Abstract
Purpose
An evolutionary action scoring algorithm (EAp53) based on phylogenetic sequence variations stratifies patients with head and neck squamous cell carcinoma (HNSCC) bearing TP53 missense mutations as high-risk, associated with poor outcomes, or low-risk, with similar outcomes as TP53 wild-type, and has been validated as a reliable prognostic marker. We performed this study to further validate prior findings demonstrating that EAp53 is a prognostic marker for patients with locally advanced HNSCC and explored its predictive value for treatment outcomes to adjuvant bio-chemoradiotherapy.
Methods and Materials
Eighty-one resection samples from patients treated surgically for stage III or IV human papillomavirus-negative HNSCC with high-risk pathologic features, who received either radiation therapy + cetuximab + cisplatin (cisplatin) or radiation therapy + cetuximab + docetaxel (docetaxel) as adjuvant treatment in a phase 2 study were subjected to TP53 targeted sequencing and EAp53 scoring to correlate with clinical outcomes. Due to the limited sample size, patients were combined into 2 EAp53 groups: (1) wild-type or low-risk; and (2) high-risk or other.
Results
At a median follow-up of 9.8 years, there was a significant interaction between EAp53 group and treatment for overall survival (P = .008), disease-free survival (P = .05), and distant metastasis (DM; P = .004). In wild-type or low-risk group, the docetaxel arm showed significantly better overall survival (hazard ratio [HR] 0.11, [0.03-0.36]), disease-free survival (HR 0.24, [0.09-0.61]), and less DM (HR 0.04, [0.01-0.31]) than the cisplatin arm. In high-risk or other group, differences between treatments were not statistically significant.
Conclusions
The docetaxel arm was associated with better survival than the cisplatin arm for patients with wild-type or low-risk EAp53. These benefits appear to be largely driven by a reduction in DM.
Introduction
Head and neck squamous cell carcinoma (HNSCC) is a common cause of cancer-related deaths worldwide.1 Surgery followed by radiation therapy (RT) and concurrent administration of high-dose cisplatin is a current global standard therapy for those patients who have operable HNSCC with either extranodal extension (ENE) or positive margin upon pathologic review.2, 3, 4, 5, 6 Subsequent studies have been conducted to determine whether more effective and less toxic postoperative chemoradiotherapeutic regimens can be used in this disease setting. One such study is NRG/RTOG 0234, a phase 2 randomized clinical trial in which patients treated surgically for stage III or IV HNSCC with high-risk pathologic features received either RT + cetuximab + cisplatin (cisplatin) or RT + cetuximab + docetaxel (docetaxel) as adjuvant treatment.7 The primary endpoint of this trial was published and demonstrated that both arms had improved outcomes with better overall survival (OS) and disease-free survival (DFS) rates than in the historical standard, which is the RT + cisplatin arm of RTOG 9501, but the largest improvement was noted for the docetaxel arm.7
Whereas most treatment decisions for patients with HNSCC are based on clinical staging and pathologic evaluation, molecular biomarkers that could provide unique insight into tumor biology have great potential to complement imaging- and pathology-based staging. One potential candidate biomarker for HNSCC is the TP53 gene, as multiple studies have demonstrated an association between TP53 mutations and decreased survival rates.8, 9, 10, 11 Additionally, recent next-generation sequencing technology confirmed that TP53 is the most common somatically mutated gene in HNSCC.12, 13, 14 However, TP53 mutational status has yet to be incorporated into clinical practice for treatment selection. To that end, we established a novel computational scoring system named “EAp53,”15 which uses an evolutionary action (EA) score based on phylogenetic sequence variations and speciation to stratify patients with HNSCC bearing TP53 missense mutations into either a high-risk group associated with poor outcomes or a low-risk group with similar outcomes to patients with wild-type TP53. The EAp53 score has been validated as a reliable prognostic marker in several clinical cohorts.16,17 Additionally, in our preclinical study, TP53-mutated and -wild-type HNSCC cell lines treated in vitro or in vivo with cisplatin indicated that high-risk EAp53 confers cisplatin resistance not seen in cells with wild-type or low-risk EAp53,16,17 whereas we have not observed TP53 mutations to be associated with taxane resistance using same model (unpublished data).
The primary objective of this study was to determine whether EAp53 is prognostic for patients in RTOG 0234. The secondary objective was to determine whether EAp53 is predictive of treatment outcome. We hypothesized that high-risk EAp53 would show worse clinical outcomes in the entire RTOG 0234 cohort compared with low-risk EAp53 and that high-risk EAp53 would show worse clinical outcomes in the cisplatin arm compared with the docetaxel arm, and there will be no difference in outcomes between treatment arms among low-risk EAp53 groups. Lastly, for comparison purposes, we performed the same analyses with the TP53 classification method established by Poeta and colleagues,9,18 which is based on protein folding.
Methods and Materials
All studies here were approved by the institutional review board and conducted in accordance with the Declaration of Helsinki. Waiver of written informed consent was provided as part of the approval process before sample collection.
EAp53 scoring
EA scores for individual TP53 mutations were obtained from the EAp53 server at Baylor College of Medicine (http://eaction.lichtargelab.org/Eap53). Based on previous analysis,16 scores below 75 were categorized as low-risk EAp53, and those 75 or above were categorized as high-risk EAp53. Mutations that were not missense mutations were designated “other.” Patients who had both missense and other mutations were classified using the missense mutation. Those whose tumors had both low-risk and high-risk EAp53 were classified as having high-risk EAp53.
The Cancer Genome Atlas database
To monitor the EAp53 effect on survival, the clinico-pathologic and TP53 sequencing information for the latest patients with HNSCC in The Cancer Genome Atlas (TCGA) were extracted from the National Cancer Institute Genomic Data Commons (https://gdc.cancer.gov) or firebrowse.org. Patient criteria, analysis method, and TP53 mutation status are described in the Supplementary Materials and Table S1.
RTOG 0234 sample collection
Tumor samples resected from patients enrolled in RTOG 0234 were used to extract genomic DNA and determine EAp53 status. All patients had American Joint Committee on Cancer pathologic stage III or IV squamous cell carcinoma of the oral cavity, oropharynx, hypopharynx, or larynx and had undergone gross total resection. Also, patients had 1 or more pathologically high-risk factors: positive margin, ENE, or 2 or more positive nodes.
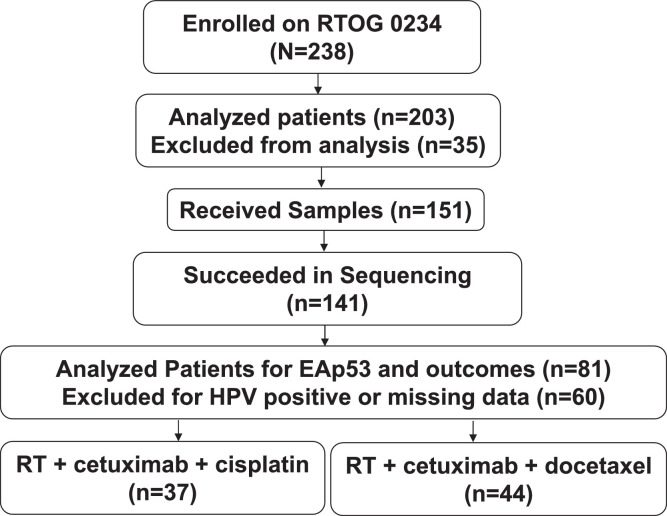
Among 203 analyzable patients, all 151 available resected tumor samples were received as 5- or 10-μm thick sections of formalin-fixed paraffin-embedded tissue samples from the NRG Oncology Biospecimen bank (Fig. 1). Samples were quality-controlled, reviewed to confirm the diagnosis (RCJ), annotated, and anonymized before being provided to our laboratory through an approval by the National Clinical Trials Network Core Correlative Sciences Committee.
Figure 1.
CONSORT diagram. Abbreviations: EAp53 =evolutionary action scoring algorithm; HPV = human papillomavirus; RT = radiation therapy.
DNA extraction and TP53 targeted sequencing
Isolated DNA was sequenced on MiSeq (Illumina, San Diego, CA). Precise information is described in the Supplementary Materials.
Statistical analysis of RTOG 0234 cohort
DFS and OS rates were estimated by the Kaplan-Meier method. Local-regional failure (LRF) and distant metastasis (DM) rates were estimated by the cumulative incidence method. Patients excluded from RTOG 0234 primary analysis were also excluded here.7 Missing data analysis was performed by comparing patients with known EAp53 against those with unknown EAp53 for DFS, OS, LRF, DM, and patient characteristics. Patient characteristics between groups were compared by Fisher's exact test or Wilcoxon rank-sum test. Automated immunohistochemistry staining and in situ hybridization were performed for detecting p16 and human papillomavirus (HPV) status, respectively.7 Due to low prevalence of TP53 mutations in HPV-positive tumors, primary analysis was limited to patients with HPV-negative tumors, defined as follows: for oropharynx primary site, both HPV-negative and p16-negative were defined as negative and HPV-positive and/or p16-positive were defined as positive; for other primary sites, only HPV status was considered. To be included in analysis, patients had to have no missing data for the following variables: assigned treatment, age, gender, race, Zubrod performance status, smoking history, primary site, T stage, N stage, ENE, positive margin, and number of positive nodes. Due to small sample sizes, patients were combined into 2 EAp53 groups: (1) wild-type or low-risk; and (2) high-risk or other. For prognostic analysis, EAp53 groups were compared by log-rank test. Hazard ratios (HRs) and 95% confidence intervals were estimated by Cox model with and without assigned treatment and additional covariates. The number of additional covariates was limited to treatment + 2 for OS, treatment + 3 for DFS, and only treatment for LRF and DM, due to low numbers of events. Model selection was performed using akaike information criterion. For predictive analysis, HRs were estimated for models including EAp53, treatment, and the interaction of EAp53 and treatment with and without additional covariates as described previously. All statistical tests were 2-sided with alpha of 0.05. With the 81 patients available for these analyses, the statistical power to detect an HR of 2.5 on OS, DFS, LRF, and DM associated with EAp53 status was 87%, 92%, 67%, and 66%, respectively, based on a Cox model. The SAS version 9.4 was used for analysis.
Poeta classification method
The same analyses for the same patients were performed replacing EAp53 with Poeta rules + splice method.9,18 Precise information is described in the Supplementary Materials.
Results
EAp53 effect on outcome in TCGA cohort
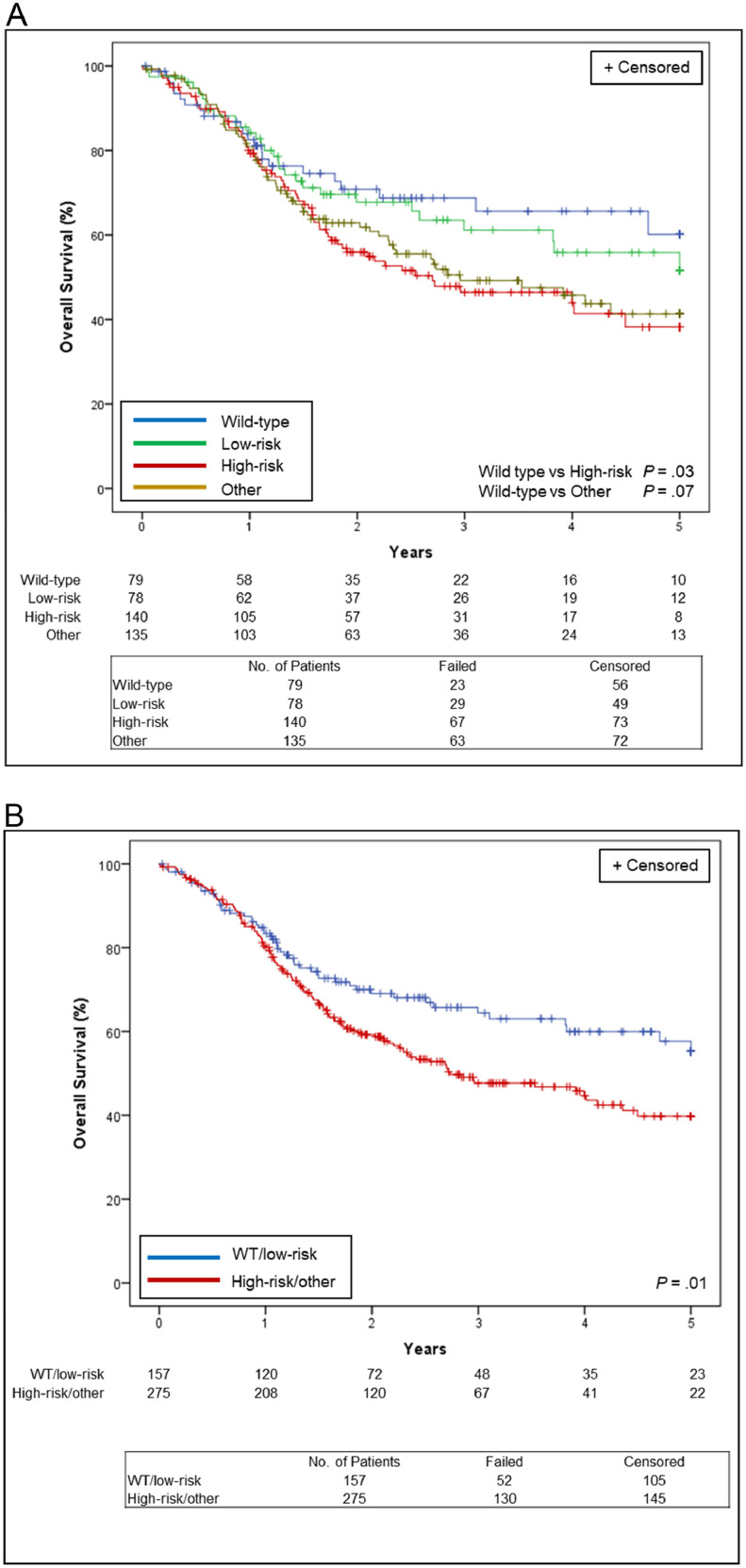
Among 432 HPV-negative patients with HNSCC in TCGA, 78 patients (18%) were categorized as low-risk, 140 (32%) were categorized as high-risk, 135 (31%) were categorized as other, with 79 (18%) wild-type for EAp53. The Kaplan-Meier survival curve in Fig. 2A shows that the 5-year OS rate was 60.2% for wild-type, 51.6% for low-risk (HR low-risk vs wild-type, 1.15 [0.67-1.99]; P = .61), 38.2% for high-risk (HR high-risk vs wild-type, 1.67 [1.04-2.69]; P = .03), and 41.3% for other EAp53 (HR other vs wild-type, 1.55 [0.96-2.51]; P = .07). Given the similarity between the outcomes for wild-type and low-risk or high-risk and other, we combined them into 2 groups: (1) wild-type or low-risk; and (2) high-risk or other, to improve the statistical power of our analysis. The 5-year OS rate was therefore 55.3% for the wild-type or low-risk group and 39.8% for the high-risk or other group (HR 1.49, [1.08-2.06]; P = .014; Fig. 2B).
Figure 2.
Overall survival estimates in 432 patients with human papillomavirus–negative head and neck squamous cell carcinoma from The Cancer Genome Atlas, (A) by each evolutionary action scoring algorithm status and (B) by evolutionary action scoring algorithm group. WT = wild-type.
TP53 targeted sequencing of 151 RTOG 0234 samples
One hundred forty-one of 151 patients had successful TP53 mutation calls (Fig. S1, Tables S2 and S3). We observed 10 sequencing failures: 9 due to low DNA yield and 1 due to low DNA quality. Seventy-nine of 141 sequenced patients (56%) had a total of 101 mutations. Of these mutations, 62 were missense mutations that could be scored by EAp53. Twenty of 79 patients (25%) had multiple mutations. Twenty-one of 141 patients (15%) were classified as low-risk, 33 (23%) were classified as high-risk, and 25 (18%) were classified as other, with 62 (44%) wild-type for EAp53.
Patient characteristics in RTOG 0234 HPV-negative cohort
The patient and tumor characteristics for 81 patients with HPV-negative tumors and complete data for covariates are summarized in Table 1. There was no imbalance in all covariates between treatment arms for EAp53 subset of patients. Sixty-eight of 81 patients (84%) had TP53 mutations; low-risk were 19 (23%), high-risk were 27 (33%), and other mutations were 22 (27%), with 13 (16%) wild-type. Overall, 52% had Zubrod performance status 1, 58% had ENE, and 37% had positive margins. Primary sites were oral cavity (65%), oropharynx (14%), larynx (14%), and hypopharynx (7%). In wild-type and low-risk group, 34% of patients had Zubrod 1, compared with 63% in the high-risk or other group. Seventy-five percent and 59% of patients in the wild-type or low-risk and high-risk or other groups, respectively, had oral cavity tumors. In the wild-type or low-risk group, 47% of patients had ENE, compared with 65% in the high-risk other group. Fifty percent and 29% in the wild-type or low-risk and high-risk or other groups, respectively, had positive margins.
Table 1.
Patient and tumor characteristics by evolutionary action scoring algorithm and assigned treatment in RTOG 0234 human papillomavirus–negative cohort
| Wild-type or low-risk |
High-risk or other |
|||||
|---|---|---|---|---|---|---|
| Cisplatin | Docetaxel | Total | Cisplatin | Docetaxel | Total | |
| Characteristic | (n = 12) | (n = 20) | (n = 32) | (n = 25) | (n = 24) | (n = 49) |
| Age (years) | P = .39* | P = .94* | ||||
| Mean | 58.2 | 55.1 | 56.2 | 55 | 53.6 | 54.3 |
| Standard deviation | 9.73 | 11.57 | 10.86 | 11.43 | 13.71 | 12.49 |
| Median | 57.5 | 58.5 | 58 | 57 | 56.5 | 57 |
| Min - max | 38-69 | 25-77 | 25-77 | 27-74 | 21-79 | 21-79 |
| First - third quartiles | 53.5-66.5 | 50.5-62.5 | 52-63 | 49-62 | 47-62 | 49-62 |
| Sex | P = .21† | P = .77† | ||||
| Male | 11 (92%) | 14 (70%) | 25 (78%) | 17 (68%) | 15 (63%) | 32 (65%) |
| Female | 1 (8%) | 6 (30%) | 7 (22%) | 8 (32%) | 9 (38%) | 17 (35%) |
| Race | P = .62† | P = .61† | ||||
| White | 10 (83%) | 18 (90%) | 28 (88%) | 22 (88%) | 23 (96%) | 45 (92%) |
| Nonwhite | 2 (17%) | 2 (10%) | 4 (13%) | 3 (12%) | 1 (4%) | 4 (8%) |
| Zubrod performance status | P = 1.00† | P = 1.00† | ||||
| 0 | 8 (67%) | 13 (65%) | 21 (66%) | 9 (36%) | 9 (38%) | 18 (37%) |
| 1 | 4 (33%) | 7 (35%) | 11 (34%) | 16 (64%) | 15 (63%) | 31 (63%) |
| Smoking history | P = .18* | P = .79* | ||||
| Never | 2 (17%) | 7 (35%) | 9 (28%) | 4 (16%) | 5 (21%) | 9 (18%) |
| Former | 7 (58%) | 11 (55%) | 18 (56%) | 18 (72%) | 16 (67%) | 34 (69%) |
| Current | 3 (25%) | 2 (10%) | 5 (16%) | 3 (12%) | 3 (13%) | 6 (12%) |
| Primary site | P = .68† | P = 1.00† | ||||
| Oral cavity | 10 (83%) | 14 (70%) | 24 (75%) | 15 (60%) | 14 (58%) | 29 (59%) |
| Oropharynx | 1 (8%) | 2 (10%) | 3 (9%) | 4 (16%) | 4 (17%) | 8 (16%) |
| Hypopharynx | 0 | 1 (5%) | 1 (3%) | 2 (8%) | 3 (13%) | 5 (10%) |
| Larynx | 1 (8%) | 3 (15%) | 4 (13%) | 4 (16%) | 3 (13%) | 7 (14%) |
| Surgical-pathologic T stage | P = .54* | P = .44* | ||||
| T1 | 2 (17%) | 5 (25%) | 7 (22%) | 1 (4%) | 5 (21%) | 6 (12%) |
| T2 | 5 (42%) | 6 (30%) | 11 (34%) | 7 (28%) | 4 (17%) | 11 (22%) |
| T3 | 1 (8%) | 7 (35%) | 8 (25%) | 5 (20%) | 5 (21%) | 10 (20%) |
| T4 | 4 (33%) | 2 (10%) | 6 (19%) | 12 (48%) | 10 (42%) | 22 (45%) |
| Surgical-pathologic N stage | P = .55* | P = .15* | ||||
| N0 | 0 | 2 (10%) | 2 (6%) | 2 (8%) | 0 | 2 (4%) |
| N1 | 2 (17%) | 2 (10%) | 4 (13%) | 4 (16%) | 2 (8%) | 6 (12%) |
| N2a | 1 (8%) | 1 (5%) | 2 (6%) | 0 | 0 | 0 |
| N2b | 7 (58%) | 14 (70%) | 21 (66%) | 13 (52%) | 14 (58%) | 27 (55%) |
| N2c | 2 (17%) | 1 (5%) | 3 (9%) | 6 (24%) | 6 (25%) | 12 (24%) |
| N3 | 0 | 0 | 0 | 0 | 2 (8%) | 2 (4%) |
| Surgical-pathologic AJCC stage | P = 1.00† | P = .67† | ||||
| III | 1 (8%) | 3 (15%) | 4 (13%) | 4 (16%) | 2 (8%) | 6 (12%) |
| IV | 11 (92%) | 17 (85%) | 28 (88%) | 21 (84%) | 22 (92%) | 43 (88%) |
| Extranodal extension | P = 1.00† | P = .77† | ||||
| No | 6 (50%) | 11 (55%) | 17 (53%) | 8 (32%) | 9 (38%) | 17 (35%) |
| Yes | 6 (50%) | 9 (45%) | 15 (47%) | 17 (68%) | 15 (63%) | 32 (65%) |
| Positive margin | P = 1.00† | P = .75† | ||||
| No | 6 (50%) | 10 (50%) | 16 (50%) | 17 (68%) | 18 (75%) | 35 (71%) |
| Yes | 6 (50%) | 10 (50%) | 16 (50%) | 8 (32%) | 6 (25%) | 14 (29%) |
| Two or more positive nodes | P = .37† | P = .70† | ||||
| No | 1 (8%) | 5 (25%) | 6 (19%) | 5 (20%) | 3 (13%) | 8 (16%) |
| Yes | 11 (92%) | 15 (75%) | 26 (81%) | 20 (80%) | 21 (88%) | 41 (84%) |
Abbreviation: AJCC = American Joint Committee on Cancer.
Wilcoxon rank-sum test.
Fisher's exact test; primary site was tested as oral cavity versus others.
EAp53 as a prognostic biomarker in RTOG 0234 HPV-negative cohort
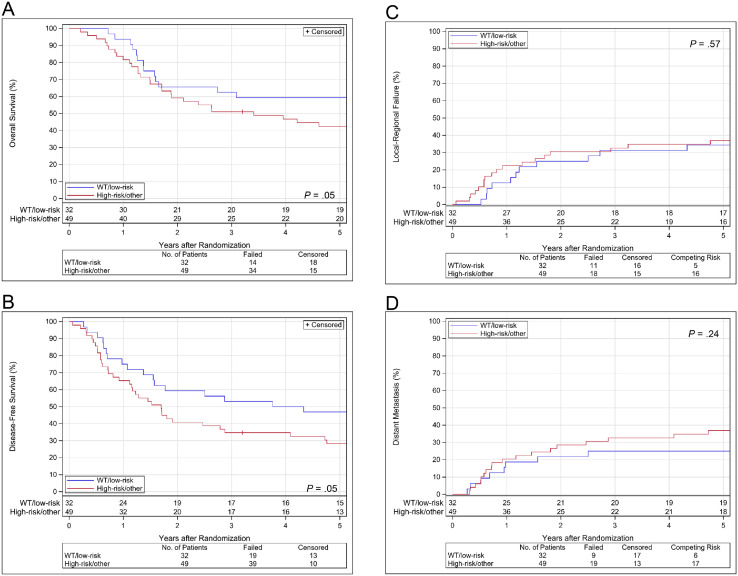
OS, DFS, LRF, and DM estimates by EAp53 group are shown in Fig. 3. Before adjustment for treatment or covariates, the high-risk or other group had significantly worse OS and DFS than the wild-type or low-risk group (HR 1.87, [1.00-3.48]; P = .05 and HR 1.74, [1.00-3.01]; P = .05, respectively). However, after adjustment, the difference was no longer significant (HR 1.56, [0.80-3.04]; P = .19 for OS and HR 1.55, [0.85-2.81]; P = .15 for DFS). In LRF, we observed no significant difference between the EAp53 groups in analysis both before and after adjustment for treatment; the HRs (high-risk or other vs wild-type or low-risk) were 1.25 (0.59-2.64; P = .57) and 1.17 (0.55-2.52; P = .68), respectively. In DM, we also observed no significant difference between the EAp53 groups in analyses both before and after adjustment for treatment; the HRs (high-risk or other vs wild-type or low-risk) were 1.62 (0.73-3.59; P = .24) and 1.31 (0.58-2.97; P = .52), respectively. Lung was the first distant metastatic site with or without other sites in 22 of 28 patients (79%).
Figure 3.
(A) Overall survival, (B) disease-free survival, (C) local-regional failure, and (D) distant metastasis estimates by evolutionary action scoring algorithm group with treatment arms combined in RTOG 0234 human papillomavirus–negative cohort. Abbreviations: WT = wild-type.
EAp53 as a predictive biomarker in RTOG 0234 HPV-negative cohort
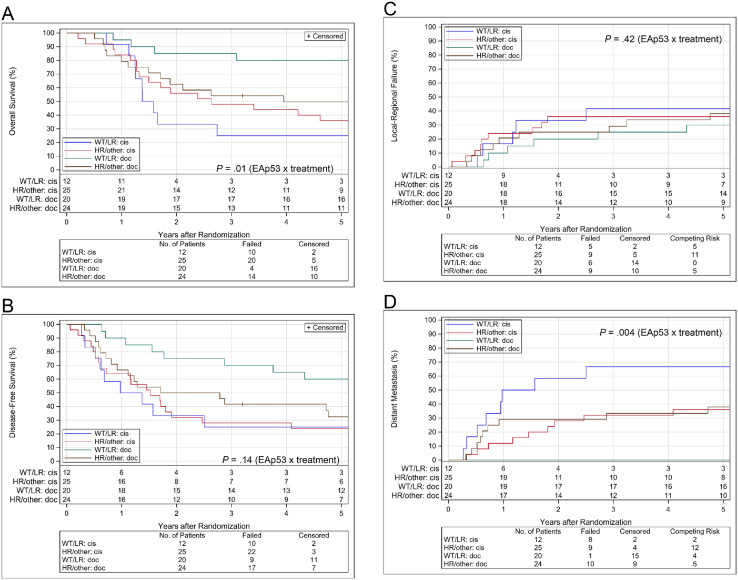
OS, DFS, LRF, and DM estimates by EAp53 group and treatment assignment are shown in Fig. 4. Final multivariate models are shown in Table 2. For OS, before adjustment for covariates, we found a significant interaction between EAp53 group and assigned treatment (P = .01); the HR comparing the docetaxel to the cisplatin arm was 0.12 (0.04-0.39) in the wild-type or low-risk group and 0.67 (0.34-1.33) in the high-risk or other group. After adjustment for covariates, we again found a significant interaction between EAp53 group and assigned treatment (P = .008); the HR comparing the docetaxel to the cisplatin arm was 0.11 (0.03-0.36) in the wild-type or low-risk group and 0.71 (0.36-1.40) in the high-risk or other group. For DFS, before adjustment for covariates, we did not observe a significant interaction between EAp53 group and assigned treatment (P = .14); the HR comparing the docetaxel to the cisplatin arm was 0.29 (0.12-0.71) in the wild-type or low-risk and 0.67 (0.35-1.26) in the high-risk other group. However, after adjustment for covariates, we found a significant interaction between EAp53 group and assigned treatment (P = .05); the HR comparing the docetaxel to the cisplatin arm was 0.24 (0.09-0.61) in the wild-type or low-risk and 0.74 (0.39-1.39) in the high-risk or other group. For LRF, we did not see a significant interaction between EAp53 group and assigned treatment without adjustment for covariates (P = .42); the HR comparing the docetaxel to the cisplatin arm was 0.49 (0.15-1.62) in the wild-type or low-risk and 0.92 (0.36-2.31) in the high-risk or other group. For DM, we found a significant interaction between EAp53 group and assigned treatment without adjustment for covariates (P = .004); the HR comparing the docetaxel to the cisplatin arm was 0.04 (0.01-0.31) in the wild-type or low-risk and 1.05 (0.42-2.59) in the high-risk or other group. There were too few events to adjust for covariates for both LRF and DM.
Figure 4.
(A) Overall survival, (B) disease-free survival, (C) local-regional failure, and (D) distant metastasis estimates by EAp53 group and treatment arm in RTOG 0234 human papillomavirus–negative cohort. Abbreviations: cis = cisplatin; doc = docetaxel; EAp53 = evolutionary action scoring algorithm; HR = high-risk; LR = low-risk; WT = wild-type.
Table 2.
Multivariable analysis of EAp53 as a predictive biomarker in RTOG 0234 human papillomavirus–negative cohort
| Endpoint | ||
|---|---|---|
| Variable | Hazard ratio | |
| subgroup | (95% confidence interval) | P value |
| Overall survival (n = 81; 48 events) | ||
| EAp53 X assigned treatment interaction | .008 | |
| EAp53 (high-risk or other vs wild-type or low-risk) | ||
| If cisplatin arm | 0.73 (0.34-1.60) | |
| If docetaxel arm | 4.69 (1.52-14.50) | |
| Assigned treatment (docetaxel vs cisplatin) | ||
| If wild-type or low risk | 0.11 (0.03-0.36) | |
| If high-risk or other | 0.71 (0.36-1.40) | |
| Sex (male vs female) | 2.52 (1.21-5.25) | .01 |
| Zubrod performance status (1 vs 0) | 1.93 (1.05-3.55) | .03 |
| Disease-free survival (n = 81; 58 events) | ||
| EAp53 X assigned treatment interaction | .05 | |
| EAp53 (high-risk or other vs wild-type or low-risk) | ||
| If cisplatin arm | 0.87 (0.40-1.91) | |
| If docetaxel arm | 2.69 (1.16-6.21) | |
| Assigned treatment (docetaxel vs cisplatin) | ||
| If wild-type or low-risk | 0.24 (0.09-0.61) | |
| If high-risk or other | 0.74 (0.39-1.39) | |
| Sex (male vs female) | 2.15 (1.13-4.08) | .02 |
| T stage (T3 and T4 vs T1 and T2) | 1.73 (0.99-3.01) | .05 |
| Extranodal extension (yes vs no) | 2.23 (1.26-3.95) | .006 |
| Local-regional failure (n = 81; 29 events) | ||
| EAp53 X assigned treatment interaction | .42 | |
| EAp53 (high-risk or other vs wild-type or low-risk) | ||
| If cisplatin arm | 0.83 (0.28-2.49) | |
| If docetaxel arm | 1.56 (0.55-4.38) | |
| Assigned treatment (docetaxel vs cisplatin) | ||
| If wild-type or low-risk | 0.49 (0.15-1.62) | |
| If high-risk or other | 0.92 (0.36-2.31) | |
| Distant metastasis (n = 81; 28 events) | ||
| EAp53 X assigned treatment interaction | .004 | |
| EAp53 (high-risk or other vs wild-type or low-risk) | ||
| If cisplatin arm | 0.42 (0.16-1.10) | |
| If docetaxel arm | 11.71 (1.50-91.68) | |
| Assigned treatment (docetaxel vs cisplatin) | ||
| If wild-type or low-risk | 0.04 (0.01-0.31) | |
| If high-risk or other | 1.05 (0.42-2.59) |
Abbreviations EAp53 = evolutionary action scoring algorithm.
Assessment of Poeta rules + splice method
In both TCGA and RTOG 0234 HPV-negative cohorts, disruptive mutation showed no discriminatory power of prognosis compared with nondisruptive mutation (Figs. S2 and S3). As a predictive biomarker to treatment outcome in RTOG 0234 HPV-negative cohort, we found a significant interaction between Poeta rules + splice group and assigned treatment after adjusting for covariates for OS (P = .04). However, regarding other outcomes, we did not see any significance (Fig. S4 and Table S4).
Discussion
We confirmed that EAp53 status is a prognostic marker in HNSCC in the HPV-negative TCGA cohort; however, EAp53 was not prognostic in RTOG 0234 HPV-negative cohort (n = 81) when adjusting for additional covariates. This difference may be due to the poor prognostic features used as inclusion criteria in RTOG 0234, the small sample size, or potential baseline covariate imbalance between biomarker subgroups, as suggested by the adjusted analysis (Table S5). Interestingly, EAp53 was also not prognostic in HPV-negative TCGA data when the HNSCC cohort was filtered with the same criteria as RTOG 0234 (Fig. S5). Overall, these results might suggest that the importance of EAp53 as a prognostic biomarker could vary depending on the disease setting. For example, it may not be prognostic in locally advanced tumors that are treated with cisplatin-based therapy but could be prognostic in lower stage tumors. These hypotheses should be confirmed with well-designed studies involving a larger number of patients for each disease setting.
We next explored the use of EAp53 as a predictive biomarker. We found EAp53 status to be predictive of outcome for cisplatin- and docetaxel-based combination bio-chemoradiotherapy in patients with pathologically high-risk HPV-negative HNSCC. In the wild-type or low-risk group, the docetaxel arm was associated with better OS, DFS, and lower DM rates than the cisplatin arm. We found a significant treatment effect favoring docetaxel over cisplatin in the wild-type or low-risk group, but there was no evidence of difference in clinical outcomes in the high-risk or other group by treatment arm. We therefore concluded that these observed differences in outcome were driven by differential response to the treatments in the wild-type or low-risk group.
These results are not consistent with our original hypothesis, especially for the wild-type or low-risk group. Our preclinical findings in HPV-negative HNSCC cell lines demonstrated relative cisplatin resistance in cell lines with high-risk EAp53 compared with those with wild-type or low-risk EAp53.16,17 Besides, multiple clinical cohorts have shown that tumors bearing TP53 mutations are relatively resistant to cisplatin-based treatment compared with those with wild-type TP53.19,20 Conversely, both in vitro and in vivo, docetaxel has shown no difference in response based on TP53 mutation status, or slightly better effect in TP53 mutant cell lines than in wild-type (unpublished data).21,22 Because we observed a difference in DM but not LRF, we hypothesized that tumors with wild-type or low-risk EAp53 might not only be resistant to the cisplatin-containing treatment but more aggressive after the treatment. Our most current experiments partially support this hypothesis, as the TP53 wild-type HPV-negative HNSCC cell line selected for cisplatin resistance retains TP53 wild-type status, and when being injected into the tongues of mice, it develops distant metastases whereas the parental cell line does not (unpublished data).
How much the inclusion of cetuximab may have affected the observed differences in outcomes between the 2 arms is not clear. The phase 3 clinical trial RTOG 0522 determined that the addition of cetuximab did not show significantly different outcomes compared with RT + cisplatin alone for 891 patients with stage III or IV HNSCC, although these patients didn't undergo surgery.23 The lack of difference could have been caused by the substantial rates of incomplete treatment in the cetuximab arm due to severe toxicity. In RTOG 0234, both arms were shown to be feasible with predictive toxicity resulting in better survival rates relative to historical control, RT + cisplatin. However, the docetaxel arm showed most favorable outcomes compared with the cisplatin arm with regard to OS and DFS; therefore, only this arm has commenced formal testing in the following randomized phase 2 and 3 trial NRG/RTOG 1216, which is currently accruing patients. Validation of our findings in a cohort of patients treated without cetuximab will be necessary to determine whether cetuximab plays a role in the phenotype that can result in different treatment outcomes. Fortunately, NRG/RTOG 1216 includes the appropriate arms to test this. Another ongoing phase 2 randomized EA3132 trial for patients with HNSCC with pathologic stage III or IVA (American Joint Committee on Cancer 8) T3-T4a, N0-3, M0 or T1-T2, N1-3, or M0 after total resection of the primary tumor may also be used as a validation.24,25 This study stratifies patient by TP53 mutational status in adjuvant RT alone versus RT + cisplatin. The fundamental concepts underlying this study are similar to our RTOG 0234 trial in that they are both designed with the long-term goal of potentially selecting postoperative adjuvant therapy for surgically treated patients with HNSCC based on TP53 mutational status. However, the studies differ in that the EA3132 trial determines the TP53 mutational status prospectively and uses Poeta rules + splice method. In addition, it is targeting patients with lower risk surgical pathology (no positive margins, ENE, and/or gross residual disease) than RTOG 0234 or NRG/RTOG 1216.
Our findings may have only been apparent because we analyzed pathologically advanced HNSCC, the biology of which is different from early stage HNSCC. We previously reported a strong association between high-risk and other EAp53 and ENE in patients with oral SCC in TCGA26. In the present study, 58% of the patients had ENE, and they were similarly enriched in high-risk and other EAp53. Of note, in the ENE-positive oral SCC cohort of TCGA, we also observed a worse OS rate in the wild-type or low-risk group than in high-risk or other group (Fig. S6). Adjuvant treatments in the TCGA cohort were not controlled in any way, but many of the ENE-positive patients likely received adjuvant therapy including RT + cisplatin. Therefore, the worse outcomes in wild-type and low-risk patients may have been related to both the treatment regimen and the pathologically high-risk tumor biology, particularly ENE positivity. What this means mechanistically is unclear.
A limitation of this study is that the sample size is small and validation in other cohorts is necessary. Additionally, the sequenced tissue sites included not only primary tumor but also metastatic lymph nodes. Mutations can be heterogeneous within a primary tumor and across metastatic sites, so it is possible that some detected mutations may not be representative of the whole tumor.24 This variability is inherent to most sequencing studies but could be more controlled in future studies.
Taken together, our results demonstrate that RT + cetuximab + docetaxel may be a good postoperative treatment option for locally advanced HPV-negative patients with HNSCC with wild-type and low-risk EAp53. This benefit appears to be largely driven by reduction in DM in wild-type and low-risk patients, who had better survival rates in docetaxel-based treatment. These findings need validation before changing clinical practice. The ongoing NRG/RTOG 1216 addressed which adjuvant treatment has more promising DFS by comparing RT + cisplatin, RT + docetaxel, and RT+ cetuximab + docetaxel in phase 2, and is addressing which combination has better OS by comparing RT + cisplatin, RT + cetuximab + docetaxel, and RT + cisplatin + atezolizumab (new arm) in phase 3 for pathologically high-risk HNSCC in the postoperative setting. The planned TP53 mutation analysis in that trial will answer at least 2 questions related to our findings: (1) Can we validate the good outcome for wild-type and low-risk patients treated with RT + cetuximab + docetaxel? (2) Does cetuximab play a role in different outcomes? If the results of the NRG/RTOG 1216 trial validate our findings and justify clinical use, then incorporating TP53 sequencing and EAp53 scoring into standard clinical practice would be easy, given the availability of Clinical Laboratory Improvement Amendments certified TP53 sequencing assays.
Conclusions
EAp53 status is a statistically significant predictive biomarker to adjuvant treatment outcome and was superior to the stratification by Poeta rules + splice method in this cohort. However, the Poeta method should still be analyzed in future studies, together with EAp53, because of its demonstrated utility in other previous studies.9,18
Acknowledgments
The authors thank Justin P. Windham for technical support of DNA extraction, Barbara Burtness and Christine H. Chung for advice about the Poeta rules + splice method, and Scientific Publication Services (Donald R. Norwood).
Footnotes
Sources of support: This project was supported by grants UG1CA189867 (NRG Oncology NCORP), U10CA180868 (NRG Oncology Operations), U10CA180822 (NRG Oncology SDMC), and U24CA196067 (NRG Oncology Biospecimen Bank) from the National Cancer Institute (NCI) and 5R01DE024601-05 from the National Institute of Dental & Craniofacial Research (NIDCR), Eli Lilly, and Aventis Pharmaceuticals. This project was funded, in part, under a grant with the Pennsylvania Department of Health; the department specifically disclaims responsibility for any analyses, interpretations, or conclusions. Paul M. Harari acknowledges support from the NIH (P50 DE026787 - UW Head and Neck SPORE Grant). Chieko Michikawa was supported by JSPS KAKENHI (grant number 16K11718) and supported in part by the Fellowship of Astellas Foundation for Research on Metabolic Disorders.
Disclosures: C. Michikawa, N. Silver, P.M. Harari, M.S. Kies, D.I. Rosenthal, Q. Le, D.Y. Duose, S. Mallampati, S. Trivedi, R. Luthra, A.A. Osman, O. Lichtarge, U. Parvathaneni, D.N. Hayes, and J.N. Myers have nothing to disclose. R.L. Foote reports Grants or contracts from any entity-Unrestricted research funding from endowed named professorship paid to Mayo Clinic from Hitachi, Ltd, Royalties from textbook sales and writing content, royalties from licensing of patent, paid to me from Elsevier, UpToDate, Bionix. Honoraria was paid to me from Opportunity and Progress of Proton Therapy Clinical Opportunities to advance the field of particle therapy Mayo Clinic Guangzhou, China. Patent licensed to Bionix. Royalties paid to Mayo Clinic and to my department and to myself from Patent issued for TruGuard intra-oral radiotherapy stent. R.C. Jordan reports Grants or contracts from National Cancer Institute U24CA196067, National Institute of Allergy and Infectious Diseases P30AI027763, National Institute of Dental & Craniofacial Diseases R01DE026502. Royalties or licenses from Elsevier – Oral Pathology Clinical Pathologic Correlations Ed 7. Payment for expert testimony for 2021- 2 hours Evans Dixon Law Firm. Stock or stock options-Exact Sciences 4 shares. C.R. Pickering reports All support for the present manuscript-NIH grants. P.A.T-Saavedra reports All support for the present manuscript- NRG Oncology SDMC Grant from NCI. I.I. Wistuba reports Grants or contracts from any entity- Genentech, Oncoplex, HTG Molecular, DepArray, Merck, Bristol-Myers Squibb, Medimmune, Adaptive, Adaptimmune, EMD Serono, Pfizer, Takeda, Amgen, Karus, Johnson & Johnson, Bayer, Iovance, 4D, Novartis, and Akoya. Consulting fees from Genentech/Roche, Bayer, BristolMyers Squibb, Astra Zeneca/Medimmune, Pfizer, HTG Molecular, Asuragen, Daiichi Sankyo, Merck, GlaxoSmithKline, Guardant Health, Flame, Novartis, Sanofi, Oncocyte, and MSD. Payment or honoraria for lectures, presentations, speakers bureaus, manuscript writing or educational events from Medscape, MSD, Genentech/Roche, Platform Health, Pfizer, AstraZeneca, Merck.
Data sharing statement: All original data of The Cancer Genome Atlas (TCGA) cohort are at the National Cancer Institute Genomic Data Commons (https://gdc.cancer.gov) or firebrowse.org. All published data of RTOG 0234 cohort from this paper will be available upon request in accordance with NRG Oncology's data sharing policy, which can be found at https://www.nrgoncology.org/Resources/Ancillary-Projects-Data-Sharing-Application. All data analyzed during this study are in this published article or the Supplementary Materials.
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.adro.2022.100989.
Appendix. Supplementary materials
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Cooper JS, Pajak TF, Forastiere AA, et al. Postoperative concurrent radiotherapy and chemotherapy for high-risk squamous-cell carcinoma of the head and neck. N Engl J Med. 2004;350:1937–1944. doi: 10.1056/NEJMoa032646. [DOI] [PubMed] [Google Scholar]
- 3.Bernier J, Domenge C, Ozsahin M, et al. Postoperative irradiation with or without concomitant chemotherapy for locally advanced head and neck cancer. N Engl J Med. 2004;350:1945–1952. doi: 10.1056/NEJMoa032641. [DOI] [PubMed] [Google Scholar]
- 4.Bernier J, Cooper JS, Pajak TF, et al. Defining risk levels in locally advanced head and neck cancers: a comparative analysis of concurrent postoperative radiation plus chemotherapy trials of the EORTC (#22931) and RTOG (# 9501) Head Neck. 2005;27:843–850. doi: 10.1002/hed.20279. [DOI] [PubMed] [Google Scholar]
- 5.Cooper JS, Zhang Q, Pajak TF, et al. Long-term follow-up of the RTOG 9501/intergroup phase III trial: postoperative concurrent radiation therapy and chemotherapy in high-risk squamous cell carcinoma of the head and neck. Int J Radiat Oncol Biol Phys. 2012;84:1198–1205. doi: 10.1016/j.ijrobp.2012.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.National Comprehensive Cancer Network (NCCN). Head and neck cancers (version 3.2019). Available at: https://www.nccn.org/professionals/physician_gls/pdf/head-and-neck.pdf. Accessed October 16, 2019.
- 7.Harari PM, Harris J, Kies MS, et al. Postoperative chemoradiotherapy and cetuximab for high-risk squamous cell carcinoma of the head and neck: Radiation Therapy Oncology Group RTOG-0234. J Clin Oncol. 2014;32:2486–2495. doi: 10.1200/JCO.2013.53.9163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Erber R, Conradt C, Homann N, et al. TP53 DNA contact mutations are selectively associated with allelic loss and have a strong clinical impact in head and neck cancer. Oncogene. 1998;16:1671–1679. doi: 10.1038/sj.onc.1201690. [DOI] [PubMed] [Google Scholar]
- 9.Poeta ML, Manola J, Goldwasser MA, et al. TP53 mutations and survival in squamous-cell carcinoma of the head and neck. N Engl J Med. 2007;357:2552–2561. doi: 10.1056/NEJMoa073770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lindenbergh-van der Plas M, Brakenhoff RH, Kuik DJ, et al. Prognostic significance of truncating TP53 mutations in head and neck squamous cell carcinoma. Clin Cancer Res. 2011;17:3733–3741. doi: 10.1158/1078-0432.CCR-11-0183. [DOI] [PubMed] [Google Scholar]
- 11.Sano D, Xie TX, Ow TJ, et al. Disruptive TP53 mutation is associated with aggressive disease characteristics in an orthotopic murine model of oral tongue cancer. Clin Cancer Res. 2011;17:6658–6670. doi: 10.1158/1078-0432.CCR-11-0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Agrawal N, Frederick MJ, Pickering CR, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333:1154–1157. doi: 10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stransky N, Egloff AM, Tward AD, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157–1160. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cancer Genome Atlas N: Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517:576–582. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Katsonis P, Lichtarge O. A formal perturbation equation between genotype and phenotype determines the Evolutionary Action of protein-coding variations on fitness. Genome Res. 2014;24:2050–2058. doi: 10.1101/gr.176214.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Neskey DM, Osman AA, Ow TJ, et al. Evolutionary action score of TP53 identifies high-risk mutations associated with decreased survival and increased distant metastases in head and neck cancer. Cancer Res. 2015;75:1527–1536. doi: 10.1158/0008-5472.CAN-14-2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Osman AA, Neskey DM, Katsonis P, et al. Evolutionary action score of TP53 coding variants is predictive of platinum response in head and neck cancer patients. Cancer Res. 2015;75:1205–1215. doi: 10.1158/0008-5472.CAN-14-2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Masica DL, Li S, Douville C, et al. Predicting survival in head and neck squamous cell carcinoma from TP53 mutation. Hum Genet. 2015;134:497–507. doi: 10.1007/s00439-014-1470-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Temam S, Flahault A, Perie S, et al. p53 gene status as a predictor of tumor response to induction chemotherapy of patients with locoregionally advanced squamous cell carcinomas of the head and neck. J Clin Oncol. 2000;18:385–394. doi: 10.1200/JCO.2000.18.2.385. [DOI] [PubMed] [Google Scholar]
- 20.Perrone F, Bossi P, Cortelazzi B, et al. TP53 mutations and pathologic complete response to neoadjuvant cisplatin and fluorouracil chemotherapy in resected oral cavity squamous cell carcinoma. J Clin Oncol. 2010;28:761–766. doi: 10.1200/JCO.2009.22.4170. [DOI] [PubMed] [Google Scholar]
- 21.Furuse S, Adachi M, Ijichi K, et al. Pre-radiation enhances the cytotoxicity of docetaxel in head and neck squamous cell carcinoma cells. Oncol Rep. 2010;23:1339–1343. doi: 10.3892/or_00000769. [DOI] [PubMed] [Google Scholar]
- 22.Tanaka N, Osman AA, Takahashi Y, et al. Head and neck cancer organoids established by modification of the CTOS method can be used to predict in vivo drug sensitivity. Oral Oncol. 2018;87:49–57. doi: 10.1016/j.oraloncology.2018.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ang KK, Zhang Q, Rosenthal DI, et al. Randomized phase III trial of concurrent accelerated radiation plus cisplatin with or without cetuximab for stage III to IV head and neck carcinoma: RTOG 0522. J Clin Oncol. 2014;32:2940–2950. doi: 10.1200/JCO.2013.53.5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.ECOG-ACRIN, head and neck cancer EA3132. Available at: https://ecog-acrin.org/wp-content/uploads/2021/02/EA3132-pocket-reference-card.pdf. Accessed February 22, 2022.
- 25.ClinicalTrials.gov. Available at: https://clinicaltrials.gov/ct2/show/NCT02734537. Accessed February 22, 2022.
- 26.Sandulache VC, Michikawa C, Kataria P, et al. High-Risk TP53 Mutations Are Associated with Extranodal Extension in Oral Cavity Squamous Cell Carcinoma. Clin Cancer Res. 2018;24:1727–1733. doi: 10.1158/1078-0432.CCR-17-0721. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.