Abstract
Background
Cluster headache is a highly debilitating neurological disorder with considerable inter-ethnic differences. Genome-wide association studies (GWAS) recently identified replicable genomic loci for cluster headache in Europeans, but the genetic underpinnings for cluster headache in Asians remain unclear. The objective of this study is to investigate the genetic architecture and susceptibility loci of cluster headache in Han Chinese resided in Taiwan.
Methods
We conducted a two-stage genome-wide association study in a Taiwanese cohort enrolled from 2007 through 2022 to identify the genetic variants associated with cluster headache. Diagnosis of cluster headache was retrospectively ascertained with the criteria of International Classification of Headache Disorders, third edition.
Control subjects were enrolled from the Taiwan Biobank. Genotyping was conducted with the Axiom Genome-Wide Array TWB chip, followed by whole genome imputation. A polygenic risk score was developed to differentiate patients from controls. Downstream analyses including gene-set and tissue enrichment, linkage disequilibrium score regression, and pathway analyses were performed.
Results
We enrolled 734 patients with cluster headache and 9,846 population-based controls. We identified three replicable loci, with the lead SNPs being rs1556780 in CAPN2 (odds ratio = 1.59, 95% CI 1.42‒1.78, p = 7.61 × 10–16), rs10188640 in MERTK (odds ratio = 1.52, 95% CI 1.33‒1.73, p = 8.58 × 10–13), and rs13028839 in STAB2 (odds ratio = 0.63, 95% CI 0.52‒0.78, p = 2.81 × 10–8), with the latter two replicating the findings in European populations. Several previously reported genes also showed significant associations with cluster headache in our samples. Polygenic risk score differentiated patients from controls with an area under the receiver operating characteristic curve of 0.77. Downstream analyses implicated circadian regulation and immunological processes in the pathogenesis of cluster headache.
Conclusions
This study revealed the genetic architecture and novel susceptible loci of cluster headache in Han Chinese residing in Taiwan. Our findings support the common genetic contributions of cluster headache across ethnicities and provide novel mechanistic insights into the pathogenesis of cluster headache.
Supplementary Information
The online version contains supplementary material available at 10.1186/s10194-022-01517-6.
Keywords: Cluster headache, Genome-wide association study, CAPN2, MERTK, STAB2
Background
Cluster headache is one of the most painful disorders in the world, with a prevalence of around 0.1% and a clear male predominance [1]. It is characterized by excruciating unilateral orbital, supraorbital or temporal pain attacks, with accompanying cranial autonomic symptoms or restlessness/agitation. The attacks last between 15 and 180 min but may occur up to 8 times per day and take weeks to months to remit. In some patients, the cluster headache presents as a chronic form, with attacks occurring for > 1 year without remission, or with remission periods lasting < 3 months [2]. The clinical features of cluster headache in Asian populations differ from those in Western populations, including a lower prevalence of chronic cluster headache, higher male/female ratio, and lower frequencies of restlessness or aura [3, 4]. Mechanisms underlying these ethnic differences remain unclear.
Despite the tremendous impact on the sufferers, detailed pathogenesis underlying cluster headache remains enigmatic. Because cluster headache has significant familial aggregation, attempts have been undertaken to determine its underlying genetic architectures. Hypothesis-driven approaches using candidate gene association studies, however, have not identified replicable signals. The first genome-wide association study (GWAS) investigating 99 Italian patients with cluster reported suggestive associations with genetic variants in ADCYAP1R1 (ADCYAP receptor type I) and MME (membrane metalloendopeptidase) [5]; however, these findings were not replicated subsequently. Two recent studies provided the first evidence to demonstrate genome-wide significant variants contributing to the predisposition of cluster headache in European cohorts [6, 7]. The Dutch and Norwegian study identified rs11579212 near RP11-815 M8.1, rs6541998 near MERTK (MER Proto-Oncogene, Tyrosine Kinase), rs10184573 near AC093590.1, and rs2499799 near UFL1 (UFM1 specific ligase 1)/FHL5 (four and a half LIM domains 5)[6] whereas the combined United Kingdom (UK) and Swedish cohorts identified rs113658130 near LINC01877/SATB2 (SATB homeobox 2), rs4519530 in MERTK, rs12121134 near LINC01705/DUSP10 (Dual Specificity Phosphatase 10), and rs11153082 in FHL5 to be associated with cluster headache [7]. Considering the inter-ethnic variability of clinical characteristics [3], it is uncertain whether these novel loci are replicable in other populations.
To interrogate the genetic architecture of cluster headache in Asians, we performed a two-stage GWAS in a total of 734 clinic-based patients and 9,846 population-based controls. We also established polygenic risk score (PRS) models to differentiate patients from controls and conducted downstream analyses to investigate genes and potential pathogenic mechanisms of cluster headache.
Methods
Study participants and clinical evaluations
This study was a two-stage case–control GWAS, including a discovery cohort and a replication cohort, followed by a combined analysis of both cohorts. All participants were unrelated and of Han Chinese ancestry. Patients with cluster headache were enrolled from the headache or neurological clinics of TVGH and TSGH, Taiwan, as well as the Taiwan Precision Medicine Initiative, from 2007 through 2022. Patients who were recruited after May 2012 were assigned to the discovery cohort and subjects recruited prior to that were assigned to the replication cohort. The control subjects were recruited from the Taiwan Biobank, the largest publicly available genetic database of individuals with East Asian ancestry that provides adequate coverage of genetic diversity across all Han Chinese [8]. To evaluate the genetic correlation of cluster headache and migraine, we also recruited an independent cohort of migraine patients from TVGH. The inclusion criteria for all participants include (1) age between 20 and 70 years old and (2) could fully understand the study objective and willing to provide written informed consent. Patients were eligible if they were diagnosed with cluster headache or migraine by board-certified neurologists based on the criteria proposed in the International Classification of Headache Disorders, third edition (ICHD-3) [2]. The exclusion criteria for all participants included history of major systemic illnesses or major neuropsychiatric disorders. Control subjects who have a history of moderate or severe headaches, first-degree relatives with a history of migraine or severe headaches, or any kinship relatedness with the patients were also excluded.
Single nucleotide polymorphism (SNP) genotyping
SNP genotyping was conducted with the Affymetrix Axiom Genome-Wide TWB2 Array Plate at the National Center for Genomics Medicine, Academia Sinica, Taiwan. SNP genotypes were called using the Axiom GT1 algorithm. Quality control (QC) criteria for SNPs were applied to exclude SNPs if they (a) were monomorphic in both cases and controls, (b) had a minor allele frequency < 0.01, (c) had a total call rate < 95% in cases and controls combined, (d) showed significant (P < 0.00001) deviation from Hardy–Weinberg equilibrium in case or controls, or (e) a significant difference in the genotype call rates between cases and controls (P < 0.001). For sample filtering, we excluded arrays with generated genotypes for fewer than 95% of loci. Heterozygosity of SNPs on the X-chromosome was used to verify the sex of the samples. PLINK software (version 1.90) [9] was used to identify samples with genetic relatedness, indicating that they were from the same individual (or monozygotic twins) or from first-, second- or third-degree relatives. These determinations were made based on evidence for cryptic relatedness from identity-by-descent status (pi-hat cutoff of 0.125).
Genotype imputation analysis
We conducted a genotype imputation analysis using the 1000 Genomes Phase 3 data [10] and TWB whole-genome sequencing data as reference and IMPUTE2 [11–13]. Well-imputed SNPs (information score > 0.8) were retained followed by systematic QC, as described above [14]. For sample filtering, we excluded arrays with generated genotypes for fewer than 95% of loci.
Statistical analysis
Association analyses were carried out by comparing allele or genotype frequencies between cases and controls. The Manhattan Plot and quantile–quantile (Q-Q) plots were generated using the “qqman” package in R [15]. Detection of population stratification was carried out by using principal component analysis (PCA). The genomic inflation factor (GIF) was calculated and the top 10 principal components (PCs) via Eigenstrat in typing datasets. We calculated the distribution of expected p values under the null hypothesis and genomic inflation value (λ = 1). Logistic regression was used to evaluate association of SNPs with cluster headache by adjusting age, sex, and the top 10 PCs (PC1‒PC10), and an additive genetic model was conducted using PLINK (version 1.9) [9]. P < 5 × 10–8 was designated as GWAS significance and p < 5 × 10–5 was designated as suggestive GWAS significance. The positional gene mapping and fine mapping of significant loci were generated using LocusZoom [16] and Probabilistic Identification of Causal SNPs (PICS2) [17]. The proportion of variance explained by a given SNP was calculated using Nagelkerke pseudo R2.
Gene-based analysis
We performed the Multimarker Analysis of GenoMic Annotation (MAGMA) gene-based association analysis [18] implemented in FUMA [19], which calculates a gene test-statistic (p-value) based on all SNPs located within genes, using imputation data.
Previously reported cluster headache or migraine loci
Genetic loci previously reported to be associated with cluster headache were tested for association in our samples [5–7, 20–26]. We also conducted meta-analysis of these loci by combining the results from previous studies and ours with METAL [27]. As recent GWASs suggested a potential genetic correlation of cluster headache with migraine [6, 7], we also tested the association of migraine-associated loci [28, 29] in the current sample.
Univariate LD-score regression
Linkage Disequilibrium Score Regression (LDSC v1.0.1) was used to estimate the proportion of a true polygenic signal opposed to confounding biases, and to calculate SNP-based heritability [30]. We estimated LD Scores from 1,446 samples in the Taiwan Biobank whole-genome sequencing data using an unbiased estimator of r2 with 1-cM windows, singletons excluded and no r2 cutoff. Heritability estimates were converted to the liability scale assuming a population prevalence of cluster headache of 0.1%.
Polygenic risk score derivation
Four sets of polygenic risk scores were developed to discriminate patients from controls. In the first set, we computed genome-wide PRS derived from the whole genome imputation data following a method reported previously [31]. We used the GWAS of the discovery cohort (i.e., the training set) for generating the estimates of regression coefficients, its standard error and associated p-value (, and ) for each SNP j based on the univariate logistic regression analysis. With SNPs that passed the QC in GWAS analysis, the SNPs with MAF less than 1% or Indels were excluded. To calculate PRS, we used the standard clumping and thresholding (C + T) method [32] using two hyperparameters for model building: the cut-off of correlation and p-value threshold p. The optimal PRS model was chosen from the pair of () that maximized the prediction performance, area under the receiver-operator characteristics curve (AUC), evaluated in the testing (i.e., replication) cohort. In the second set, the PRS was derived from all the previously reported loci associated with cluster headache. The prediction performance was also evaluated with AUC. The 3rd and 4th sets of PRS were derived from the significant SNPs of the two previously reported European cluster headache GWASs [6, 7] respectively.
Gene expression, tissue enrichment analysis, and pathway analysis
The tissue specific expression of the cluster headache-associated genes was explored based on the expression data from the Genotype-Tissue Expression (GTEx) project [33] using the GTEx platform or the MAGMA gene property in FUMA. We also implemented GIGSEA (Genotype Imputed Gene Set Enrichment Analysis) [34] to infer differential gene expression and interrogate gene set enrichment for the trait-associated SNPs. The gene expression levels were imputed from GWAS summary data of cluster headache by Elastic net (ENet) regression models using MetaXcan [35] or the pathway analyses of MetaXcan output, we employed the weighted linear regression model of the GIGSEA [34] with empirical p-values incorporating 1,000 permutations for Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), and Reactome (REAC) [36–38]. Following the guidance of GIGSEA, the Bayes factor was used to correct the multiple hypothesis testing [39]. Stringent thresholds for significant associations were set as the empirical p-values < 0.05 and Bayes factor [BF] > 100 in the pathway enrichment analyses in brain tissues. Of note, we selected the enriched pathways with the gene sets that contain from 15 to 500 genes as suggested by GSEA manual.
Results
Characteristics of study subjects
The two-stage GWAS included a discovery cohort consisting of 359 cluster headache patients and 4786 population-based controls and a replication cohort comprised 375 patients and 5060 population-based controls. Demographic characteristics of participants are shown in Additional File 1.
Association analysis
After applying stringent QC criteria, we obtained 549,611 SNPs with an average call rate of 99.78% ± 0.25%. After imputation, 6,630,979 loci were yielded. Q-Q plot (Additional File 2) and PCA (Additional File 3) showed no significant population stratification. In the combined analysis, the LDSC intercept was 0.9962 (SE 0.0061), indicating little inflation due to factors other than polygenic architecture. In addition, we estimated the SNP-based heritability (h2) of cluster headache at 16.51% (SE 4.42%) on the observed scale and 22.5% (SE 6.03%) on the liability scale.
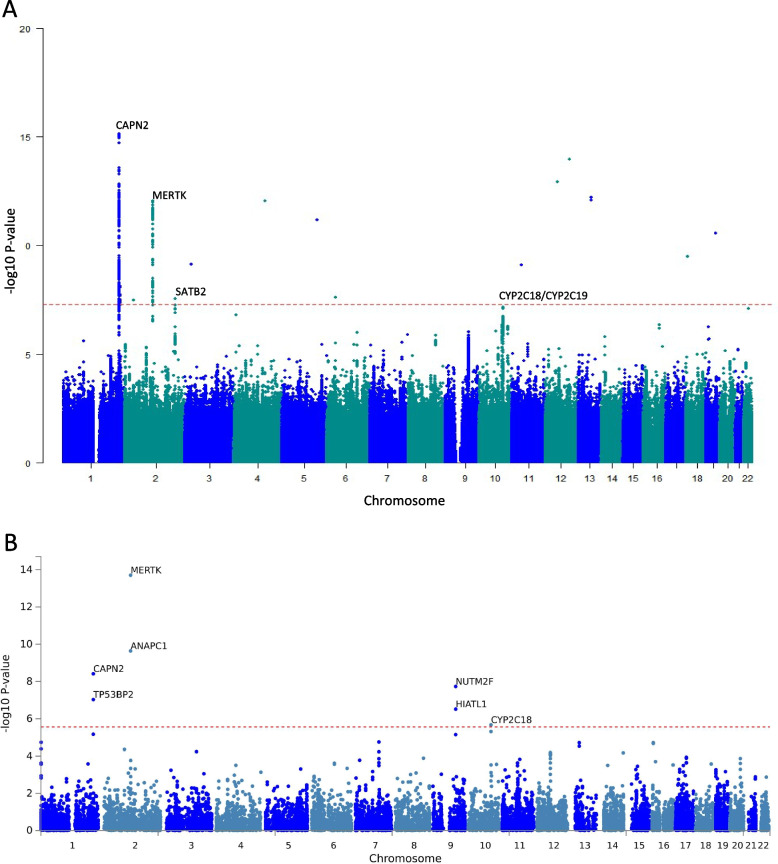
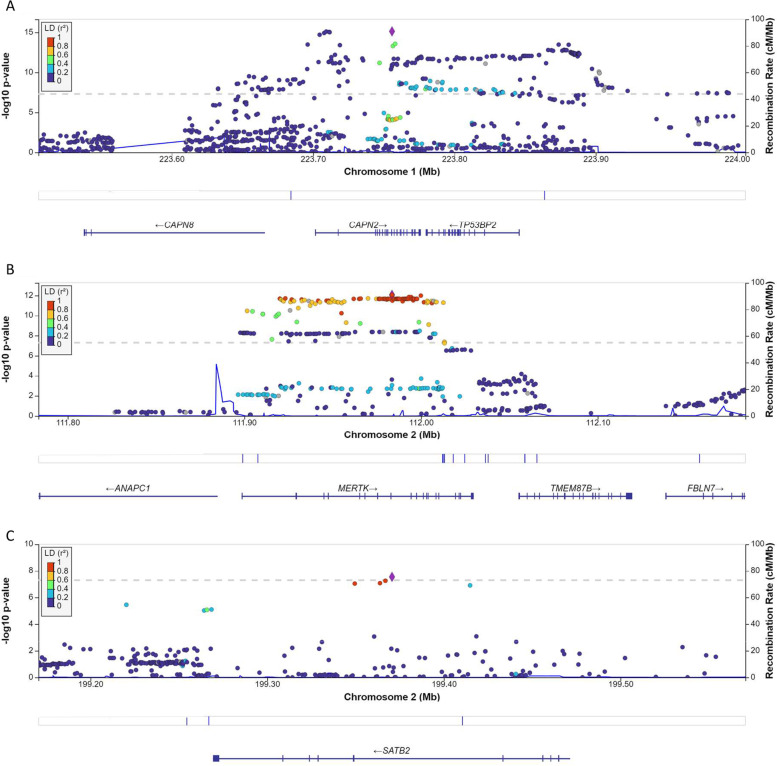
The two-stage genome-wide analysis using imputation data identified one replicable genomic locus in Calpain 2 (CAPN2) with multiple SNPs reaching GWAS significance at both discovery and replication cohorts (Fig. 1A and Additional File 4). Combined analysis found additional two loci with GWAS significance in MERTK and SATB2 (Table 1) and one locus with suggestive GWAS significance in CYP2C18/CYP2C19 (Fig. 1A). Of note, indels or singleton SNPs without significant LD with other SNPs that were deemed spurious association signals. The regional association plots for the significant loci were demonstrated in Fig. 2. In addition, fine mapping with PICS identified multiple variants with causal probability larger than 0.2, but only rs4653500 (PICS probability = 0.46) has eQTL on CANP2.
Fig. 1.
Manhattan plots for the GWAS and gene-based analysis. A Manhattan plot for the associations of SNPs with cluster headache. The horizontal axis shows the chromosomal position, and the vertical axis shows the significance of tested markers. The threshold for genome wide significance (p < 5 × 10–8) is indicated by a red dash line. B Manhattan plot of the gene-based test as computed by Multimarker Analysis of GenoMic Annotation (MAGMA). The input SNPs were mapped to 18,460 protein coding genes. The red dash line indicates the genome wide significance defined at p = 0.05/18,460 = 2.709 × 10–6
Table 1.
Basic demographics of participants
| Characteristics | N | Mean age ± SD (yr) | Male, n (%) | P value |
|---|---|---|---|---|
| Cluster headache GWAS | ||||
| Discovery cohort | ||||
| Cluster headache | 359 | 36.39 ± 10.58 | 293 (81.62%) | 0.335 |
| Controls | 4,786 | 42.98 ± 7.56 | 4,000 (83.58%) | |
| Replication cohort | ||||
| Cluster headache | 375 | 36.74 ± 10.61 | 298 (79.47%) | 0.159 |
| Controls | 5,060 | 42.84 ± 7.58 | 4,012 (79.29%) | |
| Combined analysis | ||||
| Cluster headache | 734 | 36.57 ± 10.59 | 591 (80.52%) | 0.566 |
| Controls | 9,846 | 42.91 ± 7.57 | 8,012 (81.37%) | |
| Migraine cohort for computation of genetic correlation with cluster headache | ||||
| Migraineurs | 3,173 | 38.51 ± 11.77 | 638 (20.11%) | < 0.001 |
| Controls | 24,528 | 43.31 ± 7.55 | 8,013 (32.67%) | |
P p value for sex ratio (by chi-square test)
Fig. 2.
Regional association plots of the two genome-wide significant cluster headache loci (A) CAPN2, (B) MERTK, and (C) SATB2. Each dot represents a single nucleotide polymorphism (SNP) derived from the fine-mapped imputation data. The horizontal axis gives the genomic coordinate and the vertical axis the significance level (-log10 p value). The top SNP for each locus is marked with a purple diamond (CRCh38/hg19). SNPs are colored based on their correlation (r2) with the labeled lead SNP according to the legend. The solid blue line shows the recombination rate from 1000 Genomes EAS data (right vertical axis). The gray dash line corresponds to p = 5 × 10−8. Figures were obtained from LocusZoom
Gene-based association testing
Gene-based association testing used the mean association signal from all SNPs within each gene, accounting for LD. Seven genes passed the threshold of statistical significance: MERTK, ANAPC1 (anaphase promoting complex subunit 1), CAPN2, NUTM2F (NUT Family Member 2F), TP53BP2 (Tumor Protein P53 Binding Protein 2), HIATL1 (Hippocampus Abundant Transcript-Like Protein 1), and CYP2C18 (Fig. 1B). The Q-Q plot of the gene-based test computed by MAGMA was shown in Additional File 5.
Previously reported cluster headache loci
Among the previously reported cluster headache-associated SNPs, rs4519530 in MERTK and rs6541998 near MERTK were successfully replicated in our sample using imputation data. Some SNPs were unavailable or insignificant in our sample, but the lead SNPs within or near the same genes were found to have suggestive GWAS significance (Table 2). Most of the variants, particularly those in MERTK and FHL5, were more significant in meta-analysis (Table 2); however, substantial heterogeneity was noted in some of them (Additional File 6).
Table 2.
The Association of previously reported cluster headache loci in the current sample
| Chr | Gene | Index SNP | Risk allele | Cona | OR [95%CI] | P1b | P2c | Ref No |
|---|---|---|---|---|---|---|---|---|
| 1 | near RP11-815M8.1 | rs11579212 | C | 2 | 0.97 [0.78–1.2] | 0.30 | 0.09 | 6 |
| 1 | LINC01705/DUSP10 | rs6687758 | G | 1 | 1.07 [0.94–1.23] | 0.23 | 1.58 × 10–3 | 7 |
| 1 | LINC01705/DUSP10 | rs12121134 | T | 2 | 0.61 [0.22–1.69] | 0.21 | 5.23 × 10–3 | 7 |
| 1 | LINC01705 | rs545037820 | G | 3 | 0.75 [0.50–1.12] | 0.02 | ||
| 1 | near LINC01705 | rs2034485 | T | 3 | 1.31 [1.16–1.49] | 1.16 × 10–6 | ||
| 1 | DUSP10 | rs145531779 | C | 3 | 1.43 [1.11–1.83] | 0.02 | ||
| 2 | MERTK | rs4519530 | C | 2 | 1.50 [1.32–1.72] | 4.69 × 10–12 | 9.30 × 10–27 | 7 |
| 2 | near MERTK | rs6541998 | C | 2 | 1.43 [1.25–1.63] | 1.86 × 10–8 | 6.23 × 10–15 | 6 |
| 2 | MERTK | rs10188640 rs10188642 | A | 3 | 1.52 [1.33–1.73] | 8.58 × 10–13 | ||
| 2 | near LINC01877/SATB2 | rs4675692 rs113658130 | G C | NA | 7 | |||
| 2 | LINC01877 | rs13394614 | G | 3 | 1.34 [1.18–1.52] | 5.42 × 10–7 | ||
| 2 | STAB2 | rs13028839 | A | 3 | 0.63 [0.52–0.78] | 2.81 × 10–8 | ||
| 3 | MME | rs147564881 | C | NA | 5 | |||
| 3 | MME | rs56208271 | A | 3 | 1.61 [1.17–2.22] | 4.56 × 10–3 | ||
| 3 | near MME | rs143162143 | C | 3 | 2.29 [1.56–3.36] | 1.15 × 10–4 | ||
| 4 | CLOCK | rs12649507 | G | 2 | 0.88 [0.78–1.01] | 0.07 | 1.02 × 10–3 | 21 |
| 4 | CLOCK | rs369023808 | G | 3 | 1.19 [0.76–1.88] | 0.04 | ||
| 4 | near CLOCK | rs34945396 | C | 3 | 0.78 [0.66–0.92] | 5.09 × 10–3 | ||
| 4 | ADH4 | rs1126671 | T | 1 | 1.72 [0.60–4.93] | 0.04 | 1.46 × 10–3 | 20 |
| 4 | ADH4 | rs56314548 | C | 3 | 0.77 [0.51–1.17] | 0.58 | ||
| 6 | HCRTR2 | rs3800539 | A | 2 | 1.13 [0.99–1.29] | 0.14 | 0.13 | 26 |
| 6 | HCRTR2 | rs10498801 | A | 2 | 0.88 [0.75–1.02] | 0.03 | 0.34 | 26 |
| 6 | HCRTR2 | rs2653349 | A | 2 | 0.77 [0.57–1.03] | 0.49 | 0.10 | 23, 24 |
| 6 | HCRTR2 | rs3122156 | G | NA | 25 | |||
| 6 | HCRTR2 | rs78769350 | G | 3 | 0.69 [0.46–1.06] | 0.03 | ||
| 6 | near UFL1/FHL5 | rs2499799 | T | 2 | 1.08 [0.85–1.36] | 0.74 | 4.00 × 10–3 | 6 |
| 6 | FHL5 | rs11153082 | G | 2 | 1.19 [1.04–1.35] | 1.99 × 10–4 | 7.54 × 10–11 | 7 |
| 6 | FHL5 | rs9386670 | A | 2 | 1.17 [1.02–1.34] | 9.72 × 10–4 | 1.09 × 10–9 | 7 |
| 6 | UFL1 | rs2255552 | G | 3 | 1.18 [1.04–1.35] | 1.83 × 10–4 | ||
| 6 | FHL5 | rs57425973 | A | 3 | 1.2 [1.05–1.37] | 6.21 × 10–5 | ||
| 7 | ADCYAP1R1 | rs12668955 | A | 1 | 1.07 [0.96–1.19] | 0.22 | 1.7 × 10–4 | 5 |
| 7 | ADCYAP1R1 | rs201504906 rs200124908 | C | 3 | 1.72 [1.31–2.26] | 1.01 × 10–5 | ||
| 12 | CRY1 | rs8192440 | A | 1 | 0.96 [0.66–1.41] | 0.42 | 0.09438 | 22 |
Abbreviations: Chr Chromosome, SNP Single nucleotide polymorphism, Con Condition, OR Odds ratio, CI Confidence interval, Risk allele, allele with higher frequency in cases compared to controls, NA Not available
aCondition: 1. Information obtained from real genotyping data. 2. Information derived from imputation data. 3. Most significant SNP within or near the same gene in the imputation data
bThe p values of Cochran-Armitage Trend Test
cThe p values of meta-analysis of the previous studies and the current study
Polygenic risk score
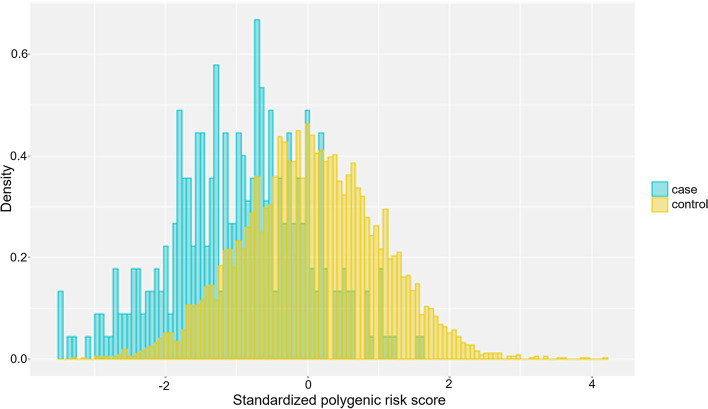
We calculated two sets of PRS. In the first set using the C + T method for genome-wide data, the distribution of the PRS for cluster headache in testing dataset was shown in Fig. 3. Grid search found that the PRS consisted of SNPs with r2 = 0.1 and p-value = 0.05 was the best model to differentiate patients from controls (AUC = 0.772, 95% confidence interval (CI) 0.747–0.797). The SNPs within the PRS model had a combined explained variance (i.e., Nagelkerke pseudo R2) of 16.97%. In the second set which used only the previously reported cluster headache loci (listed in Table 3) to derive PRS, the AUC was 0.633 (95% CI 0.604–0.662). The 3rd set of PRS derived from the Dutch and Norwegian study [6] had an AUC of 0.536 [0.504–0.567] while the 4th set from UK and Swedish study [7] had an AUC of 0.583 [0.553–0.613].
Fig. 3.
Genome-wide polygenic risk score (PRS) for cluster headache. The density distributions of polygenic score for cluster headache in testing dataset. The x-axis represents polygenic score, with values scaled to a mean of 0 and standard deviation of 1. Cases: blue; Controls: yellow
Table 3.
Lead SNPs of GWAS significant loci in the combined analysis for patients with cluster headache in Taiwan
| SNP | Chr | Position | Gene | Minor allele | Stage | MAF(Case, Control) | OR | 95% CI | P valuea | eQTL mapped genesc |
|---|---|---|---|---|---|---|---|---|---|---|
| rs1556780 | 1 | 223,755,834 | CAPN2 | G | 1 | (0.354, 0.253) | 1.637 | 1.390‒1.927 | 2.34 × 10–9 | CAPN2 |
| 2 | (0.344, 0.253) | 1.540 | 1.316‒1.801 | 5.31 × 10–8 | ||||||
| Com | (0.329, 0.253) | 1.586 | 1.416‒1.775 | 7.61 × 10–16 | ||||||
| rs10188640 | 2 | 111,983,520 | MERTK | A | 1 | (0.345, 0.257) | 1.522 | 1.297‒1.787 | 2.17 × 10–7 | AC012442.5, FBLN7, MERTK, RGPD8, TMEM87B, TTL, ZC3H8 |
| 2 | (0.343, 0.260) | 1.481 | 1.266 ‒1.733 | 8.05 × 10–7 | ||||||
| Com | (0.349, 0.258) | 1.501 | 1.342‒1.679 | 1.25 × 10–12 | ||||||
| rs13028839 | 2 | 199,370,629 | SATB2 | A | 1 | (0.096, 0.154) | 0.577 | 0.447–0.746 | 2.02 × 10–5 | C2orf69, SATB2, AC019330.1 |
| 2 | (0.099, 0.149) | 0.639 | 0.493–0.829 | 3.46 × 10–4 | ||||||
| Com | (0.102, 0.151) | 0.634 | 0.518‒0.778 | 2.81 × 10–8 |
Abbreviations: SNP Single nucleotide polymorphism, Chr Chromosome, OR Odds ratio, CI Confidence interval, MAF Minor allele frequency
aThe p values of Cochran-Armitage Trend Test The eQTL mapping was done with GTEx v.8
Genetic correlation between cluster headache and migraine
We recruited a cohort consisting of 3,173 Taiwanese migraine patients and 24,528 controls and estimated the genetic covariance between cluster headache and migraine on the liability scale was 21.0% (SE 5.7%) and the genetic correlation was 42.91% (SE 16.31%, P = 0.009). We have also examined 125 migraine-associated loci reported in previous migraine GWAS in the current sample of cluster headache.27, 28 Some SNPs or variants with high LD within or near the migraine-associated loci were found to have suggestive GWAS significance in the current cluster headache dataset (Table 4).
Table 4.
Previously reported migraine loci with suggestive GWAS significance in the current sample
| Chr | Gene | Index SNP | Risk allele | Cona | OR [95%CI] | P valueb |
|---|---|---|---|---|---|---|
| 2 | MYO3B | rs6753065 | G | 2 | 1.43 [1.20–1.71] | 4.91 × 10–5 |
| 6 | near GJA1 | rs28455731 | T | 1 | 1.32 [1.15–1.50] | 4.03 × 10–5 |
| 6 | near GJA1 | rs9482172 | C | 2 | 1.33 [1.17–1.52] | 1.98 × 10–5 |
| 6 | near PCMT1 | rs560489087 | T | 2 | 2.12 [1.49–3.03] | 2.05 × 10–5 |
| 9 | NFIB | rs10756554 | T | 2 | 0.79 [0.71–0.89] | 3.33 × 10–5 |
| 10 | PLCE1 | rs4244285 | A | 1 | 1.34 [1.20–1.49] | 1.76 × 10–7 |
| 10 | PLCE1 | rs12571421 | G | 2 | 1.34 [1.20–1.49] | 1.88 × 10–7 |
| 14 | near ITPK1 | rs150587675 | A | 1 | 1.68 [1.32–2.14] | 2.23 × 10–5 |
| 17 | HOXB3 | rs3214184 | TTAACAc | 2 | 1.33 [1.16–1.51] | 2.36 × 10–5 |
| 18 | near SKOR2 | rs62095427 | A | 2 | 0.62 [0.49–0.76] | 9.95 × 10–6 |
Abbreviations: Chr Chromosome, SNP Single nucleotide polymorphism, Con Condition, OR Odds ratio, CI Confidence interval, Risk allele, allele with higher frequency in cases compared to controls, NA Not available
aCondition: 1. Information obtained from real genotyping data. 2. Most significant SNP within or near the same gene in the imputation data
bThe p values of Cochran-Armitage Trend Test
cThe variant is an INDEL (T/TTAACA)
eQTL analysis
eQTL analysis using the GTEx online platform [33] (accessed on June 20, 2022) found that the lead SNP rs1556780 in CAPN2 and rs10188640 in MERTK had significant eQTLs on several genes (Table 1). With MetaXcan to infer the correlation between genetic variants and gene expression, we found that the genetic variants in MERTK were significantly associated with the gene expression in the pituitary gland and caudate (basal ganglia) with a false discovery rate (FDR) of less than 0.001 (Additional File 7).
Functional enrichment and pathway analysis
MAGMA gene-property analyses implemented in FUMA did not demonstrate significantly enriched tissues. GIGSEA biological pathway enrichment analyses based on the MetaXcan gene-level summary statistics found that the pathways were predominantly enriched in the pituitary gland, hippocampus, cerebellum, and basal ganglion. Top significantly associated pathways in these tissues that may be relevant to cluster headache include pathways involved in synaptic transmission, transmembrane protein kinase activity, immune responses, mitochondrial function, and metalloendopeptidase activity (Additional File 8 and Additional File 9).
Discussion
We identified three susceptibility loci, in CAPN2, MERTK, and SATB2, as well as one suggestive locus at CYP2C18/CYP2C19 at genome-wide level in patients with cluster headache in Taiwan. To the best of our knowledge, this is the first GWAS of cluster headache performed in Han Chinese and the first in Asians. While replicating the susceptibility genes recently identified in GWASs in patients of European ancestries [6, 7] suggested the validity of our study, we also identified novel genes implicating potential inter-ethnic differences. The association effect sizes are relatively large and similar to those observed in European GWASs [6, 7]. These results suggest that some phenotypes of cluster headache might be driven by these selected loci with large effect size, although further studies are needed to explore the genotype–phenotype association. In addition, several other risk loci identified in previous studies are of suggestive GWAS significance in our samples, suggesting that future studies with larger sample sizes might validate the associations of these loci with cluster headache. Moreover, the superior discriminative capacity of genome-wide PRS than the PRS composed of known cluster headache-associated loci suggests that additional loci with smaller effect sizes might also contribute to the genetic basis of cluster headache. Nevertheless, the clinical utility of PRS remains to be explored.
The novel gene CAPN2 identified in our study encodes calpain 2, a calcium-regulated non-lysosomal thiol-protease involved in cytoskeletal remodeling and signal transduction. Calpain 1/2 have been known to mediate Ca2+ influx and mediate degradation of suprachiasmatic nucleus (SCN) circadian oscillatory protein (SCOP) [40] in SCN neurons, which may contribute to coordinated regulation of circadian rhythms. In addition, calpain 1/2 play opposite roles in retinal ganglion cell (RGC) degeneration induced by ischemia/reperfusion injury [41], while degeneration of RGCs could lead to impaired circadian rhythmicity. Another potential implication of CAPN2 may be that the most commonly used preventive drug for cluster headache, the L-type calcium channel blocker verapamil, has been known to abrogate calpain activation [42]. Furthermore, calpain was found to mediate capsaicin-induced ablation of transient receptor potential vanilloid subtype 1 (TRPV1)-positive trigeminal afferent terminals [43], which may mediate the release of calcitonin gene-related peptide (CGRP) and contribute to the pathogenesis of cluster headache.
We successfully replicated MERTK and SATB2 identified in previous cluster headache GWASs [6, 7]. MERTK encodes a protein belongs to the MER/AXL/TYRO3 receptor kinase family. In addition to regulating microglia-mediated neuroinflammation and astrocyte-mediated neuronal synaptic remodeling proposed in previous studies [6, 7]. MERTK functions in the retinal pigment epithelium as a regulator of photoreceptor phagocytosis, which is a circadian-regulated process indispensable for vision [44]. Mutations of MERTK cause degeneration of photoreceptors, which in turn lead to the loss of photic and circadian control and reduced production of melanopsin mRNA in RGCs [45]. As melanopsin-expressing RGCs are responsible for circadian photoreception and project to SCN and hypothalamus [46], MERTK may thus indirectly participate in the pathogenesis of cluster headache. In addition, enrichment analysis showed significant expression of MERTK in pituitary gland, which could potentially contribute to the altered hormonal expression in cluster headache [47]. SATB2 encodes a DNA binding protein that specifically binds nuclear matrix attachment regions and involves in transcription regulation and chromatin remodeling. Previous GWAS suggested that it may be associated with hypothalamic dopaminergic neurons and structures responsible for nociceptive processing [6, 7]. SATB2 was also known as an important transcription factor of RGCs in primates [48]. In addition, both GWAS and gene-based association analysis suggested CYP2C18 as a potential novel susceptibility gene for cluster headache. Interesting, CYP2C18 is a cytochrome P450 monooxygenase involved in retinoid metabolism [49], while retinoic acid is a molecular trigger of RGC hyperactivity [50]. Taken together, the top implicated genes in our study may modulate the function of RGCs; however, how this could be involved in cluster headache pathogenesis particularly circadian rhythmicity requires further validation.
Gene-based association testing also identified several possible additional loci, among these, ANAPC1 has also been identified in another cluster headache GWAS using gene-based analysis, with enriched expression in the brain, particularly neurons [7]. In addition, among the genes that may be potentially affected by variants in MERTK, i.e., with significant eQTL expression, TMEM87B (transmembrane protein 87B) and FBLN7 (Fibulin 7) have been identified in prior GWASs via gene-based analysis or eQTL analysis [6, 7], suggesting their involvement in cluster headache. Moreover, we found evidence of suggestive association among previously suggested loci to be involved in cluster headache, including LINC01877, LINC01705, ADCYAP1R1, and MME. Although these genes are functionally plausible for the pathogenesis of cluster headache, future studies with larger sample sizes are needed to validate these findings.
We found considerable heritability of cluster headache and migraine and significant genetic correlation between these two primary headache disorders, which corroborates with the clinical observations that both disorders exhibit features of trigeminovascular activation and respond to similar treatments such as triptans or CGRP monoclonal antibodies. Some previously reported migraine loci were also found to have suggestive GWAS significance in our current sample, which may contribute to the share biology between migraine and cluster headache. In addition, pathway analyses suggested that biological processes associated with synaptic transmission or immune responses may be involved in the pathogenesis of cluster headache. In fact, immunological processes have long been considered important in the pathogenesis of migraine and we have recently found HLA class I alleles are associated with clinic-based migraine [51].
Our study has limitations. First, the sample size is relatively small. However, the successful validation of the findings of previous GWASs and replicable signals in both stages of our GWAS after stringent quality control suggest that our findings are unlikely spurious. Second, the identified variants might not be the true causal variants and it remains unknown whether the expression of the implicated genes is truly altered in patients even though eQTL analysis suggested that these variants could affect gene expression in certain tissues. Finally, although the implicated genes were plausibly relevant to the pathogenesis of cluster headache in in silico functional analysis, we were unable to validate the function of these genes in vivo at the current stage owing to the limitation of the availability of biological samples such as brain tissues or retina from the patients. Further in-depth functional analysis at molecular level, at least in a subset of the patients, are needed the increase the credibility of the findings..
Conclusions
In conclusion, our data provide the genetic architecture and mechanistic insights into cluster headache, particularly in Asians, which has been under-represented in previous genetic studies of cluster headache. Future GWAS with patients from multiple ethnicities are required to identify shared and independent genetic determinants of cluster headache across different populations and to better understand the molecular mechanisms of this debilitating disorder.
Supplementary Information
Additional file 1: Supplementary Table 1. Demographics and of headache characteristics in patients with cluster headache.
Additional file 2: Supplemental Figure 1. Quantile-quantile plot of the GWAS results of cluster headache.
Additional file 3: Supplemental Figure 2. Principal component analysis (PCA) plot. The horizontal and vertical axes are the first and second dimensions from principal component analysis based on the cluster headache GWAS samples. Blue dots indicate controls and red dots indicate patients.
Additional file 4: Supplemental Figure 3. Manhattan plot of the discovery cohort (A) and replication cohort (B).
Additional file 5: Supplemental Figure 4. Q-Q plot of the gene-based test computed by MAGMA. The horizontal axis shows -log10 p values expected under the null distribution. The vertical axis shows observed -log10 p values.
Additional file 6: Supplemental Table 2. Meta-analysis of previous studies and the current study for previously reported cluster-associated loci.
Additional file 7: Supplemental Figure 5. eQTL analysis by MetaXcan
Additional file 8: Supplemental Table 3. GIGSEA Biological Pathway Enrichment in brain tissues. The significant level of empirical P-value was 0.05 and the BayesFactor was 100. Top significantly associated pathways in these tissues that may be relevant to cluster headache were listed in the table. UsedGenes indicates number of gene used in the enrichment estimation of the GIGSEA model.
Additional file 9: Supplemental Figure 6. GIGSEA pathway analysis of the genes relevant to cluster headache GWAS variants.
Acknowledgements
We thank all study participants for their generous contribution and the Taiwan Biobank for making the data available for controls. We thank the National Center for Genome Medicine, Taiwan and Taiwan Precision Medicine Initiative (TPMI) for the technical support in the genotyping.
Abbreviations
- ADCYAP1R1
ADCYAP receptor type I
- ANAPC1
Anaphase promoting complex subunit 1
- CAPN2
Calpain 2
- CGRP
Calcitonin gene-related peptide
- DUSP10
Dual Specificity Phosphatase 10
- FHL5
Four and a half LIM domains 5
- FBLN7
Fibulin 7
- GWAS
Genome-wide association study
- HIATL1
Hippocampus Abundant Transcript-Like Protein 1
- ICHD-3
International classification of headache disorders, third edition
- MME
Membrane metalloendopeptidase
- MERTK
MER proto-oncogene, tyrosine kinase
- PRS
Polygenic risk score
- NUTM2F
NUT Family Member 2F
- SATB2
SATB homeobox 2
- SNP
Single nucleotide polymorphism
- TMEM87B
Transmembrane protein 87B
- TP53BP2
Tumor protein P53 binding protein 2
- TRPV1
Transient receptor potential vanilloid subtype 1
- UFL1
UFM1 specific ligase 1
Authors’ contributions
S.P.C, C.S-J.F., & S.J.W. conceived the study. S.P.C., Y.F.W., F.C.Y., L.H.P., J.L.F., H.C.C., & S.J.W. involved in patient recruitment and clinical data collection. S.P.C., C.L.H. T.H.C., J.H.H., Y.L.L, H.C.C, K.H.L. & Y.C.C. performed the main analyses. S.P.C., C.L.H., C.S-J.F., & S.J.W. wrote the manuscript and the other authors provided critical revision of the manuscript for important intellectual content. The authors read and approved the final manuscript.
Funding
This work was supported by the Brain Research Center, National Yang Ming Chiao Tung University from The Featured Areas Research Center Program within the framework of the Higher Education Sprout Project by the Ministry of Education (MOE) in Taiwan (to SJW & SPC); the Ministry of Science and Technology, Taiwan [MOST-108–2314-B-010 -022 -MY3 & 110–2326-B-A49A-501-MY3 (to SPC); MOST 108–2321-B-010–014-MY2, 108–2321-B-010–001-, 108–2314-B-010–023-MY3, 110–2321-B-010–005- & 111–2321-B-A49-004 (to SJW); and MOST 109–2314-B-001–006-MY2 (to CSJF)], Ministry of Health and Welfare, Taiwan [MOHW107-TDU-B-211–123001 and MOHW 108-TDU-B-211–133001] (to SJW), and Taipei Veterans General Hospital, Taiwan [VGH-106-D9-001-MY2-2, VGH-111-C-111 & VGH-111-C-006–1 (to SJW) & V110C-102, VGH-111-C-158, V109D52-001-MY3-2, VGHUST110-G1-3–1 (to SPC)]. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Availability of data and materials
Data described in the manuscript, code book, and analytic code, intended for reasonable use, will be made available upon request by contacting the corresponding authors. Anonymized data from TWB is available for application by contacting biobank@gate.sinica.edu.tw.
Declarations
Ethics approval and consent to participate
This study involves human participants. The study protocol was approved by the Institutional Review Board of Taipei Veterans General Hospital (TVGH-IRB 96–02-11A, 2013–11-001AC, and 2020–08-014A) and Tri-Service General Hospital (TSGH-IRB 2–108-05–038). All participants provided written informed consent before entering the study. All clinical investigations were conducted according to the principles expressed in the Declaration of Helsinki. All collected information was de-identified before statistical data analysis. The corresponding authors had full access to all data in the study and had final responsibility for the decision to submit the research for publication.
Consent for publication
Not applicable.
Competing interests
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Shih-Pin Chen, Email: chensp1977@gmail.com.
Chia-Lin Hsu, Email: chia.lin@ibms.sinica.edu.tw.
Yen-Feng Wang, Email: yfwang851106@gmail.com.
Fu-Chi Yang, Email: fuji-yang@yahoo.com.tw.
Ting-Huei Chen, Email: tingstat22@gmail.com.
Jia-Hsin Huang, Email: jiahsin.huang@gmail.com.
Li-Ling Hope Pan, Email: hope881212@hotmail.com.
Jong-Ling Fuh, Email: jlfuh@vghtpe.gov.tw.
Hsueh-Chen Chang, Email: hcchang1927@gmail.com.
Yi-Lun Lee, Email: soxHenry433@gmail.com.
Hung-Ching Chang, Email: rick.hcchang@gmail.com.
Ko-Han Lee, Email: kohanlee1995@gmail.com.
Yu-Chuan Chang, Email: chester75321@gmail.com.
Cathy Shen-Jang Fann, Email: csjfann@ibms.sinica.edu.tw.
Shuu-Jiun Wang, Email: sjwang@vghtpe.gov.tw.
References
- 1.May A, Schwedt TJ, Magis D, Pozo-Rosich P, Evers S, Wang SJ. Cluster headache Nat Rev Dis Primers. 2018;4:18006. doi: 10.1038/nrdp.2018.6. [DOI] [PubMed] [Google Scholar]
- 2.Headache Classification Committee of the International Headache Society (IHS) (2018) The International Classification of Headache Disorders, 3rd edition. Cephalalgia 38(1):1–211. 10.1177/0333102417738202 [DOI] [PubMed]
- 3.Peng KP, Takizawa T, Lee MJ. Cluster headache in Asian populations: Similarities, disparities, and a narrative review of the mechanisms of the chronic subtype. Cephalalgia : an international journal of headache. 2020;40(10):1104–1112. doi: 10.1177/0333102420923646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin KH, Wang PJ, Fuh JL, Lu SR, Chung CT, Tsou HK, et al. Cluster headache in the Taiwanese – a clinic-based study. Cephalalgia : an international journal of headache. 2004;24(8):631–638. doi: 10.1111/j.1468-2982.2003.00721.x. [DOI] [PubMed] [Google Scholar]
- 5.Bacchelli E, Cainazzo MM, Cameli C, Guerzoni S, Martinelli A, Zoli M, et al. A genome-wide analysis in cluster headache points to neprilysin and PACAP receptor gene variants. J Headache Pain. 2016;17(1):114. doi: 10.1186/s10194-016-0705-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harder AVE, Winsvold BS, Noordam R, Vijfhuizen LS, Børte S, Kogelman LJA, et al. Genetic Susceptibility Loci in Genomewide Association Study of Cluster Headaches. Ann Neurol. 2021 doi: 10.1002/ana.26146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O'Connor E, Fourier C, Ran C, Sivakumar P, Liesecke F, Southgate L, et al. Genome-Wide Association Study Identifies Risk Loci for Cluster Headache. Ann Neurol. 2021 doi: 10.1002/ana.26150. [DOI] [PubMed] [Google Scholar]
- 8.Wei CY, Yang JH, Yeh EC, Tsai MF, Kao HJ, Lo CZ, et al. Genetic profiles of 103,106 individuals in the Taiwan Biobank provide insights into the health and history of Han Chinese. NPJ Genom Med. 2021;6(1):10. doi: 10.1038/s41525-021-00178-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.1000 Genomes Project Consortium, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, et al (2015) A global reference for human genetic variation. Nature. 526(7571):68–74. 10.1038/nature15393 [DOI] [PMC free article] [PubMed]
- 11.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5(6):e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Howie B, Marchini J, Stephens M. Genotype imputation with thousands of genomes. G3 (Bethesda) 2011;1(6):457–70. doi: 10.1534/g3.111.001198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schizophrenia Working Group of the Psychiatric Genomics Consortium Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511(7510):421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng HF, Rong JJ, Liu M, Han F, Zhang XW, Richards JB, et al. Performance of genotype imputation for low frequency and rare variants from the 1000 genomes. PLoS ONE. 2015;10(1):e0116487. doi: 10.1371/journal.pone.0116487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. 2016.
- 16.Boughton AP, Welch RP, Flickinger M, VandeHaar P, Taliun D, Abecasis GR, et al. LocusZoom.js: interactive and embeddable visualization of genetic association study results. Bioinformatics. 2021. doi: 10.1093/bioinformatics/btab186. [DOI] [PMC free article] [PubMed]
- 17.Taylor KE, Ansel KM, Marson A, Criswell LA, Farh KK-H. PICS2: next-generation fine mapping via probabilistic identification of causal SNPs. Bioinformatics. 2021. doi: 10.1093/bioinformatics/btab122. [DOI] [PMC free article] [PubMed]
- 18.de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: Generalized Gene-Set Analysis of GWAS Data. PLoS Comput Biol. 2015;11(4):e1004219. doi: 10.1371/journal.pcbi.1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun. 2017;8(1):1826. doi: 10.1038/s41467-017-01261-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rainero I, Rubino E, Gallone S, Fenoglio P, Negro E, De Martino P, et al. Cluster headache is associated with the alcohol dehydrogenase 4 (ADH4) gene. Headache. 2010;50(1):92–98. doi: 10.1111/j.1526-4610.2009.01569.x. [DOI] [PubMed] [Google Scholar]
- 21.Fourier C, Ran C, Zinnegger M, Johansson AS, Sjöstrand C, Waldenlind E, et al. A genetic CLOCK variant associated with cluster headache causing increased mRNA levels. Cephalalgia : an international journal of headache. 2018;38(3):496–502. doi: 10.1177/0333102417698709. [DOI] [PubMed] [Google Scholar]
- 22.Fourier C, Ran C, Sjöstrand C, Waldenlind E, Steinberg A, Belin AC. The molecular clock gene cryptochrome 1 (CRY1) and its role in cluster headache. Cephalalgia. 2021;41(13):1374–1381. doi: 10.1177/03331024211024165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rainero I, Gallone S, Valfrè W, Ferrero M, Angilella G, Rivoiro C, et al. A polymorphism of the hypocretin receptor 2 gene is associated with cluster headache. Neurology. 2004;63(7):1286–1288. doi: 10.1212/01.wnl.0000142424.65251.db. [DOI] [PubMed] [Google Scholar]
- 24.Schürks M, Kurth T, Geissler I, Tessmann G, Diener HC, Rosskopf D. Cluster headache is associated with the G1246A polymorphism in the hypocretin receptor 2 gene. Neurology. 2006;66(12):1917–1919. doi: 10.1212/01.wnl.0000215852.35329.34. [DOI] [PubMed] [Google Scholar]
- 25.Fourier C, Ran C, Steinberg A, Sjöstrand C, Waldenlind E, Belin AC. Analysis of HCRTR2 Gene Variants and Cluster Headache in Sweden. Headache. 2019;59(3):410–417. doi: 10.1111/head.13462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fan Z, Hou L, Wan D, Ao R, Zhao D, Yu S. Genetic association of HCRTR2, ADH4 and CLOCK genes with cluster headache: a Chinese population-based case-control study. J Headache Pain. 2018;19(1):1. doi: 10.1186/s10194-017-0831-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26(17):2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hautakangas H, Winsvold BS, Ruotsalainen SE, Bjornsdottir G, Harder AVE, Kogelman LJA, et al. Genome-wide analysis of 102,084 migraine cases identifies 123 risk loci and subtype-specific risk alleles. Nat Genet. 2022;54(2):152–160. doi: 10.1038/s41588-021-00990-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen SP, Fuh JL, Chung MY, Lin YC, Liao YC, Wang YF, et al. Genome-wide association study identifies novel susceptibility loci for migraine in Han Chinese resided in Taiwan. Cephalalgia : an international journal of headache. 2018;38(3):466–475. doi: 10.1177/0333102417695105. [DOI] [PubMed] [Google Scholar]
- 30.Bulik-Sullivan BK, Loh PR, Finucane HK, Ripke S, Yang J, Patterson N, et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47(3):291–295. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khera AV, Chaffin M, Aragam KG, Haas ME, Roselli C, Choi SH, et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat Genet. 2018;50(9):1219–1224. doi: 10.1038/s41588-018-0183-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Purcell SM, Wray NR, Stone JL, Visscher PM, O'Donovan MC, Sullivan PF, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460(7256):748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45(6):580–5. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed]
- 34.Zhu S, Qian T, Hoshida Y, Shen Y, Yu J, Hao K. GIGSEA: genotype imputed gene set enrichment analysis using GWAS summary level data. Bioinformatics. 2018;35(1):160–163. doi: 10.1093/bioinformatics/bty529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barbeira AN, Bonazzola R, Gamazon ER, Liang Y, Park Y, Kim-Hellmuth S, et al. Exploiting the GTEx resources to decipher the mechanisms at GWAS loci. Genome Biol. 2021;22(1):49. doi: 10.1186/s13059-020-02252-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harris MA, Clark J, Ireland A, Lomax J, Ashburner M, Foulger R, et al. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32(Database issue):D258–61. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2015;44(D1):D457–D462. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Croft D, O'Kelly G, Wu G, Haw R, Gillespie M, Matthews L, et al. Reactome: a database of reactions, pathways and biological processes. Nucleic Acids Res. 2011;39((Database issue)):D691–7. doi: 10.1093/nar/gkq1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Efron B, Tibshirani R. Empirical bayes methods and false discovery rates for microarrays. Genet Epidemiol. 2002;23(1):70–86. doi: 10.1002/gepi.1124. [DOI] [PubMed] [Google Scholar]
- 40.Shimizu K, Mackenzie SM, Storm DR. SCOP/PHLPP and its functional role in the brain. Mol Biosyst. 2010;6(1):38–43. doi: 10.1039/b911410f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Y, Lopez D, Davey PG, Cameron DJ, Nguyen K, Tran J, et al. Calpain-1 and calpain-2 play opposite roles in retinal ganglion cell degeneration induced by retinal ischemia/reperfusion injury. Neurobiol Dis. 2016;93:121–128. doi: 10.1016/j.nbd.2016.05.007. [DOI] [PubMed] [Google Scholar]
- 42.Li Y, Li Y, Feng Q, Arnold M, Peng T. Calpain activation contributes to hyperglycaemia-induced apoptosis in cardiomyocytes. Cardiovasc Res. 2009;84(1):100–110. doi: 10.1093/cvr/cvp189. [DOI] [PubMed] [Google Scholar]
- 43.Wang S, Bian C, Yang J, Arora V, Gao Y, Wei F, et al. Ablation of TRPV1+ Afferent Terminals by Capsaicin Mediates Long-Lasting Analgesia for Trigeminal Neuropathic Pain. eNeuro. 2020;7(3). doi: 10.1523/eneuro.0118-20.2020. [DOI] [PMC free article] [PubMed]
- 44.Law AL, Parinot C, Chatagnon J, Gravez B, Sahel JA, Bhattacharya SS, et al. Cleavage of Mer tyrosine kinase (MerTK) from the cell surface contributes to the regulation of retinal phagocytosis. J Biol Chem. 2015;290(8):4941–4952. doi: 10.1074/jbc.M114.628297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sakamoto K, Liu C, Tosini G. Classical photoreceptors regulate melanopsin mRNA levels in the rat retina. J Neurosci. 2004;24(43):9693–9697. doi: 10.1523/jneurosci.2556-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hattar S, Kumar M, Park A, Tong P, Tung J, Yau KW, et al. Central projections of melanopsin-expressing retinal ganglion cells in the mouse. J Comp Neurol. 2006;497(3):326–349. doi: 10.1002/cne.20970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Strittmatter M, Hamann GF, Grauer M, Fischer C, Blaes F, Hoffmann KH, et al. Altered activity of the sympathetic nervous system and changes in the balance of hypophyseal, pituitary and adrenal hormones in patients with cluster headache. NeuroReport. 1996;7(7):1229–1234. doi: 10.1097/00001756-199605170-00001. [DOI] [PubMed] [Google Scholar]
- 48.Nasir-Ahmad S, Lee SCS, Martin PR, Grünert U. Identification of retinal ganglion cell types expressing the transcription factor Satb2 in three primate species. J Comp Neurol. 2021;529(10):2727–2749. doi: 10.1002/cne.25120. [DOI] [PubMed] [Google Scholar]
- 49.Marill J, Cresteil T, Lanotte M, Chabot GG. Identification of human cytochrome P450s involved in the formation of all-trans-retinoic acid principal metabolites. Mol Pharmacol. 2000;58(6):1341–1348. doi: 10.1124/mol.58.6.1341. [DOI] [PubMed] [Google Scholar]
- 50.Telias M, Sit KK, Frozenfar D, Smith B, Misra A, Goard MJ, et al. Retinoic acid inhibitors mitigate vision loss in a mouse model of retinal degeneration. Sci Adv. 2022;8(11):eabm4643. doi: 10.1126/sciadv.abm4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang C, Chen SP, Huang YH, Chen HY, Wang YF, Lee MH, et al. HLA class I alleles are associated with clinic-based migraine and increased risks of chronic migraine and medication overuse. Cephalalgia : an international journal of headache. 2020;40(5):493–502. doi: 10.1177/0333102420902228. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Supplementary Table 1. Demographics and of headache characteristics in patients with cluster headache.
Additional file 2: Supplemental Figure 1. Quantile-quantile plot of the GWAS results of cluster headache.
Additional file 3: Supplemental Figure 2. Principal component analysis (PCA) plot. The horizontal and vertical axes are the first and second dimensions from principal component analysis based on the cluster headache GWAS samples. Blue dots indicate controls and red dots indicate patients.
Additional file 4: Supplemental Figure 3. Manhattan plot of the discovery cohort (A) and replication cohort (B).
Additional file 5: Supplemental Figure 4. Q-Q plot of the gene-based test computed by MAGMA. The horizontal axis shows -log10 p values expected under the null distribution. The vertical axis shows observed -log10 p values.
Additional file 6: Supplemental Table 2. Meta-analysis of previous studies and the current study for previously reported cluster-associated loci.
Additional file 7: Supplemental Figure 5. eQTL analysis by MetaXcan
Additional file 8: Supplemental Table 3. GIGSEA Biological Pathway Enrichment in brain tissues. The significant level of empirical P-value was 0.05 and the BayesFactor was 100. Top significantly associated pathways in these tissues that may be relevant to cluster headache were listed in the table. UsedGenes indicates number of gene used in the enrichment estimation of the GIGSEA model.
Additional file 9: Supplemental Figure 6. GIGSEA pathway analysis of the genes relevant to cluster headache GWAS variants.
Data Availability Statement
Data described in the manuscript, code book, and analytic code, intended for reasonable use, will be made available upon request by contacting the corresponding authors. Anonymized data from TWB is available for application by contacting biobank@gate.sinica.edu.tw.