Abstract
To understand how distinct memories are formed and stored in the brain is an important and fundamental question in neuroscience and computational biology. A population of neurons, termed engram cells, represents the physiological manifestation of a specific memory trace and is characterized by dynamic changes in gene expression, which in turn alters the synaptic connectivity and excitability of these cells. Recent applications of single-cell RNA sequencing (scRNA-seq) and single-nucleus RNA sequencing (snRNA-seq) are promising approaches for delineating the dynamic expression profiles in these subsets of neurons, and thus understanding memory-specific genes, their combinatorial patterns and regulatory networks. The aim of this article is to review and discuss the experimental and computational procedures of sc/snRNA-seq, new studies of molecular mechanisms of memory aided by sc/snRNA-seq in human brain diseases and related mouse models, and computational challenges in understanding the regulatory mechanisms underlying long-term memory formation.
Keywords: memory formation, scRNA-seq, snRNA-seq, Alzheimer disease
Introduction
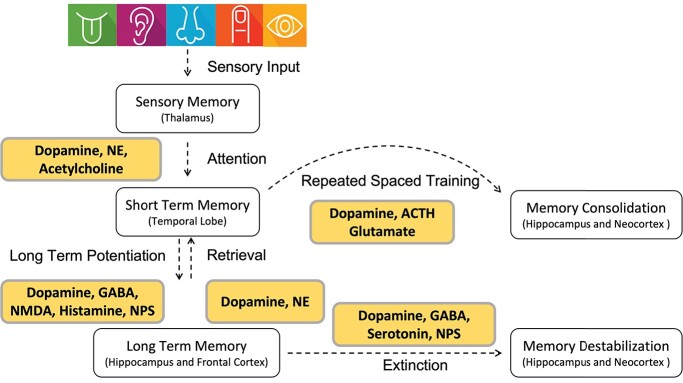
The dynamic processes of memory formation, including encoding, consolidation, storage and retrieval, are biologically essential functions of mammals to maintain information and recall it at a future time [1–3]. Many mammalian brain regions are involved in memory-related functions, including the hippocampus, the amygdala and the adjacent entorhinal, perirhinal and parahipppocampal cortices, collectively known as medial temporal lobe structures (MTL) [4]. Among MTL, a neural circuit of hippocampus, retrosplenial cortex and anterior thalamus are considered as key components involved in spatial, contextual and episodic memory [5]. The encoding process of memory begins with perception of sensory inputs along with interoceptive and emotional states (Figure 1). Networks of neurons encoding these simultaneous inputs converge, and signaling dynamics can induce initial cellular responses, such as long-term depression and long-term potentiation that encode the initial engram.
Figure 1.
Summary of different memory processes and major neurotransmitters. NE: norepinephrine, GABA: gamma-aminobutyric acid, NMDA: N-methyl-D-aspartate, NPS: neuropeptide S, ACTH: adrenocorticotropic hormone.
Memory consolidation is the process by which transient short-term memories are transformed into long-term memories, which are first stored in the MTL and then delivered to cortical areas for long-term storage. During consolidation, encoded memory traces gradually transform from an initially vulnerable state to a more permanent one by employing diverse genomic and epigenomic mechanisms. Storage is mostly based on modifications in the pattern of synaptic strength in specific groups of neurons. Alterations in synaptic connections and changes in neuronal excitability result from changes in gene transcription (and, thus, downstream translation) and are regulated by a variety of epigenetic processes [6, 7]. Recall and recognition are two main types of memory retrieval. In recall, the information is retrieved from memories. In recognition, the presentation of familiar external stimuli provides a cue that the information has been seen before. The brain uses three kinds of memory processes: sensory register, short-term memory (STM) and long-term memory (LTM) (Figure 1). In the sensory register process, the brain quickly acquires information from the environment, which usually lasts a few seconds at most. STM is the ability to hold and recall information for a short period of time, on the order of seconds to minutes, while LTM stores information for long-lasting periods, sometimes for an entire lifetime [1]. LTMs do not form quickly: they must be consolidated over time [2].
The excitability of neurons in the dentate gyrus of the hippocampus is recognized as an essential factor underlying the recruitment of a memory engram into engram cells [8, 9]. Once allocated, the successful consolidation of memory involves a dynamic time-dependent process of transcription and translation [10, 11]. During these processes, the activity of specific neuromodulators including acetylcholine (ACh), monoamines, lipids, amino acids, neuropeptides and neurotrophic factors contributes to many behaviorally relevant processes, including arousal, motivation, sleep, emotion and memory. ACh and glutamate have been broadly investigated as key neurotransmitters in learning and memory. Recent evidence further reveals that several LTM-related neurotransmitter systems, including gamma-aminobutyric acid (GABA), dopamine, serotonin and norepinephrine (NE), are also involved in the memory reconsolidation (Figure 1). Meanwhile, many of these neurotransmitters appear capable of promoting memory destabilization [12, 13], but more investigations of how these neurotransmitters work coordinately to support destabilization across different types of memory and in different brain regions are crucially needed.
Transcriptional dynamics during LTM formation is a central mechanism responding to these biological influences, maintaining long-term transcriptional stability of memory-related genes. Recent technological advancements now make it possible to assess early transcriptional variations in sparsely distributed neuronal ensembles through analysis of so-called immediate early genes which are rapidly expressed following an activity-inducing experience [14]. The enduring molecular dynamics required for encoding contextual memory within engram cells can be detected by various approaches used to detect differences in DNA and RNA at the level of bulk tissue down to single cells or nuclei. In this review, we summarize the basics of single-cell and single-nuclei RNA sequencing (sc/snRNA-seq), and describe experimental and computational workflows. We especially emphasize the novel discoveries of brain long-term memory formation facilitated by sc/snRNA-seq. Finally, we propose new strategic insights into upcoming trends, and computational challenges in the integrative analysis of single-cell sequencing datasets.
Methods to detect genomic and epigenomic changes underlying memory formation
The early introduction of DNA microarrays and bulk RNA sequencing (RNA-seq) provided the possibility of evaluating genome-wide expression patterns across heterogeneous populations of cells from essentially any tissue, bringing insights into the transcriptional programs underlying cellular identity, function and/or dysfunction for many biological systems [15, 16]. Data from several studies suggest that memory formation is linked to specific gene expression signatures that are detectable in bulk cell preparations from cortical and subcortical brain regions [17]. For example, upregulation of PRKCD, RAC1, LIMK1 and CDC42 shows strong associations with cortical memory [18–21]. On the other hand, significant changes in expression of CDK5, NLGN1, RAB3A, STX1A, SNCA, SYT1 and UNC13A are strongly associated with subcortical memory [22–25]. Genes whose changes in expression are linked to memory formation and consolidation, in both cortical and subcortical brain cells, include those involved in processes such as protein localization, transcriptional regulation and glutamate receptor signaling, cell functions previously proven to be critical for memory formation [26]. These common genes contribute to the Arp2/3 complex, and to GABA and AMPA ligand-gated ion channels which are essential for memory function [27]. Cerebral cortex-specific genes are primarily involved in DNA repair, epigenetic regulation, immunity and IFN-gamma signaling, processes also known to be related to memory [28]. Subcortex-specific genes act in neurogenesis, dendrite morphogenesis, glial cell differentiation and myelination [29].
Conditioning of fear memories in mice is an important research model that is widely investigated by bulk RNA-seq. Sustained upregulation of activity regulated cytoskeleton associated protein (Arc) mRNA is observed after fear conditioning (FC) [25], and can be used as a marker gene for time-series experiments of fear memory consolidation. Another example comes from the studies of CREB protein. A CREB-dependent network of 50 differentially expressed genes (DEGs) is recruited in engram neurons associated with fear memory after 24 h, indicating engram-specific CREB transcription is required for memory consolidation [30]. These results as well as the results of many other studies indeed demonstrate that CREB plays a critical role in the processes of memory allocation, acquisition and consolidation [31–35]. However, bulk sequencing approaches are limited to aggregate measurements across heterogeneous groups of cells. Thus, it remains unclear which subtypes of brain cells are responsible for producing the gene expression signatures of these memory functions. Furthermore, although bulk sequencing studies helped establish an outline of the gene expression signature associated with memory processing, important changes may occur only in specific cell subtypes, and these would be potentially masked by lack of change in the larger set of the cells sampled. Bulk transcriptional profiling averages expression levels across all or most of the cells sampled in any given experiment. This approach limits the ability to determine which cell types are related to any specific response or whether measured results across experimental conditions derive from changes in gene expression or changes in cellular composition of the tested samples. Overall, average expression profiles may misrepresent the signal of interest, and lead to erroneous biological interpretations.
Single-cell measurements hold the promise of specifying cellular types in measured responses, with greater integration of functional and mechanistic data relevant to cell and tissue biology. Single-cell technologies, mainly sc/snRNA-seq, offer new opportunities to address memory-related research challenges by considering the transcriptomic profile of individual cells and cell types within an engram. Since the scRNA-seq approach was first introduced [36, 37], many alternative strategies have been developed and applied to a variety of biological systems. scRNA-seq studies in the mammalian brain reveal the complexity, connectivity and functions of brain cell types [38]. So far, many sc/snRNA-seq experiments have been applied to memory-related research and provided biological insights related to memory formation.
Experimental procedures in scRNA-seq
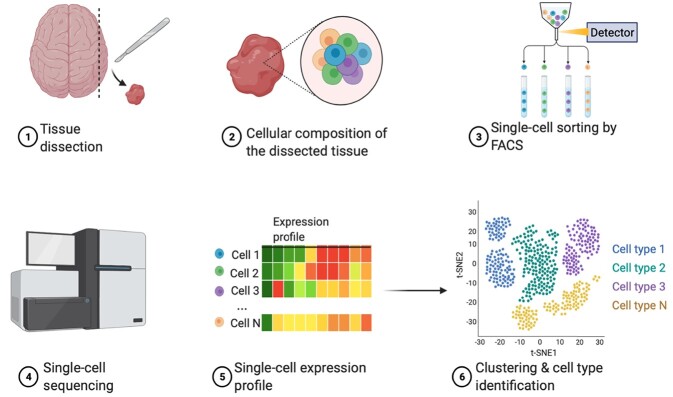
Techniques for genome-wide expression profiling in single cells were introduced in 2009 and expanded in the following years [36, 39–41]. Although early scRNA-seq techniques were limited to the study of hundreds of cells, recent approaches that combine DNA barcoding with microfluidics or combinatorial indexing provide the possibility of massively parallel scRNA-seq of up to 100 000 cells in one experiment, which may then be combined into even larger datasets [42–47]. In addition to improving statistical power via scaling, these techniques avoid the massive amplification bias and noise common in earlier approaches that did not use specific molecular identifiers. A classic scRNA-seq experimental pipeline starts with the dissociation of cells from a tissue and the isolation of single cells with special device platforms; mRNAs are then captured for reverse transcription (RT) and amplification; lastly, the synthesized complementary DNA (cDNA) molecules undergo library preparation for sequencing (Figure 2).
Figure 2.
scRNA-seq experimental workflow. 1. Tissue dissection. 2. Cellular composition of dissected tissue. Cells are usually stained by marker genes. 3. Single-cell sorting by fluorescence-activated cell sorting. For neurons, the nuclei are typically obtained for snRNA-seq. 4. Library construction and single-cell sequencing. 5. Single-cell expression profile. 6. Clustering and cell type identification.
The leading step of scRNA-seq is cell isolation, and its adequacy depends on the protocol being applied. Early techniques for single-cell isolation comprise micropipetting, micromanipulation and laser capture microdissection which are considered low throughput. A high-throughput technique is fluorescence-activated cell sorting in which fluorescently labeled antibodies are used to isolate cells that express a specific protein from a heterogeneous combinations of cells, one single cell at a time [48]. A more recent development are microfluidic devices in which a hydrodynamic flux allows isolation and processing of single cells in channels with dimensions of tens to hundreds of microns, comparable to the size of a single cell [49–53]. Instead of using transcripts from the entire cell to profile gene expression as in scRNA-seq, snRNA-seq mainly measures nuclear transcripts. snRNA-seq is most valuable in assessing the transcriptome of cells that are difficult to isolate such as neurons, adipocytes, archived frozen specimens and other preserved tissues [54–57]. snRNA-seq is especially useful for investigating the transcriptome of neurons as these cells are heterogenous in size and shape, and thus difficult to capture whole from tissues. Furthermore, isolating single cells for scRNA-seq involves extended incubations and processing, which can result in artifactual changes in gene expression [54]. On the other hand, large numbers of nuclei can be obtained quickly and easily from fresh, lightly fixed or frozen tissues [58–60]. Although nuclear transcripts comprise typically less than 50% of all the RNA in the cell [54], snRNA-seq can offer sufficient and robust markers to define broad cell classes in human and mouse brains [55, 61, 62] with resolution comparable to scRNA-seq [54, 61, 63]. Importantly, scRNA-seq and snRNA-seq identify the same cell subtype profiles in brain tissue [54]. For the purposes of clarity in this review because the technologies and workflow are quite similar, scRNA-seq will be used to refer to both sc/snRNA-seq unless specified.
Following isolation of single cells, mature mRNAs need to be captured, reverse transcribed into cDNAs and amplified. Several devices make use of genetic barcodes, which permit capturing mRNAs from multiple samples and cells, simultaneously. This procedure is called ‘multiplexing’ [64]. Another novel technique is combinatorial in situ barcoding, adopted in the single-cell combinatorial indexing RNA-seq and SPLiT-Seq method [46], where single cells follow several barcoding rounds and are uniquely labeled at the end of the procedure [37]. Usually, RT of mRNA transcripts to cDNA is performed by using an oligo-dT primer to avoid the capture of structural RNAs which account for the majority of cellular RNAs [65].
RT of mRNA to cDNA is required in scRNA-seq experiments as well as in bulk RNA-seq. RNA instability generally obviates the use of RNA-dependent polymerases. Therefore, cDNAs must be amplified to obtain the required quantities for sequencing. The major methods adopted for cDNA synthesis and amplification are template-switching coupled with polymerase chain reaction (PCR) and in vitro transcription [65–67]. The wide technical variability that arises from the combination of processing steps hinders accurate quantification of transcript abundance. Possible solutions to overcome this difficulty include the addition of quantitative standards such as RNA spike-ins or use of unique molecular identifiers (UMIs). Spike-ins are defined as artificial RNA molecules, which are added to cell lysate in specific quantities and subjected to all experimental steps after cell isolation to measure losses due to processing steps. The aim of using UMIs is to present information related to the relationship between the number of molecules of RNA input and the detected number of sequencing reads [68], and thus it is critical for removing PCR duplicates and obtaining an accurate measurement of gene expression levels.
Although most of scRNA-seq methods have similar procedures, they often differ in how they tag transcripts to their cell of origin and generate libraries for sequencing. Recent benchmarking studies provided systematic comparison for relative performance, experimental protocols and application preference [62, 67, 69]. Overall, Smart-seq2 [70] and CEL-Seq2 [71] performed similarly well among low-throughput methods, while10x Chromium was the top performer among high-throughput methods. For library preparation, Smart-seq2, CEL-Seq2 and Drop-seq [43] are most time consuming while the 10x Chromium method [45] is more automated and requires the least time. Sci-RNA-seq [46], Drop-seq, Seq-Well [72] and inDrops [73] are cost-efficient, although sci-RNA-seq would be more cost-effective with larger numbers of single cells or nuclei. Meanwhile, Smart-seq2 is the most expensive, primarily because there is no pooling during library preparation. Thus, usage preference is based on different research aims as Smart-seq2 and CEL-Seq2 are better than the high-throughput methods to obtain the highest sensitivity. In particular, Smart-seq2 can be used for calling genetic variants and alternative splicing isoforms since its reads cover the whole gene body.
Preprocessing and quality control of scRNA-seq data
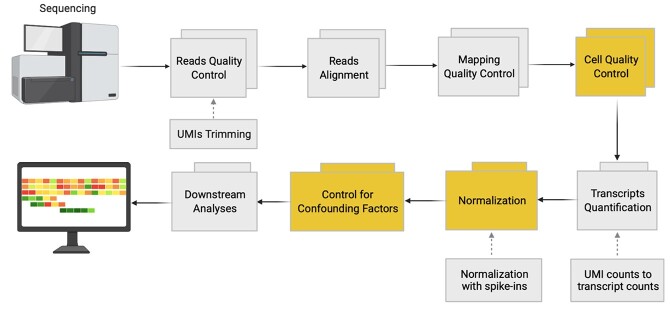
The sequencing of scRNA-seq libraries generates FASTQ files, which include thousands to millions of reads of RNA sequences and add-on sequences (e.g. UMIs). The generic scRNA-seq analysis workflow carries a further cell quality control (QC) step and the analysis of quantitative standards (Figure 3). The initial step of the scRNA-seq data analysis pipeline is the pre-processing of sequencing reads. The developed tools for QC of bulk RNA-seq data, including Falco [74], Trimmomatic [75] and Kraken [76], have also been applied for scRNA-seq [38, 77]. Later, sinQC [78] and Scater [79] were specifically designed for scRNA-seq reads QC. For the reads alignment, STAR [80], GSNAP [81], Tophat2 [82], HISAT [83] and the pseudo-aligner Kallisto [84] are most widely used. After mapping and quality checking, reads are ready to be summated to generate expression levels. This can be performed in a standard way by summing the reads mapping on every gene through use of operating tools such as HTSeq count [85], FeatureCounts [86] or maxcounts [87].
Figure 3.
scRNA-seq data quality control and pre-processing. Yellow boxes illustrate pivotal steps of scRNA-seq analysis. Boxes linked by dashed arrows indicate optional steps, depending on the scRNA-seq experimental protocol used.
In scRNA-seq analysis, every cell can be represented by a single biological system reflecting different cellular processes, such as differentiation, cellular reprogramming and disease transformations [88]. Under ideal conditions, there should be no fold difference in the levels of expression of the majority of genes for a specific cell type [89, 90]. However, read counts are usually affected by sequencing depth, and/or different protocols, introducing dropout noise in scRNA-seq data [91]. The dropout noise, reflected as a large number of zeros in the dataset, is a major bioinformatics challenge that is caused by high levels of technical variability inherent indifferent protocols [75, 77, 92], and thus will affect measurement of true gene expression levels [76, 79, 92]. The mixture of dropouts and true mRNA expression signals are usually described as zero-inflated negative binomial models by extending regular negative binomial models with mixture components to account for an excess frequency of zeros [93, 94]. To date, multiple methods have been developed to correct systematic errors, which routinely affect sequencing data. Data diversity is also due to other confounding factors, such as the intrinsic transcriptional variability and the extrinsic technical noise since they hamper the biological signal of interest from being uncovered [77]. To obtain valid analysis results, these sources of variation need to be better modeled or controlled [64].
To address the dropout noise and sparsity commonly found in scRNA-seq data, multiple imputation methods have been proposed to impute the data before downstream analysis. These imputation methods mainly employ statistical models or smoothing/diffusing strategies to smooth/insert the gene expression values in cells with similar expression profiles or deep-learning-based methods are designed to reconstruct the observed expression matrix from the estimated latent spaces. A systematic evaluation of 18 scRNA-seq imputation methods [95] shows that MAGIC [96] and SAVER [97] outperformed other evaluated methods in producing coherent datasets, improving downstream analysis. In particular, many deep-learning-based methods, such as DeepImpute [98], AutoImpute [99], stPlus [100], DCA [101], scScope [102], SAUCIE [103], scIGANs [104], scGNN [105] and GraphSCI [106], are designed to improve the accuracy of data imputation. Among them, MAGIC is highly scalable and performs well and stably with the majority of benchmark datasets, outperforming most deep learning algorithms in the analysis of human and mouse brain single-cell datasets [95, 107].
To detect genes with expression levels that have higher variability than expected from technical sources (called ‘high variable genes’), scientists modeled the relationship between gene expression level and the squared coefficient of variation, which reveals the variability in expression level of the genes associated with the mean expression level [108]. Another group of approaches has been established to account for the noise related to oscillatory behavior such as genes involved in the cell cycle. Indeed, within the cell cycle, a cell grows, duplicates its DNA and divides into daughter cells [109]. Various cells, even being of a similar type, might be at different time points of the cell cycle, therefore, having diverse gene expression profiles. Buettner and his co-workers designed a Gaussian process latent variable model, which estimates the covariance matrix allied with cell cycle alternations by factoring the expression profiles of 892 annotated cell-cycle genes [110]. They first adjust for technical variation using spike-in parameters and then adjust the variation derived from the oscillatory genes. Alternatively, the program ccRemover [109] may be used to withdraw the principal components impacted by the cell cycle. Another method, Oscope, is reported to identify genes with oscillating behavior, without a priori insight into which are the oscillatory genes, by combining a K-medoids clustering and a paired-sine model [111]. Whereas cell cycle processes may not influence the transcriptome of non-dividing cells such as neurons, other oscillatory factors may be involved such as circadian rhythms and these may require special consideration in certain studies [112].
The large number of measured genes and cells in a typical scRNA-seq experiment causes difficulties in visual presentation of data which need to be reduced to lower dimensions and cells clustered into putative subpopulations. A broadly used solution to problems rendering single-cell data is dimensionality reduction which indicates the presence of cell data in a lower-dimensional space. This approach is valuable for cell quality monitoring, data inspection before and after normalization, outlier detection and confounding effect identification [77]. Principal component analysis [113, 114], t-distributed stochastic neighbor embedding [115], UMAP [116] and diffusion maps [117] can be counted as the most widespread dimensionality reduction approaches. Meanwhile, autoencoder methods are adapted to do dimensionality reduction, e.g. DCA [101], scVI [118], scDeepCluster [119], SAUCIE [103], scGAE [120] and SCDRHA [121].
Typically, scRNA-seq profiles single cells in the transcriptome scale but will lose the spatial information of cells. As cell signaling is confined by physical location in the cellular microenvironment, it is valuable to obtain spatial information for communicating cells that tend to be spatially adjacent, particularly relevant for neuronal tissues. There are three main ways to capture and/or reconstruct spatial information from scRNA-seq data. First, a new technique, termed spatial scRNA-seq, is specifically designed to obtain transcriptomes while keeping cell position information during cell preparing and sorting. For example, targeted in situ technologies such as multiplexed error-robust fluorescence in situ hybridization (MERFISH) [122], cyclic-ouroboros single-molecule FISH (osmFISH) [123], sequential FISH (seqFISH+) [124] and spatially resolved transcript amplicon readout mapping (STARmap) [125] can achieve cellular resolution but are limited to hundreds of preselected genes. Spatial transcriptomics methods, such as Spatial Transcriptomics (ST) [126], 10X Visium and Slide-seq [127], can sequence entire transcriptomes, but with low spatial resolution (10–100 μm). The computational toolbox Giotto [128] was developed to implement a rich set of algorithms to perform common tasks for spatial omics data analysis, involving cell-type enrichment analysis, spatial pattern recognition, spatially coherent gene detection, and cell neighborhood and interaction analyses. Analysis of a variety of public datasets demonstrates that Giotto can be applied widely in conjunction with a broad range of spatial transcriptomic and proteomic approaches [128]. So far, although these methods can obtain spatial transcriptomes at the single-cell level, they are limited in either gene throughput or spatial resolution (neither transcriptome-wide in breadth nor at cellular resolution in depth). Second, if there are no available spatial transcriptomics data for specific tissues or diseases, some algorithms can reconstruct spatial locations de novo from scRNA-seq data (see more details in next section). Third, a number of integration methods were designed to reconstruct spatial information of scRNA-seq data from reference spatial data, such as Tangram [129], Cell2location [130], SpaOTsc [131], DistMap [132], SpaGE [133], SPOTlight [134], DSTG [135] and CellDART [136]. A recent paper [137] benchmarking 16 spatial and single-cell transcriptomics integration methods shows that Tangram [129] and SpaGE [133] outperformed other methods for predicting the spatial distribution of RNA transcripts. We expect that improving spatial sequencing methods will be extremely useful in mammalian memory studies as the engram cells of certain memory patterns are typically highly interconnected as local clusters.
Systematic analysis of scRNA-seq data
The downstream analyses of gene profiles include not only traditional procedures of differential gene expression analysis and functional enrichment analysis that are carried out in bulk RNA-seq analysis, but also novel analysis topics such as cell clustering analysis, trajectory inference (TI), gene regulatory inference and cell spatial localization analysis. Among all these topics, cell clustering is a core and fundamental step to reveal known or novel cell types [44, 49, 50]. There are two types of clustering methods, supervised clustering and unsupervised clustering. Supervised clustering methods usually use prior knowledge of cell-type marker genes to annotate scRNA-seq data into predefined cell types and thus achieve higher precision in clustering known cell types [138]; however, unsupervised clustering is of central importance for identifying novel cell types [44–46, 67, 68, 139]. There are many unsupervised clustering tools available, including SNN-Cliq [140], pcaReduce [141], scDEC [138], CIDR [142], SINCERA [143], GiniClust [144], RaceID [145], SIMLR [146], SC3 [147], Seurat2/3/4 [148–150] and SCANPY [151]. Recently, we designed a novel unsupervised method, SCENA, that has superior performance compared to existing methods based on large-scale validation with 13 publicly accessible scRNA-seq datasets generated from diverse biological systems [152]. SCENA has high accuracy in detecting cell populations, is robust against dropout noise and has high running speed by integrating a CPU + GPU heterogeneous parallel computing strategy. By applying it to scRNA-seq data of mouse brain cells, we detected not only known cell types but also novel cell types of interneurons that exhibit differential expression profiles of GABA receptor subunits and transporters [152].
Differential expression analysis: Applying current best practices in scRNA-seq methods and avoiding major pitfalls ultimately permits analysis of differential gene expression values from multiple experiments or conditions. Currently, scRNA-seq is allowing new insight into the dynamic molecular mechanisms underlying processes such as cell differentiation, cell-specific gene clusters and cancer driver genes [46, 77, 93, 140]. Several tools, including DESeq [153] and edgeR [154] originally developed for differential gene expression analysis on bulk RNA-seq data, are being applied to scRNA-seq data [46, 47, 51, 155]. However, due to dropout noise, low capture efficiency of RNA molecules and complicated gene expression stochastic processes in single cells, scRNA-seq data tend to exhibit more complicated distributions and huge heterogeneity compared to bulk RNA-seq data. To address these challenges, new strategies and models for differential gene expression analysis of scRNA-seq data are developed, including SCDE [156], MAST [157], Monocle2 [158], scDD [159], DEsingle [94], SigEMD [160], EMDomics [161], D3E [162] and singleCellHaystack [163]. Compared to most of the methods that use normalized transcript counts as input, Monocle2 employs census counts to better eliminate variability in different experiments. DEsingle performs well in classifying DEGs by estimating the proportion of real and drop-out zeros. Beside these model-based methods, nonparametric methods, such as SigEMD [160], EMDomics [161] and D3E [162], detect DEGs by employing a distance metric between the distributions of genes in two conditions. Recent benchmarking of 11 methods indicates that DEsingle and SigEMD achieve a better trade-off between true positive rates and precision on simulated datasets [93]. singleCellHaystack is used to find DEGs without knowing the cell labels. According to our experiments, the running time of singleCellHaystack is short, while the sensitivity is relatively low.
Trajectory inference: scRNA-seq data can provide insight into cellular dynamic processes by using TI (or pseudotime analysis) [164–166]. The basic idea of TI is to order cells along a trajectory wherein cells in similar cellular stages or differentiation lineages have high similarities in their expression patterns. TI is especially useful for studying cell cycle stages, cell differentiation, cancer development and immune responses whose gene patterns are continuously changed. The trajectories of cells can then be visualized as networks/graphs, presenting clear trajectory topologies. Although tens of methods/models of TI in scRNA-seq data have been invented and applied to divergent biological questions, a recent evaluation of 45 TI methods showed that they exhibit substantial complementarity, e.g. the optimal performance of methods depends to a large extent upon the characteristics of the dataset [164].
Gene–gene networks: An important perspective offered by scRNA-seq studies is the gene interaction landscape in which genes regulate expression of each other. scRNA-seq offers new possibilities to infer gene regulation networks (GRN) for biological processes that are time-dependent, such as the cell cycle or differentiation [167]. Precisely, by observing how gene expression differs among comparable cells subject to stochastic variations or their contribution to a dynamic process, GRN provides biologically based evidence for which transcription factors regulate which genes. As an example, a novel tool named BEELINE is designed to simulate single-cell transcriptional data from synthetic and Boolean networks [120]. It avoids certain limitations of existing methods, thus aiding the development of GRN inference algorithms [168].
Cell–cell networks: Dissociation of tissues into single cells delivers high-throughput genomics measurements; however, spatial data are generally lost. Recently, computational methods have also been advanced to infer cell–cell communications from both bulk RNA-seq and scRNA-seq data [37]. Whereas bulk cell populations often mask the critical contribution of minor subpopulations within the whole, the computational analysis of scRNA-seq data allows identification of cells that are intimately in contact. stLearn [169] and Squidpy [170] are two popular pipelines for integrating scRNA-seq and spatial transcriptomics, which both use CellPhoneDB [171] to predict ligand-receptor-mediated cell–cell interactions between identified cell clusters. Spatially optimal transporting the single cells (SpaOTsc) [131] is a method that enables mapping between scRNA-seq data and spatial data, inference of spatial distances between single cells, comparison of spatial gene expression patterns, reconstruction of spatial cell–cell communications, estimation of the spatial range of particular types of intercellular signaling and detection of gene pairs that possibly intercellularly regulate each other. Lastly, SpaOTsc is largely applicable to datasets where reasonable similarity measurements between spatial positions and single cells are attainable. Clark et al. developed a novel approach called RABID-seq for barcoded viral tracing of cell–cell interactions in central nervous system inflammation [172]. RABID-seq can simultaneously investigate cell interactions and transcriptomes of interacting cells in vivo; it successfully discovered signaling pathways controlled by the axon guidance molecules Sema4D-PlexinB1, Sema4D-PlexinB2 and Ephrin-B3/EphB3 as mediators of microglia–astrocyte interactions [122].
Applications of scRNA-seq in studying brain memory formation
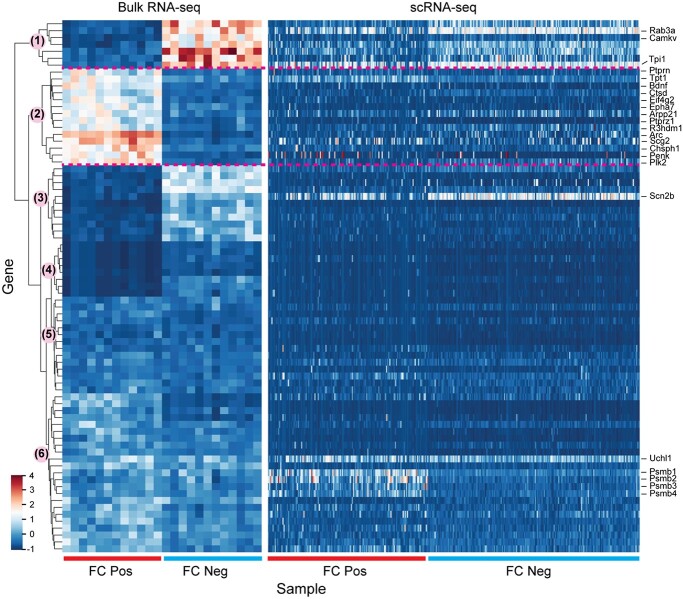
scRNA-seq studies are especially useful for revealing detailed characterization of brain cell types and developmental stages or disease grades. Advances in single-cell methods are exponentially scaling up the quantity of single cells profiled in each study, empowering not only the characterization of a wide-ranging landscape of cell types in the brain, but also the comprehensive investigation of molecular mechanisms underlying memory formation (Table 1). To monitor cells activated during a given experience (such as a fearful condition), targeted recombination in active populations (TRAP) is recognized as a promising method in which the TMX-dependent recombinase creERT2 is expressed under the control of the Arc or Fos promoter [173]. By using FosTRAP2 mice to label cells activated during memory recall in the medial prefrontal cortex, Chen et al. [155] studied the transcriptional signature 16 days after FC using scRNA-seq. Results unveiled that heterogeneous transcriptional programs specific for different neuronal and non-neuronal (e.g. astrocytes and microglia) cells are involved in remote memory retrieval [155]. Bioinformatics analysis revealed 99 DEGs in neurons by comparing FC neurons with non-neurons and non-fear conditions. Although those genes seem to be involved in multiple core biological functions related to neuronal activity, there is no significant overlap with genes reported in early memory studies. For example, among these 99 DEGs, there are 15 genes found in common in 1157 DEGs reported in conditions of fear memory consolidation in hippocampus [30], 5 genes found in common in 944 DEGs reported in conditions of associative fear-learning in temporal association cortex [174] and 10 genes found in common in 611 DEGs reported in conditions of post-visual stimulus in visual cortex [175]. To visualize expression patterns in bulk RNA-seq and scRNA-seq, we checked 77 DEGs from bulk RNA-seq data in conditions of fear memory consolidation [30] by considering smaller Fano factors (variance/mean ratio) that have higher mean scores and/or small variances (Figure 4). There are six major groups clustered by using data from bulk RNA-seq Figure 4 left [30]. In FC-positive and FC-negative conditions, the genes show smaller variances in bulk RNA-seq data, but exhibit large divergence of expression levels within cells in scRNA-seq Figure 4 right [155]. The 14 genes (cluster 2) that are highly expressed under FC in the bulk RNA-seq data seem to show no obvious differential expression in single-cell profiles, including the marker gene Arc. The inconsistency could be explained by (1) the genes/mechanisms of memory encoding [174, 175] and consolidation [30], although they are connected, are quite different; (2) the differences in the brain regions (e.g. hippocampus, cortex), the experimental conditions to which the mice were exposed, differences between Fos- and Arc-expressing neurons; and/or (3) the different experimental background and measurement limitation of scRNA-seq (e.g. dropouts for lowly expressed genes and large variances for highly expressed genes).
Table 1.
Summary of studies that characterize the single-cell transcriptome in the brain memory
| Year | Experimental model | Method | Technique | Brain region | Developmental stage | Condition/test | Number of cells | Reference (PMID) |
|---|---|---|---|---|---|---|---|---|
| 2021 | Mus musculus | snRNA-seq | 10X Genomics | Hippocampus | 8 weeks to 6 months | Barnes maze test | 15 900 | 33402532 |
| 2020 | Mus. musculus | scRNA-seq | Smart-seq2 | Hippocampus | 42–49 days | Fear Conditioning | 3691 | 33177708 |
| 2020 | M. musculus | snRNA-seq | 10X Genomics | Hippocampus | 4 to 5 months | Y-maze spatial recognition | 15 573 | 33084572 |
| 2017 | M. musculus | snRNA-seq | inDrop-seq | Cortex | 6–7 weeks | Visual stimulation | 47 209 | 29752482 |
| 2016 | M. musculus | scRNA-seq | Ovation RNA-seq | Cortex | 4 days | Fear conditioning | 165 | 27557751 |
| 2016 | Drosophila melanogaster | scRNA-seq | SMARTer | Whole brain | 24 h | Foot shock | 185 | 27160913 |
Figure 4.
Comparison of gene expression in bulk RNA-seq and scRNA-seq. A total of 77 genes with small Fano factors (variance/mean ratio) in samples under fear condition are selected from 1157 DEGs reported by bulk RNA-seq in conditions of fear memory retrieval [30]. Gene expression is normalized by z-score transformation in each cell. The hierarchical clustering is performed using the python Seaborn package based on the bulk RNA-seq levels (left). The expression levels from scRNA-seq are listed accordingly (right).
Although the different gene sets are from different studies, neuronal plasticity is found as one of the commonly enriched biological processes, confirming that it is a critical mechanism for memory formation and retrieval. To study the role of neuronal plasticity in appropriate memory recall, scientists have formulated a ‘lock-and-key’ hypothesis stating that the induction of plasticity is required but not adequate to modify motor behavior [176]. There is the further requirement that plasticity must form the dynamics of neural activity (the key) to match a temporal filter (the lock) that selectively prevents inappropriate motor responses to sensory stimuli. This idea is explored through computational analysis of two cerebellar behaviors, assessing whether deep cerebellar and vestibular nuclei neurons can filter electrical signals from Purkinje cells that would otherwise drive inappropriate motor responses. Results showed that, in different conditions, reflex acquisition requires the conditioned stimulus to precede the unconditioned stimulus. Hence, the suggested lock-and-key mechanisms connect neuronal activity and channel kinetics to recall performance and generate specific predictions of how perturbations to rebound depolarization impact motor expression [176]. More recently, Berto et al. investigated brain memory-sensitive oscillations and gene expression profiles by using both bulk and snRNA-seq techniques [177]. They identified genes correlated with oscillatory signatures of memory formation across six frequency bands and observed that isolated oscillatory signature-specific modules are highly enriched for specific classes of both excitatory and inhibitory neurons. This study establishes an experimental and analytical approach for investigating memory formation by integrating human electrophysiology, scRNA-seq and other omics techniques.
Applications of scRNA-seq in studying memory loss in Alzheimer disease
Memory dysfunction is associated with numerous brain pathologies, including tumors, epilepsy and neurodegenerative diseases such as Alzheimer disease (AD), Parkinson disease and amyotrophic lateral sclerosis. Among all these conditions, complaints of memory failure in AD patients are among the most common in clinics and hospital settings. Based on statistical data from the World Health Organization (WHO, https://www.who.int/news-room/fact-sheets/detail/dementia), currently more than 55 million people live with dementia worldwide, and there are nearly 10 million new cases every year. AD is the most common form of dementia and may contribute to 60–80% of cases. However, dementia, especially neurodegenerative dementias such as AD, has no cure because of the lack of understanding of memory formation/loss mechanisms, treatment strategies and drugs. Here we focus on reviewing the latest results of AD research by sc/snRNA-seq that have not only enhanced the understanding of the molecular pathogenesis of neurodegenerative disorders, but also helped to delineate the pathways and circuits related to memory mechanisms.
Many snRNA-seq studies have been dedicated to understanding the cell type and molecular changes in neurodegenerative diseases, especially AD [178–181]. A recent large-scale study of 169 496 nuclei from the prefrontal cortex of AD patients and healthy controls revealed the loss of neuroprotective glial cells and revealed a role of antigen presentation by angiogenic endothelial cells in AD [178]. Cell type-specific transcriptomic changes are reported to be associated with four major molecular pathways, including angiogenesis in endothelial cells, immune response in endothelial cells and microglia, myelination in oligodendrocytes, and synaptic signaling in astrocytes and neurons. The DEGs largely overlap with another independent snRNA-seq study performed by Mathys et al. [179], exhibiting concordant pathway changes of transsynaptic signaling in astrocytes, synaptic signaling in excitatory neurons, mitochondrial functions in inhibitory neurons, secretion in microglia and regulation of axonogenesis by oligodendrocytes. By comparing these results from AD research with the genes associated with remote memory of fear conditions [155], we find that the synaptic signaling and regulation of secretory vesicles in neurons are common pathways, indicating they may be key steps in memory formation.
So far, 73 scRNA-seq datasets from 10 brain regions of AD patients have been collected in the single-cell RNA-Seq database for Alzheimer’s disease (scREAD) [182]. Although general analysis of AD scRNA-seq data have focused on the development of AD pathology, it is interesting to analyze these datasets by specifically investigating how memory-related genes are altered by AD. Thus, further integrative and comparative analysis should be performed to precisely narrow the gene candidates and potential pathways of memory formation.
Computational challenges and future studies
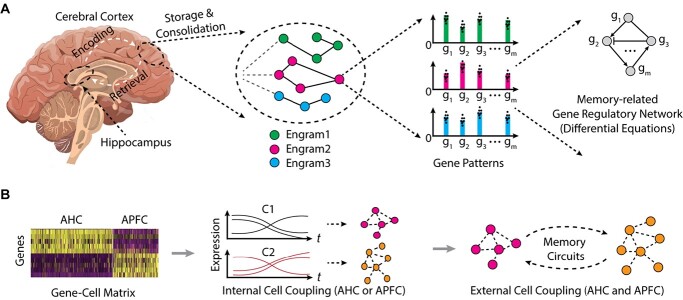
Overall, studies show scRNA-seq to be a powerful high-throughput tool for analyzing individual brain cells, enabling comprehensive and high-resolution cell type determination and identifying new cell markers. Many methods and software tools have been invented, employing diverse computational and/or statistical strategies, providing great convenience in analyzing scRNA-seq data for diverse analysis topics (Supplementary Table S1). The remarkable potential of sc/snRNA-seq is also illustrated in studies of cognitive function and memory including brain diseases in which memory is disrupted, especially AD. However, the increasing number of single cells being sequenced in current studies is starting to overcome the capacity of available data analysis approaches. Thus, new computational techniques and mathematical modeling are needed to handle even larger datasets [44, 49–51]. Here we highlight several promising topics (Figure 5).
Figure 5.
Schematic research topics from scRNA-seq data analysis to mathematical modeling of memory-related gene regulation. (A) The different engram cells are extracted, sequenced, and analyzed for gene regulatory network. (B) The cell–cell coupling/connection is to be analyzed for same cell types/regions or cross different regions. AHC: active hippocampus cell, APFC: active prefrontal cortex cell.
Specific analysis of memory-related genes: scRNA-seq allows study of the dynamic transcriptional response in memory formation. As discussed previously, most genes whose expression is altered in activated neurons in remote memory are rarely observed within those DEGs discovered in early stages of learning and memory encoding [30, 155, 174, 175]. Thus, comprehensive analysis of single-cell RNA-seq datasets is needed to detect DEGs for different memory conditions, and for different neuron and/or cell types. Current strategies to detect DEGs mainly compare transcriptomes of two or more conditions with regular significance testing [30, 155]. It is critical to further filter the DEGs with memory-specific characteristics. Since remote memory-related genes are stably regulated for long-term expression, studying LTM phenotypes or phenotypes involving oscillatory behavior may facilitate detection of DEGs with small variances in levels of expression in critical subsets of neurons (Figure 5A). Another high-level analysis of scRNA-seq data involves detecting neuronal circuits from cell–cell correlation between neurons, between neurons and non-neurons, and between brain regions (Figure 5B). The first step in studying cell–cell communication using snRNA-seq data is to calculate gene–gene correlations. For a given cell type, the conserved or different gene modules can be detected computationally for experimental and control conditions. A similar strategy can be used to detect these modules by comparing neurons and non-neurons, or across different brain regions.
Mathematical modeling: Although specific DEGs are already known to be linked to fear memory or other memory conditions, deep analysis of these genes and engram cells is lacking. Some of the main questions that need to be answered are the following: How are DEGs coordinated to encode specific memory engrams? Is there a common coding strategy at the molecular level to convert certain memory signals into specific gene expression profiles? How does establishment of a transcriptional regulation network relate to stable memory-specific connections among engram cells? To answer these questions, new mathematical models are required to delineate the dynamic processes of memory-related gene regulation and neuron–neuron communication. For example, based on the gene–gene network among DEGs, or the cell–cell network among engram cells, pseudotime methods can be established to simulate the dynamics of these interactions (Figure 5B). Ordinary differential equations and stochastic differential equations can be applied to single-cell transcriptome data of engram cells with some adjustment. For example, differential equation methods have been used to model the forgetting curves with and without anterograde amnesia and learning and forgetting curves with impaired cortical plasticity [183]. A biochemical mechanism for time-encoding memory formation within individual synapses of Purkinje cells is also studied by establishing differential equations for a set of proteins including mGluR7, G-protein, G-protein-coupled inward rectifier potassium ion channel, protein kinase A, protein phosphatase and other associated biomolecules [184]. A positive feedback loop within the molecular cascade of pCREB, C/RBP and BDNF was investigated by a differential equation-based model [185]. A potential limitation of applying differential equation methods is that current scRNA-seq data are only obtained with limited time steps of hours, days and weeks. In theory, more time points can deliver more precise information. An alternative way is to do pseudotime analysis of scRNA-seq data to achieve potential orders of cells over time. Several pseudo-temporal analysis algorithms, including Monocle [166], scTDA [186] and Waterfall [187], have been applied to study lineage relations among neurons, stem cells and complete organisms [46, 188]. New models and methods are expected to describe the dynamic processes of engram cells from the pre-memory state to the terminal condition of an established stable memory.
Integrative analysis: Current applications of divergent experimental protocols involving scRNA-seq are generating numerous brain-focused datasets. It is valuable to design methodologies and computational frameworks to integrate and compare scRNA-seq data from multiple platforms, and biological and clinical conditions. Chen et al. [155] identified 99 genes in specific cell types by comparing multiple remote memory fear conditions. Similar comparisons may be done using sc/snRNA-seq datasets from neurodegenerative diseases such as AD. Several large-scale databases of healthy reference cell atlases are publicly available, including Single Cell Expression Atlas by EMBL-EBI [189], Human Cell Atlas [190], Allen Brain Atlas [191], Mouse Cell Atlas [192], and Mouse Organs and Tissues [193]. Such resources may be used to detect memory-related genes and pathways by filtering the transcriptome background obtained from heathy brain cells. Another type of data integration involves incorporation of multiple types of single-cell omics data. The epigenome and three-dimensional (3D) genomic architecture are emerging as vital factors in the dynamic regulation of transcriptional programs essential for neuronal functions. Asaf Marco and his co-workers [194] utilized an activity-dependent tagging system in mice to determine the epigenetic state, 3D genome architecture and transcriptional landscape of engram cells within memory formation and recall. Their bulk sequencing-based discoveries revealed that memory encoding ends in an epigenetic priming event, marked by boosted accessibility of enhancers without the corresponding transcriptional variations. Memory consolidation consequently ends in spatial re-organization of large chromatin segments and promoter–enhancer interactions. Single-cell-based application of high-throughput chromosome conformation capture is required for elucidating the comprehensive transcriptional and epigenomic landscape across the lifespan of memory formation and recall in the hippocampal engram ensemble. Thus, integrating scRNA-seq with epigenomic (3D chromatin interactions and histone modifications), proteomic and metabolomic represents a potential strategy for delineating the full dynamic nature of memory engram cells, leading to a more systematic understanding of the processes determining memory formation. Overall, rapidly emerging single-cell sequencing approaches have generated big data for future investigations and are starting to reveal the high-resolution map of the brain mechanisms underlying memory function.
Key Points
The formation of memories in neurons is an important, complex and fundamental question in fully understanding mammalian brain functions.
scRNA-seq and snRNA-seq are novel and effective approaches to detect transcriptional profiles that are required for the memory formation.
Comprehensive data analysis of sc/snRNA-seq datasets is essential to detect memory-related genes and pathways.
Novel computational tools and mathematical models are urgently needed for elucidating the molecular mechanisms of memory formation.
Supplementary Material
Acknowledgement
We would like to thank Ruoyu Chen for preparing the scRNA-seq data and illustrating figures.
Atlas M. Sardoo is a PhD candidate in the Department of Biological and Biomedical Sciences at Rowan University. Her research interests focus on deep learning and single-cell/ single-nucleus RNA sequencing data analysis.
Shaoqiang Zhang is a Professor in the College of Computer and Information Engineering, Tianjin Normal University, China. His research interests include bioinformatics, combinatorial optimization and high-performance computing. He is a member of IEEE and The China Computer Federation.
Thomas N. Ferraro is a Professor in the Department of Biomedical Sciences at Cooper Medical School of Rowan University (CMSRU). His current research interests focus on human and genetic mouse models of epilepsy, opioid addiction, antidepressant treatments and fear–memory formation.
Thomas Keck is Associate Professor and Chair of the Department of Biological and Biomedical Sciences at Rowan University, with a joint appointment to the Department of Chemistry and Biochemistry. His current research interests focus in developing new medications for neuropsychiatric disorders, including Alzheimer’s disease, schizophrenia, Attention-deficit/hyperactivity disorder (ADHD), pain, anxiety and a particular interest in substance use disorders. Novel pharmacotherapies that affect memory formation can affect each of these conditions and provide new insights into their pathophysiology.
Yong Chen is an Assistant Professor in the Department of Biological and Biomedical Sciences at Rowan University. Dr Chen is a bioinformatician and computational biologist and has published over 40 peer-reviewed scientific research papers. His research interests include single-cell omics data analysis, deep learning, cancer epigenetics and neuroscience.
Contributor Information
Atlas M Sardoo, Department of Biological & Biomedical Sciences, Rowan University, Glassboro, NJ 08028, USA.
Shaoqiang Zhang, College of Computer and Information Engineering, Tianjin Normal University, Tianjin 300387, China.
Thomas N Ferraro, Department of Biomedical Sciences, Cooper Medical School of Rowan University, Camden, NJ 08103, USA.
Thomas M Keck, Department of Biological & Biomedical Sciences, Rowan University, Glassboro, NJ 08028, USA; Department of Chemistry & Biochemistry, Rowan University, Glassboro, NJ 08028, USA.
Yong Chen, Department of Biological & Biomedical Sciences, Rowan University, Glassboro, NJ 08028, USA.
Data availability
The data underlying this article are available in the article and in its online supplementary material.
Abbreviations
scRNA-seq, single-cell RNA-sequencing; snRNA-seq, single-nucleus RNA-sequencing; RNA-seq, RNA sequencing; cDNA, complementary DNA; RT, reverse transcription; PCR, polymerase chain reaction; UMI, unique molecular identifier; PD, Parkinson’s disease; AD, Alzheimer’s disease; NMDA, N-methyl-D-aspartate; NE, norepinephrine; GABA, gamma-aminobutyric acid; ACh, acetylcholine; LTM, long-term memory; STM, short-term memory; QC, quality control; GRN, gene regulation networks; DEG, differentially expressed genes
Funding
This work was supported by Rowan University Startup grant (2019) for Y.C. and a key project of the Natural Science Funds of Tianjin Municipal Science and Technology Bureau (19JCZDJC35100) for S.Z.
References
- 1. Bisaz R, Travaglia A, Alberini CM. The neurobiological bases of memory formation: from physiological conditions to psychopathology. Psychopathology 2014;47(6):347–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Squire LR. Mechanisms of memory. Science 1986;232(4758):1612–9. [DOI] [PubMed] [Google Scholar]
- 3. Kandel ER, Dudai Y, Mayford MR. The molecular and systems biology of memory. Cell 2014;157(1):163–86. [DOI] [PubMed] [Google Scholar]
- 4. Squire LR, Stark CE, Clark RE. The medial temporal lobe. Annu Rev Neurosci 2004;27:279–306. [DOI] [PubMed] [Google Scholar]
- 5. Smith DM, Yang YY, Subramanian DL, et al. The limbic memory circuit and the neural basis of contextual memory. Neurobiol Learn Mem 2022;187:107557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tonegawa S, Morrissey MD, Kitamura T. The role of engram cells in the systems consolidation of memory. Nat Rev Neurosci 2018;19(8):485–98. [DOI] [PubMed] [Google Scholar]
- 7. Han DH, Park P, Choi DI, et al. The essence of the engram: cellular or synaptic? Semin Cell Dev Biol 2022;125:122–35. [DOI] [PubMed] [Google Scholar]
- 8. Park S, Kramer EE, Mercaldo V, et al. Neuronal allocation to a hippocampal engram. Neuropsychopharmacology 2016;41(13):2987–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Silva AJ, Zhou Y, Rogerson T, et al. Molecular and cellular approaches to memory allocation in neural circuits. Science 2009;326(5951):391–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Alberini CM, Kandel ER. The regulation of transcription in memory consolidation. Cold Spring Harb Perspect Biol 2014;7(1):a021741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hernandez PJ, Abel T. The role of protein synthesis in memory consolidation: progress amid decades of debate. Neurobiol Learn Mem 2008;89(3):293–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wideman CE, Jardine KH, Winters BD. Involvement of classical neurotransmitter systems in memory reconsolidation: focus on destabilization. Neurobiol Learn Mem 2018;156:68–79. [DOI] [PubMed] [Google Scholar]
- 13. Kida S. Function and mechanisms of memory destabilization and reconsolidation after retrieval. Proc Jpn Acad Ser B Phys Biol Sci 2020;96(3):95–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lacar B, Linker SB, Jaeger BN, et al. Nuclear RNA-seq of single neurons reveals molecular signatures of activation. Nat Commun 2016;7:11022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schena M, Shalon D, Davis RW, et al. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 1995;270(5235):467–70. [DOI] [PubMed] [Google Scholar]
- 16. Mortazavi A, Williams BA, McCue K, et al. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 2008;5(7):621–8. [DOI] [PubMed] [Google Scholar]
- 17. Tan PK, Ananyev E, Hsieh PJ. Distinct genetic signatures of cortical and subcortical regions associated with human memory. eNeuro 2019;6(6):ENEURO.0283-19.2019. doi: 10.1523/ENEURO.0283-19.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Etcheberrigaray R, Tan M, Dewachter I, et al. Therapeutic effects of PKC activators in Alzheimer's disease transgenic mice. Proc Natl Acad Sci U S A 2004;101(30):11141–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Haditsch U, Leone DP, Farinelli M, et al. A central role for the small GTPase Rac1 in hippocampal plasticity and spatial learning and memory. Mol Cell Neurosci 2009;41(4):409–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Todorovski Z, Asrar S, Liu J, et al. LIMK1 regulates long-term memory and synaptic plasticity via the transcriptional factor CREB. Mol Cell Biol 2015;35(8):1316–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang X, Li Q, Wang L, et al. Cdc42-dependent forgetting regulates repetition effect in prolonging memory retention. Cell Rep 2016;16(3):817–25. [DOI] [PubMed] [Google Scholar]
- 22. Bohme MA, McCarthy AW, Grasskamp AT, et al. Rapid active zone remodeling consolidates presynaptic potentiation. Nat Commun 2019;10(1):1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mishiba T, Tanaka M, Mita N, et al. Cdk5/p35 functions as a crucial regulator of spatial learning and memory. Mol Brain 2014;7:82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bie B, Wu J, Yang H, et al. Epigenetic suppression of neuroligin 1 underlies amyloid-induced memory deficiency. Nat Neurosci 2014;17(2):223–31. [DOI] [PubMed] [Google Scholar]
- 25. Kokhan VS, Afanasyeva MA, Van'kin GI. Alpha-synuclein knockout mice have cognitive impairments. Behav Brain Res 2012;231(1):226–30. [DOI] [PubMed] [Google Scholar]
- 26. Arnatkeviciute A, Fulcher BD, Fornito A. A practical guide to linking brain-wide gene expression and neuroimaging data. Neuroimage 2019;189:353–67. [DOI] [PubMed] [Google Scholar]
- 27. Takemoto K, Iwanari H, Tada H, et al. Optical inactivation of synaptic AMPA receptors erases fear memory. Nat Biotechnol 2017;35(1):38–47. [DOI] [PubMed] [Google Scholar]
- 28. Hou Y, et al. NAD(+) supplementation normalizes key Alzheimer's features and DNA damage responses in a new AD mouse model with introduced DNA repair deficiency. Proc Natl Acad Sci U S A 2018;115(8):E1876–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hertz L, Chen Y. Editorial: all 3 types of glial cells are important for memory formation. Front Integr Neurosci 2016;10:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rao-Ruiz P, Couey JJ, Marcelo IM, et al. Engram-specific transcriptome profiling of contextual memory consolidation. Nat Commun 2019;10(1):2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhou Y, Won J, Karlsson MG, et al. CREB regulates excitability and the allocation of memory to subsets of neurons in the amygdala. Nat Neurosci 2009;12(11):1438–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yiu AP, Mercaldo V, Yan C, et al. Neurons are recruited to a memory trace based on relative neuronal excitability immediately before training. Neuron 2014;83(3):722–35. [DOI] [PubMed] [Google Scholar]
- 33. Peters M, Bletsch M, Catapano R, et al. RNA interference in hippocampus demonstrates opposing roles for CREB and PP1alpha in contextual and temporal long-term memory. Genes Brain Behav 2009;8(3):320–9. [DOI] [PubMed] [Google Scholar]
- 34. Suzuki A, Fukushima H, Mukawa T, et al. Upregulation of CREB-mediated transcription enhances both short- and long-term memory. J Neurosci 2011;31(24):8786–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Trifilieff P, Herry C, Vanhoutte P, et al. Foreground contextual fear memory consolidation requires two independent phases of hippocampal ERK/CREB activation. Learn Mem 2006;13(3):349–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tang F, Barbacioru C, Wang Y, et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods 2009;6(5):377–82. [DOI] [PubMed] [Google Scholar]
- 37. Svensson V, Vento-Tormo R, Teichmann SA. Exponential scaling of single-cell RNA-seq in the past decade. Nat Protoc 2018;13(4):599–604. [DOI] [PubMed] [Google Scholar]
- 38. Poulin JF, Tasic B, Hjerling-Leffler J, et al. Disentangling neural cell diversity using single-cell transcriptomics. Nat Neurosci 2016;19(9):1131–41. [DOI] [PubMed] [Google Scholar]
- 39. Dalerba P, Kalisky T, Sahoo D, et al. Single-cell dissection of transcriptional heterogeneity in human colon tumors. Nat Biotechnol 2011;29(12):1120–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ramskold D, Luo S, Wang Y-C, et al. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat Biotechnol 2012;30(8):777–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hashimshony T, Wagner F, Sher N, et al. CEL-Seq: single-cell RNA-Seq by multiplexed linear amplification. Cell Rep 2012;2(3):666–73. [DOI] [PubMed] [Google Scholar]
- 42. Bose S, Wan Z, Carr A, et al. Scalable microfluidics for single-cell RNA printing and sequencing. Genome Biol 2015;16:120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Macosko EZ, Basu A, Satija R, et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 2015;161(5):1202–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Klein AM, Mazutis L, Akartuna I, et al. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell 2015;161(5):1187–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zheng GX, Terry JM, Belgrader P, et al. Massively parallel digital transcriptional profiling of single cells. Nat Commun 2017;8:14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cao J, Packer JS, Ramani V, et al. Comprehensive single-cell transcriptional profiling of a multicellular organism. Science 2017;357(6352):661–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rosenberg AB, Roco CM, Muscat RA, et al. Single-cell profiling of the developing mouse brain and spinal cord with split-pool barcoding. Science 2018;360(6385):176–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Porter JR, Telford WG, Batchelor E. Single-cell gene expression profiling using FACS and qPCR with internal standards. J Vis Exp 2017;120:55219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kolodziejczyk AA, Kim JK, Svensson V, et al. The technology and biology of single-cell RNA sequencing. Mol Cell 2015;58(4):610–20. [DOI] [PubMed] [Google Scholar]
- 50. Saliba AE, Westermann AJ, Gorski SA, et al. Single-cell RNA-seq: advances and future challenges. Nucleic Acids Res 2014;42(14):8845–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Liang J, Cai W, Sun Z. Single-cell sequencing technologies: current and future. J Genet Genomics 2014;41(10):513–28. [DOI] [PubMed] [Google Scholar]
- 52. Luni C, Giulitti S, Serena E, et al. High-efficiency cellular reprogramming with microfluidics. Nat Methods 2016;13(5):446–52. [DOI] [PubMed] [Google Scholar]
- 53. Grun D, Oudenaarden A. Design and analysis of single-cell sequencing experiments. Cell 2015;163(4):799–810. [DOI] [PubMed] [Google Scholar]
- 54. Bakken TE, Hodge RD, Miller JA, et al. Single-nucleus and single-cell transcriptomes compared in matched cortical cell types. PLoS One 2018;13(12):e0209648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lake BB, Codeluppi S, Yung YC, et al. A comparative strategy for single-nucleus and single-cell transcriptomes confirms accuracy in predicted cell-type expression from nuclear RNA. Sci Rep 2017;7(1):6031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Krishnaswami SR, Grindberg RV, Novotny M, et al. Using single nuclei for RNA-seq to capture the transcriptome of postmortem neurons. Nat Protoc 2016;11(3):499–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Slyper M, Porter CBM, Ashenberg O, et al. A single-cell and single-nucleus RNA-Seq toolbox for fresh and frozen human tumors. Nat Med 2020;26(5):792–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Habib N, Avraham-Davidi I, Basu A, et al. Massively parallel single-nucleus RNA-seq with DroNc-seq. Nat Methods 2017;14(10):955–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wu H, Kirita Y, Donnelly EL, et al. Advantages of single-nucleus over single-cell RNA sequencing of adult kidney: rare cell types and novel cell states revealed in fibrosis. J Am Soc Nephrol 2019;30(1):23–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wolfien M, Galow AM, Müller P, et al. Single-nucleus sequencing of an entire mammalian heart: cell type composition and velocity. Cell 2020;9(2):318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Armand EJ, Li J, Xie F, et al. Single-cell sequencing of brain cell transcriptomes and epigenomes. Neuron 2021;109(1):11–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ding J, Adiconis X, Simmons SK, et al. Systematic comparison of single-cell and single-nucleus RNA-sequencing methods. Nat Biotechnol 2020;38(6):737–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Bakken TE, van Velthoven CTJ, Menon V, et al. Single-cell and single-nucleus RNA-seq uncovers shared and distinct axes of variation in dorsal LGN neurons in mice, non-human primates, and humans. Elife 2021;10:e64875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Islam S, Kjällquist U, Moliner A, et al. Characterization of the single-cell transcriptional landscape by highly multiplex RNA-seq. Genome Res 2011;21(7):1160–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hebenstreit D. Methods, challenges and potentials of single cell RNA-seq. Biology (Basel) 2012;1(3):658–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Picelli S. Single-cell RNA-sequencing: the future of genome biology is now. RNA Biol 2017;14(5):637–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ziegenhain C, Vieth B, Parekh S, et al. Comparative analysis of single-cell RNA sequencing methods. Mol Cell 2017;65(4):631–643.e4. [DOI] [PubMed] [Google Scholar]
- 68. Baker SC, Bauer SR, Beyer RP, et al. The external RNA controls consortium: a progress report. Nat Methods 2005;2(10):731–4. [DOI] [PubMed] [Google Scholar]
- 69. Natarajan KN, Miao Z, Jiang M, et al. Comparative analysis of sequencing technologies for single-cell transcriptomics. Genome Biol 2019;20(1):70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Picelli S, Faridani OR, Björklund ÅK, et al. Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc 2014;9(1):171–81. [DOI] [PubMed] [Google Scholar]
- 71. Muraro MJ, Dharmadhikari G, Grün D, et al. A single-cell transcriptome atlas of the human pancreas. Cell Syst 2016;3(4):385–394.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Gierahn TM, WadsworthMH, II, Hughes TK, et al. Seq-Well: portable, low-cost RNA sequencing of single cells at high throughput. Nat Methods 2017;14(4):395–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Zilionis R, Nainys J, Veres A, et al. Single-cell barcoding and sequencing using droplet microfluidics. Nat Protoc 2017;12(1):44–73. [DOI] [PubMed] [Google Scholar]
- 74. Sena Brandine G, Smith AD. Falco: high-speed FastQC emulation for quality control of sequencing data. F1000Res 2019;8:1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014;30(15):2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Davis MP, van Dongen S, Abreu-Goodger C, et al. Kraken: a set of tools for quality control and analysis of high-throughput sequence data. Methods 2013;63(1):41–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Stegle O, Teichmann SA, Marioni JC. Computational and analytical challenges in single-cell transcriptomics. Nat Rev Genet 2015;16(3):133–45. [DOI] [PubMed] [Google Scholar]
- 78. Jiang P, Thomson JA, Stewart R. Quality control of single-cell RNA-seq by SinQC. Bioinformatics 2016;32(16):2514–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. McCarthy DJ, Campbell KR, Lun AT, et al. Scater: pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R. Bioinformatics 2017;33(8):1179–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29(1):15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Wu TD, Reeder J, Lawrence M, et al. GMAP and GSNAP for genomic sequence alignment: enhancements to speed, accuracy, and functionality. Methods Mol Biol 2016;1418:283–334. [DOI] [PubMed] [Google Scholar]
- 82. Kim D, Pertea G, Trapnell C, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 2013;14(4):R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods 2015;12(4):357–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Bray NL, Pimentel H, Melsted P, et al. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol 2016;34(5):525–7. [DOI] [PubMed] [Google Scholar]
- 85. Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 2015;31(2):166–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014;30(7):923–30. [DOI] [PubMed] [Google Scholar]
- 87. Finotello F, Lavezzo E, Bianco L, et al. Reducing bias in RNA sequencing data: a novel approach to compute counts. BMC Bioinformatics 2014;15(Suppl 1):S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Armond JW, Saha K, Rana AA, et al. A stochastic model dissects cell states in biological transition processes. Sci Rep 2014;4:3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Katayama S, Töhönen V, Linnarsson S, et al. SAMstrt: statistical test for differential expression in single-cell transcriptome with spike-in normalization. Bioinformatics 2013;29(22):2943–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Finotello F, Di Camillo B. Measuring differential gene expression with RNA-seq: challenges and strategies for data analysis. Brief Funct Genomics 2015;14(2):130–42. [DOI] [PubMed] [Google Scholar]
- 91. Bacher R, Kendziorski C. Design and computational analysis of single-cell RNA-sequencing experiments. Genome Biol 2016;17:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Poirion OB, Zhu X, Ching T, et al. Single-cell transcriptomics bioinformatics and computational challenges. Front Genet 2016;7:163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Wang T, Li B, Nelson CE, et al. Comparative analysis of differential gene expression analysis tools for single-cell RNA sequencing data. BMC Bioinformatics 2019;20(1):40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Miao Z, Deng K, Wang X, et al. DEsingle for detecting three types of differential expression in single-cell RNA-seq data. Bioinformatics 2018;34(18):3223–4. [DOI] [PubMed] [Google Scholar]
- 95. Hou W, Ji Z, Ji H, et al. A systematic evaluation of single-cell RNA-sequencing imputation methods. Genome Biol 2020;21(1):218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Dijk D, Sharma R, Nainys J, et al. Recovering gene interactions from single-cell data using data diffusion. Cell 2018;174(3):716–729.e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Huang M, Wang J, Torre E, et al. SAVER: gene expression recovery for single-cell RNA sequencing. Nat Methods 2018;15(7):539–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Arisdakessian C, Poirion O, Yunits B, et al. DeepImpute: an accurate, fast, and scalable deep neural network method to impute single-cell RNA-seq data. Genome Biol 2019;20(1):211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Talwar D, Mongia A, Sengupta D, et al. AutoImpute: autoencoder based imputation of single-cell RNA-seq data. Sci Rep 2018;8(1):16329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Shengquan C, Boheng Z, Xiaoyang C, et al. stPlus: a reference-based method for the accurate enhancement of spatial transcriptomics. Bioinformatics 2021;37(Suppl_1):i299–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Eraslan G, Simon LM, Mircea M, et al. Single-cell RNA-seq denoising using a deep count autoencoder. Nat Commun 2019;10(1):390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Deng Y, Bao F, Dai Q, et al. Scalable analysis of cell-type composition from single-cell transcriptomics using deep recurrent learning. Nat Methods 2019;16(4):311–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Amodio M, van Dijk D, Srinivasan K, et al. Exploring single-cell data with deep multitasking neural networks. Nat Methods 2019;16(11):1139–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Xu Y, Zhang Z, You L, et al. scIGANs: single-cell RNA-seq imputation using generative adversarial networks. Nucleic Acids Res 2020;48(15):e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Wang J, Ma A, Chang Y, et al. scGNN is a novel graph neural network framework for single-cell RNA-Seq analyses. Nat Commun 2021;12(1):1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Rao J, Zhou X, Lu Y, et al. Imputing single-cell RNA-seq data by combining graph convolution and autoencoder neural networks. iScience 2021;24(5):102393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Dai C, Jiang Y, Yin C, et al. scIMC: a platform for benchmarking comparison and visualization analysis of scRNA-seq data imputation methods. Nucleic Acids Res 2022;50(9):4877–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Brennecke P, Anders S, Kim JK, et al. Accounting for technical noise in single-cell RNA-seq experiments. Nat Methods 2013;10(11):1093–5. [DOI] [PubMed] [Google Scholar]
- 109. Barron M, Li J. Identifying and removing the cell-cycle effect from single-cell RNA-sequencing data. Sci Rep 2016;6:33892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Buettner, F., Natarajan, K., Casale, F. et al. Computational analysis of cell-to-cell heterogeneity in single-cell RNA-sequencing data reveals hidden subpopulations of cells. Nat Biotechnol 2015;33:155–160. [DOI] [PubMed] [Google Scholar]
- 111. Leng N, Chu LF, Barry C, et al. Oscope identifies oscillatory genes in unsynchronized single-cell RNA-seq experiments. Nat Methods 2015;12(10):947–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Morris EL, Patton AP, Chesham JE, et al. Single-cell transcriptomics of suprachiasmatic nuclei reveal a Prokineticin-driven circadian network. EMBO J 2021;40(20):e108614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Gogolewski K, Sykulski M, Chung NC, et al. Truncated robust principal component analysis and noise reduction for single cell RNA sequencing data. J Comput Biol 2019;26(8):782–93. [DOI] [PubMed] [Google Scholar]
- 114. Tsuyuzaki K, Sato H, Sato K, et al. Benchmarking principal component analysis for large-scale single-cell RNA-sequencing. Genome Biol 2020;21(1):9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Kobak D, Berens P. The art of using t-SNE for single-cell transcriptomics. Nat Commun 2019;10(1):5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Becht E, McInnes L, Healy J, et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat Biotechnol 2018;37:38–44. [DOI] [PubMed] [Google Scholar]
- 117. Haghverdi L, Buettner F, Theis FJ. Diffusion maps for high-dimensional single-cell analysis of differentiation data. Bioinformatics 2015;31(18):2989–98. [DOI] [PubMed] [Google Scholar]
- 118. Lopez R, Regier J, Cole MB, et al. Deep generative modeling for single-cell transcriptomics. Nat Methods 2018;15(12):1053–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Tian T, Wan J, Song Q, et al. Clustering single-cell RNA-seq data with a model-based deep learning approach. Nat Mach Intell 2019;1(4):191–8. [Google Scholar]
- 120. Luo Z, Xu C, Zhang Z, et al. A topology-preserving dimensionality reduction method for single-cell RNA-seq data using graph autoencoder. Sci Rep 2021;11(1):20028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Zhao J, Wang N, Wang H, et al. SCDRHA: a scRNA-Seq data dimensionality reduction algorithm based on hierarchical autoencoder. Front Genet 2021;12:733906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Chen KH, Boettiger AN, Moffitt JR, et al. RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science 2015;348(6233):aaa6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Codeluppi S, Borm LE, Zeisel A, et al. Spatial organization of the somatosensory cortex revealed by osmFISH. Nat Methods 2018;15(11):932–5. [DOI] [PubMed] [Google Scholar]
- 124. Eng CL, Lawson M, Zhu Q, et al. Transcriptome-scale super-resolved imaging in tissues by RNA seqFISH. Nature 2019;568(7751):235–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Wang X, Allen WE, Wright MA, et al. Three-dimensional intact-tissue sequencing of single-cell transcriptional states. Science 2018;361(6400):eaat5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Stahl PL, Salmén F, Vickovic S, et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science 2016;353(6294):78–82. [DOI] [PubMed] [Google Scholar]
- 127. Rodriques SG, Stickels RR, Goeva A, et al. Slide-seq: a scalable technology for measuring genome-wide expression at high spatial resolution. Science 2019;363(6434):1463–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Dries R, Zhu Q, Dong R, et al. Giotto: a toolbox for integrative analysis and visualization of spatial expression data. Genome Biol 2021;22(1):78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Biancalani T, Scalia G, Buffoni L, et al. Deep learning and alignment of spatially resolved single-cell transcriptomes with Tangram. Nat Methods 2021;18(11):1352–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Kleshchevnikov V, Shmatko A, Dann E, et al. Cell2location maps fine-grained cell types in spatial transcriptomics. Nat Biotechnol 2022;40(5):661–71. [DOI] [PubMed] [Google Scholar]
- 131. Cang Z, Nie Q. Inferring spatial and signaling relationships between cells from single cell transcriptomic data. Nat Commun 2020;11(1):2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Karaiskos N, Wahle P, Alles J, et al. The Drosophila embryo at single-cell transcriptome resolution. Science 2017;358(6360):194–9. [DOI] [PubMed] [Google Scholar]
- 133. Abdelaal T, Mourragui S, Mahfouz A, et al. SpaGE: spatial gene enhancement using scRNA-seq. Nucleic Acids Res 2020;48(18):e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Elosua-Bayes M, Nieto P, Mereu E, et al. SPOTlight: seeded NMF regression to deconvolute spatial transcriptomics spots with single-cell transcriptomes. Nucleic Acids Res 2021;49(9):e50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Song Q, Su J. DSTG: deconvoluting spatial transcriptomics data through graph-based artificial intelligence. Brief Bioinform 2021;22(5):bbaa414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Bae S, Na KJ, Koh J, et al. CellDART: cell type inference by domain adaptation of single-cell and spatial transcriptomic data. Nucleic Acids Res 2022;50(10):e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Li B, Zhang W, Guo C, et al. Benchmarking spatial and single-cell transcriptomics integration methods for transcript distribution prediction and cell type deconvolution. Nat Methods 2022;19(6):662–70. [DOI] [PubMed] [Google Scholar]
- 138. Liu Q, Chen S, Jiang R, et al. Simultaneous deep generative modeling and clustering of single cell genomic data. Nat Mach Intell 2021;3(6):536–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Krzak M, Raykov Y, Boukouvalas A, et al. Benchmark and parameter sensitivity analysis of single-cell RNA sequencing clustering methods. Front Genet 2019;10:1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Xu C, Su Z. Identification of cell types from single-cell transcriptomes using a novel clustering method. Bioinformatics 2015;31(12):1974–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Zurauskiene J, Yau C. pcaReduce: hierarchical clustering of single cell transcriptional profiles. BMC Bioinformatics 2016;17:140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Lin P, Troup M, Ho JW. CIDR: ultrafast and accurate clustering through imputation for single-cell RNA-seq data. Genome Biol 2017;18(1):59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Guo M, Wang H, Potter SS, et al. SINCERA: a pipeline for single-cell RNA-Seq profiling analysis. PLoS Comput Biol 2015;11(11):e1004575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Jiang L, Chen H, Pinello L, et al. GiniClust: detecting rare cell types from single-cell gene expression data with Gini index. Genome Biol 2016;17(1):144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Peyvandipour A, Shafi A, Saberian N, et al. Identification of cell types from single cell data using stable clustering. Sci Rep 2020;10(1):12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Wang B, Ramazzotti D, de Sano L, et al. SIMLR: a tool for large-scale genomic analyses by multi-kernel learning. Proteomics 2018;18(2). doi: 10.1002/pmic.201700232. [DOI] [PubMed] [Google Scholar]
- 147. Kiselev VY, Kirschner K, Schaub MT, et al. SC3: consensus clustering of single-cell RNA-seq data. Nat Methods 2017;14(5):483–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Stuart T, Butler A, Hoffman P, et al. Comprehensive integration of single-cell data. Cell 2019;177(7):1888–1902.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Butler A, Hoffman P, Smibert P, et al. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 2018;36(5):411–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Hao Y, Hao S, Andersen-Nissen E, et al. Integrated analysis of multimodal single-cell data. Cell 2021;184(13):3573–3587.e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Wolf FA, Angerer P, Theis FJ. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol 2018;19(1):15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Cui Y, Zhang S, Liang Y, et al. Consensus clustering of single-cell RNA-seq data by enhancing network affinity. Brief Bioinform 2021;22:bbab236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol 2010;11(10):R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26(1):139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Chen MB, Jiang X, Quake SR, et al. Persistent transcriptional programmes are associated with remote memory. Nature 2020;587(7834):437–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Kharchenko PV, Silberstein L, Scadden DT. Bayesian approach to single-cell differential expression analysis. Nat Methods 2014;11(7):740–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Finak G, McDavid A, Yajima M, et al. MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol 2015;16:278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Qiu X, Hill A, Packer J, et al. Single-cell mRNA quantification and differential analysis with census. Nat Methods 2017;14(3):309–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Korthauer KD, Chu LF, Newton MA, et al. A statistical approach for identifying differential distributions in single-cell RNA-seq experiments. Genome Biol 2016;17(1):222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Wang T, Nabavi S. SigEMD: a powerful method for differential gene expression analysis in single-cell RNA sequencing data. Methods 2018;145:25–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Nabavi S, Schmolze D, Maitituoheti M, et al. EMDomics: a robust and powerful method for the identification of genes differentially expressed between heterogeneous classes. Bioinformatics 2016;32(4):533–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Delmans M, Hemberg M. Discrete distributional differential expression (D3E)--a tool for gene expression analysis of single-cell RNA-seq data. BMC Bioinformatics 2016;17:110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Vandenbon A, Diez D. A clustering-independent method for finding differentially expressed genes in single-cell transcriptome data. Nat Commun 2020;11(1):4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Saelens W, Cannoodt R, Todorov H, et al. A comparison of single-cell trajectory inference methods. Nat Biotechnol 2019;37(5):547–54. [DOI] [PubMed] [Google Scholar]
- 165. Lahnemann D, Köster J, Szczurek E, et al. Eleven grand challenges in single-cell data science. Genome Biol 2020;21(1):31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Trapnell C, Cacchiarelli D, Grimsby J, et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol 2014;32(4):381–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Aubin-Frankowski PC, Vert JP. Gene regulation inference from single-cell RNA-seq data with linear differential equations and velocity inference. Bioinformatics 2020;36(18):4774–80. [DOI] [PubMed] [Google Scholar]
- 168. Pratapa A, Jalihal AP, Law JN, et al. Benchmarking algorithms for gene regulatory network inference from single-cell transcriptomic data. Nat Methods 2020;17(2):147–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Pham D, Tan X, Xu J, et al. stLearn: integrating spatial location, tissue morphology and gene expression to find cell types, cell-cell interactions and spatial trajectories within undissociated tissues . bioRxiv. 2020; 2020.05.31.125658.
- 170. Palla G, Spitzer H, Klein M, et al. Squidpy: a scalable framework for spatial omics analysis. Nat Methods 2022;19(2):171–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Efremova M, Vento-Tormo M, Teichmann SA, et al. CellPhoneDB: inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat Protoc 2020;15(4):1484–506. [DOI] [PubMed] [Google Scholar]
- 172. Clark IC, Gutiérrez-Vázquez C, Wheeler MA, et al. Barcoded viral tracing of single-cell interactions in central nervous system inflammation. Science 2021;372(6540):eabf1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Guenthner CJ, Miyamichi K, Yang HH, et al. Permanent genetic access to transiently active neurons via TRAP: targeted recombination in active populations. Neuron 2013;78(5):773–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Cho JH, Huang BS, Gray JM. RNA sequencing from neural ensembles activated during fear conditioning in the mouse temporal association cortex. Sci Rep 2016;6:31753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Hrvatin S, Hochbaum DR, Nagy MA, et al. Single-cell analysis of experience-dependent transcriptomic states in the mouse visual cortex. Nat Neurosci 2018;21(1):120–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Wetmore DZ, Mukamel EA, Schnitzer MJ. Lock-and-key mechanisms of cerebellar memory recall based on rebound currents. J Neurophysiol 2008;100(4):2328–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Berto S, Fontenot MR, Seger S, et al. Gene-expression correlates of the oscillatory signatures supporting human episodic memory encoding. Nat Neurosci 2021;24(4):554–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Lau SF, Cao H, Fu AKY, et al. Single-nucleus transcriptome analysis reveals dysregulation of angiogenic endothelial cells and neuroprotective glia in Alzheimer's disease. Proc Natl Acad Sci U S A 2020;117(41):25800–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Mathys H, Davila-Velderrain J, Peng Z, et al. Single-cell transcriptomic analysis of Alzheimer's disease. Nature 2019;570(7761):332–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Grubman A, Chew G, Ouyang JF, et al. A single-cell atlas of entorhinal cortex from individuals with Alzheimer's disease reveals cell-type-specific gene expression regulation. Nat Neurosci 2019;22(12):2087–97. [DOI] [PubMed] [Google Scholar]
- 181. Olah M, Menon V, Habib N, et al. Single cell RNA sequencing of human microglia uncovers a subset associated with Alzheimer's disease. Nat Commun 2020;11(1):6129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Jiang J, Wang C, Qi R, et al. scREAD: a single-cell RNA-Seq database for Alzheimer's disease. iScience 2020;23(11):101769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Murre JM, Chessa AG, Meeter M. A mathematical model of forgetting and amnesia. Front Psychol 2013;4:76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Mandwal A, Orlandi JG, Simon C, et al. A biochemical mechanism for time-encoding memory formation within individual synapses of Purkinje cells. PLoS One 2021;16(5):e0251172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Zhang Y, Smolen P, Alberini CM, et al. Computational model of a positive BDNF feedback loop in hippocampal neurons following inhibitory avoidance training. Learn Mem 2016;23(12):714–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Rizvi AH, Camara PG, Kandror EK, et al. Single-cell topological RNA-seq analysis reveals insights into cellular differentiation and development. Nat Biotechnol 2017;35(6):551–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Shin J, Berg DA, Zhu Y, et al. Single-cell RNA-Seq with waterfall reveals molecular cascades underlying adult neurogenesis. Cell Stem Cell 2015;17(3):360–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Sebe-Pedros A, Saudemont B, Chomsky E, et al. Cnidarian cell type diversity and regulation revealed by whole-organism single-cell RNA-Seq. Cell 2018;173(6):1520–1534.e20. [DOI] [PubMed] [Google Scholar]
- 189. Papatheodorou I, Moreno P, Manning J, et al. Expression atlas update: from tissues to single cells. Nucleic Acids Res 2020;48(D1):D77–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Regev A, et al. The human cell atlas. Elife 2017;6:e27041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Sunkin SM, Ng L, Lau C, et al. Allen Brain Atlas: an integrated spatio-temporal portal for exploring the central nervous system. Nucleic Acids Res 2013;41(Database issue):D996–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Han X, Wang R, Zhou Y, et al. Mapping the mouse cell atlas by microwell-Seq. Cell 2018;172(5):1091–1107.e17. [DOI] [PubMed] [Google Scholar]
- 193. Tabula Muris Consortium, Overall coordination, Logistical coordination, et al. Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature 2018;562(7727):367–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Marco A, Meharena HS, Dileep V, et al. Mapping the epigenomic and transcriptomic interplay during memory formation and recall in the hippocampal engram ensemble. Nat Neurosci 2020;23(12):1606–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in the article and in its online supplementary material.