Abstract
Background:
In light of advancements in machine learning techniques, many studies have implemented machine learning approaches combined with data measures to predict and classify Alzheimer’s disease. Studies that predicted cognitive status with longitudinal follow-up of amyloid-positive individuals remain scarce, however.
Objective:
We developed models based on voxel-wise functional connectivity (FC) density mapping and the presence of the ApoE4 genotype to predict whether amyloid-positive individuals would experience cognitive decline after 1 year.
Methods:
We divided 122 participants into cognitive decline and stable cognition groups based on the participants’ change rates in Mini-Mental State Examination scores. In addition, we included 68 participants from Alzheimer’s Disease Neuroimaging Initiative (ADNI) database as an external validation data set. Subsequently, we developed two classification models: the first model included 99 voxels, and the second model included 99 voxels and the ApoE4 genotype as features to train the models by Wide Neural Network algorithm with fivefold cross-validation and to predict the classes in the hold-out test and ADNI data sets.
Results:
The results revealed that both models demonstrated high accuracy in classifying the two groups in the hold-out test data set. The model for FC demonstrated good performance, with a mean F1-score of 0.86. The model for FC combined with the ApoE4 genotype achieved superior performance, with a mean F1-score of 0.90. In the ADNI data set, the two models demonstrated stable performances, with mean F1-scores of 0.77 in the first and second models.
Conclusion:
Our findings suggest that the proposed models exhibited promising accuracy for predicting cognitive status after 1 year in amyloid-positive individuals. Notably, the combination of FC and the ApoE4 genotype increased prediction accuracy. These findings can assist clinicians in predicting changes in cognitive status in individuals with a high risk of Alzheimer’s disease and can assist future studies in developing precise treatment and prevention strategies.
Keywords: Alzheimer’s disease, amyloid, ApoE genotype, cognition, functional connectivity, machine learning
Introduction
Alzheimer’s disease (AD) is a neurodegenerative disease that is a worldwide health concern. To date, no effective treatment to prevent or cure AD has been developed, however. Aggregation of extracellular amyloid-beta species and intracellular hyperphosphorylated tau protein neurofibrillary tangles have been observed in the brains of patients with AD.1 In addition, these patients often experience cerebral atrophy, deterioration of cognitive function, and mental and behavioral changes. These conditions ultimately make a patient dependent on others for accomplishing the activities of daily living, thus imposing a major burden on patients, caregivers, and clinical practitioners.1 Patients with AD typically experience memory impairment – the major cognitive deficit of the disease – at the early stage of the disease.2,3 Deterioration of cognitive function may be caused by the accumulation of amyloid-beta species and hyperphosphorylated tau protein, which can lead to neuronal loss and cerebral atrophy.4,5 These pathologies of AD can also be found in individuals with mild cognitive impairment (MCI) or even individuals with normal cognitive function (i.e. the preclinical states of AD).2,6,7 Therefore, it is important to investigate and predict the process of transition from normal cognitive function to cognitive decline in individuals with associated pathologies.
Previous functional magnetic resonance imaging (fMRI) studies have demonstrated functional connectivity (FC) impairment in several brain regions in patients with AD at the early or preclinical stage.8–10 In addition, the apolipoprotein E4 (ApoE4) allele is the genetic risk factor most associated with the development of AD.11 Previous studies have indicated that, before abnormalities in the brain structure, in cognitive function, or neurobiology develop (i.e. at the asymptomatic or preclinical stages), an ApoE4 carrier may already have abnormal FC and amyloid-beta deposition.12–15 These findings indicate that, in the early or preclinical stage of AD, individuals with AD or who carry the ApoE4 allele might experience FC impairment, which is related to cognitive deterioration. Therefore, FC and the ApoE4 genotype might play key roles in the progression of AD, especially at the preclinical phase.
In light of advancements in machine learning techniques, many studies have implemented machine learning approaches combined with data measures to predict and classify AD; machine learning has been paired with, for example, neuroimaging, clinical examination, and evaluation of demographic characteristics.13,16–21 Few studies have combined FC and the ApoE4 genotype to predict cognitive decline or classify the prognosis of patients with AD. Only one magnetoencephalography study demonstrated that the combination of resting-state oscillatory connectivity and the ApoE4 genotype enabled researchers to distinguish patients with AD from older control patients and ApoE4 carriers from ApoE4 noncarriers with moderate success.13 Most studies have focused on distinguishing between AD, MCI, and normal cognitive function,13,16–21 but others have focused on distinguishing between AD and vascular dementia.22,23 Studies predicting cognitive status with longitudinal follow-up of individuals with AD remain scarce, however. Therefore, we propose a machine learning approach that examines FC and the ApoE4 genotype to predict cognitive status in the longitudinal follow-up of amyloid-positive individuals. This study included amyloid-positive individuals who received [18F]AV-45 positron emission tomography (PET) scans. This approach can assist clinical practitioners in developing a diagnosis, treatment, and prognosis for individuals with abnormal amyloid-beta biomarkers, even those with AD.
In this study, we aimed to (1) develop voxel-wise FC density mapping and identify voxels associated with change rates in Mini-Mental State Examination (MMSE) scores in the training data set to develop the first machine learning model (i.e. a model with FC), (2) combine FC density mapping and the ApoE4 genotype (ApoE4 carriers versus ApoE4 noncarriers) as features to train the second machine learning model (i.e. a model of FC combined with ApoE4 genotype), (3) apply the models to the independent hold-out test data set to predict whether amyloid-positive individuals would experience cognitive decline after 1 year, and (4) increase Alzheimer’s Disease Neuroimaging Initiative (ADNI) data set as an external validation data set to assess our model performances.
Materials and methods
Participants
This study was conducted in accordance with the Declaration of Helsinki and was approved by the Institutional Review Board of Chang Gung Memorial Hospital. The study participants were treated at the Cognition and Aging Center of the Department of General Neurology at Kaohsiung Chang Gung Memorial Hospital. The team of researchers comprised behavioral neurologists, psychiatrists, neuropsychologists, neuroradiologists, and experts in nuclear medicine. All participants received [18F]AV-45 PET scan to identify the amyloid deposition, and the tracer was synthesized at the cyclotron facility of Chang Gung Memorial Hospital according to the previously described method.24 Briefly, we obtained helical computed tomography images for attenuation correction at 40 min. Two 5-min dynamic frames composed each PET acquisition, which was acquired at around 1 h after injection in three-dimensional (3D) mode from the Biograph mCT PET/computed tomography (CT) system (Siemens Medical Solutions, Malvern, PA, USA). The scanning protocol was according to previous studies.25,26 Finally, all enrolled participants who showed amyloid pathology were confirmed by two independent nuclear medicine raters. All participants received acetylcholine esterase inhibitor treatment once they had an MMSE score <27 from time of diagnosis. The exclusion criteria were a past history of clinical stroke or depression, a negative amyloid scan, and a modified Hachinski ischemic score >4. A total of 122 participants were recruited in this study. At baseline, all participants received MRI scans and clinical dementia rating (CDR) and MMSE assessments. All participants underwent an MMSE assessment at least 8 months after the pretest (ranging from 8 to 159 months). Because the interval between pre-test and post-test MMSE were different for each participant, we calculated the annual change rate in MMSE scores for each participant according to the following formula
When the change rate in the MMSE score was negative, the participant was placed in the cognitive decline group (n = 86). By contrast, if the change rate in the MMSE score was greater than or equal to zero, the participant was placed in the stable cognition group (n = 36). A neurologist retrieved the medical information – including diagnoses of hypertension, diabetes mellitus, and hyperlipidemia – of all participants from their medical records. The Institutional Review Committee on Human Research approved this study, and all participants provided written informed consent.
We also included the data used as external validation from the ADNI database (https://adni.loni.usc.edu/), which was launched and led by Principal Investigator Michael W. Weiner, MD, in 2003. The ADNI database was established to test the feasibility of a combination of neuroimaging studies (e.g. MRI and PET), biological markers, and clinical assessment for evaluating the progression of MCI and AD. The inclusion criteria were as follows: (1) participants had to have 3T T1-weighted images and resting-state fMRI with a repetition time of 3000 ms and an echo time of 30 ms from 3T scanners; (2) participants had to have [18F]AV-45 PET scan to identify amyloid pathology; (3) the age of participants ranged from 55 to 95 years; (4) participant had to have at least 8 months follow-up for the MMSE assessment; (5) participants had to have demographic information (e.g. age and sex) and clinical assessments (e.g. MMSE, CDR, ApoE genotype). We finally downloaded the data of 68 participants used as the external validation. We divided the data set into the ADNI-cognitive decline group (n = 32) and ADNI-stable cognition group (n = 36) using the change rate in MMSE score mentioned above.
Genotyping
Single nucleotide polymorphism (SNP) genotyping was performed using the MassARRAY system with iPLEX Gold chemistry (Agena Bioscience, San Diego, CA, USA). The polymerase chain reaction (PCR) primers and single-base extension primers were designed using Assay Design Suite v2.0. The genotyping analysis was performed using an iPLEX Gold Reagent Kit according to the manufacturer’s instructions. For the analysis, 1 µl of DNA sample (10 ng/µl) was subjected to 5 µl of PCR reaction containing 0.2 units of Taq polymerase, 2.5 pmol of multiple PCR primers, and 25 mM deoxynucleoside triphosphate (dNTPs). Thermocycling started at 94°C for 2 min and was followed by 45 cycles of 94°C for 30 s, 56°C for 30 s, and 72°C for 1 min. A final extension was performed at 72°C for 1 min. Unincorporated dNTPs were dephosphorylated using 0.3 U of shrimp alkaline phosphatase. Purified amplicons were then subjected to primer extension using the iPLEX Gold Reagent Kit. Primer extension was performed with a cycling program of 94°C for 30 s, followed by 40 cycles of 94°C for 5 s and five cycles of 52°C for 5 s and of 80°C for 5 s. This was followed by a final extension at 72°C for 3 min. The extended reaction products were purified through cation-exchange resins and then spotted onto a 384-format SpectroCHIP II array (Agena Bioscience, Hamburg, Germany) using a MassARRAY Nanodispenser RS1000 (Agena Bioscience, Hamburg, Germany). Mass determination was done using a MassARRAY Compact Analyzer (Agena Bioscience, Hamburg, Germany). The resulting spectra were processed, and alleles were called using the MassARRAY Typer 4.0 with model-based cluster analysis to analyze the genotypes of the SNPs. The ApoE genotype was determined using rs7412 and rs429358. ApoE4 carriers were defined as participants with one or two E4 alleles.
Image acquisition and preprocessing
All participants underwent an MRI scan (duration: approximately 40 min), which was performed using a GE Signa Excite 3.0T scanner (GE Medical Systems, Milwaukee, WI, USA). The T1-weighted MRI and resting-state fMRI scanning protocols were consistent with those performed in our previous studies.27,28
Raw resting-state fMRI data were analyzed using DPABI toolbox (version 4.2_190919)29 and Statistical Parametric Mapping 12 (https://www.fil.ion.ucl.ac.uk/spm/) run in MATLAB R2021a (MathWorks, Natick, MA, USA). Analysis entailed the following steps: removing the first five data points in any blood-oxygen-level-dependent time series, correcting slice-timing, realigning and then manually reorienting T1-weighted and resting-state fMRI images, coregistering T1-weighted and resting-state fMRI images, normalizing the images into an MNI152 standard space, resampling to a 3-mm cubic voxel, regressing out covariates (i.e. time courses of six head motions, white matter, and cerebrospinal fluid), and performing temporal low-pass filtering (0.01–0.1 Hz).
FC density mapping
We performed Pearson’s correlation to map the intensity of FC density mapping based on participants’ preprocessed resting-state fMRI data. As performed in previous studies,30–33 we calculated the number of global functional connections with the threshold r > 0.6 between voxel xi and all other voxels in the brain (total voxels = 55,749) and set that number as the global FC density for a given voxel. We repeated this calculation for each voxel and created each participant’s FC density mapping.
Feature selection and data set assignment
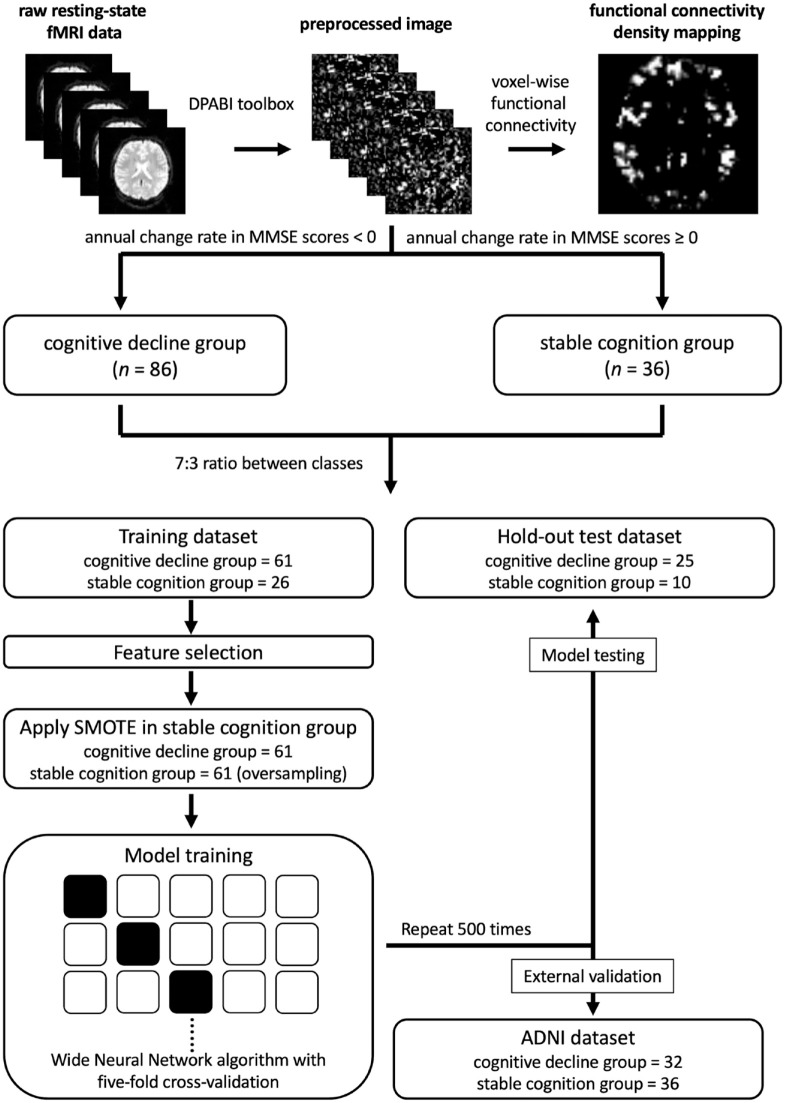
Both groups were randomly divided into a training data set (cognitive decline and stable cognition groups contained data of 61 and 26 participants, respectively) and an independent hold-out test data set (cognitive decline and stable cognition groups contained data of 25 and 10 participants, respectively) at a ratio of 7:3. In addition, we collected 68 participants from the ADNI database used as the external validation, with 32 participants assigned to the ADNI-cognitive decline group and 36 participants assigned to the ADNI-stable cognition group.
To identify a set of key voxels in the training data set, the FC of which correlated with cognitive decline, we implemented random sampling to reduce the number of voxels with spurious correlation, which is common in neuroimaging report.32 First, we randomly selected half of the individuals in the training data set and calculated Pearson’s correlation between the change rate in their MMSE scores and FC density mapping. This step was repeated 50 times for subsequent analysis. Second, in each trial, we ranked the correlation coefficients of the voxels from highest to lowest and selected the 40% of voxels with the highest correlation coefficients as the key voxels. These key voxels were further refined through analysis of the overall intersection of 50 trials. We performed the aforementioned two steps 100 times to derive 100 sets of key voxels. The more frequently a voxel was selected, the more important the voxel was considered to be in relation to cognitive decline. Therefore, the key voxels that were selected more than 10 times were considered the key features for classification in this study. All feature selection steps were performed only for the training data set to prevent information leakage.
We employed the Synthetic Minority Over-sampling Technique (SMOTE)34 to oversample the stable cognition group in the training data set and increase the number in the group from 26 to 61 to reduce classification bias due to class imbalance. Therefore, a total of 157 samples (122 in the training data set and 35 in the hold-out test data set) were included in this study (Figure 1).
Figure 1.
Schematic illustrating construction process of prediction models.
ADNI, Alzheimer’s Disease Neuroimaging Initiative; SMOTE, Synthetic Minority Over-sampling Technique.
Statistical analysis and machine learning analysis
We performed independent t test and Chi-square test to examine the demographic and clinical characteristics of continuous and categorical variables. The significance level was set at 0.05. Statistical analyses were performed using SPSS 25.0 (SPSS Inc., Chicago, IL, USA).
In this study, we developed two classification models. The first model (i.e. the model for FC) was developed using selected key voxels with FC density mapping as features. We also combined FC density mapping and the ApoE4 genotype to serve as features for training the second model (i.e. the model for FC combined with the ApoE4 genotype).
In the machine learning analysis, we performed the Classification Learner Tool in MATLAB R2021a (MathWorks) to train our classification models. First, we performed 24 different algorithms with fivefold cross-validation to train our two models. Subsequently, we chose the algorithm that best fitted our models and repeated the whole cross-validation procedure 500 times in the training, test, and ADNI data sets (Figure 1). Finally, model performance was assessed based on mean accuracy, mean recall, mean precision, and mean F1-score.
Results
Participants’ demographic and clinical characteristics
A total of 122 amyloid-positive participants (group with cognitive decline = 86; stable cognition group = 36) were recruited in this study. The two groups were balanced in terms of sex, education level, CDR at baseline, MMSE score at baseline (cognitive decline group: 24.34 ± 3.38, range: 15–30; stable cognition group: 24.56 ± 2.98, range: 18–29), prevalence of hypertension, prevalence of diabetes mellitus, and prevalence of hyperlipidemia. The cognitive decline group had a higher average age and worse MMSE score at follow-up than the stable cognition group did. Compared with the stable cognition group, the cognitive decline group had a higher proportion of ApoE4 carriers. The ADNI data set showed no significant differences between the two groups regarding age, sex, CDR at baseline, MMSE score at baseline, and proportion of ApoE4 carriers. The results are shown in Table 1.
Table 1.
Demographic and clinical characteristics of the groups.
| Characteristics | Cognitive decline group (n = 86) | Stable cognition group (n = 36) | Statistic (t or Chi-square) | p value | ADNI-cognitive decline group (n = 32) | ADNI-stable cognition group (n = 36) | Statistic (t or Chi-square) | p value |
|---|---|---|---|---|---|---|---|---|
| Age, year | 68.28 ± 7.12 | 65.19 ± 7.88 | 2.11 | 0.04* | 75.59 ± 8.45 | 72.79 ± 6.36 | 1.55 | 0.13* |
| Sex (male/female) | 40/46 | 19/17 | 0.19 | 0.67** | 12/20 | 18/18 | 0.63 | 0.30** |
| Education level, year | 9.26 ± 4.69 | 9.53 ± 3.98 | −0.31 | 0.76* | – | – | – | – |
| CDR score at baseline (n) | ||||||||
| 0 | 8 | 7 | 11 | 14 | ||||
| 0.5 | 74 | 27 | 2.54 | 0.28** | 17 | 21 | 2.35 | 0.31** |
| 1 | 4 | 2 | 4 | 1 | ||||
| MMSE score at baseline | 24.34 ± 3.38 | 24.56 ± 2.98 | −0.34 | 0.74* | 27.59 ± 2.59 | 27.78 ± 2.87 | −0.28 | 0.78* |
| MMSE score at follow-up | 17.52 ± 8.19 | 25.81 ± 2.73 | −8.33 | <0.01* | 25.31 ± 3.47 | 28.83 ± 2.18 | −4.94 | <0.01* |
| Change rate in MMSE scores | −1.62 ± 1.50 | 0.63 ± 0.80 | −8.53 | <0.01* | −1.69 ± 1.23 | 0.77 ± 0.75 | −10.05 | <0.01* |
| Hypertension, n (%) | 37 (43.02%) | 10 (27.78%) | 1.89 | 0.17** | – | – | – | – |
| Diabetes mellitus, n (%) | 17 (19.77%) | 7 (19.44%) | <0.01 | >0.99** | – | – | – | – |
| Hyperlipidemia, n (%) | 25 (29.07%) | 4 (11.11%) | 3.58 | 0.06** | – | – | – | – |
| ApoE4 carrier, n (%) | 30 (34.88%) | 1 (2.78%) | 12.16 | <0.01** | 16 (50%) | 19 (53%) | 0.05 | 0.82** |
ADNI, Alzheimer’s Disease Neuroimaging Initiative; CDR, clinical dementia rating; MMSE, Mini-Mental State Examination; ApoE4, apolipoprotein E4.
Data are mean value ± SD or n (%) unless specified otherwise.
Independent t test, significance level = 0.05.
Chi-square test, significance level = 0.05.
Model performances in the training data set
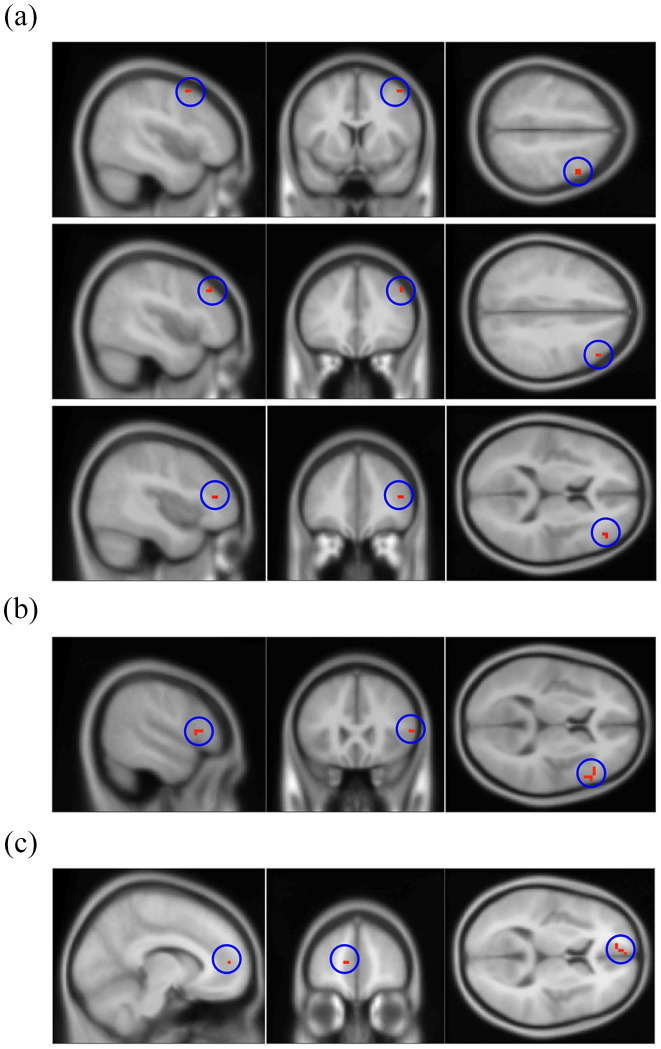
Ninety-nine key voxels were selected as final key features, which were associated with cognitive decline in this study (Figure 2 and Supplementary Table 1). These areas were mainly located in the right middle frontal gyrus (20.20%), the triangular part of the right inferior frontal gyrus (15.15%), and the medial part of the left superior frontal gyrus (9.09%).
Figure 2.
Three key brain regions associated with cognitive deterioration. (a) Right middle frontal gyrus (20 voxels, 20.20%). (b) Right inferior frontal gyrus, triangular part (15 voxels, 15.15%). (c) Left superior frontal gyrus, medial part (9 voxels, 9.09%). Supplementary Table 1 lists the details of the 99 key voxels selected as features.
Among the 24 different algorithms, the Wide Neural Network algorithm with fivefold cross-validation performed the best in the two models (model for FC: accuracy, recall, precision, and F1-score of 0.83, 0.75, 0.88, and 0.81, respectively; model for FC combined with ApoE4 genotype: accuracy, recall, precision, and F1-score of 0.84, 0.74, 0.92, and 0.82, respectively). Results are shown in Tables 2 and 3. Next, we chose the Wide Neural Network algorithm with fivefold cross-validation to train our models and repeat the whole procedure five hundred times in the training, test, and ADNI data sets. The default values were used as model hyperparameters with the following: number of fully connected layers: 1; first layer size: 100; activation: rectified linear unit (ReLU); iteration limit: 1000; regularization strength (Lambda): 0; standardize data: yes.
Table 2.
Model performance of 24 classification algorithms in training data set (model for functional connectivity).
| Algorithm | Accuracy | Recall | Precision | F1-score |
|---|---|---|---|---|
| Fine tree | 0.70 | 0.66 | 0.71 | 0.68 |
| Medium tree | 0.70 | 0.66 | 0.71 | 0.68 |
| Coarse tree | 0.66 | 0.57 | 0.70 | 0.63 |
| Linear discriminant | 0.75 | 0.54 | 0.94 | 0.69 |
| Logistic regression | 0.77 | 0.57 | 0.95 | 0.71 |
| Kernel Naïve Bayes | 0.68 | 0.67 | 0.68 | 0.67 |
| Linear SVM | 0.68 | 0.49 | 0.79 | 0.60 |
| Quadratic SVM | 0.75 | 0.62 | 0.84 | 0.71 |
| Cubic SVM | 0.80 | 0.67 | 0.89 | 0.76 |
| Fine KNN | 0.82 | 0.67 | 0.95 | 0.79 |
| Medium KNN | 0.66 | 0.39 | 0.86 | 0.54 |
| Coarse KNN | 0.49 | 0.79 | 0.49 | 0.60 |
| Cosine KNN | 0.69 | 0.84 | 0.65 | 0.73 |
| Cubic KNN | 0.72 | 0.38 | 0.92 | 0.54 |
| Weighted KNN | 0.69 | 0.38 | 1.00 | 0.55 |
| Boosted trees | 0.49 | 0.79 | 0.49 | 0.60 |
| Bagged trees | 0.76 | 0.74 | 0.78 | 0.76 |
| Subspace discriminant | 0.73 | 0.54 | 0.87 | 0.67 |
| Subspace KNN | 0.71 | 0.48 | 0.88 | 0.62 |
| Narrow neural network | 0.79 | 0.70 | 0.84 | 0.76 |
| Medium neural network | 0.79 | 0.69 | 0.86 | 0.77 |
| Wide neural network | 0.83 | 0.75 | 0.88 | 0.81 |
| Bilayered neural network | 0.82 | 0.75 | 0.87 | 0.81 |
| Trilayered neural network | 0.77 | 0.66 | 0.85 | 0.74 |
The bold values represent the best-performing algorithm among the 24 classification algorithms in the training data set (model for functional connectivity).
KNN, K-nearest neighbor; SVM, support vector machine.
Table 3.
Model performance of 24 classification algorithms in training data set (model for functional connectivity combined with the ApoE4 genotype).
| Algorithm | Accuracy | Recall | Precision | F1-score |
|---|---|---|---|---|
| Fine tree | 0.70 | 0.71 | 0.69 | 0.70 |
| Medium tree | 0.70 | 0.70 | 0.69 | 0.69 |
| Coarse tree | 0.64 | 0.61 | 0.65 | 0.63 |
| Linear discriminant | 0.79 | 0.61 | 0.95 | 0.74 |
| Logistic regression | 0.62 | 0.30 | 0.82 | 0.44 |
| Kernel Naïve Bayes | 0.70 | 0.70 | 0.69 | 0.69 |
| Linear SVM | 0.66 | 0.64 | 0.67 | 0.65 |
| Quadratic SVM | 0.75 | 0.67 | 0.80 | 0.73 |
| Cubic SVM | 0.75 | 0.57 | 0.88 | 0.69 |
| Fine KNN | 0.80 | 0.66 | 0.93 | 0.77 |
| Medium KNN | 0.66 | 0.43 | 0.81 | 0.56 |
| Coarse KNN | 0.49 | 0.79 | 0.49 | 0.60 |
| Cosine KNN | 0.64 | 0.87 | 0.60 | 0.71 |
| Cubic KNN | 0.71 | 0.48 | 0.91 | 0.63 |
| Weighted KNN | 0.71 | 0.41 | 1.00 | 0.58 |
| Boosted trees | 0.49 | 0.79 | 0.49 | 0.60 |
| Bagged trees | 0.76 | 0.72 | 0.79 | 0.75 |
| Subspace discriminant | 0.75 | 0.61 | 0.84 | 0.71 |
| Subspace KNN | 0.78 | 0.62 | 0.90 | 0.73 |
| Narrow neural network | 0.81 | 0.74 | 0.87 | 0.80 |
| Medium neural network | 0.83 | 0.72 | 0.92 | 0.81 |
| Wide neural network | 0.84 | 0.74 | 0.92 | 0.82 |
| Bilayered neural network | 0.75 | 0.69 | 0.79 | 0.74 |
| Trilayered neural network | 0.75 | 0.59 | 0.86 | 0.70 |
The bold values represent the best-performing algorithm among the 24 classification algorithms in the training data set (model for functional connectivity combined with the APOE4 genotype).
ApoE4, apolipoprotein E4; KNN, K-nearest neighbor; SVM, support vector machine.
Both of our models could distinguish between the cognitive decline and the stable cognition groups, achieving high accuracy in the training data set. The model for FC with 99 voxels as features achieved good performance (mean accuracy, mean recall, mean precision, and mean F1-score of 0.80 ± 0.02, 0.70 ± 0.03, 0.88 ± 0.03, and 0.78 ± 0.02, respectively) in the training data set. Moreover, the model for FC combined with ApoE4 genotype had superior performance in classifying the two groups, with a mean accuracy, mean recall, mean precision, and mean F1-score of 0.85 ± 0.01, 0.78 ± 0.03, 0.90 ± 0.02, and 0.83 ± 0.02, respectively, in the training data set (Table 4).
Table 4.
Performance of the models in the training data set.
| Prediction scores | ||
|---|---|---|
| Model with functional connectivity | Model for functional connectivity combined with the ApoE4 genotype | |
| Accuracy | 0.80 ± 0.02 | 0.85 ± 0.01 |
| Recall | 0.70 ± 0.03 | 0.78 ± 0.03 |
| Precision | 0.88 ± 0.03 | 0.90 ± 0.02 |
| F1-score | 0.78 ± 0.02 | 0.83 ± 0.02 |
ApoE4, apolipoprotein E4.
The model for functional connectivity included 99 voxels, and the model for functional connectivity combined with the ApoE4 genotype included 99 voxels and ApoE4 genotypes.
Model performances in the independent hold-out test data set
In the hold-out test data set, both models demonstrated high accuracy in predicting the cognitive decline and stable cognition groups. The model for FC demonstrated good performance in the hold-out test data set (mean accuracy, mean recall, mean precision, and mean F1-score of 0.80 ± 0.03, 0.87 ± 0.04, 0.86 ± 0.02, and 0.86 ± 0.02, respectively). The model with FC combined with the ApoE4 genotype achieved higher performance in distinguishing between the cognitive decline and stable cognition groups, with a mean accuracy, mean recall, mean precision, and mean F1-score of 0.86 ± 0.02, 0.90 ± 0.03, 0.91 ± 0.01, and 0.90 ± 0.02, respectively, in the hold-out test data set (Table 5).
Table 5.
Performance of the models in the hold-out test data set.
| Prediction scores | ||
|---|---|---|
| Model for functional connectivity | Model for functional connectivity combined with the ApoE4 genotype | |
| Accuracy | 0.80 ± 0.03 | 0.86 ± 0.02 |
| Recall | 0.87 ± 0.04 | 0.90 ± 0.03 |
| Precision | 0.86 ± 0.02 | 0.91 ± 0.01 |
| F1-score | 0.86 ± 0.02 | 0.90 ± 0.02 |
ApoE4, apolipoprotein E4.
The model for functional connectivity included 99 voxels, and the model for functional connectivity combined with the ApoE4 genotype included 99 voxels and ApoE4 genotypes.
Model performances in the ADNI data set
Our results showed that the two models demonstrated stable performances in the ADNI data set (model for FC: mean accuracy, mean recall, mean precision, and mean F1-score of 0.77 ± 0.02, 0.79 ± 0.04, 0.74 ± 0.03, and 0.77 ± 0.02, respectively; model for FC combined with ApoE4 genotype: mean accuracy, mean recall, mean precision, and mean F1-score of 0.78 ± 0.02, 0.80 ± 0.03, 0.75 ± 0.03, and 0.77 ± 0.02, respectively). Results are shown in Table 6.
Table 6.
Performance of the models in the ADNI data set.
| Prediction scores | ||
|---|---|---|
| Model for functional connectivity | Model for functional connectivity combined with the ApoE4 genotype | |
| Accuracy | 0.77 ± 0.02 | 0.78 ± 0.02 |
| Recall | 0.79 ± 0.04 | 0.80 ± 0.03 |
| Precision | 0.74 ± 0.03 | 0.75 ± 0.03 |
| F1-score | 0.77 ± 0.02 | 0.77 ± 0.02 |
ADNI, Alzheimer’s Disease Neuroimaging Initiative; ApoE4, apolipoprotein E4.
The model for functional connectivity included 99 voxels, and the model for functional connectivity combined with the ApoE4 genotype included 99 voxels and ApoE4 genotypes.
Discussion
In this study, we developed machine learning models using voxel-wise FC density mapping and the ApoE4 genotype to predict whether amyloid-positive individuals would experience cognitive decline after 1 year. We selected 99 voxels, which were highly correlated with change rates in MMSE scores, to train the model for FC. In addition, we developed a model for FC combined with the ApoE4 genotype comprising 99 voxels and the ApoE4 genotype to improve the accuracy of the model. The model for FC had good performance, with a mean F1-score of 0.86. Furthermore, the model for FC combined with the ApoE4 genotype achieved an excellent mean F1-score of 0.90 for predicting whether the MMSE score would decrease after 1 year for amyloid-positive individuals. Previous studies have mainly focused on differentiating between AD, MCI, and healthy individuals or differentiating between types of dementia.16–18,22,23 Furthermore, most studies have mainly focused on the progression of MCI to AD, and few studies have examined the progression from the preclinical stage to AD.35 More than 30% of cognitively normal older adults have a moderate to high accumulation of amyloid-beta species and are more likely to develop AD.2 Therefore, the strength of this study is that we recruited amyloid-positive individuals at different stages of cognitive decline (MMSE scores ranging from 15 to 30 and CDR scores ranging from 0 to 1). Moreover, we included a longitudinal MMSE assessment, FC density mapping, and ApoE4 genotype data to develop the machine learning models. Both models achieved a mean F1-score of at least 0.86, which suggests our models can predict participants’ cognitive deterioration after 1 year using only the resting-state fMRI scan. This study can provide clinical practitioners with methods and data to enable earlier recognition and prevention of AD in patients at high risk. In the ADNI data set, stable performances were found in the two models with the mean F1-score of 0.77, which were lower than those of the hold-out test data set. We speculated that the possible reason why the ADNI database did not have the similar classification performance is related to ethnic differences. Our database only included Asians, but the ADNI database included different ethnicity. Li and colleagues suggested that the ethnicity of the training data set in machine learning analysis played a crucial role in examining the accuracy of the test data set.36 In addition, a recent fMRI study found that ApoE4 had different effects on FC in different ethnic populations.37 Although the models may have a potential bias when applied to the ADNI data set, the two models we developed still had good classification performance to distinguish between the cognitive decline group and the stable cognition group in the ADNI data set.
Ninety-nine voxels, which were highly associated with change rates in MMSE scores, were selected as key features through FC density mapping. These areas in the brain were primarily located in the right middle frontal gyrus, the triangular part of the right inferior frontal gyrus, and the medial part of the left superior frontal gyrus. A recent study found that increased FC within the middle frontal gyrus was associated with severity of symptoms in patients with AD.38 Cha et al.39 demonstrated that FC in the right middle frontal gyrus was significantly higher in patients with AD than in MCI and healthy groups; they further uncovered a negative correlation between the FC of the right middle frontal gyrus and right inferior frontal gyrus and MMSE scores. One activation likelihood estimation meta-analysis revealed that, compared with healthy controls, patients with MCI had an abnormal alteration of FC in the right precentral, right middle frontal, right inferior frontal, bilateral medial frontal, bilateral superior frontal, and bilateral cingulate gyri.40 These results of previous reports are similar to our own, suggesting that the aforementioned brain regions play a key role in the progression of cognitive decline.
Compared with the first model for FC, the model with FC combined with the ApoE4 genotype achieved superior performance for every predictive score. An ApoE4 carrier might have alterations in FC and excessive deposition of amyloid-beta species at the preclinical stage before abnormalities in brain structure, cognition, or neurobiology develop.12–15 Therefore, a combination of voxel-wise FC density mapping and the ApoE4 genotype can successfully predict cognitive deterioration in amyloid-positive individuals. In addition, of those in the cognitive decline group, 34.88% were ApoE4 carriers, whereas in the stable cognition group, only 2.78% were ApoE4 carriers in this study. Previous studies have also revealed that the ApoE4 genotype increases the risk of progression from MCI to AD.41,42 In the results of the ADNI data set, the mean F1-scores were the same in the two models. These findings might result from a similar proportion of ApoE4 carriers in the ADNI-cognitive decline group (50%) and the ADNI-stable cognition group (53%). We, however, could still observe a tendency of increased accuracy, recall, and precision in the model for FC combined with the ApoE4 genotype compared with the model for FC. Hence, our findings suggest that the ApoE4 genotype is a biomarker for predicting cognitive degeneration in amyloid-positive individuals.
The strength of this study is the recruitment of amyloid-positive individuals at various stages of cognition. All participants received an MMSE assessment at baseline and follow-up. In addition, we developed a voxel-wise FC density mapping model. We selected the voxels in the FC density mapping as features, which were highly correlated with longitudinal MMSE scores. This approach enabled us to develop highly accurate models for predicting the cognitive function in amyloid-positive individuals after 1 year. This study, however, had some limitations. First, the data sets in the study were imbalanced, which may cause classification bias. Although we used SMOTE to increase the data of the stable cognition group, we cannot rule out the possibility of the data imbalance influencing model training. Second, the time between baseline and follow-up MMSE assessment of the participants in this study varied due to clinical or personal reasons. To counter this problem, we calculated a change rate for MMSE scores. In addition, we further reviewed the predicted results of our hold-out test data set with the timing of the follow-up visits less than 36 months (16 participants). Both models demonstrated high accuracy in predicting two groups, with a mean F1-score of 0.81 in the model for FC and 0.88 in the model for FC combined with ApoE4 genotype. In addition, the results of the hold-out test data set with the timing of the follow-up visits more than 36 months (19 participants) also revealed good accuracy in classifying the two groups with a mean F1-score of 0.91 in the first model and 0.91 in the second model. Although the timing of the follow-up visits in this study differed, our models could classify both groups effectively, indicating that the model performance of this study was stable and reliable.
In conclusion, the machine learning models we developed in this study demonstrated promisingly high accuracy in predicting cognitive status after 1 year in an amyloid-positive population. Notably, the combination of resting-state fMRI results and clinical information regarding the ApoE4 genotype could increase the accuracy of predictions. Furthermore, the voxel-wise FC density mapping used in the study may provide precise information regarding the brain areas related to cognitive deterioration. These findings can assist clinicians in predicting changes in cognitive status in individuals with abnormal amyloid-beta biomarkers, even those with AD and can assist future studies in developing precise treatment and prevention strategies.
Supplemental Material
Supplemental material, sj-docx-1-tan-10.1177_17562864221138154 for Functional MRI and ApoE4 genotype for predicting cognitive decline in amyloid-positive individuals by Jun-Ding Zhu, Chi-Wei Huang, Hsin-I Chang, Shih-Jen Tsai, Shu-Hua Huang, Shih-Wei Hsu, Chen-Chang Lee, Hong-Jie Chen, Chiung-Chih Chang and Albert C. Yang in Therapeutic Advances in Neurological Disorders
Acknowledgments
The authors thank the participants and their families for their generosity. The authors thank the Genomics Center for Clinical and Biotechnological Applications of National Core Facility for Biopharmaceuticals, Taiwan (MOST 109-2740-B-010-002) for genotyping.
Footnotes
ORCID iD: Jun-Ding Zhu  https://orcid.org/0000-0003-0718-4087
https://orcid.org/0000-0003-0718-4087
Supplemental material: Supplemental material for this article is available online.
Contributor Information
Jun-Ding Zhu, Institute of Brain Science, National Yang Ming Chiao Tung University, Taipei, Taiwan.
Chi-Wei Huang, Department of Neurology, Cognition and Aging Center, Institute for Translational Research in Biomedicine, Kaohsiung Chang Gung Memorial Hospital, Chang Gung University College of Medicine, Kaohsiung, Taiwan.
Hsin-I Chang, Department of Neurology, Cognition and Aging Center, Institute for Translational Research in Biomedicine, Kaohsiung Chang Gung Memorial Hospital, Chang Gung University College of Medicine, Kaohsiung, Taiwan.
Shih-Jen Tsai, Institute of Brain Science, National Yang Ming Chiao Tung University, Taipei, Taiwan; Department of Psychiatry, Taipei Veterans General Hospital, Taipei, Taiwan; Division of Psychiatry, School of Medicine, National Yang Ming Chiao Tung University, Taipei, Taiwan.
Shu-Hua Huang, Department of Nuclear Medicine, Kaohsiung Chang Gung Memorial Hospital, Chang Gung University College of Medicine, Kaohsiung, Taiwan.
Shih-Wei Hsu, Department of NeuroRadiology, Kaohsiung Chang Gung Memorial Hospital, Chang Gung University College of Medicine, Kaohsiung, Taiwan.
Chen-Chang Lee, Department of NeuroRadiology, Kaohsiung Chang Gung Memorial Hospital, Chang Gung University College of Medicine, Kaohsiung, Taiwan.
Hong-Jie Chen, Department of Nuclear Medicine, Kaohsiung Chang Gung Memorial Hospital, Chang Gung University College of Medicine, Kaohsiung, Taiwan.
Chiung-Chih Chang, Cognition and Aging Center, Institute for Translational Research in Biomedicine, Department of Neurology, Kaohsiung Chang Gung Memorial Hospital, Chang Gung University College of Medicine, No. 123 Ta-Pei Road, Niau-Sung District, Kaohsiung 833, Taiwan.
Albert C. Yang, Institute of Brain Science/Digital Medicine and Smart Healthcare Research Center, National Yang Ming Chiao Tung University, No. 155, Sec. 2, Linong Street, Beitou District, Taipei 112, Taiwan; Department of Medical Research, Taipei Veterans General Hospital, Taipei, Taiwan.
Declarations
Ethics approval and consent to participate: The study received approval from regional ethical committees at the Chang Gung Memorial Hospital and followed the tenets of the Helsinki Declaration (institutional review board (IRB) numbers: for Amyloid PET: 201801833A0C6602 and 201901949A0, for genetic of ApoE4: 201801829A3C601, and for resting-state fMRI and clinical data: 201801827A3C601 and 202001125B0). Written informed consent was obtained from all participants.
Consent for publication: Not applicable.
Author contributions: Jun-Ding Zhu: Formal analysis; Funding acquisition; Methodology; Visualization; Writing – original draft; Writing – review & editing.
Chi-Wei Huang: Data curation; Resources; Writing – review & editing.
Hsin-I Chang: Data curation; Resources; Writing – review & editing.
Shih-Jen Tsai: Funding acquisition; Investigation; Supervision; Writing – review & editing.
Shu-Hua Huang: Data curation; Resources; Writing – review & editing.
Shih-Wei Hsu: Data curation; Resources; Writing – review & editing.
Chen-Chang Lee: Data curation; Resources; Writing – review & editing.
Hong-Jie Chen: Data curation; Resources; Writing – review & editing.
Chiung-Chih Chang: Conceptualization; Funding acquisition; Project administration; Supervision; Writing – original draft; Writing – review & editing.
Albert C. Yang: Conceptualization; Formal analysis; Funding acquisition; Investigation; Methodology; Project administration; Supervision; Writing – review & editing.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by grants from the National Science and Technology Council, Taiwan (grant nos 110-2321-B-A49A-502, 110-2628-B-A49A-509, and 110-2634-F-075-001 to A.C.Y. and S.-J.T.; 110-2314-B-182A-145 to C.-C.C.). Dr A.C.Y. was also supported by the Mt. Jade Young Scholarship Award from the Ministry of Education, Taiwan, as well as Brain Research Center, National Yang Ming Chiao Tung University, and the Ministry of Education (Aim for the Top University Plan), Taipei, Taiwan. This work was also supported by grants CMRPG8J0523, CMRPG8J0842, and CMRPG8K1531 from Chang Gung Memorial Hospital, Taiwan. Mr J.-D.Z. was supported by the scholarship (108-2926-I-010-001-MY4) from the National Science and Technology Council, Taiwan.
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Availability of data and materials: The data presented here are not available due to a confidentiality agreement required by the institutional review board. The MATLAB codes in this study are available from the corresponding author upon request.
References
- 1. Scheltens P, Blennow K, Breteler MM, et al. Alzheimer’s disease. Lancet 2016; 388: 505–517. [DOI] [PubMed] [Google Scholar]
- 2. McKhann GM, Knopman DS, Chertkow H, et al. The diagnosis of dementia due to Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement 2011; 7: 263–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Joe E, Ringman JM. Cognitive symptoms of Alzheimer’s disease: clinical management and prevention. BMJ 2019; 367: l6217. [DOI] [PubMed] [Google Scholar]
- 4. Huber CM, Yee C, May T, et al. Cognitive decline in preclinical Alzheimer’s disease: amyloid-beta versus tauopathy. J Alzheimers Dis 2018; 61: 265–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Spires-Jones TL, Hyman BT. The intersection of amyloid beta and tau at synapses in Alzheimer’s disease. Neuron 2014; 82: 756–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Braak H, Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol 1991; 82: 239–259. [DOI] [PubMed] [Google Scholar]
- 7. Jack CR, Jr, Knopman DS, Jagust WJ, et al. Hypothetical model of dynamic biomarkers of the Alzheimer’s pathological cascade. Lancet Neurol 2010; 9: 119–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Berron D, van Westen D, Ossenkoppele R, et al. Medial temporal lobe connectivity and its associations with cognition in early Alzheimer’s disease. Brain 2020; 143: 1233–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lu J, Testa N, Jordan R, et al. Functional connectivity between the resting-state olfactory network and the hippocampus in Alzheimer’s disease. Brain Sci 2019; 9: 338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhou B, Yao H, Wang P, et al. Aberrant functional connectivity architecture in Alzheimer’s disease and mild cognitive impairment: a whole-brain, data-driven analysis. Biomed Res Int 2015; 2015: 495375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Serrano-Pozo A, Das S, Hyman BT. APOE and Alzheimer’s disease: advances in genetics, pathophysiology, and therapeutic approaches. Lancet Neurol 2021; 20: 68–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nuriel T, Angulo SL, Khan U, et al. Neuronal hyperactivity due to loss of inhibitory tone in APOE4 mice lacking Alzheimer’s disease-like pathology. Nat Commun 2017; 8: 1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Koelewijn L, Lancaster TM, Linden D, et al. Oscillatory hyperactivity and hyperconnectivity in young APOE-ɛ4 carriers and hypoconnectivity in Alzheimer’s disease. Elife 2019; 8: e36011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Stargardt A, Swaab DF, Bossers K. The storm before the quiet: neuronal hyperactivity and Aβ in the presymptomatic stages of Alzheimer’s disease. Neurobiol Aging 2015; 36: 1–11. [DOI] [PubMed] [Google Scholar]
- 15. Hampel H, Wilcock G, Andrieu S, et al. Biomarkers for Alzheimer’s disease therapeutic trials. Prog Neurobiol 2011; 95: 579–593. [DOI] [PubMed] [Google Scholar]
- 16. Fisher CK, Smith AM, Walsh JR. Machine learning for comprehensive forecasting of Alzheimer’s disease progression. Sci Rep 2019; 9: 13622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pellegrini E, Ballerini L, Hernandez MDCV, et al. Machine learning of neuroimaging for assisted diagnosis of cognitive impairment and dementia: a systematic review. Alzheimers Dement 2018; 10: 519–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jo T, Nho K, Saykin AJ. Deep learning in Alzheimer’s disease: diagnostic classification and prognostic prediction using neuroimaging data. Front Aging Neurosci 2019; 11: 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Meszlényi RJ, Buza K, Vidnyánszky Z. Resting state fMRI functional connectivity-based classification using a convolutional neural network architecture. Front Neuroinform 2017; 11: 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Onoda K, Yada N, Ozasa K, et al. Can a resting-state functional connectivity index identify patients with Alzheimer’s disease and mild cognitive impairment across multiple sites? Brain Connect 2017; 7: 391–400. [DOI] [PubMed] [Google Scholar]
- 21. Liu J, Li M, Lan W, et al. Classification of Alzheimer’s disease using whole brain hierarchical network. IEEE/ACM Trans Comput Biol Bioinform 2018; 15: 624–632. [DOI] [PubMed] [Google Scholar]
- 22. Castellazzi G, Cuzzoni MG, Cotta Ramusino M, et al. A machine learning approach for the differential diagnosis of Alzheimer and vascular dementia fed by MRI selected features. Front Neuroinform 2020; 14: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zheng Y, Guo H, Zhang L, et al. Machine learning-based framework for differential diagnosis between vascular dementia and Alzheimer’s disease using structural MRI features. Front Neurol 2019; 10: 1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yao CH, Lin KJ, Weng CC, et al. GMP-compliant automated synthesis of [(18)F]AV-45 (Florbetapir F 18) for imaging beta-amyloid plaques in human brain. Appl Radiat Isot 2010; 68: 2293–2297. [DOI] [PubMed] [Google Scholar]
- 25. Chang YT, Huang CW, Chang WN, et al. Altered functional network affects amyloid and structural covariance in Alzheimer’s disease. Biomed Res Int 2018; 2018: 8565620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Huang KL, Lin KJ, Hsiao IT, et al. Regional amyloid deposition in amnestic mild cognitive impairment and Alzheimer’s disease evaluated by [18F]AV-45 positron emission tomography in Chinese population. PLoS ONE 2013; 8: e58974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chang YT, Mori E, Suzuki M, et al. APOE-MS4A genetic interactions are associated with executive dysfunction and network abnormality in clinically mild Alzheimer’s disease. Neuroimage Clin 2019; 21: 101621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chang YT, Hsu SW, Huang SH, et al. ABCA7 polymorphisms correlate with memory impairment and default mode network in patients with APOEε4-associated Alzheimer’s disease. Alzheimers Res Ther 2019; 11: 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yan CG, Wang XD, Zuo XN, et al. DPABI: data processing & analysis for (resting-state) brain imaging. Neuroinformatics 2016; 14: 339–351. [DOI] [PubMed] [Google Scholar]
- 30. Tomasi D, Volkow ND. Aging and functional brain networks. Mol Psychiatry 2012; 17: 471549–471458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yang AC, Tsai SJ, Liu ME, et al. The association of aging with white matter integrity and functional connectivity hubs. Front Aging Neurosci 2016; 8: 143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tomasi D, Volkow ND. Functional connectivity density mapping. Proc Natl Acad Sci U S A 2010; 107: 9885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tomasi D, Volkow ND. Association between functional connectivity hubs and brain networks. Cereb Cortex 2011; 21: 2003–2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chawla NV, Bowyer KW, Hall LO, et al. SMOTE: synthetic minority over-sampling technique. J Artif Intell Res 2002; 16: 321–357. [Google Scholar]
- 35. Liu X, Chen K, Wu T, et al. Use of multimodality imaging and artificial intelligence for diagnosis and prognosis of early stages of Alzheimer’s disease. Transl Res 2018; 194: 56–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li J, Bzdok D, Chen J, et al. Cross-ethnicity/race generalization failure of behavioral prediction from resting-state functional connectivity. Sci Adv 2022; 8: eabj1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Turney IC, Chesebro AG, Rentería MA, et al. APOE ε4 and resting-state functional connectivity in racially/ethnically diverse older adults. Alzheimers Dement 2020; 12: e12094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sendi MSE, Zendehrouh E, Fu Z, et al. Disrupted dynamic functional network connectivity among cognitive control networks in the progression of Alzheimer’s disease. Brain Connect. Epub ahead of print 7 September 2021. DOI: 10.1089/brain.2020.0847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cha J, Jo HJ, Kim HJ, et al. Functional alteration patterns of default mode networks: comparisons of normal aging, amnestic mild cognitive impairment and Alzheimer’s disease. Eur J Neurosci 2013; 37: 1916–1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Xu W, Chen S, Xue C, et al. Functional MRI-specific alterations in executive control network in mild cognitive impairment: an ALE meta-analysis. Front Aging Neurosci 2020; 12: 578863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shu ZY, Mao DW, Xu YY, et al. Prediction of the progression from mild cognitive impairment to Alzheimer’s disease using a radiomics-integrated model. Ther Adv Neurol Disord 2021; 14: 17562864211029551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Oviedo DC, Lezcano H, Perez AR, et al. Vascular biomarkers and ApoE4 expression in mild cognitive impairment and Alzheimer’s disease. AIMS Neurosci 2018; 5: 148–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-docx-1-tan-10.1177_17562864221138154 for Functional MRI and ApoE4 genotype for predicting cognitive decline in amyloid-positive individuals by Jun-Ding Zhu, Chi-Wei Huang, Hsin-I Chang, Shih-Jen Tsai, Shu-Hua Huang, Shih-Wei Hsu, Chen-Chang Lee, Hong-Jie Chen, Chiung-Chih Chang and Albert C. Yang in Therapeutic Advances in Neurological Disorders