Significance
High-elevation species may face increased ultraviolet (UV) exposure as a consequence of climate change. By comparing the highest-elevation frog, Nanorana parkeri, with its low-elevation relatives, we showed higher UV tolerance in N. parkeri after a time course UV exposure. Combined a chromosome-level reference genome of N. parkeri with large-scale transcriptomic and microRNA (miRNA) profiling, we showed a temporal expression pattern of UV defense genes in N. parkeri. Some miRNAs correlate with both cell cycle and immune genes in expression, suggesting their regulatory role involved in UV defense. Moreover, multiple temporal-expressed genes also exhibit positive selection with function-enhancing substitutions in N. parkeri, such as TYR. This work provides a comprehensive genetic framework of UV defense, which will guide future study of skin cancer and evolution of defense.
Keywords: UV defense, melanin production, gene substitution, expression shift
Abstract
Defense against ultraviolet (UV) radiation exposure is essential for survival, especially in high-elevation species. Although some specific genes involved in UV response have been reported, the full view of UV defense mechanisms remains largely unexplored. Herein, we used integrated approaches to analyze UV responses in the highest-elevation frog, Nanorana parkeri. We show less damage and more efficient antioxidant activity in skin of this frog than those of its lower-elevation relatives after UV exposure. We also reveal genes related to UV defense and a corresponding temporal expression pattern in N. parkeri. Genomic and metabolomic analysis along with large-scale transcriptomic profiling revealed a time-dependent coordinated defense mechanism in N. parkeri. We also identified several microRNAs that play important regulatory roles, especially in decreasing the expression levels of cell cycle genes. Moreover, multiple defense genes (i.e., TYR for melanogenesis) exhibit positive selection with function-enhancing substitutions. Thus, both expression shifts and gene mutations contribute to UV adaptation in N. parkeri. Our work demonstrates a genetic framework for evolution of UV defense in a natural environment.
Human activities have significant effects on climate change, including increased ultraviolet (UV) radiation received at higher elevations (1). Excessive UV radiation is likely to have profound adverse impacts on human health as well as biodiversity (1). Exposure to intense UV can induce damage to DNA and oxidative stress through reactive oxygen species (ROS) in cells, which can lead to melanomas and other cutaneous diseases (2). UV exposure is also likely to be a factor that makes amphibians more susceptible to pathogen infections (e.g., chytridiomycosis) and thus may result in significant population declines (3, 4). Therefore, ongoing increases in UV levels may pose a serious challenge to the health of organisms and maintenance of biodiversity. For these reasons, it is important to understand the genetic and molecular mechanisms used by organisms to cope with threats of increased UV.
Organisms have evolved a series of phenotypic and behavioral adaptations to counter the harmful effects of UV radiation. For example, bird feather colors, as well as human skin pigmentation, have evolved at least partly as adaptations to protect against UV radiation (5, 6). The adaptation of human skin pigmentation is related to the production and distribution of melanin, which can block or absorb light and thereby prevent it from penetrating the skin (7). There are differences in the degree of UV resistance between different species and populations, and high-elevation species typically have higher capability to block UV than low-elevation species (8, 9); such differences can be reflected at the molecular level (10). However, most recent molecular research on this problem has been restricted to UV exposure experiments on mouse or human cells in the laboratory. These studies have identified specific genes involved in antioxidant metabolism (11, 12) and DNA repair (13, 14) pathways that can prevent or reduce UV-induced damage. Moreover, heat-shock proteins, such as hsp70, can bind to nucleic acid repair proteins after UV exposure in yeast, which is presumed to enhance DNA repair activity (15, 16). Nevertheless, we still lack a comprehensive understanding of UV defense mechanisms in vivo, including the diversity of defense pathways and genes and their dynamic evolution.
The Tibetan Plateau (TP) (17) is one of the areas with the most serious ozone layer deletion, which leads to higher UV radiation intensity in this area than in the lowlands (18). High-elevation species, such as the hot-springs snake and snub-nosed monkey, have evolved substitutions in their DNA repair genes that appear to be associated with long-term UV stress (10, 19). Compared with mammals, birds, and reptiles, high-elevation frogs have much thinner skin and lack of protective fur, feathers, or scales, making them potentially more sensitive to UV exposure. As a representative of high-elevation frogs, the genus Nanorana (Dicroglossidae) comprises 22 species distributed on the TP, Himalayas, and surrounding region. The TP subclade has colonized the highest elevations up to 5,000 m over the past 8–9 million years, and its sister lineage (Himalayan subclade) diverged from the TP subclade about 19 million years ago (20). The genus Quasipaa is a sister genus of Nanorana, and the divergence time between the two genera is dated at ∼27 million years ago (20). One of the species in the TP subclade, N. parkeri, is the world’s highest-elevation frog (∼4,500 m), which means in some circumstances it is exposed to much higher UV radiation than lower-elevation species. During the breeding season, it is active in the daytime beside small exposed rocks in streams. According to our observations, individuals of N. parkeri experience much stronger UV radiation (∼3.5 mW/cm2) than do individuals of lower-elevation relatives, such as N. phrynoides (∼1.6 mW/cm2 at ∼1,700 m) and Quasipaa spinosa (∼0.4 mW/cm2 at ∼118 m). Millions of years of selection in N. parkeri have resulted in a series of phenotypic adaptations in this species to potential UV defense, including increased skin pigmentation, compared with its lower-elevation relatives (21). For these reasons, N. parkeri is an ideal model organism for studying the long-term adaptive strategies and the underlying genetic mechanisms related to UV tolerance.
In this study, we compare the Tibetan frog (N. parkeri) with its lower-elevation relatives (N. phrynoides and Q. spinosa), using an integrated systems biology approach to study how various defense systems work together in UV defense of an organism. We examine phenotypic changes, metabolomics, and expression profile of genes over a time course of UV exposure. We then identify the positive selection signals across the genome of N. parkeri and explore how the expression and sequences of these genes have evolved in response to adaptation to high elevation, including strong UV radiation.
Results
Strong UV Resistance Is Associated with Efficient Antioxidant Activity in Tibetan Frog Skin.
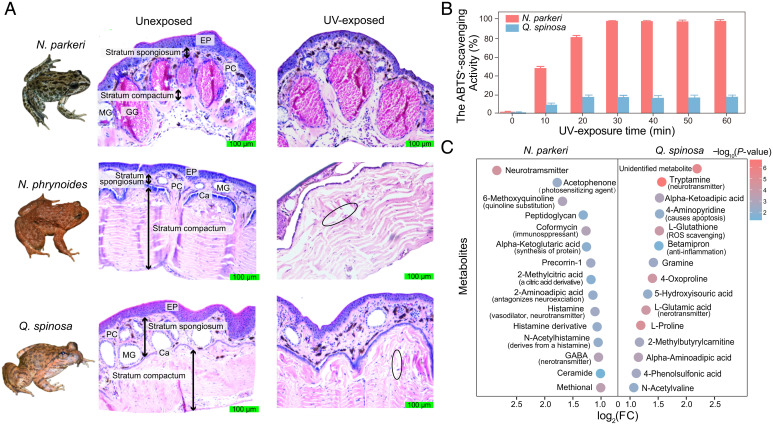
To clarify the response to extremely high UV exposure, we separately exposed N. parkeri and its lower-elevation relatives (N. phrynoides and Q. spinosa) to a time course of artificial UV exposure at an irradiance of ∼3.7 mW/cm2 (0, 0.5, 1, 2, 4, 6, and 8 h). The irradiance used was slightly higher than what N. parkeri typically experiences in the field, and the maximal exposure time was based on tolerances of individuals in pilot assessment (see Materials and Methods). Three to four individuals of each species were used per time point. To observe morphological characteristics, a dorsal skin sample after UV exposure was taken from each individual for preparing a histological section. In total, at least 54 images were captured from each species to statistically quantify the skin structures and the degrees of damage to the skin morphology. Comparisons between the UV-exposed and unexposed skins further revealed that unexposed N. parkeri had a thin dermis with tightly connected cell layers; furthermore, quantification of degree of damage in skin (see Materials and Methods) showed that the structures of the glands and the dense connective tissues remained relatively compact and stratified in shape overall along with UV exposure (Fig. 1A; comparison of degree of damage throughout UV exposure in N. parkeri,1 h vs. 0 h: false discovery rate [FDR] = 0.12, >1 h vs. 0 h: FDR < 0.05 in SI Appendix, Fig. S1, two-tailed t test). In contrast, observations on histological sections showed Q. spinosa (∼118 m) possessed a less compact skin structure (21), and the corresponding structures displayed considerable damage, including the destruction of glands, disruption of the calcified layer, and necrosis of the dermis, after UV exposure (Fig. 1A; comparison of degree of damage throughout UV exposure in Q. spinosa, 1 h vs. 0 h: FDR = 0.007, >1 h vs. 0 h: FDR < 0.0005 in SI Appendix, Fig. S1, two-tailed t test). Similar skin morphology was also seen in another lower-elevation frog, N. phrynoides (∼1,700 m) (Fig. 1A and SI Appendix, Fig. S1). These observations suggest that the skin of N. parkeri is more resistant to UV-induced necrosis compared to that of lower-elevation frogs.
Fig. 1.
Histological cross-sections and identification of antioxidant capacity and metabolites in frog skin. (A) Images separately show the basic skin structures of the unexposed group (Middle) and the UV-exposed group (Right; with ∼3.7 mW/cm2 UV exposure for 8 h) in N. parkeri, N. phrynoides, and Q. spinosa. The region marked with an ellipse is one example of damage on skin sections (Ca, calcified layer; EP, epidermis; GG, granular gland; MG, mucous gland; PC, pigment cell). (B) The ABTS+ scavenging tests for each species were performed with a 10-minute interval and the rates of ABTS+ scavenging activity (%) were calculated as (Ablank − Asample) × 100/Ablank. Each bar is based on mean ± SEM of n = 9 independent ABTS+ scavenging tests. (C) Examples of metabolites that were significantly up-regulated in the UV-exposed group compared with the unexposed group. Color indicates the significance of the difference, with P values calculated via two-tailed t tests, and log2(FC) represents the degree of change. Samples of N. phrynoides were not performed with the ABTS+ scavenging test and metabolites identification owing to deficiency.
We further examined the ROS scavenging activity of skin secretions in N. parkeri and Q. spinosa, as antioxidant capacity was implicated in preventing oxidative damage from ROS after UV exposure (2, 9). The secretions mainly from skin were separately collected from individuals with UV exposure every 10 min and then repeated with a 2, 2’-azino-bis (3-ethylbenzothiazoline-6-sulfonic acid) (ABTS+) scavenging test. N. parkeri showed a significant increase in the ABTS+ scavenging activity after UV exposure and reached nearly 100% of the scavenging activity after ∼30 min (Fig. 1B). In contrast, Q. spinosa exhibited a much smaller increase than N. parkeri after UV exposure and reached a maximum of only ∼20% of the activity detected in N. parkeri (Fig. 1B).
Metabolomics analysis of skin samples (see Materials and Methods) provided more evidence for the comparatively efficient antioxidant activity in N. parkeri. After UV exposure, we identified intermediates of UV-screening melanin and antioxidant compounds (such as quinolinate and vitamins) (22, 23) in the skin of N. parkeri that were absent in Q. spinosa (Fig. 1C and SI Appendix, Fig. S2 and Dataset S2). This result and sustained higher melanin content in N. parkeri during UV exposure (SI Appendix, Fig. S3) were consistent with the UV-absorbing function of melanin (4). Moreover, after UV exposure, individuals of N. parkeri secreted more (log2[fold change] >1, FDR <0.05) diverse neurotransmitters related to anti-inflammation and wound repair compared to unexposed frogs (Fig. 1C and SI Appendix, Fig. S2 and Dataset S2). In contrast, we did not observe a similar change in Q. spinosa associated with UV exposure. These neurotransmitters included histamine (involved in wound repair and cell apoptosis) (24), ethoxyquin (an antioxidant) (25), and gamma-aminobutyric acid (an anti-inflammatory compound) (26). These results suggest that higher ROS scavenging activity observed in N. parkeri relates to compound function in immune modulation, anti-inflammatory effects, and repair in skin, which together increase UV tolerance in N. parkeri.
Genome Assembly of the Tibetan Frog.
We next explored the underlying genetic mechanisms of UV tolerance in N. parkeri. As a high-quality genome was needed for this analysis, we updated the genome assembly of N. parkeri, with integrated datasets generated by long-range PacBio RS II (21.3 million reads), short-read Illumina sequencing (328.62 Gb), chromatin conformation capture (Hi-C) (126 Gb), and genetic mapping (SI Appendix, Tables S1–S5 and Fig. S4). The assembled genome (∼2.46 Gb) has a high continuity with a contig N50 of 2.3 Mb and a scaffold N50 of 269 Mb, both of which are more than 250 times better than the previous assembly (27) and exhibited the highest contig N50 among sequenced genomes of anurans (SI Appendix, Table S3). The contigs were anchored and oriented into 11 long pseudomolecules through a hierarchical clustering strategy (SI Appendix, Table S4). Furthermore, we identified 2,272 (87.9%, SI Appendix, Table S5) of the conserved vertebrate BUSCO genes, indicating that the N. parkeri assembled genome is of high integrity and accuracy.
We annotated 22,884 nonredundant protein-coding genes based on an integration of methods (see Materials and Methods; SI Appendix, Tables S6 and S7). Furthermore, we annotated more than 1.3 Gb (55.2% of the assembly) transposon elements and other noncoding RNAs throughout the genome assembly, including microRNA (miRNA) (SI Appendix, Tables S8 and S9).
Temporal Expression Patterns Related to UV Defense Systems in Tibetan Frog Skin.
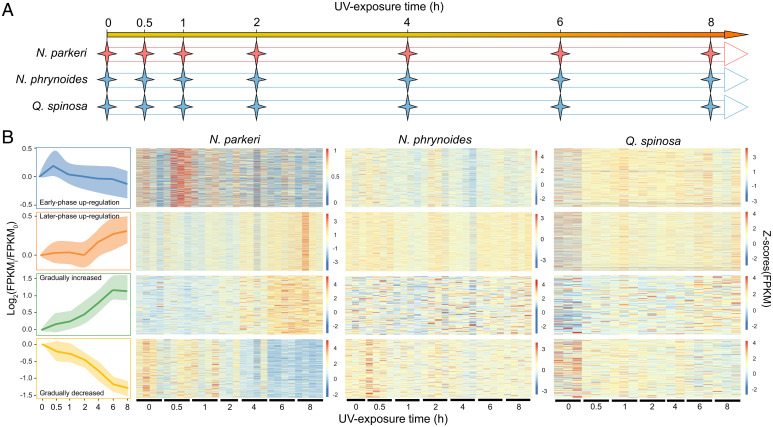
All the individuals of N. parkeri, as well as the lower-elevation species (N. phrynoides and Q. spinosa) from the time course UV exposure experiment (Fig. 2A), were separately submitted for RNA sequencing (RNA-seq) and transcriptomic analysis. We separately clustered the temporal expression profile of each species throughout the time course using the Short Time-series Expression Miner (STEM) clustering method (28). The clustering results suggest distinct patterns of gene expression in N. parkeri, including genes with continuous change during UV exposure (profiles #39 and #10 in SI Appendix, Fig. S6) and those up-regulated at a specific time point (e.g., Profiles #44, #43, and #27 in SI Appendix, Fig. S6). To precisely identify the coexpressed genes that showed up-regulation at a specific time point in the STEM results, we further performed weighted correlation network analysis (WGCNA) (29) analysis on the expression matrix of all genes (SI Appendix, Fig. S7). Combining results of STEM and WGCNA, we obtained four groups of genes in N. parkeri that are characterized by time sequence in expression, including groups of early-phase (0.5 h) up-regulation, later-phase (6–8 h) up-regulation, gradual increase throughout the UV exposure period (genes in Profile #39 of STEM), and gradual decrease throughout the UV exposure period (genes in Profile #10 of STEM). We then conducted Gene Ontology (GO) functional enrichment analyses for the coexpressed genes in each temporal group.
Fig. 2.
Experimental proposal and expression heatmaps of genes in four temporal groups throughout UV exposure. (A) Schematic shows the experimental procedures of UV exposure in a time course across each species. (B) Scaled profiles (Left) were derived from log2-normalized expression changes (FPKMih/FPKM0h) of temporal gene groups in N. parkeri, including early-phase up-regulation, later-phase up-regulation, gradually increased expression, and gradually decreased expression. The solid line denotes the median values of expression level, and the shaded areas correspond to the range of 25–75%. The profiles are indicated by a color code used for subsequent analyses. For the expression heatmap of N. parkeri, N. phrynoides, and Q. spinosa, each row represents an expression level of a gene and color represents an FPKM with Z-score standardization.
In the early-phase up-regulation group, genes associated with the immune system and skin moisturizing, such as F2RL1, IL12B, DEGS2, and PGLYRP3 (Dataset S4), were up-regulated in N. parkeri, followed by a moderate decrease in expression to their initial expression levels (Fig. 2B and SI Appendix, Fig. S8). These immune genes are involved in complex cascades including cytokine production, IL-1/6/8/17 secretion, T cell and mast cell activation, the nucleotide-binding oligomerization domain–like signaling pathway, and the nuclear factor kappa-light-chain-enhancer of activated B cells signaling pathway (Dataset S4). These results suggest that the initial activation of immunity protects N. parkeri from UV-induced immunosuppression, which would otherwise be a source of damage during early UV exposure. Genes related to the regulation of moisturization also showed increased expression during this period (Dataset S4). These included genes involved in the biosynthesis of ceramide, which functions in constructing a moisturizing screen by working with saturated fatty acids and promotes profound cell proapoptosis by regulating relative pathways in response to UV-induced oxidative stress in the hyperplasia of keratinocytes (30). In contrast, some orthologs in the early-phase up-regulation group of N. parkeri actually exhibit decreasing expression in N. phrynoides (∼1,700 m), and most orthologous genes in Q. spinosa (∼118 m) did not exhibit obvious changes in expression (Fig. 2B and Dataset S4).
For the later-phase up-regulation group, coexpressed genes that are involved in inflammation and apoptosis regulation displayed significant up-regulation in N. parkeri, especially among genes related to histamine secretion, autophagy, and leukocyte activation (Fig. 2B and SI Appendix, Fig. S8 and Dataset S5). The up-regulation of these pathways is closely related to the increased expression of immune genes in early-phase group, which involve in chemokine secretion. Increases in the expression of histamine genes in the later-phase group, including SNAP23, SLC22A3, and SNX6, potentially explained the observed higher abundance of histamine in the metabolome of N. parkeri compared with Q. spinosa after long UV exposure (Fig. 1C). The other coexpressed genes in this group of N. parkeri are related to antioxidant metabolism, including genes involved in the production of melanin and nicotinamide salvaging (SI Appendix, Fig. S8 and Dataset S5). This is reminiscent of the observed retention of high pigment content (SI Appendix, Fig. S3) and increased antioxidant production in the metabolome of N. parkeri after long UV exposure (Fig. 1C and Dataset S2). However, these orthologs displayed a slight decrease in N. phrynoides or up-regulation just in the early phase of UV exposure in Q. spinosa (Fig. 2B and Dataset S5).
Genes responsible for stress response (such as those encoding heat-shock proteins HSP90AA1 and HSF3) exhibited gradually increased expression in N. parkeri skin with ongoing exposure to UV, whereas the orthologous genes of the lower-elevation relatives exhibited no obvious change in expression (Fig. 2B and SI Appendix, Fig. S8). Given that heat-shock genes function in relieving stress and regulating adenosine triphosphatase to protect proteins from unfolding (13, 14), they are important for promoting repair of organisms. The other set of gradually up-regulated genes found in N. parkeri, such as ADCYAP1, SSTR2, and CHRNA6, are relevant to regulation of the peripheral nervous system (the neuropeptide signaling pathway and neurotransmitter biosynthesis) (Dataset S6). This finding is in accord with the abundant number of neurotransmitter metabolites identified in N. parkeri (Fig. 1C and Dataset S2). Neuropeptides are involved in cellular response to harmful stimuli and mediating UV-induced suppression of immune cells responses (31). However, the orthologs of these genes were separately decreased in expression in N. phrynoides and up-regulated in later phase of UV exposure in Q. spinosa (Fig. 2B).
The coexpressed genes in the gradually decreased expression group of N. parkeri are mostly involved in cell cycle regulation (Fig. 2B and SI Appendix, Fig. S8). In contrast, the orthologs of these genes in the lower-elevation relatives showed little change in their patterns of expression (Fig. 2B and Dataset S7). These genes in N. parkeri are down-regulated during UV exposure, which may prevent the cells from erroneously moving through the cell cycle during apoptosis (32). This has also been reported in local UV adaptation in diverse human populations, in which the repressed expression of cell cycle genes occurs in parallel with apoptosis (33).
miRNAs Serve as Regulatory Modulators of Gene Expressions in UV Defense Systems.
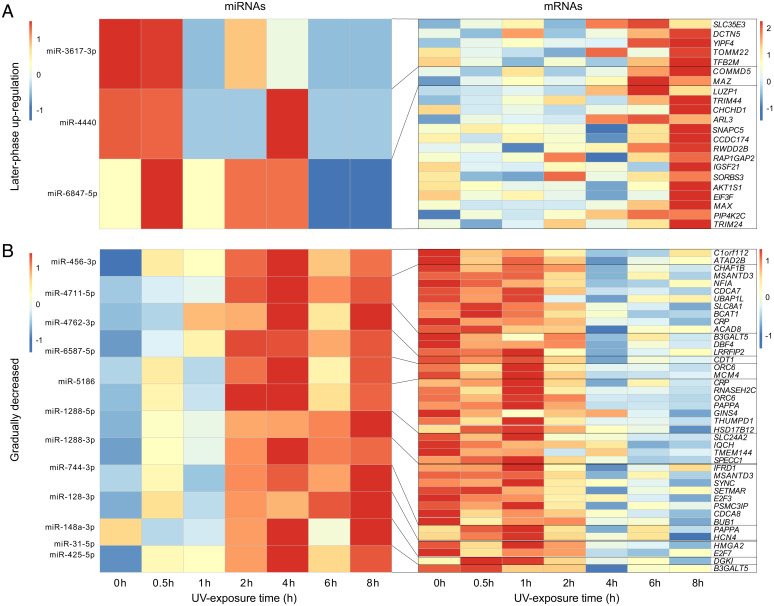
To further explore the noncoding regulatory modulators that might induce the observed temporal expression, we performed miRNA-seq for the same individuals of N. parkeri and its lower-elevation relatives (N. phrynoides and Q. spinosa) from the UV exposure time course experiment. miRNAs are commonly implicated in the regulation of stress responses mainly by silencing messenger RNA (mRNA) expression. Combining predictions from the N. parkeri genome and miRNA-seq data, we identified 515 miRNAs in N. parkeri, 76 in Q. spinosa (∼118 m), and 116 in N. phrynoides (∼1,700 m) (SI Appendix, Fig. S9 and Dataset S8). We predicted the target genes and the miRNA candidates that were coexpressed with target genes in each of the four temporal groups. Some unique miRNAs found only in N. parkeri (Fig. 3A and Dataset S8), such as miR-3617-3p and miR-4440, were associated with target genes belonging to the later-phase up-regulation group (module membership [MM = –0.67 and raw P = 0.0016 for miR-3617-3p; MM = –0.6 and raw P = 0.006 for miR-4440 in SI Appendix, Fig. S10). These miRNAs did not show conspicuous changes in expression until 8 h after UV exposure and were followed by a decrease, which was the opposite pattern of the up-regulation of inflammation and immune regulation genes in the same phase. Other N. parkeri unique coexpressed miRNAs, such as miR-1288-3p and miR-6587-5p, are predicted to drive the reduced expression of cell cycle genes (Fig. 3B and SI Appendix, Fig. S11 and Table S11). The expression of these miRNAs gradually increased and showed a significant negative correlation with that of their predicted target genes (NFIL3, RNF212, CLEC10A, and CRP for miR-1288-3p and CDT1 and LRRFIP2 for miR-6587-5p) (Fig. 3B and SI Appendix, Table S11).
Fig. 3.
Heatmaps of expression levels of coexpressed miRNA-mRNAs in (A) the later-phase group and (B) the gradually decreased expression group of N. parkeri. miRNAs (Left) exhibit a significant negative relationship with corresponding mRNA candidates (Right) in expression. The results of Pearson correlation analysis are shown in SI Appendix, Table S11.
Some miRNAs that can be found in both N. parkeri and its lower-elevation relatives showed distinctly different expression patterns in N. parkeri. For instance, miR-4762-3p and miR-5186 are coexpressed miRNAs associated with genes in the gradually decreasing expression group of N. parkeri, whereas miR-4762-3p expressed only around 1 h and 2 h of UV exposure in N. phrynoides and Q. spinosa, respectively. miR-5186 showed no clear expression patterns in both lower-elevation relatives (SI Appendix, Fig. S12). Our results indicated that miRNAs contribute to the temporal expression of UV defense genes, especially for cell-cycle related genes in N. parkeri.
UV Pressure Has Driven the Positive Selection of Defense Genes in N. parkeri.
We examined differences in genes and protein sequences themselves to determine how they might affect functional changes. Genes with an increase in the ratio of nonsynonymous to synonymous substitutions are often associated with selection pressure (such as from UV stress). Based on the phylogenetic relationships between the studied frogs (34), we applied the improved branch-site likelihood-based test (35) on their orthologs to detect positively selected genes (PSGs) in N. parkeri. We identified a total of 441 PSGs in N. parkeri after FDR correction (Dataset S10). These PSGs were significantly enriched in functional terms related to the UV defense systems identified above, including the immune regulation, apoptosis, collagen formation, cellular response to stress, melanin metabolism, biological oxidations, and DNA repair (Table 1 and Dataset S10). Some of these PSGs also belong to the specific temporal expression groups described above (Table 1), further confirming that they not only are closely related to response to UV exposure in expression but also play an important role in UV adaptation through sequence evolution.
Table 1.
Examples of PSGs and enriched functional terms in expression profiles of N. parkeri
| GO | Description | Log(P value) | Enrichment | PSGs |
|---|---|---|---|---|
| GO:w0002274 | Myeloid leukocyte activation | −7.79473 | 2.88192 | CD200*|CD63†|AGPAT2†| FGL2*|SERPINB1|HEXB|IFNGR1| PSMD1||ADAM9|HEBP2 |
| GO:1902166 | Negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class | −3.70563 | 13.12877 | CD74|CD44|ZNF385A|ATAD5 |
| GO:0046514 | Ceramide catabolic process | −2.21143 | 7.87726 | HEXB| ACER3 * |GBA2 |
| R-HSA-1474290 | Collagen formation | −2.77837 | 4.08450 | COL6A3 * |LOXL1 * | COL2A1|COL6A1| LAMC2|PLOD1 |
| GO:0002694 | Regulation of leukocyte activation | −2.329 | 1.8755 | CYLD*|FLOT2†|PGLYRP3*|CD200*| FGL2*|CASP3|CD40|CD74|CLPTM1| IGHE|LGALS9|XCL1|SFRP1 |
| GO:0010212 | Response to ionizing radiation | −3.32193 | 3.62173 | RRM1 § |EGR1 * |MAP3K20 * |CASP3| CRYAB|HSF1|SFRP1|TRIM13| TLK2|COP1 |
| GO:1900034 | Regulation of cellular response to heat | −2.40038 | 3.98848 | NUP153 * | BAG1|CREBBP|CRYAB| GSK3B|HSF1 |
| GO:0046148 | Pigment biosynthetic process | −2.27779 | 4.45043 | FXN † | TYR ‡ | SLC25A38 † |APPL1|SLC24A5 |
| hsa00760 | Nicotinate and nicotinamide metabolism | −2.67498 | 7.24346 | AOX1 † | NUDT12 † |NADK|NADK2 |
| hsa00350 | Tyrosine metabolism | −2.37062 | 6.00172 | TYR ‡ | AOX1 † |ADH1A|COMT |
| GO:0033280 | Response to vitamin D | −2.32595 | 5.83500 | TYR ‡ | KANK2 † |AQP3|SFRP1 |
Bold fonts highlight important PSGs with expression shifts in temporal phases of UV exposure.
*Genes in the early-phase up-regulation group; †Genes in the later-phase up-regulation group; ‡Genes in the gradually increased group; §Genes in the gradually decreased group.
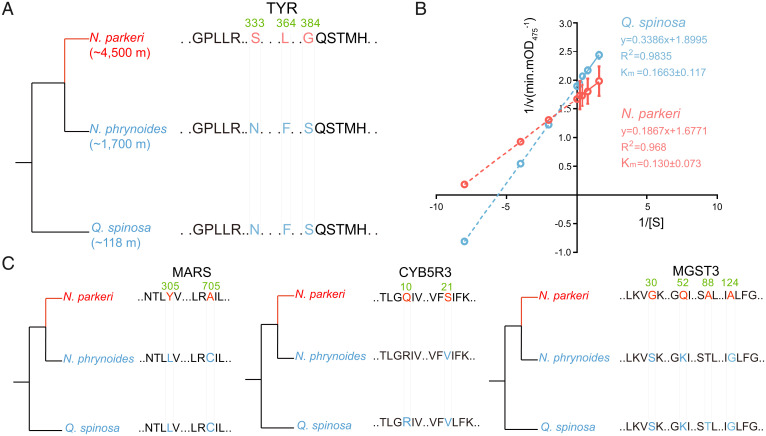
TYR, one of the PSGs, encodes a rate-limiting tyrosinase involved in melanin biosynthesis (36). The protein encoded by this gene (TYR) acquired three significant amino acid replacements (N333S, F364L, and S384G) along the N. parkeri lineage (Fig. 4A and SI Appendix, Fig. S13). Among the three replacements, S384G resulted in a change in the polarity of the encoded amino acid that is located at a functional domain of TYR, located outside the cell membrane. We thus hypothesized that the modified TYR in N. parkeri may have higher affinity with its substrate levodopa (L-DOPA). To test this hypothesis, we separately synthesized the protein sequence of TYR of both N. parkeri and Q. spinosa in vitro, then performed enzyme kinetic analyses for these proteins with the substrate L-DOPA and measured at 1-min intervals. These analyses revealed a lower Km (Michaelis constant) value (0.130 ± 0.073 mM/L) of the amino acid-replacement-bearing tyrosinase in N. parkeri compared to that in Q. spinosa (0.1663 ± 0.117 mM/L) (Fig. 4B), indicating that the identified replacements confer a higher enzymatic affinity on the TYR protein. In parallel, the increase in TYR expression was sustained over the course of UV exposure, in coordination with the up-regulated expression of other genes (e.g., SLC45A2) that transport substances required for melanin production. This suggests that the TYR gene of N. parkeri, with its function-enhancing substitutions, plays an important role in adaptation of constitutive darker coloration and persistently high melanin production in this species during UV exposure. In addition to TYR, other PSGs in N. parkeri also showed distinct sequence substitutions compared to its lower-elevation relatives (N. phrynoides, ∼1,700 m; Q. spinosa, ∼118 m), including MARS, CYB5R3, and MGST3 (P-value < 0.05, FDR > 0.05; Fig. 4C). These genes are involved in glutathione-dependent peroxidase activity and cellular redox reaction, and their orthologs in another lower-elevation relative N. medogensis (∼2,900 m) showed consistent substitutions with those in N. phrynoides and Q. spinosa (SI Appendix, Fig. S14), further confirming the unique substitutions in N. parkeri. These results suggest these PSGs that evolved with rapid rates may play an important role in long-term adaptation of N. parkeri to high elevation.
Fig. 4.
Amino acid replacements of PSGs and enzymatic activity identification of tyrosinase (TYR) across N. parkeri and Q. spinosa. (A) Phylogenetic relationships and schematic alignment of TYR amino acid sequences in N. parkeri (red) and its lower-elevation relatives (N. phrynoides and Q. spinosa; blue). Amino acid replacements unique to N. parkeri shown in red were found to be under positive selection. The reference alignment numbers refer to the amino acid sequence of N. parkeri. (B) In vitro enzymological kinetic analyses of TYR in N. parkeri and Q. spinosa. Km (Michaelis constant) is used to describe the rate of enzymatic reactions. It is numerically equal to the substrate concentration [S] at which the reaction rate is half of the maximum rate [v] achieved by enzyme. The smaller Km represents the stronger affinity the enzyme has for the substrate. Each Km value is derived from eight independent experiments, which are indicated with circles. R2 represents the correlation coefficient of the kinetic equation that fits the reaction. (C) Schematic amino acid alignments of MARS, CYB5R3, and MGST3 in N. parkeri (red) and its lower-elevation relatives (blue). Single-letter abbreviations for the amino acid residues are as follows: F, Phe; G, Gly; H, His; L, Leu; M, Met; N, Asn; P, Pro; Q, Gln; S, Ser; T, Thr; V, Val.
Discussion
Many species, including humans, are facing increased surface UV exposure as a result of anthropogenic environmental change. There has been accumulating evidence showing that the excessive UV exposure may influence local biodiversity composition and result in human skin cancers (2). Thus, it is crucial to understand the genetic basis underlying organismal adaptations to UV exposure. Most investigations have used skin cells of model species that are not well-adjusted to UV radiation to explore genetic responses. This has intrinsically set limitations to our understanding of how genetic modulations in an organism could effectively protect itself from UV-induced damages.
N. parkeri, an endemic frog of the high-elevation TP, has evolved adaptations to perennial strong UV radiation at high elevations and hence provides an excellent model system for studying UV defense. UV adaptations of N. parkeri include various types of phenotypic changes, especially increased melanin accumulation in skin (SI Appendix, Fig. S3) and UV resistance (Fig. 1), as demonstrated in this study. Compared to other highland vertebrates, N. parkeri shows some unique characteristics in skin, including much thinner skin and lack of protective scales, fur, or feathers, making this species more susceptible to UV exposure. When facing strong UV stress, unlike some other highland frogs (e.g., Odorrana andersonii), N. parkeri secretes few macromolecule peptides, but it does secrete some micromolecule antioxidant compounds that help reduce UV-induced damage. Genomic and transcriptomic analyses further showed that its response to strong UV may result from accumulation of gene sequence changes (including positive selection on the UV-defense related genes), as well as by gene regulation changes in defense pathways, especially when this species is facing long UV exposure.
Some of UV defense pathways in N. parkeri are conserved with other highland species, including melanin production in Tibetan wild boars (37) and Tibetan cashmere goats (38), as well as DNA repair in snub-nosed monkeys (10), hot-spring snakes (19), and snowfinches (39). A few genes of these pathways (e.g., CCNT2, DMAP1, CDT1, and RTEL1) have been identified as being under selection in highland species through comparative genomics. In this study, we identified additional genes involved in UV defense with consistent functions to those previously reported and also some genes with functions that have never been reported in other highland species, including immune regulation, moisturization, and neurotransmitter and neuropeptide metabolism. Neurotransmitter and heat shock proteins are typical in skin exposed to detrimental stressors. Neuropeptides (released from cutaneous nerves and immune cells in response to stimuli) are important for fine-tuned regulation of skin maintenance and repair (40, 41). The functions of these genes are consistent with changing expression patterns induced throughout UV exposure in skin of N. parkeri. Among these UV-defense related pathways, melanogenesis is an evolutionarily conserved pathway that reduces UV-induced damage in many species, from N. parkeri to humans. Besides some melanogenesis genes (e.g., MC1R, KITLG) reported in other alpine species, the expression and specific function-enhancing substitutions of TYR in N. parkeri has been verified by transcriptome and enzyme kinetic assay.
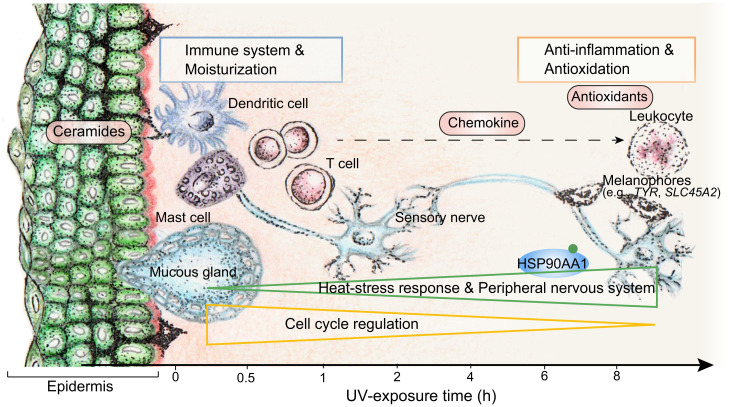
During UV defense, the immune system and ceramide production were activated in the early phase of UV exposure in skin, followed by the response of the antioxidation system (Fig. 5). The anti-inflammation system was also induced during UV exposure, probably by chemokines secreted by the activated immune cells in early phase of exposure (Fig. 5). Genes associated with the cellular stress response and the peripheral nervous system gradually increased in expression in response to accumulating damage and to form skin barrier with reinforcement of immune defenses against potential infection; meanwhile, cell cycle–associated genes gradually decreased in expression, involved in suppressing replication of damaged cells in the apoptosis process (Fig. 5). In contrast to the many observed changes in gene regulation during UV exposure in N. parkeri, we observed much less temporal expression regulation in orthologs of lower-elevation relatives, such as N. phrynoides (∼1,700 m) and Q. spinosa (∼118 m) (Fig. 2B and Dataset S4). Some of the genes we identified as involved in UV defense in N. parkeri have been reported as involved in UV adaptation of other high-elevation species, such as CCNT2 and DMAP1 in the Tibetan hot-spring snake (19), as well as CDT1 and RTEL1 in snub-nosed monkeys (10). Our results further showed the dynamic regulation of these genes in N. parkeri in response to UV exposure. For example, in N. parkeri, CCNT2 and DMAP1 are up-regulated for repressing transcription and temporal coordination of protein synthesis in the cell cycle, whereas CDT1 and RTEL1 (involved in DNA unwinding and replication) are down-regulated (SI Appendix, Fig. S15).
Fig. 5.
Molecular dynamics of temporal defense systems against UV exposure in N. parkeri skin. The defense systems and corresponding representative organelles are displayed along the timeline in relationship to their temporal up-regulation. Genes in early-phase group are related to immune system and moisturization (blue rectangle) and up-regulated at early phase of UV exposure. Immune cells secrete chemokines that induced up-regulation of anti-inflammation related genes as well as antioxidative genes (red rectangle) at the later phase of UV exposure. Genes in the gradually increased group are related to heat stress response and peripheral nervous system (green triangle) and are gradually up-regulated in expression. Genes in the gradually decreased group function in cell cycle regulation (golden triangle) and are gradually down-regulated in expression.
Previous studies based on mammalian and yeast cells have separately identified single genes or exogenous chemical compounds that are involved in mitigating UV-induced damage, including those related to melanogenesis (36, 42), vitamin synthesis (43), and DNA repair (44). However, UV defense is determined not by a single gene but by a suite of genetic adaptations acting in concert. Here, we used N. parkeri to systematically demonstrate how various defense genes, including PSGs and temporal expression genes, have evolved to work together in an organism to protect its skin from UV damage. Although some functions of these individual defense genes have been identified by previous studies, we demonstrate that UV defense involves the cooperation between many different systems and genes. For example, the gene TYR has been reported to play a key role in melanogenesis and skin disease. In this study, we observed that TYR and other PSGs (GNAS, EP300, GSK3B, PLCB) are involved in melanogenesis and evolved rapidly in N. parkeri (Dataset S10). TYR is up-regulated gradually during UV exposure and works with up-regulated SLC45A2 to induce melanin generation in melanosomes. This process may also trigger observed up-regulation of the TC-NER master organizing protein gene ERCC6 [involved in DNA repair (45)] and CRTC3 [involved in cell cycle regulation and melanogenesis (46)] (SI Appendix, Fig. S16).
Our results revealed that in addition to substitutions in genes of N. parkeri, temporal expression changes of some genes are regulated by miRNAs, especially in cell cycle genes. In cell cycle signaling of N. parkeri, PSGs such as CREBBP and EP300 induce cell apoptosis. BUB1 and another PSG, SMC1A, regulate the cohesion formation, which is critical for DNA duplication. PSG GSK3B works with downstream genes with a gradual decrease in expression, which directly affects DNA biosynthesis. MiRNAs were involved in regulating expression of these downstream genes, including miR-744-3p for BUB1 and E2F3, miR-5186 for ORC6 and MCM4, and miR-4762-3p for DBF4. All these miRNAs showed gradual increase in expression and inhibited their target genes during UV exposure.
The UV adaptations of high-elevation N. parkeri provide compelling insights into how some species may adapt to environmental changes such as UV exposure. The mechanisms and pathways that we have identified may also be useful in elucidating the regulatory mechanisms underlying skin physiology and complex diseases, such as melanoma. This study contributes to our understanding of UV defense and provides a model for the study of defense against other environmental stresses.
Materials and Methods
Sampling Information and Ethics Statement.
Collection localities and treatments for all samples are given in SI Appendix, Table S1. The collecting permit and all study protocols were approved by the Animal Care and Ethics Committee of Kunming Institute of Zoology, Chinese Academy of Sciences, and were conducted in strict accordance with the guidelines for Animal Care and Use at the Kunming Institute of Zoology (SMKX-20160301-03).
We collected male adults except one female used for genome sequencing during their reproductive periods. Captured frogs were housed in artificial ponds at the site of collection and then were used in experiments. After the frogs were euthanized using benzocaine (SMKX-20160301-03), their dorsal skin was isolated rapidly for subsequent analyses.
UV Exposure Experiments and Skin Sample Collection.
To set the UV irradiance used in the experiments, we obtained illumination photometry measurements of local habitats of the studied species by using an illuminometer (TES-1339; TES Electrical Electronic Corp., Taiwan). We used an irradiance of ∼3.7 mW/cm2 UVB exposure with artificial lamps (Philips G8T5, 313 nm) in experiments, which is slightly higher than the irradiance of local conditions that N. parkeri experiences at daytime (∼3.5 mW/cm2). In the pilot assesment of tolerance to UV, we exposed individuals to a continuous ∼3.7 mW/cm2 UVB exposure up to 8 h, based on previous research (11). After 8 h of UVB exposure, we observed that skin of N. parkeri showed no obvious change, whereas stratum corneum started showing damage in N. phrynoides and Q. spinosa.
In the UVB exposure experiment, each three or four individuals of studied frogs was washed with double-distilled water (ddH2O) and set in a plastic box (20 × 15 × 15 cm) filled with 200 mL of ddH2O. Then we exposed these individuals (a total of 27 individuals for N. parkeri and same for N. phrynoides, 20 for Q. spinosa) to separate UVB lights at an irradiance of ∼3.7 mW/cm2 following a time course (0, 0.5, 1, 2, 4, 6, and 8 h). During UVB exposure, the plastic boxes were surrounded with ice packs to keep the temperature of water baths that frogs were sitting in around room temperature. Skin secretions and dorsal skin samples were collected separately from each individual for the subsequent analyses.
Statistical Analysis of Changes to Skin Histology.
We collected dorsal skin samples near the foramen magnum in a 1 cm × 1 cm square immediately after the frogs were killed and subjected them to hematoxylin–eosin staining procedures (11). The skin tissues were fixed in 10% buffered formalin overnight and subsequently preserved in 70% ethanol for storage under ambient conditions, then embedded in paraffin. Histological sections 6 μm thick were mounted on positively charged slides and then were stained with the HE solution. The histological sections were photographed with a Leica DM4000B microscope (Leica, Heidelberg, Germany) at ×100 or ×200 magnification. All the morphological characteristics were quantified based on images of hematoxylin–eosin stained sections with Image-Pro Plus version 6.0 (https://www.mediacy.com/imageproplus). All the data used for images are given in Dataset S1.
After images of histological sections were captured at ×200 magnification, at least 54 images (from six time points of the UV exposure experiment) were randomly selected from each species to quantify damage and melanin content. For each histological image, the proportion of hollowed-out areas within the overall epithelial area was calculated as the degree of damage (%). A two-tailed t test was used for significance testing, and FDR was used for correction of P-value. The mean integrated optical density (47) was measured to represent melanin content in skin.
Measurement of ABTS+ Free Radical Scavenging Activities.
Skin secretions were collected with sterile swabs and dissolved in ddH2O. The secretion solutions were centrifuged at 12,000 rpm, 4 °C for 10 min, and then the supernatants were lyophilized and stored at −80 °C until use. An ABTS scavenging test (48) with some modification was adopted to evaluate the antioxidant activity of the samples. Briefly, a stock solution of ABTS radical (Sigma-Aldrich, USA) was prepared by incubating 2.8 mM potassium persulfate (Sigma-Aldrich, USA) with 7 mM ABTS in water for at least 6 h in the dark, after which it was used immediately. The stock solution was diluted 50-fold with ddH2O, and then 50-μL samples were added to the diluted stock solutions and kept from light for 30 min, with a blank control of the same volume of ddH2O. Vitamin C (Sigma-Aldrich, USA) dissolved in H2O was used as the positive control. The decrease in absorbance at 415 nm indicated the antioxidant activity of the samples. The rate of free radical scavenging (%) was calculated as follows: (Ablank − Asample) × 100/Ablank.
Metabolite Identification and Quantification.
Each skin sample (100 mg) was ground with liquid nitrogen, and homogenate was resuspended with prechilled 80% methanol and 0.1% formic acid. The samples were incubated on ice for 5 min and then were centrifuged at 15,000 g, 4 °C for 20 min. Some of supernatant was diluted to a final concentration containing 53% methanol by liquid chromatography mass spectrometry–grade water. Finally, the supernatant was injected into the ultra-high-performance liquid chromatography–tandem mass spectrometry system analysis (49).
The raw data files generated by ultra-high-performance liquid chromatography–tandem mass spectrometry were processed in Compound Discoverer 3.1 (ThermoFisher) to perform peak alignment, peak picking, and quantitation for each metabolite. After that, peak intensities were normalized to the total spectral intensity. Then the peaks were matched against the mzCloud (https://www.mzcloud.org/), mzVault, and Mass List databases to obtain the accurate qualitative and relative quantitative results. Statistical analyses were performed in the statistical software R (version R-3.4.3). The metabolites were annotated with the Kyoto Encyclopedia of Genes and Genomes database (50), HMDB database (https://hmdb.ca/ metabolites), and LIPID MAPS database (https://www.lipidmaps.org/). Two-tailed t test was applied to calculate the statistical significance (P value). The metabolites with FDR <0.05 and fold change ≥2 or ≤0.5 were considered to be differentially expressed metabolites.
Genome Sequencing, Assembly, and Annotation.
One female N. parkeri from Lhasa, Tibet in China (3,697 m) was prepared for DNA isolation. After euthanasia, liver, muscle, heart, lung, brain, skin, kidney, and ovary tissues were deprived and stored in liquid nitrogen. Total RNA was extracted from each of tissues, and genomic DNA was extracted from liver. For long-read sequencing, SMRTbell libraries with fragment size of 20 kb were constructed and sequenced by the PacBio Sequel system. These sequences were de novo assembled with Falcon (51). To perform error correction of the assembled genome, short-read sequencing data were obtained based on a paired-end library in the BGISEQ-500 platform. Then, the Hi-C technique was applied to capture chromatin interactions. The Hi-C reads were evaluated and further filtered by HiC-Pro (52).
We refined the primary assembly by using both PacBio long-read data and BGISEQ short-read data with Pilon (53). Then, methods based on de novo prediction, homology, and RNA sequence were used to annotate the positions and structures of protein coding genes. Databases of SwissProt and NR (National Center for Biotechnology Information nonredundant protein sequence database) (54) were selected for gene functional annotation of N. parkeri in BLAST+ (55). We annotated the repetitive sequences in the N. parkeri genome in RepeatMasker (version 4.08) and RepeatProteinMask (version 4.08) (56) based on the RepBase TE library (https://www.girinst.org/repbase/). Then, noncoding RNAs (including miRNAs) were identified through the Rfam database (57) with default parameters. The raw sequences for the whole genome assembly and annotation information have been deposited in the Genome Sequence Archive (GSA) of National Genomics Data Center (NGDC) (58) at https://ngdc.cncb.ac.cn/gsa/ (accession: CRA004362). The whole-genome assembly data have been deposited in the Genome Warehouse of NGDC (59) at https://bigd.big.ac.cn/gwh under accession number GWHBCKU00000000. The annotation files can be found in Figshare at https://figshare.com/projects/Genomic_data_of_Nanorana_parkeri/116061.
Transcriptome Sequencing and Assemblies.
Total RNA was extracted with TRIzol reagent (Takara) and an RNeasy Mini Kit (Qiagen, Valencia, CA). Sequencing libraries were generated with a NEBNext Ultra RNA Library Prep Kit for Illumina (#E7530L; NEB, USA). After cluster generation, the libraries were sequenced on an Illumina HiSeq PE platform, and 150 bp paired-end reads were generated. Reads that contained low-quality, adaptor-polluted, or high content of unknown base reads were removed.
For N. phrynoides and Q. spinosa, of which reference genomes were lacking, clean reads from the unexposed group were merged for de novo assembly in Trinity (60) with default settings (–min_contig_length 300 –min_kmer_cov 3 –min_glue 3 –bfly_opts ‘-V 5 –edge-thr = 0.1 –stderr). Then the reference transcript was produced by eliminating redundant contigs with CD-HIT (61). To produce longer and more complete consensus sequences, the TIGR Gene Indices Clustering Tools pipeline was used to cluster and assemble sequences with CAP3 (62). The quality of the reference transcript based on common statistical indexes was evaluated (SI Appendix, Table S10). The clean reads of mRNA-seq of studied species in this article are given in the GSA of NGDC under accession number CRA004830.
Ortholog Determination.
A modified reciprocal best hit method (version 0.6.9 “crb-blast”) (63) was used to identify orthologous genes between the transcript assemblies (N. phrynoides and Q. spinosa) and the genome of N. parkeri. The coding region of each assembled contig was determined in TransDecoder (version 3.0.1 with “-m 50” option) (64). If more than one open reading frame was predicted, the one with greatest similarity to the sequence in N. parkeri was chosen. Prank (65) and Gblocks (66) were used to align each gene into a consensus 1:1 orthologous gene set (Dataset S3) for subsequent evolutionary analyses. To reduce false positives, alignments with a final length of less than 120 bp were discarded. Because a high mean identity ratio is assumed to indicate a accurate orthology prediction, we estimated the mean identity ratio for each alignment to evaluate the quality of our orthologous gene set.
Expression Abundance Calculation.
For N. parkeri, all the clean reads obtained from mRNA-seq were mapped to the reference genome in HISAT2 (67) (–phred33 –sensitive –no-discordant –no-mixed -I 1 -X 1000). Then, StringTie (68) (-f 0.3 -j 3 -c 5 -g 100 -s 10000 -p 8) was used to calculate transcript expression levels. A Python script (prepDE.py) was used to extract read count information from the files generated by StringTie. For N. phrynoides and Q. spinosa, clean reads were mapped to the corresponding reference transcript in Bowtie2 (69) (-q –phred64 –sensitive –dpad 0 –gbar 99999999 –mp 1,1 –np 1 –score-min L,0,-0.1 -I 1 -X 1000 –no-mixed –no-discordant -p l -k 200). We calculated the expression (fragments per kilobase per million mapped fragments [FPKM]) of each transcript with RSEM (70) for each individual. A heatmap of gene expression was drawn in eGPS (71) based on Z-scores and normalized FPKM values.
Temporal Clustering of Genes from Time-Course Expression Profiles.
To trace the changing pattern of gene expression, we used STEM software (version 1.3.12) (28) to classify significant temporal groups. Before implementing STEM, we calculated the median gene expression value for each time point and normalized the median value with the log2 function. The normalized expression profile was used as the input for STEM. We selected the default parameters with “Log normalize data” as the data info and “STEM Clustering Method” as the clustering method. The clustering results from STEM are shown in SI Appendix, Fig. S6.
Identification of Coexpressed Genes in Temporal Groups.
The WGCNA package (29) was used to identify coexpressed genes that were strongly related to specific temporal groups (with options “unsigned correlation” and “minimum cluster size = 30”). The Benjamini–Hochberg method was used to correct for multiple testing when calculating the correlation between modules and time points (SI Appendix, Fig. S7). The genes that were highly correlated with a module (MM > 0.6, P < 0.05) were considered as coexpressed genes in the temporal groups. GO enrichment analyses on temporal group genes were performed in Metascape (72). We converted gene symbols into orthologs in Homo sapiens and then analyzed the associated functional enrichment terms. The gene list and correlation with modules, as well as respective GO terms in each temporal group, can be found in SI Appendix, Tables S3–S6.
miRNA-seq, Annotation, and Expression Calculation.
We performed miRNA-seq analysis on the same individuals (n = 19 for N. parkeri, n = 24 for N. phrynoides, n = 14 for Q. spinosa) that were used for mRNA-seq. RNA extraction and quality assessment were performed according to the above protocols. Small RNA (18–30 nt) was extracted and prepared for library construction according to the method and process of the Small RNA Sample Preparation Kit (Illumina, RS-200-0048). The qualified libraries were sequenced with an Illumina platform to generate 50-bp single-end reads. Reads that contained low-quality, adaptor-polluted, high content of unknown base reads and those with a length of less than 15 bp were filtrated. The clean reads of miRNA sequencing of studied species in this article are given in the GSA of NGDC under accession number CRA005235.
The filtered reads were mapped to the corresponding genome (N. parkeri) or reference transcripts (N. phrynoides and Q. spinosa) in Bowtie (73) (-best -strata -v 0 -k 100) and then analyzed in miRDeep2 (74) to annotate mature miRNAs and precursors. The annotation was based on known miRNAs (from the available well-annotated genomes including Xenopus laevis, X. tropicalis, Anolis carolinensis, Gallus gallus, and Homo sapiens) curated in miRBase (release 22) (75) with no mismatches allowed. miRNAs with a read counts <5 and a true positive probability <60% were excluded from downstream analyses. If a miRNA expressed in at least two of the three (or four) individuals in one time point of UV exposure, we considered it reliable. The numbers of miRNAs identified in each species are listed in Dataset S8. The abundances of miRNAs were normalized by dividing the reads quantified by miRDeep2, with the total miRNA reads quantified by Bowtie and then log2 transformed (Dataset S9).
miRNA Target Prediction.
mRNA and miRNA expression data from the same individual were combined as the input data for WGCNA to identify coexpressed miRNAs and mRNAs in the early-phase temporal group and later-phase temporal group. miRNAs that showed a significant negative correlation (MM < 0, P < 0.05) with a temporal group were selected as coexpressed miRNA candidates (SI Appendix, Fig. S10). To identify coexpressed miRNAs in the gradually increasing group and gradually decreasing group, expression profiles of the total miRNAs were further clustered in STEM (SI Appendix, Fig. S11). The mRNA targets of all the coexpressed miRNAs were predicted with TargetScan (version 7.1) (76) with the parameter “Total context++ score < -0.1.” The correlation of miRNAs and target mRNA in expression was qualified by Spearman’s rank correlation coefficient test in R (SI Appendix, Table S11). Only miRNAs that showed a significant (P < 0.05) negative correlation with predicted mRNA targets were retained as candidates.
Evolutionary Analyses.
Based on the phylogenetic relationships of the studied species (34), codeml was used to estimate dN, dS, and dN/dS values with the free ratio model (“model=1”). An improved branch site model (null hypothesis: model = 2, NSsites = 2, fix_omega = 1, omega =1; alternative hypothesis: model = 2, NSsites = 2, fix_omega = 0, omega = 1) in PAML (35) was used to identify genes showing positive selection along the N. parkeri branch, which was assigned as the foreground branch. A likelihood ratio test was used to compare the observed positive selection to a null model. The corresponding P value was calculated and corrected by FDR. Positively selected sites were deduced through Bayesian empirical Bayes analysis.
In Vitro Synthesis of Tyrosinase and Confirmation of Its Activity.
We separately synthesized DNA sequences encoding the tyrosinases of N. parkeri and Q. spinosa in vitro (see SI Appendix, Fig. S13 for expressions of synthesized proteins). The synthesized tyrosinase sequences were inserted into PET-32a bacterial plasmids, which were then transferred into BL21 competent cells (TSV-A09; Tsingke, Kunming, China). The bacterial strains transfected with the recombinant plasmid were inoculated in Luria–Bertani liquid medium resistant to ampicillin (A610028; Sangon Biotech, Shanghai, China) and cultured at 37 °C with agitation 180 rpm to an optical density at 600 nm of 0.6. The recombinant fusion tyrosinases were isolated and purified at 4 °C. Then, the tyrosinase was expressed with 200 mM CuSO4 (A600063-0500; Sangon Biotech, Shanghai, China) and 100 mM isopropylthio-β-galactoside (I6148; Macklin, Shanghai, China) induction at 28 °C with shaking (100 rpm) for 10 h and purified at 4 °C by Ni-NTA agarose column (30230; Qiagen, Valencia, CA), according to the instructions. With the buffer (15 μM CuSO4 50 mM Na2HPO4, pH 6.2), the tyrosinase solution was combined with the substrate L-DOPA (D9628-5G; Sigma-Aldrich, St. Louis, MO) at concentrations of 0.625 mM, 1.5 mM, 2.5 mM, 5 mM, and 10 mM. The absorbance at 475 nm was measured with an Epoch Microplate Reader (BioTek, VT, American) at 1-min intervals and monitored for 1 h. The dynamics of the tyrosinase catalysis are showed in a fitted curve generated with the Lineweaver–Burk equation.
Supplementary Material
Acknowledgments
We thank Yong-Bin Chen, Bao-Wei Jiao, Ju-Min Zhou, Zhen Liu, Bo Zhao, and Jing Chai for helpful discussions; Bao-Lin Zhang, Yong Shao, and Wei Xu for assistance in collections; Hong-Man Chen for technical support in the laboratory. This research was supported by Chinese Academy of Sciences, XDPB17, J.C.; the Second Tibetan Plateau Scientific Expedition and Research Program, 2019QZKK0501, J.C.; Chinese Academy of Sciences, XDA20050201, J.C.; Chinese Academy of Sciences, Animal Branch of the Germplasm Bank of Wild Species, J.C.; Yunnan Province in China, Spring City Plan: the high-level talent promotion and training project of Kunming, Y.-P.Z.; Natural Science Foundation of China, 31671326, Y.-B.S.; Yunnan Province in China, 202101AV070009, Y.-B.S.; Natural Science Foundation of China, 81760648, X.W.Y.; State Key Laboratory of Genetic Resources and Evolution, GREKF20-02, D.M.H. and J.C.; Chinese Academy of Sciences, 2018VBA0039, D.M.H.; Chinese Academy of Sciences, 2017VBA0003, R.W.M.; National Sciences and Engineering Research Council of Canada, A3148, R.W.M.; and China Scholarship Council, 201904910862, T.-T.F.
Footnotes
Reviewers: F.L., Institute of Zoology Chinese Academy of Sciences; and J.L., Lanzhou University.
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2120393119/-/DCSupplemental.
Data, Materials, and Software Availability
Raw nucleotide sequence data have been deposited in Genome Sequence Archive (GSA) in National Genomics Data Center (NGDC), China National Center for Bioinformation/Beijing Institute of Genomics (CNCB/BIG), Chinese Academy of Sciences (CAS) (CRA004362 (77), CRA004830 (78), and CRA005235 (79)) .
References
- 1.Bais A. F., et al. , Ozone-climate interactions and effects on solar ultraviolet radiation. Photochem. Photobiol. Sci. 18, 602–640 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Cadet J., Douki T., Ravanat J. L., Oxidatively generated damage to cellular DNA by UVB and UVA radiation. Photochem. Photobiol. 91, 140–155 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Garcia T. S., Romansic J. M., Blaustein A. R., Survival of three species of anuran metamorphs exposed to UV-B radiation and the pathogenic fungus Batrachochytrium dendrobatidis. Dis. Aquat. Organ. 72, 163–169 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Alton A. L., Franklin E. C., Drivers of amphibian declines: Effects of ultraviolet radiation and interactions with other environmental factors. Clim. Change Responses 4, 6 (2017). [Google Scholar]
- 5.Dyck J., Structure and colour-production of the blue barbs of Agapornis roseicollis and Cotinga maynana. Z. Zellforsch. Mikrosk. Anat. 115, 17–29 (1971). [DOI] [PubMed] [Google Scholar]
- 6.Jablonski N. G., Chaplin G., Colloquium paper: Human skin pigmentation as an adaptation to UV radiation. Proc. Natl. Acad. Sci. U.S.A. 107 (suppl. 2), 8962–8968 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin J. Y., Fisher D. E., Melanocyte biology and skin pigmentation. Nature 445, 843–850 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Porter W. P., Norris K. S., Lizard reflectivity change and its effect on light transmission through body wall. Science 163, 482–484 (1969). [DOI] [PubMed] [Google Scholar]
- 9.Yang X., Wang Y., Zhang Y., Lee W. H., Zhang Y., Rich diversity and potency of skin antioxidant peptides revealed a novel molecular basis for high-altitude adaptation of amphibians. Sci. Rep. 6, 19866 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu L., et al. , Genomic analysis of snub-nosed monkeys (Rhinopithecus) identifies genes and processes related to high-altitude adaptation. Nat. Genet. 48, 947–952 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Poljsak B., Dahmane R., Godic A., Skin and antioxidants. J. Cosmet. Laser Ther. 15, 107–113 (2013). [DOI] [PubMed] [Google Scholar]
- 12.Olin-Sandoval V., et al. , Lysine harvesting is an antioxidant strategy and triggers underground polyamine metabolism. Nature 572, 249–253 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nassour J., et al. , Defective DNA single-strand break repair is responsible for senescence and neoplastic escape of epithelial cells. Nat. Commun. 7, 10399 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rastogi P. R., Ashok Kumar R., Tyagi M. B., Sinha R. P., Molecular mechanisms of ultraviolet radiation-induced DNA damage and repair. DNA Damage, Mutagenesis, and DNA Repair 2010, 592080 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mendez F., et al. , Heat-shock proteins associated with base excision repair enzymes in HeLa cells. Radiat. Res. 153, 186–195 (2000). [DOI] [PubMed] [Google Scholar]
- 16.Jarosz D. F., Lindquist S., Hsp90 and environmental stress transform the adaptive value of natural genetic variation. Science 330, 1820–1824 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jie L., et al. , Name and scale matter: Clarifying the geography of Tibetan Plateau and adjacent mountain regions. Global Planet. Change 215, 103893 (2022). [Google Scholar]
- 18.Peng S., Du Q., Wang L., Lin A., Hu B., Long-term variations of ultraviolet radiation in Tibetan Plateau from observation and estimation. Int. J. Climatol. 35, 1245–1253 (2015). [Google Scholar]
- 19.Li J. T., et al. , Comparative genomic investigation of high-elevation adaptation in ectothermic snakes. Proc. Natl. Acad. Sci. U.S.A. 115, 8406–8411 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Che J., et al. , Spiny frogs (Paini) illuminate the history of the Himalayan region and Southeast Asia. Proc. Natl. Acad. Sci. U.S.A. 107, 13765–13770 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang C. H., et al. , Comparative skin histology of frogs reveals high-elevation adaptation of the Tibetan Nanorana parkeri. Asian Herpetol. Res. 10, 79–85 (2019). [Google Scholar]
- 22.Verdin E., NAD+ in aging, metabolism, and neurodegeneration. Science 350, 1208–1213 (2015). [DOI] [PubMed] [Google Scholar]
- 23.Stuyvesant V. W., Jolley W. B., Anti-inflammatory activity of d-α-tocopherol (vitamin E) and linoleic acid. Nature 216, 585–586 (1967). [DOI] [PubMed] [Google Scholar]
- 24.Marieb N. E., Human Anatomy & Physiology (Benjamin Cummings, San Francisco, 2001). [Google Scholar]
- 25.Morgan V. R., Handbook of Small Animal Practice (Elsevier, ed. 5, 2008). [Google Scholar]
- 26.McIntire S. L., Reimer R. J., Schuske K., Edwards R. H., Jorgensen E. M., Identification and characterization of the vesicular GABA transporter. Nature 389, 870–876 (1997). [DOI] [PubMed] [Google Scholar]
- 27.Sun Y. B., et al. , Whole-genome sequence of the Tibetan frog Nanorana parkeri and the comparative evolution of tetrapod genomes. Proc. Natl. Acad. Sci. U.S.A. 112, E1257–E1262 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ernst J., Bar-Joseph Z., STEM: A tool for the analysis of short time series gene expression data. BMC Bioinformatics 7, 191–201 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Langfelder P., Horvath S., WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics 9, 559–572 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morad S. A., Cabot M. C., Ceramide-orchestrated signalling in cancer cells. Nat. Rev. Cancer 13, 51–65 (2013). [DOI] [PubMed] [Google Scholar]
- 31.Jia Q., Sieburth D., Mitochondrial hydrogen peroxide positively regulates neuropeptide secretion during diet-induced activation of the oxidative stress response. Nat. Commun. 12, 2304 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gentile M., Latonen L., Laiho M., Cell cycle arrest and apoptosis provoked by UV radiation-induced DNA damage are transcriptionally highly divergent responses. Nucleic Acids Res. 31, 4779–4790 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fraser H. B., Gene expression drives local adaptation in humans. Genome Res. 23, 1089–1096 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun Y. B., et al. , Species groups distributed across elevational gradients reveal convergent and continuous genetic adaptation to high elevations. Proc. Natl. Acad. Sci. U.S.A. 115, E10634–E10641 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang Z., PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591 (2007). [DOI] [PubMed] [Google Scholar]
- 36.Körner A., Pawelek J., Mammalian tyrosinase catalyzes three reactions in the biosynthesis of melanin. Science 217, 1163–1165 (1982). [DOI] [PubMed] [Google Scholar]
- 37.Li M., et al. , Genomic analyses identify distinct patterns of selection in domesticated pigs and Tibetan wild boars. Nat. Genet. 45, 1431–1438 (2013). [DOI] [PubMed] [Google Scholar]
- 38.Guo J., et al. , Whole-genome sequencing reveals selection signatures associated with important traits in six goat breeds. Sci. Rep. 8, 10405 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qu Y., et al. , The evolution of ancestral and species-specific adaptations in snowfinches at the Qinghai-Tibet Plateau. Proc. Natl. Acad. Sci. U.S.A. 118, E2012398118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Farage M. A., Miller K. W., Maibach H. I., Textbook of Aging Skin (Springer Science & Business Media, Heidelberg, 2015). [Google Scholar]
- 41.Peters E. M. J., et al. , Neuropeptide control mechanisms in cutaneous biology: Physiological and clinical significance. J. Invest. Dermatol. 126, 1937–1947 (2006). [DOI] [PubMed] [Google Scholar]
- 42.Cui R., et al. , Central role of p53 in the suntan response and pathologic hyperpigmentation. Cell 128, 853–864 (2007). [DOI] [PubMed] [Google Scholar]
- 43.Dunaway S., et al. , Natural antioxidants: Multiple mechanisms to protect skin from solar radiation. Front. Pharmacol. 9, 392 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Robu M., et al. , Role of poly(ADP-ribose) polymerase-1 in the removal of UV-induced DNA lesions by nucleotide excision repair. Proc. Natl. Acad. Sci. U.S.A. 110, 1658–1663 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schwertman P., et al. , UV-sensitive syndrome protein UVSSA recruits USP7 to regulate transcription-coupled repair. Nat. Genet. 44, 598–602 (2012). [DOI] [PubMed] [Google Scholar]
- 46.Yoo H., et al. , CRTC3, a sensor and key regulator for melanogenesis, as a tunable therapeutic target for pigmentary disorders. Theranostics 11, 9918–9936 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Greenberg N. M., et al. , Prostate cancer in a transgenic mouse. Proc. Natl. Acad. Sci. U.S.A. 92, 3439–3443 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Akerström B., Maghzal G. J., Winterbourn C. C., Kettle A. J., The lipocalin alpha1-microglobulin has radical scavenging activity. J. Biol. Chem. 282, 31493–31503 (2007). [DOI] [PubMed] [Google Scholar]
- 49.Want E. J., et al. , Global metabolic profiling of animal and human tissues via UPLC-MS. Nat. Protoc. 8, 17–32 (2013). [DOI] [PubMed] [Google Scholar]
- 50.Ogata H., et al. , KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 27, 29–34 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chin C. S., et al. , Phased diploid genome assembly with single-molecule real-time sequencing. Nat. Methods 13, 1050–1054 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Servant N., et al. , HiC-Pro: An optimized and flexible pipeline for Hi-C data processing. Genome Biol. 16, 259–270 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Walker B. J., et al. , Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9, e112963 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pruitt K. D., Tatusova T., Maglott D. R., NCBI reference sequences (RefSeq): A curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 35, D61–D65 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McGinnis S., Madden T. L., BLAST: At the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. 32, W20–W25 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tarailo-Graovac M., Chen N., Using RepeatMasker to identify repetitive elements in genomic sequences. Curr. Protoc. Bioinformatics Chapter 4, Unit 4.10 (2009). [DOI] [PubMed] [Google Scholar]
- 57.Kalvari I., et al. , Rfam 13.0: Shifting to a genome-centric resource for non-coding RNA families. Nucleic Acids Res. 46, D335–D342 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen T., et al. , The Genome Sequence Archive family: Toward explosive data growth and diverse data types. Genomics Proteomics Bioinformatics 19, 578–583 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.CNCB-NGDC Members and Partners, Database resources of the National Genomics Data Center, China National Center for bioinformation in 2021. Nucleic Acids Res. 49, D18–D28 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Grabherr M. G., et al. , Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fu L., Niu B., Zhu Z., Wu S., Li W., CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 28, 3150–3152 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huang X., Madan A., CAP3: A DNA sequence assembly program. Genome Res. 9, 868–877 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Aubry S., Kelly S., Kümpers B. M., Smith-Unna R. D., Hibberd J. M., Deep evolutionary comparison of gene expression identifies parallel recruitment of trans-factors in two independent origins of C4 photosynthesis. PLoS Genet. 10, e1004365 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Oakley T. H., et al. , Osiris: Accessible and reproducible phylogenetic and phylogenomic analyses within the Galaxy workflow management system. BMC Bioinformatics 15, 230–239 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Löytynoja A., Phylogeny-aware alignment with PRANK. Methods Mol. Biol. 1079, 155–170 (2014). [DOI] [PubMed] [Google Scholar]
- 66.Talavera G., Castresana J., Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 56, 564–577 (2007). [DOI] [PubMed] [Google Scholar]
- 67.Kim D., Langmead B., Salzberg S. L., HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pertea M., et al. , StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li B., Dewey C. N., RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323–339 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yu D., et al. , eGPS 1.0: Comprehensive software for multi-omic and evolutionary analyses. Natl. Sci. Rev. 6, 867–869 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhou Y., et al. , Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 10, 1523–1533 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Langmead B., Trapnell C., Pop M., Salzberg S. L., Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25.1–R25.10 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Friedländer M. R., Mackowiak S. D., Li N., Chen W., Rajewsky N., miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 40, 37–52 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kozomara A., Birgaoanu M., Griffiths-Jones S., miRBase: From microRNA sequences to function. Nucleic Acids Res. 47, D155–D162 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Agarwal V., Bell G. W., Nam J.-W., Bartel D. P., Predicting effective microRNA target sites in mammalian mRNAs. eLife 4, e05005 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ting-Ting Fu et al., Genome data of Nanorana parkeri. GSA. https://ngdc.cncb.ac.cn/gsa/browse/CRA004362. Deposited 18 October 2022. [Google Scholar]
- 78.Ting-Ting Fu et al., Transcriptomes of skin in Nanorana species. GSA. https://ngdc.cncb.ac.cn/gsa/browse/CRA004830. Deposited 18 October 2022. [Google Scholar]
- 79.Ting-Ting Fu et al., miRNA reads of skin in Nanorana species. GSA. https://ngdc.cncb.ac.cn/gsa/browse/CRA005235. Deposited 18 October 2022. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw nucleotide sequence data have been deposited in Genome Sequence Archive (GSA) in National Genomics Data Center (NGDC), China National Center for Bioinformation/Beijing Institute of Genomics (CNCB/BIG), Chinese Academy of Sciences (CAS) (CRA004362 (77), CRA004830 (78), and CRA005235 (79)) .