ABSTRACT
Imbalance of gut microbiota homeostasis is related to the occurrence of ulcerative colitis (UC), and probiotics are thought to modulate immune microenvironment and repair barrier function. Here, in order to reveal the interaction between UC and gut microbiota, we screened a new probiotic strain by 16S rRNA sequencing from Dextran Sulfate Sodium (DSS)-induced colitis mice, and explored the mechanism and clinical relevance. Lactobacillus johnsonii (L. johnsonii), as a potential anti-inflammatory bacterium was decreased colonization in colitis mice. Gavage L. johnsonii could alleviate colitis by specifically increasing the proportion of intestinal macrophages and the secretion of Il-10 with macrophages depleted model and in Il10−/− mice. We identified this subset of immune cells activated by L. johnsonii as CD206+ macrophagesIL−10. Mechanistically, L. johnsonii supplementation enhanced the mobilization of CD206+ macrophagesIL−10 through the activation of STAT3 in vivo and in vitro. In addition, we revealed that TLR1/2 was essential for the activation of STAT3 and the recognition of L. johnsonii by macrophages. Clinically, there was positive correlation between the abundance of L. johnsonii and the expression level of MRC1, IL10 and TLR1/2 in UC tissues. L. johnsonii could activate native macrophages into CD206+ macrophages and release IL-10 through TLR1/2-STAT3 pathway to relieve experimental colitis. L. johnsonii may serve as an immunomodulator and anti-inflammatory therapeutic target for UC.
KEYWORDS: Lactobacillus johnsonii, Ulcerative colitis, Macrophages, STAT3, IL-10
Background
Ulcerative colitis (UC) is a subtype of inflammatory bowel disease (IBD), characterized by chronic nonspecific inflammation of the rectum and colon.1,2 Both adaptive and innate immunity are considered to be involved in this chronic inflammation process.3 Gut microbiota played vital roles in the occurrence and development of IBD.4,5 Proteus mirabilis as a key bacterium for Crohn’s disease could trigger inflammatory responses.6 Extracellular vesicles of Fusobacterium nucleatum compromise intestinal barrier in UC through targeting RIPK1-mediated cell death pathway.7 Gut microbiota had been discovered with the deepening of sequencing technology, while the mechanism remains to be elucidated. Lactobacillus and Bifidobacterium are two recognized genera of probiotics.8,9 We have previously reported the anti-inflammatory effect of Bifidobacterium adolescentis in UC.10 Lactobacillus johnsonii (L. johnsonii) is classified in the genus Lactobacillus. Study has shown that L. johnsonii as a potential beneficial bacteria could improve memory impairment through the brain-gut axis11 and regulate metabolic-related diseases.12,13
The regulatory role of the immune system is considered to be a bridge linking the gut microbiota and the host.14,15 A large number of immune cells such as macrophages, dendritic cells (DCs), T cells, B cells, and NK cells are colonized in the intestinal lamina propria. Macrophages, as a member of natural immunity, are widely involved in tissue repair, regeneration, and inflammation.16 In addition to the conventional use of M1-like and M2-like to differentiate macrophages, the updated recommendation is to use the marker or cytokine identifying the macrophages rather than a phenotypic designation.17 The mannose receptors (CD206+) macrophages, as surface markers of anti-inflammatory macrophages, have been reported to distinguish colonic macrophage populations between health and IBD.18
In this study, we uncovered that the abundance of L. johnsonii was lessened in colitis by 16S rRNA sequencing. Supplement with L. johnsonii alleviated DSS-induced colitis. L. johnsonii could stimulate the activation of CD206+ macrophages through the TLR1/2-STAT3 pathway and promote the secretion of the anti-inflammatory cytokine IL-10. The abundance of L. johnsonii in UC patients was positively correlated with the level of IL10 and TLR1/2. Our findings provide insights for the gut microbiota to regulate the host immune system. L. johnsonii, as a probiotic, could be a potential strategy for UC treatment.
Results
L. johnsonii relieved
In order to explore the potential gut microbiota associated with UC, we constructed a DSS-induced chronic colitis model, and performed the fecal 16S rRNA sequencing. Fecal bacterial symbiotic richness and diversity (α-diversity) were significantly decreased, which was evident in the Sobs, Ace, and Chao index in colitis mice (Figure S1A-C). Furthermore, we performed principal component analysis (PCA) on the OTU level, which showed differences in microbial structure (β-diversity) between normal and inflammatory environments (Figure S1D). We then focused on the potential beneficial bacteria by the comparison with normal and inflammatory states, which indicated the abundance of L. johnsonii was depleted in colitis mice (Figure 1a, Figure S1E). The qPCR results confirmed the lower abundance of intestinal colonized L. johnsonii in both chronic (Figure S1F) and acute (Figure S1G) colitis.
Figure 1.
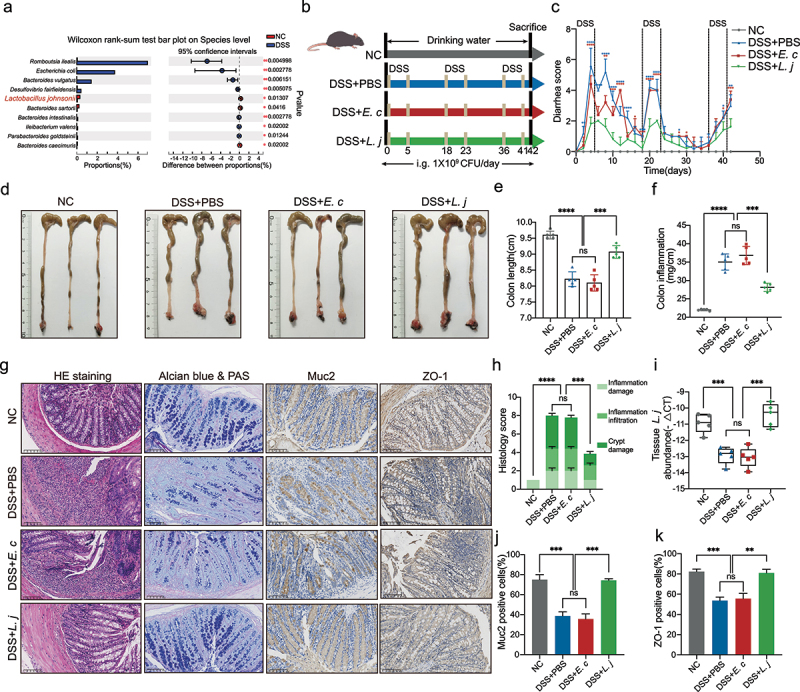
L. johnsonii relieved DSS-induced colitis and maintained intestinal mucosal barrier.
a, Analysis of differences between NC and DSS groups on the species level from 16S rRNA sequencing. b, Schematic diagram showing that the process of DSS-induced chronic colitis. c, Diarrhea score was recorded according to rectal bleeding and diarrhea. d, Representative colon images of the groups with free drinking water (NC), gavaging PBS (DSS+PBS), gavaging E. coli MG1655 (DSS+E. c) and gavaging L. johnsonii (DSS+L. j). e, Colon length was analyzed in four groups. f, Colon inflammation was analyzed in four groups10. g, Representative pictures of HE staining, Alcian blue & PAS staining, the Muc2 and ZO-1 immunohistochemistry. Scale bar, 100 µm. h, Cumulative scores including inflammation damage, inflammation infiltration and crypt damage were measured. i, Bacteria genomic DNA was extracted from feces of mice with DSS-induced chronic colitis. The abundance of L. johnsonii was tested by RT-qPCR. j-k, The relative intensities of IHC (Figure 1g) were quantified by ImageJ software in 5 random fields in each group. Data are presented as mean ± SD, n = 5. **, P < .01; ***, P < .001; ****, P < .0001; ns no significant. Wilcoxon rank-sum test (a), ANOVA test (e-f, h-k).
To determine the role of L. johnsonii in the development of colitis, we constructed DSS-induced chronic colitis and gavaged with PBS (DSS+PBS), E. coli MG1655 (DSS+E. c) or L. johnsonii (DSS+L. j) (Figure 1b). Results demonstrated that L. johnsonii alleviated the severity of diarrhea in colitis mice as compared with PBS control or E. coli group (Figure 1c). In addition, L. johnsonii could reduce the degree of intestinal inflammation and subsequent intestinal shrinkage (Figure 1d-f). The weight of the spleen in the DSS+L. j group was also decreased (Figure S1H-I). Hematoxylin-Eosin (HE) staining showed lower cumulative scores characterized by loss of epithelium, infiltration of inflammatory cells, depletion of goblet cells and damage of crypt in L. johnsonii fed mice (Figure 1g-h). The mRNA expression levels of inflammatory factors including Nos2, Tnfα, Il1β, and Il12a were reduced in DSS+L. j group (Figure S1J).
To explore the effect of L. johnsonii inoculation on the intestinal microbiota homeostasis, we performed fecal 16S rRNA sequencing from DSS+PBS, DSS+E. c and DSS+L. j groups. PCA revealed that daily L. johnsonii supplementation induced significant changes in the composition of the gut microbiota (Figure S1K). The relative abundance of bacteria on the phylum level showed that Bacteroidota was enriched after L. johnsonii gavaged, whereas Firmicutes and Actinobacteriota were decreased (Figure S1L, N). The imbalance of Bacteroidetes/Firmicutes (B/F) ratio, which reflected intestinal homeostasis, was associated with diseases such as obesity and IBD.19 Gavage L. johnsonii increased the B/F ratio (Figure S1M). The relative abundance of bacteria on the family level showed that L. johnsonii increased the levels of Prevotellaceae20 and order Clostridia,21 which were closely related to the secretion of short chain fatty acids (SCFAs), and decreased the levels of Lachnospiraceae, Oscillospiraceae and Erysipelotrichaceae (Figure S1O). We also determined gavage L. johnsonii could colonize in the colon tissues by qPCR analysis (Figure 1i).
Mucin secreted by goblet cells is an important part of colonic mucosal barrier.22 Goblet cells were less damaged in the DSS+L. j group with staining of Alcian blue & PAS and Muc2 (Figure 1g, j).23 We also found that the intestinal mucosa barrier stained by ZO-1 was protected with L. johnsonii (Figure 1g, k). Collectively, the results indicated that L. johnsonii could relieve chronic colitis and maintain the colonic mucosal barrier.
Macrophages immune response was involved in the anti-inflammatory effect by L. johnsonii in colitis
When intestinal barrier was damaged by inflammation, the microbiota entered the lamina propria and interacted with the immune cells colonized.24,25 Therefore, we examined immune cells response in colon tissues by flow cytometry (Figure S2A-B), and the cell compositions by tSNE dimensionality reduction assay (Figure S2C). The cell compositions were separated into different cell clusters (Figure 2a). The number of macrophages was significantly increased in the DSS+L. j group (Figure S2D) by the gating strategy (Figure 2b). Flow cytometry and immunofluorescence analysis confirmed that L. johnsonii supplementation could increase the proportion of colonized macrophages in the intestine (Figure 2e-f). However, no significant changes had been observed in other types of immune cells including DCs, B cells, NK cells, CD4+ T cells, Th1, Th2, Th17, Treg or CD8+ T cells (Figure 2c-d). To further confirm the anti-inflammatory effect of L. johnsonii mediated by macrophages, we constructed a DSS-induced acute colitis model (Figure S3A). Consistent results showed that L. johnsonii supplementation could reduce the degree of intestinal inflammation, cumulative scores by HE staining and the expression of inflammatory factors (Figure S3B-F), while increased the number of macrophages colonized in the intestine (Figure S3G).
Figure 2.
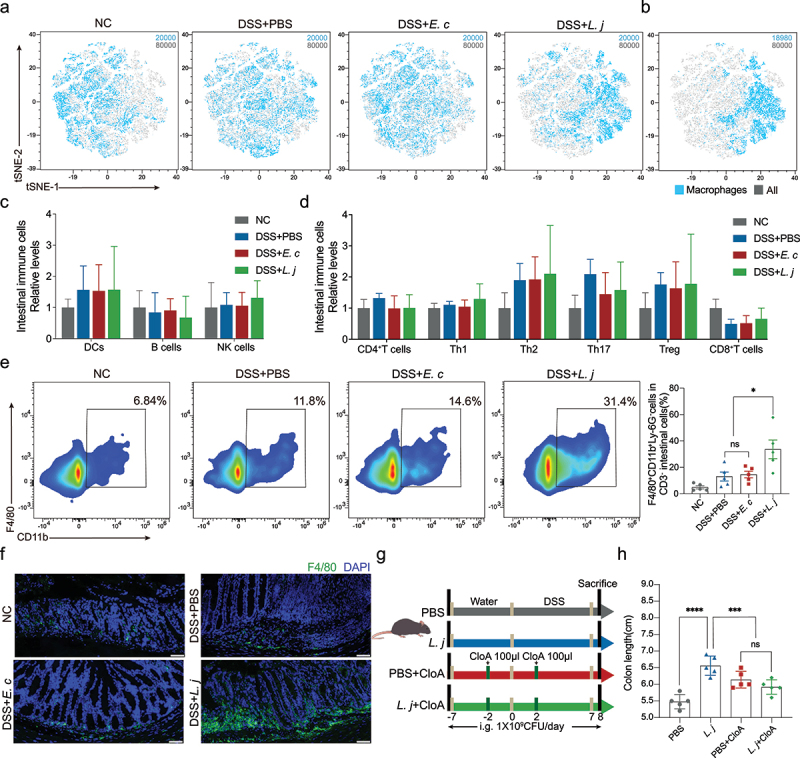
Macrophages immune response was involved in the anti-inflammatory effect by L. johnsonii in colitis.
a, tSNE dimensionality reduction analysis of CD45+ immune cells. 20000 cells were randomly selected from each group, a total of 80000 cells. b, tSNE dimensionality reduction analysis of macrophages cluster (18980 cells) in CD45+ immune cells (80000 cells). c, Frequencies of DCs, B cells, NK cells from colon lamina propria in four groups were tested by multicolor flow cytometry. d, Frequencies of CD4+T cells, Th1, Th2, Th17, Treg and CD8+T cells from colon lamina propria in four groups were tested by multicolor flow cytometry. e, Flow cytometry representation of macrophages from colon lamina propria in four groups. f, Representative images of immunofluorescence staining of macrophages (dual F4/80+) in chronic colitis mice. Scale bars, 50 µm. g, Schematic diagram showing that the process of macrophages clearance model. h, Colon length was analyzed in four groups. Data are presented as mean ± SD, n = 5. *, P < .05; ***, P < .001; ****, P < .0001; ns no significant. ANOVA test (c-e, i).
In order to explore whether macrophages were contributed to the abrogation of intestinal inflammation, we used clodronate liposomes to deplete phagocytic gut macrophages as previously reported26–28 (Figure 2g). The depletion of macrophages (Figure S4B-C) could attenuate the anti-inflammatory and colon length prolongation effect by L. johnsonii (Figure 2h, Figure S4A). Supplement with L. johnsonii could not protect the infiltration of inflammation and the damage of crypts when chemical clearance of macrophages (Figure S4D-E). Collectively, macrophages were involved in the anti-inflammatory process by L. johnsonii in colitis.
L. johnsonii promoted CD206+macrophagesIL-10 activation in vivo and in vitro
Macrophages activation and polarization was involved in inflammation.29 Flow cytometry and immunofluorescence analysis showed that L. johnsonii could upregulate intestinal CD206+ macrophages in chronic colitis mice (Figure 3a-b), but had no obvious effect on CD11c+ macrophages (Figure S5B). Similar result was observed in acute colitis (Figure S5A, C). The activation of CD206+ macrophages in bone marrow-derived macrophages (BMDMs) and mouse macrophages cell line Raw264.7 were also observed after co-cultured with L. johnsonii in vitro (Figure S5D-F). These results indicated that L. johnsonii could activate CD206+ macrophages in vivo and in vitro.
Figure 3.
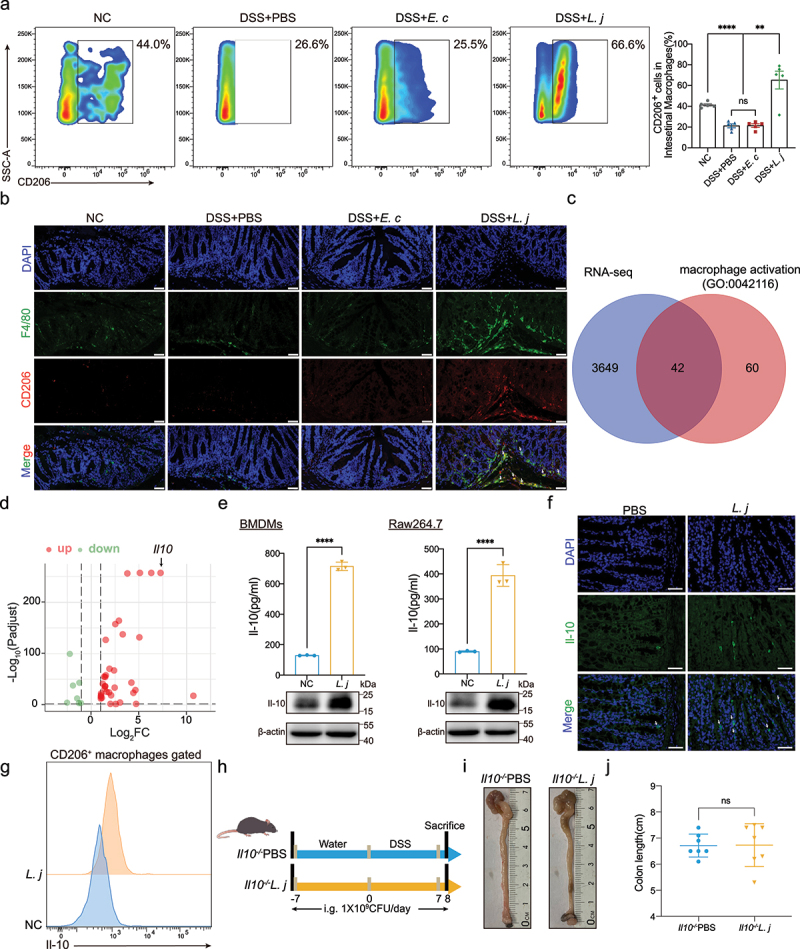
L. johnsonii promoted CD206+ macrophagesIL-10 activation in vivo and in vitro.
a, Flow cytometry representation of CD206+ macrophages from colon lamina propria in chronic colitis model. b, Representative images of immunofluorescence staining of CD206+ macrophages (dual F4/80+ CD206+) in chronic colitis mice. Scale bars, 50 µm. The white arrows indicated the positive stained cells. c, Venn diagram showed the intersecting of the RNA sequencing results and the macrophage activation gene set (GO:0042116). d, Volcano map of the differential gene expression patterns (Figure 3c) (n = 3 per group, fold change>2, logCPM>2, FDR<0.05). e, The protein expression of Il-10 of PBS control (NC) or L. johnsonii (L. j) treated BMDMs and Raw264.7 cells were examined by ELISA and western blot. f, Representative images of immunofluorescence staining of Il-10 in colon tissues of PBS control (PBS) or L. johnsonii (L. j) treated chronic colitis mice. Scale bars, 100 µm. The white arrows indicated the positive stained cells. g, Representative histograms of Il-10 expression by CD206+ macrophages were shown. h, Schematic diagram showing that the process of DSS-induced acute colitis in Il10−/− mice. i, Representative colon images of the groups with gavaging PBS (Il10−/−PBS), gavaging L. johnsonii (Il10−/−L. j). j, Colon length was analyzed in two groups. Data are presented as mean ± SD, n = 5–7. **, P < .01; ****, P < .0001; ns no significant. ANOVA test (a), unpaired Student’s t test (e, j).
To explore the effector molecules underlying L. johnsonii relieving inflammation through CD206+ macrophages, we performed RNA sequencing in BMDMs after incubated with L. johnsonii or PBS for 24 hours. Results showed that 1846 genes in BMDMs were significantly upregulated, while 1845 genes were downregulated after incubated with L. johnsonii (Figure S6A). Then we intersected this RNA sequencing results with the macrophage activation gene set (GO:0042116) (Figure 3c). In line of this, 37 genes were significantly up-regulated and 7 genes were down-regulated in BMDMs after treatment with L. johnsonii (Figure S6B). The volcano map showed that Il10 was the second up-regulated gene (Figure 3d). IL-10 played a regulatory role in IBD.29 L. johnsonii supplementation could increase the secretion of Il-10 in both BMDMs and Raw264.7 cells in vitro by ELISA and western blot assays (Figure 3e). Immunofluorescence detection showed that the content of Il-10 in colitis tissues was increased after L. johnsonii treatment (figure 3f). In particular, L. johnsonii could increase the expression of Il-10 in CD206+ macrophages in BMDMs (Figure 3g).
We then determined whether L. johnsonii exerts its anti-inflammatory role in Il10 knockout mice. L. johnsonii could not alleviate colon shortening and inflammation in Il10−/− mice treated by DSS as expected (Figure 3h-j, Figure S6C-D). We defined this macrophages subtype induced by L. johnsonii as CD206+ macrophages.IL-10 Taken together, L. johnsonii could activate intestinal CD206+ macrophages, thus promoting the secretion of IL-10 to relieve colitis.
STAT3 signaling was essential for L. johnsonii activated CD206+macrophagesIL-10
To explore the underlying mechanism of L. johnsonii promoting the activation of CD206+ macrophages,IL-10 we performed KEGG enrichment analysis in the macrophages-related gene sets from the RNA sequencing (Figure S6A), and found that the JAK-STAT signaling was significantly enriched (Figure 4a). Then we verified the target genes related to the JAK-STAT3 pathway in both BMDMs and Raw264.7 cells. The mRNA levels of Pim1, Socs3, Cish, and Socs1 were significantly increased after L. johnsonii treatment (Figure S7A-B). L. johnsonii could upregulate the phosphorylation level of STAT3 in BMDMs and Raw264.7 cells (Figure 4b-c). After treatment with Stattic, an STAT3 inhibitor,30–32 the expression of CD206 and Il-10 was downregulated (Figure 4d-e). We further constructed a STAT3 inhibition model with Stattic in acute colitis, and found that the inactivation of STAT3 abrogated the anti-inflammatory effect by L. johnsonii (figure 4f-g, Figure S7C-E). There was no significant difference in the number of CD206+ macrophagesIL-10 after Stattic was administered (Figure 4h-i, Figure S7F-H). These results showed that STAT3 pathway was involved in the activation of CD206+ macrophagesIL-10 by L. johnsonii.
Figure 4.
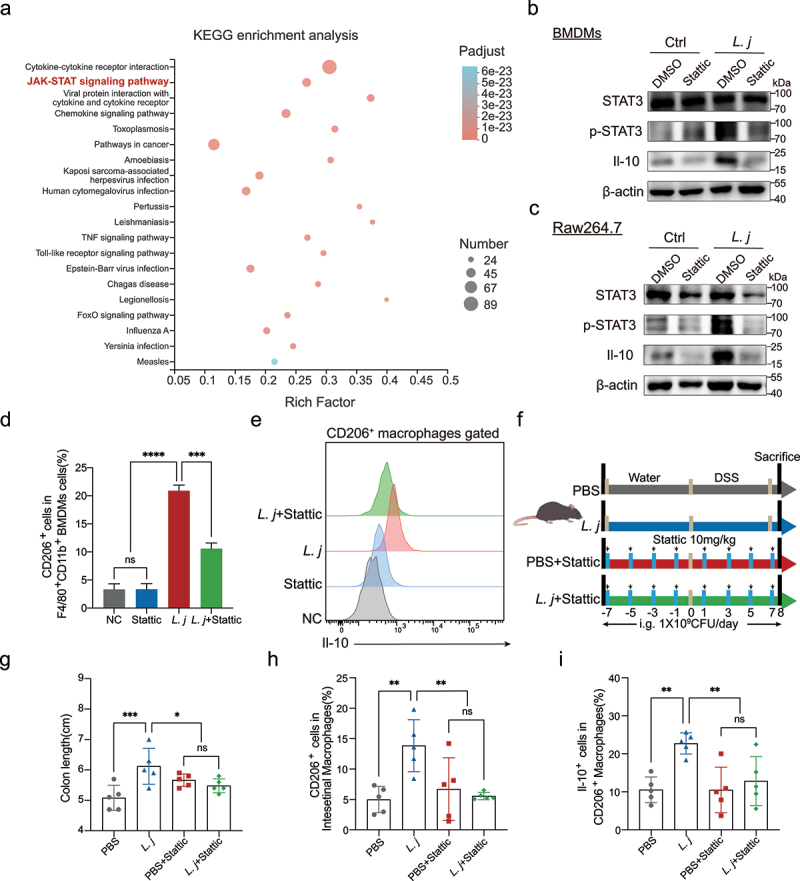
STAT3 signaling was essential for L. johnsonii activated CD206+macrophagesIL-10.
a, KEGG enrichment analysis of macrophages related gene sets. The degree of color represented p adjust value and the size of node represented the gene number in this item. b-c, The protein expression of STAT3, p-STAT3, Il-10 was tested by western blot in both BMDMs and Raw264.7 cells. d, Percentage of CD206+ macrophages were analyzed by flow cytometry after BMDMs co-cultured with PBS control (NC), Stattic, L. johnsonii (L. j) or Stattic and L. johnsonii (L. j+ Stattic) (MOI = 100:1) for 24 hours. e, Representative histograms of Il-10 expression by CD206+ macrophages were shown after BMDMs co-cultured with PBS control (NC), Stattic, L. johnsonii (L. j) or Stattic and L. johnsonii (L. j+ Stattic) (MOI = 100:1) for 24 hours. f, Schematic diagram showing that the process of STAT3 inactivation model in vivo. g, Colon length was analyzed in four groups. h, Percentage of CD206+ macrophages were analyzed from colon lamina propria in STAT3 inactivation model. i, Percentage of Il-10+ cells were analyzed from CD206+ macrophages in STAT3 inactivation model. Data are presented as mean ± SD, n = 3–5. *, P < .05; **, P < .01; ***, P < .001; ****, P < .0001; ns no significant. ANOVA test (d, g-i).
TLR1/2 participated in the recognition of L. johnsonii by CD206+macrophagesIL-10
Toll-like receptor family (TLRs) was closely involved in the interaction between microbiota and immune cells.33,34 In order to explore the pattern recognition receptors (PRRs) which macrophages recognized L. johnsonii, we filtered the relative expression of TLRs genes and found that Tlr1 and Tlr2 were significantly up-regulated in BMDMs and Raw264.7 cells after L. johnsonii stimulation (Figure 5a-b). There were no significant changes of Tlr3, Tlr4, Tlr6, Tlr7, Tlr8 and Tlr9 (Figure 5a-b).
Figure 5.
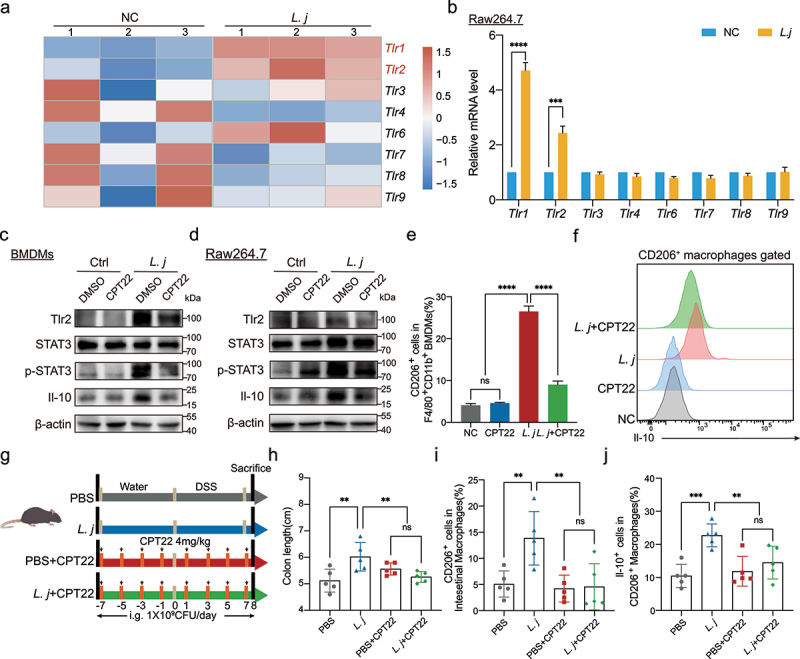
TLR1/2 participated in the recognition of L. johnsonii by CD206+macrophagesIL-10.
a, Heat map representing the TLRs genes expression patterns between L. johnsonii-treated or PBS-treated BMDMs by Microarrays (n = 3 per group, fold change>2, logCPM>2, FDR<0.05). b, The mRNA levels of the TLRs Tlr1, Tlr2, Tlr3, Tlr4, Tlr6, Tlr7, Tlr8 and Tlr9 were evaluated in Raw264.7 cells after L. johnsonii treatment. c-d, The protein expression of Tlr2, STAT3, p-STAT3, Il-10 was tested by western blot in both BMDMs (c) and Raw264.7 cells (d). e, Percentage of CD206+ macrophages were analyzed by flow cytometry after BMDMs co-cultured with PBS control (NC), CPT22, L. johnsonii (L. j) or CPT22 and L. johnsonii (L. j+ CPT22) (MOI = 100:1) for 24 hours. f, Representative histograms of Il-10 expression by CD206+ macrophages were shown after BMDMs co-cultured with PBS control (NC), CPT22, L. johnsonii (L. j) or CPT22 and L. johnsonii (L. j+ CPT22) (MOI = 100:1) for 24 hours. g, Schematic diagram showing that the process of TLR1/2 inhibition model in vivo. h, Colon length was analyzed in four groups. i, Percentage of CD206+ macrophages were analyzed from colon lamina propria in TLR1/2 inhibition model. j, Percentage of Il-10+ cells were analyzed from CD206+ macrophages in TLR1/2 inhibition model. Data are presented as mean ± SD, n = 3–5. **, P < .01; ***, P < .001; ****, P < .0001; ns no significant. unpaired Student’s t test (b), ANOVA test (e, h-j).
It has been reported that TLR1 and TLR2, as a heterodimer, was involved in the identification of microorganisms.35–37 Therefore, we used CPT22, a TLR1/2 complex inhibitor,38,39 to evaluate its effect on the activation of macrophages. Results showed that the inhibition of TLR1/2 could block the activation of STAT3 and production of Il-10 in both BMDMs (Figure 5c) and Raw264.7 macrophages cells (Figure 5d) by L. johnsonii. We further explored the effect of TLR1/2 on CD206+ macrophagesIL-10 activation by flow cytometry. Pretreatment with CPT22 significantly inhibited the expression of CD206 and the secretion of Il-10 (Figure 5e-f). The downstream targets of JAK-STAT3 pathway including Pim1, Socs3, Cish and Socs1 were suppressed by pretreatment with TLR1/2 inhibitor (Figure S8A-B). Similarly, blocking of TLR1/2 attenuated the anti-inflammatory effect by L. johnsonii in DSS-induced acute colitis (Figure 5g-h, Figure S8C-E). There was no significant difference in the number of CD206+ macrophagesIL-10 (Figure 5i-j, Figure S8F-G). These results indicated that TLR1/2 participated in the recognition of L. johnsonii by macrophages, and which was required for L. johnsonii promoting the activation of CD206+ macrophagesIL-10.
The abundance of L. johnsonii was positively correlated with MRC1, IL10 and TLR1/2 in UC patients
To uncover the clinical relevance of L. johnsonii and UC, we assessed the abundance of L. johnsonii in UC tissues (n = 13) and normal tissues (n = 22) (Figure 6a) and found that the abundance of L. johnsonii was downregulated in UC patients (Figure 6b). Consistent observations were observed in colitis mice (Figure 1i, Figure S1F-G). Furthermore, we found that the expression of human Mannose receptor C-type 1(MRC1), the homologous genes to CD206 in mouse, was decreased in UC patients (Figure 6c).
Figure 6.
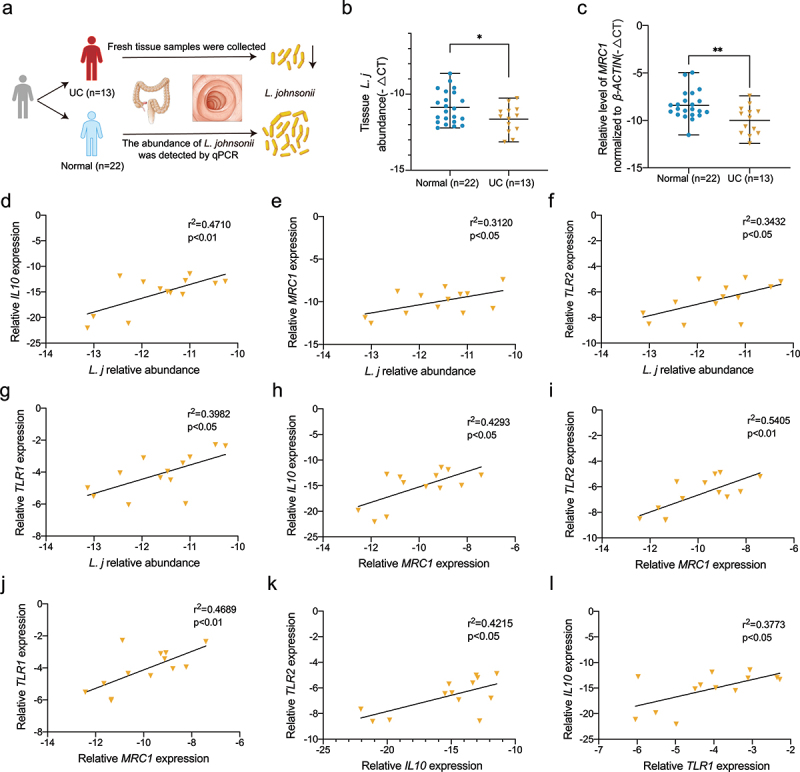
The abundance of L. johnsonii was positively correlated with MRC1, IL10 and TLR1/2 in UC patients.
a, Schematic diagram showing that tissue samples from 13 patients with ulcerative colitis and 22 normal people were collected. b, Bacteria genomic DNA was extracted from colon tissues of UC patients and healthy people. The abundance of L. johnsonii was tested by RT-qPCR. c, The mRNA expression level of MRC1 were evaluated in UC patients and healthy people. d, The correlation of L. johnsonii and IL10 were analyzed. e, The correlation of L. johnsonii and MRC1 were analyzed. f, The correlation of L. johnsonii and TLR2 were analyzed. g, The correlation of L. johnsonii and TLR1 were analyzed. h, The correlation of MRC1 and IL10 were analyzed. i, The correlation of MRC1 and TLR2 were analyzed. j, The correlation of MRC1 and TLR1 were analyzed. k, The correlation of IL10 and TLR2 were analyzed. i, The correlation of TLR1 and IL10 were analyzed. Data are presented as mean ± SD, n = 13–22. *, P < .05; **, P < .01. Mann Whitney test (a-b), Spearman correlation analysis (d-l).
Moreover, the abundance of L. johnsonii was positively correlated with the expression of IL10, MRC1, and TLR1/2 (Figure 6d-g). There were positive relationships among the expression of MRC1, IL10 and TLR1/2 (Figure 6h-l). Summarily, our results indicated that L. johnsonii colonization was reduced in UC patients, and its abundance was positively correlated with MRC1, IL10 and TLR1/2.
Discussion
In this study, we reported that L. johnsonii, as a member of the Lactobacillus, was depleted in inflammatory intestine. L. johnsonii supplementation could alleviate DSS-induced acute and chronic colitis. This view was testified by Charlet et al., who found that mixed gavage of L. johnsonii and B. thetaiotaomicron alleviated DSS-induced acute colitis.40 Recent study reported that L. johnsonii reduced Citrobacter rodentium-induced colitis by regulating inflammation and endoplasmic reticulum stress.41 However, the interaction between L. johnsonii and the host immune system have not been clarified.
We found that the anti-inflammatory effects of L. johnsonii were mediated by macrophages. We then defined the L. johnsonii induced population of anti-inflammatory macrophages as CD206+ macrophagesIL-10 based on the surface marker and secreted cytokine. As a traditional surface marker of M2-like macrophages, CD206 is participated in the anti-inflammatory process.42 We further demonstrated the importance of IL-10 in L. johnsonii induced inflammation suppression by RNA sequencing and Il10 knock out mice. IL-10 could directly affect the mucus production and properties of goblet cells.43 Il10−/− mice were thought to be defective in colonic Muc2 synthesis.44 This is also consistent with our findings that L. johnsonii supplementation could restore barrier function by increasing the secretion of IL-10.
The activation effect of L. johnsonii on CD206+ macrophagesIL-10 was mediated by STAT3 in our observations. The activation of STAT3 is crucial for mucosal repair in innate immunity45 and the activation of macrophages.46 The JAK-STAT3 pathway is often considered to be a pro-inflammatory signaling pathway in IBD,47 and inhibitors of JAK2 have been approved for clinical treatment of UC.48 JAK-STAT3 signaling may play diverse function depending on the microenvironment conditions. We found that the activation of STAT3 was involved in the anti-inflammatory effect by promoting the CD206+ macrophagesIL-10 activation. TLRs were related with the recognition of gut microbiota by immune cells. L. johnsonii was reduced in mice with Toll-like receptor 7 agonist imiquimod treatment.49 L. johnsonii N6.2 stimulated the innate immune response through Toll-like receptor 9 in Caco-2 cells.50 These suggests the connection between L. johnsonii and TLRs. We found that the activation of TLR1/2 was essential for the recognition of L. johnsonii by macrophages. TLR2 has been widely reported as a common pattern recognition receptor, and it often forms heterodimers with TLR1 or TLR6 to complete the recognition process together.35–37 In addition, we found that L. johnsonii colonization on colonic tissues from clinical UC patients was reduced. The abundance of L. johnsonii was positively correlated with the expression of TLR1/2.
In conclusion, we identified that L. johnsonii could promote the activation of CD206+ macrophagesIL-10 through TLR1/2-STAT3 pathway, and relieved experimental colitis (Figure 7a).
Figure 7.
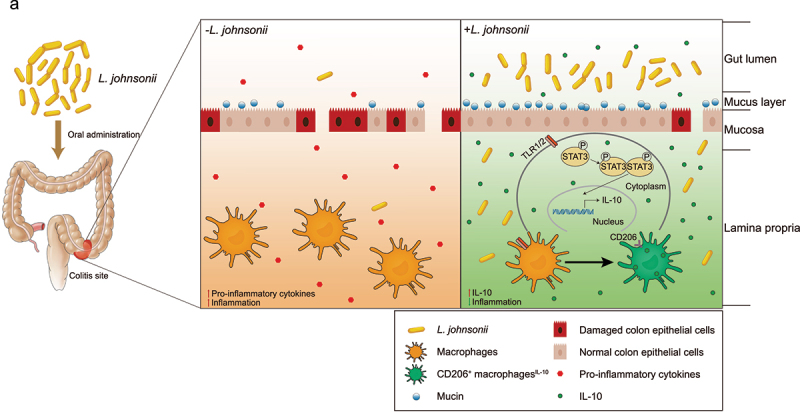
The schematic diagram of L. johnsonii relieving colitis.
a, Schematic diagram showing that the resident bacteria of L. johnsonii activates native macrophages into CD206+ macrophages and release IL-10 through TLR1/2-STAT3 pathway to relieve colitis.
Methods
Human sample collection
Tissue samples from 13 patients with ulcerative colitis and 22 normal people were obtained from Sir Run Run Shaw Hospital, Zhejiang University School of Medicine. Fresh tissue samples were collected during the health examination and immediately frozen in liquid nitrogen, stored in a − 80°C freezer. Ulcerative colitis patients were colonoscopically and pathologically diagnosed. The patients with ulcerative colitis were in active stage, without immunotherapy, and without antibiotics and probiotics in the past one month. Control individuals had no history of diarrhea or the use of antibiotics and probiotics in the past one month.
DNA extraction and bacteria quantification
Bacterial DNA from mice feces was extracted using TIANGEN stool kits (DP328-02, TIANGEN, Beijing), and bacterial genomic DNA from human tissues or mice colon tissues were extracted using TIANGEN DNA kits (DP304-02, TIANGEN, Beijing). RT-qPCR was performed in ROCHE LightCycler®480 System (Rotor gene 6000 Software, Sydney, Australia). Each reaction was performed in triplicate with qPCR SYBR Green Master Mix (11198ES08, YEASON, China), primers and template DNA. Relative abundance was calculated using -ΔCt method. Universal Eubacteria 16S was used as an internal reference gene for stool samples.
L. johnsonii: Forward: 5ʹ- TCGAGCGAGCTTGCCTAGATGA-3ʹ,
Reverse: 5ʹ- TCCGGACAACGCTTGCCACC-3ʹ;
Universal Eubacteria 16S: Forward: 5ʹ- CGGCAACGAGCGCAACCC-3ʹ,
Reverse: 5ʹ- CCATTGTAGCACGTGTGTAGCC-3.51
Bacteria strain and culture
L. johnsonii was independently isolated from pig feces by Zhejiang Academy of Agricultural Sciences and confirmed on the species level by 16S ribosomal RNA sequencing (V4 sequences). Bacterium were cultured in De Man, Rogosa and Sharpe (MRS) Medium (HB0384-5, hopebio, China) under an atmosphere of 10% H2, 10% CO2 and 80% N2 (AW500SG anaerobic workstations; ELECTROTEK, England) for 24 hours at 37°C. The nonpathogenic commensal intestinal bacteria, E. coli strain MG1655 (Biobw, China) was used as a negative control52 and was cultured in Luria-Bertani (LB) Medium (A507002 Sangon Biotech, China) at 37°C. The cultures were resuspended at a final concentration of 1 × 109 CFU/ 200 µl.
Animal models
Male C57BL/6 mice (6–8 weeks of age) were purchased from Shanghai SLAC Laboratory Animal, China. Male Il10−/− mice (6–8 weeks of age) were provided by GemPharmatech Co., Ltd. All mice were housed in Sir Run Run Shaw Hospital, Zhejiang University School of Medicine under specific pathogen-free (SPF) conditions. Barrier environment maintains a 12-hour circadian rhythm, constant temperature and humidity, sufficient water and food. Before bacterial gavage, 2 mg/mL streptomycin was added to drinking water for 3 days to ensure the consistency of routine microbiota, as reported in a previous study.53
For chronic colitis model, Mice were randomized. Each group of mice was given 2% DSS (160110, MP Biomedicals) for 5 days and then drank normal water for 14 days, this process was repeated for three cycles. The mice in the NC group drank normal water freely throughout the modeling process. Mice in DSS+L. j and DSS+E. c groups were orally gavaged with 200 µl of L. johnsonii or E. coli MG1655 containing 1 × 109 CFU per day, while mice in DSS+PBS group were given the same volume of sterile PBS.
For acute colitis model, Mice were randomized. Each group of mice was given 3% DSS (160110, MP Biomedicals) for 7 days and oral gavage lasted 14 days.
For clodronate liposomes-mediated macrophage deletion model,26 based on the acute colitis model, each mouse was injected intraperitoneally with 1 mg of clophosome-A (CloA), an anionic liposomal clodronate (F70101C-A, FormuMax, USA), in a volume of 100 µl 2 days prior to (day −2) and 2 days after (day 2) DSS exposure.28 Flow cytometry was used to detect the removal of macrophages residing in the intestine.
For STAT3 inactivation model in vivo, based on the acute colitis model, each mouse was injected intraperitoneally with 4 mg/kg Stattic (S7024, Selleckchem, Houston, TX, USA) once every two days.54,55
For TLR1/2 inhibition model in vivo, based on the acute colitis model, each mouse was injected intraperitoneally with 3 mg/kg CPT22 (S8677, Selleckchem, Houston, TX, USA) once every two days.56,57
Colorectal and spleen tissues were photographed and measured in length or weigh at the end of the experiment. Diarrhea scores were recorded daily, scoring bloody stools and diarrhea. Bloody stools: 0 = no bleeding, 2 = light bleeding, 4 = heavy bleeding; Diarrhea: 0 = well-shaped stools, 2 = soft and mushy stools, 4 = watery stools. The assessment of colonic inflammation was performed by normalization of colon weight (mg) by its length (cm).10
Histopathological analysis
Colon tissues were formalin fixed overnight at room temperature and then embedded in paraffin. 5 µm sections were stained with hematoxylin and eosin (HE) for pathological analysis. The pathological score was assessed by the severity of inflammation, the degree of inflammatory involvement, and the degree of epithelial/crypt damage as previously reported.58 Calculate each parameter and sum to get the total score.
For immunohistochemistry, paraffin-embedded tissue sections were stained with anti-MUC2 antibody (GB14110, 1:1000, Servicebio, China), anti-ZO-1 antibody (GB111402, 1:1000, Servicebio, China) and visualized by DAB staining according to manufacturer’s instructions.
For immunofluorescence, paraffin-embedded tissues were stained with anti-F4/80 antibody (GB11027, 1:1000, Servicebio, China), anti-Il-10 antibody (GB11108, 1:1000, Servicebio, China) and anti-CD206 antibody (GB113497, 1:500, Servicebio, China). The slides were independently assessed by two researchers who were unaware of the nature of the samples.
Isolation of colon lamina propria cells
As previously reported,59 residual lymphoid and adipose tissues were removed from colorectal tissue. Colon tissue was then cut into small pieces and placed in D-Hank’s buffer (MA0039, Meilunbio, China) containing 1 mM DTT (MB3047-1, Meilunbio, China) and 5 mM EDTA (MB2514, Meilunbio, China) at 37°C shaker 150 g for 30 minutes. Mucus-depleted colon tissues were cut into 1 mm pieces and further digested in Hank’s buffer (MA0041, Meilunbio, China) supplemented with 1 mg/ml type IV collagenase (A005318, Sangon, China) for 30 minutes at 37°C 150 g shaker. After complete digestion, the cell suspension was passed through a 200-mesh filter and then centrifuged at 700 g for 5 minutes. Isolated colonic lamina propria cells were collected for further flow cytometry analysis.
Flow cytometry analysis
After the colonic lamina propria cells were counted, Fc receptors were blocked, 100 μl of the cell suspension was added with 1 μg of purified CD16/32 antibody (101320, Biolegend) and incubated on ice for 10 minutes. Then, cells were stained for live cells using Fixable viability Stain 510 (564406, BD Biosciences) for 30 minutes.
After termination, epifluorescence staining was performed using antibodies as described below: Alexa Fluor 700-CD45 (560510, BD Biosciences); Brilliant Violet 605-CD11b (101257, Biolegend); Brilliant Violet 421-F4/80 (565411, BD Biosciences); PE-I-A/I-E (557000, BD Biosciences); APC-CD206 (141708, Biolegend); PE-CY7-CD11c (117318, Biolegend); BUV395-Ly-6G (563978, BD Biosciences); Brilliant Violet 711-NK1.1 (108745, Biolegend); Brilliant Violet 785-CD45R/B220 (103246, Biolegend); PECP-CY5.5-CD3 (551163, BD Biosciences); Brilliant Violet 605-CD4 (100548, Biolegend); APC-CY7-CD8 (561967, BD Biosciences). Incubated in a refrigerator at 4°C for 30 minutes in the dark.
Then, cells were fixed in the dark at room temperature for 30 minutes (422101, Biolegend), centrifuged, and then the membranes were ruptured (421002, Biolegend). Intracellular staining was performed using antibodies as described below: PE-CY7-Foxp3 (560408, BD Biosciences); Brilliant Violet 421-Gata3 (563349, BD Biosciences); Alexa Fluor 647-T-bet (561267, BD Biosciences); PE-RORγt (IC6006P-025, R&D). The cells were incubated in the dark at room temperature for 60 minutes.
For the detection of Il-10 cytokines, after resuspending the cells with culture medium, added 2 μl PMA/Ionomycin, Activation Cocktails (423303, Biolegend) to 1 ml of cell suspension, incubated for 6 hours in a 37°C cell incubator, and then performed the above flow staining.
Samples were analyzed using Flow Cytometer (BD Biosciences). Subsequent analysis was performed with FlowJo software (Tree Star Inc., San Carlos, CA). tSNEs were calculated in FlowJo using the default settings for opt-SNE. For each set of analyses, individual cell subsets (CD45+ immune cells) from each group (NC, DSS+PBS, DSS+E. c, DSS+L. j) were randomly down-sampled to an equal number of events (20000 events).
Bone marrow derived macrophages (BMDMs) and cell culture
After 8-week-old mice were sacrificed by cervical dislocation, they were disinfected by soaking in 75% alcohol for 15 minutes. The marrow cavities of the tibia and femur were opened and rinsed repeatedly with RPMI-1640 medium (Genom, China) supplemented with 10% FBS (Gibco) and 1% penicillin/streptomycin. The collected cells were cultured in RPMI-1640 medium containing mouse macrophage colony-stimulating factor (M-CSF, 20 ng/ml, Novoprotein, China), and the medium was renewed every 2 days. One week later, BMDMs were confirmed by flow cytometry for the expression of F4/80 (565411, BD Biosciences). The Raw264.7 mouse macrophage cell line was purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). Raw264.7 cells were cultured in RPMI-1640 medium (Genom, China) supplemented with 10% FBS (Gibco) and 1% penicillin/streptomycin. All cell lines were maintained at 37°C in a humidified 5% CO2 atmosphere.
BMDMs or Raw264.7 cells were seeded overnight in 6 well plates at 1 × 106 cells per well and then cultured in antibiotic-free medium. BMDMs and Raw264.7 cells were incubated with L. johnsonii or E. coli at an MOI of 100:1 for 24 hours. For STAT3 inhibition experiments, Stattic (10 μM) was administered per six-well plate and incubated for 24 hours. For TLR1/2 inhibition experiments, CPT22 (10 μM) was administered per 6 well plate and incubated for 24 hours. Finally, BMDMs and Raw264.7 cells were digested for further flow cytometry analysis.
RNA sequencing
After 24 hours of incubation with L. johnsonii (MOI = 100:1) or PBS, RNA from BMDMs were extracted and reverse transcribed into cDNA. RNA sequencing was paired-end at Shanghai MajorBio (China). Expression levels across the sample were expressed in RPKM (reads per kilobase per million). All differentially expressed genes were plotted with the “pheatmap” and “ggplot2” R packages. For KEGG enrichment analysis, p-value <0.05 was used as a threshold to determine significant enrichment for gene sets with the “clusterProfiler” R package.
16S rRNA sequencing
The fresh feces were collected for 16S rRNA sequencing on the 42nd day (before the mice were sacrificed). Bacterial genomic DNA was extracted from mouse feces as described above. Measured DNA concentration and checked DNA quality by 1% agarose gel electrophoresis. The V3-V4 hypervariable regions of the 16S ribosomal RNA gene was amplified by PCR system (GeneAmp 9700, ABI, USA) using primers 338 F and 806 R. PCR amplicons were purified by the AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, USA) and quantified by QuantiFluor™-ST (Promega, USA). Purified amplicons were sequenced on the Illumina MiSeq platform (Illumina, San Diego, USA) according to the standard protocols of Shanghai MajorBio (China). The raw data were decomposed and quality filtered by Trimmomatic and merged by FLASH. Operational taxonomic units (OTUs) were clustered using UPARSE (http://drive5.com/uparse/). We used the Ace, Chao, and Sobs diversity index to measure the species richness (α-diversity). The β-diversity was calculated using UniFrac distances and visualized using principal component analysis (PCA).
RNA extraction and quantitative real-time PCR
RNA from mouse colon tissues or cells was extracted using Trizol reagent (Takara, Japan). PrimeScript™ RT reagent Kit (Takara, Japan) was used for reverse transcription. RT-qPCR was performed in Light Cycler®480 Real-Time PCR System (Roche) using SYBR Premix Ex Taq (Takara, Japan). cDNA was amplified by PCR under the following conditions: 95°C for 2 min, followed by 50 cycles of 95°C for 15s and 60°C for 30s. The mRNA expression of genes was analyzed using the specific primers listed in (Table S1). The relative mRNA expression was calculated using -ΔCt method or the comparative cycle method (2−ΔΔCt). β-actin was used as internal control.
Western blot analysis
Colon tissues or cells were lysed using RIPA extraction buffer (R0010, Solarbio, China). Protein concentrations were quantified using BCA protein assay kit (PC0020, Solarbio, China). Proteins were separated on 10% SDS polyacrylamide gels and then transferred to PVDF membranes. Membranes were blocked with quickblock solution (P0231, Beyotime, China) for 15 minutes, and then blocked with antibodies against STAT3 (4904, CST), p-STAT3 (9145, CST), Il-10 (A12255, ABclonal) and Tlr2 (13744, CST) at 4°C overnight. The membranes were then incubated with secondary antibody-conjugated HRP (1:10000, Abcam) for 1 hour at room temperature, and bands were visualized using an ECL kit (FD8000, Fdbio science, China). β-actin served as a loading control.
Statistical analysis
All statistical analyses were performed using GraphPad Prism 9.0 software (GraphPad Software, Inc., La Jolla, CA, USA). The experiments were set up to use 3–7 samples/repeats for each experiment/group. Representative images for immunofluorescence staining, immunohistochemistry and immunoblot were shown. Each of these experiments was independently repeated over three times. Data was expressed as mean ± standard deviation (SD) or standard error of mean (SEM). Differences between groups were analyzed by unpaired Student’s t-test, one-way ANOVA test, Kruskal-Wallis test, Mann Whitney test or Wilcoxon matched-pairs signed-rank test. Spearman correlation analysis was used for correlation analysis. P value < .05 was considered statistically significant.
Supplementary Material
Acknowledgments
We thank Prof. Wei Zhuo (Department of Cell Biology and Department of Gastroenterology, Zhejiang University School of Medicine) for useful suggestions. We also appreciate Dr. Pin Wu (Cancer Institute, Second Affiliated Hospital, Zhejiang University School of Medicine) for helpful discussion and revisiting the immune phenotypes.
Funding Statement
This project was financially supported by the National Foundation of Natural Science of China (82072623 to LJ Wang), Zhejiang Province Public Welfare Technology Research Project (LGD21H160002, to SJ Chen), Zhejiang Provincial Medical and Health Science and Technology Project (2019RC043 to YF Fang, 2021RC070 to L Wang) and Natural Science Foundation of Zhejiang (LQ19H030006, to HQ He).
Authors’ contributions
LJ Wang, SJ Chen and W Liu designed and supervised the study. DJC Jia, QW Wang, YY Hu and JM He performed most experiments with the assistance of QW Ge, LY Chen, Y Zhang, LN Fan, YF Lin, Y Sun and Y Jiang. YD Qi performed most bioinformatic analysis. YY Hu, L Wang, YF Fang, HQ He and XE Pi collected human samples. DJC Jia, QW Wang, YY Hu, JM He, W Liu, SJ Chen and LJ Wang analyzed and interpreted the data. DJC Jia drafted the manuscript, and LJ Wang, SJ Chen, W Liu critically revised the manuscript. All authors read and approved the final manuscript.
Authors’ information
These authors contributed equally: DJC Jia, QW Wang, YY Hu and JM He.
Corresponding Authors: Correspondence to LJ Wang, SJ Chen, W Liu.
Ethics approval and consent to participate
Written informed consent was provided before tissue samples were collected from all participants, and the study protocol was approved by the Clinical Research Ethics Committee of Sir Run Run Shaw Hospital, Zhejiang University School of Medicine (20211103-35). All animal studies were performed in accordance with the guidelines of the Animal Experimentation Ethics Committee of Zhejiang University.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary Material:
Supplemental data for this article can be accessed online at https://doi.org/10.1080/19490976.2022.2145843
Availability of data and materials
The RNA-sequencing dataset has been deposited in the Sequence Read Archive (SRA) accession number: PRJNA780203. Source data and reagents are available from the corresponding author on reasonable request.
Competing interests
We have no conflicts of interest to declare.
References
- 1.Danese S, Fiocchi C.. Ulcerative colitis. N Engl J Med. 2011;365(18):1713–18. doi: 10.1056/NEJMra1102942. [DOI] [PubMed] [Google Scholar]
- 2.Ordás I, Eckmann L, Talamini M, Baumgart DC, Sandborn WJ. Ulcerative colitis. Lancet. 2012;380(9853):1606–1619. doi: 10.1016/s0140-6736(12)60150-0. [DOI] [PubMed] [Google Scholar]
- 3.de Souza Hs, Fiocchi C, de Souza HSP. Immunopathogenesis of IBD: current state of the art. Nat Rev Gastroenterol Hepatol. 2016;13(1):13–27. doi: 10.1038/nrgastro.2015.186. [DOI] [PubMed] [Google Scholar]
- 4.Wlodarska M, Kostic AD, Xavier RJ. An integrative view of microbiome-host interactions in inflammatory bowel diseases. Cell Host Microbe. 2015;17(5):577–591. doi: 10.1016/j.chom.2015.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Imhann F, Vich Vila A, Bonder MJ, Fu J, Gevers D, Visschedijk MC, Spekhorst LM, Alberts R, Franke L, van Dullemen HM, et al. Interplay of host genetics and gut microbiota underlying the onset and clinical presentation of inflammatory bowel disease. Gut. 2018;67(1):108–119. doi: 10.1136/gutjnl-2016-312135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang J, Hoedt EC, Liu Q, Berendsen E, Teh JJ, Hamilton A, O’ Brien AW, Ching JYL, Wei H, Yang K, et al. Elucidation of Proteus mirabilis as a Key Bacterium in Crohn’s Disease Inflammation. Gastroenterology. 2021;160(1):317–330.e11. doi: 10.1053/j.gastro.2020.09.036. [DOI] [PubMed] [Google Scholar]
- 7.Liu L, Liang L, Yang C, Zhou Y, Chen Y. Extracellular vesicles of Fusobacterium nucleatum compromise intestinal barrier through targeting RIPK1-mediated cell death pathway. Gut Microbes. 2021;13(1):1–20. doi: 10.1080/19490976.2021.1902718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nowak A, Paliwoda A, Błasiak J, Salas A, Hernandez-Rocha C, Duijvestein M, Faubion W, McGovern D, Vermeire S, Vetrano S. Anti-proliferative, pro-apoptotic and anti-oxidative activity of Lactobacillus and Bifidobacterium strains: a review of mechanisms and therapeutic perspectives. Crit Rev Food Sci Nutr. 2019;59(21):3456–3467. doi: 10.1080/10408398.2018.1494539. [DOI] [PubMed] [Google Scholar]
- 9.Shanahan F, Dinan TG, Ross P, et al. Probiotics in transition. Clin Gastroenterol Hepatol. 2012;10(11):1419–4. doi: 10.1016/j.cgh.2012.09.020. [DOI] [PubMed] [Google Scholar]
- 10.Fan L, Qi Y, Qu S, Chen X, Li A, Hendi M, Xu C, Wang L, Hou T, Si J, et al. B. adolescentis ameliorates chronic colitis by regulating Treg/Th2 response and gut microbiota remodeling. Gut Microbes. 2021;13(1):1–17. doi: 10.1080/19490976.2020.1826746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang H, Sun Y, Xin J, Zhang T, Sun N, Ni X, Zeng D, Bai Y. Lactobacillus johnsonii BS15 Prevents Psychological Stress–Induced Memory Dysfunction in Mice by Modulating the Gut–Brain Axis. Front Microbiol. 2020;11:1941. doi: 10.3389/fmicb.2020.01941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang G, Hong E, Oh S, Kim E. Non-Viable Lactobacillus johnsonii JNU3402 Protects against Diet-Induced Obesity. Foods. 2020;9(10):1494. doi: 10.3390/foods9101494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yoon H, Lee Y, Park H, Kang H-J, Ji Y, Holzapfel WH. Lactobacillus johnsonii BFE6154 Ameliorates Diet-Induced Hypercholesterolemia. Probiotics Antimicrob Proteins. 2021. doi: 10.1007/s12602-021-09859-4. [DOI] [PubMed] [Google Scholar]
- 14.Xavier RJ, Podolsky DK. Unravelling the pathogenesis of inflammatory bowel disease. Nature. 2007;448(7152):427–434. doi: 10.1038/nature06005. [DOI] [PubMed] [Google Scholar]
- 15.Amoroso C, Perillo F, Strati F, Fantini M, Caprioli F, Facciotti F. The Role of Gut Microbiota Biomodulators on Mucosal Immunity and Intestinal Inflammation. Cells. 2020;9(5):1234. doi: 10.3390/cells9051234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith PD, Ochsenbauer-Jambor C, Smythies LE. Intestinal macrophages: unique effector cells of the innate immune system. Immunol Rev. 2005;206(1):149–159. doi: 10.1111/j.0105-2896.2005.00288.x. [DOI] [PubMed] [Google Scholar]
- 17.Murray PJ, Allen JE, Biswas SK, Fisher E, Gilroy D, Goerdt S, Gordon S, Hamilton J, Ivashkiv L, Lawrence T, et al. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity. 2014;41(1):14–20. doi: 10.1016/j.immuni.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wright PB, McDonald E, Bravo-Blas A, Baer HM, Heawood A, Bain CC, Mowat AM, Clay SL, Robertson EV, Morton F, et al. The mannose receptor (CD206) identifies a population of colonic macrophages in health and inflammatory bowel disease. Sci Rep. 2021;11(1):19616. doi: 10.1038/s41598-021-98611-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stojanov S, Berlec A, Štrukelj B. The Influence of Probiotics on the Firmicutes/Bacteroidetes Ratio in the Treatment of Obesity and Inflammatory Bowel disease. Microorganisms. 2020:8. doi: 10.3390/microorganisms8111715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peng J, Li X, Zheng L, Duan L, Gao Z, Hu D, Li J, Li X, Shen X, Xiao H, et al. Ban-Lan-Gen Granule Alleviates Dextran Sulfate Sodium-Induced Chronic Relapsing Colitis in Mice via Regulating Gut Microbiota and Restoring Gut SCFA Derived-GLP-1 Production. J Inflamm Res. 2022;15:1457–1470. doi: 10.2147/jir.S352863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee J, d’Aigle J, Atadja L, Quaicoe V, Honarpisheh P, Ganesh BP, Hassan A, Graf J, Petrosino J, Putluri N, et al. Gut Microbiota–Derived Short-Chain Fatty Acids Promote Poststroke Recovery in Aged Mice. Circ Res. 2020;127(4):453–465. doi: 10.1161/circresaha.119.316448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paone P, Cani PD. Mucus barrier, mucins and gut microbiota: the expected slimy partners? Gut. 2020;69(12):2232–2243. doi: 10.1136/gutjnl-2020-322260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rupani B, Caputo FJ, Watkins AC, Vega D, Magnotti LJ, Lu Q, Xu DZ, Deitch EA. Relationship between disruption of the unstirred mucus layer and intestinal restitution in loss of gut barrier function after trauma hemorrhagic shock. Surgery. 2007;141(4):481–489. doi: 10.1016/j.surg.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 24.Glassner KL, Abraham BP, Quigley EMM. The microbiome and inflammatory bowel disease. J Allergy Clin Immunol. 2020;145(1):16–27. doi: 10.1016/j.jaci.2019.11.003. [DOI] [PubMed] [Google Scholar]
- 25.Ni J, Wu GD, Albenberg L, Tomov VT. Gut microbiota and IBD: causation or correlation? Nat Rev Gastroenterol Hepatol. 2017;14(10):573–584. doi: 10.1038/nrgastro.2017.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Rooijen N, Hendrikx E. Liposomes for specific depletion of macrophages from organs and tissues. Methods Mol Biol. 2010;605:189–203. doi: 10.1007/978-1-60327-360-2_13. [DOI] [PubMed] [Google Scholar]
- 27.Riehl TE, Alvarado D, Ee X, Zuckerman A, Foster L, Kapoor V, Thotala D, Ciorba MA, Stenson WF. Lactobacillus rhamnosus GG protects the intestinal epithelium from radiation injury through release of lipoteichoic acid, macrophage activation and the migration of mesenchymal stem cells. Gut. 2019;68(6):1003–1013. doi: 10.1136/gutjnl-2018-316226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Z, Hao C, Zhuang Q, Zhan B, Sun X, Huang J, Cheng Y, Zhu X. Excretory/Secretory Products From Trichinella spiralis Adult Worms Attenuated DSS-Induced Colitis in Mice by Driving PD-1-Mediated M2 Macrophage Polarization. Front Immunol. 2020;11:563784. doi: 10.3389/fimmu.2020.563784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murray PJ. Macrophage Polarization. Annu Rev Physiol. 2017;79(1):541–566. doi: 10.1146/annurev-physiol-022516-034339. [DOI] [PubMed] [Google Scholar]
- 30.Schust J, Sperl B, Hollis A, Mayer TU, Berg T. Stattic: a small-molecule inhibitor of STAT3 activation and dimerization. Chem Biol. 2006;13(11):1235–1242. doi: 10.1016/j.chembiol.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 31.Bouhaddou M, Memon D, Meyer B, White KM, Rezelj VV, Correa Marrero M, Polacco BJ, Melnyk JE, Ulferts S, Kaake RM, et al. The Global Phosphorylation Landscape of SARS-CoV-2 Infection. Cell. 2020;182(3):685–712.e19. doi: 10.1016/j.cell.2020.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu T-J, Liu -Y-Y, Li X-G, Lian B, Lu -X-X, Jin X, Shao Z-M, Hu X, Di G-H, Jiang Y-Z, et al. PDSS1-Mediated Activation of CAMK2A-STAT3 Signaling Promotes Metastasis in Triple-Negative Breast Cancer. Cancer Res. 2021;81(21):5491–5505. doi: 10.1158/0008-5472.Can-21-0747. [DOI] [PubMed] [Google Scholar]
- 33.Fitzgerald KA, Kagan JC. Toll-like Receptors and the Control of Immunity. Cell. 2020;180:1044–1066. doi: 10.1016/j.cell.2020.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Akira S, Takeda K. Toll-like receptor signalling. Nat Rev Immunol. 2004;4(7):499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- 35.Jin MS, Lee J-O. Structures of the toll-like receptor family and its ligand complexes. Immunity. 2008;29(2):182–191. doi: 10.1016/j.immuni.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 36.Takeuchi O, Sato S, Horiuchi T, Hoshino K, Takeda K, Dong Z, Modlin RL, Akira S. Cutting edge: role of Toll-like receptor 1 in mediating immune response to microbial lipoproteins. J Immunol. 2002;169(1):10–14. doi: 10.4049/jimmunol.169.1.10. [DOI] [PubMed] [Google Scholar]
- 37.Jin MS, Kim SE, Heo JY, Lee ME, Kim HM, Paik S-G, Lee H, Lee J-O. Crystal structure of the TLR1-TLR2 heterodimer induced by binding of a tri-acylated lipopeptide. Cell. 2007;130(6):1071–1082. doi: 10.1016/j.cell.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 38.Cheng K, Wang X, Zhang S, Yin H. Discovery of small-molecule inhibitors of the TLR1/TLR2 complex. Angew Chem Int Ed Engl. 2012;51(49):12246–12249. doi: 10.1002/anie.201204910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lan F, Zhang N, Holtappels G, De Ruyck N, Krysko O, Van Crombruggen K, Braun H, Johnston SL, Papadopoulos NG, Zhang L, et al. Staphylococcus aureus Induces a Mucosal Type 2 Immune Response via Epithelial Cell–derived Cytokines. Am J Respir Crit Care Med. 2018;198(4):452–463. doi: 10.1164/rccm.201710-2112OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Charlet R, Bortolus C, Sendid B, et al. Bacteroides thetaiotaomicron and Lactobacillus johnsonii modulate intestinal inflammation and eliminate fungi via enzymatic hydrolysis of the fungal cell wall. Sci Rep. 2020;10(1):11510. doi: 10.1038/s41598-020-68214-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Y, Mu T, Yang Y, Zhang J, Ren F, Wu Z. Lactobacillus johnsonii Attenuates Citrobacter rodentium –Induced Colitis by Regulating Inflammatory Responses and Endoplasmic Reticulum Stress in Mice. J Nutr. 2021;151(11):3391–3399. doi: 10.1093/jn/nxab250. [DOI] [PubMed] [Google Scholar]
- 42.Wang L-X, Zhang S-X, Wu H-J, et al. M2b macrophage polarization and its roles in diseases. J Leukoc Biol. 2019;106(2):345–358. doi: 10.1002/jlb.3ru1018-378rr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pelaseyed T, Bergström JH, Gustafsson JK, Ermund A, Birchenough GMH, Schütte A, van der Post S, Svensson F, Rodríguez-Piñeiro AM, Nyström EEL, et al. The mucus and mucins of the goblet cells and enterocytes provide the first defense line of the gastrointestinal tract and interact with the immune system. Immunol Rev. 2014;260(1):8–20. doi: 10.1111/imr.12182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schwerbrock NM, Makkink MK, van der Sluis M, Büller HA, Einerhand AWC, Sartor RB, Dekker J. Interleukin 10-deficient mice exhibit defective colonic Muc2 synthesis before and after induction of colitis by commensal bacteria. Inflamm Bowel Dis. 2004;10(6):811–823. doi: 10.1097/00054725-200411000-00016. [DOI] [PubMed] [Google Scholar]
- 45.Sugimoto K. Role of STAT3 in inflammatory bowel disease. World J Gastroenterol. 2008;14(33):5110–5114. doi: 10.3748/wjg.14.5110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yuan F, Fu X, Shi H, Chen G, Dong P, Zhang W. Induction of murine macrophage M2 polarization by cigarette smoke extract via the JAK2/STAT3 pathway. PLoS One. 2014;9(9):e107063. doi: 10.1371/journal.pone.0107063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Salas A, Hernandez-Rocha C, Duijvestein M, Faubion W, McGovern D, Vermeire S, Vetrano S, Vande Casteele N. JAK–STAT pathway targeting for the treatment of inflammatory bowel disease. Nat Rev Gastroenterol Hepatol. 2020;17(6):323–337. doi: 10.1038/s41575-020-0273-0. [DOI] [PubMed] [Google Scholar]
- 48.Baumgart DC, Le Berre C, Ingelfinger JR. Newer Biologic and Small-Molecule Therapies for Inflammatory Bowel Disease. N Engl J Med. 2021;385(14):1302–1315. doi: 10.1056/NEJMra1907607. [DOI] [PubMed] [Google Scholar]
- 49.Kiyohara H, Sujino T, Teratani T, Miyamoto K, Arai MM, Nomura E, Harada Y, Aoki R, Koda Y, Mikami Y, et al. Toll-Like Receptor 7 Agonist–Induced Dermatitis Causes Severe Dextran Sulfate Sodium Colitis by Altering the Gut Microbiome and Immune Cells. Cell Mol Gastroenterol Hepatol. 2019;7(1):135–156. doi: 10.1016/j.jcmgh.2018.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kingma SD, Li N, Sun F, Valladares RB, Neu J, Lorca GL. Lactobacillus johnsonii N6.2 stimulates the innate immune response through Toll-like receptor 9 in Caco-2 cells and increases intestinal crypt Paneth cell number in biobreeding diabetes-prone rats. J Nutr. 2011;141(6):1023–1028. doi: 10.3945/jn.110.135517. [DOI] [PubMed] [Google Scholar]
- 51.Denman SE, McSweeney CS. Development of a real-time PCR assay for monitoring anaerobic fungal and cellulolytic bacterial populations within the rumen. FEMS Microbiol Ecol. 2006;58(3):572–582. doi: 10.1111/j.1574-6941.2006.00190.x. [DOI] [PubMed] [Google Scholar]
- 52.Tsoi H, Chu ESH, Zhang X, Sheng J, Nakatsu G, Ng SC, Chan AWH, Chan FKL, Sung JJY, Yu J, et al. Peptostreptococcus anaerobius Induces Intracellular Cholesterol Biosynthesis in Colon Cells to Induce Proliferation and Causes Dysplasia in Mice. Gastroenterology. 2017;152(6):1419–1433.e5. doi: 10.1053/j.gastro.2017.01.009. [DOI] [PubMed] [Google Scholar]
- 53.Yang Y, Weng W, Peng J, Hong L, Yang L, Toiyama Y, Gao R, Liu M, Yin M, Pan C, et al. Fusobacterium nucleatum Increases Proliferation of Colorectal Cancer Cells and Tumor Development in Mice by Activating Toll-Like Receptor 4 Signaling to Nuclear Factor−κB, and Up-regulating Expression of MicroRNA-21. Gastroenterology. 2017;152(4):851–866.e24. doi: 10.1053/j.gastro.2016.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu Y, Gao Z, Hu R, et al. PD-L2 glycosylation promotes immune evasion and predicts anti-EGFR efficacy. J Immunother Cancer. 2021;9. doi: 10.1136/jitc-2021-002699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen -Y-Y, Ge J-Y, Zhu S-Y, Shao Z-M, Yu K-D. Copy number amplification of ENSA promotes the progression of triple-negative breast cancer via cholesterol biosynthesis. Nat Commun. 2022;13(1):791. doi: 10.1038/s41467-022-28452-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xing W, Huang P, Lu Y, Zeng W, Zuo Z. Amantadine attenuates sepsis-induced cognitive dysfunction possibly not through inhibiting toll-like receptor 2. J Mol Med (Berl). 2018;96(5):391–402. doi: 10.1007/s00109-018-1631-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Choi M, Kim MO, Lee J, Jeong J, Sung Y, Park S, Kwon W, Jang S, Park SJ, Kim H-S, et al. Hepatic serum amyloid A1 upregulates interleukin-17 (IL-17) in γδ T cells through Toll-like receptor 2 and is associated with psoriatic symptoms in transgenic mice. Scand J Immunol. 2019;89(6):e12764. doi: 10.1111/sji.12764. [DOI] [PubMed] [Google Scholar]
- 58.Jang YJ, Kim W-K, Han DH, Lee K, Ko G. Lactobacillus fermentum species ameliorate dextran sulfate sodium-induced colitis by regulating the immune response and altering gut microbiota. Gut Microbes. 2019;10(6):696–711. doi: 10.1080/19490976.2019.1589281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Smillie CS, Biton M, Ordovas-Montanes J, Sullivan KM, Burgin G, Graham DB, Herbst RH, Rogel N, Slyper M, Waldman J, et al. Intra- and Inter-cellular Rewiring of the Human Colon during Ulcerative Colitis. Cell. 2019;178(3):714–730. e22. doi: 10.1016/j.cell.2019.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-sequencing dataset has been deposited in the Sequence Read Archive (SRA) accession number: PRJNA780203. Source data and reagents are available from the corresponding author on reasonable request.


