Abstract
Phytohormones are major signaling components that contribute to nearly all aspects of plant life. They constitute an interconnected communication network to fine-tune growth and development in response to the ever-changing environment. To this end, they have to coordinate with other signaling components, such as reactive oxygen species and calcium signals. On the one hand, the two endosymbiotic organelles, plastids and mitochondria, control various aspects of phytohormone signaling and harbor important steps of hormone precursor biosynthesis. On the other hand, phytohormones have feedback actions on organellar functions. In addition, organelles and phytohormones often act in parallel in a coordinated matter to regulate cellular functions. Therefore, linking organelle functions with increasing knowledge of phytohormone biosynthesis, perception, and signaling will reveal new aspects of plant stress tolerance. In this review, we highlight recent work on organelle–phytohormone interactions focusing on the major stress-related hormones abscisic acid, jasmonates, salicylic acid, and ethylene.
Keywords: Abscisic acid (ABA), chloroplast, ethylene, jasmonates, mitochondria, plant organelles, phytohormones, plastids, salicylic acid (SA), retrograde signaling, stress signaling
A review of organelle–phytohormone interactions and retrograde signals, highlighting recent work and open questions and focusing on the stress-related hormones abscisic acid, jasmonates, salicylic acid, and ethylene.
Introduction
Organelles have essential functions in most cellular processes including growth and development of plants. As a consequence of their endosymbiotic origin, mitochondria and chloroplasts carry their own genomes. However, the majority of organellar proteins are now nucleus encoded (Zimorski et al., 2014). Development and plant fitness require the coordination of organellar functions, including coordinated expression of genes encoded in the three genomes of the plant cell. Therefore, organelles constantly transmit their physiological state as intracellular signals to the nucleus for the coordination of gene expression. The discovery of stress-related organellar signals led to the assumption that plant organelles are one of the primary sites for sensing environmental changes (reviewed in Kleine and Leister, 2016; Kmiecik et al., 2016; Crawford et al., 2018; Dopp et al., 2021; Li and Kim, 2022). Work has revealed different stress-related organelle signaling components such as the carotenoid oxidation byproducts (Ramel et al., 2012), phosphoadenosine 5ʹ-phosphate (PAP) (Estavillo et al., 2011), executer-mediated response to singlet oxygen (Lee Keun et al., 2007), and the isoprenoid-precursor methylerythritol cyclodiphosphate (Xiao et al., 2012).
In addition to their role as environmental sensors, organelles (especially chloroplasts) are the main hub for the metabolism of several phytohormone precursors, as illustrated in Fig. 1 (for a recent review see Fàbregas and Fernie, 2021). Phytohormones are associated with various physiological and metabolic processes (Cackett et al., 2022), and it is becoming clear that they constitute a network that is interconnected at multiple levels. Antagonistic and synergistic interactions between different hormones have been described during biotic (Berens et al., 2017; Aerts et al., 2021) and abiotic stress. For example, synergistic effects of jasmonic acid (JA) with salicylic acid (SA) were found in the high-light response (D’Alessandro et al., 2020), or between abscisic acid (ABA) and JA in the drought-stress response (Liu et al., 2016), while antagonistic effects of ABA and cytokinins have been reported under drought stress (Huang et al., 2018). Finally, ethylene was shown to modulate the activity of SA in the pathogen response (Ramšak et al., 2018).
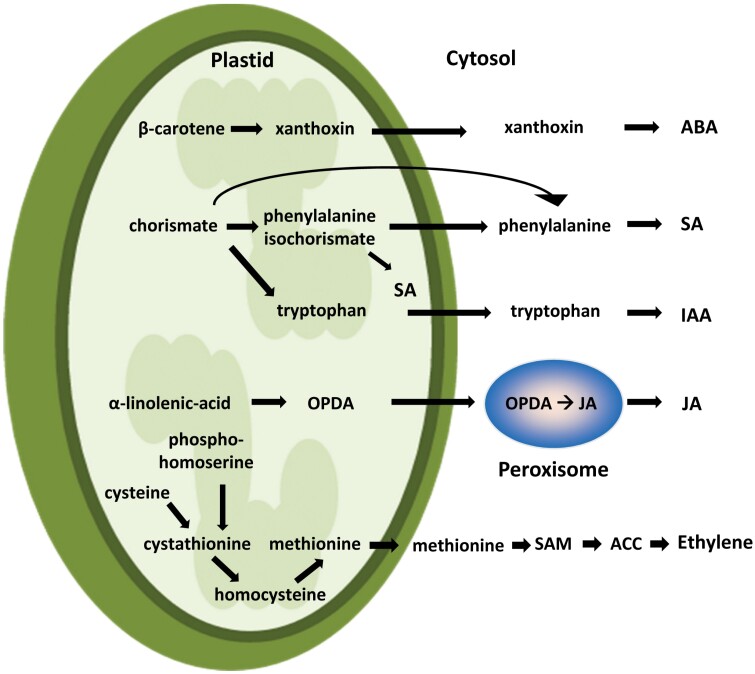
Fig. 1.
The chloroplast as metabolic hub for phytohormone precursors. Numerous phytohormone pathways start with secondary metabolism in plastids: xanthoxine biosynthesis via the xanthophyll cycle—the precursor of ABA; chorismate biosynthesis—the precursor of SA and auxin (IAA); oxidized lipids such as linolenic acid—the precursor of jasmonates (via JA); cystathionine—the precursor for methionine and thus ethylene. ACC: 1-aminocyclopropane 1-carboxylate; IAA, indole-3-acetic acid; OPDA, oxophytodienoic acid; SAM, S-adenosyl-l-methionine.
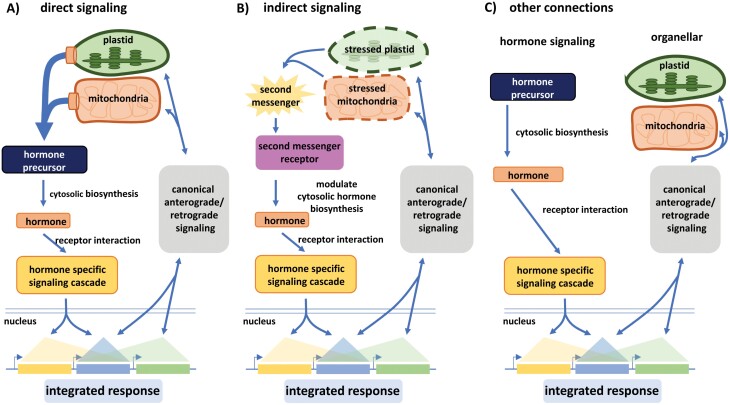
Classically, hormone signaling is discussed in a historical context where specific hormones are often viewed in a particular developmental or stress context. For this review, however, it seemed more reasonable for us to structure organelle–hormone signals to cover (i) direct signaling connections, (ii) indirect signals, and (iii) other, more complex signaling interactions (Fig. 2). Direct signals include metabolites of exclusive organellar origin that serve as key precursors for the final steps of plant hormone biosynthesis. The active forms of the hormones present the source of the signal, triggering a receptor-based signaling cascade leading to an integrated transcriptional response. The regulated genes could be exclusively under the control of a certain hormonal signaling cascade, controlled by retrograde signals from organelles, or controlled by both input pathways. Alternatively, in an indirect mechanism, the second messenger metabolites of organellar origin are perceived by receptors elsewhere in the cytoplasm and thereby act as signals leading to a modulation of biosynthetic activities of enzymes involved in hormone biosynthesis. The elicitation of organellar second messengers may be triggered via anterograde signals. Similar to the direct signaling pathways, the resulting integrated transcriptional response is a combination of exclusively hormone-derived transcriptional changes, transcriptional changes elicited by organellar retrograde signals, and a combination of both. Finally, transcriptional changes may require a functional hormonal signaling cascade and a functional organellar retrograde signaling cascade at the same time to provide an integrated transcriptional response in order to react to an environmental change, thus illustrating more complex signaling interactions (Fig. 2).
Fig. 2.
Direct/indirect signals in regulation of gene expression. Different types of connections between organelles and hormone signaling affect gene expression. (A) Direct signals: hormone levels, depending on precursor metabolites for hormone biosynthesis of exclusive organellar origin, regulate a subset of genes (yellow) while another subset of genes is controlled by retrograde signals from organelles (green), and a third subset of genes is regulated by both input pathways (blue). (B) Indirect signals: hormone levels regulating a subset of genes are indirectly altered via second messenger molecules of organellar origin, modulating the enzymes relevant for hormone biosynthesis. (C) Other signaling connections: transcriptional changes that do not fall into (A, B) but require a functional hormonal signaling cascade and a functional organellar retrograde signaling cascade at the same time.
In this review, we summarize recent work on organelle–phytohormone interactions to highlight and address open questions within these interwoven signaling cascades, concentrating on the stress-related hormones ABA, jasmonates, SA, and ethylene. However, other phytohormones also have tight relations to plant organelles, and the impact of auxin, cytokinins, gibberellins, and strigolactones on photosynthesis and photoprotection was covered in a recent review by Müller and Munné-Bosch (2021).
Abscisic acid
ABA was identified in the 1960s as an endogenous signal (Ohkuma et al., 1963). Since then, numerous functions of ABA in plant development and adaptation have been described, including reprogramming of gene expression, protection of photosynthesis, stomatal closure, maturation of seeds, and primary root growth (for recent reviews on ABA see Ma et al., 2018; Seo and Marion-Poll, 2019; Cardoso et al., 2020). Besides these diverse functions, meta-analysis of microarray studies under different operational retrograde signaling conditions, such as transition from low light to high light intensities, abolishment of the tetrapyrrole pathway by gabaculine treatment, or manipulation of electron flow via mutation of the Ser/Thr protein kinase STN7, identified ABA as a major component within the core network of inter-organelle signaling of Arabidopsis (Gläßer et al., 2014). Many nuclear genes that encode organellar proteins related to photosynthesis carry ABA responsive elements in their promoter (Koussevitzky et al., 2007), and exogenously applied ABA represses the transcription of almost all genes encoded by the plastome in Arabidopsis and barley (Yamburenko et al., 2013; Yamburenko et al., 2015). Therefore, it is no surprise that ABA is important in coordinating the response of the organelle that produces the intermediates for its own biosynthesis (the chloroplast) and that ABA signaling is linked with inter-organelle communication via direct and indirect connections.
Direct connection of chloroplast functions and ABA biosynthesis
The biosynthetic pathway of ABA is closely associated with plastid functions and starts with the hydroxylation of β-carotene to zeaxanthin, followed by subsequent conversion to violaxanthin through the xanthophyll cycle (Fig. 3; for details see Seo and Marion-Poll, 2019). Violaxanthin is then cleaved by 9-cis-epoxycarotenoid dioxygenase (NCED), which leads to xanthoxin. The oxidative cleavage via NCED is the first irreversible conversion step towards ABA and well accepted as a bottleneck in ABA biosynthesis (Endo et al., 2008). Finally, xanthoxin is transferred to the cytosol by an unknown process, where it is converted first to abscisic aldehyde and then to ABA. The ABA precursors xanthoxin and abscisic aldehyde can be found in numerous plant tissues, but reciprocal grafting of ABA biosynthetic mutants with wild type plants showed that leaves are the predominant site for the final steps of ABA biosynthesis (McAdam et al., 2016).
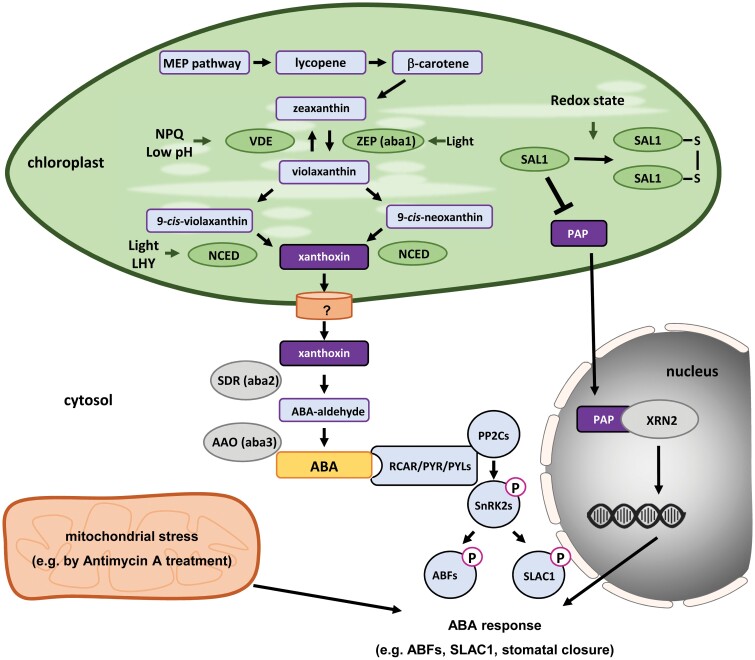
Fig. 3.
The role of organelles in abscisic acid (ABA) signaling. ABA biosynthesis is initiated in chloroplasts by the non-mevalonate (MEP−) pathway and continuous via phytoene, lycopene, β-carotene, and zeaxanthin biosynthesis. The xanthophyll cycle converts zeaxanthin into violaxanthin, which is converted into xanthoxin. In the cytosol, xanthoxin is the substrate for the synthesis of ABA aldehyde and finally ABA. ABA is perceived through receptors from the RCAR/PYR/PYL family and PP2C co-receptors, which in turn activate SnRK2s and ABFs by phosphorylation. Chloroplast signals and functions, such as the SAL–PAP pathway and NPQ, control ABA biosynthesis and signaling. AAO, ABA-aldehyde oxidase; ABF, ABA responsive element-binding factor; LHY, late hypocotyl elongation factor; NCED, 9-cis-epoxycarotenoid dioxygenase; NPQ, non-photochemical quenching; PAP, 3ʹ-phosphoadenosine-5ʹ-phosphate; PP2C, protein phosphatase 2C; SAL1, dinucleotide phosphatase/inositol phosphate phosphatase; SDR, short chain dehydrogenase; SLAC1, slow anion channel-associated 1; SnRK2s, SNF1-related protein kinases; VDE, violaxanthin de-epoxidase; XRN, exoribonucleases; ZEP, zeaxanthin epoxidase.
Several components of the ABA biosynthesis pathway are directly connected to chloroplast function and signaling. For instance, one of the most rapid components of the photo-oxidative stress response is decreased pH in the thylakoid lumen via non-photochemical quenching to rebalance photosynthesis (Ruban et al., 1992). The low pH affects the xanthophyll cycle by activating violaxanthin de-epoxidase, which converts violaxanthin into zeaxanthin and thereby counteracts the first steps of ABA biosynthesis (Pastori et al., 2003).
Indirect connection of chloroplast and ABA signaling
Besides the direct control of ABA precursor availability by chloroplast functions, ABA biosynthesis is also under the indirect control by pathways that are modulated by organelles. One example is the impact of the circadian clock, which is under the control of chloroplast functions. Genome-wide binding analysis in Arabidopsis revealed that several genes involved in ABA biosynthesis are controlled by the LATE HYPOCOTYL ELONGATION FACTOR (LHY), an important component of the molecular clock in plants (Adams et al., 2018). Subsequent analysis with LHY overexpression and loss of function lines revealed that LHY negatively effects transcription of NCED3, the gene for the most abundant NCED in Arabidopsis (Adams et al., 2018). In addition, LHY overexpression represses the accumulation of ABA during drought. Perturbation of chloroplast-related functions by chemicals such as 3-(3,4-dichlorophenyl)-1,1-dimethylurea, paraquat, and ascorbate modifies the pace of nuclear-driven circadian oscillations (Philippou et al., 2020), which led to the hypothesis that the circadian rhythm could act as an additional feedback mechanism from chloroplasts on ABA biosynthesis, due to connection of chloroplast functions and the circadian clock.
In addition to ABA biosynthesis, ABA signaling is widely interconnected with retrograde signals from chloroplasts. For instance, PAP signaling supports the ABA response pathway during stomatal closure and seed maturation via activation of the slow anion channel, SLAC1 (Pornsiriwong et al., 2017). PAP is a 3ʹ-phosphorylated-nucleotide that is maintained at very low levels by the action of the phosphatase SAL1 (also called ALX8, FIERY1, and HOS2). During photo-oxidative stress, SAL1 is inactivated by oxidation, which in turn leads to an accumulation of PAP and triggers several chloroplast stress signals, such as high expression of the H2O2 scavenger ascorbate peroxidase 2 under excess light (Estavillo et al., 2011). ABA is known as a major player in stomatal closure, as several ABA-insensitive mutants showed abolished stomatal closure under drought conditions (Chater et al., 2015). Pornsiriwong et al. (2017) showed that SAL1 inactivation/PAP accumulation can restore the guard cell responsiveness in the ABA-insensitive mutant lines ost1-2 and abi1-1 via interaction with the ABA signaling pathway downstream of these genes by the phosphorylation and activation of SLAC1.
Promoter and transcript analysis of 2-Cys PEROXIREDOXIN A (2CPA), encoding a highly abundant reactive oxygen species (ROS) scavenger in chloroplasts, revealed that its expression is controlled by ABA together with the cellular redox state (Baier et al., 2004). External application of ABA decreased the expression of 2CPA and consequently several ABA-insensitive mutants showed an increased expression of 2CPA (Baier et al., 2004). To what extent ABA-controlled expression of 2CPA could potentially feed back on the redox-responsive SAL1 inactivation/PAP accumulation is an open question.
Complex interactions: ABA signaling, plastid development, and nuclear interaction
Besides the direct connection of chloroplast functions with ABA biosynthesis and the indirect connection of ABA with operational retrograde signals, ABA itself can also shape plastid development and properties, which can be hypothesized as being an additional, more complex layer of ABA control within retrograde and anterograde signaling.
Transient expression of redox biosensors in the cytosol, the chloroplast stroma, and the nucleus revealed that photosynthesis-derived H2O2 in the chloroplast directly alters the H2O2 content of the nucleus, without affecting the cytosolic redox pool (Exposito-Rodriguez et al., 2017). It is proposed that the transfer of H2O2 occurs as a retrograde signal via the tubular structures of the chloroplast known as stromules. Gray et al. (2012) showed that ABA triggers the formation of chloroplast–nucleus complexes via stromules in tobacco and wheat seedlings. Vice versa, treatment with inhibitors of ABA biosynthesis prevents stromule formation under stress conditions. Recent work with strigolactone mutant lines and strigolactone biosynthesis inhibitors furthermore showed that ABA-induced stromule formation depends on active strigolactone biosynthesis in plastids, indicating crosstalk between ABA and strigolactones in this process (Vismans et al., 2016).
Besides signals from mature chloroplasts in response to environmental constraints (referred to as operational retrograde signals), retrograde signaling pathways also act during chloroplast development to control transcription of nucleus-encoded genes related to photosynthesis (referred to as biogenic retrograde signals). These biogenic signals are mainly studied by inhibition of carotenoid biosynthesis with norflourazon and by inhibition of the translation machinery in plastids with lincomycin (Susek et al., 1993). Screens for mutant lines that are insensitive to norflourazon and lincomycin revealed six gene loci, called GENOME UNCOUPLED 1–6 (GUN1–6). The majority of photosynthesis-related nucleus-encoded genes are repressed by ABA treatments (Yamburenko et al., 2015). Due to a certain overlap of misregulated genes between gun1-1 and abi4 knockout plants and suppression of the gun1-1 phenotype in ABA INSENSITIVE 4 (ABI4) overexpression plants, it was proposed that ABA signaling via the transcription factor ABI4 is an important downstream component of GUN1-mediated chloroplast biogenesis (Koussevitzky et al., 2007). However, recent work has re-evaluated the role of the ABA signaling component ABI4 in chloroplast biogenesis. Analysis of the phenotype of four different ABI4 alleles (abi4-102, abi4-1, abi4-2, and abi4-4) questions a role of ABI4 in GUN1-mediated biogenic control of photosynthesis-related, nucleus-encoded genes (Kacprzak et al., 2019). On the other hand, Zhu et al. (2020) showed recently that ABI5 overexpression suppresses chloroplast development and expression of photosynthesis-related genes in potato (Solanum tuberosum). Overall, the exact way that ABA is integrated in the biogenic control of chloroplast morphogenesis is still an open question.
ABA signaling and mitochondrial stress response
Inhibition of mitochondrial electron transport, for example by the chemical antimycin A, can trigger the transcription of mitochondrial stress-responsive genes. The best-known example is the increased transcript abundance of the nucleus-encoded ALTERNATIVE OXIDASE 1A (AOX1a) after perturbation of mitochondrial electron transport (Clifton et al., 2006). A forward genetic screen identified several REGULATORS OF AOX1A (RAOs) genes and thereby components of mitochondrial retrograde signaling. One of the first identified RAOs is the DREB transcription factor ABI4 (Giraud et al., 2009). Application of ABA leads to suppression of ABI4 expression and by that releases the repression of AOX1a, suggesting a positive role of ABA in mitochondrial retrograde signaling (Yao et al., 2015). It has also been reported that the induction of ABI4 and AOX1A transcription depends on RETARDED-ROOT GROWTH-LIKE FACTOR 1 (RRL1), a mitochondria-localized protein involved in mitochondrial retrograde signaling (Yao et al., 2015). Nevertheless, the full impact of ABA on mitochondrial function is still under investigation.
Jasmonates
Direct connection of organellar functions and jasmonate biosynthesis
Jasmonate is a collective term used for jasmonic acid (JA) and a diverse set of precursors and derivatives. They regulate a plethora of processes ranging from defense against stresses to the regulation of plant growth and development (reviewed in Wasternack and Hause, 2013; Koo, 2018). The first committed precursors of JA, 12-oxophytodienoic acid (cis-OPDA) and dinor-12-oxo-10,15(Z)-phytodienoic acid (dnOPDA), are made in chloroplasts (Fig. 4). As they are oxylipin derivatives, their biosynthesis starts with free fatty acids released from membrane lipids by the action of various lipases. In the so-called octadecanoid pathway, the initial substrate of cis-OPDA is α-linolenic acid (C18:3) released from the chloroplast galactolipids monogalactosyldiacylglycerol (MGDG) and digalactosyldiacylglycerol (DGDG). By contrast, dnOPDA is synthesized via a parallel hexadecanoid pathway from the less abundant 16:3 fatty acid roughanic acid (Bannenberg et al., 2009). Jasmonate synthesis seems to be closely interconnected with the MDGD/DGDG ratio since a mutant defective in DGDG production displays an increased production of cis-OPDA, as well as JA and jasmonoyl isoleucine (JA-Ile) (Lin et al., 2016).
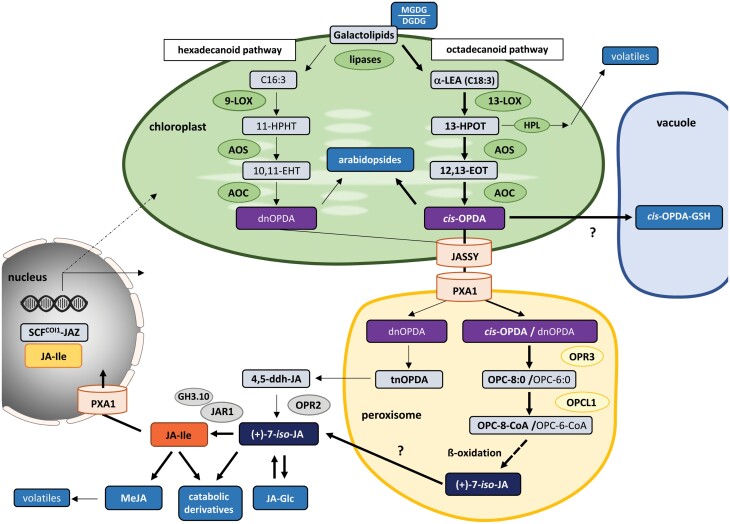
Fig. 4.
The role of organelles in jasmonate signaling. The term jasmonate comprises JA and JA-Ile as well as several precursors and catabolic derivatives some of which also possess bioactivity. Jasmonate biosynthesis is initiated in chloroplasts by oxidation of C18:3 (octadecanoid pathway) and C16:3 (hexadecanoid pathway) fatty acids derived from galactolipids, which are converted in several steps to the first committed precursor, OPDA. The MDGD/DGDG ratio, but also conjugation of OPDA to GSH or esterification to galactolipids (arabidopsides), affects OPDA homeostasis. Biosynthesis of JA continues in peroxisomes by β-oxidation of OPC-8:0 and OPC-6:0. A minor, less well described bypass pathway of JA formation involves tnOPDA and 4,5-ddh-JA. JA-Ile, the most bioactive of the jasmonates, is finally synthesized in the cytosol from JA and isoleucine, the latter being derived from methionine also made in chloroplasts. Ultimately, JA-Ile exerts its action in the nucleus by promoting the formation of SCFCOI1–JAZ co-receptor complexes and thus releasing JAZ-dependent gene suppression. 4,5ddh-JA, 4,5-didehydro-jasmonate; 10,11-EHT, 10,11(S)-epoxy-hexadecatrienoic acid; 11-HPHT, 11(S)-hydroperoxy-hexadecatrienoic acid; 12,13-EOT, 12,13(S)-epoxy-octadecatrienoic acid; 13-HPOT, 13(S)-hydroperoxylinolenic acid; α-LEA, α-linolenic acid; AOC, allene oxide cyclase; AOS, allene oxide synthase; DGDG, digalactosyl-diacylglycerol; GH3.10, glycoside hydrolase 3 gene family 10; HPL, hydroperoxide lyase; JA, jasmonic acid; JA-Glc, glycosylated jasmonate; JAR1, jasmonate-resistant 1; JASSY, chloroplast jasmonate transporter; LOX, lipoxygenases; MeJA, methyl jasmonate; MGDG, monogalactosyldiacylglycerol; OPC, 3-oxo-2-(20-[Z]-pentenyl)-cyclopentane-1-octanoic acid; OPCL1, OPC-8:0 CoA ligase 1; OPDA, oxophytodienoic acid; OPR, OPDA reductase; PXA1, peroxisomal ABC-transporter 1.
Both OPDA variants can be found esterified to galactolipids (reviewed in Genva et al., 2019). First identified in Arabidopsis and called arabidopsides, they have now also been detected in other plants. Due to their rapid increase upon certain biotic stresses, a role in plant defense was suggested. Oxylipins also have been found as glutathione (GSH) conjugates whose amount increases upon pathogen attack (Davoine et al., 2005). cis-OPDA–GSH conjugates are transported into the vacuole for either sequestration or degradation (Ohkama-Ohtsu et al., 2011). While the exact function of cis-OPDA–GSH and the arabidopsides remains elusive, they could be involved in the removal of a stimulus-induced excess of OPDA, or vice versa be an additional source for rapid jasmonate formation (Böttcher and Weiler, 2007).
Formation of JA from cis- and dnOPDA is continued in the peroxisome and the cytosol by two different routes (Fig. 4). Conjugation of JA to Ile by JASMONATE-RESISTANT 1 (JAR1), and to a lesser extent by other GH3 family proteins such as GH3.10, generates JA-Ile (Staswick and Tiryaki, 2004; Delfin et al., 2022). Isoleucine is derived from threonine and methionine made in chloroplasts. Thus, JA-Ile formation is independently connected twice to chloroplast metabolism. Most studies suggest that JA-Ile is the major biologically active JA derivative because it can promote the formation of SCF–COI1–JAZ co-receptor complexes involved in jasmonate-mediated transcriptional regulation (reviewed in Koo, 2018).
JA and JA-Ile are both substrates for further derivatization. Although these compounds are mainly considered as catabolic intermediates, bioactivity has nevertheless been suggested for some of them (reviewed in Koo, 2018; Wasternack and Hause, 2013). A notable derivative is methyl jasmonate (MeJA), a volatile communication molecule, present for example in essential oils of the jasmine flowers, where it was the first jasmonate ever identified (Demole and Stoll, 1962). After uptake into the cell, MeJA is converted to JA (Tamogami et al., 2008), thereby facilitating the external induction of jasmonate signaling. Interestingly, an intermediate of cis-OPDA synthesis, 13(S)-hydroperoxylinenic acid (13-HPOT), also gives rise to various volatiles important in the odors of fruits and vegetables (Matsui, 2006). It is not known, however, how the formation of these volatiles is interconnected with jasmonate biosynthesis and/or jasmonate signaling.
Jasmonate-mediated transcriptional changes are known to affect a wide range of cellular processes including organellar functions and expression of nucleus-encoded organellar proteins. However, comparatively little is known about organellar processes directly regulated by jasmonate signaling. Several studies showed differential effects of external jasmonate application, JAR1 overexpression, or jasmonate signaling suppression on the expression of nucleus-encoded photosynthesis-related genes (Attaran et al., 2014; Sirhindi et al., 2020; Mahmud et al., 2022). Recent findings on Arabidopsis also showed enhanced expression of most of the chloroplast-encoded genes by JA treatment (Zander et al., 2020) or JAR1 overexpression (Mahmud et al., 2022). It remains unclear whether this is a direct effect on the transcription of these genes within the organelle.
The effect of jasmonate on chlorophyll degradation has been known for a very long time (Ueda and Kato, 1980). Several studies have since reported that JA signaling influences leaf senescence by induction of SENESCENCE-ASSOCIATED GENES (SAGs) and CHLOROPHYLL CATABOLIC GENES (CCGs) involved in chlorophyll breakdown (Shan et al., 2011; Zhu et al., 2015). Regulation occurs via increase in the expression of JA-Ile-dependent MYC type transcription factors, which in turn modulate the expression of genes that facilitate chlorophyll breakdown (Zhu et al., 2015). Regulation also occurs more indirectly by MYC-mediated increase in expression of some NAC type transcription factors that were found to regulate leaf senescence. Indeed, a partially overlapping set of genes seems to be regulated by MYC2/3/4 and NAC019/055/072. By contrast, expression of RUBISCO ACTIVASE (RCA) was down-regulated by jasmonate in a COI1-dependent manner (Shan et al., 2011). Since the rca1 mutant displays typical senescence-related features and several senescence-promoting genes show up-regulation in the mutant, RCA could play an important role in jasmonate-induced de-greening (Shan et al., 2011).
Indirect mechanisms of jasmonate function
Jasmonate signaling is best described for the direct effect of JA-Ile on gene transcription via the SCF–COI1–JAZ co-receptor complex. Very little is known about indirect jasmonate signaling or other functions related to chloroplasts or mitochondria as depicted in Fig. 2B, C.
Besides JA-Ile, the most bioactive jasmonate, biological functions have been suggested for the chloroplast-derived precursor OPDA (recently reviewed in Maynard et al., 2018; Liu and Park, 2021). OPDA is found in marine algae, terrestrial algae, mosses, and ferns, but the full jasmonate synthesis pathway is absent in these organisms. Recent work suggested that dnOPDA is the evolutionary precursor of JA-Ile and can interact with the COI receptor orthologs of mosses (Monte et al. 2018; Inagaki et al., 2021). These findings indicate that the origin of jasmonate signaling can be directly connected to the chloroplast.
OPDA signaling seems to act both independently of and in concert with JA-Ile and it moreover affects the transcription of both COI1-dependent and -independent genes (Taki et al., 2005; Stinzi et al., 2001). Indeed, a signaling function of OPDA independent of JA was already suggested in the 1990s (reviewed in Bӧttcher and Pollmann, 2009). Many aspects of this OPDA-mediated signaling are not well understood but there is evidence for an action by binding to proteins such as 13-lipoxygenase, cyclophilin 20-3, thioredoxin (TRX)-m4, and peroxiredoxin (Dückershoff et al., 2008). Park et al. (2013) subsequently described that reversible binding of OPDA to cyclophilin 20-3 promotes the formation of a complex between cysteine synthase and O-acetylserine (thiol) lyase B. This leads to an increase in thiol metabolites and the cellular redox potential (Park et al., 2013). This redox change then plays a role in the expression of OPDA-dependent genes. By contrast, OPDAylation describes the covalent binding of OPDA to protein thiols (Maynard et al., 2021). Such thiols are targets of multiple post-translational modifications as part of what is called the thiol switch, and OPDAylation could be another player in this regulatory system. Recently, Knieper et al. (2022) analysed the in vitro interaction of several redox transmitters with OPDA under physiological OPDA concentrations. They obtained evidence for OPDAylation of both the cytosolic TRX-h3 and TRX-h5 and the chloroplastic TRX-f1 and TRX-m4. The in vivo relevance of this finding needs to be further analysed, but overall these works substantiate the signaling role of OPDA and suggest a close connection to redox-mediated processes.
Extracellular ATP (eATP) acts as a ‘damage-associated molecular pattern’ signal and is associated with a number of secondary messengers. Wounding, which induces ATP release, also induces expression of jasmonate-dependent genes. Tripathi et al. (2018) recently demonstrated that eATP activates these genes not via jasmonate biosynthesis but by direct enhancement of COI1–JAZ1 interaction followed by JAZ1 protein degradation. Moreover, this effect likely involves the secondary messengers Ca2+, NO, and ROS induced by eATP (Tripathi et al., 2018).
Proteomic studies suggested that the effect of MeJA application on root growth includes a reduction of proteins involved in ATP synthesis, thus affecting mitochondrial energy metabolism (Cho et al., 2007; Ruiz-May et al., 2011). Early on it was suggested that jasmonate signaling induces H2O2 production upon herbivory (Orozco-Cárdenas et al., 2001), and similarly jasmonate-induced ROS production in mitochondria was suggested to play a role in defense-related programmed cell death (Zhang and Xing, 2008). Also, MeJA-induced inhibition of root growth at least in hairy-root cultures is believed to involve cell death. Loyola-Vargas et al. (2012) found that treatment of hairy roots with MeJA enhances mitochondrial H2O2 accumulation together with an increased activity of corresponding anti-oxidant enzymes, and concluded that jasmonate signaling induced the oxidative burst that subsequently alters the mitochondrial proteome and mitochondrial function.
Mechanisms not directly related to jasmonate-mediated transcriptional regulation also seem to be involved in the initiation of jasmonate biosynthesis. Recently, Kimberlin et al. (2022) suggested that post-transcriptional processes, such as protein stability, play a role in the regulation of lipases that initiate the fast release of α-linolenic acid from chloroplast lipids upon wounding. Indeed, they suggest that positive feedforward mechanisms, i.e. jasmonate-mediated transcriptional activation of jasmonate biosynthesis pathway enzymes, have a fine-tuning role and exclude not only the lipases but also JAR1. However, these non-transcriptional regulations involved in regulating OPDA formation are not well understood.
Ethylene
Ethylene (IUPAC name: ethene) plays a central regulatory role throughout the whole plant life cycle from germination to integration of environmental changes to fruit ripening. Given its importance, the role of ethylene in plant growth, its biosynthesis pathways, and its downstream signaling pathways have been under constant investigation since its discovery as a plant growth factor by Neljubow (1901). That research led to a wealth of knowledge in plant ethylene signaling, which is reviewed in recent publications (Bakshi et al., 2015; Binder, 2020; Pattyn et al., 2021). Currently, ethylene is increasingly recognized also as an important mediator of stress signaling, with connections to plastid as well as mitochondrial signaling.
Direct signaling connections
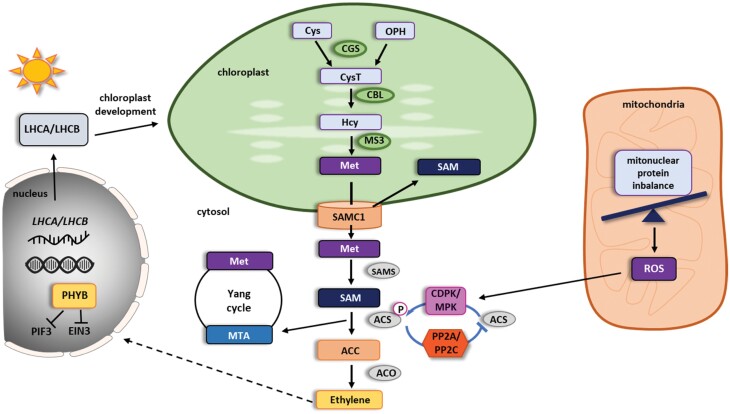
Ethylene biosynthesis is usually described as a linear two step reaction in the cytosol where aminocyclopropane 1-carboxylate synthase (ACS) converts S-adenosyl-l-methionine (SAM) to 1-aminocyclopropane 1-carboxylate (ACC) and 5ʹ-methylthioadenosine (MTA) (Adams and Yang, 1979; Boller et al., 1979). Subsequently, 1-aminocyclopropane 1-carboxylate oxidase (ACO) converts ACC to ethylene, H2O, HCN, and CO2 in the presence of oxygen (Hamilton et al., 1991; Ververidis and John, 1991; Bannenberg et al., 2009) (Fig. 5). However, SAM homeostasis is directly linked to the plastid via the methionine synthesis pathway. Also, a putative plastidial SAM importer was found to bind Ca2+, indicating stress-adaptive regulation (Stael et al., 2011). Plastidial de novo synthesis of methionine requires the formation of cystathionine from cysteine and phospho-homoserine by catalysis through cystathionine γ-synthase (CGS). Cystathionine β-lyase (CBL) then catalyses the conversion of cystathionine to homocysteine. Finally, methionine synthase (MS) methylates homocysteine, using 5-methyltetrahydrofolate as a methyl donor, into methionine (Hesse and Hoefgen, 2003), which is then exported to the cytosol. While there are three MS isoenzymes in Arabidopsis with AtMS3 localized in the plastid and AtMS1 and 2 localized in the cytosol (Hesse and Hoefgen, 2003; Ravanel et al., 2004), CGS and CBL are exclusively localized in the plastid and were shown to be embedded in a complex regulatory network with other plastid localized biochemical pathways (Ravanel et al., 1998; Watanabe et al., 2021).
Fig. 5.
The role of organelles in ethylene signaling. The chloroplast-localized enzyme cystathionine γ-synthase (CGS) catalyses the formation of cystathionine (CysT) from cysteine (Cys) and O-phosphohomoserine (OPH). Cystathionine is further transformed into homocysteine (Hcy) by cystathionine β-lyase (CBL). In the next step, the chloroplast-localized isoform of the methionine synthase 3 (MS3) forms methionine (Met), which in turn is transported out of the chloroplasts by SAMC1. In the cytoplasm Met is directly transformed into S-adenosylmethionine (SAM) by S-adenosylmethionine synthase (SAMS). SAM as main methyl donor is transported also back to the chloroplast by SAMC1. In the cytoplasm, SAM is converted into 1-aminocyclopropane 1-carboxylate (ACC) by the rate limiting ACC synthases (ACSs). After phosphorylation by calcium-dependent kinases (CDPK) and/or MAP kinases (MPK), ACSs are stabilized and therefore activated. Dephosphorylation of ACSs by different protein phosphatases (PP2A, PP2C) destabilizes the protein and leads to an immediate loss of ACS activity. The by-product of the reaction conducted by ACS, 5ʹ-methylthioadenosine (MTA), enters the Yang cycle to be detoxified and recycled into Met. ACC oxidase (ACO) catalyses the final step from ACC to ethylene. The mitonuclear protein imbalance leads to increase of mitochondrial reactive oxygen species (ROS) level. As a consequence, the mitochondrial unfolded protein response (UPRmt) is initiated. The elevated ROS level activates MPK6, which in turn promotes ET production by two ways: by phosphorylation of ACS6 and increase in transcription of the ACS6 gene. Additionally, nuclear ETHYLENE INSENSITIVE 3 (EIN3), a major ethylene responsive transcription factor, also plays a part in anterograde signaling (dashed line) in the chloroplast. After dark to light transition, PHYB promotes EIN3 and PIF3 degradation leading to LHCA and LHCB expression and chloroplast development.
A wealth of data support the notion that SAM levels influence ethylene production in the cytoplasm, and the cytoplasmic Yang cycle seems to have a primary impact on the SAM levels and subsequent ethylene biosynthesis. Yang cycle-mediated MTA recycling is especially important for plants naturally producing a high ethylene level, like ripening tomato and rice, which is demonstrated by Yang cycle gene up-regulation. In Arabidopsis, Yang cycle genes are not ethylene dependent; however, presumably during increased demand for ET, elevated ethylene biosynthesis induces MTA recycling, in order not to inhibit ACS activity (for a recent review see Pattyn et al., 2021). Despite the importance of cytoplasmic SAM and ACC levels for ethylene signaling, there are data that suggest at a direct role of the plastid in ethylene precursor production under certain conditions. In tomato, Barry et al. (2012) described a lutescent 2 loss of function mutant that displays slower chloroplast development and altered fruit ripening compared with wild type. Lutescent 2 was found to be the ortholog of the Arabidopsis EGY1 gene, which encodes a plastid thylakoid membrane-localized zinc-dependent M50 type metalloprotease (Chen et al., 2005). Similar to leaf tissue, also in tomato fruits the plastid development was impaired in the lutescent 2 mutant indicated by reduced photosynthetic rates or by whitish fruits instead of green ones at the onset of fruit formation. Ethylene levels in the fruits of lutescent 2 plants were reduced about 30% compared with the wild type, and application of exogenous ethylene was enough to alleviate the delayed onset of the ripening phenotype of lutescent 2 fruits (Barry et al., 2012). Interestingly, characterization of the OrrDs mutant, a dominant transposon-tagged tomato mutant deficient in the plastidial NADH dehydrogenase (Ndh) subunit NDH-M, revealed a delay in the onset of the fruit ripening phenotype as well as ~50% reduced ethylene levels emitted from fruits compared with the wild type (Nashilevitz et al., 2010). Based on the observation that precursors of methionine biosynthesis are reduced by ~30% in the OrrDS mutants, the authors concluded that the reduced ethylene levels are likely the result of reduced plastid-dependent ethylene production (Nashilevitz et al., 2010). In summary, the data on plastid development-retarded mutants such as egy1 in Arabidopsis (Guo et al., 2008), lutescent 2 (Barry et al., 2012), and OrrDs (Nashilevitz et al., 2010) show that in none of these mutants does the triple response for etiolated seedlings seem to be altered, and ethylene-caused phenotypes can be rescued by application of exogenous ethylene. This indicates that ethylene signaling seems to be functional and suggests that the ethylene-dependent phenotypes in these mutants are caused by reduced ethylene biosynthesis. A third indication for a role of the plastid in ethylene biosynthesis is that CGS transcript levels were found to be positively correlated with ethylene levels in ripening tomato fruits in order to sustain a high level of methionine biosynthesis (Katz et al., 2006). However, in order to determine that indeed plastid-derived metabolites are the limiting factor in the ethylene biosynthesis pathway, the possibility of indirect regulation of the core ethylene biosynthesis pathway ACCs and ACOs has to be tested in these mutants. Therefore, given current state of knowledge, the direct signaling connection originating from the metabolic capacity of the chloroplast and ethylene signaling remains hypothetical.
Indirect signaling connections
In contrast to the direct role of the chloroplast in ethylene signaling in a retrograde fashion, signaling via the ethylene responsive nuclear transcription factor ETHYLENE INSENSITIVE 3 (EIN3) has an anterograde function in chloroplast biogenesis itself. Liu et al. (2017) demonstrated that EIN3 interacts with PHYTOCHROME INTERACTING FACTOR3 (PIF3), a darkness-stabilized bHLH transcription factor, to repress the expression of most light harvesting complex (LHC) genes in the dark, thereby repressing expression of these genes and blocking the transition from etioplasts to chloroplasts. Upon exposure to light, phytochrome B-dependent degradation of EIN3 and PIF3 leads to LHCA and LHCB transcription, promoting chloroplast development (Liu et al., 2017).
Plastids and mitochondria are also connected indirectly to ethylene signaling via second messenger-related mechanisms. The best described example for this is probably the mitochondrial unfolded protein response. In an elegant study using mutants and chemical treatments leading to impaired mitochondrial translation, Wang and Auwerx (2017) discovered that the mitochondrial unfolded protein response (UPRmt) leads to a transient ROS burst within the first 60 min of triggered stress in Arabidopsis. That correlated with an increased activating phosphorylation of MITOGEN ACTIVATED PROTEIN KINASE 6 (MAPK6) in the same time frame. MAPK6 activity led to an increase in transcription of ACS6, thereby activating the ethylene response leading to a part of the transcriptional changes observed for the initially elicited UPRmt, ultimately restoring mitochondrial protein homeostasis (Wang and Auwerx, 2017).
With respect to plastid protein translational stress caused by lincomycin treatment Gommers et al. (2021) report a substantial overlap in gene expression between ACC-treated plants and lincomycin-treated plants as well as in their respective phenotypes of cotyledon opening and hypocotyl length. Somewhat surprisingly, neither an increase in ethylene nor prolonged stabilization of EIN3 was reported after 3 d of lincomycin treatment, prompting the authors to hypothesize an ethylene-independent response for activating classical ethylene-dependent genes downstream of EIN3/EIL1. Genetic analysis excluded GUN1-dependent retrograde signaling for activation of the observed gene expression and cotyledon opening patterns. However, at this point it should be mentioned that Gommers et al. (2021) analysed ethylene and EIN3 levels on constitutively lincomycin-treated plants and the data presented in Wang and Auwerx (2017) suggest a short transient ethylene signal rather than a prolonged constitutive one. Therefore, it still cannot be excluded that the actual ethylene signal might arise earlier in the lincomycin-treated plants.
An interesting link between plastid-derived ROS and the ethylene response was observed in potato plants expressing cyanobacterial plastid-targeted flavodoxin (fld) (Arce et al., 2022). In their meta-analysis of transcriptional profiles from fld-expressing Solanaceae, the authors detected common cis-elements in the promoter regions of genes involved in ethylene metabolism as a main target group of down-regulated transcripts due to fld expression and presumably reduced ROS levels in these plants (Arce et al., 2022). Unfortunately, the study lacked resolution to draw further conclusions for plastid-derived ROS and ethylene signaling, but in our opinion it is an additional reason to further investigate the relation between these.
Parallel signaling connection
This third category of plastid/mitochondria/ethylene signaling is intriguing because it requires both a functional ethylene response and functional organellar signaling but both of them are seemingly independent of each other in the first place. The Arabidopsis ETHYLENE-DEPENDENT GRAVOTROPISM-DEFICIENT and YELLOW-GREEN1 (EGY1) gene encodes a thylakoid membrane-localized protease involved in chloroplast development in leaf mesophyll cells that was isolated as an ethylene-dependent gravitropism mutant (Chen et al., 2005). Surprisingly it turned out that the egy1 mutant displayed a wild type like triple response indicating that the ethylene signaling cascade is intact in these plants (Guo et al., 2008). Through careful observation the authors noticed that egy1 shows a severe delay in chloroplast development in general. With respect to hypocotyl gravitropism the authors observed that the number, size, and starch levels of endodermal plastids were reduced compared with the wild type. Through application of exogenous sucrose, starch fill levels of endodermal plastids were restored and in the presence of ethylene also the gravitropic response of the egy1 mutant (Guo et al., 2008). From these data we can conclude that in the case of ethylene-dependent hypocotyl gravitropism, ethylene signaling is necessary to keep cells in a state that allows them to bend, whereas the endodermal plastid allows the plant to sense the gravity vector in these hypocotyl cells.
Salicylic acid
Of all the hormones addressed, SA is the least characterized. At present, indirect or complex mechanisms are still under investigation. Accumulation of SA has several layers of effects, triggering changes in expression of multiple genes, including nucleus-encoded proteins that are translocated to mitochondria and chloroplasts as well as through direct interaction with several proteins and biphasic accumulation of ROS in different subcellular compartments (Lu and Tsuda, 2020).
Direct connection of organellar functions in salicylic acid synthesis
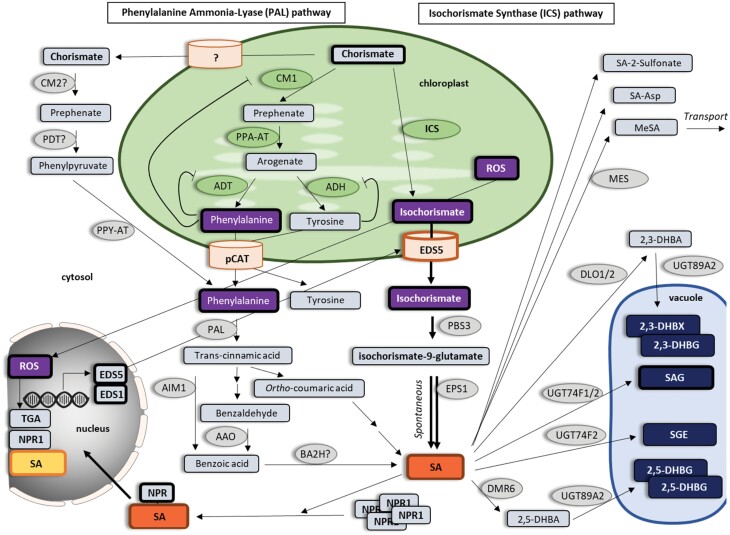
It is known that plants possess both an isochorismate synthase (ICS) and phenylalanine ammonia-lyase (PAL) pathway to synthesize SA, both starting from chorismate precursor (Zhang and Li, 2019) (Fig. 6). The ICS pathway is the primary route for SA production in Arabidopsis. However, the contribution of each pathway to the biosynthesis differs between plant species (Lefevere et al., 2020). In the well-studied ICS pathway, isochorismate is produced from chorismate in chloroplasts and transported to the cytosol, where SA is synthesized (Rekhter et al., 2019). Compartmentalization of the proteins responsible for the synthesis and transport control unidirectional forward flux to protect the pathway against evolutionary forces and pathogen perturbations (Rekhter et al., 2019). On the other hand, it is unclear whether the steps of the PAL pathway leading to the SA precursor phenylalanine take place in the chloroplast, cytosol, or both simultaneously (Lefevere et al., 2020). However, in the next steps of the PAL pathway, cytosol and peroxisomes are involved in SA synthesis (Murphy et al., 2020). Once synthesized, SA may undergo a number of chemical modifications including glucosylation, methylation, sulfonation, hydroxylation, and amino acid conjugation (D’Maris Amick et al., 2011). Some modifications inactivate SA in its regulatory roles to fine-tune SA activity, whereas others serve as temporary pools for its storage, such as SA glucoside, a vacuole-localized SA reserve (Ding and Ding, 2020).
Fig. 6.
The role of organelles in salicylic acid signaling. Overview of salicylic acid (SA) synthesis via the isochorismate synthase (ICS) and phenylalanine ammonia-lyase (PAL) pathways starting from chorismate. ICS converts chorismate into isochorismate (IC) in plastids. EDS5 exports IC from the plastid into the cytosol where PBS3 converts it into isochorismoyl-9-glutamate and further into SA by EPS1. The PAL pathway converts chorismate into prephenate by CM1 or CM2. Prephenate is converted into arogenate by PPA-AT and further to tyrosine by ADH or phenylalanine by ADT. CM1, ADH, and ADT are negatively regulated by their corresponding amino acid products. Tyrosine and phenylalanine are transported into the cytosol where PPY-AT converts them to phenylalanine. Phenylalanine produced from both plastidal and cytosolic pathways is further converted into trans-cinnamic acid by PAL and then into SA via ortho-coumaric intermediate or benzaldehyde and benzoic acid. SA can be converted into functional or non-functional metabolites such as SA-2-sulfonate, SA-Asp and MeSA, or can be stored in the vacuole as SAG, SGE, 2,3-DHBX, 2,3-DHBG, 2,5-DHBX and 2,5-DHBG. Higher SA levels induce monomerization of NPR1, translocation into the nucleus and NPR1-dependent gene expression through direct interactions with TGA transcription factors. The genes explicitly mentioned in the text are highlighted (boxed with bold line). AAO, aldehyde oxidase 4; ADH, arogenate dehydrogenase; ADT, arogenate dehydratase; AIM1, abnormal inflorescence meristem1; Asp, aspartic acid; BA2H, benzoic acid 2-hydroxylase; CM, chorismate mutase 1; DHBA, dihydroxy-benzaic acid; DHBG, dihydroxybenzoic acid glucoside; DHBX, dihydroxybenzoic acid xyloside; DLO, DMR6-like oxygenase; DMR6, SA-5 hydrolase; EDS5, enhanced disease susceptibility 5; EPS1, enhanced Pseudomonas susceptibility 1; ICS, isochorismate synthase; MES, methylesterases; NPR1, Nonexpresser of PR gene 1; PAL, phenylalanine ammonia-lyase; PBS3, avrPphB susceptible3; PDT, prephenate dehydratase; PPA-AT, plant prephenate aminotransferases; PPY-AT, phenylpyruvate aminotransferase; SA, salicylic acid; SAG, SA 2-O-β-d-glucoside; SGE, salicylate glucose ester; UGT89A2, uridine diphosphate (UDP)-glucosyltransferase.
Direct signaling connections: multiple regulatory feedback loops linking chloroplasts and SA signaling
SA signaling is known to be tightly interconnected with ROS signaling, which is also involved in several stress responses including pathogen attack (Bleau and Spoel, 2021). The transient phase of ROS accumulation occurs within minutes after infection and is mostly apoplastic and tightly linked to the activities of RBOHs, plasma membrane NADPH oxidases (Kadota et al., 2014; Li et al., 2014). In potato, RBOHD and SA regulate the immune response through a complex regulatory feedback loop, as RBOHD is required for the spatial accumulation of SA, and conversely, RBOHD is under the transcriptional regulation of SA (Lukan et al., 2020). RBOHD-dependent ROS production is also regulated by SA in Arabidopsis (Liu and He, 2016). The second, more sustained phase of ROS accumulation occurs in different compartments, including the apoplast, chloroplasts, mitochondria, and peroxisomes (Shapiguzov et al., 2012), and is associated with the establishment of defense responses and signaling for and/or execution of hypersensitive response cell death in incompatible interactions (Lu and Yao, 2018). Whether SA plays a role in this signaling has not been fully deciphered yet. Assumptions that SA signaling is not involved in cell death execution (Ochsenbein et al., 2006; Yao and Greenberg, 2006; Zurbriggen et al., 2009) were later questioned by Straus et al. (2010). They suggested that chloroplastic ROS acts as a flexible spatiotemporal integration point leading to opposite SA signaling reactions in infected and surrounding tissue to control the propagation of cell death. Interestingly, in potato hypersensitive response, ROS generated in the chloroplasts around the cell death zone are involved in two different SA-dependent and SA-independent plant response processes, which are spatially regulated. Strongly oxidized disordered chloroplasts in the cells on the edge of the cell death zone play a role in SA-independent hypersensitive response programmed cell death signaling, while the cells with moderately oxidized chloroplasts farther away from the cell death zone evidence SA-dependent signal transmission to neighboring tissue at the transcriptional level (Lukan et al., 2020). Chloroplastic ROS is involved in the up-regulation of genes upstream of SA accumulation through retrograde signaling, such as ENHANCED DISEASE SUSCEPTIBILITY1 (EDS1) (Ochsenbein et al., 2006) and ENHANCED DISEASE SUSCEPTIBILITY5 (EDS5) (Nomura et al., 2012).
Indirect mechanisms: SA binds to proteins from different subcellular compartments
Recently, multiple SA-binding proteins (SABPs) with different structures, localizations, and functions have been identified (Pokotylo et al., 2019). However, a direct function in plant physiology is currently not known for them all. Slaymaker et al. (2002) identified SALICYLIC ACID-BINDING PROTEIN 3 (SABP3) as a chloroplast β-carbonic anhydrase, which exhibits antioxidant activity in the soluble fraction of purified tobacco leaf chloroplasts. They proposed it has role in plant defense, perhaps through antioxidant activity (Slaymaker et al., 2002; DiMario et al., 2017). Treatment with SA altered the localization of the protein, which entered the cytoplasm, where interaction with SA and NONEXPRESSOR OF PATHOGENESIS-RELATED GENES 1 (NPR1) occurs (Medina-Puche et al., 2017). Another protein with antioxidative activity in plant immunity and identified as a SABP is glutathione S-transferase (GSTF) (Tian et al., 2012) with roles in the metabolism of antimicrobial compounds and detoxification of mycotoxins (Gullner et al., 2018). Dixon et al. (2009) characterized AtGSTF2 and AtGSTF8 as chloroplastic proteins and AtGSTF10 as vacuolar. SA binding inhibits GSTF activity, which modulates glutathione homeostasis and thus the cell redox state (Nazar et al., 2017). Another redox-related protein that binds SA is chloroplastic TRX-m1 (Manohar et al., 2015). The effect of SA binding on TRX-m1 activity has not yet been established; however, the activity of cytosolic TRX-h3 and TRX-h5 is required for NPR1 monomerization (Tada et al., 2008). Other chloroplastic proteins that bind SA are glyceraldehyde 3-phosphate dehydrogenase (GAPDH) isoforms AtGAPA-1 and AtGAPA-2, which play a role in the Calvin cycle (Pokotylo et al., 2019). In humans, SA binding suppresses GAPDH translocation to the nucleus, which plays a role in the regulation of replication of some viruses (Choi et al., 2015), while the role of this interaction in plants has not yet been deciphered. Another interaction with a potential role in plant antivirus defenses is suppression of mitochondrial α-ketoglutarate dehydrogenase (KGDE2) by SA in tomato, which occurs through direct binding (Liao et al., 2015). In contrast to viral resistance, the results of a recent study in tomato showed that KGDE2 plays a negative role in plant basal defense against Pseudomonas syringae in association with the SA defense pathway (Ma et al., 2020). Whether this occurs through direct binding needs to be elucidated. The presence of SABPs exhibiting a wide range of affinities for SA, combined with the varying SA levels found in specific subcellular compartments, in different tissues, at different developmental stages, or during responses to environmental cues, provides tremendous flexibility and multiple mechanisms through which SA effects can be utilized in plants.
Outlook
From the data published up to now, it can be concluded that phytohormone and organellar signaling works in concert at various levels. However, the studies discussed in this review also make clear that the connections between organellar and phytohormone signaling depend on complex factors such as tissue type, developmental status, and environmental stress conditions (Aerts et al., 2021; Cackett et al., 2022). A fact that further makes comparisons between studies difficult is that often, although arising from the same signaling pathway, different components are monitored. Subsequently, from those singular datasets conclusions for the whole pathway are implied. However, with the wealth of knowledge we currently have on processes like phytohormone biosynthesis, perception, and signaling as well as their interconnections to other signaling pathways at various points and levels (Ludwików et al., 2014; Blázquez et al., 2020; Binder, 2020; Depaepe et al., 2021; Müller, 2021; Müller and Munné-Bosch, 2021; Cackett et al., 2022), it becomes clear that taking single elements of these pathways does not generally permit predictions for the whole interconnected signaling process. These facts are increasingly being taken into consideration in current research and they will provide a more holistic framework for how organelles such as mitochondria and plastids are connected to the core phytohormone responses. Linking hormonal signaling to intracellular organellar communication brings new perspectives to and better understanding of plant signaling networks. In particular, it shows how developmental processes and responses to stress can be modulated through the functional state of organelles. Taking this perspective into account, future experiments can be designed to capture direct and indirect connections at the metabolic, transcriptional, and physiological level. In a long-term perspective, this knowledge will guide future functional studies and lead to improved crop breeding strategies for stress resilience.
Contributor Information
Andras Bittner, Plant Cell Biology, Institute of Cellular and Molecular Botany, University of Bonn, Kirschallee 1, 53115 Bonn, Germany.
Agata Cieśla, Laboratory of Biotechnology, Faculty of Biology, Adam Mickiewicz University, Uniwersytetu Poznańskiego 6, 61-614 Poznań, Poland.
Kristina Gruden, Department of Biotechnology and Systems Biology, National Institute of Biology, Večna pot 111, 1000 Ljubljana, Slovenia.
Tjaša Lukan, Department of Biotechnology and Systems Biology, National Institute of Biology, Večna pot 111, 1000 Ljubljana, Slovenia.
Sakil Mahmud, Plant Cell Biology, Institute of Cellular and Molecular Botany, University of Bonn, Kirschallee 1, 53115 Bonn, Germany.
Markus Teige, Department of Functional & Evolutionary Ecology, University of Vienna, Djerassiplatz 1, 1030 Vienna, Austria.
Ute C Vothknecht, Plant Cell Biology, Institute of Cellular and Molecular Botany, University of Bonn, Kirschallee 1, 53115 Bonn, Germany.
Bernhard Wurzinger, Department of Functional & Evolutionary Ecology, University of Vienna, Djerassiplatz 1, 1030 Vienna, Austria.
Author contributions
AB (ABA), SM (jasmonate), BW and AC (ethylene), TL (SA): writing—original draft and literature review; AB, AC, TL, MT, UV, BW: preparation of figures; KG, MT, UV: writing—review and editing. All authors contributed equally to the article and approved the submitted version.
Conflict of interest
The authors declare that no conflict of interest exists.
Funding
We acknowledge funding by the EU Horizon 2020 research and innovation project ADAPT (GA 2020 862-858), National Science Center Poland (Miniatura 4 2020/04/X/NZ3/00541), the German Academic Exchange Service (DAAD funding program no. 57299294), and the Slovenian research agency program P4-0165 (projects J4-1777 and Z4-3217).
References
- Adams DO, Yang SF.. 1979. Ethylene biosynthesis: Identification of 1-aminocyclopropane-1-carboxylic acid as an intermediate in the conversion of methionine to ethylene. Proceedings of the National Academy of Sciences, USA 76, 170–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams S, Grundy J, Veflingstad SR, Dyer NP, Hannah MA, Ott S, Carré IA.. 2018. Circadian control of abscisic acid biosynthesis and signaling pathways revealed by genome-wide analysis of LHY binding targets. New Phytologist 220, 893–907. [DOI] [PubMed] [Google Scholar]
- Aerts N, Pereira Mendes M, Van Wees SCM.. 2021. Multiple levels of crosstalk in hormone networks regulating plant defense. The Plant Journal 105, 489–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arce RC, Carrillo N, Pierella Karlusich JJ.. 2022. The chloroplast redox-responsive transcriptome of Solanaceous plants reveals significant nuclear gene regulatory motifs associated to stress acclimation. Plant Molecular Biology 108, 513–530. [DOI] [PubMed] [Google Scholar]
- Attaran E, Major IT, Cruz JA, Rosa BA, Koo AJ, Chen J, Kramer DM, He SY, Howe GA.. 2014. Temporal dynamics of growth and photosynthesis suppression in response to jasmonate signaling. Plant Physiology 165, 1302–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baier M, Ströher E, Dietz K-J.. 2004. The acceptor availability at photosystem I and ABA control nuclear expression of 2-Cys peroxiredoxin-A in Arabidopsis thaliana. Plant & Cell Physiology 45, 997–1006. [DOI] [PubMed] [Google Scholar]
- Bakshi A, Shemansky JM, Chang CR, Binder BM.. 2015. History of research on the plant hormone ethylene. Journal of Plant Growth Regulation 34, 809–827. [Google Scholar]
- Blázquez M, Nelson DC, Weijers D.. 2020. Evolution of plant hormone response pathways. Annual Review of Plant Biology 71, 327–353. [DOI] [PubMed] [Google Scholar]
- Bannenberg G, Martinez M, Hamberg M, Castresana C.. 2009. Diversity of the enzymatic activity in the lipoxygenase gene family of Arabidopsis thaliana. Lipids 44, 85–95. [DOI] [PubMed] [Google Scholar]
- Barry CS, Aldridge GM, Herzog G, Ma Q, McQuinn RP, Hirschberg J, Giovannoni JJ.. 2012. Altered chloroplast development and delayed fruit ripening caused by mutations in a zinc metalloprotease at the lutescent2 locus of tomato. Plant Physiology 159, 1086–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berens ML, Berry HM, Mine A, Argueso CT, Tsuda K.. 2017. Evolution of hormone signaling networks in plant defense. Annual Review of Phytopathology 55, 401–425. [DOI] [PubMed] [Google Scholar]
- Binder BM. 2020. Ethylene signaling in plants. Journal of Biological Chemistry 295, 7710–7725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleau JR, Spoel SH.. 2021. Selective redox signaling shapes plant–pathogen interactions. Plant Physiology 186, 53–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller T, Herner RC, Kende H.. 1979. Assay for and enzymatic formation of an ethylene precursor, 1-aminocyclopropane-1-carboxylic acid. Planta 145, 293–303. [DOI] [PubMed] [Google Scholar]
- Bӧttcher C, Pollmann S.. 2009. Plant oxylipins: Plant responses to 12-oxo-phytodienoic acid are governed by its specific structural and functional properties. FEBS Journal 17, 4693–4704. [DOI] [PubMed] [Google Scholar]
- Böttcher C, Weiler EW.. 2007. cyclo-Oxylipin-galactolipids in plants: occurrence and dynamics. Planta 226, 629–637. [DOI] [PubMed] [Google Scholar]
- Cackett L, Luginbuehl LH, Schreier TB, Lopez-Juez E, Hibberd JM.. 2022. Chloroplast development in green plant tissues: the interplay between light, hormone, and transcriptional regulation. New Phytologist 233, 2000–2016. [DOI] [PubMed] [Google Scholar]
- Cardoso AA, Gori A, Da-Silva CJ, Brunetti C.. 2020. Abscisic acid biosynthesis and signaling in plants: key targets to improve water use efficiency and drought tolerance. Applied Sciences 10, 6322. [Google Scholar]
- Chater C, Peng K, Movahedi M, et al. 2015. Elevated CO2-induced responses in stomata require ABA and ABA signaling. Current Biology 25, 2709–2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Bi YR, Li N.. 2005. EGY1 encodes a membrane-associated and ATP-independent metalloprotease that is required for chloroplast development. The Plant Journal 41, 364–375. [DOI] [PubMed] [Google Scholar]
- Cho K, Agrawal GK, Shibato J, et al. 2007. Survey of differentially expressed proteins and genes in jasmonic acid treated rice seedling shoot and root at the proteomics and transcriptomics levels. Journal of Proteome Research 6, 3581–3603. [DOI] [PubMed] [Google Scholar]
- Choi HW, Tian M, Manohar M, Harraz MM, Park S-W, Schroeder FC, Snyder SH, Klessig DF.. 2015. Human GAPDH is a target of aspirin’s primary metabolite salicylic acid and its derivatives. PLoS One 10, e0143447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifton R, Millar AH, Whelan J.. 2006. Alternative oxidases in Arabidopsis: A comparative analysis of differential expression in the gene family provides new insights into function of non-phosphorylating bypasses. Biochimica et Biophysica Acta – Bioenergetics 1757, 730–741. [DOI] [PubMed] [Google Scholar]
- Crawford T, Lehotai N, Strand A.. 2018. The role of retrograde signals during plant stress responses. Journal of Experimental Botany 69, 2783–2795. [DOI] [PubMed] [Google Scholar]
- D’Alessandro S, Beaugelin I, Havaux M.. 2020. Tanned or sunburned: how excessive light triggers plant cell death. Molecular Plant 13, 1545–1555. [DOI] [PubMed] [Google Scholar]
- Davoine C, Douki T, Iacazio G, Montillet JL, Triantaphylides C.. 2005. Conjugation of keto fatty acids to glutathione in plant tissues. Characterization and quantification by HPLC-tandem mass spectrometry. Analytical Chemistry 77, 7366–7372. [DOI] [PubMed] [Google Scholar]
- Delfin JC, Kanno Y, Seo M, Kitaoka N, Matsuura H, Tohge T, Shimizu T.. 2022. AtGH3.10 is another jasmonic acid-amido synthetase in Arabidopsis thaliana. The Plant Journal 110, 1082–1096. [DOI] [PubMed] [Google Scholar]
- Demole E, Stoll M.. 1962. Synthèses du D,L-jasmonate de méthyle (cis-pentène-2ʹ-yl-2-oxo-3-cyclopentylacétate de méthyle) et de deux isomères. Helvetica Chimica Acta 45, 692–703. [Google Scholar]
- Depaepe T, Hendrix S, Janse van Rensburg HC, Van den Ende W, Cuypers A, Van Der Straeten D.. 2021. At the crossroads of survival and death: The reactive oxygen species-ethylene-sugar triad and the unfolded protein response. Trends in Plant Science 26, 338–351. [DOI] [PubMed] [Google Scholar]
- DiMario RJ, Clayton H, Mukherjee A, Ludwig M, Moroney JV.. 2017. Plant carbonic anhydrases: structures, locations, evolution, and physiological roles. Molecular Plant 10, 30–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding P, Ding Y.. 2020. Stories of salicylic acid: A plant defense hormone. Trends in Plant Science 25, 549–565. [DOI] [PubMed] [Google Scholar]
- Dixon DP, Hawkins T, Hussey PJ, Edwards R.. 2009. Enzyme activities and subcellular localization of members of the Arabidopsis glutathione transferase superfamily. Journal of Experimental Botany 60, 1207–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Maris Amick D, Vlot AC, Mary CW, Daniel FK.. 2011. Salicylic acid biosynthesis and metabolism. The Arabidopsis Book 9, e0156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dopp IJ, Yang X, Mackenzie SA.. 2021. A new take on organelle-mediated stress sensing in plants. New Phytologist 230, 2148–2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dückershoff K, Mueller S, Mueller MJ, Reinders J.. 2008. Impact of cyclopentenone-oxylipins on the proteome of Arabidopsis thaliana. Biochimica et Biophysica Acta 1784, 1975–1985. [DOI] [PubMed] [Google Scholar]
- Endo A, Sawada Y, Takahashi H, et al. 2008. Drought induction of Arabidopsis 9-cis-epoxycarotenoid dioxygenase occurs in vascular parenchyma cells. Plant Physiology 147, 1984–1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estavillo GM, Crisp PA, Pornsiriwong W, et al. 2011. Evidence for a SAL1-PAP chloroplast retrograde pathway that functions in drought and high light signaling in Arabidopsis. The Plant Cell 23, 3992–4012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exposito-Rodriguez M, Laissue PP, Yvon-Durocher G, Smirnoff N, Mullineaux PM.. 2017. Photosynthesis-dependent H2O2 transfer from chloroplasts to nuclei provides a high-light signaling mechanism. Nature Communications 8, 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fàbregas N, Fernie AR.. 2021. The interface of central metabolism with hormone signaling in plants. Current Biology 31, R1535–R1548. [DOI] [PubMed] [Google Scholar]
- Genva M, Obounou Akong F, Andersson MX, Deleu M, Lins L, Fauconnier M-L.. 2019. New insights into the biosynthesis of esterified oxylipins and their involvement in plant defense and developmental mechanisms. Phytochemistry Reviews 18, 343–358. [Google Scholar]
- Giraud E, Van Aken O, Ho LHM, Whelan J.. 2009. The transcription factor ABI4 is a regulator of mitochondrial retrograde expression of ALTERNATIVE OXIDASE1a. Plant Physiology 150, 1286–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gläßer C, Haberer G, Finkemeier I, Pfannschmidt T, Kleine T, Leister D, Dietz K-J, Häusler RE, Grimm B, Mayer KFX.. 2014. Meta-analysis of retrograde signaling in Arabidopsis thaliana reveals a core module of genes embedded in complex cellular signaling networks. Molecular Plant 7, 1167–1190. [DOI] [PubMed] [Google Scholar]
- Gommers CMM, Ruiz-Sola MA, Ayats A, Pereira L, Pujol M, Monte E.. 2021. GENOMES UNCOUPLED1-independent retrograde signaling targets the ethylene pathway to repress photomorphogenesis. Plant Physiology 185, 67–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray JC, Hansen MR, Shaw DJ, Graham K, Dale R, Smallman P, Natesan SKA, Newell CA.. 2012. Plastid stromules are induced by stress treatments acting through abscisic acid. The Plant Journal 69, 387–398. [DOI] [PubMed] [Google Scholar]
- Gullner G, Komives T, Király L, Schröder P.. 2018. Glutathione S-transferase enzymes in plant-pathogen interactions. Frontiers in Plant Science 9, 1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo D, Gao XR, Li H, Zhang T, Chen G, Huang PB, An LJ, Li N.. 2008. EGY1 plays a role in regulation of endodermal plastid size and number that are involved in ethylene-dependent gravitropism of light-grown Arabidopsis hypocotyls. Plant Molecular Biology 66, 345–360. [DOI] [PubMed] [Google Scholar]
- Hamilton AJ, Bouzayen M, Grierson D.. 1991. Identification of a tomato gene for the ethylene-forming enzyme by expression in yeast. Proceedings of the National Academy of Sciences, USA 88, 7434–7437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesse H, Hoefgen R.. 2003. Molecular aspects of methionine biosynthesis. Trends in Plant Science 8, 259–262. [DOI] [PubMed] [Google Scholar]
- Huang X, Hou L, Meng J, You H, Li Z, Gong Z, Yang S, Shi Y.. 2018. The antagonistic action of abscisic acid and cytokinin signaling mediates drought stress response in Arabidopsis. Molecular Plant 11, 970–982. [DOI] [PubMed] [Google Scholar]
- Inagaki H, Miyamoto K, Ando N, et al. 2021. Deciphering OPDA signaling components in the momilactone-producing moss Calohypnum plumiforme. Frontiers in Plant Science 12, 688565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kacprzak SM, Mochizuki N, Naranjo B, Xu D, Leister D, Kleine T, Okamoto H, Terry MJ.. 2019. Plastid-to-nucleus retrograde signaling during chloroplast biogenesis does not require ABI4. Plant Physiology 179, 18–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadota Y, Sklenar J, Derbyshire P, et al. 2014. Direct regulation of the NADPH oxidase RBOHD by the PRR-associated kinase BIK1 during plant immunity. Molecular Cell 54, 43–55. [DOI] [PubMed] [Google Scholar]
- Katz YS, Galili G, Amir R.. 2006. Regulatory role of cystathionine-γ-synthase and de novo synthesis of methionine in ethylene production during tomato fruit ripening. Plant Molecular Biology 61, 255–268. [DOI] [PubMed] [Google Scholar]
- Kimberlin AN, Holtsclaw RE, Zhang T, Mulaudzi T, Koo AJ.. 2022. On the initiation of jasmonate biosynthesis in wounded leaves. Plant Physiology 189, 1925–1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine T, Leister D.. 2016. Retrograde signaling: Organelles go networking. Biochimica et Biophysica Acta – Bioenergetics 1857, 1313–1325. [DOI] [PubMed] [Google Scholar]
- Kmiecik P, Leonardelli M, Teige M.. 2016. Novel connections in plant organellar signaling link different stress responses and signaling pathways. Journal of Experimental Botany 67, 3793–3807. [DOI] [PubMed] [Google Scholar]
- Knieper M, Vogelsang L, Guntelmann T, Sproß J, Gröger H, Viehhauser A, Dietz KJ.. 2022. OPDAylation of thiols of the redox regulatory network in vitro. Antioxidants 11, 855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo AJ. 2018. Metabolism of the plant hormone jasmonate: a sentinel for tissue damage and master regulator of stress response. Phytochemistry Reviews 17, 51–80. [Google Scholar]
- Koussevitzky S, Nott A, Mockler TC, Hong F, Sachetto-Martins G, Surpin M, Lim J, Mittler R, Chory J.. 2007. Signals from chloroplasts converge to regulate nuclear gene expression. Science 316, 715–719. [PubMed] [Google Scholar]
- Lee Keun P, Kim C, Landgraf F, Apel K.. 2007. EXECUTER1- and EXECUTER2-dependent transfer of stress-related signals from the plastid to the nucleus of Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA 104, 10270–10275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefevere H, Bauters L, Gheysen G.. 2020. Salicylic acid biosynthesis in plants. Frontiers in Plant Science 11, 338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Li M, Yu L, et al. 2014. The FLS2-associated kinase BIK1 directly phosphorylates the NADPH oxidase RbohD to control plant immunity. Cell Host & Microbes 15, 329–338. [DOI] [PubMed] [Google Scholar]
- Li M, Kim C.. 2022. Chloroplast ROS and stress signaling. Plant Communications 3, 100264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Tian M, Zhang H, et al. 2015. Salicylic acid binding of mitochondrial alpha-ketoglutarate dehydrogenase E2 affects mitochondrial oxidative phosphorylation and electron transport chain components and plays a role in basal defense against tobacco mosaic virus in tomato. New Phytologist 205, 1296–1307. [DOI] [PubMed] [Google Scholar]
- Lin YT, Chen LJ, Herrfurth C, Feussner I, Li HM.. 2016. Reduced biosynthesis of digalactosyldiacylglycerol, a major chloroplast membrane lipid, leads to oxylipin overproduction and phloem cap lignification in Arabidopsis. The Plant Cell 28, 219–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Staswick PE, Avramova Z.. 2016. Memory responses of jasmonic acid-associated Arabidopsis genes to a repeated dehydration stress. Plant Cell and Environment 39, 2515–2529. [DOI] [PubMed] [Google Scholar]
- Liu W, Park S-W.. 2021. 12-oxo-Phytodienoic acid: a fuse and/or switch of plant growth and defense responses? Frontiers in Plant Science 12, 724079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Liu R, Li Y, Shen X, Zhong S, Shi H.. 2017. EIN3 and PIF3 form an interdependent module that represses chloroplast development in buried seedlings. The Plant Cell 29, 3051–3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, He C.. 2016. Regulation of plant reactive oxygen species (ROS) in stress responses: learning from AtRBOHD. Plant Cell Reports 35, 995–1007. [DOI] [PubMed] [Google Scholar]
- Loyola-Vargas V, Ruíz-May E, Galaz-Ávalos R, De-la-Peña C.. 2012. The role of jasmonic acid in root mitochondria disruption. Plant Signaling & Behavior 7, 611–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Tsuda K.. 2020. Intimate association of PRR- and NLR-mediated signaling in plant immunity. Molecular Plant-Microbe Interactions 34, 3–14. [DOI] [PubMed] [Google Scholar]
- Lu Y, Yao J.. 2018. Chloroplasts at the crossroad of photosynthesis, pathogen infection and plant defense. International Journal of Molecular Sciences 19, 3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwików A, Cieśla A, Kasprowicz-Maluśki A, et al. 2014. Arabidopsis protein phosphatase 2C ABI1 interacts with type I ACC synthases and is involved in the regulation of ozone-induced ethylene biosynthesis. Molecular Plant 7, 960–976. [DOI] [PubMed] [Google Scholar]
- Lukan T, Pompe-Novak M, Baebler S, et al. 2020. Precision transcriptomics of viral foci reveals the spatial regulation of immune-signaling genes and identifies RBOHD as an important player in the incompatible interaction between potato virus Y and potato. The Plant Journal 104, 645–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q, Liu Y, Fang H, Wang P, Ahammed GJ, Zai W, Shi K.. 2020. An essential role of mitochondrial α-ketoglutarate dehydrogenase E2 in the basal immune response against bacterial pathogens in tomato. Frontiers in Plant Science 11, 579772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Cao J, He J, Chen Q, Li X, Yang Y.. 2018. Molecular mechanism for the regulation of ABA homeostasis during plant development and stress responses. International Journal of Molecular Sciences 19, 3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmud S, Ullah C, Kortz A, Bhattacharyya S, Yu P, Gershenzon J, Vothknecht UC.. 2022. Constitutive expression of JASMONATE RESISTANT 1 induces molecular changes that prime the plants to better withstand drought. Plant Cell and Environment 45, 2906–2922. [DOI] [PubMed] [Google Scholar]
- Manohar M, Tian M, Moreau M, et al. 2015. Identification of multiple salicylic acid-binding proteins using two high throughput screens. Frontiers in Plant Science 5, 777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui K. 2006. Green leaf volatiles: hydroperoxide lyase pathway of oxylipin metabolism. Current Opinion in Plant Biology 9, 274–280. [DOI] [PubMed] [Google Scholar]
- Maynard D, Gröger H, Dierks T, Dietz K-J.. 2018. The function of the oxylipin 12-oxophytodienoic acid in cell signaling, stress acclimation, and development. Journal of Experimental Botany 69, 5341–5354. [DOI] [PubMed] [Google Scholar]
- Maynard D, Viehhauser A, Knieper M, Dreyer A, Manea G, Telman W, Butter F, Chibani K, Scheibe R, Dietz KJ.. 2021. The in vitro interaction of 12-oxophytodienoic acid and related conjugated carbonyl compounds with thiol antioxidants. Biomolecules 11, 457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAdam SA, Brodribb TJ, Ross JJ.. 2016. Shoot-derived abscisic acid promotes root growth. Plant Cell and Environment 39, 652–659. [DOI] [PubMed] [Google Scholar]
- Medina-Puche L, Castelló MJ, Canet JV, Lamilla J, Colombo ML, Tornero P.. 2017. β-Carbonic anhydrases play a role in salicylic acid perception in Arabidopsis. PLoS One 12, e0181820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monte I, Ishida S, Zamarreño AM, et al. 2018. Ligand-receptor co-evolution shaped the jasmonate pathway in land plants. Nature Chemical Biology 14, 480–488. [DOI] [PubMed] [Google Scholar]
- Müller M. 2021. Foes or friends: ABA and ethylene interaction under abiotic stress. Plants 10, 448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller M, Munné-Bosch S.. 2021. Hormonal impact on photosynthesis and photoprotection in plants. Plant Physiology 185, 1500–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy AM, Zhou T, Carr JP.. 2020. An update on salicylic acid biosynthesis, its induction and potential exploitation by plant viruses. Current Opinion in Virology 42, 8–17. [DOI] [PubMed] [Google Scholar]
- Nashilevitz S, Melamed-Bessudo C, Izkovich Y, et al. 2010. An orange ripening mutant links plastid NAD(P)H dehydrogenase complex activity to central and specialized metabolism during tomato fruit maturation. The Plant Cell 22, 1977–1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazar R, Iqbal N, Umar S.. 2017. Heat stress tolerance in plants: action of salicylic acid. In: Nazar R, Iqbal N, Khan NA, eds. Salicylic acid: a multifaceted hormone. Singapore: Springer Singapore, 145–161. [Google Scholar]
- Neljubow D. 1901. Ueber die horizontale Nutation der Stengel von Pisum sativum und einiger anderen Pflanzen. Beihefte zum botanischen Zentralblatt 10, 128–139. [Google Scholar]
- Nomura H, Komori T, Uemura S, et al. 2012. Chloroplast-mediated activation of plant immune signaling in Arabidopsis. Nature Communications 3, 926. [DOI] [PubMed] [Google Scholar]
- Ochsenbein C, Przybyla D, Danon A, Landgraf F, Göbel C, Imboden A, Feussner I, Apel K.. 2006. The role of EDS1 (enhanced disease susceptibility) during singlet oxygen-mediated stress responses of Arabidopsis. The Plant Journal 47, 445–456. [DOI] [PubMed] [Google Scholar]
- Ohkama-Ohtsu N, Sasaki-Sekimoto Y, Oikawa A, et al. 2011. 12-Oxo-phytodienoic acid-glutathione conjugate is transported into the vacuole in Arabidopsis. Plant & Cell Physiology 52, 205–209. [DOI] [PubMed] [Google Scholar]
- Ohkuma K, Lyon JL, Addicott FT, Smith OE.. 1963. Abscisin II, an abscission-accelerating substance from young cotton fruit. Science 142, 1592–1593. [DOI] [PubMed] [Google Scholar]
- Orozco-Cárdenas ML, Narváez-Vásquez J, Ryan CA.. 2001. Hydrogen peroxide acts as a second messenger for the induction of defense genes in tomato plants in response to wounding, systemin, and methyl jasmonate. The Plant Cell 13, 179–191. [PMC free article] [PubMed] [Google Scholar]
- Park SW, Li W, Viehhauser A, et al. 2013. Leaf vitamin C contents modulate plant defense transcripts and regulate genes that control development through hormone signaling. The Plant Cell 15, 939–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastori GM, Kiddle G, Antoniw J, Bernard S, Veljovic-Jovanovic S, Verrier PJ, Noctor G, Foyer CH.. 2003. Leaf vitamin C contents modulate plant defense transcripts and regulate genes that control development through hormone signaling. The Plant Cell 15, 939–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattyn J, Vaughan-Hirsch J, Van de Poel B.. 2021. The regulation of ethylene biosynthesis: a complex multilevel control circuitry. New Phytologist 229, 770–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippou K, Davis AM, Davis SJ, Sánchez-Villarreal A.. 2020. Chemical perturbation of chloroplast-related processes affects circadian rhythms of gene expression in Arabidopsis: salicylic acid application can entrain the clock. Frontiers in Physiology 11, 429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokotylo I, Kravets V, Ruelland E.. 2019. Salicylic acid binding proteins (SABPs): The hidden forefront of salicylic acid signaling. International Journal of Molecular Sciences 20, 4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pornsiriwong W, Estavillo GM, Chan KX, et al. 2017. A chloroplast retrograde signal, 3ʹ-phosphoadenosine 5ʹ-phosphate, acts as a secondary messenger in abscisic acid signaling in stomatal closure and germination. eLife 6, e23361. [Google Scholar]
- Ramel F, Birtic S, Cuiné S, Triantaphylidès C, Ravanat J-L, Havaux M.. 2012. Chemical quenching of singlet oxygen by carotenoids in plants. Plant Physiology 158, 1267–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramšak Ž, Coll A, Stare T, Tzfadia O, Baebler Š, Van de Peer Y, Gruden K.. 2018. Network modeling unravels mechanisms of crosstalk between ethylene and salicylate signaling in potato. Plant Physiology 178, 488–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravanel S, Block MA, Rippert P, Jabrin S, Curien G, Rebeille F, Douce R.. 2004. Methionine metabolism in plants: Chloroplasts are autonomous for de novo methionine synthesis and can import S-adenosylmethionine from the cytosol. Journal of Biological Chemistry 279, 22548–22557. [DOI] [PubMed] [Google Scholar]
- Ravanel S, Gakière B, Job D, Douce R.. 1998. The specific features of methionine biosynthesis and metabolism in plants. Proceedings of the National Academy of Sciences, USA 95, 7805–7812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rekhter D, Lüdke D, Ding Y, Feussner K, Zienkiewicz K, Lipka V, Wiermer M, Zhang Y, Feussner I.. 2019. Isochorismate-derived biosynthesis of the plant stress hormone salicylic acid. Science 365, 498–502. [DOI] [PubMed] [Google Scholar]
- Ruban AV, Walters RG, Horton P.. 1992. The molecular mechanism of the control of excitation energy dissipation in chloroplast membranes Inhibition of ΔpH-dependent quenching of chlorophyll fluorescence by dicyclohexylcarbodiimide. FEBS Letters 309, 175–179. [DOI] [PubMed] [Google Scholar]
- Ruiz-May E, De-la-Peña C, Galaz-Ávalos RM, Lei Z, Watson BS, Sumner LW, Loyola-Vargas VM.. 2011. Methyl jasmonate induces ATP biosynthesis deficiency and accumulation of proteins related to secondary metabolism in Catharanthus roseus (L.) G. hairy roots. Plant and Cell Physiology 52, 1401–1421. [DOI] [PubMed] [Google Scholar]
- Seo M, Marion-Poll A.. 2019. Abscisic acid metabolism and transport. In: Seo M, Marion-Poll A, eds. Advances in Botanical Research 92. Academic Press, 1–49. [Google Scholar]
- Shan X, Wang J, Chua L, Jiang D, Peng W, Xie D.. 2011. The role of arabidopsis rubisco activase in jasmonate-induced leaf senescence. Plant Physiology 155, 751–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiguzov A, Vainonen J, Wrzaczek M, Kangasjärvi J.. 2012. ROS-talk – how the apoplast, the chloroplast, and the nucleus get the message through. Frontiers in Plant Science 3, 292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirhindi G, Mushtaq R, Gill SS, Sharma P, Abd Allah EF, Ahmad P.. 2020. Jasmonic acid and methyl jasmonate modulate growth, photosynthetic activity and expression of photosystem II subunit genes in Brassica oleracea L. Scientific Reports 10, 9322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaymaker DH, Navarre DA, Clark D, del Pozo O, Martin GB, Klessig DF.. 2002. The tobacco salicylic acid-binding protein 3 (SABP3) is the chloroplast carbonic anhydrase, which exhibits antioxidant activity and plays a role in the hypersensitive defense response. Proceedings of the National Academy of Sciences, USA 99, 11640–11645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stael S, Rocha AG, Robinson AJ, Kmiecik P, Vothknecht UC, Teige M.. 2011. Arabidopsis calcium-binding mitochondrial carrier proteins as potential facilitators of mitochondrial ATP-import and plastid SAM-import. FEBS Letters 585, 3935–3940. [DOI] [PubMed] [Google Scholar]
- Staswick PE, Tiryaki I.. 2004. The oxylipin signal jasmonic acid is activated by an enzyme that conjugates it to isoleucine in Arabidopsis. The Plant Cell 16, 2117–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinzi A, Weber H, Reymond P, Browse J, Farmer EE.. 2001. Plant defense in the absence of jasmonic acid: the role of cyclopentenones. Proceedings of the National Academy of Sciences, USA 98, 12837–12842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straus MR, Rietz S, Ver Loren van Themaat E, Bartsch M, Parker JE.. 2010. Salicylic acid antagonism of EDS1-driven cell death is important for immune and oxidative stress responses in Arabidopsis. The Plant Journal 62, 628–640. [DOI] [PubMed] [Google Scholar]
- Susek RE, Ausubel FM, Chory J.. 1993. Signal transduction mutants of arabidopsis uncouple nuclear CAB and RBCS gene expression from chloroplast development. Cell 74, 787–799. [DOI] [PubMed] [Google Scholar]
- Tada Y, Spoel SH, Pajerowska-Mukhtar K, Mou Z, Song J, Wang C, Zuo J, Dong X.. 2008. Plant immunity requires conformational changes of NPR1 via S-nitrosylation and thioredoxins. Science 321, 952–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taki N, Sasaki-Sekimoto Y, Obayashi T, et al. 2005. 12-Oxo-phytodienoic acid triggers expression of a distinct set of genes and plays a role in wound-induced gene expression in Arabidopsis. Plant Physiology 139, 1268–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamogami S, Rakwal R, Agrawal GK.. 2008. Interplant communication: airborne methyl jasmonate is essentially converted into JA and JA-Ile activating jasmonate signaling pathway and VOCs emission. Biochemical Biophysical Research Communications 376, 723–727. [DOI] [PubMed] [Google Scholar]
- Tian M, von Dahl CC, Liu P-P, Friso G, van Wijk KJ, Klessig DF.. 2012. The combined use of photoaffinity labeling and surface plasmon resonance-based technology identifies multiple salicylic acid-binding proteins. The Plant Journal 72, 1027–1038. [DOI] [PubMed] [Google Scholar]
- Tripathi T, Zhang T, Koo AJ, Stacey G, Tanaka K.. 2018. Extracellular ATP acts on jasmonate signaling to reinforce plant defense. Plant Physiology 176, 511–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda J, Kato J.. 1980. Isolation and identification of a senescence-promoting substance from wormwood (Artemisia absinthium L.). Plant Physiology 66, 246–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ververidis P, John P.. 1991. Complete recovery in vitro of ethylene-forming enzyme activity. Phytochemistry 30, 725–727. [Google Scholar]
- Vismans G, van der Meer T, Langevoort O, Schreuder M, Bouwmeester H, Peisker H, Dörman P, Ketelaar T, van der Krol A.. 2016. Low-phosphate induction of plastidal stromules is dependent on strigolactones but not on the canonical strigolactone signaling component MAX2. Plant Physiology 172, 2235–2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Auwerx J.. 2017. Systems phytohormone responses to mitochondrial proteotoxic stress. Molecular Cell 68, 540–551.e5. [DOI] [PubMed] [Google Scholar]
- Wasternack C, Hause B.. 2013. Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Annals of Botany 111, 1021–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M, Chiba Y, Hirai MY.. 2021. Metabolism and regulatory functions of O-acetylserine, S-adenosylmethionine, homocysteine, and serine in plant development and environmental responses. Frontiers in Plant Sciences 12, 643403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Savchenko T, Baidoo EE, Chehab WE, Hayden DM, Tolstikov V, Corwin JA, Kliebenstein DJ, Keasling JD, Dehesh K.. 2012. Retrograde signaling by the plastidial metabolite MEcPP regulates expression of nuclear stress-response genes. Cell 149, 1525–1535. [DOI] [PubMed] [Google Scholar]
- Yamburenko MV, Zubo YO, Börner T.. 2015. Abscisic acid affects transcription of chloroplast genes via protein phosphatase 2C-dependent activation of nuclear genes: repression by guanosine-3ʹ-5ʹ-bisdiphosphate and activation by sigma factor 5. The Plant Journal 82, 1030–1041. [DOI] [PubMed] [Google Scholar]
- Yamburenko MV, Zubo YO, Vanková R, Kusnetsov VV, Kulaeva ON, Börner T.. 2013. Abscisic acid represses the transcription of chloroplast genes. Journal of Experimental Botany 64, 4491–4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao N, Greenberg JT.. 2006. Arabidopsis ACCELERATED CELL DEATH2 modulates programmed cell death. The Plant Cell 18, 397–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X, Li J, Liu J, Liu K.. 2015. An Arabidopsis mitochondria-localized RRL protein mediates abscisic acid signal transduction through mitochondrial retrograde regulation involving ABI4. Journal of Experimental Botany 66, 6431–6445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zander M, Lewsey MG, Clark NM, et al. 2020. Integrated multi-omics framework of the plant response to jasmonic acid. Nature Plants 6, 290–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Xing D.. 2008. Methyl jasmonate induces production of reactive oxygen species and alterations in mitochondrial dynamics that precede photosynthetic dysfunction and subsequent cell death. Plant and Cell Physiology 49, 1092–1111. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Li X.. 2019. Salicylic acid: biosynthesis, perception, and contributions to plant immunity. Current Opinion in Plant Biology 50, 29–36. [DOI] [PubMed] [Google Scholar]
- Zhu T, Li L, Feng L, Ren M.. 2020. StABI5 involved in the regulation of chloroplast development and photosynthesis in potato. International Journal of Molecular Sciences 21, 1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Chen J, Xie Z, Gao J, Ren G, Gao S, Zhou X, Kuai B.. 2015. Jasmonic acid promotes degreening via MYC2/3/4- and ANAC019/055/072-mediated regulation of major chlorophyll catabolic genes. The Plant Journal 84, 597–610. [DOI] [PubMed] [Google Scholar]
- Zimorski V, Ku C, Martin WF, Gould SB.. 2014. Endosymbiotic theory for organelle origins. Current Opinion in Microbiology 22, 38–48. [DOI] [PubMed] [Google Scholar]
- Zurbriggen MD, Carrillo N, Tognetti VB, Melzer M, Peisker M, Hause B, Hajirezaei M-R.. 2009. Chloroplast-generated reactive oxygen species play a major role in localized cell death during the non-host interaction between tobacco and Xanthomonas campestris pv. vesicatoria. The Plant Journal 60, 962–973. [DOI] [PubMed] [Google Scholar]