Abstract
C1q/TNF-related protein (CTRP) family comprises fifteen highly conserved secretory proteins with diverse central and peripheral functions. In zebrafish, mouse, and human, CTRP4 is most highly expressed in the brain. We previously showed that CTRP4 is a metabolically responsive regulator of food intake and energy balance, and mice lacking CTRP4 exhibit sexually dimorphic changes in ingestive behaviors and systemic metabolism. Recent single-cell RNA sequencing also revealed Ctrp4/C1qtnf4 expression in diverse neuronal cell types across distinct anatomical brain regions, hinting at additional roles in the central nervous system not previously characterized. To uncover additional central functions of CTRP4, we subjected Ctrp4 knockout (KO) mice to a battery of behavioral tests. Relative to wild-type (WT) littermates, loss of CTRP4 does not alter exploratory, anxiety-, or depressive-like behaviors, motor function and balance, sensorimotor gating, novel object recognition, and spatial memory. While pain-sensing mechanisms in response to thermal stress and mild shock are intact, both male and female Ctrp4 KO mice have increased sensitivity to pain induced by higher-level shock, suggesting altered nociceptive function. Importantly, CTRP4 deficiency impairs hippocampal-dependent associative learning and memory as assessed by trace fear conditioning paradigm. This deficit is sex-dependent, affects only female mice, and is associated with altered expression of learning and memory genes (Arc, c-fos, and Pde4d) in the hippocampus and cortex. Altogether, our behavioral and gene expression analyses have uncovered novel aspects of the CTRP4 function and provided a physiological context to further investigate its mechanism of action in the central and peripheral nervous system.
Keywords: C1QTNF4, cortex, hippocampus, learning and memory, mouse behavior, secreted hormone
1 ∣. INTRODUCTION
Since the discovery of immune complement C1q,1,2 over 30 additional secretory proteins that comprise the vertebrate C1q family have been characterized in humans, mice, and zebrafish.3-5 The key feature that unites all family members is the presence of a globular C1q domain located at the C-terminus of each protein.3 These secretory proteins have important and diverse biological functions, ranging from immune response,6 to blood pressure control and platelet hemostasis,7 metabolism,8 cardiovascular function,9 myoblast fusion,10 and nervous system functions.11
Among the C1q family members are the fifteen proteins referred to as the C1q/TNF-related proteins (CTRP1-15), which we have identified and characterized as part of an ongoing effort to understand how secreted hormones facilitate inter-organ communications to maintain metabolic homeostasis.12-19 Recent studies by us and others have highlighted the salutary roles of CTRPs in the peripheral tissues, most notably related to metabolic and cardiovascular functions.20-28 Since the unexpected role of complement C1q in synaptic pruning was described,29,30 the central nervous system (CNS) functions for several brain-enriched C1q family members have also been uncovered. These include the important roles of cerebellins (Cbln1-4) in hippocampus, cerebellum, and midbrain,31-35 as well as the functions of C1ql3/CTRP1336 and C1ql1/CTRP1437,38 in cerebellum and forebrain.
The distinguishing feature of CTRP4 (encoded by C1QTNF4 gene) is that it is the only vertebrate C1q family member with two tandem globular C1q domains connected by a short linker.12,39 In divergent vertebrate species such as human, mouse, and zebrafish, CTRP4 expression is predominantly detected in the CNS.39 One other tissue with a relatively high expression of CTRP4 transcript in humans is the adipose tissue.39 We first demonstrated its in vivo role in reducing food intake and altering energy balance via central delivery of recombinant CTRP4 into the lateral ventricle.39 Subsequently, we and others have further confirmed and extended its CNS role in food intake and energy metabolism using overexpression40 and knockout41 mouse models. Interestingly, anti-inflammatory properties of CTRP4 have recently been noted in humans and rodents. Transgenic mice with elevated plasma CTRP4 have significantly reduced gastrointestinal inflammation and disease severity index when subjected to dextran sulfate sodium-induced colitis.42 Conversely, its deficiency in Ctrp4 KO mice results in heightened inflammation and mortality induced by endotoxin (LPS) shock or sepsis.43 Exome sequencing of individuals with a severe form of the autoimmune disease systemic lupus erythematosus (SLE) also identified a rare variant of CTRP4/C1QTNF4 (H198Q) which encodes a protein that can reduce TNF-α-mediated NF-κB activation and cell death in vitro.44 Whether this rare variant, also identified in a separate cohort of SLE patients,45 is causally linked to SLE remains to be established. In other brain inflammatory condition due to infection, such as herpes simplex encephalitis, expression of CTRP4 was shown to be upregulated.46 More recently, nucleolin (a soluble intracellular protein) was suggested as a potential cell surface docking protein that interacts with CTRP4 on monocytes, B-cells (to a much lesser extent), and dead cells.47 Collectively, these studies demonstrated the potential central and peripheral functions of CTRP4.
Aside from the few published studies highlighted, the contribution of CTRP4 to other aspects of physiology remains largely unknown and unexplored. Recent single-cell RNA sequencing has provided an unprecedented and comprehensive view of Ctrp4 expression in different brain regions of the mouse at remarkable cellular resolution.48 We hypothesized that since Ctrp4 is expressed predominantly in the brain39 in a variety of neuronal cell types that spread across many different anatomical regions,48 CTRP4 may likely have additional CNS roles beyond regulating neuropeptide gene expression in the hypothalamus to control food intake. To test this hypothesis and explore additional functions of CTRP4 in the brain as hinted by its widespread expression, we subjected the Ctrp4 KO mice to a battery of behavioral tests. We reasoned that any behavioral deficits seen in the Ctrp4 KO mice would provide a clue to its role in the CNS. We discovered that CTRP4 deficiency impairs associative learning and memory in a sex-dependent manner, and this deficit correlates with altered learning- and memory-associated gene expression in the hippocampus and cortex. We also showed that loss of CTRP4 modestly enhances sensitivity to shock, highlighting a potential nociceptive function. Given how little is known about CTRP4 biology, the novel CNS role we uncovered will provide a context for future studies to interrogate its mechanisms of action and cellular targets within the brain.
2 ∣. MATERIALS AND METHODS
2.1 ∣. Mouse model
The CTRP4 (c1qtnf4tm1a(KOMP)Wtsi) knockout (KO) mouse strain used was previously published.41 The CTRP4-null mice were generated and maintained on a C57BL/6N genetic background. Genotyping primers for wild-type (WT) allele were forward 5′-CTCAGGGCCTTTGGAAGGGCA-3′ and reverse 5′-GGGAG GGCTTATCACAGC ACC-3′. The size of the WT band was 333 bp. Genotyping primers for the Ctrp4 KO allele were forward 5′-GGTAA ACTGGCTCGGATTAGGG-3′ and reverse 5′-TTGACTGTAGCGGCTGATGTTG-3′. The size of the KO band was 211 bp. The genotyping PCR parameters were as follows: 94°C for 5 min, followed by 10 cycles of (94°C for 15 s, 65°C for 30 s, 72°C for 1 min), then 35 cycles of (94°C for 15 s, 55°C for 30 s, 72°C for 30 s), and lastly 72°C for 5 min. All mice were generated by intercrossing Ctrp4 heterozygous (+/−) mice. Ctrp4 KO (−/−) and WT (+/+) littermate controls were housed in polycarbonate cages on a 12-h light–dark photocycle with ad libitum access to water and food. Mice were fed a standard chow (Envigo; 2018SX). At termination of the study, mice were fasted for 2 h and euthanized. Brain tissues were dissected, snap-frozen in liquid nitrogen, and kept at −80°C until analysis. All mouse experiments (protocol # MO19M481) and mouse behavioral tests (protocol # SP20M311) were approved by the Institutional Animal Care and Use Committee of the Johns Hopkins University School of Medicine. All animal experiments were conducted in accordance with the National Institute of Health guidelines and followed the standards established by the Animal Welfare Acts.
2.2 ∣. Tissue collection
Cortex and hippocampus were immediately dissected from euthanized mice, flash-frozen in liquid nitrogen and kept at −80°C until analysis.
2.3 ∣. Quantitative real-time PCR analysis
Total RNA was isolated from micro-dissected brain regions using Trizol reagent (Thermo Fisher Scientific) according to the manufacturer's instructions. Purified RNA was reverse transcribed using an iScript cDNA Synthesis Kit (Bio-rad). Real-time quantitative PCR analysis was performed on a CFX Connect Real-Time System (Bio-rad) using SsoAdvanced Universal SYBR Green Supermix (Bio-rad) per manufactuer's instructions. Data were normalized to 18S rRNA and expressed as relative mRNA levels using the ΔΔCt method.49 Fold change data were log transformed to ensure normal distribution and statistics were performed. Real-time qPCR primers used in this study are included in Table S1.
2.4 ∣. Open field
Locomotor activity was assessed over 30 min in a 40 cm × 40 cm activity chamber with infrared beams (San Diego Instruments Inc., San Diego, CA, USA). Horizontal activity, as well as time spent in the center or periphery of the chamber was automatically recorded. All behavioral tests described in this study were conducted by experimenters blinded to genotypes, and all tests were performed during the active (dark cycle) phase of the mouse circadian cycle.
2.5 ∣. Elevated plus maze
Anxiety related behavior was evaluated using the elevated plus maze test. Mice were placed in the center of a 54 cm high maze consisting of two open and two closed (30.5 cm long) arms for 5 min. Distance travelled and time spent in the open and closed arms was automatically recorded using Anymaze Tracking Software (Stoelting, Co., Wood Dale, IL, USA).
2.6 ∣. Y-maze spontaneous alternation
Working memory was assessed in the Y-maze spontaneous alternation task. Mice were placed at the end of one arm of a Y-maze consisting of three 38 cm long arms (San Diego Instruments Inc., San Diego, CA, USA) and allowed to explore the maze for five minutes. Distance travelled and entries into the arms were automatically recorded using Anymaze Tracking Software (Stoelting, Co., Wood Dale, IL, USA). The percent alternation was calculated using the equation: % Alternation = (Correct alternations/(Total arm entries – 2)) × 100.
2.7 ∣. Y-maze spatial recognition
Spatial memory was assessed using the Y-maze spatial recognition test. This test consists of a “Training” phase and a “Test” phase. During the “Training” phase, one arm of a Y-maze consisting of three 38 cm long arms (San Diego Instruments) was blocked off. Mice were placed at the end of one of two open arms and allowed to explore for five minutes. After a 30-min inter-trial interval, the “Test” phase began. The blockade was removed, and mice were allowed to explore all three arms of the maze for five minutes. Distance travelled and time spent in each arm was automatically recorded using Anymaze Tracking Software (Stoelting, Co., Wood Dale, IL, USA). Data from the first two minutes of the “Test” phase were used to evaluate percent time spent in the novel arm.
2.8 ∣. Novel object recognition test
The novel object recognition test (NORT) was used to evaluate short term memory. Briefly, mice were allowed to habituate to an empty 20 cm × 20 cm box for 10 min. Twenty-four hours following habituation, mice were placed in the box with two identical objects for a 10-min training period. Thirty minutes following the training period, one object was replaced with a novel object, and the mouse was allowed to explore the objects for five minutes. Time spent sniffing the novel and familiar objects was measured using CleverSys Topscan tracking software (Cleverysy Inc., Reston, VA, USA).
2.9 ∣. Rotarod
Motor coordination and learning was evaluated using the accelerating rotarod test. Mice were placed on the rotarod (Columbus Instruments, Columbus, OH, USA) with a starting speed of 4 rpm, with an acceleration of 6 rpm/minute. The time at which each mouse dropped from the rotating rotarod was recorded. Each mouse was given three trials per day with a 2-min inter-trial interval, over three days.
2.10 ∣. Hotplate
Mice were placed on an 11 cm × 11 cm black anodized aluminum plate heated to 52°C (IITC Life Science, Woodland Hills, CA). The latency for each mouse to withdraw one of the hind limbs from the plate was recorded, and the mouse was returned to the home cage. If a mouse did not withdraw the hind limb after 30 s, it was removed from the hotplate and placed back in the home cage.
2.11 ∣. Hargreaves plantar test
Nociception was evaluated using a Plantar Analgesia Meter Model 390G (IITC Life Science, Woodland Hills, CA) as previously described.50 Mice were placed on a glass platform heated to 32°C and habituated to the platform for 30 min prior to testing. A focused thermal heat stimulus was delivered from a light source to the plantar surface of the hind paw for up to 30 s. A full leg raises at the site where the heat stimulus was directed was considered a reaction to the thermal stimulus. The average of six trials was used as the latency for each mouse, alternating paws between trials.
2.12 ∣. Morris water maze
The Morris water maze test (MWM) was based on the protocol previously described.51 The maze consists of a circular stainless-steel tank 4 m in diameter filled with room temperature water made opaque with white tempera paint for the training, probe, and reversal trials. During the hidden platform training phase, a 10 cm platform was placed in a fixed location in the maze, with the top of the platform hidden beneath 1 cm of water. Mice were placed into the maze around the perimeter in one of four start positions used in a semi-random fashion throughout six trials each day for three days. Mice were allowed to search for the platform for 60 s and after finding the platform remained there for 10 s. Escape latency was recorded for each trial. 24 h following the hidden platform training phase, mice were tested in one 60-s probe trial. Latency to cross into the platform area, number of crossings in the platform area, and time spent in the platform quadrant were measured using Anymaze tracking software (Stoelting Co., Wood Dale, IL, USA). During the reversal phase, the hidden platform was placed in a different location of the MWM than that used during the hidden platform training phase. Like the hidden platform training phase, mice were placed into the maze around the perimeter in one of four start positions used in a semi-random fashion throughout six trials each day for two days. Mice were allowed to search for the platform for 60 s and after finding the platform remained there for 10 s. Escape latency was recorded for each trial.
2.13 ∣. Trace fear conditioning
Trace fear conditioning was conducted as previously described.52 Briefly, trace fear conditioning consisted of a habituation day, a training day, and a test day over three consecutive days. On the habituation day, mice were exposed to the shock box (Med Associates, Inc., Fairfax, VT, USA) for ten minutes. On the training day, mice were placed in the shock box and give a two-minute habituation, after which a 20-s tone (80 db, 2000 Hz) was delivered. Twenty seconds following the termination of the tone, a scrambled two-second 0.5 mA shock was delivered. The tone-shock pairing was repeated three additional times. On the test day, mice were placed in the shock box for three minutes to measure freezing in response to the context. Mice were then placed in a separate context and freezing in response to the 20-s tone was measured. Freezing behavior was automatically scored using Med Associates, Inc. Video Freeze Software (Med Associates, Inc., Fairfax, VT, USA).
2.14 ∣. Shock Sensitivity
Sensitivity to shock was evaluated using the method previously described.53 Mice were placed in a shock box (Coulbourn Instruments, Whitehall, PA) and allowed to habituate for two minutes. Following habituation mice were exposed to a 1 s shock which increased in increments of 0.05 mA every 30 s from 0.1 to 0.5 mA. The shock level at which each mouse first flinched, vocalized, and jumped was recorded.
2.15 ∣. Prepulse inhibition PPI
PPI was assessed as described.51 The experimental session consisted of a 5-min acclimatization period to a 70-dB background noise (continuous throughout the session), followed by the presentation of 10 40-ms 120-dB white noise stimuli at a 20-s inter-stimulus interval (the habituation session). Upon the completion of the habituation session, each mouse was left in the enclosure for 5 min without presentations of any startle stimuli. Immediately after, the pre-pulse inhibition (PPI) session was begun. During each PPI session, a mouse was exposed to the following types of trials: pulse-alone trial (a 120-dB, 100-ms, broadband burst); the omission of stimuli (no-stimulus trial); and five pre-pulse–pulse combinations (pre-pulse–pulse trials) consisting of a 20-ms broadband burst used as a pre-pulse and presented 80 ms before the pulse using one of the five pre-pulse intensities: 74; 78; 82; 86 and 90 dB. Each session consisted of six presentations of each type of the trial presented in a pseudorandom order. PPI was assessed as the percentage scores of PPI (%PPI): 100 × (mean startle amplitude on pulse-alone trials – mean startle amplitude on prepulse–pulse trials/ mean startle amplitude on pulse-alone trials) for each animal separately. The percentage of PPI for each animal was used as the dependent variable in statistical analysis.
2.16 ∣. Tail-suspension test
The tail-suspension test was performed as previously described.54 Each mouse was suspended individually by its tail in a styrofoam box (28 cm × 30 cm × 25 cm) in a quiet room. The tip of the mouse tail was fixed on the top part inside the styrofoam box using adhesive tape. The duration of the test was 6 min, and the immobility time of the tail-suspended mice was measured during the last 4 min of the test. The immobility time was determined using a stopwatch by an observer blinded to the genotype.
2.17 ∣. Statistical analyses
All results are expressed as mean ± standard error of the mean (SEM). Statistical analysis was performed with Prism 8 software (GraphPad Software, San Diego, CA). All real-time PCR data were log2 transformed to ensure linear distribution. Data were analyzed with two-tailed Student’s t-tests or by repeated measures ANOVA. For two-way ANOVA, we performed Bonferroni post hoc tests. p < .05 was considered statistically significant. All data are presented as mean ± SEM (standard error of the mean).
3 ∣. RESULTS
3.1 ∣. Cell-level expression patterns of Ctrp4 across distinct anatomical regions of the mouse brain
Our previous study indicated predominant and widespread expression of Ctrp4 transcript in the mouse brain.39 Recently single-cell RNA sequencing, however, has afforded a unique opportunity to re-examine Ctrp4 transcript expression across different mouse brain regions at single-cell resolution.48 In the mouse central and peripheral nervous system, there are a total of 23 distinct anatomical regions that express the Ctrp4 transcript (Figure 1). Within these anatomical regions, there are a total of 32 functional cell types and 175 single-cell clusters that express Ctrp4 (Figure 1). The top six anatomical regions with the highest expression level of Ctrp4 are the spinal cord, midbrain, hindbrain, hypothalamus, cerebral cortex, and hippocampus; together, they account for >73% of the total Ctrp4 transcript in the brain (Figure 1B). Within different brain regions, the Ctrp4 transcript can be found in a variety of functional cell types that include excitatory, inhibitory, dopaminergic, glutamatergic, cholinergic, peptidergic, and nitregic neurons (Figure 1). The expression patterns of Ctrp4 within the central and peripheral nervous system suggest potentially undiscovered CNS role of CTRP4 in regulating physiology and behavior.
FIGURE 1.
Mouse Ctrp4 transcript is widely expressed in the central and peripheral nervous system. (A) Summary of unique mouse anatomical regions, functional cell types, and single-cell clusters that express Ctrp4 based on single-cell RNA sequencing data generated by the Linnarsson lab; Details of methods and cell type clustering as performed by the Linnarsson's lab can be found in the published work.48 The data can be accessed through the mousebrain.org website. (B) Data organized by total Ctrp4 transcript expression per anatomical region. Expression values for each single-cell cluster as defined are obtained from mousebrain.org.48 The cumulative Ctrp4 expression of all single-cell clusters expressing Ctrp4 is considered the total possible Ctrp4 and represents 100% of the expression. Single-cell clusters expressing Ctrp4 were organized by their anatomical location. The Ctrp4 expression levels of each single-cell cluster within a particular anatomical region were summed and the percent of the total possible Ctrp4 expressed is indicated. The data are organized from anatomical regions expressing the most Ctrp4 to those expressing the least. For example, the spinal cord is the region of the nervous system that expressed the greatest amount of Ctrp4 mRNA. We then grouped the single cell populations by functional cell type (excitatory neuron, cholinergic interneuron, etc) and summed the amount of Ctrp4 expressed by each functional cell type. This allows us to see within a particular anatomical region: (1) which functional cell types are expressing Ctrp4 and (2) which of those are expressing the most and least Ctrp4. The next column shows the number of single-cell clusters that express Ctrp4 of that particular functional cell type (ie how many different single cell clusters are excitatory neurons that express Ctrp4 within that anatomical region). The final column shows the percent total Ctrp4 expression in a particular group of functional cell types located within a given anatomical region
3.2 ∣. Loss of CTRP4 does not alter exploratory, anxiety- or depressive-like behaviors
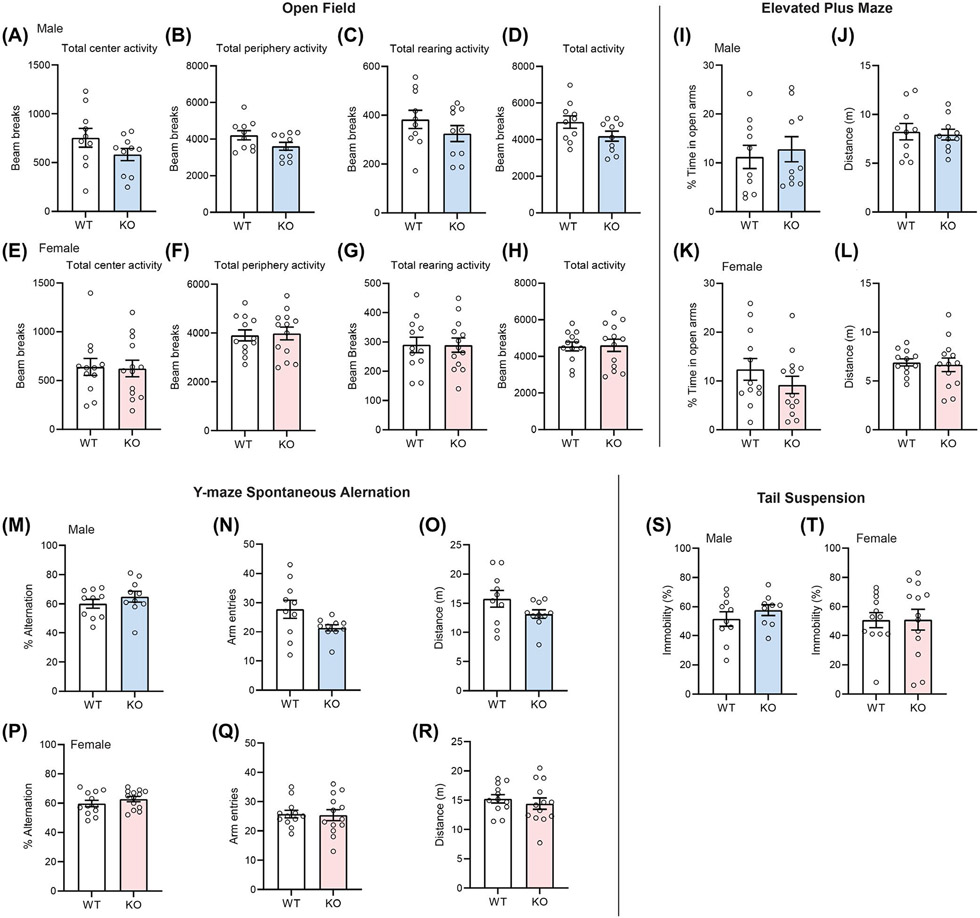
We first assessed the impact of CTRP4 deficiency on general exploratory behaviors. In an open field test for anxiety-like behavior, Ctrp4 KO mice of either sex spent similar amount of time as the WT littermates in the center and periphery of the open field, as well as having similar numbers of rearing activity (Figure 2A-H). In the elevated plus maze that also measures anxiety-like behavior, Ctrp4 KO mice of either sex also spent a similar proportion of time exploring the open arms when compared to the WT littermates (Figure 2I-L). In the Y-maze spontaneous alternation test, a measure of working memory, the number of arm entries and % correct alternation between the three arms were similar between genotypes of either sex (Figure 2M-R). Finally, in the tail suspension test, which measures potential depressive-like behavior, we also did not observe a genotype effect between WT and KO male and female mice, as indicated by the comparable percent time spent in the state of immobility (Figure 2S-T). These data, together, indicate that CTRP4 is dispensable for general exploratory, as well as anxiety- and depressive-like behaviors.
FIGURE 2.
Loss of CTRP4 does not affect exploratory, anxiety or depressive-like behaviors. (A–H) Open field tests for male (A–D) and female (E–H) wild-type (WT) and Ctrp4 knockout (KO) mice. The amount of total activity spent at the center and periphery of the open field, as well as rearing activity, were not significantly different between genotypes of either sex. (I–L) Elevated plus maze tests of male (I,J) and female (K,L) mice. The percent of time spent in the open arms and the total distance travelled were not significantly different between genotypes of either sex. (M–R) Y-maze spontaneous alternation tests of male (M–O) and female (P–R) mice. The percent alternation, arm entries, and total distance travelled were not significantly different between genotypes of either sex. (S,T) Tail suspension tests for depressive-like behavior of male (U) and female (V) mice. The % of time spent in the state of immobility was not significantly different between genotypes of either sex. All data are presented as mean ± SEM Male mice (WT, n = 10; KO, n = 10); Female mice (WT, n = 12; KO, n = 13)
3.3 ∣. CTRP4-null mice have normal motor function and prepulse inhibition
Since Ctrp4 transcript is expressed in the cerebellum that regulates motor function (Figure 1B), we assessed neuromuscular function with the rotarod test. Over a 3-day test period, both WT and Ctrp4 KO mice of either sex gradually improved their ability to stay on the rotarod for longer periods of time (ie, increased latency to drop), indicating that CTRP4 is not required for motor coordination and balance (Figure 3A,B). Next, we examined whether CTRP4 deficiency results in neuropsychiatric-like behaviors. Humans with neuropsychiatric disorders (eg, schizophrenia) and rodents with schizophrenia-like phenotypes are known to have reduced prepulse inhibition of acoustic startle response.55,56 In response to different prepulse intensities, both WT and Ctrp4 KO mice of either sex showed a similar degree of inhibition to subsequent acoustic startle response (Figure 3C,D), indicating that CTRP4 is not required for sensorimotor gating.
FIGURE 3.
CTRP4 deficiency does not affect motor coordination and balance, as well as prepulse inhibition of acoustic startle response. (A,B) Rotarod tests of wild-type (WT) and Ctrp4 knockout (KO) male and female mice. Latency to drop from the rotatod over a three-day period of testing was not significantly different between genotypes of either sex. (C,D) Prepulse inhibition of acoustic startle response in male (C) and female (D) mice. Percent of inhibition in acoustic startle response following different prepulse (74–90 decibels) were not different between genotypes of either sex. All data are presented as mean ± SEM
3.4 ∣. CTRP4 deficiency enhances sensitivity to pain induced by shock
Given Ctrp4 transcript expression in the spinal cord and dorsal root ganglia (Figure 1), we examined the potential contribution of CTRP4 to somatosensory function, focusing on sensitivity to pain elicited by thermal heat or shock. In both hotplate and Hargreaves tests, the latency to lift the hind legs in response to thermal stress was not significantly different between genotypes of either sex (Figure 4A-D), indicating that CTRP4 is dispensable for pain sensation induced by heat. We next tested whether another pain modality, such as that induced by shock, is different between genotypes. We observed the low-level shock intensity needed to elicit a flinch or vocalization was not different between genotypes of either sex (Figure 4E,F,I,J), indicating that Ctrp4 KO mice have intact shock sensing mechanisms. Greater shock will elicit a jump in mice. We observed that the shock intensity needed to elicit the first hop was significantly lower in both male and female Ctrp4 KO mice relative to WT littermates (Figure 4H,L), suggesting enhanced sensitivity to pain induced by higher shock level in the CTRP4-null animals. The shock intensity needed to elicit a full jump (where all four feet left the ground), however, was also lower in Ctrp4 KO mice relative to WT littermates but fell short of being significant (Figure 4G,K).
FIGURE 4.
Loss of CTRP4 modestly enhances pain sensitivity to shock. (A,B) Hotplate tests of wild-type (WT) and Ctrp4 knockout (KO) male and female mice. Latency time to withdraw one of the hind limbs from the plate was not significantly different between genotypes of either sex. (C,D) Hargreaves tests of male (C) and female (D) mice. The latency time for a full leg raise at the site of thermal stimulus was not significantly different between genotypes of either sex. (E–L) Shock sensitivity tests of male (E–H) and female (I–L) mice. The shock intensity (mA) at which mice flinched, vocalized, jump, and hop are indicated. Relative to WT littermates, Ctrp4 KO male and female mice required a lower shock intensity to elicit the first full jump. All data are presented as mean ± SEM *p < .05. Male mice (WT, n = 8–10; KO, n = 10–12); Female mice (WT, n = 8–12; KO, n = 10–13)
3.5 ∣. Loss of CTRP4 impairs associative learning and memory in female mice
The prominent expression of Ctrp4 transcript in the cortex and hippocampus (Figure 1) led us to next ask if CTRP4 plays a role in learning and memory. We first assessed spatial memory using two different tests, the Y-maze spatial recognition test and Morris water maze. In the former, the total distance traveled and the percent of time spent in familiar and novel arms were not significantly different between genotypes of either sex (Figure 5A-D). In the novel object recognition test, a measure of short-term memory, the percent preference for the novel object was not significantly different between WT and KO mice of either sex (Figure 5E,F). And in the more complex Morris water maze, the latency time to find the hidden platform was also comparable between WT and Ctrp4 KO female mice (Figure 5G,H). Male mice, however, were not assessed. These results indicate that CTRP4 is dispensable for spatial learning and memory. We next performed the trace fear conditioning test to evaluate complex associative learning and memory in these animals. During training and the subsequent context and cue tests, male Ctrp4 KO mice performed just as well as the WT littermates (Figure 6A-C), indicating no deficit in associative learning and memory. Ctrp4 KO female mice, however, did not learn as well during the training session (Figure 6D), as indicated by reduced freezing in response to paired tone/shock. Although Ctrp4 KO female mice performed similarly well as the WT littermates during subsequent context tests (Figure 6E), they froze significantly less in response to cue tests (Figure 6F), indicating impaired associative memory.
FIGURE 5.
CTRP4 deficiency does not impair short-term memory and long-term spatial memory. (A–D) Y-maze spatial recognition tests of male (A,B) and female (C,D) mice. Percent time spent in familiar and novel arm, as well as total distance travelled, were not significantly different between genotypes of either sex. (E,F) Novel object recognition tests of male (E) and female (F) mice. The percent preference for novel object was not significantly different between genotypes of either sex. (G,H) Morris water maze tests of WT and Ctrp4 KO female mice. The latency time to discover the platform was not significantly different between genotypes
FIGURE 6.
Loss of CTRP4 impairs associative learning and memory. (A–F) Trace fear conditioning tests of male (A–C) and female (D–F) mice. Percent freezing during training, as well as context and cue tests, were not significantly different between WT and KO male mice. Ctrp4 KO female mice, however, had reduced freezing during training and in the cue tests. Arrows point to time when shock was administered. All data are presented as mean ± SEM **p < .01; ***p < .001
3.6 ∣. CTRP4 deficiency alters learning and memory gene expression in the hippocampus and cortex
Associative learning and memory involve multiple brain regions, including the hippocampus and cortex. Consequently, we performed targeted expression analyses on genes known to be important for learning and memory57 (Table S1). Because a deficit in associative learning and memory was only seen in female Ctrp4 KO mice, we therefore focused our gene expression analyses in female mice. As expected, the expression of Ctrp4 transcript in the hippocampus and cortex was completely abolished in the CTRP4-null mice (Figure 7A,D). Of the 44 selected genes analyzed, Arc (Activity regulated cytoskeleton associated protein) expression was significantly reduced in both hippocampus and cortex (Figure 7B,E). In addition, Pde4d (Phosphodiesterase 4D) expression was significantly upregulated in the hippocampus (Figure 7C), and c-fos (Fos proto-oncogene) expression was significantly downregulated in the cortex (Figure 7F), of Ctrp4 KO female mice relative to WT controls. The average expression values (log2 fold change) of all the 44 genes analyzed by real-time PCR were shown on a heatmap (Figure 7G). Although the expression of most genes fell short of being significant (Figure S1), there were many more genes whose expression tended to be altered in the hippocampus than in the cortex of Ctrp4 KO female mice relative to WT controls (Figure 7C). Some of these genes whose expression trended to be upregulated in the Ctrp4 KO hippocampus include Fkbp1a (fkbp12; +59%, p = .06), Paip2 (+57%, p = .06), and Hdac1 (+65%, p = .08). Combined, these data suggest a possible link between altered gene expression in the hippocampus and cortex and the observed deficit in associative learning and memory in Ctrp4 KO female mice.
FIGURE 7.
Altered expression of learning and memory genes in the hippocampus and cortex of WT and Ctrp4 KO female mice. (A–C) Altered expression of Ctrp4, Arc, and Pde4d in the hippocampus of wild-type (WT) and Ctrp4 knockout (KO) female mice. (D–F) Altered expression of Ctrp4, Arc, and c-fos in the cortex of WT and KO female mice. All data are presented as mean ± SEM *p < .05; ***p < .001; ****p < .0001. (G) Heatmap of average Log2(fold change) expression data in hippocampus and cortex of WT and KO female mice based on real-time qPCR analyses. WT, n = 12; KO, n = 12
4 ∣. DISCUSSION
Despite widespread expression of Ctrp4 transcript in many different neuronal cell types across distinct anatomical brain regions, a battery of behavioral tests indicated that CTRP4 is dispensable for CNS functions governing general exploratory behaviors, motor function and balance, sensorimotor gating, as well as anxiety- and depressive-like behaviors. Loss of CTRP4, however, enhances pain sensitivity elicited by shock in both male and female mice lacking CTRP4, implying altered nociceptive function; this effect is modest since CTRP4 deficiency lowers the shock intensity needed to elicit the first hop. The shock intensity needed to induce the first jump (all four legs are off the ground), however, was not significantly different between genotypes of either sex, although it trended to be lower in Ctrp4 KO female mice relative to WT controls. Given the expression of Ctrp4 transcript in dorsal root ganglia (DRG) and our behavioral data, CTRP4 likely has a role in nociception, but that additional confirmation using different pain modality paradigms (eg, acute or chronic pain stimuli induced by chemical or mechanical damage to nerve cells) are needed to fully establish if and what forms of nociception are regulated by CTRP4.
While spatial learning and memory was preserved in the Ctrp4 KO mice, associative learning and memory was impaired in female, but not male, Ctrp4 KO mice. Sexual dimorphism in learning and memory is not entirely unexpected, as this has been documented in humans and animal models.58-60 The underlying mechanisms accounting for sex differences in learning and memory are complex and not well understood, and likely involve sex hormones. Although estrogen is generally thought to account for sexual dimorphism in memory formation,61,62 there is no simple relationship between female sex hormone and memory formation.60 It should be noted that sex differences in the activation of synaptic kinases (eg, CaMKKα, CaMKKβ, ERK), transcription factors (eg, CREB, Nf-κB), and gene transcription (eg, BDNF, GAA1, SRp20) have also been documented60; it is unclear, however, whether any of these molecules are differentially altered in Ctrp4 KO female relative to male mice. At least at the transcript level, we observed no differences in the expression of Camk4, Creb, and Bdnf between WT and Ctrp4 KO female mice. Additional studies are needed to clarify the sex differences in associative learning and memory in CTRP4 deficient animals.
Associative learning and memory, as assessed by the trace fear conditioning paradigm, is dependent on multiple brain regions, including the hippocampus, amygdala, and cortex.63-66 Altered expression of genes in these brain regions can affect learning and memory. Indeed, when we examined 44 select genes known to play roles in learning and memory in mice (based on gain- or loss-of-function mouse models),57 the expression of Arc and c-fos were significantly downregulated in the hippocampus and/or cortex of Ctrp4 KO female mice relative to WT littermates. Arc/Arg3.1 and c-fos are known to play important roles in learning and memory,67,68 and mice lacking Arc or c-Fos in the CNS have a marked deficit in long-term memory.69,70 Thus, reduced expression of Arc and c-fos in the hippocampus and cortex may contribute to the deficit in associative learning and memory as seen in the Ctrp4 KO female mice. In addition to impaired associative learning and memory (as judged by trace fear conditioning paradigm), loss of Arc also impaired novel object recognition and spatial memory (as indicated by the Morris water maze test).69 In contrast, reduced expression of Arc in the hippocampus and cortex of Ctrp4 KO female mice appears to adversely affect associative learning and memory but has no impact on short-term memory for object recognition and complex spatial (Morris water maze) memory. The difference may be attributable to the complete absence of Arc in the Arc KO mice versus 30%–40% reduction in Arc expression seen in the Ctrp4 KO female mice.
Interestingly, we also observed increased expression of Pde4d in the hippocampus of the Ctrp4 KO female mice. Pde4d encodes phosphodiesterase 4D, an enzyme that hydrolyzes cAMP and plays an important role in modulating cAMP/PKA/CREB pathway in the hippocampus critical for learning and memory.71 Increased neuronal cAMP level in the hippocampus via modulation of a G-protein coupled receptor enhances hippocampal synaptic plasticity and improves short and long-term memory for contextual fear conditioning.72 Similarly, increasing cellular cAMP level by pharmacologic inhibition of Pde4d73,74 or genetic inactivation of Pde4d75 enhances memory. Thus, elevated Pde4d expression in the hippocampus of Ctrp4 KO female mice would presumably reduce hippocampal cAMP level and signaling, thereby contributing to impaired associative learning and memory seen in these animals. Complete deletion of Pde4d gene or miRNA-mediated downregulation of Pde4d have also been shown to improve short-term memory of object recognition and long-term spatial memory (Morris water maze)75; neither of these two types of memory was affected in Ctrp4 KO female mice with an elevated hippocampal expression of Pde4d. Since Pde4d is expressed in multiple brain regions (including hippocampus and cortex), the complete loss of Pde4d or its downregulation by miRNA would likely affect multiple neuronal cell types throughout the brain. Thus, in these loss-of-function mouse models, both short-term and long-term memory are improved, whereas the modest (~65%) upregulation of Pde4d seen in the hippocampus (but not cortex) of Ctrp4 KO female mice may not be sufficient to negatively impact short-term memory and spatial memory.
Our expression analyses also uncovered multiple genes that trended to be upregulated in the hippocampus of Ctrp4 KO female mice relative to WT controls; among these are Fkbp1a/fkbp12, Paip2a, and Hdac1. The potential relevance of these upregulated genes is highlighted by enhanced hippocampal-dependent memory in KO mouse models lacking FK506-binding protein 1a (Fkbp1a) or Poly(A) binding protein interacting protein 2 (Paip2a).76,77 Similarly, pharmacologic inhibition of class I histone deacetylase (Hdac1-3, −8) also improves hippocampal-dependent memory in contextual fear conditioning.78 Thus, the combined trended upregulation of Fkbp1a, Paip2a, and Hdac1 in the hippocampus of Ctrp4 KO female mice could also potentially contribute to impaired associative learning and memory seen in these animals.
Several limitations of the study are noted. Our behavioral tests were not exhaustive, even though we had assessed many of the mouse behaviors commonly used to evaluate altered brain function and memory deficit. Additional behavioral tests, in particular those related to somatosensory functions, may reveal a previously unrecognized role of CTRP4 in the central or peripheral nervous system. Although CTRP4 deficiency altered learning and memory gene expression in the hippocampus and cortex, we do not know the mechanism that links CTRP4 deficiency to impaired associative memory in a sex-dependent manner. To date, no plasma membrane receptor has been identified for CTRP4. Although nucleolin, an intracellular protein, was recently shown to be a docking protein that can bind CTRP4, such interaction has only been demonstrated in monocytes, B-cells, and dead cells.47 The relevance of this finding to CTRP4 action in the brain is presently unclear.
In summary, our behavioral and gene expression analyses have highlighted a novel and sexually dimorphic CNS role of CTRP4 in regulating hippocampal-dependent memory. Given the dearth of knowledge concerning the biological function of this highly conserved secretory protein, our study provides important and valuable information needed to further investigate its mechanism of action in the central and peripheral nervous system.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by the National Institutes of Health (DK084171 to GWW).
Funding information
HHS ∣ NIH ∣ National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), Grant/Award Number: DK084171
Abbreviations:
- Arc
activity regulated cytoskeleton associated protein
- Camk4
calcium/calmodulin dependent protein kinase IV
- cAMP
cyclic adenosine monophosphate
- c-fos
Fos proto-oncogene
- CNS
central nervous system
- CREB
cAMP response element binding protein
- CTRP
C1q/TNF-related protein
- Fkbp1a
FK506-binding protein 1a
- Hdac1
histone deacetylase 1
- KO
knockout
- Paip2
Poly(A) binding protein interacting protein 2
- PKA
protein kinase A
- WT
wildtype
Footnotes
DISCLOSURES
The authors declare no conflicts of interest.
SUPPORTING INFORMATION
Additional Supporting Information may be found online in the Supporting Information section.
REFERENCES
- 1.Kishore U, Reid KB. C1q: structure, function, and receptors. Immunopharmacology. 2000;49:159–170. [DOI] [PubMed] [Google Scholar]
- 2.Reid KB, Porter RR. Subunit composition and structure of subcomponent C1q of the first component of human complement. Biochem J. 1976;155:19–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ghai R, Waters P, Roumenina LT, et al. C1q and its growing family. Immunobiology. 2007;212:253–266. [DOI] [PubMed] [Google Scholar]
- 4.Tom Tang Y, Hu T, Arterburn M, et al. The complete complement of C1q-domain-containing proteins in Homo sapiens. Genomics. 2005;86:100–111. [DOI] [PubMed] [Google Scholar]
- 5.Mei J, Gui J. Bioinformatic identification of genes encoding C1q-domain-containing proteins in zebrafish. J Genet Genomics. 2008;35:17–24. [DOI] [PubMed] [Google Scholar]
- 6.Thielens NM, Tedesco F, Bohlson SS, Gaboriaud C, Tenner AJ. C1q: a fresh look upon an old molecule. Mol Immunol. 2017;89:73–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colombatti A, Spessotto P, Doliana R, Mongiat M, Bressan GM, Esposito G. The EMILIN/Multimerin family. Front Immunol. 2011;2:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seldin MM, Tan SY, Wong GW. Metabolic function of the CTRP family of hormones. Rev Endocr Metab Disord. 2014;15:111–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lau WB, Ohashi K, Wang Y, et al. Role of adipokines in cardiovascular disease. Circ J. 2017;81:920–928. [DOI] [PubMed] [Google Scholar]
- 10.Hamoud N, Tran V, Aimi T, et al. Spatiotemporal regulation of the GPCR activity of BAI3 by C1qL4 and Stabilin-2 controls myoblast fusion. Nat Commun. 2018;9:4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yuzaki M. The C1q complement family of synaptic organizers: not just complementary. Curr Opin Neurobiol. 2017;45:9–15. [DOI] [PubMed] [Google Scholar]
- 12.Wong GW, Wang J, Hug C, Tsao TS, Lodish HF. A family of Acrp30/adiponectin structural and functional paralogs. Proc Natl Acad Sci U S A. 2004;101:10302–10307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wei Z, Seldin MM, Natarajan N, Djemal DC, Peterson JM, Wong GW. C1q/tumor necrosis factor-related protein 11 (CTRP11), a novel adipose stroma-derived regulator of adipogenesis. J Biol Chem. 2013;288:10214–10229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wong GW, Krawczyk SA, Kitidis-Mitrokostas C, et al. Identification and characterization of CTRP9, a novel secreted glycoprotein, from adipose tissue that reduces serum glucose in mice and forms heterotrimers with adiponectin. FASEB J. 2009;23:241–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wong GW, Krawczyk SA, Kitidis-Mitrokostas C, Revett T, Gimeno R, Lodish HF. Molecular, biochemical and functional characterizations of C1q/TNF family members: adipose-tissue-selective expression patterns, regulation by PPAR-gamma agonist, cysteine-mediated oligomerizations, combinatorial associations and metabolic functions. Biochem J. 2008;416:161–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wei Z, Peterson JM, Wong GW. Metabolic regulation by C1q/TNF-related protein-13 (CTRP13): activation of AMP-activated protein kinase and suppression of fatty acid-induced JNK signaling. J Biol Chem. 2011;286:15652–15665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seldin MM, Peterson JM, Byerly MS, Wei Z, Wong GW. Myonectin (CTRP15), a novel myokine that links skeletal muscle to systemic lipid homeostasis. J Biol Chem. 2012;287:11968–11980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peterson JM, Wei Z, Wong GW. C1q/TNF-related protein-3 (CTRP3), a novel adipokine that regulates hepatic glucose output. J Biol Chem. 2010;285:39691–39701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peterson JM, Aja S, Wei Z, Wong GW. C1q/TNF-related protein-1 (CTRP1) enhances fatty acid oxidation via AMPK activation and ACC inhibition. J Biol Chem. 2012;287:1576–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lei X, Rodriguez S, Petersen PS, et al. Loss of CTRP5 improves insulin action and hepatic steatosis. Am J Physiol Endocrinol Metab. 2016;310:E1036–E1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Little HC, Rodriguez S, Lei X, et al. Myonectin deletion promotes adipose fat storage and reduces liver steatosis. FASEB J. 2019;33:8666–8687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Petersen PS, Lei X, Wolf RM, et al. CTRP7 deletion attenuates obesity-linked glucose intolerance, adipose tissue inflammation, and hepatic stress. Am J Physiol Endocrinol Metab. 2017;312:E309–E325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peterson JM, Seldin MM, Wei Z, Aja S, Wong GW. CTRP3 attenuates diet-induced hepatic steatosis by regulating triglyceride metabolism. Am J Physiol Gastrointest Liver Physiol. 2013;305:G214–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peterson JM, Wei Z, Seldin MM, Byerly MS, Aja S, Wong GW. CTRP9 transgenic mice are protected from diet-induced obesity and metabolic dysfunction. Am J Physiol Regul Integr Comp Physiol. 2013;305:R522–R533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rodriguez S, Lei X, Petersen PS, Tan SY, Little HC, Wong GW. Loss of CTRP1 disrupts glucose and lipid homeostasis. Am J Physiol Endocrinol Metab. 2016;311:E678–E697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tan SY, Lei X, Little HC, et al. CTRP12 ablation differentially affects energy expenditure, body weight, and insulin sensitivity in male and female mice. Am J Physiol Endocrinol Metab. 2020;319:E146–E162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wei Z, Lei X, Petersen PS, Aja S, Wong GW. Targeted deletion of C1q/TNF-related protein 9 increases food intake, decreases insulin sensitivity, and promotes hepatic steatosis in mice. Am J Physiol Endocrinol Metab. 2014;306:E779–E790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wolf RM, Lei X, Yang ZC, Nyandjo M, Tan SY, Wong GW. CTRP3 deficiency reduces liver size and alters IL-6 and TGFbeta levels in obese mice. Am J Physiol Endocrinol Metab. 2016;310:E332–E345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stevens B, Allen NJ, Vazquez LE, et al. The classical complement cascade mediates CNS synapse elimination. Cell. 2007;131:1164–1178. [DOI] [PubMed] [Google Scholar]
- 30.Chu Y, Jin X, Parada I, et al. Enhanced synaptic connectivity and epilepsy in C1q knockout mice. Proc Natl Acad Sci U S A. 2010;107:7975–7980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hirai H, Pang Z, Bao D, et al. Cbln1 is essential for synaptic integrity and plasticity in the cerebellum. Nat Neurosci. 2005;8:1534–1541. [DOI] [PubMed] [Google Scholar]
- 32.Uemura T, Lee SJ, Yasumura M, et al. Trans-synaptic interaction of GluRdelta2 and Neurexin through Cbln1 mediates synapse formation in the cerebellum. Cell. 2010;141:1068–1079. [DOI] [PubMed] [Google Scholar]
- 33.Seigneur E, Südhof TC. Genetic ablation of all cerebellins reveals synapse organizer functions in multiple regions throughout the brain. J Neurosci. 2018;38:4774–4790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krishnan V, Stoppel DC, Nong Y, et al. Autism gene Ube3a and seizures impair sociability by repressing VTA Cbln1. Nature. 2017;543:507–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matsuda K, Miura E, Miyazaki T, et al. Cbln1 is a ligand for an orphan glutamate receptor delta2, a bidirectional synapse organizer. Science. 2010;328:363–368. [DOI] [PubMed] [Google Scholar]
- 36.Martinelli DC, Chew KS, Rohlmann A, et al. Expression of C1ql3 in discrete neuronal populations controls efferent synapse numbers and diverse behaviors. Neuron. 2016;91:1034–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kakegawa W, Mitakidis N, Miura E, et al. Anterograde C1ql1 signaling is required in order to determine and maintain a single-winner climbing fiber in the mouse cerebellum. Neuron. 2015;85:316–329. [DOI] [PubMed] [Google Scholar]
- 38.Sigoillot SM, Iyer K, Binda F, et al. The secreted protein C1QL1 and Its receptor BAI3 control the synaptic connectivity of excitatory inputs converging on cerebellar purkinje cells. Cell Rep. 2015;10:820–832. [DOI] [PubMed] [Google Scholar]
- 39.Byerly MS, Petersen PS, Ramamurthy S, et al. C1q/TNF-related protein 4 (CTRP4) is a unique secreted protein with two tandem C1q domains that functions in the hypothalamus to modulate food intake and body weight. J Biol Chem. 2014;289:4055–4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li Y, Ye L, Jia G, Chen H, Yu L, Wu D. C1q/TNF-related protein 4 induces signal transducer and activator of transcription 3 pathway and modulates food intake. Neuroscience. 2020;429:1–9. [DOI] [PubMed] [Google Scholar]
- 41.Sarver DC, Stewart AN, Rodriguez S, Little HC, Aja S, Wong GW. Loss of CTRP4 alters adiposity and food intake behaviors in obese mice. Am J Physiol Endocrinol Metab. 2020;319:E1084–E1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luo Y, Wu X, Ma Z, et al. Expression of the novel adipokine C1qTNF-related protein 4 (CTRP4) suppresses colitis and colitis-associated colorectal cancer in mice. Cell Mol Immunol. 2016;13:688–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cao L, Tan W, Chen W, et al. CTRP4 acts as an anti-inflammatory factor in macrophages and protects against endotoxic shock. Eur J Immunol. 2021;51:380–392. [DOI] [PubMed] [Google Scholar]
- 44.Pullabhatla V, Roberts AL, Lewis MJ, et al. De novo mutations implicate novel genes in systemic lupus erythematosus. Hum Mol Genet. 2018;27:421–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pakzad B, Shirpour R, Mousavi M, et al. C1QTNF4 gene p.His198Gln mutation is correlated with early-onset systemic lupus erythematosus in Iranian patients. Int J Rheum Dis. 2020;23:1594–1598. [DOI] [PubMed] [Google Scholar]
- 46.Xu W, Zhou H, Li X, et al. C1Q/TNF-related protein 4 expression correlates with herpes simplex encephalitis progression. Ann Transl Med. 2019;7:235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vester SK, Beavil RL, Lynham S, et al. Nucleolin acts as the receptor for C1QTNF4 and supports C1QTNF4-mediated innate immunity modulation. J Biol Chem. 2021;296:100513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zeisel A, Hochgerner H, Lonnerberg P, et al. Molecular architecture of the mouse nervous system. Cell. 2018;174:999–1014.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. [DOI] [PubMed] [Google Scholar]
- 50.Ballinger MD, Saito A, Abazyan B, et al. Adolescent cannabis exposure interacts with mutant DISC1 to produce impaired adult emotional memory. Neurobiol Dis. 2015;82:176–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pletnikov MV, Ayhan Y, Nikolskaia O, et al. Inducible expression of mutant human DISC1 in mice is associated with brain and behavioral abnormalities reminiscent of schizophrenia. Mol Psychiatry. 2008;13:173–186, 115. [DOI] [PubMed] [Google Scholar]
- 52.Terrillion CE, Abazyan B, Yang Z, et al. DISC1 in astrocytes influences adult neurogenesis and hippocampus-dependent behaviors in mice. Neuropsychopharmacology. 2017;42:2242–2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kovacsics CE, Gould TD. Shock-induced aggression in mice is modified by lithium. Pharmacol Biochem Behav. 2010;94:380–386. [DOI] [PubMed] [Google Scholar]
- 54.Bilkei-Gorzo A, Racz I, Michel K, Zimmer A. Diminished anxiety- and depression-related behaviors in mice with selective deletion of the Tac1 gene. J Neurosci. 2002;22:10046–10052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Braff DL, Geyer MA. Sensorimotor gating and schizophrenia. Human and animal model studies. Arch Gen Psychiatry. 1990;47:181–188. [DOI] [PubMed] [Google Scholar]
- 56.Powell SB, Zhou X, Geyer MA. Prepulse inhibition and genetic mouse models of schizophrenia. Behav Brain Res. 2009;204:282–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee YS. Genes and signaling pathways involved in memory enhancement in mutant mice. Mol Brain. 2014;7:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Andreano JM, Cahill L. Sex influences on the neurobiology of learning and memory. Learn Mem. 2009;16:248–266. [DOI] [PubMed] [Google Scholar]
- 59.Wang W, Le AA, Hou B, et al. Memory-related synaptic plasticity is sexually dimorphic in rodent hippocampus. J Neurosci. 2018;38:7935–7951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mizuno K, Giese KP. Towards a molecular understanding of sex differences in memory formation. Trends Neurosci. 2010;33:285–291. [DOI] [PubMed] [Google Scholar]
- 61.Romeo RD, Waters EM, McEwen BS. Steroid-induced hippocampal synaptic plasticity: sex differences and similarities. Neuron Glia Biol. 2004;1:219–229. [DOI] [PubMed] [Google Scholar]
- 62.McEwen B. Estrogen actions throughout the brain. Recent Prog Horm Res. 2002;57:357–384. [DOI] [PubMed] [Google Scholar]
- 63.Kochli DE, Thompson EC, Fricke EA, Postle AF, Quinn JJ. The amygdala is critical for trace, delay, and contextual fear conditioning. Learn Mem. 2015;22:92–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gilmartin MR, Helmstetter FJ. Trace and contextual fear conditioning require neural activity and NMDA receptor-dependent transmission in the medial prefrontal cortex. Learn Mem. 2010;17:289–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Beeman CL, Bauer PS, Pierson JL, Quinn JJ. Hippocampus and medial prefrontal cortex contributions to trace and contextual fear memory expression over time. Learn Mem. 2013;20:336–343. [DOI] [PubMed] [Google Scholar]
- 66.Esclassan F, Coutureau E, Di Scala G, Marchand AR. A cholinergic-dependent role for the entorhinal cortex in trace fear conditioning. J Neurosci. 2009;29:8087–8093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gallo FT, Katche C, Morici JF, Medina JH, Weisstaub NV. Immediate early genes, memory and psychiatric disorders: focus on c-Fos, Egr1 and Arc. Front Behav Neurosci. 2018;12:79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bramham CR, Worley PF, Moore MJ, Guzowski JF. The immediate early gene arc/arg3.1: regulation, mechanisms, and function. J Neurosci. 2008;28:11760–11767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Plath N, Ohana O, Dammermann B, et al. Arc/Arg3.1 is essential for the consolidation of synaptic plasticity and memories. Neuron. 2006;52:437–444. [DOI] [PubMed] [Google Scholar]
- 70.Fleischmann A, Hvalby O, Jensen V, et al. Impaired long-term memory and NR2A-type NMDA receptor-dependent synaptic plasticity in mice lacking c-Fos in the CNS. J Neurosci. 2003;23:9116–9122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ricciarelli R, Fedele E. Phosphodiesterase 4D: an enzyme to remember. Br J Pharmacol. 2015;172:4785–4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Isiegas C, McDonough C, Huang T, et al. A novel conditional genetic system reveals that increasing neuronal cAMP enhances memory and retrieval. J Neurosci. 2008;28:6220–6230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Burgin AB, Magnusson OT, Singh J, et al. Design of phosphodiesterase 4D (PDE4D) allosteric modulators for enhancing cognition with improved safety. Nat Biotechnol. 2010;28:63–70. [DOI] [PubMed] [Google Scholar]
- 74.Barad M, Bourtchouladze R, Winder DG, Golan H, Kandel E. Rolipram, a type IV-specific phosphodiesterase inhibitor, facilitates the establishment of long-lasting long-term potentiation and improves memory. Proc Natl Acad Sci U S A. 1998;95:15020–15025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li YF, Cheng YF, Huang Y, et al. Phosphodiesterase-4D knockout and RNA interference-mediated knock-down enhance memory and increase hippocampal neurogenesis via increased cAMP signaling. J Neurosci. 2011;31:172–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hoeffer CA, Tang W, Wong H, et al. Removal of FKBP12 enhances mTOR-Raptor interactions, LTP, memory, and perseverative/repetitive behavior. Neuron. 2008;60:832–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Khoutorsky A, Yanagiya A, Gkogkas CG, et al. Control of synaptic plasticity and memory via suppression of poly(A)-binding protein. Neuron. 2013;78:298–311. [DOI] [PubMed] [Google Scholar]
- 78.Zhao WN, Ghosh B, Tyler M, et al. Class I histone deacetylase inhibition by tianeptinaline modulates neuroplasticity and enhances memory. ACS Chem Neurosci. 2018;9:2262–2273. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.