Abstract
The ability to respond to injury is essential for the survival of an organism and involves analogous mechanisms in animals and plants. Such mechanisms integrate coordinated genetic and metabolic reprogramming events requiring regulation by small RNAs for adequate healing of the wounded area. We have previously reported that the response to injury of the filamentous fungus Trichoderma atroviride involves molecular mechanisms closely resembling those of plants and animals that lead to the formation of new hyphae (regeneration) and the development of asexual reproduction structures (conidiophores). However, the involvement of microRNAs in this process has not been investigated in fungi. In this work, we explore the participation of microRNA-like RNAs (milRNAs) molecules by sequencing messenger and small RNAs during the injury response of the WT strain and RNAi mutants. We found that Dcr2 appears to play an important role in hyphal regeneration and is required to produce the majority of sRNAs in T. atroviride. We also determined that the three main milRNAs produced via Dcr2 are induced during the damage-triggered developmental process. Importantly, elimination of a single milRNA phenocopied the main defects observed in the dcr2 mutant. Our results demonstrate the essential role of milRNAs in hyphal regeneration and asexual development by post-transcriptionally regulating cellular signalling processes involving phosphorylation events. These observations allow us to conclude that fungi, like plants and animals, in response to damage activate fine-tuning regulatory mechanisms.
Keywords: conidiation, filamentous fungi, hyphal regeneration, milRNAs, RNAi machinery, RNA-seq, small RNAs, signaling, transcriptome
Data Summary
The datasets generated and/or analysed during the current study are available in the Gene Expression Omnibus repository, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE123330. Six supplementary figures, three supplementary tables and 12 supplementary datasets are available with the online version of this article. Supplementary Material can be accessed in the Microbiology Society’s data repository Figshare at https://doi.org/10.6084/m9.figshare.20054486.v1 [1].
Impact Statement.
The ability to respond to injury is a biological process essential for the survival of an organism. This ability that may even result in the regeneration of a whole-body part or lost structure is shared by organisms of different kingdoms and has always fascinated humans. Filamentous fungi, due to their immobility and because they represent a nutrient source rich in amino acids and sugars, are consumed by predators and therefore continuously exposed to mechanical damage. Here we describe how the filamentous fungus Trichoderma atroviride fine-tunes its transcriptional response to injury through small non-coding RNAs (sRNAs), a process that had never been studied at this level. We describe how Dcr2 plays a major role in hyphal regeneration. We also describe the participation of microRNA-like molecules (milRNAs) and small interfering RNAs during the injury response making use of mutants in the RNAi machinery. This work represents one of the few reports describing the role of a milRNA in the biology of filamentous fungi. We found that elimination of a single milRNA phenocopied the main defects observed in a dcr2 mutant. These observations allowed us to conclude that fungi share with plants and animals fine tuning regulatory mechanisms key for their development and survival.
Introduction
The ability to rapidly and precisely respond and, in some cases, regenerate a damaged area after an injury is essential for the survival of an organism [2]. Plants and animals display analogous, sophisticated mechanisms, requiring coordinated genetic and metabolic reprogramming events that will define a new molecular programme to direct proliferation and differentiation to replace the lost cells and prevent further damage [2, 3].
In eukaryotes, post-transcriptional regulation by small non-coding RNAs (sRNAs) can allow rapid changes in the transcriptional landscape in response to environmental stimuli [4] or fine-tuning of the transcriptional profile during development [5], through processes known as RNA interference (RNAi). There are two main classes of cytoplasmic regulatory sRNAs, namely small interfering RNAs (siRNAs) and microRNAs (miRNAs), which are distinguished based on their biosynthetic pathway. Biogenesis of siRNAs depends on RNA-dependent RNA polymerases (RdRPs), Dicer and Argonaute proteins, and they directly regulate the expression level of coding transcripts [6]. siRNAs negatively regulate their targets and were initially proposed to be involved in genome protection against viral infections and transposons, yet siRNAs also play important roles in response to environmental stimuli [7, 8].
On the other hand, biosynthesis of miRNAs does not depend on RdRPs but requires the transcription of a hairpin structure by canonical RNA polymerases [9]. These primary transcripts, or primary miRNAs (pri-miRNAs), are subsequently processed in the nucleus by Dicer-like proteins in the case of plants or Drosha in the case of animals, to excise the hairpin structure (precursor miRNA or pre-miRNA), which is further processed by Dicer to generate single-stranded ~20–22 bp microRNAs that are loaded onto Argonautes in the cytoplasm [10, 11]. miRNAs have been extensively described in plants and animals as negative regulators of translation and transcript stability, and play important roles during development, response to environmental stresses, and are also implicated in several diseases [4, 10, 12, 13].
In animals, miRNAs are involved in tissue and organ regeneration [14], as in the case of miR-133b that represses the small GTPase RhoA after spinal cord injury in zebrafish, enhancing spinal cord regeneration [15]. Also in zebrafish, miR-99/100 and let-7a/c regulate cardiomyocyte dedifferentiation and heart regeneration [16]. Furthermore, local injection of double-stranded miR-1, miR-133 and miR-206 can accelerate muscle regeneration in a rat skeletal muscle injury model [17]. Therefore, strategies for organ regeneration, based on miRNAs, emerge as an interesting therapeutic alternative. Similarly, in plants, miRNAs and siRNAs are involved in the response to wounding and systemic defence against herbivory [18, 19]. In fungi, ex-siRNAs (siRNAs derived from exons) have been implicated in the regulation of gene expression in response to antibiotics [20].
As a nutrient-rich source and due to their immobility, filamentous fungi are prey to a variety of predators, including fungivorous nematodes and arthropods, and are therefore constantly injured. In ascomycetes, when a hypha is damaged the septal pore nearest to the point of injury is sealed to prevent excessive cytoplasmic leakage and cell death [21]. Thereafter, new hyphal tips are formed, resulting in the resumption of growth and reconnection of hyphae [22]. Interestingly, this process is very similar at the genetic level to regeneration in other, phylogenetically distant, organisms [22].
The filamentous fungus Trichoderma atroviride responds to mycelial injury, regenerating broken hyphae and forming asexual reproduction structures [23]. This response to injury has many commonalities with that of higher eukaryotes, likely due to convergent evolution. The mechanisms used by T. atroviride involve the detection of extracellular ATP released from damaged cells, which leads to a rapid and transient increase in cytosolic calcium [24]. In parallel, reactive oxygen species (ROS) are produced by the NADPH oxidase complex [24]. Extracellular ATP activates two mitogen-activated protein kinase (MAPK) pathways [24]. The MAPK Tmk1 is required for hyphal regeneration and is responsible for the transcriptional activation of genes involved in cell cycle, DNA repair and programmed cell death, which are essential processes for regeneration, while Tmk3 is necessary for conidiation [23]. Despite these findings and the knowledge accumulated in plants and animals, the role of sRNAs in the fungal response to injury has never been studied. Regulation of gene expression by sRNAs in fungi is only starting to be understood.
In Neurospora crassa, microRNA-like RNAs (milRNAs) were identified and, like canonical miRNAs, are capable of post-transcriptionally repressing gene expression [25]. In Mucor circinelloides it has been suggested that the regulation of gene expression by exonic siRNAs (ex-siRNAs) is important during development [26, 27] and allows this fungus to resist the antifungal drug FK506 [20]. Finally, in Fusarium graminearum and Metarhizium robertsii, milRNAs have been correlated with the regulation of sexual and asexual reproduction, and putative target genes have been predicted [28, 29]. This evidence suggests that, like other eukaryotes, fungi can fine-tune gene expression through sRNAs, during growth and development. However, in contrast with what has been observed in plants and animals, no fungal milRNA appears to be conserved, few loss-of-function experiments have been carried out, and to date, no defect has been observed in sexual or asexual reproduction [30]. Furthermore, several clades of fungi have naturally lost their RNAi machinery, likely because of adaptation to killer viruses, without further negative consequences to their survival [31]. Therefore, it is unclear if fungal microRNAs (milRNAs) are central regulators of biological processes like they are in other eukaryotes [7].
We previously showed that in T. atroviride the RNAi machinery is important for colony growth and that light-induced conidiation was impaired in dcr2 (Dicer2) null mutants [32]. Here we show that the Δdcr2 and Δrdr3 mutants are affected in injury-induced conidiation. By performing RNA-seq, we determined that the RNAi machinery regulates the expression of genes involved in oxidation-reduction processes and amino acid metabolism in the first minutes after injury. Interestingly, analysis of hyphal regeneration showed that dcr2 plays an important role in the regeneration of damaged hyphae, but rdr3 is not required. We identified three main Dicer2-dependent milRNAs induced during asexual development triggered by mechanical damage, suggesting that they play an important role even during the formation of conidia. Finally, we demonstrate that a single T. atroviride milRNA plays a key role during damage-induced conidiation and regeneration.
Methods
Strains and culture conditions
We used T. atroviride IMI206040 as a wild-type strain. All RNAi mutants used for this investigation have been described previously [32]. The strains were cultivated on potato dextrose agar PDA (Difco) at 28 °C.
Growth rate
Radial growth of the strains was measured in 14 cm in diameter Petri dishes at 12, 24 36, 48 and 60 h post-inoculation, at 28 °C in the dark.
Injury induction and conidia quantification
Mycelial plugs of the different T. atroviride strains were inoculated in PDA plates and allowed to grow for 36 h in the dark at 28 °C, mycelia were cut using a scalpel or a cookie mould under a red safety light [22]. To observe and quantify conidiation, the strains were incubated for 36 additional hours under the same conditions. Conidia quantification was performed using a Zeiss 542 Axiostar Plus Microscope in a hemocytometer [32].
Regeneration assay
The WT and Δdcr2, Δrdr3, dcr2C, ΔmilRNA2 mutants of T. atroviride were grown in half-strength PDB with 1.5 % agar medium on glass slides (Corning), incubated for 24 h at 28 °C. Mycelia were then cut with a scalpel, incubated for three additional hours, and hyphae stained with lactophenol cotton blue for 10 min, visualized in a Leica CTR6000-B microscope (40×), and photographed. To determine the percentage of regeneration, 150 broken hyphae per colony were examined and from these, we counted how many formed a new hypha. Three biological replicates were made per experiment and a one-way ANOVA and Tukey’s test was performed to evaluate the significance of the differences between the strains.
RNA-seq and differential expression analysis
The mycelium of the strains analysed in the transcriptomic experiments was collected and frozen immediately after growing for 36 h in the dark for the control condition or 30 min after damage. Total RNA was extracted using the TRIzol protocol (Invitrogen). Libraries were prepared with the TruSeq RNA Sample Preparation Kit v2 (Illumina). Library size and quality were determined using a Bioanalyzer (Agilent Technologies). Three biological replicates were sequenced for all conditions. The libraries of the Δdcr2 experiment and its WT control were sequenced on an Illumina HiSeq 2 500 with the 1×100 format, with an average of 17 million reads. Libraries of the Δrdr3 and ΔmilRNA2 mutants and the WT control for each experiment were sequenced on an Illumina NextSeq 500 with the 1×75 format, with an average of 27 and 15 million reads, respectively (Table S1, available in the online version of this article). FastQC was used for quality control (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). The reads obtained were pseudoaligned to the T. atroviride V2 transcriptome (Tatroviridev2_FrozenGeneCatalog_20100319.transcripts.fasta.gz) using Kallisto to quantify transcript abundance [33].
For differential expression analysis, only those genes that had at least three reads per million in at least ten libraries were considered. All analyses were carried out using the edgeR package [34]. For determining differential expression between the comparisons, we used the generalized linear model (GLM) likelihood ratio test. A common dispersion between replicates of 0.0194 was calculated and a tagwise dispersion was also calculated. False discovery rates (FDR) were calculated and genes with an FDR <0.05 were considered differentially expressed.
The sequencing data generated and analysed during the current study are available in the GEO database with accession GSE123330 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE123330). All other data generated or analysed during this study are included in this published article and its supplementary information files.
GO enrichment analysis
Annotation of all the genes of T. atroviride was performed using Blast2GO [35]. GO-terms assigned to less than four genes or more than 1000 were not considered and only five levels below the root terms were used. These filters were performed with R v3.5.0 (www.r-project.org). Enrichment analyses for cellular components and biological processes were performed using the GOseq package [36]. GO terms with FDR ≤0.05 were considered significantly enriched in each comparison. We present this analysis in a clustered heatmap that highlights the significant categories with asterisks, where ** represents FDR <0.01 and * FDR <0.05.
Small RNA-seq and read annotation
sRNA-seq of WT and Δdcr2 was performed on an Illumina MiSeq and libraries prepared using the TruSeq Small RNA Library Preparation Kits. Three biological replicates were made with an average of 745 000 reads (Table S2). Reaper, part of the Kraken toolset [37], was used to remove adapters, and ShortStack v3.8.5 to predict sRNA-producing clusters generated by Dicer activity, as well as microRNAs [38]. All mapping was performed with Bowtie v1 [39].
For the genomic annotation of these sRNAs, we used the frozen GFF gene catalogue 20 100 319 and the unmasked assembly v2 from the JGI website (https://genome.jgi.doe.gov/portal/Triat2/Triat2.download.ftp.html). A prediction of UTR regions considering 500 bp upstream and downstream of the genes was made using the function flank of GenomicRanges [40]. Rfam 11.0 and Infernal 1.0 [41, 42] were used to annotate non-coding RNAs. Reads that mapped to regions with no annotation were considered intergenic. To analyse the general functional classification of each library, we also annotated reads of each possible length (11–31 nucleotides).
Visualization of the accumulation of small RNA reads in genomic regions was performed using IGV [43, 44].
milRNA target prediction
Using the milRNAs predicted by ShortStack, we predicted the target genes for the three most abundant microRNAs; milRNA1: UGAAACCCCGGACAAACUUGCG, milRNA2: UUUUGCGAUGCCCAAUAUCUGU and milRNA3: UGUGAAGCUAAUCACUCGGUAUC, using TargetFinder [45] against all genes in the T. atroviride genome (https://genome.jgi.doe.gov/portal/Triat2/Triat2.download.ftp.html). A penalty score cutoff value of 8 was imposed for this analysis.
Stem-loop RT-PCR and qPCR
Primers for stem-loop Reverse Transcription-Polymerase Chain Reaction (RT-PCR) were designed to produce amplicons of 72 bp [46]. cDNA was synthesized following the manufacturers' recommendations of the RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific). The reverse transcription (RT) reaction was performed under the following conditions: 30 min at 16 °C, then 60 cycles {30 s at 30 °C, 30 s at 42 °C, 1 s at 50 °C} and finally 5 min at 85 °C. The PCR was carried out according to the GoTaq Green Master Mix kit for the semi-quantitative RTs and for the qPCRs the SYBR Green PCR Master Mix protocol (Applied Biosystems) was followed. As an internal control, we used a constitutive tRNA encoded in the contig_21 : 1 402 684–1 402 704 region of the genome (Table S3).
Generation of ΔmilRNA2 mutants
Contig_17 : 1 121 016–1 121 375 of the genome assembled scaffolds (unmasked), with a length of 349 bp, was replaced by a hygromycin-resistance cassette. The primers used to generate the gene replacement construct are listed in Table S2. The construct was directly used for PEG-mediated protoplast transformation of the WT strain as previously described [32] and transformants were subjected to five rounds of single-spore isolation using hygromycin as a selection marker. Ten putative transformants were analysed by PCR using the Nested oligos to confirm the mutation (Table S3). Also, a Southern-blot analysis was performed following standard techniques. The NcoI restriction enzyme was used to obtain the appropriate banding pattern and downstream fragment of pre-milRNA2 was P labelled and used as a probe for hybridization [32].
Hyphal fusion complementation
To rescue the phenotype of the milRNA2 mutant, we took advantage of our model. Firstly, Trichoderma atroviride can fuse its hyphae [47] and produce heterokaryons, which allows complementation by dikaryon formation. Secondly, the Δdcr2 mutant is resistant to benomyl while the ΔmilRNA2 mutant has the hygromycin resistance cassette. These characteristics allowed us to carry out a hyphal fusion complementation experiment.
Results
The RNAi machinery is involved in the response to injury in T. atroviride
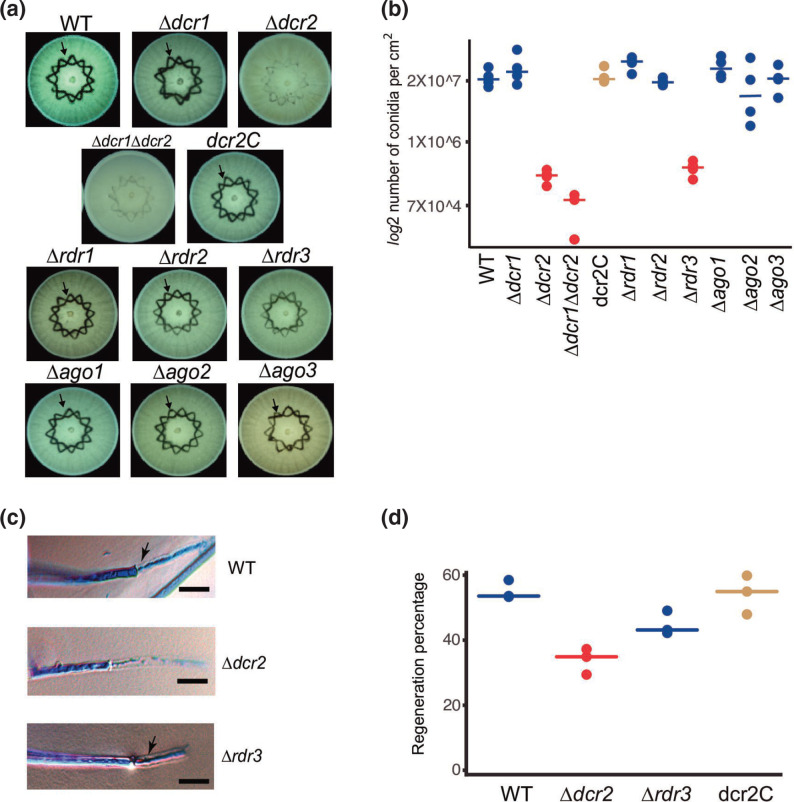
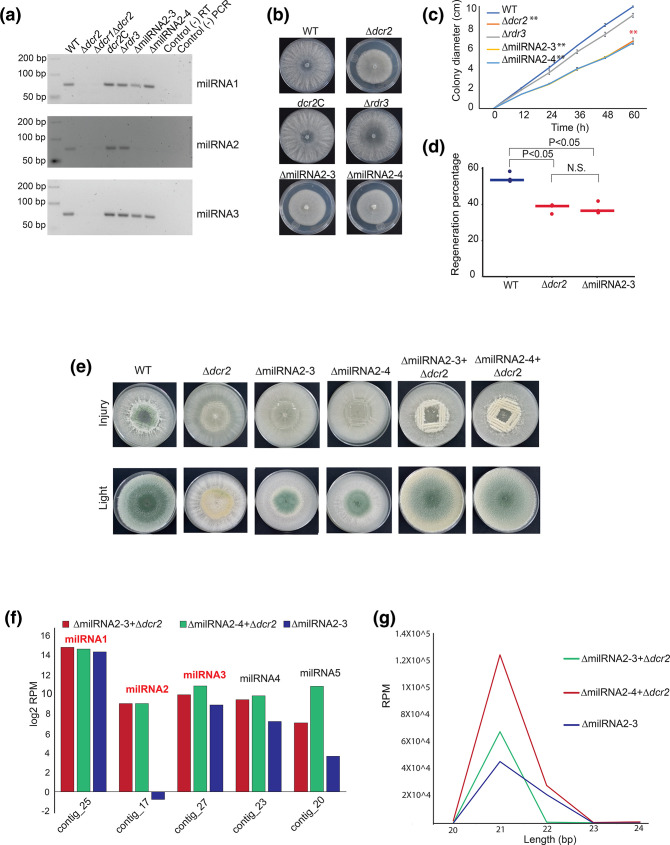
Based on the capacity of T. atroviride to regenerate and form conidia in response to injury, we decided to evaluate the phenotype of single mutants of all components of the RNAi machinery, as well as that of a double Δdcr1Δdcr2 mutant. We observed a highly significant decrease in the production of conidia in response to injury in the Δdcr2, Δdcr1Δdcr2 and Δrdr3 mutants (P <0.01), with a 99, 99.5 and 98 % reduction compared to the WT, respectively (Fig. 1a and b). These results indicate that dcr2 and rdr3 are required for injury-induced conidiation. Conidiation in the Δdcr2 mutant was fully restored when re-transformed with the dcr2 gene (dcr2C; Fig. 1b), consistent with what is observed in response to light [32]. To determine if there was an early defect in the Δdcr2 and Δrdr3 mutants that could explain their strongly reduced capacity to produce conidia, we evaluated their hyphal regeneration capacity upon injury. Microscopical examination showed that regeneration of the Δdcr2 but not that of the Δrdr3 mutant was affected (Fig. 1c). There was a significant decrease in the regenerative capacity of the Δdcr2 mutant: only 33 % of the injured hyphae regenerated, compared with 55 and 54 % of the wild-type and dcr2C strains, respectively (P <0.01; Fig. 1d). However, the difference in the regenerative capacity of the Δrdr3 mutant (45 %) compared to the WT was not statistically significant (Fig. 1d). Thus, Dcr2 has a role in hyphal regeneration, potentially producing sRNAs that help regulate this process.
Fig. 1.
The response to mechanical damage is affected in RNAi mutants. (a) Conidiation response to injury of the WT strain and RNAi mutants. Colonies of the indicated strains growing on PDA were damaged with a cookie mould and photographed 36 h later. The arrow indicates the areas of accumulation of conidia in response to injury (dark). (b) The graph shows the yield of conidia in response to injury of four biological replicates. (c) Hyphal regeneration 1 h after injury. Hyphae were stained with lactophenol cotton blue and examined by light microscopy. Arrows point to the new hyphae. Scale bar=10 µM. (d) Percentage of hyphae that regenerate after injury for the indicated strains. Three biological replicates were used, counting 50 hyphae per strain. In (b) and (d), a one-way ANOVA test, followed by Tukey honest significant differences was applied. Highlights in red indicate that there was a statistically significant difference when comparing the indicated strains with the WT (P<0.05).
The RNAi machinery regulates genes for the response to stress and metabolism
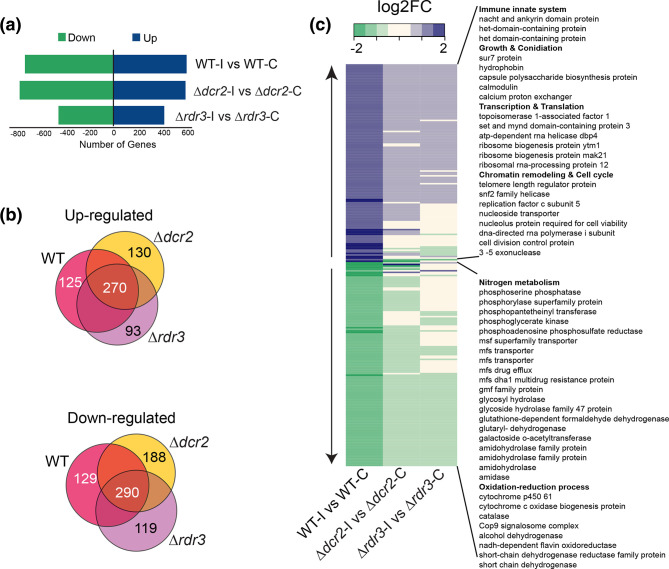
Given the major changes in the transcriptional profile that take place early after injury [22, 23], we carried out high-throughput RNA sequencing experiments for the WT, Δdcr2 and Δrdr3 strains 30 min after injury (Table S1). We identified 1346 (Dataset S1), 1384 (Dataset S2) and 884 (Dataset S3) differentially expressed genes, respectively, compared to undamaged controls (Fig. 2a). Of these, 125 induced and 129 repressed genes were exclusive to the WT strain after injury (Fig. 2b). Among the 125 induced genes that no longer respond properly in the mutants, some are involved in processes such as innate immunity response, regulation of transcription/translation, and chromatin remodelling. Within this set, we found several strongly affected genes such as three subunits of DNA-dependent RNA polymerase I, two DNA repair genes (rhm52 and a 3´−5´ exonuclease), the replication factor C subunit 5, and five helicases, including two DEAD/DEAH-box helicases (ID: 292 633, ID: 35745) highly affected in the mutants (Fig. 2c and Dataset S4).
Fig. 2.
Genes involved in nitrogen metabolism and redox balance are regulated by RNAi. (a) Number of differentially expressed genes in the WT, Δdcr2 and Δrdr3 subjected to injury, compared to their corresponding undamaged control (FDR <0.05, fold-change >1). (b) Venn diagrams of the overexpressed and repressed genes in the different comparisons (blue=WT, yellow = Δdcr2 and green = Δrdr3), indicating the number of exclusive genes for each strain and those shared by all strains. (c) Heat map of the expression profile of the 254 genes differentially expressed only in the WT strain (125 up-regulated and 129 down-regulated genes), and their behavior in the Δdcr2 and Δrdr3 strains.
The genes that were repressed in response to injury only in the WT strain were mostly related to nitrogen metabolism. In addition, the set of repressed genes contained five mfs transporters, which participate in the traffic of simple monosaccharides, oligosaccharides, amino acids, and peptides, among other molecules. These observations suggest that there is sRNA-guided repression of amino acid and peptide metabolism. We also found that genes that participate in the metabolism of ROS, such as cytochromes and a catalase, were repressed in the WT background, but not in the mutants. Within this set of genes, we also found cop9, a negative regulator of oxylipin biosynthesis. In this regard, oxylipins have been suggested as important secondary messengers during the damage response in fungi, as well as in plants and animals [22].
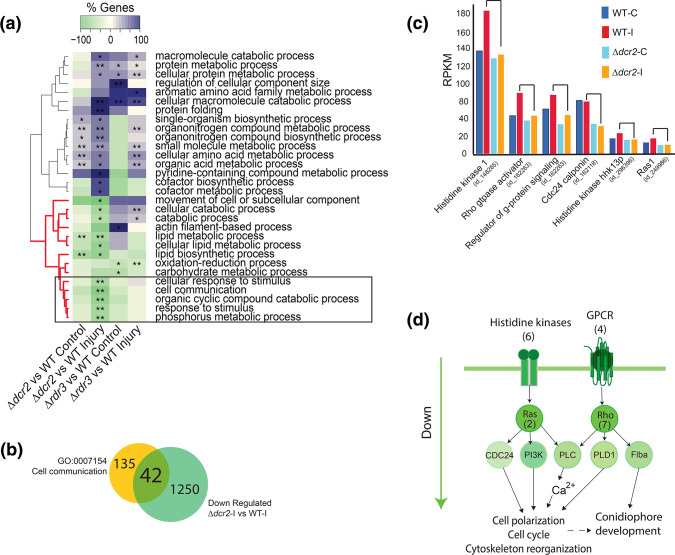
Dcr2 regulates cell communication
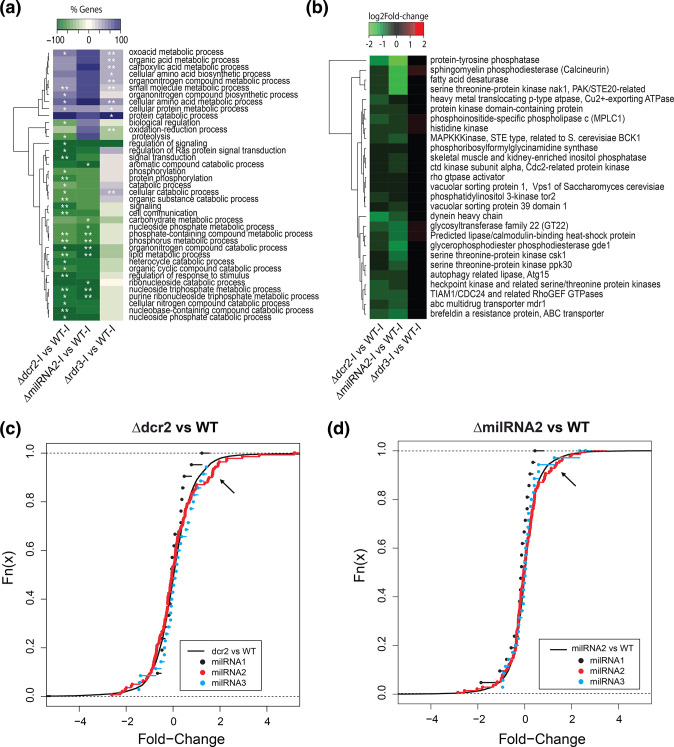
Since Δdcr2 showed regeneration defects that were not observed in the Δrdr3 strain (Fig. 1c), we separately compared the transcriptional profiles of the individual mutants in each treatment to that of the WT strain (Datasets S6–S7). This analysis revealed that the Δdcr2 strain under injury is defective in phosphorus and lipid metabolism, as well as in cellular communication (Fig. 3a, highlighted in a black box). Of the 1292 genes repressed in the Δdcr2 compared to the WT in response to injury, 42 participate in cell communication, including genes involved in phosphorus and lipid metabolism (Fig. 3b and Dataset S8). When we focused on the expression level in six of these genes with obvious changes (Fig. 3c), we observed that those encoding proteins vital for cell signalling such as six histidine kinases, four GPCRs, seven Rho and two Ras GTPases, do not increase their expression in response to injury in the Δdcr2 mutant as they do in the WT. Based on our transcriptomic data, we propose a model of the pathways that are affected in the mutant in response to injury (Fig. 3d). Briefly, two major signalling pathways involving phosphorylation cascades, linked by genes related to signalling by lipids, are no longer activated upon injury in the Δdcr2 mutant. Thus, it appears that Dcr2 plays an important role in the regulation of cellular communication independently of Rdr3.
Fig. 3.
dcr2 regulates signaling processes independently of rdr3. (a) Clustering of GO terms-enrichment analysis belonging to Biological Process (FDR <0.01 **; FDR <0.05 *) in a paired comparison of the Δdcr2 and Δdcr3 mutants vs the WT strain in the control and injury conditions. (b) Overlap between the genes contained in the cell communication GO-term and the genes repressed specifically in the Δdcr2 in response to injury compared to the WT strain. (c) Expression profile of genes involved in signaling in the WT strain and the Δdcr2 mutant, both in the control condition and in response to injury. (d) Model of signaling pathways repressed at the transcriptional level in the dcr2 mutant.
milRNAs are the main class of small RNAs produced by Dcr2
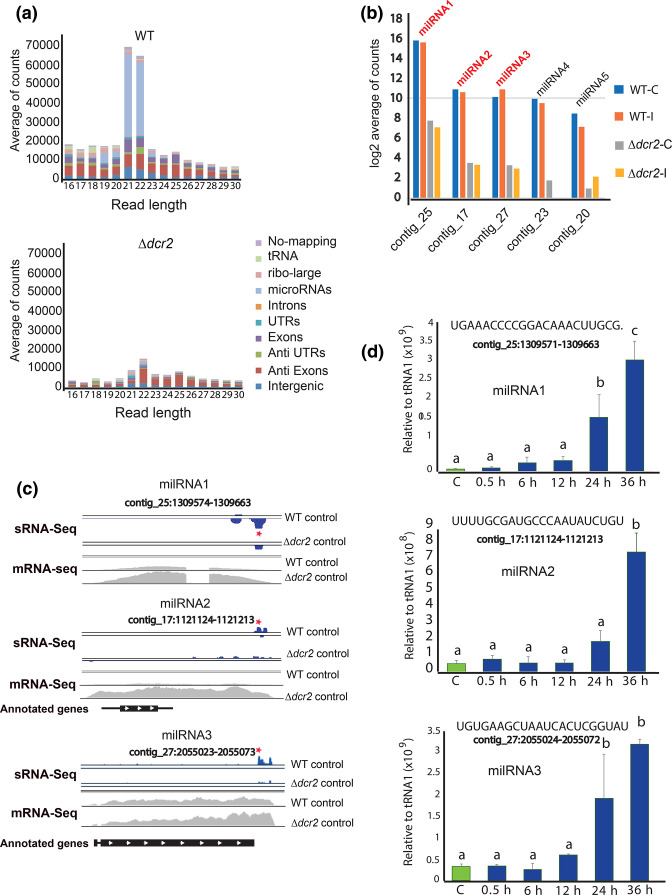
To identify the Dcr2-dependent small RNAs that could regulate gene expression during vegetative growth and in response to injury in T. atroviride, we performed small RNA sequencing (sRNA-seq) of the WT and Δdcr2 strains. The most drastic change in the sRNA profiles of the Δdcr2 strain was a reduction of 21-22nt reads (Fig. 4a, S1). Using ShortStack we predicted five milRNAs (Fig. 4b), of which three (milRNA1, milRNA2 and milRNA3) account for ~90 % of all milRNA reads in the WT (Fig. 4a and b), with a read-depth profile like that of plant and animal miRNAs [38], following the structure of the predicted hairpin (Fig. 4c), while milRNA4 and milRNA5 do not accumulate reads in the 5P region (Fig. S2). We also observed an increase in the expression of the potential primary transcripts encoding these milRNAs (pri-milRNAs) in the Δdcr2 mutant, which suggests that these transcripts are processed by Dcr2 to produce the mature milRNAs but are stabilized in its absence (Fig. 4c).
Fig. 4.
Biogenesis of milRNAs depends on dicer-2 in T. atroviride. (a) Genomic annotation and size distribution of the sRNA reads, read counts are the average across three replicates of the control condition. (b) Expression profile 30 min after injury of the five predicted milRNAs counted using ShortStack. The y-axis scale shows the average of reads in logarithm base 2 comparing WT vs Δdcr2 (FDR <0.05). (c) Visualization of the genomic loci for milRNAs 1, 2 and 3. In blue, milRNAs (sRNA-Seq) and in grey likely pri-milRNA transcription (mRNA-Seq). Red stars indicate the regions of the mature milRNAs that were used for the stem-loop RT-qPCR. The black bars at the bottom represent potential coding regions and the white arrows show the direction of transcription (annotated genes). (d) Time course of the expression of milRNA1, milRNA2 and milRNA3 during injury-induced conidiation by stem-loop RT-qPCR, green bars show control expression without injury, blue bars show expression after injury (sequences of the milRNAs are shown at the top). The expression level is expressed relative to that of tRNA1, which does not change under these conditions. Three biological and five technical replicates were performed. Error bars represent ±sem. A one-way ANOVA test, followed by Tukey honest significant differences was used, different letters indicate significant differences (P<0.05).
milRNAs increase their expression during development
To determine if the main milRNAs produced by Dcr2 participate during the response to injury, we evaluate their expression pattern during injury-induced conidiation by stem-loop RT-qPCR (Fig. 4d). We observed that the three main milRNAs are expressed during vegetative growth, but their expression strongly increases when conidiophores are being produced (24 to 36 h after injury). Their drastic increase during differentiation and conidia formation suggests that these milRNAs could play a role during development in response to injury.
A milRNA plays a key role in asexual reproduction and hyphal regeneration
To determine the role of milRNAs in response to damage in T. atroviride, we attempted to generate knockout mutants for the three most abundant milRNAs. However, we only succeeded in the case of milRNA2, where the genomic region that gives rise to its precursor was replaced by a hygromycin resistance cassette. Two independent mutants were selected and confirmed by Southern blot (Fig. S3). We performed stem-loop RT-PCR of milRNA1, milRNA2 and milRNA3, in the WT, Δdcr2, Δdcr1Δdcr2, Δrdr3, dcr2C, as well as in the milRNA2 mutant strains (Fig. 5a). As expected, we detected no expression for any of the milRNAs in the Δdcr2 and Δdcr1Δdcr2 strains, but their expression was re-established in the dcr2C strain. The expression of the three milRNAs was unaltered in the Δrdr3 mutant, indicating that, as expected, RdRP3 does not participate in their biogenesis. Only expression of miRNA2 was lost in the two replacement mutants, indicating that milRNA2 is not involved in the regulation of the expression of milRNA1 or milRNA3.
Fig. 5.
Mutation of milRNA2 blocks asexual development in response to injury. (a) Gel electrophoresis analysis of stem-loop RT-PCR products of the three most abundant milRNAs in the indicated RNAi mutants and two independent milRNA2 mutants. (b) Photographs show colonies of the indicated strains after 72 h of growth. (c) Time course of colony growth of the indicated strains. (b) and (c) Colonies were grown on PDA in darkness. Error bars represent ±sem. A one-way ANOVA test, followed by Tukey honest significant differences was used (**P <0.01). (d) The graph shows the percentage of hyphae that regenerate after injury for the indicated strains. 50 hyphae in total for each replicate were examined and three biological replicates (points) were performed per strain, lines indicate the average regeneration percentage. A one-way ANOVA test, followed by Tukey honest significant differences was used. Highlights in red indicate that there is a statistically significant difference when comparing the indicated strain with the WT (P <0.05). (e) Photographs show the conidiation phenotypes in response to injury of the WT strain, Δdcr2, ΔmilRNA2, and the dikaryon resulting from the fusion of Δdcr2/ΔmilRNAs. The experiment was performed on PDA and in complete darkness. (f) Expression level (reads per million) of the five milRNAs predicted in both the mutant and the dikaryons of milRNA2. (g) Length distribution of sRNA reads between 20 and 24 nt in the milRNA2 mutant, as well as the two dykarions.
We further characterized the Δdcr2, dcr2C, Δrdr3, and the two milRNA2-mutants, and showed that there is a similar defect in vegetative growth of the milRNA2 and the Δdcr2 mutants (Fig. 5b). We observed a highly significant difference between the growth rate of the Δdcr2 and ΔmilRNA2 compared to the WT strains and the Δrdr3, even within the first 12 h of growth (Fig. 5c). Interestingly, the growth rate of the Δdcr2 and ΔmilRNA2 mutants was statistically indistinguishable, while that of the Δrdr3 mutant was equivalent to that of the WT. Subsequently, we decided to evaluate the regenerative capacity as a specific response to injury. In the Δdcr2 and ΔmilRNA2 mutants, the capacity to regenerate was affected similarly, showing a significant reduction, compared to the WT strain (P <0.05; Fig. 5d).
To determine whether the presence of milRNA2 can re-establish the WT phenotype in the mutant, we obtained heterokaryons of the Δdcr2 mutant and, ΔmilRNA2-3 and ΔmilRNA2-4 (Fig. S4). Interestingly, the heterokaryons of the milRNA2-3 and milRNA2-4 mutants with the Δdcr2 mutant can produce conidia in response to injury and light (Fig. 5e).
To determine that there is no alteration in the expression of the other milRNAs and to observe the expression of milRNA2, we performed an sRNA-seq experiment of the ΔmilRNA2-3 mutant, and the two strains fused with Δdcr2 (ΔmilRNA2-3+Δdcr2 and ΔmilRNA2-4+Δdcr2). Clearly, both fused strains specifically recovered milRNA2 expression (Fig. 5f, Fig. S5). When evaluating the number of reads in the five predicted milRNAs, it seems that there is a slight increase in the accumulations of milRNA3, 4 and 5 in the ΔmilRNA2-4+Δdcr2 heterokaryon (Fig. 5f). We observed that the ΔmilRNA2-4+Δdcr2 fusion has a higher cumulative RPM number of approximately 6×104 compared to the ΔmilRNA2-3+Δdcr2 fusion. These results also showed that the two fused strains seem to have an increase in the population of 21 nt-sRNAs in comparison to ΔmilRNA2-3 mutant, suggesting that these sRNAs come from the milRNA2 region (Fig. 5g).
milRNA2 positively regulates signalling and the cell cycle
We performed a transcriptome analysis on the ΔmilRNA2 mutant in response to injury. It was clear that there was an increase in the primary transcript in the ΔmilRNA2 mutant, similarly to what was observed in the Δdcr2 mutant, and not observed in either the Δrdr3 mutant or the WT (Fig. S6).
To get a clearer picture of the global changes caused by the lack of milRNA2, we analysed the biological processes enriched amongst the deregulated genes in our transcriptomes. In many cases, the same processes were repressed in the Δdcr2 and ΔmilRNA2 mutants, but not in Δrdr3. This indicates that these changes are mainly due to loss of regulation by a single milRNA (Fig. 6a). Within these processes, we found phosphorus metabolism, phospholipid metabolism and organonitrogen compound catabolism, as well as metabolism of nucleoside triphosphate; changes strongly related to signalling via phosphorylation cascades. Interestingly, genes encoding master regulators of the cell cycle and proteins involved in signalling such as bck1, cdc24, cdc2, ppk30, calcineurin, and others were subtly but significantly repressed in the Δdcr2 and ΔmilRNA2 mutants but not in Δrdr3 (Fig. 6b).
Fig. 6.
Transcriptome analysis of milRNA2 mutant in response to injury and microRNA-target prediction. (a) Clustering of GO-enrichment analysis belonging to biological process (FDR <0.01 **, FDR <0.05 *) in the general comparison WT vs Δdcr2, ΔmilRNA2 and Δrdr3 mutants. (b) Heatmap of the expression profile of the genes affected by the mutation of milRNA2 and dcr2 (FDR <0.05). (c) Cumulative frequency curve of the expression profile of target genes of the three main milRNAs in the comparison Δdcr2 vs WT, the x-axis shows the log2-Fold-change, the black curve represents the expression profile of all the genes. (d) Cumulative frequency curve of the expression profile of target genes of the three main milRNAs in the comparison ΔmilRNA2 vs WT, the x-axis shows the log2-Fold-change, the black curve represents the expression profile of all the genes. The black arrow indicates the tendency of a group of milRNA2 target genes to be induced with respect to the average distribution of all the genes in the transcriptome.
Target genes of the milRNAs
To determine the target genes of the milRNAs in T. atroviride we first compared the expression profiles (in Δdcr2-Injury vs WT-Injury) of the 246 target genes for the three most abundant milRNAs predicted by TargetFinder. We observed that a subset of milRNA2-target genes increased their expression in the Δdcr2 strain compared to the WT strain, an effect that was also observed in the targets for milRNA3, but not clearly for milRNA1 (Fig. 6c).
To corroborate the observed change in abundance of predicted milRNA2 targets, we carried out an RNA-seq experiment for the ΔmilRNA2 mutant in response to injury, obtaining results like those of the Δdcr2 mutant, but in the ΔmilRNA2 we did not observe the overexpression of milRNA3 targets genes, suggesting a specific effect on the target genes of milRNA2 (Fig. 6d). We found that the target genes that were up-regulated in both the ΔmilRNA2 and Δdcr2 mutant backgrounds are related to phosphorylation signalling and the regulation of gene expression (Fig. S7). This result suggests that these genes are negatively regulated by milRNA2.
Discussion
When T. atroviride is injured, it quickly responds by activating phosphorylation-dependent signalling cascades and adjusting its transcriptional profile, resulting in hyphal regeneration and activation of asexual development [22, 24]. The observation in T. atroviride that the dcr2 and rdr3 mutant strains do not conidiate properly in response to injury suggests that there are genes that must be silenced by small RNAs to respond to this stimulus. In the transcriptomic analysis presented here, we found that the dcr2 and rdr3 genes participate in negatively controlling nitrogen metabolism and the cellular redox state after mechanical damage and it is likely that this metabolic dysregulation affects the conidiation process at later stages. We believe that future in-depth research work is necessary to describe the role of ex-siRNAs in the conidiation process in response to injury.
We have previously reported that during asexual development triggered by exposure to constant light the RNAi machinery regulates conidiation specific genes orthologous to the Neurospora crassa ccg6, con10 and con13, and flbA, medA, and wetA that encode developmental regulators in Aspergillus nidulans [32]. Therefore, in addition to an early defect in the expression pattern, it appears that both stimuli converge in the regulation of conidiation genes, and this process depends on RNAi regulation.
Remarkably, the Δdcr2 mutant is affected in its regeneration capacity. In T. atroviride hyphal regeneration initiates 1 h after injury, but the transcriptional changes crucial for this process occur even before, after 30 min [23]. By performing the transcriptional analysis at 30 min post-injury in the RNAi mutants, we were able to establish that mutation of dcr2 affects cellular communication and metabolism of phosphorus and lipids (Fig. 3a), mainly the histidine kinase, GPCR, Rho, and Ras GTPase pathways. Several components of these pathways are repressed in the dcr2 mutant, including the cdc24, pi3k, plc, pld1 and flba1 genes. Rho and Ras proteins activate Phospholipase C (plc) and Phosphatidylinositol 3-kinase (PI3K) [48]. The latter in turn catalyses the hydrolysis of phosphatidylinositol 4,5-bisphosphate into diacylglycerol and inositol (1,4,5)-trisphosphate. These phospholipids function as second messengers that activate protein kinase C and cause mobilization of intracellular Ca2+, respectively [49]. It has been reported that PLC and PI3K may regulate cytoskeleton reorganization, cell growth, membrane trafficking and cytokinesis [50]. Moreover, it has recently been reported that major cytoskeleton reorganization occurs during hyphal regeneration in T. atroviride and N. crassa [51]. These processes are decisive for tip growth, and polarized cell movement in filamentous fungi [49, 50]. The changes in the expression of these genes also appear to be responsible for a decrease in the regenerative capacity of hyphae after mechanical damage in the absence of dcr2. It could be argued that the defect in regeneration capacity observed in the dcr2 mutant may be just a defect in growth. In our view, these two processes are necessarily linked to some extent since regeneration requires growth. However, it must be taken into consideration that regeneration implies the emergence of a completely new structure that allows the re-establishment of growth. Our microscopic observations indicate that in mutants affected in regeneration, hyphae considered not regenerating show no indication of the production of the new structure, which could be expected even if growing at a slower rate. Furthermore, during regeneration, there is a highly specific transcriptional profile [23], different from that observed during normal growth, which is affected by dcr2, indicating that the RNAi machinery plays a specific role in regeneration.
Recently, we reported a detailed study of the genes and mechanisms essential for hyphal regeneration in T. atroviride [23] and were able to determine that the Tmk-1 signalling pathway participates in regeneration. However, the expression of the corresponding gene is not altered in the RNAi mutant backgrounds, suggesting that the regulation of signalling by sRNAs occurs downstream of this phosphorylation cascade. We also reported the mobilization of Ca2+ from intracellular pools in response to injury and that blocking this mobilization results in decreased regeneration and conidiation. In this sense, we also found calcineurin expression repressed in the Δdcr2 (−2.36-Fold change) which regulates genes in response to calcium increases [52]. In N. crassa calcineurin is required for normal growth, asexual development and female fertility [53]. This observation suggests that it is in this signalling pathway that Dicer2-dependent small RNAs may be playing an important role.
We also found that genes encoding proteins with HET, NACHT domains and DEAD/DEAH-box helicases (Dataset S4), related to cell death and innate immunity, are strongly induced after injury in the WT strain, but not in the RNAi mutants. These genes act as DNA/RNA sensors and regulate gene expression [54–56]. In this regard, it has been shown that genes related to programmed cell death and the immune system are involved in the ability to respond to damage and regeneration, in Hydra and zebrafish [57, 58]. These genes may play an important role in maintaining genome integrity, and appear to be important to initiate regeneration, since they are induced immediately after the injury in T. atroviride [23].
In Fusarium graminearum the RNAi machinery participates in the regulation of sexual reproduction through ex-siRNAs [28]. Likewise, in Mucor circinelloides several processes including resistance to antibiotics by epimutations depend on ex-siRNAs [20, 26]. However, even though filamentous fungi express milRNAs that seem to be involved in various processes, it is not clear yet what is the role of this class of small RNAs in development, and very little is known about the mechanism by which they regulate their target genes. Here, we determined that the main small RNAs present in T. atroviride are milRNAs of 21 to 22 nucleotides. Interestingly, these milRNAs depend entirely on Dcr2 for their synthesis, which implies that the drastic phenotype observed in this mutant may be due to the lack of milRNA regulation of certain genes. Interestingly, no drastic changes were observed in the Δdcr2 mutant in the accumulation of anti-ex-siRNAs compared to the WT strain, which may suggest a minor participation of Dcr2 in the formation of these sRNAs.
Three different types of evidence lead us to propose milRNAs as regulators of the processes affected by the dcr2 mutation during the response to injury, including hyphal regeneration. milRNAs are highly produced in T. atroviride, and in the absence of dcr2, they are no longer synthesized. On the other hand, after injury, these milRNAs tend to increase their expression during development, and finally, mutation of only one of them causes a profound modification in the phenotype of T. atroviride. This suggests that milRNAs play an important role in several steps of development in these fungi. Strikingly, removing a single milRNA in T. atroviride (milRNA2) in two independent mutants led to defects in growth, conidiation and regeneration after injury that closely resembled the Δdcr2 phenotype. An interesting observation was that in response to light, conidiation was not as affected as in response to injury, suggesting a role for specific milRNA2 in injury-triggered conidiation.
Heterokaryons formed from the fusion of hyphae between the ΔmilRNA2 mutants and a Δdcr2 mutant led to the recovery of the WT conidiation phenotype. Sequencing of the total population of small RNAs in the ΔmilRNA2-3 mutant and in heterokaryons clearly showed that milRNA2 mutation is specific and that when fused with Δdcr2 it recovers milRNA2 expression. This correlates with the phenotype and strongly suggests that this milRNA is necessary for regeneration and development in this fungus. Even though complementation using heterokaryons could be considered not as direct as retransformation with the knocked-out gene, the recovery of the capacity to conidiate and synthesize milRNA2 using two independent mutants fused with the same Δdcr2 mutant, strongly indicates that the observed phenotype is due to the lack of milRNA2 since the participation of other, undetected, mutations that could have been produced during transformation would be extremely unlikely.
Consistently, the analysis of the transcriptomic profile of the milRNA2 mutant showed that like in dcr2, phosphorylation-dependent signalling processes and cell cycle are also affected in this mutant. Although these genes are not direct targets of this milRNA, this evidence strongly suggests that the elimination of a single milRNA, perhaps by allowing an increase in the expression of one or more genes, results in a decrease in the expression of the signalling system necessary to activate the response.
Interestingly, the genes predicted as milRNA2-targets are related to phosphorylation signalling, such as histidine phosphotransferases, and serine and threonine-protein kinases (Dataset S8). One of these kinases is a homolog of the mammalian chk2 (ID: 252945) that regulates cell cycle checkpoint arrest through phosphorylation of CDC25 [59] implying that the defect in this process is due to the absence of negative control by milRNA2 (Fig. S5). In addition, we found an RNA-directed RNA polymerase, different from the three RdrP previously identified as part of the RNAi [32] This enzyme might be involved in the formation of small RNAs in a non-canonical pathway, acting as a repressor of gene expression that is regulated by milRNA2. However, determining its physiological role remains to be investigated.
In animals and plants, microRNAs play essential roles in development. In Fusarium graminearum it was suggested that milRNAs regulate genes important for sexual development, but no direct evidence for the involvement of specific milRNAs was presented [60]. However, Shao et al. reported in Cordyceps militarist that mutation of milR4 and milR16 results in defects in fruiting body development. Besides, the milR16 mutant presents a pale-yellow fruiting body primordium, covered with abnormal primordium. Thus, the authors conclude that milRNAs play vital roles in sexual development in C. militaris [61]. Although no target genes were defined, these results are consistent with our findings. Together these data provide evidence that, as in higher eukaryotes, fungi can use key milRNAs as central regulators of development and regeneration.
Conclusions
Regeneration is a widely studied process in animals due to the possibility of mining their genetics to improve the human regenerative capacity. However, to date, we have rarely looked at micro-organisms that can regenerate. Our evidence demonstrates that, like in animals, regulation by microRNAs is essential for the correct regeneration of hyphae in T. atroviride, which highlights the relevance of filamentous fungi as interesting microbial models to understand the conserved processes of regulation in response to injury.
Supplementary Data
Funding information
This work was supported by CONACYT grant - Investigación en Fronteras de la Ciencia (FON.INST./117/2016) to AHE https://www.conacyt.gob.mx/.The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Acknowledgements
We wish to thank Dr Edgar Balcazar-López and Dr Elizabeth Medina-Castellanos for providing input and assistance at the early stages of this work. We thank Dr José Luis Reyes-Taboada for his help in performing the stem-loop RT-PCR technique. J.M.V-E is indebted to Conacyt for a doctoral scholarship (262371) and was supported as a postdoctoral fellow by Conacyt project CB-284884 to CA-G.
Author contributions
Conceptualization: A.H.E., J.M.V.E. Formal analysis: J.M.V.E., P.M.G.D.l.R., R.P.F., N.C.V. Funding acquisition: A.H.E. Investigation: J.M.V.E. Methodology: P.M.H., C.A.G., A.H.E. Project administration: A.H.E. Supervision: C.A.G., A.H.E. Writing – original draft: J.M.V.E., C.A.G., and A.H.E.
Conflicts of interest
The authors declare that they have no conflicts of interests.
Footnotes
Abbreviations: FDR, false discovery rate; GLM, generalized linear model; GPCRs, G-protein coupled receptors; MAPK, mitogen-activated protein kinase; milRNAs, micro-RNA-like RNAs; miRNAs, microRNAs; PDA, potato dextrose agar; pre-miRNAs, precursor miRNA; pri-miRNAs, primary miRNAs; qPCR, quantitative polymerase chain reaction; RdRPs, RNA-dependent RNA polymerases; RNAi, RNA interference; ROS, reactive oxygen species; RPM, reads per million; RT-qPCR, reverse transcription quantitative polymerase chain reaction test; siRNAs, small interfering RNAs; sRNAs, small non-coding RNAs; tRNA, transfer RNA; UTR, untranslated region.
All supporting data, code and protocols have been provided within the article or through supplementary data files. Seven supplementary figures, three supplementary tables and 12 supplementary dataset are available with the online version of this article.
References
- 1.Villalobos-Escobedo JM, Martínez-Hernández JP, Pelagio-Flores R, González-De la Rosa PM, Carreras-Villaseñor N, et al. 2022. Trichoderma atroviride hyphal regeneration and conidiation depend on cell-signaling processes regulated by a microRNA?like RNA. Figshare. [DOI] [PMC free article] [PubMed]
- 2.Ricci L, Srivastava M. Wound-induced cell proliferation during animal regeneration. Wiley Interdiscip Rev Dev Biol. 2018;7:e321. doi: 10.1002/wdev.321. [DOI] [PubMed] [Google Scholar]
- 3.Birnbaum KD, Sánchez Alvarado A. Slicing across kingdoms: regeneration in plants and animals. Cell. 2008;132:697–710. doi: 10.1016/j.cell.2008.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biggar KK, Storey KB. Insight into post-transcriptional gene regulation: stress-responsive microRNAs and their role in the environmental stress survival of tolerant animals. J Exp Biol. 2015;218:1281–1289. doi: 10.1242/jeb.104828. [DOI] [PubMed] [Google Scholar]
- 5.Follert P, Cremer H, Béclin C. MicroRNAs in brain development and function: a matter of flexibility and stability. Front Mol Neurosci. 2014;7:5. doi: 10.3389/fnmol.2014.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hannon GJ. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- 7.Villalobos-Escobedo JM, Herrera-Estrella A, Carreras-Villaseñor N. The interaction of fungi with the environment orchestrated by RNAi. Mycologia. 2016;108:556–571. doi: 10.3852/15-246. [DOI] [PubMed] [Google Scholar]
- 8.Khraiwesh B, Zhu JK, Zhu J. Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. Biochim Biophys Acta. 2012;1819:137–148. doi: 10.1016/j.bbagrm.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 10.Hammond SM. An overview of microRNAs. Adv Drug Deliv Rev. 2015;87:3–14. doi: 10.1016/j.addr.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cui J, You C, Chen X. The evolution of microRNAs in plants. Curr Opin Plant Biol. 2017;35:61–67. doi: 10.1016/j.pbi.2016.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martin RC, Liu PP, Goloviznina NA, Nonogaki H. microRNA, seeds, and Darwin?: diverse function of miRNA in seed biology and plant responses to stress. J Exp Bot. 2010;61:2229–2234. doi: 10.1093/jxb/erq063. [DOI] [PubMed] [Google Scholar]
- 13.Li Y, Kowdley KV. MicroRNAs in common human diseases. Genomics Proteomics Bioinformatics. 2012;10:246–253. doi: 10.1016/j.gpb.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frith JE, Porrello ER, Cooper-White JJ. Concise review: new frontiers in microRNA-based tissue regeneration. Stem Cells Transl Med. 2014;3:969–976. doi: 10.5966/sctm.2014-0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu YM, Gibbs KM, Davila J, Campbell N, Sung S, et al. MicroRNA miR-133b is essential for functional recovery after spinal cord injury in adult zebrafish. Eur J Neurosci. 2011;33:1587–1597. doi: 10.1111/j.1460-9568.2011.07643.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aguirre A, Montserrat N, Zacchigna S, Nivet E, Hishida T, et al. In vivo activation of a conserved microRNA program induces mammalian heart regeneration. Cell Stem Cell. 2014;15:589–604. doi: 10.1016/j.stem.2014.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakasa T, Ishikawa M, Shi M, Shibuya H, Adachi N, et al. Acceleration of muscle regeneration by local injection of muscle-specific microRNAs in rat skeletal muscle injury model. J Cell Mol Med. 2010;14:2495–2505. doi: 10.1111/j.1582-4934.2009.00898.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pandey SP, Shahi P, Gase K, Baldwin IT. Herbivory-induced changes in the small-RNA transcriptome and phytohormone signaling in Nicotiana attenuata . Proc Natl Acad Sci U S A. 2008;105:4559–4564. doi: 10.1073/pnas.0711363105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin JS, Lin CC, Lin HH, Chen YC, Jeng ST. MicroR828 regulates lignin and H2O2 accumulation in sweet potato on wounding. New Phytol. 2012;196:427–440. doi: 10.1111/j.1469-8137.2012.04277.x. [DOI] [PubMed] [Google Scholar]
- 20.Calo S, Shertz-Wall C, Lee SC, Bastidas RJ, Nicolás FE, et al. Antifungal drug resistance evoked via RNAi-dependent epimutations. Nature. 2014;513:555–558. doi: 10.1038/nature13575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hernández-Oñate MA, Herrera-Estrella A. Damage response involves mechanisms conserved across plants, animals and fungi. Curr Genet. 2015;61:359–372. doi: 10.1007/s00294-014-0467-5. [DOI] [PubMed] [Google Scholar]
- 22.Hernández-Oñate MA, Esquivel-Naranjo EU, Mendoza-Mendoza A, Stewart A, Herrera-Estrella AH. An injury-response mechanism conserved across kingdoms determines entry of the fungus Trichoderma atroviride into development. Proc Natl Acad Sci U S A. 2012;109:14918–14923. doi: 10.1073/pnas.1209396109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Medina-Castellanos E, Villalobos-Escobedo JM, Riquelme M, Read ND, Abreu-Goodger C, et al. Danger signals activate a putative innate immune system during regeneration in a filamentous fungus. PLoS Genet. 2018;14:e1007390. doi: 10.1371/journal.pgen.1007390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Medina-Castellanos E, Esquivel-Naranjo EU, Heil M, Herrera-Estrella A. Extracellular ATP activates MAPK and ROS signaling during injury response in the fungus Trichoderma atroviride . Front Plant Sci. 2014;5:659. doi: 10.3389/fpls.2014.00659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee HC, Li L, Gu W, Xue Z, Crosthwaite SK, et al. Diverse pathways generate microRNA-like RNAs and Dicer-independent small interfering RNAs in fungi. Mol Cell. 2010;38:803–814. doi: 10.1016/j.molcel.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nicolas FE, Moxon S, de Haro JP, Calo S, Grigoriev IV, et al. Endogenous short RNAs generated by Dicer 2 and RNA-dependent RNA polymerase 1 regulate mRNAs in the basal fungus Mucor circinelloides. Nucleic Acids Res. 2010;38:5535–5541. doi: 10.1093/nar/gkq301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nicolás FE, Vila A, Moxon S, Cascales MD, Torres-Martínez S, et al. The RNAi machinery controls distinct responses to environmental signals in the basal fungus Mucor circinelloides. BMC Genomics. 2015;16:237. doi: 10.1186/s12864-015-1443-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Son H, Park AR, Lim JY, Shin C, Lee YW. Genome-wide exonic small interference RNA-mediated gene silencing regulates sexual reproduction in the homothallic fungus Fusarium graminearum . PLoS Genet. 2017;13:e1006595. doi: 10.1371/journal.pgen.1006595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meng H, Wang Z, Wang Y, Zhu H, Huang B. Dicer and argonaute genes involved in RNA interference in the Entomopathogenic Fungus Metarhizium robertsii . Appl Environ Microbiol. 2017;83:e03230-16. doi: 10.1128/AEM.03230-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang Q, Li L, Xue Z, Ye Q, Zhang L, et al. Transcription of the major neurospora crassa microRNA-like small RNAs relies on RNA polymerase III. PLoS Genet. 2013;9:e1003227. doi: 10.1371/journal.pgen.1003227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Drinnenberg IA, Fink GR, Bartel DP. Compatibility with killer explains the rise of RNAi-deficient fungi. Science. 2011;333:1592. doi: 10.1126/science.1209575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carreras-Villaseñor N, Esquivel-Naranjo EU, Villalobos-Escobedo JM, Abreu-Goodger C, Herrera-Estrella A. The RNAi machinery regulates growth and development in the filamentous fungus Trichoderma atroviride. Mol Microbiol. 2013;89:96–112. doi: 10.1111/mmi.12261. [DOI] [PubMed] [Google Scholar]
- 33.Bray NL, Pimentel H, Melsted P, Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol. 2016;34:525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- 34.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Götz S, García-Gómez JM, Terol J, Williams TD, Nagaraj SH, et al. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008;36:3420–3435. doi: 10.1093/nar/gkn176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Young MD, Wakefield MJ, Smyth GK, Oshlack A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 2010;11:R14. doi: 10.1186/gb-2010-11-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Davis MPA, van Dongen S, Abreu-Goodger C, Bartonicek N, Enright AJ. Kraken: a set of tools for quality control and analysis of high-throughput sequence data. Methods. 2013;63:41–49. doi: 10.1016/j.ymeth.2013.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Axtell MJ. ShortStack: comprehensive annotation and quantification of small RNA genes. RNA. 2013;19:740–751. doi: 10.1261/rna.035279.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lawrence M, Huber W, Pagès H, Aboyoun P, Carlson M, et al. Software for computing and annotating genomic ranges. PLoS Comput Biol. 2013;9:e1003118. doi: 10.1371/journal.pcbi.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nawrocki EP, Kolbe DL, Eddy SR. Infernal 1.0: inference of RNA alignments. Bioinformatics. 2009;25:1335–1337. doi: 10.1093/bioinformatics/btp157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Burge SW, Daub J, Eberhardt R, Tate J, Barquist L, et al. Rfam 11.0: 10 years of RNA families. Nucleic Acids Res. 2013;41:D226–32. doi: 10.1093/nar/gks1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative genomics viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fahlgren N, Carrington JC. Plant MicroRNAs. Humana Press; 2010. miRNA target prediction in plants; pp. 51–57. [DOI] [PubMed] [Google Scholar]
- 46.Varkonyi-Gasic E, Wu R, Wood M, Walton EF, Hellens RP. Protocol: a highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods. 2007;3:12. doi: 10.1186/1746-4811-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gruber S, Zeilinger S. The transcription factor Ste12 mediates the regulatory role of the Tmk1 MAP kinase in mycoparasitism and vegetative hyphal fusion in the filamentous fungus Trichoderma atroviride . PLoS One. 2014;9:e111636. doi: 10.1371/journal.pone.0111636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Seifert JP, Zhou Y, Hicks SN, Sondek J, Harden TK. Dual activation of phospholipase C-epsilon by Rho and Ras GTPases. J Biol Chem. 2008;283:29690–29698. doi: 10.1074/jbc.M805038200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fukami K, Inanobe S, Kanemaru K, Nakamura Y. Phospholipase C is a key enzyme regulating intracellular calcium and modulating the phosphoinositide balance. Prog Lipid Res. 2010;49:429–437. doi: 10.1016/j.plipres.2010.06.001. [DOI] [PubMed] [Google Scholar]
- 50.Castellano E, Downward J. RAS Interaction with PI3K: more than just another effector pathway. Genes Cancer. 2011;2:261–274. doi: 10.1177/1947601911408079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Garduño-Rosales M, Callejas-Negrete OA, Medina-Castellanos E, Bartnicki-García S, Herrera-Estrella A, et al. F-actin dynamics following mechanical injury of Trichoderma atroviride and Neurospora crassa hyphae. Fungal Genet Biol. 2022;159:103672. doi: 10.1016/j.fgb.2022.103672. [DOI] [PubMed] [Google Scholar]
- 52.Connolly S, Quasi-Woode D, Waldron L, Eberly C, Waters K, et al. Calcineurin regulatory subunit calcium-binding domains differentially contribute to calcineurin signaling in Saccharomyces cerevisiae . Genetics. 2018;209:801–813. doi: 10.1534/genetics.118.300911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tamuli R, Deka R, Borkovich KA. Calcineurin subunits A and B interact to regulate growth and asexual and sexual development in Neurospora crassa . PLoS One. 2016;11:e0151867. doi: 10.1371/journal.pone.0151867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Glass NL, Dementhon K. Non-self recognition and programmed cell death in filamentous fungi. Curr Opin Microbiol. 2006;9:553–558. doi: 10.1016/j.mib.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 55.Paoletti M, Saupe SJ. Fungal incompatibility: evolutionary origin in pathogen defense? Bioessays. 2009;31:1201–1210. doi: 10.1002/bies.200900085. [DOI] [PubMed] [Google Scholar]
- 56.Perčulija V, Ouyang S. Helicases from All Domains of Life. Academic Press; 2019. Diverse roles of DEAD/DEAH-Box helicases in innate immunity and diseases; pp. 141–171. [DOI] [Google Scholar]
- 57.Wenger Y, Buzgariu W, Reiter S, Galliot B. Injury-induced immune responses in Hydra. Semin Immunol. 2014;26:277–294. doi: 10.1016/j.smim.2014.06.004. [DOI] [PubMed] [Google Scholar]
- 58.Lai SL, Marín-Juez R, Moura PL, Kuenne C, Lai JKH, et al. Reciprocal analyses in zebrafish and medaka reveal that harnessing the immune response promotes cardiac regeneration. Elife. 2017;6:e25605. doi: 10.7554/eLife.25605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zannini L, Delia D, Buscemi G. CHK2 kinase in the DNA damage response and beyond. J Mol Cell Biol. 2014;6:442–457. doi: 10.1093/jmcb/mju045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zeng W, Wang J, Wang Y, Lin J, Fu Y, et al. Dicer-like proteins regulate sexual development via the biogenesis of perithecium-specific microRNAs in a plant pathogenic ungus Fusarium graminearum . Front Microbiol. 2018;9:818. doi: 10.3389/fmicb.2018.00818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shao Y, Tang J, Chen S, Wu Y, Wang K, et al. milR4 and milR16 mediated fruiting body development in the medicinal fungus Cordyceps militaris . Front Microbiol. 2019;10:83. doi: 10.3389/fmicb.2019.00083. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Villalobos-Escobedo JM, Martínez-Hernández JP, Pelagio-Flores R, González-De la Rosa PM, Carreras-Villaseñor N, et al. 2022. Trichoderma atroviride hyphal regeneration and conidiation depend on cell-signaling processes regulated by a microRNA?like RNA. Figshare. [DOI] [PMC free article] [PubMed]