Abstract
For proper function of proteins, their subcellular localization needs to be monitored and regulated in response to the changes in cellular demands. In this regard, dysregulation in the nucleocytoplasmic transport (NCT) of proteins is closely associated with the pathogenesis of various neurodegenerative diseases. However, it remains unclear whether there exists an intrinsic regulatory pathway(s) that controls NCT of proteins either in a commonly shared manner or in a target-selectively different manner. To dissect between these possibilities, in the current study, we investigated the molecular mechanism regulating NCT of truncated ataxin-3 (ATXN3) proteins of which genetic mutation leads to a type of polyglutamine (polyQ) diseases, in comparison with that of TDP-43. In Drosophila dendritic arborization (da) neurons, we observed dynamic changes in the subcellular localization of truncated ATXN3 proteins between the nucleus and the cytosol during development. Moreover, ectopic neuronal toxicity was induced by truncated ATXN3 proteins upon their nuclear accumulation. Consistent with a previous study showing intracellular calcium-dependent NCT of TDP-43, NCT of ATXN3 was also regulated by intracellular calcium level and involves Importin α3 (Imp α3). Interestingly, NCT of ATXN3, but not TDP-43, was primarily mediated by CBP. We further showed that acetyltransferase activity of CBP is important for NCT of ATXN3, which may acetylate Imp α3 to regulate NCT of ATXN3. These findings demonstrate that CBP-dependent acetylation of Imp α3 is crucial for intracellular calcium-dependent NCT of ATXN3 proteins, different from that of TDP-43, in Drosophila neurons.
Keywords: acetylation, ATXN3, calcium, CBP, Importin α, nucleocytoplasmic transport
INTRODUCTION
Proper subcellular localization of proteins is essential for their functioning and/or interactions with binding partners, of which dysregulation often leads to diverse cellular defects (Bauer et al., 2015). In line with this, disruption of nucleocytoplasmic transport (NCT) of proteins is known to cause diverse neurodegenerative diseases (NDs), such as amyotrophic lateral sclerosis (ALS) and polyglutamine (polyQ) diseases, through aberrant accumulation of toxic disease-related proteins (e.g., TDP-43 in ALS and polyQ proteins in polyQ diseases) in afflicted neurons (Chung et al., 2018). However, our understanding on the molecular and cellular basis underlying NCT dysregulation in these NDs remains incomplete. Recently, we showed that NCT of TDP-43 is controlled by the intracellular calcium-Calpain-A (CalpA)-Importin α3 (Imp α3) signaling pathway in Drosophila neurons, implicating that dysregulation of this intrinsic regulatory mechanism may lead to mislocalization of TDP-43 in ALS (Park et al., 2020a). Given that a limited number of karyopherins take charge of NCT of numerous proteins containing nuclear localization signal (NLS) (Wing et al., 2022), it can be speculated that the identified calcium-dependent intrinsic regulatory mechanism for NCT of TDP-43 may be, at least partly, involved in the NCT of other disease-related proteins.
As one of ND-related proteins known to mislocalize in the disease context, ataxin-3 (ATXN3), also known as SCA3, is a deubiquitinating enzyme that normally localizes in the cytosol (Nijman et al., 2005). Upon genetic mutation of CAG repeats encoding polyQ repeats, mutated ATXN3 proteins aberrantly translocate into the nucleus, thereby inducing nuclear proteotoxicity in neurons (Bichelmeier et al., 2007; Evert et al., 1999; Lee et al., 2020; 2011). Mutated ATXN3 proteins are known to undergo proteolytic cleavage into smaller toxic fragments having higher toxicity (Breuer et al., 2010; Haacke et al., 2006; Simoes et al., 2022). Generally, previous studies have focused only on the proteotoxicity of mutated ATXN3 proteins in neurons with using ATXN3 proteins containing normal length of polyQ repeats as a control (Peng et al., 2022; Sowa et al., 2022). Given that the unmutated ATXN3 proteins are known to occasionally translocated into the nucleus (Fujigasaki et al., 2000; Perez et al., 1999; Reina et al., 2010; Tait et al., 1998), the proper localization of unmutated ATXN3 can be dynamically regulated in a cellular context-dependent manner, as was shown for TDP-43 proteins (Park et al., 2020a). It is reported that unmutated ATXN3 proteins can also undergo proteolytic cleavage (Simoes et al., 2022). Notably, we observed that unmutated ATXN3 proteins can confer aberrant protein toxicity, when truncated, leading to defective neuronal remodeling during metamorphosis of Drosophila (Chung et al., 2017). In the current study, we examined whether the identified intrinsic regulatory mechanism for NCT of TDP-43 is also involved in the NCT of unmutated ATXN3 proteins that are truncated (ATXN3tr-27Q). We found that NCT of ATXN3tr-27Q is regulated by intracellular calcium level similar to the NCT of TDP-43, but involves CBP as a key mediator linking calcium and Imp α3 unlike the NCT of TDP-43 (Park et al., 2020a).
MATERIALS AND METHODS
Drosophila stocks and gene switch-mediated expression
All flies were maintained at 25°C and 60% humidity. The following lines were obtained from Bloomington Drosophila Stock Center (BL) (USA): w1118 (BL 3605), elav-GAL4 (BL 8760), ppk-GAL4 (32078), UAS-ATXN3tr-27Q (BL 8149), UAS-ATXN3tr-78Q (BL 8141), UAS-NFAT RNAi (BL 51422), UAS-CrebB RNAi (BL 63681), UAS-Cam RNAi (BL 34609), UAS-CanA-14F RNAi (BL 58249), UAS-CanB RNAi (BL 27307), UAS-CaMKII RNAi (BL 35362), UAS-Pka-C1 RNAi (BL 31599), UAS-Pkc53E RNAi (BL 55864), UAS-CalpA RNAi (BL 29455), UAS-CalpB RNAi (BL 25963), 109(2)80-GAL4 (BL 8769), UAS-CBP.wt-v5 (BL 32573), UAS-CBP.F2161A-v5 (BL 32574), UAS-HDAC1 RNAi (BL 33725), UAS-HDAC3 RNAi (BL 34778), UAS-HDAC4 RNAi (BL 34744), and UAS-RedStinger (UAS-NLS-DsRed, BL 8546). UAS-Imp α3 RNAi (106249KK), UAS-IP3R RNAi (106982KK), UAS-CBP RNAi (102885KK), and UAS-CaMKI RNAi (101380KK) were obtained from Vienna Drosophila RNAi Centre (VDRC) (Austria). UAS-CD4-tdGFP, ppk1a-GAL4, Gene-Switch (GS)-ppk1a-GAL4, UAS-Htt-152Q-eGFP, and ppk-CD4-tdTom were provided by Yuh Nung Jan (University of California, San Francisco, USA). To activate gene switch-mediated transcription of GAL4, adult flies were fed food with RU486 (100 μM RU486 dissolved in 80% ethanol), according to each experimental condition.
Generation of Drosophila transgenic construct
UAS-2x Flag-Imp α3.K17R transgene was generated by using the LD13917 (Flybase ID: FBcl0163088) with a point mutation resulting in the replacement of K17 with R within the “FKNKGK” sequence of the encoded proteins. UAS-nuclear export signal (NES)-NLS-GFP was generated by using the sequences from: NES (LALKLAGLDI), NLS (PKKKRKVD), and green fluorescent protein (GFP) (Cat. No. 632370; TaKaRa Bio, Japan). These transgenes were incorporated into pACU2 vectors, and each transgenic fly line was generated by BestGene (USA).
Immunohistochemistry
Immunohistochemistry was performed as previously described (Chung et al., 2017; Kim et al., 2021; Kwon et al., 2018; Park et al., 2020b). larvae (120 h after egg laying [120 h AEL]), pupae (18 h after puparium formation [18 h APF]), and adult flies were dissected in Schneider’s Insect Medium (Cat. No. S0146; Sigma, USA) to obtain fillet or brain samples for immunohistochemical analyses. Samples were fixed in 4% Paraformaldehyde for 20 min, washed in 0.3% PBST (Triton-X100 0.3% in phosphate-buffered saline [PBS]) for 10 min (repeated three times), and blocked in blocking buffer (5% Normal donkey serum or normal goat serum in 0.3% PBST) for 45 min at room temperature. The samples were incubated at 4°C with primary antibody. The following primary antibodies were used in this study: mouse anti-Flag (1E6, 1:400 dilution; Wako, Japan) for detecting the Flag epitope; rat anti-HA (3F10, 1:200 dilution; Roche, Switzerland) for detecting the HA epitope; and goat anti-HRP Alexa Fluor 488 (1:400 dilution; Jackson Immunoresearch Laboratories, USA) and anti-HRP Alexa Fluor 647 (1:400 dilution; Jackson Immunoresearch Laboratories) for detecting neuronal plasma membrane. To detect primary antibodies, the following secondary antibodies were used: goat anti-mouse Alexa Fluor 594 (1:200 dilution; Invitrogen, USA) and goat anti-mouse Alexa Fluor 488 (1:400 dilution; Invitrogen).
Confocal microscopy
All images were acquired using LSM 700, 800 confocal microscope (Zeiss, Germany) and Zen software (Zeiss). All images of samples used after immunohistochemistry experiments or immediately were taken at 200× and 400× magnifications using 20× and 40× objective lens, respectively. Images of the Drosophila class IV dendritic arborization (C4da) neurons were obtained from the abdominal segments A2-A6, where anterior is to the left and dorsal is up.
Quantification of dendrite remodeling defects in C4da neurons
To perform quantification of dendrite remodeling defects in C4da neurons, we used a method as previously described (Wolterhoff et al., 2020), we determines the number of neurons that still have dendrites attached to soma to reflect the phenotypic penetrance, and these data were analyzed using a two-tailed Fisher’s exact test.
Lifespan assays
The lifespan assay was performed as previously described (Han et al., 2020). Adult flies were collected under CO2 anesthesia and housed at a density of 20 flies per vial. Flies were passed every other day, and the number of dead flies was recorded. At least 80 flies were used for each experimental group tested.
Quantification and statistical analysis
The cytoplasm-to-nucleus (Cyt/Nuc) ratios of immunostained proteins (HA-ATXN3tr-27Q, HA-ATXN3tr-78Q, 2xFlag-Imp α3, 2xFlag-Imp α3.K17R) was analyzed using a method previously described (Park et al., 2020a), the mean pixel intensities of them in the nucleus and the cytosol were measured using ImageJ (National Institutes of Health, USA). Statistical analysis was performed using Prism 8 (GraphPad Software, USA), with Student’s unpaired t-test, one-way ANOVA (with Tukey post hoc test), or two-way ANOVA (with Tukey post hoc test). In all figures, not significant, *, **, ***, and **** represent P > 0.05, P < 0.05, P < 0.01, P < 0.001, and P < 0.0001, respectively. Values are presented as mean ± SD.
RESULTS
ATXN3tr-27Q undergoes context-dependent nucleocytoplasmic translocation in Drosophila neurons and induces neuronal toxicity upon nuclear accumulation
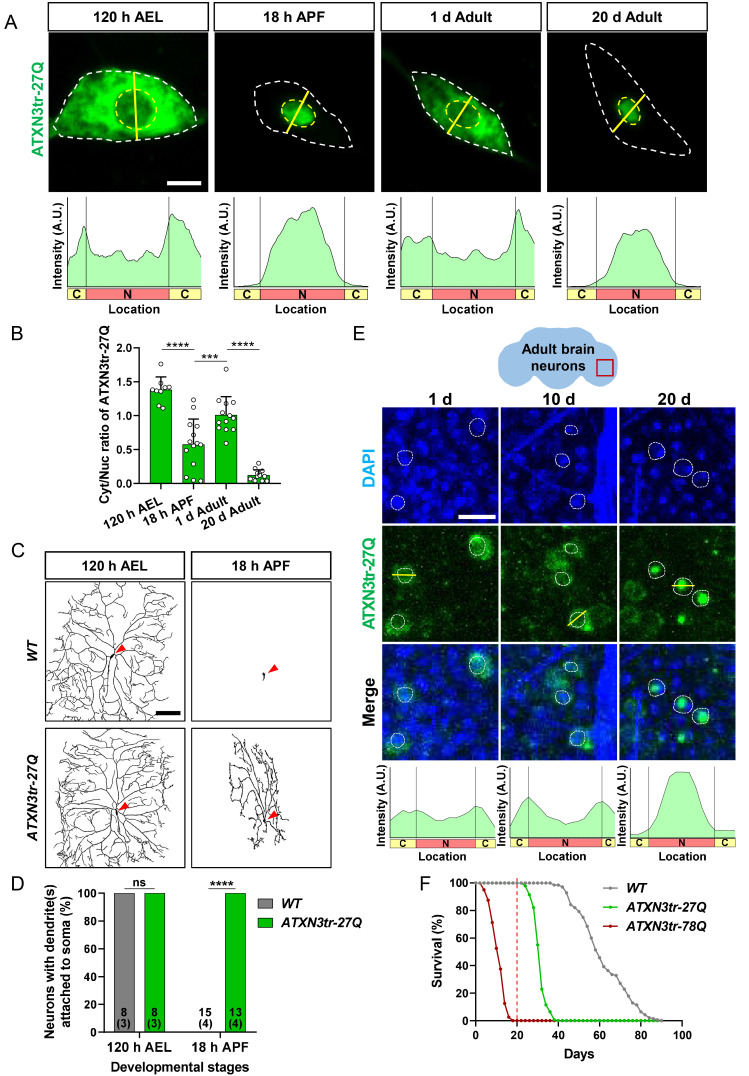
We first explored whether additional disease-related proteins other than TDP-43 undergo context-dependent nucleocytoplasmic translocation similar to that shown in TDP-43 (Park et al., 2020a) in Drosophila C4da neurons, one of well-established neuronal model system to easily monitor neuronal phenotypes and/or protein localization in a single cell level (Chung et al., 2017; Han et al., 2020; Kwon et al., 2018; Lee et al., 2011; Park et al., 2020a). To this end, we focused on unmutated ATXN3 proteins that is truncated (ATXN3tr-27Q) and found that ATXN3tr-27Q undergoes dynamic changes in its nucleocytoplasmic localization during development (Figs. 1A and 1B) similar to TDP-43 in C4da neurons (Park et al., 2020a). Consistent with previous studies reporting cytosolic localization of unmutated and truncated ATXN3 (Kwon et al., 2018; Warrick et al., 2005), ATXN3tr-27Q showed predominant localization in the cytosol of C4da neurons during larval stage or in young adults. Interestingly, nuclear translocation of ATXN3tr-27Q observed in C4da neurons during pupal stage or in late adults (Fig. 1A). Quantitative analysis of ATXN3tr-27Q localization between the nucleus and the cytosol (Cyt/Nuc) revealed dynamic changes of ATXN3tr-27Q localization during development (Fig. 1B). On the other hand, we found that the pathogenic form of ATXN3, ATXN3tr-78Q, showed constant nuclear localization in C4da neurons during development (Supplementary Fig. S1), suggesting that a pathogenic mutation of ATXN3 leads to the extreme shift in NCT of the protein toward nuclear import.
Fig. 1. ATXN3tr-27Q undergoes dynamic nucleocytoplasmic translocation in a cellular context-dependent manner and induces neuronal toxicity upon nuclear accumulation in Drosophila neurons.
(A) Subcellular localization of overexpressed HA-ATXN3tr-27Q proteins in C4da neurons during development stages (120 h AEL, 18 h APF, 1-day adult, and 20-day adult) [+/+;ppk1a-GAL4>UAS-CD4-tdGFP/UAS-HA-ATXN3tr-27Q]. Outer and inner dashed lines indicate the borders of cell bodies and nuclei, respectively. Scale bar = 5 μm. The intensity profile of fluorescent signals representing ATXN3tr-27Q proteins across cell bodies along yellow lines are presented at the bottom. (B) Quantification of cytosol/nuclear (Cyt/Nuc) ratio of HA-ATXN3tr-27Q proteins during development stages. Values are presented as mean ± SD. ****P < 0.0001, ***P = 0.0005 by one-way ANOVA with Tukey post hoc test; n = 9 for 120 h AEL, n = 14 for 18 h APF, n = 13 for 1 d adult, n = 13 for 20 d adult. (C) Skeletonized dendrite images of C4da neurons of 120 h AEL and 18 h APF [WT, +/+;ppk1a-GAL4>UAS-CD4-tdGFP/+, HA-ATXN3tr-27Q, +/+; ppk1a-GAL4>UAS-CD4-tdGFP/UAS-HA-ATXN3tr-27Q]. Red-colored arrowheads indicate cell bodies of C4da neurons. Scale bar = 100 μm. (D) Penetrance of dendrite defects at 120 h AEL and 18 h APF. Values are presented as mean ± SD. ****P < 0.0001 by Fisher’s exact test; ns, not significant. The number of neurons for each genotype is shown in the figure, and the number of animals is shown below in parentheses. (E) Subcellular localization of overexpressed HA-ATXN3tr-27Q proteins in adult brain neurons along aging (1 day, 10 days, and 20 days adults) [+/+;elav-GAL4/UAS-HA-ATXN3tr-27Q]. Red-colored box indicates imaging area of adult brain neurons. (F) Comparison of survival rates over time between control flies and flies pan-neuronally expressing HA-ATXN3tr-27Q and HA-ATXN3tr-78Q [WT, +/+;elav-GAL4/+, ATXN3tr-27Q, +/+;elav-GAL4/UAS-HA-ATXN3tr-27Q, ATXN3tr-78Q, +/+;elav-GAL4/UAS-HA-ATXN3tr-78Q].
Given that ATXN3tr-78Q conferring strong neuronal toxicity predominantly accumulates in the nucleus of neurons (Kwon et al., 2018; Lee et al., 2011; Park et al., 2020a), we suspected that nuclear accumulated ATXN3tr-27Q may also induce ectopic neuronal toxicity. Supporting this possibility, a previous study reported that overexpression of ATXN3tr-27Q in C4da neurons induced defects in neuronal remodeling during pupal stage when ATXN3tr-27Q translocates into the nucleus (Chung et al., 2017). To confirm this, we examined dendrite phenotypes of C4da neurons expressing ATXN3tr-27Q during larval and pupal stages. We confirmed that overexpression of ATXN3tr-27Q induced obvious defects in dendrite pruning at 18 h after pupal formation (18 h APF) (Figs. 1C and 1D), while overexpression of ATXN3tr-27Q did not induced any noticeable changes in dendrite arborization at the larval stage (Figs. 1C and 1D) as previously reported (Chung et al., 2017). Then, we also examined dendrite phenotypes of C4da neurons expressing ATXN3tr-27Q at adult stage. Because overexpression of ATXN3tr-27Q during pupal stage induced obvious defects in neuronal remodeling, we used inducible expression system (Supplementary Fig. S2A) to check the effect of nuclear translocation of ATXN3tr-27Q only during adult stage on dendrites with excluding the preceding effects during pupal stage. We confirmed that inducibly expressed ATXN3tr-27Q also translocated into the nucleus of C4da neurons in adult flies along aging (Supplementary Fig. S2B), similar to constantly expressed ATXN3tr-27Q through development (Fig. 1A). We found that ATXN3tr-27Q can induce obvious changes in dendrite morphology of C4da neurons in aged adult (45 days after hatching from pupa) upon nuclear accumulation, compared to the control (Supplementary Fig. S2C). These results indicate that ATXN3tr-27Q can induce ectopic neuronal toxicity in C4da neurons upon nuclear accumulation.
Next, we asked whether the observed dynamic NCT of ATXN3tr-27Q in C4da neurons also occurs in other neuronal cell types in Drosophila. To this end, we overexpressed ATXN3tr-27Q in whole neuronal cells and examined subcellular localization of ATXN3tr-27Q in adult fly brains at three different time points (1 day, 10 days, and 20 days after eclosion from pupa). Consistent with the findings in C4da neurons (Fig. 1A), ATXN3tr-27Q primarily localized in the cytosol of neurons of young adult brains (1 day and 10 days) and translocated into the nucleus along aging (20 days) (Fig. 1E). These results indicate that ATXN3tr-27Q undergoes context-dependent NCT generally in Drosophila neurons. Then, we wondered whether dynamic changes of ATXN3tr-27Q NCT in whole neurons of adult brains can also induce obvious phenotypes, as was shown for dendrite pruning defects in C4da neurons (Fig. 1C). To this end, we examined survival of adult flies overexpressing ATXN3tr-27Q or the pathogenic form of ATXN3, ATXN3tr-78Q and compared the results with those of controls. We found that pan-neuronal overexpression of ATXN3tr-27Q significantly affected the survival of flies compared to the control (Fig. 1F). However, the degree of survival defects caused by ATXN3tr-27Q was relatively milder than that caused by ATXN3tr-78Q (Fig. 1F). Notably, the survival curve of flies expressing ATXN3tr-27Q abruptly declines around 20 days when ATXN3tr-27Q translocates into the nucleus (Fig. 1F), further supporting that ATXN3tr-27Q induces ectopic neuronal toxicity upon nuclear accumulation in Drosophila. Taken together, these results demonstrate that ATXN3tr-27Q undergoes context-dependent nucleocytoplasmic translocation in Drosophila neurons similar to TDP-43 (Park et al., 2020a) and induces neuronal toxicity upon nuclear accumulation.
Context-dependent NCT of ATXN3tr-27Q is regulated by the level of intracellular calcium and involves Imp α3 in Drosophila neurons
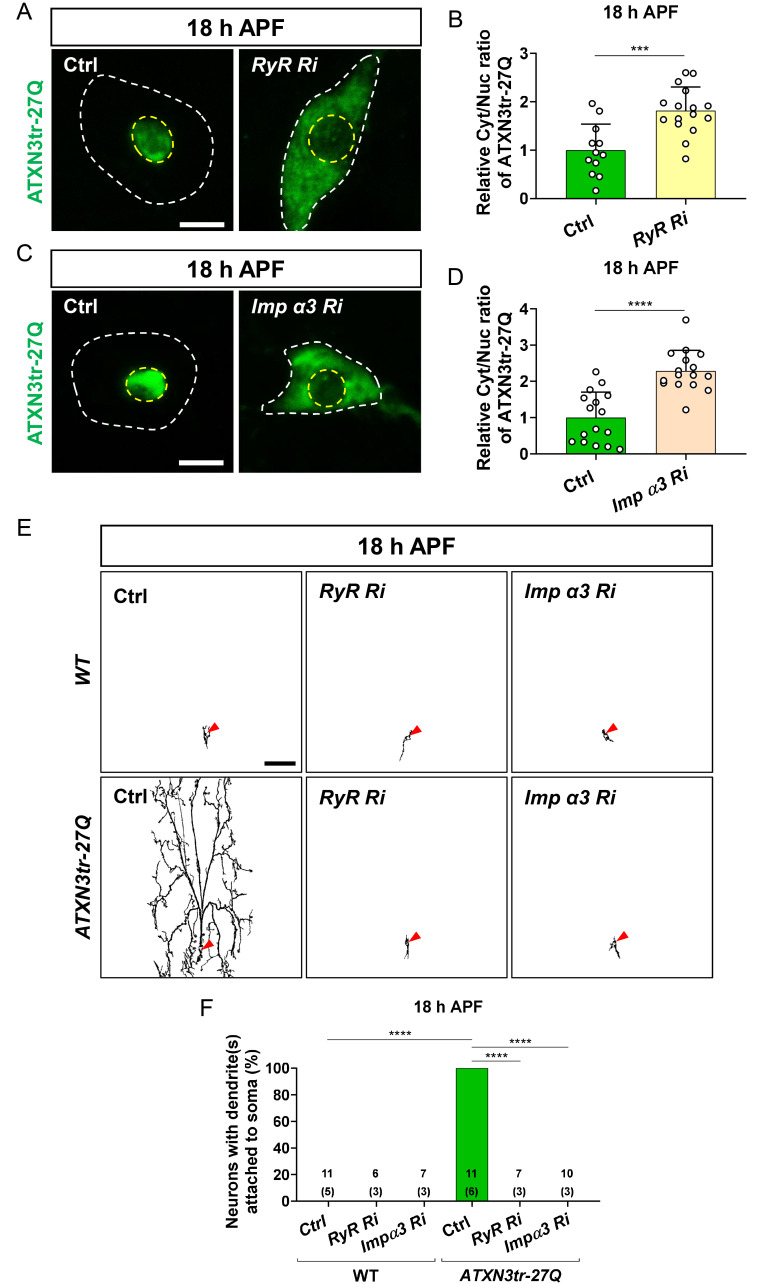
In a previous study, we showed that intracellular calcium level is a critical regulator of NCT of TDP-43 in C4da neurons (Park et al., 2020a). Based on this, we explored whether the level of intracellular calcium is also crucial for NCT of ATXN3tr-27Q in C4da neurons. To this end, we genetically manipulated intracellular calcium level in C4da neurons expressing ATXN3tr-27Q and then examined subcellular localization of ATXN3tr-27Q as well as neuronal phenotypes. We used RNAi of ryanodine receptor (RyR), one of major calcium-releasing channel in the endoplasmic reticulum (ER), to lower intracellular calcium level of C4da neurons. Knockdown (KD) of RyR inhibited nuclear translocation of ATXN3tr-27Q during pupal stage (Figs. 2A and 2B), implicating that intracellular calcium level may be also critical for NCT of ATXN3tr-27Q in C4da neurons similar to TDP-43 (Park et al., 2020a). To support these data, we knocked down Inositol 1,4,5-trisphosphate receptor (IP3R), calcium-releasing channel in the ER, in C4da neurons expressing ATXN3tr-27Q and observed significant prevention of nuclear translocation of ATXN3tr-27Q compared to control during pupal stage (Supplementary Figs. S3A and S3B). In addition, we employed chemical manipulation of intracellular calcium level through feeding of 1 mM 1, 2-bis (o-aminophenoxy) ethane-N,N,N′,N′-tetraacetic acid (BAPTA) for chelating calcium ions. BAPTA feeding (i.e., decreasing intracellular calcium level) led to inhibition of nuclear translocation of ATXN3tr-27Q during pupal stage (Supplementary Fig. S3C), consistent with our above results obtained with genetic manipulation of intracellular calcium (Fig. 2A).
Fig. 2. Nuclear accumulation of ATXN3tr-27Q inducing dendrite remodeling defects during metamorphosis is regulated by the intracellular calcium level and involves Imp α3 in neurons.
(A) Subcellular localization of overexpressed HA-ATXN3tr-27Q proteins in C4da neurons of control (Ctrl) or expressing RyR Ri at 18 h APF [Ctrl, +/+;ppk1a-GAL4/UAS-HA-ATXN3tr-27Q, RyR Ri, UAS-RyR RNAi/+;ppk1a-GAL4/UAS-HA-ATXN3tr-27Q]. Outer and inner dashed lines indicate borders of cell bodies and nuclei, respectively. Scale bar = 5 μm. (B) Quantification of Cyt/Nuc ratio of HA-ATXN3tr-27Q proteins in C4da neurons of Ctrl or expressing RyR Ri at 18 h APF. Values are presented as mean ± SD. ***P = 0.0003 by two-tailed t-test; n = 12 neurons for Ctrl, n = 16 neurons for for RyR Ri. (C) Subcellular localization of overexpressed HA-ATXN3tr-27Q in C4da neurons of Ctrl or expressing Imp α3 Ri at 18 h APF [Ctrl, +/+;ppk1a-GAL4/UAS-HA-ATXN3tr-27Q, Imp α3 Ri, UAS-Imp α3 RNAi/+;ppk1a-GAL4/UAS-HA-ATXN3tr-27Q]. Outer and inner dashed lines indicate borders of cell bodies and nuclei, respectively. Scale bar = 5 μm. (D) Quantification of Cyt/Nuc ratio of HA-ATXN3tr-27Q proteins in C4da neurons of Ctrl or expressing Imp α3 Ri at 18 h APF. Values are presented as mean ± SD. ****P < 0.0001 by two-tailed t-test; n = 19 neurons for Ctrl, n = 19 neuron for Imp α3 Ri. (E) Skeletonized dendrite images of C4da neurons of 18 h APF [Ctrl in WT, +/+;ppk1a-GAL4>UAS-CD4-tdGFP/+, RyR Ri in WT, UAS-RyR RNAi/+;ppk1a-GAL4>UAS-CD4-tdGFP/+, Imp α3 Ri in WT, UAS-Imp α3 RNAi/+;ppk1a-GAL4>UAS-CD4-tdGFP/+, Ctrl in ATXN3tr-27Q, +/+;ppk1a-GAL4>UAS-CD4-tdGFP/UAS-HA-ATXN3tr-27Q, RyR Ri in ATXN3tr-27Q, UAS-RyR RNAi/+;ppk1a-GAL4>UAS-CD4-tdGFP/UAS-HA-ATXN3tr-27Q, Imp α3 Ri in ATXN3tr-27Q, UAS-Imp α3 RNAi/+;ppk1a-GAL4>UAS-CD4-tdGFP/UAS-HA-ATXN3tr-27Q]. Scale bar = 50 μm. (F) Penetrance of dendrite defects at 18 h APF. Values are presented as mean ± SD. ****P < 0.0001 by Fisher’s exact test. The number of neurons for each genotype is shown in the figure, and the number of animals is shown below in parentheses.
Next, we examined whether NCT of ATXN3tr-27Q involves Imp α3 that has been identified as a key karyopherin mediating nuclear translocation of TDP-43 (Park et al., 2020a). To this end, we knocked down Imp α3 in C4da neurons expressing ATXN3tr-27Q and examined subcellular localization of ATXN3tr-27Q during pupal stage. KD of Imp α3 inhibited nuclear translocation of ATXN3tr-27Q during pupal stage (Figs. 2C and 2D). These results collectively indicate that NCT of ATXN3tr-27Q is regulated by the level of intracellular calcium and involves Imp α3 in C4da neurons.
Given that ATXN3tr-27Q induces ectopic neuronal toxicity upon nuclear accumulation (Fig. 1), we then investigated whether inhibition of ATXN3tr-27Q nuclear translocation during pupal stage can protect neurons from pruning defects. Inhibition of ATXN3tr-27Q nuclear translocation by KD of either RyR or Imp α3 showed dramatic effects in the prevention of pruning defects during pupal stage (Figs. 2E and 2F). These results suggest that ectopic neuronal toxicity of ATXN3tr-27Q can be prevented or suppressed by inhibition of its nuclear translocation. Taken together, these results demonstrate that ATXN3tr-27Q share, at least partially, the regulatory mechanism of NCT with TDP-43 (Park et al., 2020a) in Drosophila neurons.
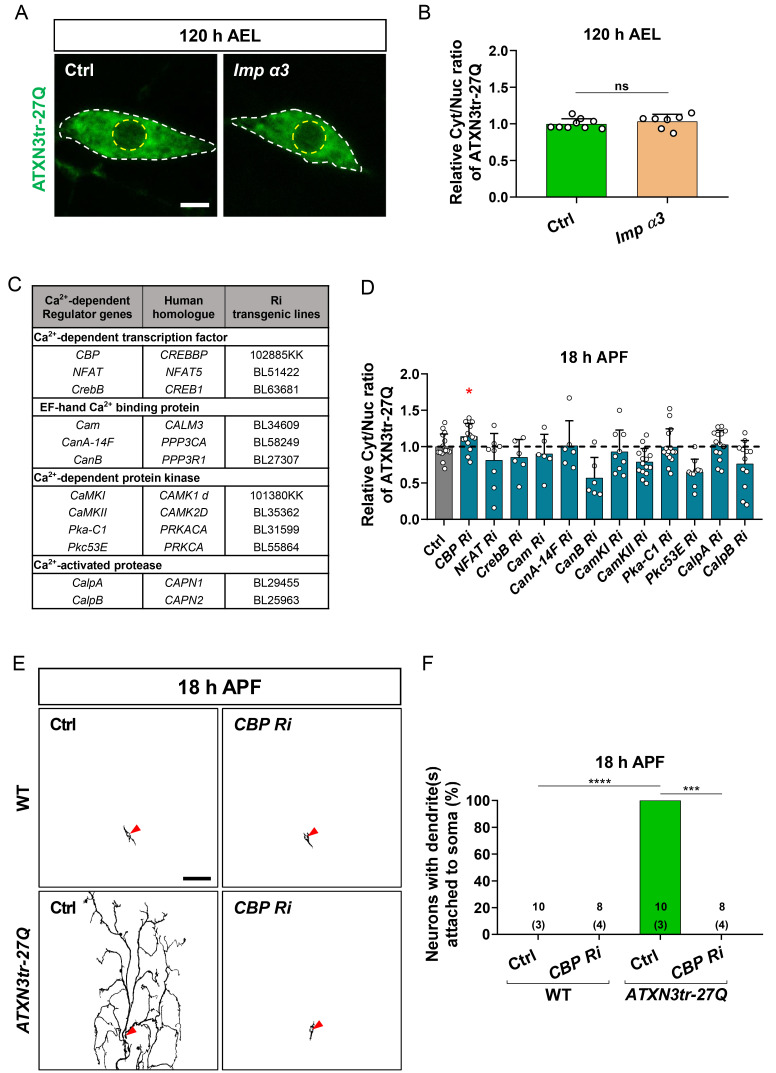
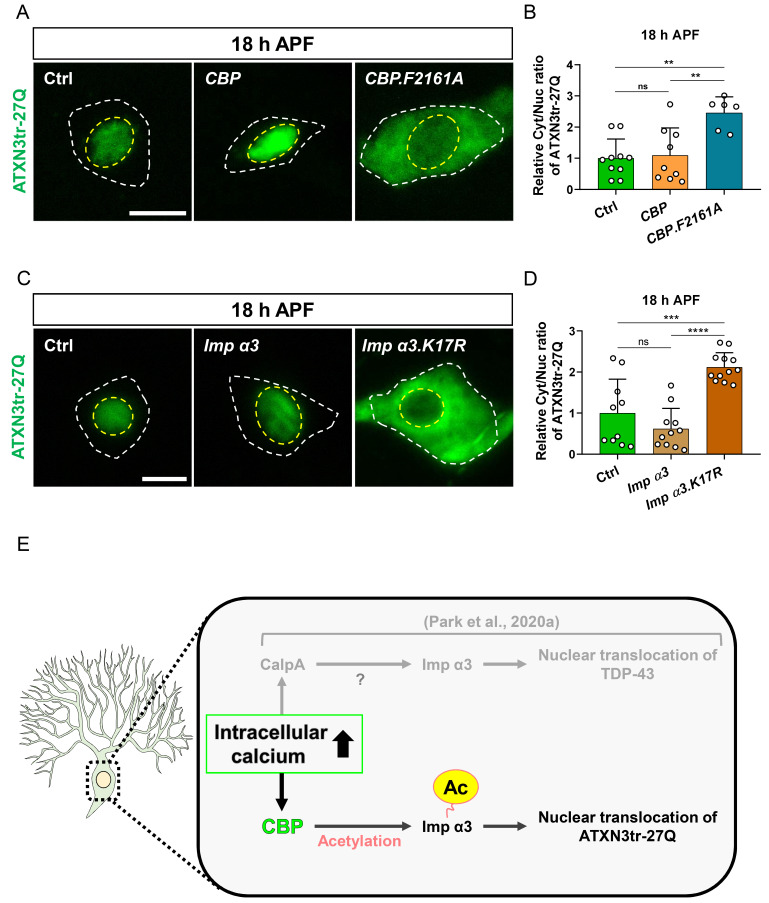
CBP mediates intracellular calcium-dependent NCT of ATXN3tr-27Q in Drosophila neurons
Given the involvement of Imp α3 in NCT of ATXN3tr-27Q, we speculated that overexpression of Imp α3 may facilitate nuclear translocation of ATXN3tr-27Q during larval stage, as shown for TDP-43 in a previous study (Park et al., 2020a). Interestingly, we found that overexpression of Imp α3 did not induce nuclear translocation of ATXN3tr-27Q during larval stage different from TDP-43 (Figs. 3A and 3B), implicating that NCT of ATXN3tr-27Q may involve certain mediators between intracellular calcium and Imp α3 that are different from those involved in NCT of TDP-43. Thus, we screened available calcium-dependent regulators using RNAi lines, as previously described (Park et al., 2020a), to identify specific mediators of NCT of ATXN3tr-27Q (Fig. 3C). We expressed RNAi line of calcium-dependent regulators in C4da neurons expressing ATXN3tr-27Q and examined whether nuclear accumulation of ATXN3tr-27Q is affected by KD of these regulators during pupal stage. For NCT of TDP-43, a previous study identified CalpA as one of critical mediators between intracellular calcium and Imp α3 (Park et al., 2020a). Notably, our genetic screening identified that CBP is primarily involved in NCT of ATXN3tr-27Q (Fig. 3D). Then, we examined whether genetic manipulation of CBP can alter neuronal phenotypes caused by nuclear accumulation of ATXN3tr-27Q. KD of CBP prevented pruning defects caused by ATXN3tr-27Q nuclear accumulation during pupal stage (Figs. 3E and 3F). Taken together, we identified CBP as a new calcium-dependent regulator that specifically mediates NCT of ATXN3tr-27Q in Drosophila neurons.
Fig. 3. Calcium-dependent NCT of ATXN3tr-27Q is mediated by CBP in neurons.
(A) Subcellular localization of overexpressed HA-ATXN3tr-27Q proteins in C4da neurons of control (Ctrl) or expressing Imp α3 at 120 h AEL [Ctrl, +/+;ppk1a-GAL4/UAS-HA-ATXN3tr-27Q, Imp α3, UAS-2xFlag-Imp α3/+;ppk1a-GAL4/UAS-HA-ATXN3tr-27Q]. Outer and inner dashed lines indicate borders of cell bodies and nuclei, respectively. Scale bar = 5 μm. (B) Quantification of Cyt/Nuc ratio of HA-ATXN3tr-27Q proteins in C4da neurons of Ctrl or expressing Imp α3 at 120 h AEL. Values are presented as mean ± SD. P > 0.05 (P = 0.4235) by two-tailed t-test; ns, not significant; n = 9 for Ctrl, n = 7 for Imp α3. (C) List of intracellular calcium-dependent regulators screened in this study. (D) Quantification of Cyt/Nuc ratio of HA-ATXN3tr-27Q proteins in C4da neurons of Ctrl or expressing denoted transgenes described in C. [Ctrl, ppk-Gal4/+;UAS-HA-ATXN3tr-27Q, CBP Ri, ppk-Gal4/UAS-CBP RNAi;UAS-HA-ATXN3tr-27Q, NFAT Ri, ppk-Gal4/+;UAS-HA-ATXN3tr-27Q/UAS-NFAT RNAi, CrebB Ri, ppk-Gal4/UAS-CrebB RNAi;UAS-HA-ATXN3tr-27Q/+, Cam Ri, ppk-Gal4/+;UAS-HA-ATXN3tr-27Q/UAS-Cam RNAi, CanA-14F Ri, ppk-Gal4/CanA-14F;UAS-HA-ATXN3tr-27Q/+, CanB Ri, ppk-Gal4/+;UAS-HA-ATXN3tr-27Q/UAS-CanB RNAi, CaMKI Ri, ppk-Gal4/UAS-CaMKI RNAi;UAS-HA-ATXN3tr-27Q/+, CaMKII Ri, ppk-Gal4/+;UAS-HA-ATXN3tr-27Q/UAS-CamKII RNAi, Pka-C1 Ri, ppk-Gal4/+;UAS-HA-ATXN3tr-27Q/UAS-Pka-C1 RNAi, Pkc53E Ri, ppk-Gal4/+;UAS-HA-ATXN3tr-27Q/UAS-Pkc53E RNAi, CalpA Ri, ppk-Gal4/+;UAS-HA-ATXN3tr-27Q/CalpA Ri, CalpB Ri, ppk-Gal4/+; UAS-HA-ATXN3tr-27Q/UAS-CalpB RNAi]. Values are presented as mean ± SD. *P = 0.0358 by two-tailed t-test; n = 15 for Ctrl, n = 15 for CBP Ri, n = 8 for NFAT Ri, n = 6 for CrebB Ri, n = 6 for Cam Ri, n = 6 for CanA-14F Ri, n = 6 for CanB Ri, n = 9 for CamKI Ri, n = 15 for CamKII Ri, n = 15 for Pka-C1 Ri, n = 10 for Pkc53E Ri, n= 15 for CalpA Ri, n = 12 for CalpB Ri. (E) Skeletonized dendrite images of C4da neurons of 18 h APF [Ctrl in WT, +/+;ppk1a-GAL4>UAS-CD4-tdGFP/+, CBP Ri in WT, UAS-CBP RNAi/+; ppk1a-GAL4>UAS-CD4-tdGFP/+, Ctrl in ATXN3tr-27Q, +/+;ppk1a-GAL4>UAS-CD4-tdGFP/UAS-HA-ATXN3tr-27Q, CBP Ri in ATXN3tr-27Q, UAS-CBP RNAi/+;ppk1a-GAL4>UAS-CD4-tdGFP/UAS-HA-ATXN3tr-27Q]. Scale bar = 50 μm. (F) Penetrance of dendrite defects at 18 h APF. Values are presented as mean ± SD. ****P < 0.0001 by Fisher’s exact test. The number of neurons for each genotype is shown in the figure, and the number of animals is shown below in parentheses.
CBP-dependent acetylation of Imp α3 is crucial for NCT of ATXN3tr-27Q in Drosophila neurons
CBP is known as a HAT (histone acetyltransferase) that regulates expression profiles of genome through post-translational modification of histones (Bannister and Kouzarides, 1996). Notably, it is also known that CBP can acetylate target proteins other than histones, such as a subset of Importins (Importin α7 and Rch1) (Bannister et al., 2000) and HNF4 (hepatocyte nuclear factor 4) (Soutoglou et al., 2000). To understand how CBP mediates calcium-dependent NCT of ATXN3tr-27Q in C4da neurons, we first asked whether CBP-dependent acetylation of histones is involved in this process. To this end, we knocked down several histone deacetylases (HDACs) that counteract function of CBP on histone modification (Gaub et al., 2010; Saha and Pahan, 2006) and examined subcellular localization of ATXN3tr-27Q at the larval stage. We found that KD of HDACs such as HDAC1, HDAC3, and HDAC4 did not induce noticeable changes in subcellular localization of ATXN3tr-27Q in C4da neurons at the larval stage (Supplementary Fig. S4), indicating that CBP-dependent acetylation of histones is not involved in NCT of ATXN3tr-27Q.
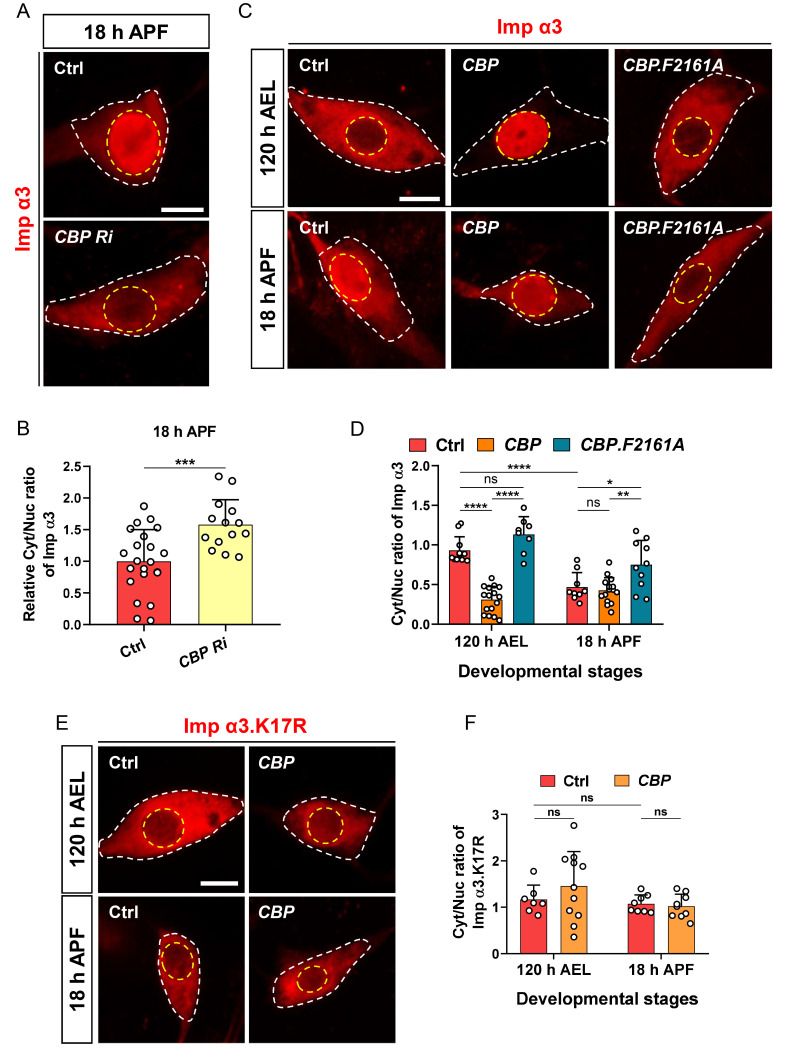
Then, to determine whether CBP has a regulatory function on Imp α3, we examined the effect of CBP KD on the subcellular localization of Imp α3 during development. Drosophila Imp α3 primarily localized in the cytosol during larval stage and then highly translocated into the nucleus during pupal stage in C4da neurons (Park et al., 2020a). Based on this study, we first knocked down CBP in C4da neurons expressing Imp α3 and examined nucleocytoplasmic localization of Imp α3 during pupal stage. KD of CBP significantly prevented nuclear translocation of Imp α3 during pupal stage (Figs. 4A and 4B). We next asked whether enzyme activity of CBP is critical for the regulatory role of CBP on Imp α3. To address this, we overexpressed both wild-type and an enzymatic-dead form of CBP, CBP.F2161A, in C4da neurons expressing Imp α3 and examined whether NCT of Imp α3 is regulated by enzymatic activity of CBP. At larval stage, CBP.F2161A overexpression failed to induce nuclear translocation of Imp α3 in C4da neurons, while CBP overexpression induced its nuclear translocation (Figs. 4C and 4D). At pupal stage, CBP.F2161A overexpression significantly prevented nuclear translocation of Imp α3 (Figs. 4C and 4D), implicating that CBP.F2161A acts as a dominant-negative form of CBP. These results demonstrate that the acetyltransferase activity of CBP is crucial for NCT of Imp α3 in Drosophila neurons, which may be involved in the calcium-dependent NCT of ATXN3tr-27Q as a cargo of Imp α3.
Fig. 4. CBP regulates NCT of Drosophila Imp α3 via acetyltransferase activity in neurons.
(A) Subcellular localization of overexpressed Flag-Imp α3 proteins in C4da neurons of control (Ctrl) or expressing CBP Ri at 18 h APF [Ctrl, UAS-2xFlag-Imp α3/+;ppk1a-Gal4/+, CBP Ri, UAS-2xFlag-Imp α3/UAS-CBP RNAi;ppk1a-Gal4/+]. Outer and inner dashed lines indicate borders of cell bodies and nuclei, respectively. Scale bar = 5 μm. (B) Quantification of Cyt/Nuc ratio of Flag-Imp α3 proteins in C4da neurons of Ctrl or expressing CBP Ri at 18 h APF. Values are presented as mean ± SD. ***P = 0.0009 by two-tailed t-test; n = 22 for Ctrl, n = 14 for CBP Ri. (C) Subcellular localization of overexpressed Flag-Imp α3 proteins in C4da neurons of Ctrl or expressing CBP or CBP.F2161A at 120 h AEL and 18 h APF [Ctrl, UAS-2xFlag-Imp α3/+;ppk1a-Gal4/+, CBP, UAS-2xFlag-Imp α3/+;ppk1a-Gal4/UAS-CBP.wt-V5, CBP.F2161A, UAS-2xFlag-Imp α3/UAS-CBP.F2161A-V5;ppk1a-Gal4/+]. Outer and inner dashed lines indicate borders of cell bodies and nuclei, respectively. Scale bar = 5 μm. (D) Quantification of Cyt/Nuc ratio of Flag-Imp α3 proteins in C4da neurons of Ctrl or expressing CBP or CBP.F2161A at 120 h AEL and 18 h APF. Values are presented as mean ± SD. P = 0.28, ****P < 0.0001, **P = 0.0025, *P = 0.0377 by two-way ANOVA with Tukey’s post hoc test; ns, not significant; n = 11 for Ctrl at 120 h AEL, n = 16 for CBP at 120 h AEL, n = 8 for CBP.F2161A at 120 h AEL, n = 9 for Ctrl at 18 h APF, n = 15 for CBP at 18 h APF, n = 10 for CBP.F2161A at 18 h APF. (E) Subcellular localization of overexpressed Flag-Imp α3.K17R proteins in C4da neurons of Ctrl or expressing CBP at 120 h AEL and 18 h APF [Ctrl, UAS-2xFlag-Imp α3.K17R/+;ppk1a-Gal4/+, CBP, UAS-2xFlag-Imp α3.K17R/+;ppk1a-Gal4/UAS-CBP.wt-V5]. Outer and inner dashed lines indicate borders of cell bodies and nuclei, respectively. Scale bar = 5 μm. (F) Quantification of Cyt/Nuc ratio of Flag-Imp α3.K17R proteins in C4da neurons of Ctrl or expressing CBP at 120 h AEL and 18 h APF. Values are presented as mean ± SD. P > 0.3 by two-way ANOVA with Tukey’s post hoc test; ns, not significant; n = 7 for Ctrl at 120 h AEL, n = 11 for CBP at 120 h AEL, n = 8 for Ctrl at 18 h APF, n = 9 for CBP at 18 h APF.
Notably, a previous study reported that CBP acetylates a subset of Imp α in cultured mammalian cells (Bannister et al., 2000). Although a previous study reported that CBP does not acetylate the mammalian homolog of Drosophila Imp α3 (Bannister et al., 2000), we found that the mammalian Imp α3 does not contain “FKNKGK” sequence known to be acetylated by CBP (Bannister et al., 2000). Interestingly, the “FKNKGK” sequence is present in the Importin-β (Imp β)-binding (IBB) domain of Drosophila Imp α3 (Supplementary Fig. S5A), similar to mammalian Rch1 that is known to be acetylated by CBP (Bannister et al., 2000), suggesting that CBP can acetylate Drosophila Imp α3 to mediate NCT of ATXN3tr-27Q. To address this possibility, we generated a new transgene of Drosophila Imp α3 (Imp α3.K17R) that contains a point mutation resulting in the replacement of K17 with R within “FKNKGK” sequence of the encoded proteins (Supplementary Fig. S5B), thereby making the encoded proteins unable to be acetylated by CBP (Bannister et al., 2000). Using this new transgene, we examined whether CBP-dependent regulation of NCT of Drosophila Imp α3 is abolished by the K17R point mutation. At the larval stage, CBP overexpression failed to induce nuclear translocation of Imp α3.K17R (Figs. 4E and 4F), unlike Imp α3 (Figs. 4C and 4D). Moreover, Imp α3.K17R did not undergo nuclear translocation during pupal stage (Figs. 4E and 4F), different from Imp α3 (Figs. 4C and 4D), where overexpression of CBP failed to induce nuclear translocation of Imp α3.K17R (Figs. 4E and 4F). These results collectively suggest that CBP-dependent acetylation of Imp α3 is crucial for its nuclear import in Drosophila neurons.
Next, we decided to verify that CBP-mediated acetylation of Imp α3 described above (Fig. 4) is involved in the calcium-dependent NCT of ATXN3tr-27Q in C4da neurons. To this end, we first overexpressed either CBP or CBP.F2161A in C4da neurons expressing ATXN3tr-27Q and examined whether enzymatic activity of CBP is important for the subcellular localization of ATXN3tr-27Q. Overexpression of CBP.F2161A acting as a dominant-negative form indeed prevented nuclear translocation of ATXN3tr-27Q during pupal stage, while overexpression of CBP induced nuclear translocation of ATXN3tr-27Q (Figs. 5A and 5B). Then, we explored whether the status of CBP-mediated acetylation in Imp α3 is associated with NCT of ATXN3tr-27Q by comparing the effects of overexpressed Imp α3 with overexpressed Imp α3.K17R on subcellular localization of ATXN3tr-27Q in C4da neurons. We found that overexpression of Imp α3.K17R prevented nuclear translocation of ATXN3tr-27Q during pupal stage (Figs. 5C and 5D), consistent with the above results using overexpression of CBP.F2161A (Figs. 5A and 5B). Taken together, these results demonstrate that CBP-mediated acetylation of Drosophila Imp α3, which contains the conserved “FKNKGK” sequence, is crucial for intracellular calcium-dependent NCT of ATXN3tr-27Q, which distinguish NCT of ATXN3tr-27Q from that of TDP-43 (Park et al., 2020a) (Fig. 5E).
Fig. 5. CBP-mediated acetylation of Imp α3 is crucial for NCT of ATXN3tr-27Q in neurons.
(A) Subcellular localization of overexpressed HA-ATXN3tr-27Q proteins in C4da neurons of control (Ctrl) or expressing CBP or CBP.F2161A at 18 h APF [Ctrl, ppk-Gal4/+; UAS-HA-ATXN3tr-27Q/+, CBP, ppk-gal4/+;UAS-HA-ATXN3tr-27Q/UAS-CBP.wt-V5, CBP.F2161A, ppk-Gal4/UAS-CBP.F2161A-V5;HA-ATXN3tr-27Q/+]. Outer and inner dashed lines indicate borders of cell bodies and nuclei, respectively. Scale bar = 5 μm. (B) Quantification of Cyt/Nuc ratio of HA-ATXN3tr-27Q proteins in C4da neurons of Ctrl or expressing CBP or CBP.F2161A at 18 h APF. Values are presented as mean ± SD. P = 0.9498, **P < 0.004 by one-way ANOVA with Tukey’s post hoc test; ns, not significant; n = 10 for Ctrl, n = 9 for CBP, n = 6 for CBP.F2161A. (C) Subcellular localization of overexpressed HA-ATXN3tr-27Q proteins in C4da neurons of Ctrl or expressing Flag-Imp α3 or Flag-Imp α3.K17R at 18 h APF [Ctrl, +/+;ppk1a-Gal4/UAS-HA-ATXN3tr-27Q, Imp α3, UAS-2xFlag-Imp α3/+;ppk1a-Gal4/HA-ATXN3tr-27Q, Imp α3.K17R, UAS-2xFlag-Imp α3.K17R/+;ppk1a-Gal4/UAS-HA-ATXN3tr-27Q]. Outer and inner dashed lines indicate borders of cell bodies and nuclei, respectively. Scale bar = 5 μm. (D) Quantification of Cyt/Nuc ratio of HA-ATXN3tr-27Q proteins in C4da neurons of Ctrl or expressing Flag-Imp α3 or Flag-Imp α3.K17R at 18 h APF. Values are presented as mean ± SD. P = 0.2972, ***P = 0.0002, ****P < 0.0001 by one-way ANOVA with Tukey’s post hoc test; ns, not significant; n = 10 for Ctrl, n = 11 for Imp α3, n = 12 for Imp α3.K17R. (E) A schematic illustration for the molecular mechanism regulating NCT of ATXN3tr-27Q proteins.
DISCUSSION
In this study, we demonstrate that CBP-mediated acetylation of Imp α3 is crucial for intracellular calcium-dependent NCT of ATXN3tr-27Q in Drosophila neurons. Interestingly, the regulatory mechanism of NCT of ATXN3tr-27Q appears to be partly different from that of TDP-43 (Park et al., 2020a). For NCT of ATXN3tr-27Q, CBP-mediated acetylation of Imp α3 is critical, while for NCT of TDP-43, CalpA and several other calcium-dependent regulators, except for CBP, mediate calcium-dependent regulation of Imp α3, potentially through an uncharacterized mechanism(s) different from acetylation (Park et al., 2020a). Since NCT of both ATXN3tr-27Q and TDP-43 (Park et al., 2020a) in Drosophila neurons is critically regulated by intracellular calcium level and involves Imp α3 mediating nuclear transport of a broad range of proteins containing NLS sequences, it can be speculated that NCT of many other proteins, particularly disease-related proteins for NDs, may be similarly regulated by intracellular calcium level. Given that different sets of mediators are engaged in NCT of ATXN3tr-27Q and TDP-43 (Park et al., 2020a), there may exist a certain target-selective mechanism(s) at the downstream of intracellular calcium. Although the current study could not characterize how these two targets sharing classical NLS (cNLS) sequence are differentially recognized, our findings suggest that certain molecular features of proteins may predispose at least one of multiple regulatory mechanisms for their NCT. Notably, we found that KD of CBP also affected NCT of NLS-tagged DsRed proteins (Supplementary Fig. S6A), suggesting that CBP-dependent acetylation of Imp α3 is involved in NCT of other proteins containing cNLS sequence beside ATXN3tr-27Q in Drosophila neurons. In addition, we also found that inducible overexpression of Imp α3.K17R in C4da neurons of adult flies prevented nuclear translocation of NES-NLS-tagged GFP (Supplementary Fig. S6B), further supporting the notion that identified regulatory mechanism for NCT of proteins can be generalized, to a certain extent, in Drosophila neurons. These additional findings implicate that at least CBP-mediated acetylation of Imp α can serve as one default mode-of-regulatory mechanism shared for NCT of a group of proteins in Drosophila neurons. Further studies focused on the exact nature of a target-selective mechanism(s) will provide invaluable insights into the understanding of delicate and dynamic control of protein function and localization in response to the changes in cellular context.
To figure out the target-selective nature of calcium-dependent regulatory mechanisms underlying NCT of these disease-related proteins, we need to also understand how CBP-dependent acetylation affects molecular features of Imp α. IBB domain containing the known acetylation site of Imp α is required for the interaction with Imp β (Bannister et al., 2000). According to a previous study, the acetylation of Imp α is known to increase the binding affinity to Imp β. From these previous results, we speculate that the enhanced formation of heterodimer between Imp α and Imp β may be responsible for target selectivity distinguishing between TDP-43 and ATXN3. Future studies on the molecular details underlying the target selectivity of CBP-dependent acetylation of Imp α are highly demanded.
In this study, we demonstrate that nuclear accumulation of ATXN3tr-27Q can induce ectopic neuronal toxicity (e.g., aberrant neuronal remodeling during metamorphosis and morphological changes of dendrites at the adult stage) in Drosophila neurons. ATXN3tr-27Q was generally used as a control to study protein toxicity caused by a mutated polyQ form of ATXN3 (ATXN3tr-78Q) (Chung et al., 2017; Kwon et al., 2018; Saitoh et al., 2015; Warrick et al., 1998). Our finding has potential implication that context-dependent changes in NCT of ATXN3 proteins may result in ectopic toxicity unrelated to the pathogenic mutation of ATXN3 proteins during development and neuronal aging. Thus, it seems possible that aging-dependent nuclear accumulation of ATXN3 proteins, potentially together with other mislocalized proteins regulated by same NCT mechanism, can contribute to the aging-dependent impairment of neuronal functions. However, in the physiological condition, the endogenous protein level of truncated ATXN3 may be far less than that of overexpressed ATXN3tr-27Q used in our study. Thus, it seems more likely that the toxic effect of nuclear accumulated truncated ATXN3 by itself may be not enough to induce specific disease in humans. Instead, our additional data (Supplementary Figs. S6C and S6D) raise an alternative possibility that calcium-mediated NCT of ATXN3tr-27Q can have certain pathological roles in polyQ diseases. Co-overexpression of ATXN3tr-27Q with expanded polyQ forms of truncated ATXN3 (ATXN3tr-78Q) or truncated huntingtin (Htt-152Q) induced ectopic translocation of ATXN3tr-27Q into the protein aggregates in a dorsal cluster of da neurons during larval stage (Supplementary Figs. S6C and S6D), suggesting that nuclear accumulated ATXN3tr-27Q can participate in the multimerization of toxic polyQ proteins. Given that multimerization of mutated polyQ proteins is associated with polyQ toxicity, we surmise that calcium-mediated nuclear accumulation of truncated ATXN3 may contribute to the protein toxicity of nuclear accumulated mutated polyQ proteins by facilitating their multimerization in the nucleus of neurons. Thus, we speculate that calcium-mediated nuclear accumulation of truncated ATXN3 may have limited pathological roles likely through contributing to the protein toxicity primarily attributed to mutated polyQ proteins, rather than inducing ectopic toxicity enough to elicit specific diseases by itself in humans.
We showed here that reducing intracellular calcium concentration can prevent ectopic neuronal toxicity induced by nuclear accumulation of ATXN3tr-27Q. In our study, we primarily used the genetic manipulation of intracellular calcium level (Figs. 2A and 2B), but we also showed that the use of BAPTA can lead to the effects in the regulation of NCT of ATXN3tr-27Q (Supplementary Fig. S2C) comparable to the effects of genetic manipulation. This implies that pharmacochemical approach to prevent ectopic neuronal toxicity induced by mislocalized proteins under the control of intracellular calcium level may be feasible. For the functional homeostasis of neurons, there are distinct mechanisms regulating intracellular calcium levels that involve concerted actions of plasma membrane calcium channels and intracellular calcium reservoirs such as ER and mitochondria (Bagur and Hajnoczky, 2017; Bootman and Bultynck, 2020). Given that ectopic neuronal toxicity associated with calcium-dependent protein mislocalization can be a contributing factor for either the neuronal disorders or aging-dependent neuronal impairments in humans, diverse chemicals regulating intracellular calcium level through different mode-of-actions need to be tested for their effectiveness and side effects. For example, there exist mitochondria-specific chemicals (MIT-001/NecroX-7 and NecroX-5) that show a strong regulatory effect on the intracellular calcium level without inducing serious side effects (Hwang et al., 2018; Park et al., 2017; Thu et al., 2012), which can be good candidates for the potential future application to humans. We believe that our study will pave a new avenue toward understanding the exact nature of calcium-dependent NCT of proteins in neurons and be useful to develop an effective therapeutic intervention targeting neuronal toxicity associated with dysregulated NCT of proteins.
Supplemental Materials
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
ACKNOWLEDGMENTS
This work was supported by Basic Science Research Program through the National Research Foundation of Korea (2022R1A4A2000703 and 2021R1A2C1003817) and the Korea Brain Research Institute (KBRI) Research Program (22-BR-03-02), funded by the Ministry of Science and ICT, Republic of Korea, and the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI) and Korea Dementia Research Center (KDRC), funded by the Ministry of Health & Welfare and Ministry of Science and ICT, Republic of Korea (HU21C0027).
Footnotes
AUTHOR CONTRIBUTIONS
J.H.C., E.S.K., and S.B.L. wrote the manuscript. J.H.C., M.G.J., and N.Y.L. performed experiments. J.H.C., M.G.J., and N.Y.L. analyzed the data. E.S.K., J.H.P., C.G.C., S.H.K., and S.B.L. provided expertise and feedback. S.B.L. supervised the research.
CONFLICT OF INTEREST
The authors have no potential conflicts of interest to disclose.
REFERENCES
- Bagur R., Hajnoczky G. Intracellular Ca2+ sensing: its role in calcium homeostasis and signaling. Mol. Cell. 2017;66:780–788. doi: 10.1016/j.molcel.2017.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister A.J., Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- Bannister A.J., Miska E.A., Gorlich D., Kouzarides T. Acetylation of importin-alpha nuclear import factors by CBP/p300. Curr. Biol. 2000;10:467–470. doi: 10.1016/S0960-9822(00)00445-0. [DOI] [PubMed] [Google Scholar]
- Bauer N.C., Doetsch P.W., Corbett A.H. Mechanisms regulating protein localization. Traffic. 2015;16:1039–1061. doi: 10.1111/tra.12310. [DOI] [PubMed] [Google Scholar]
- Bichelmeier U., Schmidt T., Hubener J., Boy J., Ruttiger L., Habig K., Poths S., Bonin M., Knipper M., Schmidt W.J., et al. Nuclear localization of ataxin-3 is required for the manifestation of symptoms in SCA3: in vivo evidence. J. Neurosci. 2007;27:7418–7428. doi: 10.1523/JNEUROSCI.4540-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bootman M.D., Bultynck G. Fundamentals of cellular calcium signaling: a primer. Cold Spring Harb. Perspect. Biol. 2020;12:a038802. doi: 10.1101/cshperspect.a038802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuer P., Haacke A., Evert B.O., Wullner U. Nuclear aggregation of polyglutamine-expanded ataxin-3: fragments escape the cytoplasmic quality control. J. Biol. Chem. 2010;285:6532–6537. doi: 10.1074/jbc.M109.036335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung C.G., Kwon M.J., Jeon K.H., Hyeon D.Y., Han M.H., Park J.H., Cha I.J., Cho J.H., Kim K., Rho S., et al. Golgi outpost synthesis impaired by toxic polyglutamine proteins contributes to dendritic pathology in neurons. Cell Rep. 2017;20:356–369. doi: 10.1016/j.celrep.2017.06.059. [DOI] [PubMed] [Google Scholar]
- Chung C.G., Lee H., Lee S.B. Mechanisms of protein toxicity in neurodegenerative diseases. Cell. Mol. Life Sci. 2018;75:3159–3180. doi: 10.1007/s00018-018-2854-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evert B.O., Wullner U., Schulz J.B., Weller M., Groscurth P., Trottier Y., Brice A., Klockgether T. High level expression of expanded full-length ataxin-3 in vitro causes cell death and formation of intranuclear inclusions in neuronal cells. Hum. Mol. Genet. 1999;8:1169–1176. doi: 10.1093/hmg/8.7.1169. [DOI] [PubMed] [Google Scholar]
- Fujigasaki H., Uchihara T., Koyano S., Iwabuchi K., Yagishita S., Makifuchi T., Nakamura A., Ishida K., Toru S., Hirai S., et al. Ataxin-3 is translocated into the nucleus for the formation of intranuclear inclusions in normal and Machado-Joseph disease brains. Exp. Neurol. 2000;165:248–256. doi: 10.1006/exnr.2000.7479. [DOI] [PubMed] [Google Scholar]
- Gaub P., Tedeschi A., Puttagunta R., Nguyen T., Schmandke A., Di Giovanni S. HDAC inhibition promotes neuronal outgrowth and counteracts growth cone collapse through CBP/p300 and P/CAF-dependent p53 acetylation. Cell Death Differ. 2010;17:1392–1408. doi: 10.1038/cdd.2009.216. [DOI] [PubMed] [Google Scholar]
- Haacke A., Broadley S.A., Boteva R., Tzvetkov N., Hartl F.U., Breuer P. Proteolytic cleavage of polyglutamine-expanded ataxin-3 is critical for aggregation and sequestration of non-expanded ataxin-3. Hum. Mol. Genet. 2006;15:555–568. doi: 10.1093/hmg/ddi472. [DOI] [PubMed] [Google Scholar]
- Han M.H., Kwon M.J., Ko B.S., Hyeon D.Y., Lee D., Kim H.J., Hwang D., Lee S.B. NF-kappa B disinhibition contributes to dendrite defects in fly models of neurodegenerative diseases. J. Cell Biol. 2020;219:e202004107. doi: 10.1083/jcb.202004107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I.C., Kim J.Y., Kim J.H., Lee J.E., Seo J.Y., Lee J.W., Park J., Yang H.M., Kim S.H., Cho H.J., et al. Therapeutic potential of a novel necrosis inhibitor, 7-amino-indole, in myocardial ischemia-reperfusion injury. Hypertension. 2018;71:1143–1155. doi: 10.1161/HYPERTENSIONAHA.117.09405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E.S., Chung C.G., Park J.H., Ko B.S., Park S.S., Kim Y.H., Cha I.J., Kim J., Ha C.M., Kim H.J., et al. C9orf72-associated arginine-rich dipeptide repeats induce RNA-dependent nuclear accumulation of Staufen in neurons. Hum. Mol. Genet. 2021;30:1084–1100. doi: 10.1093/hmg/ddab089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon M.J., Han M.H., Bagley J.A., Hyeon D.Y., Ko B.S., Lee Y.M., Cha I.J., Kim S.Y., Kim D.Y., Kim H.M., et al. Coiled-coil structure-dependent interactions between polyQ proteins and Foxo lead to dendrite pathology and behavioral defects (vol 115, pg E10748, 2018) Proc. Natl. Acad. Sci. U. S. A. 2018;115:E11563. doi: 10.1073/pnas.1807206115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D., Lee Y.I., Lee Y.S., Lee S.B. The mechanisms of nuclear proteotoxicity in polyglutamine spinocerebellar ataxias. Front. Neurosci. 2020;14:489. doi: 10.3389/fnins.2020.00489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.B., Bagley J.A., Lee H.Y., Jan L.Y., Jan Y.N. Pathogenic polyglutamine proteins cause dendrite defects associated with specific actin cytoskeletal alterations in Drosophila. Proc. Natl. Acad. Sci. U. S. A. 2011;108:16795–16800. doi: 10.1073/pnas.1113573108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijman S.M., Luna-Vargas M.P., Velds A., Brummelkamp T.R., Dirac A.M., Sixma T.K., Bernards R. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- Park J.H., Chung C.G., Park S.S., Lee D., Kim K.M., Jeong Y., Kim E.S., Cho J.H., Jeon Y.M., Shen C.J., et al. Cytosolic calcium regulates cytoplasmic accumulation of TDP-43 through Calpain-A and Importin alpha3. Elife. 2020a;9:e60132. doi: 10.7554/eLife.60132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.H., Chung C.G., Seo J., Lee B.H., Lee Y.S., Kweon J.H., Lee S.B. C9orf72-associated arginine-rich dipeptide repeat proteins reduce the number of Golgi outposts and dendritic branches in Drosophila neurons. Mol. Cells. 2020b;43:821–830. doi: 10.14348/molcells.2020.0130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.H., Kim H.K., Jung H., Kim K.H., Kang M.S., Hong J.H., Yu B.C., Parks S., Seo S.K., Choi I.W., et al. NecroX-5 prevents breast cancer metastasis by AKT inhibition via reducing intracellular calcium levels. Int. J. Oncol. 2017;50:185–192. doi: 10.3892/ijo.2016.3789. [DOI] [PubMed] [Google Scholar]
- Peng L., Peng Y., Chen Z., Wang C., Long Z., Peng H., Shi Y., Shen L., Xia K., Leotti V.B., et al. The progression rate of spinocerebellar ataxia type 3 varies with disease stage. J. Transl. Med. 2022;20:226. doi: 10.1186/s12967-022-03428-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez M.K., Paulson H.L., Pittman R.N. Ataxin-3 with an altered conformation that exposes the polyglutamine domain is associated with the nuclear matrix. Hum. Mol. Genet. 1999;8:2377–2385. doi: 10.1093/hmg/8.13.2377. [DOI] [PubMed] [Google Scholar]
- Reina C.P., Zhong X.Y., Pittman R.N. Proteotoxic stress increases nuclear localization of ataxin-3. Hum. Mol. Genet. 2010;19:235–249. doi: 10.1093/hmg/ddp482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha R.N., Pahan K. HATs and HDACs in neurodegeneration: a tale of disconcerted acetylation homeostasis. Cell Death Differ. 2006;13:539–550. doi: 10.1038/sj.cdd.4401769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh Y., Fujikake N., Okamoto Y., Popiel H.A., Hatanaka Y., Ueyama M., Suzuki M., Gaumer S., Murata M., Wada K., et al. p62 plays a protective role in the autophagic degradation of polyglutamine protein oligomers in polyglutamine disease model flies. J. Biol. Chem. 2015;290:1442–1453. doi: 10.1074/jbc.M114.590281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoes A.T., Carmona V., Duarte-Neves J., Cunha-Santos J., de Almeida L.P. Identification of the calpain-generated toxic fragment of ataxin-3 protein provides new avenues for therapy of Machado-Joseph disease| Spinocerebellar ataxia type 3. Neuropathol. Appl. Neurobiol. 2022;48:e12748. doi: 10.1111/nan.12748. [DOI] [PubMed] [Google Scholar]
- Soutoglou E., Katrakili N., Talianidis I. Acetylation regulates transcription factor activity at multiple levels. Mol. Cell. 2000;5:745–751. doi: 10.1016/S1097-2765(00)80253-1. [DOI] [PubMed] [Google Scholar]
- Sowa A.S., Haas E., Hubener-Schmid J., Lorentz A. Ataxin-3, the spinocerebellar ataxia type 3 neurodegenerative disorder protein, affects mast cell functions. Front. Immunol. 2022;13:870966. doi: 10.3389/fimmu.2022.870966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tait D., Riccio M., Sittler A., Scherzinger E., Santi S., Ognibene A., Maraldi N.M., Lehrach H., Wanker E.E. Ataxin-3 is transported into the nucleus and associates with the nuclear matrix. Hum. Mol. Genet. 1998;7:991–997. doi: 10.1093/hmg/7.6.991. [DOI] [PubMed] [Google Scholar]
- Thu V.T., Kim H.K., Long L.T., Lee S.R., Hanh T.M., Ko T.H., Heo H.J., Kim N., Kim S.H., Ko K.S., et al. NecroX-5 prevents hypoxia/reoxygenation injury by inhibiting the mitochondrial calcium uniporter (vol 94, pg 342, 2012) Cardiovasc. Res. 2012;95:134. doi: 10.1093/cvr/cvs122. [DOI] [PubMed] [Google Scholar]
- Warrick J.M., Morabito L.M., Bilen J., Gordesky-Gold B., Faust L.Z., Paulson H.L., Bonini N.M. Ataxin-3 suppresses polyglutamine neurodegeneration in Drosophila by a ubiquitin-associated mechanism. Mol. Cell. 2005;18:37–48. doi: 10.1016/j.molcel.2005.02.030. [DOI] [PubMed] [Google Scholar]
- Warrick J.M., Paulson H.L., Gray-Board G.L., Bui Q.T., Fischbeck K.H., Pittman R.N., Bonini N.M. Expanded polyglutamine protein forms nuclear inclusions and causes neural degeneration in Drosophila. Cell. 1998;93:939–949. doi: 10.1016/S0092-8674(00)81200-3. [DOI] [PubMed] [Google Scholar]
- Wing C.E., Fung H.Y.J., Chook Y.M. Karyopherin-mediated nucleocytoplasmic transport. Nat. Rev. Mol. Cell Biol. 2022;23:307–328. doi: 10.1038/s41580-021-00446-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolterhoff N., Gigengack U., Rumpf S. PP2A phosphatase is required for dendrite pruning via actin regulation in Drosophila. EMBO Rep. 2020;21:e48870. doi: 10.15252/embr.201948870. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.