Abstract
INTRODUCTION:
Before the 1800s, dogs were probably primarily selected for functional roles such as hunting, guarding, and herding. Modern dog breeds are a recent invention defined by conformation to a physical ideal and purity of lineage. Breeds are commonly ascribed temperaments and behavioral proclivities based on the purported function of the ancestral source population. By extension, the breed ancestry of individual dogs is assumed to be predictive of temperament and behavior. Through our community science project Darwin’s Ark (darwinsark.org), we enrolled a diverse cohort of pet dogs to explore how genetics shapes complex behavioral traits in this exceptional natural model.
RATIONALE:
Dogs are a natural system for investigating the genetics of complex traits. Millions of pet dogs live in human homes, sharing our environment, and receive sophisticated medical care. Behavioral disorders are treated with human psychiatric drugs, achieving similar response rates, and genetic studies suggest shared etiology with some human psychiatric conditions.
We developed Darwin’s Ark as an open data resource for collecting owner-reported phenotypes and genetic data and invited any dog owner to enroll their dog. We paired this with low-pass sequencing to capture nearly all common variation in this outbred population. Our inclusive approach achieved the large samples needed to investigate complex traits.
RESULTS:
We surveyed owners of 18,385 dogs (49% purebred) and sequenced the DNA of 2155 dogs. Most behavioral traits are heritable [heritability (h2) > 25%], but behavior only subtly differentiates breeds. Breed offers little predictive value for individuals, explaining just 9% of variation in behavior. For more heritable, more breed-differentiated traits, like biddability (responsiveness to direction and commands), knowing breed ancestry can make behavioral predictions somewhat more accurate (see the figure). For less heritable, less breed-differentiated traits, like agonistic threshold (how easily a dog is provoked by frightening or uncomfortable stimuli), breed is almost uninformative.
We used dogs of mixed breed ancestry to test the genetic effect of breed ancestry on behavior and compared that to survey responses from purebred dog owners. For some traits, like biddability and border collie ancestry, we confirm a genetic effect of breed that aligns with survey responses. For others, like human sociability and Labrador retriever ancestry, we found no significant effect.
Through genome-wide association, we found 11 regions that are significantly associated with behavior, including howling frequency and human sociability, and 136 suggestive regions. Regions associated with aesthetic traits are unusually differentiated in breeds, consistent with a history of selection, but those associated with behavior are not.
CONCLUSION:
In our ancestrally diverse cohort, we show that behavioral characteristics ascribed to modern breeds are polygenic, environmentally influenced, and found, at varying prevalence, in all breeds. We propose that behaviors perceived as characteristic of modern breeds derive from thousands of years of polygenic adaptation that predates breed formation, with modern breeds distinguished primarily by aesthetic traits. By embracing the full diversity of dogs—including purebred dogs, mixed-breed dogs, purpose-bred working dogs, and village dogs—we can fully realize dogs’ long-recognized potential as a natural model for genetic discovery.
Graphical Abstract
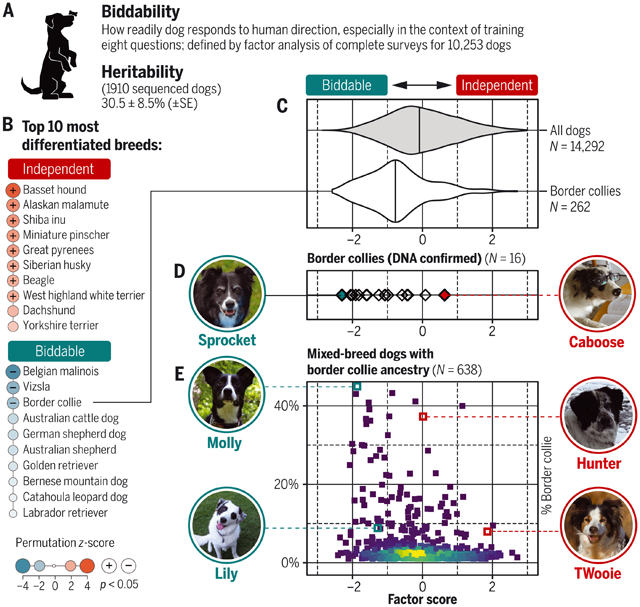
Effect of breed on behavior. (A) Biddability is among eight behavioral factors defined from surveys. SE, standard error. (B) Dogs in some breeds tend to score unusually high or low for this factor compared with dogs overall. (C and D) Border collies score lower on average for biddability (vertical line at median) but vary widely (C), including genetically confirmed border collies (D). (E) In mixed-breed dogs, border collie ancestry has a small genetic effect on biddability. [Photo credits: K. Wirka (Sprocket); M. Wisniewski (Caboose); B. Hoadley (Molly); M. Logsdon (Hunter); A. Macias (Lily); S. Staples (TWooie)]
Behavioral genetics in dogs has focused on modern breeds, which are isolated subgroups with distinctive physical and, purportedly, behavioral characteristics. We interrogated breed stereotypes by surveying owners of 18,385 purebred and mixed-breed dogs and genotyping 2155 dogs. Most behavioral traits are heritable [heritability (h2) > 25%], and admixture patterns in mixed-breed dogs reveal breed propensities. Breed explains just 9% of behavioral variation in individuals. Genome-wide association analyses identify 11 loci that are significantly associated with behavior, and characteristic breed behaviors exhibit genetic complexity. Behavioral loci are not unusually differentiated in breeds, but breed propensities align, albeit weakly, with ancestral function. We propose that behaviors perceived as characteristic of modern breeds derive from thousands of years of polygenic adaptation that predates breed formation, with modern breeds distinguished primarily by aesthetic traits.
What is your dog like?
Modern domestic dog breeds are only ~160 years old and are the result of selection for specific cosmetic traits. To investigate how genetics aligns with breed characteristics, Morrill et al. sequenced the DNA of more than 2000 purebred and mixed-breed dogs. These data, coupled with owner surveys, were used to map genes associated with behavioral and physical traits. Although many physical traits were associated with breeds, behavior was much more variable among individual dogs. In general, physical trait heritability was a greater predictor of breed but was not necessarily a predictor of breed ancestry in mutts. Among behavioral traits, biddability—how well dogs respond to human direction—was the most heritable by breed but varied significantly among individual dogs. Thus, dog breed is generally a poor predictor of individual behavior and should not be used to inform decisions relating to selection of a pet dog. —LMZ
Modern dog breeds are less than 160 years old (~50 to 80 generations), a blink in evolutionary history compared with the origin of dogs more than 10,000 years ago (1, 2) (Fig. 1A). Prehistoric wolves likely adapted to use human refuse through changes in morphology, behavior, metabolism, and reproduction (3–7). Early humans may have given favored dogs increased access to limited resources, but there is little evidence of humans intentionally breeding dogs until 2000 years ago (8, 9). By contrast, the modern dog breed, emphasizing conformation to a physical ideal and purity of lineage, is a Victorian invention (10). Before the 1800s, dogs were primarily selected for functional roles such as hunting, guarding, and herding (11)—heritable behaviors derived from the wolf predatory sequence (12). Modern breeds retain these component motor patterns, but their contexts, sequences, and thresholds vary (12, 13). The extent to which ancient behavioral propensities persist in modern breeds, defined primarily by aesthetics and often disconnected from functional behavioral selection, is unclear.
Fig. 1. The ancestry-inclusive Darwin’s Ark project collected surveys and genetic data from a diverse cohort of dogs.
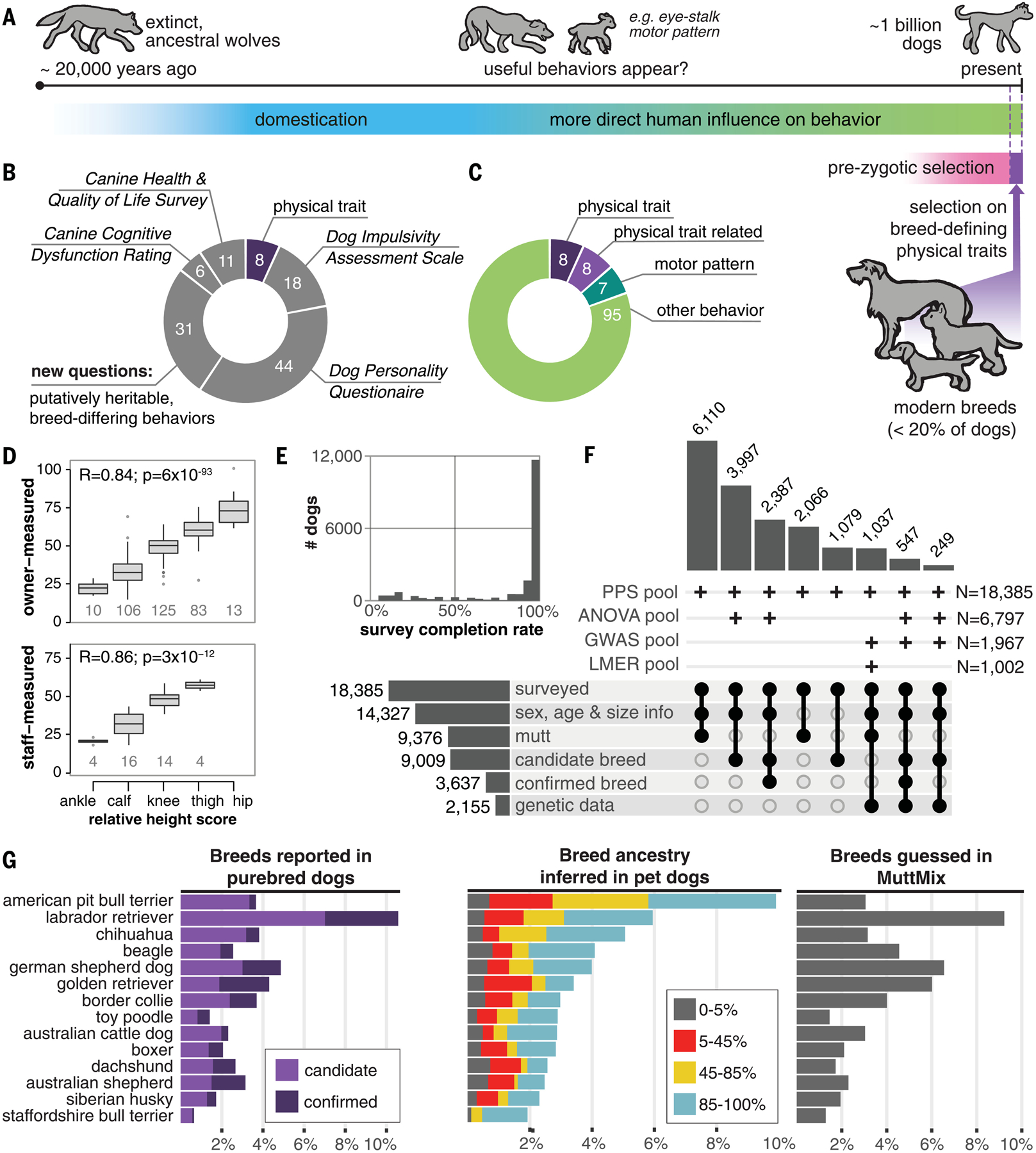
(A) Selection on behavior in dogs predates modern breeds, which have existed for ~1% of dog history (10). (B and C) Surveys include (B) 79 published behavior questions (34–37) and 39 new questions about heritable, easy-to-identify traits that fall into (C) four categories that potentially vary in heritability, including physical traits (fig. S3). (D) Owner responses to the size question (fig. S2) were highly correlated with measured size in 375 dogs (Pearson correlation). Boxes and whiskers represent 25% quartile, 75% quartile, minimum, and maximum, with horizontal line at median. (E) Owners of most dogs answered >95% of questions. (F) Upset plot visualizing the dataset. Six horizontal bars show the number of dogs subsetted by information type or breed category (“subsets”). Each column is a possible intersection of subsets, with black circles indicating the inclusion of a subset and vertical bars showing the number of dogs intersecting. Four rows (“pools”) represent the dogs used in four major analyses. Plus signs denote the inclusion of an intersection, with total number of dogs (N) on the right. (G) The frequency of breeds among purebred dogs in Darwin’s Ark (left), compared with the 14 most common breed ancestries we detected in all dogs through global ancestry inference (middle). More popular breeds tended to be guessed more frequently by MuttMix participants (right).
Dogs with ancestry from a single modern breed (purebred dogs) predominate in genetic studies, which capitalize on their unusual population history and limited genetic diversity (14–16), but are a minority of all dogs (3, 17). More than 80% of the nearly 1 billion dogs on Earth are free-living, free-breeding, and not under human control (e.g., village dogs) (3). Even in countries with large purebred populations, dogs with ancestry from more than one breed are common [~50% in the United States (18)]. Herein, we use the word “mutt” to describe a dog with ancestry from more than one breed, and potentially from non-breed populations.
Modern breeds are commonly ascribed characteristic temperaments (e.g., bold, affectionate, or trainable), and behavioral proclivities on the basis of their purported ancestral function (e.g., herding or hunting) (19, 20). By extension, the breed ancestry of an individual dog is assumed to be predictive of temperament and behavior (21), with dog DNA tests marketed as tools for learning about a dog’s personality and training needs (22). Studies, however, found that within-breed behavioral variation approaches levels similar to the variation between breeds (23, 24), suggesting that such predictions are error prone even in purebred dogs.
Behavioral traits in dogs are also a potentially powerful natural model for human neuropsychiatric disease. Pet dogs are regularly treated with human psychiatric drugs, including selective serotonin reuptake inhibitors, and have similar response rates, and genetic studies suggest shared etiology (25–29). Dog behavioral traits are polygenic, driven by many small effect loci and the environment (30, 31). Given this genetic complexity, the success of survey-based phenotypes for mapping complex human diseases (32, 33), and the availability of well-validated dog-owner surveys (34–37), a large-scale, survey-based study design is ideal for investigating the genetics of canine behavior.
Through our community science project Darwin’s Ark (darwinsark.org), we enrolled a diverse cohort of pet dogs to explore the complicated, and sometimes unexpectedly weak, relationship between breed and behavior. We show that using ancestrally diverse dog cohorts enables more powerful studies of behavioral genetics in this notable natural model.
Results
Survey data
We developed Darwin’s Ark as an open data resource for collecting owner-reported phenotypes and genetic data in dogs. Dog owners were asked to complete 12 short surveys (117 questions) on behavioral and physical traits (Fig. 1, B and C; figs. S1, S2A, and S3; and table S1). Darwin’s Ark surveys, each with no more than 10 questions, are designed to be easy to complete. Owner survey responses are susceptible to rater bias, including the influence of breed stereotypes.
To ascertain size, we asked whether the dog is ankle-high, calf-high, knee-high, thigh-high, or hip-high rather than requiring owners to measure their dog (fig. S2A). This simple question proved effective when validated in three ways: (i) We measured dogs [N = 38 dogs; Pearson correlation coefficient (Rpearson) = 0.86; p = 3 × 10−12]; (ii) owners measured their dogs (N = 337 dogs; Rpearson = 0.84; p = 6 × 10−93); and (iii) we tested correlation with breed average height (38) in the subset of dogs that were purebred (N = 2025; Rpearson = 0.85; p < 2.2 × 10−16) (Fig. 1D, fig. S2, and data S1).
Owners answered, on average, 100 ± 34 (±SD) questions per dog; 70% of dogs have answers for more than 95% of questions (22) (Fig. 1, E and F). For the 48 questions drawn from the Dog Personality Questionnaire, between-question correlations matched published results (22, 37) [Mantel’s correlation coefficient (r) = 0.95, p = 1 × 10−7; fig. S1].
Overall, the Darwin’s Ark cohort (N = 18,385; 85% from the United States) is broadly similar to the US dog population. Half (49.2%) are reported as purebred (18), and breed frequencies (Fig. 1G) correlate with US breed popularity (39) (Rpearson = 0.88; p = 1.48 × 10−32) (table S2).
To reduce the dimensionality of the survey data, we performed exploratory factor analysis (22), defined eight factors that explain a cumulative 24.3% of variance in behavior (fig. S4 and tables S3 and S4), and scored 16,522 dogs. We named each factor for the facet of behavior captured and established standard terms for describing low- and high-scoring dogs (Fig. 2A). In subsequent analyses, we examined the influence of breed and genetics on each behavioral factor (Fig. 2B).
Fig. 2. Behavioral traits do not define breeds the way aesthetic traits like size do.
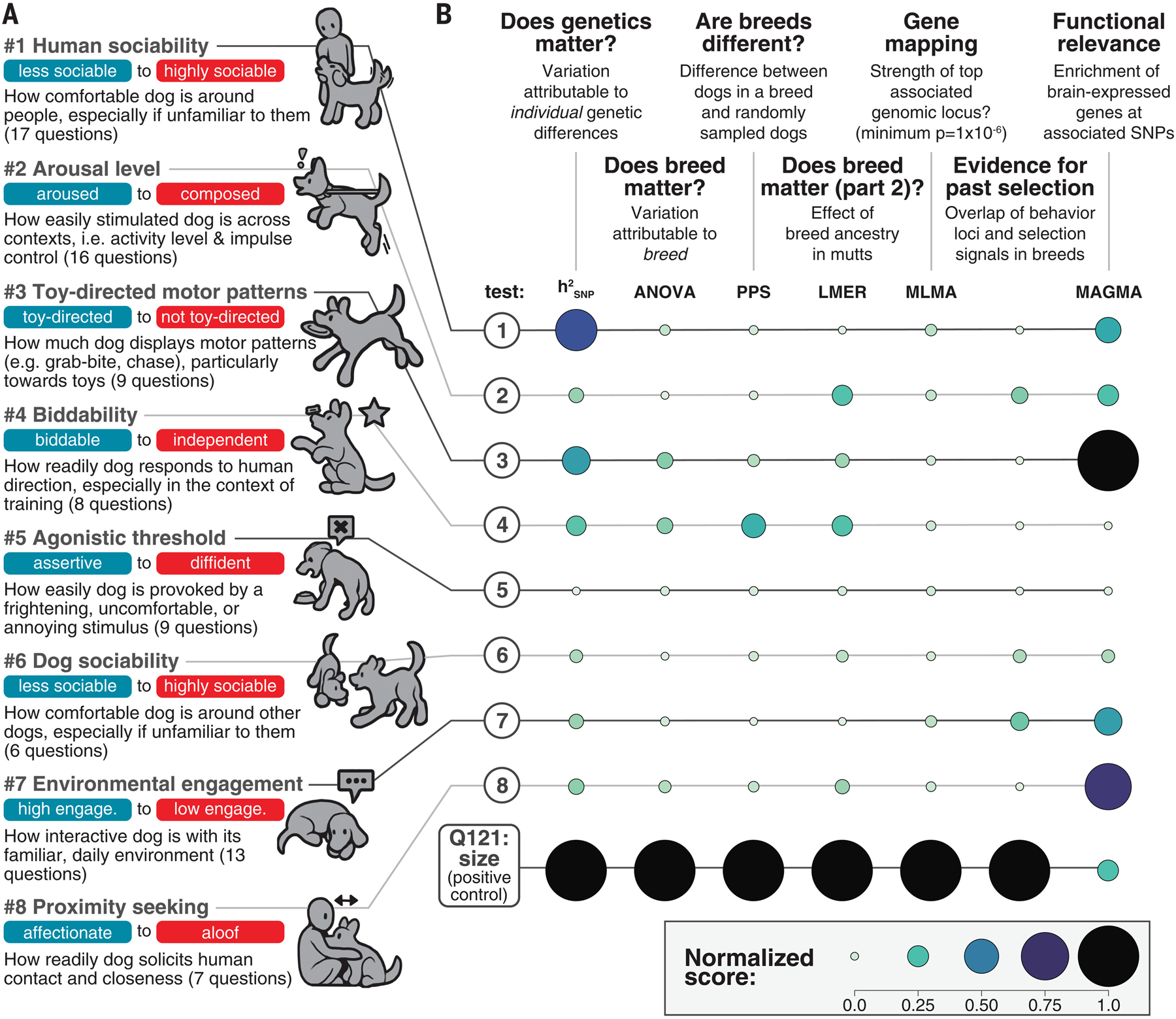
(A) Exploratory factor analysis bins behavioral questions into eight inferred factors, which correspond to latent behavioral propensities (blue, negative score; red, positive score). (B) In a series of seven analyses, we explored how behavior relates to breed in the context of size, which is a strongly breed-differentiated trait. For each column, circle size is proportional to the minimum-maximum normalized values of (i) LD-corrected h2SNP, (ii) effect size of breed in ANOVA (confirmed breed), (iii) standard deviation of PPS (candidate breeds), (iv) standard deviation of LMER t scores, (v) −log10 (minimum p) for MLMA, (vi) fraction of breeds with significant overlap (pFDR < 0.05) between PBS and GWAS, and (vii) maximum MAGMA log10(p) for 13 brain regions in GTEx (85).
Genetic data
Nearly half of the Darwin’s Ark cohort are mutts, an understudied population with un-characterized genomic diversity. We whole-genome sequenced (mean coverage = 46×; range 30× to 67×) 27 pet dogs of unknown breed ancestry (the “Mendel’s Mutts” cohort), including 26 from the United States (one originally from Mauritius and two from St. Kitts) and one from Ireland (data S2). We compared jointly called variant records in this cohort with published whole genomes for 530 purebred dogs from 128 breeds (22, 40).
Sequencing mutts efficiently captures common variants in the dog population, including variation not detected by sequencing large numbers of purebred dogs. Sequencing an additional mutt yields nearly as many new variants as sequencing a purebred dog, even when each new purebred dog is from a different breed (Fig. 3A). In the 27 mutts, we discovered 11,974,853 biallelic single-nucleotide polymorphisms (SNPs) total, including 375,474 variants not found in the 128 breeds (530 dogs).
Fig. 3. Mutts have complex ancestry from many breeds.
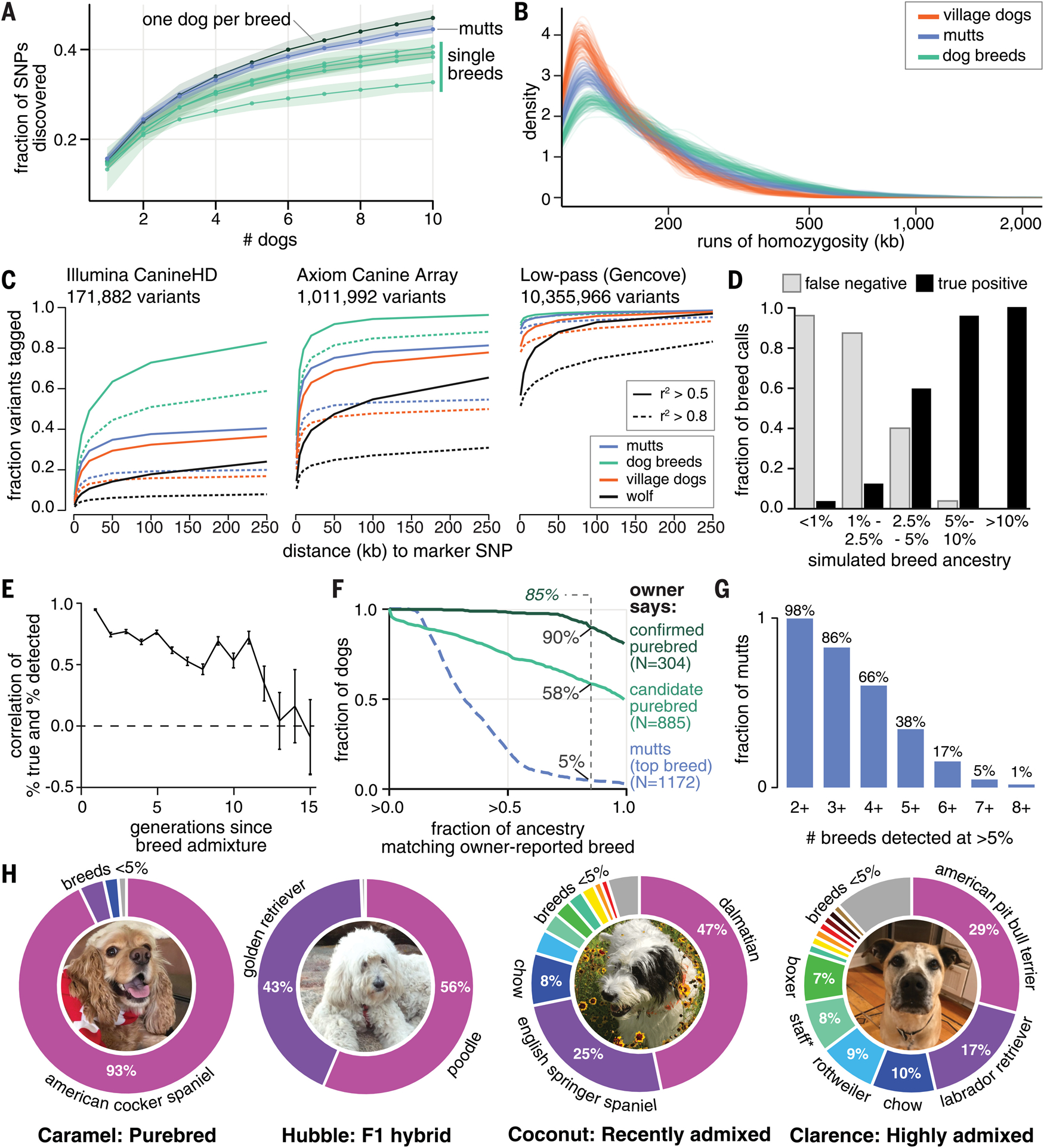
(A) Sequencing mutts discovers new variants at nearly the same rate as sequencing a series of purebreds of different breeds and faster than sequencing multiple individuals within any one breed (green; from top down: golden retriever, Labrador retriever, Yorkshire terrier, and Leonberger). Shaded regions indicate 95% confidence intervals from random reordering within each cohort. (B) Runs of homozygosity are shorter in mutts than in purebred dogs but are slightly longer than in outbred village dogs. (C) Fraction of variants tagged (out of a random sample of 20,000 autosomal SNPs) by a marker SNP is lower for the Illumina and Axiom genotyping arrays compared with low-pass sequencing with imputation, particularly in outbred populations like mutts. (D and E) Global breed ancestry inference pipeline (assessed using simulated breed admixture) (D) calls breeds comprising >5% ancestry accurately but misses lower-frequency breeds and (E) can discern admixture occurring up to ~12 generations ago (~24 to 36 years). Error bars in (E) represent standard deviation across 10 simulations of 100 admixed genomes. (F) For dogs categorized as confirmed purebred based on owner reports, breed calling assigns >85% ancestry (vertical dashed line) to the owner-reported breed in 90% of dogs. For candidate purebred dogs, 58% meet this criterion (4.4% had no detectable ancestry from the owner-reported breed). Just 5% of dogs categorized as mutts have >85% ancestry from their most common breed (blue dashed line). (G) Most mutts have ancestry detected (>5%) from more than three breeds (1205 dogs total). (H) Examples of breed calling in four dogs with different ancestry types: Caramel, a purebred dog, who has 93% of her ancestry assigned to her owner-reported breed; Hubble, an F1 goldendoodle; Coconut, who has apparent mutt ancestry mixed with dalmatian; and Clarence, a mutt with <45% ancestry from any one breed (*Staffordshire bull terrier). [Photo credits: M. Movassagh (Caramel); J. Luban (Hubble); A. Pensarosa (Coconut); R. Skloot (Clarence)]
We also confirm that genetic variation private to a single breed is rare [38,097 ± 13,206 (±SD) SNPs per breed after excluding Tibetan mastiffs, a more distantly related lineage (41) with 651,551 private SNPs]. Breeds are not distinguished by a small number of “breed-defining” variants (table S5). After analyzing all 13 breeds with more than 10 sequenced representatives, we found just 332 SNPs (298 autosomal) exclusive to, and fixed in, a single breed (data S3) out of 16,702,091 SNPs total (0.002%). Tibetan mastiffs account for 142 SNPs (121 autosomal), with just 16 ± 31 (±SD) SNPs (15 ± 29 autosomal) in each of the nine other breeds. These variants are unlikely to affect phenotype. Annotation with SNPEff classifies 98.2% (326) as occurring at loci without obvious function.
Mendel’s Mutts have shorter runs of homozygosity than purebred dogs (Fig. 3B) and linkage disequilibrium (LD) that decays more rapidly (fig. S5). Thus, genotyping arrays designed with sufficient marker density for purebred dog studies miss much of the genetic variation in mutts. Average squared correlation coefficient (r2) between SNPs drops below 0.2 at 9.8 kb, which is slightly longer than in village dogs (6.2 kb) but 5- to 10-fold shorter than in breeds (fig. S5). Because of the short LD, the markers included on the Illumina Canine HD Beadchip (N = 171,882) and the Axiom Canine Genotyping Array Sets A and B (N = 1,011,992) tag only 19 and 53% of genomic variation in mutts, respectively, compared with 51 and 85% in breeds (Fig. 3C).
We adopted a low-pass sequencing and imputation approach (42–46), using a reference panel of 435 deeply sequenced dogs and other canids (data S4). We validated by resequencing 11 mutts with high-coverage whole-genome sequencing (WGS) at low coverage [1.0× ± 0.6× (±SD)]. We imputed, on average, 32,438,672 SNPs and 13,910,371 insertions and/or deletions (indels) per dog, or 19.8 ± 6.9 (±SD) variants per kilobase (~40× denser than the Axiom array), which was sufficient to tag nearly all the common variants (94% tagged by a marker within 100 kb and 87% within 1 kb) (Fig. 3C). Concordance between low-pass and 30× sequencing was 98.3 ± 0.7% (±SD) (N = 11 dogs; ~7.7 million common SNPs), which was slightly lower than that between the Axiom array and 30× (99.3 ± 0.1%; N = 10 dogs; 0.83 million SNPs) but better than that between imputed array calls and 30× (97.3 ± 0.3%; 7.6 million SNPs), including higher concordance for heterozygous genotypes (98.9 versus 98.3%) (data S5).
Our final genetic dataset comprises 1715 Darwin’s Ark dogs sequenced at 0.6× ± 0.3× (±SD) coverage, each genotyped for 32,213,747 ± 141,060 (±SD) SNPs, and 440 dogs genotyped on the Axiom array and imputed using the same pipeline for 32,006,290 ± 157,307 (±SD) SNP genotypes, for a total of 2155 dogs (data S6). We selected dogs for sequencing on the basis of the enrollment date and survey completion rate (7.4% of dogs had sequencing funded by the owner’s donation).
Breed ancestry assignment
In our genetic data, owner-reported breed is a reasonable proxy for predominant genetic ancestry. We developed a breed-calling pipeline using the software ADMIXTURE (22, 47) to infer ancestry using a supervised analysis and a reference panel of 101 breeds (12 dogs per breed; 688,060 SNPs) collated from public and Darwin’s Ark data (Fig. 3, D and E; figs. S6 and S7A; and table S2). Genetically inferred breed ancestry across dogs correlated well with the proportions of dogs registered to breeds in the American Kennel Club (93 breeds, Rpearson = 0.74; p = 2.9 × 10−17) (Fig. 1G).
The top breed that was called matched the owner-reported breed in 98.7% of dogs described as registered purebred (N = 304) and in 85.8% of all dogs for which owners report just a single breed (N = 885) (table S6). Dogs described as registered purebreds vary somewhat in the percent ancestry assigned to the owner-reported breed (potentially because the reference data are not representative of all diversity in the breed or because of shared ancestry between breeds). We empirically set the threshold for defining a dog as genetically “purebred” as when 85% of ancestry is inferred to come from a single breed because 90% of dogs described as registered purebred by owners fell above this threshold (Fig. 3F).
We designated three classifications of breed ancestry: (i) “confirmed purebred dogs” were either described as registered purebred by the owner or confirmed by sequencing (3637 dogs), (ii) “candidate purebred dogs” included all confirmed purebred dogs and dogs with owner-reported ancestry from one breed (9009 dogs), and (iii) “mutts” were all other dogs (9376 dogs) (Fig. 1F). Genetically inferred ancestry superseded owner-reported breed when both were available, although discrepancies were rare (15 out of 556 candidate purebreds, 3 out of 323 confirmed purebred) and were primarily nominal variations on the same breed (e.g., Landseer versus Newfoundland). Extrapolating from the subset of dogs with genetic data, 89.7% of registered purebred and 58.2% of dogs with owner-reported ancestry from one breed would, if sequenced, have >85% ancestry called from their owner-reported breed (Fig. 3F). Because confirmed purebred dogs have a substantially higher percentage of their ancestry assigned to their owner-reported breed, in subsequent analyses, we prioritized the confirmed set or, when the larger and more diverse candidate set was useful, validated findings in the confirmed set.
Mutts are rarely (17%) mixes of just two breeds. Most (66%) carry >5% ancestry from four or more breeds (Fig. 3G). We find that 1071 dogs (70%) are highly admixed, carrying under 45% ancestry from any one breed (Fig. 3H). The most common breed ancestry (data S7) is American pit bull terrier (9.9%) followed by Labrador retriever (6.0%), Chihuahua (5.1%), beagle (4.1%), and German shepherd dog (4.0%) (Fig. 1G and fig. S7C), varying by geographic region (fig. S7D). Purebred dogs had higher coefficients of inbreeding, as estimated from the proportion of the genome in runs of homozygosity [FROH = 0.06 ± 0.04 (±SD); N = 633] than mutts (FROH = 0.02 ± 0.02; N = 1221) [for Student’s t test p value (pt-test) = 1.7 × 10−122; t = 28.4, degrees of freedom (df) = 776.8] (fig. S7B).
Heritability of surveyed traits
Combining genetic and survey data for 1967 dogs, we found that genetic variation explains more than 25% of the variation in factor scores for human sociability, toy-directed motor patterns, and biddability (responsiveness to commands), as well as in responses to 38 of 83 (46%) behavioral questions and eight physical traits. We estimated SNP-based heritability (h2SNP) with standard errors using restricted maximum likelihood (48) and LD score correction (excluding 27 questions for which more than half of LD-stratified variance components were constrained) on genetic relationship matrices calculated for dogs with genetic data [8,518,951 autosomal SNPs with minor allele frequency (MAF) >2%] (data S8). Physical traits are exceptionally heritable, with five out of eight exceeding 85% heritability. Retrieving is the most heritable behavioral trait [52.5 ± 9.2% (±SE)], and human sociability is the most heritable factor (factor 1, 67.3 ± 13.0%). Behaviors related to intrinsic motor patterns and physical traits are more heritable than other behaviors (Fig. 4A). To assess whether these heritabilities are overestimated because of correlation between traits and breed ancestry, we recalculated them by incorporating the top 10 principal component eigenvectors of genetic variance (49, 50). For the most part, we saw little change [median fold change of +0.02, 25% quartile = −0.049 and 75% quartile = +0.140] between estimates (Rpearson = 0.97, p = 1.1 × 10−58). Heritability decreases the most for biddability (factor 4, drops from 30.5 ± 8.5% to 20.0 ± 8.8%) and “circles before pooping” [question 64 (Q64), drops from 25.1 ± 8.1% to 8.0 ± 7.6%] (fig. S8).
Fig. 4. Breed and age (and not size) have subtle effects on the behavior traits surveyed.
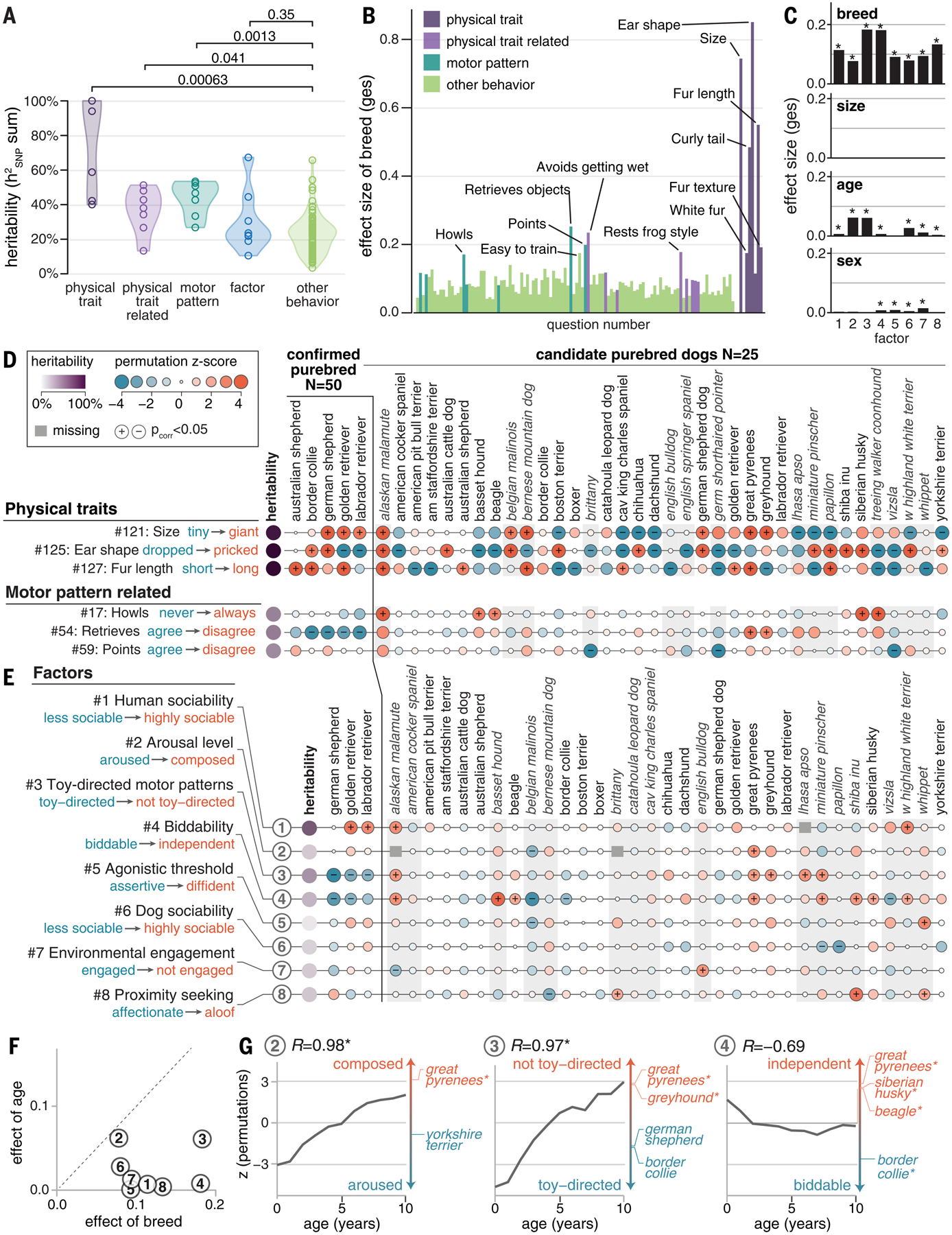
(A) LD-corrected SNP-based heritability is much higher for physical traits, and somewhat higher for motor patterns, compared with other behavioral traits (significance measured using Student’s t test; Benjamini-Hochberg (BH)–adjusted p value is shown). Shaded regions indicate the probability density. (B and C) ANOVA in confirmed purebred dogs shows that (B) the effect size of breed exceeds 15% for 6/7 physical traits and some behavioral questions (labeled bars) and that (C) for factors, breed explains more variation in scores than age and sex, and size has no significant effects. In (C), asterisks indicate statistical significance (BH-adjusted p < 0.05). (D and E) Permutations comparing breeds to randomly sampled dogs show that (D) many breeds are significantly differentiated on physical traits but that differentiation is much rarer for behavioral traits, including intrinsic motor patterns and (E) the eight behavioral factors. The selection of the most popular and/or most differentiated breeds is shown, with full results in fig. S12. Breeds in italics are represented by fewer than 50 individuals and skew toward more extreme z-scores (gray background; difference in mean score = 0.29; pt-test = 2 × 10−71). (F) The effect of age, compared with the effect of breed, in the ANOVA shows that age explains nearly as much variance as breed for factor 2. (G) The PPS for arousal level and toy-directed motor patterns are significantly correlated with age, whereas biddability is more breed driven. Asterisks indicate significant results.
Breed explains some behavior variance
In the owner surveys, breed explains a larger fraction of the variance in behavior phenotypes (110 questions and eight factors) than size, sex, or age, but the effect is relatively small (Fig. 4, B and C; fig. S9A; and data S9). In an analysis of variance (ANOVA) of confirmed purebred dogs representing 78 breeds, the breed effect, measured as generalized eta squared (ges) (51), averages 0.089 ± 0.039 (±SD) (range 0.034 to 0.253), correlates with heritability (Rpearson = 0.89; p = 7.9 × 10−44) (fig. S9B), and is about fivefold higher for the physical traits characteristic of breeds than for behavioral traits (fig. S9C). The same analysis using the less stringent “candidate purebred” breed definition is nearly perfectly correlated with the confirmed purebred analysis (Rpearson = 0.99, p = 5.2 × 10−102; N = 125), with ges values ~30% lower (mean ratio = 0.70 ± 0.11) (fig. S9D).
Age explains little of the variation [0.018 ± 0.035 (±SD)] overall, but for a subset of traits it exceeds 0.05, including two factors (arousal level and toy-directed motor patterns) and nine questions, which include five designed to assess aging-related traits (36) (Fig. 4C and fig. S9E). Sex has little effect (0.009 ± 0.044), except for “lifts leg to urinate” (Q66; ges = 0.48). Size has virtually no effect (6.6 × 10−4 ± 8.6 × 10−4; range 2.5 × 10−7 to 0.006).
Breed is not a reliable predictor of individual behavior
For several factors, score distributions for individual breeds differ from the distribution of all dogs (fig. S10), with at least a few breeds over- or underrepresented in the highest-scoring quartile (fig. S11 and data S10). These distributions are based on owner survey data that may be influenced by breed stereotypes and other factors, and differences are not necessarily genetic in origin. For example, for human sociability (factor 1), an individual Labrador retriever (1.4-fold), golden retriever (1.6-fold), American pit bull terrier (1.4-fold), or Siberian husky (1.7-fold) was more likely to score in the highest quartile than a randomly selected dog, whereas a German shepherd dog (0.78-fold), Chihuahua (0.72-fold), or dachshund (0.56-fold) was less likely. Even so, in every breed represented by 25 or more dogs, the majority scored within one SD of the Darwin’s Ark cohort mean (67.2 ± 7.5% within one SD and 95.4 ± 3.0% within two SD for confirmed purebred dogs). Behavioral factors show high variability within breeds, suggesting that although breed may affect the likelihood of a particular behavior to occur, breed alone is not, contrary to popular belief, informative enough to predict an individual’s disposition.
We developed an interactive dashboard (https://darwinsark.org/muttomics) to illustrate the value offered by breed for predicting behavior in any individual dog. For example, the chance that an owner scores an individual dog in the highest quartile for human sociability increases from 22% for a mutt to 40% if that dog is a golden retriever (fig. S11). Users can select one or a combination of characteristics, and the site dynamically updates to show the frequency in 23 breeds and in mutts.
Measuring breed peculiarity
We developed a permutation-based approach to measure when dogs of a particular breed are described by owners as having behavioral characteristics that significantly differentiate them from other dogs. For each phenotype, we compared dogs within each breed to dogs sampled randomly from the full cohort, producing a “population peculiarity score” (PPS) (22). We tested both “confirmed” breeds (sample n = 50; up to six breeds) and “candidate” breeds (sample n = 25; up to 62 breeds) (Fig. 4, D and E; figs. S12 and S13A; and data S11).
Overall, breeds were only subtly differentiated on behavioral phenotypes. In the confirmed purebreds, only 5.1% (30/583) of breed-phenotype pairs were significantly differentiated for behavioral questions, compared with 41.5% (17/41 pairs) for physical traits. Scores for behavioral questions were not more correlated with each other than were scores for physical questions (table S7). Intrinsic motor patterns and physical traits tend to be slightly more breed differentiated (fig. S13B).
No behaviors are exclusive to any breed (fig. S14). Even in the breed with the lowest howling propensity, confirmed Labrador retrievers (Q17; N = 241; 78.4% never howl), 8% of owners report that their Labrador howls sometimes, often, or always. Although 90% (53/57) of confirmed greyhounds are reported to never bury their toys (Q29), owners described three dogs as frequent buriers.
We used the same permutation approach to measure how behavior changes as dogs age (fig. S13C). Most questions (72%) and factors (63%) are correlated with age [false discovery rate (FDR) p value (pFDR) < 0.05] (figs. S15 and S16). Older dogs score nearly as composed in their arousal level (factor 2) as the most composed breed (Great Pyrenees), and puppies are far more toy-directed in their motor patterns (factor 3) than one of the most toy-directed breeds (German shepherd dog) (Fig. 4, F and G).
Testing breed stereotypes
PPSs aligned, to a limited degree, with behavioral stereotypes described in the breed standards (data S12). The American Kennel Club (AKC) describes each breed with a three-word phrase (e.g., border collies are “affectionate, smart, energetic” and beagles are “friendly, curious, merry”) (52) (table S2). Breeds described with particular words are not behaviorally distinct from other breeds; however, “charming” tends to describe breeds that are less toy directed (factor 3; pFDR = 0.039) (fig. S17). Grouping breeds by their proposed historic working role, as captured by AKC show groups (53), finds that four out of six show groups are peculiar on at least one factor (Fig. 5A). Herding breeds are more toy directed, more biddable, more engaged, and more aloof, whereas toy breeds are more independent and less dog social. Sporting breeds are more toy directed, and working breeds are more dog social (Fig. 5B and fig. S18A).
Fig. 5. Breed stereotypes can be assessed in mutts, where environmental effects (e.g., owner perception) are mitigated by the difficulty of accurately discerning the breed.
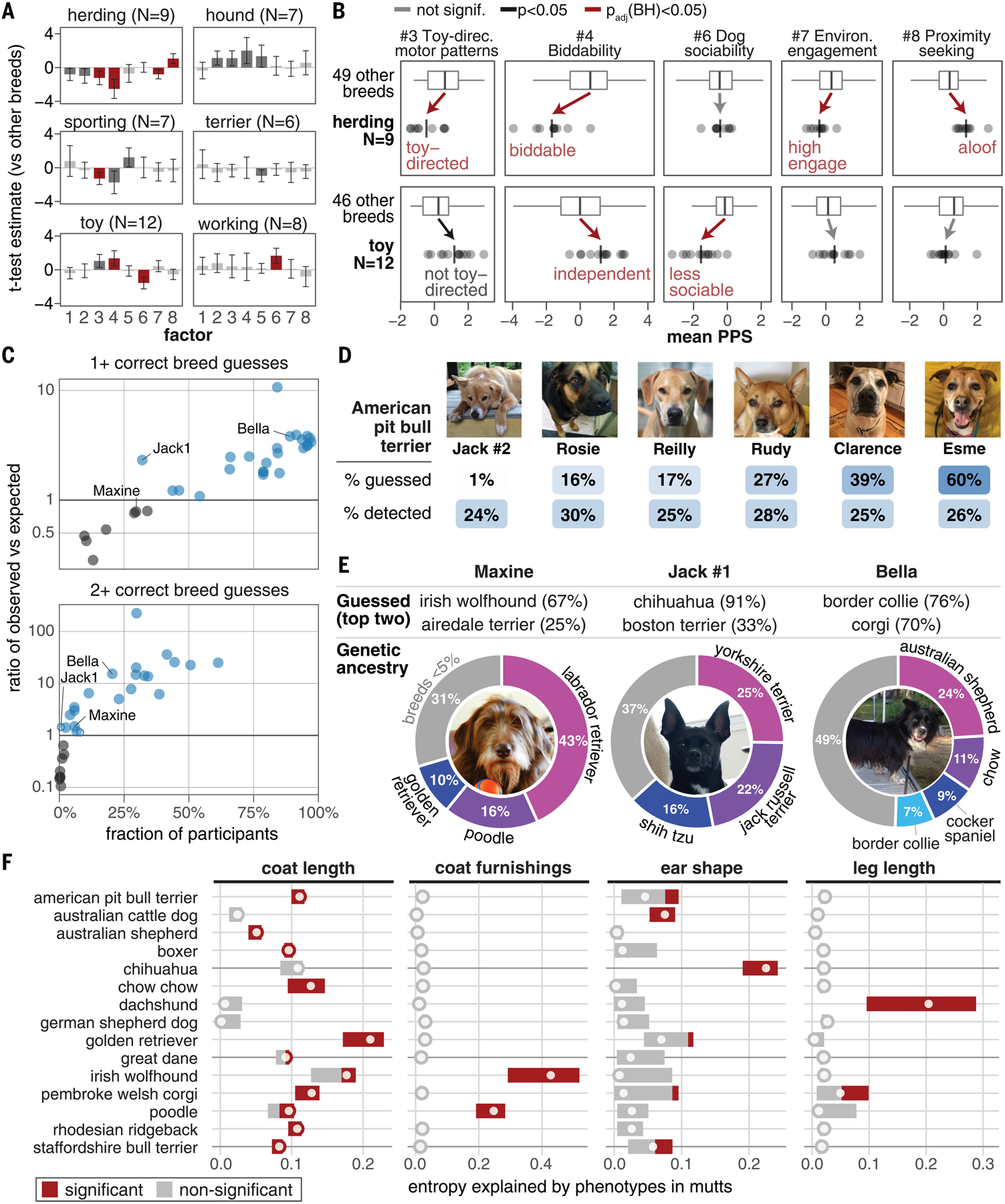
(A) Breeds grouped by purported historic working roles are more differentiated (measured by mean PPS) on some factors than other breeds (58 breeds total), measured as a t statistic using Student’s t test. Error bars represent 95% confidence intervals. (B) Toy and herding breeds illustrate groups with shifts in opposite directions (full results in fig. S18). Points are PPS for breed, vertical lines are mean PPS for group, boxes enclose the 25 to 75% quartiles, and horizontal lines extend from 1.5 times the interquartile range (IQR) below the 25% quartile to 1.5 times the IQR above the 75% quartile. Arrows indicate the direction of change in means, and words show the favored behavioral propensity. Herding breeds are Australian cattle dog, Australian shepherd, Belgian malinois, border collie, Catahoula leopard dog, collie, German shepherd dog, Pembroke Welsh corgi, and Shetland sheepdog. Toy breeds are bichon frise, Cavalier King Charles spaniel, Chihuahua, Havanese, Maltese, miniature pinscher, papillon, Pomeranian, pug, shih tzu, toy poodle, and Yorkshire terrier. (C) For each dog in the MuttMix survey, the ratio of observed:expected correct guesses for (i) one or more or (ii) two or more of the highest-content breed ancestries (blue indicates a ratio >1; open circles are not significant). (D) For six dogs that have a similar amount of genetic ancestry detected from American pit bull terrier (~25 to 30%), participants guessed this breed at rates ranging from 1 to 60%. [Photo credits: J. O’Donnell (Jack); T. Fortier (Rosie); A. Phelps (Reilly); L. Moses (Rudy); R. Skloot (Clarence); M. Bishop (Esme)] (E) For three individual mutts, the most guessed breeds (top) differ from the genetically inferred breed ancestry (bottom). [Photo credits: E. Winchester (Maxine); R. Bacon (Jack); E. Stackpole (Bella)] (F) The dogs in (E) illustrate how a mutt’s physical characteristics influence participant breed guesses. Points show entropy explained by traits using guesses for all mutts (22), and bars span values from a leave-one-out analysis (full results in fig. S18). For example, 67% of participants likely guessed Irish wolfhound for Maxine because of her coat furnishings.
We found more support for breed behavioral stereotypes when comparing the PPS results to quantitative rankings from the Encyclopedia of Dog Breeds for each breed on 10 behavioral characteristics (19) (fig. S17B). Nine of the 10 correlated significantly with PPS for at least one factor. Breeds ranked high on “ease of training” tended to be more biddable (factor 4) and more toy directed (factor 3). Breeds ranked as low on “energy level” scored as more composed, more dog social, and less environmentally engaged (factors 2, 6, and 7). “Watchdog ability” and “friendliness towards strangers” both correlated with human sociability (factor 1), but in opposite directions.
Overall, when comparing breeds to all pet dogs, breed differences based on owner reports align with some breed behavioral stereotypes, with one major caveat. Using survey data alone, we cannot distinguish environmental effects, including the effects of the stereotypes themselves (e.g., by influencing owner’s perception of their dog’s behavior), from genetic effects.
Human perception of breed in mutts
Half of the Darwin’s Ark cohort were mutts, offering an opportunity to test whether breed stereotypes have a genetic etiology. In purebred dogs, cultural breed stereotypes affect the perception of a dog’s behavior and thus may alter a dog’s environment (54, 55) or introduce rater bias into owner survey responses. If breed ancestry is not readily discernible in mutts, these nongenetic factors would be mitigated, allowing us to discern the genetic effects of breed from human perception and other environmental factors.
To measure how accurately breed can be discerned from physical characteristics in mutts, we ran a 2-month community science project, MuttMix (muttmix.org) that recruited 26,639 participants (Fig. 5C). For 30 mutts with complex genetic ancestry (i.e., no first-generation crosses) (22), we asked participants to guess their top three breeds. Between 13,662 and 14,160 participants submitted responses for each dog. They accurately identified, on average, 20.9 ± 20.4% (±SD) of each dog’s breed ancestry (fig. S19A). Breeds comprising a smaller proportion of a dog’s ancestry were especially challenging to identify (Rpearson = 0.61; p = 3.3 × 10−10) (fig. S19B). Thus, any effect of perceived ancestry on survey responses is likely to be substantially mitigated in mutts.
The physical characteristics associated with a breed, like short fur (American pit bull terrier), short legs (dachshund), or pricked ears (Chihuahua), influenced how participants guessed, but this is an error-prone approach (Fig. 5, D to F, and fig. S19C). Dogs with ancestry from more popular breeds had more correct guesses because participants tended to guess popular breeds more frequently (with the exception of the underguessed American pit bull terrier) (Fig. 1G and fig. S20). To control for this, we calculated how often we would expect to see each possible combination of breed guesses if breed guess rates matched the population frequencies in Darwin’s Ark (table S2) and compared the observed rate of correct guesses to the expected rate. Participants guessed correctly more often than expected for 73% of dogs (Fig. 5C and fig. S21A). The number of correctly guessed breeds by each participant for each dog was slightly higher for self-described dog professionals [N = 84,918; mean = 0.93 (SD 0.74)] than nonprofessionals [N = 333,614; mean = 0.81 (SD 0.72)] (fig. S21B).
Effect of breed ancestry in mutts
We measured whether breed influences behavior through genetics by examining only mutts with <45% ancestry from any single breed. This analysis illustrates the power of including complex mixes in behavioral studies, because any influence of breed stereotypes is mitigated when true breed ancestry is not readily discernible from appearance. We built linear mixed-effects regression (LMER) models for all factors and questions (breed ancestries as fixed effects; age and pairwise genetic relatedness as random effects) (22) (Fig. 6, A to C; fig. S22; and data S13). The proportion of variance in factor scores captured by genetic breed ancestry (marginal R2) averaged 9 ± 3% (±SD), suggesting a weak but discernible genetic effect of breed on disposition (fig. S23B and data S14). Breed ancestry explained, on average, 20 ± 12% (±SD) of variance for physical traits in mutts.
Fig. 6. Genetics of aesthetic and behavioral traits in dogs and influence of breed ancestry in mutts.
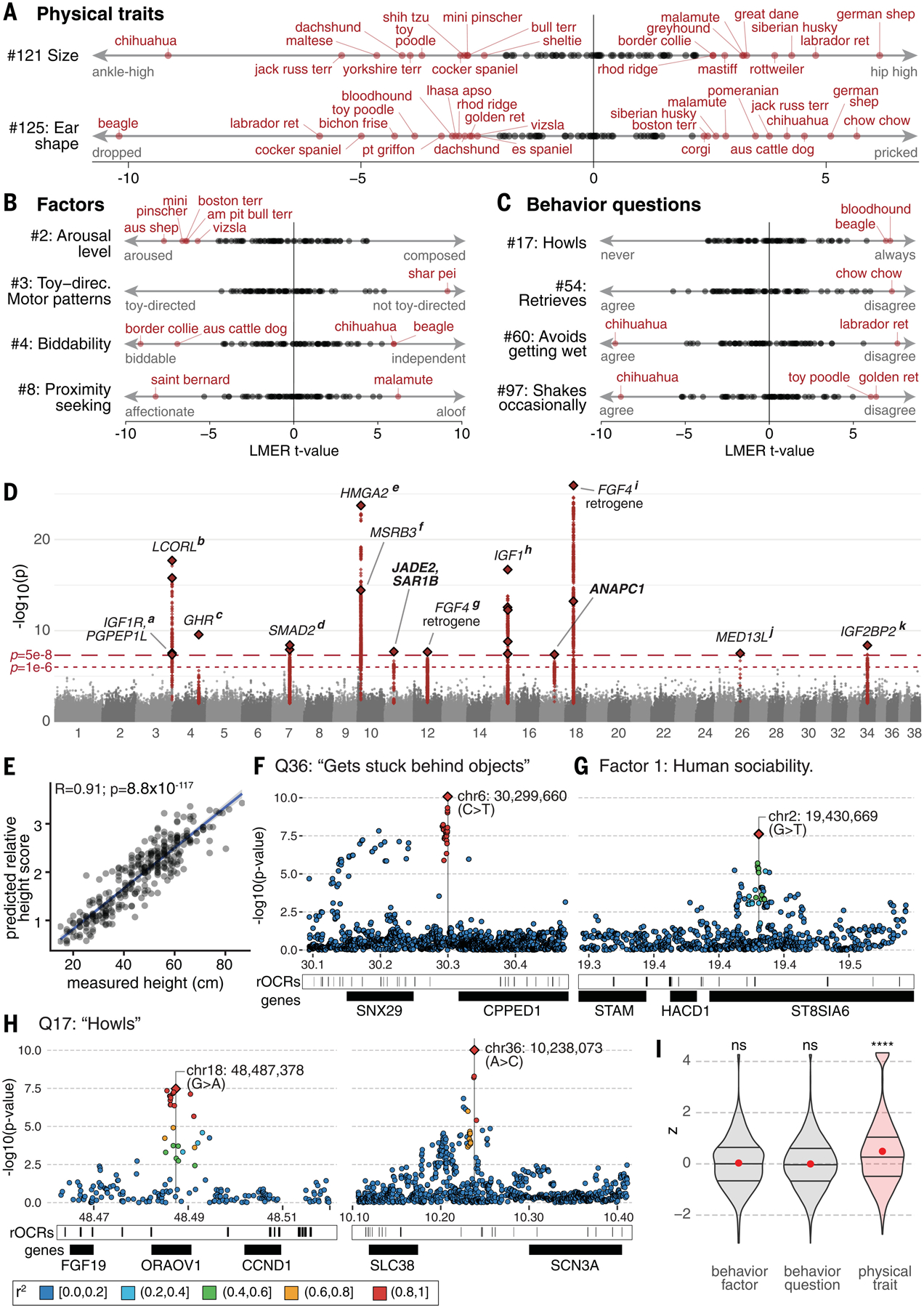
(A to C) In highly admixed dogs with no breed ancestry over 45%, the fixed effects of breed ancestry on (A) physical traits, (B) behavioral factor scores, and (C) individual behavioral question scores are shown. (D) Manhattan plot for the GWAS of surveyed height (Q121) from 1951 dogs, including covariates for age and sex. Linkage blocks (r2 > 0.2) associated (p < 5 × 10−8) with stature align with previous associations for body size in (a) IGF1R (68), (b) LCORL (40), (c) GHR (69), (d) SMAD2 (84), (e) HMGA2 (69) and the nearby (f) MSRB3, (g) a chromosome 12 retrogene insertion of FGF4 (70), (h) IGF1 (64–66), (i) another FGF4 retrogene on chromosome 18 (67), (j) MED13L (40), and (k) IGF2BP2 (40). Two previously unknown associations were found spanning JADE2 and SAR1B, and in ANAPC1. (E) Random forest models based on size-associated SNPs (p < 1 × 10−5) accurately predict body size and correlate strongly (N = 310 dogs; Rpearson = 0.91, p = 8.8 × 10−117, t = 37.451, df = 308) with real measurements in those dogs. (F to H) Regional association plots for (F) scores on Q36 “gets stuck behind objects,” (G) human sociability, and (H) frequency of howling from Q17. In addition to protein coding genes (black boxes), we also show representative open chromatin regions (rOCRs; narrow vertical lines). We annotated rOCRs genome-wide using ENCODE methods (101) applied to canine ATAC-seq (assay for transposase-accessible chromatin using sequencing) data from 14 tissues (102) and mammalian sequence constraint (103). (I) Breeds show high genetic differentiation, measured as the population branch statistic, overlapping physical trait associated loci compared with ~100,000 randomly permuted regions (N = 1232, mean z = 0.49, p = 7.3 × 10−33). Regions associated with behavioral factors (N = 512, mean z = 0.03, p = 0.224) and question scores (N = 9317, mean z = 0.00, p = 0.603) do not show such differentiation in breeds. Red circles indicate mean, with horizontal lines at the 25% quartile, median, and 75% quartile. The shaded area is the probability density, with significant differentiation in red. ns, not significant; ****p < 0.0001 (Student’s t test).
We validated the LMER method by confirming that the ancestry effects for physical traits matched the breed standards for physical appearance (38). For example, ancestry from nine breeds (six with long fur and three with short fur) had a significant effect on fur length in mutts, and for each breed, the direction of effect matches the breed standard. In total, we assessed 51 breed-trait pairs for four traits (size, white coat color, ear shape, and fur length). The direction of effect matched in 50 out of 51 (table S8). For the size question (Q121), the LMER score is strongly correlated with the breed average height (Rpearson = 0.86; p = 2.3 × 10−6; N = 19).
Correlation between the LMER results and the PPSs confirm that some behavioral differences in mutts derive from differences in breed ancestry (N = 6333; Rpearson = 0.28; p = 1.8 × 10−111) (fig. S24 and data S15). For example, mutts with more border collie ancestry tend to be more biddable (factor 4; LMER t = −4.6; pFDR = 0.0002), consistent with survey data for confirmed border collies [PPS z = −4.6; corrected p value (pcorr) = 2 × 10−6]. Similarly, mutts with more Labrador retriever ancestry tend not to avoid getting wet (Q60; LMER t = 3.8; pFDR = 0.003), like many confirmed Labrador retrievers (PPS z = 4.3; pcorr = 0.003).
Discordance between the LMER results and breed differentiation measured by PPS may capture nongenetic influences on survey responses such as breed stereotypes. Owners of confirmed golden retrievers, for example, tend to disagree that their dog is fearful of unfamiliar people (Q46; PPS z = 4.6; pcorr = 0.002), which fits the breed stereotype that golden retrievers are friendly to strangers (19). In mutts, however, golden retriever ancestry had no effect on this question (LMER t = 0.3; pFDR = 0.88), suggesting that the reported propensity may not be driven by genetics (fig. S23A). Similarly, whereas owners of confirmed Labrador retrievers tend to describe their dogs as more human social (factor 1; PPS z = 3.3; pcorr = 0.006), in line with the breed stereotype (“friendly, active, and outgoing”), in mutts, Labrador retriever ancestry has little effect (LMER t = 0.4; pFDR = 0.90). Owners of confirmed border collies tend to score their dogs higher on “wants to play” (Q2; PPS z = −3.6; pcorr = 0.04); this is consistent with the stereotype that border collies are “affectionate, smart, and energetic” but discordant with the LMER results, which find no effect of border collie ancestry (LMER t = 0.089; pFDR = 0.97)
By analyzing the effect of ancestry on behavior in mutts, we can anticipate the likely behavioral propensities of breeds that are not well represented in our survey data (table S9). For example, Saint Bernard ancestry correlates with being more affectionate (factor 8, LMER t = −4.1; pFDR = 0.002) and Shar-Pei ancestry with being less toy directed (factor 3; LMER t = 4.6; pFDR = 0.0002) (Fig. 6B). Ancestry from Chesapeake Bay retrievers increases with propensity to damage doors (Q40; LMER t = 4.2; p = 0.001) and escape from enclosures (Q35; LMER t = 3.5; pFDR = 0.02).
Genome-wide association studies in all dogs
We first investigated breed-defining physical traits with known large-effect loci using a mixed linear model–based approach for genome-wide association (56) across 8,518,951 SNPs of >2% MAF. We controlled for population and family structure and cryptic relatedness in our complex cohort (600 purebred dogs, representing 88 breeds, and 1496 mutts) using a genetic relationship matrix in a mixed-model framework. None of the genome-wide association studies (GWASs) had unusual genomic inflation [mean inflation factor (λGC0.5) = 0.985 ± 0.016 (±SD); range 0.960 to 1.03; N = 14], suggesting that the mixed-model framework controls for confounding due to population structure and other factors (57).
We successfully replicated 17 published associations for physical traits other than size (table S10), including for genes MITF (14) with white spotting [p = 2.89 × 10−37; SNP effect size (b) = −0.78], FGF5 and RSPO2 (58) with coat length (p = 5.46 × 10−54; b = +0.37) and texture (p = 6.35 × 10−9; b = +0.11), USH2A (59) with roan and/or ticking (p = 5.31 × 10−16; b = +0.20), RUNX3 (60) with pheomelanin intensity (p = 4.11 × 10−8; b = −0.20), and the β-defensin region (61–63) with brindle coat patterns (p = 2.50 × 10−107; b = +0.35) (fig. S25, D to I, and data S16).
For size, a quantitative trait, we replicated 10 previously published associations (40, 64–71) (Fig. 6D, fig. S25A, and table S10) and found new associations to SAR1B [p = 2.01 × 10−8; b = +0.12; metabolic disorders (72, 73)] and ANAPC1 [p = 4.11 × 10−8; b = +0.15; short stature in Rothmund-Thomson syndrome (74)]. By comparing giant dogs (N = 55) and then tiny dogs (N = 55) to average dogs (N = 1841), we distinguished variants associated with gigantism (fig. S25B) and dwarfism (fig. S25C) specifically. The FGF4 retrogene locus, previously associated with chondrodysplasia (67), is more strongly associated in the tiny GWAS (pall = 1.16 × 10−26; ptiny = 1.15 × 10−29), dwarfing all other loci.
The height associations were robust even in the absence of purebred dogs, suggesting that the all-mutt GWAS might offer equivalent power to one that includes purebred dogs. In dogs carrying less than 45% ancestry from any breed, a cohort with about half as many dogs (970 versus 1951), we identified all the major stature-associated loci (Rpearson = 0.91; p < 1 × 10−8) as well as a new association in LRIG3 (p = 7.29 × 10−10; b = −0.31), a gene involved in bone morphogenetic protein–mediated body-size regulation (75) (fig. S25J).
Genomic predictions for height based on the GWAS-identified variants perform well in both purebred dogs and mutts, reflecting the strong selection on size among dog breeds. For a random forest regression model built using 1730 dogs and 2733 size-associated SNPs (p < 1 × 10−5) (22), predictions carried a mean squared error of 0.3 (fig. S26) and 66% of predictions fell within ±0.5 units of the relative size score (fig. S2) (Rpearson = 0.77, p = 3.90 × 10−305), with no drop in accuracy for predictions made on mutts [predicted and true values differed by 0.46 ± 0.35 (±SD) in purebreds versus 0.43 ± 0.36 (±SD) in mutts; pt-test = 0.08, t = 1.75, df = 832]. Randomly selected SNPs, by comparison, performed poorly, with a mean squared error of 0.5 (45% of predictions within ±0.5 units). Predictions for relative stature validated well against more precise measurements taken in person (N = 310 dogs; Rpearson = 0.91, p = 8.8 × 10−117) (Fig. 6E and fig. S27).
Behavioral GWASs
Applying the same GWAS approach to the behavioral phenotypes identified 11 genome-wide significant (p < 5 × 10−8) (76) and 136 suggestive (p < 1 × 10−6) associations (data S16). As with physical traits, the behavioral GWAS had minimal genomic inflation [mean λGC0.5 = 0.995 ± 0.0087 (±SD); range 0.976 to 1.05; N = 118]. The associations for behavioral traits were weaker, consistent with a more complex genetic architecture. They have not yet been independently replicated. The most significant association, to “gets stuck behind objects” (Q36), mapped to a 380-kb region (p = 8.36 × 10−11; b = +0.54) (Fig. 6F and fig. S25K) containing SNX29, a gene associated with cognitive performance in human GWASs (77–79). “Dog howls” (Q17) mapped to an intergenic region (p = 9.63 × 10−11; b = +0.54) (Fig. 6G and fig. S25L) between SLC38A11 and SCN3A, a voltage-gated sodium channel involved in the development of speech and language (80). The top association to a behavioral factor was for human sociability (factor 1), downstream of the gene HACD1 (p = 2.41 × 10−8; b = −0.36) (Fig. 6H and fig. S25M), a regulator of long-term memory (81) that is also associated with centronuclear myopathies (82).
In our diverse cohort with dense genotyping data, associated regions are smaller than those discovered using intrabreed GWASs with sparser marker sets. We compared our behavior-associated regions to those found in an earlier study of a different complex trait (osteosarcoma) at the same linkage threshold (r2 > 0.8). In the Darwin’s Ark GWAS, associated regions extend to a median 5.6 kb (25 to 75% quartile = 2.0 to 14 kb, mean 16.8 kb) around suggestive (p < 1 × 10−6) behavioral loci and 5.7 kb at physical trait loci (1.4 to 22 kb, mean 26.2 kb). By contrast, intrabreed GWASs of osteosarcoma in three breeds with diverse population structures mapped at median ranges of 86 kb (25 to 75% quartile = 57 to 162 kb) in racing greyhounds, 54 kb (21 to 409 kb) in rottweilers, and 1 Mb (743 kb to 1.4 Mb) in Irish wolfhounds (83). This increased resolution may facilitate the search for causal variants. In the Darwin’s Ark GWAS, we can distinguish a region on chromosome 10 that is associated with stature (76.2 kb at r2 > 0.8; HMGA2; p = 1.84 × 10−24; b = −0.31) from one associated with ear shape (118.7 kb at r2 > 0.8; MSRB2; p = 6.02 × 10−23; b = −0.33) that were previously linked in interbreed GWASs (71, 84) (fig. S28).
The mixed-model association approach may not fully control for spurious association that arises when a trait differs between breeds. An association for “focused in distracting situations” (Q21) (chr32:4,512,005; p = 1.0 × 10−8; b = −0.22) (fig. S25N) mapped to a locus containing FGF5, a gene associated with long-coated breeds (58). This association was lost when we conditioned on the top coat length–associated SNP (chr32:4,509,367; p = 0.0001; b = −0.15) (fig. S29), which is linked to the top focus-associated SNP (r2 = 0.33). The original association likely reflected the spurious difference in focus scores between dogs with shorter and longer coats (pt-test = 0.00023; 2012 dogs). Pleiotropy is unlikely because fur length explains almost no variation in focus scores (ANOVA ges = 0.0004; p = 0.35; N = 2456). Consistent with this, the focus association on chromosome 32 weakens (chr32:4,512,005; p = 1.2 × 10−6; b = −0.18) when we include the top 10 SNP-based principal components in the mixed model (fig. S30).
To assess whether spurious breed-trait correlations are a major confounder in our analyses, we reran all GWASs and included the top 10 principal components in the mixed model. Only 6% (3/48) of our genome-wide significant associations were lost (p > 1 × 10−6) (data S16). We also tested whether the top 75 regions associated with dog size (ahighly breed-differentiated trait) were enriched for SNPs associated in any of the 119 behavioral GWASs (fig. S31). Only one GWAS (Q66, “lifts leg to urinate”) was significant [adjusted p value (padj) = 0.013]. Thus, although spurious associations due to aesthetic traits are a concern in multibreed GWASs, they are likely rare in the GWAS run on our diverse cohort.
Unattributed heritability
A large proportion of the genetic and environmental contributions to behavior remains undiscovered. The SNPs that we found to be associated with heritable (h2SNP > 0.1) behavioral traits account for a smaller proportion of overall heritability than do aesthetic trait associations (22), consistent with a complex genetic architecture. For the 14 physical traits, 53.0 ± 30.2% (±SD) of heritability is attributable to associated SNPs (p < 1 × 10−6), but for the eight behavioral factors and 73 questions, this drops to 21.0 ± 12.8% and 27.9 ± 20%, respectively. The six associated loci accounted for 42.7% of the genetic component of dog sociability (h2SNP = 14.8 ± 6.1%), whereas just 4.3% of highly heritable human sociability (h2SNP = 41.5 ± 9.1%) could be explained by its single associated region.
Brain-expressed genes enriched in behavior GWASs
Regions associated with dog behavioral phenotypes are enriched in brain-expressed genes. We cataloged the genes expressed in 38 tissue types, including 13 brain regions, using human GTEx data (85), an approach used previously in dogs (86). We also collated genes from curated lists for obsessive-compulsive disorder (OCD) (87), autism-spectrum disorders (88), and schizophrenia (89, 90). Using MAGMA (22, 91), we tested all GWASs for enrichment (data S17). Regions associated with toy-directed motor patterns (factor 3) had the strongest enrichments, which were for genes expressed in the hippocampus and in the basal ganglia of the nucleus accumbens, caudate, and putamen. Associations for “not keen on new situations” (Q84) were enriched for hypothalamus-expressed genes (fig. S30). Overall, enrichments in genes associated with neuropsychiatric conditions were weak, peaking for the enrichment of human OCD genes in Q84-associated regions (p = 0.0012; padj = 0.24).
Aesthetic selection predominates in breeds
Associations to physical traits, but not behaviors, tend to overlap signals of genetic differentiation in modern breeds, suggesting that aesthetics, and not behavior, has been the focus of selection. We tested for sites with excess differentiation in each breed with >12 dogs using the population branch statistic (PBS) test (92), using all dogs (N = 3802 to 3878) and wolves (N = 48) as the two outgroups across ~27.6 million SNPs from publicly available and our genetic data (data S4 and S6). Among the top 0.1% of breed-differentiated regions (26 ± 6 regions per breed), we validated genetic signals of selection reported at EPAS1, for hypoxia tolerance, in Tibetan mastiffs (93); at CACNA1A, unknown phenotype, in two sled dog breeds (94); at ESR1, unknown phenotype, in long-legged sighthounds (40); and at ALX4, a blue eye color gene, in Siberian huskies (95) (data S18).
We used permutation (22) to test whether PBS scores are unexpectedly high in regions associated with traits (data S19) and found that, whereas physical trait–associated regions are more differentiated, those associated with behavioral traits are not (mean z = 0.491 versus −0.001; pt-test = 4 × 10−31) (Fig. 6I). Considering all moderately associated GWAS regions (p < 1 × 10−6), 25 of 65 (39%) physical trait loci are unusually differentiated, whereas only 38 of 515 (7%) behavioral trait loci are, and a subset of those are also connected to physical traits (data S20). Differentiation at physical trait loci is consistent with ongoing selection to meet strict morphometric standards in breeds (38), and the lack of overlap for most behavioral traits suggests weaker or absent selection.
The lack of differentiation at behavioral loci is not inconsistent with heritable behavioral differences in breeds, which may reflect genetic drift or selection that predates breed creation, neither of which the PBS test is designed to detect. To this point, neither of the two loci associated with howling are differentiated in either the Siberian huskies or beagles, even though ancestry from these breeds influences howling propensity.
Discussion
Behavioral traits are subtly differentiated in modern breeds (Fig. 2B). Furthermore, breed offers only modest value for predicting the behavior of individual dogs. For more heritable and more breed-differentiated traits, like biddability (factor 4), knowing breed ancestry can make behavioral predictions somewhat more accurate in purebred dogs. For less heritable, less breed-differentiated traits, like agonistic threshold (factor 5), which measures how easily a dog is provoked by frightening, uncomfortable, or annoying stimuli, breed is almost uninformative.
In our ancestrally diverse cohort, we show that behavioral characteristics ascribed to modern breeds are polygenic, environmentally influenced, and found, at varying prevalence, in all breeds. They likely naturally arose over millennia as dogs followed human migrations and adapted to new human technologies (2). The tight bottlenecks that established modern breeds captured ancient variation, at varying frequencies, with subsequent genetic drift or selection further shaping modern breeds (Fig. 3, A and B).
We found no evidence that the behavioral tendencies in breeds reflect intentional selection by breeders (Fig. 6I) but cannot exclude the possibility. Current datasets are too small to detect more subtle, recent directional selection, which requires hundreds of thousands of samples (96). In dogs, breed demographic history makes detecting selection particularly challenging (1, 97).
Canine behavioral disorders are a proposed natural model for human neuropsychiatric diseases (25, 27). Here, we show that large-scale behavior GWASs in dogs are tractable, identifying dozens of loci associated with behavioral traits in dogs. These associations explain a fraction of overall heritability, suggesting that still-larger sample sizes are needed. Our study design, combining owner-engagement with low-pass sequencing (45), makes this eminently achievable. We anticipate that this approach will be even more powerful once methods for accurately assigning local ancestry in individuals with >100 potential source populations (compared with two or three in human studies) are validated and incorporated into dog GWASs (98).
As dog studies grow in scale and complexity, it is crucial that we meet the standards of statistical rigor developed by the human genetics community and carefully account for confounding by artificial selection for aesthetic extremes in modern breeds (99), which can create misleading signals of association. One approach for studying behavior in dogs has been to compare breeds, rather than individuals, using breed-level behavioral phenotypes. The wide variability in behavior within breeds, and the potential for spurious correlations with breed-defining aesthetic traits, suggests that any discoveries made using this approach should be carefully validated using other methods.
To date, dog genetics has focused on modern breeds, which capture just a tiny fraction of global canine diversity. Although this made early genomic studies feasible (14), it limits discovery today (100). By embracing the full diversity of dogs, including purebred dogs, mutts, purpose-bred working dogs, and village dogs, we can fully realize dogs’ long-recognized potential as a natural model for genetic discovery.
Materials and methods summary
Materials and methods described in full detail can be found in the supplementary materials (22).
Survey data collection
We collected consent, profile information, and surveys for 18,385 dogs enrolled by their owners via the Darwin’s Ark platform (https://darwinsark.org) on or before 15 November 2019. Profile information included the dog’s approximate birth date, sex and sterilization status, suspected or known breed(s), purebred registration, and/or photograph. We collected 12 surveys, including 11 about behavior (10 questions each) and one about physical characteristics (eight questions), for a total of 118 survey items (table S1). All responses to survey questions were time stamped, and ages at the time of survey were calculated relative to reported birth date (22).
The 110 behavioral questions all used a five-point Likert scale: (i) 81 questions had options of strongly agree, agree, neither agree nor disagree, disagree, or strongly disagree; and (ii) 29 had options of never, rarely, sometimes, often, or always. We sourced 79 behavioral questions from published and validated surveys: (i) Dog Personality Questionnaire (DPQ/DPQL; 45 questions) (37); (ii) Canine Health-related Quality of Life Survey (CHQLS; 11 questions) (36); (iii) Dog Impulsivity Assessment Scale (DIAS; 18 questions, including one also in DPQ) (34); and (iv) Canine Cognitive Dysfunction Rating scale (CCDR; six questions) (35). We validated the performance of behavioral surveys using a Mantel’s test on the inter-item correlation distance (d = 1 − |r|) matrices between published data for 48 DPQ items (N = 2556 dogs) and our data. We included 31 new behavior questions developed with input from canine behavior professionals in the International Association of Animal Behavior Consultants. The physical characteristics survey used a variety of response types (table S1). Answers of “I’m not sure,” “I don’t know,” “not sure,” and “surgically cropped ears” (Q125) were excluded.
Dog size was measured through Q121: “When DOG is standing next to someone of average height, how high are HIS shoulders?” This question was validated in three ways (fig. S2 and data S1): (i) owners were provided with a measuring tape by mail and instructed to measure the height from their dog’s shoulder to the ground using the provided measuring tape (337 dogs); (ii) dogs were measured (height to withers) by professionals during the 2017 Somerville Dog Festival in Somerville, MA (38 dogs); and (iii) owner-reported size was compared with average breed height (2025 purebred dogs).
We performed exploratory factor analysis on the behavioral surveys (10,253 dogs with responses for all 110 questions) and extracted the optimal number of factors as estimated by the Horn’s parallel analysis and optimal coordinates heuristic methods (20 factors; table S3). A varimax orthogonal rotation was applied to generate a structure matrix with factor loadings for each item, and items with low pattern or structure loadings (less than ±0.3) were removed. We generated factor scores for 6269 additional dogs with responses to >80% of questions by populating missing responses through random sampling. The dog’s age for each factor is the mean age for all responses to included questions.
Sample collection
Animal study protocols for saliva and blood collection from dogs were approved by the UMass Chan Medical School Institutional Animal Care and Use Committee (IACUC) (no. A-2520–18). We sent or gave owners saliva collection kits (DNA Genotek PG-100 saliva swabs) for sampling. For a subset of dogs, owners provided blood collected by their veterinarian. We selected dogs for sequencing primarily based on survey completeness and enrollment date. Of 1715 samples submitted for low-coverage DNA sequencing, 159 samples (7.4% of 2155 dogs in the genetic dataset) were funded by owner donations to the Darwin’s Ark Foundation, a 501(c)(3) nonprofit organization (82–3942341).
High-coverage genome sequencing and analysis
We performed high-coverage [45× ± 10× (±SD)] WGS on samples from 27 putatively mixed-breed dogs (the Mendel’s Mutts cohort) (data S2). For the initial 22 mutts sequenced, we performed joint variant calling with publicly available data for 620 other dogs and 34 canids (data S4) using the Genome Analysis Toolkit (GATK3) (22) on the CanFam3.1 reference assembly. The final variant call file contained 34,191,821 SNPs and 11,943,064 indels. For the five mutts sequenced later, genotypes were called for the same set of variants using GATK3 HaplotypeCaller.
We compared cumulative variant discovery using purebred versus mutt genomes using chromosome 13 as a random proxy for the whole genome. We tested six cohorts: one dog sampled at random per breed (N = 128 possible dogs), Mendel’s Mutts (N = 27 dogs), and the four breeds with >27 individuals sequenced (22). We computed the cumulative distribution of the fraction of 619,031 variants discovered using 557 purebred dogs versus using 10 dogs randomly chosen and ordered within each cohort and computed the 95% confidence interval using random reordering within each cohort.
We compared the lengths of detected runs of homozygosity (ROH) in mutts, dog breeds, and village dog genomes across biallelic SNPs using PLINK v1.90b6.21 with a minimum length of 100 kb and 100 SNPs, with at least 1 kb per SNP (22). We then randomly sampled n = 464 runs (the mean number of ROH detected per mutt) from the pool of ROH detected in mutts, purebred dogs, and village dogs, re-sampling N = 100 times.
We measured LD in mixed-breed dogs (Mendel’s Mutts), breeds (golden retriever, Labrador retriever, Leonberger, and Yorkshire terrier), village dogs, and wolves by randomly sampling 25 dogs from each cohort and, for 20,000 randomly sampled biallelic SNPs, measuring r2 to all SNPs within 100 kb. We assessed tagging of genetic variation using genotyping arrays by measuring r2 between the same set of random SNPs and the subset of SNPs on the array (171,882 for the Illumina HD Canine Genotyping Array and 1,011,992 for the Axiom Canine Genotyping Array Sets A and B).
Low-coverage sequencing and imputation
We piloted a low-pass sequencing and imputation approach (42–46) using a panel of reference haplotypes from high-coverage whole-genome sequences. Autosomal variant calls were inferred directly from sequencing reads through Gencove loimpute software (46) and a panel of reference haplotypes from publicly available WGS data [mean coverage 22.9× (SD 14.2×)] for 435 canids (data S4). The imputation process generated unfiltered genotypes for 32,438,672 SNPs and 13,910,371 indels with imputation genotype probability (GP) scores per genotype per dog. We validated performance by comparing low-pass sequencing and imputation [1.0× ± 0.6× (±SD)] to array data (Axiom array) and high-coverage WGS data for 11 mutts with high-coverage WGS at low coverage. We also performed down-sampling of high-coverage WGS and subsequent imputation by the same method.
We combined low-pass sequencing data for 1715 dogs [0.6× ± 0.3× (±SD)] with data for 440 dogs genotyped on the Axiom array and imputed using the same haplotype reference panel (excluding genotypes of GP < 0.7). After merging, we performed additional quality control based on MAF, call rate, and Hardy-Weinberg equilibrium and validated owner-reported sex (22). The final dataset included 8,518,951 biallelic, autosomal SNPs and 2155 dogs at a genotyping rate of 97.5% (1084 males and 1071 females).
Breed ancestry assignment
We assembled a reference panel of 101 of the most common dog breeds in the United States (table S2) using high-coverage WGS for 380 dogs of 74 breeds (data S4), low-coverage WGS for 115 dogs of 54 breeds, Axiom genotyping array data for 109 dogs of 43 breeds, and Illumina CanineHD arrays for 883 dogs of 90 breeds (22). For each breed, we selected 12 dogs for inclusion, prioritizing high-density raw data and genetic diversity within breeds. We imputed genotypes for low-density data using the 435-canid panel of reference haplotypes. We retained SNPs genotyped in more than 80% of dogs and at a MAF of at least 5%. Among ancestry-informative SNPs of Hudson’s estimator of fixation index (FST) > 0.15 between breeds, we selected a dense set of 2,468,442 markers (r2 > 0.9 within 5 kb) for admixture simulations and a sparser set of 688,060 markers (r2 > 0.5 within 50 kb) for ancestry inference.
We used a Monte Carlo approach to generate simulated admixed genomes of known ancestral haplotypes and then compared the breed ancestry composition with ancestry inferred using ADMIXTURE (22). We simulated admixed individuals through N = 15 generations of admixtures with the following procedure: N + 1 random individuals from different breeds were selected to contribute to the admixture. With each iteration, recombination was simulated to incorporate a new individual. Recombination was treated as a Poisson event that occurred, on average, once every Morgan. Simulations ran on 10 independently drawn datasets of six dogs per reference breed to create 1000 admixed individuals of known ancestry. We inferred global ancestry for simulated individuals using the supervised mode of ADMIXTURE (random seed = 43) and the reference genotypes from six dogs reserved from each breed.
We then performed supervised admixture analysis of the Darwin’s Ark genetic cohort. Genotype data from all query dogs was merged with all reference-breed data and filtered for SNPs in the global breed ancestry panel. Global ancestry from the 101 reference breeds were inferred using the supervised mode of ADMIXTURE (random seed = 43) that was supplied with reference population assignments. Population weights less than 1% were discarded from individual ancestry results.
We combined breed ancestry assignments with survey data for dogs without genetic data to define three breed sets as decribed in the results: confirmed purebred dogs, candidate purebred dogs, and mutts.
Heritability analysis
We estimated the SNP-based heritability (h2SNP) of surveyed traits using restricted maximum likelihood (REML) analysis implemented in the genome-wide complex trait analysis (GCTA, version 1.92.3 beta 3) software tool (56). We calculated LD scores in 250-kb regions using a block size of 10,000 kb with an overlap of 5000 kb between blocks. We generated a genetic relationship matrix (GRM) for the genetic cohort of 2155 dogs, as well as multiple GRMs calculated from SNPs stratified into LD score quartiles (22). The four LD-stratified GRMs were used to run REML analysis (GREML-LDMS) and estimate h2SNP with standard errors (data S8).
Population peculiarity scoring
We applied a custom permutation-based analysis (22) to test whether groups of dogs defined by breed or age differed significantly in survey responses from randomly sampled groups on any survey item or factor. We included all dogs with any survey responses. For each permutation and a given sample size N (table S14), we calculated the mean (the observed test statistic) for each normalized survey response or factor score for N dogs sampled from among dogs of each grouping. For each permutation, we also calculated the mean for a random sampling of size N from the full dataset (the permuted test statistics). We counted how often the observed test statistics for each population were higher than the permuted test statistics. We ran a total of 500,000 permutations. To obtain the PPSs, we calculated the one-tailed empirical p values and generated z-scores matching the survey directionality. We also calculated the two-tailed p values corrected for multiple testing by a maxT procedure that preserves the correlational structure between survey items (22).
Ancestry perception survey
We designed the web-based MuttMix survey (muttmix.org) to assess perceptions of breed ancestry in mixed-breed dogs by nonowner observers. Participants self-identified as either general public or dog professional (yes or no to “Do you work with dogs professionally and/or are you a breeder?”). The survey consisted of 30 mixed-breed dogs with ancestry assignments and one purebred dog. Owners provided front and side photographs and a short video. Owners reported the dog’s relative size (fig. S2F) and other physical descriptors. The images and information that were provided were shared with participants, who were asked to guess, for each dog, the three breeds detected in largest proportion (22). The survey launched on 16 April 2018 and closed on 16 June 2018, and responses were collected from 26,639 people over a 2-month period.
We compared breed guesses to genetically inferred breed ancestry (22). Any breed call below 5% was removed and only breeds offered as survey options were examined. To calculate the average total percentage of ancestry guessed correctly, we first calculated the percentage guessed correctly by each user for each dog by summing the percent genetic ancestry attributed to their top three breed guesses. To assess the accuracy of user guesses of breed ancestry, we first counted the number of breed guesses for a given dog that were among the top two or three breeds that were genetically detected.
We measured how specific physical attributes affected participants’ breed choices using entropy analysis (22). For each breed option, we calculated how well mutts’ phenotypes, defined binarily for each of eight different traits (height, leg length, ear type, coat type, coat length, coat furnishings, white spotting, and pigmentation), distinguished between participant guesses of presence versus absence of each ancestry. We applied a leave-one-out analysis, omitting guesses for each mutt in series, to assess the impact of guesses for each mutt on entropy reduction. To calculate significance, we randomized trait assignments across mutts and then asked whether entropy reductions from true traits were greater than those randomly assigned.
We calculated how often we expected to see each possible combination of breed guesses by chance, assuming the guess rate for each breed to be the overall frequency of that breed (22) (table S2). We then calculated the observed rate of guesses with 1+, 2+, and 3 breeds correct for each dog and then calculated the ratio of the observed-to-expected rate.
LMER models
To measure the relationship of genetic breed ancestry to physical and behavioral phenotypes, we constructed LMER models using all dogs with <45% ancestry from any single breed (1002 dogs total). We treated normalized question and factor scores as independent variables, breed ancestry as fixed effects, and age as random effect. For each survey item and factor, we built a model with REML to obtain unbiased estimates, standard deviations, and Wald statistics (t.val) for the fixed effects of breed on factor score and performed ANOVA to obtain the breed F statistics. To obtain the likelihood ratio for each breed, we constructed models using maximum likelihood with and without the breed and performed an ANOVA.
GWASs using mixed linear models
We performed genome-wide mixed linear model–based associations in the Darwin’s Ark genetic cohort using the “leave-one-chromosome-out” approach (MLMA-LOCO) implemented in GCTA (56) with categorical covariates for sex and data type (genotyping or low-pass sequencing) and quantitative covariates for height and age for nonmorpho-logical traits. Because LD is nearly as short in diverse dogs as in humans, we used the thresholds for genome-wide significance (p = 5 × 10−8) and suggestive associations (p = 1 × 10−6) that are conventionally used in human GWASs (1, 76).
We defined regions of association by clumping SNPs in LD (r2 > 0.2 and r2 > 0.5) and near (<250 kb) associated index SNPs using PLINK (data S16). When comparing region sizes to the earlier osteosarcoma study (83), we used the same clumping thresholds. To assess how much phenotypic variance was explained by associated regions, we derived genetic relationship matrices for regions of suggestive association (p = 1 × 10−6) with each trait and the set of all other SNPs and estimated the partitioned heritability as the proportion of total heritability unattributed by discovered associations.
We built a predictive model for height as responses to survey Q121 for 1730 dogs older than 18 months and assessed its power through 10-fold cross validation (9/10 training, 1/10 testing). At each round, we performed GWASs on the training set, selected SNPs for prediction at given p value cutoffs, built a random forest regression model, and assessed accuracy using the testing set. The reported accuracy and mean squared error are averaged across 10 rounds.
We tested for enrichment of association summary statistics in three types of gene sets (22) by applying MAGMA (version 1.09) (91), a method that accounts for region size, variant count, and LD (data S17).
Genetic differentiation of breeds
We calculated genome-wide normalized PBS scores using the Hudson estimator of fixation (FST) for each breed (NP ≥ 12 dogs, maximum 88) relative to dogs overall (ND = 3890 − NP) and wolves (NW = 48) in sliding windows of 100 kb by 10 kb (data S18) over ~27.6 million SNPs from publicly available and Darwin’s Ark genetic data (22). After dividing locus tests into physical trait, behavioral question, and behavioral factor associations, we performed a one-tailed Student’s t test to test whether the observed maximum PBS within associated loci exceeded what we expect by random chance (data S20). To test whether allele frequencies at SNPs associated with behavioral or physical traits tended to differ more in breeds, we calculated the max FST observed between one of the top 10 breeds and all other dogs and compared this to 29,903 randomly sampled SNPs using a one-sided t test.
Supplementary Material
ACKNOWLEDGMENTS
We thank all of our participants and their dogs for making this work possible, as well as the International Association of Animal Behavior Consultants community for their feedback and help with outreach; E. A. Ostrander for helpful advice and the Ostrander lab for providing the high-coverage whole-genome data (available in data S4); B. Klein and the National Entlebucher Mountain Dog Association for providing whole-genome data; R. Peters and the 360design.com team; J. Gallagher, H. Herzog, A. Karlsson, K. Lindblad-Toh, L. Moses, B. Neale, C. Painter, D. Promislow, B. Rosener, and D. Weaver for useful discussions; H. Beberman for support; B. Burton, R. Crisler, G. Fisher, S. Fraser, L. Haug, J. Hourihan, M. Mullins, C. Pachel, L. Strassberg, and M. Workman for input on survey development; C. Abrams, R. Bacon, K. Bigger, M. Bishop, J. Quintal, D. Church, J. Conner, A. Dauphin, A. Derr, T. and K. Flotte, T. Fortier, S. Garamszegi, B. R. Granger, S. Humphreys, C. Mitchell, M. Movassagh, A. Jorgensen, M. Lane, A. and M. Lek, B. Lengsfeld, J. Luban, A. Macbeth, L. McGuire, S. and J. Moeling, D. Morrissey, E. Neenan, J. O’Donnell, T. Ollier, L. Paquin, G. Peloso, A. Pensarosa, A. Phelps, S. Richardson, J. Rivera, S. Rulnick, S. Schnaffner, R. Skloot, E. Stackpole, W. Theurkauf, S. Thier, and E. Winchester for their time and thoughtful replies to our many questions about their dogs as well as permission to share photographs of their dogs.
Funding:
This project was funded by the National Institutes of Health, including NIMH R21 MH109938, NCI R01 CA255319, NCI R37 CA218570, NCI F32 CA247088, NHGRI R01 HG008742, NHGRI U24 HG009446, OD R24 OD018250, and NIA U19 AG057377. It was also supported by NSF EF-2022007, Broad Institute BroadIgnite and Next10 awards, the Darwin’s Ark Foundation, the Food Allergy Science Initiative, the Manton Foundation, and the Working Dog Project.
Footnotes
Competing interests: L.G. is a co-founder of, equity owner in, and chief technical officer at Fauna Bio Inc. H.J.N is an employee of AbbVie Inc.
View the article online
Data and materials availability:
All sequencing data have been deposited into the National Center for Biotechnology Information (NCBI) Sequence Read Archive, including high-coverage sequencing reads for the Mendel’s Mutts dataset in BioProject PRJNA683923 and low-coverage sequencing reads deposited to BioProject PRJNA675863. The Broad-UMass Canid Variants in variant call format (VCF) is available publicly via FTP at https://data.broadinstitute.org/DogData/. All survey and genetic data from dogs of the Darwin’s Ark and Mendel’s Mutts cohorts, data from the MuttMix survey, and scripts used in analyses are archived in Dryad (104) and Zenodo (105). The genome-wide association summary statistics and plots are shared on Terra (106) and Figshare (107), respectively.
REFERENCES AND NOTES
- 1.Lindblad-Toh K et al. , Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature 438, 803–819 (2005). doi: 10.1038/nature04338 [DOI] [PubMed] [Google Scholar]
- 2.Bergström A et al. , Origins and genetic legacy of prehistoric dogs. Science 370, 557–564 (2020). doi: 10.1126/science.aba9572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lord K, Feinstein M, Smith B, Coppinger R, Variation in reproductive traits of members of the genus Canis with special attention to the domestic dog (Canis familiaris). Behav. Processes 92, 131–142 (2013). doi: 10.1016/j.beproc.2012.10.009 [DOI] [PubMed] [Google Scholar]
- 4.Hansen Wheat C, van der Bijl W, Temrin H, Dogs, but not wolves, lose their sensitivity toward novelty with age. Front. Psychol 10, 2001 (2019). doi: 10.3389/fpsyg.2019.02001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moretti L, Hentrup M, Kotrschal K, Range F, The influence of relationships on neophobia and exploration in wolves and dogs. Anim. Behav 107, 159–173 (2015). doi: 10.1016/j.anbehav.2015.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arendt M, Cairns KM, Ballard JWO, Savolainen P, Axelsson E, Diet adaptation in dog reflects spread of prehistoric agriculture. Heredity 117, 301–306 (2016). doi: 10.1038/hdy.2016.48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Axelsson E et al. , The genomic signature of dog domestication reveals adaptation to a starch-rich diet. Nature 495, 360–364 (2013). doi: 10.1038/nature11837 [DOI] [PubMed] [Google Scholar]
- 8.de G. Mazzorin J, Tagliacozzo A, “Morphological and osteological changes in the dog from the Neolithic to the Roman period in Italy” in Dogs Through Time: An Archaeological Perspective (Archaeopress, 2000), pp. 141–161. [Google Scholar]
- 9.Mazzorin JDG, Tagliacozzo A, Dog remains in Italy from the Neolithic to the Roman period. Anthropozoologica 25, 429 (1997). [Google Scholar]
- 10.Worboys M, Strange J-M, Pemberton N, The Invention of the Modern Dog: Breed and Blood in Victorian Britain (JHU Press, 2018). [Google Scholar]
- 11.Ritvo H, Pride and pedigree: The evolution of the Victorian dog fancy. Vic. Stud 29, 227–253 (1986). [Google Scholar]
- 12.Lord K, Schneider RA, Coppinger R, in The Domestic Dog: Its Evolution, Behavior and Interactions with People, Serpell J, Ed. (Cambridge Univ. Press, 2016), chap. 4, pp. 42–66. [Google Scholar]
- 13.Arons CD, Shoemaker WJ, The distribution of catecholamines and β-endorphin in the brains of three behaviorally distinct breeds of dogs and their F1 hybrids. Brain Res. 594, 31–39 (1992). doi: 10.1016/0006-8993(92)91026-B [DOI] [PubMed] [Google Scholar]
- 14.Karlsson EK et al. , Efficient mapping of mendelian traits in dogs through genome-wide association. Nat. Genet 39, 1321–1328 (2007). doi: 10.1038/ng.2007.10 [DOI] [PubMed] [Google Scholar]
- 15.Karlsson EK, Lindblad-Toh K, Leader of the pack: Gene mapping in dogs and other model organisms. Nat. Rev. Genet 9, 713–725 (2008). doi: 10.1038/nrg2382 [DOI] [PubMed] [Google Scholar]
- 16.Sutter NB, Ostrander EA, Dog star rising: The canine genetic system. Nat. Rev. Genet 5, 900–910 (2004). doi: 10.1038/nrg1492 [DOI] [PubMed] [Google Scholar]
- 17.Gompper ME, Ed., Free-Ranging Dogs and Wildlife Conservation (Oxford Univ. Press, 2013). [Google Scholar]
- 18.American Veterinary Medical Association (AVMA), United States Pet Ownership and Demographics Sourcebook (AVMA, Veterinary Economics Division, 2018). [Google Scholar]
- 19.Coile DC, Encyclopedia of Dog Breeds (Barrons Educational Series, 1998). [Google Scholar]
- 20.American Kennel Club, The New Complete Dog Book: Official Breed Standards and Profiles for Over 200 Breeds (Fox Chapel Publishing, 2017). [Google Scholar]
- 21.Scott JP, Fuller JL, Genetics and the Social Behavior of the Dog (Univ. Chicago Press, 1965). [Google Scholar]
- 22. See the supplementary materials.
- 23.Mehrkam LR, Wynne CDL, Behavioral differences among breeds of domestic dogs (Canis lupus familiaris): Current status of the science. Appl. Anim. Behav. Sci 155, 12–27 (2014). doi: 10.1016/j.applanim.2014.03.005 [DOI] [Google Scholar]
- 24.Svartberg K, Breed-typical behaviour in dogs—Historical remnants or recent constructs? Appl. Anim. Behav. Sci 96, 293–313 (2006). doi: 10.1016/j.applanim.2005.06.014 [DOI] [Google Scholar]
- 25.Noh HJ et al. , Integrating evolutionary and regulatory information with a multispecies approach implicates genes and pathways in obsessive-compulsive disorder. Nat. Commun 8, 774 (2017). doi: 10.1038/s41467-017-00831-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dodman NH et al. , A canine chromosome 7 locus confers compulsive disorder susceptibility. Mol. Psychiatry 15, 8–10 (2010). doi: 10.1038/mp.2009.111 [DOI] [PubMed] [Google Scholar]
- 27.Overall KL, Natural animal models of human psychiatric conditions: Assessment of mechanism and validity. Prog. Neuropsychopharmacol. Biol. Psychiatry 24, 727–776 (2000). doi: 10.1016/S0278-5846(00)00104-4 [DOI] [PubMed] [Google Scholar]
- 28.Sarviaho R et al. , A novel genomic region on chromosome 11 associated with fearfulness in dogs. Transl. Psychiatry 10, 169 (2020). doi: 10.1038/s41398-020-0849-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sarviaho R et al. , Two novel genomic regions associated with fearfulness in dogs overlap human neuropsychiatric loci. Transl. Psychiatry 9, 18 (2019). doi: 10.1038/s41398-018-0361-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ilska J et al. , Genetic characterization of dog personality traits. Genetics 206, 1101–1111 (2017). doi: 10.1534/genetics.116.192674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tang R et al. , Candidate genes and functional noncoding variants identified in a canine model of obsessive-compulsive disorder. Genome Biol. 15, R25 (2014). doi: 10.1186/gb-2014-15-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.DeBoever C et al. , Assessing digital phenotyping to enhance genetic studies of human diseases. Am. J. Hum. Genet 106, 611–622 (2020). doi: 10.1016/j.ajhg.2020.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hyde CL et al. , Identification of 15 genetic loci associated with risk of major depression in individuals of European descent. Nat. Genet 48, 1031–1036 (2016). doi: 10.1038/ng.3623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wright HF, Mills DS, Pollux P, Development and validation of a psychometric tool for assessing impulsivity in the domestic dog, Canis familiaris. Int. J. Comp. Psychol (2011). doi: 10.1016/j.physbeh.2011.09.019; 21986321 [DOI] [Google Scholar]
- 35.Salvin HE, McGreevy PD, Sachdev PS, Valenzuela MJ, The canine cognitive dysfunction rating scale (CCDR): A data-driven and ecologically relevant assessment tool. Vet. J 188, 331–336 (2011). doi: 10.1016/j.tvjl.2010.05.014 [DOI] [PubMed] [Google Scholar]
- 36.Lavan RP, Development and validation of a survey for quality of life assessment by owners of healthy dogs. Vet. J 197, 578–582 (2013). doi: 10.1016/j.tvjl.2013.03.021 [DOI] [PubMed] [Google Scholar]
- 37.Jones AC, Development and Validation of a Dog Personality Questionnaire (Univ. Texas, 2008). [Google Scholar]
- 38.Sutter NB, Mosher DS, Gray MM, Ostrander EA, Morphometrics within dog breeds are highly reproducible and dispute Rensch’s rule. Mamm. Genome 19, 713–723 (2008). doi: 10.1007/s00335-008-9153-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yordy J et al. , Body size, inbreeding, and lifespan in domestic dogs. Conserv. Genet 21, 137–148 (2020). doi: 10.1007/s10592-019-01240-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Plassais J et al. , Whole genome sequencing of canids reveals genomic regions under selection and variants influencing morphology. Nat. Commun 10, 1489 (2019). doi: 10.1038/s41467-019-09373-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parker HG et al. , Genomic analyses reveal the influence of geographic origin, migration, and hybridization on modern dog breed development. Cell Rep. 19, 697–708 (2017). doi: 10.1016/j.celrep.2017.03.079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martin AR et al. , Low-coverage sequencing cost-effectively detects known and novel variation in underrepresented populations. Am. J. Hum. Genet 108, 656–668 (2021). doi: 10.1016/j.ajhg.2021.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rubinacci S, Ribeiro DM, Hofmeister RJ, Delaneau O, Efficient phasing and imputation of low-coverage sequencing data using large reference panels. Nat. Genet 53, 120–126 (2021). doi: 10.1038/s41588-020-00756-0 [DOI] [PubMed] [Google Scholar]
- 44.Buckley RM et al. , Best practices for analyzing imputed genotypes from low-pass sequencing in dogs. Mamm. Genome 33, 213–229 (2021). doi: 10.1007/s00335-021-09914-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li JH, Mazur CA, Berisa T, Pickrell JK, Low-pass sequencing increases the power of GWAS and decreases measurement error of polygenic risk scores compared to genotyping arrays. Genome Res. 31, 529–537 (2021). doi: 10.1101/gr.266486.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wasik K et al. , Comparing low-pass sequencing and genotyping for trait mapping in pharmacogenetics. BMC Genomics 22, 197 (2021). doi: 10.1186/s12864-021-07508-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alexander DH, Lange K, Enhancements to the ADMIXTURE algorithm for individual ancestry estimation. BMC Bioinformatics 12, 246 (2011). doi: 10.1186/1471-2105-12-246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang J et al. , Common SNPs explain a large proportion of the heritability for human height. Nat. Genet 42, 565–569 (2010). doi: 10.1038/ng.608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Athanasiadis G et al. , Estimating narrow-sense heritability using family data from admixed populations. Heredity 124, 751–762 (2020). doi: 10.1038/s41437-020-0311-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Luo S et al. , Genome-wide association study of serum metabolites in the African American Study of Kidney Disease and Hypertension. Kidney Int. 100, 430–439 (2021). doi: 10.1016/j.kint.2021.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bakeman R, Recommended effect size statistics for repeated measures designs. Behav. Res. Methods 37, 379–384 (2005). doi: 10.3758/BF03192707 [DOI] [PubMed] [Google Scholar]
- 52.American Kennel Club, Dog breeds; https://www.akc.org/dog-breeds/.
- 53.American Kennel Club, The Complete Dog Book (Ballantine Books, 2006). [Google Scholar]
- 54.Wells DL, Morrison DJ, Hepper PG, The effect of priming on perceptions of dog breed traits. Anthrozoos 25, 369–377 (2012). doi: 10.2752/175303712X13403555186370 [DOI] [Google Scholar]
- 55.Gunter LM, Barber RT, Wynne CDL, What’s in a name? Effect of breed perceptions & labeling on attractiveness, adoptions & length of stay for pit-bull-type dogs. PLOS ONE 11, e0146857 (2016). doi: 10.1371/journal.pone.0146857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang J, Lee SH, Goddard ME, Visscher PM, GCTA: A tool for genome-wide complex trait analysis. Am. J. Hum. Genet 88, 76–82 (2011). doi: 10.1016/j.ajhg.2010.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pirinen M, Donnelly P, Spencer CCA, Efficient computation with a linear mixed model on large-scale data sets with applications to genetic studies. Ann. Appl. Stat 7, 369–390 (2013). doi: 10.1214/12-AOAS586 [DOI] [Google Scholar]
- 58.Cadieu E et al. , Coat variation in the domestic dog is governed by variants in three genes. Science 326, 150–153 (2009). doi: 10.1126/science.1177808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brancalion L et al. , Roan, ticked and clear coat patterns in the canine are associated with three haplotypes near usherin on CFA38. Anim. Genet 52, 198–207 (2021). doi: 10.1111/age.13040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Slavney AJ et al. , Five genetic variants explain over 70% of hair coat pheomelanin intensity variation in purebred and mixed breed domestic dogs. PLOS ONE 16, e0250579 (2021). doi: 10.1371/journal.pone.0250579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Candille SI et al. , A β-defensin mutation causes black coat color in domestic dogs. Science 318, 1418–1423 (2007). doi: 10.1126/science.1147880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Oguro-Okano M, Honda M, Yamazaki K, Okano K, Mutations in the melanocortin 1 receptor, β-defensin103 and agouti signaling protein genes, and their association with coat color phenotypes in Akita-inu dogs. J. Vet. Med. Sci 73, 853–858 (2011). doi: 10.1292/jvms.10-0439 [DOI] [PubMed] [Google Scholar]
- 63.Kerns JA et al. , Linkage and segregation analysis of black and brindle coat color in domestic dogs. Genetics 176, 1679–1689 (2007). doi: 10.1534/genetics.107.074237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chase K et al. , Genetic basis for systems of skeletal quantitative traits: Principal component analysis of the canid skeleton. Proc. Natl. Acad. Sci. U.S.A 99, 9930–9935 (2002). doi: 10.1073/pnas.152333099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chase K, Carrier DR, Adler FR, Ostrander EA, Lark KG, Interaction between the X chromosome and an autosome regulates size sexual dimorphism in Portuguese water dogs. Genome Res. 15, 1820–1824 (2005). doi: 10.1101/gr.3712705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sutter NB et al. , A single IGF1 allele is a major determinant of small size in dogs. Science 316, 112–115 (2007). doi: 10.1126/science.1137045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Parker HG et al. , An expressed fgf4 retrogene is associated with breed-defining chondrodysplasia in domestic dogs. Science 325, 995–998 (2009). doi: 10.1126/science.1173275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hoopes BC, Rimbault M, Liebers D, Ostrander EA, Sutter NB, The insulin-like growth factor 1 receptor (IGF1R) contributes to reduced size in dogs. Mamm. Genome 23, 780–790 (2012). doi: 10.1007/s00335-012-9417-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rimbault M et al. , Derived variants at six genes explain nearly half of size reduction in dog breeds. Genome Res. 23, 1985–1995 (2013). doi: 10.1101/gr.157339.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Brown EA et al. , FGF4 retrogene on CFA12 is responsible for chondrodystrophy and intervertebral disc disease in dogs. Proc. Natl. Acad. Sci. U.S.A 114, 11476–11481 (2017). doi: 10.1073/pnas.1709082114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Webster MT et al. , Linked genetic variants on chromosome 10 control ear morphology and body mass among dog breeds. BMC Genomics 16, 474 (2015). doi: 10.1186/s12864-015-1702-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Okada T et al. , Anderson’s disease/chylomicron retention disease in a Japanese patient with uniparental disomy 7 and a normal SAR1B gene protein coding sequence. Orphanet J. Rare Dis 6, 78 (2011). doi: 10.1186/1750-1172-6-78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Levy E et al. , Sar1b transgenic male mice are more susceptible to high-fat diet-induced obesity, insulin insensitivity and intestinal chylomicron overproduction. J. Nutr. Biochem 25, 540–548 (2014). doi: 10.1016/j.jnutbio.2014.01.004 [DOI] [PubMed] [Google Scholar]
- 74.Ajeawung NF et al. , Mutations in ANAPC1, encoding a scaffold subunit of the anaphase-promoting complex, cause Rothmund-Thomson syndrome type 1. Am. J. Hum. Genet 105, 625–630 (2019). doi: 10.1016/j.ajhg.2019.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gumienny TL et al. , Caenorhabditis elegans SMA-10/LRIG is a conserved transmembrane protein that enhances bone morphogenetic protein signaling. PLOS Genet. 6, e1000963 (2010). doi: 10.1371/journal.pgen.1000963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen Z, Boehnke M, Wen X, Mukherjee B, Revisiting the genome-wide significance threshold for common variant GWAS. G3 11, jkaa056 (2021). doi: 10.1093/g3journal/jkaa056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lee JJ et al. , Gene discovery and polygenic prediction from a genome-wide association study of educational attainment in 1.1 million individuals. Nat. Genet 50, 1112–1121 (2018). doi: 10.1038/s41588-018-0147-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Savage JE et al. , Genome-wide association meta-analysis in 269,867 individuals identifies new genetic and functional links to intelligence. Nat. Genet 50, 912–919 (2018). doi: 10.1038/s41588-018-0152-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lam M et al. , Pleiotropic meta-analysis of cognition, education, and schizophrenia differentiates roles of early neurodevelopmental and adult synaptic pathways. Am. J. Hum. Genet 105, 334–350 (2019). doi: 10.1016/j.ajhg.2019.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Smith RS et al. , Sodium channel SCN3A (NaV1.3) regulation of human cerebral cortical folding and oral motor development. Neuron 99, 905–913.e7 (2018). doi: 10.1016/j.neuron.2018.07.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Widmer YF, Bilican A, Bruggmann R, Sprecher SG, Regulators of long-term memory revealed by mushroom body-specific gene expression profiling in Drosophila melanogaster. Genetics 209, 1167–1181 (2018). doi: 10.1534/genetics.118.301106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pelé M, Tiret L, Kessler J-L, Blot S, Panthier J-J, SINE exonic insertion in the PTPLA gene leads to multiple splicing defects and segregates with the autosomal recessive centronuclear myopathy in dogs. Hum. Mol. Genet 14, 1417–1427 (2005). doi: 10.1093/hmg/ddi151 [DOI] [PubMed] [Google Scholar]
- 83.Karlsson EK et al. , Genome-wide analyses implicate 33 loci in heritable dog osteosarcoma, including regulatory variants near CDKN2A/B. Genome Biol. 14, R132 (2013). doi: 10.1186/gb-2013-14-12-r132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Boyko AR et al. , A simple genetic architecture underlies morphological variation in dogs. PLOS Biol. 8, e1000451 (2010). doi: 10.1371/journal.pbio.1000451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lonsdale J et al. , The Genotype-Tissue Expression (GTEx) project. Nat. Genet 45, 580–585 (2013). doi: 10.1038/ng.2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.MacLean EL, Snyder-Mackler N, vonHoldt BM, Serpell JA, Highly heritable and functionally relevant breed differences in dog behaviour. Proc. Biol. Sci 286, 20190716 (2019). doi: 10.1098/rspb.2019.0716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Privitera AP et al. , OCDB: A database collecting genes, miRNAs and drugs for obsessive-compulsive disorder. Database 2015, bav069 (2015). doi: 10.1093/database/bav069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Abrahams BS et al. , SFARI Gene 2.0: A community-driven knowledgebase for the autism spectrum disorders (ASDs). Mol. Autism 4, 36 (2013). doi: 10.1186/2040-2392-4-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Schizophrenia Working Group of the Psychiatric Genomics Consortium, Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–427 (2014). doi: 10.1038/nature13595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pardiñas AF et al. , Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nat. Genet 50, 381–389 (2018). doi: 10.1038/s41588-018-0059-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.de Leeuw CA, Mooij JM, Heskes T, Posthuma D, MAGMA: Generalized gene-set analysis of GWAS data. PLOS Comput. Biol 11, e1004219 (2015). doi: 10.1371/journal.pcbi.1004219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yi X et al. , Sequencing of 50 human exomes reveals adaptation to high altitude. Science 329, 75–78 (2010). doi: 10.1126/science.1190371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.vonHoldt B, Fan Z, Ortega-Del Vecchyo D, Wayne RK, EPAS1 variants in high altitude Tibetan wolves were selectively introgressed into highland dogs. PeerJ 5, e3522 (2017). doi: 10.7717/peerj.3522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sinding MS et al. , Arctic-adapted dogs emerged at the Pleistocene-Holocene transition. Science 368, 1495–1499 (2020). doi: 10.1126/science.aaz8599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Deane-Coe PE, Chu ET, Slavney A, Boyko AR, Sams AJ, Direct-to-consumer DNA testing of 6,000 dogs reveals 98.6-kb duplication associated with blue eyes and heterochromia in Siberian Huskies. PLOS Genet. 14, e1007648 (2018). doi: 10.1371/journal.pgen.1007648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sanjak JS, Sidorenko J, Robinson MR, Thornton KR, Visscher PM, Evidence of directional and stabilizing selection in contemporary humans. Proc. Natl. Acad. Sci. U.S.A 115, 151–156 (2018). doi: 10.1073/pnas.1707227114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sabeti PC et al. , Positive natural selection in the human lineage. Science 312, 1614–1620 (2006). doi: 10.1126/science.1124309 [DOI] [PubMed] [Google Scholar]
- 98.Atkinson EG et al. , Tractor uses local ancestry to enable the inclusion of admixed individuals in GWAS and to boost power. Nat. Genet 53, 195–204 (2021). doi: 10.1038/s41588-020-00766-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Barton N, Hermisson J, Nordborg M, Population genetics: Why structure matters. eLife 8, e45380 (2019). doi: 10.7554/eLife.45380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wojcik GL et al. , Genetic analyses of diverse populations improves discovery for complex traits. Nature 570, 514–518 (2019). doi: 10.1038/s41586-019-1310-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Moore JE et al. , Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature 583, 699–710 (2020). doi: 10.1038/s41586-020-2493-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Megquier K et al. , BarkBase: Epigenomic annotation of canine genomes. Genes 10, 433 (2019). doi: 10.3390/genes10060433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zoonomia Consortium A comparative genomics multitool for scientific discovery and conservation. Nature 587, 240–245 (2020). doi: 10.1038/s41586-020-2876-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Morrill K et al. , Associated data files and scripts for “Ancestry-inclusive dog genomics challenges popular breed stereotypes”. Dryad (2022); doi: 10.5061/dryad.g4f4qrfr0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Morrill K et al. , Associated data files and scripts for “Ancestry-inclusive dog genomics challenges popular breed stereotypes”. Zenodo (2022); doi: 10.5281/zenodo.5808330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Morrill K, Darwin’s Ark - Data release 2022. Terra (2022); https://app.terra.bio/#workspaces/darwins-ark/Darwins%20Ark%20-%20Data%20Release%202022. [Google Scholar]
- 107.Morrill K et al. , Additional figures for “Ancestry-inclusive dog genomics challenges popular breed stereotypes”. FigShare (2022); doi: 10.6084/m9.figshare.16608793 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing data have been deposited into the National Center for Biotechnology Information (NCBI) Sequence Read Archive, including high-coverage sequencing reads for the Mendel’s Mutts dataset in BioProject PRJNA683923 and low-coverage sequencing reads deposited to BioProject PRJNA675863. The Broad-UMass Canid Variants in variant call format (VCF) is available publicly via FTP at https://data.broadinstitute.org/DogData/. All survey and genetic data from dogs of the Darwin’s Ark and Mendel’s Mutts cohorts, data from the MuttMix survey, and scripts used in analyses are archived in Dryad (104) and Zenodo (105). The genome-wide association summary statistics and plots are shared on Terra (106) and Figshare (107), respectively.


