splicing defect | Inactive | 92.9 | 86.8 | 92.9 | 93 | 90 | 71.4 | 74 | 76.5 | 66.8 | 29.8 | 79.2 | 88 |
| *6 | rs10264272 | Splicing defect | Inactive | <0.1 | 0.2 | <0.1 | <0.1 | 0 | 0 | 0 | 0 | <0.1 | 12.9 | 0.8 | 3.5 |
| *7 | rs41303343 | Frameshift | Inactive | <0.1 | <0.1 | <0.1 | 0 | 0 | <0.1 | 0 | 0 | <0.1 | 10.2 | 0.5 | 0 |
AFR African, EAS East Asian, SAS South Asian, AMR admixed Americans, FIN Finnish, AJ Ashkenazi Jewish, ME Middle Eastern, SE South European, NWE Northwest European, JP Japanese, KR Korean.
CYP3A5 activity is primarily governed by the CYP3A5*3 allele, which is defined by the presence of a variant that results in the generation of a cryptic splice site that causes a premature stop codon. The *3 allele constitutes the major allele in most populations studied (MAF = 66.8–93%) except for Africans (MAF = 29.8%; Table 5). The latter also harbor the population-specific loss-of-function frameshift allele CYP3A5*7 (MAF = 10.2%) as well as CYP3A5*6 (MAF = 12.9%), a splice variant restricted to African and Middle Eastern populations. Consequently, 53% to 94% of all CYP3A5 alleles are inactive across all major populations (Fig. 1h).
The genetic landscape of CYP variability
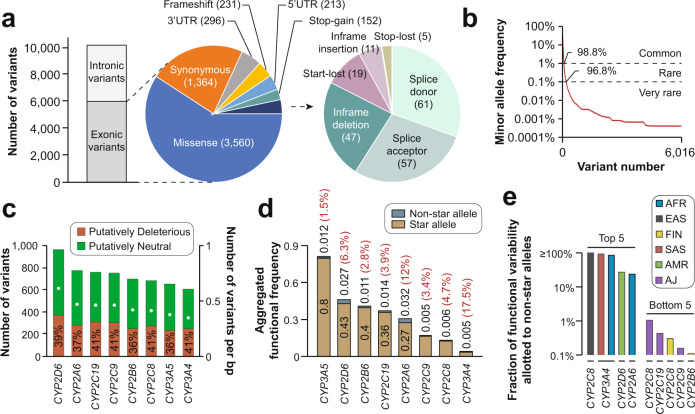
In addition to the well-characterized star alleles, CYP genes harbor a multitude of variants with unknown functional consequences. Using whole exome and whole genome sequencing data from 141,614 unrelated individuals, a total of 10,176 genetic variants were identified across the eight CYPs, of which 6016 were exonic (Fig. 2a). Notably, intronic variations are likely underreported as the majority of samples were sequencing using exome sequencing, which does not systematically cover introns. We thus we focused our further analyses exclusively on exonic variants. Among the exonic variations, missense (n = 3560; 59%) and synonymous variants (n = 1364; 23%) were most common, whereas only 19 start-lost variants, 11 in-frame insertions and 5 stop lost variants were identified. Importantly, 98.8% (n = 5891) and 96.8% (n = 5695) of the exonic variants were rare with MAFs < 1% and MAF < 0.1%, respectively (Fig. 2b). Compared to the approximately 150 variants with established functional annotations based on experimental or epidemiological data, these results indicate that the functional impact of the vast majority of variants in drug metabolizing CYPs remains to be determined.
Fig. 2. The landscape of genetic variability in major drug-metabolizing CYP genes.
a Across 141,614 individuals, a total of 10,176 genetic variants were identified 6016 of which were exonic. Pie charts indicate the distribution of exonic variants across variant classes. b The majority of exonic variants have MAFs < 1% (98.8%) or <0.1% (96.8%). c Stacked column plot showing the number of identified exonic variants for each analyzed CYP gene (left y-axis). White dots indicate the number of variants per base pair (bp; right y-axis). All variants were categorized into deleterious (red) or neutral (green) using the ADME Prediction Framework (see Methods). The fraction of putatively deleterious variants (including both reduced function and loss-of-function variants) for each CYP gene is shown. A list of all putatively deleterious variants and their estimated activity scores is provided in Supplementary Table 2. d Functional contribution of star alleles and non-star alleles to the overall functional variability of CYP genes. Black numbers indicate the aggregated frequencies of functional star alleles and non-star alleles. The ratio of the genetically encoded functional variability allotted to non-star alleles and star alleles is indicated by the red percentage values. e Functional contribution of non-star alleles to the population-specific functional variability of CYP genes. Only the top 5 and bottom 5 gene-population pairs are shown. All population-specific contributions of non-star alleles are shown in Supplementary Table 3. AFR African, EAS East Asian, SAS South Asian, AMR admixed Americans, FIN Finnish, AJ Ashkenazi Jewish, MAF minor allele frequency, UTR untranslated region.
To estimate the functional relevance of these unexplored variations at the population scale, we thus utilized computational predictions. Specifically, we used the APF algorithm (see Methods), which has been specifically developed for the interpretation of genetic variability in pharmacogenes. Overall, we identified 2175 variants that were predicted to reduce enzyme function (Supplementary Table 2). While more variants were identified in CYP2D6 (n = 962; 0.65 per bp), CYP2A6 (n = 768; 0.52 per bp) and CYP2C19 (n = 753; 0.51 per bp) and less in CYP2C8 (n = 679; 0.46 per bp), CYP3A5 (n = 654; 0.43 per bp) and CYP3A4 (n = 609; 0.4 per bp), the fraction of variants with impacts on gene function was similar across all CYPs (36–41%; Fig. 2c). Next, we aggregated the functional predictions based on computational inference with the functional variability allotted to star alleles (Fig. 2d). Expectedly, CYP3A5 harbored the overall largest fraction of altered function alleles (81.2%), followed by CYP2D6 (45.7%) and CYP2B6 (41.1%) and lowest was in CYP2C9 (16.7%), CYP2C8 (13.8%) and CYP3A4 (3.7%). Globally, star alleles accounted for majority of functionally relevant alleles with non-star alleles contributing between 1.5% for CYP3A5 and 17.5% in CYP3A4 to the total genetically encoded functional variability. However, when stratifying the analysis by population, cases were observed where the putative functional impact of non-star alleles was considerable (Fig. 2e and Supplementary Table 3). In East Asian populations, uncharacterized variants in CYP2C8 are predicted to be as relevant as the deleterious CYP2C8 star alleles *2, *3, *4, *5, *7 and *11. Similarly, non-star alleles in CYP3A4 were estimated to have similar impacts compared to star alleles in South Asian and African populations, and more than one fifth of the functional variability of CYP2D6 and CYPA6 can be attributed to non-star alleles in admixed Americans and Africans, respectively. In contrast, the functional contribution from non-star alleles in Ashkenazi Jews and Finnish are found to be marginal (Fig. 2e).
Translation of CYP genetic variability profiles into population-specific functional effects
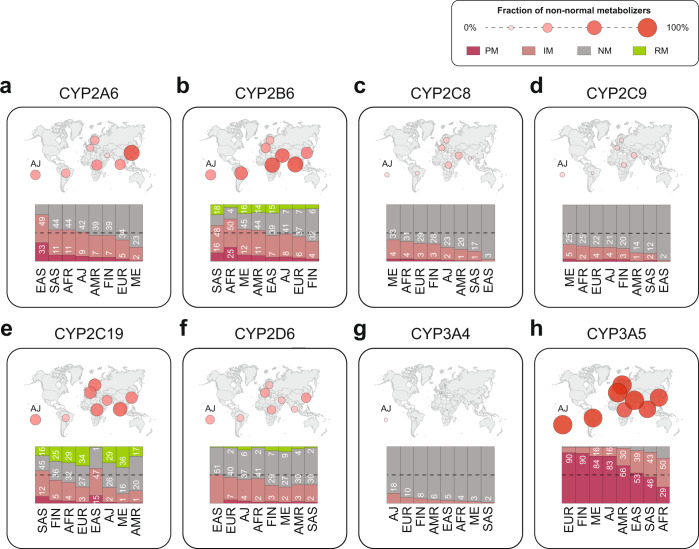
Lastly, we used the aggregated inferred functionality data of star alleles and predicted uncharacterized variants to calculate the distributions of population-specific metabolizer phenotypes (Fig. 3). Poor metabolizers (PM) and intermediate metabolizers (IM) of CYP2A6 were most common in East Asian populations (33% PM; 49% IM). In contrast, only 5% and 2% of the population were CYP2A6 PMs in Europe and the Middle East (Fig. 3a). For CYP2B6, non-normal metabolizer phenotypes were most frequent in South Asia and Africa where only 19% and 21% were classified as normalizer metabolizers (NM; Fig. 3b). By contrast, 58%, 49% and 45% of Finnish, non-Finish European and Ashkenazim individuals were inferred to be NM. Moreover, rapid metabolizers (RM) for CYP2B6 were substantially more frequent in Asian and Middle Eastern populations as well as in admixed Americans (14–18% compared to ≤7% in other populations).
Fig. 3. Global distributions of metabolic phenotypes.
Frequencies of inferred population-specific metabolic phenotypes are shown for CYP2A6 (a), CYP2B6 (b), CYP2C8 (c), CYP2C9 (d), CYP2C19 (e), CYP2D6 (f), CYP3A4 (g) and CYP3A5 (h). Numbers indicate the fraction of poor metabolizers (PM; dark red), intermediate metabolizers (IM; light red) and rapid metabolizers (RM; green). The fraction of non-normal metabolizers is indicated by the size of circles on the respective world maps. AFR African, EAS East Asian, SAS South Asian, AMR admixed Americans, FIN Finnish, AJ Ashkenazi Jewish, ME Middle Eastern, EUR non-Finnish European.
In contrast to CYP2A6 and CYP2B6, the majority of CYP2C8 and CYP2C9 classified as NM in every population analyzed (Fig. 3c, d). Impaired activity (IM or PM) of CYP2C8 and CYP2C9 was most prevalent in and Middle Easterners (38% and 30%, respectively) and Africans (35% and 27%, respectively), whereas East Asians are the most conserved among all studied populations (≤3% for both enzymes). In these calculations we considered CYP2C8*3 as a decreased function allele; however, as discussed below, functional effects of this allele are not clear and might be substrate-specific. An alternative map in which CYP2C8*2 is considered as functionally neutral is provided in Supplementary Fig. 1. For CYP2C19, South Asians were estimated to be phenotypically most variable, with 12%, 45% and 16% being classified as PM, IM and RM, respectively. In East Asians the fraction of PM (15%) and IM (47%) individuals were even higher than South Asians. However, only 1% of East Asians were CYP2C19 RM, whereas the respective frequencies in Middle Easterners (36%), Europeans (34%), Africans (29%) and Ashkenazim (29%) was substantially higher (Fig. 3e).
While CYP2D6 has a multitude of population-specific polymorphisms with functional relevance, the fraction of non-normal CYP2D6 metabolizer phenotypes does not vary substantially across different ethnogeographic groups (Fig. 3f). The fraction of CYP2D6 IMs is highest in East Asians (51% of the population), whereas PMs are very rare (<1%). The highest frequency of CYP2D6 PMs is found in Europeans (7%) and Ashkenazim (4%). Normal CYP2D6 metabolizers are most common among individuals of Southeast Asian (65%) and admixed American (63%) descent while RMs are most frequent in the Middle East (9%).
For CYP3A4, the frequency of IMs is highest in Ashkenazi Jews and Europeans with an estimated 18% and 10% of the entire population, whereas the prevalence of IMs in other populations is <10% (Fig. 3g). In contrast, the majority of individuals across all populations are classified as IMs (9–50%) or PMs (29–90%; Fig. 3h). When using the older but still widely used classification scheme of expressors (defined as individuals with at least one active CYP3A5 allele) and non-expressors (defined as individuals with two inactive CYP3A5 alleles), the range of expressors varied between 10% in Europeans and 71% in Africans.
Discussion
CYP genes are long known to be highly polymorphic with distinct genetic population differences. Comprehensive maps of interethnic differences in CYP variability have previously been presented for individual genes or alleles based on literature analyses [19–25]. Importantly however, genotyping strategies can differ between studies, which can impact allele frequency estimates particularly for genes with complex haplotype structures, such as CYP2B6 and CYP2D6. In addition, there have been multiple efforts in mapping CYP variability by genotyping individuals across populations using consistent profiling approaches [26, 27]. However, due to practical limitations, these efforts were limited to relatively small cohorts. By analyzing comprehensive sequencing data from 141,614 individuals across a total of 12 populations we here update our previous meta-analysis [11] and provide a systematic overview of the global landscape of genetic variability for the eight CYP genes of highest clinical relevance.
Among the well-characterized CYP alleles, we find considerable differences between related ethnogeographic groups. In East Asia, despite substantial admixture of Japanese, Korean and Han Chinese [28], frequencies of functionally relevant alleles can differ substantially. For instance, the reduced function allele CYP3A4*16 is common in Japanese (MAF = 3.9%), whereas it is almost absent in Koreans and other East Asian populations with important implications for the treatment with statins and immunosuppressants. Furthermore, frequencies of CYP2D6*10 and CYP2D6*41 were substantially lower in Japanese compared to other East Asian groups, suggesting that recognition of these differences might aid in the optimization of population-specific dosing recommendations for antipsychotic and antidepressant treatment [29]. Our results moreover confirm considerable differences between Finnish and other European populations. Specifically, we find substantially lower frequencies of the reduced function alleles CYP2D6*41 (3.2% in Finnish compared to 9.3% in other European populations) as well as a lack of CYP2B6*9 (0% vs. 4.4%).
The functional effects of some common alleles remain controversial. For instance, CYP2C8*3, the most frequent CYP2C8 variant allele in most populations, has been shown in vitro studies to result in reduced [30, 31], normal [32] or increased metabolism [33, 34]. Further complication is added by the fact that CYP2C8*3 is in strong linkage disequilibrium with the decreased activity allele CYP2C9*2 [35]. As both enzymes substantially overlap in their substrate specificity, this might have influenced previous clinical investigations in their interpretation that CYP2C8*3 results in decreased CYP2C8 activity [36].
Of note, phenotype assignments based on diplotypes differs between genes. For instance, for CYP2C9 NMs are defined as an activity score of 2 (i.e. individuals having two functional alleles), whereas IM status is assigned for individuals with activity scores of 1–1.5 and PMs are defined as 0–0.5 [37]. Similarly, an individual carrying a reduced function allele would be classified as IM for CYP2B6 [38] and CYP2C19 [39]. For CYP2D6 however, individuals with one reduced function and one normal function allele are classified as NMs and CYP2D6 IMs and PMs are defined as activity scores of 0–1.25 and 0, respectively [40]. Here, we followed these current established guidelines and conventions for the translation of genotypes into activity scores and metabolizer phenotypes. As a consequence of these differences in phenotype annotations, we would like to point out to the readers that individuals who carry, for instance, a normal function allele and the reduced function allele CYP2D6*10 (activity score = 0.25), which is highly common across Asia, are classified as NMs, not IMs. Similarly, homozygosity for CYP2D6*10, i.e. a diplotype activity score of 0.5, results in an IM definition, whereas the same activity would be classified as PM in other CYPs.
While well-characterized star alleles play important roles in determining CYP function, a considerable amount of heritable variability in drug response remains unexplained. For instance, elegant twin studies have shown that while 90% of the pharmacokinetic variation of metoprolol and torsemide were heritable, known genetic variants in CYP2D6 and CYP2C9 only explained 39% and 2% of the respective variation [41], raising the possibility that additional variants in these or other genes might explain at least part of the “missing heritability”. Further evidence for the potential relevance of as of yet uncharacterized variants in CYP genes comes from a retrospective study of 2087 patients taking the antidepressant escitalopram, which revealed that although common CYP2C19 alleles were well-corelated with escitalopram serum concentrations, substantial variability particularly in the *1/*1 group remained [42]. Our results indicate that, as expected, common star alleles indeed constitute the predominant genetic determinants of functional CYP variability. However, rare, as of yet uncharacterized variants are estimated to account for 1.5–17.5% of the overall genetically encoded functional variability in CYP genes. These estimates suggest that the relative contribution of rare variants in CYPs is overall lower compared to phase I and phase II enzymes or drug transporters, for which previous studies suggested that approximately 20–40% of the total variability is allotted to rare variants [43–45].
Notably, we here focused on genetic variability in coding regions of the analyzed genes. However, rare variants in regulatory or untranslated regions (UTRs) of CYP genes might also modulate gene activity. Previous studies indicated that expression of multiple CYPs, including CYP2C8, CYP2C9 and CYP2C19, was modulated by miRNAs and variants in the respective UTRs might impact these miRNA-mRNA interactions [46]. Furthermore, rare variations in regulatory regions, which are largely understudied due to the challenges associated with their functional interrogation, might contribute substantially to the missing heritability in CYP activity. Importantly, in recent years the repertoire of methodologies and algorithms to predict the functional impact of regulatory variants is rapidly increasing [47]. However, a systematic application of these tools to CYP variability in non-coding regions has not been presented and the assessment of the relative importance of such variants for inter-individual variability in drug metabolism remains an important frontier of pharmacogenomic research.
Non-star allele variants that were predicted to be deleterious by APF featured multiple population-specific variants with experimental support of reduced activity. For instance, rs181297724, causing a p.Ala161Pro amino acid exchange in CYP2C19, strongly reduced metabolic activity in vitro [48] and was found to be common in Finnish (MAF = 5.6%), whereas it was almost absent in other populations, including Europeans (MAF < 0.1%). In CYP2D6, rs79392742 (p.Ala449Asp) was detected in 3.5% of admixed American alleles and was associated with decreased activity. measured with substrates dextromethorphan and metoprolol [49]. Similarly, rs145014075 (p.Ser467X) and rs145308399 (p.Glu97Lys) in CYP2A6 are common in Ashkenazim and South Asians (MAF = 2.3%), respectively, and have been associated with decreased activity in vitro or in vivo [50, 51]. Notably, the relevance of such variants can differ substantially between populations; for instance, uncharacterized variants in CYP2C8 and CYP3A4 are estimated to be as relevant as the characterized star alleles in Asian populations, whereas the contribution of non-star allele variants in CYP2B6 and CYP2Cs in Finnish and Ashkenazi Jewish individuals is very low. These results pinpoint variants and populations in which additional functionally relevant variability in CYP genes might be discovered, which might provide useful guidance for the optimization of population-specific genotyping strategies for related drugs.
In summary, the presented analyses provide CYP allele frequencies using population-scale sequencing data broadening the data base from previous work by increasing the number of individuals from 56,945 to 141,614. We furthermore make use of improved variant calling quality as well as updated functional annotations and linkage information. In addition, we complement our previous studies by systematically considering rare variants into functional effect predictions in CYP genes based on dedicated pharmacogenomic algorithms. The presented data refines the worldwide landscape of ethnogeographic variability in CYP genes and emphasizes the importance of considering genetic differences for the optimization of population-specific pharmacotherapy and precision public health.
Supplementary information
Author contributions
YZ and VML designed the study. YZ was responsible for analyzing the data and writing the initial draft. VML was responsible for reviewing, revising the manuscript and providing feedbacks.
Funding
The work is funded by the Swedish Research Council [grant agreement numbers: 2019-01837 and 2021-02801], by the EU/EFPIA/OICR/McGill/KTH/Diamond Innovative Medicines Initiative 2 Joint Undertaking (EUbOPEN grant number 875510), and by the Robert Bosch Foundation, Stuttgart, Germany. Open access funding provided by Karolinska Institute.
Data availability
All data were available in the main tables and the supplementary information.
Competing interests
YZ and VML are co-founders and shareholders of PersoMedix AB. In addition, VML is CEO and shareholder of HepaPredict AB and discloses consultancy work for Enginzyme AB.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41397-022-00288-2.
References
- 1.Eichelbaum M, Ingelman-Sundberg M, Evans WE. Pharmacogenomics and Individualized Drug Therapy. Annu Rev Med. 2006;57:119–37. doi: 10.1146/annurev.med.56.082103.104724. [DOI] [PubMed] [Google Scholar]
- 2.Lauschke VM, Ingelman-Sundberg M. Prediction of drug response and adverse drug reactions: From twin studies to Next Generation Sequencing. Eur J Pharm Sci. 2019;130:65–77. doi: 10.1016/j.ejps.2019.01.024. [DOI] [PubMed] [Google Scholar]
- 3.Ingelman-Sundberg M. Pharmacogenetics of cytochrome P450 and its applications in drug therapy: the past, present and future. Trends Pharm Sci. 2004;25:193–200. doi: 10.1016/j.tips.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 4.Zanger UM, Schwab M. Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharm Ther. 2013;138:103–41. doi: 10.1016/j.pharmthera.2012.12.007. [DOI] [PubMed] [Google Scholar]
- 5.Zhou S-F, Liu J-P, Chowbay B. Polymorphism of human cytochrome P450 enzymes and its clinical impact. Drug Metab Rev. 2015;41:89–295. doi: 10.1080/03602530902843483. [DOI] [PubMed] [Google Scholar]
- 6.Fujikura K, Ingelman-Sundberg M, Lauschke VM. Genetic variation in the human cytochrome P450 supergene family. Pharmacogenet Genomics. 2015;25:584–94. doi: 10.1097/FPC.0000000000000172. [DOI] [PubMed] [Google Scholar]
- 7.Zhou Y, Lauschke VM. Population pharmacogenomics: an update on ethnogeographic differences and opportunities for precision public health. Hum Genet. 2021;141:1113–36. doi: 10.1007/s00439-021-02385-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nelson MR, Wegmann D, Ehm MG, Kessner D, St Jean P, Verzilli C, et al. An abundance of rare functional variants in 202 drug target genes sequenced in 14,002 people. Science. 2012;337:100–4. doi: 10.1126/science.1217876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bush WS, Crosslin DR, Owusu-Obeng A, Wallace J, Almoguera B, Basford MA, et al. Genetic variation among 82 pharmacogenes: The PGRNseq data from the eMERGE network. Clin Pharm Ther. 2016;100:160–9. doi: 10.1002/cpt.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kozyra M, Ingelman-Sundberg M, Lauschke VM. Rare genetic variants in cellular transporters, metabolic enzymes, and nuclear receptors can be important determinants of interindividual differences in drug response. Genet Med. 2017;19:20–9. doi: 10.1038/gim.2016.33. [DOI] [PubMed] [Google Scholar]
- 11.Zhou Y, Ingelman‐Sundberg M, Lauschke VM. Worldwide Distribution of Cytochrome P450 Alleles: a meta‐analysis of population‐scale sequencing projects. Clin Pharm Ther. 2017;102:688–700. doi: 10.1002/cpt.690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaedigk A, Ingelman-Sundberg M, Miller NA, Leeder JS, Whirl-Carrillo M, Klein TE, et al. The Pharmacogene Variation (PharmVar) Consortium: incorporation of the Human Cytochrome P450 (CYP) Allele Nomenclature Database. Clin Pharm Ther. 2018;103:399–401. doi: 10.1002/cpt.910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–43. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Machiela MJ, Chanock SJ. LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics. 2015;31:3555–7. doi: 10.1093/bioinformatics/btv402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Desta Z, El-Boraie A, Gong L, Somogyi AA, Lauschke VM, Dandara C, et al. PharmVar GeneFocus: CYP2B6. Clin Pharm Ther. 2021;110:82–97. doi: 10.1002/cpt.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Twist GP, Gaedigk A, Miller NA, Farrow EG, Willig LK, Dinwiddie DL, et al. Constellation: a tool for rapid, automated phenotype assignment of a highly polymorphic pharmacogene, CYP2D6, from whole-genome sequences. NPJ Genom Med. 2016;1:15007. doi: 10.1038/npjgenmed.2015.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou Y, Mkrtchian S, Kumondai M, Hiratsuka M, Lauschke VM. An optimized prediction framework to assess the functional impact of pharmacogenetic variants. Pharmacogenomics J. 2019;19:115–26. doi: 10.1038/s41397-018-0044-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yasar U, Lundgren S, Eliasson E, Bennet A, Wiman B, de Faire U, et al. Linkage between the CYP2C8 and CYP2C9 genetic polymorphisms. Biochem Biophys Res Commun. 2002;299:25–8. doi: 10.1016/S0006-291X(02)02592-5. [DOI] [PubMed] [Google Scholar]
- 19.Gaedigk A, Sangkuhl K, Whirl-Carrillo M, Klein T, Leeder JS. Prediction of CYP2D6 phenotype from genotype across world populations. Genet Med. 2017;19:69–76. doi: 10.1038/gim.2016.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fricke-Galindo I, Cespedes-Garro C, Rodrigues-Soares F, Naranjo ME, Delgado A, de Andres F, et al. Interethnic variation of CYP2C19 alleles, “predicted” phenotypes and “measured” metabolic phenotypes across world populations. Pharmacogenomics J. 2016;16:113–23. doi: 10.1038/tpj.2015.70. [DOI] [PubMed] [Google Scholar]
- 21.Petrović J, Pešić V, Lauschke VM. Frequencies of clinically important CYP2C19 and CYP2D6 alleles are graded across Europe. Eur J Hum Genet. 2020;28:88–94. doi: 10.1038/s41431-019-0480-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Céspedes-Garro C, Fricke-Galindo I, Naranjo ME, Rodrigues-Soares F, Farinas H, de Andres F, et al. Worldwide interethnic variability and geographical distribution of CYP2C9 genotypes and phenotypes. Expert Opin Drug Met. 2015;11:1893–905. doi: 10.1517/17425255.2015.1111871. [DOI] [PubMed] [Google Scholar]
- 23.Langmia IM, Just KS, Yamoune S, Brockmöller J, Masimirembwa C, Stingl JC. CYP2B6 functional variability in drug metabolism and exposure across populations—Implication for Drug Safety, Dosing, and Individualized Therapy. Front Genet. 2021;12:692234. doi: 10.3389/fgene.2021.692234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koopmans AB, Braakman MH, Vinkers DJ, Hoek HW, van Harten PN. Meta-analysis of probability estimates of worldwide variation of CYP2D6 and CYP2C19. Transl Psychiat. 2021;11:141. doi: 10.1038/s41398-020-01129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sistonen J, Sajantila A, Lao O, Corander J, Barbujani G, Fuselli S. CYP2D6 worldwide genetic variation shows high frequency of altered activity variants and no continental structure. Pharmacogenet Genomics. 2007;17:93–101. doi: 10.1097/01.fpc.0000239974.69464.f2. [DOI] [PubMed] [Google Scholar]
- 26.Mizzi C, Dalabira E, Kumuthini J, Dzimiri N, Balogh I, Basak N, et al. A European spectrum of pharmacogenomic biomarkers: implications for clinical pharmacogenomics. PLoS ONE. 2016;11:e0162866. doi: 10.1371/journal.pone.0162866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klein K, Lang T, Saussele T, Barbosa-Sicard E, Schunck WH, Eichelbaum M, et al. Genetic variability of CYP2B6 in populations of African and Asian origin: allele frequencies, novel functional variants, and possible implications for anti-HIV therapy with efavirenz. Pharmacogenet Genomics. 2005;15:861–73. doi: 10.1097/01213011-200512000-00004. [DOI] [PubMed] [Google Scholar]
- 28.Wang Y, Lu D, Chung Y-J, Xu S. Genetic structure, divergence and admixture of Han Chinese, Japanese and Korean populations. Hereditas. 2018;155:19. doi: 10.1186/s41065-018-0057-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Milosavljević F, Bukvic N, Pavlovic Z, Miljevic C, Pesic V, Molden E, et al. Association of CYP2C19 and CYP2D6 poor and intermediate metabolizer status with antidepressant and antipsychotic exposure. JAMA Psychiatry. 2021;78:270–80. doi: 10.1001/jamapsychiatry.2020.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao Y, Liu D, Wang H, Zhu J, Chen C. Functional characterization of five CYP2C8 variants and prediction of CYP2C8 genotype-dependent effects on in vitro and in vivo drug–drug interactions. Xenobiotica. 2010;40:467–75. doi: 10.3109/00498254.2010.487163. [DOI] [PubMed] [Google Scholar]
- 31.Tsukada C, Saito T, Maekawa M, Mano N, Oda A, Hirasawa N, et al. Functional characterization of 12 allelic variants of CYP2C8 by assessment of paclitaxel 6α-hydroxylation and amodiaquine N-deethylation. Drug Metab Pharmacokinet. 2015;30:366–73. doi: 10.1016/j.dmpk.2015.07.003. [DOI] [PubMed] [Google Scholar]
- 32.Rodriguez-Antona C, Niemi M, Backman JT, Kajosaari LI, Neuvonen PJ, Robledo M, et al. Characterization of novel CYP2C8 haplotypes and their contribution to paclitaxel and repaglinide metabolism. Pharmacogenomics J. 2008;8:268–77. doi: 10.1038/sj.tpj.6500482. [DOI] [PubMed] [Google Scholar]
- 33.Muschler E, Lal J, Jetter A, Rattay A, Zanger U, Zadoyan G, et al. The role of human CYP2C8 and CYP2C9 variants in pioglitazone metabolism in vitro. Basic Clin Pharm Toxicol. 2009;105:374–9. doi: 10.1111/j.1742-7843.2009.00457.x. [DOI] [PubMed] [Google Scholar]
- 34.Kaspera R, Naraharisetti SB, Tamraz B, Sahele T, Cheesman MJ, Kwok P-Y, et al. Cerivastatin in vitro metabolism by CYP2C8 variants found in patients experiencing rhabdomyolysis. Pharmacogenet Genomics. 2010;20:619–29. doi: 10.1097/FPC.0b013e32833ecace. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Speed WC, Kang SP, Tuck DP, Harris LN, Kidd KK. Global variation in CYP2C8–CYP2C9 functional haplotypes. Pharmacogenomics J. 2009;9:283–90. doi: 10.1038/tpj.2009.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martinez C, Garcia-Martin E, Blanco G, Gamito FJ, Ladero JM, Agundez JA. The effect of the cytochrome P450 CYP2C8 polymorphism on the disposition of (R)-ibuprofen enantiomer in healthy subjects. Br J Clin Pharm. 2005;59:62–9. doi: 10.1111/j.1365-2125.2004.02183.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Theken KN, Lee CR, Gong L, Caudle KE, Formea CM, Gaedigk A, et al. Clinical Pharmacogenetics Implementation Consortium Guideline (CPIC) for CYP2C9 and Nonsteroidal Anti‐Inflammatory Drugs. Clin Pharm Ther. 2020;108:191–200. doi: 10.1002/cpt.1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Desta Z, Gammal RS, Gong L, Whirl-Carrillo M, Gaur AH, Sukasem C, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for CYP2B6 and Efavirenz-Containing Antiretroviral Therapy. Clin Pharm Ther. 2019;106:726–33. doi: 10.1002/cpt.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee CR, Luzum JA, Sangkuhl K, Gammal RS, Sabatine MS, Stein CM, et al. Clinical Pharmacogenetics Implementation Consortium Guideline for CYP2C19 Genotype and Clopidogrel Therapy: 2022 Update. Clin Pharmacol Ther. 2022. 10.1002/cpt.2526 [DOI] [PMC free article] [PubMed]
- 40.Crews KR, Monte AA, Huddart R, Caudle KE, Kharasch ED, Gaedigk A, et al. Clinical Pharmacogenetics Implementation Consortium Guideline for CYP2D6, OPRM1, and COMT Genotypes and Select Opioid Therapy. Clin Pharm Ther. 2021;110:888–96. doi: 10.1002/cpt.2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Matthaei J, Brockmöller J, Tzvetkov MV, Sehrt D, Sachse-Seeboth C, Hjelmborg JB, et al. Heritability of metoprolol and torsemide pharmacokinetics. Clin Pharm Ther. 2015;98:611–21. doi: 10.1002/cpt.258. [DOI] [PubMed] [Google Scholar]
- 42.Jukić MM, Haslemo T, Molden E, Ingelman-Sundberg M. Impact of CYP2C19 Genotype on Escitalopram Exposure and Therapeutic Failure: A Retrospective Study Based on 2,087 Patients. Am J Psychiatry. 2018;175:463–70. doi: 10.1176/appi.ajp.2017.17050550. [DOI] [PubMed] [Google Scholar]
- 43.Ramsey LB, Bruun GH, Yang W, Trevino LR, Vattathil S, Scheet P, et al. Rare versus common variants in pharmacogenetics: SLCO1B1 variation and methotrexate disposition. Genome Res. 2012;22:1–8. doi: 10.1101/gr.129668.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ingelman-Sundberg M, Mkrtchian S, Zhou Y, Lauschke VM. Integrating rare genetic variants into pharmacogenetic drug response predictions. Hum Genomics. 2018;12:26. doi: 10.1186/s40246-018-0157-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xiao Q, Zhou Y, Lauschke VM. Ethnogeographic and inter-individual variability of human ABC transporters. Hum Genet. 2020;139:623–46. doi: 10.1007/s00439-020-02150-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rieger JK, Klein K, Winter S, Zanger UM. Expression variability of absorption, distribution, metabolism, excretion–related MicroRNAs in Human Liver: influence of nongenetic factors and association with gene expression. Drug Metab Dispos. 2013;41:1752–62. doi: 10.1124/dmd.113.052126. [DOI] [PubMed] [Google Scholar]
- 47.Zhou Y, Lauschke VM. Computational tools to assess the functional consequences of rare and noncoding pharmacogenetic variability. Clin Pharm Ther. 2021;110:626–36. doi: 10.1002/cpt.2289. [DOI] [PubMed] [Google Scholar]
- 48.Wang H, An N, Wang H, Gao Y, Liu D, Bian T, et al. Evaluation of the effects of 20 nonsynonymous single nucleotide polymorphisms of CYP2C19 on S-Mephenytoin 4′-Hydroxylation and Omeprazole 5′-Hydroxylation. Drug Metab Dispos. 2011;39:830–7. doi: 10.1124/dmd.110.037549. [DOI] [PubMed] [Google Scholar]
- 49.Dalton R, Lee S-B, Claw KG, Prasad B, Phillips BR, Shen DD, et al. Interrogation of CYP2D6 structural variant alleles improves the correlation between CYP2D6 genotype and CYP2D6‐mediated metabolic activity. Clin Transl Sci. 2020;13:147–56. doi: 10.1111/cts.12695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Borrego-Soto G, Perez-Paramo YX, Chen G, Satuario-Facio SK, Santos-Guzman J, Posadas-Valay R, et al. Genetic variants in CYP2A6 and UGT1A9 genes associated with urinary nicotine metabolites in young Mexican smokers. Pharmacogenomics J. 2020;20:586–94. doi: 10.1038/s41397-020-0147-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.El-Boraie A, Tanner J-A, Zhu AZ, Claw KG, Prasad B, Schuetz EG, et al. Functional characterization of novel rare CYP2A6 variants and potential implications for clinical outcomes. Clin Transl Sci. 2022;15:204–20. doi: 10.1111/cts.13135. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data were available in the main tables and the supplementary information.