Abstract
The extracellular cysteine protease from Streptococcus pyogenes is a virulence factor that plays a significant role in host-pathogen interaction. Streptococcal protease is expressed as an inactive 40-kDa precursor that is autocatalytically converted into a 28-kDa mature (active) enzyme. Replacement of the single cysteine residue involved in formation of the enzyme active site with serine (C192S mutation) abolished detectable proteolytic activity and eliminated autocatalytic processing of zymogen to the mature form. In the present study, we investigated activity of the wild-type (wt) streptococcal protease toward human fibrinogen and bovine casein. The former is involved in blood coagulation, wound healing, and other aspects of hemostasis. Treatment with streptococcal protease resulted in degradation of the COOH-terminal region of fibrinogen α chain, indicating that fibrinogen may serve as an important substrate for this enzyme during the course of human infection. Polyclonal antibodies generated against recombinant 40- and 28-kDa (r40- and r28-kDa) forms of the C192S streptococcal protease mutant exhibited high enzyme-linked immunosorbent assay titers but demonstrated different inhibition activities toward proteolytic action of the wt enzyme. Activity of the wt protease was readily inhibited when the reaction was carried out in the presence of antibodies generated against r28-kDa C192S mutant. Antibodies produced against r40-kDa C192S mutant had no significant effect on proteolysis. These data suggest that the presence of the NH2-terminal prosegment prevents generation of functionally active antibodies and indicate that inhibition activity of antibodies most likely depends on their ability to bind the active-site region epitope(s) of the protein.
The group A streptococcus (Streptococcus pyogenes) is a common human pathogen causing a wide variety of pathological conditions ranging from relatively mild diseases, such as pharyngitis and impetigo, to more serious nonsuppurative sequelae, acute rheumatic fever and glomerulonephritis. In addition, S. pyogenes can cause invasive diseases such as toxic shock syndrome and necrotizing fasciitis. S. pyogenes strains express several extracellular proteins that are involved in virulence. One of these proteins is a highly conserved extracellular cysteine protease also known as streptopain (EC 3.4.22.10) (46) or streptococcal pyrogenic exotoxin B (SpeB) (3, 17, 36). The structural gene encoding streptococcal cysteine protease is chromosomally located and is found in all natural isolates tested (48). Streptococcal protease is expressed as a 40-kDa inactive zymogen (3, 27) which upon secretion undergoes autocatalytic activation resulting in the removal of the 12-kDa NH2-terminal propeptide and formation of the mature 28-kDa active enzyme. This mechanism of conversion to active enzyme prevents unwanted protein degradation and enables spatial and temporal regulation of proteolytic activity (23). As a member of cysteine endopeptidase group of enzymes, streptococcal cysteine protease contains a Cys-His pair at the active site (26, 28, 43). Replacement of the single cysteine residue at position 192 with serine (C192S mutation) resulted in loss of detectable proteolytic activity of the enzyme and in prevention of processing of the 40-kDa zymogen to the 28-kDa mature form (14, 35).
Several lines of evidence suggest that streptococcal cysteine protease may play an important role in host-pathogen interaction. In vitro streptococcal protease has been shown to degrade extracellular matrix proteins including fibronectin and vitronectin and thus can affect the structural integrity of host tissue (20). Tissue integrity also could be damaged as a result of activation of 66-kDa human endothelial cell matrix metalloprotease by streptococcal protease, with subsequent degradation of type IV collagen (2). In addition, the protease cleaves human interleukin-1β precursor, resulting in formation of biologically active interleukin-1β and indicating an important role of this virulence factor in inflammation reaction and shock (21). Streptococcal protease also cleaves monocytic cell urokinase receptor and releases an active fragment of the receptor from the cell surface, suggesting possible involvement of this enzyme in cellular activation of plasminogen (47). In vivo data also suggest that secreted cysteine protease contributes to S. pyogenes pathogenesis. It was reported that patients with fatal invasive streptococcal infections had lower acute-phase antibody levels against cysteine protease than patients with less serious infections, indicating that anti-streptococcal protease antibody may play a protective role in humans (18). Immunization of mice with wild-type streptococcal protease conferred protection against lethal group A streptococcal infections (22), and inactivation of the structural gene encoding this enzyme significantly decreased lethality of mice following challenge with S. pyogenes (29).
In this study, we investigated the effect of the streptococcal protease on human fibrinogen and discuss pathological consequences of protease-mediated fibrinogen degradation on streptococcal infection and the wound healing process. Fibrinogen is a polyfunctional, multidomain protein involved in several aspects of hemostasis. It is mainly known as a blood clotting protein which, after thrombin-induced activation into fibrin, undergoes polymerization to prevent the loss of blood upon vascular injury. Fibrinogen, with a molecular mass of 340 kDa, consists of three pairs of nonidentical polypeptide chains, Aα, Bβ, and γ, linked together by inter- and intrachain disulfide bonds (10). The deposition at sites of trauma allows fibrin and/or fibrinogen to serve as a substrate for microbial adhesion (38). Several surface components of S. pyogenes including M (19, 16) and T (40) proteins have been identified as fibrinogen/fibrin binding proteins. Thus, both fibrin (or fibrinogen) and fibronectin binding proteins of group A streptococci may mediate initial attachment to host tissue (38), while extracellular cysteine protease that cleaves fibronectin, vitronectin, and fibrinogen may contribute to further colonization and invasion.
One of the goals of this investigation was to test whether antibodies generated against proteolytically inactive recombinant 40- and 28-kDa (r40- and r28-kDa) forms of the C192S streptococcal protease mutant can inhibit the activity of the wild-type enzyme. To address this issue, antibodies were produced in mice and rabbits, purified, and tested in inhibition assays. Human fibrinogen and resorufin-labeled bovine casein were used as substrates for monitoring proteolytic activity of the streptococcal protease and for analyzing the inhibition by the antibodies. We show that antibodies produced against the r28-kDa truncated form of the C192S mutant effectively inhibited digestion of casein or fibrinogen by cysteine protease whereas antibodies generated against the r40-kDa form of the mutant had no significant effect on proteolysis.
MATERIALS AND METHODS
Expression of r40- and r28-kDa C192S mutants of the streptococcal cysteine protease in Escherichia coli.
r40- and r28-kDa C192S streptococcal cysteine protease mutants were produced in E. coli by using the pET-28a expression vector (Novagen, Inc., Madison, Wis.). To clone designated regions of DNA encoding two forms of cysteine protease, we designed the PCR primers shown in Table 1. The forward and reverse primers contained 21 to 24 bases corresponding to the 5′- and 3′-terminal sequences of the desired coding segment. Both forward primers incorporated NcoI restriction sites immediately before the coding regions corresponding to the NH2-terminal sequence of the zymogen and mature form, respectively. A common reverse primer included a TAA stop codon immediately after the coding segment, followed by a BamHI site. The specific DNA fragments were generated by PCR using Taq DNA polymerase (Boehringer Mannheim Corp., Indianapolis, Ind.) and a template consisting of the mutated TGT→AGT streptococcal cysteine protease gene that encodes the single C192S substitution (14, 35). The amplified DNA fragments were ligated into the pCR2.1 vector (Invitrogen Corporation, Carlsbad, Calif.). After in vivo amplification, the resultant pCR2.1 plasmids were digested with NcoI and BamHI restriction enzymes (New England Biolabs, Inc., Beverly, Mass.), and 1.1- and 0.7-kb fragments were purified by agarose gel electrophoresis. These DNA fragments were ligated into pET-28a expression vector by using NcoI and BamHI restriction sites. The resulting plasmids were transformed into E. coli BL21(DE3) host cells (Novagen). For expression of the proteins, BL21(DE3) cells were grown overnight at 37°C in HSY medium (20 g of HySoy and 5 g of yeast extract per liter, 10 mM NaCl, 10 mM potassium phosphate [pH 7.2]) containing 50 μg of kanamycin per ml. Overnight cultures were diluted 1:100 with fresh HSY medium, grown to an optical density at 600 nm of 1.5, induced with 1 mM isopropyl-β-d-thiogalactopyranoside for 3 h, harvested by centrifugation, and lysed by the freeze-thaw method.
TABLE 1.
Primers used in this study
| Recombinant protein | Primera
|
|
|---|---|---|
| Orientation | Sequence | |
| 40-kDa C192S precursor of streptococcal protease (Asp 28-Pro 398) | Fwd | CC ATG GAT CAA AAC TTT GCT CGT AAC G |
| NcoI | ||
| Rev | GGA TCC TTA CTA AGG TTT GAT GCC TAC AAC AGC | |
| BamHI | ||
| 28-kDa C192S streptococcal protease (Gln 146-Pro 398) | Fwd | CC ATG GAA CCA GTT GTT AAA TCT CTC CTT G |
| NcoI | ||
| Rev | GGA TCC TTA CTA AGG TTT GAT GCC TAC AAC AGC | |
| BamHI | ||
Fwd, forward; Rev, reverse.
Purification of r40- and r28-kDa C192S streptococcal cysteine protease mutants.
The r40-kDa C192S mutant was purified from the soluble fraction of bacterial lysate. After overnight dialysis against 20 mM Tris (pH 7.2), the sample was loaded onto a DyeMatrex Red A gel (Amicon, Inc., Beverly, Mass.) affinity column equilibrated with the same buffer. Bound protein was eluted with a 0 to 2 M NaCl gradient in 20 mM Tris (pH 7.2) and dialyzed against 20 mM Tris (pH 7.4)–0.15 M NaCl (TBS [Tris-buffered saline]).
Since the r28-kDa C192S protease mutant expressed in E. coli was exclusively in an insoluble form, we prepared a mature or truncated form of protein by limited proteolysis of the r40-kDa C192S proenzyme. The r40-kDa C192S form (2 mg/ml) was digested with thermolysin (Fluka Chemical Corp., Ronkonkoma, N.Y.) or pepsin (Worthington Biochemical Corporation, Freehold, N.J.) in 20 mM Tris (pH 7.2) or 100 mM Gly (pH 3.0), respectively. Reactions were carried out at an enzyme/substrate ratio of 1:30 (wt/wt) at 25°C. Analytical digestion of the r40-kDa C192S mutant with elastase (Worthington) was performed in 20 mM Tris (pH 7.2)–500 mM NaCl buffer at 25°C. The enzyme/substrate ratio of this reaction was 1:50 (wt/wt). Time course digestion was monitored by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), performed either with the Bio-Rad electrophoresis system using 15% homogeneous gels (Bio-Rad Laboratories, Hercules, Calif.) or with the Pharmacia Phast system using 8 to 25% gradient gels (Pharmacia Biotech Inc., Piscataway, N.J.). All SDS-polyacrylamide gels in this study were stained with Coomassie brilliant blue R (Sigma Chemical Co., St. Louis, Mo.) or Coomassie R-350 (Pharmacia Biotech) solution. The r28-kDa C192S form of protease was prepared from a 5-h thermolysin or pepsin digest. Affinity chromatography on the DyeMatrex Red A Gel column was used for further purification of the r28-kDa C192S cysteine protease mutant. The thermolysin digestion mixture was directly loaded onto an affinity column equilibrated with 20 mM Tris (pH 7.2), and bound protein was subsequently eluted with a 0 to 2 M NaCl gradient in 20 mM Tris (pH 7.2). Pepsin hydrolysis was terminated by shifting the pH of the reaction mixture from acidic to neutral with 1 M Tris (pH 8.0); after overnight dialysis against 20 mM Tris (pH 7.2), the sample was loaded onto the DyeMatrex Red Gel column. The elution profiles of thermolysin and pepsin digestion mixtures exhibited single peaks, each of which contained a protein with a relative mobility corresponding to a molecular mass of 28 kDa according to SDS-PAGE analysis. Protein containing fractions were pooled and dialyzed against TBS (pH 7.4). Both r40- and r28-kDa streptococcal protease mutants were concentrated and stored frozen at −20°C.
Purification of wild-type 28-kDa streptococcal cysteine protease from S. pyogenes.
Native 28-kDa streptococcal cysteine protease was purified from the culture supernatant of S. pyogenes MGAS 1719 as described previously (21).
Protein concentrations.
Protein concentrations were determined spectrophotometrically, using extinction coefficients calculated from the amino acid composition by the following equation: E280,0.1% = (5,690W + 1,280Y)/m, where W and Y represent the number of Trp and Tyr residues and m represents molecular mass (11, 37). The following values of E280,0.1% were obtained for proteins: wild-type and recombinant 28-kDa C192S streptococcal protease, 1.52; and r40-kDa C192S streptococcal protease, 1.21.
Sequence analysis.
NH2-terminal sequence analysis was performed with an Applied Biosystems model 477A sequenator. The NH2 termini were determined by direct sequencing for 10 to 12 cycles.
Protease activity assays.
Streptococcal cysteine protease activity assay was performed with resorufin-labeled casein (Boehringer Mannheim) as described by Twining (44). The wild-type streptococcal cysteine protease was activated by 30-min incubation with 10 mM dithiothreitol at room temperature followed by overnight dialysis at 4°C against TBS (pH 7.4). To assess proteolytic activity of the enzyme, 1.25 μg of cysteine protease was incubated for various times at 37°C in the presence of 0.4% resorufin-labeled casein in 50 mM Tris (pH 7.8)–5 mM CaCl2 buffer. Undigested substrate was removed by 5% trichloroacetic acid precipitation; after centrifugation, the absorbance of released resorufin-labeled peptides in the supernatant fractions was measured spectrophotometrically at 574 nm. Alternatively, activity of streptococcal protease was analyzed by detecting digestion of human fibrinogen (91% clottable; Sigma). In this assay, 1.25 μg of streptococcal protease was incubated in the presence of 250 μg of fibrinogen, resulting in an enzyme/substrate ratio of 1:200 (wt/wt). The reaction was carried out in TBS (pH 7.4) for various times at 25°C. The incubation mixture was then transferred into solution containing 10% β-mercaptoethanol and/or 2% SDS, heated at 95°C, and analyzed by SDS-PAGE performed with the Bio-Rad electrophoresis system using precast 4 to 20% gradient gels.
Western blot analysis of the streptococcal protease-mediated digestion of human fibrinogen was performed with monoclonal mouse anti-fibrinogen Aα chain (Aα epitope 529-539) antibody NYB 1C2-2 (Accurate Chemical and Scientific Corp., Westbury, N.Y.).
The specificity of casein and fibrinogen digestion was confirmed by using the specific cysteine protease inhibitor N-[N-(l-3-trans-carboxyoxirane-2-carbonyl)-l-leucyl]-agmatine (E-64; Boehringer Mannheim). Proteolysis of casein or fibrinogen by streptococcal cysteine protease was completely blocked in the presence of 20 μM E-64.
Generation of antisera, purification of IgG fractions, and ELISA.
Female Swiss Webster mice, 6 to 8 weeks old, were immunized subcutaneously with 5 μg of r40- or r28-kDa C192S streptococcal cysteine protease mutants in the presence of 50 μg of 3-O-deacylated monophosphoryl lipid A (Ribi ImmunoChem Research, Hamilton, Mont.) and 100 μg of aluminum phosphate at weeks 0, 3, and 5. Samples of mouse sera were collected at weeks 0 (before the first immunization), 3, 5, and 7. Female New Zealand White rabbits were immunized intradermally with 50 μg of r40- or r28-kDa C192S streptococcal cysteine protease mutants in the presence of complete Freund’s adjuvant at week 0, followed by subsequent intramuscular immunizations with 100 μg of protein in the presence of incomplete Freund’s adjuvant at weeks 4 and 8. Samples of rabbit sera were collected at week 0 (before the first immunization), 6, and 10. The immunoglobulin G (IgG) fractions of the rabbit antisera were purified by affinity chromatography using a 5-ml HiTrap protein G-Sepharose column (Pharmacia Biotech). The titers of both mouse antisera and purified rabbit IgG were determined by an enzyme-linked immunosorbent assay (ELISA) using plates coated with wild-type or mutated cysteine protease species.
Antibody-mediated inhibition of the streptococcal cysteine protease.
The inhibition activities of mouse antisera and purified rabbit IgG were examined as follows. The wild-type streptococcal cysteine protease (1.25 μg) was incubated for 5 h at 37°C with 0.4% resorufin-labeled casein in 50 mM Tris (pH 7.8)–5 mM CaCl2 buffer, in the presence of increasing concentrations of preimmune or immune anti-r40-kDa and anti-r28-kDa C192S mutant mouse sera or rabbit IgG. Mouse and rabbit antisera (IgG) tested in these studies were collected at weeks 7 and 10, respectively. The total volume of each reaction mixture was 0.2 ml. Undigested substrate and sera components were removed by 5% trichloroacetic acid precipitation; after centrifugation, the absorbance of released resorufin-labeled peptides in the supernatant fractions was measured spectrophotometrically at 574 nm.
The inhibition activity of the rabbit IgG was also tested by monitoring of the streptococcal protease-mediated degradation of the human fibrinogen. In these experiments, 250 μg of fibrinogen was treated with 1.25 μg of streptococcal protease in the presence of 20 μg of IgG purified from preimmune or immune anti-r40-kDa and anti-r28-kDa C192S sera, resulting in an enzyme/antibody ratio of 1:16 (wt/wt). The reaction was carried out in TBS (pH 7.4) at 25°C. At various times, aliquots were taken and reaction was terminated by heating sample at 95°C in the presence of 10% β-mercaptoethanol and 2% SDS. All samples were analyzed by SDS-PAGE performed with the Bio-Rad electrophoresis system using precast 4 to 20% gradient gels. All experiments in this study were performed three to four times; data from one representative experiment are shown.
RESULTS
Preparation of r40- and r28 kDa C192S streptococcal protease mutants.
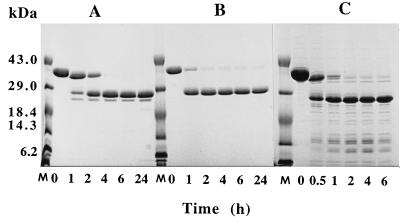
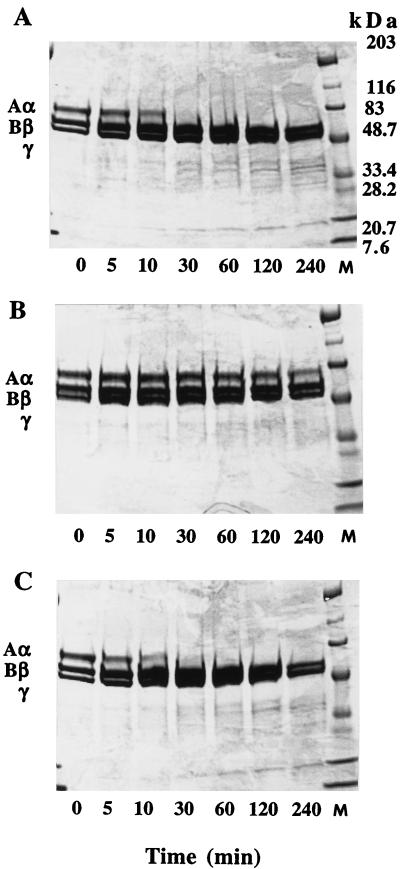
The r40- and r28-kDa C192S streptococcal protease mutants comprising residues Asp 28 through Pro 398 and Gln 146 through Pro 398, respectively, were produced in E. coli by using the pET-28a expression vector as described in Materials and Methods. Both proteins contained an NH2-terminal formylmethionyl residue that initiated translation. The r40-kDa C192S mutant was found predominantly in the insoluble fraction, and only 10 to 15% of the protein remained soluble. In all preparations described here, the total soluble fraction of the E. coli lysate was used as the starting point for purification. The final yield of purified r40-kDa C192S streptococcal protease mutant varied between 30 and 40 mg/liter of bacterial culture. The r28-kDa C192S mutant was found exclusively in the insoluble fraction of bacterial lysate; therefore, limited proteolysis of the r40-kDa C192S form of streptococcal protease was used for generation of the desired protein. To select appropriate conditions for preparation of the truncated version of the streptococcal protease mutant, we digested the protein with different proteases. The results obtained with elastase, pepsin, and thermolysin are presented in Fig. 1. In all cases, digestion of the r40-kDa C192S streptococcal protease produced a major discrete 28-kDa fragment which seems to be the terminal product of proteolysis. The 28-kDa products of limited proteolysis were purified from both pepsin and thermolysin digests, and their NH2 termini were determined by direct sequencing for several cycles. Both proteins consistently displayed a single sequence, IKQPVVKSLLD, corresponding to the NH2-terminal sequence of the mature form of streptococcal protease preceded by two extra residues, Ile 144 and Lys 145, derived from the propeptide region of the protein. Thus, the 28-kDa form of the streptococcal protease mutant can be readily generated by limited proteolysis of the r40-kDa C192S precursor with a variety of proteases. The yield of thermolysin or pepsin generated 28-kDa C192S streptococcal protease corresponded to 50% of starting protein and thus varied between 15 and 20 mg/liter of bacterial culture. The homogeneity of the purified mutants and wild-type streptococcal protease was checked by SDS-PAGE. All proteins exhibited single bands on SDS-PAGE with relative mobility consistent with their expected molecular masses (Fig. 2).
FIG. 1.
Treatment of the r40-kDa C192S precursor of the streptococcal cysteine protease with elastase (A), pepsin (B), and thermolysin (C), analyzed by SDS-PAGE (15% gel). Lanes M, molecular mass standards.
FIG. 2.
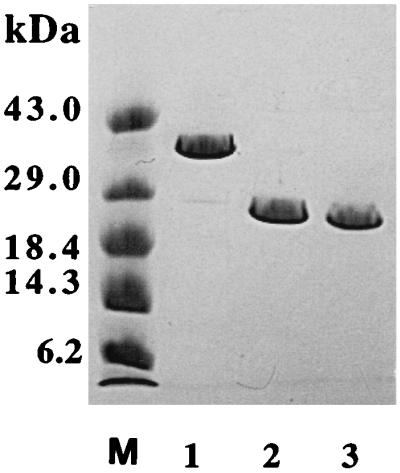
SDS-PAGE (8 to 25% gel) analysis of the purified r40-kDa C192S precursor of the streptococcal protease (lane 1), thermolysin generated r28-kDa C192S streptococcal protease (lane 2), and wild-type 28-kDa streptococcal protease (lane 3). Lane M, molecular mass standards.
Cleavage of human fibrinogen by wild-type streptococcal cysteine protease.
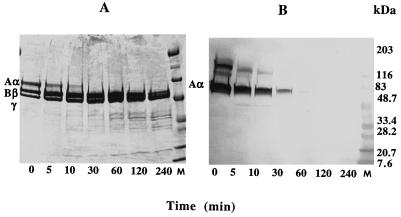
To investigate the effect of streptococcal protease on human fibrinogen, the course of digestion was studied by examining a series of SDS-gels under nonreduced and reduced conditions. Cleavage of fibrinogen at an enzyme/substrate ratio 1:200 (wt/wt) produced a single fragment which upon 4-h incubation with streptococcal protease exhibited a gradual increase of mobility on SDS-PAGE from 340 to 260 kDa (not shown). To identify the region of fibrinogen molecule attacked by streptococcal protease, the digestion mixture was analyzed by SDS-PAGE at reduced conditions. Electrophoresis of a sample corresponding to starting material showed bands in the positions expected for the intact Aα (66 kDa, 610 amino acid residues), Bβ (54 kDa, 461 amino acid residues), and γ (48 kDa, 411 amino acid residues) chains of fibrinogen. Upon incubation with streptococcal protease, the Bβ and γ chains appeared to be the same as those of fibrinogen whereas the Aα chain gradually disappeared, resulting in the formation of a truncated species of about 50 kDa (Fig. 3A). Degradation of the Aα chain was accompanied by accumulation of low-molecular-mass fragments ranging from 35 to 10 kDa.
FIG. 3.
Time course of digestion of human fibrinogen with streptococcal protease analyzed by SDS-PAGE (4 to 20% gel) (A) and Western blotting (B) under reducing conditions. Lanes M, molecular mass standards.
To localize the streptococcal protease-sensitive area within the Aα chain of fibrinogen, the digestion mixture was studied by Western blot analysis using anti-fibrinogen Aα chain monoclonal antibody NYB 1C2-2, which recognizes the COOH-terminal portion of the Aα chain (Aα epitope 529-539) (39). Figure 3B shows Western blot analysis of the same course of digestion reaction presented in Fig. 3A. The major band represents intact Aα chain that reacts with NYB 1C2-2. Incubation with streptococcal protease resulted in depletion of this band, suggesting COOH-terminal cleavage of the Aα chain and removal of the 529-539 epitope portion of the protein. After 1 h of treatment with protease, no detectable binding of NYB 1C2-2 with the Aα chain or its degradation products was observed. Thus, the above results indicate that streptococcal protease effectively degrades human fibrinogen by attacking preferentially the COOH-terminal portion of its Aα chain (αC domain), while the Bβ and γ chains remain relatively resistant to proteolysis. When 20 μM E-64 inhibitor was added to the reaction mixture, no sign of fibrinogen degradation was observed after 24 h of incubation with streptococcal protease at an enzyme/substrate ratio of 1:50 (not shown).
Digestion of casein-resorufin by streptococcal cysteine protease.
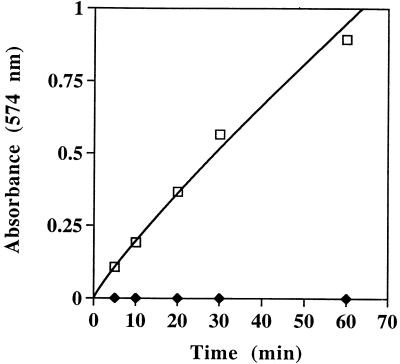
For analysis of proteolytic activity of the wild-type streptococcal protease, we also used an approach based on spectrophotometric determination of digestion of a commonly used substrate, casein. Treatment of resorufin casein with the wild-type streptococcal protease resulted in an increase in absorbance at 574 nm due to the release of resorufin-labeled peptides from casein. No increase of absorbance was observed upon incubation of casein-resorufin with the r28-kDa C192S streptococcal protease mutant (Fig. 4). These results suggest that under the tested conditions, wild-type streptococcal protease exhibited detectable proteolytic activity whereas the processed C192S mutant, lacking propeptide, was completely inactive. The results also indicate that the casein-resorufin substrate-based assay can serve as a convenient approach for evaluating the effects of antibodies generated against the r40- and r28-kDa C192S streptococcal protease mutants on activity of the wild-type enzyme.
FIG. 4.
Influence of the incubation time on the casein-resorufin hydrolysis by wild-type streptococcal protease (squares) and the r28-kDa C192S streptococcal protease mutant (diamonds).
Inhibition of proteolytic activity of the wild-type streptococcal cysteine protease by antibodies produced against r40- and r28-kDa C192S mutants.
Mouse antisera collected at week 7 and rabbit IgG purified from antisera collected at week 10 exhibited the highest ELISA titers against both r40-kDa and r28-kDa C192S streptococcal protease antigens. As shown in Table 2, in mice, the r40-kDa C192S streptococcal protease mutant elicited an antibody titer 25 times higher against itself than against the heterologous (r28-kDa C192S) form, indicating an important role of the 12-kDa NH2-terminal propeptide in the immune response. In contrast, antibodies produced in rabbits exhibited similar titers against homologous and heterologous forms of the C192S mutant. When the wild-type streptococcal protease was used as an antigen in ELISAs, antibody titers were very similar to those of the r28-kDa C192S mutant, suggesting that the single C192S amino acid substitution in streptococcal protease most likely did not affect the antigenicity of the protein.
TABLE 2.
Cross-reactivity of mouse antisera and rabbit IgG to different forms of streptococcal cysteine proteasea
| Tested antigen | ELISA titer of mouse antiserumb
|
ELISA titer of rabbit IgGc
|
||
|---|---|---|---|---|
| r40-kDa C192S streptococcal protease | r28-kDa C192S streptococcal protease | r40-kDa C192S streptococcal protease | r28-kDa C192S streptococcal protease | |
| r40-kDa C192S streptococcal protease | 1,602,749 | 154,012 | 1,493,202 | 824,487 |
| r28-kDa C192S streptococcal protease | 61,771 | 138,926 | 754,740 | 702,198 |
| Wild-type 28-kDa streptococcal protease | 56,042 | 380,779 | 1,165,038 | 1,300,976 |
The r28-kDa C192S streptococcal protease was prepared by treatment of the r40-kDa C192S streptococcal protease precursor with thermolysin.
ELISA titers are for IgG antibodies of pooled sera from five mice. Serum was collected at week 7.
ELISA titers are for purified IgG samples from individual rabbits. IgG antibodies purified from serum collected at week 10 were diluted to 1 mg/ml and used as starting material for further analysis.
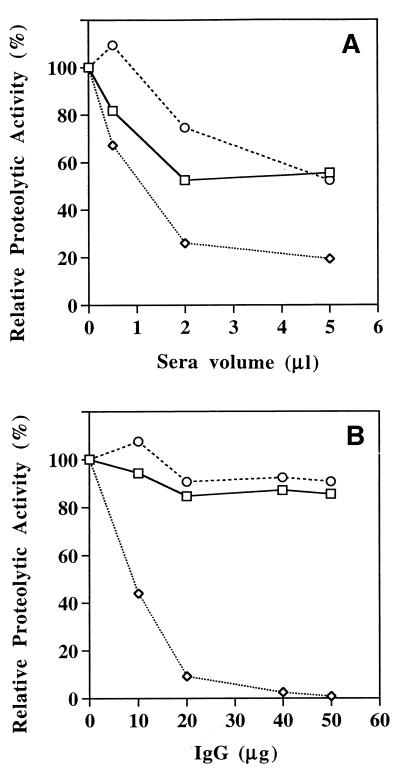
To determine whether antibodies generated against r40- and r28-kDa C192S mutants were capable of inhibiting proteolytic activity of the wild-type streptococcal protease, we studied the activity of the enzyme as a function of antibody concentration in the reaction mixture. The results in Fig. 5A show that hydrolysis of casein-resorufin is inhibited in the presence of increasing concentrations of serum from vaccinated mice relative to a control (preimmune) sera. Inhibition activity of the serum generated against the r28-kDa C192S streptococcal protease was higher than that of the serum against r40-kDa C192S form. To eliminate nonspecific inhibition effect of control (preimmune) sera on the streptococcal cysteine protease, we analyzed the caseinolytic activity of the enzyme in the presence of IgG purified from immunized and nonimmunized rabbits (Fig. 5B). Proteolytic activity of the streptococcal protease was totally inhibited with 50 μg of IgG purified from serum of a rabbit immunized with r28-kDa C192S streptococcal protease. In contrast, IgG purified from serum of the rabbit that was immunized with the r40-kDa C192S mutant had no effect on activity of the streptococcal protease and exhibited properties similar to those of the control (preimmune) IgG (Fig. 5B).
FIG. 5.
Effect of mouse antisera (A) and rabbit IgG (B) on streptococcal protease caseinolytic activity. Mouse serum and rabbit IgG from animals immunized with r40-kDa C192S precursor of the streptococcal protease (squares) or r28-kDa C192S streptococcal protease (diamonds) and from control, nonimmunized animals (circles) are indicated.
To further investigate the inhibition activity of rabbit IgG antibodies, we used an assay based on SDS-PAGE monitoring of the streptococcal protease-mediated cleavage of human fibrinogen. The time course of the digestion reaction was performed as described in Materials and Methods in the presence of 20 μg of purified IgG (Fig. 6). Within 10 min of reaction, in the presence of anti-r40-kDa C192S mutant IgG (Fig. 6A) or preimmune IgG (Fig. 6C), the Aα chain is significantly depleted. This is again accompanied by the appearance of low-molecular-mass degradation products ranging between 35 and 10 kDa. Further incubation leads to the disappearance of the band corresponding to intact Aα chain and the appearance of a higher-mobility band representing the truncated form of the Aα chain, which comigrates with the Bβ and γ chains. Figure 6B presents results obtained in the presence of anti-r28-kDa C192S mutant IgG. The band representing the Aα chain remains intact and does not undergo a shift in mobility, indicating efficient inhibition of streptococcal protease activity by this type of IgG. The above results clearly demonstrate that antibodies generated against the r28-kDa C192S streptococcal protease exhibit inhibition activity, and thus the mutant corresponding to mature enzyme provides the best option as a potential vaccine candidate.
FIG. 6.
Effect of rabbit IgG on streptococcal protease fibrinogenolytic activity analyzed by SDS-PAGE (4 to 20% gel) under reducing conditions. Time course of digestion reaction was performed in the presence of IgG purified from rabbits immunized with r40-kDa C192S precursor of the streptococcal protease (A) or r28-kDa C192S streptococcal protease (B) and from control, nonimmunized rabbit (C). Lanes M, molecular mass standards.
DISCUSSION
In vitro experiments revealed that several human proteins, including fibronectin, vitronectin (20), 66-kDa human endothelial cell matrix metalloprotease (2), interleukin-1β precursor (21), and monocytic cell urokinase receptor (47), could serve as substrates for streptococcal cysteine protease. These data show broad specificity of the streptococcal protease and its ability to interfere with a number of important physiological processes in humans. In this study, we examine streptococcal protease-mediated degradation of human fibrinogen. Our results clearly demonstrate that streptococcal protease cleaves soluble fibrinogen in a time-dependent manner, preferentially attacking the COOH-terminal regions of the Aα chains. SDS-PAGE analysis of the reaction mixture at reduced conditions revealed high susceptibility of the Aα chain to streptococcal protease digestion, while the Bβ and γ chains were less readily available to proteolysis. After 5 min of digestion at an enzyme/substrate ratio of 1:200, the band corresponding to intact Aα chain was significantly depleted, and no intact Aα chain was detected after 10 min of reaction (Fig. 3A). Progressive reduction of the intact Aα chain was also observed when the digestion mixture was studied by Western blot analysis using anti-fibrinogen Aα chain monoclonal antibody NYB 1C2-2, directed against the 529-539 epitope (Fig. 3B). Incubation of fibrinogen with protease results in complete elimination of the binding of monoclonal antibody NYB 1C2-2 with the Aα chain, suggesting the removal of the epitope-containing portion from the polypeptide chain. Interestingly, NYB 1C2-2 did not react with the degradation products of the Aα chain either. None of the low-molecular-weight fragments derived from the COOH-terminal portion of the Aα chain (Fig. 3A) interacted with NYB 1C2-2 (Fig. 3B), indicating that one of the cleavage sites is located within the 529-539 epitope region. Thus, results of the Western blotting analysis confirmed that streptococcal protease cleaves the COOH-terminal portion of the Aα chains. The high sensitivity of the COOH-terminal regions of the fibrinogen Aα chain to the action of the streptococcal cysteine protease reported in this study may explain the frequently observed Aα heterogeneity of human fibrinogen preparations (41, 42). Although the COOH-terminal regions of the Aα chains of fibrinogen (αC domains) are sensitive to a variety of proteases (9, 31), including plasmin (15, 33), trypsin (33), leukocyte (1), and platelet (25) proteases, it is still not known what causes cleavage in vivo. Sequence analysis of the Aα peptides extracted from human blood filtrate revealed that these fragments are generated at known plasmin attack sites and at several novel cleavage sites, especially at hydrophobic amino acid residues (42). Taking into account that streptococcal protease preferentially cleaves polypeptides with hydrophobic residues at P2, P1, and P1′ positions (46), it seems likely that streptococcal infections in humans and fibrinogenolytic activity of the secreted cysteine protease described in this study may be responsible for the existence of fibrinogen species with COOH-terminal truncated Aα chains.
The αC domains are involved in a number of activities within fibrinogen and fibrin. They play an important role in fibrin polymer assembly, presumably being involved in lateral aggregation of protofibrils (45). They also control activation of factor XIII (7) and serve subsequently as its substrate, becoming cross-linked to each other (5, 31) and to fibronectin (34, 30, 32). While covalent cross-linking between fibrin molecules is essential for clot structural stability, the presence of fibronectin with its multiple adhesive domains is important for the cell adhesion and migration events required for wound healing. Factor XIIIa-catalyzed cross-linking of fibronectin to αC domains is required for maximal cell adhesion to a fibronectin-fibrin matrix (4). Covalent incorporation of fibronectin into fibrin clot provides an effective substrate for attachment, spreading, and migration of fibroblasts (12, 24). Thus, the ability of streptococcal protease to cleave both fibronectin (20) and the COOH-terminal regions of fibrinogen Aα chain indicates that this virulence factor may inhibit the wound healing process. This finding is important for a better understanding of the molecular mechanisms involved in pathological conditions such as necrotizing infection of soft tissues in humans caused by S. pyogenes as a result of penetrating injuries, surgical procedures, and minor cuts. Additional studies will be required to elucidate biological properties of generated fibrinogen fragments and the effect of fibrinogen cleavage on the bacterium’s ability to cause severe, invasive infections.
One of the goals of this study was to produce antibodies against two forms of the C192S streptococcal protease mutant (40 and 28 kDa) and to investigate whether they are capable of inhibiting proteolytic activity of the wild-type enzyme. Two forms of the C192S streptococcal protease mutant, the 40-kDa proenzyme (residues 28 to 398) and truncated 28-kDa version (residues 146 to 398), were expressed with high yield in E. coli by using the pET-28a expression vector. The r28-kDa truncated form was found in the insoluble fraction of the bacterial lysate, while the r40-kDa form was present in both soluble and insoluble fractions. Interestingly, we observed similar distribution of the expressed streptococcal protease mutants with another expression vector, pTrc99A (Pharmacia Biotech). These results are consistent with multiple data suggesting that prosegments of the cysteine proteases play an important role in the folding reaction during protein synthesis (23). Although only about 10 to 15% of the r40-kDa C192S streptococcal protease mutant was recovered from the soluble fraction of E. coli lysate, it was sufficient to purify up to 40 mg of protein from 1 liter of bacterial culture. The r40-kDa precursor of the C192S streptococcal protease mutant was used as a starting material for conversion to the 28-kDa mature form of the mutant. Limited proteolysis of the r40-kDa form with a variety of enzymes, including elastase, pepsin, and thermolysin, resulted in the formation of essentially the same truncated 28-kDa form of the C192S streptococcal protease mutant (Fig. 1). These observations are consistent with the report of Liu and Elliott (27) that treatment of the wild-type streptococcal zymogen with trypsin and subtilisin produced the mature enzyme. The availability of the NH2-terminal segment of the proenzyme to proteases with different specificities and the resistance of the remaining portion of the molecule to further digestion indicates a possible mechanism which triggers autocatalytic activation of the streptococcal protease in vivo.
Substitution of a single cysteine residue at position 192 with serine in the 40-kDa streptococcal cysteine proenzyme resulted in the elimination of autocatalytic processing of the precursor (35) and the absence of detectable proteolytic activity after removal of the NH2-terminal prosegment (14). These observations were confirmed in the present study. Comparison of the caseinolytic activity of the wild-type and mutated forms of streptococcal protease revealed that the wild-type enzyme cleaves casein-resorufin, while the thermolysin-generated r28-kDa C192S mutant exhibited no detectable proteolytic activity (Fig. 4). It seems unlikely that the C192S amino acid substitution would result in the loss of structural integrity of the streptococcal protease with subsequent elimination of enzymatic activity. This conclusion is based on available crystallographic data for the Cys→Ser mutants of several cysteine proteases, including rat procathepsin B (8), human procathepsin L (6), and papain-like protease procaricain from Carica papaya (13). Three-dimensional structures of mutated enzymes clearly indicated that such an amino acid substitution did not affect overall fold of these proteins. Further evidence supporting this conclusion is provided by the results of ELISAs in this study. Comparison of the ELISA titers revealed that antibodies generated against the r28-kDa C192S streptococcal protease bind equally well to the truncated form of the mutant and to the wild-type streptococcal protease, indicating similar degrees of availability of epitopes in both proteins. Interestingly, ELISA titers of both anti-r28- and anti-r40-kDa C192S protease mouse sera or rabbit IgG toward wild-type streptococcal cysteine protease (Table 2) did not correlate with the ability of these antibodies to inhibit the enzyme (Fig. 5 and 6). Digestion of both casein and fibrinogen by streptococcal protease was readily inhibited by antibodies generated against the r28-kDa C192S streptococcal protease mutant, while antibodies produced against the r40-kDa C192S mutant had little or no effect (Fig. 5 and 6). These data suggest that only a small fraction of antibodies generated against the r28-kDa C192S streptococcal protease mutant that are capable of binding to the active site area of the protein are responsible for inhibition activity. Since the major function of the activation prosegment in the cysteine protease class of enzymes is to sterically block the active site and thus inhibit unwanted protein degradation (13, 23), the presence of the NH2-terminal prosegment in the zymogen 40-kDa form of streptococcal protease mutant makes the active site unavailable to the immune system and prevents the generation of functionally active antibodies. Its seems that the r28- and r40-kDa antigens are processed differently by the immune system, and therefore removal of the NH2-terminal prosegment from streptococcal protease mutant prior to immunization is critical for the generation of antibodies with maximum inhibition activity.
ACKNOWLEDGMENTS
This work was supported in part by Public Health Service grant AI-33119 and Texas Technology Development and Transfer grant 004949-036 to J. M. Musser.
We thank E. Bortell for protein sequence analysis and K. Belanger for performing ELISAs. Thanks also go to E. Baranyi-Thomas for help with preparation of the manuscript.
REFERENCES
- 1.Bilezikian S B, Nossel H L. Unique pattern of fibrinogen cleavage by human leukocyte proteases. Blood. 1977;50:21–28. [PubMed] [Google Scholar]
- 2.Burns E H, Marciel A M, Musser J M. Activation of a 66-kilodalton human endothelial cell matrix metalloprotease by Streptococcus pyogenes extracellular cysteine protease. Infect Immun. 1996;64:4744–4750. doi: 10.1128/iai.64.11.4744-4750.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chaussee M S, Gerlach D, Yu C E, Ferretti J J. Inactivation of the streptococcal erythrogenic toxin B gene (speB) in Streptococcus pyogenes. Infect Immun. 1993;61:3719–3723. doi: 10.1128/iai.61.9.3719-3723.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corbett S A, Lee L, Wilson C L, Schwarzbauer J E. Covalent cross-linking of fibronectin to fibrin is required for maximal cell adhesion to a fibronectin-fibrin matrix. J Biol Chem. 1997;272:24999–25005. doi: 10.1074/jbc.272.40.24999. [DOI] [PubMed] [Google Scholar]
- 5.Cottrell B A, Strong D D, Watt K W K, Doolittle R F. Amino acid sequence studies on the α chain of human fibrinogen. Exact location of cross-linking acceptor sites. Biochemistry. 1979;18:5405–5410. doi: 10.1021/bi00591a023. [DOI] [PubMed] [Google Scholar]
- 6.Coulombe R, Grochulski P, Sivaraman J, Menard R, Mort J S, Cygler M. Structure of human procathepsin L reveals the molecular basis of inhibition by the prosegment. EMBO J. 1996;15:5492–5503. [PMC free article] [PubMed] [Google Scholar]
- 7.Credo R B, Curtis C G, Lorand L. α-Chain domain of fibrinogen controls generation of fibrinoligase (coagulation factor XIIIa). Calcium Ion regulatory aspects. Biochemistry. 1981;20:3770–3778. doi: 10.1021/bi00516a016. [DOI] [PubMed] [Google Scholar]
- 8.Cygler M, Sivaraman J, Grochulski P, Coulombe R, Storer A C, Mort J S. Structure of rat procathepsin B: model for inhibition of cysteine protease activity by the proregion. Structure. 1996;4:405–416. doi: 10.1016/s0969-2126(96)00046-9. [DOI] [PubMed] [Google Scholar]
- 9.Doolittle R F, Watt K W K, Cottrell B A, Strong D D, Riley M. The amino acid sequence of the α-chain of human fibrinogen. Nature. 1979;280:464–468. doi: 10.1038/280464a0. [DOI] [PubMed] [Google Scholar]
- 10.Doolittle R F. Fibrinogen and fibrin. In: Bloom A L, Thomas D P, editors. Haemostasis and thrombosis. 2nd ed. London, England: Churchill Livingstone Co.; 1987. pp. 192–215. [Google Scholar]
- 11.Edelhoch H. Spectroscopic determination of tryptophan and tyrosine in proteins. Biochemistry. 1967;6:1948–1954. doi: 10.1021/bi00859a010. [DOI] [PubMed] [Google Scholar]
- 12.Grinnel F, Feld M, Minter D. Fibroblast adhesion to fibrinogen and fibrin substrata: requirement for cold-insoluble globulin (plasma fibronectin) Cell. 1980;19:517–525. doi: 10.1016/0092-8674(80)90526-7. [DOI] [PubMed] [Google Scholar]
- 13.Groves M R, Taylor M A J, Scott M, Cummings N J, Pickersgill R W, Jenkins J A. The prosequence of procaricain forms an α-helical domain that prevents acces to the substrate-binding cleft. Structure. 1996;4:1193–1203. doi: 10.1016/s0969-2126(96)00127-x. [DOI] [PubMed] [Google Scholar]
- 14.Gubba S, Low D E, Musser J M. Expression and characterization of group A Streptococcus extracellular cysteine protease recombinant mutant proteins and documentation of seroconversion during human invasive disease episodes. Infect Immun. 1998;66:765–770. doi: 10.1128/iai.66.2.765-770.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harfenist E J, Canfield R E. Degradation of fibrinogen by plasmin. Isolation of an early cleavage product. Biochemistry. 1975;14:4110–4117. [Google Scholar]
- 16.Havlicek J, Pokorny J, Havlickova H. Quantitative character of fibrinogen uptake by M+ and M− variants of Streptococcus pyogenes. Zentbl Bakteriol Hyg Reihe A. 1987;265:1–11. doi: 10.1016/s0176-6724(87)80147-5. [DOI] [PubMed] [Google Scholar]
- 17.Hauser A R, Schlievert P M. Nucleotide sequence of the streptococcal pyrogenic exotoxin type B gene and relationship between the toxin and the streptococcal proteinase precursor. J Bacteriol. 1990;172:4536–4541. doi: 10.1128/jb.172.8.4536-4542.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holm S E, Norrby A, Bergholm A M, Norgren M. Aspects of pathogenesis of serious group A streptococcal infections in Sweden, 1988–1989. J Infect Dis. 1992;166:31–37. doi: 10.1093/infdis/166.1.31. [DOI] [PubMed] [Google Scholar]
- 19.Kantor F S. Fibrinogen precipitation by streptococcal M protein. I. Identity of the reagents, and stoichiometry of the reaction. J Exp Med. 1965;121:849–859. doi: 10.1084/jem.121.5.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kapur V, Topouzis S, Majesky M W, Li L L, Hamrick M R, Hamill R J, Patti J M, Musser J M. A conserved Streptococcus pyogenes extracellular cysteine protease cleaves human fibronectin and degrades vitronectin. Microb Pathog. 1993;15:327–346. doi: 10.1006/mpat.1993.1083. [DOI] [PubMed] [Google Scholar]
- 21.Kapur V, Majesky M W, Li L L, Black R A, Musser J M. Cleavage of interleukin 1β (IL-1β) precursor to produce active IL-1β by a conserved extracellular cysteine protease from Streptococcus pyogenes. Proc Natl Acad Sci USA. 1993;90:7676–7680. doi: 10.1073/pnas.90.16.7676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kapur V, Maffei J T, Greer R S, Li L L, Adams G J, Musser J M. Vaccination with streptococcal extracellular cysteine protease (interleukin-1β convertase) protects mice against challenge with heterologous group A streptococci. Microb Pathog. 1994;16:443–450. doi: 10.1006/mpat.1994.1044. [DOI] [PubMed] [Google Scholar]
- 23.Khan A R, James M N G. Molecular mechanisms for the conversion of zymogens to active proteolytic enzymes. Protein Sci. 1998;7:815–836. doi: 10.1002/pro.5560070401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Knox P, Crooks S, Rimmer C S. Role of fibronectin in the migration of fibroblasts into plasma clots. J Cell Biol. 1986;102:2318–2323. doi: 10.1083/jcb.102.6.2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kunicki T J, Mosesson M W, Pidard D. Cleavage of fibrinogen by human platelet calcium-activated protease. Thromb Res. 1984;35:169–182. doi: 10.1016/0049-3848(84)90212-3. [DOI] [PubMed] [Google Scholar]
- 26.Liu T Y, Stein W H, Moore S, Elliott S D. The sequence of amino acid residues around the sulfhydryl group at the active site of streptococcal protease. J Biol Chem. 1965;240:1143–1149. [PubMed] [Google Scholar]
- 27.Liu T Y, Elliott S D. Streptococcal proteinase: the zymogen to enzyme transformation. J Biol Chem. 1965;240:1138–1142. [PubMed] [Google Scholar]
- 28.Liu T Y. Demonstration of the presence of a histidine residue at the active site of streptococcal proteinase. J Biol Chem. 1965;242:4029–4032. [PubMed] [Google Scholar]
- 29.Lukomski S, Sreevatsan S, Amberg C, Reichardt W, Wolschnik M, Podbelski A, Musser J M. Inactivation of Streptococcus pyogenes extracellular cysteine protease significantly decreases mouse lethality of serotype M3 and M49 strains. J Clin Investig. 1997;99:2574–2580. doi: 10.1172/JCI119445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matsuka Y V, Medved L V, Brew S A, Ingham K C. The NH2-terminal fibrin-binding site of fibronectin is formed by interacting fourth and fifth finger domains. J Biol Chem. 1994;269:9539–9546. [PubMed] [Google Scholar]
- 31.Matsuka Y V, Medved L V, Migliorini M M, Ingham K C. Factor XIIIa-catalyzed cross-linking of recombinant αC fragments of human fibrinogen. Biochemistry. 1996;35:5810–5816. doi: 10.1021/bi952294k. [DOI] [PubMed] [Google Scholar]
- 32.Matsuka Y V, Migliorini M M, Ingham K C. Cross-linking of fibronectin to C-terminal fragments of the fibrinogen α-chain by factor XIIIa. J Protein Chem. 1997;16:739–745. doi: 10.1023/a:1026307731751. [DOI] [PubMed] [Google Scholar]
- 33.Mihalyi E, Weinberg R M, Towne D W, Friedman M E. Proteolytic fragmentation of fibrinogen. I. Comparison of the fragmentation of human and bovine fibrinogen by trypsin or plasmin. Biochemistry. 1976;15:5372–5381. doi: 10.1021/bi00669a025. [DOI] [PubMed] [Google Scholar]
- 34.Mosher D F, Johnson R B. Specificity of fibronectin-fibrin cross-linking. Ann N Y Acad Sci. 1983;408:583–593. doi: 10.1111/j.1749-6632.1983.tb23275.x. [DOI] [PubMed] [Google Scholar]
- 35.Musser J M, Stockbauer K, Kapur V, Rudgers G W. Substitution of cysteine 192 in a highly conserved Streptococcus pyogenes extracellular cysteine protease (interleukin 1β convertase) alters proteolytic activity and ablates zymogen processing. Infect Immun. 1996;64:1913–1917. doi: 10.1128/iai.64.6.1913-1917.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ohara-Nemoto Y, Sasaki M, Kaneko M, Nemoto T, Ota M. Cysteine protease activity of streptococcal pyrogenic exotoxin B. Can J Microbiol. 1994;40:930–936. doi: 10.1139/m94-149. [DOI] [PubMed] [Google Scholar]
- 37.Pace C N, Vajdos F, Fee L, Grimsley G, Gray T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Patti J M, Allen B L, McGavin M J, Hook M. MSCRAMM-mediated adherence of microorganisms to host tissues. Annu Rev Microbiol. 1994;48:585–617. doi: 10.1146/annurev.mi.48.100194.003101. [DOI] [PubMed] [Google Scholar]
- 39.Procyk R, Bishop P D, Kudryk B. Fibrin-recombinant factor XIII A-subunit association. Thromb Res. 1993;71:127–138. doi: 10.1016/0049-3848(93)90179-r. [DOI] [PubMed] [Google Scholar]
- 40.Schmidt K H, Kohler W. T-proteins of Streptococcus pyogenes. IV. Communication: isolation of T1-protein by affinity chromatography on immobilized fibrinogen. Zentbl Bakteriol Hyg Reihe A. 1984;258:449–456. doi: 10.1016/s0176-6724(84)80021-8. [DOI] [PubMed] [Google Scholar]
- 41.Semeraro N, Collen D, Verstraete M. On the origin of the Aα chain heterogeneity of human fibrinogen. Biochim Biophys Acta. 1977;492:204–214. doi: 10.1016/0005-2795(77)90227-6. [DOI] [PubMed] [Google Scholar]
- 42.Standker L, Sillard R, Bensch K W, Ruf A, Raida M, Schulz-Knappe P, Schepky A G, Patscheke H, Forssmann W G. In vivo degradation of human fibrinogen Aα: detection of cleavage sites and release of antithrombotic peptides. Biochem Biophys Res Commun. 1995;215:896–902. doi: 10.1006/bbrc.1995.2548. [DOI] [PubMed] [Google Scholar]
- 43.Tai J Y, Kortt A A, Liu T Y, Elliott S D. Primary structure of streptococcal proteinase. J Biol Chem. 1976;251:1955–1959. [PubMed] [Google Scholar]
- 44.Twining S S. Fluorescein isothiocyanate-labeled casein assay for proteolytic enzymes. Anal Biochem. 1984;143:30–34. doi: 10.1016/0003-2697(84)90553-0. [DOI] [PubMed] [Google Scholar]
- 45.Veklich Y I, Gorkun O V, Medved L V, Nieuwenhuizen W, Wesel J W. Carboxyl-terminal portions of the α chains of fibrinogen and fibrin. J Biol Chem. 1993;268:13577–13585. [PubMed] [Google Scholar]
- 46.Webb E C, editor. Enzyme nomenclature. San Diego, Calif: Academic Press, Inc.; 1992. pp. 399–403. [Google Scholar]
- 47.Wolf B B, Gibson C A, Kapur V, Hussaini I M, Musser J M, Gonias S L. Proteolytically active streptococcal pyrogenic exotoxin B cleaves monocytic cell urokinase receptor and releases an active fragment of the receptor from cell surface. J Biol Chem. 1994;269:30682–30687. [PubMed] [Google Scholar]
- 48.Yu C E, Ferretti J J. Frequency of the erythrogenic toxin B and C genes (speB and speC) among clinical isolates of group A streptococci. Infect Immun. 1991;59:211–215. doi: 10.1128/iai.59.1.211-215.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]