Summary
Multisystem inflammatory syndrome in children (MIS-C) is a delayed-onset, COVID-19-related hyperinflammatory illness characterized by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) antigenemia, cytokine storm, and immune dysregulation. In severe COVID-19, neutrophil activation is central to hyperinflammatory complications, yet the role of neutrophils in MIS-C is undefined. Here, we collect blood from 152 children: 31 cases of MIS-C, 43 cases of acute pediatric COVID-19, and 78 pediatric controls. We find that MIS-C neutrophils display a granulocytic myeloid-derived suppressor cell (G-MDSC) signature with highly altered metabolism that is distinct from the neutrophil interferon-stimulated gene (ISG) response we observe in pediatric COVID-19. Moreover, we observe extensive spontaneous neutrophil extracellular trap (NET) formation in MIS-C, and we identify neutrophil activation and degranulation signatures. Mechanistically, we determine that SARS-CoV-2 immune complexes are sufficient to trigger NETosis. Our findings suggest that hyperinflammatory presentation during MIS-C could be mechanistically linked to persistent SARS-CoV-2 antigenemia, driven by uncontrolled neutrophil activation and NET release in the vasculature.
Keywords: neutrophil, multisystem inflammatory syndrome in children, COVID-19, pediatrics, RNA sequencing, neutrophil extracellular traps
Graphical abstract
Highlights
-
•
Neutrophils in pediatric COVID-19 express an interferon-stimulated gene response
-
•
MIS-C neutrophils display a granulocytic myeloid-derived suppressor cell signature
-
•
Neutrophil activation and extensive spontaneous NETosis are seen in MIS-C
-
•
SARS-CoV-2 spike immune complexes, especially IgA immune complexes, trigger NETs
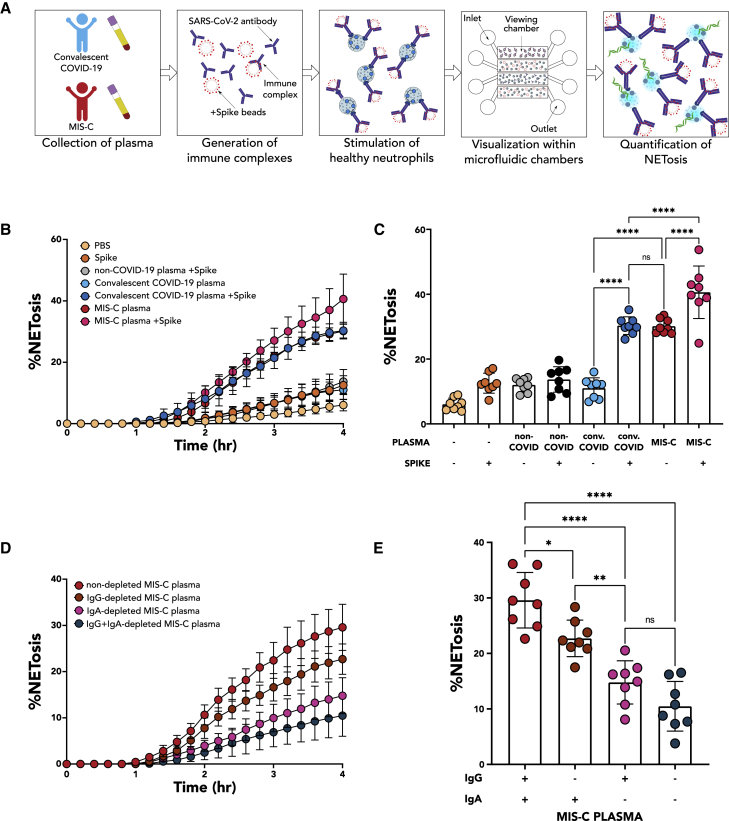
Boribong, LaSalle, et al. extensively profile gene expression, protein production, and functionality of neutrophils from children with acute COVID-19 or multisystem inflammatory syndrome in children (MIS-C) to define neutrophil responses driving distinct SARS-CoV-2 disease states. They propose a model whereby neutrophil activation elicits endothelial dysfunction and cardiovascular complications within MIS-C.
Introduction
Neutrophil activation is a central component of the immune response against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection; however, excessive neutrophil hyperactivation can contribute to severe COVID-19 in adults.1,2,3,4 The role of neutrophils in pediatric SARS-CoV-2 infections has not been defined. Children are less likely to suffer serious symptoms of acute COVID-19, but some children can develop a severe, life-threatening hyperinflammatory illness called multisystem inflammatory syndrome in children (MIS-C) weeks to months after the resolution of upper respiratory tract symptoms.5 MIS-C is defined by sepsis-like physiology with high fevers, systemic inflammation, and multiorgan involvement, likely caused by SARS-CoV-2 antigenemia originating from gastrointestinal sources of virus leaking across a permeable mucosal barrier into circulation.6 Children with MIS-C display signs of vascular injury, including elevated C-reactive protein, erythrocyte sedimentation rate, and D-dimer. 80% of children with MIS-C develop cardiac complications, including myocarditis, ventricular dysfunction, and coronary aneurysms,7,8 although mechanisms of cardiovascular injury are unclear.
Neutrophils play a significant role in innate immune responses against bacterial, fungal, and viral infections.9 However, at the same time, hyperactivation of neutrophils has been shown to be detrimental in severe diseases, including sepsis, acute respiratory distress syndrome (ARDS), and severe acute COVID-19 in adults.1,8,10,11 Not only can hyperactivation of polymorphonuclear leukocytes (PMNs) result in vascular and organ injury, but they can also contribute to hypercoagulation and thrombus formation.12 Understanding neutrophil activation in acute pediatric COVID-19 and MIS-C is essential to developing clinical guidelines and advances in treatment recommendations.
Extensive immune profiling has shown distinct hyperinflammatory patterns distinguishing acute pediatric COVID-19 from MIS-C;13,14,15 however, very little information on neutrophil activation has been included in these reports. This paucity of research on neutrophil activation in acute pediatric COVID-19 and MIS-C may be due to technical challenges with studying neutrophils: neutrophils have a very short lifespan and are extensively damaged by freezing. In contrast to peripheral blood mononuclear cells (PBMCs), neutrophils require isolation and fixation or analysis within hours of blood collection. Thus, a significant knowledge gap exists between pediatric COVID-19 and MIS-C and the contribution of neutrophils to disease pathogenesis.
Here, we isolated neutrophils immediately after blood collection from children with acute COVID-19 and MIS-C and healthy controls. We also extensively profiled gene expression, protein production, and neutrophil functionality to define neutrophil responses driving these distinct SARS-CoV-2 disease states in children.
Results
Cohort characteristics
Blood was collected from 152 children, including 43 children with COVID-19 confirmed by RT-PCR (Table S1), 31 children clinically diagnosed with MIS-C (Table S2), and 78 pediatric controls, including healthy children, children with convalescent COVID-19, and children with non-COVID-19 illness. The average ages of the acute pediatric COVID-19 cohort, the MIS-C cohort, and the pediatric control cohort were 14.2, 7.4, and 10.9 years, respectively. Gender, race, and ethnicity are shown in Table 1. Immediately after blood collection, plasma was collected, and neutrophils were enriched by negative selection. Neutrophils from each cohort were used for bulk RNA sequencing (RNA-seq; n = 48) and neutrophil extracellular trap (NET) assays (spontaneous NET, n = 21, and microfluidics, n = 24); plasma was used for proteomics (n = 55) and cell-free DNA measurements (n = 63) (Figure 1; Table 1).
Table 1.
Patient demographics and distribution of pediatric blood samples used for each assay
| Total enrolled (n = 152) | Pediatric controls (n = 78) | COVID-19 (n = 43) | MIS-C (n = 31) |
|---|---|---|---|
| Age, mean years (min, max) | 10.9 (1.2, 20) | 14.2 (0.04, 22.2) | 7.4 (0.17, 21) |
| Female gender, n (%) | 39 (50) | 14 (33) | 9 (29) |
| Male gender, n (%) | 39 (50) | 29 (67) | 22 (71) |
| Race, n (%) | |||
| White | 43 (55) | 15 (34) | 13 (42) |
| Black or African American | 3 (4) | 4 (9) | 5 (16) |
| Asian | 7 (9) | 2 (5) | 2 (6) |
| Ethnicity, n (%) | |||
| Hispanic or Latino | 28 (36) | 21 (51) | 13 (42) |
| Samples used in assays, n | |||
| RNA sequencing | 12 | 19 | 17 |
| Proteomics | 23 | 19 | 13 |
| Cell-free DNA | 21 | 23 | 19 |
| Spontaneous NETosis | 14 | 0 | 7 |
Figure 1.
Overview of cohort and experimental schematic
Whole blood samples were collected from three cohorts: (1) pediatric healthy controls, (2) pediatric acute COVID-19 infection, and (3) multisystem inflammatory syndrome in children (MIS-C). Plasma collected from whole-blood samples was used in proteomic assays to quantify levels of protein and cell-free DNA. Isolated neutrophils were used in bulk RNA sequencing and NETosis assays. Patient samples were used in individual or multiple assays.
Transcriptional profiling of neutrophils from pediatric COVID-19 and MIS-C
To confirm the successful negative selection enrichment of neutrophils, we used a digital cytometry method, CIBERSORTx, to estimate the fractions of major cell-type lineages in each bulk RNA-seq sample.16 We used the single-cell RNA-seq COVID-19 Bonn Cohort 2 reference dataset to generate cell-type-specific signatures for mature neutrophils, immature neutrophils, monocytes, T/natural killer (NK) cells, B cells, and plasmablasts.4 The total neutrophil fraction was defined as the sum of mature and immature neutrophils. High purity of neutrophils was confirmed in all samples that passed quality control (Figure S1A; Table S3), and the majority of samples had high mature-to-immature neutrophil ratios. Notably, there were no significant differences in any CIBERSORTx fractions between the patient cohorts (Figure S1B). Additionally, flow cytometry confirmed high neutrophil purity with minimal monocyte and lymphocyte contamination (Figure S1C) and no eosinophil contamination (Figure S1D).
Uniform manifold approximation and projection (UMAP) visualization of bulk RNA-seq samples did not reveal distinct groupings based on patient cohort or CIBERSORTx-estimated cell-type percentages. Though we did not perform clustering analyses due to the small number of rare samples, the UMAP landscape was broadly characterized by two major groupings, which represent distinct transcriptional profiles (Figure S1E).
Acute COVID-19 infection is associated with neutrophil interferon response signatures in children
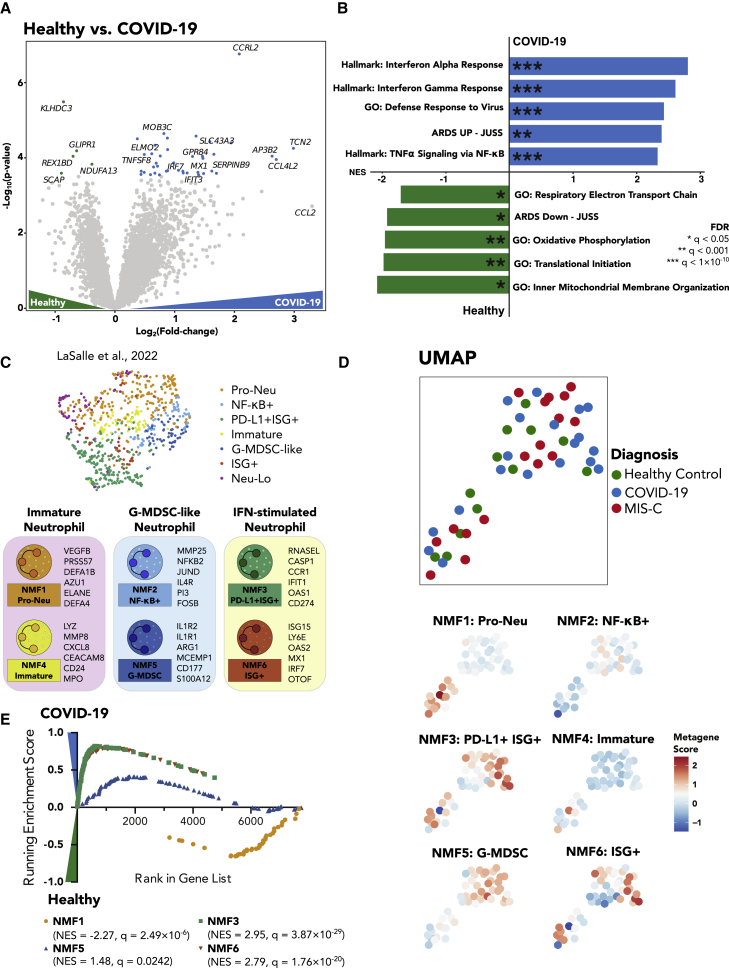
To identify differentially expressed genes and pathways in neutrophils associated with acute SARS-CoV-2 infection in children, we performed differential expression analysis and gene set enrichment analysis (GSEA) between COVID-19-positive samples and healthy samples (Table S4). The most highly upregulated gene in COVID-19-positive samples was CCRL2, a gene critical for CXCR2-dependent neutrophil recruitment and β2-integrin activation17 (Figure 2A). Following closely were other genes implicated in neutrophil chemotaxis, activation, and type I and II interferon responses such as ELMO2, GPR84, IRF7, IFIT3, and MX1. Overall, COVID-19-positive samples showed strong enrichment of pathways involved in antiviral response, including responses to interferon gamma (IFNγ) and IFNα (Figure 2B; Table S4).
Figure 2.
Acute pediatric COVID-19 neutrophils are marked by a robust interferon-stimulated gene signature
(A) Volcano plot showing differentially expressed genes between pediatric acute COVID-19 and healthy control samples. Color-coded points indicate genes that pass false discovery rate (FDR) correction with q < 0.05.
(B) Gene set enrichment analysis for the differentially expressed genes in (A). Bar lengths correspond to normalized enrichment score (NES), with positive NES values indicating enrichment in COVID-19 and negative values indicating enrichment in healthy pediatric controls. Asterisks denote significance as defined in the figure.
(C) UMAP of neutrophil bulk RNA sequencing (RNA-seq) samples from adults with acute COVID-19 and healthy controls from LaSalle et al.1 UMAP is color coded by cluster designation. Bottom, schematic illustrating several top genes associated with each neutrophil subtype. Neu-Lo, samples excluded from clustering based on estimated low neutrophil fraction by CIBERSORTx.
(D) (Top) UMAP plots of neutrophil bulk RNA-seq data. Each point is one bulk sample, and points are color coded according to disease status. (Bottom) UMAPs of all neutrophil samples that were sequenced with bulk RNA-seq. Samples are colored according to their expression score of each neutrophil state metagene.
(E) GSEA enrichment plots for the NMF1 (Pro-Neu), NMF3 (PD-L1+ ISG+), NMF5 (G-MDSC), and NMF6 (ISG+) signatures, the four NMF neutrophil signatures that passed FDR correction. Each point represents one gene in the gene signature. Positive scores denote enrichment in acute pediatric COVID-19, and negative scores denote enrichment in healthy controls.
In order to identify bulk neutrophil gene expression subtypes involved in response to SARS-CoV-2 infection in children, we utilized the bulk neutrophil gene signatures defined by an identical protocol in a large adult cohort,1 as well as previously defined neutrophil signatures from single-cell and bulk RNA-seq data.10,18 Pre-defined signatures included progenitor neutrophils (NMF1: Pro-Neu), nuclear factor κB (NF-κB)-activated neutrophils (NMF2: NF-κB+), IFN-stimulated neutrophils with high expression of PD-L1 (NMF3: PD-L1+ ISG+), immature activated neutrophils (NMF4: Immature), granulocytic myeloid-derived suppressor cell-like (G-MDSC) neutrophils (NMF5: G-MDSCs), and a second IFN-stimulated neutrophil signature with lower expression of PD-L1 and a non-overlapping set of IFN-stimulated genes (ISGs) (NMF6: ISG+)1,10,18 (Figure 2C). Since these signatures were also derived from bulk RNA-seq data, they represent transcriptional profiles of a mixture of neutrophil states rather than one specific subpopulation. When scoring each sample based on their average expression of NMF genes compared with their average expression of a control gene set (method details), we found that many of the NMF signatures produced groupings on the UMAP (Figure 2D), indicating the utility of these bulk signatures in defining neutrophil state signatures in children with COVID-19.
Next, we performed GSEAs comparing neutrophil subtype signature gene sets from children with COVID-19 and healthy controls (Table S4). Briefly, genes were ranked by signed log(p values) of the DESeq2 differential expression analysis between acute pediatric COVID-19 and healthy controls, and an enrichment score was calculated to indicate the degree to which the gene set was overrepresented at the top or bottom of the ranked list. We found significant enrichment of the NMF3: PD-L1+ ISG+ and the NMF6: ISG+ neutrophil subtypes in COVID-19 (Figure 2E), plus a weak enrichment for the NMF5: G-MDSC subtype in COVID-19. Conversely, the NMF1: Pro-Neu signature was slightly enriched in the healthy controls compared with pediatric COVID-19 (Figure 2E). Taken together, these results confirm that acute COVID-19 infection in children induces an ISG signature in neutrophils similar to early stages of infection in adults.1
MIS-C is associated with a strong G-MDSC signature having similarities to severe COVID-19 in adults and sepsis
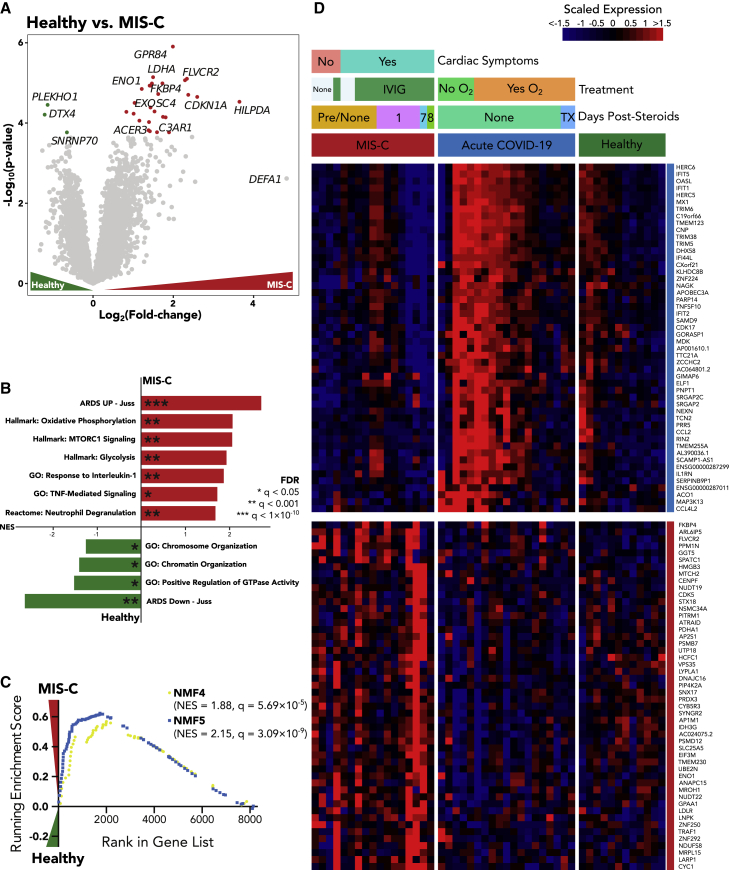
To define neutrophil activation in MIS-C, we performed differential expression analysis and GSEAs comparing MIS-C samples with healthy pediatric controls (Table S4). The most highly upregulated gene in MIS-C neutrophils was GPR84, a gene involved in reactive oxygen species (ROS) production and neutrophil activation19 (Figure 3A). Following closely was ACER3, a gene that has been shown to be upregulated in blood neutrophils of adult patients with ARDS, and several genes involved in glucose metabolism including LDHA and ENO1 and the complement C3 receptor C3AR1, which may modulate neutrophil recruitment, were also elevated in neutrophils from patients with MIS-C.20
Figure 3.
Neutrophils from patients with MIS-C display a strong G-MDSC signature and are characterized by altered metabolism
(A) Volcano plot showing differentially expressed genes between MIS-C and pediatric healthy control samples. Color-coded points indicate genes that pass FDR correction with q < 0.05.
(B) Gene set enrichment analysis for the differentially expressed genes in (A). Bar lengths correspond to NES, with positive NES values indicating enrichment in MIS-C and negative values indicating enrichment in healthy controls. Asterisks denote significance as defined in the figure.
(C) GSEA enrichment plots for the NMF4 (Immature) and NMF5 (G-MDSC) signatures, the two NMF neutrophil signatures that passed FDR correction.
(D) Heatmap displaying scaled RNA-seq expression values for gene markers distinguishing pediatric acute COVID-19 and MIS-C. The color coding on the right of the heatmap indicates whether the gene was differentially expressed in MIS-C versus COVID-19 and healthy or COVID-19 versus MIS-C or healthy. Color-coded bars above indicate disease status, cardiac involvement for patients with MIS-C, course of treatment, and days since the administration of steroids.
See also Figures S2 and S3 and Tables S3, S4, and S5.
The most highly enriched pathway in MIS-C was ARDS Up - Juss, a collection of genes that were found to be upregulated at least 3-fold in adult ARDS blood neutrophils compared with healthy controls10 (Figure 3B; Table S4). Accordingly, healthy control samples relative to MIS-C most prominently expressed the ARDS Down - Juss pathway, containing genes upregulated at least 3-fold in healthy controls over ARDS samples. In agreement with the top differentially expressed genes, the next most highly enriched pathways in MIS-C were oxidative phosphorylation, glycolysis, and mTORC1 signaling, suggesting that MIS-C neutrophils are highly metabolically active. Finally, MIS-C samples were also enriched for neutrophil degranulation, tumor necrosis factor (TNF) signaling, and interleukin-1 (IL-1) signaling, suggesting a mechanism by which neutrophils contribute to tissue damage in this highly inflammatory disease.
Next, we sought to determine which neutrophil subtypes were enriched in MIS-C. GSEA revealed a strong enrichment of the NMF5: G-MDSC subtype in MIS-C, the subtype that was most strongly associated with severe disease and death in adult patients with COVID-191,4,21 (Figure 3C; Table S4), and NMF4, suggesting that a proportion of patients with MIS-C may have high levels of a specific population of immature neutrophils (pre-neutrophils) that have been linked to increased NETosis, neutrophil degranulation, and activation.1
We performed a direct comparison of MIS-C and acute pediatric COVID-19 neutrophils to validate our findings from the individual healthy control comparisons (Table S4). Indeed, we observed a host of upregulated ISGs in acute COVID-19 that vanish in MIS-C, suggesting that the acute antiviral response has subsided in this late stage of disease, and instead, we find several markers of increased cellular metabolism consistent with neutrophil activation (Figure S2A; Table S4). In agreement with these gene markers, the ISG+ neutrophil states (NMF3 and NMF6) were found to be consistently highly enriched in acute pediatric COVID-19, while immature, NETosis-associated, and G-MDSC states (NMF1, NMF4, and NMF5, respectively) were enriched in MIS-C, albeit with more heterogeneity (Figures S2B, S2C, and S3). We confirmed the enrichment of these states in acute pediatric COVID-19 versus MIS-C using a second gene set analysis technique called sigPathway22 (Table S4; method details). Additional markers for pediatric acute COVID-19 and MIS-C are highlighted in Figure 3D.
Cardiac involvement is often associated with MIS-C but is not required for diagnosis; several patients in our cohort had either myocarditis, ventricular dysfunction, or coronary arterial aneurysm, with two patients requiring extracorporeal membrane oxygenation (ECMO). Thus, we next performed a hypothesis-generating differential expression analysis to identify neutrophilic transcriptional differences between patients with (n = 13) or without (n = 4) cardiac symptoms of MIS-C (Figure S3A). Several hallmark genes of sepsis and antigen presentation were enriched in cardiac MIS-C (Figure S3B), including several of the MHC II genes (HLA-DQB1, HLA-DRA, HLA-DMB). Other potential markers of severe cardiac MIS-C are CD163, which has been shown to be upregulated in PMNs of children with sepsis,23 ZBTB16, which is high in adults with sepsis and severe COVID-19,1,10 and OLAH, which is high in complex disease courses in sepsis.24
Neutrophils from children with MIS-C undergo high levels of spontaneous NETosis
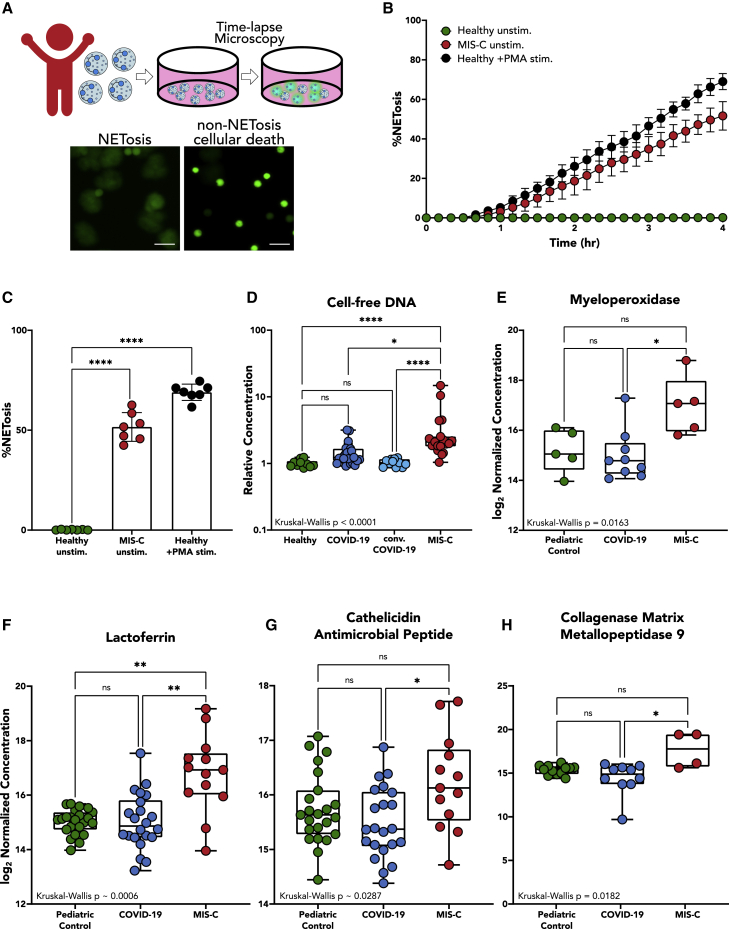
As many of the highly upregulated genes and pathways from children with MIS-C are involved in neutrophil activation and potentially destructive neutrophil effector functions such as NET formation, granule release, and ROS production, we sought to verify that the findings from the gene expression analysis corresponded to functional changes in neutrophils. We visualized neutrophil activation by studying spontaneous NET formation by neutrophils from patients with MIS-C compared with those from healthy donors. NETs were observed by microscopy using microfluidic technology; Neutrophils stimulated with 100 nM phorbol myristate acetate (PMA), a known, potent stimulator of NET formation, served as a positive control.
Neutrophils were isolated immediately after phlebotomy, plated without any added stimulants or neutrophil activators, and visualized via time-lapse microscopy for 4 h (experimental overview in Figure 4A). While neutrophils from healthy pediatric controls showed minimal spontaneous NETosis over the 4 h (Video S1, top), neutrophils isolated from children with MIS-C showed ∼500-fold higher rates of spontaneous NETosis (percentage of neutrophils undergoing NETosis: MIS-C 52% ± 7% versus healthy controls 0.17% ± 0.21%, p < 0.0001; Figures 4B and 4C; Video S1, bottom). The spontaneously formed NETs in MIS-C neutrophils were similar to PMA-stimulated NETs from healthy pediatric children, reaching markedly elevated levels of NET formation (69% ± 4% NETosis; Figures 4B and 4C).
Figure 4.
Fluorescence microscopy, cell-free DNA, and plasma protein markers implicate high levels of NETosis in MIS-C disease pathology
(A) Neutrophils were isolated from patients with MIS-C or healthy controls and plated on a 96-well plate without any stimulus to measure neutrophil activation via spontaneous NET release. Scale bar: 50 μM.
(B) Temporal dynamics of NET release in unstimulated neutrophils from healthy children (n = 7) and children with MIS-C (n = 7), as well as healthy neutrophils stimulated with 100 nM PMA (n = 7).
(C) End-point percentage of NETosis in unstimulated neutrophils from healthy children and children with MIS-C and healthy neutrophils stimulated with 100 nM PMA.
(D) Quantification of circulating cell-free DNA in plasma of healthy patients (n = 10), children with COVID-19 (n = 19), children with convalescent COVID-19 (n = 11), and children with MIS-C (n = 19). Displayed as fold increase from baseline (healthy controls).
(E–H) Peak values of myeloperoxidase (E), lactoferrin (F), cathelicidin antimicrobial peptide (G), and collagenase matrix metallopeptidase 9 (H) quantified in plasma from pediatric controls (n = 23), children with acute COVID-19 (n = 19), and children with MIS-C (n = 13).
(B) Mean values and SD are presented. (C) One-way ANOVA with multiple comparisons. Mean values and SD are presented. (D)–(H) Median values are shown with interquartile ranges; significance was determined by Kruskal-Wallis. Statistical significance is defined as ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001.
See also Video S1.
(Top) Spontaneous NETosis of neutrophils isolated from a healthy pediatric patient and (Bottom) a child with MIS-C captured by fluorescence (brightfield and FITC) microscopy. Extracellular DNA stained with SYTOX green. Scale bar = 100 μM.
To corroborate evidence of NETosis in MIS-C, we probed for markers of NET release. We found that children with MIS-C had significantly higher levels of relative cell-free DNA within their plasma compared to children with acute COVID-19, children with convalescent COVID-19 (p = 0.0160) and healthy children (p < 0.0001) (Figure 4D). Though cell-free DNA is non-specific and could come from other forms of cell death, recent studies showed that circulating cell-free DNA in severe adult COVID-19 is primarily derived from hematopoietic cells, specifically neutrophils.25 We also used mass spectrometry to quantify levels of protein markers of NET release. When comparing plasma from children with acute COVID-19, MIS-C, and non-COVID-19 illnesses, significant differences were detected in levels of myeloperoxidase (MPO) (p = 0.0163); lactoferrin (p < 0.0182); collagenase matrix metallopeptidase 9 (MMP9) (p = 0.0006); and cathelicidin anti-microbial peptide (CAMP) (p = 0.0287) (Figures 4E–4H), with children with MIS-C displaying elevated levels of each of these enzymes in circulation.
Circulating anti-SARS-CoV-2 immune complexes stimulate NETosis
Current models of MIS-C pathogenesis suggest that the characteristic immune hyperactivation of the disease is driven by persistent circulating spike antigenemia6 without viremia26 months after the initial SARS-CoV-2 infection. In one study, viral persistence in the gastrointestinal (GI) tract led to the release of zonulin, a biomarker of intestinal permeability, which subsequently led to leakage of viral antigens into the bloodstream and subsequent immune hyperactivation.6 Thus, we hypothesized that SARS-CoV-2 antigen:antibody immune complexes (ICs) could be responsible for triggering NETosis in the vasculature, contributing to cardiovascular pathology seen in MIS-C. To probe this mechanism of increased NET formation in children with MIS-C, neutrophils isolated from healthy controls were treated for 4 h with ICs generated by incubating patient-derived plasma with SARS-CoV-2 spike antigen (experimental overview, Figures 5A and S4). We observed low levels of NETosis when neutrophils were stimulated with spike antigen plus buffer (12% ± 3%; Figures 5B and 5C) or plasma from non-COVID-19 controls without spike antigen (12% ± 2%) and with spike antigen (14% ± 4%), likely due to the absence of spike-specific antibodies and ICs in these children (Figures 5B and 5C). We then stimulated neutrophils with convalescent COVID-19 plasma, both in the presence and in the absence of spike antigen, to determine if SARS-CoV-2 antibodies alone could induce NETosis, and we again observed low levels of NETs in these stimulated neutrophils (11% ± 3%; Figures 5B and 5C), indicating that neither antibodies nor antigen alone will induce NETosis.
Figure 5.
SARS-CoV-2 spike immune complexes trigger NETosis
(A) Plasma was collected from several children with non-COVID-19 illness, convalescent COVID-19, and MIS-C and pooled together. Pooled plasma was diluted at 1:10 and incubated with beads coated with spike proteins to generate immune complexes or without the presence of spike protein to measure the stimulation of plasma alone. Neutrophils were isolated from healthy children, stimulated, and visualized within four viewing channels of a microfluidic device.
(B) Temporal dynamics of NET release in neutrophils (n = 8) stimulated with PBS, PBS-treated spike beads, non-COVID-19 plasma, non-COVID-19-plasma-coated spike beads, convalescent COVID-19 plasma, convalescent COVID-19-plasma-coated spike beads, MIS-C plasma, and MIS-C-plasma-coated spike beads.
(C) End-point percentage of NET release in neutrophils (n = 8) stimulated with conditions described in (B).
(D) Temporal dynamics of NET release in neutrophils (n = 8) stimulated with non-depleted MIS-C plasma, IgG-depleted MIS-C plasma, IgA-depleted MIS-C plasma, and MIS-C plasma depleted of both IgG and IgA.
(E) End-point percentage of NET release in neutrophils (n = 8) stimulated with plasma as described in (D).
(B-E) Mean values and SD are presented. (C and D) Significance was determined by one-way ANOVA with multiple comparisons. Statistical significance is defined as ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001.
See also Figures S4 and S5 and Videos S2 and S3.
However, the addition of spike antigen to convalescent COVID-19 plasma induced significant NETosis (30% ± 3%) (Video S2, top) compared with convalescent COVID-19 plasma alone (p < 0.0001) (Figures 5B and 5C). This level of NETosis was comparable with that seen when healthy neutrophils were stimulated with MIS-C plasma (30% ± 1%; Video S2, bottom). This increase in NET formation is likely due to the formation of SARS-CoV-2-specific antibody:antigen ICs in the sample. To further verify this claim, we added additional spike antigen to MIS-C plasma, which stimulated an even greater amount of NET formation (40% ± 8%; Figures 5B and 5C). Similarly, we found that plasma from healthy, vaccinated children alone did not stimulate neutrophils but that the exogenous addition of spike beads to their vaccine-generated antibodies stimulated NETosis (29% ± 5%; Figure S4). These results provide further evidence of circulating spike antigen in patients with MIS-C, which triggers the formation of inflammatory ICs and subsequent neutrophil activation.
(Top) NETosis of isolated neutrophils isolated from a healthy patient stimulated with convalescent COVID-19 plasma-treated Spike beads and (Bottom) stimulated with MIS-C plasma captured by fluorescence (brightfield and FITC) microscopy. Extracellular DNA stained with SYTOX green. Scale bar = 100 μM.
To better understand which antibody class of anti-spike ICs was driving NETosis in MIS-C, we depleted either immunoglobulin G (IgG)-ICs, IgA-ICs, or both IgG- and IgA-ICs from MIS-C plasma and quantified NETosis. Compared with non-depleted MIS-C plasma (29% ± 5%), IgA depletion (14% ± 3%) resulted in the greatest decrease in NET formation (p < 0.0001), although IgG depletion (22% ± 3%) contributed to a lesser degree (p = 0.0145) (Figures 5D and 5E).
Together, these results suggest that SARS-CoV-2 antigenemia in the setting of a mature humoral response appears to instigate excessive neutrophil activation with hyperinflammatory and detrimental intravascular NET formation in MIS-C.
Discussion
In this study, we report the first in-depth characterization of neutrophils in pediatric acute COVID-19 and MIS-C. Utilizing neutrophil bulk RNA-seq, plasma protein expression, and functional analyses of neutrophils, we highlight major differences in the neutrophil response that help to explain the divergent presentations of these SARS-CoV-2-related diseases and provide mechanistic insight into neutrophil activation by SARS-CoV-2 antigen:antibody ICs in MIS-C.
MIS-C, though SARS-CoV-2-related, presents as a completely distinct set of symptoms and neutrophil phenotypes from acute COVID-19 in both children, shown here, and previous reports in adults.1 MIS-C occurs weeks to months after the initial infection, with a notable absence of viremia,26 in contrast to acute severe COVID-19.27 Instead, MIS-C is linked to SARS-CoV-2 antigenemia without replication-competent virus in a mature humoral response setting. Studies have demonstrated that viral RNA is still detected in the stool weeks after infection or exposure.6,28 In children who develop MIS-C, viral persistence is associated with increased gut permeability through zonulin release by gut epithelial cells,6 allowing viral antigens to escape into the bloodstream, where primed antibodies await. The formation of ICs triggers a hyperinflammatory immune response with neutrophil activation and NET formation within the vasculature (Figure 6).
Figure 6.
Schematic of the role of neutrophils in pediatric COVID-19 and MIS-C
Acute SARS-CoV-2 infection begins in the respiratory epithelium, and interferon signaling induces ISG+ neutrophils in circulation. Weeks later, viral persistence in the gut lumen results in zonulin release from gut epithelial cells, leading to loss of tight junctions and SARS-CoV-2 antigen leakage. Finally, immune complexes trigger NETosis and induce G-MDSCs, resulting in endothelial dysfunction and cardiovascular complications.
As a result of this distinct mechanism, neutrophils in MIS-C are characterized by upregulated genes and pathways associated with G-MDSCs and the formation of NETs, which is highly distinct from the antiviral gene signature seen in acute pediatric COVID-19. Neutrophils in patients with MIS-C more closely resemble those found in patients with non-viral ARDS10 or sepsis,21 rather than mild acute COVID-19, in that they appear to take on an immature, T cell-suppressive phenotype. This activated and destructive phenotype is hypothesized to cause the systemic damage seen in patients with MIS-C, with potential mechanisms being the interaction of neutrophil Fc receptors with SARS-CoV-2 antigen ICs, or TNF-ɑ signaling via NF-κB, which in excess can dysregulate T cells29 or activate further NETosis. Indeed, increased NF-κB signaling has been shown to be upregulated in monocytes and dendritic cells in MIS-C, especially in severe cardiac MIS-C,30 but prior reports have not examined neutrophils. Activated neutrophils have been implicated in the pathology of myocarditis through infiltration into the heart, with evidence of NETs found in endomyocardial biopsies from patients with myocarditis.31,32,33 As activated neutrophils and viral particles have been found in the myocardium postmortem in patients with MIS-C,34,35 increased antigen presentation by neutrophils and upregulation of septic responses by neutrophils may play a role in pathology driving severe cardiac complications of MIS-C. Importantly, this neutrophil activation can modulate B cell function, promoting autoimmune processes and contributing to immune activation in MIS-C36 and potentially long COVID-19.37 The role of the neutrophils in autoimmunity has already been established in conditions like anti-neutrophil cytoplasmic antibody-associated (ANCA-associated) vasculitis, Behcet’s disease, and systemic lupus erythematosus.38
In this study, we also provide visual evidence of high rates of spontaneous NETosis in neutrophils from children with MIS-C with imaging experiments performed within hours of fresh whole-blood collection. Furthermore, we provide mechanistic insight into the abundant NET formation by showing that SARS-CoV-2 antigen:patient-derived antibody ICs, predominantly anti-spike IgA-ICs, can trigger NETosis in healthy donor neutrophils. While IgA plays a key role in neutralizing SARS-CoV-2 at the mucosal surface,39 increased levels of IgA in circulation have been associated with severe COVID-19 in adults, with a corresponding increase in neutrophilic inflammatory responses.40,41 IgA can be a potent inducer and potentiator of NET formation42,43 but has not previously been implicated in MIS-C pathogenesis.
The observed NETosis has several implications for the disease pathology observed in MIS-C. Although NETs are effective at capturing and destroying pathogens at mucosal barriers, they can be a source of excessive inflammation and coagulation when released in circulation. First, NET components such as MPO can generate ROS and directly damage endothelium through several mechanisms, leading to tissue factor (TF) release. NETs themselves often contain TFs;44 thus, both NETs and the damaged endothelium can initiate the extrinsic coagulation cascade. Second, the abundant extracellular DNA can serve as a damage-associated molecular pattern (DAMP),45 triggering additional inflammation alongside inflammatory cytokine production, and the negative charge can initiate the intrinsic coagulation cascade.46 Taken together, these mechanisms can serve as a scaffold for platelets, induce hypercoagulability and endothelial dysfunction and lead to thrombosis, vasculitis, and cardiovascular complications, providing a uniting framework for MIS-C pathogenesis (Figure 6).
Neutrophil activation in MIS-C starkly contrasts the ISG profile seen in neutrophils from children with acute COVID-19. In children, the antiviral response to SARS-CoV-2 is sufficient to clear the acute infection without severe symptoms, as evidenced by the mild or asymptomatic presentations in most cases. While live, infectious SARS-CoV-2 can be detected in the upper airways of children,47 accounting for the antiviral response, there is little to no viremia detected in pediatric COVID-19.26 This picture may be similar to that seen in adults with mild COVID-19. However, in adults with moderate to severe disease, impaired antiviral signaling can lead to viremia in a larger fraction of cases, resulting in a more severe presentation of acute COVID-19 in these individuals.48,49,50 Thus, containment of the virus in the upper respiratory tract in children likely prevents the progression to more severe circulating neutrophil phenotypes seen in life-threatening COVID-19 in adults, and it is unlikely that circulating neutrophils alone determine disease severity in acute pediatric COVID-19.
There is an increasing appreciation of neutrophil activation in severe COVID-19 in adults, and therapies are being designed to target neutrophil hyperactivation. However, in most children, their immune system can already manage SARS-CoV-2 due to a strong IFN-mediated antiviral response.14,15 In both adults and children, SARS-CoV-2 viremia or antigenemia and the resultant ICs appear to be responsible for deleterious NET formation. While therapies blunting excessive neutrophil activation could improve clinical outcomes in severe disease, mitigating the antigenemia or viremia will be critical. Previous studies have indicated that preventing SARS-CoV-2 antigen leakage from the gut with the zonulin antagonist larazotide may be effective in preventing antigenemia-included hyperinflammation in MIS-C.6,51 While vaccination and avoidance of infection are the best prevention strategies, if SARS-CoV-2 infection does occur and MIS-C ensues, future studies may explore combination therapies that both prevent antigenemia by enhancing tight junctions in gut epithelium and simultaneously inhibiting systemic NET formation to improve upon the current treatment paradigm.
In summary, our study provides much-needed mechanistic insight into the drivers of pathology in MIS-C and its departure from mild acute SARS-CoV-2 infection in children. We link existing knowledge of gut mucosal breakdown and antigenemia in MIS-C to increased IC-dependent NETosis and a shift away from the antiviral state toward a sepsis-like G-MDSC phenotype, highlighting potential connections between neutrophil activation and the characteristic cardiovascular pathology to suggest pathways of intervention to improve the standard of care.
Limitations of the study
We acknowledge several limitations of our study. First, because of the nature of the pandemic and logistics around sample collection and processing of neutrophils, we chose to utilize bulk transcriptomics due to the technical challenges of performing single-cell RNA-seq on neutrophils, so each sample represents a mixture of neutrophils with distinct gene expression programs. However, our high-purity double isolation method left little contamination with other cell types. Second, we acknowledge the small cohort size, although it represents the largest cohort of its kind, due to the low incidence rate of the disease. Third, based on the sample size limitation, we were unable to define new gene expression states that may have been specific to pediatric neutrophils and had to rely on the adult-derived signatures. However, that work demonstrated a relatively small set of conserved neutrophil states across many different disease contexts. Relatedly, we used GSEA to determine the enrichment of these previously defined states. This method can only indicate relative enrichment and may overestimate effect sizes; however, it currently remains an integral component of RNA-seq analysis. Fourth, MIS-C can present heterogeneously, and there was a mixture of treatment regimens and disease duration among the patients, which may have resulted in greater heterogeneity in the gene expression profiles. Fifth, we acknowledge that the concentration of spike protein used to elicit NETosis may not directly align with levels of antigenemia in patients with MIS-C; however, these spike beads allowed us to test the impact of ICs on neutrophil activation. All limitations considered, numerous technical challenges and time-sensitive issues were overcome in this study to allow for deep characterization of neutrophil profiles in acute pediatric COVID-19 and MIS-C, which can guide future studies, including utilizing single-cell technologies.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| FITC anti-human CD66b | BioLegend | CAT# 305103; RRID:AB_314495 |
| Pacific Blue™ anti-human CD14 Antibody | BioLegend | CAT# 325615; RRID:AB_830688 |
| Biological samples | ||
| Patient samples used in this study are described in Tables 1, S3, and S4 | Massachusetts General Hospital | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| DRAQ5® | Cell Signaling Technology | CAT #4084 |
| SYTOX™ Green Nucleic Acid Stain | Invitrogen | CAT# S7020 |
| Phorbol 12-myristate 13-acetate | Sigma Aldrich | CAT# P1585 |
| Ficoll® Paque Plus | Cytiva | CAT# 17-1440-03 |
| Buffer TCL | Qiagen | CAT# 1031576 |
| β-mercaptoethanol | Sigma Aldrich | CAT# M6250 |
| Hoechst 33,342 Solution | Thermo Fisher Scientific | CAT# 62249 |
| Critical commercial assays | ||
| EasySep™ Direct Human Neutrophil Isolation Kit | STEMCELL Technologies | CAT# 19666 |
| HetaSep™ | STEMCELL Technologies | CAT# 07806 |
| Qubit dSDNA High Sensitivity Assay Kit | Invitrogen | CAT #Q32854 |
| High-Sensitivity DNA Bioanalyzer Kit | Agilent | CAT# 5067-4626 |
| Nextera XT Library Prep kit | Illumina | CAT# FC-131-1024 |
| Deposited data | ||
| Neutrophil bulk RNAseq analyzed data | This manuscript | GEO: GSE217370https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE217370 |
| Adult COVID-19 Neutrophil bulk RNA-seq data | LaSalle et al. 2022 | GEO: GSE212041 |
| COVID-19 Neutrophil scRNA-Seq data | Schulte-Schrepping et al. 2020 | EGA accession -EGAS00001004571 |
| Sepsis Neutrophil scRNA-seq data | Reyes et al. 2021 | https://singlecell.broadinstitute.org/single_cell/study/SCP1492/ |
| ARDS Neutrophil RNA-Seq | Juss et al. 2016 | GEO accession GSE76293 |
| Lung cancer single-cell neutrophil RNA-seq data | Zilionis et al. 2019 | GEO: GSE127465 |
| Oligonucleotides | ||
| RT primer (DNA oligo) | IDT | 5′-AAGCAGTGGTATCAACGCAGAGTACT30VN-3′ |
| TSO primer (RNA oligo with LNA) | Qiagen | 5-AAGCAGTGGT ATCAACGCAGAG TACATrGrG + G-3′ |
| ISPCR (DNA oligo) | IDT | 5′-AAGCAGTGGT ATCAACGCAGAG T-3′ |
| Software and algorithms | ||
| Code and Data from this manuscript | This manuscript |
https://doi.org/10.5281/zenodo.7277479 https://doi.org/10.5281/zenodo.7277353 |
| FIJI Is Just ImageJ | NIH | https://imagej.net/software/fiji/ |
| IntelliCyt ForeCyt (v8.1) | Sartorius | https://www.sartorius.com/en/products/flow-cytometry/flow-cytometry-software |
| INSPIRE software | EMD Millipore | https://www.emdmillipore.com/US/en/20150212_143429 |
| IDEAS software | EMD Millipore | https://www.emdmillipore.com/US/en/20150212_144049 |
| AutoCAD | Autodesk | https://www.autodesk.com/products/autocad/overview |
| GTEx-TOPMed RNA-Seq pipeline | Broad Institute | https://github.com/broadinstitute/gtexpipeline/ |
| STAR v2.5.3a | Dobin et al., 2013 | https://github.com/alexdobin/STAR/releases/tag/2.5.3a |
| RSEM v1.3.0 | Li et al., 2011 | https://github.com/deweylab/RSEM/releases/tag/v1.3.0 |
| RNA-SeQC 2 | Graubert et al. 2021 | https://github.com/getzlab/rnaseqc |
| CIBERSORTx | Newman et al., 2019 | https://cibersortx.stanford.edu |
| DESeq2 v1.30.1 | Love et al., 2014 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| Fgsea | Korotkevich et al., 2019 | http://bioconductor.org/packages/release/bioc/html/fgsea.html |
| Prism 9.3.1 (471) | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| sigPathway v1.64.0 | Lai et al., 2022 | https://bioconductor.org/packages/release/bioc/html/sigPathway.html |
| Other | ||
| FluoSpheres™ NeutrAvidin™-Labeled Microspheres, 1.0 μm, yellow-green fluorescent (505/515), 1% solids | Invitrogen | CAT #F8776 |
| CaptureSelect™ IgA Affinity Matrix | Thermo Fisher Scientific | CAT #194288010 |
| Protein G Resin | GenScript | CAT #L00209 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Lael Yonker (lyonker@mgh.harvard.edu).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
MGH patient cohort description
Biospecimens were obtained from pediatric patients at Massachusetts General Hospital (MGH) under the institutional review board (IRB) approved ‘MGH Pediatric COVID-19 Biorepository’ (no. 2020P000955). Healthy pediatric controls were collected under the IRB-approved ‘the Center for Celiac Research Pediatric Biorepository’ (no. 2016P000949). Informed consent, and assent, when appropriate, were obtained in accordance with IRB guidelines from patients and/or parents/guardians before study enrollment. Patients classified as COVID-19 positive had positive SARS-CoV-2 RT-PCR results upon enrollment. Patients classified as MIS-C were diagnosed per CDC guidelines. Healthy children were children presenting for well-child visits, a vaccine appointment, or an endoscopy (with normal pathology), and are otherwise healthy. Non-COVID-19 pediatric controls were children that were sick (hospitalized or outpatient) during the pandemic but tested negative for SARS-CoV-2.
Method details
Neutrophil isolation and lysis
Blood samples were processed as previously described.1,52 Briefly, blood samples were collected in vacutainer tubes containing EDTA anticoagulant. Whole blood was centrifuged at room temperature at 1000g for 10 min with breaks activated to allow for plasma collection. The remaining blood was mixed 1:1 with HBSS and gently layered on top of Ficoll at a 2:1 ratio. This was centrifuged at room temperature at 1000g for 30 min with the brakes off. PBMCs were then removed, and neutrophils were isolated from the remaining blood pellet via negative selection using the EasySep Direct Neutrophil Isolation Kit (STEMCELL Technologies, Cat# 19666), following manufacturer’s directions. 1x105 cells were centrifuged at 300g for 5 min at room temperature with soft brakes activated. Neutrophils were lysed with 100 μL of RNA lysis buffer (TCL) (Qiagen, Cat# 1031576) with 1% β-mercaptoethanol and frozen at −80°C.
Validation of isolated neutrophil purity
Nucleated cells were collected from whole blood via red blood cell sedimentation using HetaSep solution (STEMCELL Technologies, Cat# 07906). Neutrophils were isolated as previously described (plasma centrifuged, PBMCs isolated and collected, and neutrophils isolated from remaining red blood cell pellet). Isolated neutrophils and nucleated cells were centrifuged at 300g for 5 min at room temperature with the brakes off. The cells were resuspended in 50 μL of FACS buffer (0.5% BSA in PBS) and then fixed in 1% paraformaldehyde in PBS. All fixed cells were then stained with CD66b (1:200 dilution) (Biolegend, Cat# 305103), CD14 (1:20 dilution) (Biolegend, Cat# 325615), and DRAQ5 (1:2000 dilution) (Cell Signaling Technology, Cat# 4084S). Data was obtained through the Amnis ImageStreamX Mark II imaging flow cytometer and INSPIRE software (EMD Millipore). The accompanying IDEAS software (EMD Millipore) was used to perform data analysis. Additionally, Giemsa staining was performed on the isolated neutrophils to ensure no contamination of eosinophils. The isolated neutrophil solution was prepared in PBS at a concentration of 1x105 cells/mL. Cytospin preparations were made at 600 RPM for 5 min, and the cells allowed to air dry overnight. The next morning, cells were stained with fixative, eosin, and methylene blue (Thermo Fisher Scientific, Cat# 9990701), washed with DI water, and left to dry overnight. The cells and slide were mounted in the morning. Images were captured using a ZEISS Stemi 305 stereo microscope.
Patient matched plasma isolation
Blood samples were processed as previously described.52 Briefly, blood samples were collected in EDTA vacutainer tubes. Plasma was collected by centrifuging tubes at 1000g for 10 min at room temperature with brakes activated. Aliquoted plasma was stored at −80°C.
Smart-Seq2 cDNA preparation
cDNA was prepared from bulk populations of 1x105 neutrophils as recently described1 using a modified reverse transcription step.53 Neutrophil lysates were thawed on ice, and 20 μL per sample was added to a 96-well plate prior to brief centrifugation. Following RNA purification with Agencourt RNAClean XP SPRI beads (Beckman Coulter, Cat# A63987), samples were resuspended in 4 μL of Mix-1 [Per 1 sample: 1 μL (10 μM) RT primer (DNA oligo) 5′–AAGCAGTGGTATCAACGCAGAGTACT30VN-3′; 1 μL (10 μM) dNTPs; 1μL (10%, 4 U/μL) recombinant RNase inhibitor; 1 μL nuclease-free water], denatured at 72°C for 3 min and immediately cooled on ice for 1 min. Next, 7 μL of Mix-2 [Per 1 sample: 0.75 μL nuclease-free water; 2 μL 5X RT buffer (Thermo Fisher Scientific, Cat# EP0753); 2 μL (5 M) betaine; 0.9 μL (100 mM) MgCl2; 1 μL (10 μM) TSO primer (RNA oligo with LNA) 5′-AAGCAGTGGTATCAACGCAGAGTACATrGrG + G-3′; 0.25 μL (40 U/μL) recombinant RNase inhibitor; 0.1 μL (200 U/μL) Maxima H Minus Reverse Transcriptase] was added. Reverse transcription was performed at 50°C for 90 min, followed by a 5-min incubation at 85°C. Next, 14 μL of Mix-3 [Per 1 sample: 1 μL nuclease-free water; 0.5 μL (10 μM) ISPCR primer (DNA oligo) 5′-AAGCAGTGGTATCAACGCAGAGT-3′; 12.5 μL 2X KAPA HiFi HotStart ReadyMix] was added to each well and amplification was performed at 98°C for 3 min, followed by 16 cycles of [98°C for 15 s, 67°C for 20 s, and 72°C for 6 min], and final extension at 72°C for 5 min. Primer removal and cDNA purification was performed using AgencourtAMPureXP SPRI beads (Beckman Coulter, Cat# A63881). Concentration measurements were determined with the Qubit dsDNA high sensitivity assay kit (Invitrogen, Cat# Q32854) on the Cytation 5 Microplate Reader (BioTek), and cDNA size distribution was measured using the High-Sensitivity DNA Bioanalyzer Kit (Agilent, Cat# 5067-4626).
Library construction and sequencing
Libraries were constructed with the Nextera XT Library Prep kit (Illumina, Cat# FC-131-1024) using custom indexing adapters53 in a 384-well PCR plate. Residual primer dimers were removed with an additional cleanup step. One pooled library containing 26 samples were sequenced on a NextSeq 500 sequencer (Illumina) using paired-end 38-base reads. Following quality control and acquisition of additional samples, a second pooled library containing 40 samples was sequenced using the same method.
Spontaneous NET release fluorescence microscopy data acquisition
Blood was processed from healthy pediatric and MIS-C patients as previously described.52 An aliquot of 1x104 cells in 50 μL RPMI (no FBS) was collected from the isolated neutrophils. Neutrophils were plated in a single well of a black, clear-bottom 96-well plate. 50 μL of RPMI was added to the well for the unstimulated neutrophils. For the stimulated neutrophils, 50 μL of phorbol myristate acetate (PMA) (Sigma Aldrich, Cat# P1585) was added to a final concentration of 100 nM. 50 μL of SYTOX green (Thermo Fisher Scientific, Cat# S7020) in RPMI was added to a final concentration of 2 μM. NETosis was imaged using the Personal AUtomated Lab Assistant (PAULA) Cell Imager (Leica Microsystems) placed in a 37°C incubator with 5% CO2. Brightfield and FITC images were taken every 10 min for 4 h to allow for temporal imaging of NET formation.
Cell-free DNA quantification
cfDNA quantification was performed using the Qubit dsDNA High Sensitivity Assay Kit (Invitrogen, Cat# Q32854) according to the manufacturer’s protocol. 98 μL of Qubit DNA dye was aliquoted per well of a 96-well black clear bottom plate (Corning, Cat# 3904). 2 μL of plasma sample was added to each well of the assay plate, and fluorescence was measured on a Cytation 5 Microplate reader at 523 nm. The concentration of cell-free DNA was analyzed relative to the average concentration of the healthy controls.
Plasma protein measurement
Samples were prepared as previously described.6 100 μg were denatured, reduced, alkylated, and purified using solid-phase-enhanced sample-preparation (SP3) technology before lysc and tryptic digest.54,55 25 μg of the resulting peptides were subsequently labeled using TMTpro reagents (Thermo Fisher Scientific) according to the manufacturer’s instructions.56,57 Labeled samples were combined and fractionated using a basic reversed phase HPLC.54 The fractions were analyzed via reversed phase LC-M2/MS3 on either an Orbitrap Fusion Lumos or an Orbitrap Eclipse mass spectrometer using the Simultaneous Precursor Selection (SPS) supported MS3 method58,59 essentially as described previously60 and using Real-Time Search when using the Orbitrap Eclipse.61 MS2 spectra were assigned using a SEQUEST-based62 in-house built proteomics analysis platform63 using a target-decoy database-based search strategy to assist filtering for a false-discovery rate (FDR) of protein identifications of less than 10%.64 Searches were done using the Uniprot human protein sequence database (UP000005640)64 using an in-house-built platform.62 Search strategy included a target-decoy database-based search in order to filter against a false-discovery rate (FDR) of protein identifications of less than 1%.64 An average signal-to-noise value of larger than 10 per reporter ion as well as with an isolation specificity59 of larger than 0.75 were considered for quantification. Lysozyme, Myeloperoxidase, collagenase matrix metallopeptidase 9, Lactoferrin, and Cathelicidin antimicrobial peptide were quantified. Protein concentration data were normalized as previously described.56
Design and fabrication of the microfluidic devices
Microfluidic devices used for time-lapse imaging of NETosis were designed using AutoCAD (Autodesk) and fabricated using standard photolithographic and soft lithography approaches (Figures S5A and S5B). Briefly, a master negative mold was prepared by spin-coating a silicon wafer with negative photoresist (SU-8, Microchem) to a thickness of 100 μm. The design was patterned onto the wafer by exposure to UV light through a mylar mask (Fineline Imaging) and developing away the unexposed areas. The silicon wafer was then used as a template for casting PDMS devices. PDMS base and primer were mixed at a ratio of 10:1 and poured over the wafer, degassed for at least 1 h, then baked overnight at 65°C. Inlets and outlets were punched using a 1 mm diameter biopsy punch (Harris UniCore, Millipore Sigma), and the entire device was liberated from the PDMS block using an 8 mm diameter punch. PDMS devices were treated with oxygen plasma and bonded irreversibly to oxygen plasma-treated glass-bottom well plates (Mattek) by heating to 75°C for 10 min.
Immune complex generation
Immune complexes were generated as previously described.40 Briefly, SARS-CoV-2 Spike proteins were biotinylated, conjugated to NeutrAvidin beads, and incubated with 1:10 diluted plasma samples.
Plasma IgG and IgA depletion
IgG and IgA were depleted from MIS-C plasma samples as previously described.40 Briefly, IgG was depleted using Protein A/G Agarose (Fisher Scientific, Cat# NC1466772) and IgA using CaptureSelect IgA Affinity Matrix (Thermo Fisher Scientific, Cat# 194288010). Pierce Centrifuge Columns were washed with 1X PBS and incubated overnight with the plasma samples. Depleted plasma was collected the following day by centrifugation of the columns. Control of a non-depleted plasma sample was treated identically but without adding the affinity matrix.
NETosis assay
10 μL of RPMI was loaded into the inlet port of the microfluidic device until a droplet was formed on the outlet port. RPMI was added to the wells until the devices were submerged. The plate was placed in a 37°C, 5% CO2 incubator until the neutrophil solution was prepared to be loaded. Neutrophils were isolated from a healthy patient cohort. 1–2 mL of whole blood was collected in EDTA vacutainer tubes, and neutrophils were isolated via negative selection using the Easysep Direct Neutrophil Isolation Kit, following the manufacturer’s directions. Isolated neutrophils were stained with 32 μM Hoechst 3342 dye for 10 min at 37°C, with 5% CO2. Stained neutrophils were resuspended in RPMI (no FBS) to a concentration of 1x10 7 cells/mL. 10 μL of cells were mixed with 10 μL of stimulant and 10 μL of SYTOX green in RPMI (no FBS) to a final concentration of 2 μM. 5 μL of this solution was loaded into the inlet port of the microfluidic device using a standard pipette tip until cells were observed to exit the outlet port. Brightfield and FITC fields were imaged at 10x magnification using a fully automated fluorescent Nikon TiE inverted wide-field microscope with a bio chamber heated to 37°C with 5% CO2. Each device was imaged every 12 min for 4 h to allow for temporal imaging of NET formation (Figure S5C). An initial image was taken with brightfield and DAPI at the beginning of the experiment to count fluorescently stained neutrophils.
Quantification and statistical analysis
RNA-seq alignment
Raw FASTQ files were aligned to a custom genome using the Terra platform of the Broad Institute. Alignment was performed using STAR v2.5.3a, and expression quantification was performed using RSEM v1.3.0. The custom FASTA was generated from the Homo sapiens genome assembly GRCh38 (hg38, excluding ALT, HLA, and Decoy contigs according to the Broad Institute GTEx-TOPMed RNA-seq pipeline, https://github.com/broadinstitute/gtex-pipeline/) with an appended SARS-CoV-2 genome. GENCODE v35 with the appended SARS-CoV2 GTF was used for annotation.
Quality control
RNA-SeQC 265 (https://github.com/getzlab/rnaseqc) was used to calculate quality control metrics for each sample. Samples were excluded if they did not meet the following criteria: 1) percentage of mitochondrial reads less than 20%, 2) greater than 10,000 genes detected with at least 5 unambiguous reads, 3) median exon CV less than 1, 4) exon CV MAD less than 0.75, 5) exonic rate greater than 25%, 6) median 3′ bias less than 90%. One sample was excluded for having low neutrophil content and high B cell contamination as estimated by CIBERSORTx, described in the next section. Only one sample was kept per patient: if more than one passed quality control, the earlier or pre-treatment sample was selected. Thus, out of 55 sequencing reactions, 48 samples were carried forward for analysis. Genes were included in the analysis if they were expressed at a level of 0.1 TPM in at least 20% of samples and if there were at least 6 counts in 20% of samples. In total, 15,406 genes passed the filtration criteria.
Neutrophil fraction estimation and contamination control
CIBERSORTx16 was used to estimate total neutrophil, mature neutrophil, immature neutrophil, T/NK cells (either T or NK, due to similar gene expression profiles), B, plasmablast, and monocyte content in each sample. We generated a signature matrix from the single-cell RNA-seq data of PBMCs and neutrophils from whole blood from Cohort 2 of the Schulte-Schrepping et al. dataset (2)4 using the pseudobulking method previously described.1 CIBERSORTx was run using default parameters.
Dimensionality reduction and visualization
PCA and UMAP were performed in R using prcomp() and umap() with default parameters.
Differential expression analysis
Differential expression analyses were performed using the DESeq2 package in R66 without additional covariates.
Gene set enrichment analysis and sigPathway
Gene set enrichment analysis was performed using the fgsea package in R using MSigDB Release v7.2 pathways from the H, C5 GO BP databases. We also added custom MSigDB pathways using the keyword “neutrophil,” and the gmt file is available on Zenodo. In addition, we added gene sets defining neutrophil states in ARDS,10 blood and lung tissue of lung cancer patients,18 sepsis (20), single-cell neutrophil clusters in COVID-19 4, and bulk RNA-seq signatures of neutrophils in COVID-19 1. sigPathway was performed using the sigPathway v1.64.0 package in R using the custom neutrophil state gene sets described above with all other parameters set to default.
Sample pathway scoring
Bulk RNA-seq samples were scored according to their expression of NMF neutrophil state gene signatures according to a previously described method used to control for sample complexity.67 We defined the pathway score for each sample as the average expression of the genes in the gene set minus the average expression of genes in a control gene set. Genes were ranked according to average expression across all samples and divided into 25 bins, and for each gene in the NMF signature, 100 different genes were selected from the same bin to create a control gene set with comparable expression levels which is 100-fold larger.
Disease marker gene selection and heatmap
To identify genes uniquely associated with acute pediatric COVID-19 or MIS-C, we performed three differential expression analyses: MIS-C vs. COVID-19 and controls, COVID-19 vs. MIS-C and controls, and controls vs. MIS-C and COVID-19. We filtered the results for padj >0.05 and only selected positive markers (log(fold-change) greater than 0) and capped the lists at 50 genes. There were no significant marker genes for control neutrophils. Heatmaps of the gene markers for the two diseases were generated with the pheatmap package in R. Row (genes) are ordered according to p value, and columns are ordered according to the clinical subtypes shown in the legend.
Quantification of NETosis
Detection of NETs was performed automatically on the FITC channel using the TrackMate plugin in FIJI.68,69 As neutrophils undergo NETosis, the nucleus undergoes condensation, followed by decondensation of the chromatin. Next, the cell’s nuclear membrane breaks down, allowing for SYTOX green to stain the DNA and (Figure S5D). NETs are quickly dispersed, and the fluorescence is diffused (Figure S5E). Non-NETosis cellular death can become stained with SYTOX green as the membrane breaks down, however, the fluorescence signal is stable and long-lasting (Figure S5F). The methodology was validated using manual tracking within FIJI68 to determine the average length of time of cells undergoing NETosis against cells undergoing non-NETosis cellular death. A cutoff differentiating the two was defined at 1.5 h (Figure S4G). Thus, NETs were defined as tracked cells with a duration ≤1.5 h, and non-NETosis cellular death was defined as tracked cells with a course> 1.5 h (Movie S3). A total number of neutrophils was counted using the DAPI channel at time t = 0 68. The percentage of NETs was calculated using the number of NETs released at a specific time point divided by the total number of neutrophils.
Statistical analysis
To compare multiple groups, results were compared by 1-way ANOVA with multiple comparisons or by Kruskal-Wallis with post-hoc comparisons between the groups completed using Dunn’s test and corrected for multiple comparisons with Tukey’s method in GraphPad Prism v9. Statistical tests used are noted in the figure caption. Statistical significance is defined as ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001, unless otherwise noted in the figure.
Acknowledgments
We acknowledge and thank the study participants and their families. Additionally, we would like to acknowledge the efforts of all members of the MGH Pediatric COVID-19 Biorepository, especially Ms. Rosiane Lima, Ms. Madeleine Burns, Ms. Eva Farkas, Ms. Christina Lee, Ms. Aditi Sirsikar, and the Center for Celiac Research Pediatric Biorepository, especially Dr. Maureen M. Leonard and Ms. Victoria Kenyon, for their assistance in recruiting and processing biospecimens for the study. This work was funded by National Institute of General Medical Sciences grant T32GM007753 (T.J.L.); National Heart, Lung, and Blood Institute grant 5K08HL143183 (L.M.Y.); the Massachusetts General Hospital Executive Committee on Research; COVID-19 Clinical Trials Initiative grant (L.M.Y.); and the Department of Pediatrics at Massachusetts General Hospital for Children (L.M.Y.) The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIGMS or the NIH.
Author contributions
Conceptualization, M.S.-F. and L.M.Y.; methodology, B.P.B., T.J.L., Y.C.B., F.E., M.E.L., J.P.D., A.L.K.G., S.H., S.P., W.H., G.A., D.I., M.S.-F., and L.M.Y.; validation, B.P.B., T.J.L., Y.C.B., F.E., M.E.L., J.P.D., A.L.K.G., S.H., S.P., W.H., G.A., D.I., M.S.-F., and L.M.Y.; formal analysis, B.P.B., T.J.L., Y.CB., F.E., S.H., G.A., D.I., M.S.-F., and L.M.Y.; investigation, B.P.B., T.J.L., Y.C.B., F.E., M.E.L., J.P.D., A.L.K.G., S.H., S.P., W.H., A.G.E., A.F., G.A., D.I., M.S.-F., and L.M.Y.; resources, S.P., W.H., A.G.E., A.F., G.A., D.I., M.S.-F., and L.M.Y.; data curation, B.P.B., T.J.L., S.H., M.S.-F., and L.M.Y.; writing – original draft, B.P.B., T.J.L., M.S.-F., and L.M.Y.; writing – review & editing, B.P.B., T.J.L., Y.C.B., F.E., M.E.L., J.P.D., A.L.K.G., S.H., S.P., W.H., A.E., A.F., G.A., D.I., M.S.-F., and L.M.Y.; visualization, B.P.B., T.J.L., F.E., M.E.L., D.I., M.S.-F., and L.M.Y.; supervision, M.S.-F. and L.M.Y.
Declaration of interests
M.S.-F. receives funding from Bristol-Myers Squibb. G.A. is a founder of Seromyx Systems, Inc. A.F. is co-founder of and stockholder in Alba Therapeutics.
Inclusion and diversity
We worked to ensure gender balance in the recruitment of human subjects. We worked to ensure ethnic or other types of diversity in the recruitment of human subjects. One or more of the authors of this paper self-identifies as an underrepresented ethnic minority in their field of research or within their geographical location. One or more of the authors of this paper self-identifies as a gender minority in their field of research. One or more of the authors of this paper self-identifies as a member of the LGBTQIA+ community.
Published: November 21, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2022.100848.
Contributor Information
Moshe Sade-Feldman, Email: msade-feldman@mgh.harvard.edu.
Lael M. Yonker, Email: lyonker@mgh.harvard.edu.
Supplemental information
Data and code availability
-
•
Due to IRB consent limitations, raw sequencing data is not publicly available. However, the read count matrix and TPM matrix used in this study will be publicly available in GEO (Gene Expression Omnibus) under accession number GSE217370 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE217370) and Table S5. Additional Supplemental Items are available from Zenodo Data: https://doi.org/10.5281/zenodo.7277479. Raw sequencing data can be made available from the lead contact upon request.
-
•
All code required to run the analyses in this manuscript is deposited in Zenodo Data: https://doi.org/10.5281/zenodo.7277479 based on the associated Github repository (https://github.com/lasalletj/Pediatric_COVID_MISC_Neutrophils).
-
•
Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.
References
- 1.LaSalle T.J., Gonye A.L.K., Freeman S.S., Kaplonek P., Gushterova I., Kays K.R., Manakongtreecheep K., Tantivit J., Rojas-Lopez M., Russo B.C., et al. Longitudinal characterization of circulating neutrophils uncovers phenotypes associated with severity in hospitalized COVID-19 patients. Cell Rep. Med. 2022;3 doi: 10.1016/j.xcrm.2022.100779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aschenbrenner A.C., Mouktaroudi M., Krämer B., Oestreich M., Antonakos N., Nuesch-Germano M., Gkizeli K., Bonaguro L., Reusch N., Baßler K., et al. Disease severity-specific neutrophil signatures in blood transcriptomes stratify COVID-19 patients. Genome Med. 2021;13:7. doi: 10.1186/s13073-020-00823-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meizlish M.L., Pine A.B., Bishai J.D., Goshua G., Nadelmann E.R., Simonov M., Chang C.H., Zhang H., Shallow M., Bahel P., et al. A neutrophil activation signature predicts critical illness and mortality in COVID-19. Blood Adv. 2021;5:1164–1177. doi: 10.1182/bloodadvances.2020003568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schulte-Schrepping J., Reusch N., Paclik D., Baßler K., Schlickeiser S., Zhang B., Krämer B., Krammer T., Brumhard S., Bonaguro L., et al. Severe COVID-19 is marked by a dysregulated myeloid cell compartment. Cell. 2020;182:1419–1440.e23. doi: 10.1016/j.cell.2020.08.001. e1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.CDC . 2021. Information for Healthcare Providers about Multisystem Inflammatory Syndrome in Children (MIS-C)https://www.cdc.gov/mis/mis-c/hcp/index.html [Google Scholar]
- 6.Yonker L.M., Gilboa T., Ogata A.F., Senussi Y., Lazarovits R., Boribong B.P., Bartsch Y.C., Loiselle M., Rivas M.N., Porritt R.A., et al. Multisystem inflammatory syndrome in children is driven by zonulin-dependent loss of gut mucosal barrier. J. Clin. Invest. 2021;131:149633. doi: 10.1172/jci149633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feldstein L.R., Tenforde M.W., Friedman K.G., Newhams M., Rose E.B., Dapul H., Soma V.L., Maddux A.B., Mourani P.M., Bowens C., et al. Characteristics and outcomes of US children and adolescents with multisystem inflammatory syndrome in children (MIS-C) compared with severe acute COVID-19. JAMA. 2021;325:1074–1087. doi: 10.1001/jama.2021.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meghraoui-Kheddar A., Chousterman B.G., Guillou N., Barone S.M., Granjeaud S., Vallet H., Corneau A., Guessous K., de Roquetaillade C., Boissonnas A., et al. Two new neutrophil subsets define a discriminating sepsis signature. Am. J. Respir. Crit. Care Med. 2022;205:46–59. doi: 10.1164/rccm.202104-1027OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Y., Wang Q., Mackay C.R., Ng L.G., Kwok I. Neutrophil subsets and their differential roles in viral respiratory diseases. J. Leukoc. Biol. 2022;111:1159–1173. doi: 10.1002/jlb.1mr1221-345r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Juss J.K., House D., Amour A., Begg M., Herre J., Storisteanu D.M.L., Hoenderdos K., Bradley G., Lennon M., Summers C., et al. Acute respiratory distress syndrome neutrophils have a distinct phenotype and are resistant to phosphoinositide 3-kinase inhibition. Am. J. Respir. Crit. Care Med. 2016;194:961–973. doi: 10.1164/rccm.201509-1818OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pastorek M., Dúbrava M., Celec P. On the origin of neutrophil extracellular traps in COVID-19. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.821007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gómez-Moreno D., Adrover J.M., Hidalgo A. Neutrophils as effectors of vascular inflammation. Eur. J. Clin. Invest. 2018;48 doi: 10.1111/eci.12940. [DOI] [PubMed] [Google Scholar]
- 13.Hsieh L.E., Grifoni A., Sidney J., Shimizu C., Shike H., Ramchandar N., Moreno E., Tremoulet A.H., Burns J.C., Franco A. Characterization of SARS-CoV-2 and common cold coronavirus-specific T-cell responses in MIS-C and Kawasaki disease children. Eur. J. Immunol. 2021;52:123–137. doi: 10.1002/eji.202149556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ravichandran S., Tang J., Grubbs G., Lee Y., Pourhashemi S., Hussaini L., Lapp S.A., Jerris R.C., Singh V., Chahroudi A., et al. SARS-CoV-2 immune repertoire in MIS-C and pediatric COVID-19. Nat. Immunol. 2021;22:1452–1464. doi: 10.1038/s41590-021-01051-8. [DOI] [PubMed] [Google Scholar]
- 15.Vella L., Giles J.R., Baxter A.E., Oldridge D.A., Diorio C., Kuri-Cervantes L., Alanio C., Pampena M.B., Wu J.E., Chen Z., et al. Deep Immune Profiling of MIS-C demonstrates marked but transient immune activation compared to adult and pediatric COVID-19. medRxiv. 2020 doi: 10.1101/2020.09.25.20201863. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Newman A.M., Steen C.B., Liu C.L., Gentles A.J., Chaudhuri A.A., Scherer F., Khodadoust M.S., Esfahani M.S., Luca B.A., Steiner D., et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat. Biotechnol. 2019;37:773–782. doi: 10.1038/s41587-019-0114-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Del Prete A., Martínez-Muñoz L., Mazzon C., Toffali L., Sozio F., Za L., Bosisio D., Gazzurelli L., Salvi V., Tiberio L., et al. The atypical receptor CCRL2 is required for CXCR2-dependent neutrophil recruitment and tissue damage. Blood. 2017;130:1223–1234. doi: 10.1182/blood-2017-04-777680. [DOI] [PubMed] [Google Scholar]
- 18.Zilionis R., Engblom C., Pfirschke C., Savova V., Zemmour D., Saatcioglu H.D., Krishnan I., Maroni G., Meyerovitz C.V., Kerwin C.M., et al. Single-cell transcriptomics of human and mouse lung cancers reveals conserved myeloid populations across individuals and species. Immunity. 2019;50:1317–1334.e10. doi: 10.1016/j.immuni.2019.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sundqvist M., Christenson K., Holdfeldt A., Gabl M., Mårtensson J., Björkman L., Dieckmann R., Dahlgren C., Forsman H. Similarities and differences between the responses induced in human phagocytes through activation of the medium chain fatty acid receptor GPR84 and the short chain fatty acid receptor FFA2R. Biochim. Biophys. Acta. Mol. Cell Res. 2018;1865:695–708. doi: 10.1016/j.bbamcr.2018.02.008. [DOI] [PubMed] [Google Scholar]
- 20.Metzemaekers M., Gouwy M., Proost P. Neutrophil chemoattractant receptors in health and disease: double-edged swords. Cell. Mol. Immunol. 2020;17:433–450. doi: 10.1038/s41423-020-0412-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reyes M., Filbin M.R., Bhattacharyya R.P., Sonny A., Mehta A., Billman K., Kays K.R., Pinilla-Vera M., Benson M.E., Cosimi L.A., et al. Plasma from patients with bacterial sepsis or severe COVID-19 induces suppressive myeloid cell production from hematopoietic progenitors in vitro. Sci. Transl. Med. 2021;13:eabe9599. doi: 10.1126/scitranslmed.abe9599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tian L., Greenberg S.A., Kong S.W., Altschuler J., Kohane I.S., Park P.J. Discovering statistically significant pathways in expression profiling studies. Proc. Natl. Acad. Sci. USA. 2005;102:13544–13549. doi: 10.1073/pnas.0506577102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Groselj-Grenc M., Ihan A., Derganc M. Neutrophil and monocyte CD64 and CD163 expression in critically ill neonates and children with sepsis: comparison of fluorescence intensities and calculated indexes. Mediators Inflamm. 2008;2008 doi: 10.1155/2008/202646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Banerjee S., Mohammed A., Wong H.R., Palaniyar N., Kamaleswaran R. Machine learning identifies complicated sepsis course and subsequent mortality based on 20 genes in peripheral blood immune cells at 24 H post-ICU admission. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.592303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Andargie T.E., Tsuji N., Seifuddin F., Jang M.K., Yuen P.S., Kong H., Tunc I., Singh K., Charya A., Wilkins K., et al. Cell-free DNA maps COVID-19 tissue injury and risk of death and can cause tissue injury. JCI Insight. 2021;6:147610. doi: 10.1172/jci.insight.147610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yonker L.M., Neilan A.M., Bartsch Y., Patel A.B., Regan J., Arya P., Gootkind E., Park G., Hardcastle M., St John A., et al. Pediatric severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): clinical presentation, infectivity, and immune responses. J. Pediatr. 2020;227:45–52.e5. doi: 10.1016/j.jpeds.2020.08.037. e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fajnzylber J., Regan J., Coxen K., Corry H., Wong C., Rosenthal A., Worrall D., Giguel F., Piechocka-Trocha A., Atyeo C., et al. SARS-CoV-2 viral load is associated with increased disease severity and mortality. Nat. Commun. 2020;11:5493. doi: 10.1038/s41467-020-19057-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Natarajan A., Zlitni S., Brooks E.F., Vance S.E., Dahlen A., Hedlin H., Park R.M., Han A., Schmidtke D.T., Verma R., et al. Gastrointestinal symptoms and fecal shedding of SARS-CoV-2 RNA suggest prolonged gastrointestinal infection. Med. 2022;3:371–387.e9. doi: 10.1016/j.medj.2022.04.001. e379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mehta A.K., Gracias D.T., Croft M. TNF activity and T cells. Cytokine. 2018;101:14–18. doi: 10.1016/j.cyto.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Cevins C., Luka M., Smith N., Meynier S., Magérus A., Carbone F., García-Paredes V., Barnabei L., Batignes M., Boullé A., et al. A monocyte/dendritic cell molecular signature of SARS-CoV-2-related multisystem inflammatory syndrome in children with severe myocarditis. Med (N Y) 2021;2:1072–1092.e7. doi: 10.1016/j.medj.2021.08.002. e1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Müller I., Vogl T., Pappritz K., Miteva K., Savvatis K., Rohde D., Most P., Lassner D., Pieske B., Kühl U., et al. Pathogenic role of the damage-associated molecular patterns S100A8 and S100A9 in coxsackievirus B3-induced myocarditis. Circ. Heart Fail. 2017;10:e004125. doi: 10.1161/circheartfailure.117.004125. [DOI] [PubMed] [Google Scholar]
- 32.Tschöpe C., Ammirati E., Bozkurt B., Caforio A.L.P., Cooper L.T., Felix S.B., Hare J.M., Heidecker B., Heymans S., Hübner N., et al. Myocarditis and inflammatory cardiomyopathy: current evidence and future directions. Nat. Rev. Cardiol. 2021;18:169–193. doi: 10.1038/s41569-020-00435-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weckbach L.T., Grabmaier U., Uhl A., Gess S., Boehm F., Zehrer A., Pick R., Salvermoser M., Czermak T., Pircher J., et al. Midkine drives cardiac inflammation by promoting neutrophil trafficking and NETosis in myocarditis. J. Exp. Med. 2019;216:350–368. doi: 10.1084/jem.20181102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dolhnikoff M., Ferreira Ferranti J., de Almeida Monteiro R.A., Duarte-Neto A.N., Soares Gomes-Gouvêa M., Viu Degaspare N., Figueiredo Delgado A., Montanari Fiorita C., Nunes Leal G., Rodrigues R.M., et al. SARS-CoV-2 in cardiac tissue of a child with COVID-19-related multisystem inflammatory syndrome. Lancet. Child Adolesc. Health. 2020;4:790–794. doi: 10.1016/s2352-4642(20)30257-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Duarte-Neto A.N., Caldini E.G., Gomes-Gouvêa M.S., Kanamura C.T., de Almeida Monteiro R.A., Ferranti J.F., Ventura A.M.C., Regalio F.A., Fiorenzano D.M., Gibelli M.A.B.C., et al. An autopsy study of the spectrum of severe COVID-19 in children: from SARS to different phenotypes of MIS-C. EClinicalMedicine. 2021;35 doi: 10.1016/j.eclinm.2021.100850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Porritt R.A., Binek A., Paschold L., Rivas M.N., McArdle A., Yonker L.M., Alter G., Chandnani H.K., Lopez M., Fasano A., et al. The autoimmune signature of hyperinflammatory multisystem inflammatory syndrome in children. J. Clin. Invest. 2021;131:e151520. doi: 10.1172/jci151520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Noval Rivas M., Porritt R.A., Cheng M.H., Bahar I., Arditi M. Multisystem inflammatory syndrome in children and long COVID: the SARS-CoV-2 viral superantigen hypothesis. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.941009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thieblemont N., Wright H.L., Edwards S.W., Witko-Sarsat V. Human neutrophils in auto-immunity. Semin. Immunol. 2016;28:159–173. doi: 10.1016/j.smim.2016.03.004. [DOI] [PubMed] [Google Scholar]
- 39.Sterlin D., Mathian A., Miyara M., Mohr A., Anna F., Claër L., Quentric P., Fadlallah J., Devilliers H., Ghillani P., et al. IgA dominates the early neutralizing antibody response to SARS-CoV-2. Sci. Transl. Med. 2021;13:eabd2223. doi: 10.1126/scitranslmed.abd2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bartsch Y.C., Wang C., Zohar T., Fischinger S., Atyeo C., Burke J.S., Kang J., Edlow A.G., Fasano A., Baden L.R., et al. Humoral signatures of protective and pathological SARS-CoV-2 infection in children. Nat. Med. 2021;27:454–462. doi: 10.1038/s41591-021-01263-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hasan Ali O., Bomze D., Risch L., Brugger S.D., Paprotny M., Weber M., Thiel S., Kern L., Albrich W.C., Kohler P., et al. Severe coronavirus disease 2019 (COVID-19) is associated with elevated serum immunoglobulin (ig) A and antiphospholipid IgA antibodies. Clin. Infect. Dis. 2021;73:e2869–e2874. doi: 10.1093/cid/ciaa1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gimpel A.K., Maccataio A., Unterweger H., Sokolova M.V., Schett G., Steffen U. IgA complexes induce neutrophil extracellular trap formation more potently than IgG complexes. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.761816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aleyd E., van Hout M.W.M., Ganzevles S.H., Hoeben K.A., Everts V., Bakema J.E., van Egmond M. IgA enhances NETosis and release of neutrophil extracellular traps by polymorphonuclear cells via Fcα receptor I. J. Immunol. 2014;192:2374–2383. doi: 10.4049/jimmunol.1300261. [DOI] [PubMed] [Google Scholar]
- 44.Qi H., Yang S., Zhang L. Neutrophil extracellular traps and endothelial dysfunction in atherosclerosis and thrombosis. Front. Immunol. 2017;8:928. doi: 10.3389/fimmu.2017.00928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Denning N.L., Aziz M., Gurien S.D., Wang P. DAMPs and NETs in sepsis. Front. Immunol. 2019;10:2536. doi: 10.3389/fimmu.2019.02536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.de Bont C.M., Boelens W.C., Pruijn G.J.M. NETosis, complement, and coagulation: a triangular relationship. Cell. Mol. Immunol. 2019;16:19–27. doi: 10.1038/s41423-018-0024-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yonker L.M., Boucau J., Regan J., Choudhary M.C., Burns M.D., Young N., Farkas E.J., Davis J.P., Moschovis P.P., Bernard Kinane T., et al. Virologic features of severe acute respiratory syndrome coronavirus 2 infection in children. J. Infect. Dis. 2021;224:1821–1829. doi: 10.1093/infdis/jiab509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bastard P., Gervais A., Le Voyer T., Rosain J., Philippot Q., Manry J., Michailidis E., Hoffmann H.H., Eto S., Garcia-Prat M., et al. Autoantibodies neutralizing type I IFNs are present in ∼4% of uninfected individuals over 70 years old and account for ∼20% of COVID-19 deaths. Sci. Immunol. 2021;6:eabl4340. doi: 10.1126/sciimmunol.abl4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jacobs J.L., Bain W., Naqvi A., Staines B., Castanha P.M.S., Yang H., Boltz V.F., Barratt-Boyes S., Marques E.T.A., Mitchell S.L., et al. SARS-CoV-2 viremia is associated with COVID-19 severity and predicts clinical outcomes. Clin. Infect. Dis. 2022;74:1525–1533. doi: 10.1093/cid/ciab686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li Y., Schneider A.M., Mehta A., Sade-Feldman M., Kays K.R., Gentili M., Charland N.C., Gonye A.L., Gushterova I., Khanna H.K., et al. SARS-CoV-2 viremia is associated with distinct proteomic pathways and predicts COVID-19 outcomes. J. Clin. Invest. 2021;131:148635. doi: 10.1172/jci148635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yonker L.M., Swank Z., Gilboa T., Senussi Y., Kenyon V., Papadakis L., Boribong B.P., Carroll R.W., Walt D.R., Fasano A. Zonulin antagonist, larazotide (AT1001), as an adjuvant treatment for multisystem inflammatory syndrome in children: a case series. Crit. Care Explor. 2022;10 doi: 10.1097/cce.0000000000000641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lima R., Gootkind E.F., De la Flor D., Yockey L.J., Bordt E.A., D'Avino P., Ning S., Heath K., Harding K., Zois J., et al. Establishment of a pediatric COVID-19 biorepository: unique considerations and opportunities for studying the impact of the COVID-19 pandemic on children. BMC Med. Res. Methodol. 2020;20:228. doi: 10.1186/s12874-020-01110-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Villani A.C., Satija R., Reynolds G., Sarkizova S., Shekhar K., Fletcher J., Griesbeck M., Butler A., Zheng S., Lazo S., et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science. 2017;356:eaah4573. doi: 10.1126/science.aah4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Edwards A., Haas W. Multiplexed quantitative proteomics for high-throughput comprehensive proteome comparisons of human cell lines. Methods Mol. Biol. 2016;1394:1–13. doi: 10.1007/978-1-4939-3341-9_1. [DOI] [PubMed] [Google Scholar]
- 55.Hughes C.S., Moggridge S., Müller T., Sorensen P.H., Morin G.B., Krijgsveld J. Single-pot, solid-phase-enhanced sample preparation for proteomics experiments. Nat. Protoc. 2019;14:68–85. doi: 10.1038/s41596-018-0082-x. [DOI] [PubMed] [Google Scholar]
- 56.Lapek J.D., Jr., Greninger P., Morris R., Amzallag A., Pruteanu-Malinici I., Benes C.H., Haas W. Detection of dysregulated protein-association networks by high-throughput proteomics predicts cancer vulnerabilities. Nat. Biotechnol. 2017;35:983–989. doi: 10.1038/nbt.3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li J., Van Vranken J.G., Pontano Vaites L., Schweppe D.K., Huttlin E.L., Etienne C., Nandhikonda P., Viner R., Robitaille A.M., Thompson A.H., et al. TMTpro reagents: a set of isobaric labeling mass tags enables simultaneous proteome-wide measurements across 16 samples. Nat. Methods. 2020;17:399–404. doi: 10.1038/s41592-020-0781-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McAlister G.C., Nusinow D.P., Jedrychowski M.P., Wühr M., Huttlin E.L., Erickson B.K., Rad R., Haas W., Gygi S.P. MultiNotch MS3 enables accurate, sensitive, and multiplexed detection of differential expression across cancer cell line proteomes. Anal. Chem. 2014;86:7150–7158. doi: 10.1021/ac502040v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ting L., Rad R., Gygi S.P., Haas W. MS3 eliminates ratio distortion in isobaric multiplexed quantitative proteomics. Nat. Methods. 2011;8:937–940. doi: 10.1038/nmeth.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Erickson B.K., Jedrychowski M.P., McAlister G.C., Everley R.A., Kunz R., Gygi S.P. Evaluating multiplexed quantitative phosphopeptide analysis on a hybrid quadrupole mass filter/linear ion trap/orbitrap mass spectrometer. Anal. Chem. 2015;87:1241–1249. doi: 10.1021/ac503934f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Erickson B.K., Mintseris J., Schweppe D.K., Navarrete-Perea J., Erickson A.R., Nusinow D.P., Paulo J.A., Gygi S.P. Active instrument engagement combined with a real-time database search for improved performance of sample multiplexing workflows. J. Proteome Res. 2019;18:1299–1306. doi: 10.1021/acs.jproteome.8b00899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Eng J.K., McCormack A.L., Yates J.R. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 63.Huttlin E.L., Jedrychowski M.P., Elias J.E., Goswami T., Rad R., Beausoleil S.A., Villén J., Haas W., Sowa M.E., Gygi S.P. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell. 2010;143:1174–1189. doi: 10.1016/j.cell.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Elias J.E., Gygi S.P. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 65.Graubert A., Aguet F., Ravi A., Ardlie K.G., Getz G. RNA-SeQC 2: efficient RNA-seq quality control and quantification for large cohorts. Bioinformatics. 2021;37:3048–3050. doi: 10.1093/bioinformatics/btab135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Puram S.V., Tirosh I., Parikh A.S., Patel A.P., Yizhak K., Gillespie S., Rodman C., Luo C.L., Mroz E.A., Emerick K.S., et al. Single-cell transcriptomic analysis of primary and metastatic tumor ecosystems in head and neck cancer. Cell. 2017;171:1611–1624.e24. doi: 10.1016/j.cell.2017.10.044. e1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tinevez J.Y., Perry N., Schindelin J., Hoopes G.M., Reynolds G.D., Laplantine E., Bednarek S.Y., Shorte S.L., Eliceiri K.W. TrackMate: an open and extensible platform for single-particle tracking. Methods. 2017;115:80–90. doi: 10.1016/j.ymeth.2016.09.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(Top) Spontaneous NETosis of neutrophils isolated from a healthy pediatric patient and (Bottom) a child with MIS-C captured by fluorescence (brightfield and FITC) microscopy. Extracellular DNA stained with SYTOX green. Scale bar = 100 μM.
(Left) Neutrophil undergoing NETosis and (Right) neutrophil undergoing non-NETosis cellular death and detection by TrackMate software, seen as a magenta circle, captured by 20X fluorescence microscopy (FITC). Extracellular DNA stained with SYTOX green. Scale bar = 25 μM.
(Top) NETosis of isolated neutrophils isolated from a healthy patient stimulated with convalescent COVID-19 plasma-treated Spike beads and (Bottom) stimulated with MIS-C plasma captured by fluorescence (brightfield and FITC) microscopy. Extracellular DNA stained with SYTOX green. Scale bar = 100 μM.
Data Availability Statement
-
•
Due to IRB consent limitations, raw sequencing data is not publicly available. However, the read count matrix and TPM matrix used in this study will be publicly available in GEO (Gene Expression Omnibus) under accession number GSE217370 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE217370) and Table S5. Additional Supplemental Items are available from Zenodo Data: https://doi.org/10.5281/zenodo.7277479. Raw sequencing data can be made available from the lead contact upon request.
-
•
All code required to run the analyses in this manuscript is deposited in Zenodo Data: https://doi.org/10.5281/zenodo.7277479 based on the associated Github repository (https://github.com/lasalletj/Pediatric_COVID_MISC_Neutrophils).
-
•
Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.