Abstract
Surfactant proteins A (SP-A) and D (SP-D) are thought to play important roles in pulmonary host defense. We investigated the interactions of rat and human SP-A and SP-D with Aspergillus fumigatus conidia. Rat SP-D but not rat SP-A bound the conidia, and the binding was inhibited by EDTA, mannose, glucose, maltose, and inositol. Binding studies using a mutant recombinant rat SP-D with altered carbohydrate recognition but normal structural organization clearly established a role for the carbohydrate recognition domain in binding to conidia. However, neither rat SP-A nor SP-D increased the association of fluorescein isothiocyanate-labeled conidia with rat alveolar macrophages as determined by flow cytometry. Both human SP-A (isolated from normal and alveolar proteinosis lungs) and SP-D (recombinant protein and protein isolated from alveolar proteinosis lungs) bound the conidia. These data indicate that important differences exist between rat and human SP-A in binding to certain fungi. Human SP-A and SP-D binding to conidia was also examined in the presence of hydrophobic surfactant components (HSC), containing both the phospholipid and hydrophobic proteins of surfactant. We found that HSC inhibited but did not eliminate human SP-A binding to Aspergillus conidia. In contrast, the SP-D binding to conidia was unaffected by HSC. These findings indicate that SP-D plays a major role in the recognition of Aspergillus conidia in alveolar fluid.
Aspergillus fumigatus is a fungus found in soil, decaying organic material, and air. The organism is an important airborne pathogen capable of causing a variety of clinical conditions in humans, including allergic bronchopulmonary aspergillosis and invasive aspergillosis (34). Although this organism may cause disease in the immunosuppressed host, invasive disorders in the normal host are rare. Thus, the healthy lung typically provides a strong defense against inhaled A. fumigatus, but the mechanisms involved in this defense are not completely understood.
Pulmonary surfactant proteins A (SP-A) and D (SP-D) are members of the C-type lectin superfamily which also includes serum mannose-binding protein, conglutinin, and collectin 43 (12). SP-A and SP-D are synthesized by type II cells and Clara cells in the lung and share many structural features. Both proteins contain an amino terminal region involved in interchain disulfide bonding, a collagen-like domain, a neck region, and a C-terminal carbohydrate recognition domain (CRD) (18). SP-A and SP-D monomers oligomerize to form trimers, but differ in the organization of the trimers into higher-order structures. SP-D forms a cruciform-like 12 mer composed of four homotrimers, whereas SP-A forms a bouquet-like 18 mer of six trimers (18).
SP-A and SP-D bind many microorganisms in vitro, including viruses, bacteria, and fungi (4, 31). In many cases, their recognition patters are similar, although perhaps by different mechanisms. For example, both SP-A and SP-D tightly bind rough but not smooth gram-negative bacterial lipopolysaccharide (14, 30) and both interact with influenza A virus (3, 7). In the case of influenza A virus, SP-D binds via its CRD, whereas an N-linked oligosaccharide on SP-A is bound by the virus (8). Additionally, the host defense role played by SP-A and SP-D will likely depend on the target organism and specific surfactant protein involved. For example, Hartshorn et al. (6) showed that both SP-A and SP-D increased neutrophil uptake of bacteria and Benne et al. (2) demonstrated that SP-A but not SP-D increased alveolar macrophage phagocytosis of influenza A virus. Conversely, SP-A reduced phagocytosis of Pneumocystis carinii by alveolar macrophages (13) and inhibited uptake of serum-opsonized Candida albicans by alveolar macrophages (32).
Recently, Madan et al. (22) examined the binding of human SP-A and SP-D (hSP-A and hSP-D, respectively) to A. fumigatus conidia. In this study, both proteins bound to and enhanced killing and clearance of the organism in vitro. To more completely understand the roles played by SP-A and SP-D in host defense against Aspergillus, we have been independently examining the interactions of rat SP-A and SP-D (rSP-A and rSP-D, respectively) with A. fumigatus conidia (1). Interestingly, we found that rSP-D but not rSP-A bound the conidia. Additionally, neither rSP-A nor rSP-D caused increased association of the conidia with rat alveolar macrophage cells. When this work was extended to hSP-A and hSP-D, we found that both proteins bound the conidia, confirming the findings of the previous study (22). Because SP-A is known to bind phospholipids, we also evaluated the binding in the presence of phospholipids. We found the binding of hSP-A was inhibited by hydrophobic components of pulmonary surfactant, suggesting that the observed binding in vitro in the absence of surface active material may differ from the in vivo situation. This work also demonstrates that rSP-A differs from hSP-A in binding to certain microbes and caution must be used in extrapolating certain observations with rSP-A to the human situation.
MATERIALS AND METHODS
Preparation of A. fumigatus conidia.
A. fumigatus was cultured from a clinical isolate provided by the University of Colorado Health Sciences Center. The organism was grown on Sabouraud dextrose agar at 30°C. Conidia were harvested and fixed by gently scraping the fungal mat into 2% paraformaldehyde in phosphate-buffered saline (PBS). The conidia were passed through 20-μm nylon mesh and incubated for approximately 16 h at room temperature. Following the incubation, the conidia were washed with PBS, counted with a hemacytometer, and stored at 4°C until needed. For macrophage association analysis, the desired number of conidia were resuspended in 0.1 M carbonate buffer (pH 9.0) containing 0.3 mg of fluorescein isothiocyanate isomer I (FITC)/ml (Molecular Probes, Eugene, Oreg.). Following a 24-h incubation at room temperature, the conidia were washed with calcium binding buffer (see below). Labeling was confirmed by fluorescence microscopy.
Preparation of native rSP-A and rSP-D.
Surfactant was isolated from the bronchoalveolar lavage of Sprague-Dawley rats 28 days after intratracheal instillation of 25 mg (approximately 125 mg/kg of body weight) of silica (10). SP-A was purified from the surfactant by delipidation with butanol, mannose-sepharose affinity chromatography, elution with EDTA, and gel filtration chromatography using Bio-Gel A-15m (BioRad, Hercules, Calif.) (16). Native rSP-D was purified by centrifugation at 30,000 × g from supernatant of bronchoalveolar lavage from silica-treated rats by using a modification of a recently published method (36). The supernatant was adjusted to 5 mM CaCl2 and immediately applied to a mannose-sepharose column, which had been equilibrated with 5 mM Tris-HCl (pH 7.4)–150 mM NaCl–5 mM CaCl2. The SP-D was eluted with a solution containing 5 mM Tris-HCl (pH 7.4), 150 mM NaCl, and 100 mM MnCl2. The eluted protein was dialyzed against 5 mM Tris-HCl (pH 7.4)–150 mM NaCl and then adjusted to 5 mM in CaCl2. The dialyzed material was applied to a fresh mannose-sepharose column and eluted with 5 mM Tris-HCl (pH 7.4)–150 mM NaCl–10 mM EDTA. The eluted protein was dialyzed against 5 mM Tris-HCl (pH 7.4)–150 mM NaCl. The SP-A and SP-D preparations were judged pure by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (19), Coomassie blue staining, and Western blotting. The proteins were stored at −20°C.
Preparation of recombinant rSP-A and rSP-D.
The expression of rSP-A and rSP-D in CHO-K1 cells has previously been described (25, 27) as has the construction and characterization of E321Q/N323D and collagen domain deletion mutant (CDM) recombinant rSP-Ds (27, 28). For this work, the rSP-A cDNA was ligated into the XbaI site on a pEE 14 plasmid vector and expressed as described for rSP-D (27). CHO-K1 cells expressing either rSP-A or rSP-D were grown in glutamine-free Glasgow minimum essential medium (GMEM) (Life Technologies, Inc., Grand Island, N.Y.) containing 10% heat-inactivated and dialyzed fetal bovine serum. The medium was supplemented with 600 μM and 250 μM methionine sulfoxamine for cells expressing SP-A and SP-D, respectively. For protein purification, the cell lines were grown for 3 days in GMEM and were then transferred to serum-free EC-CELL 301 medium (JRH Biosciences, Lenexa, Kans.) and incubated for 4 days. The medium was removed and four additional harvests were performed, allowing 24 h incubation per harvest. For SP-A, the medium was adjusted to 2 mM CaCl2 and applied to a mannose-sepharose column that had been equilibrated with 5 mM Tris-HCl (pH 7.4)–2 mM CaCl2. The SP-A was eluted with 5 mM Tris-HCl (pH 7.4)–2 mM EDTA, dialyzed against 5 mM Tris-HCl (pH 7.4), and stored at −20°C. For SP-D, the medium was dialyzed against 5 mM Tris-HCl (pH 7.4)–150 mM NaCl–1 mM EDTA. Following dialysis, the medium was adjusted to 5 mM CaCl2, applied to a mannose-sepharose column equilibrated with 5 mM Tris-HCl (pH 7.4)–150 mM NaCl–5 mM CaCl2 and eluted with 5 mM Tris-HCl (pH 7.4)–150 mM NaCl–5 mM EDTA. The eluted protein was dialyzed against 5 mM Tris-HCl (pH 7.4)–150 mM NaCl and stored at −20°C. The SP-A and SP-D preparations were judged pure by SDS-PAGE, Coomassie blue staining, and Western blotting.
Preparation of hSP-A and hSP-D.
CHO-K1 cells expressing hSP-D (5) were a gift from Erika Crouch. Recombinant hSP-D was expressed and purified as described above for recombinant rSP-D except the cells were grown in 25 μM methionine sulfoxamine. The hSP-D was also isolated from the bronchoalveolar lavage of alveolar proteinosis (AP) patients by first centrifuging the lavage fluid at 30,000 × g for 16 h at 4°C. The supernatant was adjusted to 5 mM CaCl2 and applied to a mannose-sepharose column equilibrated with 5 mM Tris-HCl (pH 7.4)–150 mM NaCl–5 mM CaCl2. The SP-D was eluted with 5 mM Tris-HCl (pH 7.4)–150 mM NaCl–100 mM MnCl2 and dialyzed against 5 mM Tris-HCl (pH 7.4)–150 mM NaCl. The dialyzed protein was further purified by mannose-sepharose affinity chromatography and eluted with 5 mM Tris-HCl (pH 7.4)–150 mM NaCl–5 mM CaCl2–50 mM inositol. The eluted material was dialyzed against 5 mM Tris-HCl (pH 7.4)–150 mM NaCl–5 mM EDTA to remove inositol, and then against 5 mM Tris-HCl (pH 7.4)–150 mM NaCl. Normal hSP-A was purified as described above for native rSP-A, from the bronchoalveolar lavage of a donor lung not used for lung transplantation, except that gel filtration was not performed. The purification of hSP-A from the bronchoalveolar lavage of AP patients was performed as previously described (9). The SP-A and SP-D preparations were judged pure by SDS-PAGE, Coomassie blue staining, and Western blotting. The proteins were stored at −20°C.
Preparation of HSC.
Hydrophobic surfactant components (HSC) were isolated from the bronchoalveolar lavage of Sprague-Dawley rats 28 days after intratracheal instillation of 25 mg of silica (approximately 125 mg/kg) (10, 16). Initially, the surfactant was purified by the method of Hawgood et al. (10) using NaBr density gradient centrifugation. The purified surfactant was extracted with butanol (16) and segregated into butanol-soluble and -insoluble material. The butanol-soluble material (HSC) was recovered by drying under vacuum and resuspending in chloroform. The phospholipid content was determined by the method of Rouser et al. (33), and the mixture was stored at −20°C. Prior to use, an aliquot of HSC was dried under N2, twice resuspended in 50 μl of methanol, and dried again under a stream of N2. The lipid film was resuspended by vortex mixing in 50 mM Tris (pH 7.4). The mixture was further dispersed by probe sonicating on ice for two 1-min intervals with 1 min of cooling on ice between bursts.
Binding of surfactant proteins to A. fumigatus conidia.
Binding reactions were carried out in calcium binding buffer (pH 7.4) (CBB) which contained 130 mM NaCl, 13 mM NaN3, 5 mM KCl, 3 mM sodium phosphate buffer, 10 mM HEPES, 2 mM CaCl2, 1 mM MgSO4, and 1% heat-inactivated and dialyzed fetal bovine serum. Aliquots of 2× 106 conidia were resuspended in CBB and incubated with the appropriate surfactant protein (20 μg/ml unless otherwise noted) at 25°C for 1 h. The total reaction volume was 100 μl. The conidia were then washed three times with CBB and incubated with a 10-μg/ml concentration of the appropriate rabbit polyclonal immunoglobulin G (IgG) in CBB for 1 h at 25°C. The conidia were again washed three times with CBB and incubated with 10 μg of FITC-conjugated F(ab′)2 fragment of donkey anti-rabbit IgG (Jackson ImmunoResearch Laboratories, West Grove, Pa.)/ml for 1 h at 25°C in CBB. Finally, the conidia were washed twice with CBB and analyzed for FITC fluorescence with a Becton Dickinson FACSCalibur flow cytometer and CELLQuest software.
Macrophage association analysis.
The CBB used for macrophage association analysis was the same as used in the conidia binding experiments, but without NaN3 and serum. A rat alveolar macrophage cell line (ATCC CRL 2192) was grown in Dulbecco’s modified Eagle medium (Life Technologies, Inc.) supplemented with 10% heat-inactivated fetal bovine serum on bacteriologic plastic plates. The cells were removed by repeated pipetting, washed three times with CBB, and counted with a hemacytometer. The macrophages (3 × 105) were then incubated with 9 × 105 FITC-labeled conidia which had been washed with CBB and preincubated (37°C for 30 min) with rSP-A, rSP-D, 10% rabbit anti-conidia immune serum, or Tris-buffered saline (pH 7.4). The macrophage association was allowed to progress for 1 h at 37°C. Following the incubation, the cells were washed three times with CBB with or without 10 mM EDTA (pH 7.4) to remove conidia bound to the cell surface via SP-A or SP-D. Finally, the cells were fixed with 2% paraformaldehyde in PBS for 30 min at room temperature, washed with PBS, and analyzed for fluorescence with a Becton Dickinson FACSCalibur flow cytometer and CELLQuest software. Some samples were also examined visually by fluorescence microscopy. In other experiments, macrophages were isolated from the bronchoalveolar lavage of Sprague-Dawley rats by centrifugation at 600 × g. The macrophages were washed with CBB lacking NaN3 and serum, and association of conidia with the macrophages was performed as described above.
Other methods.
Protein concentrations were determined by using the bicinchoninic acid assay (Pierce, Rockford, Ill.) and bovine serum albumin as a standard. Polyclonal anti-rSP-A and rSP-D were raised in rabbits against recombinant rSP-A and rSP-D. Polyclonal anti-hSP-A was raised in a rabbit against SP-A purified from the bronchoalveolar lavage of AP patients. Polyclonal anti-hSP-D was raised in a rabbit against recombinant hSP-D. Immune serum against A. fumigatus was raised in a rabbit against 2% paraformaldehyde-fixed conidia.
Statistical analysis.
Data are shown as means ± standard errors. Data were compared by using the Student’s t test, and P values of <0.05 were considered significant.
RESULTS
rSP-A and rSP-D binding to A. fumigatus conidia.
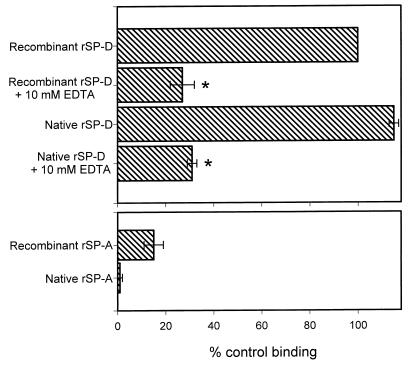
As an initial step in characterizing the interactions of rSP-A and rSP-D with A. fumigatus conidia, binding assays were performed as described in Materials and Methods using both recombinant and native proteins (Fig. 1). Preliminary experiments were performed with A. fumigatus ATCC 14110, with results (not shown) nearly identical to those shown in Fig. 1. We chose to include recombinant proteins since mutant recombinant proteins were available. Any binding activity could then be explored further by using the mutant proteins (see below). Native and recombinant rSP-D bound the conidia tightly, and this binding was inhibited by 10 mM EDTA. Significantly less binding was observed for recombinant rSP-A, and native rSP-A did not bind the conidia. Similar results were obtained for rSP-A and rSP-D binding to conidia treated with paraformaldehyde, ethanol, or high temperature (not shown). From our data, it is not possible to quantitatively compare the binding of rSP-A and rSP-D since different primary antibodies were used in binding detection. However, it is unlikely that the lack of binding seen for rSP-A was due to a failure of the primary antibody. The polyclonal rabbit anti-rSP-A or rSP-D antibodies had similar reactivity to their respective proteins in enzyme-linked immunosorbent assays (ELISAs) and Western blots. In addition, to verify that rSP-A did not bind the conidia, solutions containing 20 million conidia were incubated for 1 h at 25°C with 4 μg of native rSP-A/ml. Centrifuged supernatants of these solutions showed no SP-A loss when analyzed by quantitative ELISA. However, approximately 50% of the rSP-D was lost on the conidia in parallel experiments using that protein (data not shown).
FIG. 1.
Binding of native and recombinant rSP-A and rSP-D to A. fumigatus conidia. Aliquots of 2 × 106 conidia were incubated with 20 μg of SP-A or SP-D/ml for 1 h at 25°C followed by washing and similar incubations with primary and secondary antibodies. For inhibition studies, the proteins were preincubated with EDTA for 15 min at 25°C and the EDTA-surfactant protein mixture was then added to the conidia. Binding was detected by flow cytometry and normalized to the mean fluorescent intensity of recombinant rSP-D (taken as 100% binding). Data are the averages ± standard errors of four independent experiments. ∗, P < 0.05 compared to binding without EDTA.
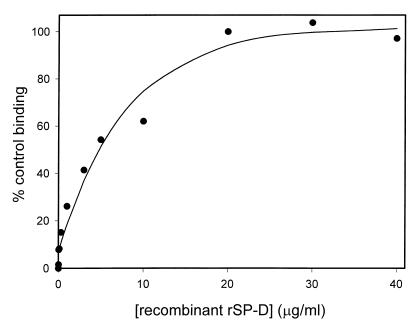
Recombinant rSP-D bound the conidia in a concentration-dependent manner, reaching maximal binding at about 20 μg/ml (Fig. 2). Although estimation of the SP-D concentration in alveolar lining fluid is difficult, Wright (39) has suggested concentrations of 36 to 216 μg of SP-D/ml in rat lung alveolar fluid. Thus, the maximal binding observed at 20 μg/ml is in the physiological range.
FIG. 2.
Dose response curve for recombinant rSP-D binding to A. fumigatus conidia. Aliquots of 2 × 106 conidia were incubated with various concentrations of SP-D for 1 h at 25°C followed by washing and similar incubations with primary and secondary antibodies. Binding was detected by flow cytometry and normalized to the mean fluorescent intensity at 20 μg/ml. Data are the averages of duplicate analysis.
Inhibition of recombinant rSP-D binding to A. fumigatus conidia.
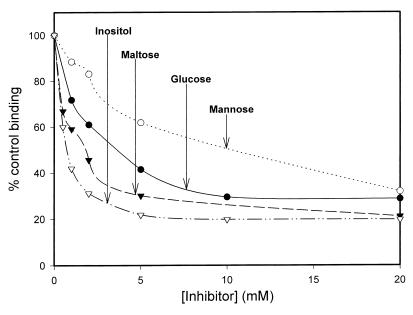
Previous work has demonstrated that SP-D binds carbohydrates via its CRD in a calcium-dependent manner (29). The observation that EDTA inhibited native and recombinant rSP-D binding to A. fumigatus conidia suggested a role for the CRD. To further establish a role for the CRD, we performed binding experiments in the presence of various carbohydrate inhibitors. As shown in Fig. 3 and Table 1 inositol, maltose, glucose, and mannose all inhibited rSP-D binding to the microorganism. This is in agreement with previous observations which showed maltose, glucose, and mannose inhibited rSP-D binding to maltosyl-bovine serum albumin (29).
FIG. 3.
Inhibition of recombinant rSP-D binding to A. fumigatus conidia. Recombinant rSP-D (20 μg/ml) was incubated with various concentrations of mannose, glucose, maltose, and inositol for 15 min at 25°C. The SP-D-inhibitor mixture was then added to 2 × 106 conidia and binding was allowed to progress for 1 h at 25°C, followed by washing and similar incubations with primary and secondary antibodies. Binding was detected by flow cytometry and normalized to the mean fluorescent intensity in the absence of inhibitor. Four independent experiments were performed for each inhibitor. The graph shows representative data for each inhibitor.
TABLE 1.
IC50s for the inhibition of recombinant rSP-D binding to A. fumigatus conidia
| Inhibitor | IC50 (mM)a |
|---|---|
| Mannose | 13.4 ± 3.3 |
| Glucose | 4.6 ± 0.5 |
| Maltose | 1.2 ± 0.1 |
| Inositol | 0.9 ± 0.1 |
Values are means ± standard errors of four experiments.
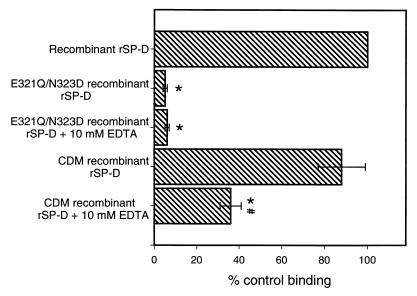
Mutant rSP-D binding to A. fumigatus conidia.
To conclusively establish a role for the CRD in binding to conidia, we performed experiments using previously constructed recombinant rSP-D mutants. One of these mutants (CDM) lacked the collagen-like domain but retained a functional CRD (28), and the other mutant (E321Q/N323D) had altered carbohydrate recognition properties (27). As shown in Fig. 4, the CDM mutant rSP-D bound the conidia and the binding was inhibited by EDTA. However, when the E321Q/N323D mutant recombinant rSP-D was tested, essentially no binding was seen (Fig. 4). This mutant is essentially identical to wild-type recombinant rSP-D in size and structural organization and retains recognition of phospholipid ligands (27). As discussed below, the fact that the E321Q/N323D mutant failed to bind conidia clearly establishes a role for the CRD in recognition of the microorganism.
FIG. 4.
Binding of mutant recombinant rSP-D to A. fumigatus conidia. Aliquots of 2 × 106 conidia were incubated with 20 μg of SP-D/ml for 1 h at 25°C followed by washing and similar incubations with primary and secondary antibodies. For inhibition studies, the proteins were preincubated with EDTA for 15 min at 25°C and the EDTA-surfactant protein mixture was then added to the conidia. Binding was detected by flow cytometry and normalized to the mean fluorescent intensity of wild-type recombinant rSP-D. ∗, P < 0.05 compared to the binding of wild-type recombinant rSP-D; #, P < 0.05 compared to mutant protein binding without EDTA. Data are the averages ± standard errors of four independent experiments. EDTA inhibition of wild-type recombinant rSP-D is shown in Fig. 1.
Association of A. fumigatus with rat alveolar macrophage cells.
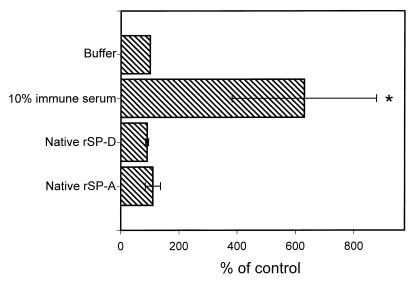
Several studies have demonstrated that SP-A and SP-D can enhance the uptake of various pathogens by phagocytic cells (2, 6, 11, 22). To determine whether rSP-A or rSP-D acted similarly, we examined the association of conidia with rat alveolar macrophages. FITC-labeled conidia were preincubated with 10 μg of native rSP-A or rSP-D/ml and then added to alveolar macrophage cells as described in Materials and Methods. A positive control for these experiments used 10% rabbit anti-conidia immune serum, and buffer alone served as a negative control. Flow cytometry was used to determine the association of conidia with the alveolar macrophages. The results are shown in Fig. 5 and clearly demonstrate that neither rSP-A or rSP-D altered the association of A. fumigatus conidia with alveolar macrophage cells. Similar results were obtained with alveolar macrophages isolated from rat lungs and when the cells were washed with buffer lacking EDTA (not shown). To visualize the interactions of SP-A, SP-D, macrophages, and conidia, parallel studies were performed with fluorescence microscopy. No discernable alterations in the association of conidia with macrophages could be attributed to SP-A or SP-D. Additionally, to verify that labeling the conidia with FITC did not alter the interactions with the macrophages, similar studies using unlabeled conidia were performed, and the association of conidia with the macrophages was examined microscopically. The results of these experiments (not shown) were similar to those shown in Fig. 5.
FIG. 5.
Association of A. fumigatus conidia with cultured rat alveolar macrophage cells. Samples of 9 × 105 FITC-labeled conidia were incubated with native rSP-A (10 μg/ml), native rSP-D (10 μg/ml), 10% rabbit immune serum, or buffer for 30 min at 37°C. Following incubation, 3 × 105 alveolar macrophage cells were added to the conidia and the mixture was incubated at 37°C for 1 h. The cells were then washed in buffer containing 10 mM EDTA and fixed with 2% paraformaldehyde in PBS for 30 min at room temperature. The cell association was determined by measuring the FITC fluorescence associated with the macrophages. The data were normalized to the buffer control and are the averages ± standard errors of four or five independent experiments. ∗, P < 0.05 compared to buffer control.
Binding of hSP-A and hSP-D to A. fumigatus conidia.
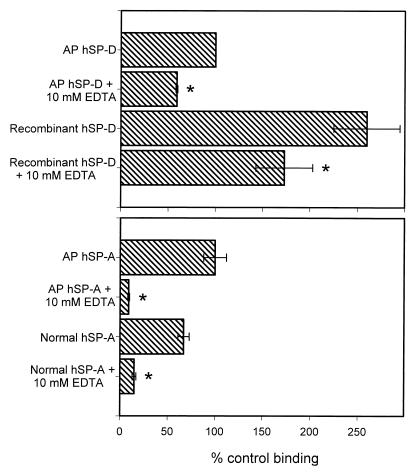
Madan et al. (22) showed that both hSP-A and hSP-D, isolated from the bronchoalveolar lavage fluid of AP patients (AP hSP-A and AP hSP-D, respectively), bound A. fumigatus conidia. Given the result that rSP-A failed to bind, we examined the interactions of hSP-A and hSP-D with conidia in our system. We performed binding experiments with AP hSP-D, recombinant hSP-D produced using a CHO-K1 cell expression system, AP hSP-A, and SP-A isolated from a normal human lung (normal hSP-A). As can be seen in Fig. 6, all forms of human SP-A and SP-D bound the conidia, and the binding was inhibited by EDTA. AP hSP-A and normal hSP-A showed similar binding and EDTA inhibition. However, the binding of recombinant hSP-D was 2.5 times greater than AP hSP-D and neither was inhibited by EDTA to the extent shown by native or recombinant rSP-D. Additionally, in preliminary experiments we found that magnesium could not substitute for calcium in hSP-D binding to the conidia. Surprisingly, however, we found that magnesium could partially substitute for calcium in hSP-A binding (not shown). We are continuing to investigate this phenomenon. As noted above for rSP-A and rSP-D, quantitative comparison between the binding of hSP-A and hSP-D cannot be made with our data since different primary antibodies were used for these proteins. However, from our data it is clear that both hSP-A and hSP-D bind the conidia.
FIG. 6.
Binding of hSP-A and hSP-D to A. fumigatus conidia. Conidia (2 × 106) were incubated with 20 μg of hSP-A or hSP-D/ml for 1 h at 25°C followed by washing and similar incubations with primary and secondary antibodies. For inhibition studies, the proteins were preincubated with EDTA for 15 min at 25°C and the EDTA-surfactant protein mixture was then added to the conidia. Binding was detected by flow cytometry and normalized to the mean fluorescent intensity of AP hSP-D. Data are the averages ± standard errors of three or four independent experiments. ∗, P < 0.05 compared to binding without EDTA.
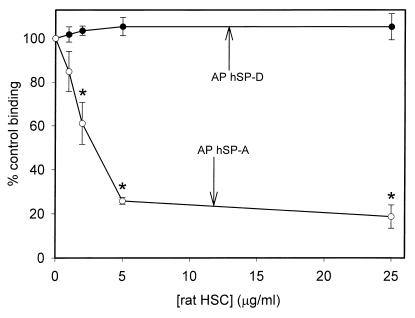
It is well know that SP-A, and to a lesser extent SP-D, associate with surfactant phospholipids (17), and SP-A must be extracted from these lipids during purification from lavage fluid. Therefore, we examined if hSP-A and hSP-D binding to A. fumigatus conidia could be affected by the presence of surfactant. We chose to use surfactant free of SP-A and SP-D. This material contains surfactant phospholipids and other hydrophobic surfactant constituents, including SP-B and SP-C. Fig. 7 shows the results of binding experiments performed in the presence HSC at various concentrations. Rat HSC inhibited AP hSP-A binding with a 50% inhibitory concentration (IC50) of approximately 3 μg of phospholipid/ml, but did not affect AP hSP-D binding. This indicates that in the lung alveolar space, SP-A may not bind A. fumigatus conidia as extensively as SP-D or purified SP-A in the absence of phospholipid.
FIG. 7.
Inhibition of AP hSP-A and AP hSP-D binding to A. fumigatus conidia by rat HSC. Purified AP hSP-A or AP-hSP-D (20 μg/ml) was incubated with various concentrations of rat HSC (based on total phospholipid content) for 15 min at 25°C. The surfactant protein-inhibitor mixture was then added to 2 × 106 conidia and binding was allowed to progress for 1 h at 25°C, followed by washing and similar incubations with primary and secondary antibodies. Binding was detected by flow cytometry and normalized to the mean fluorescent intensity in the absence of inhibitor. The data are the averages ± standard errors of three independent experiments. ∗, P < 0.05 compared to binding in the absence of HSC.
DISCUSSION
SP-A and SP-D are thought to play important roles in the host defense properties of pulmonary surfactant (4, 23, 39). Several studies have shown that SP-A and SP-D can bind to various pathogens and stimulate their uptake by phagocytic cells in vitro (2, 6, 11, 22). Additionally, mice deficient in SP-A have recently been shown to be more susceptible to group B streptococci and Pseudomonas aeruginosa when exposed by intratracheal instillation (20, 21), clearly demonstrating a host defense role in vivo.
Madan et al. (22) showed that AP hSP-A and AP hSP-D bound and increased killing of A. fumigatus in vitro. Independently, we have been studying the interactions of rSP-A and rSP-D with A. fumigatus conidia with different results (1). We have shown that rSP-D binds conidia in a calcium-dependent manner (Fig. 1) and the binding is inhibited by inositol, maltose, glucose, and mannose (Fig. 3 and Table 1). It is interesting that we found an IC50 of 13 mM for mannose inhibition of rSP-D binding to the conidia whereas Madan et al. (22) showed approximately 50% inhibition of hSP-D binding with 100 mM mannose. We performed preliminary inhibition experiments for human SP-D binding to the conidia in the presence of various concentrations of glucose and inositol (not shown). Our results suggest that inhibition of hSP-D binding may require higher inhibitor concentrations to achieve the same inhibition seen with rSP-D. For example, in one experiment, hSP-D binding was inhibited by 49% with 20 mM inositol.
CDM and E321Q/N323D mutant recombinant rSP-Ds were also tested for binding (Fig. 4). CDM rSP-D lacks the collagen-like domain but retains a functional CRD and bound the conidia. This is significant, as it shows that SP-D dodecamers are not required for binding. CDM rSP-D forms only trimers (28). Deletion of the collagen-like domain also removes the only site of N-linked glycosylation in rat SP-D (35). Thus, these data clearly demonstrate that neither the collagen-like domain or the N-linked oligosaccharide of SP-D are required for binding to conidia. In contrast, the E321Q/N323D mutant failed to bind the conidia. This latter SP-D mutant has altered carbohydrate recognition specificity and also fails to bind mannose-sepharose but retains phosphotidylinositol binding ability (27). The amino acids at positions 321 and 323 are predicted to be contact residues for Ca2+ and carbohydrate ligands within the CRD based upon homology with rat mannose-binding protein A (MBP-A). The corresponding amino acids in MBP-A (E185 and N187) are in the carbohydrate binding pocket of the protein, elucidated by solution of the crystal structure (38). Both the binding defect of the E321Q/N323D and the carbohydrate inhibition of SP-D binding to conidia demonstrate a role for the CRD in the recognition and ligation of conidia.
In contrast to the findings with rSP-D, the recombinant form of rSP-A bound weakly to conidia, and the native form of rSP-A failed to bind (Fig. 1). Despite the relatively poor activity of rSP-A, hSP-A displayed significant binding to conidia (Fig. 6). The reasons for the difference between rSP-A and hSP-A remain unclear. Both our work and that of Madan et al. (22) implicate the CRD in the binding of A. fumigatus conidia by hSP-A since the binding was inhibited by EDTA and saccharides. It is possible that differences in carbohydrate recognition between rSP-A and hSP-A account for this observation.
We have examined the CRDs of human and rat SP-A in more detail. Although the primary structures are very similar (70% amino acid identity and 79% amino acid homology), significant differences exist. Based on amino acid alignments with rat MBP-A, the region between residues 195 and 216 in SP-A appears to be critical for carbohydrate recognition and binding. This has been confirmed by mutagenesis at several critical residues in rSP-A (24, 26). When this region is examined more closely, two potentially important nonidentities are apparent; positions 197 and 199 in rSP-A are occupied by Arg and Glu, respectively, while in hSP-A the sites are Ala and Arg, respectively. Future mutagenesis experiments will be required to determine if humanizing the carbohydrate binding pocket of rSP-A alters the interaction of rSP-A with the conidia.
Neither rSP-A nor rSP-D increased the association of conidia with a rat alveolar macrophage cell line (Fig. 5) or macrophages directly isolated from rat lungs (not shown). Similar results were obtained whether or not the macrophages were washed with EDTA after incubation with the conidia-surfactant protein mixture (not shown). Madan et al. (22) showed that AP hSP-A and AP hSP-D increased the association and killing of conidia by human alveolar macrophages and neutrophils. It is important to note that the rat alveolar macrophages used in our study were either from a cultured cell line or isolated from normal rats, whereas Madan et al. (22) used human alveolar macrophages isolated from aspergillosis patients.
We repeated the binding studies of Madan et al. (22), using hSP-A and hSP-D. hSP-A isolated from bronchoalveolar lavage of both normal and AP lungs showed binding to conidia that was inhibited by EDTA (Fig. 6). This is significant, since structural differences between AP hSP-A and normal hSP-A have been described (37). Our data show that these structural differences do not cause functional differences in our system. The results obtained with recombinant hSP-D and protein isolated from bronchoalveolar lavage of AP patients were unexpected. Although both hSP-D forms bound the conidia, they showed relatively greater calcium-independent binding than other proteins tested; Madan et al. showed almost complete inhibition using 10 mM EDTA (22). At present, the reason for the high calcium-independent binding seen in our system remains unclear.
In the lung, SP-A and SP-D are components of a complex and dynamic environment. This environment contains a variety of lipids and other proteins. SP-A, and to a lesser extent SP-D, associate with extracellular alveolar lipids and several studies have examined these interactions (15, 17, 26). One study found that greater than 99% of total rat SP-A in bronchoalveolar lavage was associated with the lipid pellet from surfactant isolation, whereas less than 30% of the SP-D was found in the lipid pellet (17). To examine the effects of other surfactant components upon the activity of SP-A and SP-D, we performed binding studies in the presence of material extracted from rat surfactant (HSC). When preincubated with the proteins, HSC inhibited AP hSP-A binding to conidia but did not affect AP hSP-D binding (Fig. 7). This may be due to steric hindrance of the conidia binding site or conformational changes in SP-A brought about by the lipids.
In summary, we have shown that rSP-D, hSP-D, and hSP-A bind A. fumigatus conidia. In contrast, rSP-A does not bind the conidia. Thus, rSP-A is not a satisfactory model for studying the interactions of hSP-A with certain fungi. Additionally, while a previous study (22) demonstrated that hSP-A and hSP-D increased the phagocytosis and killing of conidia by human alveolar macrophages, we saw no rSP-A- or rSP-D-mediated increase in association of conidia with rat alveolar macrophages. Finally, we demonstrated that hSP-A binding was inhibited by surfactant material, suggesting that the binding observed in vitro may be an overestimate of that occurring in vivo.
ACKNOWLEDGMENTS
We thank Amanda Evans, Sandra Plaga, and Surapon Pattanajitvilai for providing many reagents; Rhonda Emerick for assistance with flow cytometry; and Erika Crouch for providing CHO-K1 cells expressing recombinant human SP-D. We also acknowledge Shuyu Ye for performing some preliminary experiments.
This work was supported by grants from the National Institutes of Health (HL-29891 and HL-45286) and from the Environmental Protection Agency (R825793). This research was performed in the Lord and Taylor Laboratory for Lung Biochemistry at National Jewish Medical and Research Center.
REFERENCES
- 1.Allen M, Mason R, Harbeck R, Greene K, Smith B, Voelker D. Abstracts of the 1998 International Conference of the American Thoracic Society. Am. J. Respir. Crit. Care Med. 157:A865. 1998. Binding of rat SP-A and SP-D to Aspergillus spores. [Google Scholar]
- 2.Benne C A, Benaissa-Trouw B, van Strijp J A, Kraaijeveld C A, van Iwaarden J F. Surfactant protein A, but not surfactant protein D, is an opsonin for influenza A virus phagocytosis by rat alveolar macrophages. Eur J Immunol. 1997;27:886–890. doi: 10.1002/eji.1830270413. [DOI] [PubMed] [Google Scholar]
- 3.Benne C A, Kraaijeveld C A, van Strijp J A, Brouwer E, Harmsen M, Verhoef J, van Golde L M, van Iwaarden J F. Interactions of surfactant protein A with influenza A viruses: binding and neutralization. J Infect Dis. 1995;171:335–341. doi: 10.1093/infdis/171.2.335. [DOI] [PubMed] [Google Scholar]
- 4.Crouch E C. Collectins and pulmonary host defense. Am J Respir Cell Mol Biol. 1998;19:177–201. doi: 10.1165/ajrcmb.19.2.140. [DOI] [PubMed] [Google Scholar]
- 5.Hartshorn K, Chang D, Rust K, White M, Heuser J, Crouch E. Interactions of recombinant human pulmonary surfactant protein D and SP-D multimers with influenza A. Am J Physiol. 1996;271:L753–L762. doi: 10.1152/ajplung.1996.271.5.L753. [DOI] [PubMed] [Google Scholar]
- 6.Hartshorn K L, Crouch E, White M R, Colamussi M L, Kakkanatt A, Tauber B, Shepherd V, Sastry K N. Pulmonary surfactant proteins A and D enhance neutrophil uptake of bacteria. Am J Physiol. 1998;274:L958–L969. doi: 10.1152/ajplung.1998.274.6.L958. [DOI] [PubMed] [Google Scholar]
- 7.Hartshorn K L, Crouch E C, White M R, Eggleton P, Tauber A I, Chang D, Sastry K. Evidence for a protective role of pulmonary surfactant protein D (SP-D) against influenza A viruses. J Clin Investig. 1994;94:311–319. doi: 10.1172/JCI117323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hartshorn K L, White M R, Shepherd V, Reid K, Jensenius J C, Crouch E C. Mechanisms of anti-influenza activity of surfactant proteins A and D: comparison with serum collectins. Am J Physiol. 1997;273:L1156–L1166. doi: 10.1152/ajplung.1997.273.6.L1156. [DOI] [PubMed] [Google Scholar]
- 9.Hattori A, Kuroki Y, Katoh T, Takahashi H, Shen H Q, Suzuki Y, Akino T. Surfactant protein A accumulating in the alveoli of patients with pulmonary alveolar proteinosis: oligomeric structure and interaction with lipids. Am J Respir Cell Mol Biol. 1996;14:608–619. doi: 10.1165/ajrcmb.14.6.8652189. [DOI] [PubMed] [Google Scholar]
- 10.Hawgood S, Benson B J, Hamilton R L., Jr Effects of a surfactant-associated protein and calcium ions on the structure and surface activity of lung surfactant lipids. Biochemistry. 1985;24:184–190. doi: 10.1021/bi00322a026. [DOI] [PubMed] [Google Scholar]
- 11.Hickman-Davis J M, Lindsey J R, Zhu S, Matalon S. Surfactant protein A mediates mycoplasmacidal activity of alveolar macrophages. Am J Physiol. 1998;274:L270–L277. doi: 10.1152/ajplung.1998.274.2.L270. [DOI] [PubMed] [Google Scholar]
- 12.Holmskov U, Malhotra R, Sim R B, Jensenius J C. Collectins: collagenous C-type lectins of the innate immune defense system. Immunol Today. 1994;15:67–74. doi: 10.1016/0167-5699(94)90136-8. [DOI] [PubMed] [Google Scholar]
- 13.Koziel H, Phelps D S, Fishman J A, Armstrong M Y, Richards F F, Rose R M. Surfactant protein-A reduces binding and phagocytosis of Pneumocystis carinii by human alveolar macrophages in vitro. Am J Respir Cell Mol Biol. 1998;18:834–843. doi: 10.1165/ajrcmb.18.6.3059. [DOI] [PubMed] [Google Scholar]
- 14.Kuan S F, Rust K, Crouch E. Interactions of surfactant protein D with bacterial lipopolysaccharides. Surfactant protein D is an Escherichia coli-binding protein in bronchoalveolar lavage. J Clin Investig. 1992;90:97–106. doi: 10.1172/JCI115861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuroki Y, Akino T. Pulmonary surfactant protein A (SP-A) specifically binds dipalmitoylphosphatidylcholine. J Biol Chem. 1991;266:3068–3073. [PubMed] [Google Scholar]
- 16.Kuroki Y, Mason R J, Voelker D R. Pulmonary surfactant apoprotein A structure and modulation of surfactant secretion by rat alveolar type II cells. J Biol Chem. 1988;263:3388–3394. [PubMed] [Google Scholar]
- 17.Kuroki Y, Shiratori M, Ogasawara Y, Tsuzuki A, Akino T. Characterization of pulmonary surfactant protein D: its copurification with lipids. Biochim Biophys Acta. 1991;1086:185–190. doi: 10.1016/0005-2760(91)90006-4. [DOI] [PubMed] [Google Scholar]
- 18.Kuroki Y, Voelker D R. Pulmonary surfactant proteins. J Biol Chem. 1994;269:25943–25946. [PubMed] [Google Scholar]
- 19.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 20.LeVine A M, Bruno M D, Huelsman K M, Ross G F, Whitsett J A, Korfhagen T R. Surfactant protein A-deficient mice are susceptible to group B streptococcal infection. J Immunol. 1997;158:4336–4340. [PubMed] [Google Scholar]
- 21.LeVine A M, Kurak K E, Bruno M D, Stark J M, Whitsett J A, Korfhagen T R. Surfactant protein-A-deficient mice are susceptible to Pseudomonas aeruginosa infection. Am J Respir Cell Mol Biol. 1998;19:700–708. doi: 10.1165/ajrcmb.19.4.3254. [DOI] [PubMed] [Google Scholar]
- 22.Madan T, Eggleton P, Kishore U, Strong P, Aggrawal S S, Sarma P U, Reid K B. Binding of pulmonary surfactant proteins A and D to Aspergillus fumigatus conidia enhances phagocytosis and killing by human neutrophils and alveolar macrophages. Infect Immun. 1997;65:3171–3179. doi: 10.1128/iai.65.8.3171-3179.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mason R J, Greene K, Voelker D R. Surfactant protein A and surfactant protein D in health and disease. Am J Physiol. 1998;275:L1–L13. doi: 10.1152/ajplung.1998.275.1.L1. [DOI] [PubMed] [Google Scholar]
- 24.McCormack F X, Festa A L, Andrews R P, Linke M, Walzer P D. The carbohydrate recognition domain of surfactant protein A mediates binding to the major surface glycoprotein of Pneumocystis carinii. Biochemistry. 1997;36:8092–8099. doi: 10.1021/bi970313f. [DOI] [PubMed] [Google Scholar]
- 25.McCormack F X, Fisher J H, Suwabe A, Smith D L, Shannon J M, Voelker D R. Expression and characterization of rat surfactant protein A synthesized in Chinese hamster ovary cells. Biochim Biophys Acta. 1990;1087:190–198. doi: 10.1016/0167-4781(90)90204-f. [DOI] [PubMed] [Google Scholar]
- 26.McCormack F X, Kuroki Y, Stewart J J, Mason R J, Voelker D R. Surfactant protein A amino acids Glu195 and Arg197 are essential for receptor binding, phospholipid aggregation, regulation of secretion, and the facilitated uptake of phospholipid by type II cells. J Biol Chem. 1994;269:29801–29807. [PubMed] [Google Scholar]
- 27.Ogasawara Y, Voelker D R. Altered carbohydrate recognition specificity engineered into surfactant protein D reveals different binding mechanisms for phosphatidylinositol and glucosylceramide. J Biol Chem. 1995;270:14725–14732. doi: 10.1074/jbc.270.24.14725. [DOI] [PubMed] [Google Scholar]
- 28.Ogasawara Y, Voelker D R. The role of the amino-terminal domain and the collagenous region in the structure and the function of rat surfactant protein D. J Biol Chem. 1995;270:19052–19058. doi: 10.1074/jbc.270.32.19052. [DOI] [PubMed] [Google Scholar]
- 29.Persson A, Chang D, Crouch E. Surfactant protein D is a divalent cation-dependent carbohydrate-binding protein. J Biol Chem. 1990;265:5755–5760. [PubMed] [Google Scholar]
- 30.Pikaar J C, Voorhout W F, van Golde L M, Verhoef J, Van Strijp J A, van Iwaarden J F. Opsonic activities of surfactant proteins A and D in phagocytosis of gram-negative bacteria by alveolar macrophages. J Infect Dis. 1995;172:481–489. doi: 10.1093/infdis/172.2.481. [DOI] [PubMed] [Google Scholar]
- 31.Reid K B M. Interactions of surfactant protein D with pathogens, allergens and phagocytes. Biochim Biophys Acta. 1998;1408:290–295. doi: 10.1016/s0925-4439(98)00074-x. [DOI] [PubMed] [Google Scholar]
- 32.Rosseau S, Guenther A, Seeger W, Lohmeyer J. Phagocytosis of viable Candida albicans by alveolar macrophages: lack of opsonin function of surfactant protein A. J Infect Dis. 1997;175:421–428. doi: 10.1093/infdis/175.2.421. [DOI] [PubMed] [Google Scholar]
- 33.Rouser G, Siakotos A N, Fleischer S. Quantitative analysis of phospholipids by thin-layer chromatography and phosphorus analysis of spots. Lipids. 1966;1:85–86. doi: 10.1007/BF02668129. [DOI] [PubMed] [Google Scholar]
- 34.Sharma O P, Chwogule R. Many faces of pulmonary aspergillosis. Eur Respir J. 1998;12:705–715. doi: 10.1183/09031936.98.12030705. [DOI] [PubMed] [Google Scholar]
- 35.Shimizu H, Fisher J H, Papst P, Benson B, Lau K, Mason R J, Voelker D R. Primary structure of rat pulmonary surfactant protein D. cDNA and deduced amino acid sequence. J Biol Chem. 1992;267:1853–1857. [PubMed] [Google Scholar]
- 36.Strong P, Kishore U, Morgan C, Lopez Bernal A, Singh M, Reid K B. A novel method of purifying lung surfactant proteins A and D from the lung lavage of alveolar proteinosis patients and from pooled amniotic fluid. J Immunol Methods. 1998;220:139–149. doi: 10.1016/s0022-1759(98)00160-4. [DOI] [PubMed] [Google Scholar]
- 37.Voss T, Schafer K P, Nielsen P F, Schafer A, Maier C, Hannappel E, Maassen J, Landis B, Klemm K, Przybylski M. Primary structure differences of human surfactant-associated proteins isolated from normal and proteinosis lung. Biochim Biophys Acta. 1992;1138:261–267. doi: 10.1016/0925-4439(92)90002-5. [DOI] [PubMed] [Google Scholar]
- 38.Weis W I, Drickamer K, Hendrickson W A. Structure of a C-type mannose-binding protein complexed with an oligosaccharide. Nature. 1992;360:127–134. doi: 10.1038/360127a0. [DOI] [PubMed] [Google Scholar]
- 39.Wright J R. Immunomodulatory functions of surfactant. Physiol Rev. 1997;77:931–962. doi: 10.1152/physrev.1997.77.4.931. [DOI] [PubMed] [Google Scholar]