Abstract
The field of extracellular vesicles (EVs) has expanded tremendously over the last decade. The role of cell-to-cell communication in neighboring or distant cells has been increasingly ascribed to EVs generated by various cells. Initially, EVs were thought to a means of cellular debris or disposal system of unwanted cellular materials that provided an alternative to autolysis in lysosomes. Intercellular exchange of information has been considered to be achieved by well-known systems such as hormones, cytokines, and nervous networks. However, most research in this field has searched for and found evidence to support paracrine or endocrine roles of EV, which inevitably leads to a new concept that EVs are synthesized to achieve their paracrine or endocrine purposes. Here, we attempted to verify the endocrine role of EV production and their contents, such as RNAs and bioactive proteins, from the regulation of biogenesis, secretion, and action mechanisms while discussing the current technical limitations. It will also be important to discuss how blood EV concentrations are regulated as if EVs are humoral endocrine machinery.
Keywords: adiponectin, ceramide, exosome, extracellular vesicles, syntenin, tetraspanin
EXTRACELLULAR VESICLES (EVs)
Small vesicles delimited by a lipid bilayer released from cells are now defined as EVs (Witwer and Thery, 2019). Virtually all cells, if not all, in any tissue or organ can be the source of EVs (Mathieu et al., 2019). Exosomes typically have size distributions smaller than 150 nm, whereas ectosomes have diameters larger than 150 nm.
EVs can be defined in subpopulations based on their generating mechanism as “ectosomes” or are called “microvesicles” that are shed directly from the plasma membrane, “exosomes” are released by exocytosis of multivesicular bodies (MVBs), where the limiting membrane of MVBs is inwardly budded while exosomes (intraluminal vesicles [ILVs]) are formed in the internal space of this endosome (Mathieu et al., 2019). Therefore, the term “exosomes” is defined by their intracellular biogenesis. It starts from endocytosis, which forms an early endosome. Early endosomes mature into late endosomes, in which ILVs are generated. Late endosomes containing multiple ILVs are called MVBs. Finally, exosomes are secreted from cells by exocytosis of the MVB contents.
There have been extensive reviews of the reported functions (El Andaloussi et al., 2013; Gézsi et al., 2019; Kalluri and LeBleu, 2020; Kita et al., 2019b), biogenesis mechanism (Colombo et al., 2014; Hessvik and Llorente, 2018; Jadli et al., 2020; Kita and Shimomura, 2021; Xie et al., 2022), and isolation of EVs (Poupardin et al., 2021; Théry et al., 2006); here, we briefly summarize the most current views on these specific aspects and discuss them based on the endocrine roles of EVs.
EXOSOME BIOGENESIS
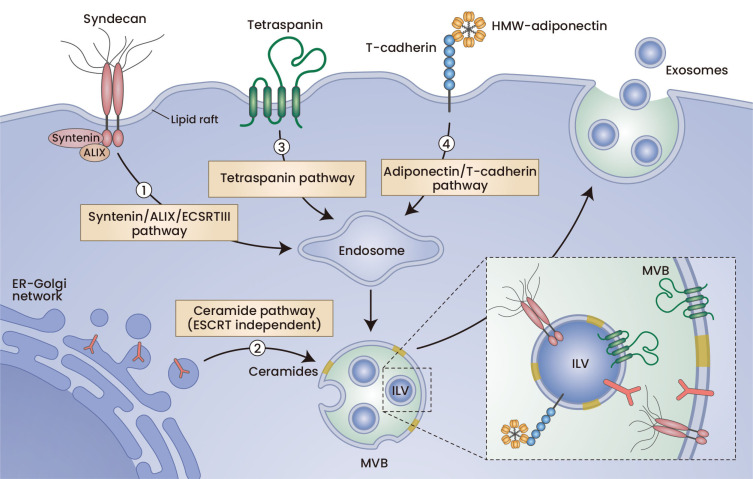
From plasma membrane endocytosis to exosome secretion, the term “exosome biogenesis” refers to ILV formation in the late endosome, MVBs. Exosome biogenesis progresses with the conserved membrane-neck-directed activities of the endosomal sorting complex required for transport (ESCRT) complexes (Xie et al., 2022). ESCRTs are composed of approximately 20 proteins assembled into 4 complexes (e.g., ESCRT-0, -I, -II, and -III) with associated proteins (e.g., VPS4, VTA1, and ALIX) and are conserved from yeast to mammals (Henne et al., 2011). ILV biogenesis starts with the formation of a nascent bud toward the interior of an MVB, which is connected via a membranous stalk to the endosomal limiting membrane (Fig. 1). The subsequent severing of this nascent ILV from the limiting membrane requires membrane fission activity of the ESCRT complex (van Niel et al., 2018). Currently, what makes a nascent bud become an MVB is obscure. Exosome biogenesis is mediated by a variety of ESCRT proteins, including Hrs, CHMP4, TSG101, STAM1, and VPS4 (Colombo et al., 2013; Hessvik and Llorente, 2018). The syntenin-Alix-ESCRT-III system is also important and has physiological significance, especially in tumor cells (Baietti et al., 2012; Ghossoub et al., 2014; Imjeti et al., 2017; Roucourt et al., 2015). ILVs are considered to form independently of ESCRTs (Stuffers et al., 2009). Sphingolipid regulation also plays a significant role in both exosome biogenesis and secretion (Verderio et al., 2018). The abundance of tetraspanins, such as CD63, also affects exosome biogenesis (Andreu and Yanez-Mo, 2014; Ghossoub et al., 2020).
Fig. 1. Exosome biogenesis pathways.
Exosomes are intracellularly generated inside MVBs. Endocytosis of the plasma membrane starts with endosome maturation, in which the limited membrane is invaginated to form ILVs. If ILVs are secreted outside the cells, they are called exosomes. Membrane protein clustering is an important mechanism by which membrane budding occurs. Syndecan clustering, tetraspanin enrichment, and high-molecular-weight multimer adiponectin-induced T-cadherin clustering are all supposed to enhance ILV biogenesis. Ceramide generation may affect membrane protein clustering in the late endosome and thereby induce ILV biogenesis. ER, endoplasmic reticulum.
Syntenin-ALIX-ESCRT-III system
The syntenin-ALIX-ESCRT-III system facilitates signaling-induced sorting of plasma membrane proteins to endosomes and intracellular biogenesis of exosomes (Baietti et al., 2012). One plausible mechanism of exosome biogenesis may be membrane budding by clustering of membrane proteins (Hurley et al., 2010). Interestingly, higher order oligomerization of proteins with membrane anchoring was shown to be sufficient for protein sorting into the exosome (Fang et al., 2007). Clustering of an extremely abundant membrane protein, syndecan, which is associated with cytosolic syntenin and in turn syntenin, binds ALIX, a component of ESCRT, and is facilitated by the enhancement of ILV biogenesis (Baietti et al., 2012). In this study, Baietti et al. (2012) demonstrated accumulations of DAB-stained ILVs in MVBs by incubating them with anti-CD63:HRP conjugates for one hour. This strategy enables visualization of newly generated ILVs in MVBs and thus enables direct evaluation of the speed of exosome biogenesis (Baietti et al., 2012). Because exosome secretion can be regulated in multiple steps, including ILV biogenesis and MVB fusion with the plasma membrane, a direct evaluation strategy of ILV genesis such as this is mandatory.
In this syntenin-ALIX system, it was also revealed that small GTPase ADP ribosylation Factor 6 (ARF6) regulates ILV biogenesis (Ghossoub et al., 2014). Because the formation of a bud from a flat membrane requires thermal energy, the involvement of a small GTPase is reasonable (Hurley et al., 2010).
It was reported that heparanase overexpression in human cancer cells (e.g., myeloma, lymphoblastoid, and breast cancer) enhances exosome secretion (Thompson et al., 2013). The addition and knockdown of heparanase affect exosome biogenesis, as revealed by the dependence on Rab27a, and the effects of heparanase depend on syndecans and interactions of syntenin with ALIX (Roucourt et al., 2015). Heparanase is the only mammalian enzyme capable of cleaving heparan sulfate internally and is upregulated in many cancers. Although heparanase can activate several signal transduction pathways independently of its enzymatic activity and can enhance cell survival and cell migration, it may promote tumor progression by increasing tumor exosome biogenesis (Roucourt et al., 2015; Thompson et al., 2013).
The syntenin-ALIX exosome biogenesis system is also regulated by a proto-oncogene, Src (Imjeti et al., 2017). Src plays important roles in cell proliferation, invasion, motility, and signal transduction induced by various external stimuli, such as growth factors and integrins. It was reported that Src directly phosphorylates syndecan and syntenin, thereby enhancing syndecan endocytosis and syntenin-syndecan endosomal budding (Imjeti et al., 2017).
Because both syntenin and ALIX are ubiquitously expressed throughout the body in fetal and adult humans (Zimmermann et al., 2001) and in mouse tissues (Jeon et al., 2013), it is expected that a loss of syntenin would affect whole-body exosome biogenesis and therefore the physiological significance of exosome biogenesis at the whole-body level. However, systemic syntenin knockout mice were reported to be viable, showed no major defects, and had normal fertility (Kashyap et al., 2021). Moreover, surprisingly, EVs secreted by the MEFs of syntenin KO mice did not differ in their numbers and sizes compared to those of WT mice (Kashyap et al., 2021). The congenital deficiency of syntenin may be compensated for by some other factors.
We found that knockdown of ALIX in human adipose-derived mesenchymal stem cells (MSCs) reduced exosomal cargo secretion by more than 95% and significantly attenuated the therapeutic effects of intravenously injected cells in a pressure-over load-induced heart failure model (Nakamura et al., 2020). ALIX deficiencies in these stem cells did not affect the cytokine profile, growth of cells, or differentiation potential to adipocytes, chondrocytes, and osteoblasts (Nakamura et al., 2020). Because “loss-of-exosome” studies are crucially important to know the roles of exosomes, transient knockdown of ALIX may be an ideal method to study the functions of exosomes in at least the cultured MSCs.
Sphingolipids and their metabolizing enzymes
Sphingolipid ceramide is composed of sphingosine and fatty acids and is contained in high concentrations within cell membranes. Using a mouse oligodendroglial cell line, Trajokovic et al. (2008) reported that ILV biogenesis into an MVB depends on ceramide, purified exosomes are enriched in ceramide, and the release of exosomes decreases after inhibition of neutral sphingomyelinases (nSMase) (Trajkovic et al., 2008), which convert sphingomyelin to ceramide and phosphocholine. This suggests proposing a lipid-based mechanism for exosome biogenesis where inward membrane budding to form ILV proceeds at sphingolipid-rich membrane microdomains through enzymatic generation of ceramides and subsequent coalescence into larger domains (Trajkovic et al., 2008). Indeed, small membrane vesicles such as exosomes need ceramides, cone-shaped structures, to stably form sharp curvatures. From this finding, many studies employed the nSMase inhibitor, GW4869, or an siRNA against nSMase2 to limit exosome biogenesis. Intraperitoneal injections of GW4869 were reported to reduce the levels of brain and serum exosomes in a mouse model of Alzheimer’s disease (Dinkins et al., 2014). The same group also reported decreased numbers of exosomes for a genetic deficiency of nSMase2 using the same mouse model (Dinkins et al., 2016). Therefore, these studies suggest that ceramide regulation of exosome biogenesis has physiological importance in vivo. Ceramide generated by nSMase2 is considered to mediate ESCRT-independent exosome biogenesis (Stuffers et al., 2009; Trajkovic et al., 2008). The sphingomyelin metabolite, sphingosine-1-phosphate (S1P), was shown to induce cargo sorting to ILVs and maturation of MVBs by continuously activating Gi-coupled S1P receptors in MVBs (Kajimoto et al., 2013). Recently, it was shown that nSMase2 inhibition induces V-ATPase complex assembly that drives MVB lumen acidification and consequently reduces exosome secretion (Choezom and Gross, 2022). Rab31 was shown to mediate flotillin sorting into ILVs and inhibit MVB fusion to the lysosome and enhance exosome secretion (Wei et al., 2021). The effects were also dependent on lipids and were inhibited by nSMase inhibitors or cholesterol biosynthesis inhibitors (Wei et al., 2021).
To date, however, it has been unreported whether such lipids and their metabolism affect exosome biogenesis in terms of the ILV biogenesis rate. Furthermore, the effects of inhibition or deficiency of sSMase2 on exosome biogenesis and secretion do not seem to be general (Colombo et al., 2013; Phuyal et al., 2014; Skotland et al., 2017; van Niel et al., 2011). We also found that knockdown of sSMase2 in cultured murine endothelial cells or human adipose-derived MSCs did not change the yield of EVs (Nakamura et al., 2020; Obata et al., 2018). Because inhibition of nSMase seems to have different effects depending on cell type despite the physicochemical basis of the ceramide, it will be important to understand what causes the differences among cell types (Verderio et al., 2018).
Tetraspanin
Tetraspanin is a four transmembrane protein superfamily that organizes membrane microdomains termed tetraspanin-enriched microdomains by forming clusters and interacts with a large variety of transmembrane and cytosolic signaling proteins (Andreu and Yanez-Mo, 2014). Tetraspanins such as CD9, CD81, and CD63 are major constituents of EVs and serve as canonical surface markers for EVs. Tetraspanin plays roles in both ESCRT-dependent and ESCRT-independent exosome production and cargo sorting. For example, the melanocyte-specific glycoprotein, PMEL ESCRT, independently enters ILVs and is secreted into exosomes upon interaction with CD63 (van Niel et al., 2011). It was also reported that exosome release from dendritic cells generated from CD9 knockout mice is reduced compared to that from wild-type dendritic cells (Chairoungdua et al., 2010). From a transmission electron microscopy analysis, substantial evidence supported the role of tetraspanin in exosome biogenesis (Bari et al., 2011). The tetraspanin, CD81, promotes microvillus formation and/or extension, whereas CD82 does not. CD81 enhances outward bending of the plasma membrane, whereas CD82 inhibits it (Bari et al., 2011). Because tetraspanins interact with various membrane proteins, the mechanism of how overexpression and/or depletion of one specific tetraspanin affects the membrane budding reaction that leads to endocytosis and forms ILVs in an MVB is not well understood. Recently, a connection between the tetraspanin system and syntenin-ALIX-ESCRT-III system was reported (Baietti et al., 2012; Ghossoub et al., 2020). TSPN6, a poorly characterized tetraspanin, supports lysosomal degradation of SDC4 and syntenin and suppresses exosome biogenesis (Ghossoub et al., 2020). Tetraspanins, by forming a membrane protein network including syndecans and flotillin, may facilitate both ESCRT-dependent and ESCRT-independent pathways and regulate membrane homeostasis and exosome biogenesis.
Adiponectin/T-cadherin system
Adiponectin is a secretory protein produced specifically by adipocytes. It is considered a beneficial protein that decreases with obesity progression and has various organ-protecting effects, such as insulin sensitization and cardiovascular protection (Kita and Shimomura, 2021; Kita et al., 2019b). We found that adiponectin, an adipocyte-derived factor, enhances ILV biogenesis and subsequent exosome secretion by binding to cell-surface T-cadherin, a sole physiologically proven receptor for adiponectin (Fukuda et al., 2017; Kita et al., 2019a; Obata et al., 2018) (Fig. 1). Adiponectin increases the numbers of DAB-stained ILVs counted by immune electron microscopy in an MVB during a one-hour incubation period with a labeled antibody against CD63 following transfection with siRNA against Rab7 and Rab27a to damn up MVB fusion with lysosomes or the plasma membrane (Obata et al., 2018). Moreover, the adiponectin/T-cadherin system strongly affects the total levels of small EVs in the plasma of mice (Obata et al., 2018). A deficiency in either adiponectin or T-cadherin similarly decreased plasma EVs in almost half of wild-type mice. Overexpression of adiponectin, in turn, increased plasma EVs. To our knowledge, adiponectin/T-cadherin is the only mechanism that is proven to regulate systemic concentrations of EVs by regulating ILV biogenesis in T-cadherin-expressing cells, including endothelial cells (Obata et al., 2018), skeletal and heart muscle cells (Tanaka et al., 2019), and tissue residential stem/stromal cells (Kita and Shimomura, 2021; Kita et al., 2019b; Nakamura et al., 2020; Tsugawa-Shimizu et al., 2021). Interestingly, this system also affects cellular ceramide contents (Obata et al., 2018). The addition of adiponectin decreases cellular ceramide contents and increases secreted ceramides associated with EVs, which could be sedimented by ultracentrifugation (Obata et al., 2018). However, as mentioned above, exosome production was not decreased by nSMase inhibition or silencing in cultured endothelial cells (Obata et al., 2018).
The molecular mechanism of the adiponectin/T-cadherin system is not fully understood. Balatskaya et al. recently demonstrated clustering of T-cadherin by incubation with high-molecular-weight adiponectin (Balatskaya et al., 2019).
EXOSOME RELEASE
The numbers of released exosomes per hour, namely, the exosome production speed, was determined by both exosome biogenesis as discussed above and by MVB fusion rates to the plasma membrane (Fig. 2). In this chapter, we will discuss regulation of the release of already matured MVBs in which ILVs are formed. MVB movement and fusion to the plasma membrane are conducted by the Rab GTPase family and SNARE family proteins (Fader et al., 2009). Intracellular calcium concentrations are a key factor affecting these steps (Savina et al., 2003). Additionally, the ability of the lysosome to degrade the cargos of MVBs determines MVB fates and therefore affects the exosome secretion rate.
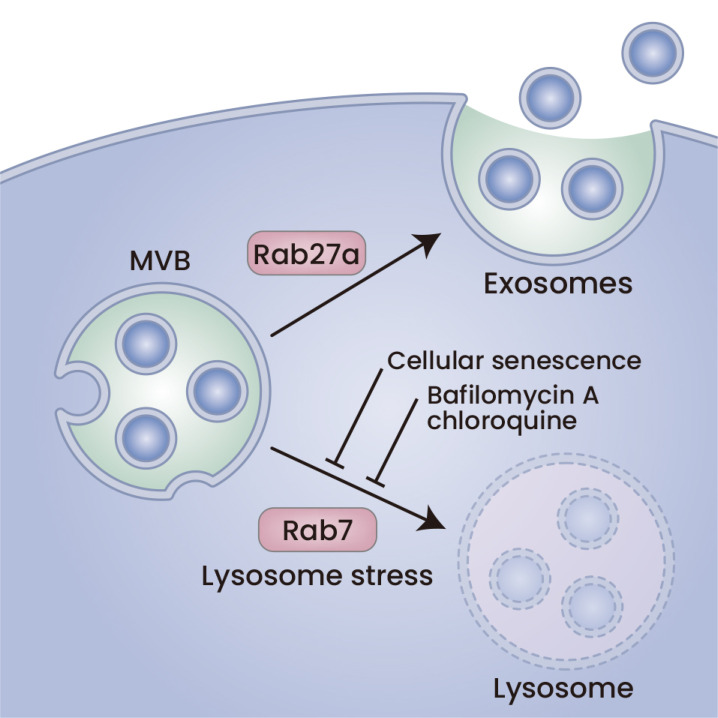
Fig. 2. MVB fate determines exosome secretion.
MVB can fuse with both lysosomes and the plasma membrane. The latter secretes exosomes. MVB fusion with lysosomes can be affected by senescence, similar to treatments with bafilomycin A or CQ. Lysosome stress is a hallmark of aging-related cellular dysfunction and may regulate exosome secretion.
MVB fate
MVB is one of the late endosomes that matures from early endosomes. If MVB fuses with lysosomes, then the MVB contents are degraded in lysosomal acidic and strongly proteolytic circumstances. If an MVB fuses with the plasma membrane, the ILVs in this MVB are released to form exosomes. Release of exosomes into the extracellular milieu is driven by proteins of the Rab GTPase family, including RAB27 (Ostrowski et al., 2010) and SNARE family proteins (Fader et al., 2009). Currently, it is thought that there are different subgroups of MVBs in cells: one is utilized for lysosomal degradation, and the other is utilized for exosomal secretion. The molecular mechanism of this regulatory event is unclear (Gurunathan et al., 2021). On the other hand, it is well known that lysosome status affects exosome secretion (Eitan et al., 2016). It has been shown that inhibition of lysosomes with different alkaline agents increases EV secretion. Treating cells expressing α-synuclein, as a cargo of ILVs, with bafilomycin A, a lysosome inhibitor, increases the levels of α-synuclein that are released in EVs (Alvarez-Erviti et al., 2011). Lysosome inhibition by bafilomycin A not only increases EV cargos but also increases the EV numbers that are secreted into the media, as determined by nanoparticle tracking analysis (NTA) (Miao et al., 2015). We also found that bafilomycin A increases multiple exosome cargo secretions, including T-cadherin, the binding partner of adiponectin, from endothelial cells (Obata et al., 2018).
Autophagy
Bafilomycin A specifically inhibits the enzyme activity of vacuolar-type H+-ATPase (V-ATPase), a membrane-spanning proton pump that acidifies endosomes such as lysosomes (Fedele and Proud, 2020). Because lysosome fusion and subsequent cargo degradation are the final step of autophagy, bafilomycin A and chloroquine (CQ) have been used interchangeably to block autophagy in in vitro experiments, assuming that they both block autophagy itself or its final step (Devis-Jauregui et al., 2021). According to this assumption, autophagy inhibition may increase exosome secretion. However, these compounds inhibit lysosomes, which are alternative deposits of MVBs that fuse with the plasma membrane and release exosomes (Devis-Jauregui et al., 2021) (Fig. 2). However, it is also true that the autophagy-lysosome pathway is also involved in exosome secretion. It has been reported that the ubiquitin-like ATG12 protein and its ligase, ATG3, interact with ALIX. ALIX regulates MVB fusion of autophagosomes and release of exosomes (Murrow et al., 2015). However, whether autophagosome fusion with lysosomes and MVB fusion with lysosomes are coordinately regulated in response to cellular status is not well understood.
Cellular senescence
Concerning the above notion, it is important to discuss exosome secretion from senescent cells. Senescence is associated with the secretion of factors, the so-called senescence-associated secretory phenotype (SASP). Several studies have suggested the involvement of EVs as a secretion feature of SASPs, namely, “senescence-associated EVs (Kadota et al., 2018; Misawa et al., 2020; Salminen et al., 2020; Takasugi, 2018; Urbanelli et al., 2016).” Both aspects of EV secretion are included, the alternative waste disposal method and cell-to-cell communication. Takahashi et al. (2017) reported that inhibition of exosome secretion results in nuclear DNA accumulation in the cytoplasm, thereby activating the cytoplasmic DNA sensing machinery. The authors used ALIX or Rab27a knockdown, ILV biogenesis inhibition or MVB movement, and fusion to the plasma membrane to examine the roles of exosome secretion in cellular senescence (Takahashi et al., 2017). This will imply that both waste disposal from cytosolic compartments to ILVs and from intracellular degradation through MVBs to lysosomes equally save cells from cellular senescence. Recently, the importance of lysosomal function in cellular senescence has emerged (Johmura et al., 2021; Suda et al., 2021). Two different studies targeting senescent cells found lysosome functions for “senolysis,” removal of senescent cells. It was found that cellular senescence is associated with lysosomal membrane damage causing lower intracellular pH, induction of glutaminase 1 (GLS1) expression, and glutaminolysis-induced ammonia production for neutralization. GLS1 inhibition successfully induces senolysis in vivo (Johmura et al., 2021). By searching for molecules with transmembrane domains specifically expressed in senescent cells, Suda et al. (2021) found glycoprotein nonmetastatic melanoma protein B (GPNMB) as a molecular target for senolytic vaccine development. Importantly, GPNMB is a lysosomal protein that is overexpressed due to cellular senescence and becomes located in the plasma membrane (Suda et al., 2021; 2022). GPNMB can be upregulated by lysosomal stress and maintains lysosomal integrity (Suda et al., 2022).
It was reported that DNA damage-induced cellular senescence increases sEV secretion (Hitomi et al., 2020) (Fig. 2). It upregulated nSMase2 expression and downregulated SMS2, a counter enzyme against SMase, thereby increasing ceramide synthesis (Hitomi et al., 2020). In this study, the authors also showed that lysosome inhibition by CQ or bafilomycin A1 induced SASP expression along with the p16Ink4a gene, a master regulator of cellular senescence (Hitomi et al., 2020). In both cross-sectional and longitudinal analyses of EVs in the context of age in human subjects, circulating EVs analyzed using NTA suggested that EV concentrations declined with advancing age (Eitan et al., 2017). The decreased EV concentrations in aged people were explained by the increased uptake into B cells and monocytes in the blood (Eitan et al., 2017) (Fig. 2).
ACTION MECHANISM OF EVs
RNA cargos
Based on the discovery of ribonucleic acids in small EVs (Valadi et al., 2007), numerous reports have demonstrated the importance of RNA cargos such as microRNA and long noncoding RNA (lncRNA) and messenger RNA (mRNA) in EVs by having paracrine and endocrine roles (O’Brien et al., 2020).
Genetic information is stored in genomic DNA in the nucleus. RNA is transcribed using this DNA as a template, and proteins are translated based on RNA. That is the central dogma. It turns out that there are only approximately 30,000 genes in the human genome, but the genes mentioned here are those that are transcribed into RNA to become mRNA and translated into proteins. The total human genome is very long at 3.1 billion base pairs, but even if all parts that eventually become proteins are added, these form less than 2% of the total genome length (Yao et al., 2019). More than 98% of this region of the genome encodes at least thousands of noncoding RNAs (ncRNAs), and in fact, approximately 80% of genomic DNA has been revealed to encode ncRNAs. Of course, tRNA (transfer RNA) carries amino acids at the translation stage, and rRNA (ribosomal RNA), which is the location translation occurs, is also a type of ncRNA. On the other hand, microRNA is a single-stranded RNA consisting of 21-25 bases that complementarily binds to the nonprotein region of the 3' end of mRNA that is translated into a protein. It regulates the expressions and translations of various genes by suppressing protein translation and guiding RNA to degradation. Additionally, lncRNAs with two hundred bases or more have a wide range of lengths and various functions. Most lncRNAs are present in the nucleus and function in the structural regulation of chromatin, transcriptional regulation of mRNA, and regulation of stability (Yao et al., 2019). The characteristic of these ncRNAs is that one ncRNA regulates the expressions of a large number of genes. Since the roles of ncRNAs have been reported to include the processes of cell differentiation, transformation, and tumorigenesis, ncRNAs may play a role in such aspects of the coordinated regulation of the entire cell.
There have been reports on the roles of RNA cargos, including microRNAs and lncRNAs, in EVs. For example, fat-derived exosomal microRNAs regulate Fgf21 expression in the liver (Thomou et al., 2017). Obesity-associated exosomal microRNAs affect glucose and lipid metabolism (Castaño et al., 2018). The lncRNA, PUNISHER, is upregulated in patients with coronary artery disease and can induce vascular endothelial growth Factor A (VEGFA) mRNA expression and promote an angiogenic response in recipient endothelial cells (Hosen et al., 2021). Several studies have demonstrated that RNA-binding proteins (RBPs) are particularly involved in the selective loading of RNAs into EVs (Fabbiano et al., 2020; Garcia-Martin et al., 2022; Tosar et al., 2021).
Here, we attempt to argue against such a genetic communication paradigm (Fig. 3). One important question regarding this paradigm is raised from the finding that tRNAs are among the most abundant RNAs involved in EVs (Tosar et al., 2021). The levels of tRNAs are much greater than those of others, including microRNAs (Tosar et al., 2021). Moreover, the number of molecules of a particular microRNA contained in an EV is too low to exert its effect (Tosar et al., 2021). There are also issues with the experimental strategies used to support the genetic communication paradigm in EV studies (Jeppesen et al., 2019; Murillo et al., 2019). In addition, techniques such as overexpressing specific microRNAs of interest, overexpressing a molecule called an antagomir that complementarily inhibits function, or knocking out the microRNA in its encoding genomic region are employed. However, as mentioned above, microRNAs play an important role in the cells that produce them, such as changing the traits of these cells; therefore, such a method may alter the phenotype of the producing cells (Yao et al., 2019). The same thing can be said of RBPs. RBPs impact multiple aspects of RNA biology, and their silencing could affect cellular phenotypes other than RNA sorting into EVs (Tosar et al., 2021).
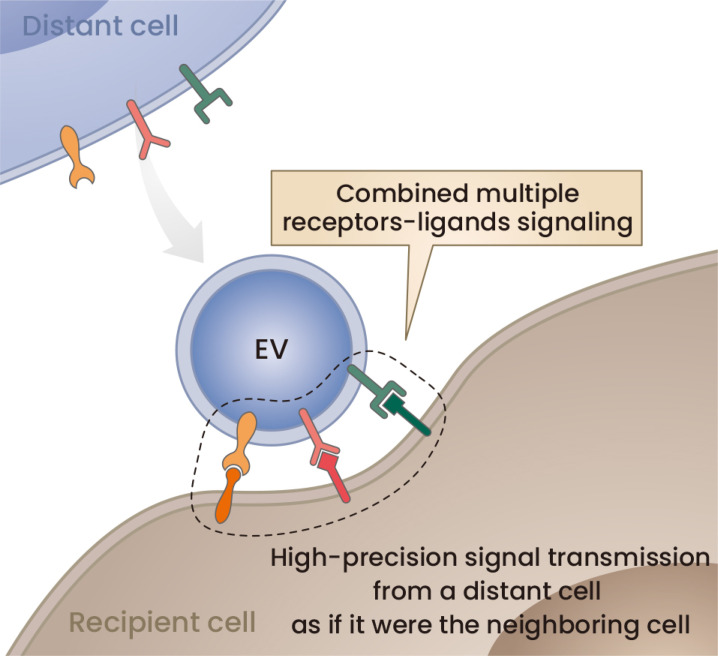
Fig. 3. Hypothesis on the role of EV surface ligands and receptors.
Because there has been a lack of influential evidence showing the importance of RNA cargos and because there have been increasing numbers of reports on the role of EV surface receptors/ligands, we propose a hypothesis in which EVs can transmit high-precision signals between distant cells by using multiple receptors and ligands as if they were the neighboring cell.
Other mechanisms
The membranes of EVs, especially those of exosomes, are derived from the plasma membrane. In particular, lipid rafts are thought to invaginate inside cells to form early endosomes. ILVs are generated by inward budding of their membranes. Because lipid rafts are thought to be specialized membranes rich in integral membrane proteins such as signaling receptors along with ceramides and cholesterol, exosomes represent membrane proteins at the surface and are important for signaling, such as PD-L1 (Chen et al., 2018), MHC class switch (Clayton et al., 2001), Notch ligand Delta-like 4 (Sheldon et al., 2010), and EphA2 (Takasugi et al., 2017). Importantly, these membrane proteins have become recognized as the functional mechanism by which EVs transfer signals to recipient cells. Similar to cell‒cell junctions, EV membrane proteins may facilitate cell‒cell communications in distant locations (Fig. 3).
Exosomes are signaling packages that integrate all of the nucleic acids, various bioactive proteins, and lipids, and these various signals must act cooperatively at various stages in recipient cells.
SYSTEMIC LEVELS OF EVs
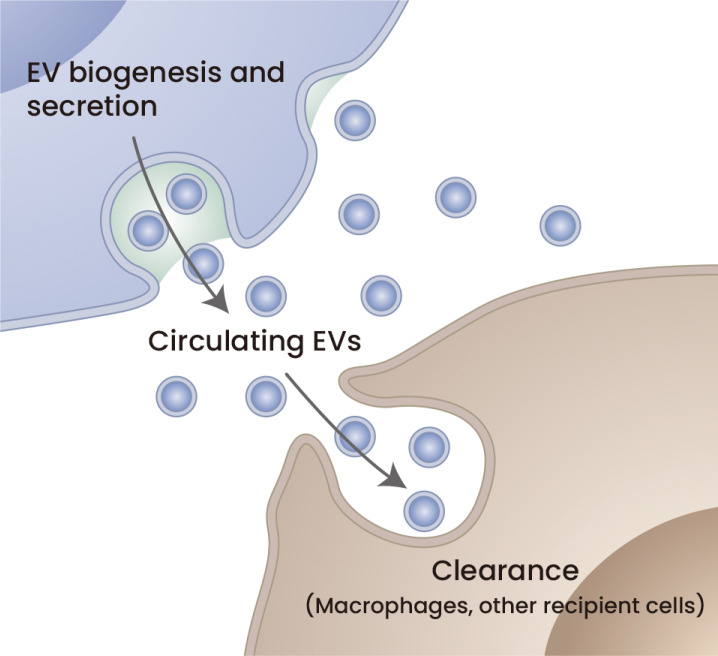
It is of great interest to know which cell type or which organ most strongly affects the total mass of EVs systemically in the blood. Currently, multiple strategies and tools have been developed to isolate EVs from biological fluids such as blood (Sidhom et al., 2020). Additionally, multiple methods have been developed to evaluate the masses of EVs. We have reported that the adiponectin/T-cadherin system affects plasma EV levels (Kita et al., 2019b; Obata et al., 2018). We employed a combined method of PEG precipitation and differential ultracentrifugation to semipurify EVs from plasma (Kawada-Horitani et al., 2022; Nakamura et al., 2020; Obata et al., 2018). A similar approach was fully validated recently (Rider et al., 2016). Because such methods cannot exclude contaminants such as aggregated proteins, we evaluated sEV-specific markers by western blotting. Various approaches have been employed to measure plasma sEV levels. PEG-based precipitation combined with NTA offered insights into the physiological changes of sEVs in plasma (Eitan et al., 2017; Freeman et al., 2018). Increased particle numbers in the fasting plasma of type 2 diabetes patients compared with euglycemic control patients were reported (Freeman et al., 2018). An age-related increase in plasma sEVs was also reported using similar methods (Eitan et al., 2017). Quantification of exosomes in biological fluids is still challenging in its methodology (Mateescu et al., 2017; Witwer et al., 2013). Recently, a knockin mouse expressing the luciferase fusion CD63 was reported (Luo et al., 2020). The strategy to enable the measurement of sEVs by engineering CD63 fusion with 19 kDa small and efficient luciferase, nanoluc, was also established at the cell culture level (Cashikar and Hanson, 2019; Hikita et al., 2018; Kojima et al., 2018). Using mice expressing CD63-nanoluc fusion protein in the desired cell type may be a simplified approach to study changes in sEV secretion in vivo. Apart from these, it was reported that plasma EVs labeled with MFG-E8-fused gLuc were cleared with a half-life of approximately 7 min in mice (Matsumoto et al., 2020). The pharmacokinetic analysis further demonstrated that the plasma sEV secretion rate was calculated as 18 μg/min in mice (Matsumoto et al., 2020) (Fig. 4).
Fig. 4. Circulating EV levels are regulated by EV secretion and clearance.
Plasma EVs labeled with MFG-E8-fused gLuc were cleared with a half-life of approximately 7 min in mice. The plasma sEV secretion rate was calculated as 18 μg/min in mice based on pharmacokinetic analysis (Matsumoto et al., 2020).
CONCLUDING REMARKS
For future studies to reveal the physiological and pharmacological significance of sEVs, exosome biogenesis, and secretion, we propose several points below.
EVs isolated from biological fluids or even produced by a single cell type are heterogeneous and contain substantial amounts of contaminants with the currently available purification technologies. A loss-of-exosome study will be required to demonstrate the importance of exosomes.
Because endocrine functions depend on the concentration changes in sEVs in biological fluids similar to other endocrine factors, such as hormones, a versatile measurement technology for sEVs will be a key point. A detailed comparison between a luciferase fusion exosome marker protein and its native protein in various measurement technologies may push this field forward.
Concerning this, an approach to measure tissue/cell-type-specific sEVs in mice will have great importance. A mouse expressing a space-temporal controllable luciferase fusion exosome marker may be desired.
Regarding the mechanism by which sEVs transmit signals to recipient cells, we propose the importance of cell-surface ligands and receptors on the sEV surface (Fig. 3). sEVs may be a tool to communicate with recipient cells at a distance by engaging their plasma membranes with each other.
Footnotes
AUTHOR CONTRIBUTIONS
S.K. wrote the original draft and contributed to the tables and figure. I.S. revised and edited the manuscript.
CONFLICT OF INTEREST
The authors have no potential conflicts of interest to disclose.
REFERENCES
- Alvarez-Erviti L., Seow Y., Schapira A.H., Gardiner C., Sargent I.L., Wood M.J., Cooper J.M. Lysosomal dysfunction increases exosome-mediated alpha-synuclein release and transmission. Neurobiol. Dis. 2011;42:360–367. doi: 10.1016/j.nbd.2011.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreu Z., Yanez-Mo M. Tetraspanins in extracellular vesicle formation and function. Front. Immunol. 2014;5:442. doi: 10.3389/fimmu.2014.00442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baietti M.F., Zhang Z., Mortier E., Melchior A., Degeest G., Geeraerts A., Ivarsson Y., Depoortere F., Coomans C., Vermeiren E., et al. Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat. Cell Biol. 2012;14:677–685. doi: 10.1038/ncb2502. [DOI] [PubMed] [Google Scholar]
- Balatskaya M.N., Sharonov G.V., Baglay A.I., Rubtsov Y.P., Tkachuk V.A. Different spatiotemporal organization of GPI-anchored T-cadherin in response to low-density lipoprotein and adiponectin. Biochim. Biophys. Acta Gen. Subj. 2019;1863:129414. doi: 10.1016/j.bbagen.2019.129414. [DOI] [PubMed] [Google Scholar]
- Bari R., Guo Q., Xia B., Zhang Y.H., Giesert E.E., Levy S., Zheng J.J., Zhang X.A. Tetraspanins regulate the protrusive activities of cell membrane. Biochem. Biophys. Res. Commun. 2011;415:619–626. doi: 10.1016/j.bbrc.2011.10.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cashikar A.G., Hanson P.I. A cell-based assay for CD63-containing extracellular vesicles. PLoS One. 2019;14:e0220007. doi: 10.1371/journal.pone.0220007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castaño C., Kalko S., Novials A., Párrizas M. Obesity-associated exosomal miRNAs modulate glucose and lipid metabolism in mice. Proc. Natl. Acad. Sci. U. S. A. 2018;115:12158–12163. doi: 10.1073/pnas.1808855115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chairoungdua A., Smith D.L., Pochard P., Hull M., Caplan M.J. Exosome release of β-catenin: a novel mechanism that antagonizes Wnt signaling. J. Cell Biol. 2010;190:1079–1091. doi: 10.1083/jcb.201002049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G., Huang A.C., Zhang W., Zhang G., Wu M., Xu W., Yu Z., Yang J., Wang B., Sun H., et al. Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature. 2018;560:382–386. doi: 10.1038/s41586-018-0392-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choezom D., Gross J.C. Neutral sphingomyelinase 2 controls exosome secretion by counteracting V-ATPase-mediated endosome acidification. J. Cell Sci. 2022;135:jcs259324. doi: 10.1242/jcs.259324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton A., Court J., Navabi H., Adams M., Mason M.D., Hobot J.A., Newman G.R., Jasani B. Analysis of antigen presenting cell derived exosomes, based on immuno-magnetic isolation and flow cytometry. J. Immunol. Methods. 2001;247:163–174. doi: 10.1016/S0022-1759(00)00321-5. [DOI] [PubMed] [Google Scholar]
- Colombo M., Moita C., van Niel G., Kowal J., Vigneron J., Benaroch P., Manel N., Moita L.F., Thery C., Raposo G. Analysis of ESCRT functions in exosome biogenesis, composition and secretion highlights the heterogeneity of extracellular vesicles. J. Cell Sci. 2013;126:5553–5565. doi: 10.1242/jcs.128868. [DOI] [PubMed] [Google Scholar]
- Colombo M., Raposo G., Théry C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 2014;30:255–289. doi: 10.1146/annurev-cellbio-101512-122326. [DOI] [PubMed] [Google Scholar]
- Devis-Jauregui L., Eritja N., Davis M.L., Matias-Guiu X., Llobet-Navàs D. Autophagy in the physiological endometrium and cancer. Autophagy. 2021;17:1077–1095. doi: 10.1080/15548627.2020.1752548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinkins M.B., Dasgupta S., Wang G., Zhu G., Bieberich E. Exosome reduction in vivo is associated with lower amyloid plaque load in the 5XFAD mouse model of Alzheimer's disease. Neurobiol. Aging. 2014;35:1792–1800. doi: 10.1016/j.neurobiolaging.2014.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinkins M.B., Enasko J., Hernandez C., Wang G., Kong J., Helwa I., Liu Y., Terry A.V., Bieberich E. Neutral sphingomyelinase-2 deficiency ameliorates Alzheimer's disease pathology and improves cognition in the 5XFAD mouse. J. Neurosci. 2016;36:8653–8667. doi: 10.1523/JNEUROSCI.1429-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eitan E., Green J., Bodogai M., Mode N.A., Bæk R., Jørgensen M.M., Freeman D.W., Witwer K.W., Zonderman A.B., Biragyn A., et al. Age-related changes in plasma extracellular vesicle characteristics and internalization by leukocytes. Sci. Rep. 2017;7:1342. doi: 10.1038/s41598-017-01386-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eitan E., Suire C., Zhang S., Mattson M.P. Impact of lysosome status on extracellular vesicle content and release. Ageing Res. Rev. 2016;32:65–74. doi: 10.1016/j.arr.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Andaloussi S., Mäger I., Breakefield X.O., Wood M.J. Extracellular vesicles: biology and emerging therapeutic opportunities. Nat. Rev. Drug Discov. 2013;12:347–357. doi: 10.1038/nrd3978. [DOI] [PubMed] [Google Scholar]
- Fabbiano F., Corsi J., Gurrieri E., Trevisan C., Notarangelo M., D'Agostino V.G. RNA packaging into extracellular vesicles: an orchestra of RNA‐binding proteins? J. Extracell. Vesicles. 2020;10:e12043. doi: 10.1002/jev2.12043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fader C.M., Sánchez D.G., Mestre M.B., Colombo M.I. TI-VAMP/VAMP7 and VAMP3/cellubrevin: two v-SNARE proteins involved in specific steps of the autophagy/multivesicular body pathways. Biochim. Biophys. Acta. 2009;1793:1901–1916. doi: 10.1016/j.bbamcr.2009.09.011. [DOI] [PubMed] [Google Scholar]
- Fang Y., Wu N., Gan X., Yan W., Morrell J.C., Gould S.J. Higher-order oligomerization targets plasma membrane proteins and HIV gag to exosomes. PLoS Biol. 2007;5:e158. doi: 10.1371/journal.pbio.0050158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedele A.O., Proud C.G. Chloroquine and bafilomycin A mimic lysosomal storage disorders and impair mTORC1 signalling. Biosci. Rep. 2020;40:BSR20200905. doi: 10.1042/BSR20200905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman D.W., Noren Hooten N., Eitan E., Green J., Mode N.A., Bodogai M., Zhang Y., Lehrmann E., Zonderman A.B., Biragyn A., et al. Altered extracellular vesicle concentration, cargo, and function in diabetes. Diabetes. 2018;67:2377–2388. doi: 10.2337/db17-1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda S., Kita S., Obata Y., Fujishima Y., Nagao H., Masuda S., Tanaka Y., Nishizawa H., Funahashi T., Takagi J., et al. The unique prodomain of T-cadherin plays a key role in adiponectin binding with the essential extracellular cadherin repeats 1 and 2. J. Biol. Chem. 2017;292:7840–7849. doi: 10.1074/jbc.M117.780734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Martin R., Wang G., Brandão B.B., Zanotto T.M., Shah S., Kumar Patel S., Schilling B., Kahn C.R. MicroRNA sequence codes for small extracellular vesicle release and cellular retention. Nature. 2022;601:446–451. doi: 10.1038/s41586-021-04234-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gézsi A., Kovács Á., Visnovitz T., Buzás E.I. Systems biology approaches to investigating the roles of extracellular vesicles in human diseases. Exp. Mol. Med. 2019;51:1–11. doi: 10.1038/s12276-019-0226-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghossoub R., Chery M., Audebert S., Leblanc R., Egea-Jimenez A.L., Lembo F., Mammar S., Le Dez F., Camoin L., Borg J.P., et al. Tetraspanin-6 negatively regulates exosome production. Proc. Natl. Acad. Sci. U. S. A. 2020;117:5913–5922. doi: 10.1073/pnas.1922447117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghossoub R., Lembo F., Rubio A., Gaillard C.B., Bouchet J., Vitale N., Slavik J., Machala M., Zimmermann P. Syntenin-ALIX exosome biogenesis and budding into multivesicular bodies are controlled by ARF6 and PLD2. Nat. Commun. 2014;5:3477. doi: 10.1038/ncomms4477. [DOI] [PubMed] [Google Scholar]
- Gurunathan S., Kang M.H., Kim J.H. A comprehensive review on factors influences biogenesis, functions, therapeutic and clinical implications of exosomes. Int. J. Nanomedicine. 2021;16:1281–1312. doi: 10.2147/IJN.S291956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henne W.M., Buchkovich N.J., Emr S.D. The ESCRT pathway. Dev. Cell. 2011;21:77–91. doi: 10.1016/j.devcel.2011.05.015. [DOI] [PubMed] [Google Scholar]
- Hessvik N.P., Llorente A. Current knowledge on exosome biogenesis and release. Cell. Mol. Life Sci. 2018;75:193–208. doi: 10.1007/s00018-017-2595-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hikita T., Miyata M., Watanabe R., Oneyama C. Sensitive and rapid quantification of exosomes by fusing luciferase to exosome marker proteins. Sci. Rep. 2018;8:14035. doi: 10.1038/s41598-018-32535-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitomi K., Okada R., Loo T.M., Miyata K., Nakamura A.J., Takahashi A. DNA damage regulates senescence-associated extracellular vesicle release via the ceramide pathway to prevent excessive inflammatory responses. Int. J. Mol. Sci. 2020;21:3720. doi: 10.3390/ijms21103720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosen M.R., Li Q., Liu Y., Zietzer A., Maus K., Goody P., Uchida S., Latz E., Werner N., Nickenig G., et al. CAD increases the long noncoding RNA PUNISHER in small extracellular vesicles and regulates endothelial cell function via vesicular shuttling. Mol. Ther. Nucleic Acids. 2021;25:388–405. doi: 10.1016/j.omtn.2021.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley J.H., Boura E., Carlson L.A., Rozycki B. Membrane budding. Cell. 2010;143:875–887. doi: 10.1016/j.cell.2010.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imjeti N.S., Menck K., Egea-Jimenez A.L., Lecointre C., Lembo F., Bouguenina H., Badache A., Ghossoub R., David G., Roche S., et al. Syntenin mediates SRC function in exosomal cell-to-cell communication. Proc. Natl. Acad. Sci. U. S. A. 2017;114:12495–12500. doi: 10.1073/pnas.1713433114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jadli A.S., Ballasy N., Edalat P., Patel V.B. Inside (sight) of tiny communicator: exosome biogenesis, secretion, and uptake. Mol. Cell. Biochem. 2020;467:77–94. doi: 10.1007/s11010-020-03703-z. [DOI] [PubMed] [Google Scholar]
- Jeon H.Y., Das S.K., Dasgupta S., Emdad L., Sarkar D., Kim S.H., Lee S.G., Fisher P.B. Expression patterns of MDA-9/syntenin during development of the mouse embryo. J. Mol. Histol. 2013;44:159–166. doi: 10.1007/s10735-012-9468-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeppesen D.K., Fenix A.M., Franklin J.L., Higginbotham J.N., Zhang Q., Zimmerman L.J., Liebler D.C., Ping J., Liu Q., Evans R., et al. Reassessment of exosome composition. Cell. 2019;177:428–445.e18. doi: 10.1016/j.cell.2019.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johmura Y., Yamanaka T., Omori S., Wang T.W., Sugiura Y., Matsumoto M., Suzuki N., Kumamoto S., Yamaguchi K., Hatakeyama S., et al. Senolysis by glutaminolysis inhibition ameliorates various age-associated disorders. Science. 2021;371:265–270. doi: 10.1126/science.abb5916. [DOI] [PubMed] [Google Scholar]
- Kadota T., Fujita Y., Yoshioka Y., Araya J., Kuwano K., Ochiya T. Emerging role of extracellular vesicles as a senescence-associated secretory phenotype: insights into the pathophysiology of lung diseases. Mol. Aspects Med. 2018;60:92–103. doi: 10.1016/j.mam.2017.11.005. [DOI] [PubMed] [Google Scholar]
- Kajimoto T., Okada T., Miya S., Zhang L., Nakamura S. Ongoing activation of sphingosine 1-phosphate receptors mediates maturation of exosomal multivesicular endosomes. Nat. Commun. 2013;4:2712. doi: 10.1038/ncomms3712. [DOI] [PubMed] [Google Scholar]
- Kalluri R., LeBleu V.S. The biology, function, and biomedical applications of exosomes. Science. 2020;367:eaau6977. doi: 10.1126/science.aau6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashyap R., Balzano M., Lechat B., Lambaerts K., Egea-Jimenez A.L., Lembo F., Fares J., Meeussen S., Kugler S., Roebroek A., et al. Syntenin-knock out reduces exosome turnover and viral transduction. Sci. Rep. 2021;11:4083. doi: 10.1038/s41598-021-81697-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawada-Horitani E., Kita S., Okita T., Nakamura Y., Nishida H., Honma Y., Fukuda S., Tsugawa-Shimizu Y., Kozawa J., Sakaue T., et al. Human adipose-derived mesenchymal stem cells prevent type 1 diabetes induced by immune checkpoint blockade. Diabetologia. 2022;65:1185–1197. doi: 10.1007/s00125-022-05708-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kita S., Fukuda S., Maeda N., Shimomura I. Native adiponectin in serum binds to mammalian cells expressing T-cadherin, but not AdipoRs or calreticulin. Elife. 2019a;8:e48675. doi: 10.7554/eLife.48675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kita S., Maeda N., Shimomura I. Interorgan communication by exosomes, adipose tissue, and adiponectin in metabolic syndrome. J. Clin. Invest. 2019b;129:4041–4049. doi: 10.1172/JCI129193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kita S., Shimomura I. Stimulation of exosome biogenesis by adiponectin, a circulating factor secreted from adipocytes. J. Biochem. 2021;169:173–179. doi: 10.1093/jb/mvaa105. [DOI] [PubMed] [Google Scholar]
- Kojima R., Bojar D., Rizzi G., Hamri G.C.E., El-Baba M.D., Saxena P., Ausländer S., Tan K.R., Fussenegger M. Designer exosomes produced by implanted cells intracerebrally deliver therapeutic cargo for Parkinson's disease treatment. Nat. Commun. 2018;9:1305. doi: 10.1038/s41467-018-03733-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W., Dai Y., Chen Z., Yue X., Andrade-Powell K.C., Chang J. Spatial and temporal tracking of cardiac exosomes in mouse using a nano-luciferase-CD63 fusion protein. Commun. Biol. 2020;3:114. doi: 10.1038/s42003-020-0830-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateescu B., Kowal E.J., van Balkom B.W., Bartel S., Bhattacharyya S.N., Buzás E.I., Buck A.H., de Candia P., Chow F.W., Das S., et al. Obstacles and opportunities in the functional analysis of extracellular vesicle RNA - an ISEV position paper. J. Extracell. Vesicles. 2017;6:1286095. doi: 10.1080/20013078.2017.1286095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu M., Martin-Jaular L., Lavieu G., Thery C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat. Cell Biol. 2019;21:9–17. doi: 10.1038/s41556-018-0250-9. [DOI] [PubMed] [Google Scholar]
- Matsumoto A., Takahashi Y., Chang H.Y., Wu Y.W., Yamamoto A., Ishihama Y., Takakura Y. Blood concentrations of small extracellular vesicles are determined by a balance between abundant secretion and rapid clearance. J. Extracell. Vesicles. 2020;9:1696517. doi: 10.1080/20013078.2019.1696517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Y., Li G., Zhang X., Xu H., Abraham S.N. A TRP channel senses lysosome neutralization by pathogens to trigger their expulsion. Cell. 2015;161:1306–1319. doi: 10.1016/j.cell.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misawa T., Tanaka Y., Okada R., Takahashi A. Biology of extracellular vesicles secreted from senescent cells as senescence‐associated secretory phenotype factors. Geriatr. Gerontol. Int. 2020;20:539–546. doi: 10.1111/ggi.13928. [DOI] [PubMed] [Google Scholar]
- Murillo O.D., Thistlethwaite W., Rozowsky J., Subramanian S.L., Lucero R., Shah N., Jackson A.R., Srinivasan S., Chung A., Laurent C.D., et al. exRNA Atlas analysis reveals distinct extracellular RNA cargo types and their carriers present across human biofluids. Cell. 2019;177:463–477.e15. doi: 10.1016/j.cell.2019.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murrow L., Malhotra R., Debnath J. ATG12-ATG3 interacts with Alix to promote basal autophagic flux and late endosome function. Nat. Cell Biol. 2015;17:300–310. doi: 10.1038/ncb3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y., Kita S., Tanaka Y., Fukuda S., Obata Y., Okita T., Nishida H., Takahashi Y., Kawachi Y., Tsugawa-Shimizu Y., et al. Adiponectin stimulates exosome release to enhance mesenchymal stem-cell-driven therapy of heart failure in mice. Mol. Ther. 2020;28:2203–2219. doi: 10.1016/j.ymthe.2020.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien K., Breyne K., Ughetto S., Laurent L.C., Breakefield X.O. RNA delivery by extracellular vesicles in mammalian cells and its applications. Nat. Rev. Mol. Cell Biol. 2020;21:585–606. doi: 10.1038/s41580-020-0251-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obata Y., Kita S., Koyama Y., Fukuda S., Takeda H., Takahashi M., Fujishima Y., Nagao H., Masuda S., Tanaka Y., et al. Adiponectin/T-cadherin system enhances exosome biogenesis and decreases cellular ceramides by exosomal release. JCI Insight. 2018;3:e99680. doi: 10.1172/jci.insight.99680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrowski M., Carmo N.B., Krumeich S., Fanget I., Raposo G., Savina A., Moita C.F., Schauer K., Hume A.N., Freitas R.P., et al. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat. Cell Biol. 2010;12:19–30. doi: 10.1038/ncb2000. sup pp 1-13. [DOI] [PubMed] [Google Scholar]
- Phuyal S., Hessvik N.P., Skotland T., Sandvig K., Llorente A. Regulation of exosome release by glycosphingolipids and flotillins. FEBS J. 2014;281:2214–2227. doi: 10.1111/febs.12775. [DOI] [PubMed] [Google Scholar]
- Poupardin R., Wolf M., Strunk D. Adherence to minimal experimental requirements for defining extracellular vesicles and their functions. Adv. Drug Deliv. Rev. 2021;176:113872. doi: 10.1016/j.addr.2021.113872. [DOI] [PubMed] [Google Scholar]
- Rider M.A., Hurwitz S.N., Meckes D.G. ExtraPEG: a polyethylene glycol-based method for enrichment of extracellular vesicles. Sci. Rep. 2016;6:23978. doi: 10.1038/srep23978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roucourt B., Meeussen S., Bao J., Zimmermann P., David G. Heparanase activates the syndecan-syntenin-ALIX exosome pathway. Cell Res. 2015;25:412–428. doi: 10.1038/cr.2015.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salminen A., Kaarniranta K., Kauppinen A. Exosomal vesicles enhance immunosuppression in chronic inflammation: impact in cellular senescence and the aging process. Cell. Signal. 2020;75:109771. doi: 10.1016/j.cellsig.2020.109771. [DOI] [PubMed] [Google Scholar]
- Savina A., Furlán M., Vidal M., Colombo M.I. Exosome release is regulated by a calcium-dependent mechanism in K562 cells. J. Biol. Chem. 2003;278:20083–20090. doi: 10.1074/jbc.M301642200. [DOI] [PubMed] [Google Scholar]
- Sheldon H., Heikamp E., Turley H., Dragovic R., Thomas P., Oon C.E., Leek R., Edelmann M., Kessler B., Sainson R.C., et al. New mechanism for Notch signaling to endothelium at a distance by Delta-like 4 incorporation into exosomes. Blood. 2010;116:2385–2394. doi: 10.1182/blood-2009-08-239228. [DOI] [PubMed] [Google Scholar]
- Sidhom K., Obi P.O., Saleem A. A review of exosomal isolation methods: is size exclusion chromatography the best option? Int. J. Mol. Sci. 2020;21:6466. doi: 10.3390/ijms21186466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skotland T., Sandvig K., Llorente A. Lipids in exosomes: current knowledge and the way forward. Prog. Lipid Res. 2017;66:30–41. doi: 10.1016/j.plipres.2017.03.001. [DOI] [PubMed] [Google Scholar]
- Stuffers S., Sem Wegner C., Stenmark H., Brech A. Multivesicular endosome biogenesis in the absence of ESCRTs. Traffic. 2009;10:925–937. doi: 10.1111/j.1600-0854.2009.00920.x. [DOI] [PubMed] [Google Scholar]
- Suda M., Shimizu I., Katsuumi G., Hsiao C.L., Yoshida Y., Matsumoto N., Yoshida Y., Katayama A., Wada J., Seki M., et al. Glycoprotein nonmetastatic melanoma protein B regulates lysosomal integrity and lifespan of senescent cells. Sci. Rep. 2022;12:6522. doi: 10.1038/s41598-022-10522-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suda M., Shimizu I., Katsuumi G., Yoshida Y., Hayashi Y., Ikegami R., Matsumoto N., Yoshida Y., Mikawa R., Katayama A., et al. Senolytic vaccination improves normal and pathological age-related phenotypes and increases lifespan in progeroid mice. Nat. Aging. 2021;1:1117–1126. doi: 10.1038/s43587-021-00151-2. [DOI] [PubMed] [Google Scholar]
- Takahashi A., Okada R., Nagao K., Kawamata Y., Hanyu A., Yoshimoto S., Takasugi M., Watanabe S., Kanemaki M.T., Obuse C., et al. Exosomes maintain cellular homeostasis by excreting harmful DNA from cells. Nat. Commun. 2017;8:15287. doi: 10.1038/ncomms15287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takasugi M. Emerging roles of extracellular vesicles in cellular senescence and aging. Aging Cell. 2018;17:e12734. doi: 10.1111/acel.12734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takasugi M., Okada R., Takahashi A., Virya Chen D., Watanabe S., Hara E. Small extracellular vesicles secreted from senescent cells promote cancer cell proliferation through EphA2. Nat. Commun. 2017;8:15729. doi: 10.1038/ncomms15728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y., Kita S., Nishizawa H., Fukuda S., Fujishima Y., Obata Y., Nagao H., Masuda S., Nakamura Y., Shimizu Y., et al. Adiponectin promotes muscle regeneration through binding to T-cadherin. Sci. Rep. 2019;9:16. doi: 10.1038/s41598-018-37115-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Théry C., Amigorena S., Raposo G., Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. Chapter. 2006;3 doi: 10.1002/0471143030.cb0322s30. Unit 3.22. [DOI] [PubMed] [Google Scholar]
- Thomou T., Mori M.A., Dreyfuss J.M., Konishi M., Sakaguchi M., Wolfrum C., Rao T.N., Winnay J.N., Garcia-Martin R., Grinspoon S.K., et al. Adipose-derived circulating miRNAs regulate gene expression in other tissues. Nature. 2017;542:450–455. doi: 10.1038/nature21365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson C.A., Purushothaman A., Ramani V.C., Vlodavsky I., Sanderson R.D. Heparanase regulates secretion, composition, and function of tumor cell-derived exosomes. J. Biol. Chem. 2013;288:10093–10099. doi: 10.1074/jbc.C112.444562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosar J.P., Witwer K., Cayota A. Revisiting extracellular RNA release, processing, and function. Trends Biochem. Sci. 2021;46:438–445. doi: 10.1016/j.tibs.2020.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trajkovic K., Hsu C., Chiantia S., Rajendran L., Wenzel D., Wieland F., Schwille P., Brügger B., Simons M. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science. 2008;319:1244–1247. doi: 10.1126/science.1153124. [DOI] [PubMed] [Google Scholar]
- Tsugawa-Shimizu Y., Fujishima Y., Kita S., Minami S., Sakaue T.A., Nakamura Y., Okita T., Kawachi Y., Fukada S., Namba-Hamano T., et al. Increased vascular permeability and severe renal tubular damage after ischemia-reperfusion injury in mice lacking adiponectin or T-cadherin. Am. J. Physiol. Endocrinol. Metab. 2021;320:E179–E190. doi: 10.1152/ajpendo.00393.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanelli L., Buratta S., Sagini K., Tancini B., Emiliani C. Extracellular vesicles as new players in cellular senescence. Int. J. Mol. Sci. 2016;17:1408. doi: 10.3390/ijms17091408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valadi H., Ekström K., Bossios A., Sjöstrand M., Lee J.J., Lötvall J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- van Niel G., Charrin S., Simoes S., Romao M., Rochin L., Saftig P., Marks M.S., Rubinstein E., Raposo G. The tetraspanin CD63 regulates ESCRT-independent and -dependent endosomal sorting during melanogenesis. Dev. Cell. 2011;21:708–721. doi: 10.1016/j.devcel.2011.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Niel G., D'Angelo G., Raposo G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2018;19:213–228. doi: 10.1038/nrm.2017.125. [DOI] [PubMed] [Google Scholar]
- Verderio C., Gabrielli M., Giussani P. Role of sphingolipids in the biogenesis and biological activity of extracellular vesicles. J. Lipid Res. 2018;59:1325–1340. doi: 10.1194/jlr.R083915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei D., Zhan W., Gao Y., Huang L., Gong R., Wang W., Zhang R., Wu Y., Gao S., Kang T. RAB31 marks and controls an ESCRT-independent exosome pathway. Cell Res. 2021;31:157–177. doi: 10.1038/s41422-020-00409-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witwer K.W., Buzás E.I., Bemis L.T., Bora A., Lässer C., Lötvall J., Nolte-'t Hoen E.N., Piper M.G., Sivaraman S., Skog J., et al. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J. Extracell. Vesicles. 2013;2 doi: 10.3402/jev.v2i0.20360. 10.3402/jev.v2i0.20360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witwer K.W., Thery C. Extracellular vesicles or exosomes? On primacy, precision, and popularity influencing a choice of nomenclature. J. Extracell. Vesicles. 2019;8:1648167. doi: 10.1080/20013078.2019.1648167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie S., Zhang Q., Jiang L. Current knowledge on exosome biogenesis, cargo-sorting mechanism and therapeutic implications. Membranes (Basel) 2022;12:498. doi: 10.3390/membranes12050498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao R.W., Wang Y., Chen L.L. Cellular functions of long noncoding RNAs. Nat. Cell Biol. 2019;21:542–551. doi: 10.1038/s41556-019-0311-8. [DOI] [PubMed] [Google Scholar]
- Zimmermann P., Tomatis D., Rosas M., Grootjans J., Leenaerts I., Degeest G., Reekmans G., Coomans C., David G. Characterization of syntenin, a syndecan-binding PDZ protein, as a component of cell adhesion sites and microfilaments. Mol. Biol. Cell. 2001;12:339–350. doi: 10.1091/mbc.12.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]