Abstract
The antibody response to Cryptococcus neoformans capsular glucuronoxylomannan (GXM) in BALB/c mice frequently expresses the 2H1 idiotype (Id) and is restricted in variable gene usage. This study examined the immunogenicity of GXM-protein conjugates, V (variable)-region usage, and 2H1 Id expression in seven mouse strains: BALB/c, C57BL/6, A/J, C3H, NZB, NZW, and (NZB × NZW)F1 (NZB/W). All mouse strains responded to vaccination with GXM conjugated to tetanus toxoid (TT), the relative magnitude of the antibody response being BALB/c ∼ C3H > C57BL/6 ∼ NZB ∼ NZW ∼ NZB/W > A/J. Analysis of serum antibody responses to GXM with polyclonal and monoclonal antibodies to the 2H1 Id revealed significant inter- and intrastrain differences in idiotype expression. Thirteen monoclonal antibodies (MAbs) (two immunoglobulin M [IgM], three IgG3, one IgG1, three IgG2a, two IgG2b, and two IgA) to GXM were generated from one NZB/W mouse and one C3H/He mouse. The MAbs from the NZB/W mouse were all 2H1 Id positive (Id+) and structurally similar to those previously generated in BALB/c mice, including the usage of a VH from the 7183 family and Vκ5.1. Administration of both 2H1 Id+ and Id− MAbs from NZB/W and C3H/H3 mice prolonged survival in a mouse model of cryptococcosis. Our results demonstrate (i) that V-region restriction as indicated by the 2H1 Id is a feature of both primary and secondary responses of several mouse strains; and (ii) that there is conservation of V-region usage and length of the third complementarity-determining region in antibodies from three mouse strains. The results suggest that V-region restriction is a result of antibody structural requirements necessary for binding an immunodominant antigen in GXM.
Cryptococcus neoformans causes life-threatening infections in immunocompromised patients, including those with AIDS and hematological malignancies (28). Cryptococcal infection in individuals with impaired immunity has high mortality, and those who survive the acute infection often have chronic infections. Given that the therapy of cryptococcosis is unsatisfactory, there is interest in vaccine development (14).
Control of C. neoformans infection is associated with cell-mediated immunity (28) and granuloma formation (23). However, there is strong evidence that humoral immunity can also be important. Antibody to the capsular glucuronoxylomannan (GXM) can prolong survival, reduce organ fungal burden, enhance granulomatous inflammation, and clear capsular polysaccharide antigen in infected mice (reviewed in references 3, 21, and 36). This suggests that vaccines which elicit high titers of protective antibodies may be useful for prevention of C. neoformans infection. GXM alone is unlikely to be an effective vaccine because it is poorly immunogenic and can be immunosuppressive (10, 26, 34). Conjugation of GXM to a protein carrier produces a highly immunogenic vaccine (6, 12, 13, 42). Mice immunized with a GXM-tetanus toxoid (GXM-TT) vaccine live longer and have lower CFU counts than controls when challenged with C. neoformans infection (12).
Molecular and idiotypic analyses of antibodies to GXM produced in response to infection or GXM-TT immunization revealed restriction in variable gene usage (5, 8, 29). Murine monoclonal antibodies (MAbs) to GXM can be classified into five groups based on molecular and idiotypic characteristics (5). Group II MAbs include several protective antibodies and are defined by a heavy-chain V (variable)-region using a VH from the 7183 gene family, a seven-amino-acid D/N segment which results in a fixed-length third complementarity-determining (CDR3) region, a light-chain V region using Vκ5.1, and reactivity with the 2H1 idiotype (Id) (5). The 2H1 Id is expressed by MAb 2H1, which is the prototypical group II antibody to GXM (5). MAb 2H1 has been demonstrated to protect against C. neoformans in murine intravenous (33), intracerebral (31), intraperitoneal (i.p.) (32), and intratracheal (20) infection. The crystal structure of MAb 2H1 has been solved with and without peptide mimetics (43). A MAb with a structure similar to that of 2H1 is in advanced preclinical development for use as adjunctive therapy in cryptococcal infections (4). However, expression of 2H1 Id is not sufficient for protection since antibodies with the same V-region usage manifest large differences in protective efficacy based on isotype (44) and fine specificity (30).
The molecular and cellular mechanisms which produce V-region-restricted responses are poorly understood. Here we studied the antibody response to a GXM conjugate vaccine in genetically different strains of mice, including three autoimmune strains. Our initial goal was to study the extent of V-region restriction in GXM-binding antibodies and generate MAbs different in specificities and molecular structure from those already available. We hypothesized that if restriction was the result of clonal deletion of self-reactive antibodies, then autoimmune mice would have a less restricted antibody response. Furthermore, we hypothesized that antibody responses in genetically different mouse strains might be quite different. This appeared to be confirmed by preliminary experiments with anti-idiotypic reagents to the 2H1 Id. However, molecular analysis of hybridomas from mice with 2H1 Id-negative (Id−) responses does not support either hypothesis since all mice studied made antibodies to GXM which are structurally similar to MAb 2H1. Instead, our findings suggest that V-region restriction in GXM-binding antibodies is a result of constrained requirements for antigen binding.
MATERIALS AND METHODS
C. neoformans strains and polysaccharide antigens.
Strains 24064 (serotype A), 24065 (serotype B), 24066 (serotype C), and 24067 (serotype D) were obtained from the American Type Culture Collection (Rockville, Md.). Strain 371 was obtained from John Bennett (Bethesda, Md.). Total cryptococcal polysaccharide was prepared by ethanol precipitation from late-log-phase cultures (24), and polysaccharide concentration was determined by the phenol-sulfuric acid method (18). GXM was purified from whole polysaccharide by cetyltrimethylammonium bromide precipitation (9).
Mice.
All mice were obtained from Jackson Laboratory (Bar Harbor, Maine). The H-2 haplotypes for A/J, BALB/c, C3H/HeJ, C57BL/6, NZB, and NZW mice are a, d, k, b, d, and z, respectively. The Igh-1 loci for BALB/c, C3H/HeJ, C57BL/6, and NZB mice are a, j, b, and e, respectively.
ELISAs for GXM.
Antibody reactivity for GXM was measured by enzyme-linked immunosorbent assay (ELISA). For serotypes A and D, the polysaccharide was added directly to polystyrene plates (Corning Glass Works, Corning, N.Y.) as described previously (7). For serotypes B and C, the polysaccharide does not bind polystyrene, and a MAb-based capture ELISA was used to immobilize the antigen (7). GXM ELISAs were done essentially as described previously (7). Goat anti-mouse alkaline phosphatase-conjugated isotype-specific antibodies (Southern Biotechnology, Birmingham, Ala.) were used as secondary antibodies. Serum responses to GXM were detected with goat anti-mouse alkaline phosphatase-conjugated antibodies to κ and λ light chains. To measure immunoglobulin G (IgG) levels in serum, IgM was dissociated by incubating serum in 0.01 M β-mercaptoethanol for 1 h at 37°C (38).
Rabbit antibody to 2H1 Id and the anti-Id ELISA.
MAb 2H1 is a protective murine IgG1 (6). MAb 2H1 protein was purified from either BALB/c ascites fluid or CellMax (Gibco BRL, Gaithersburg, Md.) supernatants by protein G column chromatography (Pierce, Rockford, Ill.). Two New Zealand White rabbits were immunized and boosted with 200 μg of 2H1 protein in Freund’s complete adjuvant (Difco, Detroit, Mich.). Rabbit bleeds were initially screened on 2H1, an irrelevant isotype-matched control, and the transfectoma 6A2 (provided by Sherie Morrison, University of California, Los Angeles), which expresses the 2H1 VH with a human IgG3 constant region and the light chain of the antiricin antibody R45. Rabbit serum with high titer to 2H1 was absorbed three times on a BALB/c total IgG-Sepharose 4B column (Pharmacia, Uppsala, Sweden). Eluent fractions containing rabbit IgG were pooled and positively affinity purified on a Sepharose 4B column containing a 2H1 mouse-human IgG3 chimeric antibody. Id-specific antibody was eluted at pH 3.0.
Generation of MAbs to the 2H1 Id.
MAb 2H1 protein was purified by protein G chromatography and conjugated to keyhole limpet hemocyanin (Pierce). Briefly, 2H1 and KLH were mixed at a weight/weight ratio of 1:1, 2% gluteraldehyde was then added dropwise, and the solution was incubated overnight at 4°C while mixing on an end-over-end rocker. The conjugate was then extensively dialyzed against sterile phosphate-buffered saline (PBS). The 2H1-keyhole limpet hemocyanin conjugate was emulsified in Freund’s complete adjuvant, and an amount corresponding to 100 μg of 2H1 was injected to five 6- to 8-week-old BALB/c female mice. Serum was screened by ELISA on TACZ1.1.4 (a mouse-human chimeric antibody expressing the 2H1 V region and the human IgG3 and kappa constant regions) captured to a polystyrene microtiter plate with a goat anti-human IgG3-specific antibody (Organon Teknika Corp., West Chester, Pa.). A mouse with high titers to the 2H1 Id was boosted by splenic immunization 3 days prior to fusion. Half of the spleen was fused to the NSO myeloma cell line, and the other half was fused to an NSO myeloma cell line transfected with bcl-2 (NSObcl-2). Hybridomas were screened by testing to a panel of MAbs which use the VH7183 and Vκ5.1 genes. Putative anti-idiotypic antibodies were tested for inhibition of 2H1 binding to GXM. Three anti-idiotypic antibodies (9A12, 26F10, and 7B8) were affinity purified from SCID mouse ascites fluid and coupled to CNBr-activated Sepharose 4B (Pharmacia). These columns were used to remove 2H1 Id-positive (Id+) antibodies from mouse sera obtained by GXM-TT immunization and to determine the amounts of 2H1 Id− antibody to GXM (see below).
Serum studies.
Two sets of female mice were immunized with GXM-TT, and their sera were analyzed for expression of the 2H1 Id. The first set consisted of 10 BALB/c, 5 NZB, 5 NZW, and 8 (NZB × NZW)F1 (NZB/W) mice. Four of the NZB/W mice were immunized at 25 to 27 weeks of age (an age after appearance of IgG to double-stranded DNA in serum), and all other mice were immunized at 6 to 8 weeks of age. The second set of mice consisted of five mice each of the BALB/c, A/J, C57BL/6, and C3H/HeJ strains. Due to experimental constraints related to the availability of conjugate, the first and second sets of mice were immunized and boosted with 3.75 and 5 μg, respectively, of GXM-TT. Sera were studied for antibody content to GXM by ELISA. Sera were assayed for (i) titer to GXM, using a mixture of anti-kappa and anti-lambda secondary antibody reagents; (ii) IgG titer to GXM (β-mercaptoethanol resistant); (iii) 2H1 Id+ titer; and (iv) 2H1 Id+ IgG titer.
Generation of MAbs to GXM from NZB/W and C3H mice.
Two female NZB/W mice were immunized with 2.5 μg of GXM conjugated to Pseudomonas aeroginosa exoprotein A (GXM-PsA) (13). The mouse with the highest titer was boosted again 3 days prior to splenocyte harvest. Since only one MAb was recovered from the first mouse due to contamination, the second mouse was boosted and its splenocytes were harvested for hybridoma generation. For the C3H fusion, mice were immunized with 5 μg of GXM-TT vaccine and boosted with the same concentration of GXM-TT in saline, and the splenocytes were fused to NSO myeloma cells by using standard protocols (11). Hybridomas were screened for binding to serotype A GXM. In the second NZB/W fusion, several plates of hybridomas were also screened by RNA dot blot hybridization with 32P-labeled oligonucleotides to VH7183 and Vκ5.1 (Table 1). When anti-idiotypic antibodies to 2H1 became available, a retrospective analysis of the supernatants from 96-well plates for 2H1 Id expression was also performed.
TABLE 1.
Oligonucleotides used in this study
| Namea | Sequence | Specificity |
|---|---|---|
| MS23 | TGGATGGTGGGAAGATG | Cκ |
| MS24 | GGGGCCAGTGGATACAC | CH1 IgG1, IgG2a, IgG2b |
| MS85 | TCTCGCAGGAGACGAGGGGGA | CH1 IgM |
| MS172 | TGTTCTTGGCATTGTCTCTG | VH7183 |
| MS193 | GAGTGTCAGTGGGTAGATGGT | CH1 IgA |
| MS194 | GAGAGCAGAAATAAACTCCC | Vκ5.1 |
| MS199 | GACGAGGGGGAAGACATT | CH1 IgM |
| MS218 | GAAGTGAAGCTGGTGGAGTCT | VH7183 |
| MS208 | AAGTAGCCTTTGACAAGGCA | CH1 IgG3 |
| MS464 | GGGAAGATGGATCCAGTTGGTGCAGCATCAGC | Cκ |
| MS465 | GGTGATATCGT(G/T)CTCAC(C/T)CA(A/G)TCTCCAGCAAT | Vκ |
| MS466 | GGTGATATC(A/T)TG(A/C)TGACCCAA(A/T)CTCCACTCTC | Vκ |
| MS467 | GAGGTGAAGCTGCAGGAGTCAGGACCTAGCCTGGTG | VH |
| MS468 | SGGT(C/G)(A/G)A(A/G)CTGCAG(C/G)AGTC(A/T)GG | VH |
| MS469 | CCAGGGGCCAGTGGATAGACAAGCTTGGGTGTCGTTTT | CH1 IgG |
Oligonucleotides MS464 to MS469 were based on sequences reported by Dubel et al. (17).
Southern blot analysis.
Genomic DNA was prepared from 5 × 106 cells by phenol-chloroform extraction. NZB/W DNA was a gift from D. Lustgarden (our laboratory). DNA was digested with restriction enzymes, using 10 U of enzyme per μg of DNA. After digestion, the DNA fragments were separated in a 1% agarose gel, transferred to a positively charged nylon membrane (Boehringer Mannheim) in 10× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), and cross-linked to the membrane in a UV Stratalinker (Stratagene, La Jolla, Calif.). Genomic probes were labeled with [α-32P]dCTP (New England Nuclear, Boston, Mass.), using random primers (Boehringer Mannheim). The labeled product was hybridized to the blot at 65°C.
V-region sequence determination.
Two methods were used for V-region sequence determination: direct sequencing of hybridoma mRNA, and DNA sequencing of amplified V regions cloned into a plasmid vector. RNA sequence determination was done as described by Geliebter et al. (22), with minor modifications (29). For DNA sequencing, cDNA was synthesized from mRNA by using oligonucleotides to the 5′ end of the heavy- and light-chain constant regions (Table 1) and then amplified by PCR with either Vent or Taq DNA polymerase. PCR products were either sequenced directly by using a Gibco BRL cycle sequencing kit or cloned into Invitrogen’s TA cloning kit and sequenced by the DNA sequencing facility at our institution. In cases where variable gene usage was unknown, PCR was performed with a set of degenerate primers designed to hybridize to most 5′ regions and all 3′ ends of murine V regions (Table 1).
Mouse protection studies.
Six- to eight-week-old female A/JCr mice were obtained from the National Cancer Institute (Bethesda, Md.). The mice were infected i.p. with 107 cells of C. neoformans 24067. A high infection inoculum was used to induce a rapidly lethal infection. Four hours before infection, mice were given 1 mg of MAbs i.p. in a total volume of 0.5 ml. The control group received either an equivalent volume of NSO myeloma ascites fluid diluted 1:1 with PBS or PBS. Mice were observed daily, and their deaths were recorded. Log rank test was used for survival analysis of lethally infected mice treated with MAbs.
RESULTS
Our studies were carried out in three phases and are presented here in the order in which they were performed. Initially, we studied MAbs to GXM generated from autoimmune mice on the premise that these MAbs would be different from previously described MAbs as a result of loss of tolerance. However, MAbs from autoimmune mice proved to be similar to the 2H1 MAb in molecular structure. This finding led us to generate anti-idiotypic antibodies to the 2H1 Id so that we could study multiple mice from autoimmune and nonautoimmune mouse strains. With these reagents, we attempted to identify mouse strains that made different antibody responses by screening immune sera to identify 2H1 Id− responses. Since studies of the 2H1 Id+ and Id− responses suggested heterogeneity in responses to GXM, we generated MAbs from a nonautoimmune mouse strain (C3H/HeJ) that responded to GXM-TT vaccination with a antibody response characterized by a predominance of 2H1 Id− antibodies. The C3H/HeJ MAbs were then studied for variable gene usage, 2H1 Id expression, and specificity. Finally, we examined the protective efficacy of several MAbs from NZB/W and C3H/HeJ in infection.
MAbs to GXM from NZB/W mice.
Female NZB/W mice develop a lupus erythematosus-like disease as they age, and we hypothesized that the immunological deficits in these mice could result in the generation of MAbs to GXM different in structure and specificity from those previously recovered from BALB/c mice (5, 29). Splenocytes were obtained from two 18- to 20-week-old female mice immunized with 2.5 μg of GXM-PsA without adjuvant (13). At this age, NZB/W mice have serological evidence of their genetic autoimmune disease but are not very ill. GXM-PsA conjugate was used because of its availability and to vary the carrier protein. The first mouse was boosted on day 14, and its splenocytes were fused 3 days later. The second mouse was boosted on day 26, and its splenocytes were fused 5 days later. From the first mouse, a single hybridoma (MAb A2F12) was recovered due to contamination of the fusion cultures. From the second mouse, 417 antigen-positive wells were identified. Isotype analysis of the MAbs in these wells revealed 313 IgM, 48 IgG1, 31 IgG3, 6 IgG2a, 7 IgG2b, and 12 IgA. Considering the numerous wells positive for reactivity with GXM, we limited our study to selected hybridomas. In particular, we tried to recover IgG2a and IgG2b isotypes since (i) pathogenic anti-DNA antibodies are usually of these isotypes and (ii) these isotypes were not recovered in previous fusions of BALB/c splenocytes (6, 8). Sixteen MAbs from NZB/W mice (1 from the first fusion and 15 from the second fusion) were cloned twice in soft agar. Isotypes and serotype reactivities of these MAbs are listed in Table 2.
TABLE 2.
GXM-binding MAbs generated in this study
| MAba | Origin | Isotype | 2H1 Id | Variable element used
|
GenBank accession no. | |||
|---|---|---|---|---|---|---|---|---|
| VH family | JH | VL | JL | |||||
| 4H9 | NZB/W | IgM | Yes | —b | — | — | — | |
| 6C3.4 | NZB/W | IgM | Yes | — | — | — | — | |
| 6A5 | NZB/W | IgG3 | Yes | — | — | — | — | |
| 9A12 | NZB/W | IgG3 | Yes | 7183 | JH1 | — | — | AF093564 |
| A2F12c | NZB/W | IgG3 | Yes | 7183 | JH1 | — | — | AF093562 |
| 6C3.3 | NZB/W | IgG1 | Yes | — | — | — | — | |
| IG5 | NZB/W | IgG2a | Yes | 7183 | JH1 | Vκ5.1 | Jκ1 | AF093561, AF093553, AF093554 |
| 7F8 | NZB/W | IgG2a | Yes | 7183 | JH1 | Vκ5.1 | Jκ1 | AF093560, AF093557, AF093558 |
| 12G6 | NZB/W | IgG2a | Yes | — | — | — | — | |
| 12A1.4 | NZB/W | IgG2b | Yes | — | — | — | — | |
| 15E6 | NZB/W | IgG2b | Yes | 7183 | JH1 | — | — | AF093559 |
| 14F8 | NZB/W | IgG2b | Yes | — | — | — | — | |
| 2G3 | NZB/W | IgA | Yes | — | — | — | — | |
| 13C8 | NZB/W | IgA | Yes | 7183 | JH1 | Vκ5.1 | Jκ1 | AF093563, AF093555, AF093556 |
| 16 | C3H | IgG1 | No | 7183 | JH2 | Vκ5.1 | Jκ1 | AF093549, AF128260 |
| 23 | C3H | IgG1 | No | 7188 | JH2 | Vκ5.1 | Jκ1 | AF093550, AF128261 |
| 25 | C3H | IgG1 | Yes | 7183 | JH2 | Vκ5.1 | Jκ1 | AF093551, AF128262 |
| 29 | C3H | IgG1 | No | 7183 | JH2 | Vκ5.1 | Jκ1 | AF093552, AF128263 |
All were reactive to serotypes A to D.
—, not done.
Sole MAb from the first fused NZB/W mouse.
The clonal relationship between the individual hybridomas was inferred from Southern blot analysis. Five different VH gene and three clearly different VL gene rearrangements were observed (data not shown). The VH sequences of five MAbs were determined; all used a member of the 7183 VH gene family rearranged to JH1, resulting in a CDR3 region of seven amino acids with conserved charge and hydrophobic residues similar to those in previously described MAbs generated in BALB/c mice (GenBank accession numbers in Table 2). Since all MAbs had the same VL rearrangement by Southern blot analysis, we limited our sequence analysis to only three, each of which expressed the combination Vκ5.1 and Jκ1 (Table 2). Hence, vaccination of at least one NZB/W mouse elicited antibodies similar in specificity, idiotype, and molecular structure to those previously generated from BALB/c mice immunized with GXM-TT. To determine whether this was a general phenomenon, we proceeded to compare the serum antibody responses in other mouse strains.
Serum responses to GXM-TT in normal and autoimmune mouse strains.
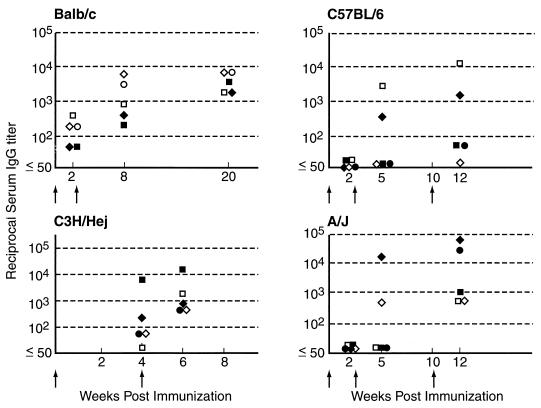
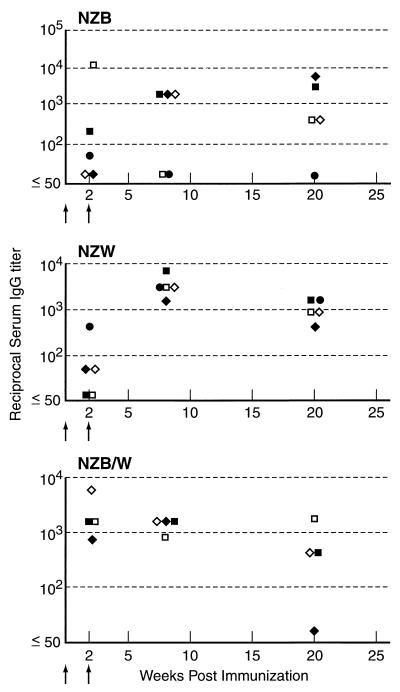
BALB/c, C3H/HeJ, C57BL/6, A/J, NZB, NZW, and NZB/W mice each made serum antibody responses to GXM after vaccination with GXM-TT (Fig. 1 and 2). Among those mice in which little or no IgG was detected, most had serum IgM. For example, each of the three A/J mice with IgG titers of only 1:50 to 1:100 had IgM titers ranging from 1:1,600 to 1:3,200. Among the mouse strains studied, the best responders were BALB/c mice, which mounted an IgG response after one vaccination and increased their IgM and IgG titers to GXM after the second immunization. Other mouse strains varied in the ability to respond to GXM-TT. Some C57BL/6 and A/J mice had no measurable serum IgG to GXM until the third dose was administered. However, even after a third dose, only two of five C57BL/6 mice had IgG titers above 1:100. GXM-TT was immunogenic in the NZB, NZW, and NZB/W mouse strains predisposed to autoimmune disorders (Fig. 2). Hence, all mouse strains studied responded to GXM-TT vaccination with high-titer responses, but the number of immunizations required to elicit an IgG response varied with the strain.
FIG. 1.
IgG antibody responses to GXM-TT vaccination for BALB/c, C57BL/6, C3H/Hej, and A/J mouse strains. The y axis denotes inverse titers. For each strain, five animals were studied. Arrows indicate the timing of vaccination. Each symbol represents a different mouse.
FIG. 2.
IgG responses to GXM vaccination for NZB, NZW, and NZB/W mouse strains. The y axis denotes inverse titers. For each strain, five animals were studied. Arrows indicate the timing of vaccination. Each symbol represents a different mouse.
To determine the prevalence of antibodies to GXM expressing the 2H1 Id in sera of mice immunized with GXM-TT, we generated rabbit polyclonal anti-idiotypic serum. In addition, five IgG1 murine MAbs to the 2H1 Id were generated, of which two (6A10 and 7B8) were 2H1-binding-site specific. The relative expression of the 2H1 Id in the polyclonal response of mice immunized with GXM-TT was assayed by using the affinity-purified rabbit polyclonal anti-Id reagent. Total (e.g., IgM plus IgG) and β-mercaptoethanol-resistant (e.g., IgG) antibodies to GXM were assayed and found to be similar with respect to predominance of the 2H1 Id (data not shown). Figure 3 shows 2H1 Id expression in serum IgG to GXM for several mouse strains immunized with GXM-TT. All ELISAs were normalized with MAb 2H1 to produce a titer with the anti-idiotypic reagents that was roughly equal to that measured in serum with the anti-isotypic reagent. That is, we adjusted the anti-idiotypic and anti-IgG ELISAs so that each gave the same absorbance for the same amount of purified 2H1. No major differences were observed for 2H1 Id expression at other times after immunization in the immune response to GXM-TT (data not shown).
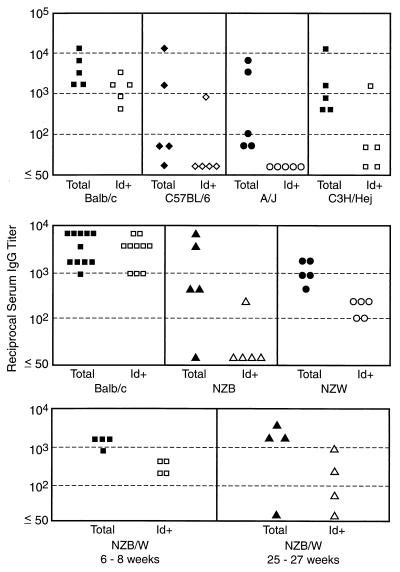
FIG. 3.
Total and 2H1 Id+ antibody responses to GXM vaccination in seven mouse strains. For the BALB/c (upper panel), NZB, NZW, and younger NZB/W mice, the titers were obtained 12 weeks after secondary vaccination; for C57BL/6, A/J, C3H/HeJ, BALB/c (middle panel), and the older NZB/W mice, the titers were measured 2 weeks after secondary vaccination. The y axis denotes inverse titers. There are two BALB/c panels because this strain was used as the positive control since it reliably produced high titers of GXM-binding antibodies which were 2H1 Id+ and the vaccination experiments for the autoimmune and nonautoimmune mice were done at different times. Serum samples for this experiment are from the same experiments as shown in Fig. 1 and 2. Some variation in titer of antibody to GXM is evident when the values in this figure are compared to those in Fig. 1 and 2, which is consistent with the experimental variation of the serological method used. The differences in the times that sera were analyzed are not important because analysis of 2H1 Id content at other times after vaccination produced similar results (data not shown).
Analysis of immune sera for 2H1 Id expression revealed strain-related differences in the relative proportion of GXM-binding antibodies expressing this idiotype (Fig. 3). In BALB/c mice, the antibody response to GXM was dominated by the 2H1 Id, and the total antibody and 2H1 Id titers to GXM varied by only one (and rarely two) dilutions in one direction or the other (data not shown). Since this comparison is semiquantitative and titer differences of one to two dilutions are within the error of the measurement, we removed the 2H1 Id+ antibodies by affinity chromatography with anti-idiotypic antibodies. When this was done for one BALB/c serum, all anti-GXM reactivity was removed, indicating that all GXM-binding antibodies were 2H1 Id+. However, occasional BALB/c mice had discordance between their total titer to GXM and the 2H1 Id titer to GXM, indicating the presence of antibodies not expressing the 2H1 Id. For the other mouse strains, the 2H1 Id constituted a significantly lower proportion of the total antibody response. 2H1 expression was not detectable in sera from the A/J mice that responded to GXM-TT (Fig. 3). Of the two C57BL/6 mice that mounted high titers in response to GXM-TT, one had a serum antibody response to GXM that was negative for the 2H1 Id and the other had antibodies expressing the 2H1 Id. C3H/HeJ mice made antibody responses to GXM after GXM-TT immunization, but for four of five mice the GXM-binding antibodies were not recognized by the reagents to the 2H1 Id.
Each of the three autoimmune mouse strains expressed significantly less 2H1 Id in IgG to GXM than did BALB/c mice (Fig. 3). Of the four NZB mice that responded to immunization, one expressed the 2H1 Id at a low level (1:200 2H1 Id titer, compared to 1:3,200 for total titer of antibody to GXM). Removal of 2H1 Id+ antibodies by column chromatography resulted in virtually no loss of GXM-binding antibody. All five NZW mice expressed the 2H1 Id, but it represented only a small portion of the total response. In a third experiment, we vaccinated four NZB/W mice with GXM-TT before the onset of autoimmune disease and four after the onset of disease (Fig. 3). Both sets of NZB/W mice responded to vaccination, but a major portion of their antibodies were 2H1 Id−. This is in contrast to the one NZB/W mouse that was used to generate hybridomas (see above). A retrospective analysis of supernatants from 96 of the positive wells from that fusion revealed that all MAbs were 2H1 Id+ (data not shown). All of the MAbs generated from the NZB/W mouse were 2H1 Id+ (Table 2).
MAbs to GXM from C3H mice.
To examine the molecular structure of 2H1 Id+ and Id− antibodies to GXM, we generated MAbs from a C3H mouse with an antibody titer to GXM of 1:11,200 and a 2H1 Id titer to GXM of 1:1,600. Hybridoma supernatants were simultaneously screened for IgG isotype and 2H1 Id binding on GXM plates. The initial screen revealed 20 hybridomas, of which 5 were IgG 2H1 Id+, 13 were IgG 2H1 Id−, and 2 were IgM isotype 2H1 Id+. Four MAbs, 16, 23, 25, and 29 (Table 2), were selected for study. These four MAbs bound to all four C. neoformans serotypes with relative affinity D > A > B > C. MAbs 16, 23, and 29 were 2H1 Id−, and MAb 25 was 2H1 Id+. Sequence analysis of heavy- and light-chain genes revealed that all utilized a VH with high homology to VH7183 and that the VL was >95% homologous with the Vκ5.1 of MAb 2H1. Hence, both 2H1 Id+ and Id− MAbs had variable regions with high homology to that of MAb 2H1.
Protective efficacy of antibodies from NZB/W and C3H mice.
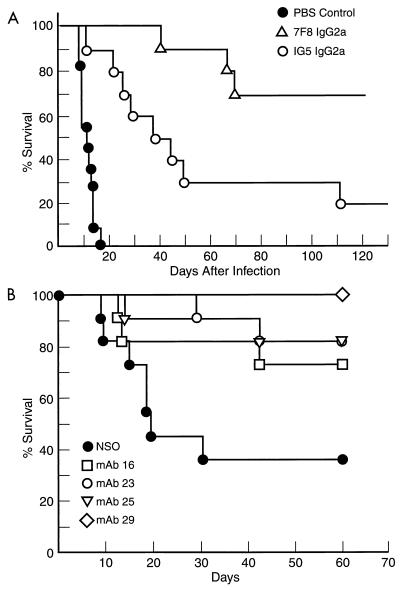
To investigate whether MAbs from autoimmune and C3H mice were protective, several were tested in passive protection experiments. Administration of either of two IgG2a MAbs from an NZB/W mouse significantly prolonged survival in lethally infected mice (Fig. 4A). The nine mice alive on day 133 of infection (two in MAb 1G5 group and seven in MAb 7F8 group) were killed, and their lungs and brains were cultured for CFU. No colonies were recovered from the organs of the two mice receiving 1G5. However, the seven mice receiving 7F8 were chronically infected with C. neoformans, indicating that antibody administration had converted a rapidly lethal infection into a chronic infection. Administration of the four MAbs from a C3H mouse also resulted in a significant prolongation in the average survival time of lethally infected mice (Fig. 4B). Hence, 2H1 Id+ MAbs from an NZB/W mouse and 2H1 Id+ and Id− MAbs from C3H mice each prolonged survival in lethally infected mice.
FIG. 4.
Survival curves for A/J mice given MAbs from either NZB/W or C3H/HeJ mice and lethally infected with C. neoformans. (A) Survival curves after administration of NZB/W-derived IgG2a MAbs 7F8 and IG5. (B) Survival curves after administration of C3H/HeJ-derived MAbs 16, 23, 25, and 29. Mice receiving antibody survived significantly longer than control mice in both experiments (P < 0.05, log rank analysis). There were 10 mice in each group. Control animals received either PBS (A) or NSO myeloma cells (B), since these negative controls had been used in prior experiments (32).
DISCUSSION
The antibody response to C. neoformans GXM in mice is under genetic control (16) and is highly restricted in variable gene usage (5, 6, 8, 29, 37). A comparative analysis of antibodies elicited in response to polysaccharide and GXM-TT revealed that both types of antibodies used the same variable gene elements but differed in affinity and isotype distribution, with the vaccine eliciting higher-affinity IgG type antibodies (29). In BALB/c mice immunized with GXM-TT, the antibody response to C. neoformans polysaccharide is dominated by the 2H1 Id, which includes both protective and nonprotective antibodies. In this study, we attempted to generate different types of antibody responses to GXM by utilizing conjugates with different carrier proteins, vaccinating autoimmune mice, vaccinating four genetically different mouse strains, and examining some mice which produced antibody responses without the 2H1 Id.
The 14 MAbs recovered from two NZB/W immunized with GXM-PsA each bound to the four C. neoformans serotypes and all were 2H1 Id+, indicating that these MAbs were serologically similar to previous MAbs generated from BALB/c mice immunized with GXM-TT. Molecular analysis of five VH and three VL revealed that the NZB/W MAbs used a 7183 variable gene element and Vκ5.1. Somatic mutations were distributed throughout the VH, with the highest concentration in CDR2, a region that may be involved in determining GXM specificity (30). As in BALB/c sequences, there were fewer somatic mutations in the VL. Nine of the thirteen VH CDR2 positions which were mutated among the NZB/W MAbs are identical to bases mutated among the 29 BALB/c MAbs studied earlier (29), and in all cases the same base changes were found. The VH of each MAb had a seven-amino-acid D/N region which has been shown to be a common feature of class II MAbs to GXM (5). However, unlike the majority of antibodies from BALB/c mice, the VH was rearranged to JH1 instead of JH2. Although JH1 is normally two amino acids longer than JH2, the VH for each of these MAbs recombined six bases into the JH1 gene element, resulting in a deletion of the first two amino acids, which conserved the length for the CDR3 relative to previously described MAbs to GXM (5). A similar event was previously noted for one BALB/c MAb that used JH4 (29). This arrangement results in conservation of both CDR3 length and the arginine-aspartic acid sequence at the beginning of CDR3. Molecular modeling of a 2H1 Id+ antibody revealed that this arginine is positioned in the GXM binding site such that it can interact with the acidic backbone of GXM (35). Crystallographic analysis of the 2H1 binding site indicates that a seven-amino-acid D/N segment is critical for forming the floor of the GXM binding site and that the RD motif is in the antigen binding site (43). The length, charge, and polarity of the VH CDR3 region is conserved in MAbs from both BALB/c and NZB/W mice. It is noteworthy that MAbs A2F12 and 1G5 were generated from different NZB/W mice yet had identical VH CDR3 sequences, a finding consistent with the importance of this region for GXM binding. The analysis of MAbs from this autoimmune mouse provides additional evidence of specific antibody structural requirements for binding GXM.
Southern blot analysis of VH genes in the 14 hybridomas from one NZB/W mouse indicates origin from at least seven precursors. Assuming that 0.001 to 0.0001% of activated B cells are captured as hybridomas in splenocyte-myeloma fusions, this mouse had between 4.1 × 105 to 4.1 × 106 activated B cells producing antibodies to GXM, an estimate consistent with the polyclonal activation found in lupus-prone NZB/W mice. Despite polyclonal activation and an autoimmune disorder, all MAbs to GXM from NZB/W utilized the same genetic elements and were similar to BALB/c MAbs. This finding argues against an explanation for variable gene restriction based solely on clonal deletion of self-reactive antibodies or anergy of antibodies to GXM which cross-react with self. Instead, the variable gene restriction observed in antibodies to GXM may be determined more by a structural requirement to bind an immunodominant epitope than by repertoire limitations imposed by the host to prevent self-reactivity.
To investigate the dependence of 2H1 Id expression on mouse genetic background, we studied the serum response to GXM-TT in four nonautoimmune strains of mice differing in H-2 and Igh haplotypes. One and two doses of GXM-TT vaccination elicited stronger antibody responses in BALB/c and C3H mice than in C57BL/6 and A/J mice. This observation with GXM-TT vaccination is consistent with previous studies of vaccination with GXM alone, which showed that BALB/c and C3H were high responders, C57BL/6 were intermediate responders, and A/J mice were low responders (16). However, after a third immunization, A/J mice had higher titers than C57BL/6 mice, suggesting additional strain differences in response after multiple doses of vaccine. Administration of GXM-TT to the three autoimmune mouse strains, NZB, NZW, and NZB/W elicited strong antibody responses to GXM, but occasional individual mice within a strain (i.e., one of five NZB and one of eight NZB/W) failed to respond to vaccination. These results indicate that GXM-TT is immunogenic in mice with diverse genetic backgrounds but there are significant mouse-to-mouse differences in the magnitude of the GXM response. Individual mouse variation in the antibody response to GXM has also been observed in C. neoformans infection (8). Although the mechanism for individual variation is not understood, it may reflect acquired immunological differences as a result of differences in antigenic exposure in utero and in the environment. Similar individual variation has been observed in the antibody response to many haptens (27, 39).
Rabbit polyclonal and mouse MAbs to the 2H1 Id were used to characterize the total antibody response to GXM in the sera of the four normal and three autoimmune mouse strains. The most striking result of the idiotypic study is that the 2H1 Id almost completely dominates the antibody response to GXM in BALB/c mice. In 14 of 15 BALB/c mice studied, the 2H1 Id accounted for most if not all of the IgM and IgG to GXM elicited by GXM-TT vaccination. The near-complete degree of 2H1 Id restriction in BALB/c mice contrasts with other restricted responses where additional V regions are utilized or appear after secondary immunization. For example, the response to phophorylcholine is dominated by the T15 Id in the primary response, but the secondary response involves the use of two additional light chains (19). The response to phenyloxazolone is dominated by two major idiotypes in 12 mouse strains, but depending on the strain the idiotypes are responsible for as little as ∼50% of the response (39). The combination of VHJ606 and Vκ11 dominates the primary response to bacterial levan, but this VH-VL pairing is only a minor component of the secondary response (2). In contrast to these examples, the 2H1 Id dominates the BALB/c response in both primary and secondary vaccination with GXM-TT and accounts for most if not all the antigen-specific response in that strain.
The six other mouse strains expressed the 2H1 Id to various degrees, and the magnitude of the response to GXM was unrelated to the ability to produce antibodies of the 2H1 Id. In the autoimmune strains, the 2H1 Id was consistently expressed in NZW mice but nearly absent in NZB mice. The fact that the NZB/W progeny show an Id expression pattern similar to that of the NZW parent when immunized at a young age suggests that the 2H1 Id is preferentially selected from several competing idiotypes. NZB/W mice vaccinated after the onset of active autoimmune disease (25 to 27 weeks) showed a more varied expression of the 2H1 Id, which may relate to the autoimmune process or differences in 2H1 Id expression with age. To compare the molecular structures of 2H1 Id+ and Id− antibodies elicited by GXM-TT, we generated MAbs in C3H mice and determined their V-region usage. Analysis of 2H1 Id+ and Id− GXM-binding MAbs from a C3H mouse revealed that all used a VH7183 gene element and Vκ5.1, which is the same gene combination described for the class II antibodies, of which 2H1 is the prototype. The negative reactivity of the three C3H MAbs with the 2H1 Id reagents presumably reflects amino acid differences in V-region genes which abolish binding with the anti-idiotypic reagents despite similar V-region usage. Hence, we cannot exclude that the 2H1 Id− component of the serum response in the various mouse strains also used a VH7183 gene element and Vκ5.1. In this regard, it is noteworthy that antibodies to GXM which are 2H1 Id− and differ in V-region usage (5) have been generated by Spiropulu et al. (40) and Dromer and collaborators (15) by using different immunization strategies. MAb variable region structure correlates with serological properties (1) and translates into differences in their complement-activating properties upon binding C. neoformans capsules (25). MAbs from both NZB/W and C3H mice prolonged survival in lethally infected mice, an indication that these MAbs are biologically active against C. neoformans.
The results of this study have been presented in the order in which they were performed. We began with the premise that antibody responses in autoimmune mice would produce antibodies different from those described previously because of their humoral immune disorder. When this proved not to be the case, we developed reagents to the 2H1 Id and studied the serological response to GXM-TT in seven mouse strains to identify those which made 2H1 Id− responses. Then we proceeded to make MAbs from a CH3/HeJ mouse with high titers of 2H1 Id− antibodies to GXM but found that these were also very similar to MAb 2H1. In retrospect, one can argue that the serological study should have preceded the generation of MAbs in autoimmune mice. However, the findings with the CH3/HeJ mouse indicates that selection of mice with 2H1 Id− responses does not exclude a predominance of antibodies very similar in molecular structure to MAb 2H1. Our experience suggests that to generate antibodies different than MAb 2H1 will require other strategies, possibly using different antigens for immunization.
In summary, our results confirm and significantly extend previous findings that the antibody response to GXM is highly restricted with regard to variable gene usage. We are unable to support or invalidate the original hypothesis that V-region restriction was related to the deletion of anti-GXM antibodies that reactive with self. Two 2H1 Id+ MAbs (one from a BALB/c mouse and the other from an NZB/W mouse) bound double-stranded DNA in a sensitive ELISA (unpublished data), suggesting that the VH7183-Vκ5.1 combination has the potential to react with self when some somatic mutations are present. The mechanism(s) responsible for V-region restriction remains unsolved; it is likely that several factors contribute to this phenomenon and that the mechanism varies with the antigen. Genes outside the H-2 locus may be involved in the regulation of V-region genes (41). The restriction in V-region usage for GXM-binding antibodies combined with conservation of the CDR3 length suggests that a specific antibody conformation is necessary for binding an immunodominant epitope shared by the four C. neoformans serotypes.
ACKNOWLEDGMENTS
A.C. and M.D.S. share senior authorship.
We thank John Robbins and Rachel Schneerson for the generous gifts of GXM-protein conjugates. We thank Antonio Nakouzi for depositing all of the sequences in GenBank.
A.C. is supported by Public Health Service awards AI33774, AI113342, and HL59842 and by a Burroughs Wellcome Fund Developmental Therapeutics Award. M.D.S. is supported by grants CA-39838, AR44192, AI42297, and AI43937 and the Harry Eagle Chair provided by the Women’s Division of the Albert Einstein College of Medicine.
REFERENCES
- 1.Belay T, Cherniak R, Kozel T R, Casadevall A. Reactivity patterns and epitope specificities of anti-Cryptococcus neoformans monoclonal antibodies by enzyme-linked immunosorbent assay and dot enzyme assay. Infect Immun. 1997;65:718–728. doi: 10.1128/iai.65.2.718-728.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boswell C M, Irwin D C, Goodnight J, Stein K E. Strain-dependent restricted VH and VL usage by antibacterial levan monoclonal antibodies. J Immunol. 1992;148:3864–3872. [PubMed] [Google Scholar]
- 3.Casadevall A. Antibody immunity and invasive fungal infections. Infect Immun. 1995;63:4211–4218. doi: 10.1128/iai.63.11.4211-4218.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Casadevall A, Cleare W, Feldmesser M, Glatman-Freedman A, Goldman D L, Kozel T R, Lendvai N, Mukherjee J, Pirofski L, Rivera J, Rosas A L, Scharff M D, Valadon P, Westin K, Zhong Z. Characterization of a murine monoclonal antibody to Cryptocococcus neoformans polysaccharide that is a candidate for human therapeutic studies. Antimicrob Agents Chemother. 1998;42:1437–1446. doi: 10.1128/aac.42.6.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Casadevall A, DeShaw M, Fan M, Dromer F, Kozel T R, Pirofski L. Molecular and idiotypic analysis of antibodies to Cryptococcus neoformans glucuronoxylomannan. Infect Immun. 1994;62:3864–3872. doi: 10.1128/iai.62.9.3864-3872.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Casadevall A, Mukherjee J, Devi S J N, Schneerson R, Robbins J B, Scharff M D. Antibodies elicited by a Cryptococcus neoformans glucuronoxylomannan-tetanus toxoid conjugate vaccine have the same specificity as those elicited in infection. J Infect Dis. 1992;65:1086–1093. doi: 10.1093/infdis/165.6.1086. [DOI] [PubMed] [Google Scholar]
- 7.Casadevall A, Mukherjee J, Scharff M D. Monoclonal antibody ELISAs for cryptococcal polysaccharide. J Immunol Methods. 1992;154:27–35. doi: 10.1016/0022-1759(92)90209-c. [DOI] [PubMed] [Google Scholar]
- 8.Casadevall A, Scharff M D. The mouse antibody response to infection with Cryptococcus neoformans: VH and VL usage in polysaccharide binding antibodies. J Exp Med. 1991;174:151–160. doi: 10.1084/jem.174.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cherniak R, Morris L C, Anderson B C, Meyer S A. Facilitated isolation, purification, and analysis of glucuronoxylomannan of Cryptococcus neoformans. Infect Immun. 1991;59:59–64. doi: 10.1128/iai.59.1.59-64.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cherniak R, Sundstrom J B. Polysaccharide antigens of the capsule of Cryptococcus neoformans. Infect Immun. 1994;62:1507–1512. doi: 10.1128/iai.62.5.1507-1512.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de St. Groth S F, Scheidegger D. Production of monoclonal antibodies: strategy and tactics. J Immunol Methods. 1980;35:1–21. doi: 10.1016/0022-1759(80)90146-5. [DOI] [PubMed] [Google Scholar]
- 12.Devi S J N. Preclinical efficacy of a glucuronoxylomannan-tetanus toxoid conjugate vaccine of Cryptococcus neoformans in a murine model. Vaccine. 1996;14:841–842. doi: 10.1016/0264-410x(95)00256-z. [DOI] [PubMed] [Google Scholar]
- 13.Devi S J N, Schneerson R, Egan W, Ulrich T J, Bryla D, Robbins J B, Bennett J E. Cryptococcus neoformans serotype A glucuronoxylomannan-protein conjugate vaccines: synthesis, characterization, and immunogenicity. Infect Immun. 1991;59:3700–3707. doi: 10.1128/iai.59.10.3700-3707.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dixon D M, Casadevall A, Klein B, Travassos L R, Deepe G. Development of vaccines and their use in the prevention of fungal infections. Med Mycol. 1998;36(Suppl.):57–67. [PubMed] [Google Scholar]
- 15.Dromer F, Salamero J, Contrepois A, Carbon C, Yeni P. Production, characterization, and antibody specificity of a mouse monoclonal antibody reactive with Cryptococcus neoformans capsular polysaccharide. Infect Immun. 1987;55:742–748. doi: 10.1128/iai.55.3.742-748.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dromer F, Yeni P, Charreire J. Genetic control of the humoral response to cryptococcal capsular polysaccharide in mice. Immunogenetics. 1988;28:417–424. doi: 10.1007/BF00355373. [DOI] [PubMed] [Google Scholar]
- 17.Dubel S, Breitling F, Fuchs P, Zewe M, Gotter S, Welschof M, Moldenhauer G, Little M. Isolation of IgG antibody Fv-DNA from various mouse and rat hybridoma cell lines using the polymerase chain reaction with a simple set of primers. J Immunol Methods. 1994;175:89–95. doi: 10.1016/0022-1759(94)90334-4. [DOI] [PubMed] [Google Scholar]
- 18.Dubois M, Gilles K A, Hamilton J K, Rebers P A, Smith F. Colorimetric method for determination of sugars and related substances. Anal Chem. 1956;28:350–357. [Google Scholar]
- 19.Feeney A J, Clarke S H, Mosier D E. Specific H chain junctional diversity may be required for non-T15 antibodies to bind phosphorylcholine. J Immunol. 1988;141:1267–1272. [PubMed] [Google Scholar]
- 20.Feldmesser M, Casadevall A. Effect of serum IgG1 against murine pulmonary infection with Cryptococcus neoformans. J Immunol. 1997;158:790–799. [PubMed] [Google Scholar]
- 21.Feldmesser M, Casadevall A. Mechanism of action of antibody to capsular polysaccharide in Cryptococcus neoformans infection. Front Biosci. 1998;3:136–151. doi: 10.2741/a270. [DOI] [PubMed] [Google Scholar]
- 22.Geliebter J, Zeff R A, Melvold R W, Nathanson S G. Mitotic recombination in germ cells generated two major histocompatibility complex mutant genes shown to be identical by RNA sequence analysis: Kbm9 and Kmb6. Proc Natl Acad Sci USA. 1986;83:3371–3375. doi: 10.1073/pnas.83.10.3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goldman D, Lee S C, Casadevall A. Pathogenesis of pulmonary Cryptococcus neoformans infection in the rat. Infect Immun. 1994;62:4755–4761. doi: 10.1128/iai.62.11.4755-4761.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kozel T R, Cazin J., Jr Immune response to Cryptococcus neoformans soluble polysaccharide. Infect Immun. 1972;5:35–41. doi: 10.1128/iai.5.1.35-41.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kozel T R, deJong B C H, Grinsell M M, MacGill R S, Wall K K. Characterization of anticapsular monoclonal antibodies that regulate activation of the complement system by Cryptococcus neoformans. Infect Immun. 1998;66:1538–1546. doi: 10.1128/iai.66.4.1538-1546.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kozel T R, Gulley W F, Cazin J J. Immune response to Cryptococcus neoformans soluble polysaccharide: immunological unresponsiveness. Infect Immun. 1977;18:701–707. doi: 10.1128/iai.18.3.701-707.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Loh D Y, Bothwell A L M, White-Scharf M E, Imanishi-Kari T, Baltimore D. Molecular basis of a mouse strain-specific anti-hapten response. Cell. 1983;33:85–93. doi: 10.1016/0092-8674(83)90337-9. [DOI] [PubMed] [Google Scholar]
- 28.Mitchell T G, Perfect J R. Cryptococcosis in the era of AIDS—100 years after the discovery of Cryptococcus neoformans. Clin Microbiol Rev. 1995;8:515–548. doi: 10.1128/cmr.8.4.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mukherjee J, Casadevall A, Scharff M D. Molecular characterization of the antibody responses to Cryptococcus neoformans infection and glucuronoxylomannan-tetanus toxoid conjugate immunization. J Exp Med. 1993;177:1105–1106. doi: 10.1084/jem.177.4.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mukherjee J, Nussbaum G, Scharff M D, Casadevall A. Protective and non-protective monoclonal antibodies to Cryptococcus neoformans originating from one B-cell. J Exp Med. 1995;181:405–409. doi: 10.1084/jem.181.1.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mukherjee J, Pirofski L, Scharff M D, Casadevall A. Antibody mediated protection in mice with lethal intracerebral Cryptococcus neoformans infection. Proc Natl Acad Sci USA. 1993;90:3636–3640. doi: 10.1073/pnas.90.8.3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mukherjee J, Scharff M D, Casadevall A. Protective murine monoclonal antibodies to Cryptococcus neoformans. Infect Immun. 1992;60:4534–4541. doi: 10.1128/iai.60.11.4534-4541.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mukherjee S, Lee S, Mukherjee J, Scharff M D, Casadevall A. Monoclonal antibodies to Cryptococcus neoformans capsular polysaccharide modify the course of intravenous infection in mice. Infect Immun. 1994;62:1079–1088. doi: 10.1128/iai.62.3.1079-1088.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murphy J W, Cozad G C. Immunological unresponsiveness induced by cryptococcal polysaccharide assayed by the hemolytic plaque technique. Infect Immun. 1972;5:896–901. doi: 10.1128/iai.5.6.896-901.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Otteson E W, Welch W H, Kozel T R. Protein-polysaccharide interactions. A monoclonal antibody specific for the capsular polysaccharide of Cryptococcus neoformans. J Biol Chem. 1994;269:1858–1864. [PubMed] [Google Scholar]
- 36.Pirofski L, Casadevall A. Antibody immunity to Cryptococcus neoformans: paradigm for antibody immunity to the fungi? Zentbl Bakteriol. 1996;284:475–495. doi: 10.1016/s0934-8840(96)80001-6. [DOI] [PubMed] [Google Scholar]
- 37.Pirofski L, Lui R, DeShaw M, Kressel A B, Zhong Z. Analysis of human monoclonal antibodies elicited by vaccination with a Cryptococcus neoformans glucuronoxylomannan capsular polysaccharide vaccine. Infect Immun. 1995;63:3005–3014. doi: 10.1128/iai.63.8.3005-3014.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scott D W, Gershon R K. Determination of total and mercaptoethanol resistant antibody in the same serum sample. Clin Exp Immunol. 1970;6:313–316. [PMC free article] [PubMed] [Google Scholar]
- 39.Solin M L, Kaartinen M, Makela O. The same few V genes account for a majority of oxalozone antibodies in most mouse strains. Mol Immunol. 1992;29:1357–1362. doi: 10.1016/0161-5890(92)90172-t. [DOI] [PubMed] [Google Scholar]
- 40.Spiropulu C, Eppard R A, Otteson E, Kozel T R. Antigenic variation within serotypes of Cryptococcus neoformans detected by monoclonal antibodies specific for the capsular polysaccharide. Infect Immun. 1989;57:3240–3242. doi: 10.1128/iai.57.10.3240-3242.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stein K E, Bona C A, Lieberman R, Chien C C, Paul W E. Regulation of anti-inulin antibody response by a nonallotype-linked gene. J Exp Med. 1998;151:1089–1102. doi: 10.1084/jem.151.5.1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Todaro-Luck F, Reiss E, Cherniak R, Kaufman L. Characterization of Cryptococcus neoformans capsular glucuronoxylomannan polysaccharide with monoclonal antibodies. Infect Immun. 1989;57:3882–3887. doi: 10.1128/iai.57.12.3882-3887.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Young A C M, Valadon P, Casadevall A, Scharff M D, Sacchettini J C. The three-dimensional structures of a polysaccharide binding antibody to Cryptococcus neoformans and its complex with a peptide from a phage display library: implication for the identification of peptide mimotopes. J Mol Biol. 1997;274:622–634. doi: 10.1006/jmbi.1997.1407. [DOI] [PubMed] [Google Scholar]
- 44.Yuan R, Casadevall A, Spira G, Scharff M D. Isotype switching from IgG3 to IgG1 converts a non-protective murine antibody to C. neoformans into a protective antibody. J Immunol. 1995;154:1810–1816. [PubMed] [Google Scholar]