Abstract
Purpose
The aim was to describe the prevalence, molecular epidemiology and clinical manifestations of human bocavirus (HBoV) in patients attended at a tertiary hospital in Barcelona, Spain.
Methods
From October 2014 to May 2017, respiratory specimens from paediatric patients were collected for respiratory viruses’ laboratory-confirmation. Phylogenetic analyses from partial VP1 sequences were performed from all HBoV laboratory-confirmed specimens. Clinical features were retrospectively studied.
Results
178/10271 cases were HBoV laboratory-confirmed. The median age was 1.53 (IQR 1.0–2.3). Co-detection was highly reported (136; 76%). All viruses belonged into HBoV1 genotype but one into HBoV2. Non-reported mutations were observed and two sites were suggestive to be under negative selection. 61% (109/178) cases had lower RTI (LRTI), of whom 84 had co-detections (77%) and 76 had comorbidities (70%). LRTI was the cause of hospitalization in 85 out of 109 cases (78%), and no differences were found regarding severity factors during hospitalization between co- and single-detections, except for median length of respiratory support, which was longer in cases with co-detections.
Conclusions
Close monitoring of predominant HBoV1 showed a high similarity between viruses. The presence of comorbidities might explain the high prevalence of LRTI. Symptomatology in HBoV single-detected cases suggest that HBoV is a true pathogen.
Keywords: Human bocavirus, Surveillance, Clinical impact, Molecular characterization
Introduction
Human bocavirus (HBoV) is a causative viral agent of respiratory and gastrointestinal tract infections (RTI and GTI, respectively) [1]. Since its first identification in 2005 [2], it has been detected worldwide predominantly during winter and spring, with a prevalence as high as 6% in both RTI and GTI [1]. Children under the age of 2 years are the most susceptible cohort of patients, being its detection in adults and the elderly considered rare [1].
HBoV is a non-enveloped virus with a linear, single-stranded DNA molecule, either positive- or negative-sensed, of about 5 kb in length [1]. Four different viral genotypes (HBoV1-4) have been described to date based on the divergence of the VP1 gene, and have been grouped into species Primate bocaparvovirus 1 (HBoV1 and HBoV3) and Primate bocaparvovirus 2 (HBoV2, subdivided into A, B and C, and HBoV4), both belonging to Parvoviridae family [3, 4]. Moreover, HBoV1 genotype classification is subdivided into ST1 and ST2 lineages [5–7].
HBoV1 is the most commonly found genotype in the respiratory tract. The most usual infection-associated clinical manifestations are cough, fever, bronchiolitis and acute wheezing [8, 9]. The remaining three genotypes (HBoV2-4) are mostly detected in the gastrointestinal tract [9] and associated with GTI [8, 9]. However, this virus has been long discussed whether it is a true pathogen, due to the high rate of co-detections and long viral excretion it presents [1, 10, 11].
This study aimed to describe the prevalence, molecular epidemiology and clinical manifestations of HBoV detected in respiratory specimens from paediatric patients attended at a tertiary hospital in Barcelona (Catalonia, Spain) from the season 2014–2015 to the season 2016–2017.
Methods
Sample collection
From October 2014 (week 40/2014) to May 2017 (week 20/2017), respiratory specimens from upper or lower respiratory tract (nasopharyngeal aspirates, nasal and pharyngeal swabs, bronchoalveolar washes and tracheal aspirates) were received for the laboratory-confirmation of respiratory viruses from paediatric patients (< 18 years old) with suspicion of acute RTI attended at a tertiary hospital in Barcelona (Catalonia, Spain).
Laboratory-confirmation of respiratory viruses
Total nucleic acids were extracted using NucliSENS easyMAG (BioMérieux, France). Rapid real-time multiplex reverse transcription PCR (RT-PCR) (GeneXpert Flu or Flu/RSV XC, Cepheid, USA) or immunofluorescent (Sofia RSV FIA, Quidel, USA) assays were performed during epidemics of human respiratory syncytial virus (HRSV) and influenza A and B viruses (FLUAV and FLUBV, respectively) to detect these respiratory viruses. In case of negative result and during the rest of the year, the detection of respiratory viruses was performed either using immunofluorescence (D3 Ultra 8™ DFA Respiratory Virus Screening & Identification kit, Diagnostic HYBRIDS, USA; this assay targets FLUAV, FLUBV, HRSV, human adenovirus [HAdV], human parainfluenza virus [HPIV] 1, 2 and 3, and human metapneumovirus [HMPV]) or real-time multiplex RT-PCR assays (Anyplex™ II RV16 Detection or Allplex™ Respiratory Panel assays, Seegene, South Korea; these assays can detect FLUAV, differentiating H1, H1pdm09 and H3 subtypes, FLUBV, HRSV A and B, HAdV, human enterovirus [EV], HMPV, HPIV-1, HPIV-2, HPIV-3, and HPIV-4, HBoV, human coronaviruses [HCoV] 229E, NL63 and OC43, and human rhinovirus [RV]).
Inclusion criteria for this study included a laboratory-confirmation of HBoV and the patient had to be under 18 years of age and considered to have a respiratory episode.
Molecular characterization of HBoV
VP1 gene was partially sequenced from laboratory-confirmed HBoV samples through a nested PCR using GoTaq®G2 Green Master Mix (Promega Corporation, USA) with the primers previously described [12] and the following PCR conditions: 94 °C × 10 min—35 cycles (94 °C × 30 s—50 °C × 20 s—72 °C × 45 s)—72 °C × 10 min. Inner PCR products were purified using Exo-SAP-IT (USB, Affymetrix Inc., Cleveland, USA) and sequenced by the ABI Prism BigDye Terminator v3.1 on the ABI PRISM 3130XL sequencer (Thermo Fisher Scientific, USA). Nucleotide sequences were edited and assembled using MEGA v6.0 [13]. Afterwards, sequences were collapsed to haplotypes with the ALTER server [14]. The best fit substitution model was also determined by MEGA v6.0, and the model with the lowest Bayesian information criterion score was used to construct the phylogenetic tree, which was further evaluated with 1000 bootstrap resamplings.
Genetic HBoV1 diversity was calculated with MEGA v6.0 software. Positively and negatively selected sites were identified through Datamonkey Webserver [15] with 4 different algorithms: single likelihood ancestor counting (SLAC) [16], fixed effects likelihood (FEL) [16], random effects likelihood (REL) [16] and mixed effects model of evolution (MEME) [17]. Each site was considered under positive or negative selection only when two or more algorithms agreed with statistical significance (p < 0.1 or Bayes Factor > 50).
Clinical impact
Clinical features and demographic data were retrospectively gathered from hospital electronic records, as well as additional microbiological data. Only patients hospitalized due to HBoV lower RTI (LRTI) were included to study the disease severity, and the following factors were considered: length of hospital stay, oxygen requirements, length of oxygen requirements, paediatric intensive care unit (PICU) admission and treatments received.
Data were analysed using the SPSS Statistics v22 software (IBM Analytics, USA). Medians, interquartile range (IQR), proportions and percentages, U Mann–Whitney, Chi-square and Fisher’s exact test were performed when appropriate. Statistical significance was taken at the p < 0.05 value.
Institutional approval
This study was considered appropriate from the ethic and scientific standpoints by the Clinical Research Ethics Committee.
Results
Respiratory viruses’ detection
A total of 10,271 specimens from 6991 patients were collected (63% nasopharyngeal aspirates, 12% nasal swabs, 14% pharyngeal swabs, 8% bronchoalveolar washes and 3% tracheal aspirates), of which 6189 (60%) were laboratory-confirmed for at least one respiratory virus. HBoV was laboratory-confirmed in 239 samples (2.3%) from 212 patients (3.0%), but only 178 samples from 168 patients were finally considered as single respiratory episodes due to clinical criteria. Median age was 1.53 years old (IQR 1.0–2.3) and 60% (99/168) were male. Importantly, 61% (102/168) of patients presented underlying diseases such as pneumopathies, immunodeficiencies or prematurity.
Other respiratory viruses were also detected as follows (in descendant order): RV (1,816; 18%), HRSV (1,361; 13%), FLUAV (689; 7%), HAdV (601; 6%), FLUBV (379; 4%), EV (279; 3%), HMPV (228; 2%), HPIV-3 (183; 2%), HCoV OC43 (143; 1%), HCoV NL63 (86; 1%), HPIV-4 (70; 1%), HCoV 229E (63; 1%), HPIV-1 (41; < 1%) and HPIV-2 (24; < 1%).
Molecular epidemiology and characterization of HBoV strains
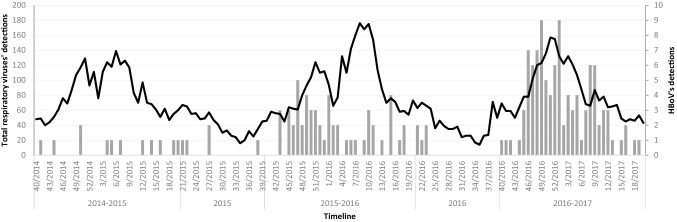
The weekly distribution of HBoV laboratory-confirmed cases showed a circulation pattern, being mostly detected during the autumn and winter months without well-defined epidemic peaks (Fig. 1). In most cases (136/178; 76%), HBoV was co-detected with one or more respiratory viruses or bacteria, most frequently RV (30%), HAdV (22%) and HRSV (14%) (Table 1). HBoV was the only pathogen detected in 42 (24%) cases.
Fig. 1.
Weekly distribution of the HBoV laboratory-confirmed samples during the period of October 2014 to May 2017. The X-axis is a timeline of the study period, separated by seasons (week 40 to week 20 of the following year) and inter-seasonal periods (week 20 to week 40). The left Y-axis marks the number of received samples from all paediatric patients attended at HUVH, which is represented by the black line. The right Y-axis marks the number of HBoV laboratory-confirmed samples, represented by the grey bars
Table 1.
HBoV detections and co-detections
| HBoV detections | Number (n, %) | |
|---|---|---|
| Single detections | 42 | 24% |
| Co-detections | 136 | 76% |
| Co-detections with 1 virus | 78 | 60.0% |
| Co-detections with 2 viruses | 45 | 33.8% |
| Co-detections with 3 viruses | 6 | 5.4% |
| Co-detections with 4 viruses | 1 | 0.8% |
| Co-detections with bacteria | 6 | |
| Number of co-detections per target | ||
| Rhinovirus | 57 | 30% |
| Human adenovirus | 42 | 22% |
| Human respiratory syncytial virus | 27 | 14% |
| Human enterovirus | 26 | 14% |
| Human metapneumovirus | 14 | 7% |
| Human parainfluenza 4 | 5 | 3% |
| Influenza A virus | 5 | 2% |
| Human coronavirus NL63 | 4 | 2% |
| Human parainfluenza 3 | 3 | 2% |
| Human coronavirus OC43 | 3 | 2% |
| Human parainfluenza 2 | 2 | 1% |
| Human coronavirus 229E | 1 | < 1% |
| Human parainfluenza 1 | 1 | < 1% |
| Influenza B virus | 0 | 0% |
The total number of episodes (178) has been considered instead of the number of patients, as some patients showed different episodes. The different viruses found co-detecting with HBoV are displayed above
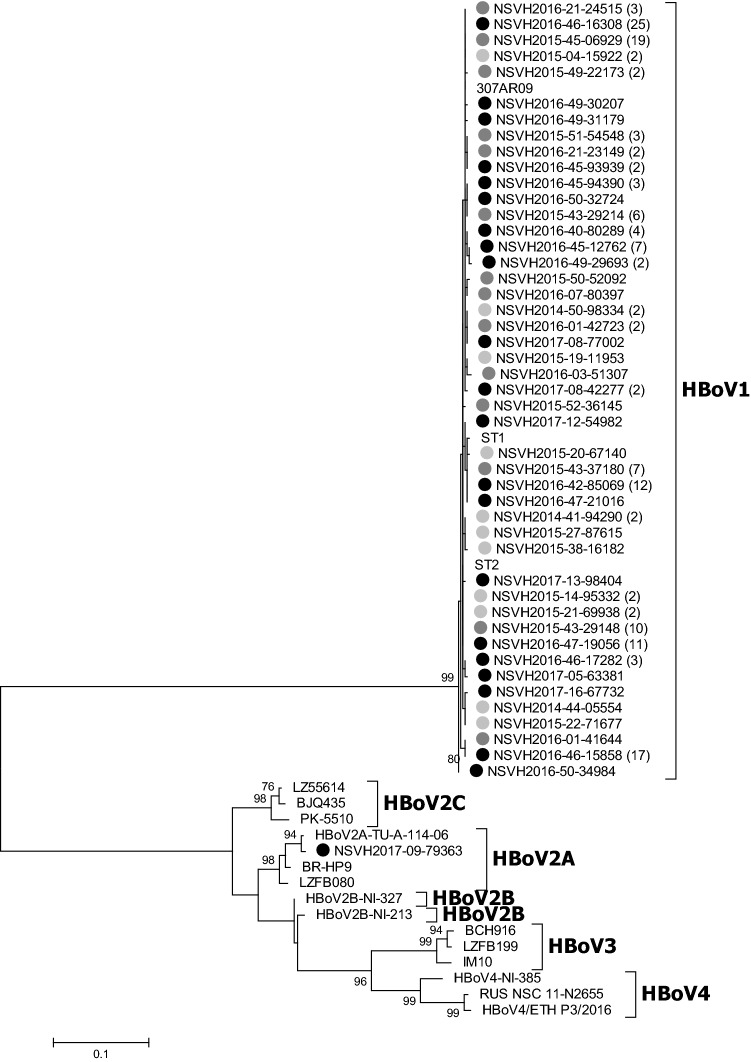
The evolutionary history was inferred with the maximum likelihood method based on Hasegawa-Kishino-Yano model. Phylogenetic analysis of 174 (98%) partial VP1 sequences revealed that all strains belonged to HBoV1 genotype, except one HBoV2A (Fig. 2). HBoV1 viruses could not be distinguished between ST1 and ST2 lineages due to low bootstrap values. None HBoV3 and HBoV4 viruses were detected. Four (2%) HBoV viruses could not be characterized due to low viral load.
Fig. 2.
Phylogenetic reconstruction of partial VP1 nucleotide sequences of detected HBoV. Phylogenetic tree of partial VP1 gene sequences from 174 HBoV viruses together with reference sequences from previous epidemiological studies available at GenBank. The analyses were performed on sequences from nucleotide position 3271 to 3817 relative to ST1 strain (accession number DQ000495). Only bootstrap values over 70% are shown. Numbers in brackets at the end of the name of the sequences indicate the number of haplotypes which are represented. Sequences marked with a black solid circle correspond to the 2016–2017 season; sequences with a dark grey solid circle correspond to the 2015–2016 season and 2016 inter-seasonal period; sequences with a light grey solid circle correspond to the 2014–2015 season and 2015 inter-seasonal period
The estimated overall nucleotide and amino acid similarities between HBoV1 strains were 99.4% and 99.8%, respectively. Nucleotide sequence comparison relative to reference ST1 sequence (accession number DQ000495), revealed up to 21 nucleotide substitutions, four of which were non-synonymous (A115T, G126E, T149A and R251K). No positive selection pressure sites were found, and codons 147 and 196 were suggestive to be under negative selection (Table 2).
Table 2.
Codons under negative selection
| Amino acid change | Nucleotide mutation | HBoV1 presenting the mutation (n, %) | SLAC | FEL | REL | Consensus | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| dN-dS | p value | dN-dS | p value | dN-dS | Bayes Factor | ||||||
| A145A | A435G | 1 (0.6%) | − 11.328 | 0.333 | − 198.922 | 0.062 | − 3.215 | 0.000 | – | ||
| Q147Q | G441A | 59 (34.1%) | − 82.751 | 0.004 | − 640.969 | 0.003 | − 20.716 | 0.000 | – | – | |
| S150S | A450T | 4 (2.3%) | − 11.328 | 0.343 | − 208.514 | 0.093 | − 3.380 | 0.000 | – | ||
| G159G | A477G | 12 (6.9%) | − 22.655 | 0.112 | − 429.518 | 0.004 | − 16.433 | 0.000 | – | ||
| V184V | T552C | 18 (10.4%) | − 11.328 | 0.333 | − 103.078 | 0.099 | − 1.275 | 0.000 | – | ||
| Q196Q | G588A | 26 (15.0%) | − 46.951 | 0.046 | − 266.130 | 0.042 | − 19.535 | 0.000 | – | – | |
| T220T | A660C | 1 (0.6%) | − 11.328 | 0.333 | − 218.146 | 0.073 | − 3.532 | 0.000 | – | ||
| K258K | G774A | 16 (9.2%) | − 23.484 | 0.106 | − 126.544 | 0.067 | − 3.163 | 0.000 | – | ||
Codon positions have been calculated using the ST1 virus originally discovered in Sweden (with accession number DQ000495). All codons with a significant value are represented
SLAC Single-Likelihood Ancestor Counting, FEL Fixed Effects Likelihood, REL Random Effects Likelihood
Sequences of the 50 different haplotypes were submitted to GeneBank Database with accession numbers MK211507-MK211556.
Clinical features
The most common HBoV-related syndromes observed were LRTI (109, 61%), followed by upper RTI (URTI) (69, 39%). There were no significant differences in the clinical features between co- and single-detections (Table 3).
Table 3.
Demographic and clinical features of patients
| Factor | Cases with co-detections | Cases with only HBoV | p value |
|---|---|---|---|
| Age | 1.5 (IQR 1–2.4) | 1.6 (IQR 1.2–2.3) | 0.4 |
| Sex | M 77; F 52 | M 22; F 17 | 0.7 |
| No co-morbidities | 51 | 15 | 1 |
| LRTI/URTI | 84/52 | 25/17 | 0.6 |
| Fever | 93 | 27 | 0.7 |
Cases are differentiated whether there were co-detections found or not along the HBoV laboratory-confirmation, and the statistical significance was calculated for each item. Age and clinical syndromes have been calculated from the 178 clinical episodes, while sex and co-morbidities have been calculated from the 168 patients
IQR Interquartile range, M Male, F Female, LRTI lower respiratory tract infection, URTI upper respiratory tract infection
The 78% (85) of the 109 LRTI cases required hospital admission, of which 21 (25%) were single-HBoV infection, while 64 (75%) presented co-detections with other pathogens. While no differences were found regarding severity factors during hospitalization (Table 4), the length of respiratory support was longer (p 0.012) in the patient group with co-detection (median length 3 days) in comparison to the patient group without co-detections (median length 1 day). The only patient admitted to PICU was previously healthy and did not require non-invasive mechanical ventilation.
Table 4.
Comparison between severity factors of patients with a co-detection and single HBoV infection
| Severity factor | Cases with co-detections (n = 64) | Cases with single HBoV detection (n = 21) | p value |
|---|---|---|---|
| Co-morbidities | 48 cases (75%) | 13 cases (62%) | 0.25 |
| Median length of stay | 4 days (IQR 2–6) | 3 days (IQR 2–4) | 0.11 |
| Respiratory support requirement | 53 cases (83%) | 19 cases (91%) | 0.4 |
| Nasal oxygen cannula | 39 (74%) | 17* (90%) | |
| High-flow nasal cannula oxygen | 8 (14%) | 1 (5%) | |
| Non-invasive mechanical ventilation | 4 (7%) | 1 (5%) | |
| Invasive mechanical ventilation | 2 (4%)† | 0 | |
| Median length of respiratory support | 3 days (IQR 2–5) | 1 day (IQR 1–3) | 0.012 |
| PICU admission | 6 cases | 1 case | – |
| Median length of stay at PICU | 5 days (IQR 3–17) | 4 days | 0.6 |
| Treatment with bronchodilators | 56 cases (87%) | 18 cases (86%) | 0.8 |
| Antibiotic treatment | 29 cases (45%) | 9 cases (43%) | 0.6 |
Only those patients manifesting a LRTI (85) have been taken into consideration for this comparison. Cases are differentiated whether there were co-detections found or not along the HBoV laboratory-confirmation, and the statistical significance was calculated for each item
IQR interquartile range, PICU paediatric intensive care unit
*One had a previous pneumopathy, requiring conventional oxygen at home
†Two episodes of the same patient, a child with a previous pneumopathy and receiving mechanical ventilation at home through tracheotomy
The only HBoV2 strain was co-detected with HCoV-NL63 in a haematological cell transplant recipient with URTI and no gastrointestinal symptoms.
Discussion
This study presents data on prevalence, genetic diversity and clinical features of respiratory HBoV at a tertiary hospital in Barcelona, Spain, during the 2014–2017 seasons.
The overall prevalence of HBoV in this study was similar to global estimations (1.4–4.45%) reported previously [1, 18–23], with a prevalence similar to that shown by EV or HMPV, and higher than other typical respiratory viruses such as HPIV and seasonal HCoV. This prevalence, though, might be underestimated for two reasons. First, since the 2015–2016 season, the hospital implemented more molecular methods in detriment of antigen-based assays to improve the sensitivity and range of respiratory viruses detected. HBoV target was not detected with the antigen-based detection assays used before, hence, an understimation of HBoV prevalence in the first season and the first half of the second one is expected. Second, during HRSV and influenza epidemics, a great number of samples are only tested through specific HRSV or influenza rapid detection assays, and in case of positive result, no additional tests are performed according to the laboratory protocols.
Specially during the second half of the study, when molecular methods for respiratory viruses detection were more widely used, the weekly distribution of detected cases showed a pattern of seasonality, in particular during the winter and autumn months. This seasonality coincides with that of other co-detected respiratory viruses [24], some of them more related to severe respiratory disease such as HRSV and influenza viruses. Hence, this virus has been suggested to be an opportunistic entity [25], whose infection only occurs when other conditions lower immunological defences, such as the prior infection by other respiratory viruses. However, HBoV has been reported to cause the illness on its own. Moreover, in the present study no differences in the seasonality pattern were found between single HBoV detections and co-detections with other respiratory viruses (data not shown), indicating an own seasonality and thus, a possible pathogenicity independent of other agents.
A large number of HBoV samples were reported to be in co-detection with other respiratory viruses. However, those with influenza or RSV might have been missed by the use of specific point-of-care tests for rapid detection of these viruses without further testing, probably being underestimated. Long viral excretion of HBoV has been previously reported, remaining detectable in serial respiratory samples from immune-competent patients up to 6 months after infection [1], which might explain such a high prevalence of co-detections. Interestingly, HBoV infection has been suggested to be triggered in its severity by other viruses, modifying its replication strategy to adapt itself to these co-detecting viruses [26] causing a symptomatology that could have remained undetectable otherwise. Contrarily, another study demonstrated that it can act as a helper virus for other viral infections [27]. Whatsoever, both of these findings justify the fact that it is commonly found co-detecting with other viruses.
Since the present study is only based on the detection of HBoV in respiratory tract, the fact that almost all viruses belonged to HBoV1 genotype was expected. The only HBoV2A strain was a case with no gastrointestinal manifestation, though respiratory symptoms of this case might be related to the co-detection with HCoV NL63.
In previous studies [5–7], two genetic lineages (ST1 and ST2) were described within HBoV1 genotype. However, in the present study, the VP1 sequences together with reference sequences showed a high genetic similarity and did not cluster in different genetic groups. In addition, in the first report describing ST1 and ST2 lineages [2] up to 18 nucleotide substitutions distinguishing between them were found, of which three were non-synonymous. In the present study, one of these amino acid changes specific of ST2-like strains was found in all HBoV1 sequences, together with other three amino acid substitutions, which is not enough genetic divergence to distinguish into two different genetic clades. These data suggests that ST1 and ST2 may not be genuine lineages within HBoV1 type, but viral variants belonging to a single clade. Nevertheless, other regions were studied in these reports [5–7], using other parts of the VP1 gene, the whole length of the gene or the whole genome. Hence, whether ST1 and ST2 are real lineages or viral variants belonging to a single clade should be further studied by whole genome sequencing.
No positive selection sites were found in the partial VP1 sequences obtained in the present study, which is an unexpected feature for an external capsid protein. Since the host immune system recognised some of the antigenic epitopes, this region is theoretically under continuous selective pressure and more susceptible to acquire changes on antigenic epitopes to evade the host immune response. In fact, a study calculated that almost 50% of the residues of this protein had antigenic properties [28]. The lack of positive selection sites might be due to the short amino acid sequences studied here (182 out of the 672 amino acids of the whole protein), and the short period of study. However, two amino acid positions were negatively selected, which means they might accomplish an important function within the protein either for maintaining the structure or for a hypothetical role in pathogenesis. The fact that they are negatively selected could be interesting for the selection of a target in the development of synthetic monoclonal antibodies, for either prophylaxis or treatment. Further studies should be done to situate both codons on the 3D structure of the protein to test the viability of their use as monoclonal antibodies’ target, as its structure is not available on the PDB database [29].
Clinical severity factors were also studied from the cohort of patients requiring hospitalization due to LRTI. The evidence that almost all severity factors were similar between cases with and without co-detections suggests that HBoV is a true pathogen capable of causing a respiratory disease on its own. This study showed a high number of cases with disease directly attributed to HBoV1 on its own (single detection), though not as severe as the disease caused by HBoV1 together with other viruses. Since the first detection of HBoV, many studies have raised the question of whether this virus can really establish a respiratory or gastrointestinal infection in humans. The high rate of HBoV co-detections with other viral respiratory pathogens and the high presence of HBoV in asymptomatic patients make difficult to draw a valid conclusion [1, 10, 11, 30], though both facts might also be explained by its long viral shedding period. This study presents the clinical impact that HBoV has by itself, without other pathogens in co-detection, suggesting that not only HBoV is a real pathogen, but it is also a concerning virus that needs to be under surveillance. Though causing a more benign disease than other respiratory viruses, since it is evidenced that this virus can cause respiratory disease, HBoV infection should be monitored as it is already done with other respiratory viruses such as HRSV, FLUAV and FLUBV. Neglected respiratory viral infections have been proved to be important to be included in viral surveillance, such as EV [31] or HMPV [32], as new viral variants can emerge, with changes in the virus tropism or in the clinical impact.
Nevertheless, this study presented some limitations. First, the active infection of HBoV was not assessed, as no viral isolation by cell culture was performed, though this might not be the best approximation due to the difficulty of isolation of HBoV). In this study, those cases presenting with LRTI and having HBoV as its only laboratory-confirmation were assumed to have an active infection by HBoV, whereas those presenting co-detections could not be assumed to have an active infection, especially since HBoV has a long period of viral shedding. Thus, comparing the different severity factors of LRTI such as length of hospital stay, oxygen requirements, length of oxygen requirements, and paediatric intensive care unit admission, might bring to light the real pathogenicity of HBoV. Nevertheless, other pathogens undetected might be participating in the clinical impact that these patients are presenting, as no metagenomics have been performed and we have only relied on the diagnostic assays we have in our hospital, but this should not happen in a significant number of patients. Hence, though it is a limitation, it does not compromise the veracity of the study. Other limitation is that nearly three quarters of the population presented co-morbidities. This is understandable, as the present study was conducted in one of the largest hospital complexes in Spain, with more than 300,000 attended patients per year only in the Maternity and Children’s Hospital. Therefore, severity of HBoV might be overestimated in this study.
Conclusions
HBoV has been proved in the present study to be considered as a true pathogen due to its capacity of triggering a respiratory symptomatology when infecting on its own, particularly during the cold months. Though having a high prevalence in the paediatric population, the disease caused by this virus was mostly clinically mild-moderate, but severe in a few cases, where antiviral treatments are not available. Further studies are needed to contribute with more clinical and molecular features, not only in hospital population but also in the community, to reveal the exact HBoV’s role on the human pathogenesis.
Acknowledgements
This study was supported by the Spanish Ministry of Economy and Competitiveness, Instituto de Salud Carlos III, co-financed by the European Development Regional Fund (ERDF) "A way to achieve Europe", Spanish Network for Research in Infectious Diseases (REIPI RD16/0016/0003), and by CIBER -Consorcio Centro de Investigación Biomédica en Red- (CB 2021), Instituto de Salud Carlos III, Ministerio de Ciencia e Innovación and Unión Europea – NextGenerationEU.
Author contributions
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by Maria Piñana, Jorgina Vila, Cristina Andrés, Jordi Saura, Alejandra González-Sánchez, and Andrés Antón. The first draft of the manuscript was written by Maria Piñana, Jorgina Vila, Cristina Andrés, Jordi Saura, Alejandra González-Sánchez, and Andrés Antón and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Contributor Information
Tomàs Pumarola, Email: virusrespiratoris@vallhebron.cat.
Andrés Antón, Email: andres.anton@vallhebron.cat.
References
- 1.Guido M, Tumolo MR, Verri T, Romano A, Serio F, De Giorgi M, et al. Human bocavirus: current knowledge and future challenges. World J Gastroenterol. 2016;22:8684. doi: 10.3748/wjg.v22.i39.8684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allander T, Tammi MT, Eriksson M, Bjerkner A, Tiveljung-Lindell A, Andersson B. Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proc Natl Acad Sci USA. 2005;102:12891–12896. doi: 10.1073/pnas.0504666102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cotmore SF, Agbandje-McKenna M, Chiorini JA, Gatherer D, Mukha D V, Pintel DJ, et al. Rationalization and extension of the taxonomy of the family Parvoviridae. ICTV. 2013. 1–18.
- 4.Kapoor A, Slikas E, Simmonds P, Chieochansin T, Shaukat S, Alam MM, et al. A new bocavirus species in human stool. J Infect Dis. 2009;199:196–200. doi: 10.1086/595831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pogka V, Moutousi A, Kossyvakis A, Kalliaropoulos A, Sgouras DN, Giannaki M, et al. Genetic variability of human metapneumo- and bocaviruses in children with respiratory tract infections. Influenza Other Respi Viruses. 2014;8:107–115. doi: 10.1111/irv.12185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kenmoe S, Vernet M-A, Njankouo-Ripa M, Penlap VB, Vabret A, Njouom R. Phylogenic analysis of human bocavirus detected in children with acute respiratory infection in Yaounde, Cameroon. BMC Res Notes. BioMed Central. 2017;10:293. doi: 10.1186/s13104-017-2620-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moesker FM, van Kampen JJA, van der Eijk AA, van Rossum AMC, de Hoog M, Schutten M, et al. Human bocavirus infection as a cause of severe acute respiratory tract infection in children. Clin Microbiol Infect. Elsevier Ltd. 2015;21:964.e1–8. doi: 10.1016/j.cmi.2015.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mietzsch M, Kailasan S, Garrison J, Ilyas M, Chipman P, Kantola K, et al. Structural insights into human bocaparvoviruses. J Virol. 2017;91. [DOI] [PMC free article] [PubMed]
- 9.Qiu J, Söderlund-Venermo M, Young NS. Human parvoviruses. Clin Microbiol Rev. 2017;30:43–113. doi: 10.1128/CMR.00040-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jartti T, Hedman K, Jartti L, Ruuskanen O, Allander T, Söderlund-Venermo M. Human bocavirus—the first 5 years. Rev Med Virol. 2012;22:46–64. doi: 10.1002/rmv.720. [DOI] [PubMed] [Google Scholar]
- 11.Martin ET, Fairchok MP, Kuypers J, Magaret A, Zerr DM, Wald A, et al. Frequent and prolonged shedding of bocavirus in young children attending daycare. J Infect Dis. 2010;201:1625–1632. doi: 10.1086/652405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kapoor A, Simmonds P, Slikas B, Li L, Bodhidatta L, Sethabutr O, et al. Humn bocaviruses are highly diverse, dispersed, recombination prone, and prevalent enteric infections. J Infect Dis. 2010;201:1633–1643. doi: 10.1086/652416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glez-Peña D, Gómez-Blanco D, Reboiro-Jato M, Fdez-Riverola F, Posada D. ALTER: Program-oriented conversion of DNA and protein alignments. Nucleic Acids Res. 2010;38:14–18. doi: 10.1093/nar/gkq321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delport W, Poon AFY, Frost SDW, Kosakovsky Pond SL. Datamonkey 2010: a suite of phylogenetic analysis tools for evolutionary biology. Bioinformatics. 2010;26:2455–2457. doi: 10.1093/bioinformatics/btq429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kosakovsky Pond SL, Frost SDWW. Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol Biol Evol. 2005;22:1208–1222. doi: 10.1093/molbev/msi105. [DOI] [PubMed] [Google Scholar]
- 17.Murrell B, Wertheim JO, Moola S, Weighill T, Scheffler K, Kosakovsky Pond SL. Detecting individual sites subject to episodic diversifying selection. Malik HS, editor. PLoS Genet. 2012;8:e1002764. doi: 10.1371/journal.pgen.1002764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dina J, Vabret A, Gouarin S, Petitjean J, Lecoq J, Brouard J, et al. Detection of human bocavirus in hospitalised children. J Paediatr Child Health. 2009;45:149–153. doi: 10.1111/j.1440-1754.2008.01442.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bastien N, Brandt K, Dust K, Ward D, Li Y. Human bocavirus infection. Canada Emerg Infect Dis. 2006;12:848–850. doi: 10.3201/eid1205.051424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shen J, Zhu Q, Zeng M, Yu H. Detection and genome analysis of human bocavirus 1–4 from hospitalized children with acute lower respiratory tract infection and symptoms of wheezing in Shanghai. Int J Mol Med. 2013;32:1415–1420. doi: 10.3892/ijmm.2013.1512. [DOI] [PubMed] [Google Scholar]
- 21.Kesebir D, Vazquez M, Weibel C, Shapiro ED, Ferguson D, Landry ML, et al. Human bocavirus infection in young children in the United States: molecular epidemiological profile and clinical characteristics of a newly emerging respiratory virus. J Infect Dis. 2006;194:1276–1282. doi: 10.1086/508213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moreno B, Abrego L, Carrera J-P, Franco D, Gaitán M, Castillo J, et al. Detection of human bocavirus type 1 infection in Panamanian children with respiratory illness. J Med Virol. 2016;88:389–394. doi: 10.1002/jmv.24346. [DOI] [PubMed] [Google Scholar]
- 23.Arnott A, Vong S, Rith S, Naughtin M, Ly S, Guillard B, et al. Human bocavirus amongst an all-ages population hospitalised with acute lower respiratory infections in Cambodia. Influenza Other Respi Viruses. 2013;7:201–210. doi: 10.1111/j.1750-2659.2012.00369.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Antón A, Marcos M, Torner N, Isanta R, Camps M, Martínez A, et al. Virological surveillance of influenza and other respiratory viruses during six consecutive seasons from 2006 to 2012 in Catalonia. Spain Clin Microbiol Infect Elsevier Ltd. 2016;22:1–9. doi: 10.1016/j.cmi.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schildgen O, Müller A, Allander T, Mackay IM, Völz S, Kupfer B, et al. Human bocavirus: passenger or pathogen in acute respiratory tract infections? Clin Microbiol Rev. 2008;21:291–304. doi: 10.1128/CMR.00030-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Streiter M, Malecki M, Prokop A, Schildgen V, Lüsebrink J, Guggemos A, et al. Does human bocavirus infection depend on helper viruses? A challenging case report. Virol J. BioMed Central Ltd. 2011;8:417. doi: 10.1186/1743-422X-8-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Z, Deng X, Zou W, Engelhardt JF, Yan Z, Qiu J. Human bocavirus 1 is a novel helper for adeno-associated virus replication. J Virol. 2017;91:1–18. doi: 10.1128/JVI.00710-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kalyanaraman N. In silico prediction of potential vaccine candidates on capsid protein of human bocavirus 1. Mol Immunol Elsevier. 2017;93:193–205. doi: 10.1016/j.molimm.2017.11.024. [DOI] [PubMed] [Google Scholar]
- 29.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Williams JV. Déjà Vu All Over Again: Koch’s Postulates and Virology in the 21st Century. J Infect Dis. 2010;201:1611–1614. doi: 10.1086/652406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Andrés C, Vila J, Creus-Costa A, Piñana M, González-Sánchez A, Esperalba J, et al. Enterovirus D68 in Hospitalized Children, Barcelona, Spain, 2014–2021. Emerg Infect Dis [Internet]. 2022;28:1327–31. https://wwwnc.cdc.gov/eid/article/28/7/22-0264_article.htm [DOI] [PMC free article] [PubMed]
- 32.Piñana M, Vila J, Maldonado C, Galano-Frutos JJ, Valls M, Sancho J, et al. Insights into immune evasion of human metapneumovirus: novel 180- and 111-nucleotide duplications within viral G gene throughout 2014–2017 seasons in Barcelona. Spain J Clin Virol. 2020;132:104590. doi: 10.1016/j.jcv.2020.104590. [DOI] [PMC free article] [PubMed] [Google Scholar]