Abstract
Protective immunity to mycobacterial infection is incompletely understood but probably involves the coordinated interaction of multiple cell types and cytokines. With the aim of developing assays that might provide a surrogate measure of protective immunity, we have investigated the use of recombinant mycobacteria carrying luciferase reporter enzymes to assess the effectiveness of antimycobacterial immunity in model systems. Measurement of luminescence was shown to provide a rapid and simple alternative to the counting of CFU as a means of monitoring mycobacterial viability. We describe optimization of a luciferase reporter strain of Mycobacterium tuberculosis and demonstrate its application for the study of mycobacterial interactions with host cells in tissue culture and the rapid assessment of vaccine efficacy in a murine model.
It is estimated that as much as one-third of the world’s population is currently infected with Mycobacterium tuberculosis, resulting in up to three million deaths per year. The recent increase in cases of tuberculosis, mainly due to an association with human immunodeficiency virus, poor living conditions, and the emergence of drug-resistant strains, has been described as a “global emergency” by the World Health Organization. Around 10% of infected individuals will go on to develop clinical tuberculosis in later life, a risk that rises to 10% annually in the case of individuals coinfected with human immunodeficiency virus (24). For the vast majority of individuals, the normal immune response is sufficient to prevent progression to clinical disease, and boosting of immunity in the susceptible minority represents a potentially powerful approach to disease control. The Mycobacterium bovis BCG (bacillus Calmette-Guérin) vaccine confers protection against childhood forms of tuberculosis but has shown efficacy against the predominant adult disease only in selected geographical locations (9). In addition, antibiotic treatment requires at least 6 months (short-course chemotherapy) with a cocktail of drugs, and resistance to these drugs is on the increase (22). The complete genome of M. tuberculosis H37Rv is now available, describing a plethora of potential new drug and vaccine targets for investigation (6). However, progress in developing improved vaccines is hampered by a limited understanding of the protective immune mechanisms that function to control mycobacterial infection.
Macrophages with microbicidal activities, activated by exposure to cytokines such as gamma interferon (IFN-γ), play a central role in the control of mycobacterial infection. Mice with defects in components of the IFN-γ pathway are impaired in their ability to control infection by mycobacteria (7), and a rare genetic defect affecting the human IFN-γ receptor is associated with enhanced susceptibility to mycobacterial disease (19). The killing of M. tuberculosis by activated macrophages can be demonstrated in vitro by using murine cell cultures, but this has proven difficult to reproduce with human macrophages (26). Several lines of evidence suggest that additional cellular interactions may contribute to mycobacterial immunity. Hypersusceptibility to M. tuberculosis infection in mice with an impairment in expression of the class I major histocompatibility complex suggests the involvement of cytotoxic T lymphocytes (10), and additional minor T-cell subsets are also activated during mycobacterial infection (17). It is probable that protection is a complex phenomenon, involving the coordinated activation of multiple cell types. The absence of a simple immunological parameter that could be used to assess the effectiveness of an individual’s immune response to mycobacterial infection presents a formidable barrier to the design and testing of new vaccines for tuberculosis.
Ultimately, the effectiveness of the immune response is expressed in its ability to control mycobacterial growth, and measurement of its effect on the physiological status of infecting mycobacteria represents one potential route to establishment of a correlate of protection. The benefit to mycobacterial pathogens of slow growth (M. tuberculosis requires 4 to 6 weeks to form colonies on agar plates with a doubling time of 18 to 24 h) is not fully understood. However, this fact and the rigorous containment facilities required are hindrances to the study of the organism. Determination of mycobacterial viability by using conventional microbiology is therefore technically cumbersome. The ability to construct recombinant mycobacterial strains carrying reporter genes provides a strategy for the simplification of such studies. The goal of this project was to evaluate the use of luciferase reporter strains as a way of assessing immune status in models of infection.
Bioluminescence may be produced by fireflies or by bacteria living in symbiotic relationships with other organisms. Deep-sea fish harbor luminescent bacteria, including Vibrio harveyi, and a parasitic nematode carries the bacterium Photorhabdus (Xhenorhabdus) luminescens (18). Such luminescence has been employed as a reporter in a number of systems to detect the presence of low levels of bacterial contamination or infection and as a promoter probe to measure specific gene expression (14). Emission of light is dependent on the presence of a cofactor, ATP, or reduced flavin mononucleotide (FMNH2), which is found only in living cells (18). Dead cells are no longer able to produce cofactor; thus, a corresponding decline in luminescence follows. Mycobacteriophages carrying the luciferase gene (luc) from the American firefly (Photinus pyralis) have been used as a rapid means of testing for drug-resistant isolates of M. tuberculosis (5, 16, 25). Bioluminescent strains of M. tuberculosis and M. bovis BCG have been constructed with firefly luciferase carried on plasmid vectors, and these strains were employed in high-throughput drug screens in both in vitro cultures (2, 3) and infected-animal models (13). Experiments were also carried out to evaluate antimycobacterial immunity in mice by using such luminescent BCG (13). A bioluminescent strain of Mycobacterium smegmatis has been constructed with luxA and luxB genes from V. harveyi and been used for the screening of antibiotics and biocides (1). In this study, we describe a comparison of different luciferase reporter strains and their use for the assessment of immune status in whole-animal and tissue culture models using M. tuberculosis as well as nonpathogenic mycobacteria.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Escherichia coli DH5α (GIBCO BRL) was maintained on Luria agar and grown in Luria broth. Transformed E. coli bacteria were selected on 250 μg of hygromycin ml−1. M. smegmatis 1-2c (an electrocompetent derivative of mc26 [30]), M. bovis BCG Montreal (ATCC 35735), and M. tuberculosis H37Rv (ATCC 27294) were grown in Middlebrook 7H9 (Difco) supplemented with albumin, dextrose, catalase (or 2% glucose for M. smegmatis) and 50 μg of hygromycin ml−1. Strains were maintained on Middlebrook 7H11 containing enrichment oleic acid, albumin, dextrose, and catalase (2% glucose for M. smegmatis), 50 μg of hygromycin ml−1, and 50 μg of amphotericin B ml−1 and grown for 3 days (M. smegmatis) or 3 weeks (BCG and M. tuberculosis) at 37°C in bags to prevent drying. Time course experiments were carried out in volumes of 50 to 100 ml in 500-ml flasks aerated at 37°C and were inoculated by diluting log-phase cultures prepared from −80°C glycerol stocks.
Plasmid construction.
Molecular biology techniques were carried out according to standard procedures (27). The luxA and luxB genes of V. harveyi were cloned under the control of the BCG hsp60 promoter, on an XbaI-NheI fragment from pPA3 (1), into pOLYG (20), a modification of p16R1 (12), to generate pSMT1 (Fig. 1). This plasmid is a shuttle vector in E. coli and mycobacteria and employs hygromycin as a selectable marker. Based on this plasmid, additional constructs were produced, including pSMT5, containing the X. luminescens luxA and luxB genes (11), and pSMT6, which consists of the firefly luc gene (8) under the control of the mycobacterial hsp60 promoter (Table 1). Further constructs where the V. harveyi luxA and luxB genes were driven by other mycobacterial putative promoter sequences are described in Table 1. The integrating vector, pLINT560, is based on a DNA segment carrying the attachment site (attP) and the integrase (int) gene from the mycobacteriophage L5 (28). Plasmid DNA was prepared from recombinant E. coli DH5α by using Qiagen columns according to manufacturer’s recommendations and was used to electroporate mycobacterial species (12). Recombinant colonies were tested for luminescence and were maintained at −80°C in 25% glycerol.
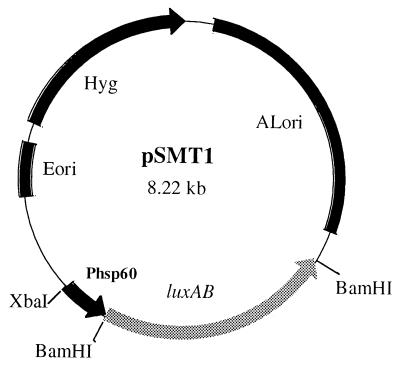
FIG. 1.
The construct pSMT1. Light production may be correlated with CFU. The reporter plasmid pSMT1 is a shuttle vector containing both an origin of replication for mycobacteria (ALori) and an E. coli origin of replication (Eori). Hygromycin (Hyg) is employed for antibiotic selection. The V. harveyi luxAB genes are under the control of the BCG hsp60 promoter (Phsp60). XbaI and BamHI cleavage sites are indicated.
TABLE 1.
Plasmid vectors constructed for this study
| Construct name | Origin of luciferase gene(s) | Promotera | Cofactor | Ratio of RLUs to CFU |
|---|---|---|---|---|
| pSMT1 | V. harveyi luxAB | hsp60 | FMNH2 | 10 |
| pSMT5 | X. luminescens luxAB | hsp60 | FMNH2 | 10 |
| pSMT6 | P. pyralis luc | hsp60 | ATP | 0.01 |
| pLINT560 | V. harveyi luxAB | hsp60 | FMNH2 | 0.1 |
| pSMT16 | V. harveyi luxAB | hsp20 | FMNH2 | 1 |
| pSMT8.1 | V. harveyi luxAB | gcvB | FMNH2 | 1 |
| pSMTBBS | V. harveyi luxAB | sodA | FMNH2 | 5 |
| pSMTCZS | V. harveyi luxAB | sodC | FMNH2 | 1 |
Promoter fragments employed were as follows: BCG heat shock protein 60 (hsp60), 404 bp of the 5′ region, including 18 bp of coding sequence (28); M. tuberculosis 16-kDa α-crystallin (hsp20), 543 bp upstream of Rv2299c, including 6 bp of coding sequence; glycine decarboxylase (gcvB), 440 bp upstream of Rv1832, finishing 19 bp upstream of the initiation codon; sodA, 111 bp upstream of Rv3846 plus 59 bp of coding sequence (30); sodC, 440 bp of the 5′ region finishing at the initiation codon of Rv0432 (6). In each V. harveyi construct, 89 bp of upstream sequence lies between the promoter element and the luxA initiation codon, including a ribosome binding site (AAGAA) 5 bp upstream of the luxA initiation codon. All constructs are transcriptional fusions.
Bioluminescence detection and assay.
Luminescence was measured in either a Berthold AutoLumat LB953 tube luminometer or a class I biosafety cabinet with a Turner Design 20/20 tube luminometer. The Turner Design machine gives a reading 1,000-fold lower than the Berthold luminometer, with proportionately lower background readings. For this study, we have defined 1 relative light unit (RLU) as 1 Berthold unit, equivalent to 0.001 Turner Design unit. A calibration curve with M. smegmatis/pSMT1 confirmed linearity between the machines. The substrate, 0.1 ml of 1% n-decyl aldehyde (Sigma) in ethanol for bacterial luciferase or luciferin for firefly luciferase (3), was injected automatically at 0.1 ml per tube with a final volume of 1 ml in phosphate-buffered saline. Raw data were collected in duplicate or triplicate over 20 s, and the mean of these totals was calculated. Samples were diluted by 10- and 100-fold to minimize any reduction of luminescence (quenching) in viscous or opaque samples. Mean RLU readings and standard errors were calculated using KaleidaGraph (Abelbeck software).
CFU.
Serial dilutions were carried out, and 0.1-ml volumes were plated for three dilutions in duplicate and were incubated in bags at 37°C. The mean number of CFU per time point and standard errors were calculated with KaleidaGraph software.
In vitro infection model.
The J774A.1 (ATCC TIB-67) murine macrophage-like cell line was seeded overnight at 5 × 105 cells per well in NUNC 24-well plates at 37°C and 5% CO2 in Hi-Glucose Dulbecco’s modified Eagle’s medium (GIBCO BRL), supplemented with 10% heat-inactivated fetal calf serum, 5 mM glutamine, and 81 μg of nonessential amino acids (Sigma) per liter. All manipulations were carried out in a class II microbiological safety cabinet. Log-phase cultures of mycobacteria prepared from frozen stocks were washed three times in phosphate-buffered saline by centrifuging at 2,200 × g for 10 min. Bacteria were resuspended in tissue culture medium to obtain the required multiplicity of infection (MOI). Cells were infected by incubation with an MOI of between 1 and 5 bacteria per cell for 1 h at 37°C and then washed three times in Hanks balanced salt solution (GIBCO BRL). The total number of cell-associated mycobacteria was determined at the first time point, and remaining cells were treated with amikacin for 2 h at a bacteriocidal concentration of 200 μg ml−1 in Dulbecco’s modified Eagle’s medium to eliminate extracellular mycobacteria. Eukaryotic cells were lysed by the addition of 1 ml of sterile distilled water containing 0.1% Triton X-100 per well. Samples were taken from duplicate or triplicate wells at different time points over a 3-day period in medium containing a bacteriostatic concentration of amikacin (20 μg ml−1) to prevent growth of extracellular bacteria released through macrophage lysis. RLUs and CFU were determined at each time point.
Murine infection model.
Female C57BL/6 mice (Harlan), aged 6 to 8 weeks, were maintained at category III containment in isolators under negative pressure. Aliquots of M. tuberculosis H37Rv/pSMT1, stored in glycerol at −80°C, were used to provide an inoculum of approximately 5 × 105 CFU in sterile saline per animal, injected intravenously (i.v.) into the tail vein. All manipulations were carried out in class I safety cabinets. For experiments to evaluate immune status, mice were immunized subcutaneously at the base of the tail with 105 BCG (Montreal) cells, prepared from frozen aliquots, 8 weeks prior to challenge. Samples were taken (from three mice per group) at 24 h and 1 to 8 weeks postinfection. Three separate experiments were undertaken to confirm the reproducibility of this model. Organs were aseptically removed and weighed. Homogenates were prepared directly from lung, liver, and spleen in 2-ml aliquots of sterile water with a Seward stomacher in a class I cabinet. These homogenates were diluted at 10- or 100-fold for direct readings of luminescence and numbers of CFU. Readings were corrected to represent whole-organ totals.
RESULTS
Comparison of luciferase constructs in M. smegmatis.
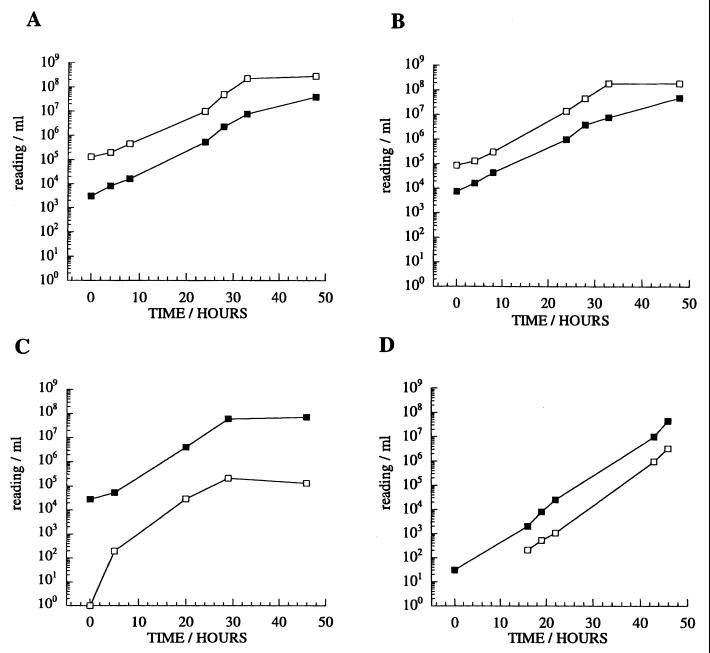
Plasmids were based on the mycobacterium-E. coli shuttle vector p16R1, and hygromycin was used for selection (12). Three luciferase enzymes were compared as reporters in recombinant mycobacteria. Bacterial luciferases from V. harveyi and X. luminescens are dimers, encoded by luxA and luxB genes, and utilize reduced flavin mononucleotide as a cofactor (18). Firefly luciferase is encoded by a single gene, luc, and utilizes ATP (16). Genes encoding each enzyme were cloned under the control of the M. tuberculosis hsp60 promoter and were tested for luminescence in the rapid-growing mycobacterium M. smegmatis (Fig. 2). The two bacterial enzymes generated similar levels of luminescence, corresponding to approximately 10 RLUs for each CFU. Luminescence generated by the firefly construct was approximately 100-fold lower than that observed from the bacterial enzymes in our system. In spite of a higher thermal stability, more appropriate to mycobacterial growth at 37°C (18), the X. luminescens luciferase showed no obvious advantage over the V. harveyi enzyme in terms of activity or stability. Bacterial luciferase (V. harveyi) was also introduced into M. smegmatis by using a vector (pLINT) capable of integration at the attB site on the mycobacterial chromosome. This construct showed lower luminescent activity, consistent with the reduced copy number of the gene in comparison to the plasmid construct, but again showed no obvious advantage in terms of stability (Fig. 2D). In fact, luciferase expression was remarkably stable, with no detectable plasmid loss after subculture or prolonged growth. However, attempts to construct a mycobacterial strain expressing the entire lux operon (encoding luciferase together with enzymes required for the production of endogenous substrate) (18) proved unstable in the mycobacteria species tested.
FIG. 2.
Luminescence activities of M. smegmatis strains containing luciferase constructs. Liquid cultures were monitored over a time course of 48 h. (A) pSMT1 (V. harveyi luxA and luxB), (B) pSMT5 (X. luminescens luxA and luxB), (C) pSMT6 (firefly luc), (D) pLINT560 (V. harveyi luxA and luxB integrated in the chromosome; in this case, the RLU value at time point 0 was below the detection limits [approximately 100 RLUs]). Open symbols, RLUs per milliliter; closed symbols, CFU per milliliter.
The V. harveyi luxA and luxB genes were also cloned in mycobacterial shuttle plasmids with promoter fragments from alternative genes. The results of the luciferase activity of these constructs in M. smegmatis are summarized in Table 1. In each case, luminescence was lower than that observed in the hsp60-driven pSMT1 construct. Based upon stability and high activity, pSMT1 was taken forward for further evaluation.
Luciferase activity in slow-growing mycobacteria.
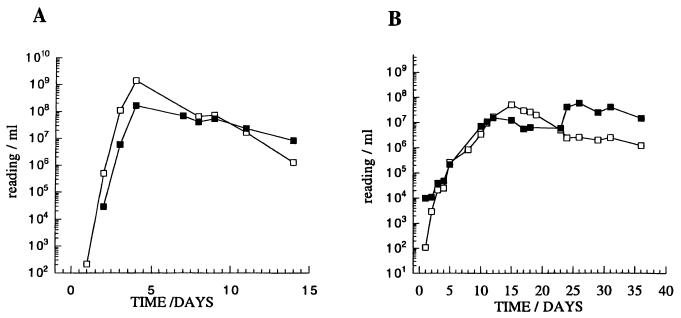
Expression from the pSMT1 construct in M. smegmatis is compared to expression from the slow-growing mycobacterial strain M. tuberculosis H37Rv in Fig. 3. A similar level of expression was observed in both organisms, with the ratio of RLUs to CFU maintained throughout the exponential phase of growth. In M. tuberculosis, a relative decline in RLUs was observed when the cultures reached stationary phase. A similar pattern was observed with BCG (data not shown). Reinoculation of stationary-phase cultures into fresh medium resulted in the rapid restoration of the original ratio of RLUs to CFU, demonstrating that the decline in RLUs is not caused by instability. Degradation of the luciferase enzyme, or a drop in the level of FMNH2 cofactor in stationary phase cells, could account for the reduced luminescence.
FIG. 3.
Comparison of luciferase expression in M. smegmatis (A) and M. tuberculosis H37Rv (B) strains containing the plasmid pSMT1. RLU and CFU readings were taken during log-phase and stationary-phase growth. Background readings obtained from wild-type mycobacteria and mycobacteria transfected with a control plasmid were routinely approximately 100 RLUs. Open symbols, RLUs per milliliter; closed symbols, CFU per milliliter.
Mycobacterial killing in a macrophage cell culture model.
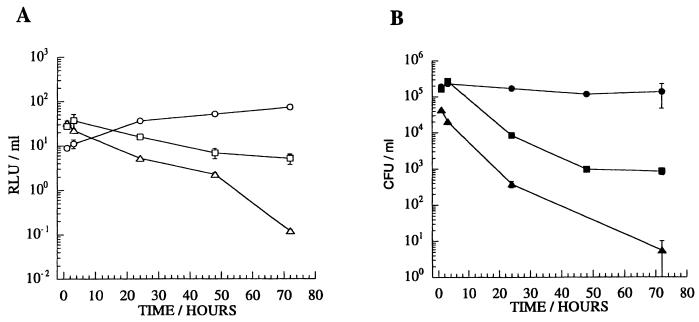
We evaluated the use of the luciferase reporter strains to monitor the mycobacteriocidal activity of the murine macrophage-like cell line J774A.1. In this model, macrophage monolayers were infected with mycobacteria at an approximate MOI of 1. After 1 h, cultures were washed, and fresh medium containing amikacin was added to prevent the growth of remaining extracellular mycobacteria. The fate of the mycobacteria was then followed over a period of 3 days by measuring luminescence and by counting CFU. Similar trends were observed with both measurements. The level of M. tuberculosis infection remained relatively stable and the level of BCG infection dropped slowly, but M. smegmatis was eliminated more rapidly (Fig. 4). However, the constant ratio of RLUs to CFU was not maintained during macrophage incubation; a relative increase in luminescence was observed in each case. This may be due to differences in the temporal relationship between the two parameters, whereby the ability to form colonies on plates declines in advance of a drop in luminescence. Thus, although changes in the number of CFU are reflected in changes in luminescence, the linear relationship between the two parameters observed in exponentially growing cultures is not necessarily retained when mycobacteria are being killed. Indeed, this may reflect the metabolic status of the population at the time of reading more accurately than does conventional CFU counting.
FIG. 4.
Mycobacterial infection of J774A.1 cells. Viability was determined by RLU (A) and CFU (B) readings. MOI was approximately 1. Circles, M. tuberculosis H37Rv/pSMT1; squares, BCG/pSMT1; triangles, M. smegmatis/pSMT1. Such infections generally yield between 1 and 10% infected cells, with approximately 3 to 5 bacteria per infected cell. The means and standard deviations (error bars) are shown for three wells at each time point.
Murine infection model.
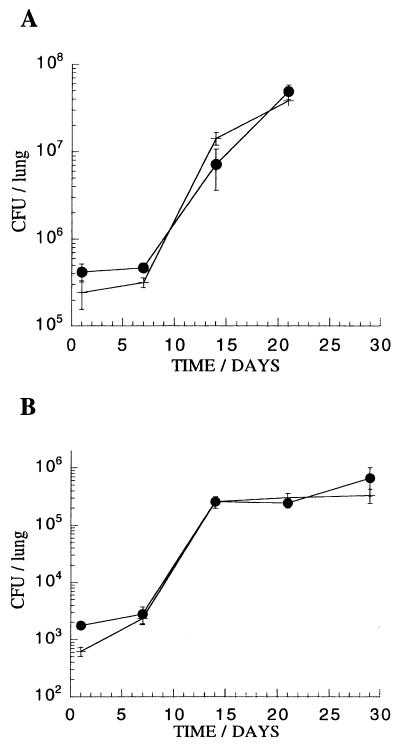
Evaluation of potential vaccine candidates currently relies on the measurement of their ability to protect against challenge with M. tuberculosis in murine or guinea pig models. To evaluate the use of luciferase reporter strains in assessing the immune status in intact animals, we have characterized the course of infection in C57BL/6 mice with a luminescent strain of M. tuberculosis H37Rv. A comparison of the in vivo growth rate of M. tuberculosis H37Rv/pSMT1 with M. tuberculosis H37Rv transformed with a vector control, pSMT3 (an identical plasmid without the luxAB genes), was undertaken (Fig. 5). The course of infection was observed over a period of 4 weeks by conventional monitoring of CFU levels. In this experiment, freshly grown log-phase cultures of M. tuberculosis were employed for the i.v. challenge, leading to higher bacterial loads than were seen with our infections using frozen stocks.
FIG. 5.
Comparison of growth rate in C57BL/6 mice of M. tuberculosis H37Rv/pSMT1 (closed circles) with M. tuberculosis H37Rv/pSMT3, a control plasmid without the luciferase luxAB genes (cross). (A) High inoculum (3.4 × 105 and 3.6 × 105 bacteria, respectively, per mouse) and (B) low inoculum (3.4 × 103 and 3.6 × 103 bacteria, respectively, per mouse) following i.v. challenge. The means and standard deviations (error bars) are shown for groups of three mice at each time point.
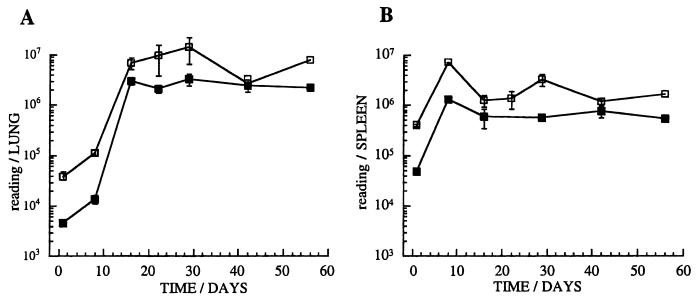
Figure 6 illustrates the measurement of luminescence in crude organ homogenates, compared to the quantification of CFU following infection with 5 × 105 CFU per mouse, over 8 weeks. A close correlation was observed between luminescence and CFU early in the infection. In spite of the absence of antibiotic selection in the mice, the pSMT1 plasmid was stably maintained, with more than 90% of the colonies isolated from infected tissues retaining antibiotic resistance (data not shown). In long-term infection the RLU to CFU ratio fluctuates, again possibly indicating a differing metabolic state of the bacterial population. Interestingly, the marked reduction in the ratio of RLUs to CFU characteristic of stationary-phase cultures was not observed during the chronic murine infection.
FIG. 6.
A murine model of infection with luminescent M. tuberculosis. C57BL/6 mice were infected with 5 × 105 M. tuberculosis H37Rv/pSMT1 bacteria. The relationship between RLUs (open squares) and CFU (closed squares) is illustrated for homogenates of lung (A) and spleen (B) in this experiment (three mice per group). RLU readings of uninfected splenocytes prepared in the same way showed background luminescence (100 RLUs). The means and standard deviations (error bars) are shown for groups of three mice at each time point.
Vaccination model.
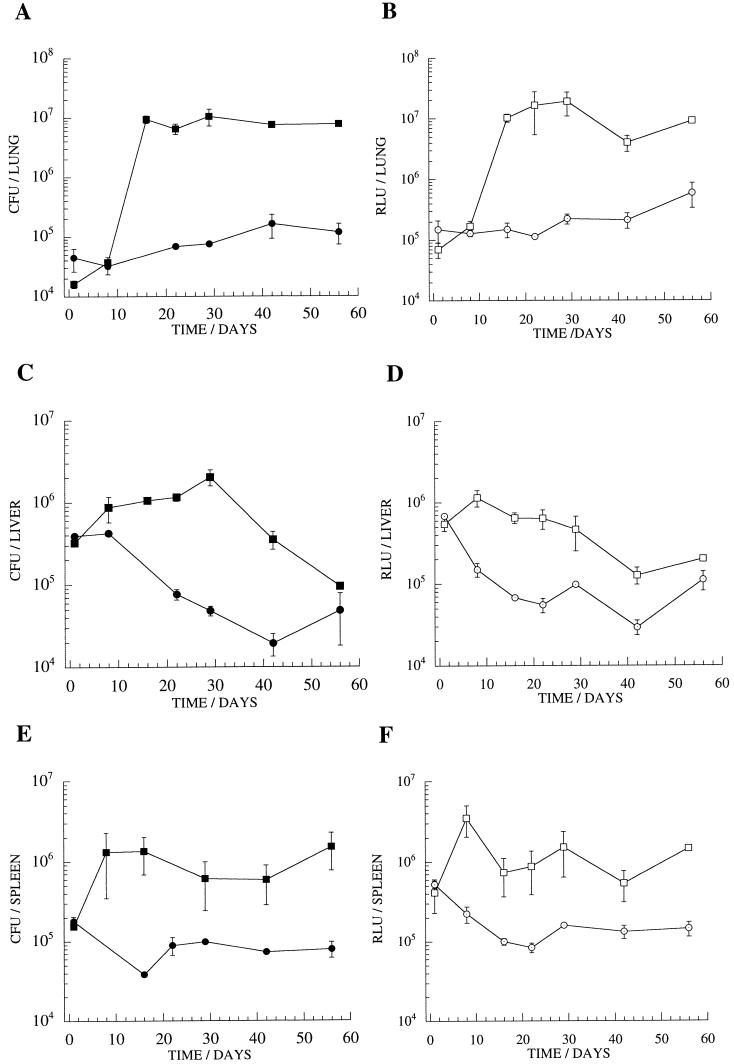
BCG-vaccinated mice were challenged in order to assess the capacity to differentiate between naive and vaccinated animals with the M. tuberculosis luciferase reporter strain. A clear difference was observed, with 10- to 100-fold lower luminescence detected in lung homogenates prepared from immunized mice 3 to 4 weeks after challenge (Fig. 7). An approximate reduction in luminescence of 1 log unit was also detectable in liver and spleen homogenates over the 2-month time course, corresponding to a drop in the number of CFU. The effect of BCG vaccination, and the correlation between RLU and CFU measurements, was also observed in mice receiving a low-dose i.v. challenge with M. tuberculosis/pSMT1 (3.5 × 103 bacteria per mouse) and in guinea pigs receiving a low-dose aerosol challenge (29a). Again, there was no evidence of any instability of the reporter construct under these conditions.
FIG. 7.
A vaccination model of murine infection with luminescent M. tuberculosis. The course of M. tuberculosis infection was monitored by measurements of CFU and RLUs in homogenates of lung (A and B), liver (C and D), and spleen (E and F) of mice immunized by BCG vaccination (circles) and of naive control mice (squares). The means and standard deviations (error bars) are shown for groups of three mice at each time point.
DISCUSSION
Measurement of luminescence is sensitive, stable, and simple to carry out, and a linear relationship generally exists between mycobacterial viability (measured in CFU) and bioluminescence in M. smegmatis, BCG, and M. tuberculosis. The utility of luminescence as a measure of viable bacteria has been confirmed during infection and killing of mycobacteria both within a macrophage-like cell line and a murine model of infection. We have employed this reporter construct to assess the effect of BCG vaccination in a murine model of M. tuberculosis infection monitored via luminescence. Approximate reductions in bacterial load of 1 to 2 log units may be observed in vaccinated animals compared to controls.
Our initial studies have focused on the pSMT1 reporter construct, selected on the basis of its strong luminescence and stability. Luciferase expression in this construct is under the control of the promoter region from the 65-kDa mycobacteria heat shock protein (HSP60). This promoter is expressed constitutively at a high level in M. tuberculosis, with a further induction in response to heat shock (28). However, in agreement with previous publications, we have not observed any significant stress-induced changes in expression when this promoter is carried on multicopy plasmid constructs. Reporter strains in which luciferase expression is placed under the control of differentially expressed promoter sequences may provide more powerful tools for assessment of immune activity. Application of the luciferase reporter approach to monitor metabolic status may provide important insights into the physiology of these persisting bacteria.
The activity of potential tuberculosis vaccine candidates is evaluated in currently available animal models, although this activity is restricted by a slow turnaround time and by the need for prolonged use of containment facilities. In this study, the possible application of reporter mycobacteria for assessment of immune status has been evaluated in a murine model. This technique has the potential for adaptation to high-throughput screens for vaccine candidates, as well as being more rapid than conventional CFU counting. Elucidation of the entire complement of almost 4,000 genes in M. tuberculosis (6) provides the foundation for a whole-genome approach to the identification of protective antigens from mycobacteria by using techniques such as DNA vaccination (4, 15, 29). In addition, the recent development of improved techniques for mutagenesis of slow-growing mycobacteria will allow the generation of a broad panel of novel, live, attenuated tuberculosis vaccine candidates (23). The potential for exploitation of these new opportunities in vaccine development will be significantly enhanced by the simplification of initial screening in animal models using the luciferase reporter strain. Demonstration of the sensitivity and stability of a virulent M. tuberculosis reporter strain in the present study represents a significant advance on the previous demonstration of resistance to a luciferase reporter strain in a short-term BCG challenge model (13).
From the liquid culture and cell culture systems, it appears that luciferase not only provides rapid enumeration of viable bacteria but may also indicate the metabolic status of the bacterial population. In the murine infection model described here, under the conditions used to infect C57BL/6 mice, a proportion of mycobacteria remain viable up to 8 weeks postchallenge with M. tuberculosis. These bacteria are detectable by luminescence, suggesting that they maintain metabolic activity. Persisting bacilli may result in a fatal infection of older animals (21), and ideally, any novel vaccine or therapy would act upon this population. This system may thus provide an inexpensive and rapid method, providing a primary screen to identify candidate antigens relevant to acute or persistent infection for further characterization.
Vaccine activity in animal models, while providing useful preliminary information, may prove an unreliable guide to efficacy in humans. The process of vaccine development would be significantly enhanced by the availability of in vitro assays for measuring immune status. The results presented here are encouraging in that they demonstrate that measurement of luciferase activity can provide a rapid and technically simple assay that provides a reliable assessment of mycobacterial viability in a range of biological systems from defined bacterial culture to intact animal models. We are also developing a splenocyte model, intended to reproduce the protection conferred by BCG vaccination of whole animals in an in vitro system. Such an in vitro screen may provide an effective correlate of protective immunity in humans. We are currently using a similar approach to evaluate the survival of a luminescent BCG reporter strain in blood cultures as a potential correlate for immune protection in humans following either BCG vaccination or exposure to M. tuberculosis.
ACKNOWLEDGMENTS
Valerie Snewin and Marie-Pierre Gares contributed equally to this study and should be considered as joint first authors.
Karen Bunting constructed plasmid pSMTCZS. We thank Peter Andrew for kindly providing us with plasmid pPA3, Kenneth Nealson for providing plasmid pCGLS1, and David Ow for providing plasmid pED32A. We are grateful to all the staff of the Huggett laboratory.
This work was supported by the Wellcome Trust.
REFERENCES
- 1.Andrew P W, Roberts I S. Construction of a bioluminescent mycobacterium and its use for assay of antimycobacterial agents. J Clin Microbiol. 1993;31:2251–2254. doi: 10.1128/jcm.31.9.2251-2254.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arain T M, Resconi A E, Hickey M J, Stover K. Bioluminescence screening in vitro (Bio-siv) assays for high-volume antimycobacterial drug discovery. Antimicrob Agents Chemother. 1996;40:1536–1541. doi: 10.1128/aac.40.6.1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arain T M, Resconi A E, Singh D C, Stover K. Reporter gene technology to assess activity of antimycobacterial agents in macrophages. Antimicrob Agents Chemother. 1996;40:1542–1544. doi: 10.1128/aac.40.6.1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barry M A, Lai W C, Johnston S A. Protection against mycoplasma infection using expression-library immunization. Nature. 1995;377:632–635. doi: 10.1038/377632a0. [DOI] [PubMed] [Google Scholar]
- 5.Carriere C, Riska P, Zimhony O, Kriakov J, Bardarov S, Burns J, Chan J, Jacobs W R., Jr Conditionally replicating luciferase reporter phages: improved sensitivity for rapid detection and assessment of drug susceptibility of Mycobacterium tuberculosis. J Clin Microbiol. 1997;35:3232–3239. doi: 10.1128/jcm.35.12.3232-3239.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S V, Eiglmeier K, Gas S, Barry III C E, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Quail M A, Rajandream M-A, Rogers J, Rutter S, Seeger K, Skelton J, Squares R, Squares S, Sulston J E, Taylor K, Whitehead S, Barrell B G. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1988;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 7.Cooper A, Dalton D, Stewart T, Griffin J, Russell D, Orme I. Disseminated tuberculosis in interferon gamma gene-disrupted mice. J Exp Med. 1993;178:2243–2247. doi: 10.1084/jem.178.6.2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dale E C, Ow D W. Intra- and intermolecular site-specific recombination in plant cells mediated by bacteriophage P1 recombinase. Gene. 1990;91:79–85. doi: 10.1016/0378-1119(90)90165-n. [DOI] [PubMed] [Google Scholar]
- 9.Davis P D O, editor. Clinical tuberculosis. 2nd ed. London, England: Chapman & Hall Medical; 1998. [Google Scholar]
- 10.Flynn J, Goldstein K L, Triebold K, Koller B, Bloom B. Major histocompatibility complex class I-restricted T cells are required for resistance to Mycobacterium tuberculosis infection. Proc Natl Acad Sci USA. 1992;89:12013–12017. doi: 10.1073/pnas.89.24.12013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frackman S, Anhalt M, Nealson K H. Cloning, organization, and expression of the bioluminescence genes of Xenorhabdus luminescens. J Bacteriol. 1990;172:5767–5773. doi: 10.1128/jb.172.10.5767-5773.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garbe T R, Barathi J, Barnini S, Zhang Y, Abou-Zeid C, Tang D, Mukherjee R, Young D B. Transformation of mycobacterial species using hygromycin resistance as selectable marker. Microbiology. 1994;140:133–138. doi: 10.1099/13500872-140-1-133. [DOI] [PubMed] [Google Scholar]
- 13.Hickey M A, Arain T M, Shawar R M, Humble D J, Langhorne M H, Morgenroth J N, Stover C K. Luciferase in vivo expression technology: use of recombinant mycobacterial reporter strains to evaluate antimycobacterial activity in mice. Antimicrob Agents Chemother. 1996;40:400–407. doi: 10.1128/aac.40.2.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hill P, Stewart G. Use of lux genes in applied biochemistry. J Biolumin Chemilumin. 1994;9:211–215. doi: 10.1002/bio.1170090315. [DOI] [PubMed] [Google Scholar]
- 15.Huygen K, Content J, Denis O, Montgomery D, Yawman A, Deck R, DeWitt C, Orme I, Baldwin S, D’Souza C, Drowart A, Lozes E, Vandenbussche P, Van Vooren J, Liu M, Ulmer J. Immunogenicity and protective efficacy of a tuberculosis DNA vaccine. Nat Med. 1996;2:893–898. doi: 10.1038/nm0896-893. [DOI] [PubMed] [Google Scholar]
- 16.Jacobs W, Jr, Barletta R G, Udani R, Chan J, Kalkut G, Sosne G, Kieser T, Sarkis G, Hatfull G, Bloom B. Rapid assessment of drug susceptibilities of Mycobacterium tuberculosis by means of luciferase reporter phages. Science. 1993;260:819–822. doi: 10.1126/science.8484123. [DOI] [PubMed] [Google Scholar]
- 17.Janis E, Kaufmann S, Schwartz R, Pardoll D. Activation of gamma delta T cells in the primary immune response to Mycobacterium tuberculosis. Science. 1989;244:713–716. doi: 10.1126/science.2524098. [DOI] [PubMed] [Google Scholar]
- 18.Meighen E. Molecular biology of bacterial bioluminescence. Microbiol Rev. 1991;55:123–142. doi: 10.1128/mr.55.1.123-142.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Newport M J, Huxley C M, Huston S, Hawrylowicz C M, Oostra B A, Williamson R, Levine M. A mutation in the interferon-gamma-receptor gene and susceptibility to mycobacterial infection. N Engl J Med. 1996;335:1941–1949. doi: 10.1056/NEJM199612263352602. [DOI] [PubMed] [Google Scholar]
- 20.Ó Gaora P, Barnini S, Hayward C, Filley E, Rook G, Young D, Thole J. Mycobacteria as immunogens: development of expression vectors for use in multiple mycobacterial species. Med Princ Pract. 1997;6:91–96. [Google Scholar]
- 21.Orme I. Aging and immunity to tuberculosis: increased susceptibility of old mice reflects a decreased capacity to generate mediator T lymphocytes. J Immunol. 1987;138:4414–4418. [PubMed] [Google Scholar]
- 22.Pablos-Mendez A, Raviglione M, Laszlo A, Binkin N, Rieder H, Bustreo F, Cohn D, Lambregts-van Weezenbeek C, Kim S, Chaulet P, Nunn P T. Global surveillance for antituberculosis-drug resistance, 1994–1997. World Health Organization-International Union against Tuberculosis and Lung Disease Working Group on Anti-Tuberculosis Drug Resistance Surveillance. N Engl J Med. 1998;338:1641–1649. doi: 10.1056/NEJM199806043382301. [DOI] [PubMed] [Google Scholar]
- 23.Pelicic V, Jackson M, Reyrat J M, Jacobs W R, Jr, Gicquel B, Guilhot C. Efficient allelic exchange and transposon mutagenesis in Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 1997;94:10955–10960. doi: 10.1073/pnas.94.20.10955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raviglione M, Snider J D, Kochi K A. Global epidemiology of tuberculosis: morbidity and mortality of a worldwide epidemic. JAMA. 1995;273:220–226. [PubMed] [Google Scholar]
- 25.Riska P, Jacobs W R, Jr, Bloom B, McKitrick J, Chan J. Specific identification of Mycobacterium tuberculosis with the luciferase reporter mycobacteriophage: use of p-nitro-alpha-acetylamino-beta-hydroxy propiophenone. J Clin Microbiol. 1997;35:3225–3231. doi: 10.1128/jcm.35.12.3225-3231.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rook G, Bloom B. Mechanisms of pathogenesis in tuberculosis. In: Bloom B, editor. Tuberculosis: pathogenesis, protection and control. Washington, D.C: ASM Press; 1994. pp. 485–501. [Google Scholar]
- 27.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 28.Stover C K, de la Cruz V F, Fuerst T R, Burlein J E, Benson L A, Bennett L T, Bansal G P, Young J F, Lee M H, Hatfull G F, Snapper S B, Barletta R G, Jacobs W R, Jr, Bloom B R. New use of BCG for recombinant vaccines. Nature. 1991;351:456–460. doi: 10.1038/351456a0. [DOI] [PubMed] [Google Scholar]
- 29.Tascon R E, Colston M J, Ragno S, Stavropoulos E, Gregory D, Lowrie D B. Vaccination against tuberculosis by DNA injection. Nat Med. 1996;2:888–892. doi: 10.1038/nm0896-888. [DOI] [PubMed] [Google Scholar]
- 29a.Williams, A., V. A. Snewin, I. N. Brown, and D. B. Young. Unpublished data.
- 30.Zhang Y, Lathigra R, Garbe T, Catty D, Young D. Genetic analysis of superoxide dismutase, the 23 kilodalton antigen of Mycobacterium tuberculosis. Mol Microbiol. 1991;5:381–391. doi: 10.1111/j.1365-2958.1991.tb02120.x. [DOI] [PubMed] [Google Scholar]