Abstract
Altered miRNA expressions are assigned pathogenic properties in several cancers including mycosis fungoides and could play a role in the early onset of the disease. The aim of this study was to examine disease-specific miRNA expression in early-stage mycosis fungoides patch and plaque lesions. A quantitative real-time PCR platform of 384 human miRNAs was used to study miRNA expression in 154 diagnostic mycosis fungoides biopsies. A total of 110 miRNAs were significantly differentially expressed (>2-fold, p < 0.05) between plaque lesions and healthy controls, and 90 miRNAs (>2-fold, p < 0.05) differed between patch lesions and healthy controls. Moreover, 13 miRNAs differed in expression between patch and plaque lesions. Early-stage mycosis fungoides exhibited miRNA features that overlapped with those of psoriasis. However, 39 miRNAs, including miR-142-3p, miR-150 and miR-146b, were specific to mycosis fungoides. In conclusion, early-stage mycosis fungoides expresses a distinct miRNA profile, indicating that miRNAs could play a role in the early development of mycosis fungoides.
Key words: mycosis fungoides, cutaneous T-cell lymphoma, microRNA, cancer
Cutaneous T-cell lymphomas (CTCLs) are a group of extranodal, non-Hodgkin’s lymphomas, defined by the presence of skin-homing malignant T-cells in a chronic inflammatory site. Mycosis fungoides (MF) is the most common subtype of CTCL, accounting for approximately 72% of cases of CTCL, with an incidence of 4.1/1,000,000 person-years (1).
Mildly erythematous patches are the earliest clinical presentation of MF, and present as flat, slightly scaling lesions, often localized in non-sun exposed skin (2). Patches can evolve into infiltrated plaques, that are also erythematous, and palpable with a well-defined border. Both patches and plaques are considered early-stage MF (stage IA–IIA) (3). Establishment of a diagnosis of early-stage MF is often challenging, since both the clinical presentation and histological pattern resemble benign inflammatory skin disorders, such as psoriasis or eczema (4). The prognosis of MF is largely determined by the clinical disease stage (5). Most cases of early-stage MF have an indolent disease course. Progression to the advanced disease stages is thus reported in one-third of patients, where the skin lesions evolve into tumours or erythroderma with a risk of nodal involvement (5). Mortality is significantly increased in these patients (5).
SIGNIFICANCE
miRNAs are small sequences of RNA that are involved in several central biological processes. However, altered expression of miRNA is also associated with malignant development. It is unknown whether this applies to early-stage mycosis fungoides. This study showed that early-stage mycosis fungoides express a disease-specific miRNA profile. These findings may improve our understanding of the development of mycosis fungoides.
Despite several investigations, the aetiology of MF remains largely unknown. Infections were previously hypothesized to initiate MF (6, 7) and, in particular, Staphylococcus aureus infections may be a key driver of disease progression (8–12). Moreover, evidence of heredity in CTCL was not identified in a recent cohort study of twins (13) and, although genetic alterations have been identified in CTCL, single aetiological or predisposing genetic factors have not yet been discovered (14, 15). Epigenetic alterations have also been investigated in CTCL and, in particular, deregulation of miRNAs may be involved in the pathogenesis of CTCL (16–21).
miRNAs are an abundant group of small, non-coding, single-stranded RNAs, involved in post-transcriptional gene regulation (22). Their ability to base-pair with target regions on messenger-RNA allows them to inhibit translation. They are involved in many crucial biological processes, such as cell differentiation, proliferation and apoptosis (22). However, recently it has become clear that deregulation of miRNAs can have an oncogenic nature (23). Thus miRNAs are of great interest in cancer research (23). miRNAs have also been shown to have prognostic value in MF, where a three miRNA-classifier makes it possible to predict the risk of disease progression and survival at the time of diagnosis (18). In addition, miRNAs may also serve as a diagnostic tool in MF (24). However, little is known about how miRNAs are involved in the early development of MF.
The aim of the current study was to investigate the miRNA profile of early MF skin lesions in order to identify miRNAs that may play a role in MF disease initiation. Identification of disease-initiating miRNAs may lead to development of new targeted therapies for patients with MF. The current analysis was based on samples from 154 Danish patients with MF. The samples were obtained from early-stage patch and plaque lesions that were initially used for the histological diagnosis of MF.
MATERIALS AND METHODS
Patient characteristics
The first formalin-fixed paraffin-embedded (FFPE) skin biopsy that established the early-stage MF diagnosis in a previously described cohort of 154 patients with MF was identified and obtained from the Departments of Pathology in Denmark for the period 1981–2013 (18). Thus, the biopsy used for analysis was the first biopsy to establish a diagnosis of early-stage MF in each patient. Consequently, only 1 biopsy was included per patient. For patients who had both patch and plaque lesions, the most infiltrated plaque lesion was chosen as the biopsy site and thus used for analysis in the current study. The diagnosis of MF was typically established based on the presence of characteristic histopathological and immunopathological findings combined with a clinical presentation consistent with early-stage MF. The patients were staged according to the proposal by the International Society for Cutaneous Lymphomas/European Organisation of Research and Treatment of Cancer (25), and additional patient characteristics were collected from the patient’s medical records (Table I). As reference, the study aimed to include patients with a clear diagnosis and disease burden, who were also untreated, in order to minimize possible biases. Therefore, the study included FFPE skin biopsies from 16 patients with untreated psoriasis with Psoriasis Area Severity Index (PASI) >10. In addition, biopsies from 20 age- and sex-matched healthy controls were included as reference samples. For patients with MF the miRNA signature was assessed according to the type of skin lesion (patch or plaque) compared with the expression in psoriasis and healthy controls.
Table I.
Patient characteristics
| Patients n = 154 | |
|---|---|
| Sex, n (%) | |
| Male | 92 (60) |
| Female | 62 (40) |
| Age, n (%) | |
| < 60 years | 56 (36) |
| ≥ 60 years | 98 (64) |
| Skin lesion, n (%) | |
| Patch | 33 (21) |
| Plaque | 121 (79) |
| Clinical stage, n (%) | |
| IA | 71 (46) |
| IB | 81 (53) |
| IIA | 2 (1) |
| T-stage, n (%) | |
| T1 | 71 (46) |
| T1a | 20 |
| T1b | 51 |
| T2 | 83 (54) |
| T2a | 5 |
| T2b | 78 |
| Treatment, n (%) | |
| No treatment | 116 (75) |
| Topical treatment | 38 (25) |
RNA extraction
RNA was extracted from 10-mm sections of the FFPE skin biopsies using the RecoverAll Total Nucleic Acid Isolation Kit (ThermoFisher Scientific/Applied Biosystems, USA) in accordance with the manufacturer’s instructions. RNA yield and quality were measured on a NanoDrop-1000 spectrophotometer (ThermoFisher Scientific).
qRT-PCR miRNA profiling
Common methods to investigate miRNA expression include quantitative real-time PCR (qRT-PCR) and microarray. Microarray is a popular tool in miRNA research, due to its ability to examine the expression of thousands of genes simultaneously. However, differences in platforms and procedures reduce reproducibility. Furthermore, reliable normalization and detection of low abundance genes, such as several miRNAs, is difficult to achieve. qRT-PCR is a robust, reproducible method for examining miRNA expression in skin biopsies. Due to its high sensitivity and specificity qRT-PCR is considered the gold standard for miRNA quantification (26).
Fifty nanograms of the extracted RNA was used for qRT-PCR-based miRNA profiling of 384 human miRNAs. The Universal cDNA synthesis kit (Exiqon A/S,Vedbaek, Denmark) was used for reverse transcription of RNA from each patient into complementary DNA (cDNA). Next, the cDNA was diluted ×100. Using a pipetting robot, ExiLENT SYBR Green master mix (Qiagen, Germany) was transferred to quantitative PCT panels preloaded with primers, and a LightCycler 480 Real-Time PCR System (Roche, Switzerland) was used for amplification. Assessment of raw crossing point (Cp) values and melting points were performed using the Roche LC software (Roche). Reactions with melting points that deviated from assay specifications, with several melting points, and with an amplification efficacy below 1.6 were excluded from analysis. Reactions with Cp values within 5 Cp values of the negative control reaction or Cp values >37 were also excluded. The data were normalized to the mean of assays detected in all samples (global mean), which was identified as the best normalizer using NormFinder. After removal of miRNA signals with Cp > 37 in most samples, 281 miRNAs were left for further analysis.
Statistical analysis
Fold-change was calculated as the difference between mean normalized Cp values for a given miRNA between 2 subgroups e.g. patch compared with plaque. Student’s t-test was used to assess the significance of differences in miRNA expression levels. p-values ≤ 0.05 were considered significant. Multiple testing adjustment was performed by estimation of the false discovery rate (q-value). The threshold for miRNAs considered top regulated was selected depending on the type of samples compared.
Heatmaps and semi-supervised hierarchical clustering was performed in Qlucore Omics Explorer v.3.4 (Qlucore AB, Lund Sweden).
Target prediction and downstream pathway analysis were performed using DIANA-miRPath v. 3. DIANA-miRPath provides statistics on predicted miRNA targets and downstream gene regulation.
RESULTS
Initial diagnostic mycosis fungoides plaque lesions display a distinct miRNA profile compared with healthy control skin
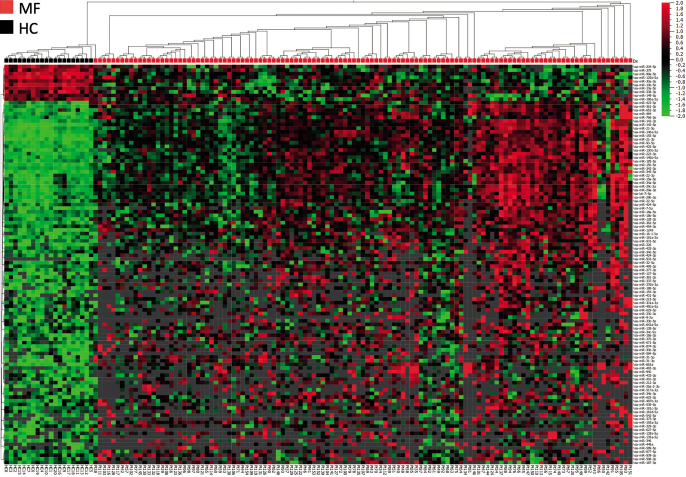
In order to assess which miRNAs were differentially expressed in plaque lesions from MF patients at the time of diagnosis, the miRNA expression profiles in MF plaque lesions compared with 20 healthy controls were assessed. A total of 110 miRNAs were found that significantly differed between patients and controls with at least a 2-fold-change. Out of these, 100 miRNAs were up-regulated in the MF plaque lesions and 10 were down-regulated. Unsupervised hierarchical clustering completely separated the patients with MF from controls (Fig. 1). Among the up-regulated miRNAs a subset of previously discovered miRNAs associated with MF were identified. These included miR-142-5p, miR-21-3p and miR-155-5p.
Fig. 1.
Heatmap and 2-way semi-supervised hierarchical clustering based on 110 differentially expressed miRNAs (fold-change > 2 and p < 0.05) between initial diagnostic mycosis fungoides (MF) plaque lesions from 121 patients (red) and 20 healthy controls (black). Expression levels are z-scaled and range from high (red) to low (green). Grey spots indicate missing (below lower level of detection) values.
Initial diagnostic mycosis fungoides patch lesions differ in their miRNA signature compared with healthy control skin
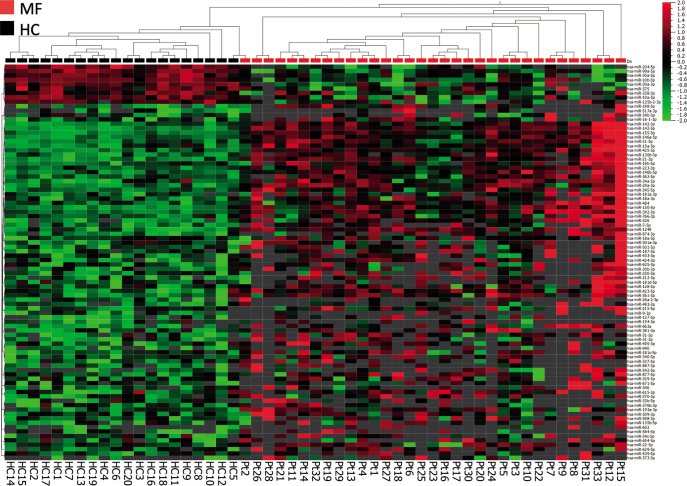
Next, the study investigated whether initial diagnostic MF patch lesions from 33 patients with MF also displayed a distinct miRNA expression pattern compared with the 20 healthy controls. A total of 90 miRNAs were significantly (p < 0.05, fold-change > 2) differentially expressed in patch lesions compared with healthy controls. A total of 81 miRNAs were up-regulated and 9 miRNAs were down-regulated. Hierarchical clustering separated the MF patients from the healthy controls (Fig. 2), except for 1 MF-patch sample that clustered with the healthy controls. Interestingly, miRNA-142-5p, miR-21-3p and miR-155-5p were up-regulated in the early MF patch lesions, as also observed for early plaque lesions.
Fig. 2.
Heatmap and 2-way semi-supervised hierarchical clustering based on 90 differentially expressed miRNAs (fold-change >2 and p < 0.05) between initial diagnostic mycosis fungoides (MF) patch lesions from 33 patients (red) and 20 healthy controls (black). Expression levels are z-scaled and range from high (red) to low (green). Grey spots indicate missing (below lower level of detection) values.
Different miRNA expression in initial diagnostic mycosis fungoides patch vs plaque lesions
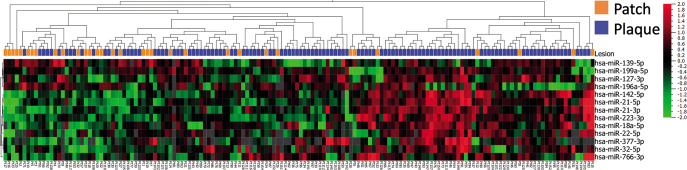
To identify miRNAs with possible involvement in early development of MF, the miRNA expression in initial diagnostic MF patch vs plaque lesions was assessed. A total of 13 miRNAs were identified that significantly differed in expression between early patch and plaque lesions, with a fold-change >1.4. Seven of these miRNAs (miR-18a-5p, miR-223-3p, miR-142-5p, miR-21-3p, miR-21-5p, miR-766-3p, miR-127-5p) were differentially expressed in both patch and plaque lesions compared with healthy controls. Eleven miRNAs, including miR-142, miR-21 and miR-22, were higher expressed in infiltrated plaques than in patch lesions, whereas the expression of miR-155 and miR-146 were similar in patch and plaque lesions. As expected, hierarchical clustering did not completely separate early patch from plaque lesions (Fig. 3).
Fig. 3.
Heatmap and 2-way semi-supervised hierarchical clustering based on 13 differentially expressed miRNAs (fold-change >1.4 and p < 0.05, q=0.29) between 33 patients with mycosis fungoides (MF) with initial diagnostic patch (orange) lesions and 121 MF patients with initial diagnostic plaque (blue) lesions. Expression levels are z-scaled and range from high (red) to low (green). Grey spots indicate missing (below lower level of detection) values. Note, incomplete separation of patch from plaque lesions.
Predicted function of the differently expressed miRNAs between patch and plaque skin lesions
To explore predicted associated differences in downstream pathway activation between initial MF patch vs plaque lesions, a DIANA-miRPath analysis based on the identified 13 differentially expressed miRNAs was performed. Pathways in cancer, transcriptional dysregulation in cancer and specific pathways associated with CTCL pathogenesis, such as the PI3K-AKT, mTOR, FoxO and TGF-beta signalling pathways, were identified (Table II).
Table II.
Selected down-stream activated Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways associated with the 13 most differentially expressed miRNAs between initial diagnostic MF patch and plaque skin lesions
| KEGG pathway | p-value | Regulated genes n | miRNAs involved n |
|---|---|---|---|
| TGF-beta signalling pathway | 0.0002 | 29 | 10 |
| FoxO signalling pathway | 0.0003 | 53 | 12 |
| mTOR signalling pathway | 0.006 | 27 | 10 |
| Pathways in cancer | 0,029 | 122 | 12 |
| PI3K-Akt signalling pathway | 0.043 | 97 | 12 |
| Transcriptional misregulation in cancer | 0.044 | 55 | 11 |
KEGG: Kyoto Encyclopedia of Genes and Genomes; TGF: transforming growth factor; FoxO: forkhead box O; mTOR: echanistic target of rapamycin; PI3K-Akt: phosphatidylinositol 3-kinase – serine/threonine kinase.
Differentially expressed miRNAs in initial diagnostic mycosis fungoides patch and plaque skin lesions compared with lesional psoriatic skin
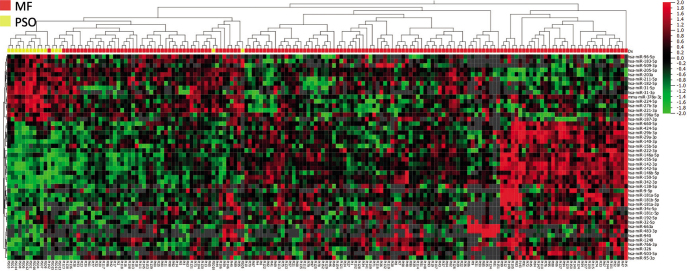
miRNA expression in MF was compared with lesional skin from 16 patients with psoriasis. Forty-six miRNAs differed significantly between patients with MF and psoriasis (p < 0.05 and fold-change > 1.5). Unsupervised hierarchical clustering showed separation between MF and psoriasis samples; however, with some overlap (Fig. 4). We found differential expression of miR-155, miR-142 and miR-146b in patients with MF. miR-21 and miR-22 were highly expressed in both MF and psoriasis skin and therefore not differentially expressed, which confirms previous findings (19). When the miRNA expression was assessed separately for MF patch and plaque lesions vs psoriasis, 35 miRNAs were significantly differentially regulated in MF patches, whereas 51 miRNAs were differentially expressed in MF plaques (Figs S1 and S2).
Fig. 4.
Heatmap and 2-way semi-supervised hierarchical clustering based on 46 differentially expressed miRNAs (fold-change >1.5 and p < 0.05, q=0.15) between initial diagnostic skin biopsies from 154 patients with early-stage mycosis fungoides (MF) (red) and skin biopsies from 16 patients with untreated psoriasis (yellow). Expression levels are z-scaled and range from high (red) to low (green). Grey spots indicate missing (below lower level of detection) values. Note: incomplete separation of MF from psoriasis lesions.
Next, the study investigated whether the distinctive miRNA signature in initial diagnostic MF skin lesions was MF specific or a sign of skin inflammation. Differentially expressed miRNAs specific for MF was identified by comparison of MF vs healthy controls (HC) and psoriasis vs HC (p < 0.05 and fold-change > 2) in a Venn diagram, (Fig. 5 and Table SI). Out of 109 differentially expressed miRNAs between MF and HC, 39 miRNAs were specific for MF, including miR-142-3p, miR-150 and miR-146b, whereas 70 of the differentially expressed miRNAs, including miR-155, miR-142-5p, miR-21 and miR-22, were also differentially expressed in psoriatic skin lesions.
Fig. 5.
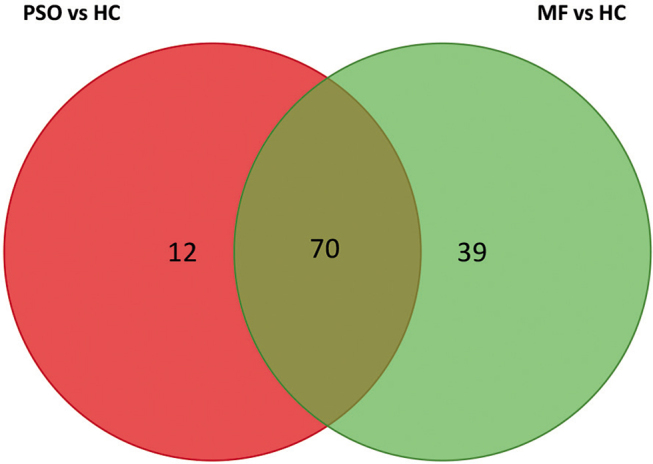
Venn diagram of differentially expressed miRNAs in psoriatic (PSO) skin (n = 82, fold-change >2, p < 0.05) and early-stage mycosis fungoides (MF) lesions (n = 109, fold-change >2, p < 0.05) compared with healthy controls (HC).
DISCUSSION
This study used a unique set of initial diagnostic skin biopsies from 33 patch lesions and 121 plaque lesions, to investigate miRNA expression in early MF. Plaque lesions were found to express a distinctive miRNA profile different from both healthy and psoriasis controls. Notably, the analysis of lesions from MF patches also showed a distinctive miRNA profile, which could indicate that dysregulation of miRNAs is associated with even the early onset of MF. miRNAs might thus play a role in the development and early progression of the disease.
When this study compared initial diagnostic patches with plaque lesions, a higher expression of 11 miRNAs in plaques was found. This indicates that the expression level of these specific miRNAs, including miR-142, miR-21 and miR-22, increases during evolution of MF, suggesting a pivotal role in MF development. In addition, associated downstream Kyoto Encyclopedia of Genes and Genomes (KEGG) activation analyses revealed that differentially expressed miRNAs between patch and plaque lesions regulate cancer- and CTCL-associated pathways, supporting that these miRNAs may be involved in early development of MF.
The results of the current study extend those of earlier research. A subset of previous CTCL-related miRNAs were found among the miRNAs that were significantly differentially expressed in plaque lesions compared with patch lesions. These included miR-142, miR-21 and miR-22. miR-142 is a previously recognized oncomiR and is deregulated in several different cancers (27–29). The miR can be produced by malignant as well as non-malignant cells and transferred to cancer cells through intercellular contacts, such as exosomes (30). An over-expression of miR-142 in cancer cells promotes cell proliferation, migration, and invasion (28). miR-142 has previously been found enhanced in MF lesions (31) and is associated with progressive MF (24). The known oncogenic properties of miR-142 taken together with our findings, suggest that miR-142 is essential in the pathogenesis of MF.
miR-21 is one of the most extensively investigated miRNAs and is found to be up-regulated in several malignancies including CTCL (16), where it exerts anti-apoptotic properties by targeting PTEN in the neoplastic T-cells (32). Furthermore, miR-21 has been linked to the epigenetic switch from inflammation to cancer (33). Recently, the expression of miR-21 was found to be regulated by IL-2 and IL-15 through the STAT5 pathway (16). IL-2 and IL-15 are important growth factors in CTCL (16), and their linkage to miR-21 through STAT5 indicates that miR-21 is associated with disease progression. miR-21 is expressed by malignant CTCL cells and stromal cells in the tumour microenviroment (16). Enhanced miR-21 expression in non-malignant cells, such as fibroblasts, is associated with tumour cell invasion and disease progression (34, 35). Thus, miR-21 might promote MF-progression by altering the function of cancer associated cells to facilitate malignant growth. The up-regulation of miR-21 found in early-stage MF compared with healthy controls in the current study and its linkage to IL-2, implies that miR-21 could play a role in the initiation of the disease.
miR-22 was up-regulated in both patch and plaque lesions compared with healthy controls. The enhanced miR-22 expression in early disease as shown here apparently contrasts with the decreased expression in malignant T cells and cell lines obtained from blood of patients with MF with advanced, systemic disease and patients with Sézary syndrome, a leukaemic and aggressive variant of CTCL (17, 21). Interestingly, miR-22 has been implicated in both oncogenic and tumour suppressive mechanisms as it targets signalling molecules with both pro- and anti-cancer properties (36–38). Accordingly, we hypothesize that the shift in miR-22 expression between early and advanced disease may reflect changes in the molecular setup during cancer progression. For instance, miR-22 targets the tumour-suppressor PTEN, suggesting that miR-22 may play a role repressing PTEN in early lymphomagenesis. In contrast, PTEN repression is mediated by miR-22-independent mechanisms in malignant T cells in advanced disease (17), suggesting that miR-22 is dispensable in relation to PTEN repression in advanced disease (17). Instead, down-regulation of miR-22 may provide an advantage to malignant T cells in advanced disease stages, because it releases the brake mediated by miR-22 on putative oncogenes in MF, such as MAX, MYCBP, HDAC4, HDAC6, CDK6, and NCoA1 (17), supporting the notion that the role of miR-22 in relation to CTCL depends on the cellular context (39).
When comparing MF lesions with psoriasis lesions, differential expression of miR-155, miR-142 and miR-146b was found in patients with MF, indicating, that these miRNAs may serve as active players in the development of MF rather than promoters or markers of inflammation. In addition, the current study confirmed a number of differentially expressed miRNAs, which can separate MF from benign inflammatory skin conditions, including an up-regulation of miR-155 and down-regulation of miR-203 and miR-205, as reported previously (19, 24). Notably, 39 miRNAs were differentially expressed in early diagnostic MF skin lesions, without being regulated in psoriatic skin, indicating that miRNAs may play a pivotal role in development of MF. In addition, the current study identified miRNAs that were both regulated in MF and psoriasis vs healthy controls, indicating that other miRNAs could have a regulatory role in both the inflammatory microenvironment and initiation of tumour cell proliferation in MF.
Differences in methodology, controls and sample quality must be considered when comparing current results with existing knowledge from the literature. Microarray studies might be prone to a higher rate of false-positive and false-negative results than qRT-PCR studies alone. Furthermore, most studies are not externally validated. Comparison of biologically very similar skin lesions provides effect sizes, measured by fold-change, which de novo will be less pronounced than in the comparison with control samples. An overview of the methodology and main results from studies referred to in this paper is shown in Table SII.
The study is strengthened by the large sample size and by the Danish registry system that enabled identification of the first biopsy to establish the diagnosis of early-stage MF for each patient. miRNAs are stable in FFPE-preserved tissue, largely independently of storage duration and time of fixation, so the miRNA quality correlates with miRNAs extracted from fresh-frozen tissue (40). Therefore, this study enabled a unique opportunity to identify miRNAs involved in the early initiation of the disease. Thus, the observed significant differences in miRNA expression within early-stage MF skin lesions (patches vs plaques) provide key biological information regarding the early process in MF.
In conclusion, this study provides evidence of a disease-specific miRNA profile in initial early patch and plaque skin lesions from patients with MF, highlighting miR-142, miR-21 and miR-22 as potential oncogenic players in the early development of MF. The current study has improved the understanding of part of the pathogenesis of MF, which may provide the basis for development of more targeted therapies.
ACKNOWLEDGEMENT
Conflicts of interest: TL is employed both by Copenhagen University and by LEO Pharma A/S. NØ has received advisory consultant honoraria from Micreos human Health B.V. The remaining authors have no conflicts of interest to declare.
REFERENCES
- 1.Bradford PT, Devesa SS, Anderson WF, Toro JR. Cutaneous lymphoma incidence patterns in the United States: a population-based study of 3884 cases. Blood 2009; 113: 5064–5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahn CS, A AL, Sangueza OP. Mycosis fungoides: an updated review of clinicopathologic variants. Am J Dermatopathol 2014; 36: 933–948; quiz 949–951. [DOI] [PubMed] [Google Scholar]
- 3.Willemze R, Jaffe ES, Burg G, Cerroni L, Berti E, Swerdlow SH, et al. WHO-EORTC classification for cutaneous lymphomas. Blood 2005; 105: 3768–3785. [DOI] [PubMed] [Google Scholar]
- 4.Pimpinelli N, Olsen EA, Santucci M, Vonderheid E, Haeffner AC, Stevens S, et al. Defining early mycosis fungoides. J Am Acad Dermatol 2005; 53: 1053–1063. [DOI] [PubMed] [Google Scholar]
- 5.Agar NS, Wedgeworth E, Crichton S, Mitchell TJ, Cox M, Ferreira S, et al. Survival outcomes and prognostic factors in mycosis fungoides/Sezary syndrome: validation of the revised International Society for Cutaneous Lymphomas/European Organisation for Research and Treatment of Cancer staging proposal. J Clin Oncol 2010; 28: 4730–4739. [DOI] [PubMed] [Google Scholar]
- 6.MacKie RM. Initial event in mycosis fungoides of the skin is viral infection of epidermal Langerhans cells. Lancet 1981; 2: 283–285. [DOI] [PubMed] [Google Scholar]
- 7.Willerslev-Olsen A, Krejsgaard T, Lindahl LM, Bonefeld CM, Wasik MA, Koralov SB, et al. Bacterial toxins fuel disease progression in cutaneous T-cell lymphoma. Toxins (Basel) 2013; 5: 1402–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Willerslev-Olsen A, Krejsgaard T, Lindahl LM, Litvinov IV, Fredholm S, Petersen DL, et al. Staphylococcal enterotoxin A (SEA) stimulates STAT3 activation and IL-17 expression in cutaneous T-cell lymphoma. Blood 2016; 127: 1287–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krejsgaard T, Willerslev-Olsen A, Lindahl LM, Bonefeld CM, Koralov SB, Geisler C, et al. Staphylococcal enterotoxins stimulate lymphoma-associated immune dysregulation. Blood 2014; 124: 761–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jackow CM, Cather JC, Hearne V, Asano AT, Musser JM, Duvic M. Association of erythrodermic cutaneous T-cell lymphoma, superantigen-positive Staphylococcus aureus, and oligoclonal T-cell receptor V beta gene expansion. Blood 1997; 89: 32–40. [PubMed] [Google Scholar]
- 11.Talpur R, Bassett R, Duvic M. Prevalence and treatment of Staphylococcus aureus colonization in patients with mycosis fungoides and Sezary syndrome. Br J Dermatol 2008; 159: 105–112. [DOI] [PubMed] [Google Scholar]
- 12.Lindahl LM, Willerslev-Olsen A, Gjerdrum LMR, Nielsen PR, Blumel E, Rittig AH, et al. Antibiotics inhibit tumor and disease activity in cutaneous T-cell lymphoma. Blood 2019; 134: 1072–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Odum N, Lindahl LM, Wod M, Krejsgaard T, Skytthe A, Woetmann A, et al. Investigating heredity in cutaneous T-cell lymphoma in a unique cohort of Danish twins. Blood Cancer J 2017; 7: e517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Litvinov IV, Tetzlaff MT, Thibault P, Gangar P, Moreau L, Watters AK, et al. Gene expression analysis in cutaneous T-cell lymphomas (CTCL) highlights disease heterogeneity and potential diagnostic and prognostic indicators. Oncoimmunology 2017; 6: e1306618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choi J, Goh G, Walradt T, Hong BS, Bunick CG, Chen K, et al. Genomic landscape of cutaneous T cell lymphoma. Nat Genet 2015; 47: 1011–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lindahl LM, Fredholm S, Joseph C, Nielsen BS, Jonson L, Willerslev-Olsen A, et al. STAT5 induces miR-21 expression in cutaneous T cell lymphoma. Oncotarget 2016; 7: 45730–45744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sibbesen NA, Kopp KL, Litvinov IV, Jonson L, Willerslev-Olsen A, Fredholm S, et al. Jak3, STAT3, and STAT5 inhibit expression of miR-22, a novel tumor suppressor microRNA, in cutaneous T-Cell lymphoma. Oncotarget 2015; 6: 20555–20569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lindahl LM, Besenbacher S, Rittig AH, Celis P, Willerslev-Olsen A, Gjerdrum LMR, et al. Prognostic miRNA classifier in early-stage mycosis fungoides: development and validation in a Danish nationwide study. Blood 2018; 131: 759–770. [DOI] [PubMed] [Google Scholar]
- 19.Ralfkiaer U, Hagedorn PH, Bangsgaard N, Lovendorf MB, Ahler CB, Svensson L, et al. Diagnostic microRNA profiling in cutaneous T-cell lymphoma (CTCL). Blood 2011; 118: 5891–5900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Kester MS, Ballabio E, Benner MF, Chen XH, Saunders NJ, van der Fits L, et al. miRNA expression profiling of mycosis fungoides. Mol Oncol 2011; 5: 273–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ballabio E, Mitchell T, van Kester MS, Taylor S, Dunlop HM, Chi J, et al. MicroRNA expression in Sezary syndrome: identification, function, and diagnostic potential. Blood 2010; 116: 1105–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116: 281–297. [DOI] [PubMed] [Google Scholar]
- 23.Esquela-Kerscher A, Slack FJ. Oncomirs – microRNAs with a role in cancer. Nat Rev Cancer 2006; 6: 259–269. [DOI] [PubMed] [Google Scholar]
- 24.Ralfkiaer U, Lindahl LM, Litman T, Gjerdrum LM, Ahler CB, Gniadecki R, et al. MicroRNA expression in early mycosis fungoides is distinctly different from atopic dermatitis and advanced cutaneous T-cell lymphoma. Anticancer Res 2014; 34: 7207–7217. [PubMed] [Google Scholar]
- 25.Olsen E, Vonderheid E, Pimpinelli N, Willemze R, Kim Y, Knobler R, et al. Revisions to the staging and classification of mycosis fungoides and Sezary syndrome: a proposal of the International Society for Cutaneous Lymphomas (ISCL) and the cutaneous lymphoma task force of the European Organization of Research and Treatment of Cancer (EORTC). Blood 2007; 110: 1713–1722. [DOI] [PubMed] [Google Scholar]
- 26.Pritchard CC, Cheng HH, Tewari M. MicroRNA profiling: approaches and considerations. Nat Rev Genet 2012; 13: 358–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Islam F, Gopalan V, Vider J, Lu CT, Lam AK. MiR-142-5p act as an oncogenic microRNA in colorectal cancer: clinicopathological and functional insights. Exp Mol Pathol 2018; 104: 98–107. [DOI] [PubMed] [Google Scholar]
- 28.Xu W, Wang W. MicroRNA1425p modulates breast cancer cell proliferation and apoptosis by targeting phosphatase and tensin homolog. Mol Med Rep 2018; 17: 7529–7536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu L, Liu S, Duan Q, Chen L, Wu T, Qian H, et al. MicroRNA-142-5p promotes cell growth and migration in renal cell carcinoma by targeting BTG3. Am J Transl Res 2017; 9: 2394–2402. [PMC free article] [PubMed] [Google Scholar]
- 30.Aucher A, Rudnicka D, Davis DM. MicroRNAs transfer from human macrophages to hepato-carcinoma cells and inhibit proliferation. J Immunol 2013; 191: 6250–6260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sandoval J, Diaz-Lagares A, Salgado R, Servitje O, Climent F, Ortiz-Romero PL, et al. MicroRNA expression profiling and DNA methylation signature for deregulated microRNA in cutaneous T-cell lymphoma. J Invest Dermatol 2015; 135: 1128–1137. [DOI] [PubMed] [Google Scholar]
- 32.Cristofoletti C, Picchio MC, Lazzeri C, Tocco V, Pagani E, Bresin A, et al. Comprehensive analysis of PTEN status in Sezary syndrome. Blood 2013; 122: 3511–3520. [DOI] [PubMed] [Google Scholar]
- 33.Iliopoulos D, Jaeger SA, Hirsch HA, Bulyk ML, Struhl K. STAT3 activation of miR-21 and miR-181b-1 via PTEN and CYLD are part of the epigenetic switch linking inflammation to cancer. Mol Cell 2010; 39: 493–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kadera BE, Li L, Toste PA, Wu N, Adams C, Dawson DW, et al. MicroRNA-21 in pancreatic ductal adenocarcinoma tumor-associated fibroblasts promotes metastasis. PLoS One 2013; 8: e71978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bullock MD, Pickard KM, Nielsen BS, Sayan AE, Jenei V, Mellone M, et al. Pleiotropic actions of miR-21 highlight the critical role of deregulated stromal microRNAs during colorectal cancer progression. Cell Death Dis 2013; 4: e684–e684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang J, Li Y, Ding M, Zhang H, Xu X, Tang J. Molecular mechanisms and clinical applications of miR-22 in regulating malignant progression in human cancer (Review). Int J Oncol 2017; 50: 345–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Song SJ, Pandolfi PP. MicroRNAs in the pathogenesis of myelodysplastic syndromes and myeloid leukaemia. Curr Opin Hematol 2014; 21: 276–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu H, Liu J, Zhang Y, Li Q, Wang Q, Gu Z. miR-22 suppresses cell viability and EMT of ovarian cancer cells via NLRP3 and inhibits PI3K/AKT signaling pathway. Clin Transl Oncol 2021; 23: 257–264. [DOI] [PubMed] [Google Scholar]
- 39.Gluud M, Willerslev-Olsen A, Gjerdrum LMR, Lindahl LM, Buus TB, Andersen MH, et al. MicroRNAs in the pathogenesis, diagnosis, prognosis and targeted treatment of cutaneous T-cell lymphomas. Cancers (Basel) 2020; 12: 1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Løvendorf MB, Zibert JR, Hagedorn PH, Glue C, Ødum N, Røpke MA, et al. Comparison of microRNA expression using different preservation methods of matched psoriatic skin samples. Exp Dermatol 2012; 21: 299–301. [DOI] [PubMed] [Google Scholar]