Abstract
Caenorhabditis elegans has been used as a major model organism to identify genetic factors that regulate organismal aging and longevity. Insulin/insulin-like growth factor 1 (IGF-1) signaling (IIS) regulates aging in many species, ranging from nematodes to humans. C. elegans is a nonpathogenic genetic nematode model, which has been extensively utilized to identify molecular and cellular components that function in organismal aging and longevity. Here, we review the recent progress in the role of IIS in aging and longevity, which involves direct regulation of protein and RNA homeostasis, stress resistance, metabolism and the activities of the endocrine system. We also discuss recently identified genetic factors that interact with canonical IIS components to regulate aging and health span in C. elegans. We expect this review to provide valuable insights into understanding animal aging, which could eventually help develop anti-aging drugs for humans.
Keywords: aging, Caenorhabditis elegans, health span, homeostasis, insulin/IGF-1 signaling, longevity
INTRODUCTION
Aging is accompanied by gradual time-dependent functional and structural changes at the molecular and cellular levels, usually leading to impaired health, age-related diseases and increased vulnerability to death in organisms (Lee et al., 2021a; 2015c; Lopez-Otin et al., 2013; Melzer et al., 2020; Son et al., 2019). The nematode Caenorhabditis elegans is a widely used model organism for aging research because of its relatively short lifespan (2-3 weeks) and genetic tractability. In addition, 83% of the C. elegans genes are homologous to human genes (Lai et al., 2000). Many studies have identified genes and pathways that affect aging and longevity using C. elegans. One of the most crucial breakthrough findings in the research field of aging is that missense mutations in daf-2 double the lifespan of C. elegans (Kenyon et al., 1993). Subsequent studies have shown that daf-2 encodes an insulin/insulin-like growth factor-1 (IGF-1) receptor homolog (Kimura et al., 1997) and that insulin/IGF-1 signaling (IIS) affects aging in diverse species, ranging from C. elegans to mammals, including humans (reviewed in Altintas et al., 2016; An et al., 2017; Kenyon, 2010). IIS is initiated by binding of agonistic or antagonistic insulin-like peptides (ILPs) to the DAF-2/insulin/IGF-1 receptor in C. elegans. The binding of an agonistic ILP to DAF-2 leads to the activation of phosphoinositide-3 kinase (PI3K) cascade composed of kinases, including AGE-1/PI3K, phosphoinositide-dependent kinase-1 (PDK-1)/3-phosphoinositide-dependent protein kinase 1, and AKT-1,2/protein kinase B (PKB). The activation of IIS subsequently inhibits multiple transcription factors, such as DAF-16/Forkhead box O (FOXO), heat shock factor-1 (HSF-1), and SKN-1/nuclear factor erythroid 2-related factor (NRF), thereby repressing their target genes that promote longevity. Conversely, reduction-of-function mutations in daf-2 decrease the activity of the downstream kinase cascade, activating those transcription factors and inducing their target genes, and result in increased lifespan. The mechanisms by which reduced IIS promotes longevity have been attributed to diverse cellular and molecular aspects, including enhanced protein homeostasis, also known as proteostasis, and resistance to various stresses.
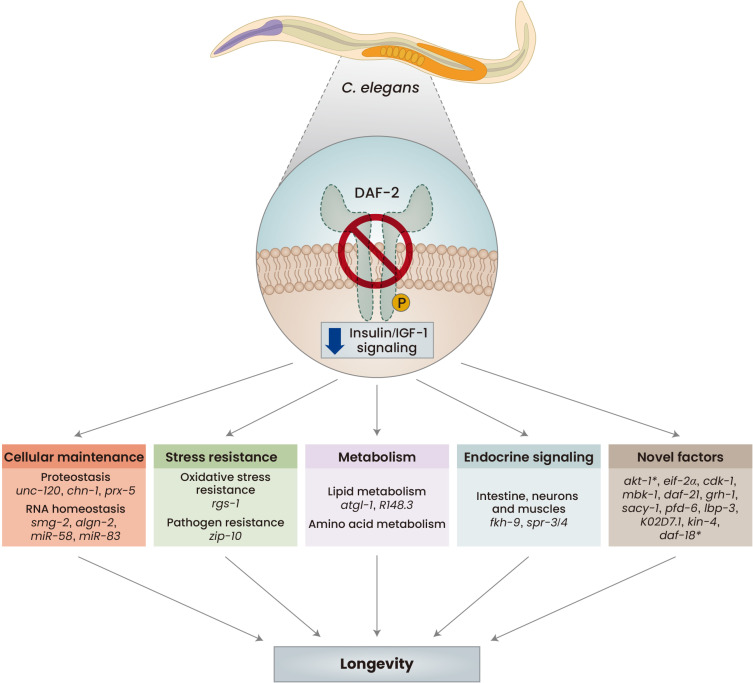
Here we review recent progress in studies regarding aging regulation by IIS using C. elegans, focusing on relevant works reported after our previous review paper covered a similar topic in 2016 (Altintas et al., 2016). We discuss findings showing that IIS affects aging by regulating cellular maintenance systems, including proteostasis and RNA homeostasis, stress resistance, metabolism, and the endocrine system (Fig. 1). We also review novel genetic factors that interact with canonical IIS components that influence longevity in C. elegans. Overall, the current review will provide useful information for our better understanding of the conserved aging-regulating roles of IIS in humans, thereby devising therapeutic strategies for developing medications and treating age-related diseases.
Fig. 1. Summary of recent progress in the roles of insulin/IGF-1 signaling (IIS) in C. elegans longevity.
Reduced IIS promotes longevity by enhancing cellular maintenance, including proteostasis, via increasing autophagy and lysosome activities, RNA homeostasis, through regulating nonsense-mediated mRNA decay (NMD) and microRNAs (miRNAs), and oxidative stress and pathogen resistance. IIS also regulates aging by modulating lipid and amino acid metabolism, endocrine signaling among several tissues, including the intestine, neurons and muscles, and the activities of novel factors that interact with classical IIS components. These processes cooperatively contribute to longevity. The roles of the genes in aging regulation shown in this figure are discussed in the corresponding sections of the text more in detail. unc-120/serum response factor, chn-1/C-term of Hsp70-interacting protein (CHIP), prx-5/peroxisomal biogenesis factor 5 (PEX5), smg-2/UPF1, algn-2/alpha-1,3/1,6-mannosyltransferase (ALG2), rgs-1/regulator of G protein signaling 20 (RGS20), zip-10/bZIP transcription factor, adipose triglyceride lipase-1 (atgl-1), forkhead transcription factor-9 (fkh-9), spr-3/4/repressor element-1 silencing transcription factor (REST), initiation factor (EIF)-2α (eif-2α), cyclin-dependent kinase 1 (cdk-1), mbk-1/human dual specificity tyrosine phosphorylation regulated kinase 1A (DYRK1A), daf-21/Hsp90, prefoldin 6 (pfd-6), suppressor of ACY-4 sterility 1 (sacy-1), lbp-3/fatty acid-binding protein, K02D7.1/purine nucleoside phosphorylase, kin-4/microtubule-associated serine/threonine kinase, and daf-18/phosphatase and tensin homolog (PTEN). Asterisk (*), new alleles.
MAIN BODY
Reduced IIS promotes longevity by enhancing cellular maintenance
The error catastrophe theory of aging proposes that aging results from errors in mRNA translation, increasing inaccurate protein synthesis (Orgel, 1963). This inaccurate protein synthesis accumulates misfolded or aggregated proteins, common features of age-associated diseases such as neurodegenerative diseases (Soto and Pritzkow, 2018). Therefore, to prevent harmful effects caused by the formation of erroneous proteins, organisms are equipped with protein quality control systems that monitor abnormal proteins, subsequently helping these proteins fold properly or degrading the proteins, maintaining proteostasis (Wolff et al., 2014).
Autophagy is a lysosome-dependent protein quality control system, which degrades abnormal cellular components and organelles (Levine and Kroemer, 2019; Shin, 2020). Autophagy is required for C. elegans longevity conferred by many regimens, including reduced IIS (Artan et al., 2022; Lee et al., 2015c; Nieto-Torres and Hansen, 2021). By performing autophagic flux assays in multiple tissues, autophagy activity has been shown to decrease with age in the intestine, the body wall muscles, the pharynx, and neurons (Chang et al., 2017). daf-2 mutants display increased autophagic activities throughout aging in several tissues, including the intestine and muscles, suggesting that reduced IIS enhances proteostasis by upregulating autophagy in these tissues. Additionally, the myogenic transcription factor, UNC-120/serum response factor, activates gene expression in the muscles during aging (Mergoud Dit Lamarche et al., 2018). UNC-120 contributes to maintaining autophagic activities in the muscles of daf-2 mutants. In addition to autophagy, the functional and anatomical features of lysosomes decline with age in C. elegans (Sun et al., 2020). daf-2 mutations suppress age-associated increases in lysosome volume and decreases in lysosomal motility, acidity and degradation activity, in a DAF-16/FOXO- and SKN-1/NRF-dependent manner. Furthermore, lysosome activity is required for the clearance of protein aggregation and longevity in daf-2 mutants. Thus, maintaining protein homeostasis by autophagy and lysosome activity during aging is pivotal for longevity conferred by reduced IIS.
Proper protein turnover contributes to proteostasis by removing damaged proteins. Several studies have reported the effect of IIS on age-associated proteome-wide changes in the protein abundance and turnover rates of C. elegans (Depuydt et al., 2016; Dhondt et al., 2016; Narayan et al., 2016; Visscher et al., 2016). Quantitative mass spectrometry indicates that the abundance of proteins involved in cellular stress responses increases in daf-2 mutants (Narayan et al., 2016). In contrast, proteins acting in cellular metabolic pathways, including those regulating lipid and amino acid metabolism, tend to be downregulated. Among the proteins exhibiting age-dependent downregulation, the decreased level of PRX-5/peroxisomal biogenesis factor 5 (PEX5) results in dysfunction of peroxisomal protein import. Surprisingly, most age-dependently altered proteins do not scale with delayed biological aging caused by daf-2 mutations. Thus, changes in the level or the activity of a small number of essential proteins appear to regulate longevity and biological aging by IIS. Interestingly, daf-2 mutations decrease the turnover rates of approximately 56% of total proteins in a young age, in particular, the components of translation machinery, including ribosomal proteins and translation factors (Dhondt et al., 2016), likely by decreasing protein synthesis and degradation rates (Depuydt et al., 2016). In contrast, old daf-2 mutant animals exhibit increased proteome-wide protein turnover rates (Visscher et al., 2016). Whether these opposite effects of daf-2 mutations on protein turnover rates in young and old animals play causal roles in aging remains undetermined. Nevertheless, these results propose the intriguing possibility that altering proteome turnover rates during aging by reducing IIS can tune protein levels to promote homeostasis, consequently leading to longevity.
In addition to proteome-wide changes caused by daf-2 mutations, the turnover rate of DAF-2 itself is regulated by the CHN-1/C-term of Hsp70-interacting protein (CHIP), an E4 ubiquitin chain elongation factor (Tawo et al., 2017). CHN-1/CHIP regulates the turnover of DAF-2 by targeting for monoubiquitylation, causing its degradation via the endocytic-lysosomal pathway. Hence, the genetic inhibition of chn-1 stabilizes DAF-2 proteins, leading to a short lifespan. Additionally, CHN-1/CHIP contributes to protein homeostasis by ubiquitylating age-dependently accumulated damaged proteins, which limit the capacity of CHN-1/CHIP to degrade DAF-2. Thus, CHN-1/CHIP appears to integrate protein homeostasis and aging regulation by IIS in C. elegans. The activity of DAF-16/FOXO increases during aging, likely by impaired proteostasis, ameliorating aging-induced perturbation of transcriptome (Li et al., 2019). These findings suggest that IIS affects proteome-wide abundance by regulating gene expression and protein turnover to promote longevity. Future studies investigating the function of upstream processes of protein synthesis during organismal aging will be an exciting research avenue.
In addition to proteostasis, IIS regulates RNA homeostasis in C. elegans. For instance, nonsense-mediated mRNA decay (NMD) is a key process for maintaining RNA quality (Kim and Maquat, 2019; Lykke-Andersen and Jensen, 2015). NMD detects and degrades transcripts containing premature termination codons, which produce truncated proteins, and mRNAs with uORFs (upstream open reading frames) and long 3´ untranslated regions (3´ UTRs). The NMD machinery comprises multiple components, including SMG-2/UPF1, a key RNA helicase for proper NMD. We have previously shown that the genetic inhibition of daf-2 enhances NMD, which is required for longevity (Son et al., 2017). Moreover, overexpression of smg-1, which encodes a protein kinase that activates SMG-2/UPF1, significantly increases lifespan. In our subsequent study that performed genome-wide RNAi and mutagenesis screens to identify NMD modulators, we reported ALGN-2/alpha-1,3/1,6-mannosyltransferase (ALG2) as a positive regulator of NMD (Kim et al., 2020b). We showed that ALGN-2 is required for the longevity of daf-2 mutants, and conversely, overexpression of algn-2 increases lifespan. These findings suggest that enhanced RNA quality control by NMD is necessary and sufficient for the longevity conferred by reduced IIS. Future research to test whether other RNA surveillance mechanisms mediate longevity caused by reduced IIS will be interesting.
IIS regulates the expression of many microRNAs (miRNAs) that affect longevity (Kim and Lee, 2019; Kinser and Pincus, 2020). For example, daf-2 mutations increase the miR-58 family of miRNA levels, in a DAF-16/FOXO-dependent fashion (Zhang et al., 2018). Moreover, the expression of miR-58 is required for the lifespan extension of daf-2 mutants. The target mRNAs of miR-58 include heat shock protein-90 (hsp-90/daf-21), ILP-1 (ins-1), and yeast imitation SWI (ISW) homolog-1 (isw-1), of which post-transcriptional regulation by the miRNA may contribute to longevity. Another miRNA, miR-83 is downregulated in daf-2 mutants, and a loss-of-function mutation in miR-83 extends lifespan in a DAF-16/FOXO-dependent manner, suggesting that miR-83 modulates longevity by acting with IIS (Dzakah et al., 2018). In addition, intestinal miR-83 is transported to the body wall muscles to downregulate autophagy by suppressing cup-5, which encodes a lysosomal calcium channel (Zhou et al., 2019). Thus, miR-83 appears to act as an endocrine factor that tissue-nonautonomously influences aging by targeting a specific player in autophagy. Overall, these findings provide insights into the interaction between protein and RNA homeostasis and IIS-controlled aging.
Enhanced stress resistance by reduced IIS is closely associated with longevity
Organisms are equipped with stress-responsive systems to cope with external stressors, including heat shock, oxidative stress, and bacterial pathogens (Park et al., 2017; Rodriguez et al., 2013). Many mutations in genes that extend lifespan, including those that reduce IIS, are associated with enhanced resistance to multiple stresses (Zhou et al., 2011). For example, RGS-1, the C. elegans homolog of the regulator of G protein signaling 20 (RGS20), acts with IIS to modulate oxidative stress resistance (Wu et al., 2017). Loss-of-function mutations in rgs-1 extend lifespan and enhance oxidative stress resistance in a DAF-16/FOXO-dependent manner. In long-lived C. elegans with mildly inhibited mitochondria (Hwang et al., 2012), increased reactive oxygen species (ROS) levels activate DAF-16/FOXO to extend lifespan (Senchuk et al., 2018). Along with previous studies demonstrating the longevity-promoting roles of mitochondrial ROS that act through AMPK (AMP-activated protein kinase) and HIF-1 (hypoxia-inducible factor-1) (Lee et al., 2010; Hwang and Lee, 2011; Hwang et al., 2014), these studies corroborated the positive role of ROS and resistance against oxidative stress in longevity conferred by reduced IIS. However, glycogen, which contributes to the lifespan-shortening effects of dietary glucose on daf-2 mutants (Gusarov et al., 2017), paradoxically increases oxidative stress resistance (Zecic et al., 2022); this is in line with several recent studies that reported uncoupling between stress resistance and longevity (Amrit et al., 2019; Dues et al., 2017). As longevity often correlates with adverse physiological outputs such as impaired reproduction and growth (Kirkwood, 1977; Lee et al., 2016; Williams, 2001), glycogen may increase stress resistance while decreasing longevity as a tradeoff. These studies provide insights into the relationship between resistance to oxidative stress and reduced IIS-mediated longevity.
IIS also regulates resistance to pathogenic bacteria. For example, loss-of-function mutations in daf-2 increase the resistance of C. elegans to bacterial colonization, the main cause of the death of aged animals, leading to extended survival in a decrepit state in old C. elegans (Podshivalova et al., 2017). Surprisingly, reduced IIS by daf-2 mutations further enhances immunocompetence in aged C. elegans, compared with young animals, in a DAF-16/FOXO- and HSF-1-dependent manner (Lee et al., 2021b). Moreover, the upregulation of DAF-16/FOXO and HSF-1 decreases the expression of ZIP-10/bZIP transcription factor, which further decreases IIS by reducing the expression of an agonistic ILP, ILP-7 (INS-7). Thus, a feedforward loop consisting of DAF-2, DAF-16/FOXO, HSF-1, ZIP-10/bZIP, and INS-7 regulates immune aging in C. elegans, and this can be exploited for reversing immunosenescence. Overall, these findings suggest that longevity conferred by reduced IIS is generally coupled with stress resistance in C. elegans. Future studies for testing whether IIS affects stress resistance in mammals will provide insights into the conserved roles of IIS in stress resistance and longevity.
Proper modulation of metabolism contributes to longevity caused by reduced IIS
Excessive dietary glucose shortens the lifespan of C. elegans by affecting biological processes (reviewed in Lee et al., 2015a; 2017), including glycerol metabolism by downregulating DAF-16/FOXO (Lee et al., 2009), and by affecting lipid metabolism (Jung et al., 2020; Lee et al., 2015b). Other metabolic processes also contribute to the lifespan extension caused by reduced IIS. A combination of anti-aging drugs targeting multiple longevity pathways, including IIS, increases the level of monounsaturated fatty acids in a sterol regulatory element-binding protein (SREBP)-dependent manner (Admasu et al., 2018). The expression of adipose triglyceride lipase-1 (ATGL-1) increases in daf-2 mutants in a DAF-16/FOXO-dependent manner, establishing it a common factor for the longevity conferred by both reduced IIS and dietary restriction (Zaarur et al., 2019). Similarly, R148.3, encoding a secreted protein in the body wall muscles and neurons, prevents abnormal accumulation of triglyceride and is critical for daf-2 mutation-induced longevity (Roy-Bellavance et al., 2017). These findings are consistent with studies demonstrating the connection between lipid metabolism and longevity in C. elegans subjected to glucose-rich diets (Jung et al., 2020; Lee et al., 2015b). Transcriptomic changes caused by daf-2 mutations and dietary restriction include the increased expression of lipid metabolism-related genes and decreased expression of macromolecule biosynthesis-related genes (Gao et al., 2018). In addition, metabolomic analyses revealed that daf-2 mutants and dietary restricted animals exhibit reduced total amounts of amino acids, adenine, and xanthine. Thus, reduced IIS affects amino acid and nucleotide metabolism, likely playing a key role in lifespan extension. Future research to identify biological factors that regulate these metabolic processes by IIS will provide important clues regarding the systematic connection between metabolic processes and longevity.
Endocrine signaling among tissues is a key feature for longevity conferred by reduced IIS
Studies have shown that the inter-tissue regulation of IIS between neurons and the intestine is pivotal for lifespan extension in C. elegans (Altintas et al., 2016; Kenyon, 2010), by transmitting environmental cues to physiological processes (Artan et al., 2016; Donato et al., 2017; Jeong et al., 2012; Kim et al., 2020a; Park et al., 2021b). For example, an agonistic ILP, INS-7, downregulates the activity of DAF-16/FOXO from the intestine to other tissues in a cell non-autonomous manner (Lee et al., 2009; Murphy et al., 2003; 2007). The expression of the intestinal ins-7 is also cell-nonautonomously downregulated by DAF-16/FOXO, indicating the feedback regulation of DAF-16/FOXO to DAF-16/FOXO. In addition to intestinal factors, the nervous tissue plays a central role in regulating longevity by affecting IIS (Altintas et al., 2016; Kenyon, 2010). A recent study using CRISPR/Cas9-mediated genome editing confirmed that the genetic depletion of daf-2 in neurons is sufficient to increase the activity of DAF-16/FOXO in the intestine and promotes longevity (Uno et al., 2021). Neuronal IIS also regulates mitochondrial function in the muscles in a DAF-16/FOXO-dependent manner, suggesting the role of neuroendocrine signaling by IIS in regulating muscle aging (Wang et al., 2019). Therefore, the inter-tissue regulation of IIS between neurons and other tissues may provide valuable information for developing therapeutics for diseases such as amyotrophic lateral sclerosis, which causes progressive loss of motor neurons, resulting in muscle defects. Previously, a study has identified FKH-9, a forkhead transcription factor, as a target of DAF-16/FOXO in neurons, by isolating adult C. elegans neurons and characterizing the neuronal transcriptome of daf-2 mutants (Kaletsky et al., 2016). FKH-9 enhances axon regeneration, short-term associative memory and longevity in daf-2 mutants. Thus, the transcriptional network that initiates from IIS and DAF-16/FOXO to neuronal FKH-9 appears to coordinate diverse behavioral and physiological processes in the whole body. Another study demonstrated that neuronal excitation increases with age in humans, mice, and C. elegans, and the transcription repressor, C. elegans spr-3/-4/repressor element-1 silencing transcription factor (REST), promotes longevity by downregulating the overall neural excitation (Zullo et al., 2019). Suppression of neural excitation by spr-3/-4 upregulates DAF-16/FOXO, leading to longevity. Thus, neural excitation and DAF-16/FOXO activation is an evolutionarily conserved mechanism by which the nervous system affects aging. Together, these recent studies pave ways to address the question regarding how the modulation of IIS factors in one tissue such as neurons and the intestine can generate a pivotal impact on the aging rate and longevity of an entire organism.
Genetic factors that interact with canonical IIS components modulate aging and longevity
A recent study that conducted large-scale quantitative phosphoproteomic analyses of wild-type and daf-2 mutant C. elegans identified phosphorylation sites of proteins that regulate longevity (Li et al., 2021b); these include novel phosphorylation sites in AKT-1, which is critical for its activity to inhibit DAF-16/FOXO, initiation factor (EIF)-2α, which contributes to inhibiting translation to promote longevity, and cyclin-dependent kinase 1 (CDK-1), which increases germ cell proliferation and limits longevity. Another study reported that C. elegans MBK-1, the ortholog of human dual specificity tyrosine phosphorylation regulated kinase 1A (DYRK1A) that phosphorylates FOXO1, contributes to longevity in daf-2 mutants, presumably by regulating the phosphorylation of DAF-16/FOXO (Mack et al., 2017). Given that IIS is a kinase cascade signaling, these phospho-protein studies provide strong evidence for the role of protein phosphorylation in IIS-mediated longevity. DAF-21/Hsp90 is required for the nuclear localization of DAF-16/FOXO isoform A and the longevity of daf-2 mutants (Somogyvari et al., 2018). Grainyhead-like transcription factor 1 (GRH-1) is necessary and sufficient for longevity conferred by reduced IIS by acting with DAF-16/FOXO (Grigolon et al., 2022). We reported that an RNA helicase, suppressor of ACY-4 sterility 1 (SACY-1), an ortholog of human DEAD-box helicase 41 (DDX41), is required for the longevity of daf-2 mutants, acting in a DAF-16/FOXO-dependent manner (Seo et al., 2016). SACY-1 functions as a general factor for multiple longevity regimens, different from another RNA helicase HEL-1, which we showed to bind and to activate DAF-16/FOXO specifically for the longevity of daf-2 mutants (Seo et al., 2015). In another report, we demonstrated that HSF-1 upregulates prefoldin 6 (PFD-6) that activates DAF-16/FOXO through physical interaction (Son et al., 2018), suggesting that PFD-6 mediates longevity response from HSF-1 to DAF-16/FOXO in animals with reduced IIS. Overall, these studies corroborated the central role of DAF-16/FOXO in the longevity caused by reduced IIS.
Global cysteine-reactivity profiling identified proteins whose activities change in the daf-2 mutants compared with those in short-lived daf-16; daf-2 double mutants (Martell et al., 2016). Specifically, the LBP-3/fatty acid-binding protein and K02D7.1/purine nucleoside phosphorylase exhibit decreased activities in daf-2 mutants in a DAF-16/FOXO-dependent manner. These decreased activities appear to contribute to longevity, as knockdown of lbp-3 or K02D7.1 further increases the long lifespan of daf-2 mutants. Thus, cysteine-reactivity profiling can identify novel mediators of C. elegans longevity, in addition to transcriptomic and proteomic analyses.
Our group reported novel regulatory mechanisms regarding DAF-18/phosphatase and tensin homolog (PTEN), which dephosphorylates phosphatidylinositol (3, 4, 5)-triphosphate (PIP3) to phosphatidylinositol (4, 5)-biphosphate (PIP2) for downregulating the downstream kinase cascade in IIS (An et al., 2019; Park et al., 2021a). We found that KIN-4, a microtubule-associated serine/threonine kinase, binds to DAF-18/PTEN through its PDZ domain and this physical interaction is required for the longevity of daf-2 mutants (An et al., 2019). Furthermore, we showed that a missense mutation in daf-18/PTEN that alters the cysteine 150 to tyrosine in DAF-18/PTEN decreases the lipid phosphatase activity, while partially retaining its protein phosphatase activity (Park et al., 2021a). This daf-18/PTEN mutant allele restores reduced motility in young daf-2 mutants and extends the health span of the animals by maintaining the partial activity of DAF-16/FOXO, while preventing the harmful hyperactivation of SKN-1/NRF. Along with these findings, AID (auxin-inducible degradation)-mediated depletion of DAF-2 after reproduction is sufficient to promote longevity without the adverse effects of daf-2 mutation on fertility and development (Venz et al., 2021). These studies suggest that the proper control of IIS components such as DAF-2 and DAF-18/PTEN can lead to increased health span by selectively promoting longevity and defying impaired fitness.
CONCLUSIONS AND PERSPECTIVES
After discovering the critical role of the IIS pathway in C. elegans longevity, researchers determined whether the mammalian homologs of IIS components influence human longevity. Minor alleles of the IGF1R and FOXO3A, C. elegans daf-2 and daf-16 homologs, respectively, are linked with human longevity (Kenyon, 2010; Tazearslan et al., 2012). In addition, growth hormone receptor deficiency in humans, which leads to reduced IGF1 levels, is associated with prevention against age-related diseases, including cancer and diabetes (Guevara-Aguirre et al., 2011). These findings suggest that the IIS pathway influences longevity and the pathophysiology of age-associated diseases in humans. A recent study reported that the chronic hyperinsulinemia induces cellular senescence in mature adipocytes (Li et al., 2021a), which may accelerate organismal aging (Kim and Kim, 2021). These findings indicate that the aging-regulatory roles of IIS are conserved across phyla, ranging from C. elegans to humans. Thus, research on aging using C. elegans is essential to identify novel anti-aging drug targets and/or to develop therapeutic interventions, thereby promoting healthy human longevity.
One of the most critical questions in the research field of aging is how to extend the healthy periods throughout life time, instead of simply extending the maximal lifespan. Thus, identifying factors that induce healthy organismal longevity is a key step for obtaining benefits from aging research. Recent studies suggest that elaborately modulating IIS can substantially improve healthy longevity. Future research to identify novel factors and mechanisms enhancing fitness and extending health span using C. elegans will help achieve healthy human longevity, the ultimate goal of aging research.
ACKNOWLEDGMENTS
We thank all Lee laboratory members for helpful comments and discussion. This research was supported by the KAIST Key Research Institutes Project (Interdisciplinary Research Group) to S.J.V.L.
Footnotes
AUTHOR CONTRIBUTIONS
H.L. and S.J.V.L. wrote the paper.
CONFLICT OF INTEREST
The authors have no potential conflicts of interest to disclose.
REFERENCES
- Admasu T.D., Chaithanya Batchu K., Barardo D., Ng L.F., Lam V.Y.M., Xiao L., Cazenave-Gassiot A., Wenk M.R., Tolwinski N.S., Gruber J. Drug synergy slows aging and improves healthspan through IGF and SREBP lipid signaling. Dev. Cell. 2018;47:67–79.e5. doi: 10.1016/j.devcel.2018.09.001. [DOI] [PubMed] [Google Scholar]
- Altintas O., Park S., Lee S.J. The role of insulin/IGF-1 signaling in the longevity of model invertebrates, C. elegans and D. melanogaster. BMB Rep. 2016;49:81–92. doi: 10.5483/BMBRep.2016.49.2.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amrit F.R.G., Naim N., Ratnappan R., Loose J., Mason C., Steenberge L., McClendon B.T., Wang G., Driscoll M., Yanowitz J.L., et al. The longevity-promoting factor, TCER-1, widely represses stress resistance and innate immunity. Nat. Commun. 2019;10:3042. doi: 10.1038/s41467-019-10759-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An S.W.A., Artan M., Park S., Altintas O., Lee S.J.V. Longevity regulation by insulin/IGF-1 signalling. In: Olsen A., Gill M., editors. Ageing: Lessons from C. elegans. Springer; Cham, Switzerland: 2017. pp. 63–81. [DOI] [Google Scholar]
- An S.W.A., Choi E.S., Hwang W., Son H.G., Yang J.S., Seo K., Nam H.J., Nguyen N.T.H., Kim E.J.E., Suh B.K., et al. KIN-4/MAST kinase promotes PTEN-mediated longevity of Caenorhabditis elegans via binding through a PDZ domain. Aging Cell. 2019;18:e12906. doi: 10.1111/acel.12906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artan M., Jeong D.E., Lee D., Kim Y.I., Son H.G., Husain Z., Kim J., Altintas O., Kim K., Alcedo J., et al. Food-derived sensory cues modulate longevity via distinct neuroendocrine insulin-like peptides. Genes Dev. 2016;30:1047–1057. doi: 10.1101/gad.279448.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artan M., Sohn J., Lee C., Park S.Y., Lee S.V. MON-2, a Golgi protein, promotes longevity by upregulating autophagy through mediating inter-organelle communications. Autophagy. 2022;18:1208–1210. doi: 10.1080/15548627.2022.2039523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J.T., Kumsta C., Hellman A.B., Adams L.M., Hansen M. Spatiotemporal regulation of autophagy during Caenorhabditis elegans aging. Elife. 2017;6:e18459. doi: 10.7554/eLife.18459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depuydt G., Shanmugam N., Rasulova M., Dhondt I., Braeckman B.P. Increased protein stability and decreased protein turnover in the Caenorhabditis elegans Ins/IGF-1 daf-2 mutant. J. Gerontol. A Biol. Sci. Med. Sci. 2016;71:1553–1559. doi: 10.1093/gerona/glv221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhondt I., Petyuk V.A., Cai H., Vandemeulebroucke L., Vierstraete A., Smith R.D., Depuydt G., Braeckman B.P. FOXO/DAF-16 activation slows down turnover of the majority of proteins in C. elegans. Cell Rep. 2016;16:3028–3040. doi: 10.1016/j.celrep.2016.07.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donato V., Ayala F.R., Cogliati S., Bauman C., Costa J.G., Lenini C., Grau R. Bacillus subtilis biofilm extends Caenorhabditis elegans longevity through downregulation of the insulin-like signalling pathway. Nat. Commun. 2017;8:14332. doi: 10.1038/ncomms14332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dues D.J., Schaar C.E., Johnson B.K., Bowman M.J., Winn M.E., Senchuk M.M., Van Raamsdonk J.M. Uncoupling of oxidative stress resistance and lifespan in long-lived isp-1 mitochondrial mutants in Caenorhabditis elegans. Free Radic. Biol. Med. 2017;108:362–373. doi: 10.1016/j.freeradbiomed.2017.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzakah E.E., Waqas A., Wei S., Yu B., Wang X., Fu T., Liu L., Shan G. Loss of miR-83 extends lifespan and affects target gene expression in an age-dependent manner in Caenorhabditis elegans. J. Genet. Genomics. 2018;45:651–662. doi: 10.1016/j.jgg.2018.11.003. [DOI] [PubMed] [Google Scholar]
- Gao A.W., Smith R.L., van Weeghel M., Kamble R., Janssens G.E., Houtkooper R.H. Identification of key pathways and metabolic fingerprints of longevity in C. elegans. Exp. Gerontol. 2018;113:128–140. doi: 10.1016/j.exger.2018.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigolon G., Araldi E., Erni R., Wu J.Y., Thomas C., La Fortezza M., Laube B., Pohlmann D., Stoffel M., Zarse K., et al. Grainyhead 1 acts as a drug-inducible conserved transcriptional regulator linked to insulin signaling and lifespan. Nat. Commun. 2022;13:107. doi: 10.1038/s41467-021-27732-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guevara-Aguirre J., Balasubramanian P., Guevara-Aguirre M., Wei M., Madia F., Cheng C.W., Hwang D., Martin-Montalvo A., Saavedra J., Ingles S., et al. Growth hormone receptor deficiency is associated with a major reduction in pro-aging signaling, cancer, and diabetes in humans. Sci. Transl. Med. 2011;3:70ra13. doi: 10.1126/scitranslmed.3001845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusarov I., Pani B., Gautier L., Smolentseva O., Eremina S., Shamovsky I., Katkova-Zhukotskaya O., Mironov A., Nudler E. Glycogen controls Caenorhabditis elegans lifespan and resistance to oxidative stress. Nat. Commun. 2017;8:15868. doi: 10.1038/ncomms15868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang A.B., Jeong D.E., Lee S.J. Mitochondria and organismal longevity. Curr. Genomics. 2012;13:519–532. doi: 10.2174/138920212803251427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang A.B., Lee S.J. Regulation of life span by mitochondrial respiration: the HIF-1 and ROS connection. Aging (Albany N.Y.) 2011;3:304–310. doi: 10.18632/aging.100292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang A.B., Ryu E.A., Artan M., Chang H.W., Kabir M.H., Nam H.J., Lee D., Yang J.S., Kim S., Mair W.B., et al. Feedback regulation via AMPK and HIF-1 mediates ROS-dependent longevity in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U. S. A. 2014;111:E4458–E4467. doi: 10.1073/pnas.1411199111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong D.E., Artan M., Seo K., Lee S.J. Regulation of lifespan by chemosensory and thermosensory systems: findings in invertebrates and their implications in mammalian aging. Front. Genet. 2012;3:218. doi: 10.3389/fgene.2012.00218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung Y., Kwon S., Ham S., Lee D., Park H.H., Yamaoka Y., Jeong D.E., Artan M., Altintas O., Park S., et al. Caenorhabditis elegans Lipin 1 moderates the lifespan-shortening effects of dietary glucose by maintaining omega-6 polyunsaturated fatty acids. Aging Cell. 2020;19:e13150. doi: 10.1111/acel.13150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaletsky R., Lakhina V., Arey R., Williams A., Landis J., Ashraf J., Murphy C.T. The C. elegans adult neuronal IIS/FOXO transcriptome reveals adult phenotype regulators. Nature. 2016;529:92–96. doi: 10.1038/nature16483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon C., Chang J., Gensch E., Rudner A., Tabtiang R. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- Kenyon C.J. The genetics of ageing. Nature. 2010;464:504–512. doi: 10.1038/nature08980. [DOI] [PubMed] [Google Scholar]
- Kim B., Lee J., Kim Y., Lee S.V. Regulatory systems that mediate the effects of temperature on the lifespan of Caenorhabditis elegans. J. Neurogenet. 2020a;34:518–526. doi: 10.1080/01677063.2020.1781849. [DOI] [PubMed] [Google Scholar]
- Kim E.J.E., Son H.G., Park H.H., Jung Y., Kwon S., Lee S.V. Caenorhabditis elegans algn-2 is critical for longevity conferred by enhanced nonsense-mediated mRNA decay. Science. 2020b;23:101713. doi: 10.1016/j.isci.2020.101713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Kim C. Transcriptomic analysis of cellular senescence: one step closer to senescence atlas. Mol. Cells. 2021;44:136–145. doi: 10.14348/molcells.2021.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.S., Lee S.V. Non-coding RNAs in Caenorhabditis elegans aging. Mol. Cells. 2019;42:379–385. doi: 10.14348/molcells.2019.0077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.K., Maquat L.E. UPFront and center in RNA decay: UPF1 in nonsense-mediated mRNA decay and beyond. RNA. 2019;25:407–422. doi: 10.1261/rna.070136.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura K.D., Tissenbaum H.A., Liu Y., Ruvkun G. daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science. 1997;277:942–946. doi: 10.1126/science.277.5328.942. [DOI] [PubMed] [Google Scholar]
- Kinser H.E., Pincus Z. MicroRNAs as modulators of longevity and the aging process. Hum. Genet. 2020;139:291–308. doi: 10.1007/s00439-019-02046-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkwood T.B. Evolution of ageing. Nature. 1977;270:301–304. doi: 10.1038/270301a0. [DOI] [PubMed] [Google Scholar]
- Lai C.H., Chou C.Y., Ch'ang L.Y., Liu C.S., Lin W. Identification of novel human genes evolutionarily conserved in Caenorhabditis elegans by comparative proteomics. Genome Res. 2000;10:703–713. doi: 10.1101/gr.10.5.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D., Hwang W., Artan M., Jeong D.E., Lee S.J. Effects of nutritional components on aging. Aging Cell. 2015a;14:8–16. doi: 10.1111/acel.12277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D., Jeong D.E., Son H.G., Yamaoka Y., Kim H., Seo K., Khan A.A., Roh T.Y., Moon D.W., Lee Y., et al. SREBP and MDT-15 protect C. elegans from glucose-induced accelerated aging by preventing accumulation of saturated fat. Genes Dev. 2015b;29:2490–2503. doi: 10.1101/gad.266304.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D., Son H.G., Jung Y., Lee S.V. The role of dietary carbohydrates in organismal aging. Cell. Mol. Life Sci. 2017;74:1793–1803. doi: 10.1007/s00018-016-2432-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G.Y., Sohn J., Lee S.V. Combinatorial approach using Caenorhabditis elegans and mammalian systems for aging research. Mol. Cells. 2021a;44:425–432. doi: 10.14348/molcells.2021.0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.J., Hwang A.B., Kenyon C. Inhibition of respiration extends C. elegans life span via reactive oxygen species that increase HIF-1 activity. Curr. Biol. 2010;20:2131–2136. doi: 10.1016/j.cub.2010.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.J., Murphy C.T., Kenyon C. Glucose shortens the life span of C. elegans by downregulating DAF-16/FOXO activity and aquaporin gene expression. Cell Metab. 2009;10:379–391. doi: 10.1016/j.cmet.2009.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., An S.W.A., Artan M., Seo M., Hwang A.B., Jeong D.E., Son H.G., Hwang W., Lee D., Seo K., et al. Genes and pathways that influence longevity in Caenorhabditis elegans. In: Mori N., Mook-Jung I., editors. In Aging Mechanisms: Longevity, Metabolism, and Brain Aging. Springer; Tokyo, Japan: 2015c. pp. 123–169. [DOI] [Google Scholar]
- Lee Y., Hwang W., Jung J., Park S., Cabatbat J.J., Kim P.J., Lee S.J. Inverse correlation between longevity and developmental rate among wild C. elegans strains. Aging (Albany N.Y.) 2016;8:986–999. doi: 10.18632/aging.100960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., Jung Y., Jeong D.E., Hwang W., Ham S., Park H.H., Kwon S., Ashraf J.M., Murphy C.T., Lee S.V. Reduced insulin/IGF1 signaling prevents immune aging via ZIP-10/bZIP-mediated feedforward loop. J. Cell Biol. 2021b;220:e202006174. doi: 10.1083/jcb.202006174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B., Kroemer G. Biological functions of autophagy genes: a disease perspective. Cell. 2019;176:11–42. doi: 10.1016/j.cell.2018.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Hagberg C.E., Silva Cascales H., Lang S., Hyvonen M.T., Salehzadeh F., Chen P., Alexandersson I., Terezaki E., Harms M.J., et al. Obesity and hyperinsulinemia drive adipocytes to activate a cell cycle program and senesce. Nat. Med. 2021a;27:1941–1953. doi: 10.1038/s41591-021-01501-8. [DOI] [PubMed] [Google Scholar]
- Li S.T., Zhao H.Q., Zhang P., Liang C.Y., Zhang Y.P., Hsu A.L., Dong M.Q. DAF-16 stabilizes the aging transcriptome and is activated in mid-aged Caenorhabditis elegans to cope with internal stress. Aging Cell. 2019;18:e12896. doi: 10.1111/acel.12896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W.J., Wang C.W., Tao L., Yan Y.H., Zhang M.J., Liu Z.X., Li Y.X., Zhao H.Q., Li X.M., He X.D., et al. Insulin signaling regulates longevity through protein phosphorylation in Caenorhabditis elegans. Nat. Commun. 2021b;12:4568. doi: 10.1038/s41467-021-24816-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Otin C., Blasco M.A., Partridge L., Serrano M., Kroemer G. The hallmarks of aging. Cell. 2013;153:1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen S., Jensen T.H. Nonsense-mediated mRNA decay: an intricate machinery that shapes transcriptomes. Nat. Rev. Mol. Cell Biol. 2015;16:665–677. doi: 10.1038/nrm4063. [DOI] [PubMed] [Google Scholar]
- Mack H.I.D., Zhang P., Fonslow B.R., Yates J.R. The protein kinase MBK-1 contributes to lifespan extension in daf-2 mutant and germline-deficient Caenorhabditis elegans. Aging (Albany N.Y.) 2017;9:1414–1432. doi: 10.18632/aging.101244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martell J., Seo Y., Bak D.W., Kingsley S.F., Tissenbaum H.A., Weerapana E. Global cysteine-reactivity profiling during impaired insulin/IGF-1 signaling in C. elegans identifies uncharacterized mediators of longevity. Cell Chem. Biol. 2016;23:955–966. doi: 10.1016/j.chembiol.2016.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melzer D., Pilling L.C., Ferrucci L. The genetics of human ageing. Nat. Rev. Genet. 2020;21:88–101. doi: 10.1038/s41576-019-0183-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mergoud Dit Lamarche A., Molin L., Pierson L., Mariol M.C., Bessereau J.L., Gieseler K., Solari F. UNC-120/SRF independently controls muscle aging and lifespan in Caenorhabditis elegans. Aging Cell. 2018;17:e12713. doi: 10.1111/acel.12713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy C.T., Lee S.J., Kenyon C. Tissue entrainment by feedback regulation of insulin gene expression in the endoderm of Caenorhabditis elegans. Proc. Natl. Acad. Sci. U. S. A. 2007;104:19046–19050. doi: 10.1073/pnas.0709613104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy C.T., McCarroll S.A., Bargmann C.I., Fraser A., Kamath R.S., Ahringer J., Li H., Kenyon C. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature. 2003;424:277–283. doi: 10.1038/nature01789. [DOI] [PubMed] [Google Scholar]
- Narayan V., Ly T., Pourkarimi E., Murillo A.B., Gartner A., Lamond A.I., Kenyon C. Deep proteome analysis identifies age-related processes in C. elegans. Cell Syst. 2016;3:144–159. doi: 10.1016/j.cels.2016.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto-Torres J.L., Hansen M. Macroautophagy and aging: the impact of cellular recycling on health and longevity. Mol. Aspects Med. 2021;82:101020. doi: 10.1016/j.mam.2021.101020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orgel L.E. The maintenance of the accuracy of protein synthesis and its relevance to ageing. Proc. Natl. Acad. Sci. U. S. A. 1963;49:517–521. doi: 10.1073/pnas.49.4.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H.H., Hwang W., Ham S., Kim E., Altintas O., Park S., Son H.G., Lee Y., Lee D., Heo W.D., et al. A PTEN variant uncouples longevity from impaired fitness in Caenorhabditis elegans with reduced insulin/IGF-1 signaling. Nat. Commun. 2021a;12:5631. doi: 10.1038/s41467-021-25920-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H.H., Jung Y., Lee S.V. Survival assays using Caenorhabditis elegans. Mol. Cells. 2017;40:90–99. doi: 10.14348/molcells.2017.0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S., Artan M., Jeong D.E., Park H.H., Son H.G., Kim S.S., Jung Y., Choi Y., Lee J.I., Kim K., et al. Diacetyl odor shortens longevity conferred by food deprivation in C. elegans via downregulation of DAF-16/FOXO. Aging Cell. 2021b;20:e13300. doi: 10.1111/acel.13300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podshivalova K., Kerr R.A., Kenyon C. How a mutation that slows aging can also disproportionately extend end-of-life decrepitude. Cell Rep. 2017;19:441–450. doi: 10.1016/j.celrep.2017.03.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez M., Snoek L.B., De Bono M., Kammenga J.E. Worms under stress: C. elegans stress response and its relevance to complex human disease and aging. Trends Genet. 2013;29:367–374. doi: 10.1016/j.tig.2013.01.010. [DOI] [PubMed] [Google Scholar]
- Roy-Bellavance C., Grants J.M., Miard S., Lee K., Rondeau E., Guillemette C., Simard M.J., Taubert S., Picard F. The R148.3 gene modulates Caenorhabditis elegans lifespan and fat metabolism. G3 (Bethesda) 2017;7:2739–2747. doi: 10.1534/g3.117.041681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senchuk M.M., Dues D.J., Schaar C.E., Johnson B.K., Madaj Z.B., Bowman M.J., Winn M.E., Van Raamsdonk J.M. Activation of DAF-16/FOXO by reactive oxygen species contributes to longevity in long-lived mitochondrial mutants in Caenorhabditis elegans. PLoS Genet. 2018;14:e1007268. doi: 10.1371/journal.pgen.1007268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo M., Park S., Nam H.G., Lee S.J. RNA helicase SACY-1 is required for longevity caused by various genetic perturbations in Caenorhabditis elegans. Cell Cycle. 2016;15:1821–1829. doi: 10.1080/15384101.2016.1183845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo M., Seo K., Hwang W., Koo H.J., Hahm J.H., Yang J.S., Han S.K., Hwang D., Kim S., Jang S.K., et al. RNA helicase HEL-1 promotes longevity by specifically activating DAF-16/FOXO transcription factor signaling in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U. S. A. 2015;112:E4246–E4255. doi: 10.1073/pnas.1505451112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin D.W. Lipophagy: molecular mechanisms and implications in metabolic disorders. Mol. Cells. 2020;43:686–693. doi: 10.14348/molcells.2020.0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somogyvari M., Gecse E., Soti C. DAF-21/Hsp90 is required for C. elegans longevity by ensuring DAF-16/FOXO isoform A function. Sci. Rep. 2018;8:12048. doi: 10.1038/s41598-018-30592-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son H.G., Altintas O., Kim E.J.E., Kwon S., Lee S.V. Age-dependent changes and biomarkers of aging in Caenorhabditis elegans. Aging Cell. 2019;18:e12853. doi: 10.1111/acel.12853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son H.G., Seo K., Seo M., Park S., Ham S., An S.W.A., Choi E.S., Lee Y., Baek H., Kim E., et al. Prefoldin 6 mediates longevity response from heat shock factor 1 to FOXO in C. elegans. Genes Dev. 2018;32:1562–1575. doi: 10.1101/gad.317362.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son H.G., Seo M., Ham S., Hwang W., Lee D., An S.W., Artan M., Seo K., Kaletsky R., Arey R.N., et al. RNA surveillance via nonsense-mediated mRNA decay is crucial for longevity in daf-2/insulin/IGF-1 mutant C. elegans. Nat. Commun. 2017;8:14749. doi: 10.1038/ncomms14749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto C., Pritzkow S. Protein misfolding, aggregation, and conformational strains in neurodegenerative diseases. Nat. Neurosci. 2018;21:1332–1340. doi: 10.1038/s41593-018-0235-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Li M., Zhao D., Li X., Yang C., Wang X. Lysosome activity is modulated by multiple longevity pathways and is important for lifespan extension in C. elegans. Elife. 2020;9:e55745. doi: 10.7554/eLife.55745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tawo R., Pokrzywa W., Kevei E., Akyuz M.E., Balaji V., Adrian S., Hohfeld J., Hoppe T. The ubiquitin ligase CHIP integrates proteostasis and aging by regulation of insulin receptor turnover. Cell. 2017;169:470–482.e13. doi: 10.1016/j.cell.2017.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tazearslan C., Cho M., Suh Y. Discovery of functional gene variants associated with human longevity: opportunities and challenges. J. Gerontol. A Biol. Sci. Med. Sci. 2012;67:376–383. doi: 10.1093/gerona/glr200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uno M., Tani Y., Nono M., Okabe E., Kishimoto S., Takahashi C., Abe R., Kurihara T., Nishida E. Neuronal DAF-16-to-intestinal DAF-16 communication underlies organismal lifespan extension in C. elegans. iScience. 2021;24:102706. doi: 10.1016/j.isci.2021.102706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venz R., Pekec T., Katic I., Ciosk R., Ewald C.Y. End-of-life targeted degradation of DAF-2 insulin/IGF-1 receptor promotes longevity free from growth-related pathologies. Elife. 2021;10:e71335. doi: 10.7554/eLife.71335.sa2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visscher M., De Henau S., Wildschut M.H.E., van Es R.M., Dhondt I., Michels H., Kemmeren P., Nollen E.A., Braeckman B.P., Burgering B.M.T., et al. Proteome-wide changes in protein turnover rates in C. elegans models of longevity and age-related disease. Cell Rep. 2016;16:3041–3051. doi: 10.1016/j.celrep.2016.08.025. [DOI] [PubMed] [Google Scholar]
- Wang H., Webster P., Chen L., Fisher A.L. Cell-autonomous and non-autonomous roles of daf-16 in muscle function and mitochondrial capacity in aging C. elegans. Aging (Albany N.Y.) 2019;11:2295–2311. doi: 10.18632/aging.101914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams G.C. Pleiotropy, natural selection, and the evolution of senescence: Evolution 11, 398-411 (1957) Sci. Aging Knowledge Environ. 2001;2001:cp13. doi: 10.1111/j.1558-5646.1957.tb02911.x. [DOI] [Google Scholar]
- Wolff S., Weissman J.S., Dillin A. Differential scales of protein quality control. Cell. 2014;157:52–64. doi: 10.1016/j.cell.2014.03.007. [DOI] [PubMed] [Google Scholar]
- Wu M., Kang X., Wang Q., Zhou C., Mohan C., Peng A. Regulator of G protein signaling-1 modulates paraquat-induced oxidative stress and longevity via the insulin like signaling pathway in Caenorhabditis elegans. Toxicol. Lett. 2017;273:97–105. doi: 10.1016/j.toxlet.2017.03.027. [DOI] [PubMed] [Google Scholar]
- Zaarur N., Desevin K., Mackenzie J., Lord A., Grishok A., Kandror K.V. ATGL-1 mediates the effect of dietary restriction and the insulin/IGF-1 signaling pathway on longevity in C. elegans. Mol. Metab. 2019;27:75–82. doi: 10.1016/j.molmet.2019.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zecic A., Dhondt I., Braeckman B.P. Accumulation of glycogen and upregulation of LEA-1 in C. elegans daf-2(e1370) support stress resistance, not longevity. Cells. 2022;11:245. doi: 10.3390/cells11020245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Zhang W., Dong M. The miR-58 microRNA family is regulated by insulin signaling and contributes to lifespan regulation in Caenorhabditis elegans. Sci. China Life Sci. 2018;61:1060–1070. doi: 10.1007/s11427-018-9308-8. [DOI] [PubMed] [Google Scholar]
- Zhou K.I., Pincus Z., Slack F.J. Longevity and stress in Caenorhabditis elegans. Aging (Albany N.Y.) 2011;3:733–753. doi: 10.18632/aging.100367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Wang X., Song M., He Z., Cui G., Peng G., Dieterich C., Antebi A., Jing N., Shen Y. A secreted microRNA disrupts autophagy in distinct tissues of Caenorhabditis elegans upon ageing. Nat. Commun. 2019;10:4827. doi: 10.1038/s41467-019-12821-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zullo J.M., Drake D., Aron L., O'Hern P., Dhamne S.C., Davidsohn N., Mao C.A., Klein W.H., Rotenberg A., Bennett D.A., et al. Regulation of lifespan by neural excitation and REST. Nature. 2019;574:359–364. doi: 10.1038/s41586-019-1647-8. [DOI] [PMC free article] [PubMed] [Google Scholar]