Abstract
Purpose
To explore a multidomain fusion model of radiomics and deep learning features based on 18F-fluorodeoxyglucose positron emission tomography/computed tomography (18F-FDG PET/CT) images to distinguish pancreatic ductal adenocarcinoma (PDAC) and autoimmune pancreatitis (AIP), which could effectively improve the accuracy of diseases diagnosis.
Materials and methods
This retrospective study included 48 patients with AIP (mean age, 65 ± 12.0 years; range, 37–90 years) and 64 patients with PDAC patients (mean age, 66 ± 11.3 years; range, 32–88 years). Three different methods were discussed to identify PDAC and AIP based on 18F-FDG PET/CT images, including the radiomics model (RAD_model), the deep learning model (DL_model), and the multidomain fusion model (MF_model). We also compared the classification results of PET/CT, PET, and CT images in these three models. In addition, we explored the attributes of deep learning abstract features by analyzing the correlation between radiomics and deep learning features. Five-fold cross-validation was used to calculate receiver operating characteristic (ROC), area under the roc curve (AUC), accuracy (Acc), sensitivity (Sen), and specificity (Spe) to quantitatively evaluate the performance of different classification models.
Results
The experimental results showed that the multidomain fusion model had the best comprehensive performance compared with radiomics and deep learning models, and the AUC, accuracy, sensitivity, specificity were 96.4% (95% CI 95.4–97.3%), 90.1% (95% CI 88.7–91.5%), 87.5% (95% CI 84.3–90.6%), and 93.0% (95% CI 90.3–95.6%), respectively. And our study proved that the multimodal features of PET/CT were superior to using either PET or CT features alone. First-order features of radiomics provided valuable complementary information for the deep learning model.
Conclusion
The preliminary results of this paper demonstrated that our proposed multidomain fusion model fully exploits the value of radiomics and deep learning features based on 18F-FDG PET/CT images, which provided competitive accuracy for the discrimination of PDAC and AIP.
Keywords: Positron emission tomography/computerized tomography (PET/CT); Carcinoma, pancreatic ductal; Autoimmune pancreatitis; Deep learning; Radiomics
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is a common digestive system malignancy, accounting for about 90% of pancreatic cancers [1], and the five-year relative survival rate of PDAC is only 10.8% [2]. The statistical data in 2021 [3] showed that pancreatic cancer ranked fourth in the mortality rate of malignant tumors; the research [4] estimated that pancreatic cancer may become the second leading cause of death among malignant tumors in 2030. Autoimmune pancreatitis (AIP) is a unique form of chronic pancreatitis, and the typical characteristics include sausage-like swelling of the focal and irregular stenosis of the main pancreatic duct [5, 6]. PDAC is cured by invasive surgical resection [7], and patients with AIP are treated with corticosteroids or rituximab [8]. However, the clinical symptoms and imaging manifestations of PDAC and AIP are very similar [9]. A systematic review showed that nearly one third (29.7%, 95% CI 18.1%–42.8%) of AIP patients undergo unnecessary pancreatectomy due to suspected malignancy in China [10]. PDAC patients will also be misdiagnosed as AIP which delays the best opportunity for surgical treatment. Therefore, the crucial issue is how to avoid the error diagnosis of these two diseases as much as possible and distinguish them accurately and noninvasively.
The clinic's common PDAC and AIP diagnostic methods are divided into three categories: serum examination, imaging examination, and histopathological examination. The detection and analysis method of serum markers [11, 12] have not yet formed standard laboratory parameters for distinguishing these two types of diseases, and the correlation between markers and diseases is still controversial [13, 14]. For invasive histological examination [15], this diagnostic method of pancreatic lesions has limitations [16, 17] when insufficient histological samples are available. In imaging examination, the studies of computer tomography (CT), magnetic resonance imaging (MRI), and contrast-enhanced ultrasound (CEUS) provided valuable distinction information for the identification of PDAC and AIP [18–21]. Compared to traditional imaging methods, 18F-fluorodeoxyglucose positron emission tomography/computer tomography (18F-FDG PET/CT) imaging can not only display anatomical information such as morphology and density of lesions but also provides functional information like metabolism and blood flow of lesions. We expected that multimodal features based on PET/CT images combining the advantages of PET and CT images could further improve the accuracy of PDAC and AIP classification.
Radiomics can obtain statistical features from clinical medical images through high-throughput computing and convert medical digital images into quantitative high dimension data that can be mined, thereby revealing subtle traces of the disease. Driven by precision medicine in recent years, many analysis methods based on radiomics have been applied to some clinical decision-making tasks, including diagnosing coronavirus, lung cancer, and breast cancer [22–24]. Radiomics is the most commonly used modeling method in PDAC and AIP identification [25–27]. However, the radiomics method relies on the accurate description of the lesion area. The different details of the edge will affect the prediction results, so we attempted to introduce the deep learning method to make up for this limitation. Deep Convolutional Neural Networks (CNN) is a method of learning deep nonlinear mapping of data through layer-by-layer training, and it can obtain digital features from images to reveal the complex abstract information contained in massive data. Deep learning algorithms are widely used in image segmentation, classification, and recognition, and have made outstanding achievements in various fields of medicine [28–31], and have become an indispensable tool for developing clinical medical research. Previous studies based on medical imaging have shown that deep learning could provide new research ideas and breakthrough diagnostic information for identifying PDAC and AIP [32–34].
In recent years, combining radiomics and deep learning methods has become a research hotspot in clinical medicine. This hybrid approach has been used to classify gastrointestinal stromal tumors or cervical lymph nodes [35, 36], and to predict IDH status in gliomas [37], but most of the previous papers discussed feature fusion based on the single-modality features of CT or MRI. Therefore, we intend to propose a multidomain fusion model based on PET/CT images, which draws on the complementary advantages of multidomain information formed by deep learning and radiomics, and the model can effectively improve the diagnosis accuracy of DPAC and AIP diseases. This method is expected to reduce the misjudgment of the disease, strive for the treatment opportunity for PDAC patients, and reduce unnecessary invasive surgical operations for AIP patients.
Materials and methods
Dataset
The initial database search identified 159 patients, and those with absence of PET or CT, incomplete clinical information records, or the history of surgery were excluded. The remaining 112 patients were included in this retrospective experiment, and the screening process is shown in Fig. 1. 64 patients with PDAC and 48 patients with AIP underwent 18F-FDG PET/CT examinations at the hospital from February 2011 to June 2019. All AIP patients were diagnosed according to the established criteria of the 14th International Pancreatology Conference: 25 cases were confirmed by histological or cytological examination, and 23 were confirmed by noninvasive means such as medical imaging, serological indicators, and medical history. All PDAC patients were diagnosed by histology or cytology. Detailed clinical statistics of 112 patients are shown in Table 1.
Fig. 1.
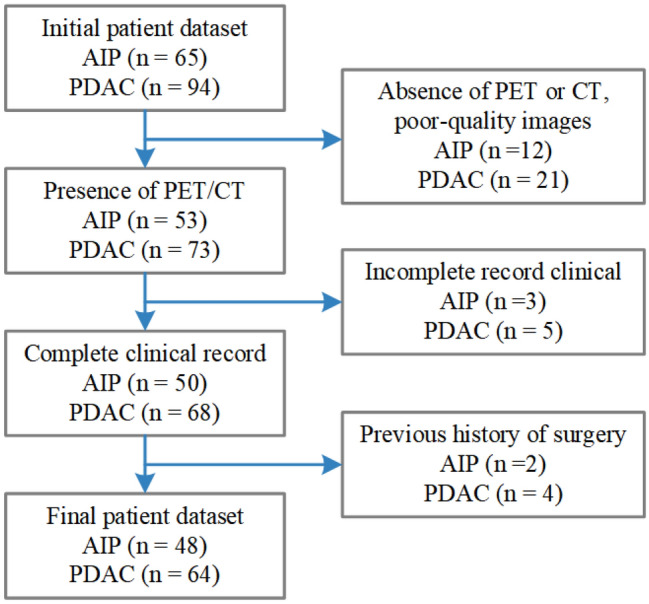
Study flowchart for selection criteria
Table 1.
Detailed clinical statistics of patients with PDAC and AIP
| Variable | PDAC (n = 64) | AIP (n = 48) |
|---|---|---|
| Patients’ sex (male/female) | 35/29 | 43/5 |
| Patients’ age (min/max/mean ± SD) | 32/88/66 ± 11.3 | 37/90/65 ± 12.0 |
| Uptake status (focal/diffuse) | 36/28 | 19/29 |
| SUV (min/max/median) | 2.71/24.62/9.41 | 1.34/6.11/3.58 |
AIP autoimmune pancreatitis, PDAC pancreatic ductal adenocarcinoma, SUV standardized uptake values
Image acquisition and processing
18F-FDG PET/CT images of all patients were collected on a Siemens Biograph64 PET/CT scanner. Before PET/CT scanning, patients should fast for at least 6 h, and 18F-FDG (3.70–5.55 MBq/kg) was intravenously injected when blood glucose < 11.1 mmol/L. PET/CT imaging was performed after sufferers rested quietly in the lounge for about 60 min. The body topogram was acquired using an electric current of 30 mA at a voltage of 120 kV. Next, whole-body CT scans were performed with a scan time of 18.67–21.93 s. Then whole-body PET scans were performed covering 5–6 bed positions with a total acquisition time of 10.0–15.0 min. The TrueX iterative algorithm was used to reconstruct the PET images and CT values for attenuation correction. The intra-layer spatial resolutions of the PET and CT images are 4.07 mm and 0.98 mm, and the sizes are 168 × 168 pixels and 512 × 512 pixels. The scanning parameters included tube voltage 120 kV, tube current 170 mA, layer thickness 3 mm.
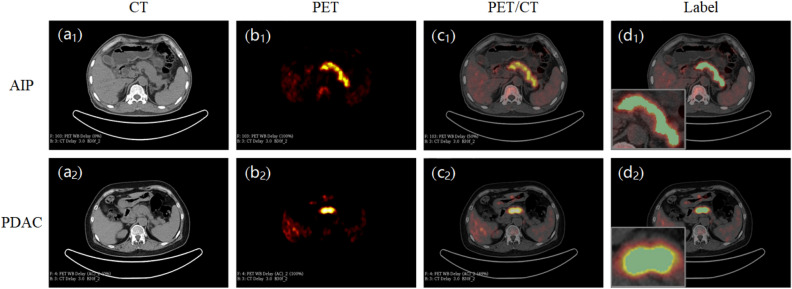
The 3D slicer software manually segments the pancreatic lesion area and obtains the region of interest (ROI) from PET/CT images (Fig. 2). To reduce patient variability, the ROI was delineated by a nuclear medicine physician and rechecked by another nuclear doctor. During the review process, if there were controversial revision opinions, a third senior nuclear medicine scientist would be invited to participate in the discussion and confirm the output. All physicians involved in ROI confirmation work have more than 10 years of experience diagnosing pancreatic diseases.
Fig. 2.
Manually delineated ROI of the pancreatic lesions on the 3D slicer software
To ensure a balanced distribution of experimental sample data, we selected slice sequences containing lesions in the 3D image of each patient to generate a new PET/CT dataset (AIP: 612, PDAC: 577). Our preprocessing pipeline for sample images is shown in Fig. 3. First, CT and PET images’ pixel values were converted into Hounsfield unit (HU) and standardized uptake value (SUV). Second, we set the pixel value threshold range of CT images (− 10 ≤ HU ≤ 100) to reduce the interference of fat, bone tissue, and other factors on texture features. Then PET images were resampled using bilinear interpolation to keep the spatial resolution consistent with CT images. Finally, we took the centroid of the ROI as the midpoint and cropped the CT, PET, and lesion label images into 64 × 64 image patches as the input of the classification model; this way not only reduces the over-reliance on the ROI but also preserves the surrounding relevant details.
Fig. 3.
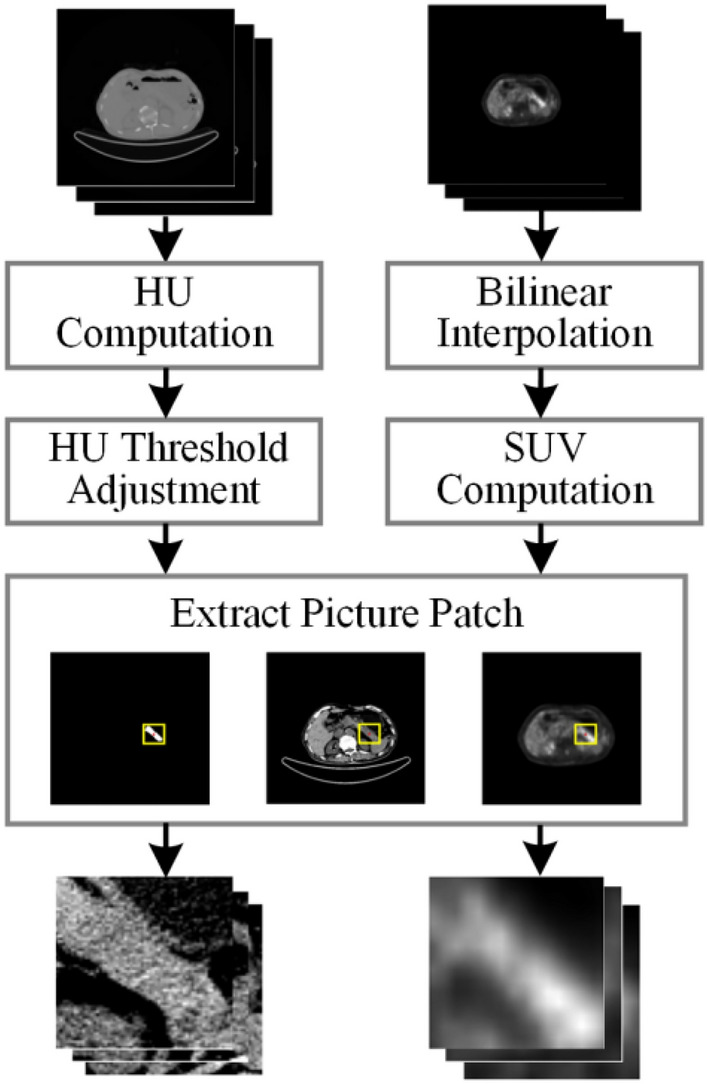
PET and CT images are preprocessing workflow
Model architecture and implementation
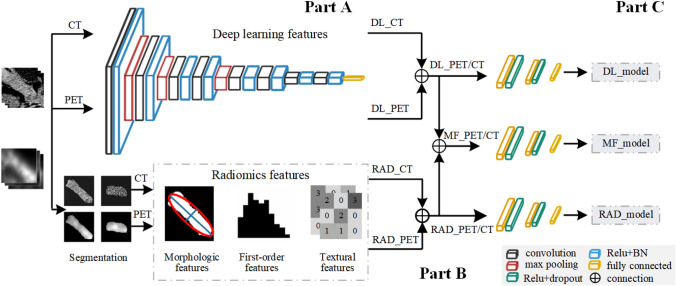
The model design process of this research is shown in Fig. 4, which consists of three parts, namely features extraction (part A), features fusion (part B), and classification prediction (part C).
Fig. 4.
Overall flowchart of our proposed multidomain features fusion classification model. A features extraction part; B features fusion part; C classification prediction part. RAD_PET/CT, radiomics features; DL_PET/CT, deep learning features; MF_PET/CT, multidomain fusion features
In the part of feature extraction, we planned to extract two groups of features: radiomics features and deep learning features. The Pyradiomics [38] open-source code in python was used to extract statistical features from PET and CT images. We chose the network framework from VGG11 [39] to extract features, which contains five blocks and the convolution layer with 3 × 3 kernel in each block. We use the VGG11 network model to train PET and CT images simultaneously to obtain high-level semantic features.
In the features fusion section, we combined the features of PET and CT images to form the multimodal features of PET/CT images, then fused radiomics and deep learning features at the decision layer to obtain multidomain feature sets of PET/CT images. Thus, we got three feature sets, namely radiomics features, deep learning features, and multidomain fusion features.
According to the features extraction process described above, we established three classification models, radiomics classification model (RAD_model), deep learning classification model (DL_model), and multidomain fusion classification model (MF_model):
RAD_model: Radiomics features include texture features (75), histogram features (18), and morphological features (9). The radiomics features of PET and CT images were connected to obtain the PET/CT multimodal features (195), which were sent to the fully connected layer (the morphological features of PET and CT images were the same, only one type was retained in the multimodal features).
DL_model: The VGG11 network was used to extract the deep learning features of PET and CT images, respectively, then the multimodal features of PET/CT (8192) were obtained through the feature fusion of the fully connected layer. The parameters of the feature extraction layer of the network were fixed, and then the linear block was adjusted to complete the binary classification task.
MF_model: We integrated radiomics and deep learning features to form multidomain features of PET/CT (8387), then input them into the full connection layer to classify PDAC and AIP. The model expected to capture valuable information from the new feature set and took this complementary advantage to identify.
Feature correlation analysis
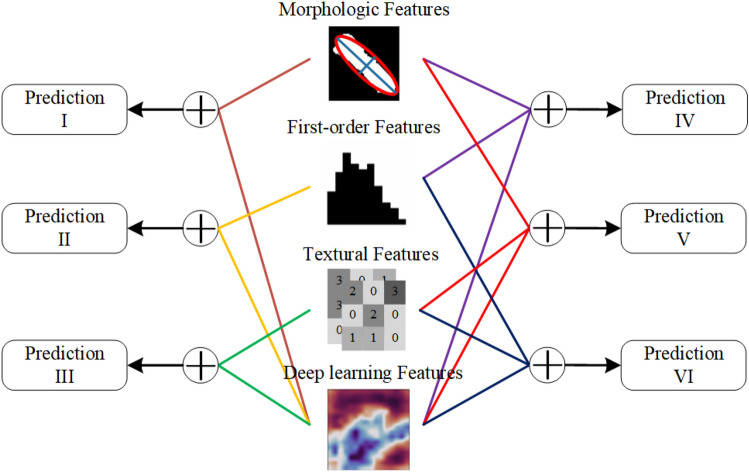
Radiomics features can be divided into morphological features, first-order features, and texture features, which represent different statistical significance [40]. Morphological features describe the geometric features of the ROI, such as volume, surface area, the surface-to-volume ratio. First-order features are sometimes called intensity features; they reflect voxel statistical variables and global properties on the ROI. Texture features focus on the voxel statistical relationship between neighboring regions, and perceive the spatial variation of voxel intensity levels. Deep learning can capture image differences that cannot be noticed by human eyes, and the diversity of feature maps makes the acquired abstract features extremely rich.
To analyze the correlation and complementarity between radiomics features with different statistical properties and deep learning features, we tried to use the morphological features, first-order features, texture features, and deep learning features to form new feature sets through different combination methods. As shown in Fig. 5, different feature sets were put into the fully connected layer for classification, and six prediction results were obtained. We hoped to infer the information categories of deep learning features by comparing these classification results.
Fig. 5.
Fused deep learning and different statistical features of radiomics
Statistical analysis
Continuous data were described as mean ± SD, and discrete data and qualitative variables were expressed as natural number or percentages. The five-fold cross-validation strategy was used to ensure the stability of the model and reduce the results contingency caused by data distribution. We evaluated model performance by calculating the mean of these quantitative indicators through five-fold cross-validation, such as accuracy (Acc), sensitivity (Sen), specificity (Spe), and area under the curve (AUC).
The DeLong test on Medcalc software (version 18.11.3) was used to verify the statistical significance of the AUC values between different models based on labels and prediction scores. Sensitivity, specificity, and accuracies were compared statistically using McNemar's tests, and p value < 0.05 was considered statistically significant difference. Models training and evaluation were run on PyTorch (version 2021.2.3), using an NVIDIA GeForce RTX 3080ti GPU with 64 GB memory.
Results
Classification performance of fusion model
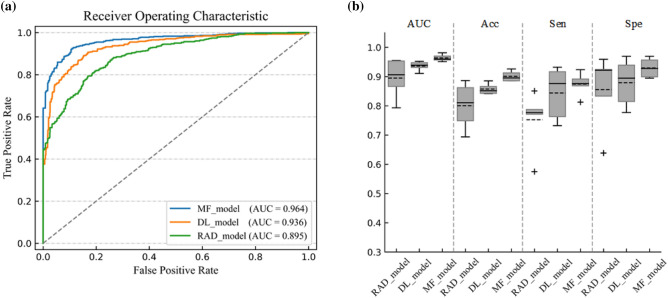
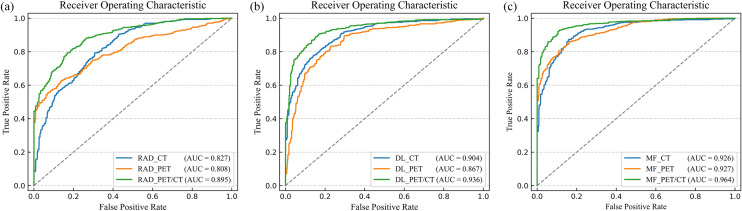
We got the classification results of multidomain features, radiomics features, and deep learning features. Figure 6a shows that the average ROC curve of MF_model was the best in the three feature classifications. The AUC of MF_model was higher than RAD_model (AUC: 96.4 vs 89.5%, p < 0.0001, DeLong test) and DL_model (AUC: 96.4 vs 93.6%, p < 0.0001, DeLong test), It is found in Fig. 6b that Acc, Sen, and Spe of MF_model were the highest among all models results at 90.1%, 87.5%, and 93.0% respectively. Table 2 summarizes the results of all methods.
Fig. 6.
Performance comparison of three classification models based on PET/CT images. a The ROC curves and AUC values of RAD_model, DL_model, and MF_model. b Performance of RAD_model, DL_model, and MF_model in terms of AUC, Acc, Spe, and Sen
Table 2.
Results of three models: AUC value, accuracy, sensitivity, and specificity
| Methods | Medical image | Performance | |||
|---|---|---|---|---|---|
| AUC (95% CI) | Accuracy (95% CI) | Sensitivity (95% CI) | Specificity (95% CI) | ||
| RAD_model | CT | 82.7% ± 7.3% (76.2–89.1%) | 73.0% ± 5.6% (68.1–77.9%) | 76.2% ± 7.6% (69.5–82.9%) | 70.4% ± 10.7% (61.0–79.8%) |
| PET | 80.8% ± 6.1% (75.4–86.1%) | 73.4% ± 7.1% (67.2–79.6%) | 66.4% ± 9.5% (58.1–74.8%) | 81.2% ± 13.2% (69.7–92.8%) | |
| PET/CT | 89.5% ± 6.1% (84.2–94.8%) | 80.0% ± 7.1% (73.8–86.3%) | 75.2% ± 9.3% (67.1–83.4%) | 85.5% ± 11.6% (75.4–95.7%) | |
| DL_model | CT | 90.4% ± 1.0% (89.6–91.3%) | 81.0% ± 1.4% (79.8–82.3%) | 79.2% ± 5.3% (74.6–83.8%) | 83.2% ± 4.5% (79.2–87.2%) |
| PET | 86.7% ± 4.1% (83.2–90.3%) | 79.7% ± 5.0% (75.3–84.0%) | 75.6% ± 10.4% (66.5–84.7%) | 84.0% ± 5.3% (79.4–88.6%) | |
| PET/CT | 93.6% ± 1.5% (92.3–94.9%) | 85.8% ± 1.7% (84.3–87.2%) | 84.4% ± 8.1% (77.3–91.5%) | 87.9% ± 7.3% (81.5–94.3%) | |
| MF_model | CT | 92.6% ± 1.6% (91.2–94.1%) | 84.3% ± 2.5% (82.1–86.4%) | 85.4% ± 9.3% (77.3–93.5%) | 83.2% ± 8.3% (75.9–90.5%) |
| PET | 92.7% ± 2.5% (90.5–94.9%) | 82.8% ± 6.0% (77.5–88.1%) | 73.8% ± 15.0% (60.6–86.9%) | 91.6% ± 4.8% (87.4–95.8%) | |
| PET/CT | 96.4% ± 1.1% (95.4–97.3%) | 90.1% ± 1.6% (88.7–91.5%) | 87.5% ± 3.6% (84.3–90.6%) | 93.0% ± 3.0% (90.3–95.6%) | |
Results of fusion features with different radiomics
We successively fused high-level semantic features from deep learning with different radiomics. As shown in Table 3, the AUC value of the multidomain feature set containing first-order features (Prediction II) was improved compared with only deep learning features (AUC: 96.2% vs 93.6%, p < 0.0001, DeLong test), and the prediction results of the feature sets that integrate deep learning features and morphological features (Prediction I) or texture features (Prediction III) of radiomics had poor performance (AUC: 91.7% vs 92.2% vs 93.6%, p < 0.05, p < 0.05, DeLong test). The comparison of the six prediction results with the deep learning method is shown in Fig. 7.
Table 3.
Evaluation of six feature sets: AUC value, accuracy, sensitivity, and specificity
| Models | Performance | ||||
|---|---|---|---|---|---|
| AUC (95% CI) | Accuracy (95% CI) | Sensitivity (95% CI) | Specificity (95% CI) | P value | |
| Mor + dl features (prediction I) | 91.7% ± 1.4% (90.4–92.9%) | 83.2% ± 2.8% (80.7–85.6%) | 82.5% ± 5.1% (78.0–87.0%) | 84.4% ± 8.6% (76.8–91.9%) | 0.0061 |
| Fir + dl features (prediction II) | 96.2% ± 1.1% (95.2–97.1%) | 89.4% ± 1.8% (87.9–91.0%) | 89.9% ± 4.0% (86.4–93.3%) | 89.7% ± 6.1% (84.4–95.0%) | 0.0006 |
| Tex + dl features (prediction III) | 92.2% ± 1.3% (91.1–93.3%) | 83.4% ± 3.4% (80.5–86.4%) | 84.0% ± 5.9% (78.9–89.2%) | 83.8% ± 10.0% (75.0–92.6%) | 0.0144 |
| Mor + Fir + dl features (prediction IV) | 96.0% ± 1.1% (95.0–97.0%) | 88.1% ± 3.1% (85.4–90.8%) | 87.3% ± 5.7% (82.3–92.3%) | 89.0% ± 9.7% (80.5–97.5%) | 0.0271 |
| Mor + Tex + dl features (prediction V) | 92.4% ± 1.1% (91.5–93.4%) | 84.4% ± 3.5% (81.3–87.5%) | 88.0% ± 2.9% (85.4–90.6%) | 81.7% ± 8.5% (74.3–89.1%) | 0.0474 |
| Fir + Tex + dl features (prediction VI) | 96.4% ± 1.0% (95.5–97.3%) | 90.5% ± 1.4% (89.4–91.7%) | 88.7% ± 4.7% (84.61–92.7%) | 92.6% ± 4.4% (88.7–96.4%) | < 0.0001 |
Mor morphological features of radiomics, Fir first-order features of radiomics, Tex texture features of radiomics, dl deep learning features; p value, McNemar test
Fig. 7.
Difference of AUC values between the six different prediction results and the deep learning model
Evaluation of multimodal features
As revealed in Fig. 8, the average ROC curve of PET/CT multimodal features was higher than only CT or PET features in three classification models. In RAD_model, the average AUC of PET/CT features was 89.5% better than CT features (AUC: 89.5 vs 82.7%, p < 0.0001, DeLong test) and PET features (AUC: 89.5 vs 80.8%, p < 0.0001, DeLong test), the Acc increased to 80.0%. DL_model classification results showed that the average AUC of PET/CT features was 93.6%, superior to CT features and PET features. There was no crossover phenomenon in the ROC curve to identify the models’ performance easily, and the Acc of PET/CT was 85.8%. In MF_model, the average AUC of PET/CT features was 96.4% higher than CT features (AUC: 96.4 vs 92.6%, p < 0.0001, DeLong test) and PET features (AUC: 96.4 vs 92.7%, p < 0.0001, DeLong test). Although the AUC value of CT and PET was very similar, the Acc of CT was 84.3% and is better than PET images.
Fig. 8.
Comparison between the performance of CT features, PET features, and PET/CT multimodal features in the three models. a Radiomics classification model. b Deep learning classification model. c Multidomain fusion classification model
Discussion
The purpose of this study was to explore an effective method for noninvasive identification of PDAC and AIP diseases. We integrated radiomics and deep learning features to establish the multidomain features fusion model (MF_model) based on 18F-FDG PET/CT images, and the better comprehensive performance of MF_model reflected the value of multidomain features in distinguishing PDAC and AIP. We found that the first-order features contribute most to improving the deep learning model by verifying the results of different feature sets.
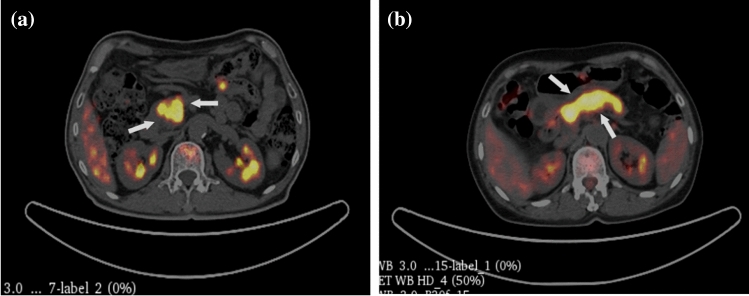
Radiomics is based on statistics which use specific functions to capture the visible information of images. The performance of the radiomics model depends on the segmentation area of the lesion, indicating that the experience and diagnostic ability of clinicians involved in ROI delineation are essential factors in the result of the model. The features extraction process of radiomics is relatively fixed and ignores the individual differences of patients. We hoped to find a more flexible way to capture the details of the area around the lesion to improve the model’s accuracy further. The method of deep learning uses image patches for features extraction which can reduce the dependence of prediction results on accurate ROI description, and the convolutional layers generate spatial features from the images that integrate local images information from the initial and deep layers. The fusion of radiomics and deep learning features to form a multidomain feature set has been applied to diagnosing and treating different diseases. PDAC prognosis and Parkinson's disease diagnosis research have proved the multidomain features' application value [41, 42]. According to our survey, the number of research papers on PDAC and AIP disease screening is limited, and fewer studies discuss the fusion classification model of radiomics and deep learning features. Therefore, we proposed to combine the advantages of radiomics and deep learning to establish a fusion classification model of PDAC and AIP based on PET/CT images. This method improves the diagnostic accuracy of these two diseases by breaking through the limitations of radiomics methods and the shortcomings of subjective differences of doctors. Figure 9 illustrates image slices in which clinical diagnosis misdiagnosed and MF_model diagnosis is correct. Under the condition of similar datasets, the Acc of our proposed method was improved by about 5% compared with the published radiomics methods [43].
Fig. 9.
Representative PET/CT slices (white arrows pointing to the lesion) which clinical diagnosis misdiagnosed and MF_model diagnosis is correct. a the slice of a 73-year-old man with AIP misdiagnosed as PDAC. b the slice of a 67 year-old woman with PDAC misdiagnosed as AIP
The high-level semantic features extracted by deep learning were difficult to define in biological or morphological terms, but we combined radiomics features into six different feature sets, fused them with deep learning features for classification training, and conjectured the information attributes of high-level semantic features by comparing the model results. It was found that the first-order features had positive impacts on the classification model, while the morphological features and texture features had negative effects by observing the experimental data. We suspect that the high-level semantic features extracted by CNN contain information related to morphological and texture features. The connection of these features would lead to information redundancy or negative correlation, which could reduce model results. The first-order features reflect the attenuation distribution of ROI voxels and reveal the homogeneity of the images, combining them with high-level semantic features to make the information complementarity to each other. Therefore, the fusion method (Prediction III) of deep learning features and first-order features not only captures abstract features that cannot be discerned by the naked eye but also considers the description information of the voxel intensity distribution in the lesion area. Hence, the feature sets showed excellent analytical performance.
In addition, we explored the advantages of PET/CT images in identifying PDAC and AIP diseases, then compared the performance differences between multimodal and single-type features. Among the three groups of models (RAD_model, DL_model, and MF_model), the PET/CT multimodal features prediction results were better than only CT and PET features. The resolution of CT images is higher than PET images, and the CT images can provide the contour information between the lesion area and the surrounding blood vessels, making the anatomical information more discriminative. PET images reflect the metabolic level of lesions and can serve as valuable supplementary information in disease classification. PET/CT images combine the advantages of CT and PET images to form diversified information, so multimodal features got the best results in experiments. This conclusion was consistent with published research findings. Xing [44] used XGBoost to establish the pathological grading prediction model of PDAC, and then found that PET/CT images had more analysis advantages than only CT and PET images. Zhang [43] used the support vector machine (SVM) algorithm to establish a classification model for PDAC and AIP diseases based on PET/CT images, and then concluded that the PET/CT multimodal features classification results were better than single-type features.
There were two main limitations of this study worth discussing. On the one hand, we used five-fold cross-validation to reduce the risk of model overfitting due to small datasets. However, we lack the generalization ability of external datasets for validating the model. We are already coordinating the collection of multi-center data to validate the reliability of the model and improve the applicability of the method in the clinic. It will take some time to achieve this goal. On the other hand, deep learning captures unique image features invisible to the human eyes, and the abstract nature of features increases the difficulty of model interpretability. Although we explored correlations between features, the impact of the deep learning process on results is still unclear. Some studies have used occlusion heat map analysis and concept attribution strategies [33, 45] to explain the “black box” decision, but these methods are still controversial and limited. Improving the interpretability of deep learning results is still a research direction we need to work on in the future.
Conclusion
We established a novel multidomain fusion model of radiomics and deep learning features based on 18F-FDG PET/CT images which demonstrated the superior diagnostic performance of multidomain features for noninvasively discriminating PDAC and AIP. This method will have the potential to become a clinical auxiliary tool to improve the performance of disease diagnosis. Moreover, we speculate that first-order features play a vital role in improving deep learning models through comparative experiments with different feature sets.
Funding
This study was funded by the National Natural Science Foundation of China (No. 62101551, 62001471, 81871390, 82001867); “234 Discipline Climbing Plan” of the First Affiliated Hospital of Naval Medical University (No. 2019YPT002, 2020YPT002).
Declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This retrospective study was approved by the Institutional Review Board of The First Affiliated Hospital of Naval Medical University (approval code: CHEC2019-089). Informed consent was waived.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Chao Cheng and Zhaobang Liu are both co-corresponding authors.
Contributor Information
Chao Cheng, Email: chao_cheng_1999@163.com.
Zhaobang Liu, Email: liuzb@sibet.ac.cn.
References
- 1.Cascinu S, Falconi M, Valentini V, Jelic S, Group EGW Pancreatic cancer: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2010;21(suppl5):v55–v58. doi: 10.1093/annonc/mdq165. [DOI] [PubMed] [Google Scholar]
- 2.Natália Alves MS, Litjens G, Bosma JS, Hermans J, Huisman H. Fully automatic deep learning framework for pancreatic ductal adenocarcinoma detection on computed tomography. Cancers (Basel) 2021;14(2):376. doi: 10.3390/cancers14020376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin. 2021;71(1):7–33. doi: 10.3322/caac.21654. [DOI] [PubMed] [Google Scholar]
- 4.Anderson EM, Thomassian S, Gong J, Hendifar A, Osipov A. Advances in pancreatic ductal adenocarcinoma treatment. Cancers (Basel). 2021;13(21):5510. doi: 10.3390/cancers13215510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ogawa H, Takehara Y, Naganawa S. Imaging diagnosis of autoimmune pancreatitis: computed tomography and magnetic resonance imaging. J Med Ultrason. 2021;48(4):565–571. doi: 10.1007/s10396-021-01145-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takahashi M, Fujinaga Y, Notohara K, Koyama T, Inoue D, Irie H, et al. Diagnostic imaging guide for autoimmune pancreatitis. Jpn J Radiol. 2020;38(7):591–612. doi: 10.1007/s11604-020-00971-z. [DOI] [PubMed] [Google Scholar]
- 7.Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74(11):2913–2921. doi: 10.1158/0008-5472.CAN-14-0155. [DOI] [PubMed] [Google Scholar]
- 8.Matsubayashi H, Ishiwatari H, Imai K, Kishida Y, Ito S, Hotta K, et al. Steroid Therapy and Steroid Response in Autoimmune Pancreatitis. Int J Mol Sci. 2019;21(1):257. doi: 10.3390/ijms21010257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dickerson LD, Farooq A, Bano F, Kleeff J, Baron R, Raraty M, et al. Differentiation of Autoimmune Pancreatitis from Pancreatic Cancer Remains Challenging. World J Surg. 2019;43(6):1604–1611. doi: 10.1007/s00268-019-04928-w. [DOI] [PubMed] [Google Scholar]
- 10.Meng Q, Xin L, Liu W, Lin H, Tian B, Wang L, et al. Diagnosis and treatment of autoimmune pancreatitis in china: a systematic review. PLoS ONE. 2015;10(6):e0130466. doi: 10.1371/journal.pone.0130466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Katherine E, Poruk DZG, Brown K, Mulvihill JD, Boucher KM, Scaife CL, Firpo MA, Mulvihill SJ. The Clinical utility of ca 19–9 in pancreatic adenocarcinoma: diagnostic and prognostic updates. Curr Mol Med. 2013;13:340–351. doi: 10.2174/1566524011313030003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shih HC, Chang MC, Chen CH, Tsai IL, Wang SY, Kuo YP, et al. High accuracy differentiating autoimmune pancreatitis from pancreatic ductal adenocarcinoma by immunoglobulin G glycosylation. Clin Proteomics. 2019 doi: 10.1186/s12014-018-9221-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pak LM, Schattner MA, Balachandran V, D'Angelica MI, DeMatteo RP, Kingham TP, et al. The clinical utility of immunoglobulin G4 in the evaluation of autoimmune pancreatitis and pancreatic adenocarcinoma. HPB (Oxford) 2018;20(2):182–187. doi: 10.1016/j.hpb.2017.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hasan S, Jacob R, Manne U, Paluri R. Advances in pancreatic cancer biomarkers. Oncol Rev. 2019;13(1):410. doi: 10.4081/oncol.2019.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Notohara K, Kamisawa T, Fukushima N, Furukawa T, Tajiri T, Yamaguchi H, et al. Guidance for diagnosing autoimmune pancreatitis with biopsy tissues. Pathol Int. 2020;70(10):699–711. doi: 10.1111/pin.12994. [DOI] [PubMed] [Google Scholar]
- 16.Zhang X, Liu X, Joseph L, Zhao L, Hart J, Xiao SY. Pancreatic ductal adenocarcinoma with autoimmune pancreatitis-like histologic and immunohistochemical features. Hum Pathol. 2014;45(3):621–627. doi: 10.1016/j.humpath.2013.08.027. [DOI] [PubMed] [Google Scholar]
- 17.Notohara K, Kamisawa T, Kanno A, Naitoh I, Iwasaki E, Shimizu K, et al. Efficacy and limitations of the histological diagnosis of type 1 autoimmune pancreatitis with endoscopic ultrasound-guided fine needle biopsy with large tissue amounts. Pancreatology. 2020;20(5):834–843. doi: 10.1016/j.pan.2020.05.026. [DOI] [PubMed] [Google Scholar]
- 18.Ha J, Choi SH, Byun JH, Kim KW, Kim SY, Kim JH, et al. Meta-analysis of CT and MRI for differentiation of autoimmune pancreatitis from pancreatic adenocarcinoma. Eur Radiol. 2021;31(5):3427–3438. doi: 10.1007/s00330-020-07416-1. [DOI] [PubMed] [Google Scholar]
- 19.Jia H, Li J, Huang W, Lin G. Multimodel magnetic resonance imaging of mass-forming autoimmune pancreatitis: differential diagnosis with pancreatic ductal adenocarcinoma. BMC Med Imaging. 2021;21(1):149. doi: 10.1186/s12880-021-00679-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dong Y, D'Onofrio M, Hocke M, Jenssen C, Potthoff A, Atkinson N, et al. Autoimmune pancreatitis: imaging features. Endosc Ultrasound. 2018;7(3):196–203. doi: 10.4103/eus.eus_23_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li J, Liu F, Fang X, Cao K, Meng Y, Zhang H, et al. CT radiomics features in differentiation of focal-type autoimmune pancreatitis from pancreatic ductal adenocarcinoma: a propensity score analysis. Acad Radiol. 2022;29(3):358–366. doi: 10.1016/j.acra.2021.04.014. [DOI] [PubMed] [Google Scholar]
- 22.Fang M, He B, Li L, Dong D, Yang X, Li C, et al. CT radiomics can help screen the Coronavirus disease 2019 (COVID-19): a preliminary study. Sci China Inform Sci. 2020 doi: 10.1007/s11432-020-2849-3. [DOI] [Google Scholar]
- 23.Chen BT, Jin T, Ye N, Mambetsariev I, Daniel E, Wang T, et al. Radiomic prediction of mutation status based on MR imaging of lung cancer brain metastases. Magn Reson Imaging. 2020;69:49–56. doi: 10.1016/j.mri.2020.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parekh VS, Jacobs MA. Multiparametric radiomics methods for breast cancer tissue characterization using radiological imaging. Breast Cancer Res Treat. 2020;180(2):407–421. doi: 10.1007/s10549-020-05533-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu Z, Li M, Zuo C, Yang Z, Yang X, Ren S, et al. Radiomics model of dual-time 2-[(18)F]FDG PET/CT imaging to distinguish between pancreatic ductal adenocarcinoma and autoimmune pancreatitis. Eur Radiol. 2021;31(9):6983–6991. doi: 10.1007/s00330-021-07778-0. [DOI] [PubMed] [Google Scholar]
- 26.Park S, Chu LC, Hruban RH, Vogelstein B, Kinzler KW, Yuille AL, et al. Differentiating autoimmune pancreatitis from pancreatic ductal adenocarcinoma with CT radiomics features. Diagn Interv Imaging. 2020;101(9):555–564. doi: 10.1016/j.diii.2020.03.002. [DOI] [PubMed] [Google Scholar]
- 27.Linning E, Yan Xu, Zhifeng Wu, Li Li, Zhang N. Differentiation of focal-type autoimmune pancreatitis from pancreatic ductal adenocarcinoma using radiomics based on multiphasic computed tomography. J Comput Assist Tomogr. 2020;44(4):511–518. doi: 10.1097/RCT.0000000000001049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Song KD. Current status of deep learning applications in abdominal ultrasonography. Ultrasonography. 2021;40(2):177–182. doi: 10.14366/usg.20085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Debelee TG, Kebede SR, Schwenker F, Shewarega ZM. Deep Learning in Selected Cancers' Image Analysis-A Survey. J Imaging. 2020;6(11):121. doi: 10.3390/jimaging6110121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang T, Lei Y, Fu Y, Wynne JF, Curran WJ, Liu T, et al. A review on medical imaging synthesis using deep learning and its clinical applications. J Appl Clin Med Phys. 2021;22(1):11–36. doi: 10.1002/acm2.13121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakaura T, Higaki T, Awai K, Ikeda O, Yamashita Y. A primer for understanding radiology articles about machine learning and deep learning. Diagn Interv Imaging. 2020;101(12):765–770. doi: 10.1016/j.diii.2020.10.001. [DOI] [PubMed] [Google Scholar]
- 32.Ziegelmayer S, Kaissis G, Harder F, Jungmann F, Muller T, Makowski M, et al. Deep convolutional neural network-assisted feature extraction for diagnostic discrimination and feature visualization in pancreatic ductal adenocarcinoma (PDAC) versus autoimmune pancreatitis (AIP) J Clin Med. 2020;9(12):4013. doi: 10.3390/jcm9124013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marya NB, Powers PD, Chari ST, Gleeson FC, Leggett CL, Abu Dayyeh BK, et al. Utilisation of artificial intelligence for the development of an EUS-convolutional neural network model trained to enhance the diagnosis of autoimmune pancreatitis. Gut. 2021;70(7):1335–1344. doi: 10.1136/gutjnl-2020-322821. [DOI] [PubMed] [Google Scholar]
- 34.Anai K, Hayashida Y, Ueda I, Hozuki E, Yoshimatsu Y, Tsukamoto J, et al. The effect of CT texture-based analysis using machine learning approaches on radiologists' performance in differentiating focal-type autoimmune pancreatitis and pancreatic duct carcinoma. Jpn J Radiol. 2022;40(11):1156–1165. doi: 10.1007/s11604-022-01298-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ning Z, Luo J, Li Y, Han S, Feng Q, Xu Y, et al. Pattern classification for gastrointestinal stromal tumors by integration of radiomics and deep convolutional features. IEEE J Biomed Health Inform. 2019;23(3):1181–1191. doi: 10.1109/JBHI.2018.2841992. [DOI] [PubMed] [Google Scholar]
- 36.Zhang W, Peng J, Zhao S, Wu W, Yang J, Ye J, et al. Deep learning combined with radiomics for the classification of enlarged cervical lymph nodes. J Cancer Res Clin Oncol. 2022;143(10):2773–2780. doi: 10.1007/s00432-022-04047-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Choi YS, Bae S, Chang JH, Kang SG, Kim SH, Kim J, et al. Fully automated hybrid approach to predict the IDH mutation status of gliomas via deep learning and radiomics. Neuro Oncol. 2021;23(2):304–313. doi: 10.1093/neuonc/noaa177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Griethuysen JJM, Fedorov A, Parmar C, Hosny A, Aucoin N, Narayan V, et al. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 2017;77(21):e104–e107. doi: 10.1158/0008-5472.CAN-17-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Simonyan K, Zisserman A (2014) Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556v6
- 40.Zhang X, Zhang Y, Zhang G, Qiu X, Tan W, Yin X, et al. Deep learning with radiomics for disease diagnosis and treatment: challenges and potential. Front Oncol. 2022;12:773840. doi: 10.3389/fonc.2022.773840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Y, Lobo-Mueller EM, Karanicolas P, Gallinger S, Haider MA, Khalvati F. Improving prognostic performance in resectable pancreatic ductal adenocarcinoma using radiomics and deep learning features fusion in CT images. Sci Rep. 2021;11(1):1378. doi: 10.1038/s41598-021-80998-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xiao B, He N, Wang Q, Cheng Z, Jiao Y, Haacke EM, et al. Quantitative susceptibility mapping based hybrid feature extraction for diagnosis of Parkinson's disease. Neuroimage Clin. 2019;24:102070. doi: 10.1016/j.nicl.2019.102070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Y, Cheng C, Liu Z, Wang L, Pan G, Sun G, et al. Radiomics analysis for the differentiation of autoimmune pancreatitis and pancreatic ductal adenocarcinoma in (18) F-FDG PET/CT. Med Phys. 2019;46(10):4520–4530. doi: 10.1002/mp.13733. [DOI] [PubMed] [Google Scholar]
- 44.Xing H, Hao Z, Zhu W, Sun D, Ding J, Zhang H, et al. Preoperative prediction of pathological grade in pancreatic ductal adenocarcinoma based on (18)F-FDG PET/CT radiomics. EJNMMI Res. 2021;11(1):19. doi: 10.1186/s13550-021-00760-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Graziani M, Andrearczyk V, Marchand-Maillet S, Müller H. Concept attribution: Explaining CNN decisions to physicians. Comput Biol Med. 2020;123:103865. doi: 10.1016/j.compbiomed.2020.103865. [DOI] [PubMed] [Google Scholar]