Abstract
Newly emerging variants of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) are continuously posing high global public health concerns and panic resulting in waves of coronavirus disease 2019 (COVID-19) pandemic. Depending on the extent of genomic variations, mutations and adaptation, few of the variants gain the ability to spread quickly across many countries, acquire higher virulency and ability to cause severe disease, morbidity and mortality. These variants have been implicated in lessening the efficacy of the current COVID-19 vaccines and immunotherapies resulting in break-through viral infections in vaccinated individuals and recovered patients. Altogether, these could hinder the protective herd immunity to be achieved through the ongoing progressive COVID-19 vaccination. Currently, the only variant of interest of SARS-CoV-2 is Omicron that was first identified in South Africa. In this review, we present the overview on the emerging SARS-CoV-2 variants with a special focus on the Omicron variant, its lineages and hybrid variants. We discuss the hypotheses of the origin, genetic change and underlying molecular mechanism behind higher transmissibility and immune escape of Omicron variant. Major concerns related to Omicron including the efficacy of the current available immunotherapeutics and vaccines, transmissibility, disease severity, and mortality are discussed. In the last part, challenges and strategies to counter Omicron variant, its lineages and hybrid variants amid the ongoing COVID-19 pandemic are presented.
Keywords: SARS-CoV-2, COVID-19, Omicron, Emerging variance, Variant of concern
1. Introduction
The pandemic of coronavirus disease 2019 (COVID-19), caused by the novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has resulted in over 617 million cases and over 6.5 million deaths globally as of October 9, 2022 [1] with significant persistent symptoms [2], [3]. Ongoing wave along with previous waves have posed significant health concerns and socio-economic threats to the world, the worst scenario ever after the 1918 Spanish Flu pandemic [1], [4], [5], [6], [7]. COVID-19 closely resembles earlier cases of the severe acute respiratory syndrome (SARS) (2002) and Middle East respiratory syndrome (MERS) (2012); however, SARS-CoV-2 gained higher transmissibility and greater disease severity, affecting multiple organs along with additional public health threats imposed by various new developing SARS-CoV-2 variants and mutants [8], [9], [10], [11], [12], [13], [14].
Face masks, hand hygiene, appropriate sanitation, contact tracing as well as lock downs with quick confirmatory diagnosis and strengthening of medical facilities altogether have aided in restricting the rapid spread of SARS-CoV-2 to some extent. However, the virus could not be controlled yet despite active vaccination drive is being in progress worldwide for rendering protection and developing herd immunity to prevent the spread of infection. This is mainly owing to the emerging new variants and mutants of SARS-CoV-2 from time to time resulting multiple waves as well as posing current huge influx in COVID-19 cases as the fourth wave that mainly due to the recently emerging Omicron variant [1], [4], [5], [12], [15], [16], [17], [18], [19], [20]. Several antiviral drugs and therapies have been proposed for use in emergency settings to improve the clinical of COVID-19 and to reduce mortality. However, any effective choice of drugs and medicines is still awaited, and for this purpose, various research and clinical trials are on the way to find out a solution for this pandemic virus which is threatening the live of millions of people globally [19], [21], [22], [23], [24], [25], [26].
Vaccination against several pathogens has saved live of millions of people in the past decades and centuries. High research efforts have paved the ways to the development of currently available COVID-19 vaccines, though with varied efficacy and potency to provide 65–95% protection levels in vaccinated people to counter COVID-19, while several other vaccines are under development and in clinical trials [27], [28], [29]. The massive vaccination drive globally is one of best options in the current scenario of the ongoing pandemic to achieve herd immunity against SARS-CoV-2 infection. The barriers to achieve the herd immunity need to be addressed appropriately, including tackling the emerging SARS-CoV-2 variants such as Omicron that causes vaccine break-through infection in COVID-19 vaccinated and recovered individuals [1], [11], [15], [16], [17], [18], [19], [20], [30], [31], [32], [33], [34], [35], [36].
This article provides an overview on the emerging Omicron variant (its lineages and hybrid/recombinant variants) including the possible origin, mutations and molecular mechanism behind phenotypic changes and immune evasion. We also discuss the impacts of immunotherapeutic, vaccine efficacy, transmissibility, disease severity, and mortality. In addition, we highlight strategies to counter and halt the spread of these variants in the midst of the ongoing COVID-19 pandemic.
2. Emerging SARS-CoV-2 variants
Emerging variants and mutants of SARS-CoV-2 owe to their evolution and adaptation in the host, environmental factors, genomic mutations involving gene insertions or deletions, amino acid modifications, and recombination events at the virus genomic level. A few of these variants have the ability to spread quickly to many countries across the globe leading to the emergence of multiple waves of COVID-19 pandemic [5], [9], [14], [19], [37]. Depending on the extent of genomic variations and adaptation, these could cause more severe disease severity and higher mortality due to higher virulency. Moreover, these could also limit the efficacy of the presently accessible COVID-19 vaccines and immunotherapies. As the results, breakthrough SARS-CoV-2 infections in both vaccinated and recovered patients, re-infection, and impeding the protective herd immunity might occur [12], [19], [33], [38], [39], [40], [41], [42], [43], [44], [45], [46].
The emerging SARS-CoV-2 variants are classified into four categories: variants of concern (VOC), variants of interest (VOI), variants being monitoring (VBM), and variant of high consequence (VOHC). This classification is based on their transmissibility, virulency, and ability to cause severe disease. Consequently, these categories could also impact the diagnostics and efficacy of vaccines and immunotherapies [20], [45]. New SARS-CoV-2 variants have shown their higher visibility during the pandemic’s second and third waves, giving rise to a rapid increase of COVID-19 cases in many countries [19], [20], [40], [46], [47]. Classification of SARS-CoV-2 variants is changing overtime. Previously, SARS-CoV-2 VOCs include Alpha (B.1.1.7), Beta (B.1.351), Gamma (P.1), Delta (B.1.617.2), and Omicron (B.1.1.529); VOI include Lambda (C.37) and Mu (B.1.621); and VBM including AZ.5, C.1.2, B.1.617.1*, B.1.526*, B.1.525*, and B.1.630, B.1.640 [48]. As of October 9, 2022, the only VOC is Omicron [49].
3. Origin of Omicron variant and its lineages: possible theories
Omicron was the fifth VOC of SARS-CoV-2 and as is the only VOC. This variant has recently posed high global public health threats, the most mutated, highly transmissible, and is comparatively resistant to prevailing immunotherapeutics or vaccines [19], [38], [50], [51], [52]. The emergence of Omicron (B.1.1.529) variant of SARS-CoV-2 was reported to the WHO in November 2021 and termed as Omicron by WHO [53]. Apart from B.1.1.529, there are other lineages of the Omicron variant: BA.1, BA.1.1, BA.2, BA.3, BA.4 and BA.5 lineages [49], [54]. Omicron was initially detected in samples collected from Botswana and several areas of South Africa, particularly Gauteng province, in the early and mid-November 2021, respectively [55], [56]. It has been speculated that the emergence of the Omicron variant in western Europe occurred prior to its first report by South African scientists [57]. In the USA, the Omicron was confirmed in California on December 1, 2021 [58]. It soon became a global concern as nearly 150 countries, including the USA, UK, Australia, France, Germany, Denmark, Japan, Netherlands, India, and other countries are facing a surge in Omicron cases [59], [60], [61], [62].
Few theories have been hypothesized with regards to the origin and evolution of the Omicron variant [63], [64], [65], [66]. It has been suggested that the rapid spread of the Omicron variant among South Africans is due to the high presence of the nation’s immunodeficient populations [60]. Immunodeficient people, including the ones with HIV/AIDS, might suffer from prolonged non-lethal COVID-19 infection [60], [67], and therefore likely to serve as a suitable host with persistent COVID-19. This is in line with the notion that the rise of a new SARS-CoV-2 variant with multimutational signatures can occur in patients with persistent COVID-19 [68], [69], [70], [71]. Nevertheless, while this is plausible, the role of immunodeficient patients in the emergence of the Omicron variant remains difficult to be concluded due to the lack of experimental and clinical evidence. In addition, Omicron variant might have evolved in areas with poor healthcare systems and a low vaccination rate [55]. The African countries with low COVID-19 vaccination coverage may be a favorable environment for the emergence of Omicron variant [72], [73], [74].
Inter-species evolution is also has been suggested as possible mechanism of Omicron variant emergence. SARS-CoV-2 spillover event to animals might have given rise to the origin of Omicron. One of the theories supporting this fact is infections (epizootic) contracted by animals from humans where there is reintroduction of the virus to humans after its mutation under several immune pressures [19]. The virus could have jumped from humans to mice, gathered mutations facilitating infection of mice, thereafter reinfection occurred to a human host, suggesting an inter-species evolution (human-mice-human) as analyzed with the presence of mouse-adapted mutation sites and adaptation to mouse [63], [64], [65], [66].
As Omicron is not a direct descendent of Delta variant or earlier variants and has genomic differences with SARS-CoV-2, it may have diverged at initial phases from other strains [75], [76]. Metric and ultrametric methods revealed that the Omicron variant is distant from other SARS-CoV-2 variants and has formed a separate monophyletic clade. Despite multiple of sequencing have been done, researchers have missed mutations that led to the emergence of Omicron [77]. The possibility of recombination mechanism as the origin of the Omicron variant also have been proposed recently. A study has reported that the Omicron variant might be resulted from the recombination of B.35 lineage (SARS‐CoV‐2/human/IRN/Ir‐3/2019) and SARS‐CoV‐2 parent strain (SARS‐CoV‐2/human/USA/COR‐21–434196/2021) [78]. Other view is that it may have evolved from chronically affected COVID-19 patient [75], [76].
4. Genetic mutations of Omicron variant
Sequencing analysis revealed that the Omicron variant harbors a large number of mutations, about 50 mutations, in comparison to the SARS-CoV-2 originally isolated from Wuhan, in which several of them are uncommon or novel [52], [59]. More than half of the mutations were found in the sequence encoding for S protein [52], a primary antigenic target of antibodies produced by adaptive immune B cells during infection or in response to vaccination. Of all mutations in the S protein, 15 are in RBD, expressing concern that the host antibodies might not work efficiently to detect and bind the S protein of this new variant, thus leading to the host's failure to mount proper immune responses. Deletion in the S protein of (Δ69–70) in the Omicron variant is being explored as a diagnostic marker as TaqPath PCR test results negative for the S gene in Omicron and positive in other SARS-CoV-2 variants [79], [80]. Mutations have also been observed in the genomic regions that encode ORF1ab (nsp3–6, nsp12, and nsp14), envelope protein, membrane protein, and nucleocapsid protein of the Omicron variant.
Mutation of Q498R, S477N, and N501Y in Omicron are associated with increased ACE2 receptor binding and improve viral infection rates in the host cells [81]. K417N mutation can moderately increase RBD promoter activity and resistance to neutralizing mAbs [46]. An investigation has been conducted to determine the strength of binding of Omicron with ACE2 receptor found that the binding between ACE2 and the RBD of Omicron is 2.5 times stronger than with prototype SARS-CoV-2 [51]. It is interesting to note that there is suppression of the Omicron by antibodies that target ACE2 [82].
Substitutions of Ser446, Arg417, Arg493 and Arg498 in RBD of Omicron variant are responsible for interactions with ACE2 and this reduces the interactions with complementarity-determining regions (CDRs) (1−3) in the monoclonal antibodies [83]. Fusogenicity of Omicron less compared to Delta making Omicron having less pathogenicity than Delta due to rearrangement (geometric) of the S1-S2 cleavage location [84]. Omicron adopts cathepsin-dependent (E64d-sensitive) entry path unlike Delta variant that uses TMPRSS-like proteases-dependent (Camostat-sensitive) entry pathway hence difference in tissue tropism and probably treatment targets [85]. Reduced fusogenicity may contribute to its weakness [85]. Omicron S protein has a tempered proinflammatory effect [85].
5. Impacts on immune responses and immunotherapeutics
Mutations, including novel insertions and deletions, may decrease the vaccine's efficacy and antibody neutralization against Omicron as the antibodies may lack specificity to mutated S protein or may be unable to recognize the mutated conformation [50], [59], [86]. Omicron has several mutations in (or near) RBD, NTD, RBM, S2 domains and furin cleavage site, which may affect antibody binding and ACE2 binding [87], [88]. More specifically, on the RBD of Omicron, there are about 15 mutations [89]. Particularly, the RBD mutations are responsible for improving the viral binding to the ACE2 receptor. As a result of such mutations, antibodies arising from the previous infection or vaccination might be unable to bind to the Omicron variant [90]. Importantly, numerous changes around the furin cleavage site have been observed surrounding the RBD at the extreme point of the S protein [76]. The large number of mutations is the main reason why the Omicron variant is more infectious and shows vaccine resistance as compared to other VOCs. Moreover, the Omicron interacts less efficiently with neutralizing convalescent mAb [91].
An animal study compared the effectiveness of mRNA vaccines synthesized from the wild-type strain and the variant-specific (BA.1 and BA.2) in a lipid nanoparticle (LNP) [92]. The results demonstrated that the animals inoculated with the variant-specific vaccines (BA.1 LNP-mRNA) showed a 2-fold increase in the concentration of nAbs compared to those animals that have received a WT vaccine. Moreover, both heterologous and homologous boosters with the WT vaccine was responsible for a significant waning of the nAbs against BA.1 and BA.2 variants. However, a booster dose with the variant-specific vaccine was responsible for a 3-fold increase in the nAbs against the newer variants. This study confirms the urgent need for improving the available vaccines to mitigate the current as well as the future variants of SARS-CoV-2.
6. Impacts on COVID-19 vaccine effectiveness
SARS-CoV-2 reinfection is common to occur [93]. The National Institute for Communicable Diseases (NICD) in South Africa has released an early report that suggested the possibility of the Omicron variant reinfecting people who were previously contracted COVID-19 [60] or even received COVID-19 vaccines [59]. This has raised a concern that humans’ immunological memory might not work properly against Omicron variant. This has significant impacts to the worldwide current effort to provide COVID-19 protection through vaccination. Studies to examine whether the Omicron variant could evade the host's innate and adaptive immune systems are urgently required. A previous study using an engineered SARS-CoV-2 variant with similar polymutational signatures on its S protein to the Omicron variant demonstrated a resistant viral phenotype to the neutralizing antibodies [94]. While antiviral protection of neutralizing antibodies against this newly identified variant is being studied [59], the protective role of innate immune cells and adaptive T cells remains to be explored. Another piece of data from a study is suggestive of the fact that the immune response mediated by T cells in persons infected previously and most likely in persons receiving vaccines should be effective against Omicron variant [95].
Initial study revealed low levels of neutralizing antibodies against the Omicron in convalescent or fully vaccinated people [96]. Only about 20% protection from a previous infection, around 70% protection from hospitalization after two doses of COVID-19 vaccine and 90% after three doses has been reported, indicating the rise of protection due to booster vaccine shots which may be due to restoring and increasing the levels of neutralizing antibodies [70], [96]. This may be a result of increasing the breadth of humoral immunity and cross-reactivity to variants [96]. Omicron is highly contagious and gives rise to vaccine breakthrough infections; however, the current mRNA booster is still protective against the severe COVID-19-related outcomes [97]. The COVID-19 vaccine booster dose may significantly enhance the protection against the Omicron variant [98]. Neutralizing response against Omicron is generated by administering a booster dose of the BNT162b2 vaccine, but the titers have been found to be much lower than that against Delta. This indicates that this variant escapes a greater extent of the antibodies elicited by vaccination as well as monoclonal antibodies (mAbs) administered therapeutically. But the antibodies generated by a booster dose of the vaccine can neutralize the Omicron variant [99]. Another study even found that there is an increase in serum neutralizing activity against the variant significantly by a booster immunization with mRNA vaccine both in convalescent individuals as well as individuals vaccinated [100]. Their study reveals that there is a critical improvement in the humoral/antibody-mediated response against the virus by booster immunization. There are reports that following immunization (primary series) with double doses (primary) of BNT162b2 or mRNA-1273 or single dose (primary) of Ad26. COV2, a reduction of immunity induced by these vaccines was observed. Thus another dose of vaccine is recommended additionally in immunocompromised (moderate to severe) individuals. A booster vaccine dose is also recommended for persons 12 years and above [101]. The prior SARS-CoV-2 infection-induced immunity offers no protection against Omicron; however, previously COVID-19 infected and recovered people with at least a dose of mRNA vaccine may be protected from Omicron [100]. It is also interesting to note that Omicron-specific cross-neutralizing activity may be induced in convalescent patients through immunization with double doses of mRNA vaccine [102]. The persons previously infected or vaccinated may also provide significant protection against Omicron infection [103], [104], [105]. Still, the country-wise used vaccine effectiveness against Omicron should be evaluated.
Omicron can be cross-recognized by immunological T cell memory upon vaccination [106]. Alone, inactivated-virus vaccines or mRNA-based vaccines have not been found effective [107], [108]; thus application of different or multiple vaccines has been evaluated and produced considerable immunity against Omicron [108], [109], [110], [111]. A study has evaluated the effect of a second booster shot on ChAd/ChAd and ChAd/BNT vaccinated recipients [112]. The study found that the second booster enhanced the S protein-specific CD8+ and CD4+ T cells in both groups moderately. Moreover, after the second booster, the neutralizing antibody responses against Omicron stayed severely impaired. A current study found that two doses of BNT162b2 provided only partial protection against Omicron lineages (BA.1 and BA.2) while three doses had ≥ 70% protection against hospital and emergency department admissions [113]. Pfizer and BioNTech have started a vaccine trial in adults aged 18–55 year-old for the Omicron-based COVID-19 vaccine [114]. The fourth dose of the Omicron mRNA vaccine has been shown to be immunogenic, safe, and efficacious to prevent disease [115]. Initial humoral immune response and antibody titers specific to Omicron against the fourth dose showed no significant difference with peaks of the third dose [115].
Since countries having high COVID-19 vaccination rates and providing booster-dose associated with reduction of disease severity, hospitalization and death from Omicron variants [116], vaccination programs must be carried out widely and ramped up to counter such new emerging SARS-CoV-2 variants including Omicron [117]. Proper initiatives must be taken to obtain and distribute vaccines uniformly throughout the globe. Unless the entire global population is vaccinated, there will be a growing concern about the emergence of new variants from low-income countries [118]. In addition, booster doses of vaccines can help in minimizing hospitalization due to Omicron. The limited protection by ChAdOx1 nCoV-19 or BNT162b2 vaccination can be boosted by BNT162b2 or mRNA-1273 booster [119].
7. Impacts on transmission, disease severity, and mortality
The changes in the basic amino acids of the S‐RBD are responsible for enhancing the transmissibility of the Omicron variant [120]. The mutations, particularly Q498R, S477N, and N501Y improve viral infection rates in the host cells [81]. Omicron was 3.3 times more transmissible than the Delta variant and had 4.2 times effective reproduction number of the Delta variant [121].
Omicron is hyped to be milder than its predecessors SARS-CoV-2 variants and frequently affects the upper respiratory tract [122], [123] with considerable clinical symptoms but with low mortality [60], [104]. The prime predictor manifestations for this variant are the presence and absence of odynophagia and dysgeusia, respectively [124]. Omicron has been found to cause attenuated / milder disease in mice and hamsters as revealed by human clinical data [122]. In South Africa, the disease severity of Omicron was relatively low [125]. However, a Denmark-based study of 785 Omicron cases suggests that Omicron may not be less severe than the Delta variant [126].
Data extracted from early case reports in South Africa indicated that the mortality rate of Omicron-infected patients did not increase significantly, despite the rapid surge of COVID-19 cases [60], [125]. In addition, the epidemiological trend seems to indicate that young people, particularly children under 5 years old, were the most affected ones [60]. Again, it is interesting to note that in South Africa, it has been found during the Omicron wave that the comorbidities as well as hospitalization rates are less in younger patients during the early phase of the wave [125].
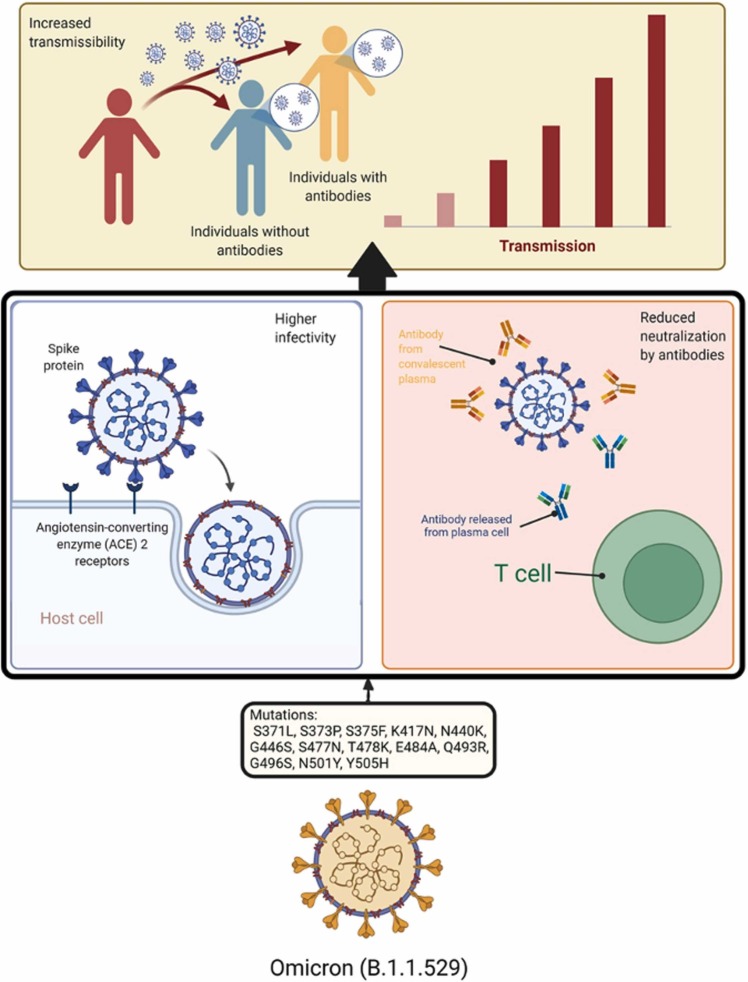
The summary of the effects of SARS-CoV-2 variants on immune responses and the transmission of the disease is provided in Fig. 1.
Fig. 1.
Mutations of spike protein amino acids of Omicron variant of SARS-CoV-2 causing in higher infectivity (by strengthening the binding with host ACE2 receptors) and reduced neutralization by antibodies (mAb, T-cell, or convalescent plasma). These changes of biological characteristics encourage higher transmissibility of the virus infecting both people with and without antibodies.
8. Multiple Omicron lineages and recombinant variants: Further challenges
Mutations are happening continuously within Omicron variant. Some lineages emerged such as BA.1, BA.1.1, BA.2, BA.3, BA.4 and BA.5. Lineage BA.1 getting replaced by BA.2 and BA.3; BA.2 has more transmissibility but without major changes than predecessor variant BA.1 [127], [128], [129], [130]. BA.2 surge is presumably due to higher transmissibility rather than enhanced immunological escape [131]. The BA.2 lineage may have cross-reactions with BA.1 as robust antibody response has been noted in persons on vaccination against BA.2 that were infected previously with BA.1 [89]. The BA.3 lineage result from mutations in BA.2 and BA 0.1 S proteins [132].
These three lineages emerged concurrently from the same location (Botswana, South Africa); BA.1 was the dominant lineage and spread faster than BA.2 initially, while BA.3 lineage spread slowly. After a few weeks, BA.2 ousted BA.1 and became a dominating variant worldwide as of late April 2022. BA.2 was shown to be more transmissible and immune-evasive than BA.1. BA.2 also reduces the efficacy of COVID-19 vaccination [129]. The BA.2 has a higher rate of secondary attack in households (39%) than BA.1 lineage (29%) [133].
Recently, multiple lineages and sub-lineages of Omicron, mainly BA.4, BA.5, BA.2.11, and BA.2.12.1, have also been identified [134]. The newly emerged Omicron lineages particularly BA.4 and BA.5, are rapidly replacing BA.2 in many countries (South Africa, Portugal) and have caused fifth wave in South Africa beginning in April 2022 [134].
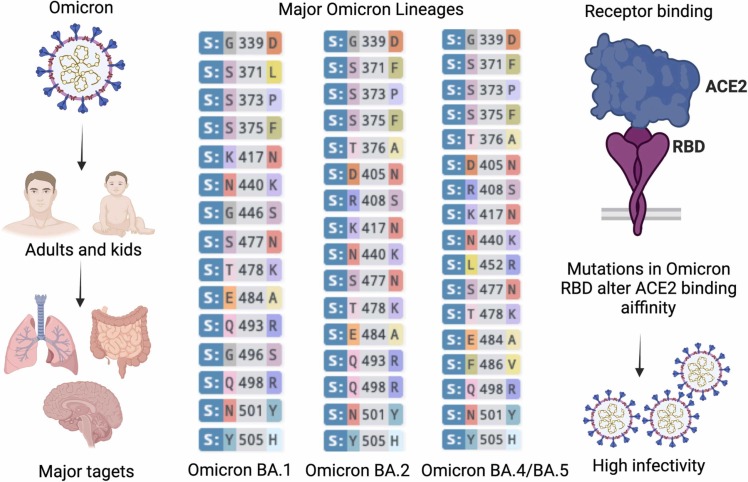
BA.4 and BA.5 lineages distinct from BA.1, such as L452R and F486V mutations in the S protein RBD [134]. A study investigated immune neutralization in vaccinated and unvaccinated individuals who had been exposed to Omicron/BA.1 [135]. Neutralizing antibodies titers against the newer BA.4 and BA.5 lineages were significantly lower (>7-fold) than the previous sub-lineage (BA.1) [135]. Hence, these newer lineages may cause symptomatic infection among the vaccinated individuals. An overview of Omicron lineages, mutations, higher infectivity and transmissibility is depicted in Fig. 2 .
Fig. 2.
Newly emerging Omicron and its lineages with increased infectivity infect both adults and kids and mainly target respiratory lung system, intestines and brain. Major Omicron lineages that spreading include BA.1, BA.2, BA4/5, with minor difference in RBD mutations in their spike. Omicron also alters and increases its binding affinity to human ACE2 to allow more efficient entry into host cells, leading higher infectivity and transmissibility.
Recently, hybrid variants of Omicron due to recombinant mechanism was also emerged. Three recombinant forms (XD, XF, and XE) of SARS-CoV-2 have been identified. The XE variant is a hybrid of BA.1 and BA.2 Omicron variant, XD is believed to be the result between Delta and BA.1 (Omicron) recombination, and XF is a hybrid between BA.1 (Omicron) and the UK Delta variant [129], [136].
Although, pathological characteristics of these hybrid variants are unknown, preliminary data found that the XE variant may be 10-fold more contagious than the BA.2 subvariant of Omicron [129]. These recombinant variants are believed to have high resistance to COVID-19 antiviral drugs or vaccines, high transmissibility, and immune evasion due to genetic diversity and viral genetic alterations, which require further explorative studies in this direction to reach a conclusive hypothesis [129], [137]. Since there is limited data on the vaccine effectiveness of the patients infected with the recombinant strains, it is too soon to comment on the specific efficacy of the ongoing vaccines. Moreover, it is speculated that the new XE variant will eventually become the dominant strain. So, it is highly recommended to monitor the progression of these recombinant variants of SARS-CoV-2 globally.
9. Rapid confirmatory diagnostic method challenges of Omicron variant
Diagnostics need to be upgraded to detect either Omicron specifically or other variants simultaneously. High-resolution melting analysis [138] and multiplex PCR and MALDI-TOF MS [139] have been evaluated for simultaneous detection of SARS-CoV-2 variants, including Omicron. These can facilitate early, rapid and timely detection. In Malaysia, scientists have used Allplex Master Assay (SARS-CoV-2 specific) (having 98.7% sensitivity) along with Variants I assay (having 100% sensitivity) for detection of the S protein mutation of Omicron and other VOCs [140]. By the combined use of single nucleotide polymorphism (SNP) PCR (targeting the S protein region) and sequencing of the viral genome, Omicron variant can be quickly detected [141]. There is a report on sequencing of the whole genome in Italy for B.1.1.529 variant identification [142]. Phylogenetic analysis of Omicron on the basis of several evolutionary substitution models including metric and ultrametric clustering methods has also been done [143].
In India, Omicron diagnostic kit called OmiSure kit has been developed by Tata MD company. It is believed to be able to detect Omicron along with other variants of SARS-CoV-2 [144]. It uses a combination of two S-gene viral targets to identify Omicron. The kit has shown 100% sensitivity and 99.2% specificity [144]. Though some tests, such as RT-PCR (NAAT), may be unaffected by Omicron, others, such as specific S-gene target failure (SGTF) may be affected. Hence there is a need for targeting multiple genomic regions to prevent the chances of false-negative tests due to changes in the targeted region [145].
10. Strategies to control Omicron
To limit the transmission of Omicron and emerging variants, proposed preventative measures are required to be implemented rigorously while keeping in mind the lessons learned from previous waves of the COVID-19 pandemic. Rapid confirmatory diagnosis along with genomic surveillance and sequencing of a larger number of SARS-CoV-2 isolates is to be carried out globally. SARS-CoV-2-host interactions, adaptation, evolutionary dynamics, mechanisms of genomic mutation or variation need deeper studies to understand higher transmissibility and pathogenicity acquired by emerging variants. This could be performed by strengthening medical research facilities and trained staff for conducting gene sequencing, identification and characterization of emerging variants, and a highly collaborative approach at the global level for updating genomic and epidemiological data repository which would altogether facilitate to tackle the emerging variants in a more effective manner [41], [43], [146], [147], [148], [149], [150], [151], [152].
Enhancing the COVID-19 mass vaccination drives with equitable global access and wider acceptance and reduce the vaccine hesitancy [153], [154] could protect the health of the worldwide population by achieving herd immunity at the earliest in coming time [29], [30], [155], [156], [157], [158], [159], but their long-term protection and achieving herd immunity is still under threats of SARS-CoV-2 adverse effects emerging variants on efficacies and protective immunity, which need to be evaluated and addressed adequately [19], [41], [46], [160], [161]. COVID-19 vaccines, such as BNT162b2, mRNA-1273, CoronaVac, Sputnik V, and AZD1222, have been evaluated as safer and more protective in preventing from developing of severe COVID-19 from infection with VOCs though with varying levels prevention and protection [14], [47], [162]. Strategic planning of strengthening vaccination with highly efficacious vaccines and booster doses of vaccination options could limit the ability of the virus to acquire mutations owing to lesser availability of susceptible population (host) compared to the slow or delayed vaccination approaches, especially in low-income countries. Special attention to speed up the development of effective and improved vaccines which could pace up against newly emerging variants of SARS-CoV-2, necessary modifications and/or updates in the presently available COVID-19 vaccines, and designing newer vaccines.
Collaborative approaches and programs of WHO, Gates Foundation, Coalition for Epidemic Preparedness Innovations (CEPI), COVID-19 Vaccines Global Access (COVAX), and Global Alliance for Vaccines and Immunizations (GAVI) need to be promoted, advanced and implemented adequately in a wider horizon for a universal and equal convenience of COVID-19 vaccines to all the countries for restraining the pandemic waves of the SARS-CoV-2 and lessening the impacts of its emerging variants [41], [155], [163]. Conducting research with regards to testing the efficacies and potencies of the currently available COVID-19 vaccines against Omicron and its lineages for evaluating breakthrough scenarios of infections are critical.
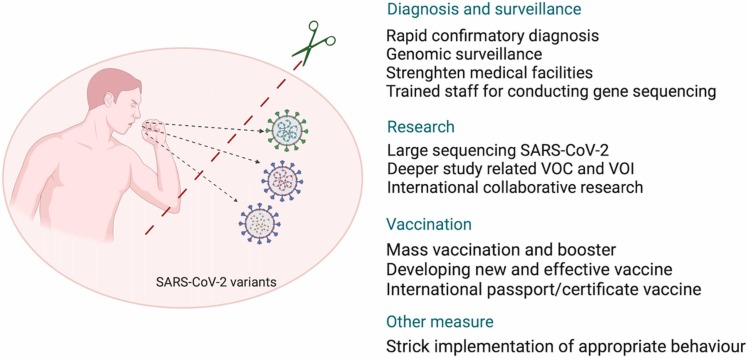
Strict implementation of recommended public health measures such as COVID-19 appropriate behaviors with following social distancing, wearing of face mask, hand hygiene, restricting movements and mass gatherings as well as sanitization and disinfection practices are the need of the hour to avoid infection with SARS-CoV-2 and variants [152], [164], [165]. Special focus on vulnerable groups, including unvaccinated, aged and those with underlying illnesses, should be laid to avoid the risk of infection and disease spread [29], [166]. Compulsory vaccination and the significance of vaccination / immunization certificates while travelling, crossing international borders, and ensuring targeted vaccination drives would act as a protective shield against community transmission of emerging variants and mutants of SARS-CoV-2 [150], [167]. Evaluating transmissibility, degree of severity, sensitivity specificity of diagnostic tests, efficacy of vaccines, and treatment effectiveness will help tackling Omicron and the future variant outbreaks [168]. The summary of the strategies to tackle SARS-CoV-2 emerging variants is provided in Fig. 3.
Fig. 3.
Strategies to tackle SARS-CoV-2 emerging variants.
11. Conclusions and future prospects
Emerging Omicron variant and its lineages have resulted a rapid and significant increase in COVID-19 cases globally while adversely impacting the protective efficacies of existing vaccines and antibodies-based therapies. Promoting equitable global access to vaccines, vaccine production, and ramping up progressive ongoing massive vaccination campaigns need to be given due priority. Necessary strategies to combat the impacts of emerging variants on vaccine efficacy and to limit the rising vaccine break-through infections are to be adopted. Appropriate mitigation and countermeasures comprising of wearing face masks, hand hygiene, social distancing, sanitation, hygiene and disinfection, and strengthening of medical facilities and infrastructure, are the top priorities for preventing SARS-CoV-2 from spreading quickly, along with formulating proactive control measures and advanced future pandemic preparedness plans.
Explorative studies are needed for investigating molecular mechanisms underlying the higher transmissibility, the evolutionary dynamics and the host adaptation of the newer variants, and understanding immunological correlation of protection and immune evasion events of previous infections with SARS-CoV-2. Advanced strategies need to be designed for modifying and updating the existing COVID-19 vaccines and vaccination schedules, adding booster shots, finding out newer vaccines with higher efficacies, and developing variant-specific vaccines, multivariant (multiple antigen-based), and mutation-proof vaccines. Additional monoclonal antibodies-based therapies, as well as efficient drugs of choice, are needed to be developed optimally to effectively treat patients with COVID-19. These advances would help in effectively countering the emerging SARS-CoV-2 variants having higher rates of infection, virulence, disease-causing ability and mortality amidst the ongoing pandemic.
CRediT authorship contribution statement
KD and HH conceptualized the manuscript. FN, AF, MIY, RKM, SC, HZ, MRD, and MI wrote the first draft. HIK, AAR, SA, AAM, JAA, MAM, AJA, HST, CC, HH reviewed and updated the manuscript. All authors approved the final manuscript.
Acknowledgements
All the authors acknowledge and thank their respective Institutes and Universities. HH supported by Lembaga Pengelola Dana Pendidikan (LPDP), managed by the Indonesian Science Fund (ISF) (Grant No: RISPRO/KI/B1/TKL/5/15448/2020).
Funding
No external funding received.
Disclosure statement
Authors have no conflict of interests.
References
- 1.WHO 2022; Available from: 〈https://covid19.who.int/?〉 Accessed on: March 2, 2022.
- 2.Fahriani M., Ilmawan M., Fajar J.K., Maliga H.A., Frediansyah A., Masyeni S., et al. Persistence of long COVID symptoms in COVID-19 survivors worldwide and its potential pathogenesis-a systematic review and meta-analysis. Narra J. 2021;1(2) doi: 10.52225/narraj.v1i2.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fajar J.K., Ilmawan M., Mamada S., Mutiawati E., Husnah M., Yusuf H., et al. Global prevalence of persistent neuromuscular symptoms and the possible pathomechanisms in COVID-19 recovered individuals: a systematic review and meta-analysis. Narra J. 2021;1(3) doi: 10.52225/narra.v1i3.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dhama K., Khan S., Tiwari R., Sircar S., Bhat S., Malik Y.S., et al. Coronavirus Disease 2019-COVID-19. Clin Microbiol Rev. 2020;33(4):e00028–20. doi: 10.1128/CMR.00028-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.El-Shabasy R.M., Nayel M.A., Taher M.M., Abdelmonem R., Shoueir K.R., Kenawy E.R. Three waves changes, new variant strains, and vaccination effect against COVID-19 pandemic. Int J Biol Macromol. 2022;204:161–168. doi: 10.1016/j.ijbiomac.2022.01.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fahriani M., Anwar S., Yufika A., Bakhtiar B., Wardani E., Winardi W., et al. Disruption of childhood vaccination during the COVID-19 pandemic in Indonesia. Narra J. 2021;1(1) doi: 10.52225/narraj.v1i1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Otolorin G.R., Oluwatobi A.I., Olufemi O.T., Esonu D.O., Dunka H.I., Adanu W.A., et al. COVID-19 pandemic and its impacts on the environment: a global perspective. Narra J. 2022;2:1. doi: 10.52225/narra.v2i1.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ganesh B., Rajakumar T., Malathi M., Manikandan N., Nagaraj J., Santhakumar A., et al. Epidemiology and pathobiology of SARS-CoV-2 (COVID-19) in comparison with SARS, MERS: an updated overview of current knowledge and future perspectives. Clin Epidemiol Glob Health. 2021;10 doi: 10.1016/j.cegh.2020.100694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thakur V., Ratho R.K., Kumar P., Bhatia S.K., Bora I., Mohi G.K., et al. Multi-organ involvement in COVID-19: beyond pulmonary manifestations. J Clin Med. 2021;10(3):446. doi: 10.3390/jcm10030446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boehm E., Kronig I., Neher R.A., Eckerle I., Vetter P., Kaiser L., et al. Novel SARS-CoV-2 variants: the pandemics within the pandemic. Clin Microbiol Infect. 2021;27(8):1109–1117. doi: 10.1016/j.cmi.2021.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farooqi T., Malik J.A., Mulla A.H., Al Hagbani T., Almansour K., Ubaid M.A., et al. An overview of SARS-COV-2 epidemiology, mutant variants, vaccines, and management strategies. J Infect Public Health. 2021;14(10):1299–1312. doi: 10.1016/j.jiph.2021.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Somerville M., Curran J.A., Dol J., Boulos L., Saxinger L., Doroshenko A., et al. Public health implications of SARS-CoV-2 variants of concern: a rapid scoping review. BMJ Open. 2021;11(12) doi: 10.1136/bmjopen-2021-055781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thye A.Y., Law J.W., Pusparajah P., Letchumanan V., Chan K.G., Lee L.H. Emerging SARS-CoV-2 variants of concern (VOCs): an impending global crisis. Biomedicines. 2021;9(10):1303. doi: 10.3390/biomedicines9101303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wahid M., Jawed A., Mandal R.K., Dailah H.G., Janahi E.M., Dhama K., et al. Variants of SARS-CoV-2, their effects on infection, transmission and neutralization by vaccine-induced antibodies. Eur Rev Med Pharmacol Sci. 2021;25(18):5857–5864. doi: 10.26355/eurrev_202109_26805. [DOI] [PubMed] [Google Scholar]
- 15.Lai C.C., Chen I.T., Chao C.M., Lee P.I., Ko W.C., Hsueh P.R. COVID-19 vaccines: concerns beyond protective efficacy and safety. Expert Rev Vaccin. 2021;20(8):1013–1025. doi: 10.1080/14760584.2021.1949293. [DOI] [PubMed] [Google Scholar]
- 16.Redwan E.M. COVID-19 pandemic and vaccination build herd immunity. Eur Rev Med Pharm Sci. 2021;25(2):577–579. doi: 10.26355/eurrev_202101_24613. [DOI] [PubMed] [Google Scholar]
- 17.Raman R., Patel K.J., Ranjan K. COVID-19: unmasking emerging SARS-CoV-2 variants, vaccines and therapeutic strategies. Biomolecules. 2021;11(7):993. doi: 10.3390/biom11070993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sharma N., Vyas S., Mohapatra A., Khanduri R., Roy P., Kumar R. Combating COVID-19 pandemic in India: Demystifying the concept of herd immunity. J Fam Med Prim Care. 2021;10(4):1515–1519. doi: 10.4103/jfmpc.jfmpc_1971_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khandia R., Singhal S., Alqahtani T., Kamal M.A., El-Shall N.A., Nainu F., et al. Emergence of SARS-CoV-2 Omicron (B.1.1.529) variant, salient features, high global health concerns and strategies to counter it amid ongoing COVID-19 pandemic. Environ Res. 2022;209 doi: 10.1016/j.envres.2022.112816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.WHO 2022; Available from: 〈https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/〉. Accessed on: March 2, 2022.
- 21.Sharun K., Tiwari R., Iqbal Yatoo M., Patel S.K., Natesan S., Dhama J., et al. Antibody-based immunotherapeutics and use of convalescent plasma to counter COVID-19: advances and prospects. Expert Opin Biol Ther. 2020;20(9):1033–1046. doi: 10.1080/14712598.2020.1796963. [DOI] [PubMed] [Google Scholar]
- 22.Ghareeb D.A., Saleh S.R., Nofal M.S., Kaddah M.M.Y., Hassan S.F., Seif I.K., et al. Potential therapeutic and pharmacological strategies for SARS-CoV2. J Pharm Investig. 2021;51(3):281–296. doi: 10.1007/s40005-021-00520-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iqbal Yatoo M., Hamid Z., Parray O.R., Wani A.H., Ul Haq A., Saxena A., et al. COVID-19 - Recent advancements in identifying novel vaccine candidates and current status of upcoming SARS-CoV-2 vaccines. Hum Vaccin Immunother. 2020;16(12):2891–2904. doi: 10.1080/21645515.2020.1788310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rabaan A.A., Al-Ahmed S.H., Sah R., Tiwari R., Yatoo M.I., Patel S.K., et al. SARS-CoV-2/COVID-19 and advances in developing potential therapeutics and vaccines to counter this emerging pandemic. Ann Clin Microbiol Antimicrob. 2020;19(1):40. doi: 10.1186/s12941-020-00384-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rabaan A.A., Al-Ahmed S.H., Muhammad J., Khan A., Sule A.A., Tirupathi R., et al. Role of inflammatory cytokines in COVID-19 patients: a review on molecular mechanisms, immune functions, immunopathology and immunomodulatory drugs to counter cytokine storm. Vaccines. 2021;9(5):436. doi: 10.3390/vaccines9050436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aleem A., Akbar S.A., Slenker A. StatPearls Publishing; Treasure Island (FL): 2022. Emerging Variants of SARS-CoV-2 And Novel Therapeutics Against Coronavirus (COVID-19) [Google Scholar]
- 27.Garcia-Montero C., Fraile-Martinez O., Bravo C., Torres-Carranza D., Sanchez-Trujillo L., Gomez-Lahoz A.M., et al. An updated review of SARS-CoV-2 Vaccines and the importance of effective vaccination programs in pandemic times. Vaccines. 2021;9(5):433. doi: 10.3390/vaccines9050433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rawat K., Kumari P., Saha L. COVID-19 vaccine: a recent update in pipeline vaccines, their design and development strategies. Eur J Pharmacol. 2021;892 doi: 10.1016/j.ejphar.2020.173751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.WHO 2022; Available from: 〈https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines〉. Accessed on: March 2, 2022.
- 30.Blasi F., Gramegna A., Sotgiu G., Saderi L., Voza A., Aliberti S., et al. SARS-CoV-2 vaccines: a critical perspective through efficacy data and barriers to herd immunity. Respir Med. 2021;180 doi: 10.1016/j.rmed.2021.106355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cihan P. Forecasting fully vaccinated people against COVID-19 and examining future vaccination rate for herd immunity in the US, Asia, Europe, Africa, South America, and the World. Appl Soft Comput. 2021;111 doi: 10.1016/j.asoc.2021.107708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garg S., Singh M.M., Deshmukh C.P., Bhatnagar N., Borle A.L., Kumar R. Critical interpretative synthesis of herd immunity for COVID-19 pandemic. J Fam Med Prim Care. 2021;10(3):1117–1123. doi: 10.4103/jfmpc.jfmpc_1127_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hirabara S.M., Serdan T.D.A., Gorjao R., Masi L.N., Pithon-Curi T.C., Covas D.T., et al. SARS-COV-2 variants: differences and potential of immune evasion. Front Cell Infect Microbiol. 2022;11 doi: 10.3389/fcimb.2021.781429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang M., Liang Y., Yu D., Du B., Cheng W., Li L., et al. A systematic review of vaccine breakthrough infections by SARS-CoV-2 Delta variant. Int J Biol Sci. 2022;18(2):889–900. doi: 10.7150/ijbs.68973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jia Z., Gong W. Will mutations in the spike protein of SARS-CoV-2 lead to the failure of COVID-19 vaccines? J Korean Med Sci. 2021;36(18) doi: 10.3346/jkms.2021.36.e124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gong W., Parkkila S., Wu X., Aspatwar A. SARS-CoV-2 variants and COVID-19 vaccines: current challenges and future strategies. Int Rev Immunol. 2022:1–22. doi: 10.1080/08830185.2022.2079642. [DOI] [PubMed] [Google Scholar]
- 37.Konings F., Perkins M.D., Kuhn J.H., Pallen M.J., Alm E.J., Archer B.N., et al. SARS-CoV-2 Variants of Interest and Concern naming scheme conducive for global discourse. Nat Microbiol. 2021;6(7):821–823. doi: 10.1038/s41564-021-00932-w. [DOI] [PubMed] [Google Scholar]
- 38.Chen J., Wang R., Gilby N.B., Wei G.W. Omicron variant (B.1.1.529): infectivity, vaccine breakthrough, and antibody resistance. J Chem Inf Model. 2022;62(2):412–422. doi: 10.1021/acs.jcim.1c01451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gupta N., Kaur H., Yadav P., Mukhopadhyay L., Sahay R.R., Kumar A., et al. Clinical characterization and Genomic analysis of COVID-19 breakthrough infections during second wave in different states of India. 2021. [DOI] [PMC free article] [PubMed]
- 40.Farinholt T., Doddapaneni H., Qin X., Menon V., Meng Q., Metcalf G., et al. Transmission event of SARS-CoV-2 delta variant reveals multiple vaccine breakthrough infections. BMC Med. 2021;19(1):255. doi: 10.1186/s12916-021-02103-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hossain M., Rahman S., Emran T., Mitra S., Islam M., Dhama K. Vol. 76. Archives of Razi Institute; 2021. pp. 1823–1830. (Recommendation and Roadmap of Mass Vaccination against COVID-19 Pandemic in Bangladesh as a Lower-Middle-Income Country (LMIC)). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin L., Liu Y., Tang X., He D. The disease severity and clinical outcomes of the SARS-CoV-2 variants of concern. Front Public Health. 2021;9 doi: 10.3389/fpubh.2021.775224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Salimi-Jeda A., Abbassi S., Mousavizadeh A., Esghaie M., Bokharaei-Salim F., Jeddi F., et al. SARS-CoV-2: current trends in emerging variants, pathogenesis, immune responses, potential therapeutic, and vaccine development strategies. Int Immunopharmacol. 2021;101(Pt A) doi: 10.1016/j.intimp.2021.108232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mascola J.R., Graham B.S., Fauci A.S. SARS-CoV-2 viral variants-tackling a moving target. J Am Med Assoc. 2021;325(13):1261–1262. doi: 10.1001/jama.2021.2088. [DOI] [PubMed] [Google Scholar]
- 45.Sharun K., Tiwari R., Dhama K., Emran T.B., Rabaan A.A., Al Mutair A. Emerging SARS-CoV-2 variants: impact on vaccine efficacy and neutralizing antibodies. Hum Vaccin Immunother. 2021;17(10):3491–3494. doi: 10.1080/21645515.2021.1923350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang R., Chen J., Hozumi Y., Yin C., Wei G.W. Emerging vaccine-breakthrough SARS-CoV-2 variants. ACS Infect Dis. 2022;8(3):546–556. doi: 10.1021/acsinfecdis.1c00557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tareq A.M., Emran T.B., Dhama K., Dhawan M., Tallei T.E. Impact of SARS-CoV-2 delta variant (B.1.617.2) in surging second wave of COVID-19 and efficacy of vaccines in tackling the ongoing pandemic. Hum Vaccin Immunother. 2021;17(11):4126–4127. doi: 10.1080/21645515.2021.1963601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.WHO. Tracking SARS-CoV-2 variants. Available from: 〈https://www.who.int/activities/tracking-SARS-CoV-2-variants/tracking-SARS-CoV-2-variants〉. 2022; Acessed on: August 1, 2022.
- 49.CDC. SARS-CoV-2 variant classifications and definitions. Available from: 〈https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-classifications.html#anchor_1632158885160〉. 2022; Accessed on October 9, 2022.
- 50.Syed A.M., Ciling A., Khalid M.M., Sreekumar B., Chen P.Y., Kumar G.R., et al. Omicron mutations enhance infectivity and reduce antibody neutralization of SARS-CoV-2 virus-like particles. medRxiv [Preprint]. 2022. [DOI] [PMC free article] [PubMed]
- 51.Shah M., Woo H.G. Omicron: a heavily mutated SARS-CoV-2 variant exhibits stronger binding to ACE2 and potently escapes approved COVID-19 therapeutic antibodies. Front Immunol. 2021;12 doi: 10.3389/fimmu.2021.830527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Torjesen I. Covid-19: Omicron may be more transmissible than other variants and partly resistant to existing vaccines, scientists fear. Br Med J. 2021;375:n2943. doi: 10.1136/bmj.n2943. [DOI] [PubMed] [Google Scholar]
- 53.Lambrou A.S., Shirk P., Steele M.K., Paul P., Paden C.R., Cadwell B., et al. Genomic surveillance for SARS-CoV-2 variants: predominance of the delta (B.1.617.2) and omicron (B.1.1.529) variants - United States, june 2021-january 2022. Morb Mortal Wkly Rep. 2022;71(6):206–211. doi: 10.15585/mmwr.mm7106a4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Desingu P.A., Nagarajan K. Omicron BA.2 lineage spreads in clusters and is concentrated in Denmark. J Med Virol. 2022 doi: 10.1002/jmv.27659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gao S.J., Guo H., Luo G. Omicron variant (B.1.1.529) of SARS-CoV-2, a global urgent public health alert! J Med Virol. 2022;94(4):1255–1256. doi: 10.1002/jmv.27491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Callaway E. Heavily mutated Omicron variant puts scientists on alert. Nature. 2021;600(7887):21. doi: 10.1038/d41586-021-03552-w. [DOI] [PubMed] [Google Scholar]
- 57.〈https://www.cbsnews.com/news/omicron-variant-covid-in-europe-netherlands-before-alert-raised/〉; Available from: 〈https://www.cbsnews.com/news/omicron-variant-covid-in-europe-netherlands-before-alert-raised/〉. Accessed on: January 5, 2022.
- 58.Jansen L., Tegomoh B., Lange K., Showalter K., Figliomeni J., Abdalhamid B., et al. Investigation of a SARS-CoV-2 B.1.1.529 (Omicron) variant cluster - Nebraska, november-december 2021. Morb Mortal Wkly Rep. 2021;70(5152):1782–1784. doi: 10.15585/mmwr.mm705152e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Callaway E., Ledford H. How bad is Omicron? What scientists know so far. Nature. 2021;600(7888):197–199. doi: 10.1038/d41586-021-03614-z. [DOI] [PubMed] [Google Scholar]
- 60.Dyer O. Covid-19: South Africa's surge in cases deepens alarm over omicron variant. Br Med J. 2021;375:n3013. doi: 10.1136/bmj.n3013. [DOI] [PubMed] [Google Scholar]
- 61.Petersen E., Ntoumi F., Hui D.S., Abubakar A., Kramer L.D., Obiero C., et al. Emergence of new SARS-CoV-2 Variant of Concern Omicron (B.1.1.529) - highlights Africa's research capabilities, but exposes major knowledge gaps, inequities of vaccine distribution, inadequacies in global COVID-19 response and control efforts. Int J Infect Dis. 2022;114:268–272. doi: 10.1016/j.ijid.2021.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.GISAID. Available from: 〈https://www.gisaid.org/hcov19-%20variants/〉. Accessed on: January 5, 2022.
- 63.Sun Y., Lin W., Dong W., Xu J. Origin and evolutionary analysis of the SARS-CoV-2 Omicron variant. J Biosaf Biosecur. 2022;4(1):33–37. doi: 10.1016/j.jobb.2021.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wei C., Shan K.J., Wang W., Zhang S., Huan Q., Qian W. Evidence for a mouse origin of the SARS-CoV-2 Omicron variant. J Genet Genom. 2021;48(12):1111–1121. doi: 10.1016/j.jgg.2021.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Du P., Gao G.F., Wang Q. The mysterious origins of the Omicron variant of SARS-CoV-2. Innovation. 2022;3(2) doi: 10.1016/j.xinn.2022.100206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lennerstrand J., Svensson L., Lundkvist Å. Hur har omikron uppstått och varför sprider den sig så snabbt? [How did Omicron evolve and why does this SARS-CoV-2 variant spread so fast?] Lakartidningen. 2022;119:21242. [PubMed] [Google Scholar]
- 67.Corey L., Beyrer C., Cohen M.S., Michael N.L., Bedford T., Rolland M. SARS-CoV-2 variants in patients with immunosuppression. N Engl J Med. 2021;385(6):562–566. doi: 10.1056/NEJMsb2104756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Avanzato V.A., Matson M.J., Seifert S.N., Pryce R., Williamson B.N., Anzick S.L., et al. Case study: prolonged infectious SARS-CoV-2 shedding from an asymptomatic immunocompromised individual with cancer. Cell. 2020;183(7):1901–1912. doi: 10.1016/j.cell.2020.10.049. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Choi B., Choudhary M.C., Regan J., Sparks J.A., Padera R.F., Qiu X., et al. Persistence and evolution of SARS-CoV-2 in an immunocompromised host. N Engl J Med. 2020;383(23):2291–2293. doi: 10.1056/NEJMc2031364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kemp S.A., Collier D.A., Datir R.P., Ferreira I., Gayed S., Jahun A., et al. SARS-CoV-2 evolution during treatment of chronic infection. Nature. 2021;592(7853):277–282. doi: 10.1038/s41586-021-03291-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Truong T.T., Ryutov A., Pandey U., Yee R., Goldberg L., Bhojwani D., et al. Increased viral variants in children and young adults with impaired humoral immunity and persistent SARS-CoV-2 infection: a consecutive case series. EBioMedicine. 2021;67 doi: 10.1016/j.ebiom.2021.103355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Data OWi 2021; Available from: 〈https://ourworldindata.org/covid-vaccinations〉. Accessed on: January 3, 2022.
- 73.Cele S., Karim F., Lustig G., San J.E., Hermanus T., Tegally H., et al. SARS-CoV-2 evolved during advanced HIV disease immunosuppression has Beta-like escape of vaccine and Delta infection elicited immunity. medRxiv [Preprint]. 2021.
- 74.Rahmani S., Rezaei N. Omicron (B.1.1.529) variant: development, dissemination, and dominance. J Med Virol. 2022;94(5):1787–1788. doi: 10.1002/jmv.27563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kupferschmidt K. Where did 'weird' Omicron come from? Science. 2021;374(6572):1179. doi: 10.1126/science.acx9738. [DOI] [PubMed] [Google Scholar]
- 76.Ren S.Y., Wang W.B., Gao R.D., Zhou A.M. Omicron variant (B.1.1.529) of SARS-CoV-2: mutation, infectivity, transmission, and vaccine resistance. World J Clin Cases. 2022;10(1):1–11. doi: 10.12998/wjcc.v10.i1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mallapaty S. Where did Omicron come from? Three key theories. Nature. 2022;602(7895):26–28. doi: 10.1038/d41586-022-00215-2. [DOI] [PubMed] [Google Scholar]
- 78.Liu X., Xiong J., Sun Z., Hu J., Thilakavathy K., Chen M., et al. Omicron: a chimera of two early SARS-CoV-2 lineages. Signal Transduct Target Ther. 2022;7(1):90. doi: 10.1038/s41392-022-00949-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Al-Tawfiq J.A., Hoang V.T., Le Bui N., Chu D.T., Memish Z.A. The emergence of the omicron (B.1.1.529) SARS-CoV-2 variant: what is the impact on the continued pandemic? J Epidemiol Glob Health. 2022:1–4. doi: 10.1007/s44197-022-00032-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Scott L., Hsiao N.Y., Moyo S., Singh L., Tegally H., Dor G., et al. Track Omicron's spread with molecular data. Science. 2021;374(6574):1454–1455. doi: 10.1126/science.abn4543. [DOI] [PubMed] [Google Scholar]
- 81.Wang L., Cheng G. Sequence analysis of the emerging SARS-CoV-2 variant Omicron in South Africa. J Med Virol. 2022;94(4):1728–1733. doi: 10.1002/jmv.27516. [DOI] [PubMed] [Google Scholar]
- 82.Ou J., Zhang Y., Wang Y., Zhang Z., Wei H., Yu J., et al. ACE2-targeting antibody suppresses SARS-CoV-2 Omicron and Delta variants. Signal Transduct Target Ther. 2022;7(1):43. doi: 10.1038/s41392-022-00913-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Khan A., Waris H., Rafique M., Suleman M., Mohammad A., Ali S.S., et al. The Omicron (B.1.1.529) variant of SARS-CoV-2 binds to the hACE2 receptor more strongly and escapes the antibody response: insights from structural and simulation data. Int J Biol Macromol. 2022;200:438–448. doi: 10.1016/j.ijbiomac.2022.01.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fantini J., Yahi N., Colson P., Chahinian H., La Scola B., Raoult D. The puzzling mutational landscape of the SARS-2-variant Omicron. J Med Virol. 2022;94(5):2019–2025. doi: 10.1002/jmv.27577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Du X., Tang H., Gao L., Wu Z., Meng F., Yan R., et al. Omicron adopts a different strategy from Delta and other variants to adapt to host. Signal Transduct Target Ther. 2022;7(1):45. doi: 10.1038/s41392-022-00903-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Saxena S.K., Kumar S., Ansari S., Paweska J.T., Maurya V.K., Tripathi A.K., et al. Characterization of the novel SARS-CoV-2 Omicron (B.1.1.529) variant of concern and its global perspective. J Med Virol. 2022;94(4):1738–1744. doi: 10.1002/jmv.27524. [DOI] [PubMed] [Google Scholar]
- 87.Harvey W.T., Carabelli A.M., Jackson B., Gupta R.K., Thomson E.C., Harrison E.M., et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat Rev Microbiol. 2021;19(7):409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sharma V., Rai H., Gautam D.N.S., Prajapati P.K., Sharma R. Emerging evidence on Omicron (B.1.1.529) SARS-CoV-2 variant. J Med Virol. 2022;94(5):1876–1885. doi: 10.1002/jmv.27626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cao Y., Wang J., Jian F., Xiao T., Song W., Yisimayi A., et al. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature. 2022;602(7898):657–663. doi: 10.1038/s41586-021-04385-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lennerstrand J., Svensson L., Lundkvist Å. How did Omicron evolve and why does this SARS-CoV-2 variant spread so fast? Lakartidningen. 2022:119. [PubMed] [Google Scholar]
- 91.Omotuyi O., Olubiyi O., Nash O., Afolabi E., Oyinloye B., Fatumo S., et al. SARS-CoV-2 Omicron spike glycoprotein receptor binding domain exhibits super-binder ability with ACE2 but not convalescent monoclonal antibody. Comput Biol Med. 2022;142 doi: 10.1016/j.compbiomed.2022.105226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fang Z., Peng L., Lin Q., Zhou L., Yang L., Feng Y., et al. Heterotypic vaccination responses against SARS-CoV-2 Omicron BA.2. bioRxiv. 2022:2022.03.22.485418. [DOI] [PMC free article] [PubMed]
- 93.Nainu F., Abidin R.S., Bahar M.A., Frediansyah A., Emran T.B., Rabaan A.A., et al. SARS-CoV-2 reinfection and implications for vaccine development. Hum Vaccin Immunother. 2020;16(12):3061–3073. doi: 10.1080/21645515.2020.1830683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Schmidt F., Weisblum Y., Rutkowska M., Poston D., DaSilva J., Zhang F., et al. High genetic barrier to SARS-CoV-2 polyclonal neutralizing antibody escape. Nature. 2021;600(7889):512–516. doi: 10.1038/s41586-021-04005-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Redd A.D., Nardin A., Kared H., Bloch E.M., Abel B., Pekosz A., et al. Minimal cross-over between mutations associated with Omicron variant of SARS-CoV-2 and CD8+ T cell epitopes identified in COVID-19 convalescent individuals. bioRxiv [Preprint]. 2021. [DOI] [PMC free article] [PubMed]
- 96.Flemming A. Omicron, the great escape artist. Nat Rev Immunol. 2022;22(2):75. doi: 10.1038/s41577-022-00676-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Young-Xu Y., Zwain G.M., Izurieta H.S., Korves C., Powell E.I., Smith J., et al. Effectiveness of mRNA COVID-19 Vaccines against Omicron among Veterans. medRxiv [Preprint]. 2022.
- 98.Tanne J.H. Covid 19: US cases rise amid omicron fears but booster shots offer protection, experts say. Br Med J. 2021;375:n3098. doi: 10.1136/bmj.n3098. [DOI] [PubMed] [Google Scholar]
- 99.Planas D., Saunders N., Maes P., Guivel-Benhassine F., Planchais C., Buchrieser J., et al. Considerable escape of SARS-CoV-2 Omicron to antibody neutralization. Nature. 2022;602(7898):671–675. doi: 10.1038/s41586-021-04389-z. [DOI] [PubMed] [Google Scholar]
- 100.Gruell H., Vanshylla K., Tober-Lau P., Hillus D., Schommers P., Lehmann C., et al. mRNA booster immunization elicits potent neutralizing serum activity against the SARS-CoV-2 Omicron variant. Nat Med. 2022;28(3):477–480. doi: 10.1038/s41591-021-01676-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Prasad N., Derado G., Nanduri S.A., Reses H.E., Dubendris H., Wong E., et al. Effectiveness of a COVID-19 additional primary or booster vaccine dose in preventing SARS-CoV-2 infection among nursing home residents during widespread circulation of the Omicron Variant - United States, february 14-march 27, 2022. Morb Mortal Wkly Rep. 2022;71(18):633–637. doi: 10.15585/mmwr.mm7118a4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kurahashi Y., Furukawa K., Sutandhio S., Tjan L.H., Iwata S., Sano S., et al. Cross-neutralizing activity against Omicron could be obtained in SARS-CoV-2 convalescent patients who received two doses of mRNA vaccination. J Infect Dis. 2022 doi: 10.1093/infdis/jiac178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Schmidt F., Muecksch F., Weisblum Y., Da Silva J., Bednarski E., Cho A., et al. Plasma neutralization of the SARS-CoV-2 Omicron variant. N Engl J Med. 2022;386(6):599–601. doi: 10.1056/NEJMc2119641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mohapatra R.K., Sarangi A.K., Kandi V., Azam M., Tiwari R., Dhama K. Omicron (B.1.1.529 variant of SARS-CoV-2); an emerging threat: current global scenario. J Med Virol. 2022;94(5):1780–1783. doi: 10.1002/jmv.27561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sohan M., Hossain M.J., Islam M.R. The SARS-CoV-2 Omicron (B.1.1.529) variant and effectiveness of existing vaccines: what we know so far. J Med Virol. 2022;94(5):1796–1798. doi: 10.1002/jmv.27574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tarke A., Coelho C.H., Zhang Z., Dan J.M., Yu E.D., Methot N., et al. SARS-CoV-2 vaccination induces immunological T cell memory able to cross-recognize variants from Alpha to Omicron. Cell. 2022;185(5):847–859. doi: 10.1016/j.cell.2022.01.015. e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Dolgin E. 2022; Available from: 〈https://www.nature.com/articles/d41586–022-00079–6〉 on January 16, 2022.
- 108.Zuo F., Abolhassani H., Du L., Piralla A., Bertoglio F., de Campos-Mata L., et al. Heterologous immunization with inactivated vaccine followed by mRNA booster elicits strong humoral and cellular immune responses against the SARS-CoV-2 Omicron variant. medRxiv [Preprint]. 2022. [DOI] [PMC free article] [PubMed]
- 109.Dejnirattisai W., Huo J., Zhou D., Zahradnik J., Supasa P., Liu C., et al. SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses. Cell. 2022;185(3):467–484. doi: 10.1016/j.cell.2021.12.046. e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Cele S., Jackson L., Khoury D.S., Khan K., Moyo-Gwete T., Tegally H., et al. Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization. Nature. 2022;602(7898):654–656. doi: 10.1038/s41586-021-04387-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Carreno J.M., Alshammary H., Tcheou J., Singh G., Raskin A.J., Kawabata H., et al. Activity of convalescent and vaccine serum against SARS-CoV-2 Omicron. Nature. 2022;602(7898):682–688. doi: 10.1038/s41586-022-04399-5. [DOI] [PubMed] [Google Scholar]
- 112.Behrens G.M.N., Barros-Martins J., Cossmann A., Ramos G.M., Stankov M.V., Odak I., et al. BNT162b2 boosted immune responses six months after heterologous or homologous ChAdOx1nCoV-19/BNT162b2 vaccination against COVID-19. medRxiv [Preprint]. 2021. [DOI] [PMC free article] [PubMed]
- 113.Tartof S.Y., Slezak J.M., Puzniak L., Hong V., Xie F., Ackerson B.K., et al. Durability of BNT162b2 vaccine against hospital and emergency department admissions due to the omicron and delta variants in a large health system in the USA: a test-negative case–control study. The Lancet Respiratory Medicine. 2022. [DOI] [PMC free article] [PubMed]
- 114.European Pharmaceutical Review 2022; Available from: 〈https://www.europeanpharmaceuticalreview.com/news/168012/studies-assess-immunogenicity-omicron-based-vaccines/〉. Accessed on: 12 June 2022.
- 115.Regev-Yochay G., Gonen T., Gilboa M., Mandelboim M., Indenbaum V., Amit S., et al. Efficacy of a fourth dose of covid-19 mRNA vaccine against Omicron. N Engl J Med. 2022;386(14):1377–1380. doi: 10.1056/NEJMc2202542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.WHO. Available from: 〈https://cdn.who.int/media/docs/default-source/immunization/covid-19/strategy-toachieve-global-covid-19-vaccination-by-mid-2022.pdf〉. Accessed on: March 5, 2022.
- 117.Andreata-Santos R., Janini L.M.R., Duraes-Carvalho R. From Alpha to Omicron SARS-CoV-2 variants: what their evolutionary signatures can tell us? J Med Virol. 2022;94(5):1773–1776. doi: 10.1002/jmv.27555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Mahmud-Al-Rafat A., Hewins B., Mannan A., Kelvin D.J., Billah M.M. COVID-19 vaccine inequity, dependency, and production capability in low-income and middle-income countries: the case of Bangladesh. Lancet Infect Dis. 2022;22(3):310–312. doi: 10.1016/S1473-3099(22)00028-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Andrews N., Stowe J., Kirsebom F., Toffa S., Rickeard T., Gallagher E., et al. Covid-19 vaccine effectiveness against the Omicron (B.1.1.529) variant. N Engl J Med. 2022;386(16):1532–1546. doi: 10.1056/NEJMoa2119451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hanai T. Quantitative in silico analysis of SARS-CoV-2 S-RBD omicron mutant transmissibility. Talanta. 2022;240 doi: 10.1016/j.talanta.2022.123206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Nishiura H., Ito K., Anzai A., Kobayashi T., Piantham C., Rodriguez-Morales A.J. Relative reproduction number of SARS-CoV-2 Omicron (B.1.1.529) compared with Delta Variant in South Africa. J Clin Med. 2021;11(1):30. doi: 10.3390/jcm11010030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Halfmann P.J., Iida S., Iwatsuki-Horimoto K., Maemura T., Kiso M., Scheaffer S.M., et al. SARS-CoV-2 Omicron virus causes attenuated disease in mice and hamsters. Nature. 2022;603(7902):687–692. doi: 10.1038/s41586-022-04441-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kozlov M. Omicron overpowers key COVID antibody treatments in early tests. Nature. 2021 doi: 10.1038/d41586-021-03829-0. [DOI] [PubMed] [Google Scholar]
- 124.Cedro-Tanda A., Gomez-Romero L., de Anda-Jauregui G., Garnica-Lopez D., Alfaro-Mora Y., Sanchez-Xochipa S., et al. Early genomic, epidemiological, and clinical description of the SARS-CoV-2 Omicron Variant in Mexico City. Viruses. 2022;14:3. doi: 10.3390/v14030545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Maslo C., Friedland R., Toubkin M., Laubscher A., Akaloo T., Kama B. Characteristics and outcomes of hospitalized patients in South Africa during the COVID-19 Omicron wave compared with previous waves. J Am Med Assoc. 2022;327(6):583–584. doi: 10.1001/jama.2021.24868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Espenhain L., Funk T., Overvad M., Edslev S.M., Fonager J., Ingham A.C., et al. Epidemiological characterisation of the first 785 SARS-CoV-2 Omicron variant cases in Denmark. Eur Surveill. 2021;26:50. doi: 10.2807/1560-7917.ES.2021.26.50.2101146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Callaway E. Why does the Omicron sub-variant spread faster than the original? Nature. 2022;602(7898):556–557. doi: 10.1038/d41586-022-00471-2. [DOI] [PubMed] [Google Scholar]
- 128.Ledford H. The next variant: three key questions about what's after Omicron. Nature. 2022;603(7900):212–213. doi: 10.1038/d41586-022-00510-y. [DOI] [PubMed] [Google Scholar]
- 129.Mohapatra R.K., Kandi V., Tuli H.S., Chakraborty C., Dhama K. The recombinant variants of SARS-CoV-2: concerns continues amid COVID-19 pandemic. J Med Virol. 2022 doi: 10.1002/jmv.27780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Viana R., Moyo S., Amoako D.G., Tegally H., Scheepers C., Althaus C.L., et al. Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in southern Africa. Nature. 2022;603(7902):679–686. doi: 10.1038/s41586-022-04411-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Yu J., Collier A.Y., Rowe M., Mardas F., Ventura J.D., Wan H., et al. Neutralization of the SARS-CoV-2 Omicron BA.1 and BA.2 Variants. N Engl J Med. 2022;386(16):1579–1580. doi: 10.1056/NEJMc2201849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Desingu P.A., Nagarajan K., Dhama K. Emergence of Omicron third lineage BA.3 and its importance. J Med Virol. 2022;94(5):1808–1810. doi: 10.1002/jmv.27601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lyngse F.P., Kirkeby C.T., Denwood M., Christiansen L.E., Mølbak K., Møller C.H., et al. Transmission of SARS-CoV-2 Omicron VOC subvariants BA.1 and BA.2: Evidence from Danish Households. medRxiv. 2022:2022.01.28.22270044.
- 134.Tegally H., Moir M., Everatt J., Giovanetti M., Scheepers C., Wilkinson E., et al. Continued Emergence and Evolution of Omicron in South Africa: New BA.4 and BA.5 lineages. medRxiv. 2022:2022.05.01.22274406.
- 135.Khan K., Karim F., Ganga Y., Bernstein M., Jule Z., Reedoy K., et al. Omicron sub-lineages BA.4/BA.5 escape BA.1 infection elicited neutralizing immunity. medRxiv. 2022:2022.04.29.22274477.
- 136.Chakraborty C., Bhattacharya M., Sharma A.R., Dhama K. Recombinant SARS-CoV-2 variants XD, XE, and XF: the emergence of recombinant variants requires an urgent call for research - Correspondence. Int J Surg. 2022;102 doi: 10.1016/j.ijsu.2022.106670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Pérez-Losada M., Arenas M., Galán J.C., Palero F., González-Candelas F. Recombination in viruses: mechanisms, methods of study, and evolutionary consequences. Infect Genet Evol. 2015;30:296–307. doi: 10.1016/j.meegid.2014.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Aoki A., Mori Y., Okamoto Y., Jinno H. Simultaneous screening of SARS-CoV-2 Omicron and Delta Variants using high-resolution melting analysis. Biol Pharm Bull. 2022;45(4):394–396. doi: 10.1248/bpb.b21-01081. [DOI] [PubMed] [Google Scholar]
- 139.Liu T., Kang L., Li Y., Huang J., Guo Z., Xu J., et al. Simultaneous detection of seven human coronaviruses by multiplex PCR and MALDI-TOF MS. Covid. 2021;2(1):5–17. [Google Scholar]
- 140.Fu J.Y.L., Chong Y.M., Sam I.C., Chan Y.F. SARS-CoV-2 multiplex RT-PCR to detect variants of concern (VOCs) in Malaysia, between January to May 2021. J Virol Methods. 2022;301 doi: 10.1016/j.jviromet.2022.114462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Sexton M.E., Waggoner J.J., Carmola L.R., Nguyen P.V., Wang E., Khosravi D., et al. Rapid detection and characterization of SARS-CoV-2 omicron variant in a returning traveler. Clin Infect Dis. 2022 doi: 10.1093/cid/ciac032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Micheli V., Bracchitta F., Rizzo A., Mancon A., Mileto D., Lombardi A., et al. First identification of the new SARS-CoV-2 Omicron variant (B.1.1.529) in Italy. Clin Infect Dis. 2022 doi: 10.1093/cid/ciab1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kandeel M., Mohamed M.E.M., Abd El-Lateef H.M., Venugopala K.N., El-Beltagi H.S. Omicron variant genome evolution and phylogenetics. J Med Virol. 2022;94(4):1627–1632. doi: 10.1002/jmv.27515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Express TI 2022; Available from: 〈https://indianexpress.com/article/india/india-test-kit-detect-omicron-icmr-7710360/〉. Accessed on: March 2, 2022.
- 145.Ferre V.M., Peiffer-Smadja N., Visseaux B., Descamps D., Ghosn J., Charpentier C. Omicron SARS-CoV-2 variant: what we know and what we don't. Anaesth Crit Care Pain Med. 2022;41(1) doi: 10.1016/j.accpm.2021.100998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Brito A.F., Semenova E., Dudas G., Hassler G.W., Kalinich C.C., Kraemer M.U.G., et al. Global disparities in SARS-CoV-2 genomic surveillance. medRxiv. 2021. [DOI] [PMC free article] [PubMed]
- 147.Burki T.K. The race between vaccination and evolution of COVID-19 variants. 2021;9(11):e109. doi: 10.1016/S2213-2600(21)00443-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Chen J., Wang R., Wei G.-W. Review of the mechanisms of SARS-CoV-2 evolution and transmission. arXiv [Preprint]. 2021.
- 149.Hossain M.J., Soma M.A., Islam M.R., Emran T.B. Urgent call for actionable measures to fight the current co-epidemic of dengue burden during the SARS-CoV-2 delta variant era in South-Asia. Ethics Med Public Health. 2021;19 doi: 10.1016/j.jemep.2021.100726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Krause P.R., Fleming T.R., Longini I.M., Peto R., Briand S., Heymann D.L., et al. SARS-CoV-2 variants and vaccines. N Engl J Med. 2021;385(2):179–186. doi: 10.1056/NEJMsr2105280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Xie X., Han J.B., Ma G., Feng X.L., Li X., Zou Q.C., et al. Emerging SARS-CoV-2 B.1.621/Mu variant is prominently resistant to inactivated vaccine-elicited antibodies. Zool Res. 2021;42(6):789–791. doi: 10.24272/j.issn.2095-8137.2021.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Asrani P., Tiwari K., Eapen M.S., Hassan M.D.I., Sohal S.S. Containment strategies for COVID-19 in India: lessons from the second wave. Expert Rev Anti Infect Ther. 2022:1–7. doi: 10.1080/14787210.2022.2036605. [DOI] [PubMed] [Google Scholar]
- 153.Hassan W., Kazmi S.K., Tahir M.J., Ullah I., Royan H.A., Fahriani M., et al. Global acceptance and hesitancy of COVID-19 vaccination: a narrative review. Narra J. 2021;1(3) doi: 10.52225/narra.v1i3.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Rosiello D., Anwar S., Yufika A., Adam R., Ismaeil M., Ismail A., et al. Acceptance of COVID-19 vaccination at different hypothetical efficacy and safety levels in ten countries in Asia, Africa, and South America. Narra J. 2021;1(3) doi: 10.52225/narra.v1i3.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Sharun K., Dhama K. COVID-19 vaccine diplomacy and equitable access to vaccines amid ongoing pandemic. Arch Med Res. 2021;52(7):761–763. doi: 10.1016/j.arcmed.2021.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Obinna D.N. Solidarity across borders: A pragmatic need for global COVID-19 vaccine equity. Int J Health Plann Manag. 2022;37(1):21–29. doi: 10.1002/hpm.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Snehota M., Vlckova J., Cizkova K., Vachutka J., Kolarova H., Klaskova E., et al. Acceptance of a vaccine against COVID-19 - a systematic review of surveys conducted worldwide. Bratisl Lek Listy. 2021;122(8):538–547. doi: 10.4149/BLL_2021_086. [DOI] [PubMed] [Google Scholar]
- 158.Wouters O.J., Shadlen K.C., Salcher-Konrad M., Pollard A.J., Larson H.J., Teerawattananon Y., et al. Challenges in ensuring global access to COVID-19 vaccines: production, affordability, allocation, and deployment. Lancet. 2021;397(10278):1023–1034. doi: 10.1016/S0140-6736(21)00306-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Yan Z.P., Yang M., Lai C.L. COVID-19 vaccines: a review of the safety and efficacy of current clinical trials. Pharmaceuticals. 2021;14(5):406. doi: 10.3390/ph14050406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Bari M.S., Hossain M.J., Akhter S., Emran T.B. Delta variant and black fungal invasion: a bidirectional assault might worsen the massive second/third stream of COVID-19 outbreak in South-Asia. Ethics Med Public Health. 2021;19 doi: 10.1016/j.jemep.2021.100722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Viveiros-Rosa S., Mendes C., Farfán-Cano G., El-Shazly M. The race for clinical trials on Omicron-based COVID-19 vaccine candidates: updates from global databases. Narra J. 2022;2(3) doi: 10.52225/narra.v2i3.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Fiolet T., Kherabi Y., MacDonald C.J., Ghosn J., Peiffer-Smadja N. Comparing COVID-19 vaccines for their characteristics, efficacy and effectiveness against SARS-CoV-2 and variants of concern: a narrative review. Clin Microbiol Infect. 2022;28(2):202–221. doi: 10.1016/j.cmi.2021.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Sk Varshney V., N K Prasanna P. Vaccine diplomacy: exploring the benefits of international collaboration. Curr Trends Biotechnol Pharm. 2020;15(1):110–114. [Google Scholar]
- 164.Rahimi F., Talebi Bezmin Abadi A. Criticality of physical/social distancing, handwashing, respiratory hygiene and face-masking during the COVID-19 pandemic and beyond. Int J Clin Pract. 2020;74(11) doi: 10.1111/ijcp.13656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Su Z., Wen J., McDonnell D., Goh E., Li X., Segalo S., et al. Vaccines are not yet a silver bullet: the imperative of continued communication about the importance of COVID-19 safety measures. Brain Behav Immun Health. 2021;12 doi: 10.1016/j.bbih.2021.100204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.ECDC 2021; Available from: 〈https://www.ecdc.europa.eu/sites/default/files/documents/threat-assessment-covid-19-emergence-sars-cov-2-variant-omicron-december-2021.pdf〉. Accessed on: March 2, 2022.
- 167.Choudhary O.P., Priyanka, Singh I. Vaccine certificate during domestic traveling: a potential initiative to prevent COVID-19 waves in India. Hum Vaccin Immunother. 2021;17(10):3487–3488. doi: 10.1080/21645515.2021.1936862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Khan A., Bibi S., Kanwal H., Umm E.K., Hussain H. Omicron: a new face of COVID-19 pandemic. Health Sci Rep. 2022;5(2) doi: 10.1002/hsr2.526. [DOI] [PMC free article] [PubMed] [Google Scholar]