Abstract
Cyclospora cayetanensis, a coccidian apicomplexan parasite, causes large outbreaks of foodborne diarrheal disease globally. Tracking the source of C. cayetanensis oocyst contamination in food items is essential to reduce, even prevent outbreaks. We previously showed that a genotyping method based on mitochondrial single nucleotide polymorphism (SNP) profiles had discriminatory power in classifying C. cayetanensis clinical isolates. In food specimens, low level contamination by oocysts and difficulties in DNA extraction present significant challenges in genotyping method development. Here, we report the development of a highly sensitive, custom-designed, targeted sequencing method based on the Illumina AmpliSeq platform; our method was capable of consistently generating near-complete mitochondrial genome sequences of C. cayetanensis from foods with low levels of contamination. To simulate environmentally observed contamination levels in foods, we seeded various food matrices, such as fresh produce and prepared dishes, with known quantities of oocysts, and isolated genomic DNA from washed food samples. Using the Ampliseq Targeted Sequencing method, we obtained near-complete mitochondrial genome sequences of C. cayetanensis from food samples seeded with as low as five to ten oocysts and used the data in downstream analysis. The flexibility of the AmpliSeq platform could potentially allow for more genomic targets to be added to achieve higher discriminatory power. This level of sensitivity in capturing high resolution genome data from contaminated food samples is a critical milestone towards the potential development of a comprehensive genotyping method for C. cayetanensis.
Keywords: Cyclospora cayetanensis, Produce, AmpliSeq, Mitochondrial genome, Targeted NGS, Genotyping, Food matrices
Cyclospora cayetanensis, produce, AmpliSeq, mitochondrial genome, targeted NGS, genotyping, food matrices.
1. Introduction
Cyclospora cayetanensis, an apicomplexan protozoan parasite, causes a foodborne and waterborne diarrheal disease called cyclosporiasis; outbreaks from this organism occur worldwide [1, 2, 3, 4, 5]. Large outbreaks of cylosporiasis have been reported in the U.S. since the mid 1990's, and are often linked to various types of imported fresh produce (e.g,. basil, cilantro, mesclun lettuce, raspberries, green onions and snow peas) [6, 7]. However, microbiological surveillance sampling studies conducted in the U.S. in 2018 and 2019 by the FDA, which targeted cilantro, parsley, and basil, identified C. cayetanensis in produce harvested in the U.S (https://www.fda.gov/food/sampling-protect-food-supply/microbiological-surveillance-sampling-fy18-19-fresh-herbs-cilantro-basil-parsley-and-processed).
Linking cyclosporiasis cases to sources of infection continues to be a public health challenge. Due to the complexity of traceback studies, only a limited number of cyclosporiasis cases during an outbreak are usually linked to specific food exposures [8]. Complex food exposure scenarios obtained via epidemiological investigations must be combined with robust traceback data aimed at identifying sources of potentially contaminated food, and if possible, potential vehicles of contamination. Identification of specific illness-causing strains connected with individual clusters of patients in an outbreak can be enhanced by the availability of epidemiologic tools based on molecular subtyping. However, designing such tools requires extensive genomic information which has been scarce, until recently, for C. cayetanensis. The lack of laboratory culture methods and limited access to the oocysts impeded the generation of such critical genomic information. With the advent of Next Generation Sequencing (NGS) technologies, nuclear and organellar genome sequences of C. cayetanensis have started to become available [9, 10, 11, 12, 13, 14, 15, 16, 17].
Analytical methods targeting either nuclear [18, 19, 20], organellar [14, 17, 21], or both nuclear and organellar genomic sequences [22, 23] of C. cayetanensis, have been published. These methods exhibit a range of discriminatory power in classifying clinical isolates of C. cayetanensis. Only one of these methods reported a classification analysis involving both clinical stool specimens from cyclosporiasis patients and a produce sample seeded with known concentrations of C.cayetanensis oocysts [17].
Food and environmental samples present unique challenges regarding the isolation of the C. cayetanensis oocysts and extraction of the genomic material [24, 25, 26]. DNA extracted from plant-origin food matrixes is metagenomic in composition and may contain a mixture of genomic material originating from plant cells, plant microflora, and other contaminants, such as human DNA, other closely-related coccidia species not pathogenic to humans such as Eimeria, as well as our detection target, the C. cayetanensis genome. Another complication is the low level of contamination in food with C. cayetanensis oocysts. We estimate that the number of C. cayetanensis oocysts in naturally contaminated produce items might be low (at 5 to 10 oocysts per 25 g of leafy greens, or per 50 g of berries), based on comparative signals from recent food surveillance studies conducted by the FDA (unpublished data). Additionally, data obtained from samples seeded with less than 10 oocysts [27] further supports this conclusion. Both the complex nature of the sample DNA and the low levels of C. cayetanensis contamination in a sample pose a great challenge by likely missing the sequencing signal from the target organism due to extreme dilution. Targeted NGS provides an effective solution to increase the sensitivity and may be applied to food and environmental samples contaminated with C. cayetanensis oocysts.
The principle of targeted NGS involves designing sequencing panels that focus on a specific part of a particular genome instead of sequencing total DNA, be it homogenous or metagenomic in composition, in a given sample. For example, to help determine the identity of all organisms present in a metagenomic sample, targeted NGS aims toward conserved regions of universal genes for prokaryotes (16S RNA genes) and eukaryotes (18S RNA genes) [28, 29, 30]. This approach has also been used for the identification of specific mutations in cancer marker genes, as a clinical test for cancer classification, and to individualize treatment decisions by physicians [31, 32, 33, 34].
C. cayetanensis mitochondrial genome is 6.3 kb linear molecule arranged in concatemeric structure with head-tail configuration. Mitochondria genomes from different isolates vary in length due to 15-mer repeat sequences located at the concatemeric junction region (9–11,15, 21). These unique structural properties along with the SNPs make this organelle genome a suitable target for genotyping method development.
Here, we report a laboratory workflow based on targeted NGS of C. cayetanensis mitochondria genome sequences within food samples with very low contamination levels of C. cayetanensis oocysts. Mitochondria genome sequences were chosen as the genomic target based on our previous work that demonstrated the use of these sequences in C. cayetanensis genotyping and classification [17]. We used an Illumina Ampliseq Custom Targeted Panel to enhance sensitivity and obtained near complete (>90 % of the 6274 bp) mitochondrial genome sequences directly from fresh or frozen food samples seeded with as low as five to ten C. cayetanensis oocysts. To our knowledge, this is the first report of C. cayetanensis genotyping in a variety of food samples with such low contamination levels.
2. Materials and methods
2.1. Sources of C. cayetanensis samples
C. cayetanensis oocysts used in this study were purified from clinical stool samples which originated from Indonesia (collection date is unknown) and Nepal (collected in 2014). We isolated C. cayetanensis oocysts from the human stool samples by a purification method previously described [10, 14]. Briefly, C. cayetanensis oocysts were recovered from sieved fecal samples by differential sucrose and cesium chloride gradient centrifugations. C. cayetanensis oocysts were counted using a haemocytometer and fluorescent microscopy using a Zeiss Axio Imager D1 microscope with an HBO mercury short arc lamp and a UV filter (350 nm excitation and 450 nm emission). This study was reviewed and approved by Institutional Review Board of FDA and identified with the files RIHSC-ID 10–095F and RIHSC-ID15-039F.
In this study we used the leftover sample DNA from seeded food samples that were used in our previous detection method development studies. Since sample DNA was not particularly prepared for this study, our samples are not in series, but provides data from various food matrixes seeded with different number of oocysts. The methods; including the ones used to prepare the DNA samples, are described below.
2.2. Seeding of fresh and frozen produce with C. cayetanensis oocysts
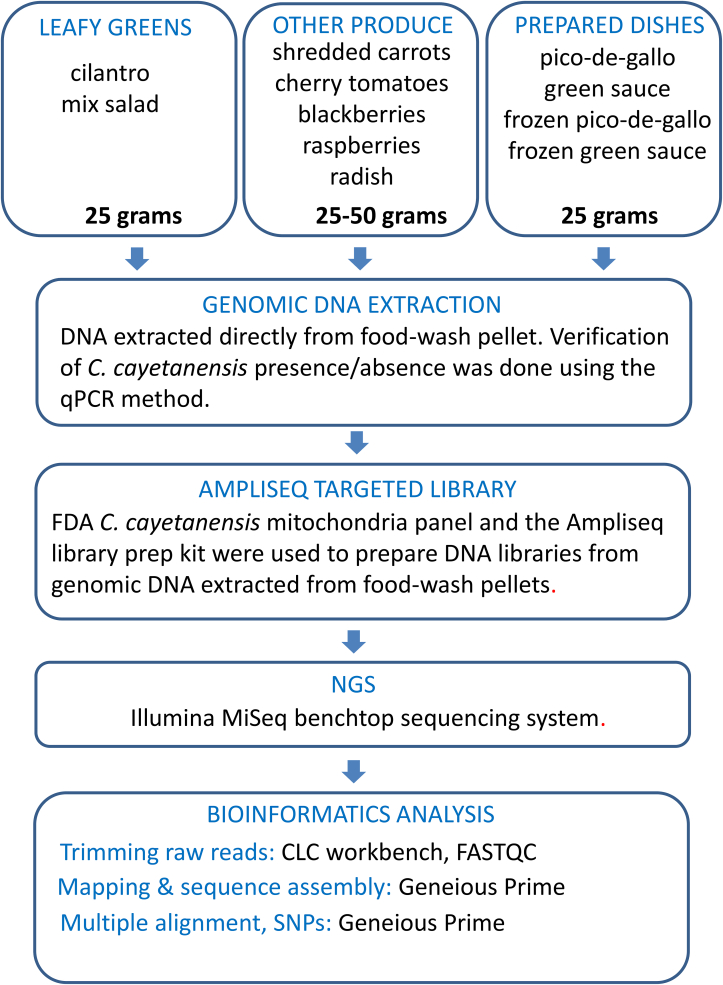
Fresh produce (cilantro, mixed salad, shredded carrots, cherry tomatoes, and blackberries, raspberries), without signs of withering, were obtained from local grocery stores and stored at 4 °C for no longer than 48 h prior to seeding experiments. Twenty-five grams of commercial fresh produce and prepared dish (pico de gallo or green sauce) test samples (total 37 samples) were prepared, with the exception of fresh berry test samples, which were prepared with 50 g for each (Figure 1), as described previously [24, 25, 35]. The samples were seeded with 200, 20, 10 or 5 oocysts originating from either Nepal or Indonesia using the FDA method BAM 19b [24]. Unseeded samples were processed together with the seeded samples as negative controls (Table 1). Samples were allowed to air dry uncovered at room temperature for approximately 2 h, transferred to BagPage filter bags (Interscience Lab Inc., Boston, MA), sealed with binder clips, and held at 4 °C for 48–72 h before initiating the produce wash step for fresh samples as described previously [24]. Frozen samples were seeded in the same manner as fresh samples and after being air dried, were held at -20 °C for 4 weeks prior to thawing at 4 °C for 24 h before initiating the produce wash step for frozen samples [26].
Figure 1.
Workflow to generate and analyze mitochondria genome assemblies from food and clinical isolates of C. cayetanensis. Wet-lab procedures and bioinformatics analysis steps are summarized in the figure.
Table 1.
Sample type, seeding and qPCR data.
| sample ID | number oocysts | food matrix | oocyst source | mean CT value (Std) |
|---|---|---|---|---|
| 1_6 | 0 | pico de gallo | N/A | und |
| 2 | 0 | shredded carrots | N/A | und |
| 2_10 | 0 | cherry tomato | N/A | und |
| 3_11 | 0 | cilantro | N/A | und |
| B2 | 5 | blackberries | Indonesia | 37.6 (0.1) |
| b1 | 5 | blackberries | Indonesia | 36.4 (1.4) |
| b2b (2) | 5 | blackberries | Indonesia | 37.3 (∗) |
| 7 | 5 | pico de gallo | Nepal | und |
| L1 | 5 | mix salad 1- | Indonesia | 37.1 (∗) |
| L6 | 5 | mix salad 1- | Indonesia | und |
| GS2-5 | 5 | frozen green sauce | Indonesia | 37.1 (∗) |
| R1 (2)-5 | 5 | raspberries | Indonesia | 37.2 (1) |
| B4 (2) | 10 | blackberries | Indonesia | 37.2 (0.7) |
| PG6 | 10 | frozen pico de gallo | Indonesia | 36.2 (0.5) |
| GS1a | 10 | green sauce | Indonesia | 37.6 (0.03) |
| GS1b(2) | 10 | green sauce | Indonesia | 37.2 (0.35) |
| 1 | 10 | shredded carrots | Nepal | 37.9 (∗) |
| 5 | 10 | shredded carrots | Nepal | und |
| GS5 | 10 | frozen green sauce | Indonesia | 37.7 (2.11) |
| R1-10 | 10 | radish | Indonesia | 36.7 (0.53) |
| R2 | 10 | radish | Indonesia | 35.3 (1) |
| C3 | 10 | cilantro | Indonesia | 37 (0.8) |
| C4 | 10 | cilantro | Indonesia | 36.3 (0.4) |
| 8 | 20 | pico de gallo | Nepal | 37.3 (∗) |
| GS10 | 200 | frozen green sauce | Indonesia | 25.6 (0.3) |
| L11 | 200 | mix salad1- | Indonesia | 31 (0.2) |
| L12 | 200 | mix salad1- | Indonesia | 31.7 (0.2) |
| C1 | 200 | cilantro | Indonesia | 32.9 (0.3) |
| C2 | 200 | cilantro | Indonesia | 33.2 (0.1) |
| B8 (2) | 200 | blackberries | Indonesia | 32.3 (0.3) |
| PG8 | 200 | frozen pico de gallo | Indonesia | 32.3 (0.1) |
| 9 | 200 | pico de gallo | Nepal | 35.2 (0.2) |
| GS1 | 200 | green sauce | Indonesia | 32.9 (0.2) |
| GS2 (2)-200 | 200 | green sauce | Indonesia | 32 (0.4) |
| 3 | 200 | shredded carrots | Nepal | 36.6 (0.8) |
| control S1 | N/A | dd H20 | N/A | N/A |
| neg-control ddH2O | N/A | dd H20 | N/A | N/A |
Und: Undetermined CT value after 45.
∗No standard deviation. One of the triplicates was positive in the qPCR reaction.
N/A: Not applicable.
2.3. Oocyst recovery and molecular detection of C. cayetanensis in food samples
The washing and molecular detection steps for both fresh and frozen berries followed the FDA's BAM Chapter 19b method [24, 35]. This method includes: 1) produce washing to recover C. cayetanensis oocysts, 2) DNA extraction from wash pellets containing concentrated oocysts, and 3) real time PCR analysis using a dual TaqMan™ method targeting the C. cayetanensis 18SrRNA, together with amplification of an internal amplification control (IAC) to monitor for reaction failure due to food matrix derived PCR inhibitors [35]. Produce wash debris pellets were stored at 4 °C for up to 24 h or frozen at -20 °C prior to DNA isolation. The DNA extraction procedure was performed using the FastDNA SPIN Kit for Soil in conjunction with a FastPrep-24 Instrument (MP Biomedicals, Santa Ana, California). The qPCR for C. cayetanensis was performed on an Applied Biosystems 7500 Fast Real time PCR System (ThermoFisher Scientific, Waltham, MA).
2.4. Targeted NGS of mitochondria genomes
A Custom AmpliSeq Panel based on the C. cayetanensis mitochondria genome (KP231180) was designed and developed by Illumina upon our request. This panel contained 35 primer sets in two primer pools to cover the mitochondria genome between 6 bp and 6266 bp (suppl. Table 1). AmpliSeq custom libraries were prepared following the protocol provided by the company (AmpliSeq for Illumina On-Demand, Custom and Community Panels Reference Guide, Document # 1000000036408 v05 October 2018) with some modifications. To quantify total DNA extracted from washed food pellets, we used the Qubit dsDNA Assay kits (Broad Range [BR] and High Sensitivity [HS]) before library construction.
Since these quantifications do not provide the actual C. cayetanensis DNA concentration due to the presence of plant and plant microbiome contamination, we used the maximum volume of DNA (7.5 μl per sample) in subsequent steps in an effort to maximize the C. cayetanensis DNA in any given sample. Sample DNA was amplified using the Amp_DNA PCR program: denaturation at 99 °C for 2 min, followed by 24 cycles of 99 °C for 15 s and 60 °C for 4 min, with a final hold at 10 °C. The primers were partially digested and phosphorylated for adapter ligation using 2 μl of the FuPa enzyme per sample at 50 °C for 10 min, 55 °C for 10 min, followed by enzyme inactivation at 62 °C for 20 min, and a final hold at 10 °C. AmpliSeq CD indexes were ligated to each sample using the Index adapter pooling guide of 2 μl of DNA Ligase at 22 °C for 30 min; this was followed by ligase inactivation at 68 °C for 5 min, and 72 °C for 5 min. The resulting library was purified using 30 μl Agencourt AMPure XP beads (Beckman Coulter, Brea, CA, USA), followed by a wash using 150 μl freshly prepared 70% ethanol. After purification, 5 μl of 10X library amplification primers and 45 μls of library amplification mix were added to the dried AMPure XP beads, and library amplification was performed at 98 °C for 2 min, followed by 7 cycles at 98 °C for 15 s and 64 °C for 1 min, with a final hold at 10 °C. The amplified amplicon library was purified using 25 μl of AMPure XP beads, to remove high-molecular weight DNA, and a second round of purification was performed with 60 μl of AMPure XP beads to remove small DNA fragments such as primers. Quantification of resulting libraries was accomplished by using the Qubit dsDNA Broad Range (BR) Assay kit. To determine the size distribution of libraries, a TapeStation 4200 D1000 chip (Agilent Technologies) was used according to manufacturer's instructions. Approximately 10–16 pmol of each library was paired-end sequenced on the MiSeq platform (Illumina) following the manufacturer's manual.
2.5. Organelle genome and 18S RNA gene copy number estimations
Copy number estimations were performed based on a comparison of NGS read coverages between a single copy nuclear gene hsp-70 (HQ216220.1) and the C. cayetanensis mitochondrial genome (KP231180), the apicoplast genome (KX189066) and the 18S RNA gene (AF111183). Raw NGS reads from sample C10 (Nepal #10) mapped to the sequences mentioned above using Geneious Prime (http://www.geneious.com/).
2.6. Generation of mitochondria genome assemblies
The CLC Genome Workbench toolkit 9.0 (Qiagen) was used for trimming the raw NGS reads based on quality and to remove the adaptor sequences as recommended. Assembly and analysis of reads were carried out in multiple steps following a combination of methods reported earlier [10, 15]. Briefly, trimmed reads were first mapped to the reference genome KP23180 using Geneious Prime. Maximum mismatch per read was set as 2%. To achieve good quality mitochondria genome assemblies with high confidence for SNP calling, a threshold of 30X was set up for mean read coverage. A reference-guided consensus sequence (‘consensus assembly’) was then extracted for the datasets, which met the read coverage threshold.
2.7. Identification of genomic variants
Each of the 27 consensus assemblies was mapped and compared with the KP231180 genome in Geneious Prime to determine SNPs.
3. Results
3.1. Framework for detection and quantification of C. cayetanensis in the seeded food samples
We designed this study to develop a genotyping method for C. cayetanensis that is applicable to produce samples collected by FDA or other regulatory bodies in the U.S. This method can be applied downstream of the FDA detection method described in BAM Chapter 19b, which is widely used in C. cayetanensis outbreak investigations, and in recent food surveillance activities performed by the FDA (Figure 1).
To simulate natural contamination events, several types of foods previously linked to C. cayetanensis outbreaks, were seeded with 5, 10, 20 and/or 200 with C. cayetanensis oocysts purified from clinical stool specimens. To establish a framework to evaluate the analytic sensitivity of the novel AmpliSeq targeted sequencing, we employed the qPCR protocol from BAM Chapter 19b. Following this protocol, samples seeded with 200 oocysts are expected to have 100% positivity by qPCR, while samples seeded with lower numbers are expected to provide fractional recoveries (ranging from 25-75% positivity for the samples seeded), a requirement for FDA validated methods [25, 35]. In the present study, this method identified all samples seeded with 200 oocysts as positive and all the unseeded samples as negative for C. cayetanensis. The samples seeded with 5 and 10 oocysts indicated the threshold level of detection for this method, and as expected, a few of the seeded samples with those oocyst numbers were negative (Table 1).
3.2. Design and application of Illumina Ampliseq C. cayetanensis mitochondria genome custom panel in targeted NGS
A custom panel targeting the complete mitochondrial genome of C. cayetanensis was designed and developed by Illumina (Methods and Suppl. File 1). We quantified the final amplification product of the libraries (Table 2) and evaluated the quality control measures of library construction (Suppl.file 2). Altogether, we were able to produce targeted AmpliSeq libraries meeting quality and quantity threshold required for Illumina NGS run protocols.
Table 2.
Targeted sequencing library and NGS data
| sample ID | Number oocysts | sample DNA (ng/μl) | library DNA (ng/μl) | size of raw sequence data (Mb) | total # of sequence reads | read coverage (mean) | % of mit. genome assembled | bp of mit.genome (total 6274 bp) assembled |
|---|---|---|---|---|---|---|---|---|
| 1_6 | 0 | 8.08 | 14.5 | 243.5 | 4 | 0 | ND∗ | ND |
| 2 | 0 | 2.95 | 3.06 | 0.6 | 24 | 1 | ND | ND |
| 2_10 | 0 | 0.037 | 0.09 | 45.7 | 6 | 0 | ND | ND |
| 3_11 | 0 | 7.74 | 17.50 | 343.2 | 6 | 0 | ND | ND |
| B2 | 5 | 22.8 | 0.67 | 296 | 657175 | 17627 | 99.20% | 6224 |
| b1 | 5 | 28.4 | 1.21 | 20 | 70709 | 1971 | 98.80% | 6198 |
| b2b (2) | 5 | 7.98 | 0.21 | 1.4 | 1227 | 21 | ND | ND |
| 7 | 5 | 50 | 0.18 | 36.4 | 64 | 2 | ND | ND |
| L1 | 5 | 105 | 5.19 | 10.4 | 13385 | 241 | 98.30% | 6170 |
| L6 | 5 | 165 | 7.75 | 5.4 | 439 | 14 | ND | ND |
| GS2-5 | 5 | 1.04 | 0.16 | 13 | 31658 | 718 | 89.80% | 5634 |
| R1 (2)-5 | 5 | 32.1 | 9.12 | 3.9 | 1350 | 39 | 98.60% | 6184 |
| B4 (2) | 10 | 4.79 | 0.74 | 3.8 | 8046 | 240 | 98.80% | 6197 |
| PG6 | 10 | 39.1 | 5.70 | 193 | 126572 | 3572 | 98.90% | 6208 |
| GS1a | 10 | 0.478 | 0.94 | 76 | 309101 | 8266 | 99.20% | 6225 |
| GS1b(2) | 10 | 0.36 | 0.19 | 2.8 | 7245 | 204 | 98.50% | 6183 |
| 1 | 10 | 25.4 | UDT | 9 | 12900 | 381 | 89.50% | 5617 |
| 5 | 10 | 3.1 | 8.23 | 2.2 | 191 | 6 | ND | ND |
| GS5 | 10 | 1.91 | 0.11 | 12.1 | 38712 | 666 | 98.30% | 6164 |
| R1-10 | 10 | 5.96 | 1.2 | 556.6 | 1101367 | 31502 | 99.40% | 6237 |
| R2 | 10 | 6.27 | 1.45 | 749.6 | 1592781 | 44675 | 99.80% | 6263 |
| C3 | 10 | 99.1 | 8.06 | 47 | 52524 | 1480 | 99.10% | 6215 |
| C4 | 10 | 115 | 2.98 | 67.5 | 82868 | 2271 | 99.10% | 6220 |
| 8 | 20 | 56 | 0.54 | 34.7 | 8956 | 265 | 98.80% | 6197 |
| GS10 | 200 | 0.535 | 18.6 | 867.9 | 1830068 | 50030 | 99.70% | 6258 |
| L11 | 200 | 280 | 24.6 | 419.1 | 865030 | 24784 | 99.30% | 6232 |
| L12 | 200 | 35.8 | 7.38 | 388.9 | 994458 | 22618 | 99.60% | 6249 |
| C1 | 200 | 85 | 5.07 | 47.5 | 96695 | 1869 | 99.10% | 6219 |
| C2 | 200 | 92.8 | 9.32 | 103.2 | 224478 | 4890 | 99.20% | 6225 |
| B8 (2) | 200 | 3.3 | 8.44 | 13.9 | 37585 | 1083 | 98.80% | 6179 |
| PG8 | 200 | 34.8 | 7.2 | 173 | 487272 | 13392 | 99.30% | 6228 |
| 9 | 200 | 60 | 0.787 | 95 | 81239 | 2355 | 99.00% | 6211 |
| GS1 | 200 | 0.125 | 0.918 | 300 | 1671319 | 45214 | 99.40% | 6238 |
| GS2 (2)-200 | 200 | 0.143 | 6.42 | 14 | 37810 | 1092 | 99.00% | 6211 |
| 3 | 200 | 12.5 | 2.05 | 4.2 | 5854 | 157 | 98.80% | 6197 |
| control S1 | N/A | N/A | 0.063 | 35 | 57 | 1 | ND | ND |
| neg-control ddH2O | N/A | N/A | 0.06 | 1.111 | 381 | 4 | ND | ND |
UDT: Under detection limit of Qubit HS assay.
N/A: Not applicable.
ND: Not done.
The AmpliSeq targeted library protocol calls for 10 ng target DNA (∼3000 copies calculated based on human genome) which may be decreased down to 1ng target DNA (∼300 copies calculated based on human genome) with suggested protocol adjustments. The total amount of DNA extracted from the wash pellet of food samples seeded with C. cayetanensis oocysts demonstrated a wide range in quantity, suggesting a variability in the amount of plant tissue in food wash pellets (Table 2). There is no feasible way to measure the exact amount of DNA coming from the oocysts spiked into the sample. However, we can estimate the number of target sequences based on the number of oocysts added to the sample. We estimated the number of mitochondria genomes in C. cayetanensis oocysts using the number of hits of NGS reads for mitochondria genomes versus the single copy gene hsp 70, used as a marker for the nuclear genome, and determined that there are approximately 67 mitochondria genome copies per haploid genome of C. cayetanensis (Suppl. file 3). Unsporulated oocyts of C. cayetanensis have two haploid genomes, while sporulated oocysts have four haploid genomes in four sporozoids. Hence, the seeding of at least 5 oocysts into food samples may represent a recoverable range of 10–20 haploid genomes in addition to 670 (67X10) to 1340 (67X20) recoverable copies of mitochondria genomes, depending on the sporulation stage of the oocysts. Although, actual copy numbers in the DNA samples are expected to be lower than these calculations, which are based on full recovery of 5 seeded oocysts, the expected range of mitochondrial genome copy numbers suggest that they may be within the detection range of targeted NGS.
Two different oocyst sources (from Nepal, and Indonesia) were used to seed the food samples. Although the main objective of this study was to obtain near-complete mitochondrial genome sequences of C. cayetanensis from seeded food samples, using the Ampliseq targeted sequencing method, and we would be able to achieve this objective using a single source, by using two sources we were able to show that method can be used to differentiate contamination sources. Also, we were able to show the performance of the method on different oocysts, since physical conditions and sporulation stages of oocysts might differ and might affect the performance of the method. Sporulation levels of these two oocyst pools were different. Oocysts originating from the Nepalese sample, Nepal 9, were mostly unsporulated, while approximately 50% of the oocysts were sporulated in the sample originated from the Indonesian sample, Indo 31. We found that performance of both qPCR and Ampliseq sequencing was lower in the samples seeded with Nepal 9 oocysts in comparison with the samples seeded with Indo 31 (Table 2); this can be explained by the presence of lower genome copies in unsporulated oocysts.
3.3. Next generation sequencing and assembly of mitochondrion genomes
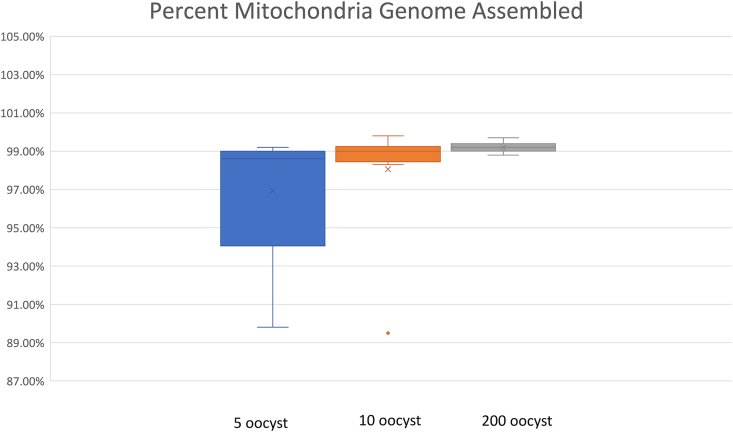
When we sequenced the Illumina Ampliseq libraries on the MiSeq (Illumina) platform, we found that size of the raw sequence data showed high variability, ranging between 0.6 Mb to 867.9 Mb, among samples. There was no correlation between the size of the raw sequence data and other factors such as the number of oocysts used, the DNA concentration of the food wash pellet, or the DNA concentration of the AmpliSeq library (Table 2). Raw sequence reads were mapped to the mitochondrial reference genome KP231180. The number of reads that mapped was between 0 and 4 among unseeded control samples, and between 2 and 50030 in seeded samples (Table 2). To achieve optimal quality mitochondria genome assemblies with high confidence for SNP calling, we set an average threshold of 30 X for read coverage. None of the unseeded controls and experimental negative controls were above the coverage threshold (Table 2). Among the samples seeded with 5 oocysts, the read coverage in 5 out of 8 samples were above the threshold. Of the 3 samples under the threshold, two samples were already undetermined by qPCR, and one showed a Ct value very close to the cut-off value (38.0). We were able to obtain mitochondria genome assemblies, the completeness of which ranged from 89.8 % to 99.2 %, in samples seeded with 5 oocysts that were above the threshold. Among the samples seeded with 10 oocysts, 10 out of 11 were above the coverage threshold and these samples yield mitochondrial genome assemblies with 89.5–99.8 % completeness. The only sample seeded with 10 oocysts below the coverage threshold was also undetermined by qPCR. We only ran one sample for the condition of 20 oocyst seeding. This sample with 20 oocysts yielded a genome assembly with 98.8 % completeness. Among the samples seeded with 200 oocysts, all 11 samples were above the coverage threshold. In these 11 samples, we were able to obtain mitochondria genome assemblies ranging between 99.0 % to 99.8 % completeness (Figure 2, Table 2). Overall, we were able to obtain mitochondria genome assemblies from 27 samples (out of 32 seeded samples) that meet the coverage threshold.
Figure 2.
C. cayetanensis mitochondria genome recovery at different seeding levels. Percent coverage of mitochondrial genome sequences obtained from samples seeded with 5, 10 and 200 oocysts.
3.4. Identification of SNPs in the mitochondria genomes of different samples
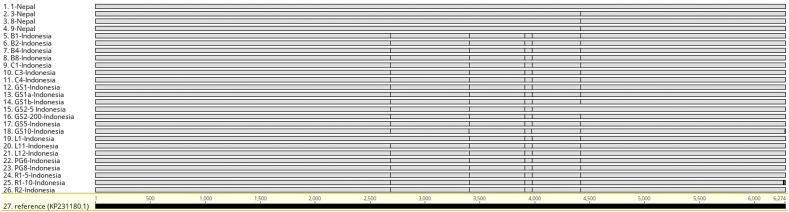
We determined distinct SNPs in 5 positions across the genome (Figure 2, Table 3). Four of these SNPs were already identified and reported [17], while a new SNP was discovered in this study. All Indonesian isolates, with the exceptions of GS2-5 and R1-5, exhibited all 5 SNPs. Although both GS2-5 and R1-5 have 4 of the 5 SNPs, low sequence coverage at the remaining SNP site did not allow reliable SNP identification. All Nepalese isolates, except sample 1, had a single SNP, which is common between the Indonesian and Nepalese isolates. Again, low read coverage at this SNP position in sample 1 prevented SNP calling (Figure 3). There was no difference identified at the15-mer repeat region between the samples seeded with Indonesian and Nepalese isolates.
Table 3.
Single nucleotide polymorphisms (SNPs) identified in the reference genome from this study.
| # | Allele | Variants | Reference genome position |
|---|---|---|---|
| 1 | A | C | 2687 |
| 2 | T | G | 3404 |
| 3 | A | C | 3910 |
| 4 | T | G | 3973 |
| 5 | C | A | 4415 |
Figure 3.
Sequence variations identified in this study. Multiple genome alignments with SNPs shown in Geneious visualization panel.
4. Discussion
We developed a highly sensitive targeted NGS method to obtain near complete mitochondrial genome sequences of C. cayetanensis from contaminated food matrices (Figure 1). Our method can be used in food samples ranging from fresh produce to prepared dishes with multiple ingredients, and frozen dishes. This method potentially opens a way to epidemiologically analyze the food items in C. cayetanensis-related outbreaks.
Analyses involving mitochondrial DNA sequences have been widely used in phylogenetics [36, 37, 38], forensic sciences [39, 40], and anthropology [41]. As genotyping targets, mitochondrial DNA has unique attributes in comparison to nuclear DNA such as maternal inheritance without recombination events, and relatively higher mutation rates [42, 43, 44, 45]. Moreover, the power of mitochondrial genome analysis builds on its ability to identify groups that descended from single source. Therefore, methods based on the mitochondrial genome sequence analysis may serve as important tools for outbreak investigations where back-tracking from individual case to outbreak source is crucial [17, 40]. Furthermore, the multi-copy nature of mitochondrial genomes in cells provides an advantage over unit-copy of nuclear sequences for amplification-based molecular detection methods such as PCR and NGS. These characteristics suggest that mitochondrial DNA sequences might be a suitable target for the development of the genotyping methods for C. cayetanensis [17].
Several methods have been developed for C. cayetanensis genotyping. Currently available methods applying Multi Locus Sequence Typing (MLST) have only been tested in clinical stool samples that contain much larger quantities of oocysts in comparison to contaminated food samples [18, 19, 20, 22, 23]. These PCR based methods might not be suitable for food samples contaminated with a few oocysts.
Conventional PCR based genotyping approaches, which were not readily applicable to food samples with low contamination levels, pose an integration problem for data from clinical isolates and food sources. As a first step to confront this problem, we previously reported a method that brings together the NGS of complete mitochondria genomes of C. cayetanensis from clinical stool samples and cilantro samples in a common platform [17]. In that paper, we were able to perform an allele-based classification scheme for clinical isolates; additionally, were able to include in the analysis the complete mitochondria genome of C. cayetanensis obtained from a cilantro sample seeded with 200 oocysts, but not at lower contamination levels. Our new method using targeted NGS sequencing brings the sensitivity level down to 5–10 oocysts per regulatory sampling for produce and prepared dish samples and provides a universal platform where both patient and food samples can be processed. This approach also opens an opportunity to integrate various genomic targets currently tested by different groups to generate informative sequence variance data from food, environmental and possibly clinical samples; the variety and number of targets should be sufficient enough to contribute to a comprehensive genotyping schema for C. cayetanensis for trace-back investigations.
Our study supports the applicability of targeted NGS technology to food samples with low level C. cayetanensis contamination. In addition to the epidemiologic analysis of food samples, this methodology (targeted NGS) could potentially be used for the surveillance and epidemiologic study of environmental samples such as water and soil, and clinical specimens. Our results re-emphasize the need for further generation of genome sequence data from clinical food and environmental samples that will clarify the course of environmental dissemination of the alleles over time and space. FDA is continuously improving any and all methodologies associated with the detection, identification, and potential genotyping of the pathogen and is currently testing additional panels containing genomic as well as mitochondrial sequences to improve sensitivity for detecting C. cayetanensis in both clinical and environmental samples. The Illumina AmpliSeq targeted sequencing approach can also be extended to other food-borne agents such as viruses, where levels are low and laboratory culture methods are limited and contamination levels in food poses difficulty in detection.
Declarations
Author contribution statement
Hediye Nese Cinar, M.D: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper. Gopal Gopinath: Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data.
Sonia Almería: Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data.
Joyce M Njoroge: Performed the experiments; Contributed reagents, materials, analysis tools or data.
Helen R. Murphy, Alexandre da Silva: Contributed reagents, materials, analysis tools or data.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Data availability statement
Data will be made available on request.
Declaration of interest's statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
C. cayetanensis oocysts from Nepal were supplied by Dr. Ynes Ortega (University of Georgia, Athens). This study was approved by the institutional review board of the FDA (RIHSC-ID 10–095F).
C. cayetanensis oocysts from Indonesia were supplied by the Centers for Disease Control and Prevention under an Interagency Agreement between FDA and CDC; IAA #224-16-2033S (RIHSC-ID 15–039F).
Appendix A. Supplementary data
The following are the supplementary data to this article:
References
- 1.Ortega Y.R., et al. Cyclospora species--a new protozoan pathogen of humans. N. Engl. J. Med. 1993;328(18):1308–1312. doi: 10.1056/NEJM199305063281804. [DOI] [PubMed] [Google Scholar]
- 2.Ortega Y.R., Gilman R.H., Sterling C.R. A new coccidian parasite (Apicomplexa: eimeriidae) from humans. J. Parasitol. 1994;80(4):625–629. [PubMed] [Google Scholar]
- 3.Ortega Y.R., Sanchez R. Update on Cyclospora cayetanensis, a food-borne and waterborne parasite. Clin. Microbiol. Rev. 2010;23(1):218–234. doi: 10.1128/CMR.00026-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sterling C.R., Ortega Y.R. Cyclospora: an enigma worth unraveling. Emerg. Infect. Dis. 1999;5(1):48–53. doi: 10.3201/eid0501.990106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Almeria S., Cinar H.N., Dubey J.P. Cyclospora cayetanensis and cyclosporiasis: an update. Microorganisms. 2019;7(9) doi: 10.3390/microorganisms7090317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Herwaldt B.L. Cyclospora cayetanensis: a review, focusing on the outbreaks of cyclosporiasis in the 1990s. Clin. Infect. Dis. 2000;31(4):1040–1057. doi: 10.1086/314051. [DOI] [PubMed] [Google Scholar]
- 7.Abanyie F., et al. 2013 Multistate outbreaks of Cyclospora cayetanensis infections associated with fresh produce: focus on the Texas investigations. Epidemiol. Infect. 2015;143(16):3451–3458. doi: 10.1017/S0950268815000370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Casillas S.M., Bennett C., Straily A. Notes from the field: multiple cyclosporiasis outbreaks - United States, 2018. MMWR Morb. Mortal. Wkly. Rep. 2018;67(39):1101–1102. doi: 10.15585/mmwr.mm6739a6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ogedengbe M.E., et al. A linear mitochondrial genome of Cyclospora cayetanensis (Eimeriidae, Eucoccidiorida, Coccidiasina, Apicomplexa) suggests the ancestral start position within mitochondrial genomes of eimeriid coccidia. Int. J. Parasitol. 2015;45(6):361–365. doi: 10.1016/j.ijpara.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cinar H.N., et al. The complete mitochondrial genome of the foodborne parasitic pathogen cyclospora cayetanensis. PLoS One. 2015;10(6):e0128645. doi: 10.1371/journal.pone.0128645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tang K., et al. Genetic similarities between Cyclospora cayetanensis and cecum-infecting avian Eimeria spp. in apicoplast and mitochondrial genomes. Parasites Vectors. 2015;8:358. doi: 10.1186/s13071-015-0966-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qvarnstrom Y., et al. Draft genome sequences from cyclospora cayetanensis oocysts purified from a human stool sample. Genome Announc. 2015;3(6) doi: 10.1128/genomeA.01324-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu S., et al. Comparative genomics reveals Cyclospora cayetanensis possesses coccidia-like metabolism and invasion components but unique surface antigens. BMC Genom. 2016;17:316. doi: 10.1186/s12864-016-2632-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cinar H.N., et al. Comparative sequence analysis of Cyclospora cayetanensis apicoplast genomes originating from diverse geographical regions. Parasites Vectors. 2016;9(1):611. doi: 10.1186/s13071-016-1896-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gopinath G.R., et al. A hybrid reference-guided de novo assembly approach for generating Cyclospora mitochondrion genomes. Gut Pathog. 2018;10:15. doi: 10.1186/s13099-018-0242-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qvarnstrom Y., et al. Purification of Cyclospora cayetanensis oocysts obtained from human stool specimens for whole genome sequencing. Gut Pathog. 2018;10:45. doi: 10.1186/s13099-018-0272-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cinar H.N., et al. Molecular typing of Cyclospora cayetanensis in produce and clinical samples using targeted enrichment of complete mitochondrial genomes and next-generation sequencing. Parasites Vectors. 2020;13(1):122. doi: 10.1186/s13071-020-3997-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo Y., et al. Multilocus sequence typing tool for cyclospora cayetanensis. Emerg. Infect. Dis. 2016;22(8):1464–1467. doi: 10.3201/eid2208.150696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo Y., et al. Population genetic characterization of Cyclospora cayetanensis from discrete geographical regions. Exp. Parasitol. 2018;184:121–127. doi: 10.1016/j.exppara.2017.12.006. [DOI] [PubMed] [Google Scholar]
- 20.Hofstetter J.N., et al. Evaluation of multilocus sequence typing of cyclospora cayetanensis based on microsatellite markers. Parasite. 2019;26:3. doi: 10.1051/parasite/2019004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nascimento F.S., et al. Mitochondrial junction region as genotyping marker for cyclospora cayetanensis. Emerg. Infect. Dis. 2019;25(7):1314–1319. doi: 10.3201/eid2507.181447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barratt J.L.N., et al. Genotyping Genetically Heterogeneous Cyclospora Cayetanensis Infections to Complement Epidemiological Case Linkage. Parasitology. 2019:1–33. doi: 10.1017/S0031182019000581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nascimento F.S., et al. Evaluation of an ensemble-based distance statistic for clustering MLST datasets using epidemiologically defined clusters of cyclosporiasis. Epidemiol. Infect. 2020;148:e172. doi: 10.1017/S0950268820001697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murphy H.R., Lee S., da Silva A.J. Evaluation of an improved U.S. Food and drug administration method for the detection of cyclospora cayetanensis in produce using real-time PCR. J. Food Protect. 2017;80(7):1133–1144. doi: 10.4315/0362-028X.JFP-16-492. [DOI] [PubMed] [Google Scholar]
- 25.Almeria S., et al. Evaluation of the U.S. Food and Drug Administration validated method for detection of Cyclospora cayetanensis in high-risk fresh produce matrices and a method modification for a prepared dish. Food Microbiol. 2018;76:497–503. doi: 10.1016/j.fm.2018.07.013. [DOI] [PubMed] [Google Scholar]
- 26.Assurian A., et al. Evaluation of the U.S. Food and Drug Administration validated molecular method for detection of Cyclospora cayetanensis oocysts on fresh and frozen berries. Food Microbiol. 2020;87 doi: 10.1016/j.fm.2019.103397. [DOI] [PubMed] [Google Scholar]
- 27.Almeria S., Assurian A., Shipley A. Modifications of the U.S. food and drug administration validated method for detection of Cyclospora cayetanensis oocysts in prepared dishes: Mexican-style salsas and guacamole. Food Microbiol. 2021;96 doi: 10.1016/j.fm.2020.103719. [DOI] [PubMed] [Google Scholar]
- 28.Dick G.J., et al. The microbiology of deep-sea hydrothermal vent plumes: ecological and biogeographic linkages to seafloor and water column habitats. Front. Microbiol. 2013;4:124. doi: 10.3389/fmicb.2013.00124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Song E.J., Lee E.S., Nam Y.D. Progress of analytical tools and techniques for human gut microbiome research. J. Microbiol. 2018;56(10):693–705. doi: 10.1007/s12275-018-8238-5. [DOI] [PubMed] [Google Scholar]
- 30.Carleton H.A., et al. Metagenomic approaches for public health surveillance of foodborne infections: opportunities and challenges. Foodb. Pathog. Dis. 2019;16(7):474–479. doi: 10.1089/fpd.2019.2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chang F., Li M.M. Clinical application of amplicon-based next-generation sequencing in cancer. Cancer Genet. 2013;206(12):413–419. doi: 10.1016/j.cancergen.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 32.Rajasagi M., et al. Systematic identification of personal tumor-specific neoantigens in chronic lymphocytic leukemia. Blood. 2014;124(3):453–462. doi: 10.1182/blood-2014-04-567933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patel N.M., et al. Enhancing next-generation sequencing-guided cancer care through cognitive computing. Oncol. 2018;23(2):179–185. doi: 10.1634/theoncologist.2017-0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bewicke-Copley F., et al. Applications and analysis of targeted genomic sequencing in cancer studies. Comput. Struct. Biotechnol. J. 2019;17:1348–1359. doi: 10.1016/j.csbj.2019.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murphy H.R., et al. Interlaboratory validation of an improved method for detection of Cyclospora cayetanensis in produce using a real-time PCR assay. Food Microbiol. 2018;69:170–178. doi: 10.1016/j.fm.2017.08.008. [DOI] [PubMed] [Google Scholar]
- 36.Brown W.M., George M., Jr., Wilson A.C. Rapid evolution of animal mitochondrial DNA. Proc. Natl. Acad. Sci. U. S. A. 1979;76(4):1967–1971. doi: 10.1073/pnas.76.4.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chikuni K., et al. Molecular phylogeny based on the kappa-casein and cytochrome b sequences in the mammalian suborder Ruminantia. J. Mol. Evol. 1995;41(6):859–866. doi: 10.1007/BF00173165. [DOI] [PubMed] [Google Scholar]
- 38.Lake J.A. Eukaryotic origins. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2015;370(1678) doi: 10.1098/rstb.2014.0321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Budowle B., et al. Forensics and mitochondrial DNA: applications, debates, and foundations. Annu. Rev. Genom. Hum. Genet. 2003;4:119–141. doi: 10.1146/annurev.genom.4.070802.110352. [DOI] [PubMed] [Google Scholar]
- 40.Grzybowski T., Rogalla U. Mitochondria in anthropology and forensic medicine. Adv. Exp. Med. Biol. 2012;942:441–453. doi: 10.1007/978-94-007-2869-1_20. [DOI] [PubMed] [Google Scholar]
- 41.Forster P., et al. Origin and evolution of Native American mtDNA variation: a reappraisal. Am. J. Hum. Genet. 1996;59(4):935–945. [PMC free article] [PubMed] [Google Scholar]
- 42.Denver D.R., et al. Abundance, distribution, and mutation rates of homopolymeric nucleotide runs in the genome of Caenorhabditis elegans. J. Mol. Evol. 2004;58(5):584–595. doi: 10.1007/s00239-004-2580-4. [DOI] [PubMed] [Google Scholar]
- 43.Denver D.R., et al. High mutation rate and predominance of insertions in the Caenorhabditis elegans nuclear genome. Nature. 2004;430(7000):679–682. doi: 10.1038/nature02697. [DOI] [PubMed] [Google Scholar]
- 44.Lynch M., Koskella B., Schaack S. Mutation pressure and the evolution of organelle genomic architecture. Science. 2006;311(5768):1727–1730. doi: 10.1126/science.1118884. [DOI] [PubMed] [Google Scholar]
- 45.Melvin R.G., Ballard J.W.O. Cellular and population level processes influence the rate, accumulation and observed frequency of inherited and somatic mtDNA mutations. Mutagenesis. 2017;32(3):323–334. doi: 10.1093/mutage/gex004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.