Abstract
Yersinia enterocolitica strains of biotype 1A are increasingly being recognized as etiological agents of gastroenteritis. However, the mechanisms by which these bacteria cause disease differ from those of highly invasive, virulence plasmid-bearing Y. enterocolitica strains and are poorly understood. We have investigated several biotype 1A strains of diverse origin for their ability to resist killing by professional phagocytes. All strains were rapidly killed by polymorphonuclear leukocytes but persisted within macrophages (activated with gamma interferon) to a significantly greater extent (survival = 40.5% ± 17.4%) than did Escherichia coli HB101 (9.3% ± 0.7%; P = 0.0001). Strains isolated from symptomatic patients were significantly more resistant to killing by macrophages (survival = 48.9% ± 19.5%) than were strains obtained from food or the environment (survival = 32.1% ± 10.3%; P = 0.04). Some strains which had been ingested by macrophages or HEp-2 epithelial cells showed a tendency to reemerge into the tissue culture medium over a period lasting several hours. This phenomenon, which we termed “escape,” was observed in 14 of 15 strains of clinical origin but in only 3 of 12 nonclinical isolates (P = 0.001). The capacity of bacteria to escape from cells was not directly related to their invasive ability. To determine if escape was due to host cell lysis, we used a variety of techniques, including lactate dehydrogenase release, trypan blue exclusion, and examination of infected cells by light and electron microscopy, to measure cell viability and lysis. These studies established that biotype 1A Y. enterocolitica strains were able to escape from macrophages or epithelial cells without causing detectable cytolysis, suggesting that escape was achieved by a process resembling exocytosis. The observations that biotype 1A Y. enterocolitica strains of clinical origin are significantly more resistant to killing by macrophages and significantly more likely to escape from host cells than are strains of nonclinical origin suggest that these properties may account for the virulence of these bacteria.
A fundamental attribute of some invasive bacterial pathogens is the ability to avoid destruction by macrophages and polymorphonuclear leukocytes (PMNs). The strategies used by various gastrointestinal pathogens to evade the bactericidal processes of these cells differ. For example, Shigella flexneri is able to escape from the phagosome of macrophages whereas Salmonella enterica serovars survive and replicate within spacious, acidic phagosomes (1, 2, 12). Both species may eventually kill murine macrophages by inducing apoptosis (24, 46).
Yersinia enterocolitica evades the host immune system via proteins termed Yops, which are encoded by an approximately 70-kb virulence plasmid (pYV). These proteins include YopH (a protein tyrosine phosphatase) and YopE (a cytotoxin), which allow the bacteria to resist phagocytosis, and YopP, which induces apoptosis of macrophages and suppresses the release of tumor necrosis factor alpha from these cells (5, 8, 22, 37, 44). YadA, another pYV-encoded protein, protects the bacteria against destruction by complement (11). The cumulative effect of these proteins enables Y. enterocolitica to persist within the extracellular compartment, although some pYV-bearing Y. enterocolitica organisms may be ingested by macrophages and PMNs (36, 38, 41).
Apart from pYV, many strains of Y. enterocolitica also carry a number of chromosomally encoded virulence factors, such as invasin, which promotes the uptake of bacteria by a variety of cells, and Ail, which promotes bacterial attachment to and invasion of epithelial cells in vitro and contributes to resistance to complement-mediated killing (7, 20, 32). In the absence of pYV, invasin induces the phagocytosis of Y. enterocolitica by macrophages but does not influence the ability of the bacteria to survive killing (13). Invasin and Ail are found only in Y. enterocolitica strains of biotypes 1B and 2 through 5, which are highly invasive in vitro and typically carry pYV. By contrast, biotype 1A strains generally lack invasin, Ail, and pYV and are far less invasive in vitro (16, 21, 31, 35, 38, 41). As a consequence, biotype 1A Y. enterocolitica was once believed to be nonpathogenic. Nevertheless, several epidemiological investigations, including one prospective case-control study, have found strains of biotype 1A to be significantly associated with diarrhea (6, 25, 28, 30, 33, 34, 39, 40). In addition, the illness experienced by patients infected with biotype 1A Y. enterocolitica is indistinguishable from that caused by pYV-bearing strains (9).
We recently reported that biotype 1A strains of Y. enterocolitica isolated from symptomatic patients are able to invade epithelial cells and to colonize the intestinal tracts of perorally inoculated mice to a significantly greater extent than are isolates obtained from the environment or food (16). These observations suggest that biotype 1A strains associated with disease may be better equipped than nonclinical strains to evade the innate immune system of the host, possibly by avoiding removal by phagocytes. The aim of this study was to investigate the interactions between biotype 1A Y. enterocolitica and professional phagocytes and thus to elucidate a possible mechanism(s) by which these bacteria persist within their hosts.
MATERIALS AND METHODS
Bacteria.
Strains of Y. enterocolitica biotype 1A were chosen to represent a variety of O serogroups isolated from widespread geographic areas. Strains of clinical origin were originally obtained from the feces of humans who displayed symptoms consistent with intestinal yersiniosis. These strains, designated clinical isolates, were presumed to be virulent by virtue of their association with disease and the results of previous studies which showed them to invade epithelial cells in significantly greater numbers than nonclinical strains (16). Clinical isolates were compared with strains of nonclinical origin, which included strains obtained from milk (five isolates), other foods (four isolates), water (two isolates), and soil (one isolate). All bacteria were maintained at −70°C in brain heart infusion (BHI) broth (Oxoid, Basingstoke, England) containing 30% (vol/vol) glycerol or for short periods on BHI agar plates at 4°C. Y. enterocolitica W22703 (biotype 2, serogroup O:9), which carries pYV, and its pYV-cured derivative, W22703c, both of which express inv and ail, were used as controls (35). Y. enterocolitica JP273 and YE2v were obtained from V. L. Miller. These strains were derived from Y. enterocolitica 8081 (biotype 1B, serogroup O:8) and contain insertional mutations in the inv and ail genes, respectively (29, 45). Derivatives of these strains cured of pYV were obtained by passaging the bacteria at 37°C on BHI agar containing 20 mM sodium oxalate and 20 mM MgCl2 and are referred to as JP273c and YE2c, respectively. Escherichia coli HB101 was used as a negative control in all assays. E. coli HB101(pVM101) and HB101(pVM103) express the inv and ail genes of Y. enterocolitica, respectively (20). For all assays, strains of Y. enterocolitica were cultivated in BHI broth with shaking at 28°C whereas strains of E. coli were grown in the same medium at 37°C.
Isolation of PMNs and macrophages.
PMNs were obtained from the peripheral blood of healthy adult volunteers by dextran sedimentation and Ficoll-Hypaque centrifugation (4). The PMNs were resuspended in a modified Krebs-Ringer buffer at approximately 2 × 107 cells per ml (38). Cell viability was >98% as determined by trypan blue exclusion.
Bone marrow-derived murine macrophages were prepared from the femurs of BALB/c mice as described previously (13, 43). Macrophages were cultured until semiconfluent in RPMI 1640 tissue culture medium (Flow Laboratories, McLean, Va.) supplemented with 15% heat-inactivated (56°C for 30 min) fetal calf serum (HIFCS; CSL, Melbourne, Australia), 30% L-cell conditioned medium (as a source of colony-stimulating factor 1), 3 mM glutamine, 50 μg of ampicillin (CSL) per ml, and 20 mM HEPES (Boehringer, Mannheim, Germany).
Epithelial cell cultures.
HEp-2 cells were cultured until almost confluent in 24-mm-diameter plastic wells (Nunc, Roskilde, Denmark) containing minimal Eagle’s medium (MEM) supplemented with 5% HIFCS and 20 mM HEPES. T84 intestinal epithelial cells were cultured and maintained in a mixture of Dulbecco’s modified Eagle’s medium and Ham’s F-12 medium supplemented with 15 mM HEPES and 5% HIFCS as described previously (14, 19). To induce cell differentiation, the cells were cultured to confluency for 7 days on 3-μm-pore-size polycarbonate membranes in 35-mm-diameter plastic wells (Nunc). The confluency of the monolayer was determined by measuring the transepithelial resistance across the monolayer with an Evom epithelial voltohmmeter (World Precision Instruments, Sarasota, Fla.). Transepithelial resistance was calculated by multiplying the measured resistance value by the area of the filter (4.2 cm2). The transepithelial resistance of cell monolayers before infection was 6,693 ± 439 ohms/cm2. Before infection with bacteria, the cells were washed twice with phosphate-buffered saline (PBS) and provided with fresh culture medium.
Assay for bactericidal activity of PMNs.
Bacteria were grown overnight with shaking in BHI broth, washed three times with PBS, and opsonized by incubation in 5% (vol/vol) pooled, heat-inactivated human serum with gentle rocking at 37°C for 15 min. The bacteria were then washed twice with modified Krebs-Ringer buffer and resuspended in the same buffer at approximately 108 CFU/ml. The assay for bacterial killing by PMNs was performed as described previously (4). Briefly, approximately 2 × 107 bacteria per ml and 107 PMNs per ml were added to wells of a microtiter plate in a final volume of 200 μl and incubated with shaking at 37°C. At predetermined times, 20-μl aliquots of the cell suspension were added to 180 μl of 0.1% (wt/vol) digitonin (Sigma, St. Louis, Mo.) to lyse the PMNs. The number of viable bacteria in the resultant suspension was then determined by measurement of growth on BHI agar. Parallel control experiments, using wells which did not contain PMNs, were performed to assess the viability of the bacteria in serum. The results were expressed as the percentage of the bacterial inoculum which resisted killing and were the mean of at least three separate determinations. The assay does not distinguish between resistance to phagocytosis and survival of ingested bacteria.
Bacterial survival within macrophages.
Quantitative assays of the ability of bacteria to survive killing by macrophages were performed as described previously (13). For these studies, a semiconfluent monolayer of bone marrow-derived macrophages was primed with 100 U of murine gamma interferon (Boehringer Mannheim) per ml for 24 h. The cells were then washed twice with PBS and maintained in fresh RPMI containing 15% HIFCS, 3 mM glutamine, and 20 mM HEPES. Approximately 2 × 107 CFU of each test strain was added to the macrophages at a multiplicity of infection of 10 bacteria per macrophage and incubated for 1 h at 37°C. The cells were then washed twice with PBS to remove nonadherent bacteria, and fresh tissue culture medium containing 100 μg of gentamicin per ml was added for 30 min to kill extracellular bacteria. The macrophages were then washed twice with PBS and lysed with 0.1% digitonin, so that the number of bacteria released from the macrophages could be determined. This value was designated the initial count (T = 0 h). In a parallel series of wells, the medium containing 100 μg of gentamicin per ml was replaced with medium containing 20 μg of gentamicin per ml and 100 U of gamma interferon per ml, after which the bacteria and tissue culture cells were incubated for 24 h. The macrophages were then washed three times with PBS and lysed with digitonin to release intracellular bacteria for enumeration as described above. This value was designated the final count (T = 24 h).
Assay for bacterial escape from macrophages and epithelial cells.
Bone marrow-derived macrophages, HEp-2, and T84 epithelial cells were cultured as described above. Bacteria were incubated with macrophages or epithelial cells for 1 or 3 h, respectively, to permit their internalization (for epithelial cells, this time was designated T = 3 h). The cells were then washed twice with PBS, and fresh tissue culture medium containing 100 μg of gentamicin per ml was added for 90 min to kill extracellular bacteria (for epithelial cells, this was T = 4.5 h). Gentamicin was removed by two washes with PBS, and the cells were reincubated in fresh tissue culture medium. The ability of bacteria to escape from cells was ascertained by the sampling culture medium and enumerating the bacteria 18 h after the removal of gentamicin (for epithelial cells, this was T = 22.5 h).
Analysis of cytotoxicity and viability of host cells.
Macrophages or HEp-2 epithelial cells were infected with bacteria as in the assay for bacterial escape, but to prevent extracellular bacterial growth, 20 μg of gentamicin per ml was included during the 18-h incubation period.
(i) Assay for release of LDH.
Tissue culture medium was removed from cells, and the lactate dehydrogenase (LDH) activity in these samples was determined colorimetrically by using Vitros LDH slides (Ortho-Clinical Diagnostics, New York, N.Y.) and a Vitros 550xrc general chemical analyzer as recommended by the manufacturer (Ortho-Clinical Diagnostics). The percentage of dead cells was calculated from the following formula:
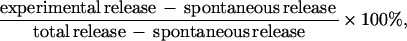 |
where spontaneous release was determined by measuring the activity of LDH in culture medium containing cells but no bacteria and total release was determined by lysing uninfected cells with 0.1% digitonin. Four wells were pooled for each strain, and each assay was performed in duplicate on at least three separate occasions. Pilot studies showed that Y. enterocolitica did not produce measurable quantities of LDH under the conditions used for this assay.
(ii) Trypan blue exclusion.
The integrity of macrophages and HEp-2 cells was also examined by their ability to exclude trypan blue. Cells were overlaid with 0.1% trypan blue for 5 min, after which at least 1,000 cells per sample were examined visually with an inverted microscope at ×400 magnification. The results are the mean of at least three separate experiments.
(iii) LIVE/DEAD assay.
The biochemical activity of macrophages was examined by using the LIVE/DEAD viability/cytotoxicity kit for animal cells (Molecular Probes, Eugene, Oreg.). Cell viability was determined by fluorescence microscopy under the conditions recommended by the manufacturer.
Electron microscopic examination of bacterium-macrophage interactions.
For transmission electron microscopy, macrophages were grown to semiconfluency in 50-mm-diameter plastic petri dishes (Becton Dickinson, Plymouth, England), after which approximately 10 bacteria per macrophage were added for 1 h at 37°C. The cells were then washed twice with PBS and fixed in 2.5% glutaraldehyde for 1 h. The cell monolayer was removed with a rubber policeman and centrifuged to form a pellet. Cells were postfixed in 2.5% osmium tetroxide for 1 h, dehydrated through a graded acetone series, and embedded in Epon-Araldite epoxy resin. Thin sections were cut and stained with 10% uranyl acetate and 2.5% lead citrate before being viewed under a Philips CM12 electron microscope at 60 kV. In some experiments, macrophages were incubated with bacteria for 1 h, washed twice with PBS, and then incubated for 18 h in fresh tissue culture medium containing 20 μg of gentamicin per ml. The cells were then washed with PBS and processed as describe above.
Statistical analysis.
Data were analyzed by the two-tailed Fisher exact test and Student’s t test as appropriate. A critical P value of 0.05 was used for all analyses.
RESULTS
Killing of Y. enterocolitica by PMNs and macrophages.
PMNs from healthy volunteers effectively killed four different biotype 1A strains, since only 20.7% ± 4.6% of bacteria (mean ± standard deviation of three independent experiments) survived a 60-min incubation with these cells (Table 1). E. coli HB101 was killed to an even greater extent (survival = 10.1% ± 2.1%; P = 0.001), suggesting that biotype 1A strains may be partly resistant to the bactericidal effects of these cells. No differences in susceptibility to killing between clinical and nonclinical strains or between the biotype 1A strains and Y. enterocolitica W22703c were observed. By contrast, Y. enterocolitica W22703(pYV) was able to avoid killing by PMNs (survival = 90.1% ± 8.3%), presumably due to pYV-encoded proteins, in particular YadA, which enable the bacteria to resist phagocytosis and inhibit the respiratory burst of PMNs and macrophages (10, 17).
TABLE 1.
Ability of Y. enterocolitica biotype 1A to evade killing by PMNs and macrophages
| Strain | % Survival of bacteria when incubated with:
|
|
|---|---|---|
| PMNsa | Macrophagesb | |
| E. coli | ||
| HB101 | 10.1 ± 2.1 | 9.3 ± 0.7c |
| Y. enterocolitica | ||
| Biotype 1A (clinical) | 20.2 ± 4.7d | 48.9 ± 19.5ce |
| Biotype 1A (nonclinical) | 21.6 ± 5.1d | 32.1 ± 10.3ce |
| W22703 (pYV+) | 90.1 ± 8.3 | NDf |
| W22703c (pYV−) | 13.3 ± 5.7 | 37.9 ± 12.5 |
The percentage of the bacterial inoculum which survived killing by human PMNs after incubation for 60 min. Results are the means ± standard deviations of at least three separate determinations.
The percentage of viable intracellular bacteria which survived incubation within murine bone marrow-derived macrophages for 24 h. The method for determining bacterial survival within macrophages is provided in Materials and Methods. Results are the means ± standard deviations of at least three separate determinations.
The difference between E. coli HB101 and strains of biotype 1A (clinical and nonclinical strains) is highly significant (P < 0.001).
Four biotype 1A strains were examined, two clinical and two nonclinical.
Sixteen strains were examined, eight clinical and eight nonclinical. The difference between clinical and nonclinical strains of biotype 1A is significant (P = 0.04).
ND, not determined due to cytolethal properties of this strain (17).
To determine the susceptibility of Y. enterocolitica biotype 1A to killing by macrophages, we incubated clinical and nonclinical isolates with macrophages that had been activated with gamma interferon. In a pilot study, we compared three sources of macrophages in this assay, namely, human peripheral blood monocyte-derived macrophages, murine peritoneal macrophages, and murine bone marrow-derived macrophages, for their ability to kill Y. enterocolitica. Since the results obtained with these different types of cells were similar (data not shown), we decided to use bone marrow-derived macrophages for all subsequent experiments. In the definitive study, eight clinical and eight nonclinical biotype 1A strains were investigated on at least three separate occasions. The number of bacteria initially phagocytosed by macrophages varied to some extent, but the differences between clinical and nonclinical isolates and between biotype 1A strains and Y. enterocolitica W22703c were not significant. W22703(pYV) was not examined due to the cytolethal properties of pYV-bearing strains when incubated with eukaryotic cells for extended periods (17, 18). E. coli HB101 was ingested to a significantly lesser extent than were the Yersinia strains, suggesting that Y. enterocolitica biotype 1A may actively invade phagocytes in a manner similar to that for invasive Y. enterocolitica strains of other biotypes (13). Moreover, only 9.3% ± 0.7% of E. coli HB101 strains phagocytosed by macrophages were recovered after 24 h, indicating effective killing of these bacteria, compared with 40.5% ± 17.4% of all biotype 1A strains (P = 0.0001) (Table 1). In addition, clinical isolates (survival = 48.9% ± 19.5%) were significantly more resistant to killing by macrophages than were nonclinical isolates (32.1% ± 10.3%) (P = 0.04).
Escape of Y. enterocolitica from macrophages and epithelial cells.
While developing the assay for the bactericidal activity of macrophages, we examined the effect of omitting gentamicin from the maintenance medium after the extracellular bacteria had been destroyed. The results of this assay were uninterpretable due to proliferation of bacteria in the culture medium. Interestingly, only some strains of Y. enterocolitica were detected in the medium during the assay, and all of these strains were clinical isolates. In contrast, most environmental strains of Y. enterocolitica and E. coli HB101 were not detected in the culture medium even after incubation for 24 h in the absence of gentamicin. These differences were not due to the differences in the ability of strains to replicate in the culture medium or to differences in their susceptibility to gentamicin (data not shown).
As a result of these observations, we postulated that some strains of Y. enterocolitica may be able to escape from macrophages, and we investigated this further by examining a wider range of biotype 1A isolates in an assay specifically designed to demonstrate bacterial escape. A characteristic of the escape assay was that at its conclusion, the culture medium was either turbid or clear. This was reflected in bacterial counts, which exceeded 107 CFU/ml for strains that had escaped from macrophages but was generally below 100 CFU/ml for strains that could not escape. Using these criteria to determine the capacity of bacteria to escape from macrophages, we found that 14 of 15 clinical isolates but only 3 of 12 nonclinical isolates were able to escape from these cells (P = 0.001) (Table 2). Strain W22703c also escaped from macrophages, indicating that Y. enterocolitica strains of other biotypes also possess this phenotype. In contrast, E. coli HB101 remained within the macrophages for the duration of the assay.
TABLE 2.
Interaction of biotype 1A strains of various origins with epithelial cells and bone marrow-derived macrophages
| Biotype 1A strain | Serotype | Invasion of Chinese hamster ovary cells (%)ab | Survival in macrophages (%)ac | Ability of bacteria to escape fromad:
|
|
|---|---|---|---|---|---|
| HEp-2 cells | Macrophages | ||||
| Clinical | |||||
| 937 | O:6 | 0.24 | 37 | 2.3 × 109 | 9.7 × 109 |
| T83 | O:5 | 0.49 | 62 | 8.6 × 108 | 2.2 × 109 |
| 61525 | O:6,30 | 1.10 | 36 | 1.2 × 109 | 4.5 × 109 |
| A4183 | O:6 | 0.26 | 89 | 4.7 × 108 | 4.1 × 109 |
| CIDC6512 | O:41,27 | 0.20 | 37 | 5.3 × 107 | 6.2 × 107 |
| 1267 | O:7,8 | 0.25 | 57 | 6.5 × 107 | 5.9 × 107 |
| A3064 | O:6 | 0.70 | 42 | 7.1 × 108 | 6.1 × 108 |
| AC12 | O:7,13 | 0.47 | 31 | 5.4 × 107 | 5.1 × 107 |
| Y125 | O:5 | 0.15 | NDe | 2.4 × 107 | 4.3 × 107 |
| A3047 | O:6 | 0.34 | ND | 5.2 × 107 | 7.9 × 107 |
| A1379 | O:6 | 0.28 | ND | 3.9 × 107 | 5.9 × 107 |
| AC22 | O:9b | 0.36 | ND | 2.1 × 107 | 2.3 × 107 |
| 1350 | O:10 | 0.29 | ND | 6.5 × 108 | 2.7 × 109 |
| CIDC6516 | O:41,27 | 0.19 | ND | 4.7 × 108 | 8.1 × 108 |
| CIDC5015 | O:46,34 | 0.20 | ND | <10 | <10 |
| Nonclinical | |||||
| AM5 | O:6 | 0.03 | 27 | <10 | <10 |
| IP2222 | O:36 | 0.01 | 25 | <10 | <10 |
| Eco88 | O:29 | 0.02 | 30 | <10 | <10 |
| CIDC5569 | O:46,34 | 0.07 | ND | 4.2 × 107 | 7.1 × 107 |
| CIDC7134 | O:6,31 | 0.50 | 53 | 2.1 × 108 | 4.3 × 108 |
| IP1477 | O:6,31 | 0.02 | 29 | <10 | <10 |
| AM2 | O:5 | 0.02 | 20 | <10 | <10 |
| AM8 | O:13,15 | 0.06 | 41 | <10 | <10 |
| IP500 | O:10,34 | 0.11 | 32 | <10 | <10 |
| 9741 | O:5 | 0.07 | ND | <10 | <10 |
| CIDC2064 | O:5 | 0.37 | ND | 3.1 × 109 | 5.4 × 109 |
| But | O:5 | 0.08 | ND | <10 | <10 |
Results are the mean of at least three independent experiments performed in duplicate.
The invasion assay was performed as described previously (16). Results are expressed as the percentage of the original bacterial inoculum.
The method for determining bacterial survival in macrophages is given in Materials and Methods. Results are expressed as the percentage of the number of bacteria recovered at T = 24 h relative to the number of bacteria at T = 0 h.
Determined by using the escape assay as described in Materials and Methods. Results are expressed as the number of CFU per milliliter of culture medium after 18 h of incubation.
ND, not determined.
To determine if the ability of Y. enterocolitica to escape from macrophages was limited to these cells, we repeated the experiments with HEp-2 cells and found that strains capable of escaping from macrophages were also able to escape from HEp-2 cells and that strains which remained intracellular in macrophages also remained within HEp-2 cells (Table 2). Since HEp-2 cells are easier to cultivate and maintain than macrophages, HEp-2 cells were subsequently used in experiments to characterize the escape phenotype.
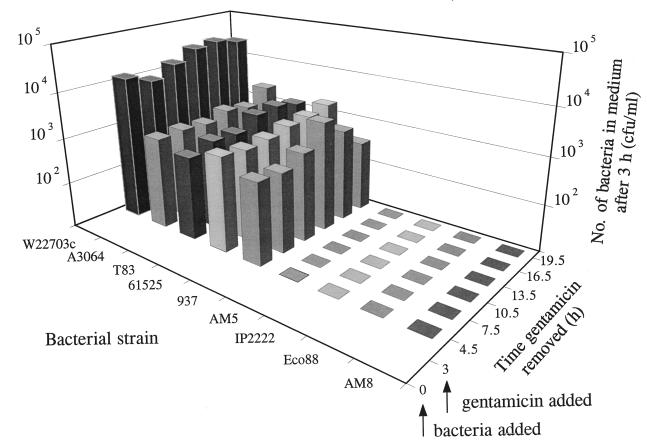
The time course of escape from cells was measured by infecting HEp-2 cells for 3 h with either Y. enterocolitica W22703c (biotype 2, serotype O:9, clinical), or eight strains of biotype 1A of clinical (four strains) or nonclinical (four strains) origin. The cells were then incubated for periods ranging from 90 min to 16.5 h in medium containing gentamicin (100 μg/ml). At the end of this incubation period, the gentamicin-containing medium was replaced with fresh medium without gentamicin and the cells were incubated for a further 3 h. This final incubation was included to allow the extracellular survival and enumeration of bacteria which had escaped from HEp-2 cells. The results showed that Y. enterocolitica W22703c and clinical isolates of biotype 1A escaped from HEp-2 cells throughout the assay whereas nonclinical isolates were not detected in significant numbers (Fig. 1).
FIG. 1.
Time course of escape of Y. enterocolitica W22703c (biotype 2), A3064 (clinical, biotype 1A), T83 (clinical, biotype 1A), 61525 (clinical, biotype 1A), 937 (clinical, biotype 1A), AM5 (nonclinical, biotype 1A), IP2222 (nonclinical, biotype 1A), Eco88 (nonclinical, biotype 1A), and AM8 (nonclinical, biotype 1A) from HEp-2 epithelial cells. Bacteria were incubated with HEp-2 cells for 3 h and then exposed to 100 μg of gentamicin per ml for periods ranging from 1.5 to 16.5 h. At the end of this period, gentamicin-containing medium was replaced with fresh medium without gentamicin. After a further 3-h incubation, the medium was removed for enumeration of viable bacteria. Data are the mean of three independent experiments.
A potential problem associated with the assay for bacterial escape is its high sensitivity. A small number of bacteria could enter the culture medium early during the course of the assay and proliferate extracellularly, giving a false impression of the ability of a strain to escape from cells. Therefore, in a variation of the assay, spectinomycin, a bacteriostatic antibiotic, was added to the culture medium during the 18-h incubation period to prevent replication of bacteria which have escaped from HEp-2 cells. The MIC of spectinomycin was determined for strains W22703c, 937, and AM5 (data not shown), and the escape assay was performed with spectinomycin at a concentration equivalent to twice the MIC. The assay, repeated on three occasions, revealed 9.0 × 104 ± 3.1 × 104 CFU of W22703c per ml and 3.2 × 102 ± 1.1 × 102 CFU of 937 per ml in the culture medium after 18 h. This was approximately 2 and 15% of the total number of intracellular bacteria at T = 3 h, respectively. By contrast, strain AM5 was not detected in the culture medium.
In a previous study, we showed that clinical isolates of Y. enterocolitica biotype 1A invaded HEp-2 cells to a significantly greater extent than did strains of nonclinical origin (16). Since clinical isolates are more likely to escape from cells than are nonclinical strains, it is conceivable that the assay for bacterial escape is biased toward strains that have a greater invasive capacity. To investigate this, we repeated the escape assay with various derivatives of Y. enterocolitica 8081 (an invasive Y. enterocolitica strain of biotype 1B, serotype O:8) and of E. coli HB101, which vary in their ability to invade HEp-2 epithelial cells. The results showed that only Y. enterocolitica strains were able to escape from these cells (Table 3). YE2c (ail mutant inv+) was isolated in similar numbers to 8081c (ail+ inv+), while JP273c (ail+ inv mutant) was isolated in smaller numbers, which corresponded to the smaller number of JP273c organisms that penetrated the HEp-2 cells initially. HB101(pVM101) (inv+), which invaded HEp-2 cells in large numbers, was not detected in the medium, confirming that the ability of a strain to escape from cells is not simply a reflection of its invasive capacity. The clear differences in the ability of the derivatives of Y. enterocolitica 8081c and E. coli HB101 to escape from cells indicates that this phenotype is chromosomally encoded and does not require the products of the inv or ail genes for its expression.
TABLE 3.
Ability of derivatives of Y. enterocolitica 8081c and E. coli HB101, which differ in their ability to invade HEp-2 cells, to escape from these cells
| Strain | No. of bacteria (CFU/ml) recovered from the medium after gentamicin had been absent for:
|
|||
|---|---|---|---|---|
| 1 h | 2 h | 3 h | 18 h | |
| E. colia | ||||
| HB101 | <10 | <10 | <10 | <10 |
| HB101(pVM103) | <10 | <10 | <10 | <10 |
| HB101(pVM101) | <10 | <10 | <10 | <10 |
| Y. enterocolitica | ||||
| W22703c | 6.2 × 103 | 1.2 × 104 | 3.1 × 104 | 3.5 × 109 |
| 8081c | 1.0 × 104 | 2.1 × 104 | 5.3 × 104 | 5.2 × 109 |
| YE2c | 1.1 × 104 | 2.0 × 104 | 4.9 × 104 | 2.8 × 109 |
| JP273c | 1.4 × 102 | 3.9 × 102 | 6.9 × 102 | 2.1 × 107 |
The number of E. coli bacteria ingested initially was 2.4 × 102, 1.9 × 104, and 3.7 × 106 CFU/ml for HB101, HB101(pVM103), and HB101(pVM101), respectively.
The number of Y. enterocolitica bacteria ingested initially was 4.6 × 106, 2.1 × 107, 2.0 × 107, and 3.2 × 103 CFU/ml for W22703c, 8081c, YE2c, and JP273c, respectively.
Interaction of Y. enterocolitica with T84 cell monolayers.
When T84 cells are cultivated as a confluent monolayer on porous membranes, they display distinct apical and basolateral poles, with a characteristic transmembrane electrical resistance (14). In this study, T84 cells were cultivated for 7 days to allow them to form a confluent monolayer as determined by measurement of the electrical resistance across the cells (6,693 ± 439 ohms/cm2). The cells were then infected with one of eight different biotype 1A strains (four clinical and four nonclinical strains) and biotype two strains W22703 and W22703c, as described for the escape assay. After 3 h, the bacteria were removed by gentle washing and by incubation with 100 μg of gentamicin per ml for 90 min. The electrical resistance across the monolayer was measured at T = 3, 4.5, 7.5, 10.5, and 22.5 h after the addition of bacteria. Monolayers infected with biotype 1A strains or W22703c showed similar electrical resistance to each other and to uninfected monolayers (7,639 ± 356 ohms/cm2 at T = 3 h) and at all time points throughout the assay (up to 22.5 h after addition of the bacteria). Nevertheless, between 1.5 × 102 and 3.0 × 103 CFU of clinical isolates of biotype 1A and W22703c per ml were isolated at T = 7.5 h from the medium bathing the apical and basolateral aspects of the monolayer, indicating that they were able to escape from T84 cells and pass through the monolayer without disrupting it. In contrast, four different nonclinical isolates of biotype 1A remained intracellular and were not isolated from either the apical or basolateral aspects of the T84 cells. These results indicate that Y. enterocolitica strains escape from T84 cells in a multidirectional manner and that the integrity of the monolayer is maintained during this process. By contrast, the pYV-bearing strain, W22703, caused a significant decrease in the potential difference of the monolayer (4,746 ohms/cm2 at T = 3 h), consistent with the known cytotoxic properties of this strains (17, 18).
Effect of Y. enterocolitica biotype 1A on host cell viability and cytoplasmic membrane integrity.
Some enteropathogenic bacteria escape from macrophages by inducing apoptosis or cytolysis (15, 23, 24). To determine if biotype 1A strains killed macrophages or epithelial cells, we infected these cells with two different escape-positive or escape-negative strains. Host cell viability and membrane integrity were measured by LDH release, trypan blue exclusion, and a commercial LIVE/DEAD assay.
LDH is a cytoplasmic enzyme that is released into the culture medium by eukaryotic cells which have lost membrane integrity, but the amount of LDH released from cells infected with biotype 1A Yersinia did not differ significantly from that released by the uninfected control, indicating that biotype 1A strains were not cytotoxic for macrophages or HEp-2 cells (Table 4). By contrast, incubation of cells with the pYV-bearing strain, W22703, resulted in significantly elevated LDH activity, presumably due to the cytotoxic effects of pYV-encoded proteins.
TABLE 4.
Membrane integrity of macrophages and HEp-2 epithelial cells incubated with strains of Y. enterocolitica
| Strain | LDH retention (%) bya:
|
Trypan blue exclusion (%) fromb:
|
||
|---|---|---|---|---|
| Macrophages | HEp-2 cells | Macrophages | HEp-2 cells | |
| W22703 (pYV+, biotype 2) | 84.6 ± 4.5 | 72.1 ± 4.4 | 78.7 ± 3.3 | 76.2 ± 4.8 |
| W22703c (pYV−, biotype 2) | 98.5 ± 0.9 | 97.8 ± 1.4 | 98.7 ± 1.3 | 99.1 ± 0.6 |
| 937 (clinical, biotype 1A) | 99.9 ± 0.1 | 99.9 ± 0.1 | 99.9 ± 0.1 | 99.9 ± 0.1 |
| AM5 (nonclinical, biotype 1A) | 99.9 ± 0.1 | 99.9 ± 0.1 | 99.9 ± 0.1 | 99.9 ± 0.1 |
The percentage of LDH retention by cells incubated with bacteria is expressed as the percentage of LDH retained by uninfected control cells. Results are expressed as mean ± standard deviation of at least three independent determinations. The assay and the equation for calculating the amount of LDH released are provided in Materials and Methods.
Data are the percentage of macrophages and HEp-2 cells incubated with bacteria that excluded trypan blue and are the mean ± standard deviation of at least three independent determinations.
The integrity of host cells was also investigated by examining their ability to exclude trypan blue. These studies showed that 99.9% ± 0.1% of macrophages or HEp-2 epithelial cells that had been incubated with biotype 1A Yersinia strains were morphologically normal and were able to exclude trypan blue (Table 4). Moreover, there was no difference between the number of dead HEp-2 cells at 3 h and at 22.5 h (data not shown), suggesting that the mechanism used by Y. enterocolitica biotype 1A and pYV-cured strains to escape from cells does not involve disruption of the cytoplasmic membrane. By contrast, a significantly greater proportion of host cells infected with the pYV-bearing strain, W22703, lost the ability to exclude trypan blue (Table 3). Moreover, almost all the cells showed a cytopathic effect.
The viability of macrophages infected with biotype 1A strains was also investigated by using the LIVE/DEAD kit, which measures the metabolic activity of eukaryotic cells. In this assay, viable macrophages appear green and dead macrophages appear red when viewed by fluorescence microscopy. The results of this assay confirmed that approximately 99.9% of macrophages infected with each of four different biotype 1A strains remained viable and showed no membrane blebbing or cell rounding (data not shown). In contrast, Y. enterocolitica W22703 killed a large number of macrophages, many of which became detached from the tissue culture tray (data not shown).
Electron microscopic examination of macrophages infected with strains of Y. enterocolitica biotype 1A.
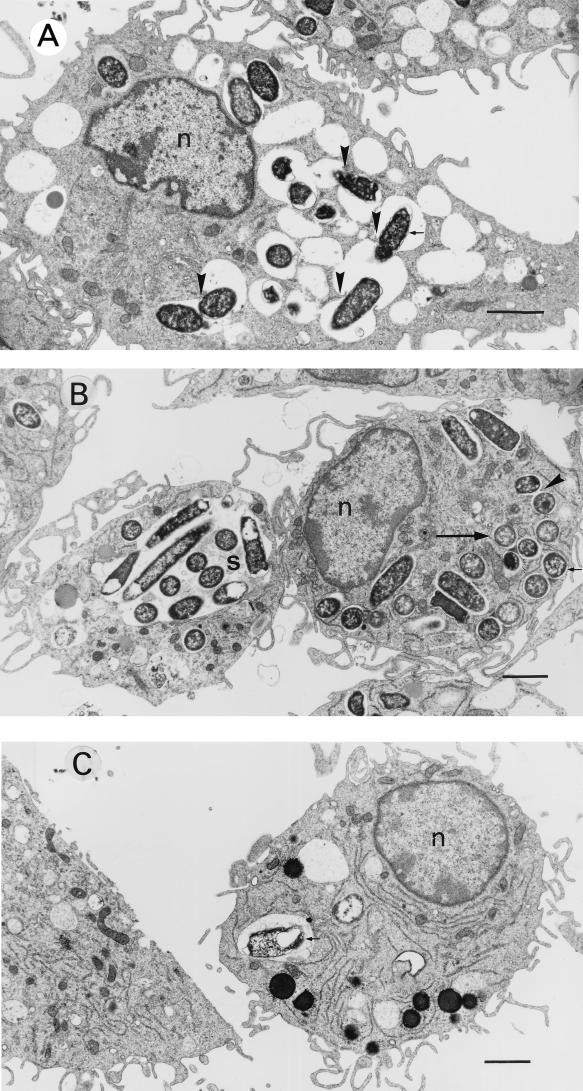
Clinical isolates of Y. enterocolitica biotype 1A incubated with macrophages were commonly observed within spacious vacuoles, many of which contained several bacteria (Fig. 2). Some bacteria appeared to induce vacuole fusion, causing the formation of large vacuoles which contained numerous bacteria. After 19 h, intact bacteria were observed within both spacious and tight vacuoles (Fig. 2B). By contrast, nonclinical isolates were observed in small numbers and, by 19 h of incubation, often appeared degraded (Fig. 2C). Despite the large number of intracellular bacteria, macrophages infected with clinical isolates of biotype 1A strains appeared healthy. Even after 19 h, the macrophage cell membrane was intact and there was no evidence of cytotoxicity.
FIG. 2.
Transmission electron micrographs of macrophages incubated for 1 h with Y. enterocolitica 937 (clinical, biotype 1A) (A), for 19 h with Y. enterocolitica 937 (B), and for 19 h with Y. enterocolitica AM5 (nonclinical, biotype 1A) (C). Findings similar to those shown in the figure were observed with three other strains of Y. enterocolitica biotypes 1A (two clinical and one nonclinical). At 1 and 19 h, strains of clinical origin appeared to induce phagosome fusion (arrowheads), and after 19 h of incubation, bacteria were observed within spacious vacuoles (s) or tight vacuoles (large arrow). In contrast, nonclinical isolates were identified in small numbers and often appeared degraded. Small arrows indicate bacteria. n, nucleus. Bar, 2 μm.
DISCUSSION
Although biotype 1A strains of Y. enterocolitica are now accepted as etiological agents of diarrhea, their pathogenic mechanisms are poorly understood. Work previously undertaken in our laboratory has shown that the ability to invade epithelial cells in vitro (in modest numbers compared to pYV-bearing strains) and to colonize the mouse intestine correlates with presumed virulence whereas the ability to produce enterotoxins does not (16). If biotype 1A Yersinia strains invade and persist within host tissues, we would expect them to be able to evade the innate immune response. However, the mechanisms which these bacteria use to evade host defenses must differ from those used by Y. enterocolitica biotypes 1B and 2 through 5, which generally rely on pYV for this purpose.
Although resistance to ingestion and killing by PMNs is associated with virulence in pYV-bearing strains of Y. enterocolitica, PMNs killed approximately 80% of each of four biotype 1A strains within 60 min. Clinical and nonclinical isolates showed a similar degree of susceptibility to killing by PMNs, confirming the impression that persistence within PMNs does not correlate with virulence in these bacteria. By contrast, biotype 1A yersiniae resisted killing by activated macrophages, with between 20 and 89% surviving for 24 h within these cells. Although strains of clinical origin were significantly more resistant to macrophage-mediated killing than were those of nonclinical origin, the difference was not pronounced, suggesting that this is not a prominent virulence attribute of these bacteria. Nevertheless, it is possible that this property contributes to the virulence of some biotype 1A strains. Since Y. enterocolitica biotype 1A is poorly equipped to survive extracellularly in host tissue by virtue of its susceptibility to killing by PMNs and complement (unpublished data), the ability to persist within macrophages may be a virulence attribute. Biotype 1A strains may also evade destruction by PMNs and complement by sequestering themselves within epithelial cells. This suggestion is supported by our previous observations that these bacteria are able to persist and replicate within epithelial cells to a greater extent than is Y. enterocolitica W22703c or E. coli HB101 (16). The finding that individual isolates in the clinical and nonclinical groups overlapped with regard to their capacity to persist within macrophages was not surprising, given that some strains isolated from nonclinical sources may be capable of causing infection and that some clinical isolates may be nonpathogens obtained from patients whose symptoms were attributable to another cause.
The mechanisms by which Y. enterocolitica resists killing by macrophages are unknown. Electron microscopic observations of macrophages infected with biotype 1A strains of clinical origin revealed large numbers of bacteria within spacious vacuoles. The significance of this observation is not known, but it suggests that survival of bacteria may partly be due to the dilution of bactericidal molecules within the vacuole, as has been suggested for Salmonella enterica (1, 2).
Some enteric pathogens resist killing by macrophages by destroying them. For example, strains of Y. enterocolitica which possess pYV may kill macrophages by secreting cytotoxins and inducing apoptosis (17, 18, 22). These properties are eliminated when the bacteria are cured of pYV, as was shown with strain W22703c in this study. Strains of biotype 1A, which inherently lack pYV, also did not appear to induce killing of macrophages or epithelial cells, as indicated by assays for LDH release and trypan blue exclusion (Table 4), measurement of electrical resistance across confluent T84 cell monolayers, and the results of the LIVE/DEAD assay. Moreover, light and electron microscopic examination of macrophages which contained large numbers of biotype 1A yersiniae revealed cells that appeared healthy even 19 h after the initial exposure to bacteria (Fig. 2). Since these techniques suggest a lack of cell death, and given the observation that up to 15% of intracellular biotype 1A bacteria may escape from cells, it seems highly unlikely that cell death would account for the bacterial escape phenotype, although a contribution from limited cytotoxicity to bacterial escape cannot be completely ruled out.
Some enteric pathogens disseminate within host cells and then destroy them before infecting new cells or reentering the intestinal lumen. A key finding of this study was that potentially pathogenic strains of Yersinia exited host cells by a mechanism that did not appear to kill the host cell or interfere with membrane integrity and thus resembled exocytosis. Nearly all biotype 1A strains isolated from symptomatic humans were able to escape from macrophages and epithelial cells. This property correlated with the ability to invade epithelial cells (Table 2). The ability to exit host cells was also exhibited by invasive, pYV-cured Y. enterocolitica strains of biotypes 1B and 2 and by mutants of the former that were defective in their cell invasion capacity. By contrast, most biotype 1A strains that were isolated from nonclinical sources invaded epithelial cells poorly and remained intracellular (Table 2), as did derivatives of E. coli HB101. The finding that the capacity to escape from cells was highly correlated with symptomatic infection and the invasion of epithelial cells suggests that this property contributes to virulence. The ability of biotype 1A yersiniae of clinical origin to escape from cells and infect new ones could explain our observation that these bacteria are able to colonize the intestines of mice for prolonged periods (16).
The mechanism used by Y. enterocolitica to escape from host cells is unknown but may be similar to exocytosis, which is used by Neisseria gonorrhoeae and biovar II strains (but not other biovars) of Chlamydia trachomatis to exit from epithelial cells (3, 26, 27, 42). Although biotype 1B and 2 strains of Y. enterocolitica also demonstrated an ability to escape from host cells, it is not known if the mechanism used by these bacteria is the same as that used by biotype 1A strains. This mechanism could be clarified in future by the isolation of the gene(s) responsible for this phenotype.
In conclusion, we have shown that Y. enterocolitica biotype 1A strains of clinical origin resisted killing by activated macrophages to a significantly greater extent than do strains of nonclinical origin. Moreover, clinical strains of biotype 1A were able to escape from macrophages and epithelial cells by a mechanism that did not appear to kill the host cell or compromise membrane integrity and thus resembled exocytosis. Because they are susceptible to killing by PMNs and complement, the ability of certain biotype 1A strains to persist within the intracellular environment of epithelial cells and macrophages may be an essential virulence determinant of these bacteria.
ACKNOWLEDGMENTS
We are indebted to V. L. Miller for the generous gift of Y. enterocolitica JP273 and YE2v and E. coli HB101(pVM101) and HB101(pVM103).
This work was supported by grants from the Australian National Health and Medical Research Council and the Royal Children’s Hospital Research Institute.
REFERENCES
- 1.Alpuche-Aranda C M, Berthiaume E P, Mock B, Swanson D S, Miller S I. Spacious phagosome formation within mouse macrophages correlates with Salmonella serotype pathogenicity and host susceptibility. Infect Immun. 1995;63:4456–4462. doi: 10.1128/iai.63.11.4456-4462.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alpuche-Aranda C M, Racoosin E L, Swanson J A, Miller S I. Salmonella stimulate macrophage macropinocytosis and persist within spacious phagosomes. J Exp Med. 1994;179:601–608. doi: 10.1084/jem.179.2.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Apicella M A, Ketterer M, Lee F K N, Zhou D, Rice P A, Blake M S. The pathogenesis of gonococcal urethritis in men: confocal and immunoelectron microscopic analysis of urethral exudates from men infected with Neisseria gonorrhoeae. J Infect Dis. 1996;173:636–646. doi: 10.1093/infdis/173.3.636. [DOI] [PubMed] [Google Scholar]
- 4.Arduino R, Murray B E, Rakita R M. Roles of antibodies and complement in phagocytic killing of enterococci. Infect Immun. 1994;62:987–993. doi: 10.1128/iai.62.3.987-993.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beuscher H U, Rödel F, Forsberg Å, Röllinghoff M. Bacterial evasion of host immune response: Yersinia enterocolitica encodes a suppressor of tumor necrosis alpha expression. Infect Immun. 1995;63:1270–1277. doi: 10.1128/iai.63.4.1270-1277.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bissett M L, Powers C, Abbott S L, Janda J M. Epidemiologic investigations of Yersinia enterocolitica and related species: sources, frequency, and serogroup distribution. J Clin Microbiol. 1990;28:910–912. doi: 10.1128/jcm.28.5.910-912.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bliska J B, Falkow S. Bacterial resistance to complement killing mediated by the Ail protein of Yersinia enterocolitica. Proc Natl Acad Sci USA. 1992;89:3561–3565. doi: 10.1073/pnas.89.8.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boland A, Sory M P, Iriarte M, Kerbourch C, Wattiau P, Cornelis G R. Status of YopM and YopN in the Yersinia Yop virulon: YopM of Y. enterocolitica is internalized inside the cytosol of PU5-1.8 macrophages by the YopB, D, N delivery apparatus. EMBO J. 1996;15:5191–5201. [PMC free article] [PubMed] [Google Scholar]
- 9.Burnens A P, Frey A, Nicolet J. Association between clinical presentation, biogroups and virulence attributes of Yersinia enterocolitica strains in human diarrhoeal disease. Epidemiol Infect. 1996;116:27–34. doi: 10.1017/s0950268800058921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.China B, N’Guyen B T, De Bruyere M, Cornelis G R. Role of YadA in resistance of Yersinia enterocolitica to phagocytosis by human polymorphonuclear leukocytes. Infect Immun. 1994;62:1275–1281. doi: 10.1128/iai.62.4.1275-1281.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.China B, Sory M P, N’Guyen B T, De Bruyere M, Cornelis G R. Role of the YadA protein in prevention of opsonization of Yersinia enterocolitica by C3b molecules. Infect Immun. 1993;61:3129–3136. doi: 10.1128/iai.61.8.3129-3136.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clerc P L, Ryter A, Mounier J, Sansonetti P J. Plasmid-mediated early killing of eucaryotic cells by Shigella flexneri as studied by infection of J774 macrophages. Infect Immun. 1987;55:521–527. doi: 10.1128/iai.55.3.521-527.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Koning-Ward T F, Grant T, Oppedisano F, Robins-Browne R. Effect of cell invasive capacity on the fate of Yersinia enterocolitica in macrophages. J Immunol Methods. 1998;215:39–44. doi: 10.1016/s0022-1759(98)00053-2. [DOI] [PubMed] [Google Scholar]
- 14.Dharmsathaphorn K, McRoberts J A, Mandel K G, Tisdale L D, Masui H. A human colonic tumor cell line that maintains vectorial electrolyte transport. Am J Physiol. 1984;246:204–208. doi: 10.1152/ajpgi.1984.246.2.G204. [DOI] [PubMed] [Google Scholar]
- 15.Fernandez-Prada C M, Hoover D L, Tall B D, Venkatesan M M. Human monocyte-derived macrophages infected with virulent Shigella flexneri in vitro undergo a rapid cytolytic event similar to oncosis but not apoptosis. Infect Immun. 1997;65:1486–1496. doi: 10.1128/iai.65.4.1486-1496.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grant T, Bennett-Wood V, Robins-Browne R M. Identification of novel virulence attributes in clinical isolates of Yersinia enterocolitica lacking classical virulence markers. Infect Immun. 1998;66:1113–1120. doi: 10.1128/iai.66.3.1113-1120.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hartland E L, Green S P, Phillips W A, Robins-Browne R M. Essential role of YopD in inhibition of the respiratory burst of macrophages by Yersinia enterocolitica. Infect Immun. 1994;62:4445–4453. doi: 10.1128/iai.62.10.4445-4453.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iriarte M, Cornelis G. YopT, a new Yersinia Yop effector protein, affects the cytoskeleton of host cells. Mol Microbiol. 1998;29:915–929. doi: 10.1046/j.1365-2958.1998.00992.x. [DOI] [PubMed] [Google Scholar]
- 19.McCormick B A, Miller S I, Carnes D, Madara J L. Transepithelial signaling to neutrophils by salmonellae: a novel virulence mechanism for gastroenteritis. Infect Immun. 1995;63:2302–2309. doi: 10.1128/iai.63.6.2302-2309.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller V L, Falkow S. Evidence for two genetic loci in Yersinia enterocolitica that can promote invasion of epithelial cells. Infect Immun. 1988;56:1242–1248. doi: 10.1128/iai.56.5.1242-1248.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller V L, Farmer III J J, Hill W E, Falkow S. The ail locus is found uniquely in Yersinia enterocolitica serotypes commonly associated with disease. Infect Immun. 1989;57:121–131. doi: 10.1128/iai.57.1.121-131.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mills S D, Boland A, Sory MP, van der Smissen P, Kerbourch C. Yersinia enterocolitica induces apoptosis in macrophages by a process requiring functional type III secretion and translocation mechanisms and involving YopP, presumably acting as an effector protein. Proc Natl Acad Sci USA. 1997;94:12638–12643. doi: 10.1073/pnas.94.23.12638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Monack D M, Mecsas J, Ghori N, Falkow S. Yersinia signals macrophages to undergo apoptosis and YopJ is necessary for this cell death. Proc Natl Acad Sci USA. 1997;94:10385–10390. doi: 10.1073/pnas.94.19.10385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Monack D M, Raupach B, Hromockyj A E, Falkow S. Salmonella typhimurium invasion induces apoptosis in infected macrophages. Proc Natl Acad Sci USA. 1996;1993:9833–9838. doi: 10.1073/pnas.93.18.9833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morris J G, Jr, Prado V, Ferreccio C, Robins-Browne R M, Bordun A-M, Cayazzo M, Kay B A, Levine M M. Yersinia enterocolitica isolated from two cohorts of young children in Santiago, Chile: incidence of and lack of correlation between illness and proposed virulence factors. J Clin Microbiol. 1991;29:2784–2788. doi: 10.1128/jcm.29.12.2784-2788.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mosleh I M, Boxberger H-J, Sessler M J, Meyer T F. Experimental infection of native human ureteral tissue with Neisseria gonorrhoeae: adhesion, invasion, intracellular fate, exocytosis, and passage through a stratified epithelium. Infect Immun. 1997;65:3391–3398. doi: 10.1128/iai.65.8.3391-3398.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moulder J W. Interaction of chlamydiae and host cells in vitro. Microbiol Rev. 1991;55:143–190. doi: 10.1128/mr.55.1.143-190.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Noble M A, Barteluk R L, Freeman H J, Subramaniam R, Hudson J B. Clinical significance of virulence-related assay of Yersinia species. J Clin Microbiol. 1987;25:802–807. doi: 10.1128/jcm.25.5.802-807.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pepe J C, Miller V L. The biological role of invasin during a Yersinia enterocolitica infection. Infect Agents Dis. 1993;2:236–241. [PubMed] [Google Scholar]
- 30.Pham J N, Bell S M, Lanzarone J Y M. Biotype and antibiotic sensitivity of 100 clinical isolates of Yersinia enterocolitica. J Antimicrob Chemother. 1991;28:13–18. doi: 10.1093/jac/28.1.13. [DOI] [PubMed] [Google Scholar]
- 31.Pierson D E, Falkow S. Nonpathogenic isolates of Yersinia enterocolitica do not contain functional inv-homologous sequences. Infect Immun. 1990;58:1059–1064. doi: 10.1128/iai.58.4.1059-1064.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pierson D E, Falkow S. The ail gene of Yersinia enterocolitica has a role in the ability of this organism to survive serum killing. Infect Immun. 1993;61:1846–1852. doi: 10.1128/iai.61.5.1846-1852.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ratnam S, Mercer E, Picco B, Parsons S, Butler R. A nosocomial outbreak of diarrheal disease due to Yersinia enterocolitica serotype O:5, biotype 1. J Infect Dis. 1982;145:242–247. doi: 10.1093/infdis/145.2.242. [DOI] [PubMed] [Google Scholar]
- 34.Robins-Browne R M, Jacobs M R, Koornhof H J, Mauff A C. Yersinia enterocolitica biotype 1 in South Africa. S Afr Med J. 1979;55:1057–1060. [PubMed] [Google Scholar]
- 35.Robins-Browne R M, Miliotis M D, Cianciosi S, Miller V L, Falkow S, Morris J G., Jr Evaluation of DNA colony hybridization and other techniques for detection of virulence in Yersinia species. J Clin Microbiol. 1989;27:644–650. doi: 10.1128/jcm.27.4.644-650.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robins-Browne R M, Tzipori S, Gonis G, Hayes J, Withers M, Prpic J K. The pathogenesis of Yersinia enterocolitica infection in gnotobiotic piglets. J Med Microbiol. 1985;19:297–308. doi: 10.1099/00222615-19-3-297. [DOI] [PubMed] [Google Scholar]
- 37.Ruckdeschel K, Roggenkamp A, Lafont V, Mangeat P, Heesemann J, Rouot B. Interaction of Yersinia enterocolitica with macrophages leads to macrophage cell death through apoptosis. Infect Immun. 1997;65:4813–4821. doi: 10.1128/iai.65.11.4813-4821.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ruckdeschel K, Roggenkamp A, Schubert S, Heesemann J. Differential contribution of Yersinia enterocolitica virulence factors to evasion of microbicidal action of neutrophils. Infect Immun. 1996;64:724–733. doi: 10.1128/iai.64.3.724-733.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stolk-Engelaar V M M, Meis J F G M, Mulder J A, Loeffen F L A, Hoogkamp-Korstanje J A A. In-vitro antimicrobial susceptibility of Yersinia enterocolitica isolates from stools of patients in The Netherlands from 1982–1991. J Antimicrob Chemother. 1995;36:839–843. doi: 10.1093/jac/36.5.839. [DOI] [PubMed] [Google Scholar]
- 40.Sulakvelidze A, Dalakishvili K, Barry E, Wauters G, Robins-Browne R, Imnadze P, Morris J G., Jr Analysis of clinical and environmental Yersinia isolates in the Republic of Georgia. J Clin Microbiol. 1996;34:2325–2327. doi: 10.1128/jcm.34.9.2325-2327.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tabrizi S N, Robins-Browne R M. Influence of a 70-kilobase virulence plasmid on the ability of Yersinia enterocolitica to survive phagocytosis in vitro. Microb Pathog. 1992;13:171–179. doi: 10.1016/0882-4010(92)90018-j. [DOI] [PubMed] [Google Scholar]
- 42.Todd W J, Caldwell H D. The interaction of Chlamydia trachomatis with host cells: ultrastructural studies of the mechanism of release of a biovar II strain from HeLa 229 cells. J Infect Dis. 1985;151:1037–1044. doi: 10.1093/infdis/151.6.1037. [DOI] [PubMed] [Google Scholar]
- 43.Vairo G, Hamilton J A. CSF-1 stimulates Na+K+-ATPase mediated 86Rb+ uptake in mouse bone marrow-derived macrophages. Biochem Biophys Res Commun. 1985;132:430–437. doi: 10.1016/0006-291x(85)91040-x. [DOI] [PubMed] [Google Scholar]
- 44.Visser L G, Annema A, Van Furth R. Role of Yops in inhibition of phagocytosis and killing of opsonized Yersinia enterocolitica by human granulocytes. Infect Immun. 1995;63:2570–2575. doi: 10.1128/iai.63.7.2570-2575.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wachtel M R, Miller V L. In vitro and in vivo characterization of an ail mutant of Yersinia enterocolitica. Infect Immun. 1995;63:2541–2548. doi: 10.1128/iai.63.7.2541-2548.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zychlinsky A, Prevost M C, Sansonetti P J. Shigella flexneri induces apoptosis in infected macrophages. Nature (London) 1992;358:167–170. doi: 10.1038/358167a0. [DOI] [PubMed] [Google Scholar]