Abstract
N-glycanase 1(NGLY1) catalyzes the removal of N-linked glycans from newly synthesized or misfolded protein. NGLY1 deficiency is a recently diagnosed rare genetic disorder. The affected individuals present a broad spectrum of clinical features. Recent studies explored several possible molecular mechanisms of NGLY1 deficiency including defects in proteostasis, mitochondrial homeostasis, innate immunity, and water/ion transport. We demonstrate abnormal accumulation of endoplasmic reticulum-associated degradation (ERAD) substrates in NGLY1-deficient cells. Global quantitative proteomics discovered elevated levels of endogenous proteins in NGLY1-defective human and mouse cells. Further biological validation assays confirmed the altered abundance of several key candidates that were subjected to isobarically labeled proteomic analysis. CCN2 was selected for further analysis due to its significant increase in different cell models of NGLY1 deficiency. Functional assays show elevated CCN2 and over-stimulated TGF-β signaling in NGLY1-deficient cells. Given the important role of CCN2 and TGF-β pathway in mediating systemic fibrosis, we propose a potential link of increased CCN2 and TGF-β signaling to microscopic liver fibrosis in NGLY1 patients.
Keywords: NGLY1, proteomics, CCN2, TGF-β signaling, fibrosis
1. Introduction
NGLY1 deficiency (OMIM 615273) is the first identified congenital disorder of deglycosylation (CDDG). This disorder is a very rare, autosomal recessive genetic disease caused by mutations in the NGLY1 gene[1]. The first NGLY1 patient was diagnosed by whole-exome sequencing in 2012, with over 70 patients identified worldwide since then [2]. Patients with NGLY1 deficiency present diverse clinical phenotypes, including systemic developmental delay, seizures, hypotonia, liver fibrosis, chronic constipation, and lack of tears (alacrima) and sweat [1, 3-5].
N-Glycanase 1 (NGLY1) is a cytoplasmic enzyme that catalyzes the removal of N-glycans from glycosylated proteins and facilitates misfolded protein degradation by endoplasmic reticulum-associated degradation (ERAD). The loss-of-function mutations in NGLY1 gene destroy the expression of NGLY1 protein and thus remove the enzyme’s deglycosylation activity[6]. Given the direct role of NGLY1 in removing glycans from misfolded proteins prior to ERAD[7], the loss of NGLY1 activity may result in the accumulation of misfolded protein causing ER stress.
In this study, we show the accumulation of ERAD substrates in NGLY1-deficient cell when compared to normal cells. We hypothesize that NGLY1 is responsible for the clearance of a subset of misfolded proteins, which may be abnormally accumulated in the absence of NGLY1. To identify these accumulated proteins, we applied a robust quantitative proteomic strategy to detect these proteins in NGLY1-defective cells. Follow-up biological validation assays confirmed the increase of several selected proteins. Among them, CCN2 displayed the highest fold change in different NGLY1-defective cells and demonstrated a critical role in the regulation of TGF-β signaling. This indicates CCN2 may be an indicator of fibrosis in NGLY1 deficiency, a potential biomarker for diagnosis and prognosis, and a potential therapeutic target for the development of NGLY1 treatments.
2. Materials and Methods
2.1. Materials
Most of the reagents were purchased from Sigma-Aldrich (St. Louis, MO). Skin fibroblasts derived from healthy controls and NGLY1 patients were obtained from Coriell Institute for Medical Research (Camden, NJ). WT, PNGase−/−, ENGase−/− and PNGase/ENGase DKO MEFs were kind gifts of Dr. Tadashi Suzuki (Institute of Physical and Chemical Research Cluster for Pioneering Research, Saitama, Japan). HeLa, MDA-MB-231, and U2Os cancer cell lines were kind gifts from Dr. Don Newmeyer (La Jolla Institute for Immunology, San Diego, CA). HEK293 cells, Dimethylsulfoxide (DMSO) and Universal Mycoplasma Detection Kit were purchased from ATCC (Manassas, VA). Dulbecco’s modified Eagle’s medium (DMEM), Dulbecco's phosphate-buffered saline (DPBS), Ultrapure water, Nunc™ Lab-Tek™ II Chamber Slide™, EDTA (0.5 M), Halt™ Protease and Phosphatase Inhibitor Cocktail, UltraPure™ SDS Solution (10%), Pierce™ Rapid Gold BCA Protein Assay Kit, TMTpro™ 16plex Label Reagent Set, TMT10plex™ Isobaric Label Reagent Set, NuPAGE™ Bis-Tris 4-12% precast gels, NuPAGE™ MOPS SDS Running Buffer (20X), NuPAGE™ MES SDS Running Buffer (20X), Restore™ PLUS Western Blot Stripping Buffer, PageRuler™ Plus Prestained 10-250kDa Protein Ladder, COL15A1 antibody, Hoechst 33324, 16% Paraformaldehyde Aqueous Solution, Lipofectamine™ RNAiMAX transfection reagent and scrambled siRNA (NC1), Instant Nonfat Dry Milk, Trypsin-EDTA (0.25%), Fetal bovine serum (FBS) and L-Glutamine (200 mM) were obtained from Thermo Fisher Scientific (Carlsbad, CA). MG132 was purchased from Tocris Bioscience (Bristol, United Kingdom). Amaxa nucleofector II device and Amaxa® Human Dermal Fibroblast Nucleofector® Kit were purchased from Lonza (Walkersville, MD). Nitrocellulose Membrane, Precision Plus Protein™ Dual Color Standards and Trans-Blot® Turbo™ RTA Midi Nitrocellulose Transfer Kit were purchased from Bio-Rad (Hercules, CA). ON-TARGETplus human NGLY1 and mouse Ccn2 siRNA Smartpool were designed and synthesized by Dharmacon (Horizon Discovery, Cambridge, UK). Autophagy Antibody Sampler kit, and antibodies against Akt1, Phospho- Akt (S473), c-Myc, Cyclin D1, GAPDH, Phospho-eEF2 (Thr56), MCAM, COL1A1, COL3A1, Phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204), p44/42 MAPK (Erk1/2), Phospho-SAPK/JNK (Thr183/Tyr185), SAPK/JNK, Phospho-p38 MAPK (Thr180/Tyr182), Phospho-SMAD2 (Ser465/Ser467), Smad2, E-Cadherin, N-Cadherin, Vimentin, Snail were obtained from Cell Signaling Technology (Danvers, MA). GFP antibody was purchased from Fitzgerald (Acton, MA). SuperFect Transfection Reagent was purchased from Qiagen (GmbH, Germany). Ubiquitin (Ub), F-box protein 6 (FBXO6), (REB-2 (ATF4), COL1A2, COL5A1, antibodies and Protein G PLUS-Agarose were purchased from Santa Cruz Biotechnology (Dallas, TX). CCN2, CCN1, TAGLN, FHL1, HSPB1, HRP donkey anti-sheep IgG antibodies were purchased from R&D Systems (Minneapolis, MN). KDEL antibody was obtained from Enzo Life Sciences (Farmingdale, NY). COL14A1 antibody was purchased from MyBioSource (San Diego, CA). Heat shock protein antibodies, including HSP 90, 70 and 40, were kind gifts from Dr. Steven Reed (the Scripps Research Institute, La Jolla, CA). Recombinant human TGF-β3 protein, Direct-Blot™ HRP anti-β-actin, HRP Goat anti-mouse IgG, HRP donkey anti-rabbit IgG, HRP mouse anti-rat IgG antibodies, Western-Ready™ ECL Substrate Kit, and antibody against p38 MAPK were purchased from BioLegend (San Diego, CA). NewBlot Stripping Buffer was purchased from LI-COR (Lincoln, NE). PNGase F was purchased from New England Biolabs (Ipswich, MA).
2.2. Cell culture and treatment
Informed consent was obtained from all human fibroblast donors, who are not identifiable and anonymous. Control and patient fibroblasts were cultured in DMEM (with 1g/L glucose) with 10% FBS. Mouse embryonic fibroblasts (MEFs), HeLa, MDA-MB-231 and U2Os cancer cells, and HEK293 cells were maintained in DMEM (with 4.5g/L glucose) supplemented with 10% FBS. All cells were maintained 37 °C and 5% CO2. The cultured cells were routinely (every 10 passages of cells) quarantined for mycoplasma contamination using PCR-based mycoplasma detection kit.
Cells were grown to 80-90% confluence followed by the treatment with 5μM MG132 for 16h, 1μM thapsgargin (TG) for 2h-24h, 100ng/mL TGF-β3 protein or 10uM SB505124 for 24h.
2.3. Comparative Proteomics
Six different cell lines, including two human skin fibroblast lines (wild type and NGLY1 patient cells) and four MEF lines (WT, PNGase−/−, ENGase−/−, and PNGase/ENGase DKO), were grown to 80-90% confluence. Cells were collected as pellet and stored at −80°C temporarily for proteomic analysis. Three triplicate cell pellets were analyzed each composed of 5 million cells. Each of frozen cell pellet was suspended in 150 μL 8M urea Tris-HCl pH8 lysis buffer with freshly added protease inhibitor cocktail. Samples were homogenized by ultrasonication 15s, 3 pulse with 15s between runs. Homogenized samples were centrifuged at 15000 g for 15 min, and the supernatants were transferred to new Eppendorf tubes. Protein concentrations of the samples were determined by BCA assay following the manufacture’s protocol.
Eighty or 40 μg of protein from each of the 12 mouse or 6 human cell samples were reduced by dithiothreitol, alkylated by iodoacetamide and precipitated by cold acetone (−20°C) overnight. Samples were centrifuged at 8000 g for 5 min at 0 °C, and the supernatants were removed. Protein pellets were air dried for 10 min and dissolved in 40 μL 50mM tri-ethyl ammonium bicarbonate (TEAB) with 1 μg sequencing grade trypsin and incubated at room temperature overnight. Twenty-five μg of peptides were taken from each sample and labeled with TMTpro Labels or TMT10plex tags according to the protocol from the manufacture’s instruction (Table S1). The 12 TMTpro and 6 TMT10plex labeled samples were then combined separately, desalted and half of the desalted sample was used for offline fractionation using a high pH reversed phase HPLC method. The samples were first separated into 40 or 32 collections for TMTpro and TMT10plex labeled samples respectively. The collections were then combined into 10 fractions for TMTpro labeled MEF samples and 8 fractions for TMT10plex labeled human dermal fibroblast (HDF) samples. The fractions were dried in a Speedvac and reconstituted in 30 μL 1% acetic acid for LCMS analysis.
The TMT-labeled peptide digest was analyzed on a ThermoFisher Scientific UltiMate 3000 UHPLC system (ThermoFisher Scientific, Bremen, Germany) interfaced with a ThermoFisher Scientific Orbitrap Fusion Lumos Tribrid mass spectrometer (Thermo Scientific, Bremen, Germany). Liquid chromatography was performed prior to MS/MS analysis for peptide separation. The HPLC column used is a Thermo Scientific™ Acclaim™ PepMap™ 100 C18 reversed-phase capillary chromatography column (Thermo Fisher Scientific, Waltham, MA) 75 μm x 15 cm, 2 μm, 100 Å. Five μL volumes of the peptide extract were injected and peptides eluted from the column by a 110-minute acetonitrile/0.1% formic acid gradient at a flow rate of 0.30 μL/min and introduced to the source of the mass spectrometer on-line. Nano electrospray ion source was operated at 2.3 kV. The digest was analyzed using the data dependent multitask capability of the instrument acquiring full scan mass spectra using a Fourier Transform (FT) Orbitrap analyzer to determine peptide molecular weights and higher-energy collisional dissociation (HCD) MS/MS product ion spectra with the Orbitrap FT analyzer at 38% normalized collision energy (NCE) to determine both the amino acid sequence and the quantities of the isobaric tags. The MS method used in this study was a data-dependent acquisition (DDA) with 3 second duty cycle. It includes one full scan at a resolution of 120,000 followed by as many MS/MS scans as possible on the most abundant ions in that full scan. The MS/MS HCD scan start at 110 m/z with a resolution of 30,000. Dynamic exclusion was enabled with a repeat count of 1 and ions within 10 ppm of the fragmented mass were excluded for duration of 60 seconds.
The data were analyzed using Proteome Discoverer V2.3 (Thermo Fisher Scientific, Waltham, MA) with the search engine Sequest-HT which is integrated in the Proteome Discoverer software. The protein sequence databases used to search the MS/MS spectra were the Uniprot mouse protein database containing 16996 entries, and the Uniprot human protein database containing 20353 entries, respectively, with automatically generated decoy databases (reversed sequences). The protease was set to full activity trypsin with a maximum of two missed cleavages. Oxidation of Methionine and acetylation of protein N-terminus were set as dynamic modifications and carbamidomethylation of Cysteine, TMT6plex of Lysine and peptide N-terminus were set as static modifications. The precursor mass tolerance for these searches was set to 10 ppm and the fragment ion mass tolerance was set to 0.02 Da. Keratins were known contaminants and were excluded from identified proteins. A false discovery rate (FDR) was set to 1% for both peptide and protein identification and calculated using the number of identified peptides/proteins from decoy database divided by the total number of identified peptides/proteins. Two peptides were required for positive protein identification to decrease the chance of false discovery by a random match.
Relative quantitation of the samples labeled by different isobaric tags was done by the Reporter Ions Quantifier node in Proteome Discoverer using the intensity of the reporter ions from MS/MS scans. The m/z tolerance of the reporter ions was set to 20 ppm, and the ion selection was set to most confident centroid. Quantitative values were normalized by the total amount of peptide in each label channel. Peptide used for quantification was set to Unique+Razor, and precursor Co-isolation threshold was set to 50%. Razor-peptides are non-unique peptides and these are assigned to the protein group containing the largest number of other peptides, according to Occam's razor principle.
2.4. Electroporation
As described by He et al [6]. pcDNA3.1-TCRα-GFP and pcDNA3.1- A1AT-NHK-GFP plasmids were kind gifts from Dr. Randal J. Kaufman (Sanford Burnham Prebys Medical Discovery Institute, La Jolla, CA). Cells were harvested 48h posterior to electroporation for either total protein extraction or separating soluble and insoluble proteins.
2.5. Transient transfection
Cells were transfected with plasmids using SuperFect Transfection reagent according to manufacturer’s protocol. Total proteins were extracted 24h-48h posterior to transfection.
2.6. IF Microscopy
As described by He et al[8]. Briefly, the transfected Cells were grown in 8-well Chamber Slide to 80%. The tells were fixed followed by counterstaining the nuclei with Hoechst 33324. The images were captured with an Olympus IX71 inverted microscope, using an Olympus ×40 objective lens and a Hamamatsu Camera ORCA-R2 (Shinjuku, Tokyo, Japan). The fluorescence was measured and digitized by software ImageJ 1.41 based on ~1,000 cells and was normalized by transfection efficiency.
2.7. Analysis of XBP1 Splicing
U- and S- isoforms of human XBP1 mRNA were amplified and analyzed as described by Castillo-Carranza, et al[9].
2.8. siRNA knockdown
As described by He et al[8]. Briefly, cells were transfected with scrambled siRNA (NC1) and increased concentrations of NGLY1 or Ccn2 siRNA. Total protein was extracted 72h after transfection. For the TGF-β3 treatment, 50 nM NC1 or Ccn2 siRNA were transfected into the WT and PNGase−/− MEFs. Forty-eight hours posterior to transfection, cells were treated with 100ng/mL TGF-β3 or 10μM SB505124 for another 24 hours prior to WB analysis.
2.9. Cell protein preparation and Western blot (WB)
The total protein extraction, protein quantitation and WB analysis were performed as described by He et al[8]. To separate soluble from insoluble proteins, cell pellets were suspended in cold RIPA buffer (25mM Tris•HCl pH 7.6, 150mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS), placed on ice for 10min, and then centrifuged at 12,000 g for 15 min. The supernatant was collected (soluble fraction) and the pellet was dissolved in the same volume of total lysis buffer (insoluble fraction).
2.10. Statistical Analysis
Data are expressed as means± standard deviation (SD). Unpaired Student t-test was used to compare means between groups. A value of P < 0.05 was considered statistically significant. GraphPad Prism 8.0 software programs were used to plot the data and perform the statistics.
3. Results and Discussion
3.1. NGLY 1 deficiency results in the accumulation of transfected ERAD model substrates.
NGLY1 deglycosylates misfolded proteins exported from the ER into the cytosol for destruction via the proteasome. Deglycosylation is a prerequisite for subsequent proteasome-mediated degradation of some, but not all, misfolded glycoproteins[10, 11]. Indeed, a patient (NG1) liver biopsy showed accumulation of an amorphous unidentified substance throughout the cytosol[12], which may be caused by abnormal accumulation of misfolded glycoproteins due to impaired cytosolic degradation. To protect the cell from the toxic effects of misfolded protein accumulation, many of these proteins are degraded by ERAD[13]. Misfolded proteins become ubiquitinated on the cytosolic side of the ER membrane by E3 ubiquitin ligases and are targeted to the proteasome for degradation[14]. If NGLY1 deficiency causes accumulation of misfolded proteins, we expect to see higher levels of ubiquitin (Ub) modification to the misfolded proteins. However, we did not observe this in NGLY1 patient cells (Figure S1A). This may result from the activity of cytosolic de-ubiquitinating enzymes.
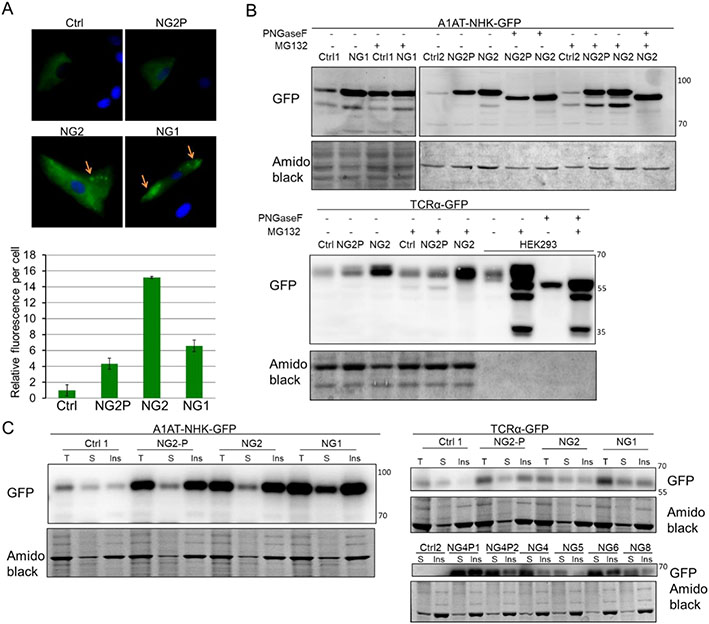
Since deglycosylation of misfolded glycoproteins precedes proteasomal degradation, we speculated that NGLY1 deficiency may result in the accumulation of ERAD substrates. T-cell receptor alpha chain (TCRα)-GFP [7, 15] and α1-antitrypsin genetic variant null Hong Kong (A1AT-NHK)-GFP[16, 17] are well-characterized ERAD substrates. Both of the substrates are N-glycosylated, but pathways directing them to dislocation sites at the ER membrane are different[18]. We transfected human skin fibroblasts with these two ERAD substrates to examine if the lack of NGLY1 will alter the expression of these transfected substrates. The transfection efficiency by electroporation was tested in control and patient fibroblast lines, and the efficiency ranged from 20% to 50% (Figure S2). The existence of those substrates can be easily detected by GFP fluorescence microscopy and WB. By microscopy, we observed a 5- to 10-fold increase in GFP fluorescence in the cytosol in patient cells transfected with A1AT-NHK-GFP (Figure 1A) as compared to controls. TCRα-GFP fluorescence was hardly detected by microscopy due to low transfection efficiency by electroporation. Moreover, A1AT-NHK-GFP and TCRα-GFP accumulation was also detected in patient cell total lysates by WB analysis (Figure 1B). We observed some punctate distribution of A1AT-NHK-GFP in the cytosol of patient fibroblasts (Figure 1A) suggesting accumulation of misfolded protein aggregates perhaps within aggresomes or inclusion bodies[19]. Since detergent-insoluble cell pellets are known to contain aggregated proteins[20], we used RIPA buffer to separate soluble (S) proteins and insoluble (Ins) protein aggregates for WB analysis. When the same amount of total protein (T) and equal volume of soluble and insoluble protein were analyzed, we found that the pronounced increase of A1AT-NHK accumulation in patient cells (T lanes) is mainly attributable to the increase of insoluble protein in those cells (Ins lanes) (Figure 1C, Figure S3). These data suggest that the primary pool of A1AT-NHK accumulation is in the protein aggregates. Unlike A1AT-NHK substrate, most of TCRα was solubilized by detergent in patient cells (Figure 1C). These data indicates that NGLY1 deficiency leads to the accumulation of some selected ERAD substrates.
Figure 1. NGLY1 deficiency causes accumulation of transfected ERAD substrates.
(A) A1AT-NHK-GFP transfection followed by fluorescence microscopy imaging. The fibroblasts were transfected with A1AT-NHK-GFP by electroporation. After cultured for 48h, the cells were fixed followed by counterstaining the nuclei with Hoechst 33324. The images were acquired with an inverted microscope, using a ×40 objective lens (Upper panel). The fluorescence was quantitated and normalized by transfection efficiency (Lower panel) as indicated in Methods. The error bars represent SD of 3 independent assays. The yellow arrows indicate protein accumulation possibly in aggresome or inclusion bodies. (B) A1AT-NHK-GFP and TCRα-GFP substrates transfection followed by WB analysis of total protein. Cells were treated with DMSO or 5μM MG132 for 16h. Equal amount (20μg) of total protein from particular lines (except for HEK293 with 1μg total protein loading) with or without MG132 treatment was also treated with PNGase F to remove the attached N-glycans. (C) WB analysis of RIPA buffer soluble (S) and insoluble (ins) ERAD substrates. Equal amount (5-20μg) of total protein (T) and equal volume of soluble and insoluble protein in each sample were analyzed. Amido black stain was used as loading control. Ctrl: control fibroblast; NG: NGLY1 patient fibroblast; NG-P (1/2): NGLY1 patient parent’s fibroblast (1, father; 2, mother).
Unexpectedly, the accumulation of ERAD substrates in NGLY1-deficient fibroblasts is not associated with any of the compensatory cellular responses, such as unfolded protein response (UPR)[21], proteostasis[22], and autophagy[23] (Figure S1, B-D). One of the possible reasons is that the role of NGLY1 in glycoprotein quality control may not be strictly conserved in evolution. Some other N-glycan releasing enzymes, such as endo-β-N-acetylglucosaminidase (ENGase) [24], may also be involved in protein quality control by deglycosylating glycoproteins, resulting in the generation of N-GlcNAc proteins[25].
3.2. Global tandem mass tags (TMT)-labeled proteomics uncovers upregulated proteins in NGLY1-deficient cells.
Accumulation of ERAD substrates in an NGLY1-dependent manner prompted us to speculate that NGLY1 deficiency may result in the abnormal buildup of endogenous proteins, which could be used as useful markers for the elucidation of pathology, diagnosis, and therapeutic targets for the disease. Hence, we adopted a robust comparative proteomic platform to compare and quantitate differentially expressed proteins between control and NGLY1-defective cells. The experiments were performed on skin fibroblast lines (normal vs. NGLY1 patient) and MEF lines, namely WT and PNGase (Ngly1 in mouse)−/−. PNGase KO caused precipitates of ERAD substrate and embryonic lethality[26], which could be partially rescued by the additional KO of ENGase gene (PNGase/ENGase double knockout [DKO])[27]. In PNGase −/− MEFs, the presence of ENGase aggravated protein aggregation by generating aggregation-prone N-Glc-NAc proteins [28]. In view of the critical roles of ENGase in NGLY1 deficiency, ENGase−/− and PNGase/ENGase DKO MEFs were also included in the proteomic analysis. Each cell line was analyzed in triplicate (Figure 2A). The samples were homogenized/lysed, trypsinized, TMT (a set of chemicals with identical mass, but yield reporter ions of different mass after fragmentation due to the varied distribution of heavy isotopes in their structure) labeled (Table S1), combined and fractionated by HPLC prior to LC/MS analysis. The data were searched against the UniProtKB databases with the program Proteome Discoverer 2.3.
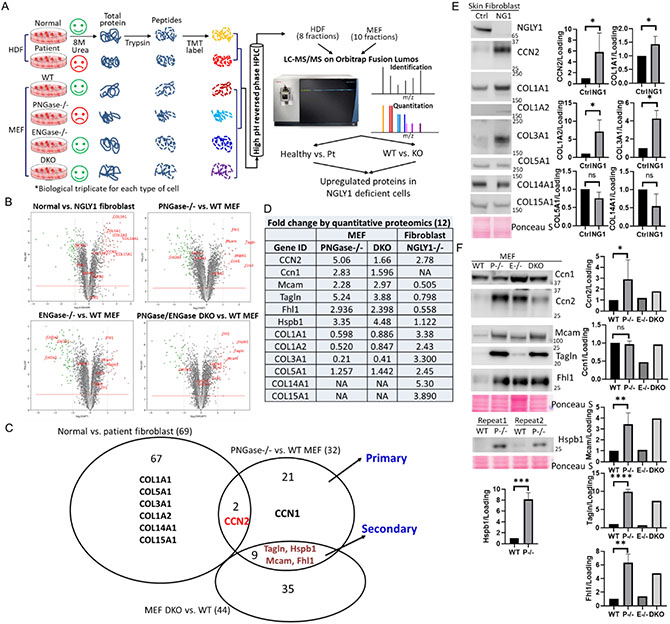
Figure 2. Quantitative proteomic analysis of upregulated proteins in NGLY1-deficient cells.
(A)Flow chart of TMT labeling proteomics. HDF, human dermal fibroblast; Pt, patient with NGLY1 deficiency. (B) Volcano plots demonstrating statistical significance (P value, y axis) versus magnitude of change (fold change, x axis). (C) Venn diagram demonstrating the upregulated proteins between cell lines. (D) Table showing the IDs and fold changes of the selected candidates. (E and F) Protein level change validated by WB analysis. Ponceau S stain was used as loading control. Data with error bars represent means ± SD from at least two independent experiments. Unpaired Student t-test was used to compare means between normal and NGLY1-deficient cells. *P < 0.05, **P < 0.01, ****P < 0.0001.
A total of 5,574 proteins (Table S2) and 5085 proteins (Table S3) were identified with quantitative information from four MEF lines and two human fibroblast lines, respectively. The goal of these experiments is to compare the abundance of proteins identified across the samples. We used two different filters to narrow down the overexpressed candidate list, including ≥2-fold change and P≤0.05 across 3 biological replicates. A total of 32 (PNGase−/− vs. WT MEF), 20 (ENGase−/− vs. WT MEF), 44 (PNGase/ENGase DKO vs. WT MEF), and 69 (normal vs. NGLY1 patient) overexpressed proteins fit these criteria and were highlighted in Table S4-7. Volcano plots are used to demonstrate the fold changes (x) and significance (y) (Figure 2B).
3.3. Biological validation
Considering the rescue effects of PNGase/ENGase DKO, we propose that the changes in protein levels seen in PNGase−/− cells but not in DKO cells reflect the primary consequences of the deficiency, whereas changes seen in both cell types most likely reflect a secondary effect of the NGLY1 deficiency. Thus, we further compared the upregulated proteins between groups (Figure 2C). We selected 12 potential targets that differed the most in these comparisons (Figure 2, B and D) and had commercially available antibodies for further WB validation. Eight out of twelve (8/12, 75%) candidate proteins were verified to have higher levels in NGLY1-defective human fibroblasts (CCN2, COL1A1, COL1A2, and COL1A3, in Figure 2E) and MEFs (Ccn2, Mcam, Tagln, Fhl1, and Hspb1, in Figure 2F) by WB analysis. It should be noted that several collagen family proteins were observed to be increased in the NGLY1 patient cells (Figure 2, B-D).
3.4. Increased CCN2 and TGF-β signaling in NGLY1-defective cells
CCN2, also known as connective tissue growth factor (CTGF), is a secreted N-glycosylated protein[29] and plays pivotal roles in cell proliferation, mobility, and cell differentiation[30, 31]. It is widely acknowledged that CCN2 is a critical downstream and synergistic mediator of TGF-β-induced epithelial-mesenchymal transition (EMT) and fibrosis[31]. The binding of secreted CCN2 to TGF-β facilitates its interaction with TGF-β receptor (TGFβR) and enhances TGF-β signaling [32]. Researchers also found TGF- β signaling alone [33, 34], or by synergizing with MLK1[35] and Hippo pathway[36], could transcriptionally enhance CCN2 expression. Hence, CCN2 could directly or indirectly amplify TGF-β signaling by establishing a positive-feedback autocrine loop[37].
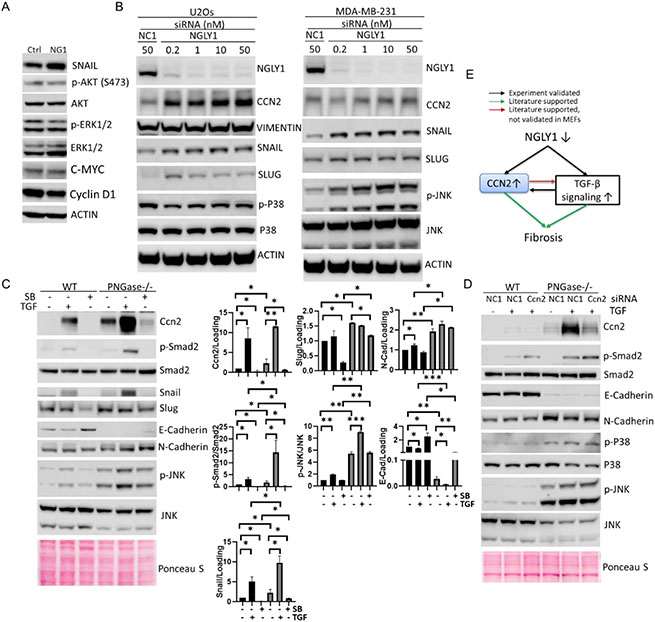
We detected markedly increased CCN2 in both NGLY1 patient cells (2.8-fold increase) and PNGase−/− MEFs (5.1-fold increase) when compared to control human fibroblast and WT MEFs respectively by comparative proteomic (Figure 2, B-D) and WB analyses (Figure 2, E and F). These results suggest CCN2 may be an indicator of NGLY1 deficiency. Since mounting evidence indicates that CCN2 may be involved with TGF-β signaling-induced systemic fibrosis as discussed above, we extensively tested the activity of different signal transducers residing in canonical (E/N-CADHERIN, SNAIL, SLUG, VIMENTIN) and non-canonical (phosphorylation of AKT, ERK1/2, JNK and P38) TGF-β signaling pathways in NGLY1-defective cells. WB analysis showed a moderate increase of SNAIL expression, but no change in AKT and ERK1/2 phosphorylation in NGLY1 patient fibroblasts (Figure 3A), indicating the constitutive over-stimulation of canonical TGF-β signaling in NGLY1 patient cells. In MDA-MB-231-LM2 and U2Os human cancer cells, siRNA knockdown of NGLY1 upregulated CCN2 expression and activate canonical (elevated levels of VIMENTIN, SNAIL and SLUG) and non-canonical (increased phosphorylation of JNK) TGF-β signal transduction (Figure 3B), suggesting a universal relationship between NGLY1 function and TGF-β signaling in human cells.
Figure 3. Elevated CCN2 and over-stimulated TGF-β signaling in NGLY1-defective cells.
(A)WB analysis of TGF-β signaling in control (Ctrl) and patient (NG1) fibroblasts. (B) siRNA knockdown of NGLY1 followed by WB analysis of the expression of NGLY1 and TGF-β signal transducers. NC1, scrambled siRNA. (C) TGF-β and TGFβR inhibitor treatment followed by WB analysis of Ccn2 and TGF-β signaling in MEFs. Cells were treated with 100ng/mL TGF-β3 (TGF) or 10μM SB505124 (SB) for 24h prior to WB analysis. Data represent means ± SD from at least two independent experiments. Unpaired Student t-test was used to compare means between two groups. *P < 0.05, **P < 0.01, ***P < 0.001. (D) siRNA knockdown of Ccn2 followed by TGF-β treatment in MEFs. Cells were transfected with 50nM siRNA for 48h and treated with 100ng/mL TGF for 24h. Actin or ponceau S stain were used as loading control. (E) Schematic model of fibrosis mediated by elevated CCN2 in NGLY1 disease.
To sort out the relationship between Ccn2 and TGF-beta signaling in NGLY1 deficiency, we further challenged MEFs with exogenous recombinant TGFβ3 protein and SB505124, a selective inhibitor of TGFβR. TGF-β3 protein treatment enhanced the phosphorylation of Smad2, JNK, and expression of Ccn2, Snail, N-Cadherin, and Slug, repressed the expression of E-cadherin, which was further enhanced by the inactivation of PNGase (Figure 3C). The effect of TGFβR inhibitor treatment was the opposite of the effect observed for treatment with TGFβ3 protein (Figure 3C). This indicates that NGLY1 deficiency could potentiate TGF-β stimulated canonical/ non-canonical signaling pathways. As Ccn2 could serve as both downstream effector and upstream regulator of TGF-β signaling[37], we further explored Ccn2’s modulation of TGF-β signaling in MEFs. We applied optimized concentration (50nM) of siRNA to knock down the expression of Ccn2 (Figure S4), followed by exposing WT and PNGase−/− MEFs to TGF-β. The mitigated Ccn2 expression did not significantly attenuate TGF-β protein stimulated TGF-β signaling (Figure 3D), indicating the increase of Ccn2 does not further amplify the over-stimulated TGF-β signaling by inactivation of PNGase.
Microscopic liver fibrosis has been noted recently in several NGLY1 patients[1, 38, 39], indicating that systemic fibrosis could be one of the important contributing factors to NGLY1 deficiency disease progression. Mounting evidence have demonstrated TGF-β mediated upregulation of CCN2 and systemic fibrosis. CCN2 is required for TGF-β-induced the overexpression of collagen[40]. It can also bind to α6β1 integrin and stimulate collagen deposition[41]. Consistently, we detected the build-up of a family of Collagens and CCN2, and found activation TGF-β signaling in NGLY1-defective cells. It suggests that NGLY1 deficiency caused the accumulation of CCN2 and the over-activation of TGF-β pathway may contribute to the organ fibrosis in NGLY1 patients (Figure 3E). Further in-depth investigation of CCN2 as a useful biomarker and TGF-β signaling dysregulation in NGLY1-defective animal models and NGLY1 patients will deepen our understanding of the molecular mechanism of NGLY1 deficiency and provide clues to discover druggable target for NGLY1 therapeutics.
In this study, we showed abnormal accumulation of engineered ERAD substrates in NGLY1 patient cells. Global quantitative proteomics further uncovered increased expression of a few endogenous proteins in NGLY1-defective cells. Biological verification studies proved the validity of those potential markers in NGLY1-deficient human and mouse cells. The functional assays showed NGLY1 deficiency caused over-stimulation of TGF-β signaling and over-expression of CCN2, suggesting critical roles of CCN2 and TGF-β pathway in pathologies of NGLY1 deficiency, especially in aspect of systemic fibrosis.
Supplementary Material
Highlights.
NGLY1 deficiency is a rare genetic disorder.
There is abnormal accumulation of ERAD substrates in NGLY1-deficient cells.
Proteomics identifies increased proteins in NGLY1-defective cells.
CCN2 as a potential marker due to role in mediating fibrosis via TGF-β signaling.
Acknowledgments:
We appreciate NGLY1 families for their cooperation. We appreciate Dr. Tadashi Suzuki (Institute of Physical and Chemical Research Cluster for Pioneering Research, Saitama, Japan) for providing WT, PNGase−/−, ENGase−/− and PNGase/ENGase DKO MEFs. We thank Dr. Don Newmeyer (La jolla Institute for Immunology, San Diego, CA) for sharing the HeLa, MDA-MB-231, and U2Os cell lines with us. We thank Dr. Steven Reed (the Scripps Research Institute, La jolla, CA) for sharing with us the heat shock protein antibodies. We also appreciate Dr. Randal J. Kaufman (Sanford Burnham Prebys Medical Discovery Institute, La Jolla, CA) for providing TCRα-GFP and A1AT-NHK-GFP plasmids. This work was supported by the Bertrand Might Research Fund, the Rocket Fund and LECOMT internal grant and Research foundation. The Fusion Lumos instrument was purchased via an NIH shared instrument grant, 1S10OD023436-01.
Footnotes
Conflicts of Interest: The authors declare that they have no competing interests.
References
- 1.Enns GM, et al. , Mutations in NGLY1 cause an inherited disorder of the endoplasmic reticulum-associated degradation pathway. Genet Med, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Might M and Might CC, What happens when N = 1 and you want plus 1? Prenat Diagn, 2017. 37(1): p. 70–72. [DOI] [PubMed] [Google Scholar]
- 3.Lam C, et al. , Prospective phenotyping of NGLY1-CDDG, the first congenital disorder of deglycosylation. Genet Med, 2017. 19(2): p. 160–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heeley J and Shinawi M, Multi-systemic involvement in NGLY1-related disorder caused by two novel mutations. Am J Med Genet A, 2015. 167A(4): p. 816–20. [DOI] [PubMed] [Google Scholar]
- 5.Abuduxikuer K, et al. , Novel NGLY1 gene variants in Chinese children with global developmental delay, microcephaly, hypotonia, hypertransaminasemia, alacrimia, and feeding difficulty. J Hum Genet, 2020. 65(4): p. 387–396. [DOI] [PubMed] [Google Scholar]
- 6.He P, et al. , A congenital disorder of deglycosylation: Biochemical characterization of N-glycanase 1 deficiency in patient fibroblasts. Glycobiology, 2015. 25(8): p. 836–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hirsch C, Blom D, and Ploegh HL, A role for N-glycanase in the cytosolic turnover of glycoproteins. EMBO J, 2003. 22(5): p. 1036–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He P, et al. , High phosphate actively induces cytotoxicity by rewiring pro-survival and pro-apoptotic signaling networks in HEK293 and HeLa cells. FASEB J, 2021. 35(1): p. e20997. [DOI] [PubMed] [Google Scholar]
- 9.Castillo-Carranza DL, et al. , Differential activation of the ER stress factor XBP1 by oligomeric assemblies Neurochem Res, 2012. 37(8): p. 1707–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blom D, et al. , A glycosylated type I membrane protein becomes cytosolic when peptide: N-glycanase is compromised. EMBO J, 2004. 23(3): p. 650–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Katiyar S, Li G, and Lennarz WJ, A complex between peptide:N-glycanase and two proteasome-linked proteins suggests a mechanism for the degradation of misfolded glycoproteins. Proc Natl Acad Sci U S A, 2004. 101(38): p. 13774–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Need AC, et al. , Clinical application of exome sequencing in undiagnosed genetic conditions. J Med Genet, 2012. 49(6): p. 353–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith MH, Ploegh HL, and Weissman JS, Road to ruin: targeting proteins for degradation in the endoplasmic reticulum. Science, 2011. 334(6059): p. 1086–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Denic V, Quan EM, and Weissman JS, A luminal surveillance complex that selects misfolded glycoproteins for ER-associated degradation. Cell, 2006. 126(2): p. 349–59. [DOI] [PubMed] [Google Scholar]
- 15.Huppa JB and Ploegh HL, The alpha chain of the T cell antigen receptor is degraded in the cytosol Immunity, 1997. 7(1): p. 113–22. [DOI] [PubMed] [Google Scholar]
- 16.Christianson JC, et al. , Defining human ERAD networks through an integrative mapping strategy. Nat Cell Biol, 2012. 14(1): p. 93–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sifers RN, et al. , A frameshift mutation results in a truncated alpha 1-antitrypsin that is retained within the rough endoplasmic reticulum. J Biol Chem, 1988. 263(15): p. 7330–5. [PubMed] [Google Scholar]
- 18.Bernasconi R and Molinari M, ERAD and ERAD tuning: disposal of cargo and of ERAD regulators from the mammalian ER. Curr Opin Cell Biol, 2011. 23(2): p. 176–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen D, et al. , Novel cell- and tissue-based assays for detecting misfolded and aggregated protein accumulation within aggresomes and inclusion bodies. Cell Biochem Biophys, 2011. 60(3): p. 173–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arslan MA, et al. , Misfolded proteins inhibit proliferation and promote stress-induced death in SV40-transformed mammalian cells. FASEB J, 2012. 26(2): p. 766–77. [DOI] [PubMed] [Google Scholar]
- 21.Walter P and Ron D, The unfolded protein response: from stress pathway to homeostatic regulation Science, 2011. 334(6059): p. 1081–6. [DOI] [PubMed] [Google Scholar]
- 22.Hartl FU, Bracher A, and Hayer-Hartl M, Molecular chaperones in protein folding and proteostasis. Nature, 2011. 475(7356): p. 324–32. [DOI] [PubMed] [Google Scholar]
- 23.Klionsky DJ, et al. , Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition)(1). Autophagy, 2021. 17(1): p. 1–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suzuki T, et al. , Endo-beta-N-acetylglucosaminidase, an enzyme involved in processing of free oligosaccharides in the cytosol. Proc Natl Acad Sci U S A, 2002. 99(15): p. 9691–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim YC, et al. , Identification and origin of N-linked beta-D-N-acetylglucosamine monosaccharide modifications on Arabidopsis proteins. Plant Physiol, 2013. 161(1): p. 455–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Asahina M, et al. , JF1/B6F1 Ngly1(−/−) mouse as an isogenic animal model of NGLY1 deficiency. Proc Jpn Acad Ser B Phys Biol Sci, 2021. 97(2): p. 89–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fujihira H, et al. , Lethality of mice bearing a knockout of the Ngly1-gene is partially rescued by the additional deletion of the Engase gene. PLoS Genet, 2017. 13(4): p. e1006696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang C, et al. , Endo-beta-N-acetylglucosaminidase forms N-GlcNAc protein aggregates during ER-associated degradation in Ngly1-defective cells. Proc Natl Acad Sci U S A, 2015. 112(5): p. 1398–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen Y, et al. , Connective tissue growth factor is secreted through the Golgi and is degraded in the endosome. Exp Cell Res, 2001. 271(1): p. 109–17. [DOI] [PubMed] [Google Scholar]
- 30.Xing X, et al. , Effects of connective tissue growth factor (CTGF/CCN2) on condylar chondrocyte proliferation, migration, maturation, differentiation and signalling pathway. Biochem Biophys Res Commun, 2018. 495(1): p. 1447–1453. [DOI] [PubMed] [Google Scholar]
- 31.Yin Q and Liu H, Connective Tissue Growth Factor and Renal Fibrosis. Adv Exp Med Biol, 2019. 1165: p. 365–380. [DOI] [PubMed] [Google Scholar]
- 32.Leask A and Abraham DJ, All in the CCN family: essential matricellular signaling modulators emerge from the bunker J Cell Sci, 2006. 119(Pt 23): p. 4803–10. [DOI] [PubMed] [Google Scholar]
- 33.Furumatsu T, et al. , Tensile strain increases expression of CCN2 and COL2A1 by activating TGF-beta-Smad2/3 pathway in chondrocytic cells. J Biomech, 2013. 46(9): p. 1508–15. [DOI] [PubMed] [Google Scholar]
- 34.Kothapalli D, et al. , Transforming growth factor beta induces anchorage-independent growth of NRK fibroblasts via a connective tissue growth factor-dependent signaling pathway. Cell Growth Differ, 1997. 8(1): p. 61–8. [PubMed] [Google Scholar]
- 35.Mao L, et al. , MKL1 mediates TGF-beta-induced CTGF transcription to promote renal fibrosis. J Cell Physiol, 2020. 235(5): p. 4790–4803. [DOI] [PubMed] [Google Scholar]
- 36.Fujii M, et al. , TGF-beta synergizes with defects in the Hippo pathway to stimulate human malignant mesothelioma growth. J Exp Med, 2012. 209(3): p. 479–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trojanowska M, Noncanonical transforming growth factor beta signaling in scleroderma fibrosis. Curr Opin Rheumatol, 2009. 21(6): p. 623–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lipinski P, et al. , Liver involvement in NGLY1 congenital disorder of deglycosylation. Pol J Pathol, 2020. 71(1): p. 66–68. [DOI] [PubMed] [Google Scholar]
- 39.Rios-Flores IM, et al. , Acute liver failure in a male patient with NGLY1-congenital disorder of deglycosylation. Eur J Med Genet, 2020. 63(8): p. 103952. [DOI] [PubMed] [Google Scholar]
- 40.Nakerakanti SS, Bujor AM, and Trojanowska M, CCN2 is required for the TGF-beta induced activation of Smad1-Erk1/2 signaling network. PLoS One, 2011. 6(7): p. e21911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heng EC, et al. , CCN2, connective tissue growth factor, stimulates collagen deposition by gingival fibroblasts via module 3 and alpha6- and beta1 integrins. J Cell Biochem, 2006. 98(2): p. 409–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.