ABSTRACT
Increasing evidence has shown that circular RNAs (circRNAs) play critical roles in various diseases, including keloid. The purpose of this study was to investigate the role of circ_0057452 and related action mechanisms during the development of keloid. The expression levels of circ_0057452, microRNA-7-5p (miR-7-5p) and GRB2 associated binding protein 1 (GAB1) mRNA were determined by quantitative real-time PCR (qRT-PCR). Cell proliferation was evaluated using methylthiazolyldiphenyl-tetrazolium bromide (MTT) and 5-Ethynyl-2’-deoxyuridine (Edu) assays. Flow cytometry analysis was utilized to determine cell cycle distribution and cell apoptosis. Western blot assay was used to measure apoptosis-related, collagen synthesis-related, and GAB1 protein levels. Cell migration and invasion were detected by wound healing assay and transwell assay. The interaction between miR-7-5p and circ_0057452 or GAB1 was confirmed by dual-luciferase reporter, RNA pull-down, and RNA Immunoprecipitation (RIP) assays. Circ_0057452 and GAB1 were upregulated in keloid tissues and keloid fibroblasts (KFs), while miR-7-5p was downregulated. Circ_0057452 knockdown or miR-7-5p overexpression inhibited the proliferation, migration, invasion, and collagen synthesis and induced cell cycle arrest and apoptosis of KFs. MiR-7-5p was targeted by circ_0057452, and its inhibition overturned the effects of circ_0057452 knockdown. In addition, GAB1 was a target of miR-7-5p, and GAB1 upregulation abolished the role of miR-7-5p overexpression and circ_0057452 knockdown in KFs. Circ_0057452 regulated the expression of GAB1 by adsorbing miR-7-5p in KFs. Circ_0057452 knockdown suppressed keloid development by regulating miR-7-5p/GAB1 axis, which might provide a promising therapeutic target for keloid.
KEYWORDS: Keloid, circ_0057452, miR-7-5p, GAB1
Introduction
Keloid is a benign dermal fibroproliferative tumor that commonly occurs after skin trauma or plastic surgery [1]. Keloid is usually defined as pathological scars caused by excessive wound healing after skin tissue injury [2]. Although keloid is a benign skin tumor, it can cause skin dysfunction via invading neighboring normal tissues, causing pain, or itching [3]. This disease possesses some features of tumor, characterized by abnormal fibroblast proliferation, aggressive invasion, and overproduction of extracellular matrix (ECM) [4,5]. Hence, the suppression of keloid fibroblast proliferation and mobility is considered as a promising therapeutic strategy for the treatment and prevention of keloid.
Circular RNAs (circRNAs), a special of noncoding RNAs (ncRNAs), possess covalently closed-loop structures with neither 5’-end cup nor 3’-end poly A tail [6]. Due to the circular structures, circRNAs are more stable and can avoid degradation by RNases [6]. Recently, multiple circRNAs were reported to play pivotal roles in diverse biological processes [7,8]. Moreover, some circRNAs have been found to be dysregulated in keloid tissues [9]. Moreover, circ_0057452 is derived from collagen type V alpha 2 chain (COL5A2) gene and upregulated in keloid tissues [10]. CircRNAs have been reported to function as competitive endogenous RNAs (ceRNAs) or microRNA (miRNA) sponges to modulate the expression of target genes and the development of disease via competitively binding to miRNA response elements [11]. Nevertheless, the exact role and the mechanism of circ_0057452 as ceRNA in keloid have not been reported.
MiRNAs, small ncRNAs, negatively regulate target gene expression via inhibiting translation levels to exert important biological functions [12,13]. Many miRNAs have been demonstrated to play crucial roles in keloid [14,15]. MiR-7-5p plays a suppressive role in keloid development [16]. Moreover, GRB2 associated binding protein 1 (GAB1) is a member of GAB family that plays an important role in keloid [17]. However, there is no relevant study about the relationships among circ_0057452, miR-7-5p, and GAB1 in keloid formation.
In this work, the levels circ_0057452, miR-7-5p, and GAB1 in keloid specimens and keloid fibroblasts (KFs) were investigated. Additionally, we explored the relationships among circ_0057452, miR-7-5p, and GAB1 and further studied their effects on the proliferation, cell cycle, apoptosis, migration, invasion, and collagen synthesis of KFs.
Materials and methods
Human tissue samples
All tissues were taken from patients who underwent operation at the First Affiliated Hospital of Zhengzhou University. After surgical excision, keloid specimens (n = 20) were acquired from keloid patients. Normal skin samples (n = 20) were provided by abdomens of healthy volunteers within plastic surgery procedures. These subjects have signed the informed consents. With approval from the Ethics Committee of the First Affiliated Hospital of Zhengzhou University, the current study was conducted.
Isolation and culture of fibroblasts
Normal fibroblasts (NFs) and primary KFs were isolated from normal skin and keloid skin tissues. Briefly, normal skin and keloid skin tissues were washed with phosphate-buffered saline (PBS; Beyotime, Jiangsu, China) and cut into 1–3 mm3 tissue blocks after removal of the epidermis and subcutaneous adipose tissues, and then digested with collagenase type I (Solarbio, Beijing, China) and trypsin (Invitrogen, Carlsbad, CA, USA). After that, NFs and KFs were cultivated in Dulbecco’s Modified Eagle Medium (DMEM; Invitrogen) containing with 10% fetal bovine serum (FBS; Invitrogen) in incubator at 37°C with 5% CO2.
Cell transfection
SiRNAs against circ_0057452 (si-circ_0057452#1, si-circ_0057452#2 and si-circ_0057452#3) and GAB1 (si-GAB1), miR-7-5p mimic (miR-7-5p), miR-7-5p inhibitor (anti-miR-7-5p), GAB1-overexpressing vector (pcDNA-GAB1), and matching controls (si-NC, miR-NC, anti-miR-NC, and pcDNA-NC) were provided by RiboBio (Guangzhou, China). KFs were transfected with the oligonucleotide or/and vector using Lipofectamine 3000 (Invitrogen).
Quantitative real-time PCR (qRT-PCR)
TRIzol (Invitrogen) was applied for extraction of total RNA. Next, cDNA was synthesized by PrimeScript RT Reagent Kit (TaKaRa, Dalian, China) or miScript II RT kit (Invitrogen). Thereafter, qRT-PCR reaction was manipulated using SYBR® Premix Ex Taq (Takara) on Bio-Rad CFX96 system (Bio-Rad, Hercules, CA, USA). Primers for genes in this study were listed as follows: circ_0057452 forward, 5’-TATGCCTGGAAAACCTGGAC-3’ and reverse, 5’-AGGGCCTTCAAGACCTTTGT-3’; COL5A2 forward, 5’- CAGGCTCCATAGGAATCAGAGG-3’ and reverse, 5’-CCAGCATTTCCTGCTTCTCCAG-3’; miR-7-5p forward, 5’-GCCGAGTGGAAGACTAGTGATT-3’ and reverse, 5’-CAGTGCGTGTCGTGGAGT-3’; GAB1 forward, 5’-GGAAACTCTTGGCATTCAGGAGG-3’ and reverse, 5’-GCAGTCTGTTTCAGAAGAGGTGG-3’; β-actin, forward, 5’- GCCGGGACCTGACTGACTAC-3’ and reverse, 5’-TCTCCTTAATGTCACGCACGAT-3’; and U6 forward, 5’-CTCGCTTCGGCAGCACATA-3’ and reverse, 5’-AACGCTTCACGAATTTGCGT-3’. The RNA levels were assessed with 2−ΔΔCt method. U6 (for miR-7-5p) and β-actin (for circ_0057452, COL5A2, and GAB1) were employed as internal controls.
CircRNA identification
The circular characteristic of circ_0057452 was confirmed through three experimental methods. For Actinomycin D (ActD) assay, cells were exposed to ActD (Sigma-Aldrich, St. Louis, MO, USA) to block transcription. Thereafter, qRT-PCR was used for measuring circ_0057452 and COL5A2 expression.
Random primers can amplify all RNAs, while oligo (dT)18 primers can only amplify RNA with poly (A) tail. Circ_0057452 and COL5A2 were amplified using random or oligo (dT)18 primers (Sangon Biotech, Shanghai, China). Next, qRT-PCR performed to verify whether circ_0057452 containing poly (A) tail.
RNase R treatment was used for degrading linear RNAs. RNA was treated with or without RNase R (Duma, Shanghai, China) for 30 min at 37°C. Next, the levels of circ_0057452 and COL5A2 were assessed by qRT-PCR.
Cell proliferation assays
Cell viability was detected by methylthiazolyldiphenyl-tetrazolium bromide (MTT) assay. In short, KFs were inoculated in a 96-well plate. After transfection, MTT reagent (Beyotime) was added to per well for 3–4 h. Next, dimethyl sulfoxide (DMSO; 200 μL, Beyotime) was added in each well after removal of cell culture medium. Finally, a microplate reader (Bio-Rad) was utilized for the detection of the optical density at a wavelength of 570 nm
For 5-Ethynyl-2’-deoxyuridine (Edu) assay, keyFluor594 Click-iT Edu detection kit (KeyGene, Nanjing, China) was used to test cell proliferation and DNA synthesis. In short, KFs were added into the 24-well plates. After transfection for 48 h, cells were incubated with Edu (50 μM) for 2 h. Next, Edu labeled cells were fixed with paraformaldehyde, followed by the addition of 0.5% Triton-X-100. Subsequently, Click-It reaction mixture was added to per well for 0.5 h in a darkness. DAPI was applied to stain the nucleic acids. Finally, stained cells were imaged by fluorescence microscope (Olympus, Tokyo, Japan) at × 200 magnification.
Flow cytometry analysis
For cell cycle analysis, KFs were collected and fixed in ethanol (Beyotime) at −20°C. After overnight fixation, the cells were harvested via centrifugation and then washed with PBS, followed by addition of propidium iodide (PI; Sangon Biotech), RNase A (Beyotime), and Triton X-100 (Sangon Biotech) in PBS for 20 min. Flow cytometry (BD Biosciences, Franklin, NJ, USA) was employed for detecting cell cycle distribution.
For apoptosis analysis, cells were harvested, washed with pre-cold PBS buffer twice, and then labeled with Annexin V-FITC and PI (Sangon Biotech). After incubation for 20 min in the darkness, the samples were measured with flow cytometry (Partec AG) for detecting cell apoptosis.
Western blot assay
Tissue samples or KFs were lysed in RIPA lysis buffer (Beyotime) to obtain total proteins. After that, the protein concentration of supernatants was tested with BCA protein assay kit (Tanon, Shanghai, China), and then proteins (about 30 μg/lane) were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) for separation. Then, the segregated proteins were transferred to PVDF membranes (Millipore, Billerica, MA, USA). Following the blockage using 5% skim milk (Beyotime), the membranes were immunoblotted with primary antibodies for 12–14 h at 4°C. The primary antibodies were all bought from Abcam (Cambridge, UK): Bax (ab53154, 1:500), Bcl-2 (ab196495, 1:2000), cleaved-caspase-3 (ab2302, 1:800), GAB1 (ab133486, 1:1000), fibronectin (ab2413, 1:1000), collagen I (ab270993, 1:1000), collagen III (ab184993, 1:1000), and β-actin (ab8227, 1:2000). Next, all membranes were maintained in secondary antibody (ab205718, 1:4000, Abcam). Finally, the visualization of protein blots was achieved by an enhanced chemiluminescence reagent (Applygen, Beijing, China).
Wound healing assay
Wound healing assay was conducted for detection of cell migration capacity. Briefly, KFs were inoculated in a 12-well plate and cultured until the cell confluence reached 80%. Thereafter, a sterile pipette tip (0.2 mL) was applied to scrape the wound on the cell monolayer. After being washed by PBS, these cells were cultured for 24 h in complete medium. Images of these scratches would be captured at 0 h and 24 h using a microscope (×40, Olympus).
Transwell assay
Transwell chamber (8 μm pore size, Costar, Corning, NY, USA) pre-coated with or without Matrigel (BD Biosciences) was used for cell invasion or migration assay. In short, KFs were re-suspended in serum-free media (0.2 mL) and added into upper chambers. The bottom chambers were supplied with the complete medium (0.6 mL). The non-migrating/invading cells from the top surface of the insert were carefully removed after 24 h incubation. After that, migrating/invading cells were fixed by paraformaldehyde (Beyotime), followed by staining with crystal violet solution (Beyotime). At last, migrating/invading cells were captured and counted with a microscope (×100, Olympus).
Dual-luciferase reporter assay
The candidate target miRNAs of circ_0057452 were predicted using circinteractome (https://circinteractome.nia.nih.gov/) and circbank (http://www.circbank.cn/). StarBase (http://starbase.sysu.edu.cn/) was utilized to predict the candidate downstream targets of miR-7-5p. The wild-type luciferase reporter vector of circ_0057452 or GAB1 (circ_0057452 WT or GAB1 3ʹUTR WT) was generated via inserting the sequence of circ_0057452 or GAB1 including the binding sequence for miR-7-5p into pmirGLO Dual-luciferase vectors (GenePharma, Shanghai, China). At the same time, mutant luciferase reporter vector of circ_0057452 or GAB1 (circ_0057452 MUT or GAB1 3ʹUTR MUT) was created via mutating the binding sites for miR-7-5p. KFs were co-transfected with the above constructs and miR-7-5p/miR-NC for 48 h. Finally, luciferase activity was tested via dual-luciferase reporter assay system (Promega, Madison, WI, USA).
RNA pull-down assay
Biotin-labeled negative control (Bio-NC), Biotin-labeled miR-7-5p (Bio-miR-7-5p), and Biotin-labeled mutant miR-7-5p (Bio-miR-7-5p MUT) were obtained from RiboBio and individually transfected into KFs. At 48 h post-transfection, Dynabeads M-280 Streptavidin (Invitrogen) was utilized to incubate cell lysates. At last, circ_0057452 and GAB1 enrichments were tested via qRT-PCR after RNA isolation from magnetic beads.
RNA immunoprecipitation (RIP) assay
EZ-Magna RIP kit (Millipore) was utilized to conduct RIP assay. Briefly, KFs were lysed in RIP lysis buffer and incubated with RIP buffer including magnetic beads coupled with antibody against immunoglobulin G (Anti-IgG) or Argonaute2 (Anti-Ago2). After incubation with Proteinase K, immunoprecipitated RNA was extracted, and circ_0057452, miR-7-5p, and GAB1 enrichment were examined via qRT-PCR.
Statistical analysis
The data were displayed as mean ± standard deviation from at least three independent experiments and analyzed by GraphPad Prism 6.0 software (GraphPad Prism, San Diego, CA, USA). The comparisons between two groups were estimated by Student’s t-test, while one-way analysis of variance (ANOVA) was applied to compare differences among multiple groups. Statistical significance was considered when P < 0.05.
Results
Circ_0057452 was upregulated in human keloid tissues and KFs
Circ_0057452 is generated from exons 35–40 of COL5A2 gene and is located on chr2:189,936,765–189,945,769, whose spliced mature sequence length is 306 bp (Figure 1a). The expression of circ_0057452 was upregulated in keloid tissues compared to normal tissues (Figure 1b). Likewise, circ_0057452 expression was higher in KFs than that in NFs (Figure 1c). Next, we analyzed the characteristics of circ_0057452 in NFs. ActD assay exhibited that the half-life of circ_0057452 transcript exceeded 24 h, while the half-life of COL5A2 mRNA was about 6 h (Figure 1d), suggesting that circ_0057452 transcript was more stable than COL5A2 transcript. The data of qRT-PCR showed that circ_0057452 expression was lower when oligo(dT)18 primers were used, than when random primers were used (Figure 1e), indicating that absence of a poly (A) tail for circ_0057452. Results from qRT-PCR confirmed that circ_0057452 was resistant to RNase R digestion (figure 1f), indicating the cyclic structure of circ_0057452. These results indicated that circ_0057452 might play a significant role in keloid progression.
Figure 1.
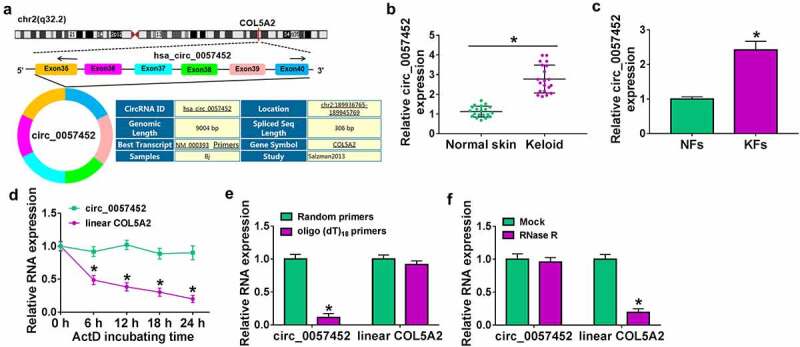
Circ_0057452 was overexpressed in human keloid tissues and KFs. (a) The information of circ_0057452 was presented. (b) The expression of circ_0057452 was determined by qRT-PCR in normal skin and keloid tissues. (c) The expression of circ_0057452 in NFs and KFs was examined by qRT-PCR. (d) The expression levels of circ_0057452 and linear COL5A2 were determined after treatment of ActD by qRT-PCR in KFs. (e) The levels of circ_0057452 and linear COL5A2 were detected after reverse transcription with random primers and oligo (dT)18 primers by qRT-PCR in KFs. (f) The levels of circ_0057452 and linear COL5A2 were determined in KFs after treatment of RNase R by qRT-PCR. *P < 0.05.
Knockdown of circ_0057452 inhibited the proliferation, migration, invasion, and collagen synthesis and induced cell cycle arrest and apoptosis in KFs
To explore the biological functions of circ_0057452 in keloid, lose-of-function assays were performed in KFs. The expression of circ_0057452 was reduced in KFs transfected with si-circ_0057452#1, si-circ_0057452#2 or si-circ_0057452#3, and the expression of circ_0057452 was the lowest in si-circ_0057452#1 group (Figure 2a). Therefore, we choose si-circ_0057452#1 for further research. MTT and Edu assay showed that circ_0057452 knockdown reduced cell viability and Edu positive cells in KFs (Figure 2 b and c), suggesting that circ_0057452 downregulation inhibited cell proliferation. Cell cycle progression and cell apoptosis were analyzed using flow cytometry. The results showed that circ_0057452 interference caused significant G0/G1 arrest and apoptosis in KFs (Figure 2 d and e). Western blot assay was sued to measure the expression of apoptosis-related proteins, including Bax (pro-apoptotic molecule), Bcl-2 (anti-apoptotic molecule), and cleaved-caspase-3 (a key executor in apoptotic process). The results revealed that knockdown of circ_0057452 increased the protein levels of Bax and cleaved-caspase-3 and reduced the protein expression of Bcl-2 (figure 2f). Wound healing assay and transwell assay showed that the migration and invasion of KFs were inhibited by downregulating circ_0057452 (Figure 2g–i). Keloid can overproduce collagens (including collagen I and collagen III) and other ECM components (including fibronectin). Western blot assay showed that circ_0057452 knockdown reduced fibronectin, collagen I and collagen III protein levels (Figure 2j), indicating inhibition of collagen synthesis ability. Taken together, circ_0057452 knockdown might inhibit the development of keloid.
Figure 2.
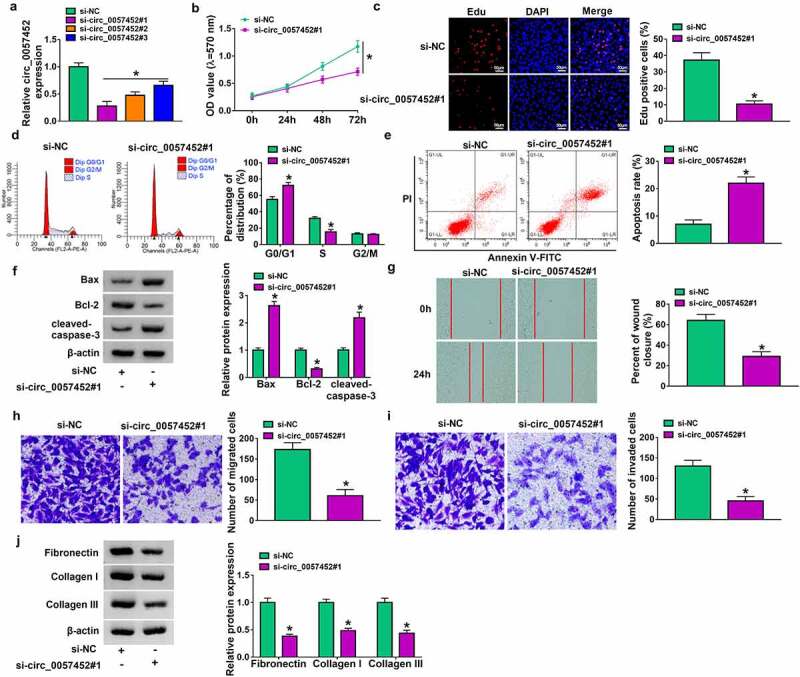
Downregulation of circ_0057452 repressed the proliferation, migration, invasion, and collagen synthesis, and induced cell cycle arrest and apoptosis in KFs. (a) The expression of circ_0057452 was measured by qRT-PCR in KFs transfected with si-NC, si-circ_0057452#1, si-circ_0057452#2, or si-circ_0057452#3. (b-i) KFs were transfected with si-NC or si-circ_0057452#1. (b) MTT assay was used for measuring cell viability. (c) DNA synthesis was determined using an Edu assay (×200). (d and e) Flow cytometry analysis was used to detect cell cycle distribution and cell apoptosis rate. (f) Western blot was performed to measure the protein levels of Bax, Bcl-2 and cleaved-caspase-3. (g) Cell migration ability was assessed by wound healing assay (×40). (h and i) Transwell assay was performed to measure cell migration and invasion abilities (×100). (j) The protein levels of fibronectin, collagen I and collagen III were analyzed using western blot analysis. *P < 0.05.
Circ_0057452 served as a sponge of miR-7-5p
Previous study reported that circRNA could act as a molecular sponge to interact with miRNA [18]. Next, we used online bioinformatics database (circinteractome and circbank) to predict the potential targets of circ_0057452. We found that five miRNAs (miR-1296-3p, miR-183-5p, miR-663b, miR-665, and miR-7-5p) were overlapped in circinteractome and circbank (Figure 3a). Moreover, we found that miR-183-5p and miR-7-5p expression were increased by transfection of si-circ_0057452#1, and the expression of miR-7-5p was higher than that of miR-183-5p expression (Figure 3b). Therefore, miR-7-5p was selected for further study. The binding sites between circ_0057452 and miR-7-5p were presented in Figure 3c. Overexpression efficiency of miR-7-5p was determined by qRT-PCR. The data showed that miR-7-5p expression was increased by transfection of miR-7-5p, indicating a high transfection efficiency (Figure 3d). Dual-luciferase reporter, RIP and RNA pull-down assays were conducted to confirm the interaction between circ_0057452 and miR-7-5p. As presented in Figure 3e, overexpression of miR-7-5p specifically inhibited the luciferase activity of circ_0057452 WT but not circ_0057452 MUT. RNA pull-down results revealed that circ_0057452 enrichment was enhanced by transfection of Bio-miR-7-5p in KFs rather than transfection of Bio-NC or Bio-miR-7-5p MUT (figure 3f). Moreover, RIP assay showed that circ_0057452 and miR-7-5p were substantially enriched in the Anti-Ago2 group compared with that in the Anti-IgG group (Figure 3g). In addition, we found that miR-7-5p expression was decreased in keloid tissues and KFs compared to normal skin tissues and NFs (Figure 3 h and i). These results suggested that miR-7-5p was a direct target of circ_0057452.
Figure 3.
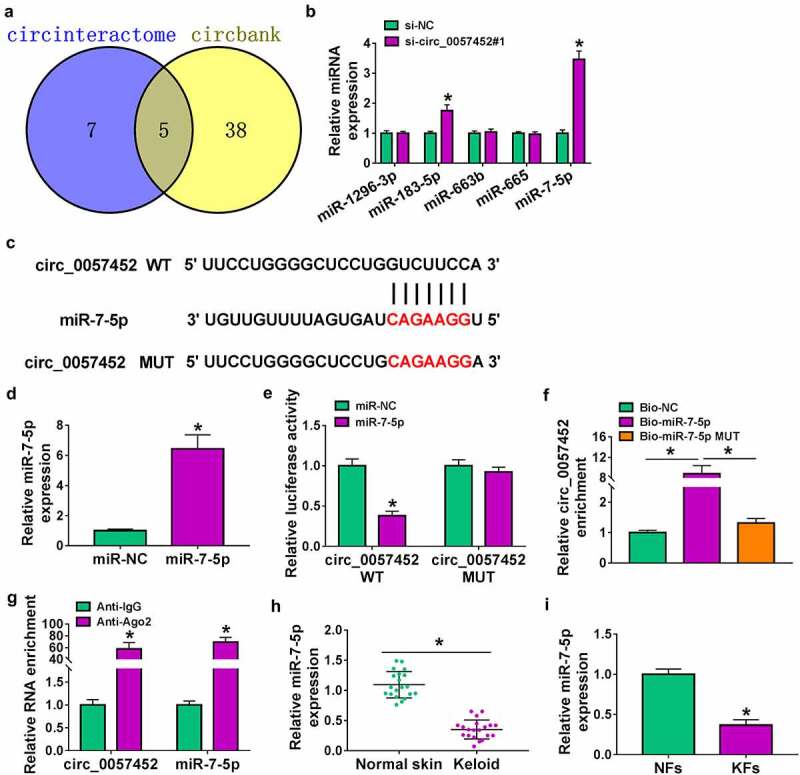
Circ_0057452 directly interacted with miR-7-5p. (a) The potential target miRNAs of circ_0057452 were predicted by circinteractome and circbank. (b) The expression levels of miR-1296-3p, miR-183-5p, miR-663b, miR-665, and miR-7-5p were determined by qRT-PCR in KFs transfected with si-NC or si-circ_0057452#1. (c) The predicted binding sites between circ_0057452 and miR-7-5p were shown. (d) MiR-7-5p expression was measured by qRT-PCR in KFs transfected with miR-NC or miR-7-5p. (e) Luciferase activity was measured in KFs co-transfected with circ_0057452 WT circ_0057452 MUT and miR-NC or miR-7-5p by dual-luciferase reporter assay. (f) The enrichment of circ_0057452 was examined in KFs transfected with Bio-NC, Bio-miR-7-5p, or Bio-miR-7-5p MUT by RNA pull-down assay. (g) The enrichment of circ_0057452 and miR-7-5p was detected in KFs by RIP assay. (h and i) MiR-7-5p expression was detected by qRT-PCR in normal skin tissues, keloid tissues, NFs, and KFs. *P < 0.05.
Inhibition of miR-7-5p reversed the regulatory functions of circ_0057452 knockdown in KFs
Transfection efficiency of anti-miR-7-5p was evaluated by qRT-PCR. The results showed that miR-7-5p expression was decreased by transfection of anti-miR-7-5p (Figure 4a). To explore whether the biological effects of circ_0057452 were mediated by miR-7-5p, rescue experiments were performed. The inhibitory effects of cell viability and DNA synthesis were abated by inhibition of miR-7-5p in KFs (Figure 4 b and c). Moreover, circ_0057452 knockdown-induced cell cycle arrest at the G0/G1 phase and cell apoptosis were relieved by the reintroduction of anti-miR-7-5p (Figure 4 d and e). Furthermore, the increased of Bax and cleaved-caspase-3 protein expression and the decreased of Bcl-2 protein expression caused by circ_0057452 depletion were neutralized by downregulating miR-7-5p in KFs (figure 4f). In addition, the suppressive effects of circ_0057452 silence on cell migration, invasion, and collagen synthesis were reversed by miR-7-5p inhibition in KFs (Figure 4g–). Together, these results indicated that circ_0057452 exerted its role in KFs by sponging miR-7-5p.
Figure 4.
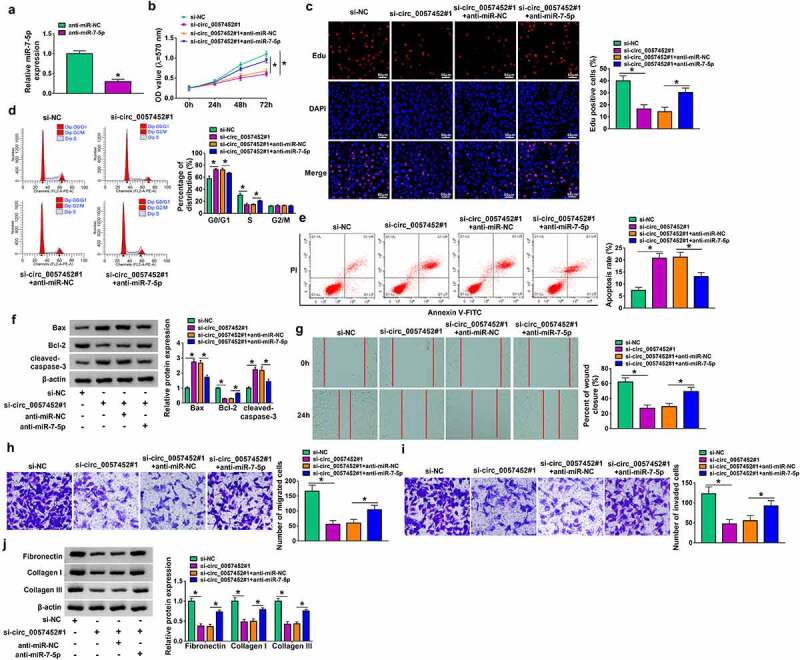
Circ_0057452 regulated the proliferation, cell cycle, migration, invasion, apoptosis, and collagen synthesis of KFs by sponging miR-7-5p. (a) The expression of miR-7-5p was examined by qRT-PCR in KFs transfected with anti-miR-NC or anti-miR-7-5p. (b-i) KFs were transfected with si-NC, si-circ_0057452#1, si-circ_0057452#1 + anti-miR-NC, or si-circ_0057452#1 + anti-miR-7-5p. (b and c) MTT assay and Edu assay were utilized to evaluate proliferation ability. (d and e) Cell cycle distribution and cell apoptosis were detected by flow cytometry analysis. (f) The protein levels of Bax, Bcl-2 and cleaved-caspase-3 were analyzed by western blot assay. (g) Cell migration was evaluated by wound healing assay (×40). (h and i) Transwell assay was used to detect cell migration and invasion (×100). (j) The protein levels of fibronectin, collagen I and collagen III were examined by western blot assay. *P < 0.05.
GAB1 was a direct target of miR-7-5p
To identify the underlying mechanism of miR-7-5p, bioinformatics analysis was performed and we found that GAB1 3ʹUTR contained binding sites for miR-7-5p (Figure 5a). To test whether GAB1 was a direct target of miR-7-5p, dual-luciferase reporter assay, RNA pull-down assay and RIP assay were carried out. As shown in Figure 5b, the luciferase activity of GAB1 3ʹUTR WT was significantly decreased after overexpression of miR-7-5p in KFs, whereas the activity of GAB1 3ʹUTR MUT was not affected. RNA pull-down assay showed that the GAB1 enrichment was increased in Bio-miR-7-5p group compared to Bio-NC and Bio-miR-7-5p MUT groups (Figure 5c). Meanwhile, GAB1 and miR-7-5p were more abundant in Ago2 pellet than in IgG pellet in KFs (Figure 5d). Next, the influence of miR-7-5p on the expression of GAB1 was detected. The data indicated that miR-7-5p overexpression inhibited the mRNA and protein expression of GAB1 in KFs (Figure 5 e and f), suggesting that GAB1 was negatively regulated by miR-7-5p. GAB1 mRNA and protein expression were higher in keloid tissues and KFs than that in normal skin tissues and NFs (Figure 5g–j). In addtion, circ_0057452 expression was negatively correlated with GAB1 mRNA expression in keloid tissues (Supplementary Figure 1A). Moreover, we found that overexpression of miR-7-5p could not significantly affect circ_0057452 expression in KFs (Supplementary Figure 1B). Subsequently, we explored whether circ_0057452 could regulate GAB1 expression via acting as a sponge of miR-7-5p in KFs. We found that GAB1 mRNA and protein expression were inhibited by knockdown of circ_0057452, which was restored by inhibiting miR-7-5p (Figure 5 k and l). These data indicated that circ_0057452 positively regulated GAB1 expression by sponging miR-7-5p.
Figure 5.
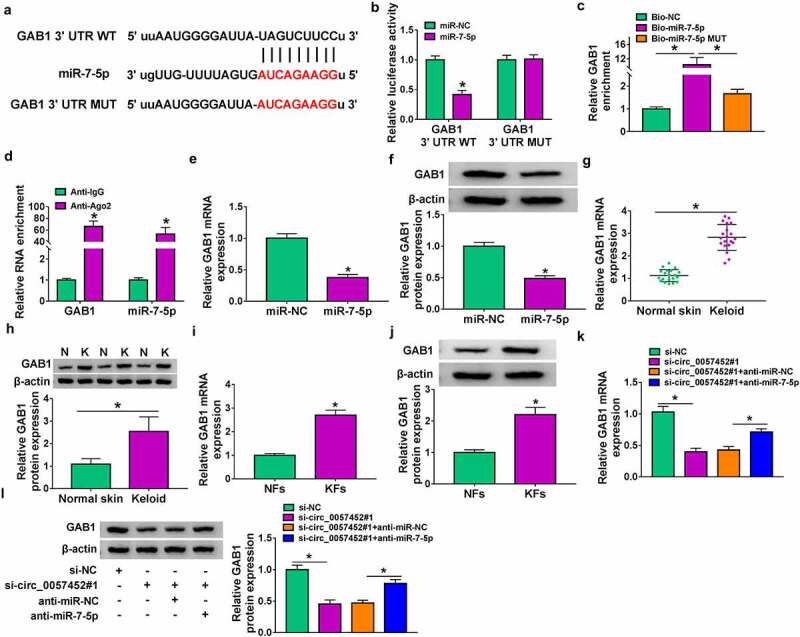
GAB1 was targeted by miR-7-5p. (a) The putative binding sites between miR-7-5p and GAB1 were predicted by starBase. (b-d) The interaction between miR-7-5p and GAB1 was confirmed by dual-luciferase reporter, RNA pull-down, and RIP assays. (e and f) The mRNA and protein expression of GAB1 in KFs transfected with miR-NC or miR-7-5p were determined by qRT-PCR and western blot, respectively. (g and h) GAB1 mRNA and protein expression in normal skin tissues and keloid tissues were detected by qRT-PCR and western blot, respectively. (i and j) The mRNA and protein levels of GAB1 in NFs and KFs were examined by qRT-PCR and western blot, respectively. (k and l) QRT-PCR and western blot were conducted to detect the mRNA and protein levels of GAB1 in KFs transfected with si-NC, si-circ_0057452#1, si-circ_0057452#1 + anti-miR-NC, or si-circ_0057452#1 + anti-miR-7-5p. *P < 0.05.
MiR-7-5p overexpress or circ_0057452 knockdown suppressed the proliferation, cell cycle, migration, invasion, and collagen synthesis and induced apoptosis in KFs by regulating GAB1
Overexpression efficiency of GAB1 was determined by western blot in KFs transfected with pcDNA-NC or pcDNA-GAB1 (Figure 6a). To explore whether GAB1 was involved in miR-7-5p/circ_0057452-mediated biological functions in KFs, rescue assays were performed. MTT and Edu assays demonstrated that enforced expression of miR-7-5p or circ_0057452 downregulation suppressed cell viability and DNA synthesis and arrested KFs in G0/G1 phase, while these effects were counteracted by overexpression of GAB1 in KFs (Figure 6b–d, supplementary Figure 2A-2C). In addition, GAB1 overexpression weakened miR-7-5p/si-circ_0057452-induced apoptosis in KFs (Figure 6e, supplementary Figure 2D). Meanwhile, miR-7-5p upregulation or circ_0057452 downregulation increased the protein levels of Bax and cleaved-caspase-3 and decreased Bcl-2 protein expression, which could be reversed by increasing GAB1 expression (figure 6f, supplementary Figure 2E). Further, the migration, invasion, and collagen synthesis abilities of KFs were suppressed by transfection of miR-7-5p or si-circ_0057452, whereas co-transfection of pcDNA-GAB1 abolished these effects (Figure 6g–j, supplementary Figure 2 F-2I). Moreover, we also detected the role of GAB1 in KFs. Transfection of si-GAB1 suppressed GAB1 protein expression in KFs (Supplementary Figure 3A), suggesting a high knockdown efficiency. In addition, GAB1 knockdown suppressed cell proliferation and induced cell cycle arrest and apoptosis (Supplementary Figure 3B-3E). GAB1 downregulation elevated the protein levels of Bax and cleaved-caspase-3 and reduced Bcl-2 protein expression (Supplementary Figure 3 F). Besides, GAB1 interference repressed the migration, invasion, and collagen synthesis abilities of KFs (Supplementary Figure 3 G-3 J). Altogether, these data indicated that miR-7-5p exerted its biological roles in KFs via regulating GAB1.
Figure 6.
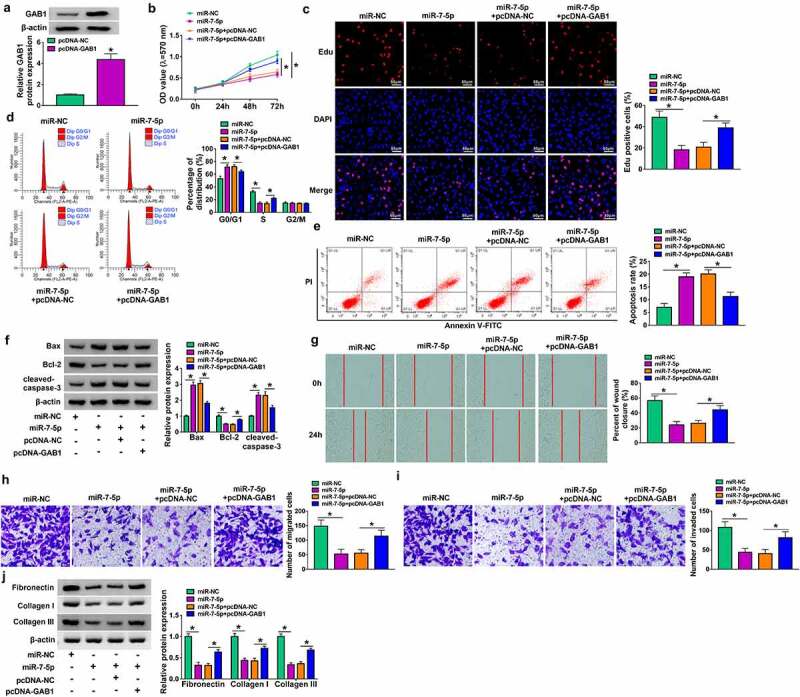
MiR-7-5p inhibited proliferation, cell cycle progression, migration, invasion, and collagen synthesis and increased apoptosis of KFs by targeting GAB1. (a) Western blot assay was carried out to detect the protein expression of GAB1 in KFs transfected with pcDNA-NC or pcDNA-GAB1. (b-i) KFs were transfected with miR-NC, miR-7-5p, miR-7-5p + pcDNA-NC, or miR-7-5p + pcDNA-GAB1. (b and c) Cell proliferation ability was evaluated by MTT assay and Edu assay. (d and e) Flow cytometry analysis was utilized to determine cell cycle distribution and cell apoptosis. (f) The protein levels of Bax, Bcl-2 and cleaved-caspase-3 were tested using western blot analysis. (g) Cell migration was assessed by wound healing assay (×40). (h and i) Cell migration and invasion were detected by transwell assay (×100). Western blot assay was carried out to detect fibronectin, collagen I and collagen III protein levels. *P < 0.05.
Discussion
Keloid is one of the pathological scars, and there is no effective treatment at present [19]. Additionally, the underlying mechanisms of keloid formation are still unclear. In the current research, we demonstrated that circ_0057452 silence repressed proliferation, mobility, and collagen synthesis as well as induced cell cycle arrest and apoptosis in KFs through modulating miR-7-5p/GAB1 axis.
Increasing evidence indicates that ncRNAs (mainly lncRNAs and miRNAs) play key roles in the pathogenesis of keloid by regulating different pathways [20–22]. Although circ_101238 and circCOL5A1 have been suggested to be implicated in the progression of keloid [16,23], the pathological mechanism of circRNAs in the progression of keloid is still poorly understood. Therefore, it is essential to better understand the roles and mechanisms of novel circRNAs for further study on the pathogenesis of keloid formation. Hence, in our research, the role and underlying mechanism of circ_0057452 in keloid progression were explored. Circ_0057452 was found to be overexpressed in keloid tissues and KFs, which was in line with a previous research [10]. Moreover, we uncovered that circ_0057452 downregulation repressed KFs proliferation, migration, invasion, and collagen synthesis and facilitated apoptosis, indicating that circ_0057452 played a promoting role in keloid progression.
It is well known that circRNAs serve as ceRNAs to bind to miRNAs, thus affecting a variety of biological processes. For instance, circ-ATP8A2 increased cervical cancer cell growth and mobility as a ceRNA to target EGFR via sponging miR-433 [24]. Moreover, circ-FOXM1 facilitated the progression of lung cancer as a ceRNA to target MACC1 and PPDPF via adsorbing miR-1304-5p [25]. Herein, circ_0057452 was demonstrated to act as a miR-7-5p sponge. It has been demonstrated that miR-7 was downregulated in keloid, and miR-7 inhibition might contribute to the pathogenesis of localized scleroderma [26]. Besides, a study by Lv et al. declared that miR-7-5p enrichment was declined in keloid specimens and KFs, and miR-7-5p inhibition could increase proliferation and miR-7-5p upregulation limited migration and augmented apoptosis via targeting Epac1 in KFs [16]. Consistently, we also found an obvious reduction in miR-7-5p expression in keloid samples and KFs. Rescue assays proved that miR-7-5p downregulation counteracted the impact of circ_0057452 interference on reduction of proliferation, migration, invasion, and collagen synthesis and enhancement of apoptosis in KFs, indicating that circ_0057452 might promote keloid progression via sponging miR-7-5p.
It has been suggested that miRNAs perform their roles through regulating the expression of target mRNAs [27]. Based on bioinformatics analysis and a series of experiments, GAB1 was demonstrated to be a downstream target for miR-7-5p. GAB1 can regulate cell differentiation and growth from many receptors, including cytokines, growth factors, and antigen receptors [28,29]. GAB1 acts as a tumor promoter in many cancers [30,31]. Importantly, Feng et al. uncovered that GAB1 was overexpressed in keloid, and GAB1 elevation neutralized the suppressing impact of miR-141-3p on keloid fibroblast proliferation and migration [17]. In line with this research, we also observed that GAB1 was upregulated in keloid specimens and KFs. Strikingly, we demonstrated that circ_0057452 could bind to miR-7-5p to modulate GAB1. Additionally, we revealed that miR-7-5p restoration repressed the growth, migration, invasion, and collagen synthesis as well as accelerated apoptosis in KFs via targeting GAB1. Hence, we concluded that circ_0057452 modulated keloid progression via regulation of miR-7-5p/GAB1 axis (Figure 7).
Figure 7.
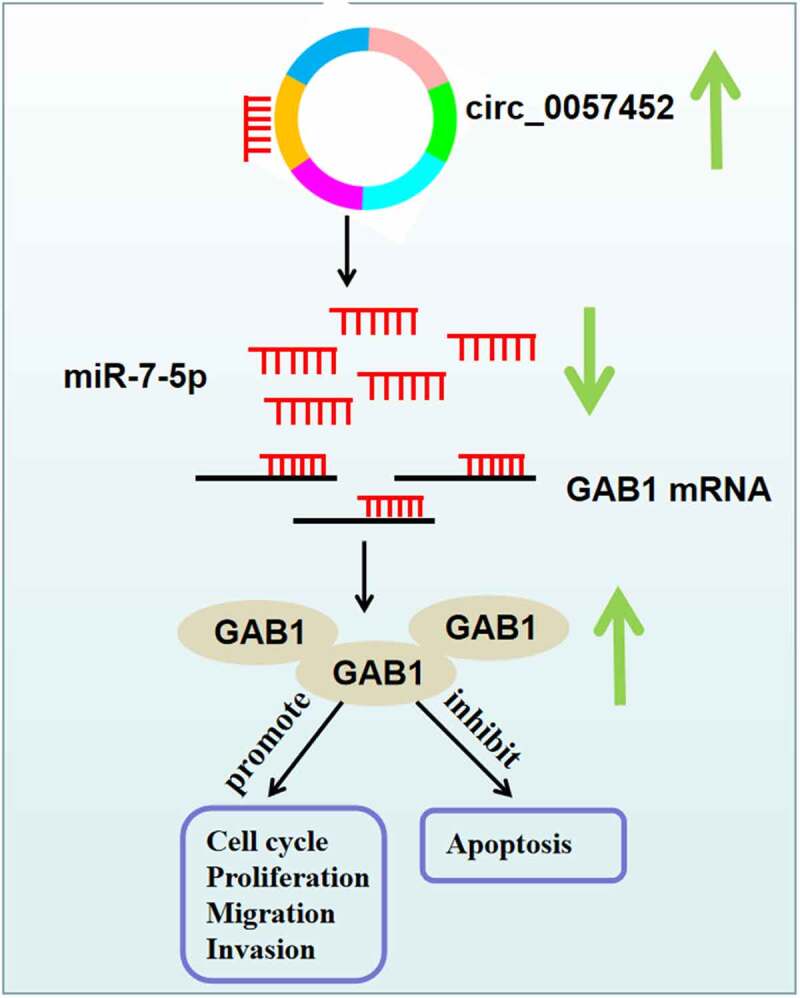
Schematic diagram of the mechanism by which the circ_0057452/miR-7-5p/GAB1 axis regulates the development of keloid.
In conclusion, circ_0057452 was upregulated in keloid. Moreover, circ_0057452 regulated the proliferation, cell cycle progression, apoptosis, migration, invasion, and collagen synthesis of KFs via regulating miR-7-5p/GAB1 axis. The circ_0057452/miR-7-5p/GAB1 regulatory network might contribute to a better understanding the progression of keloid, providing a potential therapeutic target for keloid.
Supplementary Material
Funding Statement
The author(s) reported there is no funding associated with the work featured in this article.
Data Availability Statement
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/15384101.2022.2102796
References
- [1].Alster TS, Tanzi EL.. Hypertrophic scars and keloids. Am J Clin Dermatol. 2003;4:235–243. [DOI] [PubMed] [Google Scholar]
- [2].Bock O, Schmid-Ott G, Malewski P, et al. Quality of life of patients with keloid and hypertrophic scarring. Arch Dermatol Res. 2006;297:433. [DOI] [PubMed] [Google Scholar]
- [3].Mofikoya BO, Adeyemo WL, Abdus-salam AA. Keloid and hypertrophic scars: a review of recent developments in pathogenesis and management. Nig Q J Hosp Med. 2007;17:134–139. [DOI] [PubMed] [Google Scholar]
- [4].Xue M, Jackson CJ. Extracellular matrix reorganization during wound healing and its impact on abnormal scarring. Adv Wound Care. 2015;4:119–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Unahabhokha T, Sucontphunt A, Nimmannit U, et al. Molecular signalings in keloid disease and current therapeutic approaches from natural based compounds. Pharm Biol. 2015;53:457–463. [DOI] [PubMed] [Google Scholar]
- [6].Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. [DOI] [PubMed] [Google Scholar]
- [7].Salzman J, Chen RE, Olsen MN, et al. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9:e1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Wang J, Wu H, Xiao Z, et al. Expression profiles of lncRNAs and circRNAs in Keloid. Plast Reconstr Surg Glob Open. 2019;7:e2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Shi J, Yao S, Chen P, et al. The integrative regulatory network of circRNA and microRNA in keloid scarring. Mol Biol Rep. 2020;47:201–209. [DOI] [PubMed] [Google Scholar]
- [11].Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. [DOI] [PubMed] [Google Scholar]
- [12].Ardekani AM, Naeini MM. The role of microRNAs in human diseases. Avicenna J Med Biotechnol. 2010;2:161–179. [PMC free article] [PubMed] [Google Scholar]
- [13].Jansson MD, Lund AH. MicroRNA and cancer. Mol Oncol. 2012;6:590–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Zhang Y, Guo B, Hui Q, et al. Downregulation of miR‑637 promotes proliferation and metastasis by targeting Smad3 in keloids. Mol Med Rep. 2018;18:1628–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Liu P, Hu Y, Xia L, et al. miR-4417 suppresses keloid fibrosis growth by inhibiting CyclinD1. J Biosci. 2020;45:47. [PubMed] [Google Scholar]
- [16].Lv W, Liu S, Zhang Q, et al. Circular RNA CircCOL5A1 Sponges the MiR-7-5p/Epac1 axis to promote the progression of keloids through regulating PI3K/Akt signaling pathway. Front Cell Dev Biol. 2021;9:626027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Feng J, Xue S, Pang Q, et al. miR-141-3p inhibits fibroblast proliferation and migration by targeting GAB1 in keloids. Biochem Biophys Res Commun. 2017;490:302–308. [DOI] [PubMed] [Google Scholar]
- [18].Kulcheski FR, Christoff AP, Margis R. Circular RNAs are miRNA sponges and can be used as a new class of biomarker. J Biotechnol. 2016;238:42–51. [DOI] [PubMed] [Google Scholar]
- [19].Robles DT, Berg D. Abnormal wound healing: keloids. Clin Dermatol. 2007;25:26–32. [DOI] [PubMed] [Google Scholar]
- [20].Chen Z, Tao Q, Qiao B, et al. Silencing of LINC01116 suppresses the development of oral squamous cell carcinoma by up-regulating microRNA-136 to inhibit FN1. Cancer Manag Res. 2019;11:6043–6059. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [21].Jin J, Jia Z-H, Luo X-H, et al. Long non-coding RNA HOXA11-AS accelerates the progression of keloid formation via miR-124-3p/TGFβR1 axis. Cell Cycle. 2020;19:218–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Shi K, Qiu X, Zheng W, et al. MiR-203 regulates keloid fibroblast proliferation, invasion, and extracellular matrix expression by targeting EGR1 and FGF2. Biomed Pharmacother. 2018;108:1282–1288. [DOI] [PubMed] [Google Scholar]
- [23].Yang D, Li M, Du N. Effects of the circ_101238/miR-138-5p/CDK6 axis on proliferation and apoptosis keloid fibroblasts. Exp Ther Med. 2020;20:1995–2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Ding L, Zhang H. Circ-ATP8A2 promotes cell proliferation and invasion as a ceRNA to target EGFR by sponging miR-433 in cervical cancer. Gene. 2019;705:103–108. [DOI] [PubMed] [Google Scholar]
- [25].Liu G, Shi H, Deng L, et al. Circular RNA circ-FOXM1 facilitates cell progression as ceRNA to target PPDPF and MACC1 by sponging miR-1304-5p in non-small cell lung cancer. Biochem Biophys Res Commun. 2019;513:207–212. [DOI] [PubMed] [Google Scholar]
- [26].Etoh M, Jinnin M, Makino K, et al. microRNA-7 down-regulation mediates excessive collagen expression in localized scleroderma. Arch Dermatol Res. 2013;305:9–15. [DOI] [PubMed] [Google Scholar]
- [27].Felekkis K, Touvana E, Stefanou C, et al. microRNAs: a newly described class of encoded molecules that play a role in health and disease. Hippokratia. 2010;14:236–240. [PMC free article] [PubMed] [Google Scholar]
- [28].Yamasaki S, Nishida K, Yoshida Y, et al. Gab1 is required for EGF receptor signaling and the transformation by activated ErbB2. Oncogene. 2003;22:1546–1556. [DOI] [PubMed] [Google Scholar]
- [29].Gu H, Neel BG. The ‘Gab’ in signal transduction. Trends Cell Biol. 2003;13(3):122–130. [DOI] [PubMed] [Google Scholar]
- [30].Bai R, Weng C, Dong H, et al. MicroRNA-409-3p suppresses colorectal cancer invasion and metastasis partly by targeting GAB1 expression. Int J Cancer. 2015;137:2310–2322. [DOI] [PubMed] [Google Scholar]
- [31].Wang X, Peng J, Yang Z, et al. Elevated expression of Gab1 promotes breast cancer metastasis by dissociating the PAR complex. J Exp Clin Cancer Res. 2019;38:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.


