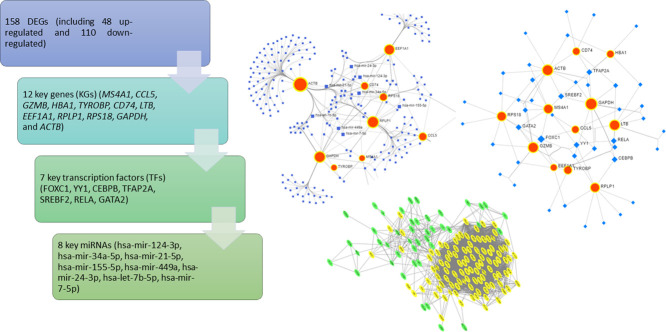
Highlights
-
•
DEGs were calculated from the publicly available scRNA seq data of NSCLC patients.
-
•
GO, KEGG, and PPI network analyses led to identification of 12 key genes.
-
•
Seven key transcription factors and 8 key miRNAs, were also identified.
-
•
Our finding might play a significant role as candidate biomarkers in NSCLC diagnosis and prognosis.
Keywords: Non-small cell lung cancer, Single-cell RNA-sequencing, Networking analysis, Gene biomarkers, Diagnosis
Abstract
Non-small cell lung cancer (NSCLC) is the most common type of lung cancer and the leading cause of cancer-related deaths worldwide. Identification of gene biomarkers and their regulatory factors and signaling pathways is very essential to reveal the molecular mechanisms of NSCLC initiation and progression. Thus, the goal of this study is to identify gene biomarkers for NSCLC diagnosis and prognosis by using scRNA-seq data through bioinformatics techniques. scRNA-seq data were obtained from the GEO database to identify DEGs. A total of 158 DEGs (including 48 upregulated and 110 downregulated) were detected after gene integration. Gene Ontology enrichment and KEGG pathway analysis of DEGs were performed by FunRich software. A PPI network of DEGs was then constructed using the STRING database and visualized by Cytoscape software. We identified 12 key genes (KGs) including MS4A1, CCL5, and GZMB, by using two topological methods based on the PPI networking results. The diagnostic, expression, and prognostic potentials of the identified 12 key genes were assessed using the receiver operating characteristics (ROC) curve and a web-based tool, SurvExpress. From the regulatory network analysis, we extracted the 7 key transcription factors (TFs) (FOXC1, YY1, CEBPB, TFAP2A, SREBF2, RELA, and GATA2), and 8 key miRNAs (hsa-miR-124-3p, hsa-miR-34a-5p, hsa-miR-21-5p, hsa-miR-155-5p, hsa-miR-449a, hsa-miR-24-3p, hsa-let-7b-5p, and hsa-miR-7-5p) associated with the KGs were evaluated. Functional enrichment and pathway analysis, survival analysis, ROC analysis, and regulatory network analysis highlighted crucial roles of the key genes. Our findings might play a significant role as candidate biomarkers in NSCLC diagnosis and prognosis.
Graphical abstract
Abbreviations
- NSCLC
Non-Small Cell Lung Cancer
- scRNA-seq
Single-cell RNA sequencing
- GEO
Gene Expression Omnibus
- DEGs
Differentially expressed genes
- GO
Gene Ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- PPI
Protein-protein interaction
- KGs
Key Genes
- ROC
Receiver Operating Characteristics
- TF
Transcription factors
- LUSC
Lung squamous cell carcinoma
- LUAD
Lung adenocarcinoma
- GAPDH
Glyceraldehyde-3 phosphate dehydrogenase
- t-SNE
t-distributed stochastic neighbor embedding
- STRING
Search Tool for the Retrieval of Interacting Genes
- AUC
Area under curve
- logFC
Log of Fold Change
- CI
Confidence Interval
- SVM
Support Vector Machine
- SCLC
Small Cell Lung Cancer
Introduction
Cancer, a heterogeneous disease, poses a serious challenge for precise treatment at the individual level. Both bulk and single-cell RNA sequencing (scRNA-seq) technologies are used for studying transcriptional profiles at the gene expression level. Several articles have identified biomarkers for NSCLC diagnosis using bulk RNAseq technology [1], [2], [3], [4], [5]. scRNA-sequencing categorizes the cell types across multiple tissues, whereas bulk RNA sequencing involves the use of a tissue or cell population [6,7]. scRNA-seq is widely used to determine tumor heterogeneity, cellular identities, novel biomarkers, and molecular and functional strategies [8]. Several scRNA-seq based studies have been performed earlier to explore tumor heterogeneity and to identify novel biomarkers for different cancers [9], [10], [11], [12].
Non-small cell lung cancer (NSCLC) is a highly heterogeneous lung cancer, accounting for approximately 85% of all the types of lung cancers, and it is strongly correlated with smoking habits [13,14]. NSCLC is mainly classified into two groups: lung squamous cell carcinoma (LUSC) and lung adenocarcinoma (LUAD). Most of the current treatment strategies for NSCLC are chemotherapies based on the histology and targeted agents for patients [15,16]. Treatment outcomes of NSCLC are quite insufficient as the post-therapy relapse rate and drug resistance remains high, and the 5-year relative survival rate of the patients is 26% [17,18]. Further exploration into the underlying mechanisms of NSCLC is thus quite urgent, which will impact the discovery of novel diagnostics and provide effective & key targets for NSCLC.
Expanding research recommends that affluent genes, miRNAs, TFs, and/or biological enriched pathways are associated with the development and progression of cancers. The altered expression of miRNAs has a strong correlation with distinct disease and carcinoma [19]. Gene-miRNAs interactions have been generally exhibited to control the convoluted molecular systems’ basic oncogenesis, advancement, intrusion, and tumor metastasis [20]. Several studies have recently shown a link of miRNA in cancer development [21]. Some studies have highlighted the importance of miRNA expression in lung tissues [22]. The deregulation of miRNA might play a role in fatal NSCLC progression [23]. Thus, several studies have identified some important TFs and miRNAs as transcriptional and post-transcriptional factors for cancer through the regulatory interaction networks [24,25].
In the last decade, several biomarkers have been reported as prognostic and predictive markers for NSCLC [2,[26], [27], [28], [29], [30]]. Song et al. identified STAT3, EGFR, PTEN, KRAS, TP53, RHOA, CTNNB1, and VEGFA, as efficient targeted genes associated with six miRNAs (hsa-miR-21-5p, hsa-miR-31-5p, hsa-miR-708-5p, hsa-miR-30a-5p, hsa-miR-451a, and has-miR-126-3p) by expression analysis and miRNA-hub gene network for NSCLC [2]. Chen et al. discovered four genes viz. CDK1, PLK1, RAD51, and RFC4 as novel biomarkers using microarray gene expression profiles that might be potential therapeutic targets in NSCLC [26]. Valk et al. identified SPAG5, POLH, KIF23, RAD54L, SGCG, NLRC4, MMRN1, and SFTPD as the novel genes that are involved in multiple pathways leading to NSCLC [31]. Puzone et al. showed that the overexpression of glyceraldehyde-3 phosphate dehydrogenase (GAPDH) correlates with poor prognosis in NSCLC patients [32]. These findings are important to understand NSCLC pathogenesis.
Although much research has already been conducted to reveal the molecular mechanism of NSCLC progression, the heterogeneity and complexity of NSCLC still poses a great challenge and need for novel and effective biomarkers. Recently, scRNA-seq technology has been used to detect tumor heterogeneity and explore the gene expression pattern in tissues that can help the researcher to detect the novel biomarkers. Here, we utilized computational models for analyzing the scRNA-seq data to reveal the tumor heterogeneity of NSCLC tissues. We identified differentially expressed genes (DEGs), their associated pathways, and PPI network to screen the key genes (KGs), key miRNAs, and key TFs for personalized diagnosis and prognosis of NSCLC and performed further analysis to validate the result. Thus, the identification of important genes, miRNAs, and TFs as well as the signaling pathways related to cancer via bioinformatics analysis, will provide worthy enlightenment in cancer research.
Materials and methods
scRNA-seq data collection and processing
The publicly available scRNA-seq data (GSE127471, data collected from the peripheral blood of a patient with NSCLC by Newman and the team) were downloaded from the gene expression omnibus (GEO) database [33]. The data were sequenced on Illumina NextSeq 500 (Human). Data processing was performed using the Seurat package V3.1.1 in R V3.6.1 [34]. For quality control check, we extracted genes with a minimum number of features 200 having non zero counts and a minimum number of cells as 3. The filtered data were normalized using log-transformation and was used for further analysis. We used two datasets associated with NSCLC from the GEO database with accession numbers GSE19188 and GSE75037 to assess the diagnostic performance of the identified KGs.
Clustering and DEG identification
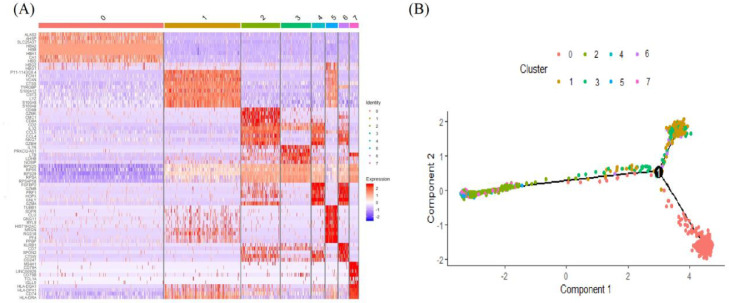
For dimensionality reduction, we performed principal component analysis (PCA) on the scaled data. The t-distributed stochastic neighbor embedding (t-SNE) was used to demonstrate two-dimensional data by first 10 principal components. The cell cluster was identified using K-means clustering based on the original Louvain algorithm. We used the Seurat's VlnPlot function to determine the expression of acquainted marker genes to assign clusters. Moreover, we constructed the trajectory analysis to reveal the tendency curve of the eight clusters using "Monocle" package [35]. The R package Seurat was used to analyze DEGs with scRNA-seq data.
Functional enrichment and pathway analysis of DEGs
We used a stand-alone software tool FunRich (version 3.1.4) for gene ontology (GO) functional enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of DEGs [36]. We considered the threshold P-value < 0.01 to obtain significant functional and pathway terms. KEGG and GO were used to annotate the enrichment analysis of bio-term classification processes and genes clusters as well as to impart a fantasy module for inference [37,38].
PPI network analysis of DEGs
Protein-protein interactions (PPIs) networks facilitate the analysis of pathogenic mechanisms and disease progression by providing knowledge on the molecular mechanism underlying cellular activity. In this study, we used the Search Tool for the Retrieval of Interacting Genes (STRING v11.0) database for constructing PPI network of DEGs [39]. Cytoscape (version 3.7.1) was used to discern the PPI networks between the DEGs [40]. We identify 12 KGs using two topological methods Betweenness and Stress in Cytoscape plugin cytoHubba [41].
Survival analysis and expression level of KGs
We used SurvExpress to check the impact of the expression pattern and survival analysis of KGs. SurvExpress (http://bioinformatica.mty.itesm.mx/SurvExpress) is an online tool for analyzing cancer gene expression data for the validation and survival analysis of multi-gene biomarkers [42]. Here we used it to verify and estimate the impact the expression pattern and prognostic value of KGs using the Kaplan-Meier curve and log-rank test.
ROC analysis of the KGs
Receiver operating characteristic (ROC) curve analysis was carried out to assess the true positive rate (Sensitivity) and false-positive rate (1− Specificity) of the identified KGs using the "pROC" package in R [43]. Area under curve (AUC) was determined and used to screen the ROC values.
Regulatory interaction network analysis of KGs
We constructed a regulatory interaction network (KGs-miRNAs and KGs-TFs) of 12 KGs using an online tool NetworkAnalyst 3.0 [44]. It is a bioinformatics tool that visualizes and deciphers the information in the association of network settings. This tool included three gene-miRNAs interaction databases TarBase [45], miRTarbase [46] and miRecords [47] and three gene-TFs interaction databases such as ENCODE [48], JASPAR [49], and ChEA [50]. We used TarBase and JASPAR databases for KGs-miRNAs and KGs-TFs interaction networks, respectively.
Results
Tumor heterogeneity and identification of DEGs
The scRNA-seq technology provided a good transcriptional detailing of cancer cells and gene expression in NSCLC patients. The 1803 cells were adopted for advanced analysis. The 1803 cells were analyzed and then classified into 8 separate clusters based on the identical gene set. Every cluster was separated by identical gene set. To make the segregation clearer among clusters, DEG analysis in cell types based on their covariance patterns and mean expression levels were evolved. We found that the identified marker gene sets were significant to ascertain cell types individually with high potentiality. Heatmap of the top ten genes showing high heterogeneity among clusters based on logFC has been illustrated in Fig. 1(A). The results of trajectory analysis showed the cells of only cluster1 may have a significant difference from other cells in NSCLC Fig. 1(B). We identified DEGs using the Wilcoxon rank sum test, based on the threshold adj.P.Val < 0.01 and logFC > 2 for up-regulated genes and adj.P.Val < 0.01 and logFC < -2 for down-regulated genes. A total of 158 DEGs, of which 48 were up-regulated and 110 down-regulated genes were identified. All DEGs (up- and down-regulated) are listed in Table 1.
Fig. 1.
(A) The heatmap of the top 10 marker genes of each cluster where each row represents genes and column represents clusters. (B) Trajectory plot differentiated by eight clusters.
Table 1.
Differentially expressed genes (DEGs) in NSCLC.
| Up-regulated DEGs | Down-regulated DEGs |
|---|---|
| TREML1, TAGLN2,LTB*,CCL5*, FTL, MNDA, LST1, SPARC, AIF1, ACRBP, AHSP,MS4A1*, CD79B, VCAN, ALAS2,CD74*, CST3, FCN1, PRF1, KLRB1,GZMB*, FGFBP2,TYROBP*, CMC1, CTSS, NRGN, RGS18, MYL9, HBD, CLU, TUBB1, SDPR, HIST1H2AC, S100A12, GNG11, CD79A, RP11-1143G9.4, CA1, LYZ, S100A9, S100A8, HBA2, GNLY, HBB, IGLL5, PF4,HBA1*, PPBP | MALAT1, RPS29, RPL39, MT-ND3, RPS27, RPS28, RPL37, RPL34, RPS26,ACTB*, RPS21, RPL23A, RPL36, MT-ND2, PTMA,EEF1A1*, MT-CO1, RPL28, RPL41, RPL38, RPL26, TMSB4X, HLA-C, RPL10, TMSB10, RPLP2, MT-CO3, MT-ND4, MT-ND1, RPL3,RPS18*, RPL27A, HA2, S100A6, RPS2, RPL27, RPS25, RPL36A, MT-ATP6, RPS8, B2M, RPS15A, HLA-B, RPL17, RPS24, RPL23,RPLP1*, MT-CO2, RPS19, RPL13A, RPL35A, MT-CYB, RPSA, RPS23, RPS3, RPL35, RPS10, RPS16, RPS15, RPS6, VIM, RPL32, RPS13, RPL13, S100A4, RPS4X, RPL30, RPL31, NBEAL1, TXNIP, TPT1, HLA-A, FAU, RPS7, NEAT1,GAPDH*, RPS3A, RPSAP58, RPS9, RPL19, RPL12, RPL15, RPL9, RPL11, RPS14, RPL6, ACTG1, H3F3A, RPL7, RPL7A, GNB2L1, RPS20, RPL5, PFN1, RPL29, RPL18, NKG7, RPL24, RPL22, TOMM7, RPL14, PABPC1, BTG1, RPL21,CFL1, RPS11, SRGN, RPL10A, RPL18A, HNRNPA1 |
N.B. Bold with star (*) indicates key genes (KGs).
Functional enrichment and pathway analysis of DEGs
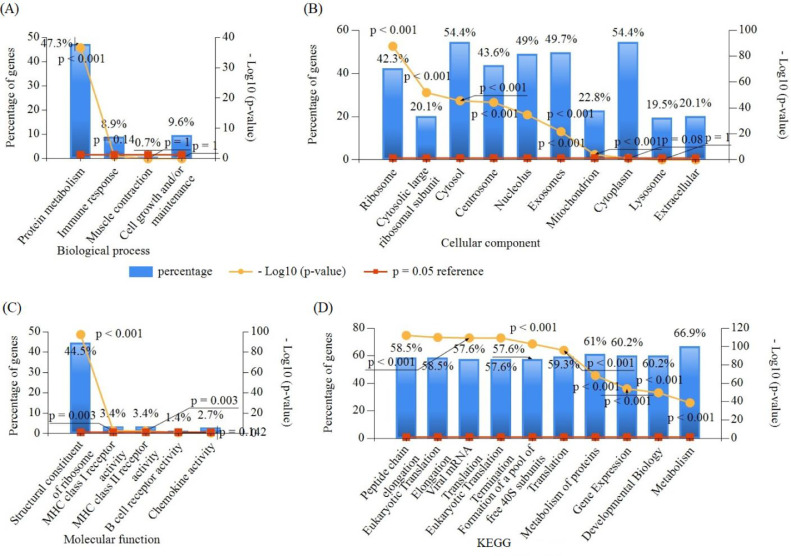
For further investigation, the GO functional and KEGG pathway enrichment analyses associated with the DEGs was performed in FunRich software. The top GO (BP, CC, and MF) and KEGG enrichment functions/terms of DEGs are shown in Fig. 2. In BP, 47.3% genes were enriched with protein metabolism, 8.90% genes were enriched in immune response, and 9.5% genes were enriched with cell growth and/or maintenance. In CC, 54.4% of genes were enriched in cytosol and cytoplasm; more than 40% of genes were enriched in exosomes, nucleolus, and ribosome; and approximately 30% of genes were enriched with cytosolic large ribosomal subunit, extracellular, lysosome, etc., where the MF enrichment was mainly correlated with structural constituent of ribosome, MHC class I receptor activity, MHC class II receptor activity, B cell receptor activity, and chemokine activity terms in Table S1. The enriched KEGG pathways for the DEGs included peptide chain elongation, eukaryotic translation elongation, eukaryotic translation termination, viral mRNA translation, 3′ -UTR-mediated translational regulation, and metabolism of proteins pathways, which are associated with lung cancer development (Table S2).
Fig. 2.
Top GO and KEGG terms enriched by DEGs ((A) Biological processes, (B) Cellular components, (C) Molecular functions, and (D) Kyoto encyclopedia of genes and genome (KEGG)).
PPI network of DEGs
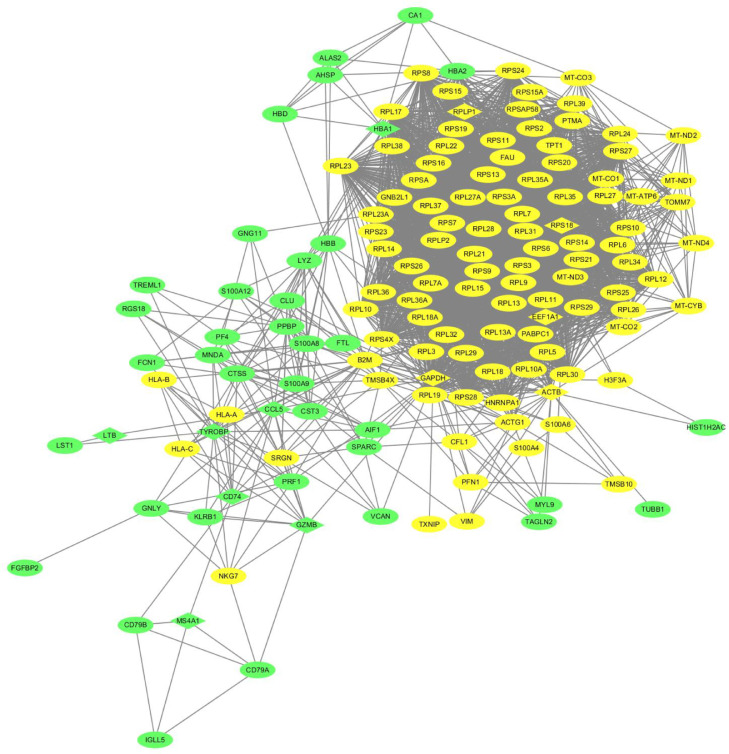
Based on the online tool STRING, a total of 158 DEGs were used in the PPI network, involving 156 nodes and 3282 edges with an average node degree 42.2 and PPI enrichment P-value < 1.0e-16 (Fig. 3). The top 15 genes were selected using each of the two network scoring methods Betweeness and Stress in Cytoscape plugin, cytoHubba. We then extracted 12 common genes from the top 15 genes of the two methods and considered them as KGs, such as MS4A1, CCL5, GZMB, HBA1, TYROBP, CD74, LTB, EEF1A1, RPLP1, RPS18, GAPDH, and ACTB, as shown in Table 2. In addition, we assessed the significance of the 12 KGs through survival analysis, cancer prediction model, and regulatory interaction network analysis.
Fig. 3.
Protein-protein interaction (PPI) network of DEGs. The green and yellow colors indicate up- and down-regulated genes respectively. The key genes (KGs) are highlighted in diamond shape.
Table 2.
The summary of the key genes (KGs) identified from the Cytoscape in NSCLC.
| S.N | Betweenness | Stress | Key Genes | ||
|---|---|---|---|---|---|
| Name | Score | Name | Score | MS4A1, CCL5, GZMB, HBA1, TYROBP, CD74, LTB, EEF1A1, RPLP1, RPS18, GAPDH, ACTB | |
| 1 | GAPDH | 6144.813 | GAPDH | 146026 | |
| 2 | MS4A1 | 2329.161 | MS4A1 | 43470 | |
| 3 | CCL5 | 1094.994 | CCL5 | 23206 | |
| 4 | ACTB | 1056.283 | HBA1 | 22566 | |
| 5 | GZMB | 830.074 | CD74 | 20158 | |
| 6 | HBA1 | 663.953 | GZMB | 20044 | |
| 7 | TYROBP | 662.6554 | ACTB | 17804 | |
| 8 | CD74 | 634.4065 | LTB | 14866 | |
| 9 | HBA2 | 551.3868 | TYROBP | 12728 | |
| 10 | EEF1A1 | 474.6351 | EEF1A1 | 11602 | |
| 11 | HBB | 462.9838 | RPS18 | 11596 | |
| 12 | RPS3 | 433.276 | RPLP1 | 10574 | |
| 13 | RPLP1 | 407.0451 | RPL13A | 10386 | |
| 14 | LTB | 399.4331 | PPBP | 10344 | |
| 15 | RPS18 | 378.9975 | RPL19 | 10194 | |
Survival analysis and expression level of KGs
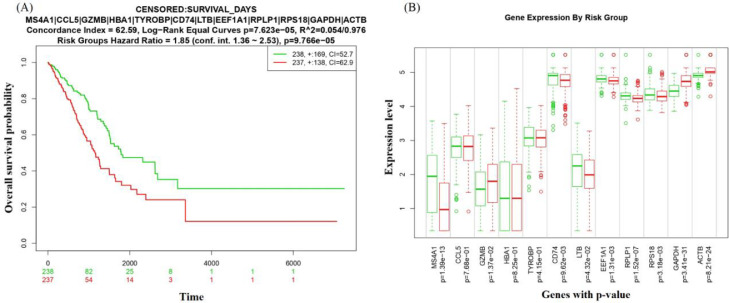
The prognostic value of the identified KGs was evaluated by fitting Cox-proportional hazards regression model between high-risk and low risk group patients in Fig. 4(A), where red indicates high-risk group patients and green indicates low-risk group patients. We observed that the overall survival probability for the high-risk group compared to the low-risk group decreased over time based on the expression level of KGs (Hazard Ratio = 1.85 at 1.36-2.53 Confidence Interval (CI), and the log-Rank p-Value = 9.766e-05) This indicates that the proposed KGs have a strong prognostic power for NSCLC. The expression patterns of the KGs by the risk group are shown in the boxplot in Fig. 4(B). The overall results showed that the KGs displayed a significant prognostic performance for NSCLC that support our original results.
Fig. 4.
(A) Kaplan-Meier plot displaying the prognostic effect of the KGs on NSCLC. (B) Boxplot displaying the expression pattern of KGs between risk groups. Red indicates high-risk group and green indicates low-risk group.
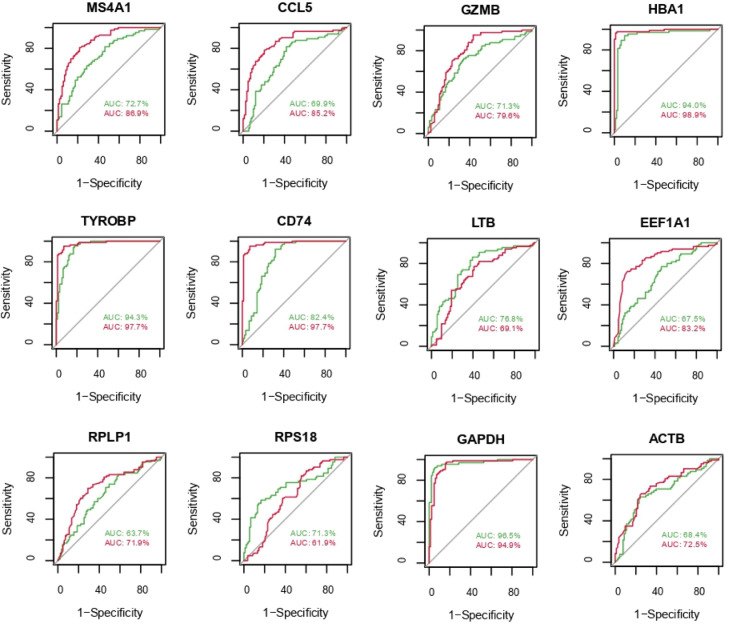
Performance measure of the identified KGs using ROC analysis
A supervised machine learning algorithm SVM classifier was considered to develop a Cancer Prediction Model for 12 KGs. We developed the cancer prediction model through the ROC curve for the training dataset with access number GSE19188 (red) and for the test dataset with access number GSE75037 (green) in Fig. 5. We observed that the AUC values range from 0.64 to 0.99 for the training dataset and 0.63 to 0.97 for the test dataset, which indicate the good prediction performance.
Fig. 5.
ROC curve evaluating the diagnostic performance of the KGs in NSCLC. Red color indicates GSE75037 dataset and green color indicates GSE19188.
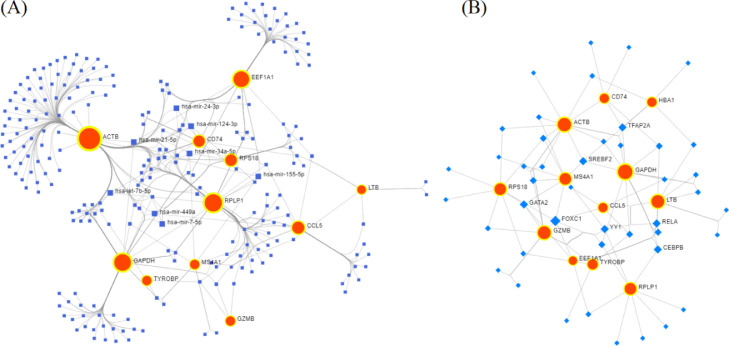
KGs-miRNAs and KGs-TFs interaction network analysis
To identify the key miRNAs associated with KGs, we constructed the interaction network between the KGs and miRNAs. We uploaded the official symbol in the gene list of human lung tissue and select the TarBase database in NetworkAnalyst to identify miRNAs that targeted the KGs (Fig. 6(A)). On the basis of degree score >= 4, we selected 8 key miRNAs (hsa-miR-124-3p, hsa-miR-34a-5p, hsa-miR-21-5p, hsa-miR-155-5p, hsa-miR-449a, hsa-miR-24-3p, hsa-let-7b-5p, and hsa-miR-7-5p) as post-transcriptional regulatory factors of KGs. From the literature review, we found that these miRNAs have a close relationship with drug resistance of lung cancer, basically NSCLC [51]. Similarly, the interaction network of KGs and TFs was constructed from the JASPAR database in NetworkAnalyst. Seven TFs (FOXC1, YY1, CEBPB, TFAP2A, SREBF2, RELA, and GATA2) with degree >= 4 were selected as transcriptional regulatory factors of KGs (Fig. 6(B)). Key miRNAs and TFs with their degree scores are listed in Table 3.
Fig. 6.
Gene-miRNAs and Gene-TFs interaction network. (A) In the Gene-miRNAs interaction network, the red bigger circles indicate genes and the blue squares indicate miRNAs. (B) In Gene-TFs interaction network, the red circles indicate genes and the blue diamond shape indicates TFs.
Table 3.
Targeted transcription factors (TFs) and miRNAs from regulatory interaction network (Gene-TFs and Gene-miRNAs).
| S.N | TFs | S.N | miRNAs | ||
|---|---|---|---|---|---|
| Label | Degree | Label | Degree | ||
| 1 | FOXC1 | 6 | 1 | hsa-miR-124-3p | 5 |
| 2 | YY1 | 4 | 2 | hsa-miR-34a-5p | 5 |
| 3 | CEBPB | 4 | 3 | hsa-miR-21-5p | 4 |
| 4 | TFAP2A | 4 | 4 | hsa-miR-155-5p | 4 |
| 5 | SREBF2 | 4 | 5 | hsa-miR-449a | 4 |
| 6 | RELA | 4 | 6 | hsa-miR-24-3p | 4 |
| 7 | GATA2 | 4 | 7 | hsa-let-7b-5p | 4 |
| 8 | hsa-miR-7-5p | 4 | |||
Discussion
NSCLC is a disease with a very high concern and is life-threatening for humans. Due to the heterogeneity of NSCLC, its treatment is very challenging. Hence, it will be helpful if NSCLC is managed by targeted treatment; however, patients with NSCLC have a lower prognosis. Hence, the identification of novel biomarkers based on the heterogeneity and the scRNA-seq data are one of the key tasks to improve the personalized and targeted medicine of NSCLC in the future.
In this study, we analyzed scRNA-seq data from the tumor tissue of peripheral blood of NSCLC patients to bioinformatically explore the cellular heterogeneity, DEGs, associated biological pathways, PPI network, key genes, miRNAs and TFs. The 1803 cells were classified into 8 explicit clusters while each cluster was mixed up with variant numbers of cells. By comparing gene expression profiles, a total 158 DEGs containing 48 up- and 110 down-regulated genes were found. To infer the biological functions and pathways associated with NSCLC, GO and KEGG pathway enrichment analyses were performed. We identified 12 KGs (MS4A1, CCL5, GZMB, HBA1, TYROBP, CD74, LTB, EEF1A1, RPLP1, RPS18, GAPDH, and ACTB) based on the two methods of Betweenness and Stress using PPI network results, and the expression pattern and survival analysis of KGs were affirmed on the basis of the TCGA data. Using the TarBase and JASPAR databases in NetworkAnalyst, we identified 8 key miRNAs (hsa-miR-124-3p, hsa-miR-34a-5p, hsa-miR-21-5p, hsa-miR-155-5p, hsa-miR-449a, hsa-miR-24-3p, hsa-let-7b-5p, hsa-miR-7-5p) and 7 key TFs (FOXC1, YY1, CEBPB, TFAP2A, SREBF2, RELA, GATA2).
All the identified 12 KGs have been supported by different studies for lung and other cancers. The deregulation of MS4A1 in lung squamous cell cancers might occur because of the expression of CD20 stromal lymphocytes [52]. Another study showed MS4A1 as a prognostic biomarker for LUAD [53]. hsa-miR-147a can inhibit the outgrowth and metastasis of NSCLC by aiming the CCL5 gene [54]. It has been found that GZMB is significantly associated with poor prognosis in SCLC [55]. Over expression of HBA1 showed low overall survival in non-smoker female lung cancer patients [56]. YAP1 promotes multidrug resistance of SCLC through signaling pathways associated with CD74 [57]. LTB-4 participates in the recruitment of neutrophils in the airways at NSCLC [58]. ACTB, EEF1A1, and RPS18 are reported to be relevant genes for qRT-PCR analysis of lung cancer also EEF1A1 is responsible for lung cancer development in smokers [59], [60], [61]. Aberrant methylation and high expression of GAPDH are associated with poor prognosis in LUAD patients [62]. TYROBP is a novel key gene with prognostic value in gastric cancer by integrated network analysis [63]. The gene biomarker RPLP1 is an anti-metastasis candidate therapeutic target with poor prognosis in triple-negative breast cancer [64]. To our knowledge TYROBP and RPLP1 are not yet been reported for lung cancer progression. Hence, we can say that these two genes (TYROBP and RPLP1) are novel with good prognostic value in our study for NSCLC.
Furthermore, hsa-miR-124-3p is reported as a tumor suppressor and inhibits the progression of several tumors, including NSCLC [65], [66], [67]. hsa-miR-34a-5p resists the brainstem glioma cell invasion [68]. hsa-miR-21-5p is one of the most important prognostic biomarkers for NSCLC [69]. hsa-miR-155-5p, hsa-miR-24-3p, and hsa-let-7a-5p were reported to be up-regulated in LUAD tissues [70]. hsa-miR-449a is reported as a genetic risk factor for gastric cancer [71]. hsa-miR-7-5p is a prognostic biomarker for small cell lung cancer [72]. Among the identified TFs, FOXC1 is one of the pioneer TF and plays an important role in the development of lung, breast, and prostate cancer [73]. The expression of FOXC1 is increased in NSCLC tissues, and it has an adverse relationship with survival [74]. TFAP2C is contributed to NSCLC tumorigenesis by downregulating numerous tumor silencers such as GADD45B, PMAIP1, and XAF1 [75].
The GO functional enrichment and KEGG pathway analysis revealed that the KGs are related to Protein metabolism, MHC class I receptor activity, B cell receptor activity, viral mRNA Translation, 3′ -UTR-mediated translational regulation etc. pathways. Most of the DEGs (47.3%) have enriched with protein metabolism (associated with KGs: EEF1A1, RPS18, RPLP1, and GZMB) term, and several studies have further claimed that NSCLC-causing genes are enriched in protein metabolism term [76,77]. The prognostic effect of the KGs with TCGA datasets in LUAD showed the worst survival rate which indicates that these KGs might be the prognostic biomarkers in LUAD. The differential expression stated the discriminating power of the KGs. Finally, the diagnostic effects of KGs were assessed by ROC analysis. The AUC values in ROC analysis indicated a comparatively good prediction performance of the KGs in NSCLC patients with higher sensitivity and specificity. Therefore, our overall analysis will provide valuable insights into NSCLC progression, KGs, key miRNAs, key TFs might be a novel diagnostic and prognostic biomarkers as well as potential regulators for the progression, diagnosis, and prognosis of NSCLC. In this study, we predicted the results through computational analysis; hence, we cannot recommend for treatment directly. We emphasize for further assessed at the molecular level by the wet-lab experiments in prior to clinical investigation.
Conclusion
The scRNA-seq data of peripheral blood cell allows the identification of distinct cell types and provides a new perspective on the pathogenesis of NSCLC. On the viewpoint of clustering analysis, we conclude that NSCLC is heterogeneous in numerous aspects. Through bioinformatics analysis, we identified 12 KGs (MS4A1, CCL5, GZMB, HBA1, TYROBP, CD74, LTB, EEF1A1, RPLP1, RPS18, GAPDH, and ACTB); among them, there were 2 novel KGs (TYROBP and RPLP1). Their targeted miRNAs and TFs were also identified, which play a significant role in NSCLC. Survival and ROC analysis showed the prognostic and diagnostic effect of KGs. Our overall findings suggested that these KGs might be the prognostic and diagnostic biomarkers for NSCLC.
Ethical Approval and Consent to participate
Not Applicable
Consent for publication
All the authors have provided consent for the publication.
Availability of supporting data
The scRNA-seq data of NSCLC were obtained from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/) with accession number GSE127471.
CRediT authorship contribution statement
Adiba Sultana: Data curation, Formal analysis, Writing – original draft, Writing – review & editing. Md Shahin Alam: Data curation, Writing – original draft, Writing – review & editing. Xingyun Liu: Formal analysis, Writing – review & editing. Rohit Sharma: Formal analysis, Writing – review & editing. Rajeev K. Singla: Formal analysis, Writing – original draft, Writing – review & editing. Rohit Gundamaraju: Formal analysis, Writing – review & editing. Bairong Shen: Conceptualization, Funding acquisition, Supervision, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgments
The authors are thankful to the National Natural Science Foundation of China for proving funding support.
Funding
This work was supported by the National Natural Science Foundation of China (Grant No. 32070671 and 32270690).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2022.101571.
Contributor Information
Rohit Sharma, Email: rohitsharma@bhu.ac.in.
Rajeev K. Singla, Email: rajeevsingla26@gmail.com, rajeevkumar@scu.edu.cn.
Bairong Shen, Email: bairong.shen@scu.edu.cn.
Appendix. Supplementary materials
References
- 1.Xu J., Nie H., He J., Wang X., Liao K., Tu L., et al. Using machine learning modeling to explore new immune-related prognostic markers in non-small cell lung cancer. Front. Oncol. 2020;10 doi: 10.3389/fonc.2020.550002. Epub 2020/11/21PubMed PMID33215029PubMed Central PMCIDPMCPMC7665579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Song F., Xuan Z., Yang X., Ye X., Pan Z. Fang Q. Identification of key microRNAs and hub genes in non-small-cell lung cancer using integrative bioinformatics and functional analyses. J. Cell. Biochem. 2020;121(3):2690–2703. doi: 10.1002/jcb.29489. Epub 2019/11/07PubMed PMID31692035. [DOI] [PubMed] [Google Scholar]
- 3.Wang Y., Huang L., Wu S., Jia Y., Yang Y., Luo L., et al. Bioinformatics analyses of the role of vascular endothelial growth factor in patients with non-small cell lung cancer. PLoS One. 2015;10(9) doi: 10.1371/journal.pone.0139285. Epub 2015/10/01PubMed PMID26422603PubMed Central PMCIDPMCPMC4589385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun Q., Li X., Xu M., Zhang L., Zuo H., Xin Y., et al. Differential expression and bioinformatics analysis of circRNA in non-small cell lung cancer. Front. Genet. 2020;11 doi: 10.3389/fgene.2020.586814. Epub 2020/12/18PubMed PMID33329727PubMed Central PMCIDPMCPMC7732606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu Q., Zhang B., Sun Y., Xu R., Hu X., Ren S., et al. Identification of novel biomarkers and candidate small molecule drugs in non-small-cell lung cancer by integrated microarray analysis. Onco Targets Ther. 2019;12:3545–3563. doi: 10.2147/OTT.S198621. Epub 2019/06/14PubMed PMID31190860PubMed Central PMCIDPMCPMC6526173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen Xi, Teichmann Sarah A., Meyer K.B. From tissues to cell types and back: single-cell gene expression analysis of tissue architecture. Annu. Rev. Biomed. Data Sci. 2018;1:29–51. doi: 10.1146/annurev-biodatasci-080917-013452. [DOI] [Google Scholar]
- 7.Olsen T.K., Baryawno N. Introduction to single-cell RNA sequencing. Curr. Protoc. Mol. Biol. 2018;122(1):e57. doi: 10.1002/cpmb.57. Epub 2018/06/01PubMed PMID29851283. [DOI] [PubMed] [Google Scholar]
- 8.Cochain C., Vafadarnejad E., Arampatzi P., Pelisek J., Winkels H., Ley K., et al. Single-Cell RNA-Seq reveals the transcriptional landscape and heterogeneity of aortic macrophages in murine atherosclerosis. Circ. Res. 2018;122(12):1661–1674. doi: 10.1161/CIRCRESAHA.117.312509. Epub 2018/03/17PubMed PMID29545365. [DOI] [PubMed] [Google Scholar]
- 9.Min J.W., Kim W.J., Han J.A., Jung Y.J., Kim K.T., Park W.Y., et al. Identification of distinct tumor subpopulations in lung adenocarcinoma via single-cell RNA-seq. PLoS One. 2015;10(8) doi: 10.1371/journal.pone.0135817. Epub 2015/08/26PubMed PMID26305796PubMed Central PMCIDPMCPMC4549254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patel A.P., Tirosh I., Trombetta J.J., Shalek A.K., Gillespie S.M., Wakimoto H., et al. Single-Cell RNA-Seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344(6190):1396–1401. doi: 10.1126/science.1254257. Epub 2014/06/14PubMed PMID24925914PubMed Central PMCIDPMCPMC4123637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gong K., Zhou H., Liu H., Xie T., Luo Y., Guo H., et al. Identification and integrate analysis of key biomarkers for diagnosis and prognosis of non-small cell lung cancer based on bioinformatics analysis. Technol. Cancer Res. Treat. 2021;20 doi: 10.1177/15330338211060202. Epub 2021/11/27PubMed PMID34825846PubMed Central PMCIDPMCPMC8649439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim J., Xu Z., Marignani P.A. Single-cell RNA sequencing for the identification of early-stage lung cancer biomarkers from circulating blood. NPJ Genom. Med. 2021;6(1):87. doi: 10.1038/s41525-021-00248-y. Epub 2021/10/17PubMed PMID34654834PubMed Central PMCIDPMCPMC8519939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen Z., Fillmore C.M., Hammerman P.S., Kim C.F., Wong K.K. Non-small-cell lung cancers: a heterogeneous set of diseases. Nat. Rev. Cancer. 2014;14(8):535–546. doi: 10.1038/nrc3775. Epub 2014/07/25PubMed PMID25056707PubMed Central PMCIDPMCPMC5712844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Warren G.W., Cummings K.M. Tobacco and lung cancer: risks, trends, and outcomes in patients with cancer. Am. Soc. Clin. Oncol. Educ. Book. 2013;33:359–364. doi: 10.14694/EdBook_AM.2013.33.359. [DOI] [PubMed] [Google Scholar]
- 15.Chan B.A., Hughes B.G. Targeted therapy for non-small cell lung cancer: current standards and the promise of the future. Transl. Lung Cancer Res. 2015;4(1):36–54. doi: 10.3978/j.issn.2218-6751.2014.05.01. Epub 2015/03/26PubMed PMID25806345PubMed Central PMCIDPMCPMC4367711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hirsch F.R., Spreafico A., Novello S., Wood M.D., Simms L., Papotti M. The prognostic and predictive role of histology in advanced non-small cell lung cancer: a literature review. J. Thorac. Oncol. 2008;3(12):1468–1481. doi: 10.1097/JTO.0b013e318189f551. Epub 2008/12/06PubMed PMID19057275. [DOI] [PubMed] [Google Scholar]
- 17.Ettinger D.S., Akerley W., Borghaei H., Chang A.C., Cheney R.T., Chirieac L.R., et al. Non-small cell lung cancer. J. Natl. Compr. Cancer Netw. 2012;10(10):1236–1271. doi: 10.6004/jnccn.2012.0130. Epub 2012/10/12PubMed PMID23054877. [DOI] [PubMed] [Google Scholar]
- 18.American Cancer Society. Lung Cancer Survival Rates 2019. Available from: https://www.cancer.org/cancer/lung-cancer/detection-diagnosis-staging/survival-rates.html.
- 19.Hayes J., Peruzzi P.P., Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol. Med. 2014;20(8):460–469. doi: 10.1016/j.molmed.2014.06.005. Epub 2014/07/17PubMed PMID25027972. [DOI] [PubMed] [Google Scholar]
- 20.Cho W.C. OncomiRs: the discovery and progress of microRNAs in cancers. Mol. Cancer. 2007;6:60. doi: 10.1186/1476-4598-6-60. Epub 2007/09/27PubMed PMID17894887PubMed Central PMCIDPMCPMC2098778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Esquela-Kerscher A., Slack F.J. Oncomirs - microRNAs with a role in cancer. Nat. Rev. Cancer. 2006;6(4):259–269. doi: 10.1038/nrc1840. Epub 2006/03/25PubMed PMID16557279. [DOI] [PubMed] [Google Scholar]
- 22.Karube Y., Tanaka H., Osada H., Tomida S., Tatematsu Y., Yanagisawa K., et al. Reduced expression of Dicer associated with poor prognosis in lung cancer patients. Cancer Sci. 2005;96(2):111–115. doi: 10.1111/j.1349-7006.2005.00015.x. Epub 2005/02/23PubMed PMID15723655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Du L., Schageman J.J., Irnov Girard L., Hammond S.M., Minna J.D., et al. MicroRNA expression distinguishes SCLC from NSCLC lung tumor cells and suggests a possible pathological relationship between SCLCs and NSCLCs. J. Exp. Clin. Cancer Res. 2010;29:75. doi: 10.1186/1756-9966-29-75. Epub 2010/07/14PubMed PMID20624269PubMed Central PMCIDPMCPMC2907339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alam M.S., Rahaman M.M., Sultana A., Wang G., Mollah M.N.H. Statistics and network-based approaches to identify molecular mechanisms that drive the progression of breast cancer. Comput. Biol. Med. 2022;145 doi: 10.1016/j.compbiomed.2022.105508. Epub 2022/04/22PubMed PMID35447458. [DOI] [PubMed] [Google Scholar]
- 25.Alam M.S., Sultana A., Reza M.S., Amanullah M., Kabir S.R., Mollah M.N.H. Integrated bioinformatics and statistical approaches to explore molecular biomarkers for breast cancer diagnosis, prognosis and therapies. PLoS One. 2022;17(5) doi: 10.1371/journal.pone.0268967. Epub 2022/05/27PubMed PMID35617355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen W., Zhu S., Zhang Y., Xiao J., Tian D. Identification of key candidate tumor biomarkers in non-small-cell lung cancer by in silico analysis. Oncol. Lett. 2020;19(1):1008–1016. doi: 10.3892/ol.2019.11169. Epub 2020/01/04PubMed PMID31897214PubMed Central PMCIDPMCPMC6924182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang M., Sun H., He J., Wang H., Yu X., Ma L., et al. Interaction of ribosomal protein L22 with casein kinase 2alpha: a novel mechanism for understanding the biology of non-small cell lung cancer. Oncol. Rep. 2014;32(1):139–144. doi: 10.3892/or.2014.3187. Epub 2014/05/21PubMed PMID24840952. [DOI] [PubMed] [Google Scholar]
- 28.Yang Z., Yang N., Ou Q., Xiang Y., Jiang T., Wu X., et al. Investigating novel resistance mechanisms to third-generation EGFR tyrosine kinase inhibitor Osimertinib in non-small cell lung cancer patients. Clin. Cancer Res. 2018;24(13):3097–3107. doi: 10.1158/1078-0432.CCR-17-2310. Epub 2018/03/07PubMed PMID29506987. [DOI] [PubMed] [Google Scholar]
- 29.Zhang X., Zhang D., Huang L., Li G., Chen L., Ma J., et al. Discovery of novel biomarkers of therapeutic responses in Han Chinese pemetrexed-based treated advanced NSCLC patients. Front. Pharmacol. 2019;10:944. doi: 10.3389/fphar.2019.00944. Epub 2019/09/12PubMed PMID31507426PubMed Central PMCIDPMCPMC6716463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shen Y., Pan X., Yang J. Gene regulation and prognostic indicators of lung squamous cell carcinoma: TCGA-derived miRNA/mRNA sequencing and DNA methylation data. J. Cell. Physiol. 2019;234(12):22896–22910. doi: 10.1002/jcp.28852. Epub 2019/06/07PubMed PMID31169310. [DOI] [PubMed] [Google Scholar]
- 31.Valk K., Vooder T., Kolde R., Reintam M.A., Petzold C., Vilo J., et al. Gene expression profiles of non-small cell lung cancer: survival prediction and new biomarkers. Oncology. 2010;79(3-4):283–292. doi: 10.1159/000322116. Epub 2011/03/18PubMed PMID21412013. [DOI] [PubMed] [Google Scholar]
- 32.Puzone R., Savarino G., Salvi S., Dal Bello M.G., Barletta G., Genova C., et al. Glyceraldehyde-3-phosphate dehydrogenase gene over expression correlates with poor prognosis in non small cell lung cancer patients. Mol. Cancer. 2013;12(1):97. doi: 10.1186/1476-4598-12-97. Epub 2013/08/31PubMed PMID23988223PubMed Central PMCIDPMCPMC3766010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Newman AM, Steen CB, Liu CL, Gentles AJ, Chaudhuri AA, Scherer F, et al. High-throughput tissue dissection and cell purification with digital cytometry [scRNA-Seq] 2019. Available from: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE127471.
- 34.Butler A., Hoffman P., Smibert P., Papalexi E., Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018;36(5):411–420. doi: 10.1038/nbt.4096. Epub 2018/04/03PubMed PMID: 29608179PubMed Central PMCIDPMCPMC6700744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qiu X., Mao Q., Tang Y., Wang L., Chawla R., Pliner H.A., et al. Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods. 2017;14(10):979–982. doi: 10.1038/nmeth.4402. Epub 2017/08/22PubMed PMID28825705PubMed Central PMCIDPMCPMC5764547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pathan M., Keerthikumar S., Ang C.S., Gangoda L., Quek C.Y., Williamson N.A., et al. FunRich: an open access standalone functional enrichment and interaction network analysis tool. Proteomics. 2015;15(15):2597–2601. doi: 10.1002/pmic.201400515. Epub 2015/04/30PubMed PMID25921073. [DOI] [PubMed] [Google Scholar]
- 37.Gene Ontology C. The Gene Ontology (GO) project in 2006. Nucleic Acids Res. 2006;34:D322. doi: 10.1093/nar/gkj021. Database issue-6Epub 2005/12/31PubMed PMID16381878PubMed Central PMCIDPMCPMC1347384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kanehisa M., Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. Epub 1999/12/11PubMed PMID10592173PubMed Central PMCIDPMCPMC102409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Szklarczyk D., Gable A.L., Lyon D., Junge A., Wyder S., Huerta-Cepas J., et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607–DD13. doi: 10.1093/nar/gky1131. Epub 2018/11/27PubMed PMID30476243PubMed Central PMCIDPMCPMC6323986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. Epub 2003/11/05PubMed PMID14597658PubMed Central PMCIDPMCPMC403769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chin C.H., Chen S.H., Wu H.H., Ho C.W., Ko M.T., Lin C.Y. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 2014;8(4):S11. doi: 10.1186/1752-0509-8-S4-S11. SupplEpub 2014/12/19PubMed PMID25521941PubMed Central PMCIDPMCPMC4290687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aguirre-Gamboa R., Gomez-Rueda H., Martinez-Ledesma E., Martinez-Torteya A., Chacolla-Huaringa R., Rodriguez-Barrientos A., et al. SurvExpress: an online biomarker validation tool and database for cancer gene expression data using survival analysis. PLoS One. 2013;8(9):e74250. doi: 10.1371/journal.pone.0074250. Epub 2013/09/26PubMed PMID24066126PubMed Central PMCIDPMCPMC3774754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Robin X., Turck N., Hainard A., Tiberti N., Lisacek F., Sanchez J.C., et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinform. 2011;12:77. doi: 10.1186/1471-2105-12-77. Epub 2011/03/19PubMed PMID21414208PubMed Central PMCIDPMCPMC3068975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou G., Soufan O., Ewald J., Hancock R.E.W, Basu N., Xia J. NetworkAnalyst 3.0: a visual analytics platform for comprehensive gene expression profiling and meta-analysis. Nucleic Acids Res. 2019;47(W1):W234–WW41. doi: 10.1093/nar/gkz240. Epub 2019/04/02PubMed PMID30931480PubMed Central PMCIDPMCPMC6602507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Karagkouni D., Paraskevopoulou M.D., Chatzopoulos S., Vlachos I.S., Tastsoglou S., Kanellos I., et al. DIANA-TarBase v8: a decade-long collection of experimentally supported miRNA-gene interactions. Nucleic Acids Res. 2018;46(D1):D239–DD45. doi: 10.1093/nar/gkx1141. Epub 2017/11/21PubMed PMID29156006PubMed Central PMCIDPMCPMC5753203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hsu S.D., Lin F.M., Wu W.Y., Liang C., Huang W.C., Chan W.L., et al. miRTarBase: a database curates experimentally validated microRNA-target interactions. Nucleic Acids Res. 2011;39:D163–D169. doi: 10.1093/nar/gkq1107. Database issueEpub 2010/11/13PubMed PMID21071411PubMed Central PMCIDPMCPMC3013699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xiao F., Zuo Z., Cai G., Kang S., Gao X., Li T. miRecords: an integrated resource for microRNA-target interactions. Nucleic Acids Res. 2009;37:D105–D110. doi: 10.1093/nar/gkn851. Database issueEpub 2008/11/11PubMed PMID18996891PubMed Central PMCIDPMCPMC2686554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Consortium E.P. The ENCODE (ENCyclopedia Of DNA Elements) project. Science. 2004;306(5696):636–640. doi: 10.1126/science.1105136. Epub 2004/10/23PubMed PMID15499007. [DOI] [PubMed] [Google Scholar]
- 49.Fornes O., Castro-Mondragon J.A., Khan A., van der Lee R., Zhang X., Richmond P.A., et al. JASPAR 2020: update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2020;48(D1):D87–D92. doi: 10.1093/nar/gkz1001. Epub 2019/11/09PubMed PMID31701148PubMed Central PMCIDPMCPMC7145627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lachmann A., Xu H., Krishnan J., Berger S.I., Mazloom A.R., ChEA Ma'ayan A. transcription factor regulation inferred from integrating genome-wide ChIP-X experiments. Bioinformatics. 2010;26(19):2438–2444. doi: 10.1093/bioinformatics/btq466. Epub 2010/08/17PubMed PMID20709693PubMed Central PMCIDPMCPMC2944209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liang A.L., Du S.L., Zhang B., Zhang J., Ma X., Wu C.Y., et al. Screening miRNAs associated with resistance gemcitabine from exosomes in A549 lung cancer cells. Cancer Manag. Res. 2019;11:6311–6321. doi: 10.2147/CMAR.S209149. Epub 2019/08/03PubMed PMID31372037PubMed Central PMCIDPMCPMC6626902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wright C.M., Savarimuthu Francis S.M., Tan M.E., Martins M.U., Winterford C., Davidson M.R., et al. MS4A1 dysregulation in asbestos-related lung squamous cell carcinoma is due to CD20 stromal lymphocyte expression. PLoS One. 2012;7(4):e34943. doi: 10.1371/journal.pone.0034943. Epub 2012/04/20PubMed PMID22514692PubMed Central PMCIDPMCPMC3325913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ma C., Luo H., Cao J., Zheng X., Zhang J., Zhang Y., et al. Identification of a novel tumor microenvironment-associated eight-gene signature for prognosis prediction in lung adenocarcinoma. Front. Mol. Biosci. 2020;7 doi: 10.3389/fmolb.2020.571641. Epub 2020/10/27PubMed PMID33102522PubMed Central PMCIDPMCPMC7546815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lu Y., Luan X.R. miR-147a suppresses the metastasis of non-small-cell lung cancer by targeting CCL5. J. Int. Med. Res. 2020;48(4) doi: 10.1177/0300060519883098. Epub 2019/12/31PubMed PMID31884861PubMed Central PMCIDPMCPMC7607764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Song Y., Sun Y., Sun T., Tang R. Comprehensive bioinformatics analysis identifies tumor microenvironment and immune-related genes in small cell lung cancer. Comb. Chem. High Throughput Screen. 2020;23(5):381–391. doi: 10.2174/1386207323666200407075004. Epub 2020/04/09PubMed PMID32264809. [DOI] [PubMed] [Google Scholar]
- 56.Shi K., Li N., Yang M., Li W. Identification of key genes and pathways in female lung cancer patients who never smoked by a bioinformatics analysis. J. Cancer. 2019;10(1):51–60. doi: 10.7150/jca.26908. Epub 2019/01/22PubMed PMID30662525PubMed Central PMCIDPMCPMC6329865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Song Y., Sun Y., Lei Y., Yang K., Tang R. YAP1 promotes multidrug resistance of small cell lung cancer by CD74-related signaling pathways. Cancer Med. 2020;9(1):259–268. doi: 10.1002/cam4.2668. Epub 2019/11/07PubMed PMID31692299PubMed Central PMCIDPMCPMC6943160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Carpagnano G.E., Palladino G.P., Lacedonia D., Koutelou A., Orlando S., Foschino-Barbaro M.P. Neutrophilic airways inflammation in lung cancer: the role of exhaled LTB-4 and IL-8. BMC Cancer. 2011;11:226. doi: 10.1186/1471-2407-11-226. Epub 2011/06/09PubMed PMID21649887PubMed Central PMCIDPMCPMC3130703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhan C., Zhang Y., Ma J., Wang L., Jiang W., Shi Y., et al. Identification of reference genes for qRT-PCR in human lung squamous-cell carcinoma by RNA-Seq. Acta Biochim. Biophys. Sin. 2014;46(4):330–337. doi: 10.1093/abbs/gmt153. Epub 2014/01/25PubMed PMID24457517. [DOI] [PubMed] [Google Scholar]
- 60.Chari R., Lonergan K.M., Pikor L.A., Coe B.P., Zhu C.Q., Chan T.H., et al. A sequence-based approach to identify reference genes for gene expression analysis. BMC Med. Genom. 2010;3:32. doi: 10.1186/1755-8794-3-32. Epub 2010/08/05PubMed PMID20682026PubMed Central PMCIDPMCPMC2928167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang Z., Zhuan B., Yan Y., Jiang S., Wang T. Identification of gene markers in the development of smoking-induced lung cancer. Gene. 2016;576(1):451–457. doi: 10.1016/j.gene.2015.10.060. Pt 3Epub 2015/11/01PubMed PMID26518718. [DOI] [PubMed] [Google Scholar]
- 62.Wang X., Shi D., Zhao D., Hu D. Aberrant methylation and differential expression of SLC2A1, TNS4, GAPDH, ATP8A2, and CASZ1 are associated with the prognosis of lung adenocarcinoma. Biomed. Res. Int. 2020;2020 doi: 10.1155/2020/1807089. Epub 2020/10/09PubMed PMID33029490PubMed Central PMCIDPMCPMC7532994. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 63.Jiang J., Ding Y., Wu M., Lyu X., Wang H., Chen Y., et al. Identification of TYROBP and C1QB as two novel key genes with prognostic value in gastric cancer by network analysis. Front. Oncol. 2020;10:1765. doi: 10.3389/fonc.2020.01765. Epub 2020/10/06PubMed PMID33014868PubMed Central PMCIDPMCPMC7516284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.He Z., Xu Q., Wang X., Wang J., Mu X., Cai Y., et al. RPLP1 promotes tumor metastasis and is associated with a poor prognosis in triple-negative breast cancer patients. Cancer Cell Int. 2018;18:170. doi: 10.1186/s12935-018-0658-0. Epub 2018/11/06PubMed PMID30386179PubMed Central PMCIDPMCPMC6203216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.He R.Q., Yang X., Liang L., Chen G., Ma J. MicroRNA-124-3p expression and its prospective functional pathways in hepatocellular carcinoma: a quantitative polymerase chain reaction, gene expression omnibus and bioinformatics study. Oncol. Lett. 2018;15(4):5517–5532. doi: 10.3892/ol.2018.8045. Epub 2018/03/20PubMed PMID29552191PubMed Central PMCIDPMCPMC5840674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sun Y., Ai X., Shen S., Lu S. NF-kappaB-mediated miR-124 suppresses metastasis of non-small-cell lung cancer by targeting MYO10. Oncotarget. 2015;6(10):8244–8254. doi: 10.18632/oncotarget.3135. Epub 2015/03/10PubMed PMID25749519PubMed Central PMCIDPMCPMC4480748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tang L.X., Chen G.H., Li H., He P., Zhang Y., Xu X.W. Long non-coding RNA OGFRP1 regulates LYPD3 expression by sponging miR-124-3p and promotes non-small cell lung cancer progression. Biochem. Biophys. Res. Commun. 2018;505(2):578–585. doi: 10.1016/j.bbrc.2018.09.146. Epub 2018/10/03PubMed PMID30274775. [DOI] [PubMed] [Google Scholar]
- 68.Chen X., Dong D., Pan C., Xu C., Sun Y., Geng Y., et al. Identification of grade-associated MicroRNAs in brainstem gliomas based on microarray data. J. Cancer. 2018;9(23):4463–4476. doi: 10.7150/jca.26417. Epub 2018/12/07PubMed PMID30519352PubMed Central PMCIDPMCPMC6277643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang K., Chen M., Wu W. Analysis of microRNA (miRNA) expression profiles reveals 11 key biomarkers associated with non-small cell lung cancer. World J. Surg. Oncol. 2017;15(1):175. doi: 10.1186/s12957-017-1244-y. Epub 2017/09/21PubMed PMID28927412PubMed Central PMCIDPMCPMC5606074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zeybek A., Oz N., Kalemci S., Edgunlu T., Kiziltug M.T., Tosun K., et al. Diagnostic value of MiR-125b as a potential biomarker for stage I lung adenocarcinoma. Curr. Mol. Med. 2019;19(3):216–227. doi: 10.2174/1566524019666190314113800. Epub 2019/03/15PubMed PMID30868951. [DOI] [PubMed] [Google Scholar]
- 71.Shi J., Liu Y., Liu J., Zhou J. Hsa-miR-449a genetic variant is associated with risk of gastric cancer in a Chinese population. Int. J. Clin. Exp. Pathol. 2015;8(10):13387–13392. Epub 2016/01/02PubMed PMID26722545PubMed Central PMCIDPMCPMC4680490. [PMC free article] [PubMed] [Google Scholar]
- 72.Li X., Ma C., Luo H., Zhang J., Wang J., Guo H. Identification of the differential expression of genes and upstream microRNAs in small cell lung cancer compared with normal lung based on bioinformatics analysis. Medicine (Baltimore). 2020;99(11):e19086. doi: 10.1097/MD.0000000000019086. Epub 2020/03/17PubMed PMID32176034PubMed Central PMCIDPMCPMC7440067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang Y., Huang Y.X., Wang D.L., Yang B., Yan H.Y., Lin L.H., et al. LncRNA DSCAM-AS1 interacts with YBX1 to promote cancer progression by forming a positive feedback loop that activates FOXA1 transcription network. Theranostics. 2020;10(23):10823–10837. doi: 10.7150/thno.47830. Epub 2020/09/16PubMed PMID32929382PubMed Central PMCIDPMCPMC7482804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cao S., Wang Z., Gao X., He W., Cai Y., Chen H., et al. FOXC1 induces cancer stem cell-like properties through upregulation of beta-catenin in NSCLC. J. Exp. Clin. Cancer Res. 2018;37(1):220. doi: 10.1186/s13046-018-0894-0. Epub 2018/09/08PubMed PMID30189871PubMed Central PMCIDPMCPMC6127900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Do H., Kim D., Kang J., Son B., Seo D., Youn H., et al. TFAP2C increases cell proliferation by downregulating GADD45B and PMAIP1 in non-small cell lung cancer cells. Biol. Res. 2019;52(1):35. doi: 10.1186/s40659-019-0244-5. Epub 2019/07/13PubMed PMID31296259PubMed Central PMCIDPMCPMC6625030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang Q., Liu J., Li R., Zhao R., Zhang M., Wei S., et al. A network pharmacology approach to investigate the anticancer mechanism and potential active ingredients of Rheum palmatum L. against lung cancer via induction of apoptosis. Front. Pharmacol. 2020;11 doi: 10.3389/fphar.2020.528308. Epub 2020/12/01PubMed PMID33250766PubMed Central PMCIDPMCPMC7672213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kim B.Y., Lee J., Kim N.S. Helveticoside is a biologically active component of the seed extract of Descurainia sophia and induces reciprocal gene regulation in A549 human lung cancer cells. BMC Genom. 2015;16:713. doi: 10.1186/s12864-015-1918-1. Epub 2015/09/20PubMed PMID26384484PubMed Central PMCIDPMCPMC4575430. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The scRNA-seq data of NSCLC were obtained from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/) with accession number GSE127471.