Abstract
Crohn’s disease (CD) is an inflammatory bowel disease in which Escherichia coli strains have been suspected of being involved. We demonstrated previously that ileal lesions of CD are colonized by E. coli strains able to adhere to intestinal Caco-2 cells but devoid of the virulence genes so far described in the pathogenic E. coli strains involved in gastrointestinal infections. In the present study we compared the invasive ability of one of these strains isolated from an ileal biopsy of a patient with CD, strain LF82, with that of reference enteroinvasive (EIEC), enteropathogenic (EPEC), enterotoxigenic (ETEC), enteraggregative (EAggEC), enterohemorrhagic (EHEC), and diffusely adhering (DAEC) E. coli strains. Gentamicin protection assays showed that E. coli LF82 was able to efficiently invade HEp-2 cells. Its invasive level was not significantly different from that of EIEC and EPEC strains (P > 0.5) but significantly higher than that of ETEC (P < 0.03), EHEC (P < 0.005), EAggEC (P < 0.004) and DAEC (P < 0.02) strains. Strain LF82 also demonstrated efficient ability to invade intestinal epithelial cultured Caco-2, Intestine-407, and HCT-8 cells. Electron microscopy examination of infected HEp-2 cells revealed the presence of numerous intracellular bacteria located in vacuoles or free in the host cell cytoplasm. In addition, the interaction of strain LF82 with epithelial cells was associated with the elongation of microvillar extensions that extruded from the host cell membranes and engulfed the bacteria. This internalization mechanism strongly resembles Salmonella- or Shigella-induced macropinocytosis. The use of cytochalasin D and colchicine showed that the uptake of strain LF82 by HEp-2 cells was mediated by both an actin microfilament-dependent mechanism and microtubule involvement. In addition, strain LF82 survived for at least 24 h in HEp-2 and Intestine-407 cells and efficiently replicated intracellularly in HEp-2 cells. PCR and hybridization experiments did not reveal the presence of any of the genetic determinants encoding EIEC, EPEC, or ETEC proteins involved in bacterial invasion. Thus, these findings show that LF82, which colonized the ileal mucosa of a patient with CD, is a true invasive E. coli strain and suggest the existence of a new potentially pathogenic group of E. coli, which we propose be designated adherent-invasive E. coli.
Escherichia coli is the predominant facultative anaerobe of the human colonic flora. Diarrheagenic E. coli strains have acquired different sets of virulence genes and thus differ from those normally resident in the colon in possessing distinct virulence factors. They are distinguished and defined on the basis of their pathogenic mechanisms and have been divided into six well-defined pathogenic groups: enteroinvasive (EIEC), enterotoxigenic (ETEC), enteropathogenic (EPEC), enterohemorrhagic (EHEC), enteroaggregative (EAggEC), and diffusely adhering (DAEC) E. coli (for a review, see reference 46). Pathogenic groups of E. coli use a common strategy of infection consisting of (i) colonization of the intestinal epithelium, (ii) evasion of host defenses, (iii) multiplication, and (iv) host damage. Although their pathogenic mechanisms are distinct, three general paradigms of virulence have been described: (i) enterotoxin production, (ii) intimate attachment with membrane signalling, and/or (iii) invasion (46).
Epithelial cell invasion is a key virulence factor only for EIEC, which may lead to a dysentery-like illness similar to that caused by Shigella flexneri (16). Like Shigella species, EIEC invades the colonic mucosa of human hosts and elicits a strong acute inflammatory response which results in abscess formation and ulceration of the colon (59). Invasion of cultured epithelial cells by Shigella spp. and EIEC is a multiple-step process consisting of (i) epithelial cell penetration by macropinocytosis, (ii) lysis of the endocytic vacuole, (iii) intracellular multiplication, (iv) movement of the bacteria within the host cell cytoplasm, and (v) spread to adjacent cells (for a review, see reference 50). Genes necessary for invasiveness are located on a 37-kb region of a large, 180- to 210-kb plasmid designated pInv (4, 41, 51, 54, 55). These genes encode secreted proteins (Ipa proteins), of which IpaB, IpaC, and IpaD are effectors of the invasion phenotype (42). Ipa proteins are secreted through a type III secretion system encoded by the mxi and spa loci located on pInv (2, 61). In addition, at least three chromosomal loci are also required for full virulence of Shigella spp.
Although not considered an invasive pathogen, EPEC is able to enter cultured epithelial cells (3, 13, 26, 44). However, the clinical significance of invasiveness in the pathogenesis due to EPEC remains unclear. EPEC pathogenesis has been described as a three-stage model consisting of (i) localized adherence to epithelial cells via the EAF plasmid-encoded bundle-forming pili, (ii) signal transduction, and (iii) intimate adherence (15). Upon adherence to the epithelial cell surface, EPEC elicits attaching and effacing lesions (45), and a subpopulation of EPEC is internalized by epithelial cells (13). EPEC intimate attachment and invasion are both mediated by a 94-kDa protein called intimin (34, 35), the product of the chromosomally encoded eae gene. A type III secretion apparatus is required for the secretion of intimin (33, 38). The intimin receptor has been recently identified as a bacterial protein called Tir (translocated intimin receptor), which is secreted via a type III secretion system and translocated through the host cell membrane (37).
The pathogenesis of ETEC is based on the action of the heat-labile and heat-stable enterotoxins, but some investigators have reported that ETEC strain H10407 is able to invade human intestinal cell lines (17). The capacity for invasion is located on two separate, chromosomally encoded loci, designated tia and tib, which direct the synthesis of a 25-kDa and a 104-kDa outer membrane protein, respectively (18, 25). However, the role of invasion during enterotoxigenic disease remains uncertain.
The other types of enterovirulent E. coli, including EHEC (48), EAggEC (5), and DAEC (29, 36, 64), can invade cultured human epithelial cells. Intracellular bacteria have been identified by both electron microscopy and the recovery of invading bacteria through gentamicin kill assays. However, the role of epithelial cell invasion in their pathogenesis has yet to be determined.
Crohn’s disease (CD) is an inflammatory bowel disease of unknown etiology that closely mimics enteric infections caused by invasive pathogens. Indeed, the inflammation observed on CD intestinal mucosa exhibits histopathologic features similar to those caused by enteric invasive bacteria such as Shigella spp. and Salmonella spp. (for a review, see reference 53). The role of luminal bacteria in the pathogenesis of CD is strongly supported by observations that patients with CD clinically improve when luminal bacterial concentrations are decreased (57, 63). E. coli strains are among the bacteria of the luminal flora suspected of being involved in the pathogenesis of CD. Tabaqchali et al. (58) showed that E. coli antibody titers are higher in patients with CD than in controls. In addition, immunocytochemistry assays revealed the presence of E. coli antigens in most intestinal resection specimens from patients with CD (9). It was recently shown that E. coli strains were abnormally predominant (between 50 and 100% of the total number of aerobes and anaerobes) in early and chronic ileal lesions of CD (12). The authors also demonstrated that most of these strains were able to adhere in vitro to differentiated Caco-2 cells, a property that could enable them to colonize the intestinal mucosa.
Subsequent studies of the interactions between these strains and cultured human epithelial cells revealed an invasive potential. The aim of the present work was to characterize the invasive ability of E. coli LF82 isolated from an ileal biopsy of a patient with CD and compare it with that of reference strains belonging to the well defined pathogenic groups of E. coli involved in gastrointestinal infections.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Strain LF82 was isolated from a chronic ileal lesion of a patient with CD. As kindly determined by Karen Krogfelt (Statens Serum Institute, Copenhagen, Denmark), strain LF82 belongs to E. coli serotype O83:H1. It was sensitive to most antibiotics but not to amoxycillin, as determined by the standard disc diffusion assay performed with antibiotic discs for agar diffusion purchased from Diagnostics Pasteur, Marnes-la-Coquette, France. It adhered strongly to Intestine-407 and Caco-2 cells even in the presence of 1% d-mannose (12).
The following E. coli strains belonging to the different pathogenic groups involved in gastrointestinal tract infections were used: EIEC strain E12860/0, kindly provided by Michael Donnenberg (University of Maryland School of Medicine, Baltimore), EPEC strain E2348/69 (40), ETEC strain H10407 (19), DAEC strain C1845 (7), EHEC strain EDL933 (47), and EAggEC strain 17-2 (62). S. flexneri SC301 (10) was used as a source of the ipaC gene. E. coli K-12 C600 was used as a noninvasive control.
All E. coli strains were grown in Luria-Bertani (LB) broth without shaking or on Mueller-Hinton agar (Institut Pasteur Production, Marnes la Coquette, France) plates overnight at 37°C.
Cell lines and cell culture.
HEp-2 cells (derived from a human larynx carcinoma) and Intestine-407 cells (derived from human intestinal embryonic jejunum and ileum) were purchased from Flow Laboratories, Inc., McLean, Va. Caco-2 and HCT-8 E11R1 cells established from human colonic adenocarcinoma were kindly provided by Alain Zweibaum (INSERM U178, Villejuif, France) and Marc Mareel (Laboratoire de Cancérologie, Université de Gent, Gent, Belgium), respectively.
HEp-2 and Intestine-407 cells were maintained in an atmosphere containing 5% CO2 at 37°C in modified Eagle medium (Seromed, Biochrom KG, Berlin, Germany) supplemented with 10% (vol/vol) heat-inactivated fetal calf serum (FCS) (Seromed), 1% nonessential amino acids (Life Technologies, Cergy-Pontoise, France), 1% l-glutamine (Life Technology), 200 U of penicillin, 50 mg of streptomycin, and 0.25 mg of amphotericin B per liter, and 1% minimal essential medium (MEM) vitamin mix X-100 (Life Technologies).
Caco-2 cells were grown in a 10% CO2 atmosphere in Dulbecco’s modified medium (Seromed) supplemented with 20% FCS, 1% nonessential amino acids, 1% l-glutamine, 200 U of penicillin, 50 mg of streptomycin, and 0.25 mg of amphotericin B per liter, and 1% MEM vitamin mix X-100.
The HCT-8 cell line was maintained in a 5% CO2 atmosphere in RPMI 1640 (Seromed) supplemented with 10% FCS, 1% nonessential amino acids, 1% l-glutamine, 200 U of penicillin, 50 mg of streptomycin, and 0.25 mg of amphotericin B per liter, and 1% MEM vitamin mix X-100.
Invasion assays.
The bacterial invasion of epithelial cells was measured as protection against gentamicin, a bactericidal antibiotic (56). The MBC (concentration that reduced the bacterial count by 99.99%) of the antibiotic for all strains included in this study was determined in tissue culture medium, and the drug was used at 10- to 100-fold the MBC (MBC ≤ 1 μg/ml). Since similar results were obtained independent of the gentamicin concentration even for prolonged periods of up to 24 h, a final concentration of 100 μg/ml was used in subsequent experiments. Monolayers were seeded in 24-well tissue culture plates (Polylabo, Strasbourg, France) with 4 × 105 cells/well for HEp-2 and Intestine-407 cell lines and incubated for 20 h. For Caco-2 and HCT-8 cell lines, monolayers were seeded with 2 × 105 cells/well and incubated for 48 h. The cell monolayers were washed twice with phosphate-buffered saline (PBS, pH 7.2). Bacteria were grown overnight in LB broth at 37°C. Each monolayer was infected in 1 ml of the cell culture medium without antibiotic at a multiplicity of infection (MOI) of either 10 or 100 bacteria per epithelial cell. After a 3-h incubation period at 37°C with 10% CO2, infected monolayers were washed three times with PBS. For measurement of invasion, fresh cell culture medium containing 100 μg of gentamicin per ml was added to kill extracellular bacteria. After incubation for an additional hour, monolayers were washed three times with PBS, and 1 ml of 1% Triton X-100 (Sigma Chemical Company, St. Louis, Mo.) in deionized water was placed in each well for 5 min to lyse the eukaryotic cells. This concentration of Triton X-100 did not affect bacterial viability for at least 30 min (data not shown). Samples were removed, diluted, and plated onto Mueller-Hinton agar plates to determine the number of CFU recovered from the lysed monolayers. Invasion levels were expressed either as the number of CFU recovered per well or as the percentage of the original inoculum resisting treatment with gentamicin.
To determine the total number of cell-associated bacteria corresponding to adherent and intracellular bacteria, the eukaryotic cells were lysed after the 3-h infection period and the bacteria were quantified as described above.
For establishment of the time course of bacterial entry, infection periods from 1 to 5 h were used, followed by 1 h of gentamicin treatment.
For measurement of intracellular multiplication, the cell monolayers were infected at an MOI of 10 for 3 h. The number of bacteria surviving the gentamicin kill assay was determined after 1, 2, 4, 6, and 24 h of gentamicin treatment.
All assays were performed at least three times in separate experiments.
Epithelial cell detachment and viability.
In parallel with each experiment of intracellular survival, a duplicate 24-well plate of epithelial cell monolayer was inoculated with bacteria and assayed as described above. At the end of each gentamicin period, the monolayers were washed three times with PBS and trypsinized. After neutralization with fresh culture medium containing 10% FCS, the cell suspension was removed and diluted. Epithelial cell viability was estimated by the blue trypan exclusion assay using a 0.4% blue trypan solution in PBS. The cells were counted to determine the number of viable epithelial cells still attached to the plates.
Effects of eukaryotic cytoskeletal inhibitors.
HEp-2 cells were preincubated for 30 min before the assay in cell culture medium devoid of antibiotics with cytochalasin D or colchicine (Sigma). The inhibitors were present throughout the 3-h infection period. The inhibitory effect of each inhibitor was evaluated against the result for a control assay without inhibitor, which was defined as 100%. All assays were performed at least three times in separate experiments.
Transmission electron microscopy (TEM).
Cross sections of HEp-2 cells were prepared as follows. The cells were fixed with 3% glutaraldehyde in 0.2 M cacodylate buffer (pH 7.4) at 4°C for 2 h and postfixed in 1% OsO4 in cacodylate buffer at 4°C for 1 h. After dehydration in a graded series of ethanol, the cultures were embedded in a 2-mm-thick Epon coating in the tissue culture well and polymerized for 3 days at 60°C. Suitable areas were reoriented either parallel or perpendicular to the cell layer surface on Epon blocks with an Epon mixture. Ultrasections were contrasted with uranyl acetate and lead citrate.
PCR and hybridization experiments.
DNA to be amplified was released from whole organisms by boiling. Bacteria were harvested from 1 ml of an overnight broth culture, suspended in 200 μl of sterile water, and incubated at 100°C for 10 min. After centrifugation of the lysate, 5 μl of the supernatant was used in the PCR assays. Oligonucleotides used for amplification of ipaC-specific, eae-specific, and tia-specific sequences were synthesized on the basis of published nucleotide sequences. The sequences of all the primers used are listed in Table 1. Thirty PCR cycles were performed. Each primer was used at 0.5 μM, with 0.4 mM each of the four deoxynucleosides triphosphates (Boehringer Mannheim, Meylan, France), 50 mM KCl, 10 mM Tris-HCl (pH 8.3), 1 mM MgCl2, and 1 U of Taq DNA polymerase (Appligène-Oncor, Illkirch, France). Ten microliters of the reaction mixture was analyzed by electrophoresis on 1.5% agarose gels, and the reaction products were visualized by staining with ethidium bromide. DNA from reference E. coli strains and a blank reagent, which contained all components except the template DNA, were included as positive and negative controls, respectively.
TABLE 1.
Oligonucleotides used and PCR product sizes
The DNA fragments obtained from each specific amplification of DNA from reference E. coli strains were used as nucleic probes for colony blot hybridization experiments. PCR fragments were purified by using Spin X centrifuge tube filters (Corning Costar Corporation, Cambridge, Mass.) and labeled by random priming with [α-32P]dATP (3,000 Ci/mmol; Amersham International plc, Amersham, United Kingdom). The 16S rRNA probe, rrnB7, was prepared from plasmid pC6 and was used as a positive control to ensure that comparable amounts of DNA were present on the blots for all strains (6). Bacteria were harvested from 1 ml of an overnight broth culture and suspended in 100 μl of PBS. Five microliters was spotted onto Hybond nylon membranes (Amersham). The membranes were transferred successively onto Whatman 3MM papers impregnated with a denaturating solution (0.5 N NaOH, 1.5 M NaCl) and a neutralizing solution (1 M Tris [pH 7.4], 1.5 M NaCl). They were then air dried and exposed for 5 min to UV.
Hybridizations were performed with rapid hybridization buffer (Amersham) overnight at 65°C with gentle agitation. The heat-denaturated probes were used at a concentration of 106 dpm/ml. The filters were washed once with 2× SSC (0.15% NaCl plus 15 mmol of sodium citrate)–0.1% (wt/vol) sodium dodecyl sulfate and twice in 0.5× SSC–0.1% (wt/vol) sodium dodecyl sulfate (SDS) at 65°C and exposed to Hyper film (Amersham) at −80°C. The films were developed according to the manufacturer’s instructions.
Statistical analysis.
The data were analyzed by the chi-square test unless the variables needed a two-tailed Fisher exact test. A P value of less than or equal to 0.05 was considered statistically significant.
RESULTS
Ability of strain LF82 to invade HEp-2 cells.
The ability of E. coli serotype O83:H1 strain LF82 to invade HEp-2 epithelial cells was investigated by using the gentamicin kill assay and compared with that of prototype E. coli strains belonging to the pathogenic groups of E. coli known to be involved in infections of the gastrointestinal tract. Infections were achieved with an MOI of either 10 or 100 bacteria per epithelial cell. Invading bacteria were recovered after a 3-h infection period followed by 1 h of treatment with gentamicin. Table 2 presents invasion efficiencies expressed as the mean number of CFU recovered per well and as the mean percentage of the inoculum resisting treatment with gentamicin. The reference strain, E. coli K-12 C600, was used as a negative control and was found to be noninvasive, since only 0.001% ± 0.001% or 0.004% ± 0.004% of the inoculum resisted gentamicin treatment with an MOI of 10 or 100, respectively.
TABLE 2.
Abilities of strain LF82 and of reference strains belonging to the different pathogenic groups of E. coli to invade HEp-2 cells
| E. coli strain | Pathogenic group | Invasion
efficiencya (mean ± SD [n ≥ 3])
|
|||
|---|---|---|---|---|---|
| MOI = 10
|
MOI = 100
|
||||
| CFU × 105/wella | % Invasionb | CFU × 105/well | % Invasion | ||
| LF82 | 160.5 ± 143.5 | 4.01 ± 3.59 | 423.6 ± 355.0 | 1.06 ± 0.89 | |
| E12860/0 | EIEC | 121.2 ± 198.1 | 3.03 ± 4.95 | 489.6 ± 204.4 | 1.22 ± 0.51 |
| E2348/69 | EPEC | 131.7 ± 168.3 | 3.29 ± 4.21 | 111.9 ± 132.5 | 0.28 ± 0.33 |
| H10407 | ETEC | 72.1 ± 57.0 | 1.80 ± 1.42 | 100.8 ± 101.4 | 0.25 ± 0.25 |
| C1845 | DAEC | 24.6 ± 30.0 | 0.61 ± 0.75 | 20.4 ± 12.7 | 0.05 ± 0.03 |
| EDL933 | EHEC | 7.4 ± 5.4 | 0.18 ± 0.13 | 16.5 ± 9.1 | 0.04 ± 0.02 |
| 17-2 | EAggEC | 3.7 ± 3.0 | 0.09 ± 0.08 | 11.5 ± 10.0 | 0.03 ± 0.03 |
| K-12 C600 | 0.04 ± 0.04 | 0.001 ± 0.001 | 1.4 ± 1.3 | 0.004 ± 0.003 | |
Number of CFU recovered from lysed monolayers.
Percentage of inoculum surviving gentamicin treatment.
When an MOI of 10 was used, E. coli LF82 efficiently entered HEp-2 cells. A mean percentage of 4.01% ± 3.59% of the original inoculum was recovered after 1 h of gentamicin treatment. The invasion level of E. coli LF82 was not significantly different (P > 0.5) from that of EPEC strain E2348/69, for which 3.29% ± 4.21% of the inoculum entered HEp-2 cells, nor from that of EIEC strain E12860/0, which gave a mean percentage of invasion of 3.03% ± 4.95%. There was daily fluctuation in the invasion efficiency of strain LF82, which varied from 0.61 to 17.25% of the original inoculum, depending on the experiment. The same pattern was observed for EIEC strain E12860/0 and EPEC strain E2348/69, for which the mean percentage of internalized bacteria varied from 0.16% to 15.80% and 0.09% to 18.10%, respectively.
The invasion level of strain LF82 was significantly higher than those of ETEC strain H10407 (P < 0.03), DAEC strain C1845 (P < 0.02), EHEC strain EDL933 (P < 0.005), and EAggEC strain 17-2 (P < 0.004).
Invasiveness at MOIs of 10 and 100.
To test the influence of the amount of inoculated bacteria on the number of internalized bacteria, cell monolayers were infected with an MOI of either 10 or 100 bacteria per epithelial cell.
With an MOI of 100, the mean percentage of invasion of strain LF82 was 1.06% ± 0.89% (Table 2). This percentage was lower than that obtained at an MOI of 10 (4.01% ± 3.59%) but not significantly different (P > 0.05). When expressed as the number of CFU recovered per well, corresponding to the number of internalized bacteria (Table 2), the results showed that a 10-fold increase in the MOI resulted only in a 2.6-fold increase in the number of internalized bacteria. Thus, strain LF82 did not enter HEp-2 cells more efficiently when infection was achieved with a 10-fold-higher inoculum. The same result was observed for all reference pathogenic E. coli strains tested. In addition, the numbers of internalized bacteria for EPEC strain E2348/69 and DAEC strain C1845 were lower at an MOI of 100 than at an MOI of 10.
To determine whether the absence of increase in the number of CFU recovered was a consequence of cell detachment or loss of cell viability, we counted the number of epithelial cells that remained attached to the cell culture plates after the assay in comparison with a noninfected monolayer. In addition, cell viability was estimated by the trypan blue exclusion assay. After a 3-h infection period, neither cell detachment nor cell death was observed in any of the E. coli strains tested (data not shown).
Internalization of strain LF82 by intestinal epithelial cells.
We compared strain LF82 and the EIEC, EPEC, and ETEC reference strains with respect to the ability to invade intestinal epithelial cells. As shown in Table 3, strain LF82 invaded the human embryonic intestinal epithelial Intestine-407 cells. Its mean invasion level (4.67% ± 1.70%) was similar to that exhibited by EIEC strain E12860/0 (5.36% ± 5.68%). Strain LF82 also entered the human colonic Caco-2 cells (mean invasion level of 4.82% ± 1.25%) as efficiently as EPEC strain E2348/69 (4.11% ± 2.64%) and more much efficiently than EIEC strain E12860/0 (0.029% ± 0.018%). With the human ileocecal epithelial HCT-8 cells, the invasive level of strain LF82 (6.02% ± 1.66%) was lower than that of EPEC strain E2348/69 (11.92% ± 9.57%) and EIEC strain E12860/0 (38.98% ± 20.19%).
TABLE 3.
Invasion of intestinal epithelial cells
| Strain | % Invasion with intestinal cell
linea (mean ± SD [n ≥ 3])
|
||
|---|---|---|---|
| Intestine-407 | Caco-2 | HCT-8 | |
| LF82 | 4.67 ± 1.70 | 4.82 ± 1.25 | 6.02 ± 1.66 |
| EIEC E12860/0 | 5.36 ± 5.68 | 0.029 ± 0.018 | 38.98 ± 20.19 |
| EPEC E2348/69 | 1.91 ± 0.40 | 4.11 ± 2.64 | 11.92 ± 9.57 |
| ETEC H10407 | 2.67 ± 0.91 | 1.53 ± 0.46 | 0.72 ± 0.44 |
| K-12 C600 | 0.001 ± 0.001 | 0.005 ± 0.003 | 0.005 ± 0.003 |
Percentage of inoculum surviving gentamicin treatment.
Search for known E. coli genetic invasive determinants.
PCR experiments were performed. Hybridization assays were also carried out to ensure that the primers chosen did not represent regions of divergence.
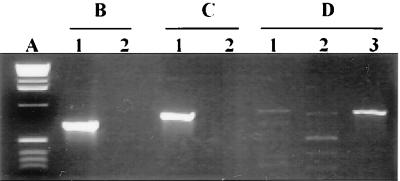
PCR experiments were performed with different sets of primers chosen to amplify an internal fragment of the ipaC gene encoding the invasin of S. flexneri, the eae gene encoding the intimin of EPEC, and the tia gene encoding a 25-kDa outer membrane protein involved in ETEC strain H10407 invasiveness (Table 1). As shown in Fig. 1, whatever the primers used, an amplification product of the predicted size was obtained with each reference strain lysate. We noticed only a slight amplification with EIEC strain E12860/0 in experiments using the primers specifying the ipaC region of S. flexneri (Fig. 1C, lane 1). However, the EIEC strain hybridized with the intragenic probe derived from the amplification product generated with S. flexneri DNA (Fig. 2). This result suggests that the ipaC gene sequence of EIEC strain E12860/0 was slightly different from that of S. flexneri. With strain LF82 DNA template, no amplification product was generated with the tia- or eae-specific primers. A weak amplification was observed with the ipaC-specific primers (Fig. 1C, lane 2). However, the amplified products did not have the predicted ipaC-specific fragment size.
FIG. 1.
PCR amplification products analysis. (A) One-kilobase DNA ladder (Life Technologies). Amplication products were generated by using primers for tia-specific sequence of ETEC strain H10407 (B), eae-specific sequence of EPEC (C), and ipaC-specific sequence of S. flexneri SC301 (D). DNA to be amplified was released by boiling from ETEC strain H10407 (B, lane 1), EPEC strain E2348/69 (C, lane 1), EIEC strain E12860/0 (D, lane 1), strain LF82 (lane 2), and S. flexneri SC301 (D, lane 3).
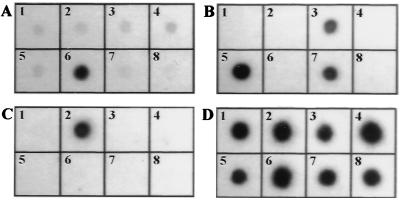
FIG. 2.
Colony blot hybridizations of intragenic tia (A), eae (B), and ipaC (C) probes corresponding to radiolabeled PCR-amplified products generated with DNA templates from ETEC strain H10407, EPEC strain E2348/69, and S. flexneri SC301, respectively. (D) Colony blot hybridization of rrnB7 probe. Blots: 1, strain LF82; 2, EIEC strain 12860/0; 3, DAEC strain C1845; 4, EAggEC strain 17-2; 5, EPEC strain E2348/69; 6, ETEC strain H10407; 7, EHEC strain EDL933; 8, E. coli K-12 C600.
The absence of known invasive determinants was verified by colony blot hybridization experiments to ensure that the primers used in PCR experiments were not specified to divergent regions of corresponding gene sequences in strain LF82. Hybridization with probe rrnB7 indicated that comparable amounts of DNA were present on the blots for all of the strains (Fig. 2D). Strain LF82 did not hybridize with the ipaC, eae, and tia intragenic probes (Fig. 2). These results indicate that the genetic determinants involved in the invasive ability of strain LF82 are not related to those of the pathogenic E. coli strains known to invade cultured epithelial cells.
Time course of bacterial entry.
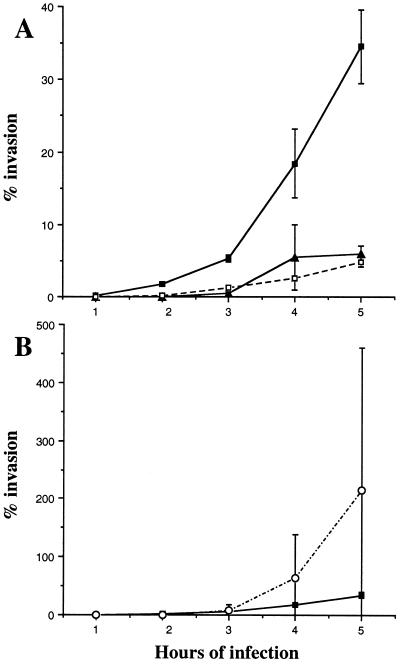
The time course of invasion was studied by determining the percentage of internalized bacteria at every hour for 5 h of infection. As shown in Fig. 3, the entry of strain LF82 into HEp-2 cells was swifter than that of EIEC strain E12860/0, EPEC strain E2348/69, and ETEC strain H10407. A mean of 1.80% ± 0.27% of the inoculum was internalized after a 2-h infection period, compared with 0.010% ± 0.007% for the EIEC strain, 0.027% ± 0.005% for the EPEC strain, and 0.23% ± 0.095% for the ETEC strain. At 3 h postinfection, the invasive level of strain LF82 was close to that of the EIEC strain (Fig. 3B). Then, like the EIEC strain, strain LF82 gradually invaded HEp-2 cells until 5 h postinfection. In contrast, the EPEC strain reached a maximum invasion level at 4 h postinfection (Fig. 3A). Although the kinetics of entry and/or multiplication of strain LF82 and EIEC strain E12860/0 exhibited similar profiles, the percentages of internalized bacteria were higher for the EIEC strain. After a 5-h infection period, the percentage of intracellular bacteria reached 34.50% ± 5.07% for strain LF82, compared to 214.50% ± 244.66% for the EIEC strain (Fig. 3B).
FIG. 3.
Time course of bacterial entry. HEp-2 cells were infected at an MOI of 10 for various infection periods and treated with gentamicin for 1 h. Percent invasion is the mean percentage of the inoculum surviving gentamicin treatment. (A) ■, strain LF82; ▴, EPEC strain E2348/69; □, ETEC strain H10407. (B) ■, strain LF82; ○, EIEC strain E12860/0. Each point is the mean of at least three separate experiments.
Survival of strain LF82 within HEp-2 and Intestine-407 cells.
The survival of strain LF82 in HEp-2 and Intestine-407 cells was estimated. After a 3-h infection period, the number of intracellular bacteria was determined at various times of gentamicin exposure and compared with that obtained with a 1-h gentamicin exposure (Table 4). To better analyze the fate of the internalized bacteria, the results were also expressed as percentages of the total bacteria recovered at 1 h of gentamicin treatment. In parallel with each experiment, epithelial cell viability was estimated by the blue trypan exclusion assay, and the detachment of the infected cells was estimated by counting the number of viable epithelial cells that remained attached to the plates after various times of gentamicin exposure (Table 4).
TABLE 4.
Survival of strain LF82 within HEp-2 and Intestine-407 cells
| Gentamicin treatment period (h) | Intracellular
survival (mean ± SD [n ≥ 3])
|
|||||
|---|---|---|---|---|---|---|
| HEp-2
|
Intestine-407
|
|||||
| CFU × 105/wella | % Relative CFUb | % Cellsc | CFU × 105/well | % Relative CFU | % Cells | |
| 1 | 690.0 ± 42.4 | 100 | 100 | 203.5 ± 60.1 | 100 | 100 |
| 2 | 1,240.0 ± 56.6 | 179.8 ± 5.0 | NDd | 283.5 ± 65.8 | 140.7 ± 9.2 | ND |
| 4 | 1,530.0 ± 183.9 | 223.0 ± 40.4 | ND | 198.0 ± 45.3 | 98.3 ± 6.8 | ND |
| 6 | 1,720.0 ± 537.4 | 247.4 ± 62.7 | 99.8 | 218.5 ± 34.6 | 109.6 ± 15.4 | 157.6 |
| 24 | 1,685.0 ± 459.6 | 242.6 ± 51.7 | 114.5 | 163.3 ± 34.9 | 81.3 ± 6.9 | 159.3 |
Number of CFU recovered from lysed monolayers.
Percent intracellular bacteria relative to the number after 1 h of gentamicin treatment, defined as 100%.
Percent epithelial cells remaining attached to the plates relative to the number after 1 h of gentamicin treatment, defined as 100%.
ND, not determined.
With HEp-2 cells, the number of LF82 bacteria recovered gradually increased over a 6-h course of exposure to gentamicin and at 6 h reached a maximum peak of 247.4% ± 62.7% of the total bacteria recovered at 1 h (Table 4). After 24 h of gentamicin treatment, the number of intracellular bacteria was not lower than at 6 h (242.6% ± 51.7%). The number of viable HEp-2 cells remaining attached did not differ between 1 and 24 h of gentamicin treatment: 99.8 and 114.5% of the HEp-2 cells counted at 1 h of gentamicin treatment were recovered at 6 and 24 h, respectively. All attached cells excluded the dye blue trypan. In addition, the culture media containing gentamicin were sterile at the various times tested, indicating that the bacteria recovered corresponded to intracellular bacteria. Thus, with HEp-2 cells, strain LF82 was able to survive for at least 24 h in the host cell cytoplasm and to efficiently replicate intracellularly, without damage to the infected cells.
When Intestine-407 cells were used, the number of intracellular bacteria recovered after 1 h of gentamicin treatment varied over a 24-h course of treatment but remained higher than 80% of the total bacteria recovered at 1 h (Table 4). It increased at 2 and 6 h of gentamicin exposure: 140.7% ± 9.2% and 109.6% ± 15.4% of the total bacteria recovered at 1 h were recovered at 2 and 6 h, respectively. Over the same period, the number of viable Intestine-407 cells remaining attached to the monolayers increased, indicating cell multiplication. Thus, strain LF82 was able to survive for at least 24 h within Intestine-407 cells without damage to the parasitized cells.
Survival of strain LF82, EPEC strain, and EIEC strain within HEp-2 cells.
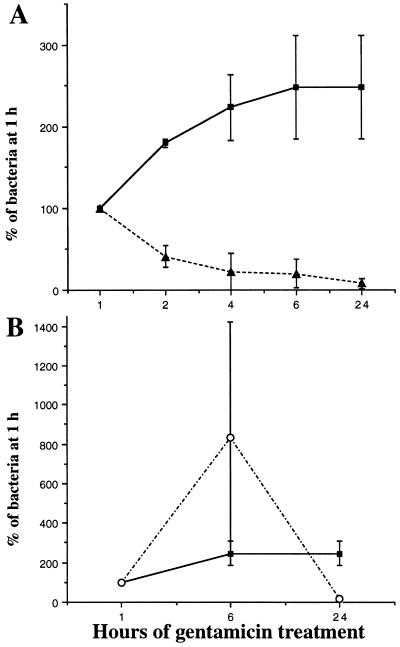
The intracellular survival of strain LF82 was compared to that of EIEC strain E12860/0 and EPEC strain E2348/69, since EIEC strains are known to extensively replicate intracellularly (52) whereas EPEC strains are reported not to survive intracellularly (46). For each strain, the number of intracellular bacteria at various times of gentamicin exposure was compared with that determined after a 1-h gentamicin exposure, which was taken as 100% (Fig. 4).
FIG. 4.
Kinetics of intracellular multiplication. HEp-2 cell monolayers were infected for 3 h, and the number of intracellular bacteria was determined after different times of gentamicin treatment. Results are expressed as the number of intracellular bacteria relative to that obtained at 1 h of gentamicin treatment, taken as 100%. (A) ■, strain LF82; ▴, EPEC strain E2348/69. (B) ■, strain LF82; ○, EIEC strain E12860/0. Each point is the mean of at least three separate experiments.
Whereas strain LF82 multiplied within invaded HEp-2 cells, the number of intracellular bacteria in EPEC strain E2348/69 decreased between 1 and 6 h of gentamicin treatment, since only 20.3% ± 17.1% of the intracellular bacteria recovered at 1 h were recovered at 6 h. After 24 h of gentamicin treatment, 8.0% ± 6.0% of the total internalized bacteria were recovered (Fig. 4A). The number of viable HEp-2 cells remaining attached did not differ between 1 and 6 h of gentamicin treatment, since 100.8% of the HEp-2 cells counted at 1 h of gentamicin treatment were recovered at 6 h. All attached cells excluded the dye blue trypan. After a 24 h of gentamicin treatment, the number of attached cells excluding the dye blue trypan decreased to 84.7%. Thus, in contrast to strain LF82, for which intracellular multiplication did not affect cell viability, the EPEC strain induced cell detachment after prolonged gentamicin periods.
The intracellular multiplication of strain LF82 was lower than that of the EIEC strain. The number of intracellular bacteria for strain E12860/0 increased 8.3-fold between 1 and 6 h of gentamicin treatment, since 833.8% ± 589.1% of the intracellular bacteria recovered at 1 h were recovered at 6 h (Fig. 4B). The number of viable HEp-2 cells remaining attached did not differ between 1 and 6 h of gentamicin treatment, since 102.9% of HEp-2 cells counted at 1 h of gentamicin treatment were recovered at 6 h. After 24 h of gentamicin treatment, 16.9% ± 8.1% of the total internalized bacteria were recovered and the number of attached cells excluding the dye blue trypan decreased to 88.8%. The observed decrease in the number of intracellular bacteria recovered was due, as reported for S. flexneri (52), to the death of the infected cells in which EIEC exhibited extensive multiplication (about 11% of the total cells of the monolayer). The bacteria would then be released and killed by gentamicin. These results indicate that even if the intracellular multiplication of strain LF82 was less efficient than that of EIEC strain E12860/0, it did not induce the cell damage evidenced in EIEC.
Effects of eukaryotic cytoskeletal inhibitors.
To examine the role of actin microfilaments and microtubules in the interactions between the bacteria and the HEp-2 epithelial cells, cell monolayers were treated with cytochalasin D and colchicine, respectively.
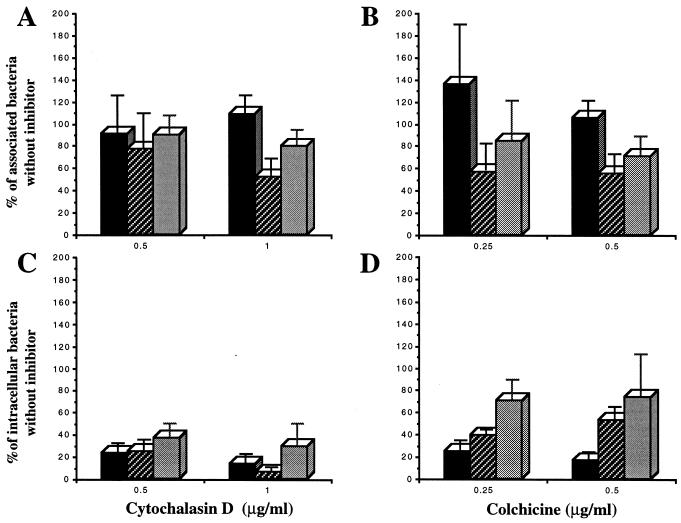
The addition of 0.5 or 1 μg of cytochalasin D per ml during the invasion assay significantly inhibited the entry of strain LF82 by 76.9% (P < 0.0001) and 86.7% (P < 0.0001), respectively (Fig. 5C). In contrast, treatment with even 1 μg of cytochalasin D per ml did not significantly (P > 0.3) modify the number of associated (adherent and internalized) bacteria (Fig. 5A). Treatment of HEp-2 cells with 0.25 or 0.5 μg of colchicine per ml also markedly inhibited the entry of strain LF82, giving inhibition percentages of 75.0 (P < 0.0001) and 83.4 (P < 0.0001), respectively (Fig. 5D), while even the higher concentration did not affect (P > 0.2) the number of cell-associated bacteria (Fig. 5B). Thus, treatment of HEp-2 cells with either cytochalasin D or colchicine efficiently decreased in a dose-dependent manner the entry of strain LF82 into the cells, without significantly affecting bacterial adhesion to these cells. These data show that the entry of E. coli LF82 into HEp-2 cells is mediated by both an actin microfilament-dependent mechanism and microtubule involvement.
FIG. 5.
Effects of eukaryotic cell inhibitors on association with (A and B) and invasion of (C and D) HEp-2 cells. HEp-2 cell monolayers were pretreated with cytochalasin D or colchicine for 30 min before the assay. Infections were performed for 3 h in the presence of the inhibitors. Cell-associated bacteria were quantified after the 3-h infection period. Invasion was determined after gentamicin treatment for an additional hour. Results are expressed as cell-associated or intracellular bacteria relative to those obtained without inhibitor, taken as 100%. ■, strain LF82; , EIEC strain E12860/0; ░⃞, EPEC strain E2348/69. Each value is the mean of at least three separate experiments.
The effects of the two cytoskeletal inhibitors on the interactions of strain LF82 with epithelial cells were compared to those obtained for EIEC strain E12860/0 and EPEC strain E2348/69. A 1-μg/ml concentration of cytochalasin D significantly reduced the cell association of EPEC strain E2348/69 compared to strain LF82 (P < 0.04) and EIEC strain E12860/0 (P < 0.02) (Fig. 5A). The inhibitory effect of the two concentrations of cytochalasin D on the bacterial entry of strain LF82 was not significantly different from that observed for EIEC strain E12860/0 and EPEC strain E2348/69 (P > 0.5) (Fig. 5C).
A 0.5-μg/ml concentration of colchicine produced an inhibitory effect on EIEC E12860/0 and EPEC E2348/69 association to epithelial cells that was not observed for strain LF82 (P < 0.05) (Fig. 5B). Moreover, the inhibitory effect of a 0.5-μg/ml concentration of colchicine on the entry of strain LF82 was significantly higher than that observed for EIEC strain E12860/0 (P < 0.005) and EPEC strain E2348/69 (P < 0.03) (Fig. 5D). All of these data indicate that the invasion process of strain LF82 is more dependent on microtubule involvement than that of EIEC strain E12860/0 and EPEC strain E2348/69.
Electron microscopic examination of LF82-infected HEp-2 cells.
To visualize the different stages of the interaction process of strain LF82 with host cells and to corroborate the data obtained with the quantitative invasion assay, TEM was performed on cell monolayers after infection for either 3 or 5 h.
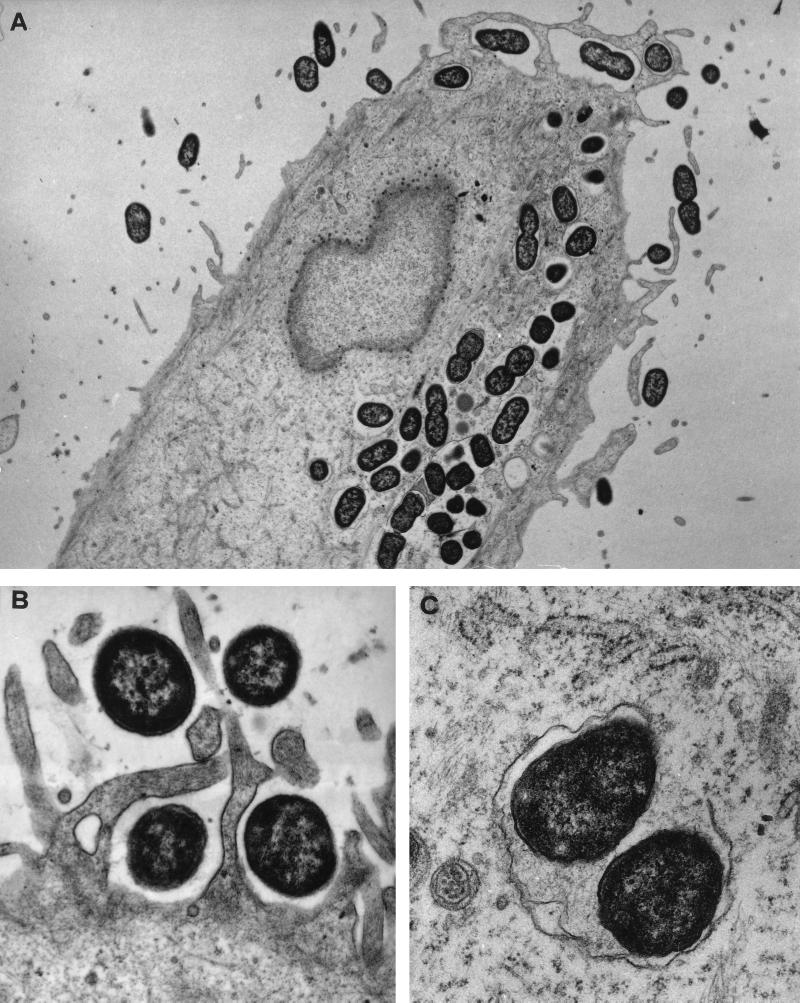
At 3 h postinfection, strain LF82 adhered closely to HEp-2 cells (Fig. 6B). The adherent bacteria strikingly induced the elongation of microvilli from the cell surface. At the site of close contact between the bacteria and the epithelial cell, the elongated microvilli surrounded the adherent bacteria. In addition, dense areas of staining, possibly related to an accumulation of cytoskeletal components, were observed beneath the sites of intimate contact.
FIG. 6.
Transmission electron micrographs of HEp-2 cells infected with strain LF82. (A) Cross section of the cell monolayer showing numerous intracellular bacteria after a 5-h infection period. (B) Micrograph showing membrane ruffling upon contact with the bacteria after a 3-h infection period. Bacteria are engulfed by elongated microvilli from the infected epithelial cells. (C) High magnification showing a partially lysed vacuolar membrane containing bacteria, indicating the ability of strain LF82 to escape from the endocytic vacuoles. Magnifications: A, ×6,200; B, ×21,600; C, ×28,800.
The photomicrograph in Fig. 6A clearly shows the presence of numerous bacteria inside the cells at 5 h postinfection. Most of the bacteria observed were enclosed by endocytic vacuoles. In addition, some bacteria were free in the cell cytoplasm, perhaps as a result of an escape from endocytic vacuoles by bacterium-induced lysis of the vacuole membrane. In addition, a high magnification (Fig. 6C) revealed the first signs of damage to the vacuole membrane containing internalized bacteria.
DISCUSSION
Invasion of eukaryotic cells is known to be an important virulence mechanism of many enteric pathogens such as Salmonella, Shigella, and Listeria species (for a review, see reference 23). All pathogenic types of E. coli involved in gastrointestinal infections are able to invade cultured epithelial cells. However, the clinical significance of invasion in the pathogenesis of all pathogenic E. coli strains except EIEC strains remains unclear (46).
In a previous study, we demonstrated that E. coli strains colonized the ileal mucosa of patients with CD (12). Further analysis revealed an invasive phenotype, and the aim of the present study was to compare the invasive properties of E. coli LF82, isolated from a chronic ileal lesion of a patient with CD, with those of reference E. coli strains involved in gastrointestinal tract infections and to analyze its invasive process.
Gentamicin kill assay showed that strain LF82 is able to efficiently invade HEp-2 cells. The number of intracellular bacteria increased only 2.6-fold when the inoculum increased from 10 to 100 bacteria per epithelial cell. The blue trypan exclusion assay showed this was due neither to cell detachment nor to cell death. Thus, all experiments reported in that study were performed with an MOI of 10.
The invasion efficiency of strain LF82 was compared with that of reference E. coli pathogenic strains. The kinetics of bacterial entry revealed that efficient internalization of strain LF82 occurs rapidly, since 1.80% of the original inoculum was internalized after a 2-h infection period. In contrast, EIEC strain E12860/0 and EPEC strain E2348/69 had not extensively invaded HEp-2 cells but achieved efficient invasion after a 3-h infection period. Strain LF82, like EIEC strain E12860/0, gradually invaded and/or multiplied within HEp-2 cells until 5 h postinfection, but without reaching the same numbers of intracellular bacteria. After a 3-h infection period and 1 h of gentamicin treatment, a mean invasion level of 4.01% (range from 0.61 to 17.25%) was found for strain LF82. This invasion level was not significantly different from what we observed with EIEC strain E12860/0 and EPEC strain E2348/69 (P > 0.5) but significantly higher than that of ETEC strain H10407 (P < 0.03), DAEC strain C1845 (P < 0.02), EHEC strain EDL933 (P < 0.005), and EAggEC strain 17-2 (P < 0.004).
It is difficult to compare our results with previously published findings on pathogenic E. coli strains because of the wide range of cell lines used and the various ways used to express the invasion results. However, EIEC strains have previously been shown to give invasion levels ranging from 0.2 to 20% of the original inoculum with HEp-2 cells, with a mean percentage of 1.87 (13). In addition, Small et al. (56) reported a mean invasion level of 2.9% of the inoculum for EIEC strain 11. Moreover, Donnenberg et al. (13) tested five EPEC strains, including strain E2348/69, and showed that 3 to 8% of the original inoculum was internalized by HEp-2 cells, with a mean level of 5.11%. The invasion levels that we observed with EIEC strain E12860/0 and EPEC strain E2348/69 were consistent with those cited in these two reports. Most of the invasion levels reported for ETEC, EHEC, and DAEC strains are consistent with those we obtained with HEp-2 cells and largely below those exhibited by strain LF82. Indeed, for ETEC strain H10407, the highest percentage of the inoculum surviving the gentamicin kill assay observed with HCT 116 intestinal cells was 1.06 (17). For EHEC strains, Oelschlaeger et al. (48) reported a maximum invasion of 1.8% ± 0.2% with HCT-8 colonic cells. In addition, Yamamoto et al. (64) found only 2.8 × 103 CFU from lysed HEp-2 cell monolayers infected with 1 ml of a 10-fold diluted overnight culture of DAEC strain D2 hybridizing with an F1845 DNA probe. Similar results were obtained for other DAEC strains with HeLa cells (29). In contrast, the invasion efficiency that we obtained for EAggEC strain 17-2 with HEp-2 cells was much lower than that reported by Benjamin et al. (5) for EAggEC strain 162 with HeLa cells.
The gentamicin kill assay has been reported to yield false-positive invasion results (11). Noninternalized bacteria may be protected from the action of the antibiotic when embedded in membrane projections extending from the cell surface. Electron microscopic examination of LF82-infected HEp-2 cells revealed numerous intracellular bacteria and corroborated the findings of the quantitative assays. Electron microscopy observation of cross sections of infected cells showed up to 45 intracellular bacteria per epithelial cell. Such high numbers of intracellular bacteria are close to those previously reported for S. flexneri (52) and Salmonella typhimurium (39).
The genetic determinants involved in pathogenic E. coli invasiveness are well documented for EIEC and EPEC. In EIEC strains, the secreted Ipa proteins, encoded by ipaB, ipaC, and ipaD plasmid genes, are effectors of the invasion phenotype (42). EPEC invasion, like intimate attachment, is mediated by intimin, the product of the chromosomally encoded eae gene (34, 35). Two chromosomal genes called tia and tib have been identified and shown to be involved in the invasive ability of ETEC strain H10407 (18, 25). To determine whether strain LF82 possessed one of the known invasive determinants, we performed PCR experiments using sets of primers specifying the ipaC gene of S. flexneri, eae of EPEC, and tia of ETEC strain H10407. No amplification product was generated when strain LF82 lysate was used as DNA template with the tia- and eae-specific primers. A weak amplification was observed with the ipaC-specific primers, but the PCR-amplified products did not have the predicted ipaC-specific fragment size. In addition, by hybridization experiments using the corresponding intragenic DNA probes, we demonstrated the absence of the ipaC, eae, and tia genes in strain LF82. These results indicate that the genetic determinants encoding the invasive properties of strain LF82 are not related to those of the known invasive E. coli strains or S. flexneri.
Most invasive bacteria activate host cell transduction pathways to promote their uptake (for a review, see reference 20) and two major mechanisms of entry into nonphagocytic cultured cells have been described for invasive bacteria. Uptake of Yersinia spp. and Listeria monocytogenes occurs via a mechanism, whereby the host cell membrane “zippers” the bacterium as it enters (32, 43). For S. flexneri and Salmonella typhimurium, the bacteria send signals to the cells that induce localized membrane rufflings and cytoskeletal rearrangements resulting in bacterial uptake by macropinocytosis (1, 22, 24, 27). Electron microscopic examination of LF82-infected HEp-2 cells revealed that the interaction of strain LF82 with HEp-2 cells was associated with the elongation of microvillar extensions extruding from the host cell membranes that surrounded the bacteria. In addition, TEM examination of LF82-infected HEp-2 cells revealed that the invading bacteria were enclosed by endocytic vacuoles. This result indicates that the localized membrane elongations, which are induced when the bacteria come into contact with the epithelial cell surface, play a key role to achieve bacterial uptake. These findings suggest that strain LF82 may trigger an active endocytic mechanism within the host cells that promotes its own internalization into nonprofessional phagocytes. Such an internalization mechanism closely resembles Salmonella- or Shigella-induced macropinocytosis (1, 22, 24, 27).
We used two eukaryotic cytoskeletal inhibitors, the actin microfilament inhibitor cytochalasin D and the microtubule inhibitor colchicine, to study the role of HEp-2 cells in the internalization process. The invasion processes of most invasive bacteria, including S. flexneri (10, 21), EIEC (8, 14), Y. enterocolitica (21), and L. monocytogenes (60), are actin microfilament but not microtubule dependent. Both microtubules and actin polymerization may be required for invasion of pathogens such as Neisseria gonorrhoeae (30), EPEC (14), DAEC (29), Campylobacter jejuni, and Citrobacter freundii (49). Strain LF82 invasion was markedly affected by the two cytoskeletal inhibitors, suggesting that this strain exploits host cell actin microfilaments but also requires microtubules to promote its own uptake into HEp-2 cells.
Some investigators claimed that as a general rule, organisms that use microtubules to invade are unable to multiply or persist within host cells (23). Intracellular multiplication of the invading microorganisms is a common feature of true intracellular pathogens and is known to be a prerequisite for virulence. Interestingly, for strain LF82, the number of intracellular bacteria increased 2.4-fold with HEp-2 cells over a 6-h period of gentamicin treatment. Furthermore, neither cell detachment nor cell death was observed until 24 h. The number of intracellular bacteria remained stable between 6 and 24 h of gentamicin treatment, indicating that the bacteria were maintained intracellularly without damage to the epithelial monolayers. Strain LF82 also demonstrated an ability to survive within Intestine-407 cells for at least 24 h without affecting cell viability and cell multiplication. Thus, strain LF82 was found to survive within Intestine-407 and HEp-2 cells for at least 24 h and to multiply within HEp-2 cells, despite the fact that its mechanism of entry requires both actin microfilaments and microtubules. However, the intracellular behavior of strain LF82 differs from that of S. flexneri, EIEC, and Salmonella typhimurium, whose intracellular growth for prolonged periods induce lysis of most of the infected cells (10, 39, 56).
Although strain LF82 had a rate of multiplication lower than those published for S. flexneri (52), EIEC (56), Salmonella typhimurium (39), and L. monocytogenes (28), it seems to be adapted for intracellular survival. In addition, transmission electron micrographs of LF82-infected HEp-2 cells revealed that the bacteria could lyse the vacuole membrane and become free in the host cell cytoplasm, evidence that they are able to escape from the phagocytic vacuoles. Such an intracellular lifestyle resembles that of S. flexneri and L. monocytogenes, which are able to escape from the endocytic vacuole through IpaB- and lysteriolysin O-mediated lysis of the endocytic vacuolar membrane, respectively (28, 31).
Our findings demonstrate that E. coli LF82 is a true invasive strain: (i) it efficiently invades cultured epithelial cells, (ii) its uptake is dependent on actin microfilaments and microtubules, (iii) it survives intracellularly for at least 24 h, and (iv) it replicates in the host cell cytoplasm after lysis of the endocytic vacuole. However, strain LF82 has none of the invasive determinants of the invasive E. coli strains known to be involved in gastrointestinal infections. The invasive strain LF82 was isolated from damaged ileal mucosa of a patient with CD, a chronic intestinal inflammatory disease. Interestingly, we found that it was able to invade Caco-2, Intestine-407, and HCT-8 intestinal cultured cells and to survive for at least 24 h within Intestine-407 cells, but its role in CD remains to be investigated.
Strain LF82 was previously reported to colonize the ileal mucosa of a patient with CD and to adhere diffusely and extensively in vitro to intestinal epithelial cells (12). These findings and the evidence presented here that strain LF82 is also a true invasive E. coli which cannot be classified in any of the pathogenic E. coli groups described to date suggest the existence of a new potentially pathogenic group of E. coli, which we propose be designated AIEC, for adherent-invasive E. coli. Cloning experiments are in progress to identify genes involved in strain LF82 invasiveness and to create molecular tools to screen the presence of such AIEC strains in patients with CD and in controls.
ACKNOWLEDGMENTS
This study was supported by grants from Association F. Aupetit, Institut de Recherche des Maladies de l’Appareil Digestif (IRMAD, Laboratoires Astra France), Fonds de Recherche de la Société Nationale Française de Gastro-Entérologie, and Ministère de la Recherche et de la Technologie (EA2148). J.B. was supported by a grant from IRMAD. We gratefully acknowledge Christel Neut and Jean-Frédéric Colombel, Groupe de Recherche sur les Maladies Inflammatoires du Tube Digestif, Lille, France, for providing E. coli strains isolated from patients with CD and for helpful discussions. We thank Karen Krogfelt for serotyping E. coli LF82. We thank Michael Donnenberg for providing EPEC strain E2348/69 and EIEC strain E12860/0, Myron M. Levine for EAggEC strain 17-2, Dolores Evans for ETEC strain H10407, Steve Moseley for DAEC strain C1845, Alison O’Brien for EHEC strain EDL933, and Philippe Sansonetti for S. flexneri SC301. We thank Alain Zweibaum and Marc Mareel for providing Caco-2 and HCT-8 E11R1 intestinal cell lines, respectively. We also thank the Microscopy Department of Michel Bourges for technical assistance with electron microscopy analysis.
REFERENCES
- 1.Adam T, Arpin M, Prevost M C, Gounon P, Sansonetti P J. Cytoskeletal rearrangements and the functional role of T-plastin during entry of Shigella flexneriinto HeLa cells. J Cell Biol. 1995;129:367–381. doi: 10.1083/jcb.129.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allaoui A, Sansonetti P J, Menard R, Barzu S, Mounier J, Phalipon A, Parsot C. MxiG, a membrane protein required for secretion of Shigellaspp. Ipa invasins: involvement in entry into epithelial cells and in intercellular dissemination. Mol Microbiol. 1995;17:461–470. doi: 10.1111/j.1365-2958.1995.mmi_17030461.x. [DOI] [PubMed] [Google Scholar]
- 3.Andrade J R, Da Veiga V F, De Santa Rosa M R, Suassuna I. An endocytic process in HEp-2 cells induced by enteropathogenic Escherichia coli. J Med Microbiol. 1989;28:49–57. doi: 10.1099/00222615-28-1-49. [DOI] [PubMed] [Google Scholar]
- 4.Baudry B, Maurelli A T, Clerc P, Sadoff J C, Sansonetti P J. Localization of plasmid DNA loci necessary for the entry of Shigella flexneriinto Hela cells, and characterization of one locus encoding four immunogenic polypeptides. J Gen Microbiol. 1987;133:3409–3413. doi: 10.1099/00221287-133-12-3403. [DOI] [PubMed] [Google Scholar]
- 5.Benjamin P, Federman M, Wanke C A. Characterization of an invasive phenotype associated with enteroaggregative Escherichia coli. Infect Immun. 1995;63:3417–3421. doi: 10.1128/iai.63.9.3417-3421.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berg K L, Squires C L, Squires C. In vivo translation of a region within the rrnB 16S rRNA gene of Escherichia coli. J Bacteriol. 1987;169:1691–1701. doi: 10.1128/jb.169.4.1691-1701.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bilge S S, Clausen C R, Lau W, Moseley S L. Molecular characterization of a fimbrial adhesin, F1845, mediating diffuse adherence of diarrhea-associated Escherichia colito HEp-2 cells. J Bacteriol. 1989;171:4281–4289. doi: 10.1128/jb.171.8.4281-4289.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bukholm G. Effect of cytochalasin B and dihydrocytochalasin B on invasiveness of entero-invasive bacteria in HEp-2 cell cultures. APMIS. 1984;92:145–219. doi: 10.1111/j.1699-0463.1984.tb02809.x. [DOI] [PubMed] [Google Scholar]
- 9.Cartun R W, Van Kruiningen H J, Pedersen C A, Berman M M. An immunocytochemical search for infectious agents in Crohn’s disease. Mod Pathol. 1993;6:212–219. [PubMed] [Google Scholar]
- 10.Clerc P, Sansonetti P J. Entry of Shigella flexneriinto HeLa cells: evidence for directed phagocytosis involving actin polymerization and myosin accumulation. Infect Immun. 1987;55:2681–2688. doi: 10.1128/iai.55.11.2681-2688.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cookson S T, Nataro J P. Characterisation of HEp-2 cell projection formation induced by diffusely adherent Escherichia coli. Microb Pathog. 1996;21:421–434. doi: 10.1006/mpat.1996.0073. [DOI] [PubMed] [Google Scholar]
- 12.Darfeuille-Michaud A, Neut C, Barnich N, Lederman E, Di Martino P, Desreumaux P, Gambiez L, Joly B, Cortot A, Colombel J F. Presence of adherent Escherichia colistrains in ileal mucosa of patients with Crohn’s disease. Gastroenterology. 1998;115:1405–1413. doi: 10.1016/s0016-5085(98)70019-8. [DOI] [PubMed] [Google Scholar]
- 13.Donnenberg M S, Donohue-Rolfe A, Keusch G T. Epithelial cell invasion: an overlooked property of enteropathogenic Escherichia coli(EPEC) associated with the EPEC adherence factor. J Infect Dis. 1989;160:452–459. doi: 10.1093/infdis/160.3.452. [DOI] [PubMed] [Google Scholar]
- 14.Donnenberg M S, Donohue-Rolfe A, Keusch G T. A comparison of HEp-2 cell invasion by enteropathogenic and enteroinvasive Escherichia coli. FEMS Microbiol Lett. 1990;69:83–86. doi: 10.1016/0378-1097(90)90417-o. [DOI] [PubMed] [Google Scholar]
- 15.Donnenberg M S, Kaper J B, Finlay B B. Interactions between enteropathogenic Escherichia coliand host epithelial cells. Trends Microbiol. 1997;5:109–114. doi: 10.1016/S0966-842X(97)01000-7. [DOI] [PubMed] [Google Scholar]
- 16.Dupont H L, Formal S B, Hornick R B, Snyder M J, Libonati J P, Sheahan D G, LaBrec E H, Kalas J P. Pathogenesis of Escherichia colidiarrhea. N Engl J Med. 1971;285:1–9. doi: 10.1056/NEJM197107012850101. [DOI] [PubMed] [Google Scholar]
- 17.Elsinghorst E A, Kopecko D J. Molecular cloning of epithelial cell invasion determinants from enterotoxigenic Escherichia coli. Infect Immun. 1992;60:2409–2417. doi: 10.1128/iai.60.6.2409-2417.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elsinghorst E A, Weitz J A. Epithelial cell invasion and adherence directed by the enterotoxigenic Escherichia coli tiblocus is associated with a 104-kilodalton outer membrane protein. Infect Immun. 1994;62:3463–3471. doi: 10.1128/iai.62.8.3463-3471.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Evans D G, Silver R P, Evans D J, Chase D G, Gorbach S L. Plasmid-controlled colonization factor associated with virulence in Escherichia colienterotoxigenic for humans. Infect Immun. 1975;12:656–667. doi: 10.1128/iai.12.3.656-667.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Finlay B B, Cossart P. Exploitation of mammalian host cell functions by bacterial pathogens. Science. 1997;276:718–725. doi: 10.1126/science.276.5313.718. [DOI] [PubMed] [Google Scholar]
- 21.Finlay B B, Falkow S. Comparison of the invasion strategies used by Salmonella cholerae-suis, Shigella flexneri and Yersinia enterocoliticato enter cultured animals cells: endosome acidification is not required for bacterial invasion or intracellular multiplication. Biochimie. 1988;70:1089–1099. doi: 10.1016/0300-9084(88)90271-4. [DOI] [PubMed] [Google Scholar]
- 22.Finlay B B, Falkow S. Salmonellainteractions with polarized human intestinal Caco-2 epithelial cells. J Infect Dis. 1990;162:1096–1106. doi: 10.1093/infdis/162.5.1096. [DOI] [PubMed] [Google Scholar]
- 23.Finlay B B, Falkow S. Common themes in microbial pathogenicity revisited. Microbiol Mol Biol Rev. 1997;61:136–169. doi: 10.1128/mmbr.61.2.136-169.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Finlay B B, Ruschkowski S, Dedhar S. Cytoskeletal rearrangements accompanying Salmonellaentry into epithelial cells. J Cell Sci. 1991;99:283–96. doi: 10.1242/jcs.99.2.283. [DOI] [PubMed] [Google Scholar]
- 25.Fleckenstein J M, Kopecko D J, Warren R L, Elsinghorst E A. Molecular characterization of the tia invasion locus from enterotoxigenic Escherichia coli. Infect Immun. 1996;64:2256–2265. doi: 10.1128/iai.64.6.2256-2265.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Francis C L, Jerse A E, Kaper J B, Falkow S. Characterization of interactions of enteropathogenic Escherichia coliO127:H6 with mammalian cells in vitro. J Infect Dis. 1991;164:693–703. doi: 10.1093/infdis/164.4.693. [DOI] [PubMed] [Google Scholar]
- 27.Francis C L, Ryan T A, Jones B D, Smith S J, Falkow S. Ruffles induced by Salmonellaand other stimuli direct macropinocytosis of bacteria. Nature. 1993;364:639–642. doi: 10.1038/364639a0. [DOI] [PubMed] [Google Scholar]
- 28.Gaillard J L, Berche P, Mounier J, Richard S, Sansonetti P. In vitro model of penetration and intracellular growth of Listeria monocytogenesin the human enterocyte-like cell line Caco-2. Infect Immun. 1987;55:2822–2829. doi: 10.1128/iai.55.11.2822-2829.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goluszko P, Vsevolod P, Selvarangan R, Nowicki S, Pham T, Nowicki B J. Dr fimbriae operon of uropathogenic Escherichia colimediate microtubule-dependent invasion to the HeLa epithelial cell line. J Infect Dis. 1997;176:158–167. doi: 10.1086/514018. [DOI] [PubMed] [Google Scholar]
- 30.Grassme H U, Ireland R M, van Putten J P. Gonococcal opacity protein promotes bacterial entry-associated rearrangements of the epithelial cell actin cytoskeleton. Infect Immun. 1996;64:1621–1630. doi: 10.1128/iai.64.5.1621-1630.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.High A, Mounier J, Prevost M C, Sansonetti P J. IpaB of Shigella flexnericauses entry into epithelial cells and escape from the phagocytic vacuole. EMBO J. 1992;11:1991–1999. doi: 10.1002/j.1460-2075.1992.tb05253.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Isberg R R, Leong J M. Multiple beta 1 chain integrins are receptors for invasin, a protein that promotes bacterial penetration into mammalian cells. Cell. 1990;60:861–871. doi: 10.1016/0092-8674(90)90099-z. [DOI] [PubMed] [Google Scholar]
- 33.Jarvis K G, Giron J A, Jerse A E, McDaniel T K, Donnenberg M S, Kaper J B. Enteropathogenic Escherichia colicontains a putative type III secretion system necessary for the export of proteins involved in attaching and effacing lesion formation. Proc Natl Acad Sci USA. 1995;92:7996–8000. doi: 10.1073/pnas.92.17.7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jerse A E, Kaper J B. The eae gene of enteropathogenic Escherichia coliencodes a 94-kilodalton membrane protein, the expression of which is influenced by the EAF plasmid. Infect Immun. 1991;59:4302–4309. doi: 10.1128/iai.59.12.4302-4309.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jerse A E, Yu J, Tall B D, Kaper J. A genetic locus of enteropathogenic Escherichia colinecessary for the production of attaching and effacing lesions on tissue culture cells. Proc Natl Acad Sci USA. 1990;87:7839–7843. doi: 10.1073/pnas.87.20.7839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jouve M, Garcia M I, Courcoux P, Labigne A, Gounon P, Le Bouguenec C. Adhesion to and invasion of HeLa cells by pathogenic Escherichia coli carrying the afa-3gene cluster are mediated by the AfaE and AfaD proteins, respectively. Infect Immun. 1998;65:4082–4089. doi: 10.1128/iai.65.10.4082-4089.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kenny B, DeVinney R, Stein M, Reinscheid D J, Frey E A, Finlay B B. Enteropathogenic E. coli(EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell. 1997;91:511–520. doi: 10.1016/s0092-8674(00)80437-7. [DOI] [PubMed] [Google Scholar]
- 38.Kenny B, Finlay B B. Protein secretion by enteropathogenic Escherichia coliis essential for transducing signals to epithelial cells. Proc Natl Acad Sci USA. 1995;92:7991–7995. doi: 10.1073/pnas.92.17.7991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leung K Y, Finlay B B. Intracellular replication is essential for the virulence of Salmonella typhimurium. Proc Natl Acad Sci USA. 1991;88:11470–11474. doi: 10.1073/pnas.88.24.11470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Levine M M, Nataro J P, Karch H, Baldini M M, Kaper J B, Black R E, Clements M L, O’Brien A D. The diarrheal response of humans to some classic serotypes of enteropathogenic Escherichia coliis dependent on a plasmid encoding an enteroadhesiveness factor. J Infect Dis. 1985;152:550–559. doi: 10.1093/infdis/152.3.550. [DOI] [PubMed] [Google Scholar]
- 41.Maurelli A T, Baudry B, d’Hauteville H, Hale T L, Sansonetti P J. Cloning of plasmid DNA sequences involved in invasion of HeLa cells by Shigella flexneri. Infect Immun. 1985;49:164–171. doi: 10.1128/iai.49.1.164-171.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Menard R, Prevost M C, Gounon P, Sansonetti P, Dehio C. The secreted Ipa complex of Shigella flexneripromotes entry into mammalian cells. Proc Natl Acad Sci USA. 1996;93:1254–1258. doi: 10.1073/pnas.93.3.1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mengaud J, Ohayon H, Gounon P, Mege R M, Cossard P. E-cadherin is the receptor for internalin, a surface protein required for entry of L. monocytogenesinto epithelial cells. Cell. 1996;84:923–932. doi: 10.1016/s0092-8674(00)81070-3. [DOI] [PubMed] [Google Scholar]
- 44.Miliotis M D, Koornhog H J, Phillips J I. Invasive potential of noncytotoxic enteropathogenic Escherichia coliin an in vitro Henle 407 cell model. Infect Immun. 1989;57:1928–1935. doi: 10.1128/iai.57.7.1928-1935.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moon H W, Whipp S C, Argenzio R A, Levine M M, Giannella R A. Attaching and effacing activities of rabbit and human enteropathogenic Escherichia coliin pig and rabbit intestines. Infect Immun. 1983;41:1340–1351. doi: 10.1128/iai.41.3.1340-1351.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nataro J P, Kaper J B. Diarrheagenic Escherichia coli. Clin Microbiol Rev. 1998;11:142–201. doi: 10.1128/cmr.11.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.O’Brien A D, Lively T A, Chen M E, Rothman S W, Formal S B. Escherichia coli O157:H7 strains associated with haemorrhagic colitis in the United States produce a Shigella dysenteriae1 (SHIGA) like cytotoxin. Lancet. 1983;i:702. doi: 10.1016/s0140-6736(83)91987-6. [DOI] [PubMed] [Google Scholar]
- 48.Oelschlaeger T A, Barrett T J, Kopecko D J. Some structures and processes of human epithelial cells involved in uptake of enterohemorragic Escherichia coliO157:H7 strains. Infect Immun. 1994;62:5142–5150. doi: 10.1128/iai.62.11.5142-5150.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oelschlaeger T A, Guerry P, Kopecko D J. Unusual microtubule-dependent endocytosis mechanisms triggered by Campylobacter jejuni and Citrobacter freundii. Proc Natl Acad Sci USA. 1993;90:6884–6888. doi: 10.1073/pnas.90.14.6884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Parsot C, Sansonetti P J. Invasion and the pathogenesis of Shigellainfections. Curr Top Microbiol Immunol. 1996;209:25–42. doi: 10.1007/978-3-642-85216-9_2. [DOI] [PubMed] [Google Scholar]
- 51.Sansonetti P J, Kopecko D J, Formal S B. Involvement of a plasmid in the invasive ability of Shigella flexneri. Infect Immun. 1982;35:852–860. doi: 10.1128/iai.35.3.852-860.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sansonetti P J, Ryter A, Clerc P, Maurelli A T, Mounier J. Multiplication of Shigella flexneriwithin HeLa cells: lysis of the phagocytic vacuole and plasmid-mediated contact hemolysis. Infect Immun. 1986;51:461–469. doi: 10.1128/iai.51.2.461-469.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sartor R B. Microbial agents in the pathogenesis, differential diagnosis, and complications of inflammatory bowel diseases. In: Blaser M J, Smith P D, Ravdin J I, Greenberg H B, Guerrant R L, editors. Infections of the Gastrointestinal Tract. New York, N.Y: Raven Press, Ltd.; 1995. pp. 435–458. [Google Scholar]
- 54.Sasakawa C, Kamata K, Sakai T, Makino S, Yamada M, Okada N, Yoshikawa M. Virulence-associated genetic regions comprising 31 kilobases of the 230-kilobase plasmid in Shigella flexneri2a. J Bacteriol. 1988;170:2480–2484. doi: 10.1128/jb.170.6.2480-2484.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Small P L C, Falkow S. Identification of regions on a 230-kilobase plasmid from enteroinvasive Escherichia colithat are required for entry into HEp-2 cells. Infect Immun. 1988;56:225–229. doi: 10.1128/iai.56.1.225-229.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Small P L C, Isberg R R, Falkow S. Comparison of the ability of enteroinvasive Escherichia coli, Salmonella typhimurium, Yersinia pseudotuberculosis, and Yersinia enterocoliticato enter and replicate within HEp-2 cells. Infect Immun. 1987;55:1674–1679. doi: 10.1128/iai.55.7.1674-1679.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sutherland L, Singleton J, Sessions J. Double blind, placebo controlled trial of metronidazole in Crohn’s disease. Gut. 1991;32:1071–1075. doi: 10.1136/gut.32.9.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tabaqchali S, O’Donoghue D P, Bettelheim K A. Escherichia coliantibodies in patients with inflammatory bowel disease. Gut. 1978;19:108–113. doi: 10.1136/gut.19.2.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Takeuchi A, Sprinz H, LaBrec E H, Formal S B. Experimental bacillary dysentery: an electron microscopic study of the response of the intestinal mucosa to bacterial invasion. Am J Pathol. 1965;47:1011–1044. [PMC free article] [PubMed] [Google Scholar]
- 60.Tilney L G, Portnoy D A. Actin filaments and the growth, movement, and spread of the intracellular parasite, Listeria monocytogenes. J Cell Biol. 1989;109:1597–1608. doi: 10.1083/jcb.109.4.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Venkatesan M M, Buysse J M, Oaks E V. Surface presentation of Shigella flexneri invasion plasmid antigens requires the products of the spalocus. J Bacteriol. 1992;174:1990–2001. doi: 10.1128/jb.174.6.1990-2001.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vial P A, Robins-Browne R, Lior H, Prado V, Kaper J B, Nataro J P, Maneval D, Elsayed A, Levine M M. Characterization of enteroadherent-aggregative Escherichia coli, a putative agent of diarrheal disease. J Infect Dis. 1988;158:70–79. doi: 10.1093/infdis/158.1.70. [DOI] [PubMed] [Google Scholar]
- 63.Wellmann W, Fink P C, Benner F. Endotoxemia in active Crohn’s disease. Treatment with whole gut irrigation and 5-aminosalicylic acid. Gut. 1986;27:814–820. doi: 10.1136/gut.27.7.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yamamoto T, Kaneko M, Changchawalit S, Serichantalergs O, Ijuin S, Echeverria P. Actin accumulation associated with clustered and localized adherence in Escherichia coliisolated from patients with diarrhea. Infect Immun. 1994;62:2917–2929. doi: 10.1128/iai.62.7.2917-2929.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]