Abstract
Burkholderia cepacia is a frequent cause of respiratory infections in cystic fibrosis patients. B. cepacia has been shown to produce at least four siderophores which may play a role in the virulence of this organism. To characterize genes involved in the synthesis of siderophores, Tn5-OT182 mutants were isolated in strain K56-2, which produces two siderophores, salicylic acid (SA) and ornibactins. Two mutants were characterized that did not produce zones on Chrome Azurol S agar in a commonly used assay to detect siderophore activity. These mutants were determined to produce sevenfold more SA than K56-2 yet did not produce detectable amounts of ornibactins. These mutants, designated I117 and T10, had a transposon insertion in genes with significant homology to pyoverdine biosynthesis genes of Pseudomonas aeruginosa. I117 contained an insertion in a pvdA homolog, the gene for the enzyme l-ornithine N5-oxygenase, which catalyzes the hydroxylation of l-ornithine. Ornibactin synthesis in this mutant was partially restored when the precursor l-N5-OH-Orn was added to the culture medium. T10 contained an insertion in a pvdD homolog, which is a peptide synthetase involved in pyoverdine synthesis. β-Galactosidase activity was iron regulated in both I117 and T10, suggesting that the transposon was inserted downstream of an iron-regulated promoter. Tn5-OT182 contains a lacZ gene that is expressed when inserted downstream of an active promoter. Both I117 and T10 were deficient in uptake of iron complexed to either ornibactins or SA, suggesting that transposon insertions in ornibactin biosynthesis genes also affected other components of the iron transport mechanism. The B. cepacia pvdA homolog was approximately 47% identical and 59% similar to l-ornithine N5-oxygenase from P. aeruginosa. Three clones were identified from a K56-2 cosmid library that partially restored ornibactin production, SA production, and SA uptake to parental levels but did not affect the rate of 59Fe-ornibactin uptake in I117. A chromosomal pvdA deletion mutant was constructed that had a phenotype similar to that of I117 except that it did not hyperproduce SA. The pvdA mutants were less virulent than the parent strain in chronic and acute models of respiratory infection. A functional pvdA gene appears to be required for effective colonization and persistence in B. cepacia lung infections.
Burkholderia cepacia is an opportunistic pathogen that can cause severe respiratory infections in patients with cystic fibrosis or chronic granulomatous disease (21). Approximately 20% of cystic fibrosis patients colonized with B. cepacia experience a rapid and often fatal pulmonary decline (21). Potential virulence factors that may contribute to the severity of B. cepacia infections include siderophores. Iron is essential for microbial growth, but its availability is very limited in mammalian hosts due to the presence of iron-binding proteins such as transferrin and lactoferrin. To acquire iron from the environment and to compete with transferrin or lactoferrin for mammalian iron, many bacteria have evolved high-affinity iron uptake systems, consisting of low-molecular-weight iron chelators, called siderophores, and specific receptor-mediated membrane-associated uptake mechanism (reviewed in references 8 and 37). B. cepacia has been reported to produce four different siderophores, salicylic acid (SA; formerly azurechelin), ornibactins, pyochelin, and cepabactin (35, 36, 53, 54, 57, 65).
We have recently determined that SA and ornibactins are the predominant siderophores produced by clinical isolates of B. cepacia (11). SA is produced by 92% of B. cepacia RAPD (randomly amplified polymorphic DNA) types from cystic fibrosis patients (11). SA, which is similar in structure to pyochelin, has also been shown to function as a siderophore in Pseudomonas aeruginosa and Pseudomonas fluorescens (65). Ornibactins are linear hydroxamate-hydroxycarboxylate siderophores that are related in their peptide structure to the pyoverdines produced by the fluorescent pseudomonads P. aeruginosa and P. fluorescens (36, 57). Ornibactins are composed of the conserved tetrapeptide l-Orn1(Nδ-OH, Nδ-acyl)-d-threo-Asp(β-OH)-l-Ser-l-Orn4(Nδ-OH,Nδ-formyl)-1,4-diaminobutane. The acyl groups of Orn1 vary in length and include 3-hydroxybutanoic acid, 3-hydroxyhexanoic acid, and 3-hydroxyoctanoic acid, forming the three different ornibactins, which are designated ornibactin-C4, ornibactin-C6, and ornibactin-C8 according to their acyl chain length (57, 58). Ornibactins are produced by 87% of B. cepacia RAPD types from cystic fibrosis patients (11).
Although the structure of ornibactins has been well defined, genes involved in their biosynthesis and transport have not yet been characterized. Because of their similarity to pyoverdines, it may be hypothesized that biosynthetic enzymes for these siderophores are conserved. Several genes have been identified which are required for the synthesis of pyoverdines. The genetic loci involved in pyoverdine biosynthesis have been mapped in P. aeruginosa to a 103-kb region located at approximately 47 min of the PAO1 chromosome (1, 62, 68). pvdA encodes the enzyme l-ornithine N5-oxygenase responsible for catalyzing the hydroxylation of l-ornithine, thus forming the hydroxamate ligands of pyoverdine (64). pvdD encodes an enzyme similar to peptide synthetases from a range of bacterial and fungal species that function in nonribosomal peptide synthesis (34). The recently characterized pvdE gene has been shown to code for an ATP-binding cassette membrane transporter protein involved in pyoverdine synthesis (31). The cytA gene has also recently been shown to be required for normal levels of pyoverdine production in P. fluorescens, in addition to being essential for cytochrome c production (17). PvdS, a novel transcriptional factor belonging to the ςE family of RNA polymerase sigma factors, is required for the transcription of pyoverdine biosynthetic genes (10, 39).
The objectives of the present study were to isolate B. cepacia mutants altered in the biosynthesis of ornibactins. Transposon mutagenesis with Tn5-OT182 was used to generate a series of mutants deficient in siderophore synthesis. Two genes were identified as directly implicated in ornibactin synthesis. The importance of ornibactin biosynthesis in the virulence of B. cepacia was also investigated.
MATERIALS AND METHODS
Strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are described in Table 1. B. cepacia K56-2 was originally isolated from the sputum of a cystic fibrosis patient. This strain produces salicylic acid and ornibactins, negligible amounts of pyochelin, and no cepabactin (11, 27). For genetic manipulations, cultures were routinely grown at 37°C in Luria-Bertani (LB) broth (Life Technologies, Burlington, Ontario, Canada) or Bacto-Terrific broth (Difco, Detroit, Mich.). Pseudomonas Isolation Agar (Difco) was used to recover B. cepacia transformants after electroporation with plasmids. Trypticase soy agar was used to quantitate bacteria in lung homogenates. When appropriate, antibiotics were added at the following concentrations: 100 μg of ampicillin, 15 μg of tetracycline, and 1.5 mg of trimethoprim per ml for Escherichia coli and 300 μg of tetracycline, 100 μg of streptomycin, and 100 μg of trimethoprim per ml for B. cepacia. A 100-mg/ml stock solution of trimethoprim was prepared in N,N-dimethyl-acetimide.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | F− φ80dlacZΔM15 Δ(lacZYZ-argF) recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 deoR U169 | Gibco BRL |
| SM10 | Mobilizing strain, RP4 tra genes integrated in chromosome, Kmr | 52 |
| XL1-Blue MR | Nonmethylating (dam dcm) cloning host used for cosmid library construction and preparation of DNA for electroporation into B. cepacia: Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 endA1 supE44 thi-1 recA1 gyrA96 relA1 lac |
Stratagene |
| B. cepacia | ||
| K56-2 | Clinical isolate, genomovar III | 11, 27 |
| K56-I117 | pvdA::Tn5-OT182 derivative of K56-2 | This study |
| K56-T10 | pvdD::Tn5-OT182 derivative of K56-2 | This study |
| K56pvdA::tp | pvdA::Tpr derivative of K56-2 | This study |
| Plasmids | ||
| pOT182 | pSUP102(GM)::Tn5-OT182; Cmr Gmr Apr Tcr | 33 |
| pNOT19 | Modified pUC19 cloning vector, Apr | 47 |
| pUCP28T | Broad-host-range vector; IncP OriT; pRO1600 ori; Tpr | 50 |
| pPDI117-2 | 9.5-kb EcoRI fragment from I117 obtained by self-cloning, Tcr | This study |
| pPDI117-3 | 11.5-kb NotI fragment from I117 obtained by self-cloning, Tcr | This study |
| pPD519 | pNOT19 with 6.1-kb SphI fragment from K56-2 with pvdA gene | This study |
| pPD520 | pUCP28T with 6.5-kb SphI fragment from pPD519 | This study |
| p34E-Tp | Source of Tp cassette; Tpr | 13 |
| pRK2013 | ColE1 Tra (RK2)+ Kmr | 16 |
| pEX18Tc | Suicide vector; sacB Tcr | 23 |
| pZ1918 | Apr Gmr; source of promoterless lacZ gentamicin resistance cassette | 48 |
| pPD520-6Z | pPD520 with 4.3-kb lacZ-Gmr cassette from pZ1918 in BglII site of pvdA gene; same orientation | This study |
| pPD520-14Z | pPD520 with 4.3-kb lacZ-Gmr cassette from pZ1918 in BglII site of pvdA gene; reverse orientation | This study |
| pUC-TP | pUC-GM (46) with the gentamicin resistance cassette replaced with a 1.1-kb Tpr gene cassette; Apr Tpr |
This study |
| pScosBC1 | Novel broad-host-range cosmid cloning vector based on pSuperCos1 (67); Apr Tpr | This study |
| pSBC-4 | pScosBC1 from K56-2 library containing the pvdA gene | This study |
| pSBC-8 | pScosBC1 from K56-2 library containing the pvdA gene | This study |
| pSBC-13 | pScosBC1 from K56-2 library containing the pvdA gene | This study |
For the analysis of salicylic acid production, cultures were grown at 32°C with maximum aeration in 50 ml of deferrated 0.5% Casamino Acids (Difco) and 0.2 mM MgCl2 (CAA medium) as previously described (53). For the detection of ornibactins, cultures were grown at 32°C for 40 h with maximum aeration in 200 ml of succinate medium supplemented with 10 mM ornithine (36). These growth conditions were used to obtain maximum yields of siderophores. All glassware was washed with acid and rinsed with deionized water to remove iron. All reagents were made with water purified by the Milli-Q System (Millipore Corp., Mississauga, Ontario, Canada). Cultures were normally grown in succinate medium plus 0.5% glucose at 32°C for siderophore-iron uptake assays.
Tn5-OT182 mutagenesis.
Tn5-OT182 (33) was introduced into K56-2 by conjugation with SM10(pOT182) as previously described (27). Transconjugants were selected on LB agar containing 300 μg of tetracycline and 100 μg of streptomycin per ml and 100 μM FeCl3 to select against wild-type B. cepacia and the E. coli donor strain. Tn5-OT182 insertion mutants were screened for mutations in genes involved in siderophore biosynthesis on Chrome Azurol S (CAS) plates (51). Mutants that did not produce detectable zones after 2 to 3 days of incubation at 32°C were selected for further characterization.
DNA manipulations.
Molecular biology techniques were performed as generally described by Sambrook et al. (44). Genomic DNA was isolated from K56-2 as described by Ausubel et al. (3). Colony hybridizations were performed as previously described (68). Recombinant plasmids were electroporated into E. coli by using a Gene Pulser (Bio-Rad, Richmond, Calif.) according to the manufacturer’s recommendations or into B. cepacia I117 as previously described (12). For the self-cloning of B. cepacia DNA from Tn5-OT182 mutants, approximately 5 μg of genomic DNA was digested with an appropriate restriction enzyme, ligated, and transformed into E. coli DH5α cells made competent by treatment with CaCl2 (28). For cloning of the parental pvdA gene, appropriate size restriction fragments were isolated by sucrose density gradient centrifugation of restriction endonuclease digested genomic DNA (20) and ligated into pNOT19 (47).
For construction of a pvdA allelic exchange mutant, the 6.1-kb SphI fragment containing pvdA was cloned into pEX18TC (23). The 0.7-kb BglII fragment was deleted and replaced with the 0.6-kb Tpr cassette from p34E-Tp (14). This plasmid was transferred from E. coli to K56-2 by triparental mating with pRK2013 (16) as a mobilizing plasmid. Tpr transconjugants were plated on 5% sucrose to select for excision of the plasmid. Insertional inactivation of pvdA was confirmed by Southern hybridization.
Construction of a cosmid library.
A novel broad-host-range cosmid vector, pScosBC1, derived from the ColE1-based cloning vector pSuperCos1 (67) (Stratagene, La Jolla, Calif.), was used to construct a cosmid library of B. cepacia K56-2. Cosmid pScosBC1 was constructed as follows. A 1.97-kb fragment encoding the kanamycin resistance gene and simian virus 40 origin of pSuperCos1 was removed from this vector by digestion with the endonucleases HpaI and PvuII, creating plasmid pScosDHP1. The 1.8-kb PstI-generated replication stabilizing fragment from pRO1614, which facilitates the stable maintenance of ColE1-based replicons in a wide range of gram-negative bacteria (46), was inserted into NsiI of pScosDHP1, creating plasmid pScosRSF.2. To enable selection of this vector in B. cepacia, a 1-kb Tpr gene cassette was added to pScosRSF.2. The cassette was derived as follows. Plasmid pCHR61 encoding the Tpr resistance gene from plasmid R388 (45) was partially digested with Sau3A1, and fragments of approximately 1 kb were ligated into the vector backbone of plasmid pUC-GM (49) isolated by restriction with BamHI. Several Tpr clones resulting from this ligation were screened, and one clone (which liberated a 1-kb Tpr gene cassette after digestion with either HindIII, PstI, SalI, SphI, or XbaI) was designated plasmid pUC-TP. A 1-kb SalI Tpr cassette isolated from pUC-TP was ligated into a single AvaI site within pSCOSRSF.2 to create the final broad-host-range cosmid vector pScosBC1.
A cosmid library of B. cepacia K56-2 was constructed in pScosBC1 as follows. Genomic DNA was extracted from strain K56-2 as previously described (30). Approximately 2 μg of K56-2 DNA partially digested with Sau3A1 (ranging from 20 to 35 kb) was combined with 2 μg of pScosBC1 linearized with BamHI, and the mixture was ligated in a 20-μl reaction mixture for 4 h at 16°C. After ligation, 4 μl of the reaction mixture was packaged into the bacteriophage lambda by using a commercial kit which selectively packages large recombinant cosmids (Gigapack III XL; Stratagene). The resulting phage extract was transfected into E. coli XL1-Blue MR. The average insert size of library clones was 33 kb, and less than 3% of the clones were nonrecombinant. A total of 1,596 cosmids (sufficient to cover the 8-Mb B. cepacia genome [7, 43] approximately six times) were picked into 96-well plates. After overnight growth, each 96-well grid was replica plated onto positively charged nylon membranes (Boehringer Mannheim, Laval, Quebec, Canada) which had been placed onto selective agar. Dimethyl sulfoxide was then added to a final concentration of 8% to each well, and the cosmid library was frozen at −70°C. After further overnight growth, DNA from each gridded clone was immobilized onto the nylon membranes as described by the manufacturer’s guidelines (Boehringer Mannheim).
Nucleotide sequencing.
Nucleotide sequencing was performed by using the ABI PRISM DyeDeoxy Termination Cycle Sequencing System with AmpliTaq DNA polymerase (Perkin-Elmer Corp.). DNA sequencing reactions were analyzed with an ABI 1373A DNA Sequencer by the University Core DNA Services (University of Calgary). The oligodeoxynucleotide OT182-LT (5′-GATCCTGGAAAACGGGAAAG-3′) was used to initiate DNA sequence reactions with plasmids obtained from Tn5-OT182 mutants by self-cloning. Custom oligonucleotides were synthesized by the University Core DNA Services or Life Technologies. Analysis of the sequence was performed with PC/Gene software (Intelligenetics, Mountain View, Calif.). The BLASTX and BLASTN programs were used to search the nonredundant sequence database for homologous sequences (29).
PFGE of genomic DNA.
Separation of intact chromosomes was carried out by pulsed-field gel electrophoresis (PFGE) by using a protocol adapted from previous methods (7, 43). Bacterial growth from 5 ml of LB overnight culture was harvested by centrifugation and resuspended to an A620 of 0.8 to 0.9 in SE buffer (75 mM NaCl, 25 mM EDTA, pH 7.4). This suspension was then concentrated eightfold by centrifugal harvesting of bacteria, followed by suspension of the pellet in one-eighth of the original volume of SE buffer. The concentrated bacterial suspension was briefly warmed to 45°C, mixed gently with an equal volume of molten 2% low-melting-point agarose (Type 7; Sigma-Aldrich Canada, Oakville, Ontario, Canada) kept at the same temperature, and poured into 70-μl disposable plug molds (Bio-Rad, Mississauga, Ontario, Canada). After cooling to 4°C for 15 min the solidified plugs (three to five per tube) were placed in 10 ml of PEN buffer (0.5 M EDTA [pH 9.6] plus 1% N-lauroyl sarcosine containing 1 mg of pronase [Boehringer Mannheim] per ml) in a 15-ml sterile tube. Plugs were incubated with gentle rocking for 48 h at 37°C and then washed thoroughly with several volume changes of TE buffer (44). Slices were then cut from plugs and loaded on a 0.8% agarose gel (14 by 13 cm) made with TAE electrophoresis buffer (44). Intact chromosomal DNA was separated by PFGE (Bio-Rad CHEF DR-II apparatus) at 3 V/cm for 64 h, with the pulse switch time ramped from 250 to 900 s.
Siderophore production assays.
Siderophore activity was measured by CAS assays (51). SA was isolated and quantitated by thin-layer chromatography of ethyl acetate-extracted culture supernatants as previously described (54). Ornibactin production was assayed as previously described (11, 27). Briefly, supernatants were lyophilized and extracted with methanol, and ornibactins were separated by Sephadex LH-20 (Pharmacia) column chromatography. Fractions containing CAS activity were pooled, and the total ornibactin concentration was estimated by using the CAS assay.
Iron uptake assays.
For iron uptake assays, cultures were grown to an A600 of 0.3, washed once, and resuspended to a final A600 of 0.3 in nitrogen-free succinate medium (36), or cultures were grown to late log phase (A600 of 1.0 to 1.4), and the cells were then washed and resuspended to a final A600 of 0.3. Similar rates of uptake were observed by using either procedure (data not shown). Ornibactins (3.6 nmol) were mixed with an equal amount of 59FeCl3 in a total volume of 100 μl and equilibrated for 10 to 30 min prior to the assay. Uptake reactions were initiated by the addition of 100 μl of the 59Fe-ornibactin mixture to a 10-ml cell suspension. Then, 1-ml samples of these reaction mixtures were removed at selected intervals, filtered through cellulose acetate 0.45-μm-pore-size filters (Sartorius, GmbH), and washed with 3 ml of 10 mM Tris (pH 7.5)–0.9% NaCl. The amount of 59Fe accumulated on the filters was measured in an LKB Compugamma counter. SA uptake assays were performed as described above except that 7.2 nmol of SA was equilibrated with 3.6 nmol of 59FeCl3 and 100 μl of 59Fe-SA was used to initiate the uptake reactions (54).
β-Galactosidase assays.
Cultures were grown to an A600 of 0.5 in TSBD-C medium (42) in the presence or absence of 50 μM FeCl3, and β-galactosidase assays were performed as previously described by Miller (38).
Effect of l-N5-hydroxyornithine on ornibactin production.
l-N5-Hydroxyornithine (l-N5-OH-Orn) was prepared by the acid hydrolysis of rhodoturulic acid as previously described (2). K56-2 and I117 were grown in succinate medium, 0.5% glucose, and 400 μM l-N5-OH-Orn (pH 7.0) for 40 h at 32°C. Ornibactin production was determined by measuring the CAS activity of culture supernatants.
Animal studies.
Chronic infection experiments were performed in the respiratory infection model in rats as described by Cash et al. (6). Groups of 16 male Sprague-Dawley rats weighing 150 to 170 g (Charles River Canada, Inc.) were tracheostomized under anesthesia and inoculated with the appropriate strain embedded in agar beads as previously described (6). On days 7 and 28 postinfection, the lungs from four animals in each group were removed aseptically and homogenized (Polytron Homogenizer; Brinkman Instruments, Westbury, N.Y.) in 3 ml of phosphate-buffered saline (0.05 M [pH 7.2] containing 0.9% saline) (PBS). Serial dilutions were plated on Trypticase soy agar or Trypticase soy agar plus the appropriate antibiotic. The lungs of four additional animals in each group were removed en bloc, fixed in 10% formalin, and examined for qualitative and quantitative pathological changes as previously described (15, 55). Infiltration of the lung with inflammatory cells and exudate was measured by the point counting method (15, 55). Briefly, with a Zeiss integrating eyepiece (Zeiss, Oberkochen, Germany), the number of points overlying the surface area of the infiltrate was divided by the total number of points counted over the entire surface area of the section of the left lobe to measure the percentage of infiltration.
Acute infections were performed in neutropenic mice infected by aerosol inoculation. Swiss Webster male mice (18 to 20 g), five per group, were made neutropenic by intraperitoneal injection of cyclophosphamide (150 mg/kg) on days 4, 2, and 0 prior to infection (9). For initiation of bacterial infections by an aerosol route, PBS dilutions of a 10-ml overnight culture were washed once in PBS and resuspended in 10 ml of PBS. Bacteria (ca. 6 × 108 CFU/ml) were aerosolized into animals by using an aerosol chamber designed for small-animal use (5). The aerosol chamber was housed in a Biosafety Cabinet in a level 2 containment facility with the airflow isolated for additional safety. The chamber is designed to accept animals ranging in size from 20 to 250 g, and it is equipped with sampling ports for an Anderson Air Sampler to allow the monitoring of the size of the particles. The aerosol is removed through HEPA-filtered adjustable vacuum flow which allows the control of flow rates. The aerosol flow rates are calibrated by using fluorescent microspheres obtained from Duke Scientific Corporation, and this was done prior to each aerosol treatment. On days 0 (ca. 2 h), 3, and 7 after bacterial inoculation by the aerosol route, animals were sacrificed, and their lungs were removed and examined for bacterial counts.
Nucleotide sequence accession number.
The nucleotide sequence for the pvdA gene has been deposited in GenBank and assigned accession no. AF013993. The nucleotide sequence of vectors pScosBC1 and pUC-TP was assembled from the published sequences of their components held within GenBank; each complete sequence has been assigned accession no. AF136442 and AF136443, respectively.
RESULTS
Isolation and characterization of B. cepacia siderophore mutants.
The objectives of this study were to isolate mutants altered in the synthesis of ornibactins. The transposon Tn5-OT182, which contains a tetracycline resistance determinant and a pBR325 origin of replication to allow the cloning of DNA adjacent to the transposon without the construction of genomic libraries, was used to generate mutants (33). This transposon also contains a promoterless lacZ reporter gene, allowing the formation of lacZ transcriptional fusions when inserted downstream from a functional promoter (33).
Approximately 5,300 Tcr Smr transconjugants from four independent mutagenesis experiments were screened for mutations in genes involved in siderophore synthesis on CAS agar (51). Colonies that produce siderophores on this medium remove iron from the blue CAS dye, and orange zones are formed around the colonies. Strains lacking siderophores produce no zones or very small zones compared to the parent strain. Nine mutants lacking zones on CAS agar were identified that contained Tn5-OT182 in a unique location, as determined by Southern hybridization (data not shown).
The region of the chromosome flanking Tn5-OT182 in each mutant was mapped by using Southern hybridization analysis of DNA digested with EcoRI, NotI, BamHI, XhoI, SalI, ClaI, HindIII, SstI, and SmaI, and a probe internal to Tn5-OT182. An appropriate restriction enzyme which would yield a fragment containing the origin of replication, the Tcr determinant, and approximately 0.5 to 5.0 kb of DNA outside the transposon was then selected for cloning the flanking region (33). Chromosomal DNA was digested with this enzyme, ligated, and transformed into E. coli DH5α. For some mutants, DNA from both the right and left sides of the transposon was cloned (Table 1). A primer specific to the end of Tn5-OT182 was used to perform a cycle sequencing reaction on the plasmids isolated by self-cloning (13). Generally, about 300 to 400 bp of sequence was obtained and used to search the nonredundant protein sequence database by using the local alignment search tool BLASTX (29) through the National Center for Biotechnology Information at the National Institutes of Health. Two of the mutants had Tn5-OT182 insertions in genes with homology to siderophore biosynthetic genes, and these mutants were selected for further characterization.
Mutants I117 and T10 contained insertions in genes homologous to P. aeruginosa genes involved in pyoverdine synthesis. The gene inactivated in I117 had homology to the pvdA gene, which is the structural gene for the enzyme l-ornithine N5-oxygenase, which catalyzes the hydroxylation of l-ornithine, an early step in the biosynthesis of the peptide moiety of pyoverdine (64). Mutants lacking this enzyme require l-N5-OH-Orn for pyoverdine synthesis (66). In the mutant T10, the transposon was inserted into a gene with homology to a peptide synthetase gene, pvdD, which has homology to peptide synthetase genes involved in nonribosomal peptide synthesis (34). P. aeruginosa pvdD encodes a 2,448-amino-acid residue protein with a predicted molecular mass of 273,061 Da. It contains two highly similar domains of ca. 1,000 amino acids each. Each domain is likely involved in the recognition, activation, and incorporation of a specific amino acid into the peptide moiety of pyoverdine (34). The insertion in T10 is located in one of the two similar domains, since the portion sequenced has homology to both domains.
Characterization of mutants I117 and T10.
Since mutants I117 and T10 were determined to have transposon insertions in genes homologous to pyoverdine biosynthetic genes which are likely similar to genes directly involved in ornibactin synthesis, these mutants were selected for further characterization. These mutants were grown in CAA medium for 24 h to stationary phase, and culture supernatants were assayed for CAS activity (Table 2). I117 and T10 had no detectable CAS activity. These mutants were also unable to grow in the presence of the iron chelator ethylenediamine-di(o-hydroxy-phenylacetic acid (EDDHA; data not shown). Ornibactins were quantitated by measuring CAS activity in methanol extracts from supernatants of cultures grown in succinate medium and chromatographed on Sephadex LH-20. I117 and T10 produced no detectable ornibactins (Table 2).
TABLE 2.
Characterization of CAS-negative mutants of B. cepacia K56-2a
| Strain | CAS activityb (A630/A600) | SA (μg/ml/A600) | Ornibactin (μg/ml/A600) | Genetic locus associated with transposon insertion and function |
|---|---|---|---|---|
| K56-2 | 1.9 | 1.6 | 7.0 | |
| I117 | 0 | 11.4 | 0 | pvdA, pyoverdin biosynthesis |
| T10 | 0 | 12.7 | 0 | pvdD, pyoverdin biosynthesis |
Values represent the average of three experiments.
Activity per 200 μl of supernatant.
To determine whether mutants were deficient in the production of both SA and ornibactins, SA was quantitated from ethyl acetate extracts of acidified supernatants chromatographed on silica gel. These mutants, surprisingly, produced approximately sevenfold-higher yields of SA than did K56-2. The SA produced by these strains was not active in the CAS assay even with the addition of 5-sulfosalicylic acid, which acts as a shuttle between the dye complex and the siderophores (51). SA isolated from these mutants, however, was able to promote iron uptake, as well as SA isolated from K56-2 or purchased commercially (BDH, Toronto, Ontario; data not shown). Therefore, these mutants were negative for the production of ornibactins, yet they hyperproduced SA, which indicates that CAS agar and the liquid CAS assay only detect the iron-binding activity of ornibactins and not SA in culture supernatants of B. cepacia.
Tn5-OT182 contains a promoterless lacZ reporter gene which can generate transcriptional fusions upon insertion into a target gene (33). The amount of β-galactosidase produced corresponds to the activity of the fused promoter upstream from the inserted lacZ gene. Since siderophore production is regulated by the amount of iron in the culture medium, the amount of β-galactosidase produced was determined in cultures grown to the same stage of growth at an A600 of 0.5 in TSBD-C and in TSBD-C plus 50 μM FeCl3. B. cepacia K56-2 produces barely detectable levels of β-galactosidase (Table 3). Mutants I117 and T10 produced β-galactosidase when grown in low-iron medium but only produced background levels of β-galactosidase in medium with 50 μM FeCl3, which was not significantly different than K56-2. There was at least a 90% decrease in β-galactosidase production in medium with iron added. This indicates that Tn5-OT182 is inserted downstream of an active iron-regulated promoter in both I117 and T10. Sequence analysis with the primer OT182-L confirmed the orientation of the pvdA and pvdD genes with respect to the lacZ gene.
TABLE 3.
Iron regulation of β-galactosidase activity in B. cepacia pvdA and pvdD mutants
| Strain | β-Galactosidase activity (Miller units), mean ± SDa
|
||
|---|---|---|---|
| 0 μM FeCl3 | 50 μM FeCl3 | P | |
| K56-2 | 1.2 ± 0.7 | 8.9 ± 2.3 | NS |
| I117 | 1,113 ± 34 | 30 ± 9.5 | <0.001 |
| T10 | 523 ± 10 | 41 ± 2.3 | <0.001 |
Means ± the standard deviations of triplicate experiments. Experiments were performed twice with similar results. NS, not significant.
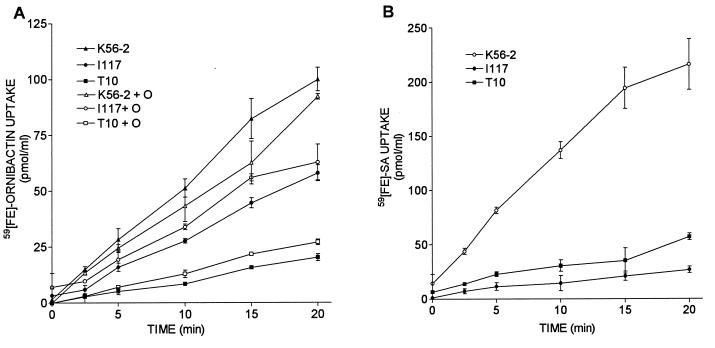
I117 and T10 were shown not to produce detectable levels of ornibactin (Table 2). To determine whether these mutants were capable of utilizing ornibactin the ability of these mutants to take up 59Fe-ornibactins was compared to the parent strain. I117 and T10 took up 59Fe-ornibactins at markedly lower rates, 60 and 20%, respectively, than K56-2 (Fig. 1A; P < 0.001). Therefore, the integration of Tn5-OT182 in the pvdA and pvdD homologs in I117 and T10 affects not only the biosynthesis of ornibactins but also the uptake of ornibactins. The expression of pyoverdine or pseudobactin receptors in P. aeruginosa and Pseudomonas putida, respectively, has previously been shown to be induced by the presence of the corresponding siderophore in the medium (18, 25). To determine whether the defect in uptake 59Fe-ornibactin uptake in T10 and I117 was due to the lack of ornibactin induction of receptor expression, 59Fe-ornibactin uptake was also examined in cultures grown in the presence of 10 μg of ornibactins per ml (Fig. 1A). The presence of ornibactins in the culture medium only slightly increased the rate of 59Fe-ornibactin uptake in T10 and I117 and did not restore uptake to parental levels. Therefore, the ability of these mutants to take up 59Fe-ornibactins is not due solely to the lack of ornibactin production and the possible subsequent induction of the uptake mechanism.
FIG. 1.
Uptake of 59Fe-siderophore complexes by B. cepacia K56-2, I117, and T10. (A) Uptake assays were initiated by the addition of 59Fe-ornibactins; 1-ml samples were removed at intervals, and the amount of 59Fe accumulated was determined. Cultures used in the assay were grown in the presence (open symbols) or absence (closed symbols) of ornibactins. (B) Uptake assays were initiated by the addition of 59Fe-SA; 1-ml samples were removed at intervals, and the amount of 59Fe accumulated was determined from a standard curve. Values represent the means ± the standard deviations of triplicate assays. These experiments were repeated at least three times with similar results.
To determine if the uptake defect was specific for ornibactins, the ability of these mutants to take up 59Fe-SA was also examined. Both I117 and T10 were also defective in 59Fe-SA uptake compared to K56-2 (Fig. 1B). I117 had a slightly higher rate of 59Fe-SA and 59Fe-ornibactin uptake than T10, but significantly less than K56-2. Therefore, despite hyperproducing SA, these mutants did not appear to be able to use this siderophore very effectively to acquire iron.
Characterization of the B. cepacia pvdA gene.
l-Ornithine N5-oxygenase is a key step in the biosynthesis of pyoverdine type siderophores since it is essential for the generation of the hydroxamate residues of these molecules (64). Therefore, the pvdA gene from B. cepacia was isolated to further characterize this essential step in the biosynthesis of ornibactins. By using a 1.2-kb SalI-BamHI fragment from pPDI117-2 as a probe in Southern hybridization, a 6.1-kb SphI fragment from K56-2 was identified which contained the entire pvdA gene. This fragment was cloned into pNOT19 (pPD519) and was used to determine the nucleotide sequence of the pvdA gene. An open reading frame of 1,332 bp was identified which began with an ATG at position 232 of the deposited sequence (Fig. 2, residue no. 1) and ended with a TGA stop codon at position 1564. There was a putative ribosome binding site, AGGGA, 9 bp upstream of the start codon (data not shown). The protein encoded by pvdA is predicted to be a 444-amino-acid peptide with a molecular mass of 50 kDa. The estimated isoelectric point (pI) is 9.1. A partial open reading frame was detected with a stop codon ending just 6 bp upstream of pvdA. This open reading frame did not have any homologous sequences in the database. No evident −35 and −10 consensus promoter sequences were identified within the 231 bp sequenced upstream of pvdA. There were no sequences with significant homology to the −10 region upstream of the T1 promoter of P. aeruginosa pvdA or to the proposed −35 consensus sequence of ςE-dependent promoters. There were no apparent consensus sequences in this upstream region which might be predicted to bind PvdS or Fur homologs (26, 41).
FIG. 2.
Computer-generated alignment of the deduced amino acid sequence of B. cepacia PvdA with P. aeruginosa PvdA (64) (accession number A49892), Ustilago maydis Sid1 (32) (accession number A47266), B. bronchiseptica AlcA (19) (accession number U32117), and E. coli IucD (22) (M18968) determined by using the programs PC/GENE CLUSTAL and Seqvu. Boxed shaded areas indicate amino acids identical in at least four of the five proteins. Unboxed shaded areas indicate regions of similarity. Gaps introduced to increase homology are indicated by dashes.
The predicted l-ornithine N5-oxygenase from B. cepacia is slightly larger than the protein from P. aeruginosa which consists of 426 amino acids and has a molecular mass of 47.7 kDa. The amino acid sequences are 47% identical and 59% similar. The deduced amino acid sequence of B. cepacia PvdA had extensive homology to three other enzymes: l-ornithine N5-oxygenase from the fungus Ustilago maydis (sid1 [32]); l-lysine N6-hydroxylase, which is required for synthesis of the siderophore aerobactin in E. coli (iucD [22]); and an enzyme from Bordetella bronchiseptica required for synthesis of the siderophore alcaligin (alcA [19]). The optimal alignment of the PvdA, Sid1, IucD, and AlcA sequences is shown in Fig. 2. B. cepacia PvdA was approximately 46% similar to Sid1 and 24% similar to AlcA and IucD. The greatest similarity was observed in the N-terminal half of the protein. The two PvdA genes and the IucD gene contain a putative flavin-binding motif (IGVG[T/F]GP) beginning at residues 10, 14, and 8, respectively (Fig. 2) (64). This sequence is not well conserved in Sid1 or AlcA.
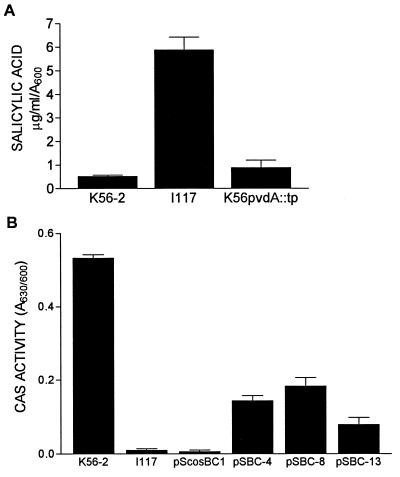
P. aeruginosa mutants which lacked l-ornithine N5-oxygenase activity were previously shown to produce pyoverdine at wild-type levels when supplemented with the biosynthetic precursor l-N5-OH-Orn (64). To determine whether provision of this precursor could restore ornibactin synthesis in I117, K56-2 and I117 were grown in succinate medium in the presence or absence of 400 μM l-N5-OH-Orn. Culture supernatants were assayed for ornibactin production by the CAS assay. Addition of l-N5-OH-Orn to the culture medium had no effect on CAS activity in K56-2 since supernatants contained 1.10 U of CAS activity in medium with l-N5-OH-Orn and 1.07 U of CAS activity in medium without the precursor. CAS activity in I117, however, was restored to approximately 30% of parental levels (0.41 U) when l-N5-OH-Orn was added to the medium. Since addition of l-N5-OH-Orn did not fully restore ornibactin synthesis to parental levels, it is possible that this compound is not taken up very well by B. cepacia, or that the transposon insertion in I117 may affect other genes involved in ornibactin synthesis in addition to pvdA.
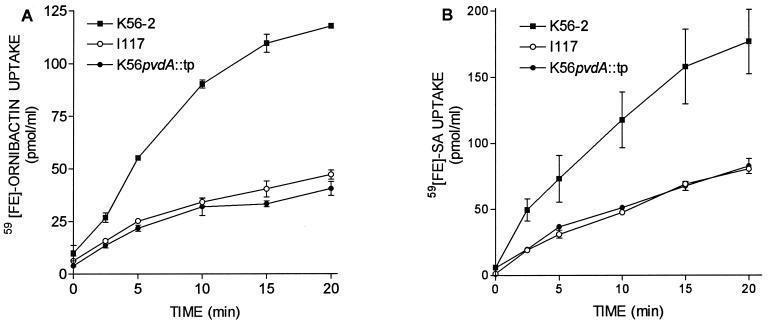
To determine whether the phenotype of I117 was due to the transposon insertion in the pvdA gene or possible polar effects, a pvdA deletion mutant was constructed. An approximately 0.7-kb BglII fragment was deleted from within the pvdA gene, and a trimethoprim resistance cassette was inserted in frame into the BglII site. The inactivated gene was inserted into the chromosome by allelic exchange and designated K56pvdA::tp. This mutant was negative for ornibactin production as measured by CAS activity. The rates of 59Fe-ornibactin and 59Fe-SA uptake in K56pvdA::tp were similar to I117 (Fig. 3). The amount of SA produced by K56pvdA::tp was only slightly greater than that produced by K56; however, the yield of SA in supernatants of I117 was approximately 10-fold greater than in supernatants from K56-2 (Fig. 4A). This suggests that the increase in SA production observed in I117 may be due to either polar effects of the transposon insertion or to a second mutation in I117.
FIG. 3.
Comparison of 59Fe-siderophore uptake by I117 and K56pvdA::tp. (A) Uptake of 59Fe-ornibactins. (B) Uptake of 59Fe-SA as described in Fig. 1. Values represent the means ± the standard deviations of triplicate assays. These experiments were repeated at least three times with similar results.
FIG. 4.
Siderophore production assays. (A) Comparison of SA production by K56-2, I117, and K56pvdA::tp. Values represent the means ± the standard deviations of triplicate assays. I117 significantly different than K56-2 and K56pvdA::tp (P < 0.001) by using analysis of variance (ANOVA). (B) Partial complementation of I117 CAS activity (ornibactin production) by cosmid clones pSBC-4, pSBC-8, and pSBC-13. Values represent the means ± the standard deviation of triplicate assays. CAS activity is per 10 μl of culture supernatant. K56-2, pSBC-4, pSBC-8, and pSBC-13 are significantly different than I117 or pScosBC1 (P < 0.001) by ANOVA.
Analysis of the nucleotide sequence flanking the ends of Tn5-OT182 revealed that the transposon had been inserted into the chromosome of mutant I117 just after amino acid residue 293, at approximately two-thirds the distance of the protein from the N terminus. To determine if the cloned pvdA gene could complement the mutation in I117 and restore ornibactin production, the 6.1-kb SphI fragment from pPD519 was ligated into the SphI site of pUCP28T (pPD520) and introduced into I117 by electroporation. I117 (pPD520) did not produce orange zones on CAS agar, nor did it demonstrate CAS activity in culture supernatants, indicating that the cloned pvdA gene was not able to complement the pvdA mutation in I117. A 4.3-kb BamHI fragment containing a lacZ-Gmr cassette was ligated into the BglII site in pvdA on pPD520 in both orientations. Neither of these constructs expressed β-galactosidase activity, indicating that the pvdA gene was not expressed from this construct and therefore was not able to complement I117.
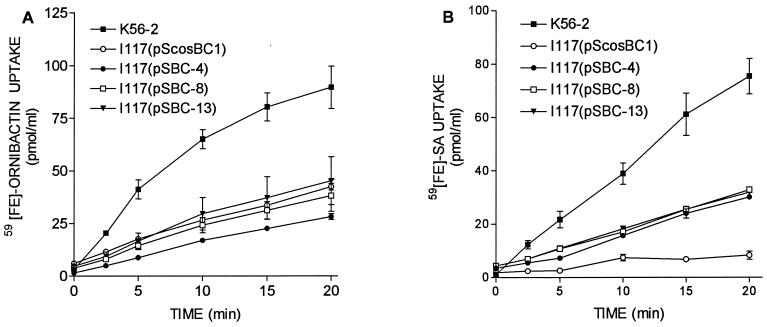
In order to isolate a larger fragment with the potential to complement I117, a cosmid library of K56-2 was constructed in the vector pScosBC1 and screened for clones containing pvdA by colony hybridization. Six positive clones were identified, and three were electroporated into I117 and analyzed for complementation. I117 containing either of these clones (pSBC-4, pSBC-8, and pSBC-13) produced zones on CAS agar. The CAS activity in culture supernatants of these strains is shown in Fig. 4B. These cosmids were able to partially restore CAS activity in I117 but not to parental levels. pSBC-4 and pSBC-8 restored CAS activity to slightly higher levels than pSBC-13. The ability of these cosmids to complement the defects in siderophore uptake by I117 was examined, and none of the cosmids had an effect on 59Fe-ornibactin uptake (Fig. 5A). However, all three of the cosmids partially restored the ability of I117 to accumulate 59Fe-SA (Fig. 5B). These cosmids also partially restored the level of SA produced by I117 to levels between that of the K56-2 and I117 containing the vector control (data not shown). The three cosmids contained overlapping inserts ranging in size from 35 to 40 kb.
FIG. 5.
Comparison of the ability of cosmid clones to restore 59Fe-ornibactin uptake (A) or 59Fe-SA uptake (B) in I117. Assays were performed as described in Fig. 1. Values represent the means ± the standard deviations of triplicate assays. These experiments were repeated at least three times, with similar results.
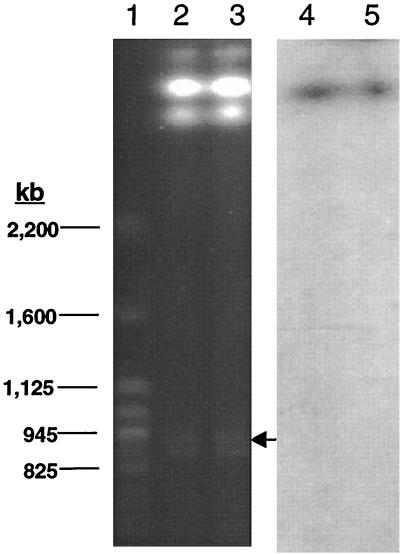
Bacteria belonging to the B. cepacia complex (63) possess large genomes which consist of multiple replicons (7, 43). Basic genomic maps have been published for strain ATCC 17616 (7), now classified as Burkholderia multivorans (63), and strain ATCC 25416 (43), now classified as B. cepacia complex genomovar I (63). Strain K56-2 belongs to the cable-pilus encoding epidemic cystic fibrosis strain lineage (11, 30) which has been classified as B. cepacia genomovar III (63). To determine whether pvdA was present on more than one replicon in K56-2, intact genomic DNA was resolved by PFGE, transferred to nylon membrane, and hybridized with a pvdA probe. Three replicons, ca. 3.5, 3.0, and 0.8 Mb in size, were detected by PFGE (Fig. 6), indicating that B. cepacia genomovar III strains possess a multireplicon genome structure in common with B. multivorans (7) and B. cepacia genomovar I (43). The pvdA probe hybridized only to the largest chromosome (Fig. 6) correlating to the presence of a single copy of the gene detected by conventional Southern hybridization described above.
FIG. 6.
(Lanes 1 to 3) Resolution of the linearized forms of the three large replicons (3.5, 3.0, and 0.8 Mb) from K56-2 by PFGE. Lane 1, Saccharomyces cerevisiae YNN295 molecular weight marker; lanes 2 and 3, K56-2 genomic DNA. (Lanes 4 and 5) Southern hybridization of PFGE in panel A hybridized to [32P]dCTP-labelled pvdA probe. The gel and autoradiogram were scanned with Hewlett-Packard ScanJet 4c and Hewlett-Packard Deskscan II software.
Role of the pvdA gene in virulence.
B. cepacia causes chronic respiratory infections, and the agar bead model of Cash et al. (6) has previously been used to assess the virulence of this organism in chronic lung infections (56, 59). Therefore, this model was selected to determine the importance of ornibactin biosynthesis in the pathogenesis of B. cepacia infections. In one experiment, rats were infected with K56-2 and I117. Quantitative bacteriology and quantitative pathology analysis were performed on days 7 and 28 postinfection (p.i.). Rats were infected with approximately 104 CFU of bacteria. On day 7 p.i., the numbers of K56-2 in the lungs had increased to 4.3 × 106 CFU/ml or 1.3 × 107 CFU/lung, whereas the numbers of I117 had decreased to 3.1 × 102 CFU/ml (Table 4). On day 28 p.i., K56-2 continued to colonize the lungs at a concentration of 3.3 × 105 CFU/ml. I117, however, was recovered from the lungs at a concentration 4 logs lower than K56-2 on both days 7 and 28 p.i. In a second experiment, K56-2 was compared to K56pvdA::tp by using the same protocol. On day 7 p.i. the number of K56-2 recovered from the lung was more than 2 logs higher than the mutant, and on day 28 p.i. K56pvdA::tp was no longer recovered from the lungs, indicating that the infection had cleared (Table 4).
TABLE 4.
Virulence of K56-2 and pvdA mutants in a chronic respiratory infection modela
| Strain | Day 7 p.i.
|
Day 28 p.i.
|
||
|---|---|---|---|---|
| CFU/ml/lung | % Pathology | CFU/ml/lung | % Pathology | |
| Expt 1 | ||||
| K56-2 | 4.3 × 106 ± 4.1 × 106 | 51.5 ± 32.9 | 3.3 × 105 ± 5.1 × 105 | 22.0 ± 9.8 |
| I117 | 3.1 × 102 ± 2.1 × 102b | 26.2 ± 7.7 | 4.0 × 101 ± 6.1 × 101b | 5.0 ± 3.1c |
| Expt 2 | ||||
| K56-2 | 1.5 × 106 ± 2.5 × 106 | 40.5 ± 4.2 | 2.4 × 105 ± 4.5 × 105 | 39.0 ± 4.7 |
| K56pvdA::tp | 7.2 × 103 ± 1.3 × 104d | 15.2 ± 5.0d | 0d | 13.5 ± 3.5d |
Values represent the means ± the standard deviations of four animals.
I117 is significantly different than K56-2 (P < 0.05) by the one-tailed Mann-Whitney t test.
I117 is significantly different than K56-2 (P < 0.05) by the unpaired t test.
K56pvdA::tp is significantly different than K56-2 (P < 0.05) by ANOVA.
The pathology of K56-2 compared to each of the mutant strains was also examined on days 7 and 28 p.i. On day 7 p.i., the degree of pathology was approximately 50% greater in rats infected with K56-2 than in rats infected with I117, and it was about 60% greater than in lungs of rats infected with K56pvdA::tp (Table 4). On day 28 p.i., the pathology observed with K56-2-infected lungs was fourfold greater than in lungs infected with I117. The extent of pathology was similar in animals infected with K56pvdA::tp on both days 7 and 28 p.i., and on both days significantly less pathology was noted than in lungs infected with the parent strain. These data indicate that the ability of B. cepacia to produce and take up ornibactin is important for colonization and persistence in lung infections and also contributes to the pathological damage that occurs during infections with B. cepacia.
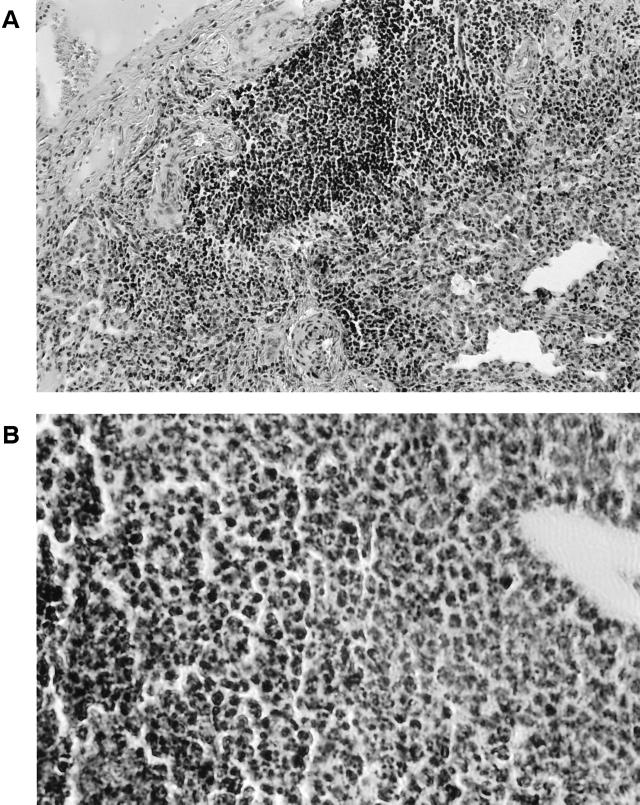
The histopathological changes associated with B. cepacia infections of murine lungs were characterized by a mixed cellular infiltrate composed predominantly of mononuclear phagocytes and lymphocytes with some polymorphonuclear leukocyte involvement (Fig. 7A). The lung architecture during B. cepacia infections remained relatively intact. There was a marked increase in lymphoid follicle size associated with the predominantly lymphocytic response (Fig. 7B). There were no observed qualitative differences in the pathology observed in lungs infected with B. cepacia K56-2 versus strains I117 and K56pvdA::tp.
FIG. 7.
Hematoxylin-and-eosin-stained sections of rat lungs infected with B. cepacia K56-2. (A) Mixed cellular infiltrate composed primarily of lymphocytes and mononuclear phagocytes. Also shown is a swollen lymphoid follicle (magnification, ×75). (B) Photomicrograph of higher-power magnification (×150) demonstrating the mononuclear nature of the infiltrate.
Establishment of infections by using the agar bead model bypasses the normal colonization mechanisms of the organism, since the bacteria are introduced directly into the lung by intratracheal inoculation. Since the results obtained from the agar bead model suggested that pvdA may also play a role in colonization of the lung, an acute respiratory infection model in mice was used to assess the importance of pvdA in colonization. Neutropenic mice were infected with K56-2 or I117 by aerosol administration. On days 0, 3, and 7 p.i., the lungs were removed from groups of five mice, and quantitative bacteriology was performed. As shown in Table 5, K56-2 was able to colonize the lungs and persist for the 7-day period. There was no significant difference between the numbers of K56-2 organisms present in the lungs on day 0, 3, or 7 p.i., which suggests that there is a balance between in vivo growth and clearance from the lung. Strain I117, however, was only transiently able to colonize the lung and was cleared from the majority of the mice by day 3. These data suggest that ornibactin biosynthesis and uptake may also play a role in the early stages of colonization.
TABLE 5.
Virulence of K56-2 and I117 in an acute respiratory infection model
| Strain | Mean CFU/ml/lunga ± SD
|
||
|---|---|---|---|
| Day 0 | Day 3 | Day 7 | |
| K56-2 | 3,926 ± 5,712 | 542 ± 923 | 697 ± 1330 |
| I117 | 792 ± 74 | 2 ± 4b | 3 ± 5b |
Values represent means ± the standard deviations of five animals.
I117 is significantly different than K56-2 (P < 0.001) by ANOVA.
DISCUSSION
B. cepacia ornibactin-deficient mutants identified by transposon insertion mutagenesis had insertions in genes homologous to genes involved in pyoverdine production in P. aeruginosa. Due to the degree of sequence homology and similar function, these genes were given the same designation as their P. aeruginosa homologs, pvdA and pvdD, respectively. K56-2, the parent strain, produces three ornibactins, designated ornibactin-C4, ornibactin-C6, and ornibactin-C8, which vary in the length of the acyl group. I117 and T10 did not produce any of the three ornibactin molecules.
The pvdA gene was cloned and sequenced from B. cepacia K56-2, and its product was determined to have significant homology with other ω-amino acid oxygenases. Therefore, enzymes involved in the hydroxylation of an ω-amino acid and its acylation are conserved steps in the synthesis of hydroxamate-like siderophores. These enzymes characterize a unique reaction that is only found in bacterial and fungal species (64) and, therefore, may represent a potential therapeutic target for the development of antimicrobial compounds.
Previously, Visca et al. (64), using a DNA probe containing the P. aeruginosa pvdA gene, demonstrated that B. cepacia and P. fluorescens DNA fragments hybridized with the probe, suggesting that l-ornithine N5-oxygenases are conserved among Pseudomonas and Burkholderia species. Tabacchioni et al. (61), however, were not able to demonstrate hybridization of B. cepacia DNA with the same P. aeruginosa pvdA probe. These conflicting reports, which included the same strain, BC TVV75, suggest that even though the l-ornithine N5-oxygenases are very similar between these two species, hybridization results with the two pvdA genes are somewhat variable. Therefore, it may have been difficult to clone the B. cepacia pvdA gene by using a hybridization strategy with a P. aeruginosa pvdA probe.
Despite the extensive similarity between the PvdA proteins of B. cepacia and P. aeruginosa, some differences have also been observed. Pyoverdine synthesis is restored to wild-type levels when a P. aeruginosa pvdA mutant is fed the precursor l-N5-OH-Orn (46). In B. cepacia, provision of the precursor l-N5-OH-Orn only restored ornibactin yields to approximately 30% of parental levels; however, there may be differences between these two organisms in their abilities to take up this compound. In P. aeruginosa, a 1.7-kb SphI fragment containing the pvdA gene was sufficient to complement a pvdA mutant and fully restore pyoverdine synthesis (64). In B. cepacia, attempts to complement a pvdA mutant with a 6.1-kb SphI fragment containing the pvdA gene were not successful in restoring ornibactin biosynthesis. Construction of a pvdA::lacZ fusion in this construct confirmed that additional sequences not contained on the 6.1-kb SphI fragment were required for pvdA expression. The vector pScosBC1 containing 35- to 40-kb inserts that included pvdA was able to restore ornibactin synthesis to approximately 30% of parental levels. Although both B. cepacia pvdA and pvdD were found to be iron regulated, as shown by the effect of iron on β-galactosidase activity in the transposon-generated lacZ fusion, no promoter regions similar to those described for P. aeruginosa pvdA (26) were identified within the 231 bp sequenced upstream of B. cepacia pvdA. These studies suggest that, although the enzymes in the biosynthetic pathways of pyoverdine and ornibactins may be highly conserved, there may be differences in the organization and regulation of genes required for the biosynthesis and uptake of these siderophores.
A probe constructed from the pvdD sequences flanking the transposon insertion did not hybridize to pPD519, indicating that pvdD is not adjacent to pvdA. This probe did hybridize to the cosmid clones, indicating that at least a portion of the pvdD gene is contained within these 35- to 40-kb fragments (data not shown).
The B. cepacia pvdA and pvdD mutants were determined to be defective in the uptake of ornibactins even when exogenous ornibactins were added to the culture medium to induce the expression of the ornibactin transport mechanism. In P. aeruginosa, the pvdD gene has been shown to be located just upstream of fpvA, the gene for the outer membrane receptor for pyoverdine (34). The two open reading frames are in the opposite orientation. Although the outer membrane receptor(s) for ferric ornibactins have not yet been described, we compared the outer membrane protein profiles of K56-2, I117, and T10 grown in low-iron and high-iron media for the absence of any iron-regulated proteins (data not shown). There were no dramatic differences observed in the outer membrane protein profiles between either of these mutants and the parent strain, suggesting that these mutants were not deficient in the expression of a specific ornibactin receptor. I117 and T10 were also deficient in the uptake of SA. These data suggest that the transposon insertions in these strains are also affecting a gene that is required for both siderophore-mediated iron transport systems. The transposon may be causing polar effects on a gene downstream of the insertion site. Mutations in tonB, exbB, or exbD homologs, for example, would likely have phenotypes similar to those of I117 and T10 in terms of their inability to accumulate iron complexed to various siderophores (4). It is also possible that there is a common enzyme required for the release of 59Fe from the siderophore that is affected by the transposon insertion. Insertion of Tn5-OT182 does not have nonspecific effects on siderophore-mediated iron uptake, as many other K56-2 isolates containing transposon insertions had normal iron uptake phenotypes (data not shown). An allelic exchange mutant created by insertion of a Tpr cassette in the pvdA gene was also deficient in uptake of either 59Fe-ornibactins or 59Fe-SA, suggesting that ornibactins are required for optimum expression of some components of siderophore-mediated iron uptake mechanisms. It is not clear why ornibactin biosynthesis would be required to induce the expression of genes involved in the SA uptake system unless these genes encoded proteins that were common to both systems.
I117 and T10 hyperproduced SA, which suggests that the genes involved in SA biosynthesis are upregulated when ornibactins are not produced. Strain K56-2 does not produce cepabactin and produces very low levels of pyochelin. No dramatic increase in the yields of pyochelin were observed in any of these mutants, and cepabactin production was not detected, suggesting that the upregulation of SA observed may be specific for this siderophore or that SA is the only siderophore that K56-2 is capable of synthesizing in greater amounts.
Previously, SA has been shown to bind iron, remove iron from transferrin in an equilibrium dialysis assay, promote growth in iron-restricted conditions, and promote iron uptake (54, 65). Purified SA has been shown to have weak activity in the CAS assay, but it requires the addition of 5-sulfosalicylic acid (54). The current study indicates that the levels of SA produced in the culture medium are not active in the CAS assay, even in the mutants that produce up to 10-fold-higher amounts of SA than the parent strain. The increased production of SA in ornibactin mutants may be a necessary response due to the apparent lower affinity for iron of SA compared to ornibactins and the reduced ability of these mutants to take up iron-SA complexes. K56pvdA::tp produced levels of SA only slightly greater than the parental levels of SA. The increase observed may be more due to the level of iron starvation of the cells than to a specific upregulation of SA biosynthesis due to this pvdA mutation. These data suggest that the increase in SA production observed in I117 and T10 may be due to polar effects of the transposon.
Since pPD519 containing approximately 1 kb of DNA upstream of the pvdA gene was not sufficient for complementation of ornibactin production in I117, a cosmid library of K56-2 was constructed. Three cosmid clones that contained pvdA complemented CAS activity in I117 and partially restored SA production and uptake to parental levels. The cosmids were not examined for their ability to complement K56pvdA::tp, since the resistance marker on both this mutant and the cosmid vector is trimethoprim. The cosmids were found to be somewhat unstable in I117 and were not well maintained in the absence of antibiotic selection. It was also observed that fragments of DNA were frequently lost from these vectors, a result possibly due to recombination. This may be the reason that siderophore production and uptake were not complemented to parental levels. The partial complementation may also be due to low levels of gene expression from these vectors in I117. It was surprising that these cosmids were able to partially restore SA-mediated iron uptake, but not Fe-ornibactin uptake. They may not be large enough to contain the appropriate genes, or the levels of ornibactin produced by I117 containing these clones may be too low to induce the required component(s) of the ornibactin uptake system.
The ability of B. cepacia to produce and take up ornibactin was demonstrated to be critical in the establishment of infections by using an acute lung model. I117 was only transiently able to colonize lungs of neutropenic mice and was almost cleared from the lungs by days 3 and 7 p.i. This suggests that iron may be required for some mechanisms involved in adherence or colonization. In the agar bead infection model, I117 and K56pvdA::tp were able to persist in the lung for at least 7 days p.i. but at a significantly lower level than was the parent strain. On day 28 p.i., most of these animals had also cleared the pvdA mutant strains from the lungs, whereas the parent strain maintained an infection level of approximately 105 CFU. The degree of pathology observed in animals infected with the pvdA mutants was only 40 to 50% of that observed in lungs of animals infected with K56-2 on day 7 p.i. and only 25 to 30% on day 28 compared to strain K56-2. These mutants were severely compromised in their ability to acquire iron since they were also at least partially deficient in their ability to take up iron complexed to SA. These strains also do not produce pyochelin, which previously has been shown to contribute to virulence of B. cepacia respiratory infections (52, 56). It would be interesting to examine the contribution of ornibactin synthesis and uptake to virulence in a strain that produces pyochelin and/or cepabactin.
It is of significant interest to note the differences in the histopathological picture associated with B. cepacia versus those observed in P. aeruginosa infections of murine lungs. The lungs of both mice and rats infected with B. cepacia presented a histopathological picture characterized by a mixed cellular infiltrate composed predominantly of mononuclear phagocytes and lymphocytes. This is contrasted with the histopathological changes associated with P. aeruginosa infections of murine lungs that are characterized by multiple microabscesses, mucus plugging, and alveolar destruction (6). The reasons for these very different histopathological pictures are not clear; however, the explanation for these differences is unlikely to be associated with iron uptake since there were no qualitative differences in the histopathology observed between B. cepacia K56-2 and the mutant strains I117 and K56pvdA::tp. Studies are ongoing to determine those B. cepacia factors that might play a role in the histopathological changes associated with lung infections due to these organisms.
ACKNOWLEDGMENTS
This study was supported by grants from the Canadian Cystic Fibrosis Foundation to P.A.S. and E.M. and from the Animal Studies Core of the Special Program in Applied Therapeutics (SPARX) from the CCFF to D.E.W.
We thank Francis Green for assistance with the interpretation of the pathology, Jocelyn Bischof for technical assistance with construction of the cosmid library, and Tod Strugnell for technical assistance with the animal experiments.
REFERENCES
- 1.Ankenbauer R, Hanne L, Cox C D. Mapping of mutations in Pseudomonas aeruginosa defective in pyoverdin production. J Bacteriol. 1986;167:7–11. doi: 10.1128/jb.167.1.7-11.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atkin C L, Neilands J B. Rhodotorulic acid, a diketopiperazine dihydroxamic acid with growth-factor activity. I. Isolation and characterization. Biochemistry. 1968;7:3734–3739. doi: 10.1021/bi00850a054. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 1. New York, N.Y: John Wiley & Sons, Inc.; 1989. [Google Scholar]
- 4.Braun V, Gunter K, Hantke K. Transport of iron across the outer membrane. Biol Metals. 1991;4:14–22. doi: 10.1007/BF01135552. [DOI] [PubMed] [Google Scholar]
- 5.Cantin, A. M., and D. E. Woods. Prolastin suppresses lung damage, inflammation and bacterial proliferation in a model of chronic Pseudomonas aeruginosa lung infection. Am. Rev. Res. Crit. Care Med., in press. [DOI] [PubMed]
- 6.Cash H A, Woods D E, McCullough B, Johanson W G, Bass J A. A rat model of chronic respiratory infection with Pseudomonas aeruginosa Am. Rev Respir Dis. 1979;119:453–459. doi: 10.1164/arrd.1979.119.3.453. [DOI] [PubMed] [Google Scholar]
- 7.Cheng H-P, Lessie T G. Multiple replicons constituting the genome of Pseudomonas cepacia 17616. J Bacteriol. 1994;176:4034–4042. doi: 10.1128/jb.176.13.4034-4042.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crosa J H. Genetics and molecular biology of siderophore-mediated iron transport in bacteria. Microbiol Rev. 1989;53:517–530. doi: 10.1128/mr.53.4.517-530.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cryz S J, Jr, Furer E, Germanier E. Simple model for the study of Pseudomonas aeruginosa infections in leukopenic mice. Infect Immun. 1983;39:1067–1071. doi: 10.1128/iai.39.3.1067-1071.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cunlliffe H E, Merriman T R, Lamont I A. Cloning and characterization of pvdS, a gene required for pyoverdine synthesis in Pseudomonas aeruginosa: PvdS is probably an alternative sigma factor. J Bacteriol. 1995;177:2744–2750. doi: 10.1128/jb.177.10.2744-2750.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Darling P, Chan M, Cox A, Sokol P A. Siderophore production by cystic fibrosis isolates of Burkholderia cepacia. Infect Immun. 1998;66:874–877. doi: 10.1128/iai.66.2.874-877.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dennis J J, Sokol P A. Electrotransformation of Pseudomonas. In: Nickoloff J A, editor. Electroporation and electrofusion of microorganisms. Clifton, N.J: Humana Press; 1995. pp. 125–133. [DOI] [PubMed] [Google Scholar]
- 13.DeShazer D, Brett P J, Carlyon R, Woods D E. Mutagenesis of Burkholderia pseudomallei with Tn5-OT182: isolation of motility mutants and molecular characterization of the flagellin structural gene. J Bacteriol. 1997;179:2116–2125. doi: 10.1128/jb.179.7.2116-2125.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeShazer D, Woods D E. Broad-host range cloning and cassette vectors based on the R388 trimethoprim resistance gene. BioTechniques. 1996;20:762–764. doi: 10.2144/96205bm05. [DOI] [PubMed] [Google Scholar]
- 15.Dunnil M S. Quantitative methods in the study of pulmonary pathology. Thorax. 1962;17:320–328. doi: 10.1136/thx.17.4.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Figuski D H, Helinski D R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gaballa A, Liedam N, Cornelis P. A cytochrome c biogenesis gene involved in pyoverdine production in Pseudomonas fluorescens ATCC 17400. Mol Microbiol. 1996;21:777–785. doi: 10.1046/j.1365-2958.1996.391399.x. [DOI] [PubMed] [Google Scholar]
- 18.Gensberg K, Hughes K, Smith A W. Siderophore-specific induction of iron uptake in Pseudomonas aeruginosa. J Gen Microbiol. 1993;138:2381–2387. doi: 10.1099/00221287-138-11-2381. [DOI] [PubMed] [Google Scholar]
- 19.Giardina P C, Foster L A, Toth S I, Roe B A, Dyer D W. Identification of alcA, a Bordetella bronchiseptica gene necessary for alcaligin production. Gene. 1995;167:133–136. doi: 10.1016/0378-1119(95)00659-1. [DOI] [PubMed] [Google Scholar]
- 20.Goldberg J B, Ohman D E. Cloning and expression in Pseudomonas aeruginosa of a gene involved in the production of alginate. J Bacteriol. 1984;158:1115–1121. doi: 10.1128/jb.158.3.1115-1121.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Govan J R W, Hughes J E, Vandamme P. Burkholderia cepacia: medical, taxonomic and ecological issues. J Med Microbiol. 1996;45:395–407. doi: 10.1099/00222615-45-6-395. [DOI] [PubMed] [Google Scholar]
- 22.Herrero M, de Lorenzo V, Neilands J B. Nucleotide sequence of the iucD gene of the pColV-K30 aerobactin operon and topology of its product studied with phoA and lacZ gene fusions. J Bacteriol. 1988;170:56–64. doi: 10.1128/jb.170.1.56-64.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoang T T, Karkhoff-Schweizer R R, Kutchma A J, Schweizer H P. A broad host-range Flp-FRT recombination system for sip-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene. 1998;212:77–86. doi: 10.1016/s0378-1119(98)00130-9. [DOI] [PubMed] [Google Scholar]
- 24.Hopfl P, Ludwig W, Schleifer K H, Larsen N. The 23S ribosomal RNA higher-order structure of Pseudomonas cepacia and other prokaryotes. Eur J Biochem. 1989;185:355–364. doi: 10.1111/j.1432-1033.1989.tb15123.x. [DOI] [PubMed] [Google Scholar]
- 25.Koster M, van de Vossenberg J, Leong J, Weisbeek P J. Identification and characterization of the pupB gene encoding an inducible ferric-pseudobactin receptor of Pseudomonas putida WCS358. Mol Microbiol. 1993;8:591–601. doi: 10.1111/j.1365-2958.1993.tb01603.x. [DOI] [PubMed] [Google Scholar]
- 26.Leoni L, Ciervo A, Orsi N, Visca P. Iron-regulated transcription of the pvdA gene in Pseudomonas aeruginosa: effect of Fur and PvdS on promoter activity. J Bacteriol. 1996;178:2299–2313. doi: 10.1128/jb.178.8.2299-2313.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lewenza S, Conway B, Greenberg E P, Sokol P A. Quorum sensing in Burkholderia cepacia: identification of the LuxRI homologs CepRI. J Bacteriol. 1999;181:748–756. doi: 10.1128/jb.181.3.748-756.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.MacLachlan P R, Sanderson K E. Transformation of Salmonella typhimurium with plasmid DNA: differences between rough and smooth strains. J Bacteriol. 1985;161:442–445. doi: 10.1128/jb.161.1.442-445.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Madden T L, Tatusov R L, Zhang J. Applications of network BLAST server. Methods Enzymol. 1996;266:131–141. doi: 10.1016/s0076-6879(96)66011-x. [DOI] [PubMed] [Google Scholar]
- 30.Mahenthiralingam E, Simpson D A, Speert D P. Identification and characterization of a novel DNA marker associated with epidemic strains of Burkholderia cepacia recovered from patients with cystic fibrosis. J Clin Microbiol. 1997;35:808–816. doi: 10.1128/jcm.35.4.808-816.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McMorran B J, Merriman M E, Rombel I T, Lamont I L. Characterization of the pvdE gene which is required for pyoverdine synthesis in Pseudomonas aeruginosa. Gene. 1996;176:55–59. doi: 10.1016/0378-1119(96)00209-0. [DOI] [PubMed] [Google Scholar]
- 32.Mei B, Budde A D, Leong S A. sid1, a gene initiating siderophore biosynthesis in Ustilago maydis: molecular characterization, regulation by iron, and role in phytopathogenicity. Proc Natl Acad Sci USA. 1993;90:903–907. doi: 10.1073/pnas.90.3.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Merriman T R, Lamont I L. Construction and use of a self-cloning promoter probe vector for gram-negative bacteria. Gene. 1993;126:17–23. doi: 10.1016/0378-1119(93)90585-q. [DOI] [PubMed] [Google Scholar]
- 34.Merriman T R, Merriman M E, Lamont I L. Nucleotide sequence of pvdD, a pyoverdine biosynthetic gene from Pseudomonas aeruginosa: PvdD has similarity to peptide synthetases. J Bacteriol. 1995;177:252–258. doi: 10.1128/jb.177.1.252-258.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meyer J M, Hohnadel D, Halle F. Cepabactin from Pseudomonas cepacia, a new type of siderophore. J Gen Microbiol. 1989;135:1479–1487. doi: 10.1099/00221287-135-6-1479. [DOI] [PubMed] [Google Scholar]
- 36.Meyer J-M, Tran Van V, Stintzi A, Berge O, Winkelmann G. Ornibactin production and transport properties in strains of Burkholderia vietnamiensis and Burkholderia cepacia (formerly Pseudomonas cepacia) Biometals. 1995;8:309–317. doi: 10.1007/BF00141604. [DOI] [PubMed] [Google Scholar]
- 37.Mietzner T A, Morse S A. The role of iron-binding proteins in the survival of pathogenic bacteria. Annu Rev Nutr. 1994;14:471–493. doi: 10.1146/annurev.nu.14.070194.002351. [DOI] [PubMed] [Google Scholar]
- 38.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 39.Miyazaki H, Kato H, Nakazawa T, Tsuda M. A positive regulatory gene, pvdS, for expression of pyoverdin biosynthetic genes in Pseudomonas aeruginosa PAO. Mol Gen Genet. 1995;248:17–24. doi: 10.1007/BF02456609. [DOI] [PubMed] [Google Scholar]
- 40.Nakayashiki T, Nishimura K, Inokuchi H. Cloning and sequencing of a previously identified gene that is involved in the biosynthesis of heme in Escherichia coli. Gene. 1995;153:67–70. doi: 10.1016/0378-1119(94)00805-3. [DOI] [PubMed] [Google Scholar]
- 41.Ochsner U A, Vasil M L. Gene repression by the ferric uptake regulator in Pseudomonas aeruginosa: cycle selection of iron regulated genes. Proc Natl Acad Sci USA. 1996;93:4409–4414. doi: 10.1073/pnas.93.9.4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ohman D E, Sadoff J, Iglewski B H. Toxin A-deficient mutants of Pseudomonas aeruginosa PA-103: isolation and characterization. Infect Immun. 1980;28:899–908. doi: 10.1128/iai.28.3.899-908.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rodley P D, Römmling U, Tümmler B. A physical genome map of the Burkholderia cepacia type strain. Mol Microbiol. 1995;17:57–67. doi: 10.1111/j.1365-2958.1995.mmi_17010057.x. [DOI] [PubMed] [Google Scholar]
- 44.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 45.Sasakawa C, Yoshikawa M. A series of Tn5 variants with various drug-resistance markers and suicide vector for transposon mutagenesis. Gene. 1987;56:283–288. doi: 10.1016/0378-1119(87)90145-4. [DOI] [PubMed] [Google Scholar]
- 46.Schweizer H P. Escherichia-Pseudomonas shuttle vectors derived from pUC18/19. Gene. 1991;97:109–112. doi: 10.1016/0378-1119(91)90016-5. [DOI] [PubMed] [Google Scholar]
- 47.Schweizer H P. Allelic exchange in Pseudomonas aeruginosa using novel ColE1-type vectors and a family of cassettes containing a portable oriT and the counter-selectable Bacillus subtilis sacB marker. Mol Microbiol. 1992;6:1195–1204. doi: 10.1111/j.1365-2958.1992.tb01558.x. [DOI] [PubMed] [Google Scholar]
- 48.Schweizer H P. Two plasmids, X1918 and Z1918, for easy recovery of the xylE and lacZ reporter genes. Gene. 1993;134:89–91. doi: 10.1016/0378-1119(93)90178-6. [DOI] [PubMed] [Google Scholar]
- 49.Schweizer H P. Small broad-host-range gentamicin resistance gene cassette for site specific insertion and deletion mutagenesis. BioTechniques. 1993;15:831–834. [PubMed] [Google Scholar]
- 50.Schweizer H P, Klassen T, Hoang T. Improved methods for gene analysis and expression in Pseudomonas spp. In: Nakazawa T, Furukawa K, Haas D, Silver S, editors. Molecular biology of pseudomonads. Washington, D.C: ASM Press; 1996. pp. 229–237. [Google Scholar]
- 51.Schwyn B, Neilands J B. Universal chemical assay for the detection and determination of siderophores. Anal Biochem. 1987;160:47–56. doi: 10.1016/0003-2697(87)90612-9. [DOI] [PubMed] [Google Scholar]
- 52.Simon R, Priefer U, Puhler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 53.Sokol P A. Production and utilization of pyochelin by clinical isolates of Pseudomonas cepacia. J Clin Microbiol. 1986;23:560–562. doi: 10.1128/jcm.23.3.560-562.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sokol P A, Lewis C J, Dennis J J. Isolation of a novel siderophore from Pseudomonas cepacia. J Med Microbiol. 1992;36:184–189. doi: 10.1099/00222615-36-3-184. [DOI] [PubMed] [Google Scholar]
- 55.Sokol P A, Woods D E. Relationship of iron and extracellular virulence factors to Pseudomonas aeruginosa lung infections. J Med Microbiol. 1984;18:125–133. doi: 10.1099/00222615-18-1-125. [DOI] [PubMed] [Google Scholar]
- 56.Sokol P A, Woods D E. Effect of pyochelin on Pseudomonas cepacia respiratory infections. Microb Pathog. 1988;5:197–205. doi: 10.1016/0882-4010(88)90022-8. [DOI] [PubMed] [Google Scholar]
- 57.Stephan H, Freund S, Beck W, Jung G, Meyer J-M, Winkelmann G. Ornibactins—a new family of siderophores from Pseudomonas. Biometals. 1993;6:93–100. doi: 10.1007/BF00140109. [DOI] [PubMed] [Google Scholar]
- 58.Stephan H, Freund S, Meyer J M, Winklemann G, Gung G. Structure elucidation of the gallium-ornibactin complex by 2D-NMR spectroscopy. Liebigs Ann Chem. 1993;1993:43–48. [Google Scholar]
- 59.Straus D C, Woods D E, Lonon M K, Garner C W. The importance of extracellular antigens in Pseudomonas cepacia infections. J Med Microbiol. 1988;26:269–280. doi: 10.1099/00222615-26-4-269. [DOI] [PubMed] [Google Scholar]
- 60.Strohmaier H, Remler P, Renner W, Högenauer G. Expression of genes kdsA and kdsB involved in 3-deoxy-d-manno-octulosonic acid metabolism and biosynthesis of enterobacterial lipopolysaccharide is growth phase regulated primarily at the transcriptional level in Escherichia coli K-12. J Bacteriol. 1995;177:4488–4500. doi: 10.1128/jb.177.15.4488-4500.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tabacchioni S, Visca P, Chiarine L, Bevivino A, DiSerio C, Facelli S, Fani R. Molecular characterization of rhizosphere and clinical isolates of Burkholderia cepacia. Res Microbiol. 1995;146:531–542. doi: 10.1016/0923-2508(96)80559-6. [DOI] [PubMed] [Google Scholar]
- 62.Tsuda M, Miyzzaki H, Nakazawa T. Genetic and physical mapping of genes involved in pyoverdin production in Pseudomonas aeruginosa PAO. J Bacteriol. 1995;177:423–431. doi: 10.1128/jb.177.2.423-431.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vandamme P, Holmes B, Vancanneyt M, Coenye T, Hoste B, Coopman R, Revets H, Lauwers S, Gillis M, Kersters K, Govan J R W. Occurrence of multiple genomovars of Burkholderia cepacia in cystic fibrosis patients and proposal of Burkholderia multivorans sp. nov. Int J Syst Bacteriol. 1997;47:1188–1200. doi: 10.1099/00207713-47-4-1188. [DOI] [PubMed] [Google Scholar]
- 64.Visca P, Ciervo A, Orsi N. Cloning and nucleotide sequence of the pvdA gene encoding the pyoverdin biosynthetic enzyme l-ornithine N5-oxygenase in Pseudomonas aeruginosa. J Bacteriol. 1994;176:1128–1140. doi: 10.1128/jb.176.4.1128-1140.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Visca P, Ciervo A, Sanfilippo V, Orsi N. Iron-regulated salicylate synthesis by Pseudomonas spp. J Gen Microbiol. 1993;139:1995–2001. doi: 10.1099/00221287-139-9-1995. [DOI] [PubMed] [Google Scholar]
- 66.Visca P, Serino L, Orsi N. Isolation and characterization of Pseudomonas aeruginosa mutants blocked in the synthesis of pyoverdin. J Bacteriol. 1992;174:5727–5731. doi: 10.1128/jb.174.17.5727-5731.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wahl G M, Lewis K A, Ruiz J C, Rothenberg B, Zhao J, Evans G A. Cosmid vectors for rapid genomic walking, restriction mapping and gene transfer. Proc Natl Acad Sci USA. 1987;84:2160–2164. doi: 10.1073/pnas.84.8.2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Woods D. Oligonucleotide screening of cDNA libraries. Focus. 1984;6:1–2. [Google Scholar]