Abstract
SARS-CoV-2 is the causative agent of the immune response-driven disease COVID-19 for which new antiviral and anti-inflammatory treatments are urgently needed to reduce recovery time, risk of death and long COVID development. Here, we demonstrate that the immunoregulatory kinase p38 MAPK is activated during viral entry, mediated by the viral spike protein, and drives the harmful virus-induced inflammatory responses. Using primary human lung explants and lung epithelial organoids, we demonstrate that targeting p38 signal transduction with the selective and clinically pre-evaluated inhibitors PH-797804 and VX-702 markedly reduced the expression of the pro-inflammatory cytokines IL6, CXCL8, CXCL10 and TNF-α during infection, while viral replication and the interferon-mediated antiviral response of the lung epithelial barrier were largely maintained. Furthermore, our results reveal a high level of drug synergism of both p38 inhibitors in co-treatments with the nucleoside analogs Remdesivir and Molnupiravir to suppress viral replication of the SARS-CoV-2 variants of concern, revealing an exciting and novel mode of synergistic action of p38 inhibition. These results open new avenues for the improvement of the current treatment strategies for COVID-19.
Keywords: SARS-CoV-2, p38 MAPK, Remdesivir, Molnupiravir, Immunomodulation, Antiviral response
1. Introduction
The newly emerged pandemic severe acute respiratory coronavirus 2 (SARS-CoV-2) infects the human respiratory tract by aerosol transmission and is the etiological agent of coronavirus disease 2019 (COVID-19) (Huang et al., 2020). Up to June 2022, SARS-CoV-2 has infected more than 532 million individuals worldwide and caused over 6.3 million deaths (WHO, June 2022). The continuous emergence of new SARS-CoV-2 variants with altered transmission and immune escape phenotypes as well as the relaxation of testing strategies and non-pharmaceutical interventions contribute to the persisting high numbers of global infections. Risk groups for the development of severe COVID-19 include people of high age (+70 Years) and individuals with comorbidities such as cancer, diabetes, chronic kidney, heart and lung diseases or chronic autoimmune diseases (Williamson et al., 2020). However, also young and immune competent individuals as well as children can develop COVID-19 and other inflammatory complications such as the multi-inflammatory syndrome in children (MIS-C)(Ahmed et al., 2020). In addition, emerging reports describe the occurrence of long-term sequelae following mild or acute COVID-19 called “long COVID” and post-acute COVID, respectively (Nalbandian et al., 2021).
The currently approved treatments for hospitalized COVID-19 patients are limited to the direct acting antiviral (DAA) drugs, such as Remdesivir, a nucleoside analog targeting the viral polymerase, with ambivalent therapeutic efficacy for severely ill patients in the later stages of the disease as well as immunocompromised patients with chronic infection (Gordon et al., 2020a; Beigel et al., 2020; Repurposed, 2020). In contrast, the recently approved DAA Molnupiravir (MK-4482, EIDD-2801, Merck) (Jayk Bernal et al., 2021; Fischer 2nd et al., 2022), also a nucleoside analog, and the viral protease inhibitor Paxlovid (PF-07321332 and Ritonavir, Pfizer) are only approved for outpatient treatment of infected individuals with a high risk to develop life threatening COVID-19 due to a weak or defective immune system or other risk factors. In addition, the JAK1/2 inhibitor Olumiant (Baricitinib, Eli Lilly) has recently been approved for anti-inflammatory applications in hospitalized patients with COVID-19 (Kalil et al., 2021).
The clinical features of COVID-19 are broad and range from asymptomatic infections to critical conditions that require treatment in intensive care units and extracorporeal membrane oxygenation but nevertheless often lead to death. Severe COVID-19 cases are associated with a dysbalanced cytokine response characterized by remarkable high blood levels of pro-inflammatory cytokines such as IL6, CXCL8, CXCL10 and TNF-α, which is associated with multiorgan failure and mortality (Chen et al., 2020; Lucas et al., 2020; Kox et al., 2020; Moore and June 2020). Virus-mediated suppression of the early interferon (IFN) response at the site of infection and imbalanced activation of immune signaling networks is suggested to govern the excessive inflammatory immune response during severe COVID-19. The involved immune regulatory signaling pathways are therefore discussed as promising targets for anti-inflammatory interventions (Blanco-Melo et al., 2020; Lei et al., 2020; Oh and Shin, 2022).
The mitogen-activated protein kinase (MAPK) p38 is a central component of the signaling pathways that regulate pro-inflammatory cytokine expression and has also been associated with virus-supportive functions (Kumar et al., 2003; Canovas and Nebreda, 2021; Cheng et al., 2020). Accumulating evidence suggests that activation of MAPK p38 is a primary factor in the induction of excessive inflammatory responses during infections with highly pathogenic influenza viruses as well as SARS-CoV-2, making it a highly attractive therapeutic target for COVID-19 treatments (Borgeling et al., 2014; Gordon et al., 2020b; Grimes and Grimes, 2020). In this study, we explore the antiviral and anti-inflammatory properties of the two clinically pre-evaluated and highly selective inhibitors of the p38 MAPK α/β isoforms, PH-797804 (PH, Pfizer) (MacNee et al., 2013) and VX-702 (VX, Vertex) (Ding, 2006), in lung epithelial cell lines, three-dimensional primary human lung explants and human lung epithelial organoids to provide a pre-clinical evaluation of their therapeutic potential to dampen cytokine responses in COVID-19. Our results demonstrated that p38 phosphorylation is induced by the spike (S) protein mediated viral entry in the absence of other SARS-CoV-2 proteins. Treatment of lung epithelial cells and human lung explants with both inhibitors efficiently ablated p38-dependent signaling and mitigated virus-induced upregulation of pro-inflammatory cytokines without affecting viral replication. In contrast, the type I IFN-dependent antiviral response in organoids resembling the lung epithelial barrier was not impeded, supporting the unique therapeutic value of p38 signaling inhibition for COVID-19 treatment. Furthermore, our results revealed that p38 inhibition by PH and VX resulted in synergistic antiviral activity during co-treatment with the nucleoside analogs and direct acting antivirals Remdesivir (Gilead Sciences) and Molnupiravir (MK-4482, EIDD-2801, Merck) against SARS-CoV-2, including the Alpha, Beta and Delta variants of concern. This was reflected in an increased reduction of viral titers by 2–3 log steps compared to mono-treatments with Remdesivir or Molnupiravir and culminated in drug synergy scores of up to 21.082 using common reference models for drug interaction. These results unveil fundamental and unexpected novel insights into the virus-specific mechanism that triggers the activation of MAPK p38 and the valuable therapeutic potential of p38 signaling inhibition for anti-inflammatory as well as antiviral (co)-treatments during COVID-19. Using primary experimental models that resemble different compartments of the human lung, our data provide the essential preclinical basis for follow up clinical investigations to explore the anti-inflammatory potential of PH and VX as well as their synergistic antiviral properties in co-treatments with the approved DAA Remdesivir and Molnupiravir to combat SARS-CoV-2.
2. Results
2.1. Spike protein-mediated viral entry triggers p38 MAPK phosphorylation and signaling
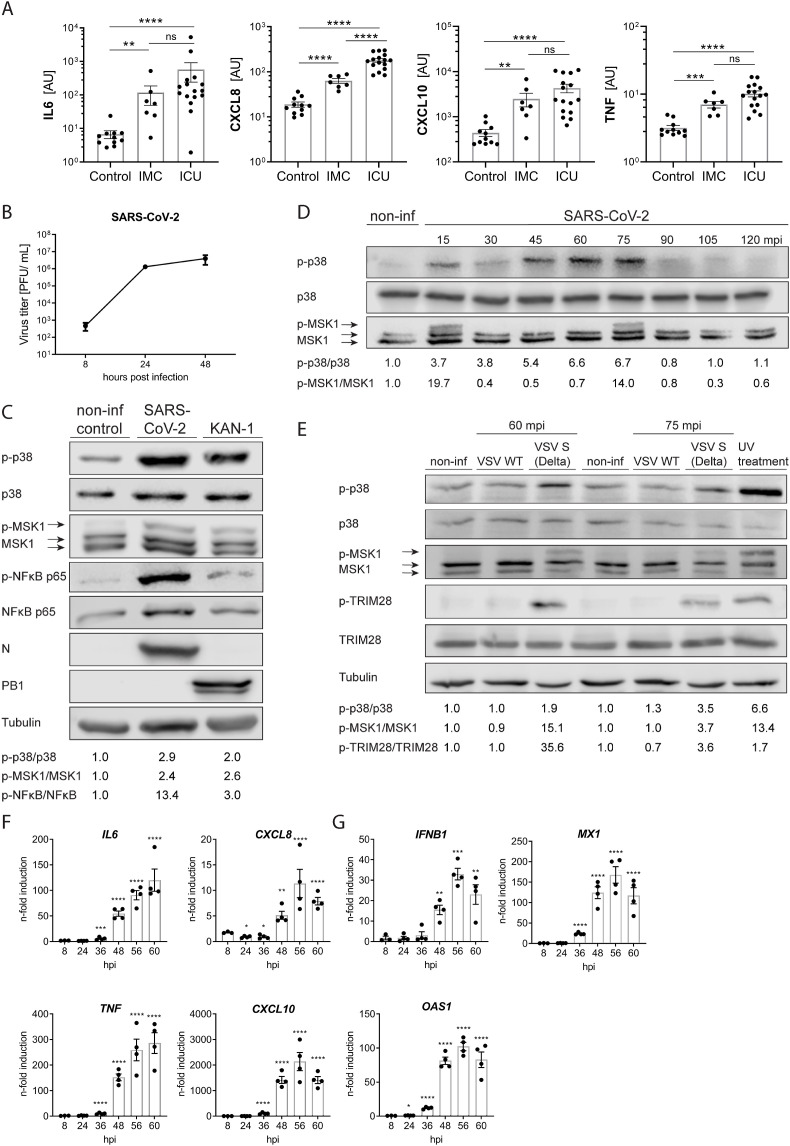
While the full spectrum of the pathophysiological mechanisms leading to tissue damage and organ failure during COVID-19 is still not fully understood, increased blood levels of pro-inflammatory cytokines are described as reliable clinical biomarkers (Kox et al., 2020). Determination of the cytokine levels from the blood of moderate to severe COVID-19 patients substantiated this correlation and revealed significantly increased levels of IL6, CXCL8, TNF-α and CXCL10 in patients that were submitted to intensive care units (ICU) compared to patients in the intensive medical ward (IMC) (Fig. 1 A). To explore a potential involvement of the p38 MAPK signaling pathway in the induction of these pro-inflammatory cytokines during infection with SARS-CoV-2, we infected human airway epithelial cells (Calu-3) cells with SARS-CoV-2 or the highly pathogenic avian influenza virus (HPAIV) A/Thailand/1(KAN-1)/2004(H5N1) (KAN-1) that was previously reported to induce a p38-driven cytokine storm under involvement of the co-repressor TRIM28 (Borgeling et al., 2014; Krischuns et al., 2018). SARS-CoV-2 demonstrated robust viral replication in Calu-3 cells (Fig. 1B) and Vero E6 cells, which was accompanied by pronounced phosphorylation of p38 at the activating phosphorylation sites T180/Y182 (Fig. 1C and suppl. Fig. S1A). In addition, we observed phosphorylation of the p38 target and downstream kinase MSK1 at T581 at 48 h p.i. as well as of the NFκB component p65 at S536. To further determine the timing of p38 activation and signal transduction after SARS-CoV-2 infection, we monitored p38 and MSK1 phosphorylation during the first 2 h of the infection using a higher viral dose. p38 phosphorylation was detectable as early as 15 min p.i. and remained up to 75 min p.i. after which it declined (Fig. 1D). Following a similar kinetic, MSK1 phosphorylation appeared at 15 and 75 min p.i. corroborating dependency on p38 signaling. To elucidate the mechanism of p38 phosphorylation during infection in more detail, we inactivated SARS-CoV-2 with either UV-light or β-propiolactone (BPL) prior to infection. However, incubation with Calu-3 cells did not result in detectable phosphorylation of p38 or MSK1 (Suppl Fig. S1B). Taking it further, we infected Vero E6 cells with pseudotyped VSV viruses encoding for either the wild type VSV-G protein (VSV-WT) or the SARS-CoV-2 S protein of the Delta variant (VSV–S (Delta)). Intriguingly, only infection with VSV-S (Delta) resulted in the phosphorylation of p38, MSK1 and the MSK1 target TRIM28 (Fig. 1E), suggesting that S protein-mediated virus entry triggers p38 phosphorylation and signal transduction in the absence of other viral proteins. Finally, we confirmed that SARS-CoV-2 infection in Calu-3 cells was accompanied by a dynamic transcriptional upregulation of the COVID-19 relevant pro-inflammatory cytokines IL6, CXCL8, CXCL10, TNF (TNF-α), IL1B (Fig. 1F) as well as IFNB1 and IFN-induced viral restriction factors, such as OAS1 and MX1 reaching maximum values after 56–60 h p.i. (Fig. 1G). These results demonstrate that the SARS-CoV-2 entry step triggers p38 phosphorylation and a pronounced IFN-dependent antiviral and inflammatory response in Calu-3 cells.
Fig. 1.
SARS-CoV-2 infection activates p38 MAPK signaling and induces a pro-inflammatory cytokine response. A) Comparisons of a priori selected blood cytokine levels in COVID-19 patients with moderate/severe (IMC, n = 7) or critical (ICU, n = 16) disease and healthy controls (n = 11). Levels of IL6, CXCL8, CXCL10 and TNF were measured by multiplex proximity extension assay. Statistical significance was determined using non-parametric Mann-Whitney-U-test. **p < 0.01, ***p < 0.001, ****p < 0.0001. AU: arbitrary units, IMC: intermediate care wards, ICU: intensive care units. B) Replication kinetic in Calu-3 cells infected with SARS-CoV-2 strain hCoV-19/Germany/FI1103201/2020 at 0.01 MOI. Virus titers are expressed as plaque forming units (PFU/ml), n = 3. C) Phosphorylation of p38, MSK1 and NF-κB p65 during SARS-CoV-2 infection. Calu-3 cells were infected with SARS-CoV-2 at MOI 0.01 for 48 h or the highly pathogenic influenza A virus KAN-1 with MOI 0.001 for 48 h. Viral infections were verified by immuno-detection of the SARS-CoV-2 Nucleoprotein (N) and the IAV polymerase basic 1 (PB1) protein. Signals for total and phosphorylated p38, MSK1 and NF-κB p65 were detected by Western blot using (phospho-) specific antibodies. Quantification of p38, MSK1 and NF-κB p65 phosphorylation is provided below the blot. D) Phosphorylation and signal transduction of p38 at early time points during SARS-CoV-2 infection. Calu-3 cells were infected with SARS-CoV-2 at MOI 2 for 2 h. Signals for total and phosphorylated p38 and MSK1 were detected by Western blot using (phospho-) specific antibodies. Quantification of p38 and MSK1 phosphorylation is shown below the blot. E) Phosphorylation of p38 and MSK1 following infection with Delta S pseudotyped VSV. Vero E6 cells were infected with VSV WT or VSV expressing the spike protein of the Delta variant with MOI 1.5 for 60 min or 75 min, respectively. As a positive control, cells were exposed to 1 kJ/m2 ultraviolet light 30 min prior to lysis. Tubulin was used as a loading control. Quantification of p38, MSK1 and TRIM28 phosphorylation is shown below the blot. F) and G) Cytokine response over time during SARS-CoV-2 infection. Calu-3 cells were infected with SARS-CoV-2 with MOI 0.01 for the indicated time points and mRNA expression of pro-inflammatory cytokines, IFNB1 and ISGs OAS1 and MX1 was determined by qRT-PCR. Statistical significance was determined using one-way ANOVA followed by Dunnett's multiple comparison test. Data are presented as n-fold gene induction over non-infected cells; bars indicate means ± SEM; n = 4.
2.2. Phosphorylation and signal transduction by p38 MAPK majorly contributes to the SARS-CoV-2 induced expression of pro-inflammatory cytokines
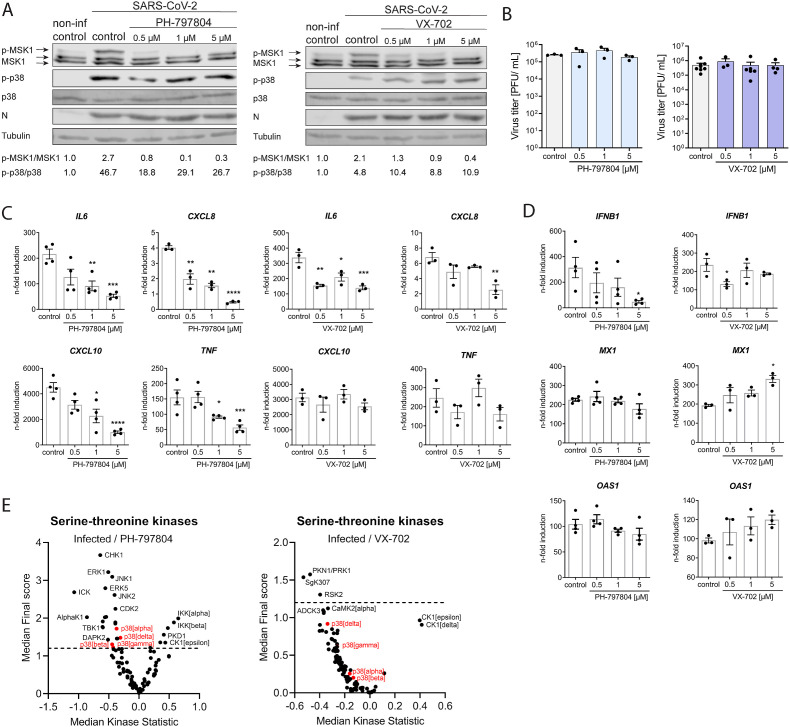
The pre-clinically evaluated p38 inhibitors PH and VX reversibly bind to the ATP-binding pocket of the p38 α/β isoforms and block further downstream signal transduction in a highly specific manner. To provide evidence that SARS-CoV-2-mediated activation of p38 signaling is responsible for the expression of pro-inflammatory cytokines we treated Calu-3 cells for 60 min prior to infection. Both inhibitors efficiently ablated phosphorylation of the p38 target MSK1 (Fig. 2 A) without cytotoxic or antiviral effects (Fig. 2B and Suppl. Figs. S2A and B). Importantly, inhibition of p38 signaling by treatment with PH and VX was accompanied by a significant reduction in the mRNA expression levels of IL6 and CXCL8. In addition, PH also significantly reduced expression of CXCL10 and TNF at 48 h p.i. (Fig. 2C). In contrast, mRNA expression of the IFN-dependent antiviral restriction factors MX1 and OAS1 was not reduced, despite significantly decreased expression of IFNB1 (Fig. 2D), suggesting that p38 inhibition by PH and VX limits the inflammatory response while the antiviral response is less affected.
Fig. 2.
p38 MAPK inhibition reduces the expression of pro-inflammatory cytokines.
Calu-3 cells were pre-treated for 1 h with DMSO (control) or the inhibitors PH-797804 and VX-702 before infection with SARS-CoV-2 at MOI 0.01 for 48 h. A) Western blot analysis to determine inhibition of p38 MAPK by the inhibitors PH-797804 and VX-702. Phosphorylation of p38 (p-p38) and the downstream kinase MSK1 (p-MSK1) was determined using phospho-specific antibodies. Quantification of p38 (p-p38/p38) and MSK1 (p-MSK1/MSK1) phosphorylation levels are depicted below the blots. B) Effect of PH-797804 and VX-702 treatment on virus replication. Calu-3 cells were treated as described above. Viral titers are displayed as PFU/ml ± SEM from 3 independent experiments and effect of C) PH-797804 and D) VX-702 treatment on cytokine expression. Calu-3 cells were treated as described above and 48 h p.i. total RNA was isolated and mRNA levels of the indicated cytokines and ISGs were determined using qRT-PCR with gene specific primers. Data are displayed as n-fold induction over non-treated and non-infected cells. Bars represent mean values ± SEM from at least 3 independent replicates. Statistical significance was determined using two-way ANOVA followed by Dunnett's multiple comparison test. E) Chip-based kinase activity profiling of SARS-CoV-2-infected and PH-797804 or VX-702 inhibitor-treated cells. Calu-3 cells were infected with SARS-CoV-2 at 0.1 MOI for 24 h in the presence of DMSO, PH-797804 or VX-702 (5 μM). Serine-threonine kinase activity was evaluated using PamGene technology and differences in kinase activity in infected inhibitor-treated compared to infected and DMSO-treated cells are depicted as median kinase statistic (negative values = lower activity, positive values = higher activity), n = 3.
In the next step, we employed kinase activity profiling in SARS-CoV-2-infected and inhibitor-treated Calu-3 cells to determine the broader impacts of p38 inhibition on the cellular kinome. As expected, the results demonstrate a reduction of the p38 kinase activity following treatment with PH and VX (Fig. 2E). Moreover, we observed reduced activity of several other inflammation-related kinases, among which we found the extracellular signal-regulated kinase 1 (ERK1), c-Jun N-terminal kinase 1/2 (JNK1/2), tank binding kinase 1 (TBK1), ribosomal S6 kinase 2 (RSK2) and serine/threonine protein kinase 1 (PKN1/PRK1) as well as increased activity of inhibitor of nuclear factor kappa B kinase subunit A/B (IKKα/β), protein kinase D1 (PKD1), janus kinase 1B (JAK1B) and others (Fig. 2E and suppl. Figs. S3A and B). These results demonstrate that p38 phosphorylation occurs in a virus-dependent manner and that p38 MAPK signaling plays a major role in the inflammatory kinase network and in expression of pro-inflammatory cytokines during SARS-CoV-2 infection.
2.3. Treatment with PH-797804 and VX-702 blocks p38 signaling and mitigates the expression of pro-inflammatory cytokines but retains the epithelial antiviral response
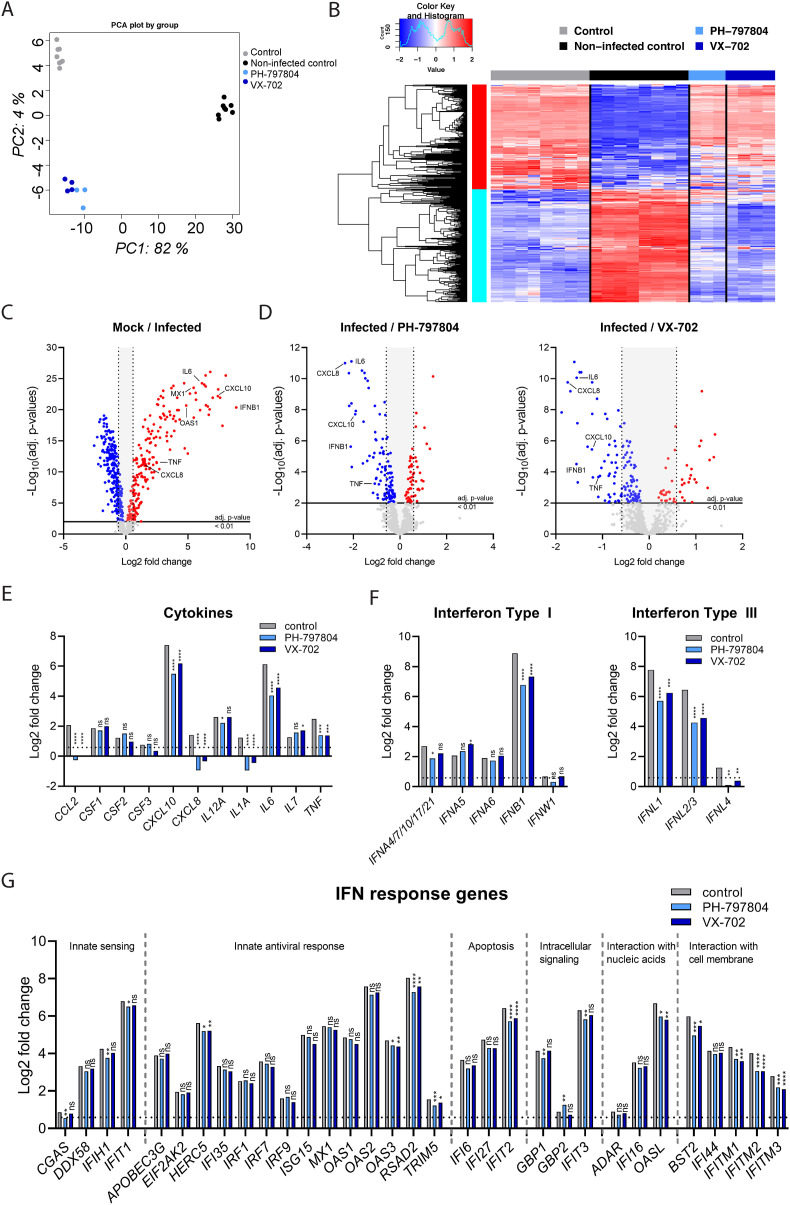
Next, we assessed the effects of p38 signaling inhibition by PH and VX on the expression of host response genes during SARS-CoV-2 infection by performing multiplexed RNA hybridization. Principal component analysis (PCA) and heatmap presentations of the total gene expression demonstrated a broadly reduced host response in the presence of PH and VX (Fig. 3 A and B). Gene specific analysis confirmed upregulation of pro-inflammatory cytokines IL6, CXCL8, CXCL10, TNF as well as the innate immune response genes IFNB1, MX1 and OAS1 in SARS-CoV-2 infected cells (Fig. 3C), whereas treatment with PH and VX significantly reduced the expression of these pro-inflammatory cytokines and IFNB1 (Fig. 3D). Beyond this, our analysis revealed that SARS-CoV-2 infection resulted in the upregulation of CCL2/3, IL12A, IL1A, IL7, IL17A and CSF1-3 (M-CSF, GM-CSF, G-CSF). Interestingly, only CCL2 and IL1A were significantly suppressed by PH and VX treatment (Fig. 3E and suppl. Fig. S4A), suggesting that the expression of cytokines and chemokines responsible for recruitment and activation of immune cells is largely maintained upon p38 signaling inhibition while the expression of immunopathological cytokines is reduced. In line with our previous results, we observed stable mRNA expression of the IFN-dependent restriction factors MX1 and OAS1 despite reduced levels of IFNB1 upon PH and VX treatment, suggesting the existence of a compensatory mechanism. Indeed, we found that the induction of IFN-α subtypes was less affected compared to IFNB1 providing a possible explanation for the stable MX1 and OAS1 levels (Fig. 3F, left panel and suppl. Fig. S4B). Like IFNB1, mRNA expression of the type III IFNs IFNL1-3 was strongly upregulated by SARS-CoV-2 infection and reduced after the treatment with both inhibitors (Fig. 3F, right panel). In contrast, we did not observe significant virus-induced upregulation of IFNG (Suppl. Fig. S4C). Broader analysis of the of IFN induced gene (ISG) landscape revealed that treatment with PH and VX did not globally affect ISG expression but rather reduced the mRNA levels of some ISGs in a gene specific manner, which included HERC5, OAS3, RSAD2, TRIM5, IFIT2, OASL, BST2 as well as IFITM1-3 (Fig. 3G and suppl. Fig. S4D).
Fig. 3.
p38 MAPK inhibitors PH-797804 and VX-702 reduce the inflammatory response to SARS-CoV-2. Calu-3 cells were treated for 1 h with 5 μM of PH-797804, 5 μM of VX-702 or DMSO (control) before infection with SARS-CoV-2 at MOI 0.001 for 48 h in the presence of the inhibitors. Non-infected, DMSO-treated cells were used as an additional control (non-infected control, mock). Gene expression was analyzed using the multiplexed RNA hybridization NanoString Human Host Response Panel and compared between non-treated and infected cells as well as infected and inhibitor-treated cells to determine differentially expressed genes (DEG). Statistical significance was determined using multiple testing and Benjamini and Hochberg correction. A) Principal component analysis (PCA) of relative gene expression levels from all samples. B) Heatmap representation of gene expression levels in mock (n = 6), solvent control (n = 6) and SARS-CoV-2 infected samples treated with PH-797804 (n = 3) and VX-702 (n = 4). C) Volcano plot of up- and down-regulated host response genes during SARS-CoV-2 infection; adj. p-value <0.01, <1.5-fold change (Log2 = 0.5849625). D) Effect of inhibitor treatment on host response genes compared to infected cells using linear regression from the package LIMMA; adj. p-value <0.01, <1.5-fold change (Log2 = 0.5849625). Comparison of mRNA expression in infected and inhibitor-treated infected cells of E) COVID-19-relevant pro-inflammatory cytokines, F) Type I, II and III interferons, G) IFN response genes normalized to the non-infected control. Data are expressed as means of Log2-fold changes; adj. p-value <0.01, <1.5-fold change (Log2 = 0.5849625). *p < 0.05; **p < 0.01, ***p < 0.001.
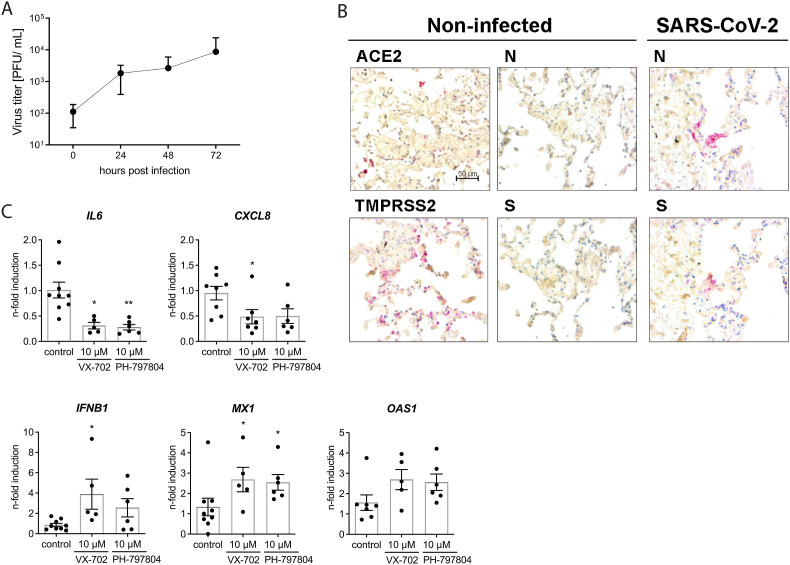
To investigate whether this cytokine-directed effect of the inhibitors PH and VX was reproducible in primary and more complex experimental models, we next employed human lung tissue explants and organoids derived from FACS-sorted human alveolar type II pneumocytes (AT-II), which resemble the natural multicellular and three-dimensional environment of the human lung as well as the unique niche of the lung epithelial barrier, respectively. Following ex vivo infection SARS-CoV-2 titers increased by 2 log10-steps over 72 h in human lung tissue and successful infection was verified by immunohistochemical detection of the viral S and N protein (Fig. 4 A and B). Gene expression analysis confirmed the induction of pro-inflammatory cytokines IL6 and CXCL8 as well as the antiviral type I IFN response exemplified by IFNB1 and ISGs MX1 and OAS1 at 48 h p.i. Application of the inhibitors PH and VX prior to viral infection resulted in significantly reduced levels of IL6 and CXCL8 mRNA expression, resembling the results in Calu-3 cells (Fig. 4C). In contrast, mRNA expression of IFNB1, MX1 and OAS1 was not reduced compared to the infected non-treated control, confirming that inhibition of p38 signaling by PH and VX targets the expression of pro-inflammatory cytokines but retains the IFN-dependent antiviral response.
Fig. 4.
p38 inhibitors reduce IL6 and CXCL8 expression in ex vivo infected human lung explants A) Human lung explants were infected with 106 PFU/well SARS-CoV-2 for 72 h (n = 5). Infectious virus in the supernatants was determined by plaque assay. Bars indicate means ± SEM. B) Visualization of SARS-CoV-2 N protein, ACE2 and TMPRSS2 in human lung explants 48 h after ex vivo infection with 106 PFU/well by immunohistochemistry. C) Gene expression in human lung explants infected for 48 h with 1 × 106 PFU/well SARS-CoV-2 in the presence or absence of the respective p38 inhibitors VX-702 and PH-797804. Expression of pro-inflammatory cytokines and ISGs was determined by qRT-PCR. Data represent means ± SEM from at least 5 patients. Statistical significance was determined with one-way-ANOVA followed by Dunnett's multiple comparison test. *p < 0.05; **p < 0.01.
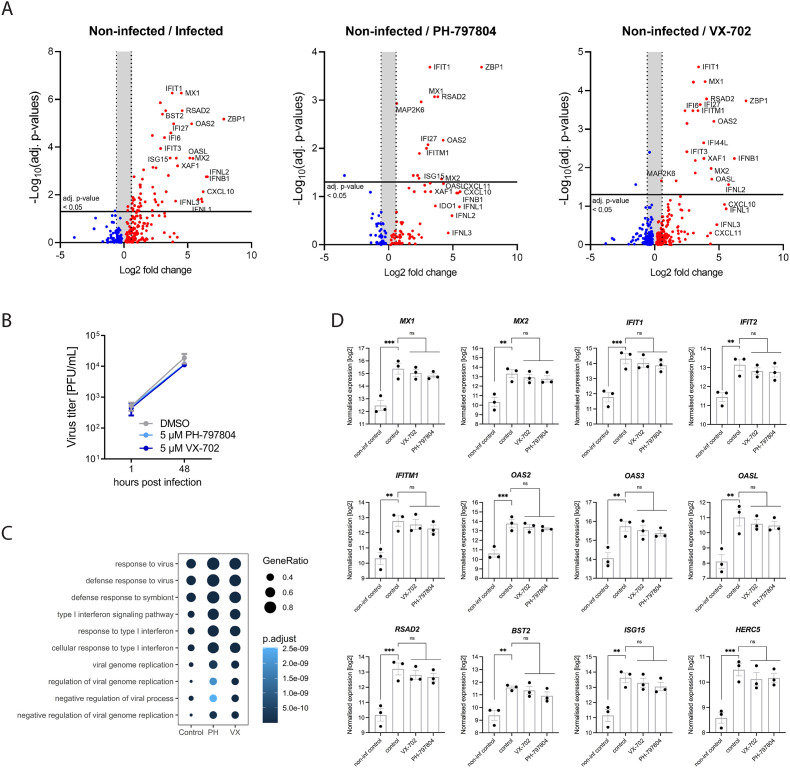
To further dissect the effect of p38 inhibition by PH and VX on the antiviral response of the lung epithelial barrier, we applied organoids from patient-derived and FACS-purified AT-II cells and performed a global transcriptome analysis. In these organoids the SARS-CoV-2-induced host response was markedly dominated by the expression of IFN-dependent genes belonging to the antiviral response such as IFIT1, MX1, BST2, RSAD2 and OAS1 (Fig. 5 A, left panel, 5B and C) while CXCL10 was the only significantly upregulated pro-inflammatory cytokine.
Fig. 5.
Treatment with p38 inhibitors PH and VX maintains the antiviral type I IFN response in SARS-CoV-2 infected human lung epithelial (AT-II) organoids. Human lung stem cell organoids from three patients were treated with DMSO (control) or the inhibitors PH and VX (5 μM) after infection with SARS-CoV-2 (MOI 1). A) SARS-CoV-2-induced immune response in non-treated cells (non-infected/infected) and inhibitor-treated infected cells (non-infected/PH-797804, non-infected/VX-702) from human lung stem cell organoids. Volcano plots displaying gene expression from RNAseq analysis; adj. p-value <0.05, <1.5-fold change (Log2 = 0.5849625). B) Replication of SARS-CoV-2 in the human lung epithelial organoids treated or non-treated with PH and VX (5 μM). C) Top 10 biological processes induced by SARS-CoV-2 infection with or without p38 inhibition. Analysis was performed using GO term enrichment for all up-regulated DEGs comparing infected control and infected treated cells with PH or VX (5 μM). D) Gene expression levels of COVID-19 relevant antiviral restriction factors. Data are displayed as n-fold induction over non-treated and non-infected organoids. Bars represent mean values ± SEM from three donors. Statistical significance was determined using one-way ANOVA followed by Dunnett's multiple comparison test. *p < 0.05; **p < 0.01, ***p < 0.001.
Importantly, inhibition of p38 signaling by PH and VX treatment did not lead to significant reduction of the IFN-mediated antiviral response (Fig. 5C and D). This further supports that p38 inhibition selectively reduces the upregulation of pro-inflammatory cytokines while the virus-induced antiviral defense in cells of the lung epithelial barrier is maintained.
2.4. Co-treatment of PH and VX with the nucleoside analogs Remdesivir and Molnupiravir results in synergistic antiviral activity
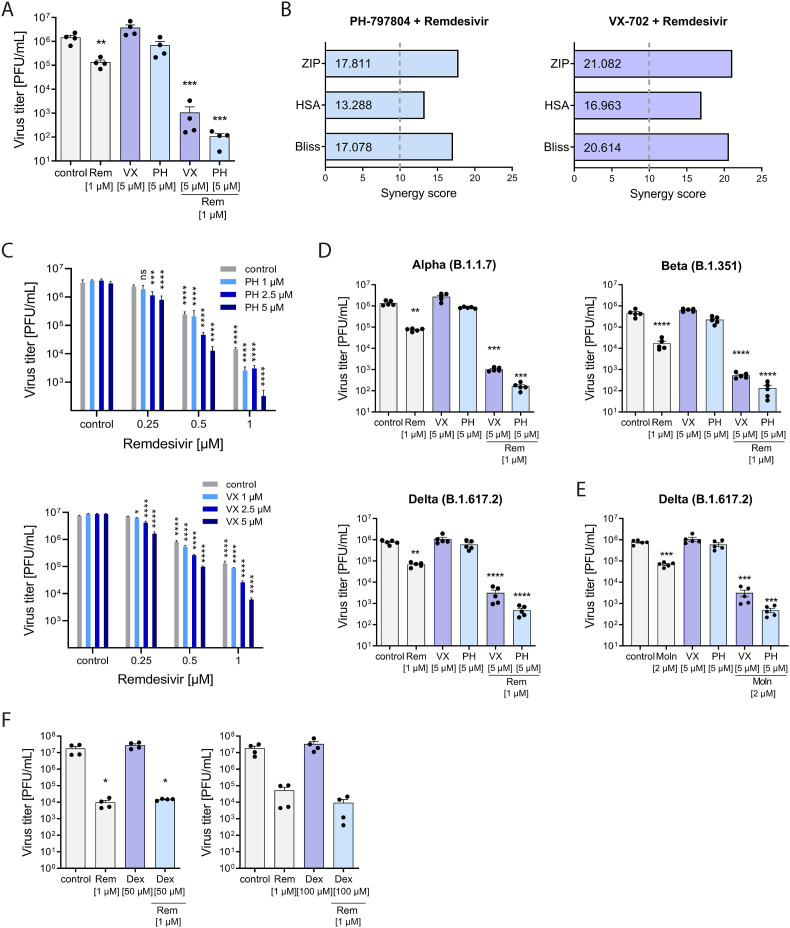
Our results demonstrated that pharmacological inhibition of the p38α/β isoforms potently suppressed the SARS-CoV-2-induced pro-inflammatory responses in the applied primary infection models, which emphasizes the high therapeutic value of PH and VX for anti-inflammatory treatment of COVID-19. To assess unfavorable drug interactions, we next analyzed the co-application of VX and PH with the approved DAA Remdesivir as a potential combinatory clinical treatment. This revealed a significantly increased reduction of SARS-CoV-2 viral titers for the drug combinations, PH/Remdesivir and VX/Remdesivir compared to Remdesivir alone (Fig. 6 A). While treatment with the p38 inhibitors (5 μM) demonstrated no antiviral activity against SARS-CoV-2 as shown before, both inhibitors increased the antiviral effect of Remdesivir mono-treatment from 1-log reduction in infectious viral titers to 3-logs (VX) and 4-logs (PH), respectively (Fig. 6A), resulting in a 100- to 1000-fold increased suppression of viral replication. As expected, the decreased viral replication under combined treatment was accompanied by statistically significant reduction in the mRNA expression of IL6, CXCL8, CXCL10 and TNF as well as reduced expression of IFNB1, MX1 and OAS1(Suppl. Fig. S5A), suggesting that the combination of PH and VX with Remdesivir provides major improvement to Remdesivir COVID-19 treatments. To assess whether the increase in the antiviral activity during co-treatment was the result of an additive or synergistic effect, we further evaluated the pharmacological interactions of PH and VX with Remdesivir in the three commonly used reference synergy models, including Bliss independence (Bliss, 1939), highest single agent (HSA)(Berenbaum, 1989), and zero interaction potency (ZIP) (Yadav et al., 2015). All three models resulted in high synergy scores of 17.811 (ZIP), 13.288 (HSA) and 17.078 (Bliss) for the drug combination PH/Remdesivir and 21.082 (ZIP), 16.963 (HSA) and 20.614 for VX/Remdesivir (Fig. 6B). Drug interaction relationships and landscape visualizations demonstrated a stronger synergy for the drug pair VX/Remdesivir than for PH/Remdesivir at lower concentration ranges of both drugs (Fig. 6C and Suppl. Figs. S5A and B). None of the combinations resulted in cell cytotoxicity at the tested or higher concentrations (Suppl. Figs. S5C and D). Most importantly, we could confirm the synergistic antiviral activities of both drug pairs during infections with the SARS-CoV-2 variants of concern (VOC), Alpha (B1.1.7), Beta (B.1.351) and Delta (B.1.617.2) (Fig. 6D). All three VOCs harbor distinct mutations in the spike protein conferring increased transmissibility and partial immune escape (reviewed in (Günl et al., 2021)). Finally, we also assessed whether PH and VX treatment increased the antiviral activity of the nucleoside analog Molnupiravir against the Delta variant and found an increased reduction in viral titers of 2–2.5 log10 steps compared to Molnupiravir alone (Fig. 6E). These results suggest that inhibition of the p38 MAPK by PH and VX potentiates the antiviral effect of nucleoside analogs against SARS-CoV-2. In contrast, we did not observe synergistic antiviral activity with dexamethosen and remdesivir (Fig. 6F), supporting that this property is specific to p38 inhibition by PH and VX.
Fig. 6.
PH and VX synergistically reduce viral replication during co-treatment with the direct acting antiviral drugs Remdesivir and Molnupiravir. Calu-3 cells were treated with the indicated concentrations of PH, VX, Dexamethasone or DMSO 1 h prior to viral infection with MOI 0.01 and until sample harvest. Remdesivir (1 μM) and Molnupiravir (1 μM) were added 1 h p.i. A) Production of infectious virus particles was determined by plaque assay and expressed as PFU/ml. Statistical significance was calculated by one way ANOVA followed by Dunnett's multiple comparison test (n = 4). B) Synergy scores for both drug pairs were calculated using the indicated reference drug interaction models. Score values above 10 are considered as synergistic. C) Production of infectious virus particles for the indicated drug combinations was determined by plaque assay and expressed as PFU/ml. Statistical significance was calculated by two-way-ANOVA followed by Dunnett's multiple comparison test (n = 4). Calu-3 cells were infected with SARS-CoV-2 variants of concern Alpha, Beta or Delta at MOI 0.01 and treated with D) Remdesivir (Rem) or E) Molnupiravir (Moln) at the indicated concentrations for 48 h. Data are expressed as PFU/ml, bars indicate means ± SEM; n = 5/treatment. Statistical significance was determined using one way ANOVA followed by Dunnett's multiple comparison test. F) Combination treatment of Dexamethasone (Dex, 50 μM and 100 μM) and Remdesivir (1 μM). Production of infectious virus particles was determined by plaque assay and expressed as PFU/ml. Statistical significance was calculated by one way ANOVA followed by Dunnett's multiple comparison test (n = 4). *p < 0.05, **p ≤ 0.05, ***p ≤ 0.001, ****p ≤ 0.0001.
3. Discussion
The ongoing evolution of SARS-CoV-2 has repeatedly given rise to new virus variants with enhanced phenotypes of transmission and immune escape from the current vaccines resulting in increased global infections and hospitalizations. The remaining low number of antiviral and also anti-inflammatory treatment options that are available for patients with severe COVID-19 remains problematic and emphasizes the urgent need for new therapeutic strategies. The p38 MAPK signaling pathway has emerged as an attractive host target for anti-inflammatory and antiviral approaches against several viruses, including HPAIV and SARS-CoV-2. The results of our study demonstrate that pharmacological inhibition of p38 MAPK with the clinically pre-evaluated inhibitors PH-797804 and VX-702, that specifically target the α/β isoforms of p38, provides remarkable high therapeutic potential for prospective anti-inflammatory treatments of COVID-19. This was evident by efficient ablation of p38 signaling, which was accompanied by significant reduction in the expression of COVID-19 relevant pro-inflammatory cytokines in the applied primary infection models. Importantly, our results revealed that despite a significant decrease in the gene expression of IFNB1 and IFNL, the reduction of the inflammatory response was not associated with a global downregulation of the IFN-dependent antiviral response in the lung epithelial barrier. Instead, we observed a distinct and gene-specific downregulation of IFITM1-3, BST2, IFIT2, HERC5, TRIM5, OAS3 and OASL in Calu-3 cells, while other genes with proven antiviral activity against SARS-CoV-2, such as OAS1, were not reduced (Zhou et al., 2021). Hence, it is uncertain whether downregulation of these viral restriction factors leads to a compromised antiviral response to SARS-CoV-2 in patients. At least for IFITM proteins antiviral but also virus-supportive functions were reported for SARS-CoV-2, suggesting that their reduced expression under PH and VX treatment could also be of therapeutic benefit (Prelli Bozzo et al., 2021; Shi et al., 2021). Importantly, we found that in primary human lung explants as well as the AT-II organoids, downregulation of IFNB1 and ISGs during PH and VX treatment was ablated, indicating that this effect is relevant in Calu-3 cells but not in tissue-derived experimental models. The importance of an intact IFN-response for the early control of viral replication and prevention of COVID-19 has been highlighted by recent reports of inborn errors of the type I IFN response and the presence of neutralizing autoantibodies against IFN-ω and IFN-α that are positively associated with the development of severe COVID-19 (Bastard et al., 2020; Asano et al., 2021). In addition, a malfunctional IFN-response would jeopardize the therapeutic efficacy by increasing the risk of secondary viral or bacterial infections and skims essential downstream priming of cellular antiviral and adaptive immune responses (Stackaruk et al., 2013; Maier et al., 2016; LeMessurier et al., 2013; Mancuso et al., 2007). Based on these important and novel insights on the effect of p38 signaling inhibition in native lung tissue, we suggest that the therapeutic application of PH and VX should be further investigated in suitable in vivo models of SARS-CoV-2 as well as in clinical studies.
In addition to the inflammation-targeted therapeutic properties of PH and VX in single treatments our experimental analysis unveiled an unexpected strong antiviral drug synergism of PH and VX during co-treatment with the nucleoside analogs Remdesivir and Molnupiravir. This synergism was evident by a 100- to 1000-fold increased suppression of SARS-CoV-2 replication in the combined treatment setting. Although antiviral activity of p38 inhibitors has been reported for several plus and negative strand RNA viruses, we did not observe antiviral activity against SARS-CoV-2 to be associated with PH and VX-treatment in this study. Nevertheless, all three used algorithms used to discriminate between additive and synergistic drug effects resulted in high synergy scores, indicating, that both PH and VX together with Remdesivir and Molnupiravir suppress SARS-CoV-2 replication in a synergistic manner during co-treatments. This novel and undescribed feature of PH and VX opens a new perspective on the use of p38 inhibitors during SARS-CoV-2 infections that could lead to major improvements of the current COVID-19 treatments. However, further experimental investigation of the underlying mechanism of action is needed. We speculate that, PH and VX confer a low level of antiviral activity, either by blocking an essential virus-supportive cellular function of p38 or by directly affecting the activity of a viral protein. While under single drug treatments, this low antiviral activity is not sufficient to achieve significant reduction in viral replication, the double-pronged attack through restriction of the viral polymerase by the nucleoside analogs as well as the inhibition of p38 signaling leads to a synergistically enhanced inhibition of viral replication. Regardless of the mechanism, the results of our study emphasize that the pre-evaluated p38 inhibitors PH and VX provide a rapid and feasible option to improve current nucleoside-based DAA treatments for hospitalized patients with severe COVID-19. With the demonstrated actions, p38 inhibitors could provide a relevant option for clinical applications. It could be envisioned that p38 inhibitors substitute for dexamethasone as the anti-inflammatory arm in the standard of care for hospitalized COVID-19. At the same time p38 inhibitors would synergistically promote the antiviral actions of remdesivir and thereby also support the antiviral arm of the treatment. Combination therapy with drugs that target relevant cellular pathways is considered a key strategy to achieve therapeutic success with lower drug doses. Especially with regard to the reported risk of drug resistance development associated with Remdesivir in immunocompromised patients (Choi et al., 2020; Martinot et al., 2020; Gandhi et al., 2022), our pre-clinical results are of fundamental importance and provide a compelling therapeutic option to increase the suppression of SARS-CoV-2 replication and reduced risk of in-host evolution. Although Molnupiravir poses a higher barrier for the development of drug resistance compared to Remdesivir, it displays a significant risk of host mutagenesis and only provides a small treatment window during the first days after symptoms onset in order to be effective against COVID-19 (Gordon et al., 2021). Here, drug combination with the described p38 inhibitors can be applied to maintain antiviral activity during treatments with reduced drug concentrations to avoid mutagenic effects and to increase the effective treatment window. Whether PH and VX also confer drug synergism with other DAA against SARS-CoV-2 such as the Mpro protease inhibitor Paxlovid remains to be investigated.
While p38 phosphorylation in SARS-CoV-2 infected cells was reported before, the viral or host triggers for p38 activation were not identified. Here, we demonstrate that p38 signaling and phosphorylation of the downstream factors MSK1 and TRIM28 already occurred within the first 15–75 min after viral infection, indicating that p38 signaling is triggered during a very early step of the viral replication cycle. Using inactivated SARS-CoV-2 or a pseudotyped VSV virus presenting the SARS-CoV-2 Delta S protein, we were further able to dissect that p38 activation was facilitated by the S protein mediated viral entry process in the absence of other viral proteins. It is possible that additional mechanisms exist that facilitate p38 activation at later stages of infection when other viral proteins are expressed, as it was reported for infections with SARS-CoV (Mizutani et al., 2004), however, this remains to be elucidated for SARS-CoV-2.
Taken together, the presented findings provide strong pre-clinical evidence for the exciting hypothesis that the parallel blockade of p38 signal transduction not only increases the antiviral potency of the nucleoside analogs Remdesivir and Molnupiravir, but could also concomitantly act against the deleterious and dysbalanced cytokine response observed in severe COVID-19 without compromising the IFN-governed protective antiviral response.
4. Materials and methods
4.1. Circulating plasma cytokine levels
Adult patients with moderate/severe (Intermediate care; n = 7) or critical (Intensive care unit; n = 16) COVID-19 disease hospitalized in the University Hospital Münster (UKM) and three local teaching hospitals (St. Franziskus-Hospital Münster, Clemens Hospital Münster, UKM-Marienhospital Steinfurt) were enrolled in a non-consecutive, prospective fashion. Eleven apparently healthy volunteers served as controls. The study was approved by the competent ethics committee (amendments of 2016–073-f-S) and was performed in accordance with the Declaration of Helsinki. Informed consent was obtained. These participants were already included in a previous study on microvascular dysfunction in COVID-19 (Rovas et al., 2021). Plasma samples were drawn and immediately centrifuged at 4 °C with 4000×g for 10 min and stored at −80 °C for further analysis. Circulating levels of IL6, CXCL8, TNF and CXCL10 were measured in plasma by multiplex proximity extension assay (Olink, Utrecht, the Netherlands). Briefly, the Olink proximity extension assay uses two specific oligonucleotide-labeled antibodies per protein (“probes”). When the two probes are in close proximity, a new PCR target sequence is formed by a proximity-dependent DNA polymerization event. The resulting sequence is subsequently detected and quantified using standard real-time quantitative PCR. Measurements were carried out in triplicate.
4.2. Cells and viruses
Calu-3 cells were maintained in a mix of Dulbeccos's modified Eagle Medium (DMEM)/Ham's Nutrient Mixture F12 (Sigma-Aldrich, St. Louis, Missouri, USA) supplemented with 10% fetal bovine serum (FBS; Capricorn Scientific, Ebsdorfergrund, Germany). Vero E6 cells were maintained in DMEM supplemented with 10% FBS. All cells were cultured at 37 °C and 5% CO2. The SARS-CoV-2 isolates hCoV-19/Germany/FI1103201/2020 (GISAID: EPI_ISL_463008), hCoV-19/Germany/NW-RKI-I-0026/2020 (GISAID: EPI_ISL_751799) (Alpha, B.1.1.7), hCoV-19/Germany/NW-RKI-I-0029/2020 (GISAID: EPI_ISL_803957) (Beta, B.1.351), hCoV-19/Germany/un-326763/2021 (GISAID: EPI_ISL_4397068) (Delta, B.1.617.2) were isolated on Vero E6 or Vero E6-TMPRSS2-overexpessing cells from swab samples. Virus titers were determined by standard plaque assay on Vero E6 cells.
Vesicular stomatitis virus (VSV) pseudotyped viruses expressing green fluorescence protein gene (VSV ΔG/GFP-Luc + S Δ21) were generated on HEK293T cells (Berger Rentsch and Zimmer, 2011). Briefly, HEK293T cells were transfected with a SARS-CoV-2 Delta spike encoding plasmid in Opti-MEM (Gibco, Waltham, Massachusetts, USA) supplemented with PEI (Sigma-Aldrich) and subsequently infected 24 h later with VSV ΔG together with G-trans-complemented virus particles in DMEM supplemented with 10% FBS. Excess VSV WT particles were neutralized using α-VSV-G antibodies, which were produced in l1-hybridoma cells (CRL-2700™, ATCC, Manassas, Virginia, USA). After 18 h, the pseudovirus containing supernatant was concentrated using the Pierce™ protein concentrator PES (100 K MWCO, #88533; Thermo Fisher, Waltham, Massachusetts, USA) and the titer was determined in Vero E6 cells by quantifying GFP-positive cells using the Axiovert 200 M microscope and the Axiovision 4 software (Zeiss, Jena, Germany).
For infection of the human lung organoids the SARS-CoV-2 isolate Munich/2020/984 (BetaCoV/Munich/BavPat1/2020) was propagated in Vero E6 cells in DMEM supplemented with 2% FCS, 4.5 g/L D-glucose, 4 mM L-glutamine, 10 mM non-essential amino acids and 1 mM sodium pyruvate. For stock production virus in supernatants was concentrated using Vivaspin 20 concentrators (Sartorius Stedim Biotech, Göttingen, Germany). A/Thailand/KAN-1/2004 (H5N1) was used with kind permission from P. Puthavathana (Bangkok, Thailand). All experiments involving infections were performed in a fully equipped biosafety level 3 (BSL3) category laboratory under consideration of the required safety measures.
4.3. Plaque assay
Vero E6 cells were washed with phosphate-buffered saline (PBS, Sigma-Aldrich) and infected with virus-containing supernatants, which were serially diluted in PBS with 0.21% bovine serum albumin (BSA, Sigma-Aldrich), 1% Penicillin/Streptomycin, 0.01% MgCl2 and CaCl2. After 30 min at 37 °C, the inoculum was removed and cells were overlayed with 2 ml of 2x MEM (2% MEM, 2% Penicillin/Streptomycin + L-glutamine, 0.42% BSA, 0.02 M HEPES pH 7.2, 0.24% NaHCO3) mixed with 2% agar (Oxoid, Wesel, Germany) and 2% FBS. The cells were incubated at 37 °C, 5% CO2, for 3 days and plaques were counted.
4.4. Lactate-dehydrogenase (LDH)-Assay
Calu-3 cells were treated with varying concentrations of the p38 inhibitors PH-797804 (Selleckchem, Houston, Texas, USA; CAS No. 586379-66-0) and VX-702 (Selleckchem, CAS No. 745833-23-2) for 24 h. Treatment with water or Triton-X-100 served as negative and positive controls, respectively. After 10 min incubation at room temperature, the supernatant of the cells was transferred to another 96-well plate, LDH Cytotoxicity Assay Reagent (Cell Biolabs, Inc., San Diego, California, USA) was added to the plate and incubated at 37 °C and 5% CO2 for 30 min. The OD was measured at 450 nm as the primary wave length using the Epoch Microplate Spectrophotometer to calculate relative cytotoxicity levels. For the drug combinations with Remdesivir (Hycultec, Beutelsbach, Germany; CAS No. 1809249-37-3), cells were treated for 48 h before analysis.
4.5. Virus infections and inhibitor treatments
Calu-3 cells were infected with SARS-CoV-2 at an MOI of 0.01 or 0.001 for 48 h p.i. in a mix of supplemented DMEM/Ham's F12 (1% sodium pyruvate, 1% P/S, 1% non-essential amino acids solution, 1 M HEPES and 2% FBS). The p38 inhibitors PH-797804 and VX-702, Remdesivir, Molnupiravir (Selleckchem CAS No. 2349386-89-4) and Dexamethasone MK-125 (Selleckchem cat #S1322) were dissolved in DMSO (Roth). For inhibitor treatments, Calu-3 cells were washed with PBS and incubated with the indicated concentrations of the p38 inhibitors or Dexamethasone for 1 h before infection. p38 inhibitors, Remdesivir, Molnupiravir or Dexamethasone were also added to the medium alone or in the indicated combinations after infection using the same concentrations. DMSO was used as solvent control. 48 h p.i., supernatants and cells were harvested for plaque titration, RNA isolation for qRT-PCR analysis and hybridization, or Western blot analysis.
4.6. Virus inactivation
Inactivation of SARS-CoV-2 particles was performed by either treating the virus containing supernatant with 1 kJ/m2 ultraviolet light or adding 0.1% β-propiolactone (BPL, Thermo Fisher) for 20 min at 37 °C/5% CO2. After incubation with BPL, the supernatants were incubated at 37 °C/5% CO2 for 2 h.
4.7. Ex vivo infection of human lung explants with SARS-CoV-2
Tumor-free human lung explants were derived from patients undergoing lung surgery at the Department of Thoracic surgery of the University Hospital Muenster according to medical recommendations. All patients gave their written consent for donation of excessive lung tissue to our experimental investigations prior to surgery. The study received ethical approval by the ethics review board of the University of Muenster and the Ärztekammer Westfalen-Lippe (2016-265-f-S). Lung tissue was kept at 4 °C in Roswell Park Memorial Institute 1640 (RPMI, Sigma-Aldrich) medium directly after surgery. Tissue was processed into entities of 100 mg and incubated overnight at 37 °C. For infection with SARS-CoV-2, 1 × 106 PFU/well were added to 2 ml RPMI-infection medium (RPMI-1940 supplemented with 1% sodium pyruvate, 1% P/S, 1% non-essential amino acids solution, 1 M HEPES and 2% FBS) and 200 μl of the virus-containing medium was injected twice into the tissue followed by incubation for 1 h at 37 °C and 5% CO2. As a negative control, patient-matched tissue was injected with medium without virus. Afterwards, the tissue was washed two times in medium to remove excess virus and incubated in fresh infection medium for the indicated times. RNA isolation from tissue for qRT-PCR analysis was performed at 48 h p.i. using the Qiagen RNeasy Plus Mini Kit following the manufacturer instructions. Briefly, tissues were homogenized in RLT + buffer (Qiagen, Hilden, Germany) supplemented with 10 μl/ml β-Mercaptoethanol using a tissue homogenizer (MP FastPrep-24; MP Biomedicals, Santa Ana, California, USA) and Lysing Matrix A (MP Biomedicals). The homogenates were centrifuged at 14 000 rpm and 4 °C for 10 min. Supernatants were added to spin columns and treated with DNAse following the manufacturer instructions of the RNase-Free DNase Set (Qiagen). The supernatants were then washed with 70% ethanol, RW1 buffer (Qiagen) and RPE buffer (Qiagen). RNA was eluted in 30 μl water.
4.8. Immunohistochemistry
Human lung explants were cut in approximately 100 mg pieces and fixed in 4% paraformaldehyde for at least 12 h at RT. After dehydration the tissue was embedded in paraffin. Paraffin section of 4 μm were submitted to heat-mediated antigen retrieval performed with 10 mM citric acid buffer (Roth). The sections were blocked with PBS including 10% FBS and 0,1% triton X-100 for 30 min. Afterwards, the slides were incubated for 1 h at room temperature with the primary antibodies targeting human ACE2 (Biolegend, San Diego, California, USA; clone A20069I, cat #375802), TMPRSS2 (Abcam, Cambridge, UK; EPR3861, ab92323), SARS-CoV-2 Nucleocapsid (SinoBiological, Beijing, China; clone 019, cat #40143-R019), SARS-CoV-2 spike glycoprotein antibody (Abcam, ab272504) followed by species-specific biotinylated secondary antibody incubation for 30 min. The Vectastain ABC-AP Kit (Vector Laboratories, cat #AK-5000) was used as described in the manufacturer's protocol. Images were taken with the Axiovert 200M fluorescence microscope (Zeiss, Jena, Germany) and the Axiovision 4 software.
4.9. Generation and infection of human lung adult stem cell organoids
For generation of adult stem cell derived lung organoids, primary cells were isolated from healthy parts of distal lung tissue obtained from lung cancer patients undergoing tumor resection surgery. The tissue was enzymatically digested for 1.5 h (cocktail containing 500 U/ml Collagenase I (Gibco, Waltham, Massachusetts, USA), 5 U/ml Dispase II (Gibco), and 1 U/ml DNase (Applichem, Darmstadt, Germany) in HBSS supplemented with 10 μM Y-27632 dihydrochloride (Tocris, Wiesbaden, Germany)), stained with anti HTII-280 (Terrace Biotech, San Francisco, California, USA; TB-27AHT2-280) and labeled with Alexa 488 (goat anti-mouse IgG (H + L) A488 antibody (Thermo Fisher, Waltham, Massachusetts, USA; A-11017)). HTII-280+ cells were FACS sorted, counted and seeded in the extracellular matrix substitute Cultrex (R&D, Wiesbaden, Germany) at a concentration of ∼1000 cells/μl. After solidification, growth medium was added containing 10% R-spondin1-conditioned medium, 1x B27 supplement (Invitrogen, Waltham, Massachusetts, USA), 1x Primocin antibiotic mix (Invivogen, Toulouse, France), 1.25 mM N-Acetylcysteine (Sigma), 5 mM Nicotinamide (Sigma-Aldrich), 0.5 μM SB202190 (Sigma), 1 μM A83-01 (Merck, Darmstadt, Germany), 100 ng/μl human Noggin (Peprotech, Hamburg, Germany), 100 ng/μl human FGF10 (Peprotech) and 25 ng/ml human FGF7 (Peprotech). Y-27632 dihydrochloride (10 μM, Tocris) was added for the first time after seeding and the GSK3 inhibitor CHIR99021 (Sigma-Aldrich, 3 μM). Organoids were kept in an incubator at 37 °C, 5% CO2. Upon growth for 2–3 weeks, the organoids were expanded by enzymatic digestion. Mature organoids were collected on ice to remove remaining matrix and broken up by repeated resuspension using a disposable syringe with needle (27G). Infections were done in ADF++ (Advanced DMEM/F12 460 (Invitrogen)) with 10 mM HEPES (Invitrogen) and 1x GlutaMax (Invitrogen) at 37 °C with 5% CO2. Organoid fragments were either mock-infected or challenged with SARS-CoV-2 isolate Munich/2020/984 (MOI 1) for 1 h at 37 °C with 5% CO2. After infection, organoids were washed with PBS and resuspended in Cultrex. After incubation at 37° and 5% CO2 for 30 min, organoid medium with 5 μM of the respective inhibitor was added. Infectious particles were quantified by plaque titration on Vero E6 cells. Briefly, cell monolayers were seeded in 24-well plates, incubated with virus-containing cell culture supernatants and overlaid with 1.2% Avicel in appropriate medium. After 72 h cells were washed with PBS and plaques were fixed and visualized by staining with crystal violet. For total RNA isolation of human lung organoids, the RNeasy Plus Mini Kit (Qiagen) was used according to the manufacturer's instructions. Organoids were released from Cultrex with cold PBS, pelleted, and resuspended by vortexing in 350 μl RLT lysis buffer supplemented with β-mercaptoethanol. RNA was eluted in 30 μl water. Quality and integrity of total RNA was controlled on TapeStation 2200 (Agilent Technologies, Santa Clara, California, USA).
Informed consent was obtained from all volunteers and the study was approved by the Charité Ethics Committee (project 451, EA2/079/13).
4.10. RNA sequencing
Sequencing was performed at the Core Facility Genomics of the Medical Faculty Muenster. PolyA + RNA was purified from 200 ng total RNA using Poly(A) mRNA Magnetic Isolation module Kit (NEB E7490L, New England Biolabs, Ipswich, Massachusetts, USA). The RNA Sequencing library was prepared with NEBNext Ultra™ II Directional RNA Library Prep Kit for Illumina (New England Biolabs). The libraries were sequenced on Illumina Nextseq 2000 using NextSeq2000 P3 Reagent Kit (200 cycles, paired end run 2x 111 bp) with an average of 74.2 M reads per RNA sample.
4.11. Quantitative real-time PCR
For RNA isolation from Calu-3 cells approximately 5 × 105 cells were lysed using 500 μl of the RNA Solv® (Omega Bio-tek, Norcross, Georgia) and RNA was extracted following the manufacturer's protocol. Total RNA was reverse transcribed using oligo d(T) primers and SuperScript II Reverse Transcriptase (Thermo Fisher). qRT-PCR was performed using the Brilliant SYBR Green Mastermix (Agilent Technologies) in a LightCycler 480 (Roche, Basel, Swiss). Target gene expression was calculated using the 2−ΔΔCT method using GAPDH as housekeeping control gene (Livak and Schmittgen, 2001). Primer sequences are provided in Supplementary Table 1.
4.12. Multiplex gene expression profiling
Relative mRNA abundance was determined by multiplexed hybridization of fluorescently labeled and barcoded gene specific probes for digital readout using the analysis system NanoString nCounter FLEX gene expression (Nanostring Technologies, Seattle, Washington, USA) with DX enablement following the manufacturer's instructions. The nCounter Human Host Response Panel was employed covering 785 genes across more than 50 pathways to evaluate the innate and adaptive immune responses to SARS-CoV-2 infection and treatment with the p38 inhibitors. 5 × 105 Calu-3 cells were solvent-treated but non-infected (non-infected control), infected with SARS-CoV-2 at MOI 0.001 in the presence of the solvent (control), or infected and inhibitor-treated (PH and VX) for 48 h. 350 ng total RNA was hybridized to the reporter and capture probes according to the CodeSet Hybridization Setup for 18 h at 65 °C. Hybridized samples were loaded into the nCounter prep station for post-hybridization processing and analyzed with the nCounter Digital analyzer. The NanoString nCounter nSolver software (version 4.0), with an additional NanoString Advanced Analysis Module (version 2.0.134) was used to perform quality control assessment, normalization and cartridge correction. Differential gene expression analysis was performed as described in statistical analysis. Expression data are shown in Supplementary Table 2.
4.13. Western blot
Calu-3 cells were infected with SARS-CoV-2 at an MOI of 0.01 in DMEM/Nutrient Mixture F-12 Ham supplemented with 1% sodium pyruvate, 1% P/S, 1% non-essential amino acids, 1 M HEPES and 2% FBS. Vero E6 cells were infected in supplemented DMEM. Cells were lysed in radioimmunoprecipitation assay (RIPA) buffer (25 mM Tris-HCl pH 8, 137 mM NaCl, 10% glycerol, 0.5% sodium deoxycholate, 0.1% SDS, 1% NP-40, 2 mM EDTA) containing the following protease and phosphatase inhibitors: 200 nM aprotinine (Roth, Karlsruhe, Germany), 10 μM leupeptin hemisulphate (Roth), 5 mM benzamidine hydrochloride (Sigma-Aldrich), 2.5 mM pefabloc® (Roth), 10 mM β-glycerophosphate (Sigma-Aldrich), 1 mM sodium orthovanadate (Sigma-Aldrich), 10 mM sodium fluoride (Roth), 2.5 mM sodium pyrophosphate (Sigma-Aldrich) at 48 h p.i. Lysate concentrations were adjusted using the Pierce BCA Protein Assay (Thermo Fisher). Protein lysates were then mixed with 5x Laemmli buffer, separated by SDS-PAGE and blotted onto nitrocellulose membranes. Primary antibodies directed against p-p38 (phospo-sites T180/Y182, clone 3D7), p-MSK1 and MSK1 detected by the same antibody (phospho-site T581, cat #9595) and p-NFκB p65 (phospho-site S536, cat #3033) were purchased from Cell Signaling Technology (Danvers, Massachusetts, USA). Antibodies directed against the SARS-CoV-2 N protein (cat #40143-R019) were purchased from Sino Biological and primary tubulin antibodies (cat #T6199) were obtained from Sigma-Aldrich. Primary antibodies directed against total p38 MAPK (cat #ab31828) were purchased from Abcam and primary antibodies directed against NFκB p65 (cat #610868) were purchased from BD Biosciences (Franklin Lakes, New Jersey, USA). Target proteins were detected via chemiluminescence, using horse radish peroxidase (HRP)-coupled secondary antibodies (Cell Signaling Technology) on the Odyssey imaging system (LI-COR Biosciences GmbH, Bad Homburg, Germany). Tubulin was used as housekeeping control.
4.14. Determination of drug synergy
Calu-3 cells were pre-treated with the solvent DMSO, p38 inhibitors PH-797804 (5 μM) and VX-702 (5 μM) or Dexamethasone (50 µM and 100 µM) diluted in DMSO for 1 h and subsequently infected with SARS-CoV-2 (hCoV-19/Germany/FI1103201/2020) or the indicated VOCs with an MOI of 0.01 at 37 °C for 1 h. Cells were washed with PBS and further cultured in DMEM/F-12 Ham's (1% sodium pyruvate, 1% P/S, 1% non-essential amino acids solution, 1 M HEPES and 2% FBS) containing the p38 inhibitors or Dexamethasone alone or combined with Remdesivir (1 μM) or Molnupiravir (1 μM). 48 hpi, supernatants were collected and titrated via plaque assay. Drug synergy scores were calculated using ZIP, HSA and Bliss algorithms.
4.15. Kinase activity profiling
Approximately 5 × 10 Calu-3 cells were pre-treated with the p38 inhibitors PH-797804 (5 μM) or VX-702 (5 μM) or DMSO, respectively, for 1 h before infection with SARS-CoV-2 at an MOI of 0.1 in supplemented DMEM/F-12 Ham mixed medium. After 24 hpi, cells were washed twice with ice cold PBS and lysed with ice cold M-PER™ Mammalian Extraction Buffer (Thermo Fisher) supplemented with Halt™ Phosphatase Inhibitor Cocktail (100x) and EDTA free Halt™ Protease Inhibitor Cocktail (100x) (Thermo Fisher). The lysates were centrifuged at 10,000 rpm for 15 min at 4 °C and the cleared supernatants were frozen at −80 °C. Protein concentrations were adjusted using Bradford assay (Bio-Rad) following the manufacturer's instructions. The activities of protein-tyrosine kinases (PTK) and serine-threonine kinases (STK) in the protein lysates were measured using PamChips (PamGene International BV,‘S-Hertogenbosch, The Netherlands) peptide arrays processed in a PamStation12 (PamGene International BV). 5 μg of protein extract was used for the PTK array protocol (v06) and 1.5 μg for the STK array protocol (v11). Kinases present in the lysates phosphorylate the peptide substrates on the chips, which is detected using fluorescently labeled antibodies. Signal intensities and correlation to kinase activity were analyzed using Bionavigator6 v. 6.3.67.0 (PamGene International BV). Briefly, Software-based image analysis integrates the signals obtained within the time course of the incubation of the kinase lysate on the chip into one single value for each peptide for each sample (exposure time scaling). Only peptides that show a steady increase of signal over time on at least a quarter of the arrays are used for further analysis (quality control). Peptides with significant alteration in phosphorylation were subsequently used for upstream kinase analysis. Processed signals are log2 transformed and in a two-group comparison between two conditions (e.g. infected and PH), the de-regulation of phosphorylation for each peptide demonstrates the effect of changes in the kinase activity. Permutation analysis results in a specificity (mapping of peptides to kinases) and a significance score (difference between treatment groups) for each kinase. The combined score was used to rank and predict top kinase hits.
4.16. Bioinformatics and statistical analysis
Statistical analysis was performed using GraphPad Prism version 9.1.1. Analysis and visualization of NanoString expression data was performed using the R software package (version 3.6.3) (R_Core_Team., 2014). The normalized, batch corrected data was log2 transformed. PCA was used to visualize groups (non-infected controls, infected controls, PH-797804-treated and VX-702-treated); no outliers were identified. For identification of differentially expressed genes (DEG), the Limma package (version 3.42.2) (Ritchie et al., 2015) was used (model.matrix(∼0 + group)). DEGs were identified based on a threshold of multiple-testing adjusted p-value of <0.05 and exhibiting more than a 1.5-fold (log2 = 0.5849625) difference in expression levels. Multiple testing adjusted p-values were calculated according to Benjamini and Hochberg (1995). Heatmaps were generated with the heatmap.2 function in the package ggplots (version 3.1.1) (gplots. Various R, 2020).
Organoid RNAseq reads were quality checked with package FastQC (FastQC. http) (version 0.11.4), then trimmed using Trimgalore (Trimgalore. https) (version 0.4.4) with default settings. Trimmed reads were mapped to human genome annotation hg38 (ENSMBL hg38 release 91) using STAR (Dobin and Gingeras, 2015) (version 2.5.2b) with overhang 150 and default settings. Number of raw reads per sample (mean of 74.2 million), trimmed reads per sample (mean of 73.9 million), uniquely mapped reads per sample (mean of 70.1 million), and percentage of mapped reads per sample (mean of 95%). Mapped reads per gene were counted using RsubRead (version 1.32.4) (Liao et al., 2019). Reads to the virus genome were mapped using STAR and the SARS-CoV-2 genome version EPI_ISL_463008_Cov2_AF_110821 and added to the human raw counts matrix. Raw counts were then normalized and log2 transformed using DESeq2 (Love et al., 2014) (version 1.16.1). Analysis and visualization of expression data was performed using the R software package (R_Core_Team, 2013) (version 3.6.3). Principal component analysis (PCA) was used to visualize groups. No outliers were identified but a strong batch effect by the source of the individual donors was found. Therefore, data was batch corrected for the donor effect using function removeBatchEffect from the Limma package (version 3.42.2). For identification of differentially expressed genes (DEG), the DESeq2 package (version 1.16.1) with the model design(dds) <- ∼ cell_prep + group was used. DEGs were identified based on a threshold of multiple-testing adjusted p-value of <0.05 and exhibiting more than a 1.5-fold (log2 = 0.5849625) difference in expression levels.
Author contributions
Conceptualization: LB, SL; Methodology: LB, SL; Experimental investigations: AF, SS, AMZ, JJ, AS, YB, BC, SK, HK, MB, MK, MT, AC, AM, PK, AR, JK, CW, PB, RW; Supervision: LB, AH, WH, MK, UR, TW; Bioinformatics and statistical analysis: KS, SS, AF, SK; Visualization: AF, SS, LB; Writing and editing: LB, AF, SS, SL.
Declaration of competing interest
LB, SL, SS, AF and UR submitted a patent application for the use of p38 inhibitors with nucleoside analogs for the treatment of COVID-19 (EU patent nr. 21183887.5). SL is a cofounder and member of the advisory board of Atriva Therapeutics GmbH, Tuebingen, Germany.
Acknowledgements
We would like to express our gratitude for support from Christoph Koenig and Vanessa Gerlt. Furthermore, we acknowledge support from the Core Facility Genomic of the Medical Faculty of the Westfaelische Wilhelms-University, especially Anika Witten for excellent technical support and advice. LB, YB, JK and SL are part of the Virus Alliance NRW (VIRAL.NRW), partner site Muenster, supported by the government of the State of North-Rhine Westphalia, Germany.
This work was supported by the German Research Foundation (DFG), grant CRU 342, project P06 to LB and SL, project P04 to PK and AM and a rotational position to AR (ZA428/18-1) as well as grants CRC 1009 B13 to SL and A06 to UR, CRC 1009 B04 to AM, CRC 1348 A11 to UR and the Federal Ministry of Education and Research (BMBF) granted to LB (CoIMMUNE, Project number 01KI20218) and to LB and SL within the NUM Organo-Strat (project 01KX2021). Further support was provided by the IZKF to LB (Bru2/015/19) and UR (Re2/022/20) as well as the Innovative Medizinische Forschung (IMF) to SS (SC121912) and AR (RO221907) of the Medical Faculty of the University of Muenster. ACH and KH were supported by DFG/SFB-TR84 (A7, B6, Z1a). ACH, KH and MK were supported by BMBF/Organo-Strat and Einstein Foundation/EC3R. TW acknowledges partial support by the BMBF through the projects NUM Organo-Strat and RAPID (01KI2006F). This work was also supported by intra-mural grants from the Helmholtz-Association (Program Infection and Immunity), a start-up grant from the University of MemphisUniversity of Memphis Tennessee Health Science Center, and NIAID Research Grants 2-U19-AI100625-06 REVISED and 5U19A|100625-07 awarded to KS.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.antiviral.2022.105475.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- Ahmed M., et al. Multisystem inflammatory syndrome in children: a systematic review. EClinicalMedicine. 2020;26 doi: 10.1016/j.eclinm.2020.100527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano T., et al. X-linked recessive TLR7 deficiency in ∼1% of men under 60 years old with life-threatening COVID-19. Sci. Immunol. 2021;6 doi: 10.1126/sciimmunol.abl4348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastard P., et al. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science. 2020;370 doi: 10.1126/science.abd4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beigel J.H., et al. Remdesivir for the treatment of covid-19 - final report. N. Engl. J. Med. 2020;383:1813–1826. doi: 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful apporach to multiple testing. J. Roy. Stat. Soc. 1995;57:289–300. [Google Scholar]
- Berenbaum M.C. What is synergy? Pharmacol. Rev. 1989;41:93–141. [PubMed] [Google Scholar]
- Berger Rentsch M., Zimmer G. A vesicular stomatitis virus replicon-based bioassay for the rapid and sensitive determination of multi-species type I interferon. PLoS One. 2011;6 doi: 10.1371/journal.pone.0025858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Melo D., et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–1045 e1039. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bliss C.I. The Toxicity of Poisons applied jointly. Ann. Appl. Biol. 1939 doi: 10.1111/j.1744-7348.1939.tb06990.x. [DOI] [Google Scholar]
- Borgeling Y., et al. Inhibition of p38 mitogen-activated protein kinase impairs influenza virus-induced primary and secondary host gene responses and protects mice from lethal H5N1 infection. J. Biol. Chem. 2014;289:13–27. doi: 10.1074/jbc.M113.469239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canovas, B. & Nebreda, A. R. Diversity and versatility of p38 kinase signalling in health and disease. Nat. Rev. Mol. Cell Biol. 22, 346-366, doi:10.1038/s41580-020-00322-w(2021). [DOI] [PMC free article] [PubMed]
- Chen Y., et al. IP-10 and MCP-1 as biomarkers associated with disease severity of COVID-19. Mol. Med. 2020;26:97. doi: 10.1186/s10020-020-00230-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y., et al. Virus-induced p38 MAPK activation facilitates viral infection. Theranostics. 2020;10:12223–12240. doi: 10.7150/thno.50992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi B., et al. Persistence and evolution of SARS-CoV-2 in an immunocompromised host. N. Engl. J. Med. 2020;383:2291–2293. doi: 10.1056/NEJMc2031364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding C. Drug evaluation: VX-702, a MAP kinase inhibitor for rheumatoid arthritis and acute coronary syndrome. Curr. Opin. Invest. Drugs. 2006;7:1020–1025. [PubMed] [Google Scholar]
- Dobin A., Gingeras T.R. Mapping RNA-seq reads with STAR. Curr Protoc Bioinformat. 2015;51:19. doi: 10.1002/0471250953.bi1114s51. 11 14 11-11 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FastQC. http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
- Fischer W.A., 2nd, et al. A phase 2a clinical trial of molnupiravir in patients with COVID-19 shows accelerated SARS-CoV-2 RNA clearance and elimination of infectious virus. Sci. Transl. Med. 2022;14 doi: 10.1126/scitranslmed.abl7430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi, S. et al. De novo emergence of a remdesivir resistance mutation during treatment of persistent SARS-CoV-2 infection in an immunocompromised patient: a case report. Nat. Commun. 13, 1547, doi:10.1038/s41467-022-29104-y(2022). [DOI] [PMC free article] [PubMed]
- Gordon C.J., et al. Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J. Biol. Chem. 2020;295:6785–6797. doi: 10.1074/jbc.RA120.013679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D.E., et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon C.J., Tchesnokov E.P., Schinazi R.F., Götte M. Molnupiravir promotes SARS-CoV-2 mutagenesis via the RNA template. J. Biol. Chem. 2021;297 doi: 10.1016/j.jbc.2021.100770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- gplots. Various R Programming tools for plotting data. github URLhttps://github.com/talgalili/gplots (2020).
- Grimes J.M., Grimes K.V. p38 MAPK inhibition: a promising therapeutic approach for COVID-19. J. Mol. Cell. Cardiol. 2020;144:63–65. doi: 10.1016/j.yjmcc.2020.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Günl F., et al. Shooting at a moving target—effectiveness and emerging challenges for SARS-CoV-2 vaccine development. Vaccines. 2021;9:1052. doi: 10.3390/vaccines9101052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayk Bernal A., et al. Molnupiravir for oral treatment of covid-19 in nonhospitalized patients. N. Engl. J. Med. 2021 doi: 10.1056/NEJMoa2116044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalil A.C., et al. Baricitinib plus remdesivir for hospitalized adults with covid-19. N. Engl. J. Med. 2021;384:795–807. doi: 10.1056/NEJMoa2031994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kox M., Waalders N.J.B., Kooistra E.J., Gerretsen J., Pickkers P. Cytokine levels in critically ill patients with COVID-19 and other conditions. JAMA. 2020;324:1565–1567. doi: 10.1001/jama.2020.17052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krischuns T., et al. Phosphorylation of TRIM28 enhances the expression of IFN-beta and proinflammatory cytokines during HPAIV infection of human lung epithelial cells. Front. Immunol. 2018;9:2229. doi: 10.3389/fimmu.2018.02229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Boehm J., Lee J.C. p38 MAP kinases: key signalling molecules as therapeutic targets for inflammatory diseases. Nat. Rev. Drug Discov. 2003;2:717–726. doi: 10.1038/nrd1177. [DOI] [PubMed] [Google Scholar]
- Lei, X. et al. Activation and evasion of type I interferon responses by SARS-CoV-2. Nat. Commun. 11, 3810, doi:10.1038/s41467-020-17665-9(2020). [DOI] [PMC free article] [PubMed]
- LeMessurier K.S., Hacker H., Chi L., Tuomanen E., Redecke V. Type I interferon protects against pneumococcal invasive disease by inhibiting bacterial transmigration across the lung. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Smyth G.K., Shi W. The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res. 2019;47:e47. doi: 10.1093/nar/gkz114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas C., et al. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature. 2020;584:463–469. doi: 10.1038/s41586-020-2588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacNee W., Allan R.J., Jones I., De Salvo M.C., Tan L.F. Efficacy and safety of the oral p38 inhibitor PH-797804 in chronic obstructive pulmonary disease: a randomised clinical trial. Thorax. 2013;68:738–745. doi: 10.1136/thoraxjnl-2012-202744. [DOI] [PubMed] [Google Scholar]
- Maier B.B., et al. Type I interferon promotes alveolar epithelial type II cell survival during pulmonary Streptococcus pneumoniae infection and sterile lung injury in mice. Eur. J. Immunol. 2016;46:2175–2186. doi: 10.1002/eji.201546201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancuso G., et al. Type I IFN signaling is crucial for host resistance against different species of pathogenic bacteria. J. Immunol. 2007;178:3126–3133. doi: 10.4049/jimmunol.178.5.3126. [DOI] [PubMed] [Google Scholar]
- Martinot M., et al. Emerging RNA-dependent RNA polymerase mutation in a remdesivir-treated B-cell immunodeficient patient with protracted coronavirus disease 2019. Clin. Infect. Dis. 2020;73:e1762–e1765. doi: 10.1093/cid/ciaa1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani T., Fukushi S., Saijo M., Kurane I., Morikawa S. Phosphorylation of p38 MAPK and its downstream targets in SARS coronavirus-infected cells. Biochem. Biophys. Res. Commun. 2004;319:1228–1234. doi: 10.1016/j.bbrc.2004.05.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore J.B., June C.H. Cytokine release syndrome in severe COVID-19. Science. 2020;368:473–474. doi: 10.1126/science.abb8925. [DOI] [PubMed] [Google Scholar]
- Nalbandian, A. et al. Post-acute COVID-19 syndrome. Nat. Med. 27, 601-615, doi:10.1038/s41591-021-01283-z(2021). [DOI] [PMC free article] [PubMed]
- Oh S.J., Shin O.S. SARS-CoV-2-mediated evasion strategies for antiviral interferon pathways. J. Microbiol. 2022 doi: 10.1007/s12275-022-1525-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prelli Bozzo, C. et al. IFITM proteins promote SARS-CoV-2 infection and are targets for virus inhibition in vitro. Nat. Commun. 12, 4584, doi:10.1038/s41467-021-24817-y(2021). [DOI] [PMC free article] [PubMed]
- R_Core_Team. (2013).
- R_Core_Team R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria URL http://www.R-project.org/(2014).
- Repurposed W.H.O. Antiviral drugs for covid-19 — interim WHO solidarity trial results. N. Engl. J. Med. 2020;384:497–511. doi: 10.1056/NEJMoa2023184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie M.E., et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovas, A. et al. Microvascular dysfunction in COVID-19: the MYSTIC study. Angiogenesis 24, 145-157, doi:10.1007/s10456-020-09753-7(2021). [DOI] [PMC free article] [PubMed]
- Shi G., et al. Opposing activities of IFITM proteins in SARS-CoV-2 infection. EMBO J. 2021;40 doi: 10.15252/embj.2020106501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stackaruk M.L., Lee A.J., Ashkar A.A. Type I interferon regulation of natural killer cell function in primary and secondary infections. Expert Rev. Vaccines. 2013;12:875–884. doi: 10.1586/14760584.2013.814871. [DOI] [PubMed] [Google Scholar]
- Trimgalore. https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/.
- Williamson E.J., et al. Factors associated with COVID-19-related death using OpenSAFELY. Nature. 2020;584:430–436. doi: 10.1038/s41586-020-2521-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav B., Wennerberg K., Aittokallio T., Tang J. Searching for drug synergy in complex dose-response landscapes using an interaction potency model. Comput. Struct. Biotechnol. J. 2015;13:504–513. doi: 10.1016/j.csbj.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, S. et al. A Neanderthal OAS1 isoform protects individuals of European ancestry against COVID-19 susceptibility and severity. Nat. Med. 27, 659-667, doi:10.1038/s41591-021-01281-1(2021). [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.