Abstract
Ets factors are members of one of the largest families of evolutionarily conserved transcription factors, regulating critical functions in normal cell homeostasis, which when perturbed contribute to tumor progression. The well-documented alterations in ETS factor expression and function during cancer progression result in pleiotropic effects manifested by the downstream effect on their target genes. Multiple ETS factors bind to the same regulatory sites present on target genes, suggesting redundant or competitive functions. The anti- and prometastatic signatures obtained by examining specific ETS regulatory networks will significantly improve our ability to accurately predict tumor progression and advance our understanding of gene regulation in cancer. Coordination of multiple ETS gene functions also mediates interactions between tumor and stromal cells and thus contributes to the cancer phenotype. As such, these new insights may provide a novel view of the ETS gene family as well as a focal point for studying the complex biological control involved in tumor progression. One of the goals of molecular biology is to elucidate the mechanisms that contribute to the development and progression of cancer. Such an understanding of the molecular basis of cancer will provide new possibilities for: (1) earlier detection, as well as better diagnosis and staging of disease; (2) detection of minimal residual disease recurrences and evaluation of response to therapy; (3) prevention; and (4) novel treatment strategies. Increased understanding of ETS-regulated biological pathways will directly impact these areas.
1. INTRODUCTION
1.1. The ETS gene family
The oncogene v-ets was discovered in 1983 as part of the transforming fusion protein (p135, gag-myb-ets) of E26, a replication-defective avian retrovirus. Both v-ets and v-myb contribute to the transformation of different lineages and cell types. The name ets is derived from E26 transforming sequence or E-twenty-six specific sequence. The v-ets oncogene transforms fibroblasts, myeloblasts, and erythroblasts in vitro and causes mixed erythroid–myeloid and lymphoid leukemia in vivo (reviewed in Blair & Athanasiou, 2000). Molecular comparisons with the predicted chicken c-Ets1 protein demonstrated that the v-ets contained three internal amino acid substitutions and unique carboxy terminal amino acids. This change resulted from the inversion of the 3′ sequences of the chicken gene during retroviral transduction (Lautenberger & Papas, 1993).
All ETS family members are defined by a conserved sequence that encodes the DNA-binding (ETS) domain (Fig. 1.1 and Table 1.1). Identification of v-ets-related genes from metazoan species has established ETS as one of the largest families of transcriptional regulators, consisting of 28 ETS genes in humans, 27 in mice, 11 in sea urchin, 10 in Caenorhabditis elegans, and 9 in Drosophila (for reviews, see Gutierrez-Hartmann, Duval, & Bradford, 2007; Hollenhorst, McIntosh, & Graves, 2011; Hsu, Trojanowska, & Watson, 2004; Seth & Watson, 2005; Turner & Watson, 2008; Watson, Turner, Scheiber, Findlay, & Watson, 2010 and references therein). The human ETS factors are classified into 12 subgroups based upon ETS domain sequence homology: ETS, ERG, PEA3, ETV, TCF, GABP, ELF1, SPI1, TEL, ERF, SPDEF, and ESE (Hollenhorst, McIntosh, et al., 2011; Seth & Watson, 2005; Watson et al., 2010; see Table 1.1 for subgroup members). In addition, a subset of four ETS family genes (ELF3, ELF5, EHF, SPDEF) has been placed in a unique subgroup based upon their restricted expression to tissues with high epithelial cell content (Feldman, Sementchenko, & Watson, 2003). In this review, the Unigene Names will be used (alternative nomenclatures are provided in Table 1.1).
Figure 1.1.
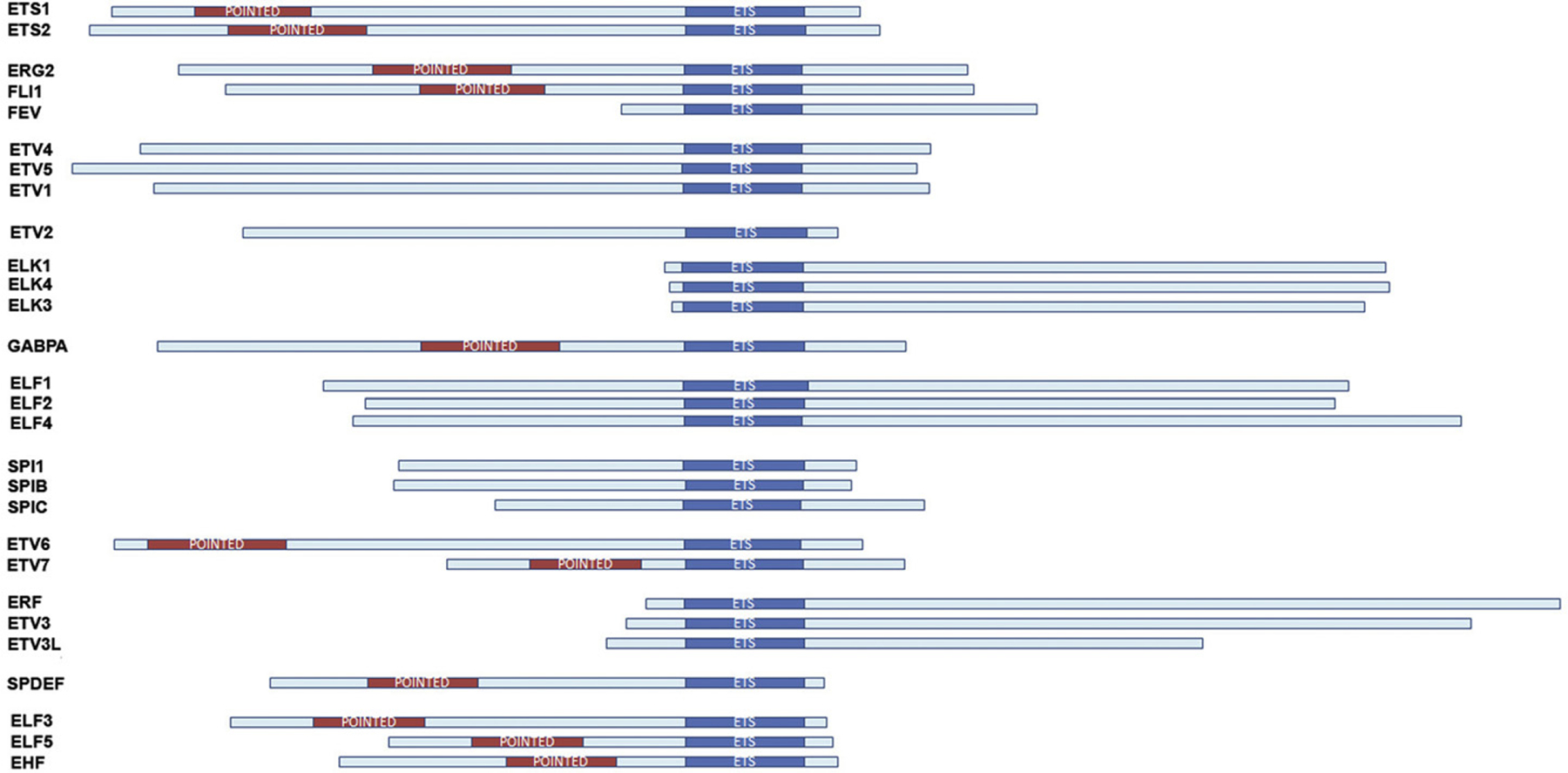
The human ETS family of transcription factors. The main structural organization of each human ETS protein by subfamily (see Table 1.1) is depicted. The ETS and Pointed domains are indicated.
Table 1.1.
The human ETS gene family
| Group | Name | Unigene name | Original name | Alternative names | Locus | Size | ETS domain | Pointed domain | |
|---|---|---|---|---|---|---|---|---|---|
| 1 | ETS | ETS1 | ETS1 | v-ets erythroblastosis virus E26 oncogene homolog 1 | EWSR1 | 11q23.3 | 441 | 331–416 | 54–135 |
| 2 | ETS2 | ETS2 | v-ets erythroblastosis virus E26 oncogene homolog 2 (avian) | 21q22.3 | 469 | 369–443 | 88–168 | ||
| 3 | ERG | ERG2 | ERG | v-ets erythroblastosis virus E26 oncogene-like (avian) | erg-3, p55 | 21q22.3 | 462 | 290–375 | 120–201 |
| 4 | FLI1 | FLI1 | Friend leukemia integration 1 transcription factor | ERGB, EWSR2, SIC-1 | 11q24.1–q24.3 | 452 | 277–361 | 115–196 | |
| 5 | FEV | FEV | Fifth Ewing variant | PET-1, HSRNAFEV | 2q23 | 238 | 43–126 | None | |
| 6 | PEA3 | PEA3 | ETV4 | Polyoma enhancer binding | E1AF, PEAS3 | 17q21 | 462 | 315–399 | None |
| 7 | ERM | ETV5 | Ets-related molecule | 3q28 | 510 | 368–449 | None | ||
| 8 | ER81 | ETV1 | Ets variant gene 1 | 7p21.3 | 458 | 314–397 | None | ||
| 9 | ETV | ER71 | ETV2 | Ets variant gene 2 | ETSRP71 | 19q13.12 | 370 | 265–350 | None |
| 10 | TCF | ELK1 | ELK1 | Xp11.2 | 428 | 7–92 | None | ||
| 11 | SAP1 | ELK4 | SRF accessory protein 1A | 1q32 | 431 | 4–89 | None | ||
| 12 | NET | ELK3 | Net transcription factor | SAP2, ERP | 12q23 | 407 | 5–85 | None | |
| 13 | GABP | GABPα | GABPA | GA binding protein transcription factor A | E4TF1, NFT2, NRF2 | 21q21.3 | 454 | 318–400 | 171–249 |
| 14 | ELF1 | ELF1 | ELF1 | E74-like factor 1 | 13q13 | 619 | 207–289 | None | |
| 15 | NERF | ELF2 | E74-like factor 2 | NERF1, NERF2, EU32 | 4q21 | 581 | 198–277 | None | |
| 16 | MEF | ELF4 | E74-like factor 4 | ELFR | Xq26 | 663 | 204–290 | None | |
| 17 | SPI1 | SPI1 | SPI1 | Spleen focus forming virus (SFFV) proviral integration oncogene spi1 | PU.1, SFPI1, SPIA | 11p11.2 | 264 | 168–240 | None |
| 18 | SPIB | SPIB | SpiB transcription factor | 19q13.3–q13.4 | 262 | 169–251 | None | ||
| 19 | SPIC | SPIC | SpiC transcription factor | 12q23.2 | 248 | 111–193 | None | ||
| 20 | TEL | TEL | ETV6 | Ets variant gene 6 | 12p13 | 452 | 340–419 | 38–119 | |
| 21 | TEL2 | ETV7 | Ets variant gene 7 | TEL-B, TREF | 6p21 | 264 | 149–228 | 49–114 | |
| 22 | ERF | ERF | ERF | Ets2 repressor factor | 19q13 | 548 | 26–106 | None | |
| 23 | ETV3 | ETV3 | Ets variant gene 3 | METS, PE1, bA110J1.4 | 1q21–q23 | 512 | 34–118 | None | |
| 24 | ETV3L | ETV3L | Ets variant gene 3 like | 1q23.1 | 361 | 38–121 | None | ||
| 25 | PDEF | PDEF | SPDEF | SAM pointed domain containing Ets transcription factor | PSE | 6p21.3 | 335 | 248–332 | 138–211 |
| 26 | ESE | ESE1 | ELF3 | Epithelium-specific Ets transcription factor 1 | ESX, JEN, ERT, EPR1 | 1q32.2 | 371 | 275–354 | 47–132 |
| 27 | ESE2 | ELF5 | Epithelium-specific Ets transcription factor 2 | 11p13–p12 | 255 | 165–243 | 46–115 | ||
| 28 | ESE3 | EHF | Epithelium-specific Ets transcription factor 3 | ESEJ | 11p12 | 300 | 209–288 | 42–112 |
List of the known human ETS genes (grouped by subfamily), including gene names and alternative nomenclature, chromosomal location, size of protein (amino acids), approximate boundaries of the Ets domain (~85 amino acids) and approximate boundaries of the Pointed domain (65–80 amino acids, if present).
1.2. ETS protein domains and DNA-binding specificity
The DNA-binding (ETS) and pointed (PNT) domains are the two most common domains present in ETS proteins and will be discussed briefly below. Other domains present in smaller subsets of ETS proteins have been described in previous reviews, and these include the OST GABPA and B-box (TCF subfamily; ELK1, ELK3, ELK4) domains (Hollenhorst, McIntosh, et al., 2011).
The Ets domain is an ~85-amino acid region that forms the winged helix-turn-helix (wHTH) DNA-binding domain composed of three alpha helices and a four-stranded antiparallel beta sheet that recognizes a core GGAA/T sequence (ETS binding site, EBS). The HTH motif is formed by helices H2 and H3. The third alpha helix makes major groove contacts with the DNA (GGAA/T core). Two invariant arginine residues present in helix H3 make contact with the two guanine residues of the EBS. Interestingly, crystallography data indicate that there are no direct contacts outside of the GGAA/T core. The DNA recognition sequence preference for several family members has been determined by in vitro selection of randomized oligonucleotides and indicates that target site recognition is dependent on sequences flanking the core motif, suggesting that DNA conformation may contribute to specificity for the flanking regions. A recent comprehensive genome-wide analysis of ETS factor binding specificities was conducted for 26 mouse and 27 human ETS genes using transcription factor DNA-binding specificity and protein-binding microarrays (Wei et al., 2010). These data support the model that ETS family DNA-binding specificities fall into four distinct classes, and identify key DNA-contact amino acids that contribute to class specificity based upon the published crystal structures for ETS1, GABPA, ELK1, ELF3, SPI1, and SPDEF. Class I contains 12 family members (ETS, ERG, PEA3, TCF, and ERF subfamilies) and is defined by an ACCGGAAGT consensus. Class II is composed of eight members (ELF, TEL, and ESE subfamilies) and the binding consensus differs in the first nucleotide, with a CCCGGAAGT sequence preference. Class III contains the three members of the SPI1 family, which bind to sites with an adenine-rich sequence 5′ to the core. Class IV contains a single family member, SPDEF, which has a GGAT core sequence rather than the GGAA. It is evident that ETS proteins often interact with EBS sequences that do not conform to the consensus binding site defined by in vitro selection experiments. Binding of ETS proteins to such subconsensus sequences is facilitated by the binding of other transacting factors to cis-elements in proximity to the EBS. Indeed, binding is often mediated by synergistic interaction with transcriptional partners on composite DNA elements. The most studied ETS synergistic interactions include those with AP1 (fos/jun), SRF, RUNX (AML), SP1, PAX5, and GATA1 (discussed further below).
DNA binding by multiple ETS factors is inhibited by two regions that flank the DNA-binding domain. Best exemplified by ETS1, this autoinhibition is stabilized by posttranslational modification (serine phosphorylation) on the region encoded by exon VII. Interestingly, exon VII undergoes alternative splicing, resulting in an isoform that binds DNA with higher affinity (Fisher et al., 1994). The alterations at the 3′ end of v-ets are functionally critical to the transforming properties of the virus, since the residues encoded by the 3′ region of c-ets have been shown to be capable of repressing the DNA-binding potential of c-ets; thus, the viral oncoprotein does not undergo autoinhibition and has higher DNA-binding activity. ETS1 autoinhibition is also reduced by interaction with transcriptional cofactors, such as RUNX1 and PAX5.
The second conserved domain found in a subset of ETS genes is the pointed (PNT) domain. This 65–85 amino acid domain belongs to the sterile alpha motif (SAM) family and is found in 11 of 28 human ETS genes and has been shown to function in protein–protein interaction and homo- and heterooligomerization. The PNT domains of several ETS factors also provide the docking site for regulation of extracellular signaling pathways. For example, ERK phosphorylation of ETS1 and ETS2 on threonine phosphor-acceptor sites increases their resultant transcriptional activity through enhanced interaction with the histone acetylase CBP/p300 (Foulds, Nelson, Blaszczak, & Graves, 2004).
In summary, the DNA consensus sequences determined for the different ETS proteins are very similar, and thus specificity is dependent on other factors including interaction with other nuclear factors. Such a dependence of lower affinity ETS binding sequences on coexpression and binding of cofactors would be anticipated to provide greater biological specificity.
2. MODULATION OF ETS FUNCTION
ETS functional activity is modulated at multiple levels. As noted above, ETS factors are dependent on interaction with other factors for precise transcriptional regulation. Indeed, maximal transcriptional activation of multiple target genes is dependent on simultaneous expression of ETS and other transcription factors. Second, specific intracellular signaling pathways and posttranslational modifications directly affect the activity of several ETS proteins by regulating subcellular compartmentalization, DNA-binding activity, and transactivation potential or stability.
2.1. Regulation by protein–protein interactions
Transcriptional regulation is dependent upon the combinatorial interactions between multiple nuclear proteins. ETS proteins form complexes with many transcription factors and such interactions may strengthen the transcriptional activity and/or define target gene specificity. Tissue-specific combination of ETS with other cofactors also provides a mechanism for proper regulation of relevant target genes in a particular cell type. Many transcription factors have their DNA-binding sites adjacent to EBS (for reviews, see Li, Pei, & Watson, 2000; Verger & Duterque-Coquillaud, 2002). As mentioned above, well-studied ETS interactions with transcriptional cofactors include those with AP1 (fos/jun), SRF, RUNX (AML), PAX5, SP1, and GATA1. Depending on the precise sequence context, binding of an ETS protein near other transcription factors results in higher affinity interaction, synergistic activation, and/or repression of specific target genes.
Among the earliest characterized protein–protein interactions was that between ETS factors and the AP1 transcriptional complex. Interaction was shown to result in synergistic transcriptional activation of promoters containing composite AP1-EBS binding sites, including MMP1, uPA, GM-CSF, maspin, and TIMP-1. In contrast, MafB, an AP1-like protein, inhibits ETS1-mediated transactivation of the AP1-EBS sites (Sieweke, Tekotte, Frampton, & Graf, 1996). ETS/AP1-binding sequences are proto-typical RAS-responsive elements and oncogenic ETS factors (ETV1, ETV4, ETV5, and ERG) have been shown to activate a RAS/MAPK transcriptional program in prostate cells in the absence of MAPK activation (Hollenhorst, Ferris, et al., 2011).
Another well-characterized interaction involves SRF and ELK1 (or ELK3, ELK4, FLI1, EWS-FLI1) that together form a ternary complex with the SRE motif present in several genes, including c-fos, Egr-1, pip92 Mcl-1, and SRF(Buchwalter, Gross, & Wasylyk, 2004).
RUNX1 and ETS1 interaction counteracts autoinhibition of DNA-binding activity (Garvie, Pufall, Graves, & Wolberger, 2002) and homotypic ETS1 interaction enhances binding to palindromic EBS (Baillat, Begue, Stehelin, & Aumercier, 2002). Interaction with PAX5 allows ETS1, as well as other family members, to bind to a nonconsensus EBS present in the early B-cell-specific mb-1 promoter (Fitzsimmons, Lutz, Wheat, Chamberlin, & Hagman, 2001).
SPI1 family proteins can function as activators or repressors of transcription and have been shown to interact with ETS factors with cell- and promoter-specific consequences. For example, functional interaction of FLI1 with SP1 or SP3 is essential for the inhibitory function of Fli1 on the collagen A2 promoter (Czuwara-Ladykowska, Shirasaki, Jackers, Watson, & Trojanowska, 2001).
FLI1 and GATA-1 act synergistically to activate gene transcription of multiple megakaryocytic genes, including gpIIb, gpVI, gpIX, gpIb, and c-mpl (reviewed in Szalai, LaRue, & Watson, 2006). We and others have demonstrated that FLI1 and GATA1 co-occupy these promoters in vivo (Jackers, Szalai, Moussa, & Watson, 2004; Moussa et al., 2010; Pang et al., 2006).
Several proteins that modulate ETS function have been identified, including Daxx (EAP1 (ETS1-associated protein 1)), EAPII, and SP100 (Li, Pei, Watson, & Papas, 2000; Pei et al., 2003; Yordy et al., 2004). The notion that loss of corepressor protein expression is relevant to cancer was demonstrated using the NCoR corepressor protein and the coregulators SRC-1 and AIB1, all of which interact with both ETS1 and ETS2 (Myers et al., 2005). The strongest clinical association in breast cancer was for NCoR downregulation in more aggressive hormone-unresponsive tumors (Myers et al., 2005).
2.2. Regulation by posttranslational modification
A common feature of many tumors is deregulation of signal transduction pathways, resulting in constitutive and often ligand-independent activation. As end effectors of these pathways, ETS factor function is significantly altered in cancer. In addition to being downstream of many RTKs (e.g., HER2/neu), ETS factors regulate the expression of multiple receptors, including HER2/neu, M-CSF receptor, MET, c-kit, and VEGF receptor (Sementchenko & Watson, 2000).
ETS factor functions are controlled by phosphorylation, acetylation, sumoylation, ubiquitinylation, and glycosylation (for reviews, see Charlot, Dubois-Pot, Serchov, Tourrette, & Wasylyk, 2010; Tootle & Rebay, 2005; Yordy & Muise-Helmericks, 2000).
One of the best-studied posttranslational modifications is phosphorylation. Phosphorylation of ETS proteins mediates effects on DNA binding, protein–protein interaction, transcriptional activation, and subcellular localization. ERK, JNK, and p38 MAP kinases are downstream components of signaling cascades. ERKs are activated in response to mitogenic signals, while JNKs and p38/SAPKs respond to stress signals. Specific ETS factors, including ETS1, ETS2, ELK1, ELK3, ELK4, GABPA, SPIB, ETV1, ETV4, and ETV5, can be phosphorylated by MAPKs, resulting in increased transcriptional activation (Charlot et al., 2010).
As noted above, phosphorylation of a mitogen-activated protein kinase (ERK) site adjacent to the PNT domain has been shown to positively regulate transcriptional activities of ETS1 and ETS2. Although MAP kinase phosphorylation of ETS1 does not affect DNA binding, calcium-induced phosphorylation of ETS1 occurs at serine residues present adjacent to the DNA-binding domain and inhibits ETS1 DNA-binding activity without affecting nuclear localization. ETS1 and ETS2 activity may also be activated by PKC in invasive breast cancer cells (Lindemann, Braig, Ballschmieter, et al., 2003; Lindemann, Braig, Hauser, Nordheim, & Dittmer, 2003). In contrast, ETV6 activity is negatively regulated by MAPK phosphorylation, which results in its nuclear export and decreased DNA-binding activity. Processes that are reversibly controlled by protein phosphorylation require a balance between protein kinase and protein phosphatase activities. Thus, it is important to assess whether specific protein phosphatases are associated with de-phosphorylation of ETS proteins.
Often associated with phosphorylation, acetylation also regulates ETS gene function. Acetylation of ETV1 enhances its DNA-binding activity and ability to transcriptionally activate target genes (Goel & Janknecht, 2003). In response to TGFβ signaling, ETS1 is acetylated and dissociated from the CBP/p300 complexes (Czuwara-Ladykowska, Sementchenko, Watson, & Trojanowska, 2002). FLI1 activity is repressed through a series of sequential posttranslational modifications (Thr312 phosphorylation and acetylation by p300/CREB binding protein-associated factor), resulting in detachment from target gene (e.g., collagen) promoters in response to TGFβ (Asano et al., 2009; Asano & Trojanowska, 2009).
ETS factors undergo ubiquitination and subsequent proteosomal degradation. ETV1, ETV4, and ETV5 each contain three potential binding motifs for the ubiquitin ligase COP1. ETV1 is degraded after being ubiquitinated by COP1. Data support the notion that COP1 functions as a tumor suppressor mediated by its negative regulation of ETV1, ETV4, and ETV5. Indeed, COP1 deficiency in mouse prostate is correlated with elevated ETV1 and increased cell proliferation, hyperplasia, and early prostate intraepithelial neoplasia (Vitari et al., 2011).
Sumoylation has been shown to affect the stability, activity, and localization of its targets. SUMO modification has been found to alter the function of several transcription factors, including ETS family members. For example, ELK1 is modified by SUMO, and this modification is reversed by ERK–MAP kinase pathway activation. Mechanistically, it has been shown that sumoylation of ELK1 facilitates recruitment of histone deacetylase 2 activity to promoters. This recruitment leads to decreased histone acetylation and altered chromatin structure, resulting in transcriptional repression at ELK1 target genes (Yang & Sharrocks, 2004). In contrast, sumoylation within the pointed domain of ETV6 inhibits ETV6-mediated repression (Chakrabarti & Nucifora, 1999; Chakrabarti et al., 1999), associated with sequestering to subnuclear compartments. Mutation of SUMO acceptor site(s) results in increased transcriptional repression, presumably because of decreased nuclear export (Wood, Irvin, Nucifora, Luce, & Hiebert, 2003). Sumoylation of ETS1, ETV4, and ETV5 leads to reduced transcriptional activity.
Future studies will help elucidate the functional impact of specific post-translational modifications on the activity of ETS transcription factors. As specific antibodies are developed, it will be possible to determine the temporal relationships between specific posttranslational events. Through such analyses, it will also be possible to determine whether specific events work cooperatively or antagonistically.
3. DEFINING AND CHARACTERIZING ETS TARGET GENES
3.1. ETS target genes
The importance of the ETS family of transcription factors in various biological and pathological processes necessitates the identification of downstream cellular target genes of specific ETS proteins. Although some overlap in the biological function of different ETS proteins may exist, the emergence of a family of closely related transcription factors suggests that individual ETS members may have evolved unique roles, manifested through the control of specific target genes. Several key areas are critical for understanding what defines a functionally important ETS target gene: First, the functional importance of the EBS must be demonstrated by mutagenesis. Second, the specific ETS factor or factors responsible for transcriptional control of specific target genes need to be identified. While extensive publications have identified functionally important EBS and thus, ETS target genes (Sementchenko & Watson, 2000), fewer investigations have identified definitive target genes for a specific ETS factor.
ETS factors are known to act as positive or negative regulators of the expression of genes, including those that control response to various signaling cascades, cellular proliferation, differentiation, hematopoiesis, apoptosis, adhesion, migration, invasion and metastasis, tissue remodeling, ECM composition, and angiogenesis (Fig. 1.2). Our earlier literature survey enabled identification of over 200 ETS target genes (Sementchenko & Watson, 2000) and to date, over 700 ETS target genes have been defined, based upon the presence of functional EBS in their regulatory regions (Watson, D.K., unpublished). While most ETS factors were initially characterized as transcriptional activators or repressors, it has become evident that several ETS factors can function as either activators or repressors, depending upon the type of promoter and cellular context.
Figure 1.2.
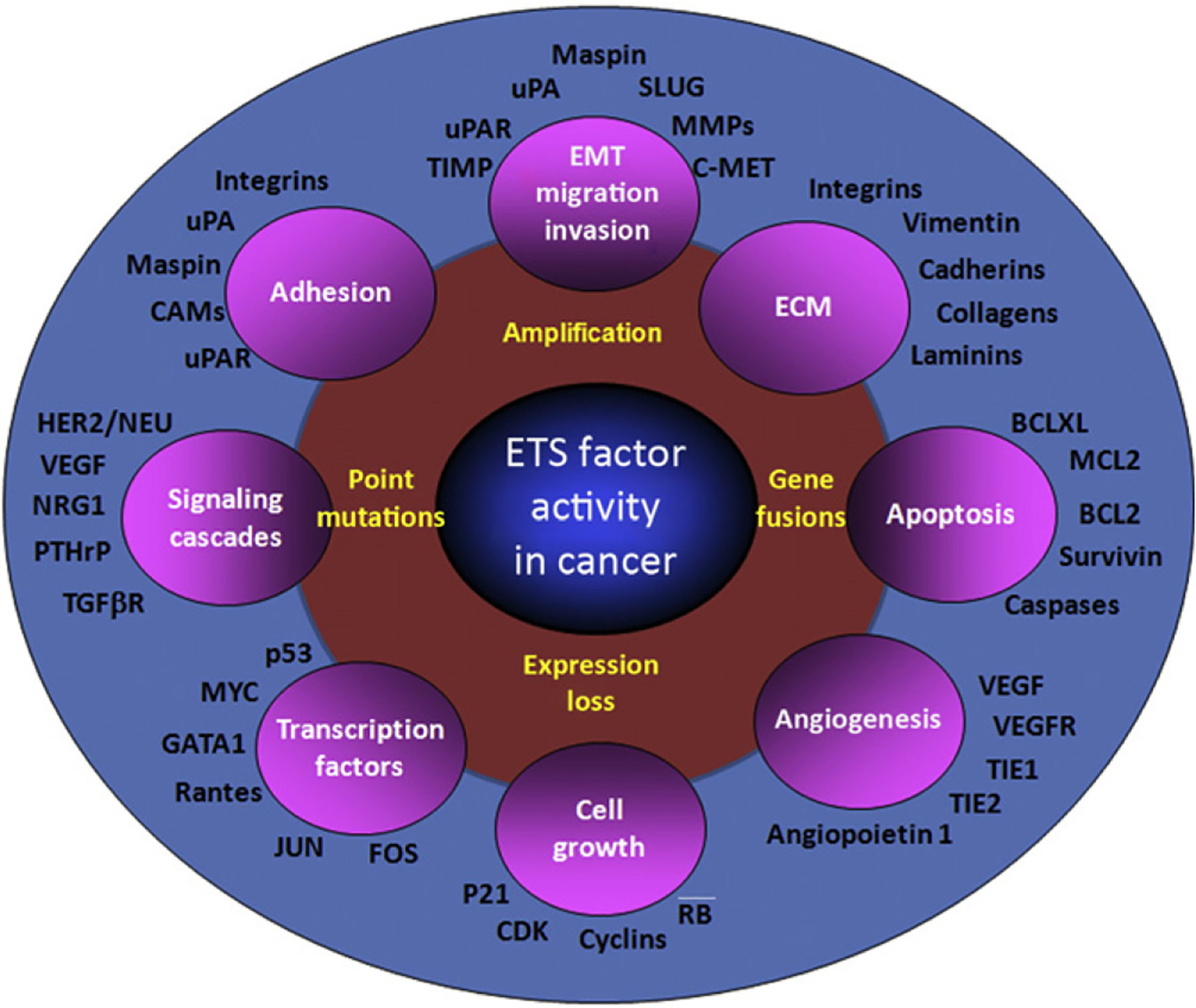
ETS factors regulate the expression of genes associated with cancer progression. Dysregulated ETS factor function leads to the altered expression of multiple target genes that are known to play critical roles in many of the processes required for cancer progression. While each of the target genes highlighted has functional EBS(s) in their regulatory regions, the role and relative affinities of specific ETS factors have only been examined in a limited subset.
During cancer progression, the oncogenic and tumor-suppressor activities of ETS factors are likely coordinated by their target genes. In the past few years, we and others have made significant strides in identifying and validating these target genes. Collectively, ETS genes have been shown to regulate the expression of genes that have important roles in malignant and metastatic processes (Fig. 1.2). Among these are those that function in control of cell proliferation (e.g., cyclins and cdks), motility (hepatocyte growth factor, HGF), invasion (uPA & uPAR, PAI, MMPs; TIMPs), extravasation (MMPs, Integrins), micro-metastasis (Osteopontin; BSP and Osteonectin), and establishment and maintenance of distant site metastasis and angiogenesis (Neovascularization and Neoangiogenesis (integrin β3, VEGF, Flt-1/KDR, Tie2; Sementchenko & Watson, 2000)). Aberrant expression of ETS factors results in the altered regulation of their target response genes. For example, upregulated ETS target genes include extracellular matrix (ECM)-degrading proteins (e.g., MMP1, MMP9, uPA), which are associated with clinical features such as lymph node status and prognosis in prostate cancer. Significantly, altered ETS expression also provides a mechanism for the downregulation of response genes that include uPA and survivin. Recent analysis of gene expression signatures allowed correlation between expression of ETS factors, ETS target genes, and prostate cancer progression (Tomlins et al., 2007).
Several studies have demonstrated that a polymorphism that generates a functional EBS within the MMP1 promoter is a negative prognostic indicator (Benbow, Tower, Wyatt, Buttice, & Brinckerhoff, 2002).
Functional studies have demonstrated that SPDEF is a negative regulator of uPA and SLUG mRNA expression. Chromatin immunoprecipitation (ChIP, discussed further below) allows definition of direct target genes for specific ETS factors. However, ChIP alone does not indicate whether the interaction is functional (e.g., causing transcriptional activation or repression). Biological rescue experiments have been used to demonstrate the importance of specific target genes (e.g., SPDEF target genes, uPA, SLUG; Findlay et al., 2011; Turner, Findlay, Kirven, Moussa, & Watson, 2008). Correlation between ChIP and gene expression can further define functional ETS targets. For example, there is an inverse correlation between SPDEF and uPA in primary colon tumors (Moussa et al., 2009).
3.2. ETS gene coexpression
Initial expression analysis supported the notion that while some ETS factors showed rather ubiquitous expression (e.g., ETS2), others had more restricted expression in specific tissues or cells (e.g., ETS1). Subsequent studies have demonstrated the simultaneous expression of 14–25 ETS mRNAs in many human tissues and cell lines. For example, studies examining ETS factor expression profiles in normal and cancerous breast cells have demonstrated that a combination of up to 25 of 28 ETS family members examined is expressed at any one time in these cells (Galang, Muller, Foos, Oshima, & Hauser, 2004; Hollenhorst, Jones, & Graves, 2004). It should be noted that mRNA expression alone does not adequately define the ETS profile, as factors including, but not limited to, alternative splicing, mRNA translation, protein stability, posttranslational modifications, and protein localization ultimately contribute to define the level of functional proteins in a cell. Complete proteomic studies need to be performed to define the relative prevalence of ETS factors in specific tissues. ETS factor function is also highly dependent upon the presence and level of specific coregulatory proteins.
3.3. Whole genome analysis: Redundant and specific binding
Multiple ETS factors bind to the same regulatory sites present on target genes, suggesting redundant or competitive functions. Furthermore, additional events contribute to, or may be necessary for, target gene regulation. As technologies have advanced, it has become possible to identify the true regulatory targets of transcription factors. ChIP has become an established method for the analysis of protein–DNA (gene regulatory elements) interactions in vivo. Sequential ChIP is an extension of the ChIP protocol, in which the immunoprecipitated chromatin is subjected to sequential immunoprecipitations with antibodies of different specificity. This provides a method of examining co-occupancy of defined promoters by multiple regulatory proteins. Furthermore, sequential ChIP provides an experimental approach to simultaneously evaluate promoter occupancy and transcriptional status (e.g., histone H3 acetylation, phosphorylated RNAPII-CTD; Jackers et al., 2004). However, ChIP and sequential ChIP methods have been restricted to the analysis of small promoter regions, the boundaries defined by the sequences of the primers designed for the PCR amplification step.
To determine the more global location of in vivo promoter binding sites of a specific protein, ChIP protocols have been combined with whole genome analysis methods to produce “ChIP-on-chip” microarrays. ChIP products are amplified and hybridized to arrays consisting of promoter regions, limiting genome coverage. ChIP sequencing (ChIP-seq) is the next generation protocol for defining Protein–DNA transcriptomes (Farnham, 2009; Schmidt et al., 2009; Visel et al., 2009). It combines ChIP with new high throughput sequencing platforms, such as Genome Analyzer (Solexa/Illumina), generating significantly more informative data (Mardis, 2007).
In the context of an ETS transcription network, ChIP-Seq analysis can potentially identify the full transcriptome for each individual ETS family member in any given scenario. Furthermore, by comparing ChIP-Seq data with mRNA expression profiles obtained following modulation of ETS expression, direct and indirect targets for each ETS factor can be ascertained. Recent genome-wide analyses of ETS-factor occupancy have identified genomic regions, both promoters and enhancers, bound by individual ETS proteins in living cells. Nine ETS proteins have been assayed for genome-wide occupancy by either promoter microarrays (ETS1, GABPA, ELF1, ELK1, EWS-FLI1, and SPI1) or high throughput sequencing (GABPA, ETS1, ERG, FLI1, EWS-ERG, EWS-FLI1, SPI1, SPDEF, ETV1, and ELF1; Hollenhorst, McIntosh, et al., 2011).
Genome-wide occupancy data for several ETS proteins have been compared and found to have a high degree of similarity. Genomic targets of ETS transcription factors can be divided into two classes (Hollenhorst et al., 2009; Hollenhorst, McIntosh, et al., 2011; Hollenhorst, Shah, Hopkins, & Graves, 2007). Class 1: redundant binding sites found in the proximal promoters of housekeeping genes. Binding sites in this class are characterized by the consensus ETS sequence (CCGGAAGT) and have the potential to bind any ETS protein with relatively high affinity. DNA regions occupied by multiple ETS proteins are frequently found a short distance (~20–40 bp) upstream of transcription start sites. Class 2: specific binding sites that are found more often in enhancer regions associated with genes that mediate the specific biological functions of an ETS family member. Specific target sites are characterized by a lower-affinity ETS sequence and are sometimes flanked by binding sites for other transcription factors. Many predicted ETS sites are not occupied in vivo and conversely, many actual sites of genomic occupancy are not predicted.
4. ETS AND MicroRNA
MicroRNAs (miRNAs) are both upstream modulators and downstream effectors of ETS transcriptional factors. miRNAs are 19–25-nucleotide RNAs that have emerged as a novel class of small, evolutionarily conserved gene regulatory molecules involved in many critical developmental and cellular functions (Wiemer, 2007). miRNAs base-pair with target mRNA sequences primarily in their 3′ untranslated region. Through specific base pairing, miRNAs induce mRNA degradation, translational repression, or both depending upon the complementarity of the miRNA to its mRNA target. Each miRNA can target numerous mRNAs, often in combination with other miRNAs, therefore controlling complex regulatory networks. It is estimated that there are ~1000 miRNAs in mammalian cells, and that approximately one third of all genes are regulated by miRNAs (Rajewsky, 2006; Shilo, Roy, Khanna, & Sen, 2007). Over 3000 identified mature miRNAs exist in species ranging from plants to humans, suggesting that miRNAs are ancient players in gene regulation (Wang & Li, 2007). Their existence and conservation throughout species support the concept that they perform critical functions in gene regulation (Wang, Stricker, Gou, & Liu, 2007). Indeed, the conserved evolution of both miRNAs and transcription factors highlights their importance in and the complexity of gene regulation (Chen & Rajewsky, 2006). In fact, one of the most widely studied miRNAs is miR-34, which has been shown to be positively and negatively regulated by the transcription factors p53 and myc, respectively (Bui & Mendell, 2010; He, He, Lowe, & Hannon, 2007).
4.1. miRNAs targeting ETS factors
A summary of the published studies that identified miR-ETS interactions is provided in Table 1.2. The majority of the studies examining miRNA-regulated ETS factors are for ETS1. miRNA 125b has been shown to be dysregulated in cancer and can act as either a tumor suppressor or oncogene, depending on cellular context. This is true for many miRNAs and adds to the complexity of targeting miRNAs therapeutically. However, in invasive breast cancer, miR-125b is downregulated and predicts poor patient survival (Zhang, Yan, et al., 2011). miR-125b expression inhibits tumor growth in vivo and has ETS1 as one of its novel direct targets. Although ETS1 protein levels were decreased by miR-125b, ETS1 mRNA levels were unchanged, suggesting a translational repression mechanism of regulation. Like miR-125b, ETS1 overexpression in invasive breast cancer predicts poor patient prognosis. Another study in hepatocellular carcinoma (HCC) identified ETS1 as a direct target of miR-193b (Xu et al., 2010). miR-193b expression inhibits tumor growth in vivo and a negative correlation between miR-193b and ETS1 mRNA levels was defined in HCC tissue samples, suggesting that miR-193b regulation of ETS1 results in mRNA degradation as opposed to translation repression; however, this was not validated in vitro. Other studies that identified ETS1 as a direct target of miRs included roles in osteoblast differentiation, inflammation, migration, angiogenesis, and megakaryopoiesis (Table 1.2). Two miRNAs, miR-204 and miR-510, were defined as direct negative regulators of SPDEF by translational repression in breast and prostate cancer (Findlay et al., 2008; Turner et al., 2011). miR-204 and miR-510 expressions are both elevated in tumor samples compared to matched normal in breast cancer and explained the apparent discordance in the literature of reports of SPDEF being elevated or downregulated in breast cancer due to the fact that SPDEF mRNA levels can be elevated in the absence of protein. miR-145 was shown to directly target FLI1 in colon cancer and in pericytes, where it was shown to block migration in response to growth factor gradients (Larsson et al., 2009; Zhang, Guo, et al., 2011). This interaction was also observed in patients with the 5q syndrome, a subtype of myelodysplastic syndrome (MDS), in which the inhibition of FLI1 by miR-145 decreases the production of megakaryocytic cells relative to erythroid cells contributing to the phenotype of the human malignancy (Kumar et al., 2011). Finally, an elegant study by the Ostrowski group showed a link between miR-320 and ETS2 in the stromal fibroblasts (Bronisz et al., 2012). Although many studies have investigated a role for miRNAs in the epithelial tumor cells, very few have focused on their regulation in the stromal compartment. This study showed that miR-320 is a critical target of PTEN in stromal fibroblasts and directly controls ETS2 expression and instructs the tumor microenvironment to suppress many of the aggressive phenotypes associated with advanced stages of breast cancer, including tumor cell invasiveness and increased angiogenic networks.
Table 1.2.
microRNAs targeting ETS factors
| microRNA | ETS factor | Function/disease | References |
|---|---|---|---|
| 569 | SPI1 | Systemic lupus erythematosis | Hikami et al. (2011) |
| 510 | SPDEF | Breast and prostate cancer | Findlay et al. (2008) and Turner et al. (2011) |
| 204 | Breast cancer | Findlay et al. (2008) | |
| 145, 214 | ELK1 | Smooth muscle cell proliferation | Park et al. (2011) |
| 7 | ERF | Lung cancer | Chou et al. (2010) |
| 196a, 196b | ERG | Acute leukemia | Coskun et al. (2011) |
| 145 | FLI1 | Megakaryocyte and erythroid differentiation | Kumar et al. (2011) |
| Colon cancer | Zhang, Guo, et al. (2011) | ||
| Migration of microvascular cells (pericytes) | Larsson et al. (2009) | ||
| 125b | ETS1 | Breast cancer | Zhang, Yan, et al. (2011) |
| 193b | Hepatocellular carcinoma | Xu et al. (2010) | |
| 370 | Osteoblast differentiation | Itoh, Ando, Tsukamasa, and Akao (2012) | |
| 155, 221/222 | Inflammation, migration of endothelial cells | Wu et al. (2011) and Zhu et al. (2011) | |
| 200b | Angiogenesis | Chun et al. (2011) | |
| 208 | Preosteoblast differentiation | Itoh, Takeda, and Akao (2010) | |
| 155 | Megakaryopoiesis | Romania et al. (2008) | |
| 320 | ETS2 | Stroma breast cancer | Bronisz et al. (2012) |
| 221 | Endothelial cell motility | Wu et al. (2011) | |
| 378 | GABPA | Metabolic shift breast cancer | Eichner et al. (2010) |
4.2. ETS factors targeting miRNAs
Only a handful of studies are published on the role of ETS factor modulated miRNAs (Table 1.3); therefore, we expect many studies in the future in this underdeveloped area of research. In ovarian cancer, a study reported that EGFR signaling leads to transcriptional repression of miR-125a through the ETS family transcription factor ETV4 (Cowden Dahl et al., 2009). It is known that overexpression of EGFR in ovarian cancer correlates with poor disease outcome and induces epithelial–mesenchymal transition (EMT) in ovarian cancer cells (Cowden Dahl et al., 2008; Nicholson, Gee, & Harper, 2001). Overexpression of miR-125a induced a conversion of highly invasive ovarian cancer cells from a mesenchymal to an epithelial morphology, suggesting that miR-125a is a negative regulator of EMT. A study to distinguish serous ovarian cancer from normal ovarian tissue using miRNA profiling identified miR-125a as downregulated (Nam et al., 2008). This correlates well with the previous study; however, miR-21 was also identified as part of the signature in this study and was reported as being upregulated. This is interesting because miR-21 was also shown to be repressed by ETV4 in colon cancer (Kern et al., 2012); therefore, this exemplifies the importance of context when studying miRNAs. Some miRNAs are regulated by the same ETS factor as illustrated by miR-126 in endothelial cells (Harris et al., 2010). miR-126 is abundantly expressed in endothelial cells, and promoter analysis showed that multiple ETS factors led to increased expression, but ETS1 and ETS2 were the most robust.
Table 1.3.
ETS factors targeting microRNAs
| ETS factor | microRNA | Action | Function/disease | References |
|---|---|---|---|---|
| SPI1 | 29b | Activation | Neutrophil differentiation (PML) | Batliner et al. (2012) |
| ETV5 | 21 | Activation | Spermatogonial stem cell self-renewal | Niu et al. (2011) |
| ETV4 | 125a | Repression | EMT in ovarian cancer | Cowden Dahl, Dahl, Kruichak, and Hudson (2009) |
| 21 | Repression | Colorectal cancer | Kern et al. (2012) | |
| ELK1 | 34a | Activation | Oncogene-induced senescence | Christoffersen et al. (2010) |
| ETS1 | 126 | Activation | Endothelial cells | Harris et al. (2010) |
| ETS2 | 126 | Activation | Endothelial cells | Harris et al. (2010) |
| 196b | Repression | Gastric cancer | Liao et al. (2012) | |
| Fusions | ||||
| TEL/AML | 494, 320a Repression | Leukemia | Diakos et al. (2010) | |
| EWS/FLI1 | 30a-5p | Activation | Ewings sarcoma | Franzetti et al. (2012) |
| let-7a | De Vito et al. (2011) | |||
| 145, 100, 125b | Repression | Ewings sarcoma | Ban et al. (2011) | |
| 22, 221/222 | McKinsey et al. (2011) | |||
| 27a, 29a, 145 | Riggi et al. (2010) | |||
Some studies have also focused on the role of ETS fusion proteins on miRNAs. In particular, one study performed a genome-wide analysis of miRNAs affected by RNAi-mediated silencing of EWS-FLI1 in Ewing’s sarcoma cell lines and identified miR-145 as the top repressed miRNA (Ban et al., 2011), which is interesting as previously mentioned miR-145 is a negative regulator of FLI1 suggesting a possible feedback loop mechanism of regulation of this fusion gene.
5. ETS MOUSE KNOCKOUT AND MUTANT MODELS
5.1. Phenotypes of mice with genetically altered Ets
To date, 23 of the 27 murine Ets genes have been genetically altered (knockouts or mutant mice; Table 1.4). Diverse biological roles of individual ETS family members are supported by the wide range of phenotypes displayed in these models. Most of these models have specific phenotypes, with the exception of Elf1 and Elk1, demonstrating nonredundant functions for the majority of Ets factors. For these, subtle phenotypes have (Elk1, minor defects in neuronal gene activation, and, Elf1, reduced NK-T cell development and function) been identified. Complete or significant embryonic and/or postnatal lethality is observed for 11 family members. Consistent with their tissue expression profiles, the majority have phenotypes that demonstrate their important functions in hematopoiesis, either exclusively or in combination with other lineage defects. There is often a wide range of phenotypes observed even within an Ets subfamily. For example, in the Spi1 subfamily, phenotypes range from Spi1, a principal regulator of myelolymphopoiesis, to SpiB, which regulates the proper function of terminally differentiated lineages and SpiC that is necessary for the function of a subset of macrophages. Ets1 and Elf4 are important regulators of T cell (T, NK, and NKT) development. Ets family members such as Fli1, Etv2, and Etv6 display functions in hematopoiesis and/or vasculo/angiogenesis. Nonhematopoietic defects were observed for Ets2 which has phenotypes related to extraembryonic development. Etv1, Etv4, and Fev each have defective neurogenesis, Etv4 and Etv5 affect male fertility. Consistent with their restricted epithelial-specific expression, the Ese and Spdef subfamilies (Elf3, Ehf, and Spdef) show tissue-specific (e.g., intestine, mammary gland) phenotypes, albeit at significantly different severities.
Table 1.4.
Mouse Ets gene knockout and mutant mice
| Gene name | Severity of mutation | Phenotype | Gene targets identified |
|---|---|---|---|
| Ets1 | Viable and fertile with 50% neonatal lethality at 4 weeks | Hematopoietic cell defects. Reduced number of B, T, and NK cells | MDM2; Cyclin G (Xu et al., 2002), Foxp3 (Mouly et al., 2010), T-bet (Nguyen et al., 2012), MMP2, 3, and 13, TIMP1, 2, and 3 (Hahne et al., 2006) |
| Ets2 | Embryonic lethal (<E8.5) | Extraembryonic tissue defect. Rescued neomorph has hair follicle defect | MMP3, MMP9, MMP13 (Yamamoto et al., 1998) |
| Erg | Embryonic lethal at E11–12.5 | Lack of definitive hematopoiesis; defective thrombopoiesis in heterozygotes | Igfr2, MMP3 (Loughran et al., 2008) |
| Fli1 | Knockout is embryonic lethal at E12.5. | Thrombocytopenia, reduced erythroid progenitors, defective vasculargenesis, and B-cell maturation | Spyropoulos et al. (2000) and Starck et al. (2010) |
| Fev | Viable with reduced numbers of homozygous knockouts | Defective development of 5-HT neurons; increased anxiety and aggression | TPH, SERT, AADC (Hendricks et al., 2003) |
| Etv4 | Viable; males infertile | Male sexual defect; lack of dendrite patterning in motor neurons | |
| Etv5 | Viable; males infertile | Defective sperm production | CXCL12, CXCL5, CCL7 (Chen et al., 2005) |
| Etv1 | Viable | Lack of motor coordination, synaptic defect, limb ataxia | |
| Etv2 | Embryonic lethal E8–9.5 | Defects in hematopoiesis and vasculogenesis. Lack of endothelial and endocardial lineages. | Tie-2 (Ferdous et al., 2009), SOX9 (DiTacchio et al., 2012) |
| Elk1 | Viable; no phenotype | Phenotypically normal, minor effects on neuronal gene activation | Cesari et al. (2004) |
| Elk4 | Viable | Defects in thymocytes and peripheral T cells | Costello, Nicolas, Watanabe, Rosewell, and Treisman (2004) |
| Elk3 | Hypomorphic mutant dies at birth | Vascular defect leading to respiratory failure | EGR-1 (Ayadi et al., 2001) |
| Gabpa | Lethal prior to implantation | Mitochondrial defects; T cells | AChRδ and AChRε; AChE; Utrophin (O’Leary et al., 2007); IL7Rα (Yu, Zhao, Jothi, & Xue, 2010) |
| Elf1 | Minor defect | Reduced NK-T cell development and function | (Choi et al., 2011) |
| Elf4 | Viable and fertile | Reduced NK and NK-T cell development and function | Perforin (Lacorazza et al., 2002) |
| Spi1 | Embryonic lethal at E18.5 | Lack of hematopoiesis; no B, T cells or monocytes | Colucci et al. (2001) |
| SpiB | Viable | Defective B-cell responses; reduced intestinal immunity | Schotte, Nagasawa, Weijer, Spits, and Blom (2004), de Lau et al. (2012), and Kanaya et al. (2012) |
| SpiC | Viable | Defect in red pulp macrophages and red blood cell recycling and iron homeostasis | Vcam1 (Kohyama et al., 2009) |
| Etv6 | Embryonic lethal E11 | Lack of embryonic angiogenesis; defective hematopoiesis | Wang et al. (1998) and Ciau-Uitz, Pinheiro, Gupta, Enver, and Patient (2010) |
| Erf | Embryonic lethal E10.5 | Defective placental development | Papadaki et al. (2007) |
| Spdef | Viable; fertile | Defective mucosal development in the gastrointestinal tract and respiratory tract. Increased gastrointestinal malignancies | Muc6,Tff (Horst et al., 2010) |
| Elf3 | 30% Embryonic lethality E11.5 | Abnormal morphogenesis and differentiation of intestinal epithelium | TGFβ (Ng et al., 2002) |
| Elf5 | Embryonic lethal E7.5 | Embryonic patterning defect; heterozygotes deficient in mammary gland development | WAP, casein (Zhou et al., 2005); Notch pathway (Chakrabarti, Wei, et al., 2012); Snai2 (Chakrabarti, Hwang, et al., 2012) |
| Ets1 −/−; Ets2A72/A72 | Embryonic lethal E11.5–15.5 | Endothelial cell defects leading to failure of vascular branching | MMP9, BCL-xL, cIAP2 (Wei et al., 2009) |
These constitutive knockout models reveal only the earliest/most distinct functions of each of these Ets family members. A better understanding of the roles and hierarchies of Ets family members in cellular differentiation and function will come with the generation of new null alleles in untargeted family members, double knockouts, ES cell differentiation and chimera rescue experiments, and tissue-specific inducible knockouts.
Analyses of ES cell differentiation and chimeric and mutant mice were used to evaluate postembryonic phenotypes observed in the constitutive Fli1 knockout mice. These studies demonstrated that Fli1 also plays an important role in multiple non-megakaryocytic hematopoietic lineages, including erythroid, granulocyte, monocyte, and lymphocyte lineages (Zhang et al., 2008). Mutant mice lacking one of two regulatory domains (Fli1ΔCTA) provide novel evidence for the importance of Fli1 in megakaryocytic differentiation and platelet function. These approaches have also established Fli1 as an important regulator of fibroblast functions (Asano et al., 2009; Asano & Trojanowska, 2009; Kubo et al., 2003).
Conditional knockouts have further allowed definition of additional phenotypes. Fli1 conditional knockout mice in combination with Tie2-Cre have shown that mice with reduced endothelial Fli1 expression have compromised vessel integrity, markedly increased vessel permeability, and impaired pericyte/vascular smooth muscle cells coverage (Asano, Stawski, et al., 2010). Conditional Fli1 deletion in the adult results in mild thrombocytopenia associated with a maturation defect of bone marrow megakaryocytes (Starck et al., 2010), as previously observed in fetal liver of constitutive Fli1 knockout mice. In addition to the decrease in megakaryocytic cells, analysis of these mice revealed increases in natural killer (NK) cells and erythrocytic cells and a decrease in granulocytic cells, in agreement with the studies with chimeric Fli1 mice.
Fewer studies have examined the phenotypes of mice with deletions or mutations of two members. Phenotypes support similar roles for ETS subgroup (Ets1 and Ets2) in endothelial cells (Wei et al., 2009), ERG subgroup (Erg and Fli1) in hematopoietic cells (Kruse et al., 2009), TCF subfamily (Elk1 and Elk4) in thymocyte development (Costello et al., 2010), SPI1 subfamily (Spi1 and SpiB) in B-cell function (Garrett-Sinha et al., 1999), and PEA3 subfamily (Etv1 and Etv5) in limb-bud development (Zhang, Verheyden, Hassell, & Sun, 2009).
5.2. Identification of ETS target genes using genetic models
One important experimental approach for identifying Ets targets is the creation of null (knockout) or mutant mice lacking the function of a single or multiple family members. Analysis of these mice provides another means for the identification of genes whose expression or repression is dependent upon an Ets family member. Specific in vivo targets for Ets genes have been identified based on the knockout mice (Table 1.4). For example, c-mpl (Kawada et al., 2001) and Tie2 (Hart et al., 2000) have reduced expression in knockout Fli1 mice, consistent with the megakaryocytic lineage and vascular defects observed.
Mutant mice lacking one of two regulatory domains (Fli1ΔCTA) are thrombocytopenic and show significantly reduced expression of multiple megakaryocytic genes, including c-mpl, platelet glycoprotein IIb (gpIIb), gpIX, and gpV. These mice also show reduced expression of genes associated with terminal differentiation of megakaryocytes to platelets. As noted above, Fli1 and GATA1 synergistically regulate gene transcription of multiple megakaryocytic genes. Transient-transfection studies indicate that only wild-type (WT) Fli1 can synergize with GATA-1, increasing promoter activity. Consistent with the failure of Fli1ΔCTA and GATA-1 to synergistically activate the c-mpl promoter-luciferase reporters in vitro, ChIP studies demonstrate that Fli1ΔCTA is not able to efficiently recruit GATA-1 to specific (c-mpl, PF4, and gpIX) promoters in vivo and further define these as Fli1 direct target genes (Moussa et al., 2010).
Fli1 has been shown to repress collagen synthesis in cultured dermal fibroblasts and mouse embryo fibroblasts and Fli1 mutant mice show significant upregulation of fibrillar collagen mRNA and altered expression of matrix-related genes, including decorin, fibromodulin, lumican, procollagen lysine, 2-oxoglutarate 5-dioxgenase 2 (PLOD2), and lysyl oxidase (Asano et al., 2009; Kubo et al., 2003).
Conditional Fli1 knockout mice with Tie2-Cre endothelial cell-specific disruption support the notion that Fli1 may function in maintenance of vascular homeostasis by directly regulating VE-cadherin, PECAM1, Tie2, MMP9, PDGF B, and S1P1 receptor (Asano, Stawski, et al., 2010).
The impact of altered expression of specific Ets response genes can be assessed by performing genetic rescue experiments. This approach is nicely represented by studies of Elf3 knockout mice (Flentjar et al., 2007; Ng et al., 2002). Elf3 knockout mice show significant embryonic and postnatal lethality, due to aberrant morphogenesis and terminal differentiation of the small intestine. Elf3 knockout mice express significantly less TGF-βRII protein. To perform a rescue experiment, transgenic mice that express human TGF-βRII specifically in the intestinal epithelium were crossed to the knockout animals. Significantly, the TGF-βRII transgenic Elf3−/− mice displayed normal small intestinal morphology.
6. ETS FACTORS AND CANCER
The hallmark features of a cancer cell consist of uncontrolled proliferation, loss of differentiation, sustained cell division, increased angiogenesis, loss of apoptosis, and a capacity to migrate and invade to other tissues and organs. All of these processes are driven by transient and/or permanent changes in gene expression profiles conferred through the activation or repression of cancer-associated genes. It is therefore clear that the role of transcriptional gene regulation in cancer progression cannot be understated and many transcription factors including ETS family members have been assigned as candidate oncogenes or tumor repressors. The importance of ETS genes in human carcinogenesis is supported by the observations that during cancer progression, ETS genes acquire point mutations (e.g., SPI1, ETS1), genomic amplification (ETS1, ETS2, ERG), increased (ETS1, ETS2, ERG) or decreased (e.g., SPDEF, EHF) expression, or rearrangements (ETV6, FLI1, ERG; Seth & Watson, 2005), resulting in altered ETS gene expression which disrupts the regulated control of many complex biological processes, promoting cellular proliferation and inhibiting apoptosis, enhancing cell migration, invasiveness, and metastasis as well as angiogenesis (Fig. 1.2).
6.1. ETS expression in cancer
Altered ETS gene expression levels are correlated with tumor progression in human neoplasias, including thyroid, pancreas, liver, prostate, colon, lung, and breast carcinomas and leukemias (Seth & Watson, 2005; Watson et al., 2010). Furthermore, in breast cancer, upregulation of multiple ETS factors, including ETS1 (Buggy et al., 2004), ETS2 (Buggy et al., 2006), ETV4 (Benz et al., 1997; Chang et al., 1997), ETV5 (Yarden & Sliwkowski, 2001), and ETV1 (Bosc, Goueli, & Janknecht, 2001), is associated with poor prognosis and metastasis. In contrast, other ETS factors including SPDEF (Doane et al., 2006; Feldman, Sementchenko, Gayed, Fraig, & Watson, 2003), ELF5 (Zhou et al., 1998), and EHF (Tugores et al., 2001) are downregulated during breast cancer progression within the same context. The impact of multiple ETS factors (e.g., ETS1, ETS2, ETV4, ELF3, SPDEF, ERG) on phenotypes and molecular regulation in cancer cells has been demonstrated through in vitro gain-of-function as well as loss-of-function experiments.
In addition to ETS-mediated transcriptional activation of multiple genes associated with cancer progression, analysis of androgen receptor (AR) genomic targets demonstrated an enrichment of ETS transcription factor family members and, more specifically, an interaction between the AR and ETS1 at a subset of the AR promoter targets was found (Massie et al., 2007). These studies support the model that ETS proteins, including ETS1, regulate genes, including androgen response genes, which contribute to prostate cancer progression.
6.2. ETS translocations
Cancer involves many chromosomal aberrations, the most studied being nonrandom chromosomal translocations resulting in recombinant chromosomes. Tumor cell formation results from the translocation associated production of FLI1 chimeric proteins as has been shown for Ewing’s sarcomas (EWS) and related primitive neuroectodermal tumors (PNET; reviewed in Arvand & Denny, 2001). In this instance, chimeric transcripts result from the fusion of the amino terminal region of the EWS gene with the carboxyl terminal DNA-binding domain of the FLI1 gene (Delattre et al., 1992; Zucman et al., 1992). The chimeric fusion protein lacks the putative RNA-binding domain of EWS and one of the transactivation domains of FLI1. It has also been shown that the EWS-FLI1 fusion is a more potent transcriptional activator than the FLI1 protein. In other Ewing’s sarcoma and PNET tumors, translocations fuse the EWS gene to other members of the ETS family, including ERG, ETV1, ETV4, and FEV.
ETV6 was originally identified by its rearrangement in specific cases of chronic myelomonocytic leukemia (CMML) presenting a t(5,12)(q33; p13) chromosomal translocation (Golub, Barker, Lovett, & Gilliland, 1994). ETV6 is rearranged in CMML, acute myelogenous leukemia (AML), acute myeloblastic leukemia (AML-M2), MDS, and acute lymphoblastic leukemia. Either the PNT domain or the ETS domain or both domains of ETV6 have been identified in over 20 different translocations observed in human leukemia and more rarely solid tumors (reviewed in Mavrothalassitis & Ghysdael, 2000). Fusions involving the PNT domain of ETV6 often lead to oligomerization that is necessary for constitutive activation of kinase activity of receptor or protein tyrosine kinases. Fusions that retain the DNA-binding domain of ETV6 are expected to result in aberrant regulation of ETS target genes.
ERG is highly expressed in over 60% of prostate tumor cells relative to benign tissues. A molecular mechanism to account for ERG overexpression in prostate cancer was subsequently provided by the identification of chromosomal rearrangements that result in the fusion between the 5′ end of the androgen-regulated, prostate-specific transmembrane serine protease TMPRSS2 gene to ERG (Soller et al., 2006; Tomlins et al., 2005). Collective studies show that the TMPRSS2-ERG fusion is present in 40–80% of prostate cancers (recently reviewed in Kumar-Sinha, Tomlins, & Chinnaiyan, 2008; Shah & Chinnaiyan, 2009; Shah & Small, 2010). TMPRSS2 gene fusions involving other ETS transcription factors ETV1, ETV4, or ETV5 have been identified in prostate cancer; however, TMPRSS2-ERG fusion and mRNA overexpression accounts for the majority of cases. Possible mechanistic insights are provided by observation that TMPRSS2-ERG fusion activates MYC and abrogates prostate epithelial cell differentiation. An 87-gene signature has been associated with TMPRSS2:ERG fusion tumors. Collective data suggest that the TMPRSS2-ERG fusions define a subset of prostate cancer and specific fusions predict poor prognosis and survival.
6.3. ETS target gene expression and function
Functional studies demonstrate the impact of such altered expression on the regulation of genes associated with proliferation, transformation, migration, invasion, anti-apoptosis, and angiogenesis (Seth & Watson, 2005) and include but are not exclusive to Her2/neu, uPA, MMPs, TIMPs, MET, Bcl2, maspin, and VEGFR (Sementchenko & Watson, 2000; Fig. 1.2).
Alterations in cell cycle control are a critical step in carcinogenesis. Cell cycle arrest at the G1–S transition by upregulation of the cyclin-dependent kinase inhibitor p21 occurs in response to DNA damage or oncogenic insult. The elevated level of p21 is known to be mediated through p53 and we have demonstrated SPDEF-mediated regulation of p21 expression is associated with inhibition of growth in vitro (Feldman, Sementchenko, Gayed, et al., 2003) and in vivo (Schaefer et al., 2010). Indeed, SPDEF-mediated inhibition of breast cancer xenograft growth can be reversed by shRNA targeting of p21 (Schaefer et al., 2010). These observations combined with ChIP demonstrate that p21 is a key direct target of SPDEF used to control cellular growth. The increased expression of the p21-activated kinase (PAK1) has been shown to be correlated with more aggressive breast cancer (Salh, Marotta, Wagey, Sayed, & Pelech, 2002). Furthermore, studies have shown that PAK1 regulates the activity of ELF3 by phosphorylation (Manavathi, Rayala, & Kumar, 2007). This novel finding raises the possibility that using a specific inhibitor to the upstream effector of ELF3 (e.g., PAK1-specific inhibitor CEP-1347) may represent a novel approach for targeting a transcription factor in breast cancer.
Migration and invasion, critical steps in the metastatic process, requires changes in cell-to-cell adhesion as well as cell adhesion to the ECM. Migration and invasion are often associated with EMT and resultant down-regulation of E-cadherin. Invasion is mediated in part by proteolytic degradation of the ECM by MMPs and uPA. Indeed, activation of the uPA system is associated with a poor prognosis in breast cancer. Significantly, we and others have shown that ETS factors are critical regulators of EMT, protease expression, and ECM (discussed further below, microenvironment). For example, studies using breast, prostate, colon, or ovarian cancer cells have demonstrated the antimigratory and antiinvasive properties of SPDEF, by negative regulation of the EMT regulator SLUG and mesenchymal genes, proteases (uPA, MMPs).
6.4. ETS conversion
To date, ETS research has mainly focused on the molecular mechanisms and functions of individual transcription factors and has produced insights into ETS factor function in both normal and cancer cells. In many cells, multiple ETS factors with similar or opposite functions are present simultaneously and the cell’s fate may depend ultimately on the balance between the activities of distinct ETS factors.
ETS factor dysregulation disturbs normal cellular homeostasis, increasing cancer growth, invasion, and metastasis. While some ETS factors are lost during cancer progression, others show increased expression: tumor suppressive and oncogenic ETS factors. We hypothesize that the balance of “tumor suppressor” and “oncogenic” ETS factors could be a marker for aggressive cancer. Taken together, accumulating evidence suggests that multiple ETS factors act in concert to positively and negatively regulate the pathways that control progression to metastatic cancer. This indicates a possible ETS conversion mechanism of gene regulation which provides the cell with an integrated mechanism by which to respond to a variety of intra- and extracellular signals efficiently (Hsu et al., 2004; Turner, Findlay, Moussa, & Watson, 2007; Watson et al., 2010). Several Ets factors are deregulated in the development of breast cancers. During cancer progression, the expression of some ETS factors (e.g., ETS1, ETS2, ETV4, ETV5, ELF3) is often increased, while the expression of other ETS factors (SPDEF, EHF) is reduced or lost (Turner, Findlay, et al., 2007; Watson et al., 2010). The ETS conversion model further hypothesizes that the change in expression pattern from what is observed in normal or benign tissues (e.g., SPDEF expression) to that observed in invasive cancer (e.g., elevated ETS1) is necessary for cancer progression to proceed.
Reciprocal ETS regulation of a metastasis-associated gene can be illustrated by the uPA promoter. ETS regulation of uPA has both positive and negative effects on cancer progression depending on the specific Ets factor expressed. ETS1 is overexpressed in invasive breast and aggressive prostate cancer and associated with increased uPA expression. In noninvasive (ETS1 negative) breast cancer cells, reexpression of ETS1 increases uPA levels leading to more aggressive tumorigenic phenotypes, including increased cell growth, migration, and invasion. In contrast, the expression of another ETS family member, SPDEF is present in noninvasive, but lost in invasive, breast cancer cells. SPDEF reexpression in invasive cells represses endogenous uPA transcription leading to an inhibition of cell migration and invasion and an antimetastatic phenotype (Feldman, Sementchenko, Gayed, et al., 2003; Turner, Moussa, Sauane, Fisher, & Watson, 2007). Significantly, a statistically significant inverse correlation between SPDEF and uPA expression is observed in colon cancer clinical specimens (Moussa et al., 2009). Intriguingly, although several potential EBS are found in the uPA promoter, both ETS1 and SPDEF have been demonstrated to bind at the same consensus EBS in vivo.
Many ETS factors (including ETS1, ETS2, ETV4) transcriptionally activate multiple MMPs, most commonly in cooperation with AP1 complexes. In contrast, SPDEF is a repressor of MMP7 (Moussa et al., 2009) and MMP9 (Johnson et al., 2010)
Another example of reciprocal regulation is provided by the maspin promoter. Maspin is a type II tumor-suppressor gene that has been shown to have antimetastatic properties when expressed in invasive breast and prostate cancer cells (Zou et al., 1994). The maspin promoter has been shown to be regulated by SPDEF (Feldman, Sementchenko, Gayed, et al., 2003; Yamada, Tamai, Miyamoto, & Nozaki, 2000). Significantly, this activation appeared to be specific for SPDEF, since neither FLI1 nor ETS1 was able to activate this promoter. Indeed, ETS1 expression inhibited SPDEF-mediated transactivation of the maspin promoter.
6.5. ETS regulatory network
Taken as a whole, this evidence strongly suggests the existence of distinct ETS expression regulatory networks that act in concert to positively or negatively regulate cancer-associated genes. Significantly, each ETS network would result in distinct patterns of target gene expression, the elucidation of which may identify prometastatic and antimetastatic signatures of gene expression that may predict the aggressive behavior of cancer cells. The ETS Regulatory Network is comprised of the ETS factors themselves, their upstream modulators, their coregulatory proteins, and their target genes (Fig. 1.3). Inflammatory cells are recruited by tumors through their secretion of chemokines, cytokines, and growth factors (1). In response, the recruited inflammatory cells and other cells of the microenvironment (e.g., fibroblasts promote tumor proliferation and progression through additional secretion of biologically active molecules). This in turn results in the activation of intracellular signaling cascades via ligand binding at the cell surface of epithelial cells (2). The activated cascades directly or indirectly (through crosstalk) result in the expression and repression of varying combinations of the 28 ETS family members (3). ETS factors can regulate their own expression and/or that of other family members (4). The composition of Ets factors defines the transcriptional regulation of their target genes, many known to be involved in cancer progression (5). The altered expression of these genes has profound consequences on many cancer-related pathways (6).
Figure 1.3.
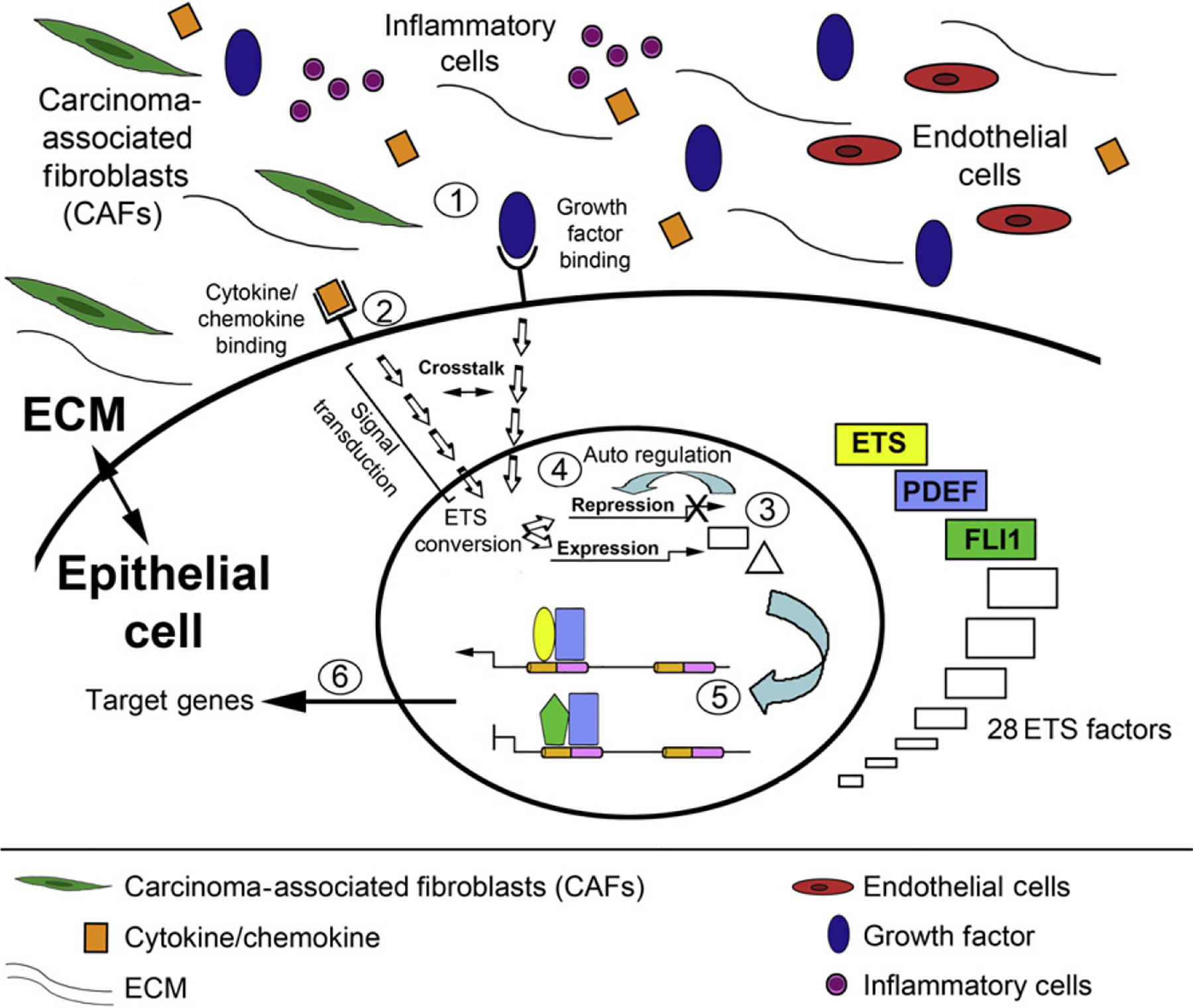
Hypothetical model of the ETS regulatory network in cancer. See text for details.
6.5.1. ETS-mediated anti- and prometastatic signatures
Gene expression signatures consist of sets of gene profiles that are known to be predictive of a disease state and/or patient response to treatment. The combined statistical analysis of multiple gene sets obtained from independent gene micro-array studies has resulted in an increased number of putative and validated “metastatic signatures” that predict the outcome of disease in cancer. In addition, comparison of gene expression profiles from primary and metastatic tumors in multiple cancer types reveals highly specific signatures that allow discrimination between primary and metastatic tumors. Similarly, by elucidating the expression networks conferred by ETS family members that elicit a prometastatic response (ETS1, etc.) and an antimetastatic response (SPDEF, etc.), improved pro- and antimetastatic signatures may be isolated which predict the aggressive behavior of cancer cells. As such, these new insights may provide a novel view of the ETS gene family as well as a focal point for studying the complex biological control involved in tumor progression.
7. THE ROLE OF ETS FACTORS IN THE MICROENVIRONMENT
The majority of cancer-related deaths are due to tumor progression, whereby cells from the primary tumor migrate, invade, and reestablish at distant metastatic sites (Guarino, Rubino, & Ballabio, 2007; Turner, Moussa, et al., 2007). The progression of solid tumors corresponds with progressive alterations in the tumor microenvironment, suggesting crosstalk between epithelium and stroma. Increasing evidence suggests that these stromal–epithelial interactions play a critical role in regulating tumor growth and progression. However, this aspect of tumorigenesis remains little understood. Previous studies have indicated that members of the ETS transcription factor family are abnormally expressed in both tumor and stromal compartments. This aberrant expression of ETS factors has been associated with cancer progression and frequently correlates with poor prognosis. For example, ETS1 is frequently overexpressed in epithelial, endothelial, and stromal cells in various tumors (Behrens, Rothe, Florin, Wellmann, & Wernert, 2001; Behrens, Rothe, Wellmann, Krischler, & Wernert, 2001; Takai, Miyazaki, Nishida, Nasu, & Miyakawa, 2002; Trojanowska, 2000). Studies have demonstrated that ETS1 is a strong independent predictor of poor prognosis in breast cancer (Myers et al., 2005; Span et al., 2002). Further, drug-resistant breast cancer cells have been shown to overexpress ETS1 (Kars, Iseri, & Gunduz, 2010), suggesting that ETS factors may play a significant role in tumor aggressiveness and contribute to failed therapies. These studies highlight the importance of understanding the mechanisms by which ETS factors function in both the epithelium and microenvironment.
The stromal compartment consists of fibroblasts, endothelial cells, perivascular cells, blood-borne cells, nerves, and intervening ECM. The fibroblasts of the tumor microenvironment, termed carcinoma-associated fibroblasts (CAFs), are thought to promote tumor progression by establishing a reactive tumor stroma, stimulating growth, sustaining angiogenesis, inhibiting the immune response, promoting the malignant phenotype, and promoting invasion and metastasis (Hanahan & Coussens, 2012; Hanahan & Weinberg, 2000, 2011; Karnoub et al., 2007; Orimo & Weinberg, 2006). While these previous studies have identified important functions for CAFs, the factors regulating the functions of these cells are undefined.
Growing evidence suggests that ETS family members are critical regulators of stromal activation. The reactive tumor stroma is characterized by excessive remodeling of the ECM via CAF production of matrix molecules (i.e., collagen-1), matrix-degrading factors (i.e., matrix metalloproteinases, MMPs), and growth factors (i.e., TGFβ). Fli1 has been established as a regulator of fibroblast function (Asano et al., 2009; Kubo et al., 2003; Truong & Ben-David, 2000; Watson et al., 1992). A hallmark of the CAF is the expression of alpha-smooth muscle actin (αSMA) and studies have shown that Fli1 reduction leads to αSMA upregulation (Nakerakanti, Kapanadze, Yamasaki, Markiewicz, & Trojanowska, 2006). Fli1 has also shown to function as a physiological transcription repressor of collagen type I gene in vivo (Asano, Bujor, & Trojanowska, 2010; Czuwara-Ladykowska et al., 2001). The absence of Fli1 correlates with elevated collagen synthesis (Kubo et al., 2003), a second hallmark of the activated tumor stroma. Fli1 has also been shown to regulate expression of tenascin-C, an additional ECM protein associated with wound healing and tumor stroma activation (Shirasaki, Makhluf, LeRoy, Watson, & Trojanowska, 1999). At least part of the Ets functions in tumor stromal cells is the regulation of ECM-degrading enzymes including MMPs, uPA, and collagenases, molecules that are crucial for the establishment of a reactive stroma and onset of metastasis (Westermarck, Seth, & Kahari, 1997). ETS1 and FLI1 have been shown to modulate MMP1 expression (Gavrilov, Kenzior, Evans, Calaluce, & Folk, 2001; Nakerakanti et al., 2006). ETS1 and ETS2 have also been implicated in uPA and MMP9 activation (Watabe et al., 1998). A recent study identified novel ETS1 target genes by subtractive hybridization in stromal fibroblasts under bFGF stimulation (Hahne, Fuchs, et al., 2011; Hahne, Okuducu, Fuchs, Florin, & Wernert, 2011). MMP1, MMP3, PAI-1, and collagen Iα2 were confirmed as ETS1 target genes. Several additional targets were identified which may play a role in generation of the activated tumor stroma: cathepsin, a lysosomal proteinase whose elevated expression is associated with several cancers; lumican, a proteoglycan that binds collagen I and II to sequester growth factors in matrix; decorin, a proteoglycan that binds collagen I during matrix assembly and interacts with fibronectin, thrombospondin, epidermal growth factor receptor, and TGFβ to affect their functions; gremlin, a secreted antagonist of BMPs that promotes cancer cell survival and proliferation and is overexpressed in stroma of many cancers; HSP-90, a heat shock protein that acts to stabilize various growth factor receptors, is required for the induction of VEGF and nitric oxide synthase, and assists MMP2 to promote invasion/metastasis. While these studies did not experimentally demonstrate effects of ETS1 on the promoters of potential ETS1 target genes identified, promoter analysis showed the presence of potential EBS in the promoter regions of each gene identified.
ETS factors have also been shown to directly regulate the expression of cytokines as well as the response to specific growth factors and chemokines (Turner et al., 2008; Turner, Moussa, et al., 2007; Turner & Watson, 2008). For example, ETS1 is a downstream effector of the stroma-derived EMT-promoting HGF and an activator of its receptor, c-Met, thereby regulating a positive feedback loop whereby HGF/c-Met affects both tumor stroma and tumor cells (Hsu et al., 2004). HGF has also been shown to induce MMP1 protein expression in cultured human dermal fibroblasts. Studies showed that the balance of ETS1 and FLI1 binding to the EBS in the MMP1 promoter regulated the effects of HGF, with ETS1 binding leading to upregulation of MMP1 and FLI1 antagonizing this expression (Jinnin, Ihn, Mimura, et al., 2005). The activities of FLI1 and ETS1 toward the expression of Tenascin-C and connective tissue growth factor (CTGF/CCN2), novel Ets target genes (Jinnin et al., 2006; Nakerakanti et al., 2006; Shirasaki et al., 1999), are modulated by acetylation in a TGFβ-dependent manner (Asano, Czuwara, & Trojanowska, 2007; Asano et al., 2009; Asano & Trojanowska, 2009). These data suggest that ETS1 and FLI1 are the effectors of the TGFβ signaling pathway through novel, previously undescribed regulatory mechanisms. Elevated ETS1 has also been shown to be an antagonist of TGF-β functions in stromal cells (Czuwara-Ladykowska et al., 2002). Significantly, ETS1 and FLI1 are targets of the TGF-β signaling pathway, the primary regulator of fibroblast maturation, activation, and function. HGF-activated ETS1 has also been shown to regulate CXCL12/CXCR4-dependent promotion of tumor cell chemoinvasion (Maroni, Bendinelli, Matteucci, & Desiderio, 2007).
Several in vivo studies have demonstrated a correlation between stromal expression of ETS factors, dysregulation of matrix factors, and tumor progression. For example, stromal upregulation of ETS1, MMP1, and MMP9 has been observed in invasive ductal and lobular breast cancers (Behrens, Rothe, Wellmann, et al., 2001) and in invasive HNPCC and sporadic colon cancer (Behrens et al., 2003). Stromal cell expression of a specific ETS target gene (MMP9) has been shown to play a critical role in angiogenesis and growth of ovarian tumors in mice (Huang et al., 2002). Together, these studies suggest that targeting Ets factors in cells of the microenvironment may be an effective antitumor therapy. To demonstrate this potential, specific inactivation of Ets2 in the CAF population in a Pten murine mammary tumor model led to a reduction in tumor size (Li, Wallace, & Ostrowski, 2010). The absence of Ets2 in fibroblasts led to decreased epithelial cell proliferation and delayed tumor progress, illustrating the ability of ETS factors to regulate crosstalk between epithelial and stromal compartments and the importance of targeting this interaction.
In addition to their role in the fibroblastic and ECM components of tumor stroma, the altered expression of several ETS factors has been suggested to regulate angiogenesis, another key step in tumor progression and metastasis. FLI1 is normally expressed in vascular cells including hematopoietic cells, perivascular cells, and endothelial cells (Jinnin, Ihn, Yamane, et al., 2005; Kubo et al., 2003; Lelievre, Lionneton, & Soncin, 2001; Lelievre, Lionneton, Soncin, & Vandenbunder, 2001; Liu, Walmsley, Rodaway, & Patient, 2008; Pimanda et al., 2007; Spyropoulos et al., 2000; Truong & Ben-David, 2000; Watson et al., 1992). Loss of Fli1 results in embryonic lethality due in part to the absence of megakaryocytes, aberrant vasculogenesis, and disruption of tissue integrity (Kawada et al., 2001; Spyropoulos et al., 2000). FLI1 expression is reduced or lost in stromal cells in epithelial tumors, suggesting that this loss of FLI1 could have a direct effect on tumor vasculogenesis. Stromal-derived VEGF can induce ETS1 expression in endothelial cells (Lavenburg, Ivey, Hsu, & Muise-Helmericks, 2003) and activated transcription of VEGFR2/Flt-1 in concert with HIF-2α (Elvert et al., 2003). In addition, expression of ERG and FLI1 has been correlated with Tie2 gene expression, which is involved in the formation and remodeling of normal vascular networks (Mattot, Vercamer, Soncin, Fafeur, & Vandenbunder, 1999).
Evolving data indicate that Fli1 plays an important role in multiple hematopoietic lineages, including erythroid, granulocyte, monocyte, and lymphocyte lineages (Hart et al., 2000; Kawada et al., 2001; Masuya et al., 2005; Nowling, Fulton, Chike-Harris, & Gilkeson, 2008; Spyropoulos et al., 2000; Zhang et al., 2008). Tumor-associated macrophages (TAMs), a cell of monocyte origin, have been implicated in tumor progression by mediating angiogenesis, invasion, and immunosuppression (Sica et al., 2008). Studies have demonstrated that ETS2 is an important downstream mediator of CSF1-R (colony stimulating factor-1, a growth factor that regulates macrophage survival, proliferation, and differentiation) signaling in TAMs. Macrophage-specific ablation of Ets2 in the PyMT tumor model resulted in significant decrease in mammary tumor metastasis to lung (Lin, Nguyen, Russell, & Pollard, 2001; Zabuawala et al., 2010). Gene expression profiling studies have demonstrated that Ets2 target genes are not only tumor specific, but compartment specific between CAFs and TAMs (reviewed in Li et al., 2010). These studies reinforce the idea that cellular context defines the direction and magnitude of response to ETS factors.
8. ETS FACTORS AND OTHER DISEASES
While less attention has been directed toward the elucidation of the roles for ETS transcription factors in diseases other than cancer, clear roles for ETS factors in autoimmune diseases have been defined; these and some other diseases will be briefly discussed below.
Transgenic mice overexpressing Fli1 develop a lupus-like disease (Zhang et al., 1995). It was also previously demonstrated that FLI1 is overexpressed in peripheral blood lymphocytes of systemic lupus erythematosis (SLE) patients compared to normal healthy controls and that NZB/NZW mice, a murine lupus model, have higher Fli1 mRNA expression in splenic lymphocytes than normal control mice (Georgiou et al., 1996). When Fli1 heterozygous mice were crossed with MRL/lpr mice, another model of SLE, Fli-1+/− MRL/lpr mice had significantly decreased serum levels of total IgG and anti-dsDNA antibodies as disease progressed. In addition, these mice had significantly increased splenic CD8+ and naive T cells and markedly decreased proteinuria and significantly lower pathologic renal scores compared to Fli1+/+ MRL/lpr mice. At 48 weeks of age, survival was significantly increased in the Fli1+/− MRL/lpr mice as 100% were alive, in contrast to only 27% of Fli1+/+ mice. Both in vivo and in vitro production of MCP-1 were significantly decreased in Fli1+/− MRL/lpr mice (Zhang et al., 2004). Similar findings were obtained in NZM2410 mice (derived from NZB X NZW F1 hybrids), where 93% of Fli1+/− NZM2410 mice survived to the age of 52 weeks compared to only 35% of WT NZM2410 mice (Mathenia et al., 2010).The primary endothelial cells isolated from the kidneys of Fli1+/− NZM2410 mice produced significantly less MCP-1. ChIP analysis demonstrated that Fli1 directly binds to the promoter of the MCP-1 gene. These data indicate that Fli1 impacts glomerulonephritis development by regulating expression of inflammatory chemokine MCP-1 and inflammatory cell infiltration in the kidneys. Together, these findings indicate that FLI1 expression is important in lupus-like disease development. The length of a GA microsatellite in the FLI1 promoter has been shown to be inversely correlated to promoter activity and is associated with SLE patients without nephritis (Morris et al., 2010). Recent genome-wide association studies have identified genetic variants of ETS1 associated with SLE (Yang et al., 2010) and Ets1 knockout mice develop lupus-like disease (high IgM and IgG autoantibodies, glomerulonephritis, and local complement activation; Wang, John, et al., 2005). It has been recently suggested that some of these phenotypes could be related to ETS1 functions, including negative regulation of Th17 and B-cell differentiation (Pan, Leng, Tao, Li, & Ye, 2011).
Systemic sclerosis (SSc) or scleroderma is an autoimmune inflammatory disease characterized by fibrosis of the skin and internal organs as well as microvessel injury. The importance of reduced FLI1 expression in the pathogenesis of SSc has been reviewed recently (Asano, Bujor, et al., 2010). Although FLI1 expression in dermal fibroblasts is relatively low, studies have shown that FLI1 plays a critical role in the regulation of ECM genes, including type I collagen (Jinnin, Ihn, Yamane, et al., 2005) and the multifunctional matricellular factor CTGF/CCN2 (Nakerakanti et al., 2006). Importantly, FLI1 has been shown to be a potent inhibitor of collagen biosynthesis in dermal fibroblasts and its aberrant expression has been implicated in the pathogenesis of cutaneous fibrosis in SSc (Kubo et al., 2003; Wang, Fan, & Kahaleh, 2006). Interestingly, MCP-1 (regulated by FLI1 in SLE) also has been shown to play a role in SSc fibrosis (Artlett, 2010). In humans, FLI1 is expressed in the healthy skin microvasculature; however, its presence is greatly reduced in endothelial and periendothelial cells in SSc skin (Kubo et al., 2003). Conditional deletion of Fli1 in the endothelium of mice results in vascular defects observed in SSc vasculature (Asano, Stawski, et al., 2010).
Jacobsen syndrome (11q-) is a rare chromosomal disorder caused by deletions in distal 11q. Individuals have thrombocytopenia with a subpopulation of cells having enlarged α-granules. In addition to platelet effects, Jacobsen syndrome patients also present with a wide spectrum of the most common congenital heart defects, including an unprecedented high frequency of hypoplastic left heart syndrome (HLHS). Both of these conditions are associated with deletions on the long arm of chromosome 11, including 11q23, where ETS1 and FLI1 are located. Thus, these patients have only one copy of these ETS genes due to a heterozygous loss of regions in Chromosome 11. FLI1 monoallelic expression combined with its hemizygous loss underlies Jacobsen thrombocytopenia (Raslova et al., 2004). Significantly, overexpression of FLI1 in patient CD34(+) cells restores the megakaryopoiesis in vitro, indicating that FLI1 hemizygous deletion contributes to the hematopoietic defects (Raslova et al., 2004). Ets1 is expressed in the endocardium and neural crest during early mouse heart development. Ets1 knockout mice show large membranous ventricular septal defects and a bifid cardiac apex, and less frequently a nonapex-forming left ventricle (one of the hallmarks of HLHS). These results implicate an important role for ETS1 in mammalian heart development and some of the most common forms of congenital heart disease (Ye et al., 2010).
The functional polymorphism in the MMP1 promoter affecting ETS binding may contribute to the pathogenesis of osteomyelitis by increased MMP1 expression (Montes et al., 2010) as well as higher disease severity in recessive dystrophic epidermolysis bullosa (Titeux et al., 2008). It has also been hypothesized that the ETS2 activation of Bcl-xL may protect glia from constitutive oxidative stress that is believed to be a key mechanism for amyotropic lateral sclerosis, an adult-onset neurodegenerative disease (Lee, Kannagi, Ferrante, Kowall, & Ryu, 2009).
Phenotypes of several of the knockout, mutant, and transgenic mice support the notion that ETS factors have roles in several diseases. For example, the importance of Fli1 and Erg in megakaryopoiesis would support a possible role for these ETS factors in other diseases affecting megakaryopoiesis (thrombocytopenia, megakaryocytopenia) or other conditions associated with thrombocytopenia (e.g., chronic liver disease, acquired immunodeficiency syndrome). Many ETS factors would be expected to have a critical role in other hematopoietic, vascular, and respiratory (e.g., asthma, cystic fibrosis, chronic obstructive pulmonary disease) diseases. ETS1 has been shown to be a mediator of inflammation and neointima formation in a model of carotid artery balloon injury (endoluminal vascular injury; Feng et al., 2010). Spdef is required for differentiation of pulmonary goblet cell and regulates genes associated with mucus production, supporting the model that Spdef plays a critical role in regulating a transcriptional network mediating the goblet cell differentiation and mucus hyperproduction associated with chronic pulmonary disorders (Park et al., 2007).
9. TARGETING THE ETS NETWORK
9.1. Therapeutic targeting of ETS transcription factors
Targeting transcription factors for therapeutic gain is the focus of intense research as being able to manipulate transcriptional expression patterns would provide a novel approach for the treatment of many human diseases. The primary limitations to targeting transcription factors are the potential for off-target effects and insufficient delivery within the cell. Overwhelming evidence suggests that the number of transcription factors whose aberrant function supports tumorigenesis is limited (Darnell, 2002). Additionally, this limited number of transcription factors function at critical focal points controlling many of the genes involved in cancer-associated processes. Therefore, targeting transcription factors has great potential for therapeutic gain.
9.2. Targeting ETS factor biology
ETS factor family members are associated with the positive and negative regulation of gene expression profiles affecting all the classic hallmarks of cancer, including sustaining proliferative signaling, evading growth suppressors, resisting apoptosis, replicative immortality, activated angiogenesis, and induced invasion and metastasis (Turner & Watson, 2008). Critically, an ever growing body of evidence demonstrates many genetic and epigenetic alterations of ETS transcription factor function and activity in cancer. ETS factors, therefore, provide potential targets for cancer therapy. Pharmacological intervention may be used to inhibit the altered expression of oncogenic ETS factors such as ETS1, ETS2, ELF3, and/or activate the expression of tumor-suppressive members such as SPDEF and ETV6 (Turner & Watson, 2008). Several of the multifaceted aspects of ETS factor biology have been explored in order to assess the potential of therapeutically targeting these proteins. Strategies have included: directly inhibiting the promoter of oncogenic ETS factors (Carbone, McGuffie, Collier, & Catapano, 2003; Carbone et al., 2004; Miwa et al., 2005; Sahin et al., 2005) or directly targeting specific ETS factor mRNA (Dohjima, Lee, Li, Ohno, & Rossi, 2003; Hu-Lieskovan, Heidel, Bartlett, Davis, & Triche, 2005; Kitange et al., 1999; Tomlins et al., 2007; Wernert et al., 1999) to prevent the expression of their target genes; directly targeting the ETS protein itself (Turner & Watson, 2008) or indirectly targeting ETS responsive promoters of transcriptional target genes (Hewett et al., 2006; Pourtier-Manzanedo et al., 2003; Sementchenko, Schweinfest, Papas, & Watson, 1998; summarized in Table 1.5; Fig. 1.4; and in Turner & Watson, 2008).
Table 1.5.
Approaches used to investigate the multifaceted aspects of ETS factor biology in order to assess the potential of therapeutically targeting these proteins
| Approach | Mode of action | Method | Molecular target | References |
|---|---|---|---|---|
| ETS promoter occupancy | Competition for binding to promoters | Decoy oligonucleotides | ETS1, ETS2 | Miwa et al. (2005) |
| Triplex-forming oligonucleotides | Carbone et al. (2003, 2004) and Sahin et al. (2005) | |||
| ETS mRNA | Prevention of expression | Antisense oligonucleotides, RNA interference | Multiple ETS factors, EWS-FLI1 | Dohjima et al. (2003), Hu-Lieskovan et al. (2005), Kitange et al. (1999), Tomlins et al. (2007), and Wernert et al. (1999) |
| ETS protein | Functional inhibition | Small-molecule inhibitor | ESE1-SER2, ETS fusions, ERG | Barber-Rotenberg et al. (2012), Rahim et al. (2011), Stegmaier et al. (2007), Turner and Watson (2008), and Patent app # 20110207675 |
| ETS responsive promoters | Prevention of ETS transcriptional regulation | Triplex-forming oligonucleotides, dominant negative | TIE1, FGF2 | Hewett et al. (2006), Pourtier-Manzanedo et al. (2003), and Sementchenko et al. (1998) |
Figure 1.4.
Therapeutic strategies for targeting ETS factor biology. Strategies have included directly inhibiting the promoter of oncogenic ETS factors; directly targeting specific ETS factor mRNA to prevent the expression of their target genes; directly targeting the ETS protein itself or indirectly targeting ETS responsive promoters of transcriptional target genes. See text for details. RNAi, RNA interference; miRNA, microRNA; GA, gambogic acid nanoparticles; ?, direct versus indirect effect; DN, dominant negative; ETS, E26 transforming sequence; ODN, decoy oligonucleotide; SMI, small-molecule inhibitor; TFO, triplex-forming oligonucleotide.
9.3. Therapeutic drugs to target ETS factor regulation
A growing body of evidence shows that many of the drugs being tested for cancer treatment alter the activity of ETS factors and provide further rationale for developing ETS targeting therapeutics. The polyphenolic flavone Luteolin demonstrates antitumor activity in several types of cancer. In prostate cancer, Luteolin treatment has been show to increase SPDEF expression while decreasing AR expression to lower the levels of prostate-specific antigen (Tsui, Chung, Feng, Chang, & Juang, 2012). Increased SPDEF expression in this model induced BTG2, NDRG1, and Maspin gene expression to inhibit proliferation and induce apoptosis. A recent study demonstrated that the therapeutic blockade of angiotensin II type1 receptor (AGTR1) inhibits CRPC through the inhibition of ETS1 (Kosaka et al., 2010). Knockdown of AGTR1 inhibits cell proliferation and influences AR expression levels in prostate cancer cells. Wang et al. has successfully used a nanoparticle drug delivery system to deliver the natural extract gambogic acid (GA) into pancreatic cancer cells (Wang, Zhang, Chen, Shi, & Chen, 2012). GA is a potent anticancer agent that inhibits cell growth and the ability for motile function in a wide variety of tumor cells. GA-treated cells show significantly decreased expression of ETS1 as well as its downstream target genes cyclin D1, uPA, and VEGF (Wang et al., 2012).
9.4. Translational ETS targeting
While these studies demonstrate the potential of targeting ETS factors for therapeutic gain, translating this success to the bedside has been limited by the challenges of directly targeting transcriptional factors in complex physiological systems. Such challenges include successfully targeting their nuclear localization and the successful design of small-molecule inhibitors given the large surface area of their DNA–protein and protein–protein interactions (Konstantinopoulos & Papavassiliou, 2011). However, the potential benefits of being able to target transcription factors have been demonstrated for the nuclear hormone-receptor family (e.g. estrogen receptor targeting by tamoxifen in breast cancer patients) as they can be targeted on the cell surface before translocation to the nucleus (Jordan, 2003). The notion that transcription factors represent undruggable targets is slowly being eroded. Technological advances in drug delivery systems and drug design provide insight into how we may overcome these challenges (Konstantinopoulos & Papavassiliou, 2011). Recently, a cell-penetrating synthetic peptide has been developed to disrupt the ERG–DNA interaction in prostate cancer. The peptide is a potent inhibitor of ERG in vitro and in vivo to inhibit DNA damage, cell growth, invasion, and metastasis. Further structure/function studies are being performed to allow further optimization of the peptide (http://www.faqs.org/patents/app/20110207675). The transcriptional activity of ETS factors themselves is regulated by multiple ligands, coregulatory proteins, transcriptional cofactors, and chromatin remodeling components, which determine not only the expression status of a target gene but also the magnitude and duration of activation or repression. The binding sites of several other key transcription factors are found adjacent to ETS binding sites (e.g. ETS/AP-1, ETS/NFκB, ETS/AR) and such composite binding sites mediate the synergistic activation or repression of target genes (Turner & Watson, 2008). The ETS protein:cofactor binding interface is therefore crucial in the regulation of DNA binding, subcellular localization, target gene specificity, and transcriptional activity. The in-depth knowledge of both the overall structure and functional domains of specific ETS family members, including SPDEF (Y. Wang, Feng, et al., 2005) and ETS1 (G. M. Lee et al., 2005), makes targeting their interactions particularly attractive for drug discovery using small-molecule inhibitors. A series of small-molecule inhibitors have been shown to inhibit the interaction between the ETS factor ELF3 and its coactivator Sur2 (a RAS-linked subunit of the mediator complex). ELF3 with its coactivator Sur2 are required for the high HER2 promoter activation observed in cancer. The ELF3-Sur2 protein interaction is mediated by one face of an eight-amino acid alpha-helical region in the ELF3 activation domain (Shimogawa et al., 2004). Screening of an indole-mimicking π-electron-rich chemical library led to the development of wrenchnolol, a small inhibitor that mimics the alpha-helical region of the ELF3 activation domain (Shimogawa et al., 2004). In vivo studies using wrenchnolol in mice have been reported to be promising (Jung, Choi, & Uesugi, 2006). Secreted alkaline phosphatase screening has also identified a fluoroquinophenoxazine derivative that also disrupts the ELF3-Sur2 binding interface. A-62176 treatment arrests the cell cycle in the G1 phase via the downregulation of cyclin D1 and the upregulation of p27Kip1 in NCI-N87 gastric cancer cells (Kim et al., 2012). Disruption of the ELF3-Sur2 binding interface in cancer cells impairs the expression of HER2, inhibits HER2-mediated phosphorylation of MAPK/AKT, and restrains the activity of topoisomerase IIa (Kim et al., 2012).
There is a growing interest in developing miRNAs as therapeutic targets due to their ability to regulate multiple genes and networks of proteins. Multiple companies have been formed to exclusively pursue this research goal. One such company is Mirna Therapeutics, Inc., a biopharmaceutical company focused on miRNA-directed oncology therapies (http://mirnatherapeutics.com). Mirna Therapeutics currently has eight lead compounds in various phases of development with their lead compound, MRX34, an miR-34-mimic, due to be the first miRNA replacement therapy in Phase I clinic trials in 2013. This is an exciting area of research and one hopes to see great advances in the very near future.
9.5. Targeting ETS fusion proteins
ETS fusion proteins offer unique therapeutic targets as they are found only in certain cancer types and not in normal cells. Ewing’s sarcoma ETS family fusion, EWS–FLI1, has been targeted using siRNA leading to more than 80% reduction in the EWS-FLI11 transcript and cell growth inhibition (Dohjima et al., 2003). Limitations to the use of siRNA approaches in vivo have been addressed by coating siRNA with a cyclodextrin-containing polycation to increase stability and by conjugating transferrin to siRNA to target to transferrin receptor-expressing tumor cells. Such modifications have successfully allowed the systemic delivery of EWS–FLI1 fusion targeting siRNA into a murine model of metastatic Ewing’s sarcoma, inhibiting tumor growth with no observed side effects (Hu-Lieskovan et al., 2005). Recent research has used a gene expression approach to identify a 14-gene expression signature for the attenuation of EWS–FLI1 expression in Ewing’s sarcoma (Stegmaier et al., 2007). The signature was then used to screen a small-molecule library highly enriched for FDA-approved drugs and identified cytosine arabinoside as a modulator of EWS–FLI1 protein expression and this compound reduced cell viability, transformation, and tumor growth in a xenograft model.
Oncogenic ETS fusion proteins are also therapeutic targets which have been targeted using small-molecule inhibitors. EWS-FLI1 fusions in Ewing’s Sarcoma have been targeted using the compound YK-4-279 which inhibits EWS-FLI1 activity, induces apoptosis in cell lines and slows down tumor growth in mouse xenograft models (Barber-Rotenberg et al., 2012; Rahim et al., 2011). The potential of YK-4-279 has also been assessed against ETS fusions found in prostate cancer. It was found to inhibit the biological activity of both ERG and ETV1 in fusion positive cell lines to decrease migration and invasion (Rahim et al., 2011).
Given the successes in targeting ETS transcription factors for therapeutic gain, an intense examination of possible additional therapeutic approaches to target these factors is warranted.
10. CONCLUDING REMARKS
The ETS family is one of a limited number of fundamentally important gene families (Hsu et al., 2004; Turner, Findlay, et al., 2007; Watson, Ascione, & Papas, 1990; Watson et al., 2010). Although considerable work has been done on individual members of this family, little effort have been made in understanding the interrelationships between the family members and thus, why they exist as a family. Progress is being made toward addressing this fundamental question. We are just beginning to define unique versus redundant ETS functions. This will require understanding which proteins interact with each family member, which signal transduction pathways are ETS family members involved in, target genes of each family member, and the roles of ETS regulatory network in oncogenesis, tumor suppression, cell proliferation/death, and differentiation/development. During normal development, ETS factor expression is tightly controlled to regulate many biological processes including cell proliferation, differentiation, hematopoiesis, apoptosis, metastasis, tissue remodeling, angiogenesis, and transformation. In cancer, aberrant ETS factor expression results in the upregulation of genes known to drive cancer and the downregulation of genes known to suppress cancer. It is becoming increasingly evident that cellular context defines the direction and magnitude of response to ETS factors. In order to advance our understanding of the ETS-dependent regulation of cancer progression and metastasis, future studies should be directed toward elucidation of the effects of simultaneous expression of multiple transcription factors on the transcriptome of nonmetastatic and metastatic cancer. Collectively, we are beginning to define the molecular mechanisms that determine which ETS family member will regulate a particular target gene and are developing appropriate approaches to determine which target genes are necessary for ETS-dependent phenotypes.
In summary, while expression and promoter arrays will allow identification of new cancer-associated target genes that are regulated by ETS transcription factors, concomitant molecular studies will increase our understanding of the mechanisms by which ETS transcription factors act as oncogenes and tumor-suppressor genes. The holy grail of any therapeutic cancer regime is the reactivation of tumor-suppressor function and/or the inhibition of oncogene activation. Direct or indirect therapeutic intervention of ETS factor function or regulation offers intriguing possibilities in order to achieve this.
ACKNOWLEDGMENTS
We apologize to those researchers whose work could not be cited because of space limitations or was only cited indirectly by referring to reviews or more recent publications.
REFERENCES
- Artlett CM (2010). Animal models of scleroderma: Fresh insights. Current Opinion in Rheumatology, 22, 677–682. [DOI] [PubMed] [Google Scholar]
- Arvand A, & Denny CT (2001). Biology of EWS/ETS fusions in Ewing’s family tumors. Oncogene, 20, 5747–5754. [DOI] [PubMed] [Google Scholar]
- Asano Y, Bujor AM, & Trojanowska M (2010). The impact of Fli1 deficiency on the pathogenesis of systemic sclerosis. Journal of Dermatological Science, 59, 153–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano Y, Czuwara J, & Trojanowska M (2007). Transforming growth factor-beta regulates DNA binding activity of transcription factor Fli1 by p300/CREB-binding protein-associated factor-dependent acetylation. Journal of Biological Chemistry, 282, 34672–34683. [DOI] [PubMed] [Google Scholar]
- Asano Y, Markiewicz M, Kubo M, Szalai G, Watson DK, & Trojanowska M (2009). Transcription factor Fli1 regulates collagen fibrillogenesis in mouse skin. Molecular and Cellular Biology, 29, 425–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano Y, Stawski L, Hant F, Highland K, Silver R, Szalai G, et al. (2010). Endothelial Fli1 deficiency impairs vascular homeostasis: A role in scleroderma vasculopathy. The American Journal of Pathology, 176, 1983–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano Y, & Trojanowska M (2009). Phosphorylation of Fli1 at threonine 312 by protein kinase C delta promotes its interaction with p300/CREB-binding protein-associated factor and subsequent acetylation in response to transforming growth factor beta. Molecular and Cellular Biology, 29, 1882–1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayadi A, Zheng H, Sobieszczuk P, Buchwalter G, Moerman P, Alitalo K, et al. (2001). Net-targeted mutant mice develop a vascular phenotype and up-regulate egr-1. EMBO Journal, 20, 5139–5152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baillat D, Begue A, Stehelin D, & Aumercier M (2002). ETS-1 transcription factor binds cooperatively to the palindromic head to head ETS-binding sites of the stromelysin-1 promoter by counteracting autoinhibition. Journal of Biological Chemistry, 277, 29386–29398. [DOI] [PubMed] [Google Scholar]
- Ban J, Jug G, Mestdagh P, Schwentner R, Kauer M, Aryee DN, et al. (2011). Hsamir-145 is the top EWS-FLI1-repressed microRNA involved in a positive feedback loop in Ewing’s sarcoma. Oncogene, 30, 2173–2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber-Rotenberg JS, Selvanathan SP, Kong Y, Erkizan HV, Snyder TM, Hong SP, et al. (2012). Single enantiomer of YK-4-279 demonstrates specificity in targeting the oncogene EWS-FLI1. Oncotarget, 3, 172–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batliner J, Buehrer E, Federzoni EA, Jenal M, Tobler A, Torbett BE, et al. (2012). Transcriptional regulation of MIR29B by PU.1 (SPI1) and MYC during neutrophil differentiation of acute promyelocytic leukaemia cells. British Journal of Haematology, 157, 270–274. [DOI] [PubMed] [Google Scholar]
- Behrens P, Mathiak M, Mangold E, Kirdorf S, Wellmann A, Fogt F, et al. (2003). Stromal expression of invasion-promoting, matrix-degrading proteases MMP-1 and -9 and the Ets 1 transcription factor in HNPCC carcinomas and sporadic colorectal cancers. International Journal of Cancer, 107, 183–188. [DOI] [PubMed] [Google Scholar]
- Behrens P, Rothe M, Florin A, Wellmann A, & Wernert N (2001). Invasive properties of serous human epithelial ovarian tumors are related to Ets-1, MMP-1 and MMP-9 expression. International Journal of Molecular Medicine, 8, 149–154. [DOI] [PubMed] [Google Scholar]
- Behrens P, Rothe M, Wellmann A, Krischler J, & Wernert N (2001). The Ets-1 transcription factor is up-regulated together with MMP 1 and MMP 9 in the stroma of pre-invasive breast cancer. The Journal of Pathology, 194, 43–50. [DOI] [PubMed] [Google Scholar]
- Benbow U, Tower GB, Wyatt CA, Buttice G, & Brinckerhoff CE (2002). High levels of MMP-1 expression in the absence of the 2G single nucleotide polymorphism is mediated by p38 and ERK1/2 mitogen-activated protein kinases in VMM5 melanoma cells. Journal of Cellular Biochemistry, 86, 307–319. [DOI] [PubMed] [Google Scholar]
- Benz CC, O’Hagan RC, Richter B, Scott GK, Chang CH, Xiong X, et al. (1997). HER2/Neu and the Ets transcription activator PEA3 are coordinately upregulated in human breast cancer. Oncogene, 15, 1513–1525. [DOI] [PubMed] [Google Scholar]
- Blair DG, & Athanasiou M (2000). Ets and retroviruses—Transduction and activation of members of the Ets oncogene family in viral oncogenesis. Oncogene, 19, 6472–6481. [DOI] [PubMed] [Google Scholar]
- Bosc DG, Goueli BS, & Janknecht R (2001). HER2/Neu-mediated activation of the ETS transcription factor ER81 and its target gene MMP-1. Oncogene, 20, 6215–6224. [DOI] [PubMed] [Google Scholar]
- Bronisz A, Godlewski J, Wallace JA, Merchant AS, Nowicki MO, Mathsyaraja H, et al. (2012). Reprogramming of the tumour microenvironment by stromal PTEN-regulated miR-320. Nature Cell Biology, 14, 159–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchwalter G, Gross C, & Wasylyk B (2004). Ets ternary complex transcription factors. Gene, 324, 1–14. [DOI] [PubMed] [Google Scholar]
- Buggy Y, Maguire TM, McDermott E, Hill AD, O’Higgins N, & Duffy MJ (2006). Ets2 transcription factor in normal and neoplastic human breast tissue. European Journal of Cancer, 42, 485–491. [DOI] [PubMed] [Google Scholar]
- Buggy Y, Maguire TM, McGreal G, McDermott E, Hill AD, O’Higgins N, et al. (2004). Overexpression of the Ets-1 transcription factor in human breast cancer. British Journal of Cancer, 91, 1308–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui TV, & Mendell JT (2010). Myc: Maestro of microRNAs. Genes & Cancer, 1, 568–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone GM, McGuffie EM, Collier A, & Catapano CV (2003). Selective inhibition of transcription of the Ets2 gene in prostate cancer cells by a triplex-forming oligonucleotide. Nucleic Acids Research, 31, 833–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone GM, Napoli S, Valentini A, Cavalli F, Watson DK, & Catapano CV (2004). Triplex DNA-mediated downregulation of Ets2 expression results in growth inhibition and apoptosis in human prostate cancer cells. Nucleic Acids Research, 32, 4358–4367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesari F, Brecht S, Vintersten K, Vuong LG, Hofmann M, Klingel K, et al. (2004). Mice deficient for the ets transcription factor elk-1 show normal immune responses and mildly impaired neuronal gene activation. Molecular and Cellular Biology, 24, 294–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti R, Hwang J, Andres Blanco M, Wei Y, Lukacisin M, Romano RA, et al. (2012). Elf5 inhibits the epithelial-mesenchymal transition in mammary gland development and breast cancer metastasis by transcriptionally repressing Snail2. Nature Cell Biology, 14, 1212–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti SR, & Nucifora G (1999). The leukemia-associated gene TEL encodes a transcription repressor which associates with SMRT and mSin3A. Biochemical and Biophysical Research Communications, 264, 871–877. [DOI] [PubMed] [Google Scholar]
- Chakrabarti SR, Sood R, Ganguly S, Bohlander S, Shen Z, & Nucifora G (1999). Modulation of TEL transcription activity by interaction with the ubiquitin-conjugating enzyme UBC9. Proceedings of the National Academy of Sciences of the United States of America, 96, 7467–7472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti R, Wei Y, Romano RA, DeCoste C, Kang Y, & Sinha S (2012). Elf5 regulates mammary gland stem/progenitor cell fate by influencing notch signaling. Stem Cells, 30, 1496–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CH, Scott GK, Kuo WL, Xiong X, Suzdaltseva Y, Park JW, et al. (1997). ESX: A structurally unique Ets overexpressed early during human breast tumorigenesis. Oncogene, 14, 1617–1622. [DOI] [PubMed] [Google Scholar]
- Charlot C, Dubois-Pot H, Serchov T, Tourrette Y, & Wasylyk B (2010). A review of post-translational modifications and subcellular localization of Ets transcription factors: Possible connection with cancer and involvement in the hypoxic response. Methods in Molecular Biology, 647, 3–30. [DOI] [PubMed] [Google Scholar]
- Chen C, Ouyang W, Grigura V, Zhou Q, Carnes K, Lim H, et al. (2005). ERM is required for transcriptional control of the spermatogonial stem cell niche. Nature, 436, 1030–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, & Rajewsky N (2006). Deep conservation of microRNA-target relationships and 3’UTR motifs in vertebrates, flies, and nematodes. Cold Spring Harbor Symposia on Quantitative Biology, 71, 149–156. [DOI] [PubMed] [Google Scholar]
- Choi HJ, Geng Y, Cho H, Li S, Giri PK, Felio K, et al. (2011). Differential requirements for the Ets transcription factor Elf-1 in the development of NKT cells and NK cells. Blood, 117, 1880–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou YT, Lin HH, Lien YC, Wang YH, Hong CF, Kao YR, et al. (2010). EGFR promotes lung tumorigenesis by activating miR-7 through a Ras/ERK/Myc pathway that targets the Ets2 transcriptional repressor ERF. Cancer Research, 70, 8822–8831. [DOI] [PubMed] [Google Scholar]
- Christoffersen NR, Shalgi R, Frankel LB, Leucci E, Lees M, Klausen M, et al. (2010). p53-independent upregulation of miR-34a during oncogene-induced senescence represses MYC. Cell Death and Differentiation, 17, 236–245. [DOI] [PubMed] [Google Scholar]
- Chun CZ, Remadevi I, Schupp MO, Samant GV, Pramanik K, Wilkinson GA, et al. (2011). Fli+ etsrp+ hemato-vascular progenitor cells proliferate at the lateral plate mesoderm during vasculogenesis in zebrafish. PLoS One, 6, e14732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciau-Uitz A, Pinheiro P, Gupta R, Enver T, & Patient R (2010). Tel1/ETV6 specifies blood stem cells through the agency of VEGF signaling. Developmental Cell, 18, 569–578. [DOI] [PubMed] [Google Scholar]
- Colucci F, Samson SI, DeKoter RP, Lantz O, Singh H, & Di Santo JP (2001). Differential requirement for the transcription factor PU.1 in the generation of natural killer cells versus B and T cells. Blood, 97, 2625–2632. [DOI] [PubMed] [Google Scholar]
- Coskun E, von der Heide EK, Schlee C, Kuhnl A, Gokbuget N, Hoelzer D, et al. (2011). The role of microRNA-196a and microRNA-196b as ERG regulators in acute myeloid leukemia and acute T-lymphoblastic leukemia. Leukemia Research, 35, 208–213. [DOI] [PubMed] [Google Scholar]
- Costello PS, Nicolas RH, Watanabe Y, Rosewell I, & Treisman R (2004). Ternary complex factor SAP-1 is required for Erk-mediated thymocyte positive selection. Nature Immunology, 5, 289–298. [DOI] [PubMed] [Google Scholar]
- Costello P, Nicolas R, Willoughby J, Wasylyk B, Nordheim A, & Treisman R (2010). Ternary complex factors SAP-1 and Elk-1, but not net, are functionally equivalent in thymocyte development. Journal of Immunology, 185, 1082–1092. [DOI] [PubMed] [Google Scholar]
- Cowden Dahl KD, Dahl R, Kruichak JN, & Hudson LG (2009). The epidermal growth factor receptor responsive miR-125a represses mesenchymal morphology in ovarian cancer cells. Neoplasia, 11, 1208–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowden Dahl KD, Symowicz J, Ning Y, Gutierrez E, Fishman DA, Adley BP, et al. (2008). Matrix metalloproteinase 9 is a mediator of epidermal growth factor-dependent e-cadherin loss in ovarian carcinoma cells. Cancer Research, 68, 4606–4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czuwara-Ladykowska J, Sementchenko VI, Watson DK, & Trojanowska M (2002). Ets1 is an effector of the transforming growth factor Beta (tgf-Beta) signaling pathway and an antagonist of the profibrotic effects of tgf-Beta. Journal of Biological Chemistry, 277, 20399–20408. [DOI] [PubMed] [Google Scholar]
- Czuwara-Ladykowska J, Shirasaki F, Jackers P, Watson DK, & Trojanowska M (2001). Fli-1 inhibits collagen type I production in dermal fibroblasts via an Sp1-dependent pathway. Journal of Biological Chemistry, 276, 20839–20848. [DOI] [PubMed] [Google Scholar]
- Darnell JE (2002). Transcription factors as targets for cancer therapy. Nature Reviews. Cancer, 2, 740–749. [DOI] [PubMed] [Google Scholar]
- Delattre O, Zucman J, Plougastel B, Desmaze C, Melot T, Peter M, et al. (1992). Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature, 359, 162–165. [DOI] [PubMed] [Google Scholar]
- de Lau W, Kujala P, Schneeberger K, Middendorp S, Li VS, Barker N, et al. (2012). Peyer’s patch M cells derived from Lgr5(+) stem cells require SpiB and are induced by RankL in cultured “miniguts” Molecular and Cellular Biology, 32, 3639–3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vito C, Riggi N, Suva ML, Janiszewska M, Horlbeck J, Baumer K, et al. (2011). Let-7a is a direct EWS-FLI-1 target implicated in Ewing’s sarcoma development. PLoS One, 6, e23592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diakos C, Zhong S, Xiao Y, Zhou M, Vasconcelos GM, Krapf G, et al. (2010). TEL-AML1 regulation of survivin and apoptosis via miRNA-494 and miRNA-320a. Blood, 116, 4885–4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiTacchio L, Bowles J, Shin S, Lim DS, Koopman P, & Janknecht R (2012). Transcription factors ER71/ETV2 and SOX9 participate in a positive feedback loop in fetal and adult mouse testis. Journal of Biological Chemistry, 287, 23657–23666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doane AS, Danso M, Lal P, Donaton M, Zhang L, Hudis C, et al. (2006). An estrogen receptor-negative breast cancer subset characterized by a hormonally regulated transcriptional program and response to androgen. Oncogene, 25, 3994–4008. [DOI] [PubMed] [Google Scholar]
- Dohjima T, Lee NS, Li H, Ohno T, & Rossi JJ (2003). Small interfering RNAs expressed from a Pol III promoter suppress the EWS/Fli-1 transcript in an Ewing sarcoma cell line. Molecular Therapy, 7, 811–816. [DOI] [PubMed] [Google Scholar]
- Eichner LJ, Perry MC, Dufour CR, Bertos N, Park M, St-Pierre J, et al. (2010). miR-378(*) mediates metabolic shift in breast cancer cells via the PGC-1beta/ERRgamma transcriptional pathway. Cell Metabolism, 12, 352–361. [DOI] [PubMed] [Google Scholar]
- Elvert G, Kappel A, Heidenreich R, Englmeier U, Lanz S, Acker T, et al. (2003). Cooperative interaction of hypoxia-inducible factor-2alpha (HIF-2alpha) and Ets-1 in the transcriptional activation of vascular endothelial growth factor receptor-2 (Flk-1). Journal of Biological Chemistry, 278, 7520–7530. [DOI] [PubMed] [Google Scholar]
- Farnham PJ (2009). Insights from genomic profiling of transcription factors. Nature Reviews. Genetics, 10, 605–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman RJ, Sementchenko VI, Gayed M, Fraig MM, & Watson DK (2003). Pdef expression in human breast cancer is correlated with invasive potential and altered gene expression. Cancer Research, 63, 4626–4631. [PubMed] [Google Scholar]
- Feldman RJ, Sementchenko VI, & Watson DK (2003). The epithelial-specific Ets factors occupy a unique position in defining epithelial proliferation, differentiation and carcinogenesis. Anticancer Research, 23, 2125–2131. [PubMed] [Google Scholar]
- Feng W, Xing D, Hua P, Zhang Y, Chen YF, Oparil S, et al. (2010). The transcription factor ETS-1 mediates proinflammatory responses and neointima formation in carotid artery endoluminal vascular injury. Hypertension, 55, 1381–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferdous A, Caprioli A, Iacovino M, Martin CM, Morris J, Richardson JA, et al. (2009). Nkx2-5 transactivates the Ets-related protein 71 gene and specifies an endothelial/endocardial fate in the developing embryo. Proceedings of the National Academy of Sciences of the United States of America, 106, 814–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findlay VJ, Turner DP, Moussa O, & Watson DK (2008). MicroRNA-mediated inhibition of prostate-derived Ets factor messenger RNA translation affects prostate-derived Ets factor regulatory networks in human breast cancer. Cancer Research, 68, 8499–8506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findlay VJ, Turner DP, Yordy JS, McCarragher B, Shriver MR, Szalai G, et al. (2011). Prostate-derived ETS factor regulates epithelial-to-mesenchymal transition through both slug-dependent and independent mechanisms. Genes & Cancer, 2, 120–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RJ, Fivash M, Casas-Finet J, Erickson JW, Kondoh A, Bladen SV, et al. (1994). Real-time DNA binding measurements of the ETS1 recombinant oncoproteins reveal significant kinetic differences between the p42 and p51 isoforms. Protein Science, 3, 257–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzsimmons D, Lutz R, Wheat W, Chamberlin HM, & Hagman J (2001). Highly conserved amino acids in Pax and Ets proteins are required for DNA binding and ternary complex assembly. Nucleic Acids Research, 29, 4154–4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flentjar N, Chu PY, Ng AY, Johnstone CN, Heath JK, Ernst M, et al. (2007). TGF-betaRII rescues development of small intestinal epithelial cells in Elf3-deficient mice. Gastroenterology, 132, 1410–1419. [DOI] [PubMed] [Google Scholar]
- Foulds CE, Nelson ML, Blaszczak AG, & Graves BJ (2004). Ras/mitogen-activated protein kinase signaling activates Ets-1 and Ets-2 by CBP/p300 recruitment. Molecular and Cellular Biology, 24, 10954–10964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzetti GA, Laud-Duval K, Bellanger D, Stern MH, Sastre-Garau X, & Delattre O (2012). MiR-30a-5p connects EWS-FLI1 and CD99, two major therapeutic targets in Ewing tumor. Oncogene, 10.1038/onc.2012.403. [DOI] [PubMed] [Google Scholar]
- Galang CK, Muller WJ, Foos G, Oshima RG, & Hauser CA (2004). Changes in the expression of many Ets family transcription factors and of potential target genes in normal mammary tissue and tumors. Journal of Biological Chemistry, 279, 11281–11292. [DOI] [PubMed] [Google Scholar]
- Garrett-Sinha LA, Su GH, Rao S, Kabak S, Hao Z, Clark MR, et al. (1999). PU.1 and Spi-B are required for normal B cell receptor-mediated signal transduction. Immunity, 10, 399–408. [DOI] [PubMed] [Google Scholar]
- Garvie CW, Pufall MA, Graves BJ, & Wolberger C (2002). Structural analysis of the autoinhibition of Ets-1 and its role in protein partnerships. Journal of Biological Chemistry, 277, 45529–45536. [DOI] [PubMed] [Google Scholar]
- Gavrilov D, Kenzior O, Evans M, Calaluce R, & Folk WR (2001). Expression of urokinase plasminogen activator and receptor in conjunction with the ets family and AP-1 complex transcription factors in high grade prostate cancers. European Journal of Cancer, 37, 1033–1040. [DOI] [PubMed] [Google Scholar]
- Georgiou P, Maroulakou IG, Green JE, Dantis P, Romano-Spica V, Kottaridis S, et al. (1996). Expression of ets family of genes in systemic lupus erythematosus and Sjogren’s syndrome. International Journal of Oncology, 9, 9–18. [PubMed] [Google Scholar]
- Goel A, & Janknecht R (2003). Acetylation-mediated transcriptional activation of the ETS protein ER81 by p300, P/CAF, and HER2/Neu. Molecular and Cellular Biology, 23, 6243–6254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golub TR, Barker GF, Lovett M, & Gilliland DG (1994). Fusion of PDGF receptor beta to a novel ets-like gene, tel, in chronic myelomonocytic leukemia with t(5; 12) chromosomal translocation. Cell, 77, 307–316. [DOI] [PubMed] [Google Scholar]
- Guarino M, Rubino B, & Ballabio G (2007). The role of epithelial-mesenchymal transition in cancer pathology. Pathology, 39, 305–318. [DOI] [PubMed] [Google Scholar]
- Gutierrez-Hartmann A, Duval DL, & Bradford AP (2007). ETS transcription factors in endocrine systems. Trends in Endocrinology and Metabolism, 18, 150–158. [DOI] [PubMed] [Google Scholar]
- Hahne JC, Fuchs T, El Mustapha H, Okuducu AF, Bories JC, & Wernert N (2006). Expression pattern of matrix metalloproteinase and TIMP genes in fibroblasts derived from Ets-1 knock-out mice compared to wild-type mouse fibroblasts. International Journal of Molecular Medicine, 18, 153–159. [PubMed] [Google Scholar]
- Hahne JC, Fuchs T, Florin A, Edwards D, Pourtier A, Soncin F, et al. (2011). Evaluation of effects caused by differentially spliced Ets-1 transcripts in fibroblasts. International Journal of Oncology, 39, 1073–1082. [DOI] [PubMed] [Google Scholar]
- Hahne JC, Okuducu AF, Fuchs T, Florin A, & Wernert N (2011). Identification of ETS-1 target genes in human fibroblasts. International Journal of Oncology, 38, 1645–1652. [DOI] [PubMed] [Google Scholar]
- Hanahan D, & Coussens LM (2012). Accessories to the crime: Functions of cells recruited to the tumor microenvironment. Cancer Cell, 21, 309–322. [DOI] [PubMed] [Google Scholar]
- Hanahan D, & Weinberg RA (2000). The hallmarks of cancer. Cell, 100, 57–70. [DOI] [PubMed] [Google Scholar]
- Hanahan D, & Weinberg RA (2011). Hallmarks of cancer: The next generation. Cell, 144, 646–674. [DOI] [PubMed] [Google Scholar]
- Harris TA, Yamakuchi M, Kondo M, Oettgen P, & Lowenstein CJ (2010). Ets-1 and Ets-2 regulate the expression of microRNA-126 in endothelial cells. Arteriosclerosis, Thrombosis, and Vascular Biology, 30, 1990–1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart A, Melet F, Grossfeld P, Chien K, Jones C, Tunnacliffe A, et al. (2000). Fli-1 is required for murine vascular and megakaryocytic development and is hemizygously deleted in patients with thrombocytopenia. Immunity, 13, 167–177. [DOI] [PubMed] [Google Scholar]
- He L, He X, Lowe SW, & Hannon GJ (2007). microRNAs join the p53 network—Another piece in the tumour-suppression puzzle. Nature Reviews. Cancer, 7, 819–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendricks TJ, Fyodorov DV, Wegman LJ, Lelutiu NB, Pehek EA, Yamamoto B, et al. (2003). Pet-1 ETS gene plays a critical role in 5-HT neuron development and is required for normal anxiety-like and aggressive behavior. Neuron, 37, 233–247. [DOI] [PubMed] [Google Scholar]
- Hewett PW, Daft EL, Laughton CA, Ahmad S, Ahmed A, & Murray JC (2006). Selective inhibition of the human tie-1 promoter with triplex-forming oligonucleotides targeted to Ets binding sites. Molecular Medicine, 12, 8–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hikami K, Kawasaki A, Ito I, Koga M, Ito S, Hayashi T, et al. (2011). Association of a functional polymorphism in the 3’-untranslated region of SPI1 with systemic lupus erythematosus. Arthritis and Rheumatism, 63, 755–763. [DOI] [PubMed] [Google Scholar]
- Hollenhorst PC, Chandler KJ, Poulsen RL, Johnson WE, Speck NA, & Graves BJ (2009). DNA specificity determinants associate with distinct transcription factor functions. PLoS Genetics, 5, e1000778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenhorst PC, Ferris MW, Hull MA, Chae H, Kim S, & Graves BJ (2011). Oncogenic ETS proteins mimic activated RAS/MAPK signaling in prostate cells. Genes & Development, 25, 2147–2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenhorst PC, Jones DA, & Graves BJ (2004). Expression profiles frame the promoter specificity dilemma of the ETS family of transcription factors. Nucleic Acids Research, 32, 5693–5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenhorst PC, McIntosh LP, & Graves BJ (2011). Genomic and biochemical insights into the specificity of ETS transcription factors. Annual Review of Biochemistry, 80, 437–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenhorst PC, Shah AA, Hopkins C, & Graves BJ (2007). Genome-wide analyses reveal properties of redundant and specific promoter occupancy within the ETS gene family. Genes & Development, 21, 1882–1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horst D, Gu X, Bhasin M, Yang Q, Verzi M, Lin D, et al. (2010). Requirement of the epithelium-specific Ets transcription factor Spdef for mucous gland cell function in the gastric antrum. Journal of Biological Chemistry, 285, 35047–35055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu T, Trojanowska M, & Watson DK (2004). Ets proteins in biological control and cancer. Journal of Cellular Biochemistry, 91, 896–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Van Arsdall M, Tedjarati S, McCarty M, Wu W, Langley R, et al. (2002). Contributions of stromal metalloproteinase-9 to angiogenesis and growth of human ovarian carcinoma in mice. Journal of the National Cancer Institute, 94, 1134–1142. [DOI] [PubMed] [Google Scholar]
- Hu-Lieskovan S, Heidel JD, Bartlett DW, Davis ME, & Triche TJ (2005). Sequence-specific knockdown of EWS-FLI1 by targeted, nonviral delivery of small interfering RNA inhibits tumor growth in a murine model of metastatic Ewing’s sarcoma. Cancer Research, 65, 8984–8992. [DOI] [PubMed] [Google Scholar]
- Itoh T, Ando M, Tsukamasa Y, & Akao Y (2012). Expression of BMP-2 and Ets1 in BMP-2-stimulated mouse pre-osteoblast differentiation is regulated by microRNA-370. FEBS Letters, 586, 1693–1701. [DOI] [PubMed] [Google Scholar]
- Itoh T, Takeda S, & Akao Y (2010). MicroRNA-208 modulates BMP-2-stimulated mouse preosteoblast differentiation by directly targeting V-ets erythroblastosis virus E26 oncogene homolog 1. Journal of Biological Chemistry, 285, 27745–27752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackers P, Szalai G, Moussa O, & Watson DK (2004). Ets-dependent regulation of target gene expression during megakaryopoiesis. Journal of Biological Chemistry, 279, 52183–52190. [DOI] [PubMed] [Google Scholar]
- Jinnin M, Ihn H, Asano Y, Yamane K, Trojanowska M, & Tamaki K (2006). Upregulation of tenascin-C expression by IL-13 in human dermal fibroblasts via the phosphoinositide 3-kinase/Akt and the protein kinase C signaling pathways. The Journal of Investigative Dermatology, 126, 551–560. [DOI] [PubMed] [Google Scholar]
- Jinnin M, Ihn H, Mimura Y, Asano Y, Yamane K, & Tamaki K (2005). Matrix metalloproteinase-1 up-regulation by hepatocyte growth factor in human dermal fibroblasts via ERK signaling pathway involves Ets1 and Fli1. Nucleic Acids Research, 33, 3540–3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinnin M, Ihn H, Yamane K, Mimura Y, Asano Y, & Tamaki K (2005). Alpha2(I) collagen gene regulation by protein kinase C signaling in human dermal fibroblasts. Nucleic Acids Research, 33, 1337–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson TR, Koul S, Kumar B, Khandrika L, Venezia S, Maroni PD, et al. (2010). Loss of PDEF, a prostate-derived Ets factor is associated with aggressive phenotype of prostate cancer: Regulation of MMP 9 by PDEF. Molecular Cancer, 9, 148. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Jordan VC (2003). Tamoxifen: A most unlikely pioneering medicine. Nature Reviews. Drug Discovery, 2, 205–213. [DOI] [PubMed] [Google Scholar]
- Jung D, Choi Y, & Uesugi M (2006). Small organic molecules that modulate gene transcription. Drug Discovery Today, 11, 452–457. [DOI] [PubMed] [Google Scholar]
- Kanaya T, Hase K, Takahashi D, Fukuda S, Hoshino K, Sasaki I, et al. (2012). The Ets transcription factor Spi-B is essential for the differentiation of intestinal microfold cells. Nature Immunology, 13, 729–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnoub AE, Dash AB, Vo AP, Sullivan A, Brooks MW, Bell GW, et al. (2007). Mesenchymal stem cells within tumour stroma promote breast cancer metastasis. Nature, 449, 557–563. [DOI] [PubMed] [Google Scholar]
- Kars MD, Iseri OD, & Gunduz U (2010). Drug resistant breast cancer cells overexpress ETS1 gene. Biomedicine & Pharmacotherapy, 64, 458–462. [DOI] [PubMed] [Google Scholar]
- Kawada H, Ito T, Pharr PN, Spyropoulos DD, Watson DK, & Ogawa M (2001). Defective megakaryopoiesis and abnormal erythroid development in Fli-1 gene-targeted mice. International Journal of Hematology, 73, 463–468. [DOI] [PubMed] [Google Scholar]
- Kern HB, Niemeyer BF, Parrish JK, Kerr CA, Yaghi NK, Prescott JD, et al. (2012). Control of MicroRNA-21 expression in colorectal cancer cells by oncogenic epidermal growth factor/Ras signaling and Ets transcription factors. DNA and Cell Biology, 31, 1403–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HL, Jeon KH, Jun KY, Choi Y, Kim DK, Na Y, et al. (2012). A-62176, a potent topoisomerase inhibitor, inhibits the expression of human epidermal growth factor receptor 2. Cancer Letters, 325, 72–79. [DOI] [PubMed] [Google Scholar]
- Kitange G, Shibata S, Tokunaga Y, Yagi N, Yasunaga A, Kishikawa M, et al. (1999). Ets-1 transcription factor-mediated urokinase-type plasminogen activator expression and invasion in glioma cells stimulated by serum and basic fibroblast growth factors. Laboratory Investigation, 79, 407–416. [PubMed] [Google Scholar]
- Kohyama M, Ise W, Edelson BT, Wilker PR, Hildner K, Mejia C, et al. (2009). Role for Spi-C in the development of red pulp macrophages and splenic iron homeostasis. Nature, 457, 318–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantinopoulos PA, & Papavassiliou AG (2011). Seeing the future of cancer-associated transcription factor drug targets. JAMA: The Journal of the American Medical Association, 305, 2349–2350. [DOI] [PubMed] [Google Scholar]
- Kosaka T, Miyajima A, Shirotake S, Kikuchi E, Hasegawa M, Mikami S, et al. (2010). Ets-1 and hypoxia inducible factor-1alpha inhibition by angiotensin II type-1 receptor blockade in hormone-refractory prostate cancer. The Prostate, 70, 162–169. [DOI] [PubMed] [Google Scholar]
- Kruse EA, Loughran SJ, Baldwin TM, Josefsson EC, Ellis S, Watson DK, et al. (2009). Dual requirement for the ETS transcription factors Fli-1 and Erg in hematopoietic stem cells and the megakaryocyte lineage. Proceedings of the National Academy of Sciences of the United States of America, 106, 13814–13819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo M, Czuwara-Ladykowska J, Moussa O, Markiewicz M, Smith E, Silver RM, et al. (2003). Persistent down-regulation of Fli1, a suppressor of collagen transcription, in fibrotic scleroderma skin. The American Journal of Pathology, 163, 571–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar MS, Narla A, Nonami A, Mullally A, Dimitrova N, Ball B, et al. (2011). Coordinate loss of a microRNA and protein-coding gene cooperate in the pathogenesis of 5q- syndrome. Blood, 118, 4666–4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar-Sinha C, Tomlins SA, & Chinnaiyan AM (2008). Recurrent gene fusions in prostate cancer. Nature Reviews. Cancer, 8, 497–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacorazza HD, Miyazaki Y, Di Cristofano A, Deblasio A, Hedvat C, Zhang J, et al. (2002). The ETS protein MEF plays a critical role in perforin gene expression and the development of natural killer and NK-T cells. Immunity, 17, 437–449. [DOI] [PubMed] [Google Scholar]
- Larsson E, Fredlund Fuchs P, Heldin J, Barkefors I, Bondjers C, Genove G, et al. (2009). Discovery of microvascular miRNAs using public gene expression data: miR-145 is expressed in pericytes and is a regulator of Fli1. Genome Medicine, 1, 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lautenberger JA, & Papas TS (1993). Inversion of a chicken ets-1 proto-oncogene segment in avian leukemia virus E26. Journal of Virology, 67, 610–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavenburg KR, Ivey J, Hsu T, & Muise-Helmericks RC (2003). Coordinated functions of Akt/PKB and ETS1 in tubule formation. The FASEB Journal, 17, 2278–2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee GM, Donaldson LW, Pufall MA, Kang HS, Pot I, Graves BJ, et al. (2005). The structural and dynamic basis of Ets-1 DNA binding autoinhibition. Journal of Biological Chemistry, 280, 7088–7099. [DOI] [PubMed] [Google Scholar]
- Lee J, Kannagi M, Ferrante RJ, Kowall NW, & Ryu H (2009). Activation of Ets-2 by oxidative stress induces Bcl-xL expression and accounts for glial survival in amyotrophic lateral sclerosis. The FASEB Journal, 23, 1739–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lelievre E, Lionneton F, & Soncin F (2001). Role of the ETS transcription factors in the control of endothelial-specific gene expression and in angiogenesis. Bulletin du Cancer, 88, 137–142. [PubMed] [Google Scholar]
- Lelievre E, Lionneton F, Soncin F, & Vandenbunder B (2001). The Ets family contains transcriptional activators and repressors involved in angiogenesis. The International Journal of Biochemistry & Cell Biology, 33, 391–407. [DOI] [PubMed] [Google Scholar]
- Li R, Pei H, & Watson DK (2000). Regulation of Ets function by protein–protein interactions. Oncogene, 19, 6514–6523. [DOI] [PubMed] [Google Scholar]
- Li R, Pei H, Watson DK, & Papas TS (2000). EAP1/Daxx interacts with ETS1 and represses transcriptional activation of ETS1 target genes. Oncogene, 19, 745–753. [DOI] [PubMed] [Google Scholar]
- Li F, Wallace JA, & Ostrowski MC (2010). ETS transcription factors in the tumor microenvironment. Open Cancer Journal, 3, 49–54. [Google Scholar]
- Liao YL, Hu LY, Tsai KW, Wu CW, Chan WC, Li SC, et al. (2012). Transcriptional regulation of miR-196b by ETS2 in gastric cancer cells. Carcinogenesis, 33, 760–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin EY, Nguyen AV, Russell RG, & Pollard JW (2001). Colony-stimulating factor 1 promotes progression of mammary tumors to malignancy. The Journal of Experimental Medicine, 193, 727–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindemann RK, Braig M, Ballschmieter P, Guise TA, Nordheim A, & Dittmer J (2003). Protein kinase Calpha regulates Ets1 transcriptional activity in invasive breast cancer cells. International Journal of Oncology, 22, 799–805. [PubMed] [Google Scholar]
- Lindemann RK, Braig M, Hauser CA, Nordheim A, & Dittmer J (2003). Ets2 and protein kinase Cepsilon are important regulators of parathyroid hormone-related protein expression in MCF-7 breast cancer cells. Biochemical Journal, 372, 787–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Walmsley M, Rodaway A, & Patient R (2008). Fli1 acts at the top of the transcriptional network driving blood and endothelial development. Current Biology, 18, 1234–1240. [DOI] [PubMed] [Google Scholar]
- Loughran SJ, Kruse EA, Hacking DF, de Graaf CA, Hyland CD, Willson TA, et al. (2008). The transcription factor Erg is essential for definitive hematopoiesis and the function of adult hematopoietic stem cells. Nature Immunology, 9, 810–819. [DOI] [PubMed] [Google Scholar]
- Manavathi B, Rayala SK, & Kumar R (2007). Phosphorylation-dependent regulation of stability and transforming potential of ETS transcriptional factor ESE-1 by p21-activated kinase 1. Journal of Biological Chemistry, 282, 19820–19830. [DOI] [PubMed] [Google Scholar]
- Mardis ER (2007). ChIP-seq: Welcome to the new frontier. Nature Methods, 4, 613–614. [DOI] [PubMed] [Google Scholar]
- Maroni P, Bendinelli P, Matteucci E, & Desiderio MA (2007). HGF induces CXCR4 and CXCL12-mediated tumor invasion through Ets1 and NF-kappaB. Carcinogenesis, 28, 267–279. [DOI] [PubMed] [Google Scholar]
- Massie CE, Adryan B, Barbosa-Morais NL, Lynch AG, Tran MG, Neal DE, et al. (2007). New androgen receptor genomic targets show an interaction with the ETS1 transcription factor. EMBO Reports, 8, 871–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuya M, Moussa O, Abe T, Deguchi T, Higuchi T, Ebihara Y, et al. (2005). Dysregulation of granulocyte, erythrocyte, and NK cell lineages in Fli-1 gene-targeted mice. Blood, 105, 95–102. [DOI] [PubMed] [Google Scholar]
- Mathenia J, Reyes-Cortes E, Williams S, Molano I, Ruiz P, Watson DK, et al. (2010). Impact of Fli-1 transcription factor on autoantibody and lupus nephritis in NZM2410 mice. Clinical and Experimental Immunology, 162, 362–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattot V, Vercamer C, Soncin F, Fafeur V, & Vandenbunder B (1999). Transcription factors of the Ets family and morphogenesis of the vascular tree. Journal de la Societe de Biologie, 193, 147–153. [PubMed] [Google Scholar]
- Mavrothalassitis G, & Ghysdael J (2000). Proteins of the ETS family with transcriptional repressor activity. Oncogene, 19, 6524–6532. [DOI] [PubMed] [Google Scholar]
- McKinsey EL, Parrish JK, Irwin AE, Niemeyer BF, Kern HB, Birks DK, et al. (2011). A novel oncogenic mechanism in Ewing sarcoma involving IGF pathway targeting by EWS/Fli1-regulated microRNAs. Oncogene, 30, 4910–4920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miwa K, Nakashima H, Aoki M, Miyake T, Kawasaki T, Iwai M, et al. (2005). Inhibition of ets, an essential transcription factor for angiogenesis, to prevent the development of abdominal aortic aneurysm in a rat model. Gene Therapy, 12, 1109–1118. [DOI] [PubMed] [Google Scholar]
- Montes AH, Valle-Garay E, Alvarez V, Pevida M, Garcia Perez E, Paz J, et al. (2010). A functional polymorphism in MMP1 could influence osteomyelitis development. Journal of Bone and Mineral Research, 25, 912–919. [DOI] [PubMed] [Google Scholar]
- Morris EE, Amria MY, Kistner-Griffin E, Svenson JL, Kamen DL, Gilkeson GS, et al. (2010). A GA microsatellite in the Fli1 promoter modulates gene expression and is associated with systemic lupus erythematosus patients without nephritis. Arthritis Research & Therapy, 12, R212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouly E, Chemin K, Nguyen HV, Chopin M, Mesnard L, Leite-de-Moraes M, et al. (2010). The Ets-1 transcription factor controls the development and function of natural regulatory T cells. The Journal of Experimental Medicine, 207, 2113–2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moussa O, LaRue AC, Abangan RS Jr., Williams CR, Zhang XK, Masuya M, et al. (2010). Thrombocytopenia in mice lacking the carboxy-terminal regulatory domain of the Ets transcription factor Fli1. Molecular and Cellular Biology, 30, 5194–5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moussa O, Turner DP, Feldman RJ, Sementchenko VI, McCarragher BD, Desouki MM, et al. (2009). PDEF is a negative regulator of colon cancer cell growth and migration. Journal of Cellular Biochemistry, 108, 1389–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers E, Hill AD, Kelly G, McDermott EW, O’Higgins NJ, Buggy Y, et al. (2005). Associations and interactions between Ets-1 and Ets-2 and coregulatory proteins, SRC-1, AIB1, and NCoR in breast cancer. Clinical Cancer Research, 11, 2111–2122. [DOI] [PubMed] [Google Scholar]
- Nakerakanti SS, Kapanadze B, Yamasaki M, Markiewicz M, & Trojanowska M (2006). Fli1 and Ets1 have distinct roles in connective tissue growth factor/CCN2 gene regulation and induction of the profibrotic gene program. Journal of Biological Chemistry, 281, 25259–25269. [DOI] [PubMed] [Google Scholar]
- Nam EJ, Yoon H, Kim SW, Kim H, Kim YT, Kim JH, et al. (2008). MicroRNA expression profiles in serous ovarian carcinoma. Clinical Cancer Research, 14, 2690–2695. [DOI] [PubMed] [Google Scholar]
- Ng AY, Waring P, Ristevski S, Wang C, Wilson T, Pritchard M, et al. (2002). Inactivation of the transcription factor Elf3 in mice results in dysmorphogenesis and altered differentiation of intestinal epithelium. Gastroenterology, 122, 1455–1466. [DOI] [PubMed] [Google Scholar]
- Nguyen HV, Mouly E, Chemin K, Luinaud R, Despres R, Fermand JP, et al. (2012). The Ets-1 transcription factor is required for Stat1-mediated T-bet expression and IgG2a class switching in mouse B cells. Blood, 119, 4174–4181. [DOI] [PubMed] [Google Scholar]
- Nicholson RI, Gee JM, & Harper ME (2001). EGFR and cancer prognosis. European Journal of Cancer, 37(Suppl. 4), S9–S15. [DOI] [PubMed] [Google Scholar]
- Niu Z, Goodyear SM, Rao S, Wu X, Tobias JW, Avarbock MR, et al. (2011). MicroRNA-21 regulates the self-renewal of mouse spermatogonial stem cells. Proceedings of the National Academy of Sciences of the United States of America, 108, 12740–12745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowling TK, Fulton JD, Chike-Harris K, & Gilkeson GS (2008). Ets factors and a newly identified polymorphism regulate Fli1 promoter activity in lymphocytes. Molecular Immunology, 45, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Leary DA, Noakes PG, Lavidis NA, Kola I, Hertzog PJ, & Ristevski S (2007). Targeting of the ETS factor GABPalpha disrupts neuromuscular junction synaptic function. Molecular and Cellular Biology, 27, 3470–3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orimo A, & Weinberg RA (2006). Stromal fibroblasts in cancer: A novel tumor-promoting cell type. Cell Cycle, 5, 1597–1601. [DOI] [PubMed] [Google Scholar]
- Pan HF, Leng RX, Tao JH, Li XP, & Ye DQ (2011). Ets-1: A new player in the pathogenesis of systemic lupus erythematosus? Lupus, 20, 227–230. [DOI] [PubMed] [Google Scholar]
- Pang L, Xue HH, Szalai G, Wang X, Wang Y, Watson DK, et al. (2006). Maturation stage-specific regulation of megakaryopoiesis by pointed-domain Ets proteins. Blood, 108, 2198–2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadaki C, Alexiou M, Cecena G, Verykokakis M, Bilitou A, Cross JC, et al. (2007). Transcriptional repressor erf determines extraembryonic ectoderm differentiation. Molecular and Cellular Biology, 27, 5201–5213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C, Hennig GW, Sanders KM, Cho JH, Hatton WJ, Redelman D, et al. (2011). Serum response factor-dependent MicroRNAs regulate gastrointestinal smooth muscle cell phenotypes. Gastroenterology, 141, 164–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park KS, Korfhagen TR, Bruno MD, Kitzmiller JA, Wan H, Wert SE, et al. (2007). SPDEF regulates goblet cell hyperplasia in the airway epithelium. The Journal of Clinical Investigation, 117, 978–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei H, Yordy JS, Leng Q, Zhao Q, Watson DK, & Li R (2003). EAPII interacts with ETS1 and modulates its transcriptional function. Oncogene, 22, 2699–2709. [DOI] [PubMed] [Google Scholar]
- Pimanda JE, Ottersbach K, Knezevic K, Kinston S, Chan WY, Wilson NK, et al. (2007). Gata2, Fli1, and Scl form a recursively wired gene-regulatory circuit during early hematopoietic development. Proceedings of the National Academy of Sciences of the United States of America, 104, 17692–17697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourtier-Manzanedo A, Vercamer C, Van Belle E, Mattot V, Mouquet F, & Vandenbunder B (2003). Expression of an Ets-1 dominant-negative mutant perturbs normal and tumor angiogenesis in a mouse ear model. Oncogene, 22, 1795–1806. [DOI] [PubMed] [Google Scholar]
- Rahim S, Beauchamp EM, Kong Y, Brown ML, Toretsky JA, & Uren A (2011). YK-4-279 inhibits ERG and ETV1 mediated prostate cancer cell invasion. PLoS One, 6, e19343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajewsky N (2006). microRNA target predictions in animals. Nature Genetics, 38(Suppl.), S8–S13. [DOI] [PubMed] [Google Scholar]
- Raslova H, Komura E, Le Couedic JP, Larbret F, Debili N, Feunteun J, et al. (2004). FLI1 monoallelic expression combined with its hemizygous loss underlies Paris-Trousseau/Jacobsen thrombopenia. The Journal of Clinical Investigation, 114, 77–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggi N, Suva ML, De Vito C, Provero P, Stehle JC, Baumer K, et al. (2010). EWS-FLI-1 modulates miRNA145 and SOX2 expression to initiate mesenchymal stem cell reprogramming toward Ewing sarcoma cancer stem cells. Genes & Development, 24, 916–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romania P, Lulli V, Pelosi E, Biffoni M, Peschle C, & Marziali G (2008). MicroRNA 155 modulates megakaryopoiesis at progenitor and precursor level by targeting Ets-1 and Meis1 transcription factors. British Journal of Haematology, 143, 570–580. [DOI] [PubMed] [Google Scholar]
- Sahin A, Velten M, Pietsch T, Knuefermann P, Okuducu AF, Hahne JC, et al. (2005). Inactivation of Ets 1 transcription factor by a specific decoy strategy reduces rat C6 glioma cell proliferation and mmp-9 expression. International Journal of Molecular Medicine, 15, 771–776. [PubMed] [Google Scholar]
- Salh B, Marotta A, Wagey R, Sayed M, & Pelech S (2002). Dysregulation of phosphatidylinositol 3-kinase and downstream effectors in human breast cancer. International Journal of Cancer, 98, 148–154. [DOI] [PubMed] [Google Scholar]
- Schaefer JS, Sabherwal Y, Shi HY, Sriraman V, Richards J, Minella A, et al. (2010). Transcriptional regulation of p21/CIP1 cell cycle inhibitor by PDEF controls cell proliferation and mammary tumor progression. Journal of Biological Chemistry, 285, 11258–11269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D, Wilson MD, Spyrou C, Brown GD, Hadfield J, & Odom DT (2009). ChIP-seq: Using high-throughput sequencing to discover protein-DNA interactions. Methods, 48, 240–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schotte R, Nagasawa M, Weijer K, Spits H, & Blom B (2004). The ETS transcription factor Spi-B is required for human plasmacytoid dendritic cell development. The Journal of Experimental Medicine, 200, 1503–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sementchenko VI, Schweinfest CW, Papas TS, & Watson DK (1998). ETS2 function is required to maintain the transformed state of human prostate cancer cells. Oncogene, 17, 2883–2888. [DOI] [PubMed] [Google Scholar]
- Sementchenko VI, & Watson DK (2000). Ets target genes: Past, present and future. Oncogene, 19, 6533–6548. [DOI] [PubMed] [Google Scholar]
- Seth A, & Watson DK (2005). ETS transcription factors and their emerging roles in human cancer. European Journal of Cancer, 41, 2462–2478. [DOI] [PubMed] [Google Scholar]
- Shah RB, & Chinnaiyan AM (2009). The discovery of common recurrent transmembrane protease serine 2 (TMPRSS2)-erythroblastosis virus E26 transforming sequence (ETS) gene fusions in prostate cancer: Significance and clinical implications. Advances in Anatomic Pathology, 16, 145–153. [DOI] [PubMed] [Google Scholar]
- Shah S, & Small E (2010). Emerging biological observations in prostate cancer. Expert Review of Anticancer Therapy, 10, 89–101. [DOI] [PubMed] [Google Scholar]
- Shilo S, Roy S, Khanna S, & Sen CK (2007). MicroRNA in cutaneous wound healing: A new paradigm. DNA and Cell Biology, 26, 227–237. [DOI] [PubMed] [Google Scholar]
- Shimogawa H, Kwon Y, Mao Q, Kawazoe Y, Choi Y, Asada S, et al. (2004). A wrench-shaped synthetic molecule that modulates a transcription factor-coactivator interaction. Journal of the American Chemical Society, 126, 3461–3471. [DOI] [PubMed] [Google Scholar]
- Shirasaki F, Makhluf HA, LeRoy C, Watson DK, & Trojanowska M (1999). Ets transcription factors cooperate with Sp1 to activate the human tenascin-C promoter. Oncogene, 18, 7755–7764. [DOI] [PubMed] [Google Scholar]
- Sica A, Larghi P, Mancino A, Rubino L, Porta C, Totaro MG, et al. (2008). Macrophage polarization in tumour progression. Seminars in Cancer Biology, 18, 349–355. [DOI] [PubMed] [Google Scholar]
- Sieweke MH, Tekotte H, Frampton J, & Graf T (1996). MafB is an interaction partner and repressor of Ets-1 that inhibits erythroid differentiation. Cell, 85, 49–69. [DOI] [PubMed] [Google Scholar]
- Soller MJ, Isaksson M, Elfving P, Soller W, Lundgren R, & Panagopoulos I (2006). Confirmation of the high frequency of the TMPRSS2/ERG fusion gene in prostate cancer. Genes, Chromosomes & Cancer, 45, 717–719. [DOI] [PubMed] [Google Scholar]
- Span PN, Manders P, Heuvel JJ, Thomas CM, Bosch RR, Beex LV, et al. (2002). Expression of the transcription factor Ets-1 is an independent prognostic marker for relapse-free survival in breast cancer. Oncogene, 21, 8506–8509. [DOI] [PubMed] [Google Scholar]
- Spyropoulos DD, Pharr PN, Lavenburg KR, Jackers P, Papas TS, Ogawa M, et al. (2000). Hemorrhage, impaired hematopoiesis, and lethality in mouse embryos carrying a targeted disruption of the Fli1 transcription factor. Molecular and Cellular Biology, 20, 5643–5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starck J, Weiss-Gayet M, Gonnet C, Guyot B, Vicat JM, & Morle F (2010). Inducible Fli-1 gene deletion in adult mice modifies several myeloid lineage commitment decisions and accelerates proliferation arrest and terminal erythrocytic differentiation. Blood, 116, 4795–4805. [DOI] [PubMed] [Google Scholar]
- Stegmaier K, Wong JS, Ross KN, Chow KT, Peck D, Wright RD, et al. (2007). Signature-based small molecule screening identifies cytosine arabinoside as an EWS/FLI modulator in Ewing sarcoma. PLoS Medicine, 4, e122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szalai G, LaRue AC, & Watson DK (2006). Molecular mechanisms of megakaryopoiesis. Cellular and Molecular Life Sciences, 63, 2460–2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai N, Miyazaki T, Nishida M, Nasu K, & Miyakawa I (2002). c-Ets1 is a promising marker in epithelial ovarian cancer. International Journal of Molecular Medicine, 9, 287–292. [DOI] [PubMed] [Google Scholar]
- Titeux M, Pendaries V, Tonasso L, Decha A, Bodemer C, & Hovnanian A (2008). A frequent functional SNP in the MMP1 promoter is associated with higher disease severity in recessive dystrophic epidermolysis bullosa. Human Mutation, 29, 267–276. [DOI] [PubMed] [Google Scholar]
- Tomlins SA, Laxman B, Dhanasekaran SM, Helgeson BE, Cao X, Morris DS, et al. (2007). Distinct classes of chromosomal rearrangements create oncogenic ETS gene fusions in prostate cancer. Nature, 448, 595–599. [DOI] [PubMed] [Google Scholar]
- Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, et al. (2005). Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science, 310, 644–648. [DOI] [PubMed] [Google Scholar]
- Tootle TL, & Rebay I (2005). Post-translational modifications influence transcription factor activity: A view from the ETS superfamily. BioEssays, 27, 285–298. [DOI] [PubMed] [Google Scholar]
- Trojanowska M (2000). Ets factors and regulation of the extracellular matrix. Oncogene, 19, 6464–6471. [DOI] [PubMed] [Google Scholar]
- Truong AH, & Ben-David Y (2000). The role of Fli-1 in normal cell function and malignant transformation. Oncogene, 19, 6482–6489. [DOI] [PubMed] [Google Scholar]
- Tsui KH, Chung LC, Feng TH, Chang PL, & Juang HH (2012). Upregulation of prostate-derived Ets factor by luteolin causes inhibition of cell proliferation and cell invasion in prostate carcinoma cells. International Journal of Cancer, 130, 2812–2823. [DOI] [PubMed] [Google Scholar]
- Tugores A, Le J, Sorokina I, Snijders AJ, Duyao M, Reddy PS, et al. (2001). The epithelium-specific ETS protein EHF/ESE-3 is a context-dependent transcriptional repressor downstream of MAPK signaling cascades. Journal of Biological Chemistry, 276, 20397–20406. [DOI] [PubMed] [Google Scholar]
- Turner DP, Findlay VJ, Kirven AD, Moussa O, & Watson DK (2008). Global gene expression analysis identifies PDEF transcriptional networks regulating cell migration during cancer progression. Molecular Biology of the Cell, 19, 3745–3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner DP, Findlay VJ, Moussa O, Semenchenko VI, Watson PM, LaRue AC, et al. (2011). Mechanisms and functional consequences of PDEF protein expression loss during prostate cancer progression. The Prostate, 71, 1723–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner DP, Findlay VJ, Moussa O, & Watson DK (2007). Defining ETS transcription regulatory networks and their contribution to breast cancer progression. Journal of Cellular Biochemistry, 102, 549–559. [DOI] [PubMed] [Google Scholar]
- Turner DP, Moussa O, Sauane M, Fisher PB, & Watson DK (2007). Prostate-derived ETS factor is a mediator of metastatic potential through the inhibition of migration and invasion in breast cancer. Cancer Research, 67, 1618–1625. [DOI] [PubMed] [Google Scholar]
- Turner DP, & Watson DK (2008). ETS transcription factors: Oncogenes and tumor suppressor genes as therapeutic targets for prostate cancer. Expert Review of Anticancer Therapy, 8, 33–42. [DOI] [PubMed] [Google Scholar]
- Verger A, & Duterque-Coquillaud M (2002). When Ets transcription factors meet their partners. BioEssays, 24, 362–370. [DOI] [PubMed] [Google Scholar]
- Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, et al. (2009). ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature, 457, 854–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitari AC, Leong KG, Newton K, Yee C, O’Rourke K, Liu J, et al. (2011). COP1 is a tumour suppressor that causes degradation of ETS transcription factors. Nature (London), 474, 403–406. [DOI] [PubMed] [Google Scholar]
- Wang Y, Fan PS, & Kahaleh B (2006). Association between enhanced type I collagen expression and epigenetic repression of the FLI1 gene in scleroderma fibroblasts. Arthritis and Rheumatism, 54, 2271–2279. [DOI] [PubMed] [Google Scholar]
- Wang Y, Feng L, Said M, Balderman S, Fayazi Z, Liu Y, et al. (2005). Analysis of the 2.0 A crystal structure of the protein-DNA complex of the human PDEF Ets domain bound to the prostate specific antigen regulatory site. Biochemistry, 44, 7095–7106. [DOI] [PubMed] [Google Scholar]
- Wang D, John SA, Clements JL, Percy DH, Barton KP, & Garrett-Sinha LA (2005). Ets-1 deficiency leads to altered B cell differentiation, hyperresponsiveness to TLR9 and autoimmune disease. International Immunology, 17, 1179–1191. [DOI] [PubMed] [Google Scholar]
- Wang C, & Li Q (2007). Identification of differentially expressed microRNAs during the development of Chinese murine mammary gland. Journal of Genetics and Genomics, 34, 966–973. [DOI] [PubMed] [Google Scholar]
- Wang Y, Stricker HM, Gou D, & Liu L (2007). MicroRNA: Past and present. Frontiers in Bioscience, 12, 2316–2329. [DOI] [PubMed] [Google Scholar]
- Wang LC, Swat W, Fujiwara Y, Davidson L, Visvader J, Kuo F, et al. (1998). The TEL/ETV6 gene is required specifically for hematopoiesis in the bone marrow. Genes & Development, 12, 2392–2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Zhang H, Chen Y, Shi F, & Chen B (2012). Gambogic acid-loaded magnetic Fe(3)O(4) nanoparticles inhibit Panc-1 pancreatic cancer cell proliferation and migration by inactivating transcription factor ETS1. International Journal of Nanomedicine, 7, 781–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watabe T, Yoshida K, Shindoh M, Kaya M, Fujikawa K, Sato H, et al. (1998). The Ets-1 and Ets-2 transcription factors activate the promoters for invasion-associated urokinase and collagenase genes in response to epidermal growth factor. International Journal of Cancer, 77, 128–137. [DOI] [PubMed] [Google Scholar]
- Watson DK, Ascione R, & Papas TS (1990). Molecular analysis of the ets genes and their products. Critical Reviews in Oncogenesis, 1, 409–436. [PubMed] [Google Scholar]
- Watson DK, Smyth FE, Thompson DM, Cheng JQ, Testa JR, Papas TS, et al. (1992). The ERGB/Fli-1 gene: Isolation and characterization of a new member of the family of human ETS transcription factors. Cell Growth & Differentiation, 3, 705–713. [PubMed] [Google Scholar]
- Watson DK, Turner DP, Scheiber MN, Findlay VJ, & Watson PM (2010). ETS transcription factor expression and conversion during prostate and breast cancer progression. Open Cancer Journal, 3, 24–39. [Google Scholar]
- Wei GH, Badis G, Berger MF, Kivioja T, Palin K, Enge M, et al. (2010). Genome-wide analysis of ETS-family DNA-binding in vitro and in vivo. EMBO Journal, 29, 2147–2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei G, Srinivasan R, Cantemir-Stone CZ, Sharma SM, Santhanam R, Weinstein M, et al. (2009). Ets1 and Ets2 are required for endothelial cell survival during embryonic angiogenesis. Blood, 114, 1123–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernert N, Stanjek A, Kiriakidis S, Hugel A, Jha HC, Mazitschek R, et al. (1999). Inhibition of angiogenesis in vivo by ets-1 antisense oligonucleotides-inhibition of Ets-1 transcription factor expression by the antibiotic fumagillin. Angewandte Chemie International Edition, 38, 3228–3231 (in English). [DOI] [PubMed] [Google Scholar]
- Westermarck J, Seth A, & Kahari VM (1997). Differential regulation of interstitial collagenase (MMP-1) gene expression by ETS transcription factors. Oncogene, 14, 2651–2660. [DOI] [PubMed] [Google Scholar]
- Wiemer EA (2007). The role of microRNAs in cancer: No small matter. European Journal of Cancer, 43, 1529–1544. [DOI] [PubMed] [Google Scholar]
- Wood LD, Irvin BJ, Nucifora G, Luce KS, & Hiebert SW (2003). Small ubiquitin-like modifier conjugation regulates nuclear export of TEL, a putative tumor suppressor. Proceedings of the National Academy of Sciences of the United States of America, 100, 3257–3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu YH, Hu TF, Chen YC, Tsai YN, Tsai YH, Cheng CC, et al. (2011). The manipulation of miRNA-gene regulatory networks by KSHV induces endothelial cell motility. Blood, 118, 2896–2905. [DOI] [PubMed] [Google Scholar]
- Xu C, Liu S, Fu H, Li S, Tie Y, Zhu J, et al. (2010). MicroRNA-193b regulates proliferation, migration and invasion in human hepatocellular carcinoma cells. European Journal of Cancer, 46, 2828–2836. [DOI] [PubMed] [Google Scholar]
- Xu D, Wilson TJ, Chan D, De Luca E, Zhou J, Hertzog PJ, et al. (2002). Ets1 is required for p53 transcriptional activity in UV-induced apoptosis in embryonic stem cells. EMBO Journal, 21, 4081–4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada N, Tamai Y, Miyamoto H, & Nozaki M (2000). Cloning and expression of the mouse Pse gene encoding a novel Ets family member. Gene, 241, 267–274. [DOI] [PubMed] [Google Scholar]
- Yamamoto H, Flannery ML, Kupriyanov S, Pearce J, McKercher SR, Henkel GW, et al. (1998). Defective trophoblast function in mice with a targeted mutation of Ets2. Genes & Development, 12, 1315–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SH, & Sharrocks AD (2004). SUMO promotes HDAC-mediated transcriptional repression. Molecular Cell, 13, 611–617. [DOI] [PubMed] [Google Scholar]
- Yang W, Shen N, Ye DQ, Liu Q, Zhang Y, Qian XX, et al. (2010). Genome-wide association study in Asian populations identifies variants in ETS1 and WDFY4 associated with systemic lupus erythematosus. PLoS Genetics, 6, e1000841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarden Y, & Sliwkowski MX (2001). Untangling the ErbB signalling network. Nature Reviews. Molecular Cell Biology, 2, 127–137. [DOI] [PubMed] [Google Scholar]
- Ye M, Coldren C, Liang X, Mattina T, Goldmuntz E, Benson DW, et al. (2010). Deletion of ETS-1, a gene in the Jacobsen syndrome critical region, causes ventricular septal defects and abnormal ventricular morphology in mice. Human Molecular Genetics, 19, 648–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yordy JS, Li R, Sementchenko VI, Pei H, Muise-Helmericks RC, & Watson DK (2004). SP100 expression modulates ETS1 transcriptional activity and inhibits cell invasion. Oncogene, 23, 6654–6665. [DOI] [PubMed] [Google Scholar]
- Yordy JS, & Muise-Helmericks RC (2000). Signal transduction and the Ets family of transcription factors. Oncogene, 19, 6503–6513. [DOI] [PubMed] [Google Scholar]
- Yu S, Zhao DM, Jothi R, & Xue HH (2010). Critical requirement of GABPalpha for normal T cell development. Journal of Biological Chemistry, 285, 10179–10188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabuawala T, Taffany DA, Sharma SM, Merchant A, Adair B, Srinivasan R, et al. (2010). An ets2-driven transcriptional program in tumor-associated macrophages promotes tumor metastasis. Cancer Research, 70, 1323–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Eddy A, Teng Y-T, Fritzler M, Kluppel M, Melet F, et al. (1995). An immunological renal disease in transgenic mice that overexpress Fli-1, a member of the ets family of transcription factor genes. Molecular and Cellular Biology, 15, 6961–6970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XK, Gallant S, Molano I, Moussa OM, Ruiz P, Spyropoulos DD, et al. (2004). Decreased expression of the Ets family transcription factor Fli-1 markedly prolongs survival and significantly reduces renal disease in MRL/lpr mice. Journal of Immunology, 173, 6481–6489. [DOI] [PubMed] [Google Scholar]
- Zhang J, Guo H, Zhang H, Wang H, Qian G, Fan X, et al. (2011). Putative tumor suppressor miR-145 inhibits colon cancer cell growth by targeting oncogene Friend leukemia virus integration 1 gene. Cancer, 117, 86–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XK, Moussa O, LaRue A, Bradshaw S, Molano I, Spyropoulos DD, et al. (2008). The transcription factor Fli-1 modulates marginal zone and follicular B cell development in mice. Journal of Immunology, 181, 1644–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Verheyden JM, Hassell JA, & Sun X (2009). FGF-regulated Etv genes are essential for repressing Shh expression in mouse limb buds. Developmental Cell, 16, 607–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Yan LX, Wu QN, Du ZM, Chen J, Liao DZ, et al. (2011). miR-125b is methylated and functions as a tumor suppressor by regulating the ETS1 proto-oncogene in human invasive breast cancer. Cancer Research, 71, 3552–3562. [DOI] [PubMed] [Google Scholar]
- Zhou J, Chehab R, Tkalcevic J, Naylor MJ, Harris J, Wilson TJ, et al. (2005). Elf5 is essential for early embryogenesis and mammary gland development during pregnancy and lactation. EMBO Journal, 24, 635–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Ng AY, Tymms MJ, Jermiin LS, Seth AK, Thomas RS, et al. (1998). A novel transcription factor, ELF5, belongs to the ELF subfamily of ETS genes and maps to human chromosome 11p13–15, a region subject to LOH and rearrangement in human carcinoma cell lines. Oncogene, 17, 2719–2732. [DOI] [PubMed] [Google Scholar]
- Zhu N, Zhang D, Chen S, Liu X, Lin L, Huang X, et al. (2011). Endothelial enriched microRNAs regulate angiotensin II-induced endothelial inflammation and migration. Atherosclerosis, 215, 286–293. [DOI] [PubMed] [Google Scholar]
- Zou Z, Anisowicz A, Hendrix MJ, Thor A, Neveu M, Sheng S, et al. (1994). Maspin, a serpin with tumor-suppressing activity in human mammary epithelial cells. Science, 263, 526–529. [DOI] [PubMed] [Google Scholar]
- Zucman J, Delattre O, Desmaze C, Plougastel B, Joubert I, Melot T, et al. (1992). Cloning and characterization of the Ewing’s sarcoma and peripheral neuroepithelioma t(11; 22) translocation breakpoints. Genes, Chromosomes & Cancer, 5, 271–277. [DOI] [PubMed] [Google Scholar]


