Graphical abstract
Keywords: Lipid nanoparticle, Analytical ultracentrifugation, Dynamic light scattering, Nanoparticle density, Particle size distribution, Vaccines, Flotation
Abstract
During the SARS-CoV2 pandemic mRNA vaccines in the form of lipid nanoparticles (LNPs) containing the mRNA, have set the stage for a new area of vaccines. Analytical methods to quantify changes in size and structure of LNPs are crucial, as changes in these parameters could have implications for potency. We investigated the application of sedimentation velocity analytical ultracentrifugation (SV-AUC) as quantitative stability-indicating method to detect structural changes of mRNA-LNP vaccines upon relevant stress factors (freeze/thaw, heat and mechanical stress), in comparison to qualitative dynamic light scattering (DLS) analysis. DLS was capable to qualitatively determine size and homogeneity of mRNA-LNPs with sufficient precision. Stress factors, in particular freeze/thaw and mechanical stress, led to increased particle size and content of larger species in DLS and SV-AUC. Changes upon heat stress at 50 °C were only detected as increased flotation rates by SV-AUC. In addition, SV-AUC was able to observe changes in particle density, which cannot be detected by DLS. In conclusion, SV-AUC can be used as a highly valuable quantitative stability-indicating method for characterization of LNPs.
1. Introduction
New types of vaccines based on mRNA technology were developed and approved during the recent SARS-CoV2 pandemic. These vaccines induce the expression of the antigen by the human body itself. Thus, the immune system responds without direct injection of the antigen or parts of it. The new approach requires that the mRNA reaches the cytoplasm, where the translation and production of the antigenic SARS-CoV2 spike protein is performed by the ribosomes. Major hurdles are the fast degradation of the mRNA by ribonucleases (RNases) and the poor transfection to the target compartment [11]. These challenges can be tackled by encapsulation of the mRNA into lipid nanoparticles (LNPs) or other vectors. Comirnaty™ and SpikeVax™ were the first vaccines using LNPs as delivery strategy to get approved by the FDA [5]. Compared to traditional antigen-based vaccines the development time can be reduced for an existing LNP platform and process. In case of new virus strains [7] the mRNA sequence can be adapted, whereas the LNP strategy and the production process remain the same [4]. Size and homogeneity of LNPs are relevant parameters reporting about the product and its reproducible manufacturing [10]. Moreover, influences of LNP size on potency were observed in siRNA-containing LNPs in model systems [3]. Therefore, reliable methods to assess size and structural properties of LNPs are required. High resolution single particle methods, like cryogenic transmission electron microscopy (cryo-TEM), are capable of this but require highly sophisticated instruments and skilled experts, which make the routine application during drug product development challenging. Dynamic light scattering (DLS) is used for quality control for the Comirnaty™ vaccine [1]. DLS determines the hydrodynamic radius with good resolution for monodisperse samples but has limitations when characterizing individual species in polydisperse samples [14]. Small-angle X-ray scattering provides another option to investigate the size and structure of LNPs [13].
SV-AUC is a quantitative, first principal method, from which structural information can be derived on the size, density and shape of the molecule under investigation. SV-AUC is applied already for the structural investigation of nanoparticles [15] along other methods [16]. It can be used to analyze polypeptides (1–10 kDa), small proteins, mAbs and large proteins (100–300 kDa) up to virus-like particles, RNA, DNA and drug delivery systems (MDa – GDa range). Regarding lipid delivery systems, drug loading of LNPs [6] and drug distribution of doxorubicin loaded liposomes in human serum have been investigated by using SV-AUC [8].
Our aim was to compare SV-AUC and DLS with respect to their capability to determine changes in structural properties based on LNP’s mass, shape or density after exposure to stress by freeze/thawing, thermal and mechanical stress.
2. Materials & methods
2.1. Sample preparation
Commercially available, recently expired (at most 2 months after expiry date) Comirnaty concentrate from Pfizer/BioNTech (1.5 mRNA mg/mL, batch 34397 TB) was used and diluted by adding 1.8 mL 0.9% NaCl as defined by the manufacturer, followed by a 10.5-fold dilution with phosphate buffered saline (10 mM HPO4 2−/H2PO4 −, 137 mM NaCl, 2.7 mM KCl, pH 7.4) to the final samples (referred to as Untreated Samples (UT)). UV-spectra from 220 to 350 nm were recorded for confirmation on a NanoDrop One device (Thermo Scientific, Waltham).
Untreated samples (UT) were continuously stored at 5 °C in 1.5-mL reaction tubes. For stressing, the diluted samples were transferred into 1.5-mL reaction tubes.
For mechanical stress, samples were vortexed 10 times for 10 s at maximum speed (VWR International Vortex, Radnor). Untreated samples and mechanical stressed samples were prepared and analyzed with n = 6.
For heat-stress the samples were exposed to 30 °C, 50 °C or 80 °C for 4 h by using an Eppendorf Thermomixer. For freeze/thaw stress, samples were exposed to three cycles of freezing in a −80 °C freezer (Thermo Scientific, Waltham) for 60 min and thawing at room temperature for 30 min. In these cases, all the samples were prepared with n = 4. After exposure to stress, each individual sample was analyzed by SV-AUC and DLS.
2.2. Dynamic light scattering (DLS)
DLS was performed with a Wyatt DynaPro Plate Reader III (Wyatt Technology Corp., Santa Barbara). 35 µL samples were filled into 384-Black COP wells in a clear film bottom well plate (Aurora Microplates, Scottsdale) and measured in triplicates at 25 °C with five acquisitions for 5 s.
Polystyrene beads with a diameter of 20 nm were used as reference standard (Thermo Fisher, Waltham). Results were exported with the Dynamics software package (Wyatt Technology Corp., Santa Barbara).
For analysis the main peak was evaluated based on predefined ranges with range intervals between 0 and 100 nm, 100 nm and 500 nm and larger ranges. Data for the main peak are taken from the range that contains the main peak. In cases where correlation functions suggest very high polydispersity (PDI > 0.4) the samples were classified as “Mulitmodal” by the Dynamics software.
2.3. Sedimentation velocity – Analytical ultracentrifugation (SV-AUC)
SV-AUC experiments were performed with an Optima AUC using an AN-50 Ti rotor and double sector centerpiece cells with sapphire windows (all Beckman Coulter). Measurements were performed at 10,000 rpm with absorption optics at 260 nm and 280 nm, scanning time of roughly 120 s and data resolution of 10 µm at 20 °C.
Data analysis was performed in UltraScan III employing the first 95 scans. Sedimentation velocity experiments were processed by using the two-dimensional spectrum analysis (2DSA) and time-invariant and radial-invariant noises were fit together with the bottom position [2]. This approach provides concentration distributions for independent sedimentation and diffusion coefficients of each solute in the mixture. To model the flotation process, a density value lower than buffer density was attributed to the LNPs.
For the final fitting, the Parametrically Constrained Spectrum Analysis was used employing a straight-line correlation in combination with a Monte Carlo (PCSA-SL-MC) simulation. Quantification of selected sedimentation coefficient ranges was performed based on a bin set, ranging from −1 S to −200 S (Main species), −200 S to −1200 S (HMWS1) and −1200 S to −2000 S (HMWS2). For these ranges, the weight averaged sedimentation coefficient, the frictional ratio (f/f0) and the content percentages were determined.
Under conditions, where density and shape factors of the LNP are well known, a conversion to size ranges might be helpful for comparison between the DLS and SV-AUC output. As our knowledge on these parameters is limited, we refrained from converting the well-reproducible, first principal sedimentation coefficient distribution, which does not require any further assumptions into the size or mass distribution, which requires assumptions regarding the density and mass of the LNP.
3. Results & discussion
3.1. Dynamic light scattering of mRNA containing LNPs
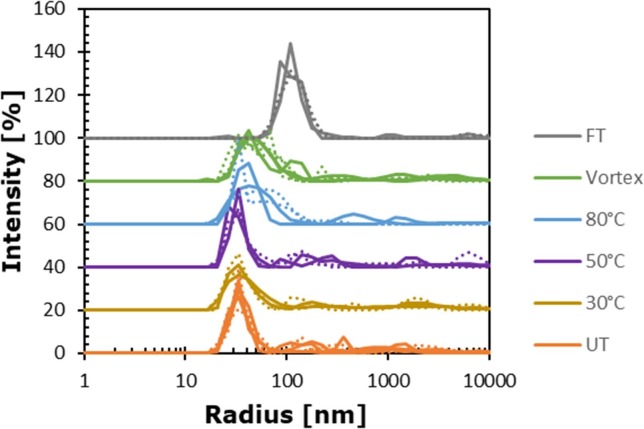
In DLS the average hydrodynamic radius of LNPs in the untreated vaccine sample was ∼66 nm with a multimodal distribution (PDI > 0.4). The size distribution by intensity (Fig. 1 ) provided information about the main mRNA-LNP species and potential additional low molecular weight species (LMWS), as well as high molecular weight species (HMWS). The main peak was detected at 36 ± 2 nm (relative intensity of about 70%) and in addition larger species between 100 nm and the upper measurement range were observed. This is in line with previous reports on LNPs investigated by DLS in a cuvette-based system [12], with the HMWS species being slightly elevated in our study.
Fig. 1.
Average DLS size distribution of UT samples (orange), 30 °C heat stress samples (ocher), 50 °C heat stress samples (purple), 80 °C heat stress samples (blue), mechanical stressed samples (green) and freeze/thaw stressed samples (grey). Dotted and solid lines represent different runs. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
To assess the stability-indicating properties of DLS we exposed the LNPs to mechanical stress (vortexing), thermal stress and freeze/thaw stress. We observed a substantial shift of the main peak to >100 nm after freeze/thaw. With the size increase at the expense of the main species, the multimodal nature of the system became attenuated as reflected by the reduced PDI of ∼0.38. After mechanical stress (vortexing) the average hydrodynamic radius of the LNP main species, increased from ∼36 nm to ∼49 nm (Table 1 ). Similarly, heat-stress at 80 °C resulted in a main peak shift to ∼47 nm whereas treatment at 30 °C and 50 °C did not lead to a substantial change (Table 1).
Table 1.
Hydrodynamic diameter, polydispersity index (PDI), radius of main peak and relative intensity of main peak of untreated and stressed samples of Comirnaty vaccine determined by DLS.
| Sample | Hydrodynamic radius [nm] | PDI | Radius of main peak [nm] | Relative intensity of main peak [%] |
|---|---|---|---|---|
| Untreated | 66 ± 9 | Multimodal | 36 ± 2 | 70 ± 6 |
| 30 °C | 77 ± 16 | Multimodal | 37 ± 4 | 65 ± 7 |
| 50 °C | 76 ± 12 | Multimodal | 35 ± 4 | 65 ± 9 |
| 80 °C | 55 ± 8 | Multimodal | 47 ± 7 | 84 ± 6 |
| Freeze-thawed | 132 ± 3 | 0.38 ± 0.07** | 139 ± 28 | 75 ± 13 |
| Vortex | 71 ± 7 | Multimodal | 49 ± 5 | 77 ± 10 |
based on 5 out of 6 measurements, as one was regarded as “multimodal” and did not result in a numerical PDI. The main peak of “freeze-thawed” was included into the larger bin as defined in Materials and Methods.
Thus, DLS can be used to monitor changes in the size and size distribution of mRNA-LNPs which may occur upon stress exposure. DLS demonstrated that especially freeze/thaw cycles can affect the size of the LNPs.
3.2. SV-AUC analysis of mRNA-LNPs’ size and polydispersity
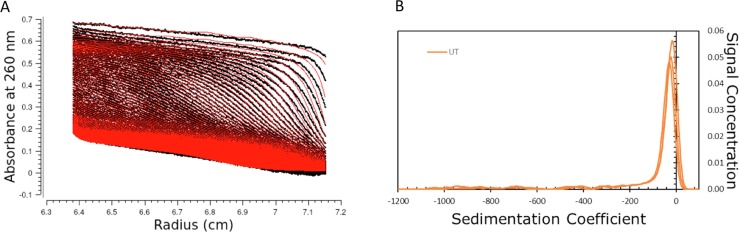
SV-AUC with UV detection at 260 nm and 280 nm was applied as an additional method to gather further information on the structural properties of mRNA-LNP samples and potential changes upon stress. Interestingly, the ComirnatyTM LNPs did not sediment in PBS, but floated and accumulated at the meniscus region of the solution column (Fig. 2 A). mRNA-LNP may sediment or float depending on their ratio of ionizable cationic lipid to nucleic acid phosphate [6]. The recorded flotation process could be modelled with UltraScan assuming an LNP density of 0.99 g/cm3 and a PBS density of 1.00567 g/cm3 at 20 °C. At both detection wavelengths the fitting process led to sufficiently low root-mean-square deviation values if sedimentation coefficient ranges between −1 and −2000 S were evaluated. The sedimentation boundary showed a main species plus additional species at higher sedimentation coefficient values. Integration of the fitted results with preset integration ranges showed that repeatable results can be obtained for the main species. In addition, two HMWS ranges were defined, which cover the entire sedimentation distribution range of the LNPs. When integrating over this distribution with defined ranges, a comparable species distribution with 83% LNP main species (s20,w of −24 ± 2.5 S) (Fig. 2B) and 17% HMWS resulted from both 260- and 280-nm data (Fig. 2A for 260 nm). Under the conditions employed in this experiment, we refrained from converting the sedimentation distribution into a size distribution due to insufficient information on density and shape of the LNPs, which is essential for the conversion. Nevertheless, based on the highly reproducible sedimentation coefficients, SV-AUC provides relevant and highly repeatable information on the polydispersity of mRNA-LNPs and on differences upon exposure to stress.
Fig. 2.
(A) SV-AUC sedimentation data of untreated sample (black lines) at 260 nm and their corresponding fit (red lines) generated by UltraScan III. (B) s20,w distribution of untreated samples (n = 3) at 260 nm plotted against the species concentrations. The dominant peak is reported as the main peak. Peaks left to the main peak represent faster floating species, peaks to the right slower moving particles, additional peaks indicate a polydisperse sample. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
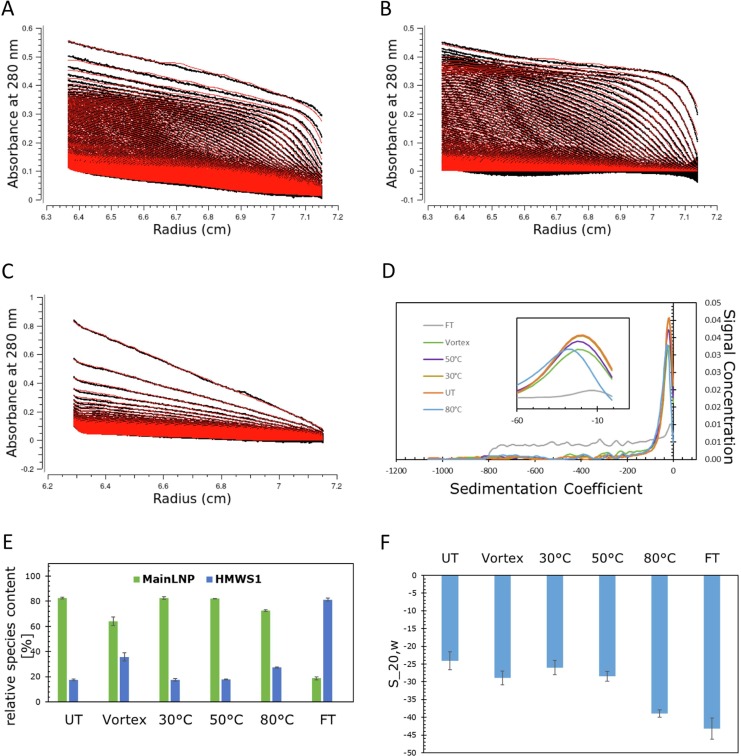
In line with DLS, the SV-AUC sedimentation process was visibly altered after mechanical and 80 °C heat stress (Fig. 3 A, 3B), while drastic changes could be observed in the freeze/thawed sample (Fig. 3C). These changes can be observed in the size distributions obtained from the fitting process (Fig. 3D), the species contents (Fig. 3E), and the sedimentation coefficients of the main species (Fig. 3F).
Fig. 3.
SV-AUC data for the mechanically (A), 80 °C (B) and freeze/thaw stressed sample (C) at 280 nm (black lines) and their corresponding fit (red lines) generated by UltraScan III. (D) S20,w distribution of untreated sample (orange), vortexed sample (green), 30 °C (ocher), 50 °C (purple), 80 °C (blue) heat stressed sample and freeze/thaw sample (grey) at 280 nm plotted against the species concentrations. (E) Average of the relative species content of the main peak (green), the HMWS1 (blue), and their standard deviation. (F) Average S20,w values of all samples and their associated standard deviation. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
In both 80 °C and mechanically stressed samples, the HMWS content was substantially increased compared to the untreated sample. In addition, the weight averaged sedimentation coefficients within this range (-1 S to -200 S) were decreased to −39 S and −29 S revealing species with a faster flotation process (Fig. 3F). This implies that either a size increase and/or a density decrease of the particles led to increased buoyancy. A slight increase in size was observed after these treatments in DLS measurements. As in DLS, heat stress at 30 °C and 50 °C did not affect the relative species content obtained from SV-AUC at either wavelength (Fig. 3E). The sedimentation coefficient was unchanged at −25 S for the untreated and the 30 °C treated LNPs, but a small shift of the s20,w to −28 S was detected after 50 °C treatment. This may indicate a small change of the LNPs, which was not evident from the DLS measurements (Table 1, Fig. 1). Reflecting on the measurement range, DLS with its logarithmic scale is providing an overview over a broader size range, while SV-AUC is more focused on the range of the main product and potential subtle changes.
As already visible from the raw data (Fig. 3C) the species contents of the LNPs changed drastically after freeze/thaw stress with an increase in HMWS to 79% reflected by the broad sedimentation coefficient distribution with only 21% main species remaining at an s20,w reduced to −43 S. Whereas DLS indicated a shifted, but fairly narrow size distribution, the broad size distribution in SV-AUC revealed a much more pronounced polydispersity.
Thus, the dependency of the sedimentation process on the particle density allows to also compare structural properties beyond the size itself by SV-AUC, which is not accessible by DLS.
4. Conclusions
DLS and SV-AUC can detect changes in size and species distributions of mRNA-LNPs after exposure to different stress factors (heat, mechanical and freeze/thaw stress) in PBS buffer. Both methods provide similar information for heat and mechanically stressed samples. Changes in the LNPs after a 50 °C heat treatment could only be detected based on and their sedimentation properties in SV-AUC.
It should be mentioned that SV-AUC as method requires higher investment in equipment, as well as more time and expertise, as compared to DLS. However, SV-AUC provides more precise particle size distributions than DLS. Furthermore, a quantification of the HMWS is only possible by SV-AUC (DLS only provides qualitative results). In addition, the sedimentation process in SV-AUC allows to get information on the particle density of different mRNA-LNP formulations, which is not accessible by DLS, but can be very relevant to decide on LNP structural integrity. In conclusion, SV-AUC can provide a more detailed, characterization of the mRNA-LNPs, which can support improving stability and quality of the resulting drug products.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Data availability
Data will be made available on request.
References
- 1.BioNTech Comirnaty, INN-COVID-19 mRNA Vaccine (nucleoside-modified) (europa.eu), 2021 [DATASET].
- 2.Brookes E., Cao W., Demeler B. A two-dimensional spectrum analysis for sedimentation velocity experiments of mixtures with heterogeneity in molecular weight and shape. Eur. Biophys. J. 2010;39(3):405–414. doi: 10.1007/s00249-009-0413-5. [DOI] [PubMed] [Google Scholar]
- 3.Chen S., Tam Y.Y.C., Lin P.J.C., Sung M.M.H., Tam Y.K., Cullis P.R. Influence of particle size on the in vivo potency of lipid nanoparticle formulations of siRNA. J. Control Release. 2016;235:236–244. doi: 10.1016/j.jconrel.2016.05.059. [DOI] [PubMed] [Google Scholar]
- 4.Corbett K.S., Edwards D.K., Leist S.R., Abiona O.M., Boyoglu-Barnum S., Gillespie R.A., Himansu S., Schäfer A., Ziwawo C.T., DiPiazza A.T., Dinnon K.H., Elbashir S.M., Shaw C.A., Woods A., Fritch E.J., Martinez D.R., Bock K.W., Minai M., Nagata B.M., Hutchinson G.B., Wu K., Henry C., Bahl K., Garcia-Dominguez D., Ma LingZhi, Renzi I., Kong W.-P., Schmidt S.D., Wang L., Zhang Y.i., Phung E., Chang L.A., Loomis R.J., Altaras N.E., Narayanan E., Metkar M., Presnyak V., Liu C., Louder M.K., Shi W., Leung K., Yang E.S., West A., Gully K.L., Stevens L.J., Wang N., Wrapp D., Doria-Rose N.A., Stewart-Jones G., Bennett H., Alvarado G.S., Nason M.C., Ruckwardt T.J., McLellan J.S., Denison M.R., Chappell J.D., Moore I.N., Morabito K.M., Mascola J.R., Baric R.S., Carfi A., Graham B.S. SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature. 2020;586(7830):567–571. doi: 10.1038/s41586-020-2622-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.FDA, Comirnaty and Pfizer-BioNTech COVID-19 vaccine, 2021.
- 6.Henrickson A., Kulkarni J.A., Zaifman J., Gorbet G.E., Cullis P.R., Demeler B. Density Matching Multi-wavelength Analytical Ultracentrifugation to Measure Drug Loading of Lipid Nanoparticle Formulations. ACS Nano. 2021;15(3):5068–5076. doi: 10.1021/acsnano.0c10069. [DOI] [PubMed] [Google Scholar]
- 7.Jackson N.A.C., Kester K.E., Casimiro D., Gurunathan S., DeRosa F. The promise of mRNA vaccines: a biotech and industrial perspective. NPJ Vaccines. 2020;5:11. doi: 10.1038/s41541-020-0159-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mehn D., Iavicoli P., Cabaleiro N., Borgos S.E., Caputo F., Geiss O., Calzolai L., Rossi F., Gilliland D. Analytical ultracentrifugation for analysis of doxorubicin loaded liposomes. Int. J. Pharm. 2017;523(1):320–326. doi: 10.1016/j.ijpharm.2017.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sato Y., Note Y., Maeki M., Kaji N., Baba Y., Tokeshi M., Harashima H. Elucidation of the physicochemical properties and potency of siRNA-loaded small-sized lipid nanoparticles for siRNA delivery. J. Control Release. 2016;229:48–57. doi: 10.1016/j.jconrel.2016.03.019. [DOI] [PubMed] [Google Scholar]
- 11.Schoenmaker L., Witzigmann D., Kulkarni J.A., Verbeke R., Kersten G., Jiskoot W., Crommelin D.J.A. mRNA-lipid nanoparticle COVID-19 vaccines: Structure and stability. Int. J. Pharm. 2021;601:120586. doi: 10.1016/j.ijpharm.2021.120586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Selmin F., Musazzi U.M., Franzè S., Scarpa E., Rizzello L., Procacci P., Minghetti P. Pre-Drawn Syringes of Comirnaty for an Efficient COVID-19 Mass Vaccination: Demonstration of Stability. Pharmaceutics. 2021;13(7):1029. doi: 10.3390/pharmaceutics13071029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Uebbing L., Ziller A., Siewert C., Schroer M.A., Blanchet C.E., Svergun D.I., Ramishetti S., Peer D., Sahin U., Haas H., Langguth P. Investigation of pH-Responsiveness inside Lipid Nanoparticles for Parenteral mRNA Application Using Small-Angle X-ray Scattering. Langmuir. 2020;36(44):13331–13341. doi: 10.1021/acs.langmuir.0c02446. [DOI] [PubMed] [Google Scholar]
- 14.Vezocnik V., Rebolj K., Sitar S., Ota K., Tusek-Znidaric M., Strus J., Sepcic K., Pahovnik D., Macek P., Zagar E. Size fractionation and size characterization of nanoemulsions of lipid droplets and large unilamellar lipid vesicles by asymmetric-flow field-flow fractionation/multi-angle light scattering and dynamic light scattering. J. Chromatogr. A. 2015;1418:185–191. doi: 10.1016/j.chroma.2015.09.048. [DOI] [PubMed] [Google Scholar]
- 15.X. Xu, H. Colfen, Ultracentrifugation Techniques for the Ordering of Nanoparticles. Nanomaterials (Basel) 11 (2021). [DOI] [PMC free article] [PubMed]
- 16.Yousefi S.R., Sobhani A., Alshamsi H.A., Salavati-Niasari M. Green sonochemical synthesis of BaDy2NiO5/Dy2O3 and BaDy2NiO5/NiO nanocomposites in the presence of core almond as a capping agent and their application as photocatalysts for the removal of organic dyes in water. RSC Adv. 2021;11:11500–11512. doi: 10.1039/d0ra10288a. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.