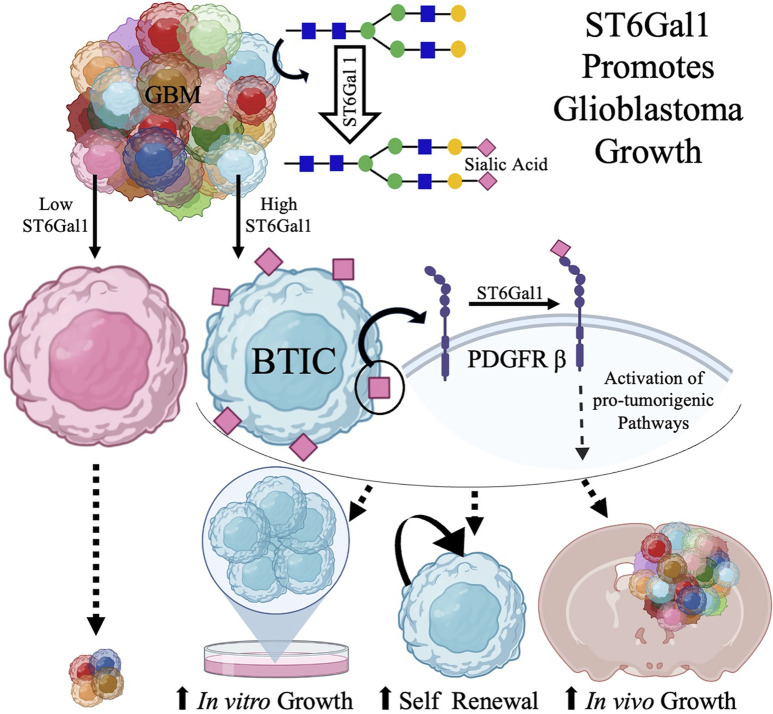
Abstract
One of the least-investigated areas of brain pathology research is glycosylation, which is a critical regulator of cell surface protein structure and function. β-Galactoside α2,6-sialyltransferase (ST6GAL1) is the primary enzyme that α2,6 sialylates N-glycosylated proteins destined for the plasma membrane or secretion, thereby modulating cell signaling and behavior. We demonstrate a potentially novel, protumorigenic role for α2,6 sialylation and ST6GAL1 in the deadly brain tumor glioblastoma (GBM). GBM cells with high α2,6 sialylation exhibited increased in vitro growth and self-renewal capacity and decreased mouse survival when orthotopically injected. α2,6 Sialylation was regulated by ST6GAL1 in GBM, and ST6GAL1 was elevated in brain tumor-initiating cells (BTICs). Knockdown of ST6GAL1 in BTICs decreased in vitro growth, self-renewal capacity, and tumorigenic potential. ST6GAL1 regulates levels of the known BTIC regulators PDGF Receptor β (PDGFRB), Activated Leukocyte Cell Adhesion Molecule, and Neuropilin, which were confirmed to bind to a lectin-recognizing α2,6 sialic acid. Loss of ST6GAL1 was confirmed to decrease PDGFRB α2,6 sialylation, total protein levels, and the induction of phosphorylation by PDGF-BB. Thus, ST6GAL1-mediated α2,6 sialylation of a select subset of cell surface receptors, including PDGFRB, increases GBM growth.
Keywords: Cell Biology, Oncology
Keywords: Brain cancer, Glycobiology
Introduction
Glioblastoma (GBM) is one of the most aggressive and fatal cancers with a median survival of less than 15 months past diagnosis (1, 2). Contributing to treatment failures and disease progression is the highly heterogeneous nature of GBMs, including a subset of GBM cells called brain tumor-initiating cells (BTICs). BTICs have similarities to neural progenitors including expression of the stem cell marker SOX2 and the ability to self-renew in neurosphere formation assays, but BTICs can form tumors when orthotopically injected (3–10). BTICs are thought to be a major cause of disease recurrence, and it is therefore imperative that the cellular mechanisms involved in BTIC maintenance are elucidated. While there are many studies dedicated to understanding the genome and proteome of GBMs and BTICs, studies of glycosylation as a post-translational modification are limited, even though altered cell surface glycosylation was one of the earliest modifications observed in malignant neoplastic progression.
Among the various glycosyltransferases present in human cells, Golgi sialyltransferase β-galactoside α2,6-sialyltransferase 1 (ST6GAL1) and 2 (ST6GAL2) add sialic acid residues in α2,6 linkage to membrane-bound and secreted N-glycosylated proteins (11). Due to the position and negative charge of sialic acid, α2,6 sialylation can alter conformation, clustering, and retention of glycoproteins (12–14). Altered glycosylation is a cancer hallmark, and ST6GAL1 is one of the main glycosyltransferases upregulated in malignancies. In epithelial cancers, ST6GAL1 has been shown to regulate α2,6 sialylation and impart tumor-initiating cell (TIC) phenotypes, including sustained proliferative capacity, upregulation of TIC markers (CD133, ALDH1), sphere formation capacity, resistance to cell death induced by chemotherapies, growth factor withdrawal, and inflammatory mediators (15–24). To date, ST6GAL1 is known to exert its biological effects in cancer cells by modulating the function of select receptors including TNF Receptor 1 (TNFR1) and EGFR, leading to the activation of transcription factors such as NF-κB (25, 26). While these pathways are known to play critical roles in brain tumors, the levels, or ability, of ST6GAL1, ST6GAL2, or α2,6 sialylation to modulate BTIC signaling or maintenance to increase glioma growth has not been investigated.
In contrast to the protumorigenic role of ST6GAL1 in pancreatic and ovarian cancers (among others), prior reports suggested that ST6GAL1 is suppressed and plays a tumor-suppressive role in GBM (27, 28). The studies used standard human glioma cell lines, primarily U373MG, propagated in media containing FBS, which is known to promote cell differentiation. Studies herein show that ST6GAL1 levels are much higher in GBM BTICs than in differentiated GBM cells. Importantly, we define a potentially novel protumorigenic role for ST6GAL1 in GBM due, in part, to the regulation of α2,6 sialylation in BTICs. We have assessed α2,6 sialylation, specifically by ST6GAL1, using patient-derived xenografts (PDXs) representing different GBM subtypes. Using a lectin that binds α2,6 sialic acids, we demonstrated that α2,6 sialylationhi GBM cells were enriched for growth in vitro and in vivo. We determined that ST6GAL1 levels were high in BTICs and, using lentivirus expressing nontargeting or ST6GAL1-directed shRNAs, we demonstrated a critical role for ST6GAL1 in α2,6 sialylation in GBMs. In this study, we have also identified specific N-glycosylated proteins that are sialylated and whose expression is regulated by ST6GAL1. Our findings define α2,6 sialylation and ST6GAL1 as central regulators of GBM growth and BTIC maintenance. These results are important because ST6GAL1 inhibitors are in development, although specific sialyltransferase inhibitors are not yet available. Furthermore, defining α2,6 sialylated proteins in GBMs or BTICs may identify new biomarkers for the disease or cellular subsets.
Results
GBM cells with elevated α2,6 sialylation have increased growth in vitro and in vivo.
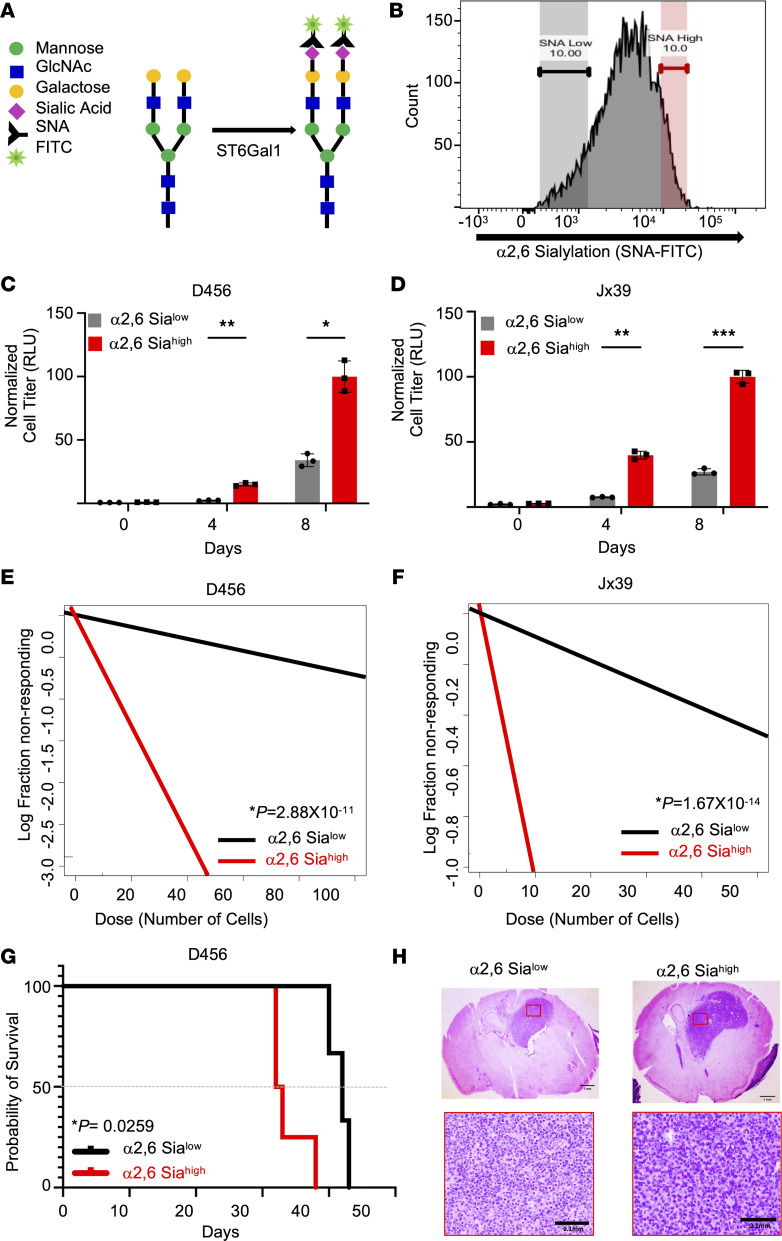
While TIC maintenance is known to require cell surface glycosylation, the interplay between cell surface protein modification/cell signaling and TIC maintenance, particularly with regard to sialylation, remains largely understudied (16, 29–31). To first determine if there were any functional consequences for α2,6 sialylation in GBM, we utilized Sambucus nigra agglutinin (SNA), a lectin that specifically binds to terminal Gal- or GalNAc-linked α2,6-linked sialic acid (Figure 1A). Using SNA conjugated to FITC with FACS, GBM cells were isolated directly from PDXs and sorted for α2,6 sialylation (Figure 1B). GBM cells in the highest tenth percentile of intensity for SNA binding were identified as α2,6 sialylationhi and those in the lowest tenth percentile of intensity for SNA binding as α2,6 sialylationlo. In 2 different xenografts, α2,6 sialylationhi GBM cells had significantly higher growth rates than α2,6 sialylationlo cells as determined via cell titer assays (Figure 1, C and D) and crystal violet staining (Supplemental Figure 1, A and B; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.158799DS1). As α2,6 sialylation has been linked to pluripotency, we next sought to determine if α2,6 sialylation could impact self-renewal and the percentages of BTICs in GBMs using neurosphere formation assays. Using cells directly isolated from 2 different PDXs and sorted for α2,6 sialylation using SNA-FITC, we determined an enrichment for neurosphere-formation capacity in α2,6 sialylationhi GBM cells (Figure 1, E and F). We next evaluated the importance of α2,6 sialylation for GBM growth in vivo. α2,6 sialylationhi or α2,6 sialylationlo GBM cells were intracranially injected into BALB/c nu/nu mice and monitored daily for the development of neurologic signs (Figure 1, G and H). Consistent with the in vitro data, the Kaplan-Meier survival curves revealed that the mice injected with α2,6 sialylationhi cells had significantly decreased survival, demonstrating that α2,6 sialylation promotes GBM growth in vivo. The presence of brain tumors in mice with neurologic signs was confirmed via H&E staining of fixed tissue sections (Figure 1H). Blinded review of these sections or those stained with Ki67 by a neuropathologist did not indicate substantial pathologic differences at this endpoint. These data defined a novel role for α2,6 sialylation in GBM growth and self-renewal.
Figure 1. α2,6 Sialylation increases GBM growth and self-renewal.
(A) Schematic of SNA, lectin with high affinity for α2,6 sialic acid, tagged with FITC as used for flow cytometry. (B) Representative histogram using SNA-FITC for FACS to sort SNAhi or α2,6 sialylationhi (highest 10% intensity) and SNAlo or α2,6 sialylationlo (lowest 10% intensity) cells. A total of 1,000 α2,6 sialylationhi versus α2,6 sialylationlo cells isolated from (C) D456 and (D) JX39 GBM PDXs were directly plated during FACS, and growth was measured over time using CellTiter-Glo 2.0 (luminescence, RLU). Individual data points are shown with the error bars as mean ± SD (n = 3). *P < 0.05; **P < 0.01; ***P < 0.001, 2-way ANOVA with Tukey’s multiple comparisons test. The experiments were repeated in 3 independent biological replicates. Data from 1 representative experiment are shown. Differences in self-renewal and BTIC frequencies were determined using in vitro limiting dilution assays with α2,6 sialylationhi versus α2,6 sialylationlo cells isolated from (E) D456 and (F) JX39 GBM PDXs. Each group was plated in decreasing number of cells (100, 50, 10, 5, and 1 cell per well). Extreme limiting dilution analysis (ELDA) was done using the software (http://bioinf.wehi.edu.au/software/elda/). P values were calculated from χ2 analysis of group comparisons. The experiments were repeated in 3 independent biological replicates. Data from 1 representative experiment are shown. (G) Kaplan-Meier survival curves for BALB/c nu/nu mice injected orthotopically with 2,500 α2,6 sialylationhi or α2,6 sialylationlo cells isolated from D456 PDX cells and euthanized upon development of neurological signs. P value was calculated using log-rank (Mantel-Cox) test. (H) Representative histological images of tumors stained with H&E support the presence of brain tumors in mice with neurological signs. Top panels: Image objective = 1.25×; scale bar: 1.0 mm. Bottom panels: Image objective = 20×; scale bar: 0.1 mm.
α2,6 Sialylation in GBM is regulated by ST6GAL1, which is increased in BTICs.
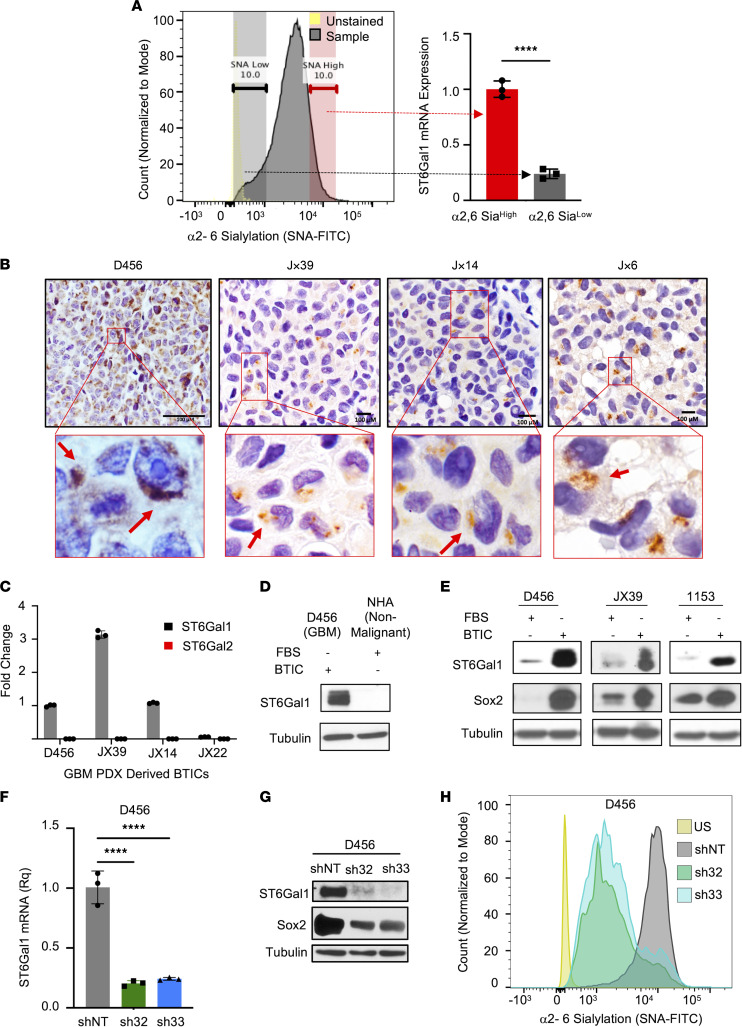
After verifying that α2,6 sialylation plays an important role in GBM, we next sought to determine the sialyltransferase mediating this effect. The primary enzyme that α2,6 sialylates N-glycosylated proteins in the secretory pathway is ST6GAL1 (11, 32). ST6GAL1 is thought to be broadly expressed with its paralog ST6GAL2, relatively restricted and at substantially lower levels, but RNA-Seq suggests that both ST6GAL1 and ST6GAL2 are expressed in brain tissue (32, 33). While ST6GAL1 is known to be important for N-glycan sialylation in the mouse brain (34), ST6GAL1/2 expression and function in the human brain or brain tumors have not been well characterized. Our analysis of data from the Human Protein Atlas RNA-Seq normal tissues project (PRJEB4337) demonstrated higher expression of ST6GAL1 mRNA compared with ST6GAL2 in normal brain tissue (Supplemental Figure 2A) (35). Both ST6GAL1 and ST6GAL2 were detected in GBMs (regardless of Isocitrate dehydrogenase status) in data from The Cancer Genome Atlas accessed via Gliovis at http://gliovis.bioinfo.cnio.es (36, 37) and/or GBMseq (38) accessed online (Supplemental Figure 2, B–G). While there was no difference in ST6GAL1 mRNA in GBM compared to nontumor tissue, ST6Gal2 was significantly decreased (Supplemental Figure 2, B and C). We further determined that higher levels of ST6GAL1 or lower levels of ST6Gal2 correlated with worse glioma patient survival, but there was no difference in survival with a similar mRNA cutoff in only GBM patients (Supplemental Figure 2, H and I; and data not shown). These data suggested ST6GAL1 as a potential mediator of α2,6 sialylation in GBM, although roles for ST6GAL2 could not be eliminated. We recognize the limitations of interpreting mRNA data from bulk tumor. However, we did verify that α2,6 sialylationhi cells isolated from GBM PDXs had higher levels of ST6GAL1 mRNA (Figure 2A). IHC using an extensively validated Ab confirmed that the typical punctate Golgi expression of ST6GAL1 was observed in sections of GBM PDXs, indicating ST6GAL1 is expressed in vivo (Figure 2B). As ST6GAL1 is highly expressed in induced pluripotent stem cells (iPSCs), has been implicated in the maintenance of epithelial cancer TICs, and is known to be regulated by the BTIC marker SOX2 (16, 39–42), we further evaluated ST6GAL1 and ST6GAL2 expression in BTICs. Quantitative real-time PCR (qRT-PCR) analysis revealed that, while heterogeneous for the extent of expression, ST6GAL1 was present in all PDX-derived BTICs tested (Figure 2C). In contrast, ST6GAL2 mRNA was not detected in the same BTICs (Figure 2C) but was confirmed to be expressed in nontumorigenic but immortalized human astrocytes (Supplemental Figure 2J). ST6GAL1 protein was higher in BTICs compared with human astrocytes (Figure 2D and Supplemental Figure 3A), and the notion that elevated ST6GAL1 expression is present in BTICs was further confirmed. When IB was used to compare BTICs and their differentiated counterparts (as determined by differential SOX2 expression), ST6GAL1 protein was consistently higher in BTICs (Figure 2E and Supplemental Figure 3, B and C). These data suggested a substantial role for ST6GAL1 in GBM that could depend on the differentiation state. To evaluate whether α2,6 sialylation is indeed regulated by ST6GAL1 in GBM, we utilized a lentiviral system to express 2 different shRNAs targeting ST6GAL1 (sh32 and sh33) or a nontargeting control (shNT) in BTICs isolated from PDXs. We confirmed knockdown (KD) of ST6GAL1 mRNA and protein using qRT-PCR (Figure 2F) and IB (Figure 2G and Supplemental Figure 3D). These IBs also revealed that targeting ST6GAL1 reduced expression of SOX2 (Figure 2G and Supplemental Figure 3E), providing the first suggestion that ST6GAL1 may be important for BTIC maintenance. Importantly, KD of ST6GAL1 reduced α2,6 sialylation as determined by FACS analysis with SNA-FITC (Figure 2H). Together, these data indicate that α2,6 sialylation in BTICs is largely imparted by ST6GAL1.
Figure 2. ST6GAL1 is expressed in GBM and elevated in BTICs to increase α2,6 sialylation.
(A) Example histogram of FACS with SNA-FITC to identify α2,6 sialylationhi and α2,6 sialylationlo cells that were lysed after sorting and ST6GAL1 mRNA levels determined using qRT-PCR. Relative quantification (Rq) is normalized to SNAhi. Individual data points are shown with the error bars as mean ± SD (n = 3). ****P < 0.0001 with 2-tailed t test. The experiments were repeated in at least 3 independent biological replicates. Data from 1 representative experiment are shown. (B) IHC of ST6GAL1 in sections of 4 different s.c. human GBM xenografts. From left, D456, JX39, Jx14, and JX6. Image objective D456 40x and JX39, Jx14, and JX6 60x oil immersion. Scale bars: 100 μm. The red box represents the section from which the magnified images were collected. The red arrows indicate punctate Golgi staining for ST6GAL1. (C) mRNA levels of ST6GAL1 or ST6GAL2 in BTICs isolated from the indicated GBM xenografts; Rq for individual PDX is normalized to D456. Individual data points are shown with the error bars as mean ± SD (n = 3). The experiments were repeated in at least 3 independent biological replicates. Data from 1 representative experiment are shown. (D) ST6GAL1 protein levels in nonmalignant brain cells (NHA) or GBM (D456) were determined via IB. NHA, normal human astrocytes. (E) ST6GAL1 protein levels in BTICs or BTICs differentiated in FBS for 96 hours were determined via IB. Differences in expression of the BTIC marker SOX2 were used as control. The experiments were repeated in at least 3 independent biological replicates. Data from 1 representative experiment are shown. (F–H) Lentivirus was used to transduce D456 cells with nontargeting control shRNA (shNT) or 2 different shRNA constructs targeting ST6GAL1 (sh32 and sh33). Cells for analysis and experiments were collected after 24 hours of lentivirus exposure and 72 hours of antibiotic selection. (F) KD of ST6GAL1 mRNA was validated using qRT-PCR. Individual data points are shown with the error bars as mean ± SD (n = 3). ****P < 0.0001 with 1-way ANOVA. (G) KD of ST6GAL1 protein was validated with IB. SOX2 expression in the shNT compared with the KD groups were determined via IB. (H) Representative histogram of FACS analysis with SNA-FITC of BTICs with and without ST6GAL1 modulation in D456 PDX cells demonstrating reduced α2,6 sialylation. The experiments were repeated in at least 3 independent biological replicates. Data from 1 representative experiment are shown.
ST6GAL1 is critical for BTIC maintenance.
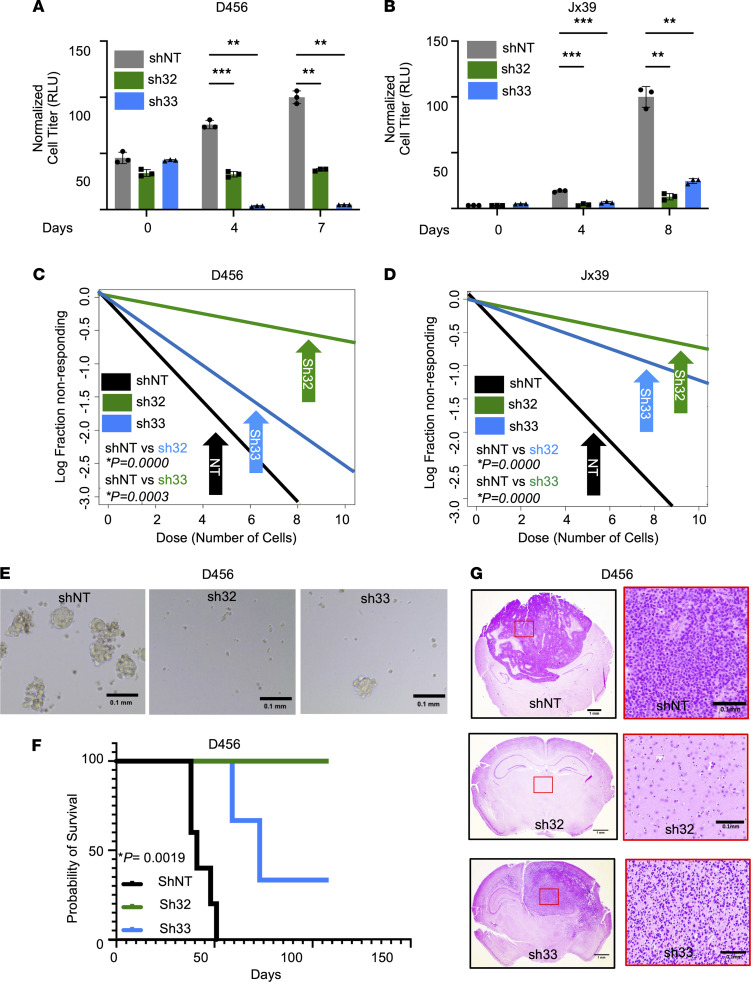
To determine if loss of ST6GAL1 could result in phenotypes similar to those in α2,6 sialylationlo cells, we utilized the lentiviral system described above (Figure 2, F–H). BTICs expressing either of 2 different ST6GAL1 shRNAs had significantly decreased in vitro growth compared with nontargeting controls as determined via cell titer assays (Figure 3, A and B) or crystal violet staining (Supplemental Figure 4, A and B). As ST6GAL1-specific inhibitors are not yet available, we next employed an analog of sialic acid to determine the impact of sialyltransferase inhibition on BTIC growth. 3Fax-Peracetyl Neu5Ac inhibits sialyltransferase via the generation of a mimetic of cytosine 5′-monophosphate N-acetylneuraminic acid (CMP-Neu5Ac), the substrate Golgi sialyltransferases use to form sialic acid (43). 3Fax-Peracetyl Neu5Ac significantly decreased the growth of BTICs derived from D456 (Supplemental Figure 4, C and D) and Jx39 (Supplemental Figure 4, E and F) PDX. BTICs were substantially more sensitive to the growth inhibitory effects of sialyltransferase inhibition than their non-BTIC counterparts (Supplemental Figure 4, D and F). Additional experiments determined impacts of loss of ST6GAL1 on BTIC self-renewal; similar to the α2,6 sialylationlo cells, ST6GAL1 KD cells have significantly decreased neurosphere formation capacity as determined in extreme limiting dilution assays (Figure 3, C–E). Furthermore, loss of ST6GAL1 significantly inhibited the ability of BTICs to initiate tumors in vivo (Figure 3, F and G). These data indicate that ST6GAL1 promotes BTIC maintenance in vitro and GBM growth in vivo and defines a novel protumorigenic role for ST6GAL1 and α2,6 sialylation in GBM.
Figure 3. Targeting ST6GAL1 decreases GBM growth and self-renewal.
Growth of (A) D456 and (B) Jx39 BTICs with (sh32, sh33) and without (nontargeting control, shNT) ST6GAL1 was measured over time using CellTiter-Glo 2.0 (luminescence, RLU). Individual data points are shown with the error bars as mean ± SD (n = 3). **P < 0.01; ***P < 0.001, 2-way ANOVA with Tukey’s multiple comparisons test. The experiments were repeated in 3 independent biological replicates. Data from 1 representative experiment are shown. BTIC frequencies were compared using in vitro limiting dilution assays with (C) D456 and (D) Jx39 BTICs with and without ST6GAL1 KD. Each group was plated in decreasing number of cells (100, 50, 10, 5, and 1 cell per well). ELDA was done using the software (http://bioinf.wehi.edu.au/software/elda/). P values were calculated from chi-square analysis of group comparisons. The experiments were repeated in at least 3 independent biological replicates. Data from 1 representative experiment are shown. (E) Representative images of D456 neurospheres at day 7 at 4× magnification. Scale bar: 0.1 mm. (F) Kaplan-Meier survival curves for BalbC nu/nu mice injected orthotopically with 5,000 shNT, sh32, or sh33 D456 BTIC cells and sacrificed upon development of neurological signs. The log-rank test was employed to calculate the indicated P value. (G) Representative histological images of tumors from F stained with H&E support the presence of brain tumors in mice with neurological signs. Left panels: Image objective = 1.25×; scale bar: 1.0 mm. Right panels: Image objective = 20×; scale bar: 0.1 mm.
ST6GAL1 regulates a select subset of cell surface proteins known to regulate TIC maintenance.
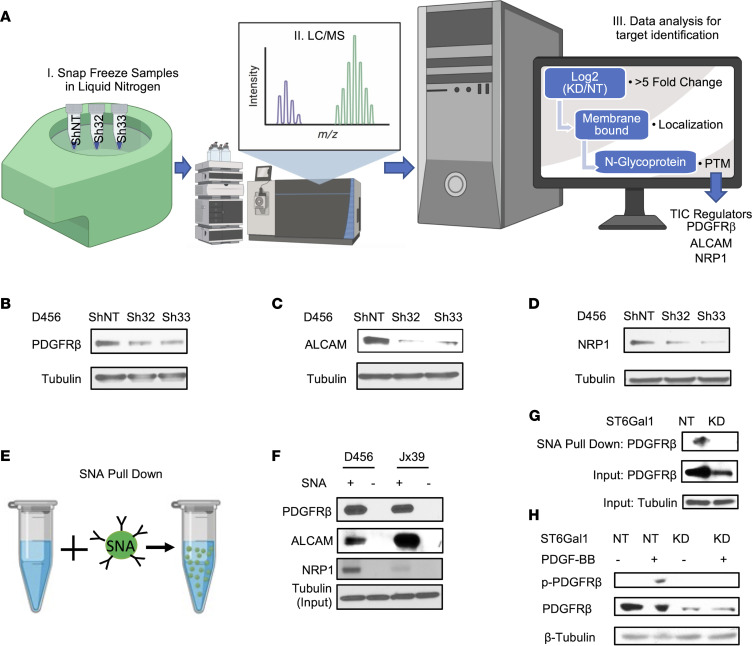
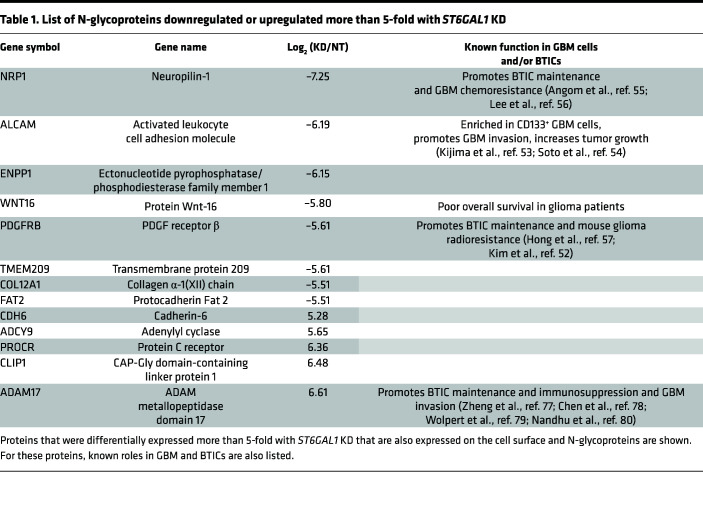
Having determined that ST6GAL1 had a protumorigenic biological role in GBM, we next sought to define the molecular mechanisms through which ST6GAL1 could increase GBM growth. Through the addition of the negatively charged sialic acid, ST6GAL1 is known to modulate many aspects of glycoprotein structure and function, including protein turnover (12–14). While molecular effects of ST6GAL1 have not been well-studied in BTICs, studies in other tumor types have demonstrated sialylation of a select group of cell surface molecules that are known to play critical roles in brain tumors. Thus, we expected that ST6GAL1 could regulate the levels of a subset of cell surface proteins and, therefore, performed proteomics of lysates from BTICs with and without ST6GAL1 KD (Figure 4A). Proteins were identified via mass spectrometry and differentially expressed proteins determined as those with 5-fold or greater positive or negative log2 fold changes (Table 1). We focused on proteins that are known to be N-glycosylated. To further prioritize targets for validation, this list was interrogated for known GBM, BTIC, and TIC regulators (Table 1). Through this process, we identified PDGF Receptor β (PDGFRB), Activated Leukocyte Cell Adhesion Molecule (ALCAM, CD166), and Neuropilin (NRP1) as top potential candidates. Regulation of these N-glycoproteins by sialyltransferases has not yet been studied, nor have these proteins been investigated as mediators of ST6GAL1 effects including those in TICs. While it is likely to be beneficial to further investigate ST6GAL1 roles in the activity of known targets with key roles in GBM, those targets, including EGFR, were not identified as differentially expressed in our analysis as described above. In samples isolated separately from those used for proteomics, we validated that KD of ST6GAL1 decreased expression of PDGFRB (Figure 4B and Supplemental Figure 5A), ALCAM (Figure 4C and Supplemental Figure 5B), and NRP1 (Figure 4D and Supplemental Figure 5C). To determine if this subset of N-glycoproteins was α2,6 sialylated, we performed pulldowns using SNA bound to agarose (Figure 4E). In lysates from BTICs isolated from 2 different PDXs, PDGFRB, ALCAM, and NRP1 associated with SNA agarose beads, but not controls, indicating that these proteins are targets for α2,6 sialylation (Figure 4F and Supplemental Figure 5, D–F). Considering the known importance for PDGFRB in GBM growth, we also further explored the impact of ST6GAL1 on PDGFRB and its phosphorylation. Using SNA pulldowns in lysates collected from nontargeting and ST6GAL1 KD cells, we confirmed that α2,6 sialylation levels of PDGFRB were diminished with loss of ST6GAL1 (Figure 4G and Supplemental Figure 5, G and H). Treatment of nontargeting and ST6GAL1 KD cells with the PDGFRB ligand PDGF-BB also demonstrated that KD of ST6GAL1 resulted in decreased phosphorylation of PDGFRB (Figure 4H and Supplemental Figure 5, I and J). As total levels of PDGFRB were decreased as expected based on our proteomics screen, we normalized phospho- to total PDGFRB and confirmed a significant decrease (Supplemental Figure 5, I and J). Thus, ST6GAL1 is a critical regulator of PDGFRB signaling whose protumorigenic role in GBM was previously unrecognized. The potentially novel finding that ST6GAL1 post-translationally modifies critical TIC regulators further implicates ST6GAL1-mediated sialylation as an important regulator of BTIC maintenance.
Figure 4. ST6GAL1 targeting decreases levels of a subset of N-glycoproteins that are known BTIC regulators.
(A) Schematic of proteomic analysis of D456 BTICs with and without ST6GAL1 KD (n = 4 for each group of shNT, sh32, and sh33). IB with samples independent of the proteomic analysis verified that successful targeting ST6GAL1 resulted in decreased (B) PDGFRB, (C) ALCAM, and (D) NRP1 protein. (E) Schematic of pulldown using SNA-bound Agarose beads. (F) SNA pulldown and protein A/G bound agarose beads as a control demonstrated that PDGFRB, ALCAM, and NRP1 were targets for α2,6 sialylation. (G) SNA pulldown of D456 PDX cells with ST6GAL1 KD compared with NT, illustrating differential pulldown of PDGFRB. (H) PDGF-BB–induced (10 minutes) activation of PDGFRB in D456 GBM PDX cells with ST6GAL1 KD compared with NT; IB for p-PDGFRB and total PDGFRB. The experiments were repeated in at least 3 independent biological replicates. Data from 1 representative experiment are shown.
Table 1. List of N-glycoproteins downregulated or upregulated more than 5-fold with ST6GAL1 KD.
Discussion
Sialylation is an important post-translational modification that is highly understudied in the brain and in brain tumors, including GBM. We find that α2,6 sialylation and ST6GAL1 are protumorigenic in GBM. Reports from the Moskal group using GBM cell lines in differentiation-promoting conditions suggested that ST6GAL1 levels were low in GBM and that loss of ST6GAL1 would be protumorigenic (27, 28). Our data confirmed that in differentiating conditions the levels of ST6GAL1 protein were low, but we determined elevated ST6GAL1 in BTICs. We also observed elevated ST6GAL1 protein in BTICs compared with nontumor brain cells. We found that enrichment for α2,6 sialylation increased GBM growth in vitro and in vivo in association with increased self-renewal as evaluated through neurosphere formation. α2,6 Sialylation was mediated by ST6GAL1 and genetic targeting of ST6GAL1 decreased GBM growth and self-renewal. While the extent of ST6GAL1 KD appeared similar between the 2 shRNAs used at the mRNA level, results often suggested a greater biological effect of shRNA 32 that correlated with changes in the extent of reduction in sialylation. For example, while both shRNA 32 and shRNA 33 significantly reduced α2,6 sialylation and tumor growth, the effects were most substantial for shRNA 32. Together, our findings demonstrate that ST6GAL1-mediated α2,6 sialylation is critical for BTIC maintenance and GBM growth and suggest the translational potential of targeting ST6GAL1 or α2,6 sialylation. While ST6GAL1-specific inhibitors are in development but are not yet available (44–50), sialyltransferase inhibitors have been identified and continue to be developed. We found that inhibiting sialyltransferase activity with 3Fax-Peracetyl Neu5Ac decreased BTIC growth at concentrations that remained ineffective in non-BTICs, which expressed lower levels of ST6GAL1. Thus, targeting of ST6GAL1 may offer benefits for GBM treatment in the future, particularly if it led to increased death of therapy resistant BTICs.
TIC phenotypes and signaling have been informed by results in non-neoplastic stem cells and iPSCs. For ST6GAL1, the literature suggests important roles in both the stem cell niche and during reprogramming (42). ST6GAL1 is present in the base of colon crypts, a well-characterized stem cell niche (29). ST6GAL1 is elevated during iPSC induction where it is critical for the acquisition of stem cell phenotypes (42). Indeed, somatic cells displayed mostly α2,3 sialylation, whereas iPSCs had high levels of α2,6 sialylation (51). Reprogramming involves the Yamanaka and neural stem cell (and BTIC) transcription factor SOX2, which we demonstrated regulates ST6GAL1 expression (40). Data in this report further demonstrate that ST6GAL1, in turn, regulates SOX2 in BTICs (Figure 2G). Thus, a SOX2-ST6GAL1 feedforward loop that regulates the glycosylation state of GBM cells may exist in BTICs. ST6GAL1 regulation of SOX2 may be an indirect outcome of an overall change in the stem cell state. However, ST6GAL1-mediated sialylation of cell surface receptors that signal to control SOX2 transcription could provide a more direct link between the 2 molecules. If true, this could be an important mechanism through which ST6GAL1 regulates a neural stem cell or neural stem cell-like state in normal and neoplastic cells, respectively.
Although ST6GAL1 is known to alter the function of a subset of cell surface proteins that have established roles in tumor biology, the molecular mechanisms through which ST6GAL1 mediates protumorigenic effects remain to be fully elucidated. Our proteomics analysis and SNA pulldowns identified PDGFRB, ALCAM, and NRP1 as potentially novel targets for ST6GAL1 that are sialylated. PDGFRB has important roles in GBM, including in BTICs, where it is elevated and promotes BTIC maintenance, invasion, and tumorigenic potential (52). Similarly, ALCAM was suggested as a BTIC marker as ALCAM was highly expressed in BTICs where it promoted neurosphere formation capacity and tumor growth while also increasing GBM invasion and metastasis to the brain (53, 54). BTICs also express NRP1 which increases BTIC marker expression, neurosphere formation capacity, migration, and tumor growth (55). These data support known protumorigenic roles for the VEGF-NRP1 signaling axis, ALCAM, and PDGFRB in GBM and other tumors, including in therapy-resistant tumor cell subsets that are likely to be enriched for TICs (56–64). Thus, the molecules we have identified as ST6GAL1 targets in GBM may mediate ST6GAL1 effects in other cancers as well.
PDGFRB, ALCAM, and NRP1 have multiple N-glycosylation sites and are membrane-bound proteins that go through modification in the secretory pathway. Considering our SNA pulldown results, these established regulators of TICs are likely targets of ST6GAL1, which post-translationally modifies proteins in the secretory pathway by adding the terminal sialic acid in trans Golgi. Certainly, the SNA pulldown results in lysates from ST6GAL1 KD cells indicates PDGFRB is a target for ST6GAL1-mediated sialylation and that ST6GAL1 regulates PDGFRB levels and phosphorylation. While additional studies outside the scope of the current report will be required to define exact mechanisms through which ST6GAL1 modulates PDGFRB, ALCAM, and NRP1 as well as other proteins, the literature does support a role for ST6GAL1 in cell surface protein turnover, including through the regulation of cell surface retention, internalization, and degradation. For example, α2,6 sialylation by ST6GAL1 increases the turnover of cell surface E-cadherin (65), affects cell surface retention of PECAM receptor by internalization and degradation (66), and regulates the internalization of the Fas death receptor (39). Through these mechanisms, ST6GAL1-mediated sialylation of integrins, growth factor receptors, and death receptors can regulate cell migration, survival, and differentiation state (11, 20, 25, 29, 39, 67). Therefore, decreases in PDGFRB, ALCAM, and NRP1 levels with ST6GAL1 KD could be due to specific changes in internalization and degradation of these targets that impact BTIC survival and maintenance. We, however, acknowledge the possibility that the effects of ST6GAL1 could be more global and other surface receptors that are differentially sialylated could transcriptionally change expression of the proteins that we identified in our screen. Furthermore, proteins that are known targets for ST6GAL1-mediated sialylation with roles in GBM (such as EGFR) would be important to investigate, even though they were not identified as priority targets in our proteomics analysis. Therefore, it is imperative to further probe sialylation and ST6GAL1 effects in GBM in future studies.
We demonstrated that inhibition of sialyltransferase activity with a small molecule inhibitor decreases BTIC growth (Supplemental Figure 4) and that ST6GAL1 sialylates PDGFRB (Figure 4). These data indicate that ST6GAL1-mediated sialylation regulates BTIC maintenance, especially as sialylation-independent roles of ST6GAL1 have not been characterized. However, it would be beneficial to determine whether a catalytically inactive mutant of ST6GAL1 would fail to rescue effects of ST6GAL1 KD. To achieve this goal, the field would benefit from human ST6GAL1 mutants that are known to alter α2,6 sialylation and impact ST6GAL1-regulated biologies, such as growth, survival, migration, and/or invasion in vitro and in vivo. In rat ST6GAL1, Meng et al. demonstrated that aa, including N230, C350, C361, H367, and Y366, are required for sialyltransferase activity as demonstrated in a biochemical assay using CMP-Neu5Ac as a donor and N-acetyllactosamine as an acceptor substrate (68). There is homology in these regions with the human sequence, so determining if similar mutations in human ST6GAL1 would alter α2,6 sialylation in BTICs or other ST6GAL1-expressing human cells would be valuable.
While our study has only explored the role of α2,6 sialylation and ST6GAL1 in BTICs in vitro and in immunocompromised mouse models, we acknowledge the potential importance of ST6GAL1 and/or ST6GAL2 in the brain tumor microenvironment. Indeed, data from GBMseq indicates strong expression of ST6GAL1 in myeloid cells, ST6GAL1, and ST6GAL2 in oligodendrocyte progenitor cells, and ST6GAL2 in astrocytes (Supplemental Figure 2, F and G). Thus, we may be underestimating the roles for ST6GAL1 and/or ST6GAL2 for GBM growth. Importantly, α2,6 sialylation and ST6GAL1 have several known functions in immunomodulation that could be relevant for GBMs or other cancers, especially as immunotherapies become increasingly used and tested (69, 70). Glycans with α2,6 sialic acids can bind to siglec2 (CD22) to inhibit B cell receptor signaling. ST6GAL1 is expressed in B cells where it is important for development and immunoglobulin levels, but sialylation of IgG can occur even when ST6GAL1 is knocked out from B cells as ST6GAL1 is secreted from the liver (22, 71, 72). Reducing extracellular ST6GAL1 via liver KO also results in a proinflammatory state linked to changes in macrophages and T cells (73). ST6GAL1 has also been linked to macrophage survival (13). Inhibition of ST6GAL1 and α2,6 sialylation was also associated with a proinflammatory state in arthritis (74). As these data may suggest, when ST6GAL1 was elevated in hepatocarcinoma cells, an immunosuppressive environment was supported: this was due, in part, to inhibition of T cell proliferation (74). ST6GAL1 is also expressed in human NK cell lines and primary cells, and activation of NK cells with IL2 results in increased α2,6 sialylation. While this increase in sialylation was not associated with an increase in ST6GAL1 mRNA levels, ST6GAL1 mRNA and protein are not always correlated and the protein expression of ST6GAL1 was not fully determined in this study (23). Thus, there are multiple mechanisms through which ST6GAL1 could impact the immuno-landscape of cancers including GBMs.
In conclusion, these data indicate the understudied importance of post-translational modifications, including sialylation and other types of glycosylation, in GBM. Considering that BTIC characterization may rely on Abs that recognize glycosylated forms of cell surface proteins (such as AC133 for CD133), it is possible that TIC enrichment is selecting for more global differences in glycosylation than currently appreciated. Taken together, our investigation defines a novel role of ST6GAL1-mediated α2,6 sialylation in the promotion of GBM growth.
Methods
Culture and maintenance of GBM PDX and BTICs.
The GBM PDXs were obtained from Yancey Gillespie and the Brain Tumor Core Facility of the University of Alabama at Birmingham (UAB), Darrel Bigner at Duke University, and Jann Sarkaria at the Mayo Clinic. CSC293T cells were produced and expanded as previously described (4, 5, 9). For dissociation, papain from Worthington Biochemical was used per the manufacturer’s instructions. For in vitro BTIC propagation, DMEM/F12 basal media (catalog 21041-025, Life Technologies) supplemented with EGF, (catalog 300-110P, GeminiBio), FGF (catalog 300-112P, GeminiBio), sodium pyruvate (catalog 11360070, Gibco), penicillin/streptomycin (catalog 15-140-122, Gibco), and GEM21 (a B27 equivalent; catalog 400-161, GeminiBio) were used. To differentiate the BTICs, 10% FBS (catalog PS-FB2, Peak Serum) was added while growth factors and GEM21 were removed.
Lentiviral gene modulation.
CSC293T cells were transiently cotransfected with psPAX2, pCMV-VSVG, and shRNA constructs using FuGENE HD Transfection Reagent (catalog PRE2312, Promega) as previously reported. Virus titer was determined using Lenti-X qRT-PCR Titration Kit (catalog 740956.50, Takara). Lentivirus-expressing ST6GAL1 shRNAs (TRCN0000035432 and TRCN0000035433) and nontargeting control shRNA (pLKO.1-TC cloning vector; catalog SHC002) were purchased from Dharmacon. Both shRNA constructs were designed against the ST6GAL1 coding sequence.
The shRNA sequences were as follows: ST6Gal-I shRNA32: 5′ CCGGCGTGTGCTACTACTACCAGAACTCGAGTTCTGGTAGTAGTAGCACACGTTTTTG 3′; ST6Gal-I shRNA33: 5′ CCGGGCGCTTCCTCAAAGACAGTTTCTCGAGAAACTGTCTTTGAGGAAGCGCTTTTTG 3′.
mRNA extraction, cDNA generation, and qRT-PCR.
Total mRNA from cells in BTIC media or 96 hours after treatment in differentiation medium was harvested using Qiagen RNeasy Mini Kit (catalog 74106) and synthesized into cDNA using the M-MLV reverse transcriptase cDNA Synthesis Kit (catalog M170A, Promega). qRT-PCR was performed on the generated cDNA with the Taq Man Fast Advanced Master Mix (catalog A44360, Thermo Fisher Scientific). The relative expression of ST6GAL1 and ST6GAL2 was measured using ST6GAL1 FAM/MGB TaqMan Primer (catalog HS00949382_m1, Life Technologies) and ST6GAL2 FAM/MGB TaqMan Primer (catalog Hs00383641_m1, Life Technologies). The data were analyzed and normalized against housekeeping gene 18S Subunit (catalog Hs99999901_s1, Life Technologies) expression to determine relative expression of target genes.
IB.
Cells in BTIC media or 96 hours after culture in differentiation medium were harvested and lysed using RIPA Lysis and Extraction Buffer (catalog 89901, Thermo Fisher Scientific). Protein concentration was determined using the BCA assay (catalog 23227, Thermo Fisher Scientific). Prior to electrophoresis on 4%–20% Tris-Glycine Mini Gels (catalog xp04200Box, Invitrogen), protein lysates were denatured with Pierce Lane Marker Reducing Sample Buffer (catalog 39000, Thermo Fisher Scientific). Protein was then transferred to PVDF membranes (catalog SLHV033RS, Thermo Fisher Scientific) and blocked using 5% nonfat milk in TBST or Pierce Protein Free Blocking Buffer (catalog 37571, Thermo Fisher Scientific). The primary Abs for Western blot were ST6GAL1 (catalog AF5924, R&D Systems), SOX2 (catalog 561469, BD Biosciences), Tubulin (catalog ab21058, Abcam), PDGFRB (catalog 3169, Cell Signaling), Phospho-PDGFRB (Tyr751) (catalog 3161, Cell Signaling), NRP1 (catalog AF3870, R&D Systems), and ALCAM (catalog AF656, R&D Systems). HRP-conjugated secondary Abs for Western blot were Anti-Goat (catalog MP-7405, Vector Labs), Anti-Rabbit (catalog A27036, Invitrogen), Anti-Mouse (catalog, A28177, Invitrogen), and Anti-Sheep (catalog HAF016, R&D Systems). SuperSignal West Dura Chemiluminescent (catalog 34076, Thermo Fisher Scientific) reagent was used for chemiluminescent reaction, which was captured using HXR Film (catalog XC6A2, Hawkins X-Ray Supply) and developed in Medical Film Processor (catalog SRX-101A, 105235078, Konika Minolta Medical and Graphic). The developed respective bands on the film were quantified using ImageJ2 (NIH) (75).
SNA pulldown.
Sambucus Nigra Agglutinin bound Agarose beads (catalog AL-1303-2, Vector Laboratories) were washed twice with ice-cold PBS (catalog 10010049, Thermo Fisher Scientific). Pierce Protein A/G Plus Agarose beads (catalog 20423, Thermo Fisher Scientific) were used as control. After washing 50 μL of SNA Agarose beads or Protein A/G Plus Agarose beads, they were incubated with 500–1,000 μg cell lysates (collected as described above) in a total of 1 mL volume for 4 hours to overnight in a dark cold room (4°C) on a rotator. Beads were washed twice with ice-cold PBS followed by incubation with Pierce Lane Marker Reducing Sample Buffer (catalog 39000, Thermo Fisher Scientific) for 5 minutes at higher than 90°C for 5 minutes. The pulled lysates were subjected to IB as described above.
Recombinant human PDGF-BB protein treatment.
Cells with indicated modifications and culture conditions were treated with PDGF-BB (catalog 220-BB-010, R&D Systems) reconstituted in 4 mM HCL per manufacturer’s instruction at a final concentration of 5 μg/mL for 10 minutes. Cells were lysed and collected for IB. The appropriate dilution of 4 mM HCL was used as control.
IHC.
The GBM PDXs propagated intracranially and s.c. were formalin-fixed and paraffin-embedded. The respective tissue samples were incubated overnight at 4°C ST6GAL1 primary Ab (1–5 μg/mL; catalog AF5924, R&D Systems) and IHC was performed as previously described (29). The images were captured using Nikon Eclipse 80i camera and ISCapture software as well as EVOS XL microscope.
FACS.
Cells from culture or directly isolated the night prior to sorting from GBM xenografts were used for flow cytometry. Cells were washed with cold DMEM:F12 (Gibco) and counted. Cells were resuspended in 90 μL of DMEM:F12 per 7 × 106 cells and incubated with or without SNA-FITC (catalog F-6802-1, EY Laboratories) and sorted by BD-FACS ARIA. Forward and side scatter and viability dyes were also used. Cells were sorted with the assistance of the Flow Cytometry Core at the UAB. The top and bottom 10% were designated as α2,6 sialylationhi and α2,6 sialylationlo populations and were directly sorted into 96-well plates in BTIC medium for experiments or sorted into flow cytometry tubes, pelleted, and lysed.
Measurement of cell growth.
First, 1 × 103 cells were seeded in 96-well plates containing 100 μL of BTIC medium. Cells were incubated for the indicated number of days at 37°C and total ATP was determined using CellTiter-Glo 2.0 kit (catalog G9243, Promega) in which ATP-driven luminescence corresponds with cell numbers. The luminescence was read using the Biotek synergy H1 microplate reader. For crystal violet growth assay, cells were seeded as described above on the Geltrex-treated plate (catalog A14133-02, Thermo Fisher Scientific) for adherence for the indicated number of days at 37°C. At endpoint, cells were washed twice with PBS (catalog 10010049, Thermo Fisher Scientific) and fixed in 10% buffered formalin (catalog 305-510, Thermo Fisher Scientific). Cells were then incubated with 0.05% crystal violet (catalog S25274B, Thermo Fisher Scientific) for 30 minutes at room temperature, extensively washed in deionized water to remove excess crystal violet and air-dried overnight. Crystal violet absorbed by cells corresponding to cell number in each group were dissolved in 50 μL of 10% acetic acid (catalog A38S-500, Thermo Fisher Scientific) for 15 minutes and absorbance was read at 590 nm using the Biotek synergy H1 microplate reader.
Neurosphere formation assay.
For in vitro limiting dilution assays, α2,6 sialylationhi and α2,6 sialylationlo or ST6GAL1 modulated cells were plated in decreasing numbers of cells per well (100, 10, 5, 2, and 1) in 96-well plates containing BTIC medium. The wells containing neurospheres were marked and counted after 14–21 days of incubation. ELDA was performed using software available at http://bioinf.wehi.edu.au/software/elda/
In vivo tumor initiation assay.
All animal procedures were performed in accordance with the UAB IACUC approved protocols. Animals were housed in a temperature-controlled vivarium with a 14-hour light/10-hour dark cycle at no more than 7 animals per cage. Viable cells were intracranially injected into female athymic nude mice 4–6 weeks of age. A total of 2,500 cells were used for experiments with FACS-sorted cells, whereas 1,000 cells were used for experiments with lentivirus-infected BTICs. Animals were maintained until development of neurological signs (for example, lethargy, ataxia, seizures, and/or paralysis), when brains were collected. Animals without neurologic signs were sacrificed at the termination of the experiment. Harvested brains were fixed in 4% paraformaldehyde and embedded in paraffin and sectioned on slides with subsequent H&E staining at the UAB Tissue Biorepository. Developed slides were imaged with a Nikon Eclipse 80i camera and ISCapture software.
In silico data analysis.
ST6GAL1 and ST6GAL2 gene expression and patient survival data were downloaded from GlioVis (http://gliovis.bioinfo.cnio.es) and plotted to assess expression and survival. The reads per kilobase of transcript per million data for ST6GAL1 and ST6GAL2 in brain from Human Protein Atlas RNA-Seq normal tissues project were downloaded from the National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/gene/6480) and (https://www.ncbi.nlm.nih.gov/gene/84620). ST6GAL1 and ST6GAL2 gene expression data in various cell types present in GBM patient samples via single-cell RNA-Seq were downloaded from GBMSeq (http://www.gbmseq.org).
Proteomics.
Lysates were prepared in M-Per in quadruplicates and peptide digests separated and analyzed with a Thermo Orbitrap Velos Pro hybrid mass spectrometer equipped with a nano-electrospray source (Thermo Fisher Scientific) similar to our prior report (76). The XCalibur RAW files were converted and mgf files searched using SEQUEST to generate peptide IDs that were filtered using Scaffold (Protein Sciences). Normalized spectral counts were used to calculate fold changes and proteins with greater than 5-fold changes among the nontargeting control and KD samples further analyzed to determine cell surface N-glycoproteins.
Statistics.
All statistics were performed with GraphPad Prism Version 7 or 9 (GraphPad Software). Both 1- and 2-way ANOVA and multiple t tests were performed with Dunnett’s or Tukey’s test for multiple comparisons, and P values indicate a confidence level of 95% and significance of 0.05. Correlation analysis was performed using Pearson’s correlation analysis with a CI of 95%. Kaplan-Meier survival curves were compared with the log-rank statistical analysis to determine significant differences in outcome.
Study approval.
All animal studies were approved by the UAB Institutional Animal Care and Use Committee.
Author contributions
SGC, SLB, and ABH conceived the project. SGC, AC, CRM, EAB, VSH, JAM, SLB, and ABH designed the experiments. SGC, KT, LR, RJ, AC, VSH, ANT, JAM, and ABH performed experiments and/or analyzed the data generated. SGC and ABH wrote the manuscript with review and approval by all authors. ABH supervised the work.
Supplementary Material
Acknowledgments
We appreciate the support of the O’Neal Comprehensive Cancer Center Mass Spectrometry/Proteomics Shared Facility, NIH grants P30AR048311 and P30AI27667 awarded to the UAB Comprehensive Flow Cytometry Core, and the UAB Tissue Biorepository Facility. This work was supported by pilot awards from the UAB Department of Cell, Developmental and Integrative Biology and O’Neal Invests of the O’Neal Comprehensive Cancer Center as well as NIH grant R01NS127424. The O’Neal Invests award included support from Clarence and Debby Pouncey. The O’Neal Comprehensive Cancer Center is supported by the O’Neal Cancer Center Support Grant through NIH award P30CA013148. Additional support to the Hjelmeland laboratory was provided via startup funds from the UAB and the Pittman Scholar Award. The Hjelmeland laboratory is supported by awards from the NIH via grants R01NS127434 and R03NS125506, as well as R01NS104339, and the Bellis Laboratory is supported via grants U01CA233581 and R01CA225177.
Version 1. 11/08/2022
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2022, GC et al. This is an open access article published under the terms of the Creative Commons Attribution 4.0 International License.
Reference information: JCI Insight. 2022;7(21):e158799.https://doi.org/10.1172/jci.insight.158799.
Contributor Information
Sajina GC, Email: sajinagc@uab.edu.
Robert Jones, Email: robert.brent.jones@emory.edu.
Asmi Chakraborty, Email: achakrab@fiu.edu.
C. Ryan Miller, Email: ryanmiller@uabmc.edu.
Elizabeth A. Beierle, Email: elizabeth.beierle@childrensal.org.
Vidya Sagar Hanumanthu, Email: vhanumanthu@uabmc.edu.
Anh N. Tran, Email: trannhatanh89@gmail.com.
References
- 1.Stupp R, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10(5):459–466. doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 2.Hottinger AF, et al. Standards of care and novel approaches in the management of glioblastoma multiforme. Chin J Cancer. 2014;33(1):32–39. doi: 10.5732/cjc.013.10207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Singh SK, et al. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63(18):5821–5828. [PubMed] [Google Scholar]
- 4.Bao S, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444(7120):756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 5.Bao S, et al. Stem cell-like glioma cells promote tumor angiogenesis through vascular endothelial growth factor. Cancer Res. 2006;66(16):7843–7848. doi: 10.1158/0008-5472.CAN-06-1010. [DOI] [PubMed] [Google Scholar]
- 6.Meacham CE, Morrison SJ. Tumour heterogeneity and cancer cell plasticity. Nature. 2013;501(7467):328–337. doi: 10.1038/nature12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eyler CE, et al. Glioma stem cell proliferation and tumor growth are promoted by nitric oxide synthase-2. Cell. 2011;146(1):53–66. doi: 10.1016/j.cell.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Z, et al. Hypoxia-inducible factors regulate tumorigenic capacity of glioma stem cells. Cancer Cell. 2009;15(6):501–513. doi: 10.1016/j.ccr.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hjelmeland AB, et al. Acidic stress promotes a glioma stem cell phenotype. Cell Death Differ. 2011;18(5):829–840. doi: 10.1038/cdd.2010.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flavahan WA, et al. Brain tumor initiating cells adapt to restricted nutrition through preferential glucose uptake. Nat Neurosci. 2013;16(10):1373–1382. doi: 10.1038/nn.3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schultz MJ, et al. Regulation of the metastatic cell phenotype by sialylated glycans. Cancer Metastasis Rev. 2012;31(3–4):501–518. doi: 10.1007/s10555-012-9359-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhuo Y, Bellis SL. Emerging role of alpha2,6-sialic acid as a negative regulator of galectin binding and function. J Biol Chem. 2011;286(8):5935–5941. doi: 10.1074/jbc.R110.191429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Z, et al. ST6Gal-I regulates macrophage apoptosis via α2-6 sialylation of the TNFR1 death receptor. J Biol Chem. 2011;286(45):39654–39662. doi: 10.1074/jbc.M111.276063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shaikh FM, et al. Tumor cell migration and invasion are regulated by expression of variant integrin glycoforms. Exp Cell Res. 2008;314(16):2941–2950. doi: 10.1016/j.yexcr.2008.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schultz MJ, et al. ST6Gal-I sialyltransferase confers cisplatin resistance in ovarian tumor cells. J Ovarian Res. 2013;6(1):25. doi: 10.1186/1757-2215-6-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schultz MJ, et al. The tumor-associated glycosyltransferase ST6Gal-I regulates stem cell transcription factors and confers a cancer stem cell phenotype. Cancer Res. 2016;76(13):3978–3988. doi: 10.1158/0008-5472.CAN-15-2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park JJ, Lee M. Increasing the α 2, 6 sialylation of glycoproteins may contribute to metastatic spread and therapeutic resistance in colorectal cancer. Gut Liver. 2013;7(6):629–641. doi: 10.5009/gnl.2013.7.6.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park JJ, et al. Sialylation of epidermal growth factor receptor regulates receptor activity and chemosensitivity to gefitinib in colon cancer cells. Biochem Pharmacol. 2012;83(7):849–857. doi: 10.1016/j.bcp.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 19.Chakraborty A, et al. ST6Gal-I sialyltransferase promotes chemoresistance in pancreatic ductal adenocarcinoma by abrogating gemcitabine-mediated DNA damage. J Biol Chem. 2018;293(3):984–994. doi: 10.1074/jbc.M117.808584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Britain CM, et al. The glycosyltransferase ST6Gal-I protects tumor cells against serum growth factor withdrawal by enhancing survival signaling and proliferative potential. J Biol Chem. 2017;292(11):4663–4673. doi: 10.1074/jbc.M116.763862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou X, et al. Sialylation of MUC4β N-glycans by ST6GAL1 orchestrates human airway epithelial cell differentiation associated with type-2 inflammation. JCI Insight. 2019;4(5):122475. doi: 10.1172/jci.insight.122475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones MB, et al. B-cell-independent sialylation of IgG. Proc Natl Acad Sci U S A. 2016;113(26):7207–7212. doi: 10.1073/pnas.1523968113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rosenstock P, et al. Sialylation of human natural killer (NK) cells is regulated by IL-2. J Clin Med. 2020;9(6):1816. doi: 10.3390/jcm9061816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alam S, et al. Altered (neo-) lacto series glycolipid biosynthesis impairs α2-6 sialylation on N-glycoproteins in ovarian cancer cells. Sci Rep. 2017;7:45367. doi: 10.1038/srep45367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holdbrooks AT, et al. ST6Gal-I sialyltransferase promotes tumor necrosis factor (TNF)-mediated cancer cell survival via sialylation of the TNF receptor 1 (TNFR1) death receptor. J Biol Chem. 2018;293(5):1610–1622. doi: 10.1074/jbc.M117.801480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Britain CM, et al. Sialylation of EGFR by the ST6Gal-I sialyltransferase promotes EGFR activation and resistance to gefitinib-mediated cell death. J Ovarian Res. 2018;11(1):12. doi: 10.1186/s13048-018-0385-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kroes RA, Moskal JR. The role of DNA methylation in ST6Gal1 expression in gliomas. Glycobiology. 2016;26(12):1271–1283. doi: 10.1093/glycob/cww058. [DOI] [PubMed] [Google Scholar]
- 28.Yamamoto H, et al. Alpha2,6 sialylation of cell-surface N-glycans inhibits glioma formation in vivo. Cancer Res. 2001;61(18):6822–6829. [PubMed] [Google Scholar]
- 29.Swindall AF, et al. ST6Gal-I protein expression is upregulated in human epithelial tumors and correlates with stem cell markers in normal tissues and colon cancer cell lines. Cancer Res. 2013;73(7):2368–2378. doi: 10.1158/0008-5472.CAN-12-3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barkeer S, et al. Glycosylation of cancer stem cells: function in stemness, tumorigenesis, and metastasis. Neoplasia. 2018;20(8):813–825. doi: 10.1016/j.neo.2018.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Glumac PM, LeBeau AM. The role of CD133 in cancer: a concise review. Clin Transl Med. 2018;7(1):18. doi: 10.1186/s40169-018-0198-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paulson JC, et al. Tissue-specific expression of sialyltransferases. J Biol Chem. 1989;264(19):10931–10934. doi: 10.1016/S0021-9258(18)60407-7. [DOI] [PubMed] [Google Scholar]
- 33.Kitagawa H, Paulson JC. Differential expression of five sialyltransferase genes in human tissues. J Biol Chem. 1994;269(27):17872–17878. doi: 10.1016/S0021-9258(17)32390-6. [DOI] [PubMed] [Google Scholar]
- 34.Ohmi Y, et al. Majority of alpha2,6 sialylated glycans in the adult mouse brain exist in O-glycans: SALSA-MS analysis for knockout mice of alpha2,6-sialyltransferase genes. Glycobiology. 2021;31(5):557–570. doi: 10.1093/glycob/cwaa105. [DOI] [PubMed] [Google Scholar]
- 35.Fagerberg L, et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteomics. 2014;13(2):397–406. doi: 10.1074/mcp.M113.035600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brennan CW, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155(2):462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bowman RL, et al. GlioVis data portal for visualization and analysis of brain tumor expression datasets. Neuro Oncol. 2017;19(1):139–141. doi: 10.1093/neuonc/now247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Darmanis S, et al. Single-cell RNA-seq analysis of infiltrating neoplastic cells at the migrating front of human glioblastoma. Cell Rep. 2017;21(5):1399–1410. doi: 10.1016/j.celrep.2017.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Swindall AF, Bellis SL. Sialylation of the Fas death receptor by ST6Gal-I provides protection against Fas-mediated apoptosis in colon carcinoma cells. J Biol Chem. 2011;286(26):22982–22990. doi: 10.1074/jbc.M110.211375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dorsett KA, et al. Sox2 promotes expression of the ST6Gal-I glycosyltransferase in ovarian cancer cells. J Ovarian Res. 2019;12(1):93. doi: 10.1186/s13048-019-0574-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tateno H, et al. Glycome diagnosis of human induced pluripotent stem cells using lectin microarray. J Biol Chem. 2011;286(23):20345–20353. doi: 10.1074/jbc.M111.231274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang YC, et al. Glycosyltransferase ST6GAL1 contributes to the regulation of pluripotency in human pluripotent stem cells. Sci Rep. 2015;5:13317. doi: 10.1038/srep13317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rillahan CD, et al. Global metabolic inhibitors of sialyl- and fucosyltransferases remodel the glycome. Nat Chem Biol. 2012;8(7):661–668. doi: 10.1038/nchembio.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bull C, et al. Targeting aberrant sialylation in cancer cells using a fluorinated sialic acid analog impairs adhesion, migration, and in vivo tumor growth. Mol Cancer Ther. 2013;12(10):1935–1946. doi: 10.1158/1535-7163.MCT-13-0279. [DOI] [PubMed] [Google Scholar]
- 45.Chang WW, et al. Soyasaponin I decreases the expression of alpha2,3-linked sialic acid on the cell surface and suppresses the metastatic potential of B16F10 melanoma cells. Biochem Biophys Res Commun. 2006;341(2):614–619. doi: 10.1016/j.bbrc.2005.12.216. [DOI] [PubMed] [Google Scholar]
- 46.Chiang CH, et al. A novel sialyltransferase inhibitor AL10 suppresses invasion and metastasis of lung cancer cells by inhibiting integrin-mediated signaling. J Cell Physiol. 2010;223(2):492–499. doi: 10.1002/jcp.22068. [DOI] [PubMed] [Google Scholar]
- 47.Natoni A, et al. Sialyltransferase inhibition leads to inhibition of tumor cell interactions with E-selectin, VCAM1, and MADCAM1, and improves survival in a human multiple myeloma mouse model. Haematologica. 2020;105(2):457–467. doi: 10.3324/haematol.2018.212266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Szabo R, Skropeta D. Advancement of sialyltransferase inhibitors: therapeutic challenges and opportunities. Med Res Rev. 2017;37(2):219–270. doi: 10.1002/med.21407. [DOI] [PubMed] [Google Scholar]
- 49.Wang L, et al. Sialyltransferase inhibition and recent advances. Biochim Biophys Acta. 2016;1864(1):143–153. doi: 10.1016/j.bbapap.2015.07.007. [DOI] [PubMed] [Google Scholar]
- 50.Montgomery AP, et al. Design, synthesis and evaluation of carbamate-linked uridyl-based inhibitors of human ST6Gal I. Bioorg Med Chem. 2020;28(14):115561. doi: 10.1016/j.bmc.2020.115561. [DOI] [PubMed] [Google Scholar]
- 51.Hasehira K, et al. Structural and quantitative evidence for dynamic glycome shift on production of induced pluripotent stem cells. Mol Cell Proteomics. 2012;11(12):1913–1923. doi: 10.1074/mcp.M112.020586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim Y, et al. Platelet-derived growth factor receptors differentially inform intertumoral and intratumoral heterogeneity. Genes Dev. 2012;26(11):1247–1262. doi: 10.1101/gad.193565.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kijima N, et al. CD166/activated leukocyte cell adhesion molecule is expressed on glioblastoma progenitor cells and involved in the regulation of tumor cell invasion. Neuro Oncol. 2012;14(10):1254–1264. doi: 10.1093/neuonc/nor202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Soto MS, et al. Functional role of endothelial adhesion molecules in the early stages of brain metastasis. Neuro Oncol. 2014;16(4):540–551. doi: 10.1093/neuonc/not222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Angom RS, et al. Ablation of neuropilin-1 improves the therapeutic response in conventional drug-resistant glioblastoma multiforme. Oncogene. 2020;39(48):7114–7126. doi: 10.1038/s41388-020-01462-1. [DOI] [PubMed] [Google Scholar]
- 56.Lee J, et al. Combined inhibition of vascular endothelial growth factor receptor signaling with temozolomide enhances cytotoxicity against human glioblastoma cells via downregulation of Neuropilin-1. J Neurooncol. 2016;128(1):29–34. doi: 10.1007/s11060-016-2091-3. [DOI] [PubMed] [Google Scholar]
- 57.Hong JD, et al. Silencing platelet-derived growth factor receptor-β enhances the radiosensitivity of C6 glioma cells in vitro and in vivo. Oncol Lett. 2017;14(1):329–336. doi: 10.3892/ol.2017.6143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim DK, et al. CD166 promotes the cancer stem-like properties of primary epithelial ovarian cancer cells. BMB Rep. 2020;53(12):622–627. doi: 10.5483/BMBRep.2020.53.12.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dalerba P, et al. Phenotypic characterization of human colorectal cancer stem cells. Proc Natl Acad Sci U S A. 2007;104(24):10158–10163. doi: 10.1073/pnas.0703478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dong Z, et al. The role of the tumor microenvironment in neuropilin 1-induced radiation resistance in lung cancer cells. J Cancer. 2019;10(17):4017–4030. doi: 10.7150/jca.28163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jimenez-Hernandez LE, et al. NRP1-positive lung cancer cells possess tumor-initiating properties. Oncol Rep. 2018;39(1):349–357. doi: 10.3892/or.2017.6089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li Q, et al. Metastasis initiating cells in primary prostate cancer tissues from transurethral resection of the prostate (TURP) predicts castration-resistant progression and survival of prostate cancer patients. Prostate. 2015;75(12):1312–1321. doi: 10.1002/pros.23011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Siegle JM, et al. SOX2 is a cancer-specific regulator of tumour initiating potential in cutaneous squamous cell carcinoma. Nat Commun. 2014;5:4511. doi: 10.1038/ncomms5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chang KK, et al. Platelet-derived growth factor receptor-α and -β promote cancer stem cell phenotypes in sarcomas. Oncogenesis. 2018;7(6):47. doi: 10.1038/s41389-018-0059-1. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 65.Engdahl C, et al. Estrogen induces St6gal1 expression and increases IgG sialylation in mice and patients with rheumatoid arthritis: a potential explanation for the increased risk of rheumatoid arthritis in postmenopausal women. Arthritis Res Ther. 2018;20(1):84. doi: 10.1186/s13075-018-1586-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kitazume S, et al. Alpha2,6-sialic acid on platelet endothelial cell adhesion molecule (PECAM) regulates its homophilic interactions and downstream antiapoptotic signaling. J Biol Chem. 2010;285(9):6515–6521. doi: 10.1074/jbc.M109.073106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Christie DR, et al. ST6Gal-I expression in ovarian cancer cells promotes an invasive phenotype by altering integrin glycosylation and function. J Ovarian Res. 2008;1(1):3. doi: 10.1186/1757-2215-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Meng L, et al. Enzymatic basis for N-glycan sialylation: structure of rat α2,6-sialyltransferase (ST6GAL1) reveals conserved and unique features for glycan sialylation. J Biol Chem. 2013;288(48):34680–34698. doi: 10.1074/jbc.M113.519041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jones MB. IgG and leukocytes: targets of immunomodulatory α2,6 sialic acids. Cell Immunol. 2018;333:58–64. doi: 10.1016/j.cellimm.2018.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Videira PA, et al. Surface alpha 2-3- and alpha 2-6-sialylation of human monocytes and derived dendritic cells and its influence on endocytosis. Glycoconj J. 2008;25(3):259–268. doi: 10.1007/s10719-007-9092-6. [DOI] [PubMed] [Google Scholar]
- 71.Irons EE, et al. Blood-borne ST6GAL1 regulates immunoglobulin production in B cells. Front Immunol. 2020;11:617. doi: 10.3389/fimmu.2020.00617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Irons EE, Lau JTY. Systemic ST6Gal-1 is a pro-survival factor for murine transitional B cells. Front Immunol. 2018;9:2150. doi: 10.3389/fimmu.2018.02150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Oswald DM, et al. Disruption of hepatocyte sialylation drives a T cell-dependent pro-inflammatory immune tone. Glycoconj J. 2020;37(3):395–407. doi: 10.1007/s10719-020-09918-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang Y, et al. Loss of α2-6 sialylation promotes the transformation of synovial fibroblasts into a pro-inflammatory phenotype in arthritis. Nat Commun. 2021;12(1):2343. doi: 10.1038/s41467-021-22365-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rueden CT, et al. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinformatics. 2017;18(1):529. doi: 10.1186/s12859-017-1934-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Moehle MS, et al. The G2019S LRRK2 mutation increases myeloid cell chemotactic responses and enhances LRRK2 binding to actin-regulatory proteins. Hum Mol Genet. 2015;24(15):4250–4267. doi: 10.1093/hmg/ddv157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zheng X, et al. Inhibition of ADAM17 reduces hypoxia-induced brain tumor cell invasiveness. Cancer Sci. 2007;98(5):674–684. doi: 10.1111/j.1349-7006.2007.00440.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chen X, et al. ADAM17 regulates self-renewal and differentiation of U87 glioblastoma stem cells. Neurosci Lett. 2013;14:537:44–49. doi: 10.1016/j.neulet.2013.01.021. [DOI] [PubMed] [Google Scholar]
- 79.Wolpert F, et al. A disintegrin and metalloproteinases 10 and 17 modulate the immunogenicity of glioblastoma-initiating cells. Neuro Oncol. 2014;16(3):382–391. doi: 10.1093/neuonc/not232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nandhu MS, et al. Tumor-derived fibulin-3 activates pro-invasive NF-κB signaling in glioblastoma cells and their microenvironment. Oncogene. 2017;36(34):4875–4886. doi: 10.1038/onc.2017.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.