Highlights
-
•
Prior exposure to metformin protects from SARS-CoV-2 in Caco2 and Calu3 cells.
-
•
Metformin treatment in SARS-CoV-2 infected cells also inhibits up to two-logs, in a dose-dependent manner.
-
•
Metformin's mode of action may be mediated by AMPK.
Keywords: SARS-CoV-2, COVID-19, Antiviral activity, Metformin, AMPK, Diabetes
Abstract
Comorbidities such as diabetes worsen COVID-19 severity and recovery. Metformin, a first-line medication for type 2 diabetes, has antiviral properties and certain studies have also indicated its prognostic potential in COVID-19. Here, we report that metformin significantly inhibits SARS-CoV-2 growth in cell culture models. First, a steady increase in AMPK phosphorylation was detected as infection progressed, suggesting its important role during viral infection. Activation of AMPK in Calu3 and Caco2 cell lines using metformin revealed that metformin suppresses SARS-CoV-2 infectious titers up to 99%, in both naïve as well as infected cells. IC50 values from dose-variation studies in infected cells were found to be 0.4 and 1.43 mM in Calu3 and Caco2 cells, respectively. Role of AMPK in metformin's antiviral suppression was further confirmed using other pharmacological compounds, AICAR and Compound C. Collectively, our study demonstrates that metformin is effective in limiting the replication of SARS-CoV-2 in cell culture and thus possibly could offer double benefits as diabetic COVID-19 patients by lowering both blood glucose levels and viral load.
1. Introduction
COVID-19 prevails and rages on in several parts of the world. While vaccination drives continue to ensure its spread is limited, newer worrying aspects of long-COVID are a cause for concern. Patients with comorbidities such as cancer, auto-immune diseases, cardiovascular conditions, and diabetes, are still highly susceptible to contracting the disease (Gómez et al., 2021; Antonelli et al., 2022). The hospitalization rate for patients with comorbidities who contracted COVID-19 was significantly higher during the first wave of the pandemic, associated with poor prognosis (Sanyaolu et al., 2020). Growing evidence of long-COVID indicates that long-term effects of the illness along with a possibility of latency of SARS-CoV-2 can drastically affect the complete recovery of COVID-19 patients (Pietsch et al., 2021; Davis et al., 2021). Evidences of SARS-CoV-2 for prolonged periods have been observed in lung (Baang et al., 2021), and intestinal epithelium (Natarajan et al., 2022). The prospect of repurposing drugs to combat prolonged presence of the virus is of utmost importance to manage disease outcomes.
Type 2 diabetes, one of the most common metabolic disorders, is universally treated using insulin and a number of other drugs, chiefly metformin (Davidson and Peters, 1997). Metformin is a biguanide compound, used as first-line antidiabetic medication worldwide. It acts primarily by increasing glucose intake and limiting gluconeogenesis in the liver, and its action is mediated in part by the energy-sensing kinase, 5’-AMP-activated protein kinase (AMPK) (Zhou et al., 2001). Metformin inhibits Complex I of the electron transport chain and suppresses ATP synthesis, which triggers AMPK activation. This results in a cascade of events that decreases anabolic processes and initiates macromolecular breakdown to reinstate homeostasis. AMPK is a trimeric complex where the α subunit harbors the kinase and activating domains. The regulatory subunit γ is where AMP or ATP binds, thereby determining the allosteric regulation of AMPK (Herzig and Shaw, 2018). Competitive binding of AMP or ATP to one of four binding domains on the γ subunit is believed to cause a conformational change that makes the kinase domain on the α subunit more accessible for phosphorylation, although this notion has been debated (Herzig and Shaw, 2018; Kim et al., 2016).
In the past year, several reports have debated the clinical use of metformin in COVID-19 (Bramante et al., 2021; Dardano and Del Prato, 2021; Ibrahim et al., 2021; Zangiabadian et al., 2021). Many case studies on metformin treatment report a decrease in hospital mortality rates for patients that were on metformin prior to admission (Dardano and Del Prato, 2021; L et al., 2020; Marmor et al., 2021). Reports suggest that high glucose levels are associated with poorer prognosis and well-controlled glucose levels were indicative of lesser complications (Lihua et al., 2020). Metformin has also been reported to show antiviral activity against other viruses (Chen et al., 2020). In this study, we aimed to investigate the effect of metformin on SARS-CoV-2, and identify the effects of AMPK perturbation on infection. With recent reports having clarified that COVID-19 is not solely a respiratory illness with evidence of SARS-CoV-2 found in feces of patients showing successful replication in enterocytes (Chen et al., 2020; Lamers et al., 2020) we hence used cell lines Calu3 and Caco2, a lung carcinoma and a colorectal carcinoma cell line, respectively. Treatment of cells with metformin prior to infection substantially lowered the viral titer. Additionally, metformin strongly restricted SARS-CoV-2 levels in previously infected cells in a dose-dependent manner. Pharmacological activation of AMPK through AICAR suppressed viral infection while its inhibition using Compound C promoted it, confirming the effect of metformin is through AMPK. Our results support the promising use of metformin as a therapeutic drug in COVID-19.
2. Materials and methods
2.1. Cell culture and reagents
Calu3 (HTB-55™; ATCC) and Vero (CCL-81™; ATCC) cells were grown in DMEM supplemented with 10% FBS and 1 × Pen Strep, and Caco2 (HTB-37™; ATCC) cells were grown in DMEM supplemented with 20% FBS and 1 × Pen-Strep, at 37 °C and 5% CO2. Rabbit monoclonal antibodies against AMPK and phospho-AMPK (T172) were procured from CST. Mouse monoclonal GAPDH, β-tubulin antibodies, and rabbit monoclonal Nucleocapsid antibodies were purchased from ThermoFisher Scientific. HRP-conjugated secondary antibodies were purchased from Jackson ImmunoResearch. Metformin, AICAR, and Compound C were procured from Merck Millipore.
2.2. Infections and treatments
All experiments involving virus culture were carried out in the biosafety level-3 laboratory at the Centre for Cellular and Molecular Biology (CCMB). SARS-CoV-2 strain B.1.1.8 (virus ID-hCoV-19/India/TG-CCMB-L1021/2020; isolate; EPI_ISL_458046) was used for all experiments at 1 MOI (Gupta et al., 2021). Cells were grown to 80% confluency and treated as described. For pre-infection treatments, cells were subjected to 10 mM metformin or 1 mM AICAR for 24 h, followed by infection with SARS-CoV-2 in serum-free medium (SFM) for 3 h in the presence of the respective compound. The inoculum was subsequently replaced with complete medium containing the compound and the cells were harvested at 24 h post-infection (hpi). In post-infection mode of metformin treatment, the cells were first infected for 3 h after which the inoculum was replaced by media containing 10 mM metformin and further incubated until 24 hpi. Dose-dependent effect of metformin was studied by subjecting cells to varying doses of metformin (5, 10, 20, and 40 mM in Caco2, and 1, 2, 5, 10, and 20 mM in Calu3 cells) similar to the post-infection treatment regimen mentioned above. Compound C treatment was carried out by infecting cells for 3 h at 1 MOI, complete media for 21 h, and then an additional 24 h with 10 µM Compound C. For all treatments, media supernatant was collected to measure extracellular viral RNA as well as infectious viral titers, and cells were processed for immunoblotting.
2.3. Virus quantification and titration
RNA from viral supernatants was isolated using Nucleospin Viral RNA isolation kit (Macherey-Nagel GmbH & Co. KG). qRT-PCR was carried out using nCOV-19 RT-PCR detection kit from Q-line Molecular to quantify SARS-CoV-2 RNA using the manufacturer's protocol on a Roche LightCycler 480. Briefly, reverse transcription was performed for 15 min, followed by initial denaturation at 94 °C for 3 min, then PCR and detection was carried out at 58 °C for 30 s for 45 cycles. The viral E gene was normalized with internal control provided in the kit, and fold changes were calculated using 2(-ΔΔCt) method.
Infectious titers of the supernatants were calculated using plaque forming assay (PFU/mL) as mentioned previously (Gupta et al., 2021). Briefly, the supernatant was serially diluted in SFM and added to a confluent monolayer of Vero cells for infection for 3 h. The medium was then replaced with a 1:1 mixture of agarose: 2 × DMEM (1% low-melting agarose (LMA) containing a final concentration of 5% FBS and 1 × Pen-Strep). Six days post-infection, cells were fixed in 4% formaldehyde prepared in 1 × PBS and subsequently washed and stained with 0.1% crystal violet to count the plaques.
2.4. Immunoblotting
Cell pellets were lysed in an NP-40 lysis buffer as described earlier (Gupta et al., 2021). Protein quantification was done using BCA method (G Biosciences). Lysates were then mixed with 6 × Laemmli buffer, and equal amounts of protein were run on SDS-PAGE, followed by transfer onto PVDF membrane. Blots were blocked in 5% BSA and incubated with specific primary antibodies at 4 °C overnight. Incubation with HRP-conjugated secondary antibodies was done for 1 h and the blots were developed on a BioRad Chemidoc MP system using ECL reagents (ThermoFisher and G Biosciences). Quantification was performed using ImageJ (Schneider et al., 2012).
2.5. MTT assay
MTT was prepared at a concentration of 5 mg/mL in 1 × PBS. To determine cell viability, 10,000 cells for Caco2 and 15,000 cells for Calu3 were seeded in a 96 well format. Cells were first infected with 1 MOI SARS-CoV-2 for 3 h followed by media change with different concentrations of metformin (0.1, 1, 2, 5, 10, 20, 40, and 80 mM) along with vehicle control, for 24 h. At 24 hpi, media containing 0.5 mg/mL MTT was added to the cells and incubated at 37 °C for 3 h. The formazan crystals formed were solubilized using DMSO on a shaking platform for 30 min, and absorbance was measured at 570 nm with a reference reading at 620 nm.
2.6. Statistical analysis
All experiments were performed in triplicate to calculate mean ± SEM. Statistical significance was calculated using two-tailed, unpaired Student's t-test and p values are represented as *, **, ***, indicating p ≤ 0.05, 0.005, and 0.0005, respectively. IC50 and IC90 were calculated from PFU data from metformin titration experiments.
3. Results
3.1. SARS-CoV-2 infection causes long-term phosphorylation of AMPK
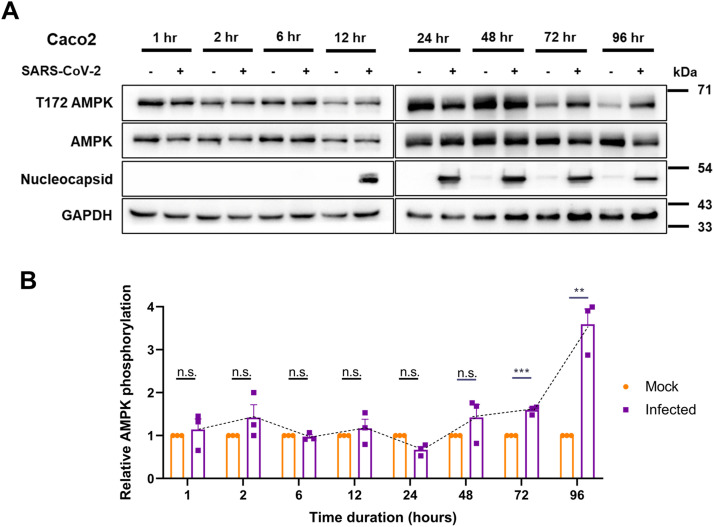
We tracked the activity and levels of AMPK during SARS-CoV-2 infection over a 96-h time course beginning from 1 h. Caco2 cells were infected with 1 MOI of SARS-CoV-2 for 1, 2, 6, 12, 24, 48, 72, and 96 h time. Though no significant change in the AMPK phosphorylation was detected until 48 h post-infection (hpi), marked increase in phosphorylation was evident from 48 hpi that further strengthened until 96 hpi, despite a drop in the abundance of the protein (Fig. 1A and B). These results indicated a major metabolic reprogramming, resulting in AMPK phosphorylation occurring after 24 hpi. Interestingly, AMPK phosphorylation coincided with the accumulation of viral proteins (Fig. 1A). This result suggests that changes in AMPK phosphorylation may affect SARS-CoV-2 replication.
Fig. 1.
SARS-CoV-2 infection induces AMPK phosphorylation.
(A) Immunoblots analyzing the phosphorylation of AMPK in SARS-CoV-2 infected Caco2 cells for early (1, 2, 6, and 12 h) and late (24, 48, 72, and 96 h) time points. Cells infected with 1 MOI of SARS-CoV-2 were harvested at various time intervals post-infection and analyzed by immunoblotting. (B) Quantitative representation of AMPK phosphorylation from three independent replicates. Densitometric values of phospho-AMPK bands were normalized against those of total-AMPK expression and GAPDH belonging to the corresponding samples and the values were plotted graphically.
3.2. Metformin protects cells from SARS-CoV-2 infection
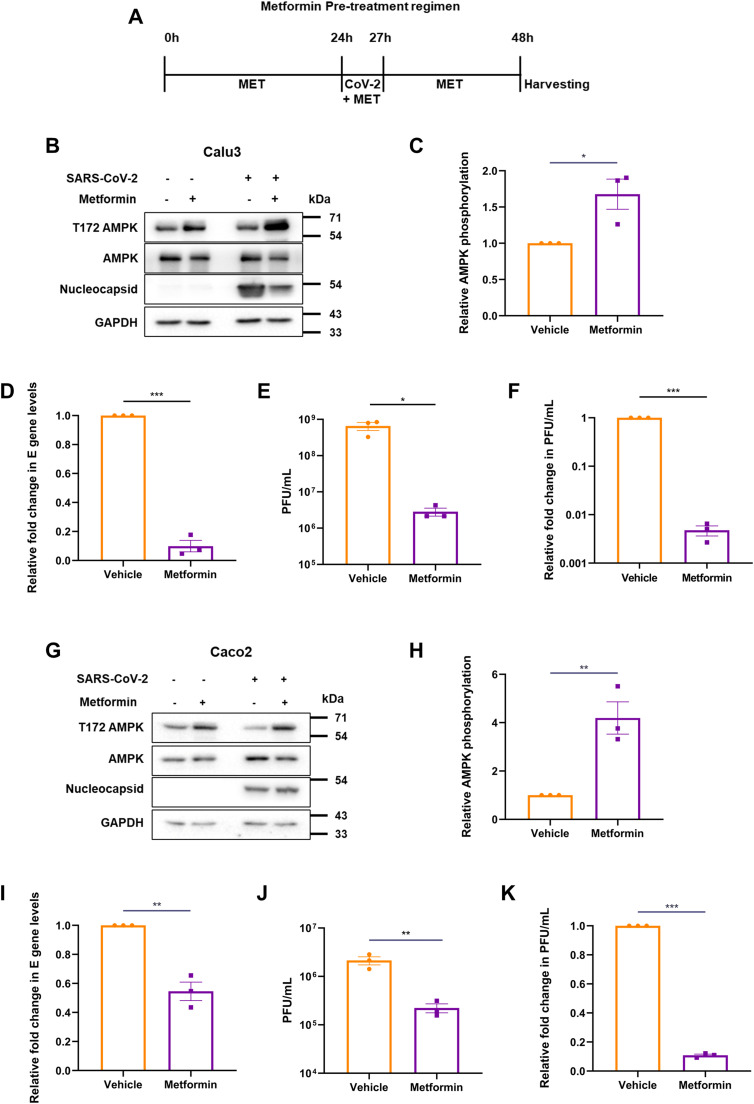
We investigated the role of AMPK activation during SARS-CoV-2 infection as previous reports have clearly established the roles played by this molecule on the outcome of viral infections (Bhutta et al., 2021). One compound of clinical relevance known to activate AMPK is metformin. With the emergence of reports suggesting a possible beneficial role of metformin in COVID-19 patients (Bramante et al., 2021; Dardano and Del Prato, 2021), we used this compound to both activate AMPK, as well as study it from a drug-repurposing point of view. In this study, we used Calu3, a respiratory epithelial cell line, and Caco2, a gut epithelial cell line, allowing us to study a larger impact of metformin and extrapolate its results in cells from different organs of relevance. Calu3 cells were pre-treated with 10 mM concentration of metformin for 24 h after which they were infected with 1 MOI of SARS-CoV-2 for 3 h in presence of metformin. Metformin at 10 mM is widely used in studies, is physiologically relevant and well reported to induce AMPK activity (Tsai et al., 2017; Cauchy et al., 2017; Alhourani et al., 2021). Subsequently, the inoculum was replaced with growth medium containing metformin and incubated until 24 hpi (Fig. 2A). Increased phosphorylation of AMPK in the drug-treated cells confirmed the effect of metformin (Fig. 2B and C). Metformin treatment resulted in nearly 90% drop in the viral RNA (Fig. 2D), and nearly two-log10 drop in the infectious viral titers (Fig. 2E and F) indicating that metformin treatment is protective against SARS-CoV-2 infection. Viral protein levels also decreased by 50%, suggesting a clear suppression of the viral life cycle by metformin (Fig. 2B). Metformin treatment performed in Caco2 cells corroborated the results from Calu3 (Fig. 2G–K), confirming that the drug has strong protective effects against SARS-CoV-2 infection in multiple cell lines.
Fig. 2.
Metformin pre-treatment inhibits SARS-CoV-2. (A) Schematic of the experimental set up for pre-treatment. Cells were infected with 1 MOI SARS-CoV-2 24 h after 10 mM metformin treatment. The vehicle control cells were also infected with the virus in parallel. (B) Immunoblots from Calu3 cells confirming AMPK phosphorylation by metformin or vehicle treatment, and SARS-CoV-2 infection. (C) Densitometric quantification of AMPK phosphorylation in infected cells. (D) SARS-CoV-2 RNA levels in the supernatant of metformin treated samples, measured by qRT-PCR of E gene. Relative fold change in the E levels between metformin and vehicle treated samples is depicted. (E) Infectious viral titers of SARS-CoV-2 in metformin treated Calu3 samples against vehicle, represented as average PFU/mL. (F) Relative fold change in the infectious viral titers of SARS-CoV-2 in metformin treated samples compared against that treated with vehicle, represented as fold change in PFU/mL. Similar experiments were carried out in Caco2 cells, with immunoblots representing AMPK phosphorylation (G), its quantification (H), viral RNA (I), average PFU/mL (J) and fold change in infectious units (K). Graphs indicate mean ± SEM, indicating the three biological replicates.
3.3. Metformin treatment post-infection causes more profound restriction of SARS-CoV-2
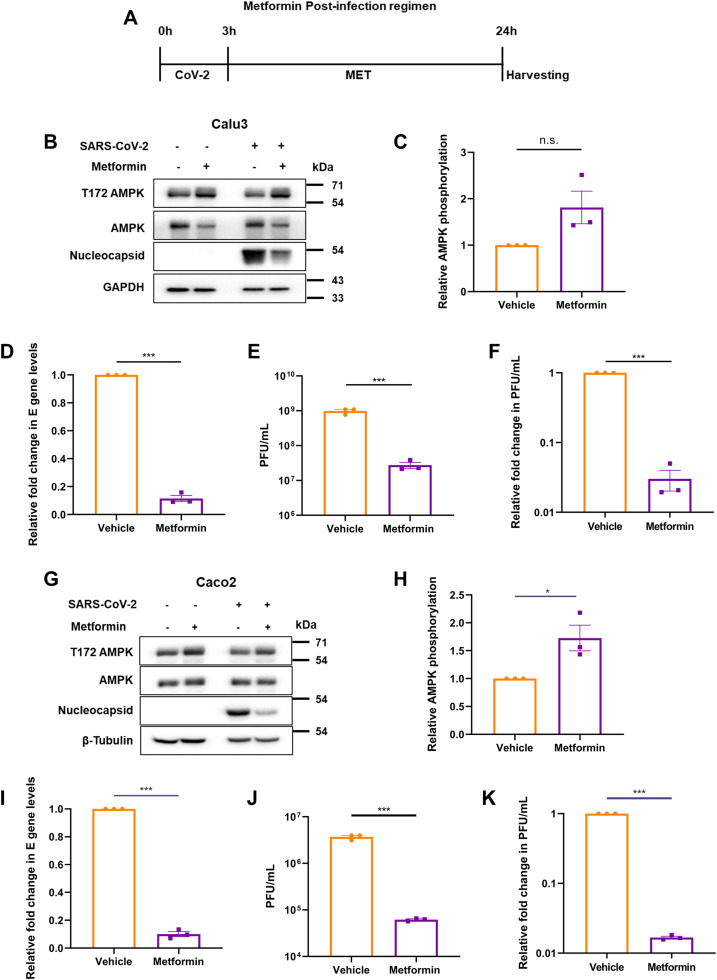
We next tested the effect of metformin in cells previously infected with SARS-CoV-2 to extrapolate its impact on the infected patients. Cells infected with 1 MOI of SARS-CoV-2 for 3 h were subsequently treated with 10 mM metformin until harvested at 24 hpi (Fig. 3A). Metformin caused higher AMPK phosphorylation in both Calu3 and Caco2 cultures infected with SARS-CoV-2 (Fig. 3B and C, F and G). Post-infection treatment with metformin resulted in a significant decrease in viral RNA and almost one-log10 drop in infectious titer in the supernatant of Calu3 cells with reduction in nucleocapsid expression as well (Fig. 3B, D–F). Similar results were observed in Caco2 cells, including a two-log10 reduction in viral titers (Fig. 3G–K), further confirming the profound restrictive effect of the drug on SARS-CoV-2. These results collectively indicate that metformin offers strong protection to both naïve as well as infected cells.
Fig. 3.
Metformin suppresses SARS-CoV-2 replication in the infected cells. (A) Schematic of the post-infection treatment by metformin. Cells were infected with 1 MOI SARS-CoV-2 for 3 h, followed by 21 h of metformin treatment before harvesting at 24 hpi. Vehicle treated cells were infected in parallel. (B) Confirmation of SARS-CoV-2 infection in Calu3 cells and AMPK phosphorylation by immunoblotting. (C) Densitometric analysis of AMPK phosphorylation in the treated, infected cells. (D) Relative fold change in SARS-CoV-2 E gene measured by qRT-PCR in the supernatants of metformin treated cells compared with the those treated with vehicle. (E) Infectious viral titers of SARS-CoV-2 in metformin treated samples against vehicle, represented as average PFU/mL. (F) Relative SARS-CoV-2 infectious titers of the supernatant from samples treated with metformin represented as fold change in PFU/mL. Similar experiments were performed in Caco2 cells. Confirmatory immunoblots (G), quantification of AMPK phosphorylation (H), RNA levels in the supernatant (I), average PFU/mL (J) and fold change in infectious units from vehicle and treated cells (K) are represented. and infectious titers from vehicle and treated cells (I), are represented. Graphs indicate mean ± SEM, indicating the three biological replicates.
3.4. Metformin treatment post-infection causes a dose-dependent suppression of SARS-CoV-2
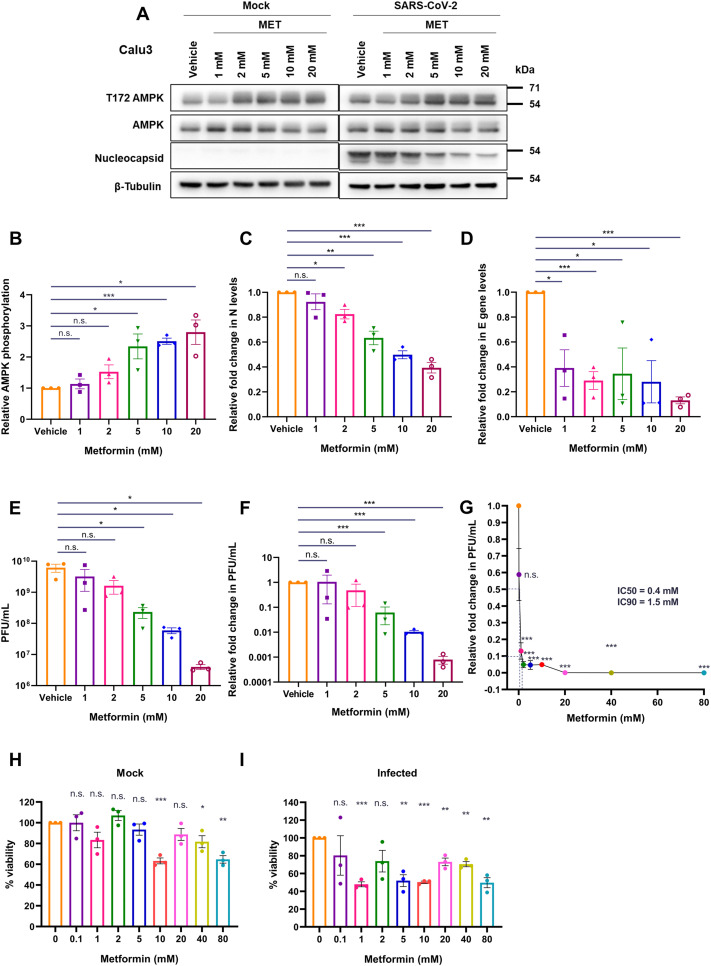
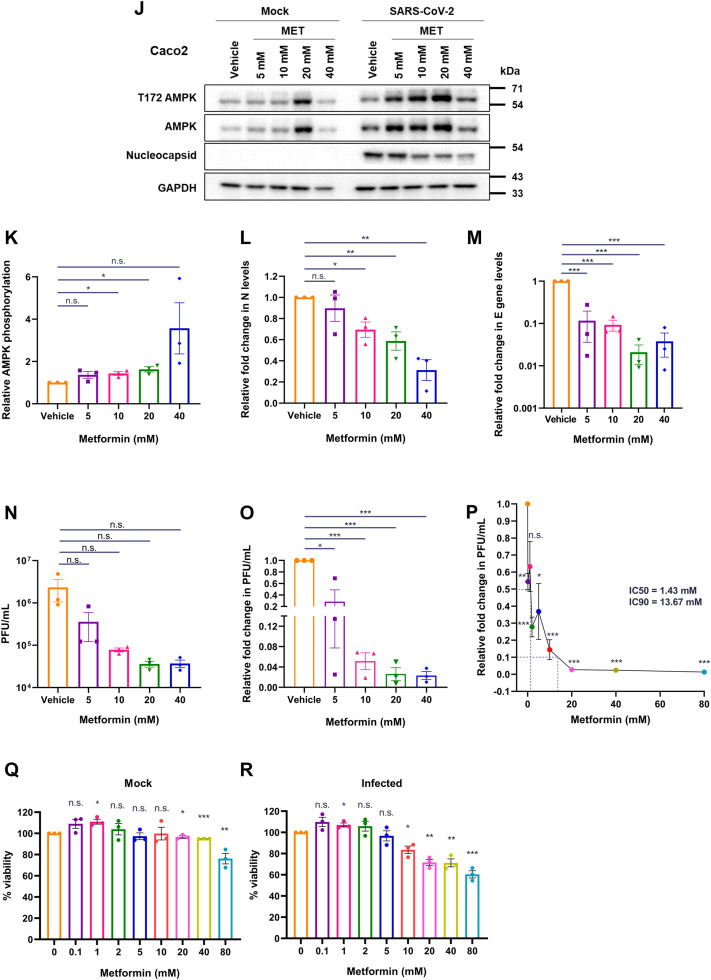
Metformin dose administered to diabetic individuals varies widely depending on a number of clinical parameters including fasting plasma glucose levels and HbA1c (Sanchez-Rangel and Inzucchi, 2017). Hence, based on our hypothesis, it is possible that individuals prescribed with different metformin doses are affected differently when infected with SARS-CoV-2. To assess whether in vitro administration of different doses of metformin affected the virus differently, we performed a dose-variation analysis. A concentration-dependent increase in AMPK phosphorylation was evident in SARS-CoV-2 infected cells from 1 to 20 mM metformin concentrations in Calu3 cells (Fig. 4A and B). A gradual and dose-dependent decrease in N levels was also evident (Fig. 4A and C). The drop in viral RNA levels was more dramatic with a very significant drop detected even at 1 mM concentration, that stabilized between 2 and 20 mM concentration (Fig. 4D), suggesting even low doses of metformin had a far greater and a more immediate impact on viral replication, possibly through alteration of cellular metabolism. Drop in viral titers was even more profound, with viral titers sharply decreasing beyond 2 mM metformin (Fig. 4E and F). The quicker response of viral RNA to metformin treatment is indicative of the drug's effect on viral RNA replication. We then expanded the range of metformin concentration in infected cells and calculated the IC50/IC90 based on plaque forming units (PFU). Metformin showed remarkably low IC50 and IC90 in Calu3 cells at 0.4 and 1.5 mM concentrations, respectively (Fig. 4G). We also measured the viability of cells using these different concentrations in uninfected and infected cells. A non-linear decrease in viability was noticed with increasing concentrations, which was further enhanced in the presence of virus, indicating that SARS-CoV-2-infected cells are more susceptible to metformin treatment (Fig. 4H and I). Similar experiments in Caco2 cells showed a comparable decrease in N, viral RNA, and infectious titers (Fig. 4J–O). A higher IC50 and IC90 were observed in Caco2 cells, 1.43 and 13.67 mM, respectively (Fig. 4P). Unlike Calu3 cultures, a gradual decrease in viability was observed with increasing metformin amounts in Caco2 cultures, with no significant loss of viability detected at these concentrations (Fig. 4Q and R). The drop in viral titer with metformin treatment was much more significant than the cell toxicity in both the cell culture models, indicating that metformin has targeted impact on viral replication. Together, these results unambiguously demonstrate a potent anti-SARS-CoV-2 effect of metformin.
Fig. 4.
Metformin treatment inhibits SARS-CoV-2 replication in a dose-dependent manner. (A-I) represent results from Calu3 cells. (A) Immunoblots confirming the infection and AMPK phosphorylation following the treatment of SARS-CoV-2 infected Calu3 cells with metformin at the doses described above the panel. (B) Relative AMPK phosphorylation in the samples treated with metformin. The graph was generated from the densitometric analysis of the immunoblots. (C) Relative abundance of N quantified from the immunoblots from the panel (A). (D) Relative fold change in SARS-CoV-2 E gene measured by qRT-PCR in the supernatants of metformin treated cells compared with the those treated with vehicle. (E) Infectious units of supernatants from the different metformin treatments against vehicle control, represented as average PFU/mL. (F) Relative SARS-CoV-2 infectious titers of the supernatant from samples treated with metformin represented as fold change in PFU/mL. (G) Measurement of IC50 and IC90 for metformin in Calu3 cells on SARS-CoV-2 infectious virus particle production. PFU/mL data for the individual samples treated with the different concentrations of metformin (0.1, 1, 2, 5, 10, 20, 40, and 80 mM) were plotted in the graph to calculate the respective values. (H and I) Viability of cells treated with varying concentrations of metformin (0.1, 1, 2, 5, 10, 20, 40, 80) assessed using MTT assay under mock (H) and infected (I) conditions, represented as % viability. (J-R) represent results from Calu3 cells. (J) Immunoblots confirming infection and AMPK phosphorylation in SARS-CoV-2 infected Caco2 cells treated with metformin at the doses mentioned. (K) Relative AMPK phosphorylation in the metformin treated samples. (L) Relative abundance of N quantified from the immunoblots from (H). (M) Relative fold change in SARS-CoV-2 E gene measured by qRT-PCR in the supernatants of metformin treated cells compared with vehicle control. (N) Infectious units of supernatants from the different metformin treatments against vehicle control, represented as average PFU/mL. (O) Relative SARS-CoV-2 infectious titers from varying metformin treatments (P) Measurement of IC50 and IC90 for metformin in Caco2 cells on SARS-CoV-2 infectious virus particle production. PFU/mL data for the individual samples treated with the different concentrations of metformin (0.1, 1, 2, 5, 10, 20, 40, and 80 mM) were plotted in the graph to calculate the respective values. (Q and R) Viability of cells treated with varying concentrations of metformin (0.1, 1, 2, 5, 10, 20, 40, 80) assessed using MTT assay under mock (Q) and infected (R) conditions, represented as % viability. Graphs indicate mean ± SEM, indicating the three biological replicates.
3.5. AMPK plays a role in restricting SARS-CoV-2
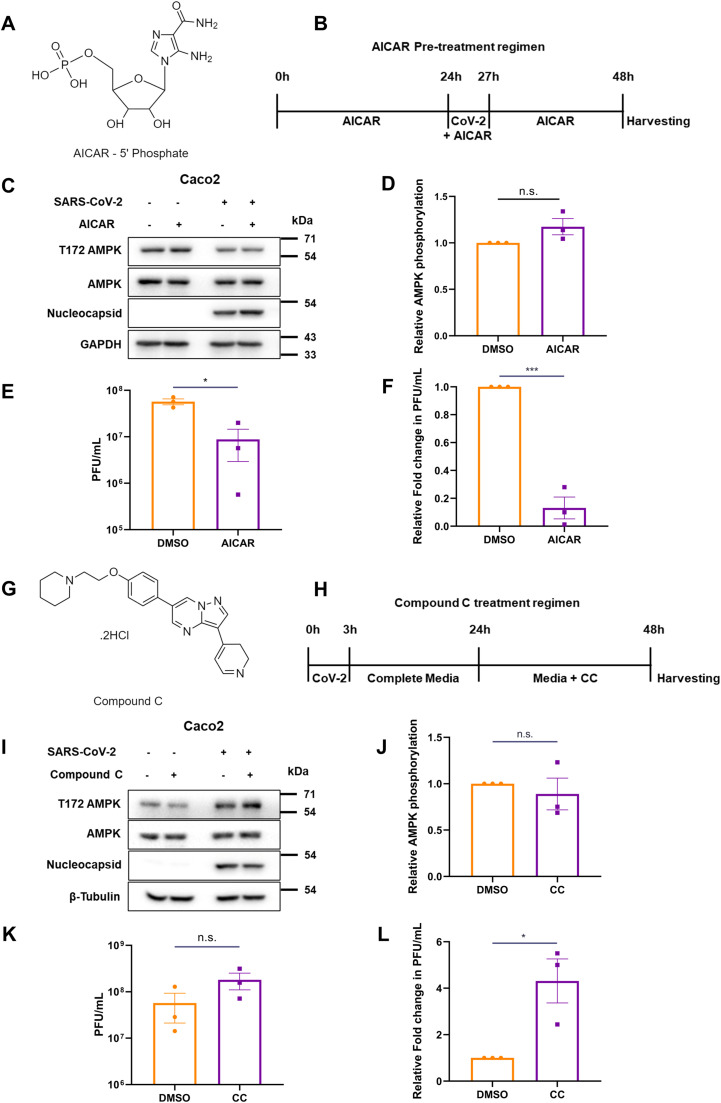
We then asked whether metformin's mode of antiviral activity could be through AMPK, a well-known mediator. To verify the same, we used pharmacological compounds that would activate or inhibit AMPK activity. AICAR, an allosteric activator, directly activates AMPK by binding to the γ subunit, mimicking AMP-binding, thereby activating downstream signaling. Cells were pre-treated with 1 mM of AICAR (Fig. 5A), for 24 h followed by infection for 3 h (Fernandes-Siqueira et al., 2018; Shen et al., 2016). The media was then replaced with AICAR-containing DMEM for another 24 h until harvesting (Fig. 5B). Although AICAR treatment did not result in appreciable change in AMPK phosphorylation (Fig. 5C and D), it resulted in a significant lowering of infectious viral titers of SARS-CoV-2 (Fig. 5E and F) by almost one-log10, as demonstrated previously with metformin (Fig. 2K). These results, along with metformin data, demonstrate that AMPK activation may be beneficial to the host cells by significantly limiting the viral titers.
Fig. 5.
AMPK activation is beneficial to the host against SARS-CoV-2. (A) Structure of AICAR 5’-phosphate. (B) Schematic of the treatment of Caco2 cells with AICAR and infection by SARS-CoV-2. (C) Immunoblots depicting AMPK phosphorylation and levels, and confirming infection. (D) Relative AMPK phosphorylation from AICAR-treated, infected cells. (E) Infectious viral titers of AICAR treated cells against DMSO control, represented as average PFU/mL. (F) Relative infectious titers of SARS-CoV-2 in samples that underwent pre-treatment with AICAR, as against the vehicle. (G) Chemical structure of Compound C. (H) Schematic of the treatment of SARS-CoV-2 infected Caco2 cells with CC. (I) Immunoblot confirmation of the infection and inhibition of AMPK activity. (J) Quantification of AMPK phosphorylation in CC treated, infected samples. (K) Infectious viral titers of Compound C-treated cells against DMSO control, represented as average PFU/mL. (L) Relative infectious titers of SARS-CoV-2 in samples that underwent CC-treatment, as against the vehicle, DMSO. Graphs indicate mean ± SEM, indicating the three biological replicates.
We further confirmed this effect by inhibiting AMPK by using Compound C (CC, Fig. 5G), a small molecule ATP-competitive inhibitor, during SARS-CoV-2 infection. Since AMPK phosphorylation peaked beyond 24 h in the time-course study (Fig. 1A), we decided to infect the cells first to activate AMPK and then inhibited AMPK with CC. Cells infected at 1 MOI were treated with 10 µM CC at 24 hpi and incubated for an additional 24 h (Fig. 5H), for a total of 48 h of infection. CC is used at this concentration widely (Zhou et al., 2001; Quan et al., 2013). Though CC treatment caused a visible drop in AMPK phosphorylation in mock cells, there was no apparent decrease observed in infected cells, indicating that the virus-induced AMPK activation overrides CC inhibition (Fig. 5I and J). As anticipated, CC treatment resulted in over four-fold higher viral titers in the supernatants as against the control sample (Fig. 5K and L). These results collectively suggest a possible protective role of AMPK in during SARS-CoV-2 infection.
4. Discussion
The anti-diabetic drug, metformin, has been projected to influence the prognosis of COVID-19 patients. As patients with comorbidities fared worse when infected with SARS-CoV-2, management of an ongoing illness alongside COVID-19 treatment became paramount. Some studies early during the pandemic identified a positive correlation between improved glucose levels in diabetic patients on metformin and better clinical outcome (L et al., 2020; Huang et al., 2020; Crouse et al., 2021; Pan et al., 2020). A number of reports proposed metformin as a possible “miracle or menace” in COVID-afflicted patients, based on retrospective data from hospitalisations, comparing the length of hospitalisation, severity of symptoms, or mortality (Marmor et al., 2021; Lui and Tan, 2021). In this study, we demonstrate that metformin profoundly lowers SARS-CoV-2 infectivity. While extrapolating these results to a clinical setup may not be appropriate, our data indicate that metformin can be effective not only as a treatment option, but as a prophylactic agent as well. With this data, we suggest that treatment of patients with metformin prior to infection with SARS-CoV-2 may have assisted in decreasing their symptoms of COVID-19. Our results also suggest that metformin could be beneficial in non-diabetic, COVID-19 patients and expand the scope of its coverage. In summary, this data lies in agreement with the numerous case studies published during the pandemic that suggested an antiviral role for this known anti-diabetic drug.
Metformin's mechanisms of action are multi-dimensional. Although the majority of reports indicate the involvement of AMPK and its substrates, other pathways are activated as well by metformin. For instance, other than blocking Complex-I activity and activating AMPK, the decrease in ATP levels also affects cAMP levels which affects Protein kinase A (PKA) activity. This in turn affects phosphofructokinase (PFK) in the glycolytic pathway, increasing glycolysis, and fructose-1,6-bisphosphatase (FBPase-1) in gluconeogenesis (Rena et al., 2017; Miller et al., 2013). Metformin also decreases hepatic lipid levels, increases skeletal muscle uptake of glucose, and in parallel helps in decreasing circulating lipids that are associated with cardiovascular risk especially in diabetic adults who are obese (Rena et al., 2017; Pernicova and Korbonits, 2014). Phosphorylation of ACC by AMPK causes inhibition of fatty acid synthesis and causes beta-oxidation that influences lipid breakdown, an important process known to be modulated by multiple viruses for the production of membranous web structures for their replication (Bhutta et al., 2021; Foster, 2012; Foretz et al., 2014). Additionally, the effects of metformin on viruses such as HBV, zika and dengue by interrupting their replication or protein synthesis, have been reported (Xun et al., 2014; Carlos Noe et al., 2021). RNA viruses in particular are known to modulate lipogenesis to steer cells to produce more vesicles to aid replication, as well as packaging and release (Pereira-Dutra et al., 2019; Herker and Ott, 2012). Certain viral infections trigger induction of lipogenic genes through activating factors such as SREBPs and PPARs, resulting in accumulation of lipid droplets (Syed and Siddiqui, 2011; Széles et al., 2007). These facilitate the establishment of key sites of replication and assembly, which has been documented for several viruses including flaviviruses (Marcelo et al., 2009; Yusuke et al., 2007; Priya et al., 2018; Steeve et al., 2007) and others (Doerflinger et al., 2017; Coffey et al., 2006; Winsome et al., 2010). This can also act as a double-edged sword as a number of antiviral molecules also assemble on lipid vesicles. Activation of interferon pathways, particularly types I and III, results in the production of interferon-stimulated genes (ISGs), some of which are reported to load onto lipid droplets, especially viperin and IFIT1 (Monson et al., 2018). A recent report on SARS-CoV-2 highlighted the possible role that lipid droplets play in its infection (Dias et al., 2020). Not only did they observe higher colocalization of viral RNA with the lipid droplets, they also demonstrated that inhibition of its formation decreased viral load as well as pro-inflammatory cytokines and apoptosis markers. These results in conjunction with ours indicate that the anti-viral effect of metformin is probably a resultant of altered lipid metabolism.
Certain RNA viruses have been found to remain in the host long after onset of symptoms. Some studies have observed the presence of SARS-CoV-2 RNA several weeks after symptoms develop, leading to prolonged illness (Baang et al., 2021). Presence of viral RNA for extended periods indicates the possibility of latency in COVID-19, though not proven yet. These factors are predicted to play a key role in long COVID-symptoms observed in a large number of individuals that have been infected (Proal and VanElzakker, 2021). From the results of this study, it is likely that the use of metformin in such cases could be an interesting proposition to combat latent COVID-19.
We sought to decipher whether metformin's antiviral effects as reported in the retrospective studies can be examined and confirmed in an in vitro setup. For this, we used concentrations of metformin that have been widely reported previously in cell-culture setups and that also fall in the range of doses used in pharmacokinetic studies (Zake et al., 2021). An important point to note is the differences observed in metformin absorption into different tissues, that we also observed between the cell lines we used. Metformin had a stronger impact on SARS-CoV-2 in lung epithelial culture than the gut epithelial cultures used in our study. It is unclear whether the lung cells had a higher absorption of the drug than the gut cells. Although a direct extrapolation to a clinical setting is not possible, we can appreciate the level of antiviral activity of metformin over a range of metformin doses.
We conducted studies in Caco2 and Calu3, two cell lines with pertinent tissue origin to our study. Metformin is absorbed in the intestine, which makes Caco2 an appropriate choice, in addition to its permissivity to SARS-CoV-2. We adopted Calu3 to study the suppressive effects of metformin in SARS-CoV-2 permissive lung epithelial cells. Previous studies quoting metformin usage in these cell lines focussed on metformin's absorption and mode of passage through the system (Nicklin et al., 1996; Proctor et al., 2008), or its anti-proliferative effects in cancer cells (Morgillo et al., 2013). A recent study performed in Calu3 cells even describes metformin's role in upregulating ACE2 expression (Albini et al., 2021). In our studies, we observed potent and impressive antiviral effect of metformin on both lung as well as gut cells, suggesting the possibility that other tissues where SARS-CoV-2 may infect would also respond to the drug similarly (Foretz et al., 2014; Chu et al., 2020; Hoffmann et al., 2020; Bojkova et al., 2020).
We speculate that the loss in infectivity of SARS-CoV-2 by metformin could also be an outcome of altered lipid metabolism mediated by AMPK. AMPK affects cellular lipid levels through a number of its substrates, such as ACC and SREBP1. AMPK also regulates macromolecular metabolism, mitochondrial homeostasis, autophagy as well as apoptosis. As its role is multifaceted and vital for maintaining energy levels, it has been reported to play key roles in many virus infections. Multiple reports show that AMPK activation can be either detrimental or beneficial for virus survival and propagation (Bhutta et al., 2021). Our results using AICAR and CC in SARS-CoV-2 infection implies an unfavorable/antiviral environment for the virus when AMPK is activated. In this context, it is interesting to note that N protein was unaffected during pharmacological activation of AMPK unlike the viral RNA and infectious titer, indicating that viral protein translation may not be inhibited during the treatments.
AICAR is an AMP analog that when phosphorylated, binds to the AMPK γ subunit. Accumulation of AICAR in the cytosol mimics increased AMP levels, skewing the AMP/ATP ratio thereby activating AMPK (Kim et al., 2016). Although a more specific activator of AMPK than metformin, AICAR does not necessarily have therapeutic potential, and hence we didn't replicate all metformin experiments with AICAR. Compound C, or Dorsomorphin, is a largely-selective small molecule AMPK inhibitor that was picked up from a kinase inhibitor screen, for delineating the mechanism of action of metformin (Zhou et al., 2001). Since we observed AMPK activation after 24 h of infection, we wanted to inhibit AMPK when it is active. In our study, AMPK activation using AICAR reiterated the results of metformin albeit to a lower extent, while Compound C exhibited the opposite effect to metformin. These results suggest that although AMPK plays a pertinent role in bringing about the effects of metformin, there are other pathways involved in inhibiting SARS-CoV-2.
5. Conclusion
Metformin suppresses SARS-CoV-2 in vitro and this data indicates both therapeutic and prophylactic potential for metformin in the management of COVID-19. AICAR and Compound C results explain AMPK's partial role in mediating metformin's antiviral effect.
Institutional biosafety
Institutional biosafety clearance was obtained for the experiments pertaining to SARS-CoV-2.
Consent for publication
All authors agree to publish the article.
Funding
The work was supported by the funding from the Council of Scientific and Industrial Research, Govt. of India (6/1/FIRST/2020-RPPBDD-TMD-SeMI).
CRediT authorship contribution statement
Haripriya Parthasarathy: Methodology, Conceptualization, Formal analysis, Investigation, Visualization, Writing – original draft, Writing – review & editing. Dixit Tandel: Visualization, Formal analysis, Investigation. Abdul Hamid Siddiqui: Investigation, Visualization. Krishnan H. Harshan: Conceptualization, Funding acquisition, Methodology, Project administration, Supervision, Writing – original draft, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no competing interests.
Acknowledgment
We thank Divya Gupta, Vishal Sah, Poojitha Sai Potharaju, and Prangya Paramita Sahoo for their help with generation of virus and for conducting experiments. We specially thank Mohan Singh Moodu and Amit Kumar for their assistance with logistics.
Data availability
No data was used for the research described in the article.
References
- Gómez C.E., Perdiguero B., Esteban M. Emerging SARS-CoV-2 variants and impact in global vaccination programs against SARS-CoV-2/COVID-19. Vaccines. 2021;9(3):1–13. doi: 10.3390/vaccines9030243. [Internet]Mar 1 [cited 2022 Jul 15]Available from: /pmc/articles/PMC7999234/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonelli M., Pujol J.C., Spector T.D., Ourselin S., Steves C.J. Risk of long COVID associated with delta versus omicron variants of SARS-CoV-2. Lancet. 2022;399(10343):2263–2264. doi: 10.1016/S0140-6736(22)00941-2. http://www.thelancet.com/article/S0140673622009412/fulltext [Internet]Jun 18 [cited 2022 Jul 15];Available from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyaolu A., Okorie C., Marinkovic A., Patidar R., Younis K., Desai P., et al. Comorbidity and its impact on patients with COVID-19. SN Compr. Clin. Med. 2020;2(8):1. doi: 10.1007/s42399-020-00363-4. [Internet]Aug [cited 2021 Sep 14]Available from: /pmc/articles/PMC7314621/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietsch H., Escher F., Aleshcheva G., Baumeier C., Morawietz L., Elsaesser A., et al. Proof of SARS-CoV-2 genomes in endomyocardial biopsy with latency after acute infection. Int. J. Infect. Dis. 2021;102:70–72. doi: 10.1016/j.ijid.2020.10.012. Jan 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis H.E., Assaf G.S., McCorkell L., Wei H., Low R.J., Re'em Y., et al. Characterizing long COVID in an international cohort: 7 months of symptoms and their impact. eClinicalMedicine. 2021;38 doi: 10.1016/j.eclinm.2021.101019. Aug 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baang J.H., Smith C., Mirabelli C., Valesano A.L., Manthei D.M., Bachman M.A., et al. Prolonged severe acute respiratory syndrome coronavirus 2 replication in an immunocompromised patient. J. Infect. Dis. 2021;223(1):23–27. doi: 10.1093/infdis/jiaa666. https://pubmed.ncbi.nlm.nih.gov/33089317/ [Internet]Jan 1 [cited 2022 Jul 11]Available from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan A., Zlitni S., Brooks E.F., Vance S.E., Dahlen A., Hedlin H., et al. Gastrointestinal symptoms and fecal shedding of SARS-CoV-2 RNA suggest prolonged gastrointestinal infection. Med. 2022;3(6):371–387. doi: 10.1016/j.medj.2022.04.001. Jun 10.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson M.B., Peters A.L. An overview of metformin in the treatment of type 2 diabetes mellitus. Am. J. Med. 1997;102:99–110. doi: 10.1016/s0002-9343(96)00353-1. [DOI] [PubMed] [Google Scholar]
- Zhou G., Myers R., Li Y., Chen Y., Shen X., Fenyk-Melody J., et al. Role of AMP-activated protein kinase in mechanism of metformin action. J. Clin. Invest. 2001;108(8):1167–1174. doi: 10.1172/JCI13505. http://www.jci.org [Internet]Oct 15 [cited 2022 Jan 19]Available from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzig S., Shaw RJ. AMPK: guardian of metabolism and mitochondrial homeostasis. Nat. Rev. Mol. Cell Biol. 2018;19(2):121–135. doi: 10.1038/nrm.2017.95. [Internet]Available from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Yang G., Kim Y., Kim J., Ha J. AMPK activators: mechanisms of action and physiological activities. Exp. Mol. Med. 2016;48(4):e224. doi: 10.1038/emm.2016.16. https://www.nature.com/articles/emm201616 2016 484 [Internet]Apr 1 [cited 2022 Jan 17]–e224Available from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramante C.T., Ingraham N.E., Murray T.A., Marmor S., Hovertsen S., Gronski J., et al. Metformin and risk of mortality in patients hospitalised with COVID-19: a retrospective cohort analysis. Lancet Healthy Longev. 2021;2(1):e34–e41. doi: 10.1016/S2666-7568(20)30033-7. [Internet]Available from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crouse A.B., Grimes T., Li P., Might M., Ovalle F., Shalev A. Metformin use is associated with reduced mortality in a diverse populationwWith COVID-19 and diabetes. Front. Endocrinol. (Lausanne) 2021;11:1081. doi: 10.3389/fendo.2020.600439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dardano A., Del Prato S. Metformin: an inexpensive and effective treatment in people with diabetes and COVID-19? Lancet Healthy Longev. 2021;2(1):e6–e7. doi: 10.1016/S2666-7568(20)30047-7. [Internet]Available from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim S., Lowe J.R., Bramante C.T., Shah S., Klatt N.R., Sherwood N., et al. Metformin and COVID-19: focused review of mechanisms and current literature suggesting benefit. Front. Endocrinol. 2021;0:625. doi: 10.3389/fendo.2021.587801. (Lausanne)Jul 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zangiabadian M., Nejadghaderi S.A., Zahmatkesh M.M., Hajikhani B., Mirsaeidi M., Nasiri M.J. The efficacy and potential mechanisms of metformin in the treatment of COVID-19 in the diabetics: a systematic review. Front. Endocrinol. 2021;12(March):1–9. doi: 10.3389/fendo.2021.645194. (Lausanne) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lihua Z., Zhi-Gang S., Xu C., Juan-Juan Q., Xiao-Jing Z., Jingjing C., et al. Association of blood glucose control and outcomes in patients with COVID-19 and pre-existing type 2 diabetes. Cell Metab. 2020;31(6):1068–1077. doi: 10.1016/j.cmet.2020.04.021. https://pubmed.ncbi.nlm.nih.gov/32369736/ [Internet]Jun 2 [cited 2021 Nov 1].e3Available from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmor S., Bramante C.T., Ingraham N.E., Murray T.A., Marmor S., Hovertsen S., et al. Metformin and risk of mortality in patients hospitalised with COVID-19: a retrospective cohort analysis. Artic Lancet Healthy Longev. 2021 doi: 10.1016/S2666-7568(20)30033-7. www.thelancet.com/ [Internet][cited 2021 Sep 20];2:34–41. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Guo H., Qiu L., Zhang C., Deng Q., Leng Q. Immunomodulatory and antiviral activity of metformin and its potential implications in treating coronavirus disease 2019 and lung injury. Front. Immunol. 2020;0:2056. doi: 10.3389/fimmu.2020.02056. Aug 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Chen L., Deng Q., Zhang G., Wu K., Ni L., et al. The presence of SARS-CoV-2 RNA in the feces of COVID-19 patients. J. Med. Virol. 2020;92(7):833–840. doi: 10.1002/jmv.25825. https://pubmed.ncbi.nlm.nih.gov/32243607/ [Internet]Jul 1 [cited 2022 Jul 11]Available from. [DOI] [PubMed] [Google Scholar]
- Lamers M.M., Beumer J., Der V.J.V., Knoops K., Puschhof J., Breugem T.I., et al. SARS-CoV-2 productively infects human gut enterocytes. Science. 2020;369(6499):50–54. doi: 10.1126/science.abc1669. https://www.science.org/doi/10.1126/science.abc1669 (80-) [Internet]Jul 3 [cited 2022 Jul 5]Available from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta D., Parthasarathy H., Sah V., Tandel D., Vedagiri D., Reddy S., et al. Inactivation of SARS-CoV-2 by β-propiolactone causes aggregation of viral particles and loss of antigenic potential. Virus Res. 2021;305 doi: 10.1016/j.virusres.2021.198555. https://linkinghub.elsevier.com/retrieve/pii/S0168170221002628 [Internet]Nov 1 [cited 2021 Sep 20]Available from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C.A., Rasband W.S., Eliceiri KW. NIH image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9(7):671–675. doi: 10.1038/nmeth.2089. https://www.nature.com/articles/nmeth.2089 2012 97 [Internet]Jun 28 [cited 2021 Nov 5]Available from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhutta M.S., Gallo E.S., Borenstein R. Multifaceted role of AMPK in viral infections. Cells. 2021;10(5):1118. doi: 10.3390/cells10051118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai H.H., Lai H.Y., Chen Y.C., Li C.F., Huang H.S., Liu H.S., et al. Metformin promotes apoptosis in hepatocellular carcinoma through the CEBPD-induced autophagy pathway. Oncotarget. 2017;8(8):13832–13845. doi: 10.18632/oncotarget.14640. https://www.oncotarget.com/article/14640/text/ [Internet]Jan 13 [cited 2022 Jan 18]Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cauchy F., Mebarki M., Leporq B., Laouirem S., Albuquerque M., Lambert S., et al. Strong antineoplastic effects of metformin in preclinical models of liver carcinogenesis. Clin. Sci. 2017;131(1):27–36. doi: 10.1042/CS20160438. https://pubmed.ncbi.nlm.nih.gov/27803295/ (Lond) [Internet][cited 2022 Jan 18]Available from: [DOI] [PubMed] [Google Scholar]
- Alhourani A.H., Tidwell T.R., Bokil A.A., Røsland G.V., Tronstad K.J., Søreide K., et al. Metformin treatment response is dependent on glucose growth conditions and metabolic phenotype in colorectal cancer cells. Sci. Rep. 2021;11(1):1–10. doi: 10.1038/s41598-021-89861-6. https://www.nature.com/articles/s41598-021-89861-6 2021 111 [Internet]May 18 [cited 2022 Jan 19]Available from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Rangel E., Inzucchi S.E. Metformin: clinical use in type 2 diabetes. Diabetologia. 2017;60(9):1586–1593. doi: 10.1007/s00125-017-4336-x. https://link.springer.com/article/10.1007/s00125-017-4336-x [Internet]Sep 1 [cited 2022 Jul 11]Available from. [DOI] [PubMed] [Google Scholar]
- Fernandes-Siqueira L.O., Zeidler J.D., Sousa B.G., Ferreira T., Da Poian A.T. Anaplerotic role of glucose in the oxidation of endogenous fatty acids during dengue virus infection. mSphere. 2018;3(1):e00458–17. doi: 10.1128/mSphere.00458-17. https://journals.asm.org/doi/abs/10.1128/mSphere.00458-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C., Ka S.O., Kim S.J., Kim J.H., Park B.H., Park J.H. Metformin and AICAR regulate NANOG expression via the JNK pathway in HepG2 cells independently of AMPK. Tumor Biol. 2016;37(8):11199–11208. doi: 10.1007/s13277-016-5007-0. https://link.springer.com/article/10.1007/s13277-016-5007-0 2016 378 [Internet]Mar 3 [cited 2022 Jan 18]Available from: [DOI] [PubMed] [Google Scholar]
- Quan H.Y., Kim D.Y., Chung S.H. Caffeine attenuates lipid accumulation via activation of AMP-activated protein kinase signaling pathway in HepG2 cells. BMB Rep. 2013;46(4):207–212. doi: 10.5483/BMBRep.2013.46.4.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. http://www.thelancet.com/article/S0140673620301835/fulltext [Internet]Feb 15 [cited 2022 May 29]Available from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan L., Lin Q., Yi L., Xiu-Lan L., Jian-Ling Z., Hui-Ying X., et al. Metformin treatment was associated with decreased mortality in COVID-19 patients with diabetes in a retrospective analysis. Am. J. Trop. Med. Hyg. 2020;103(1):69–72. doi: 10.4269/ajtmh.20-0375. https://pubmed.ncbi.nlm.nih.gov/32446312/ [Internet]Jul 1 [cited 2021 Nov 1]Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui D.T.W., Tan KCB. Is metformin a miracle or a menace in COVID-19 patients with type 2 diabetes? J. Diabetes Investig. 2021;12(4):479–481. doi: 10.1111/jdi.13484. https://onlinelibrary.wiley.com/doi/full/10.1111/jdi.13484 [Internet]Apr 1 [cited 2021 Nov 5]Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rena G., Hardie D.G., Pearson E.R. The mechanisms of action of metformin. Diabetologia. 2017;60(9):1577–1585. doi: 10.1007/s00125-017-4342-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller R.A., Chu Q., Xie J., Foretz M., Viollet B., Birnbaum M.J. Biguanides suppress hepatic glucagon signalling by decreasing production of cyclic AMP. Nature. 2013;494(7436):256–260. doi: 10.1038/nature11808. https://www.nature.com/articles/nature11808 2012 4947436 [Internet]Jan 6 [cited 2022 Oct 16]Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernicova I., Korbonits M. Metformin—mode of action and clinical implications for diabetes and cancer. Nat. Rev. Endocrinol. 2014;10(3):143–156. doi: 10.1038/nrendo.2013.256. https://www.nature.com/articles/nrendo.2013.256 2014 103 [Internet]Jan 7 [cited 2021 Sep 19]Available from. [DOI] [PubMed] [Google Scholar]
- Foster DW. Malonyl-CoA: the regulator of fatty acid synthesis and oxidation. J. Clin. Invest. 2012;122(6):1958. doi: 10.1172/JCI63967. [Internet]Jun 6 [cited 2022 Oct 16]Available from: /pmc/articles/PMC3366419/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foretz M., Guigas B., Bertrand L., Pollak M., Viollet B. Metformin: from mechanisms of action to therapies. Cell Metab. 2014;20(6):953–966. doi: 10.1016/j.cmet.2014.09.018. http://www.cell.com/article/S1550413114004410/fulltext [Internet]Dec 2 [cited 2022 Jan 18]Available from. [DOI] [PubMed] [Google Scholar]
- Xun Y.H., Zhang Y.J., Pan Q.C., Mao R.C., Qin Y.L., Liu H.Y., et al. Metformin inhibits hepatitis B virus protein production and replication in human hepatoma cells. J. Viral Hepat. 2014;21(8):597–603. doi: 10.1111/jvh.12187. https://pubmed.ncbi.nlm.nih.gov/24164660/ [Internet][cited 2022 Oct 16]Available from: [DOI] [PubMed] [Google Scholar]
- Carlos Noe F.M, Carlos Daniel C.R., Juan Fidel O.R., Irma Eloisa M.M., Luis Adrián D.J.G., José Esteban M.M., et al. The antiviral effect of metformin on zika and dengue virus infection. Sci. Rep. 2021;11(1) doi: 10.1038/s41598-021-87707-9. https://pubmed.ncbi.nlm.nih.gov/33888740/ [Internet]Dec 1 [cited 2021 Sep 20]Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira-Dutra F.S., Teixeira L., Costa MF de S., Bozza PT. Fat, fight, and beyond: The multiple roles of lipid droplets in infections and inflammation. J. Leukoc. Biol. 2019;106(3):563–580. doi: 10.1002/JLB.4MR0119-035R. https://onlinelibrary.wiley.com/doi/full/10.1002/JLB.4MR0119-035R [Internet]Sep 1 [cited 2021 Nov 1]Available from: [DOI] [PubMed] [Google Scholar]
- Herker E., Ott M. Emerging role of lipid droplets in host/pathogen interactions. J. Biol. Chem. 2012;287(4):2280–2287. doi: 10.1074/jbc.R111.300202. Jan 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed G.H., Siddiqui A. Effects of hypolipidemic agent nordihydroguaiaretic acid on lipid droplets and Hepatitis C virus. Hepatology. 2011;54(6):1936. doi: 10.1002/hep.24619. [Internet]Dec [cited 2021 Nov 1]Available from: /pmc/articles/PMC3236615/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Széles L., Töröcsik D., Nagy L. PPARγ in immunity and inflammation: cell types and diseases. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2007;1771(8):1014–1030. doi: 10.1016/j.bbalip.2007.02.005. Aug 1. [DOI] [PubMed] [Google Scholar]
- Marcelo M.S, Juan A.M, Nestor G.I., Iranaia A.M, Giselle B.L., Andrea T Da P., et al. Dengue virus capsid protein usurps lipid droplets for viral particle formation. PLoS Pathog. 2009;5(10) doi: 10.1371/journal.ppat.1000632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusuke M., Kimie A., Nobuteru U., Koichi W., Takayuki H., Margarita Z., et al. The lipid droplet is an important organelle for hepatitis C virus production. Nat. Cell Biol. 2007;9(9):1089–1097. doi: 10.1038/ncb1631. https://pubmed.ncbi.nlm.nih.gov/17721513/ [Internet]Sep [cited 2021 Nov 1]Available from. [DOI] [PubMed] [Google Scholar]
- Priya S.S, Nichole L., Gwendolyn M J., Phillip P S., T Z., DL S., et al. Comparative flavivirus-host protein interaction mapping reveals mechanisms of dengue and Zika virus pathogenesis. Cell. 2018;175(7):1931–1945. doi: 10.1016/j.cell.2018.11.028. https://pubmed.ncbi.nlm.nih.gov/30550790/ [Internet]Dec 13 [cited 2021 Nov 1]e18Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steeve B., Paul T.A., John M. Disrupting the association of hepatitis C virus core protein with lipid droplets correlates with a loss in production of infectious virus. J. Gen. Virol. 2007;88(Pt 8):2204–2213. doi: 10.1099/vir.0.82898-0. https://pubmed.ncbi.nlm.nih.gov/17622624/ [Internet]Aug [cited 2021 Nov 1]Available from: [DOI] [PubMed] [Google Scholar]
- Doerflinger S.Y., Cortese M., Romero-Brey I., Menne Z., Tubiana T., Schenk C., et al. Membrane alterations induced by nonstructural proteins of human norovirus. PLoS Pathog. 2017;13(10) doi: 10.1371/journal.ppat.1006705. https://journals.plos.org/plospathogens/article?id=10.1371/journal.ppat.1006705 [Internet]Oct 1 [cited 2021 Nov 1]Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffey C.M., Sheh A., Kim I.S., Chandran K., Nibert M.L., Parker JSL. Reovirus outer capsid protein μ1 induces apoptosis and associates with lipid droplets, endoplasmic reticulum, and mitochondria. J. Virol. 2006;80(17):8422. doi: 10.1128/JVI.02601-05. [Internet]Sep [cited 2021 Nov 1]Available from: /pmc/articles/PMC1563861/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winsome C., Michael G., Alessandro E., Clemens F.K, Nathalie C., Serge C., et al. Rotaviruses associate with cellular lipid droplet components to replicate in viroplasms, and compounds disrupting or blocking lipid droplets inhibit viroplasm formation and viral replication. J. Virol. 2010;84(13):6782–6798. doi: 10.1128/JVI.01757-09. https://pubmed.ncbi.nlm.nih.gov/20335253/ [Internet]Jul [cited 2021 Nov 1]Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monson E.A., Crosse K.M., Das M., Helbig K.J. Lipid droplet density alters the early innate immune response to viral infection. PLoS One. 2018;13(1) doi: 10.1371/journal.pone.0190597. https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0190597 [Internet]Jan 1 [cited 2021 Nov 1]Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias S.S.G., Soares V.C., Ferreira A.C., Sacramento C.Q., Fintelman-Rodrigues N., Temerozo J.R., et al. Lipid droplets fuel SARS-CoV-2 replication and production of inflammatory mediators. PLoS Pathog. 2020;16(12) doi: 10.1371/journal.ppat.1009127. https://journals.plos.org/plospathogens/article?id=10.1371/journal.ppat.1009127 [Internet]Dec 16 [cited 2021 Nov 1]Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proal A.D., VanElzakker MB. Long COVID or post-acute sequelae of COVID-19 (PASC): an overview of biological factors that may contribute to persistent symptoms. Front. Microbiol. 2021;12:1494. doi: 10.3389/fmicb.2021.698169. Jun 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zake D.M., Kurlovics J., Zaharenko L., Komasilovs V., Klovins J., Stalidzans E. Physiologically based metformin pharmacokinetics model of mice and scale-up to humans for the estimation of concentrations in various tissues. PLoS One. 2021;16(4) doi: 10.1371/journal.pone.0249594. https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0249594 [Internet]Apr 1 [cited 2022 Apr 20]Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklin P., Keates A.C., Page T., Bailey C.J. Transfer of metformin across monolayers of human intestinal Caco-2 cells and across rat intestine. Int. J. Pharm. 1996;128(1–2):155–162. Feb 29. [Google Scholar]
- Proctor W.R., Bourdet D.L., Thakker D.R. Mechanisms underlying saturable intestinal absorption of metformin. Drug Metab. Dispos. 2008;36(8):1650–1658. doi: 10.1124/dmd.107.020180. https://dmd.aspetjournals.org/content/36/8/1650 [Internet]Aug 1 [cited 2022 Nov 7]Available from. [DOI] [PubMed] [Google Scholar]
- Morgillo F., Sasso F.C., Della Corte C.M., Vitagliano D., D'Aiuto E., Troiani T., et al. Synergistic effects of metformin treatment in combination with gefitinib, a selective EGFR tyrosine kinase inhibitor, in LKB1 wild-type NSCLC cell lines. Clin. Cancer Res. 2013;19(13):3508–3519. doi: 10.1158/1078-0432.CCR-12-2777. https://pubmed.ncbi.nlm.nih.gov/23695170/ [Internet]Jul 1 [cited 2022 Nov 7]Available from. [DOI] [PubMed] [Google Scholar]
- Albini A., Calabrone L., Carlini V., Benedetto N., Lombardo M., Bruno A., et al. Preliminary evidence for IL-10-induced ACE2 mRNA expression in lung-derived and endothelial cells: implications for SARS-Cov-2 ARDS pathogenesis. Front. Immunol. 2021;12:3921. doi: 10.3389/fimmu.2021.718136. Sep 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu H., Chan J.F.W., Yuen T.T.T., Shuai H., Yuan S., Wang Y., et al. Comparative tropism, replication kinetics, and cell damage profiling of SARS-CoV-2 and SARS-CoV with implications for clinical manifestations, transmissibility, and laboratory studies of COVID-19: an observational study. Lancet Microbe. 2020;1(1):e14–e23. doi: 10.1016/S2666-5247(20)30004-5. https://pubmed.ncbi.nlm.nih.gov/32835326/ [Internet]May [cited 2022 May 28]Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280. doi: 10.1016/j.cell.2020.02.052. .e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bojkova D., Klann K., Koch B., Widera M., Krause D., Ciesek S., et al. Proteomics of SARS-CoV-2-infected host cells reveals therapy targets. Nature. 2020;583(7816):469–472. doi: 10.1038/s41586-020-2332-7. [Internet]Available from. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data was used for the research described in the article.