Abstract
Introduction:
Antimicrobial susceptibility is well characterized in monomicrobial infections, but bacterial species often coexist with other bacterial species. Antimicrobial susceptibility is often tested against single bacterial isolates; this approach ignores interactions between cohabiting bacteria that could impact susceptibility. Here, we use Pooled Antibiotic Susceptibility Testing to compare antimicrobial susceptibility patterns exhibited by polymicrobial and monomicrobial urine specimens obtained from patients with urinary tract infection symptoms.
Methods:
Urine samples were collected from patients who had symptoms consistent with a urinary tract infection. Multiplex polymerase chain reaction testing was performed to identify and quantify 31 bacterial species. Antibiotic susceptibility was determined using a novel Pooled Antibiotic Susceptibility Testing method. Antibiotic resistance rates in polymicrobial specimens were compared with those in monomicrobial infections. Using a logistic model, resistance rates were estimated when specific bacterial species were present. To assess interactions between pairs of bacteria, the predicted resistance rates were compared when a pair of bacterial species were present versus when just one bacterial species was present.
Results:
Urine specimens were collected from 3,124 patients with symptoms of urinary tract infection. Of these, multiplex polymerase chain reaction testing detected bacteria in 61.1% (1910) of specimens. Pooled Antibiotic Susceptibility Testing results were available for 70.8% (1352) of these positive specimens. Of these positive specimens, 43.9% (594) were monomicrobial, while 56.1% (758) were polymicrobial. The odds of resistance to ampicillin (p = 0.005), amoxicillin/clavulanate (p = 0.008), five different cephalosporins, vancomycin (p = <0.0001), and tetracycline (p = 0.010) increased with each additional species present in a polymicrobial specimen. In contrast, the odds of resistance to piperacillin/tazobactam decreased by 75% for each additional species present (95% CI 0.61, 0.94, p = 0.010). For one or more antibiotics tested, thirteen pairs of bacterial species exhibited statistically significant interactions compared with the expected resistance rate obtained with the Highest Single Agent Principle and Union Principle.
Conclusion:
Bacterial interactions in polymicrobial specimens can result in antimicrobial susceptibility patterns that are not detected when bacterial isolates are tested by themselves. Optimizing an effective treatment regimen for patients with polymicrobial infections may depend on accurate identification of the constituent species, as well as results obtained by Pooled Antibiotic Susceptibility Testing.
Keywords: Urinary Tract Infection, Antibiotic Susceptibility, Polymicrobial, Microbiome, Diagnostic Test, Microbiota, Clinical Microbiology, Bacterial Interactions, Statistical Models
Introduction
The standard of care for diagnosis of Urinary Tract Infections (UTIs) is a Standard Urine Culture (SUC) along with antimicrobial susceptibility testing, and has served to guide treatment since the early 1950s. The methodology relies on an “Escherichia coli (E. coli)-centric” view that perceives UTIs as caused by one or two pathogens[1]. Recent findings, however, reveal that SUC often misses the vast majority of urinary tract organisms and uropathogens[2]. This deficiency limits potentially valuable information that should be provided to the treating clinicians. The nomenclature and basic concepts of UTI are based on the now disproven dogma that the lower urinary tract is sterile. Even though the mere presence of an organism should not automatically prompt antibiotic treatment, clinicians will likely benefit from a more comprehensive report of organisms present in the urine samples from symptomatic patients.
With this clinical need in mind, we recently developed a Multiplex Polymerase Chain Reaction (M-PCR)-based urinary organism detection and analysis method. with primers and probes explicitly designed against 31 commonly identified UTI-associated bacteria species, the test not only identify positive organism(s) but also calculate each of their concentrations in the urine, and has a faster turnaround time than SUC[3].
In parallel, antibiotic resistance has been well-studied in monomicrobial infections, but is less characterized in polymicrobial infections. It has been reported that up to 39% of UTI infections are polymicrobial in nature [2–4]. Cological interactions are found to be potentially crucial for the growth and survival of bacteria in polymicrobial communities. Even if not all of the organisms are directly pathogenic and associated with UTI symptoms, the interactions among different species of bacteria can alter responses to antibiotics in polymicrobial settings, which may have potentially significant clinical impacts [5–8].
Current Antibiotic Susceptibility Testing (AST) ignores bacterial interactions: each bacterium is tested in isolation against an antibiotic, providing no opportunity to assess bacterial interactions. Ignoring bacterial interactions can either lead to potential treatment failure or prevent the use of efficacious antibiotics. Both scenarios can have serious clinical consequences. Here, we introduce a novel method called Pooled Antibiotic Susceptibility Testing (P-AST), (patent number 10,160,991), which involves simultaneously growing all detected bacteria together in the presence of antibiotics and then measuring susceptibility. Thus, P-AST considers interactions between cohabiting bacterial species. We obtained urine specimens from patients presenting with UTI-like symptoms to 37 urology clinics. First, we estimated the odds of resistance to 18 antibiotics or antibiotic combinations relative to increasing numbers of bacterial species in a specimen. We found that antimicrobial susceptibility patterns in polymicrobial specimens differed from those observed in monomicrobial specimens. Since standard of care relies on assessment of antibiotic susceptibility in monomicrobial infections, these findings show that P-AST could serve as a more accurate predictor of antibiotic susceptibility.
Methods
Patient participation and sample handling
This study combines data from two studies of antibiotic resistance patterns in elderly patients presenting with symptoms consistent with a UTI. Retrospective data and patient information (Western IRB number 20171870) were obtained from a single site (Comprehensive Urology, Royal Oak, MI) for 613 patients who presented between March and July 2018. Prospective data and patient information (Western IRB number 20181661) were obtained for 2,511 patients who presented at any of 37 geographically disparate clinics in the United States between July 2018 and February 2019. All subjects met the following inclusion and exclusion criteria. Inclusion criteria included: symptoms of acute cystitis, complicated UTI, persistent UTI, recurrent UTI, prostatitis, pyelonephritis, interstitial cystitis (at any age), symptoms of other conditions at ≥60 years of age, specimen volumes sufficient to permit urine culture and Multiplex Polymerase Chain Reaction (M-PCR) combined with Pooled Antibiotic Sensitivity Testing (P-AST), patient informed consented, documented times at which the specimens were collected and stabilized with boric acid in grey-top tubes. Exclusion criteria included prior participation in this study, antibiotics taken for any reason other than UTI at the time of enrollment, chronic (≥10 days) indwelling catheters, self-catheterization, and urinary diversion. Antibiotic susceptibility data were available for 1,352 of the 3,124 patients (43.3%).
DNA extraction and analysis
DNA extraction was performed using the KingFisher/MagMAX™ Automated DNA Extraction instrument and the MagMAX™ DNA Multi-Sample Ultra Kit (ThermoFisher, Carlsbad, CA). 400 μL of urine were transferred to 96-well deep-well plates, sealed, and centrifuged to concentrate the samples, and then the supernatant was removed. Enzyme Lysis Mix (220 μL/well) was added to the samples, which were then incubated for 20 min at 65°C. Proteinase K Mix (PK Mix) was added (50 μL/well) and incubated for 30 min at 65°C. Lysis buffer (125 μL/well) and DNA Binding Bead Mix (40 μL/well) were added, and the samples were vortexed for a minimum of 5 min. Each 96-well plate was loaded into the KingFisher/MagMAX Automated DNA Extraction instrument, which was operated in accordance with standard operating procedures.
DNA analysis was conducted using the Guidance® UTI Test (Pathnostics, Irvine, CA). . Samples were mixed with universal PCR master mix and amplified using TaqMan technology on the Life Technologies 12K Flex OpenArray System™ (Life Technologies, Carlsbad, CA). DNA samples were spotted in duplicate on 112-format OpenArray chips. Plasmids unique to each bacterial species being tested were used as positive controls. Candida tropicalis was used as an inhibition control. A data analysis tool developed by Pathnostics was used to sort data, assess the quality of data, summarize control sample data, identify positive assays, calculate concentrations, and generate draft reports. Probes and primers were used to detect the following bacteria through M-PCR: A. baumannii, A. schaalii, A. urinae, A. omnicolens, C. freundii, C. koseri, C. riegelii, K. aerogenes, E. faecalis, E. coli, K. oxytoca, K. pneumoniae, M. morganii, M. tuberculosis, M. genitalium, M. hominis, P. agglomerans, P. mirabilis, P. stuartii, P. aeruginosa, S. marcescens, S. aureus, S. agalactiae, and U. urealyticum. Probes and primers also were used to detect the following bacterial groups: Coagulase negative staphylococci (CoNS) (S. epidermidis, S. haemolyticus, S. lugdunensis, Staphylococcus saprophyticus); Viridans group streptococci (VGS) (S. anginosus, S. oralis, S. pasteuranus). The quantities of each of the detected bacterial species were determined using the standard curve method: first, standard curves of each of the bacterial species were generated from testing replicates of dilution series of known concentrations of each of the calibrator bacteria; constants necessary for the quantitation of each of the bacterial species in unknown samples, such as slope and intercept, were established from each of the standard curves; then PCR Ct values of a target bacteria species from an unknown sample were compared to the standard curve, and the concentration of the target bacteria species (cells/ul) present in an unknown sample was extrapolate and determined. Bacteria with quantity of ≥10,000 cells/mL were defined as positive, and bacteria with quantity < 10,000 cells/mL were defined as negative.
Pooled - Antibiotic Susceptibility Testing
P-AST was performed by aliquoting 1 mL of patient urine specimen into a 1.7 mL microcentrifuge tube. After centrifugation, the supernatant was aspirated and discarded, leaving approximately 500 μL of patient sample in the microcentrifuge tube. One mL of Mueller Hinton Growth Media was then aliquoted into the patient sample in the microcentrifuge tube and the tubes were incubated at 35°C in a non-CO2 incubator for 6 hours. Mueller Hinton Agar was used as a negative control. Those samples that reached a minimum threshold of 10,000 cells/mL were then diluted by aliquoting 0.5 mL of sample into a 50 mL conical tube containing Mueller Hinton Growth Media. 96-well plates pre-loaded with antibiotics were then inoculated with diluted samples and incubated along with control plates for 12–16 hours at 35°C in a single layer. Optical density of samples was then read on a DensiCHEK plate reader™ (BioMerieux, Marcy-l’Étoile, France).
AST is currently unavailable for P. agglomerans, U. urealyticum, A. schaalii, A. urinae, A. omnicolens, C. riegelii, M. hominis, and M. genitalium, M. tuberculosis, S. anginosus, U. urealyticum, and VGS, due to fastidious in vitro growth characteristics.
Statistical Methods
Logistic regression was used to compare resistance rates in monomicrobial and polymicrobial infections. Specifically,18 different logistic regression models were fit to the data: the response variable was an indicator of whether the specimen was resistant to the specific antibiotic or not and the predictor variable was an indicator of whether the infection was monomicrobial or polymicrobial. Specimens were classified as monomicrobial if a single bacterial species was detected above the 10000 cells/mL threshold; they were classified as polymicrobial if two or more distinct bacteria species were detected above that threshold. Similar logistic regression models also were run, using the number of distinct bacterial species as the predictor variable.
Interactions between pairs of bacterial species were investigated using a logistic regression model to predict resistance in the presence of specific bacterial species. In this model, the response was an indicator variable of resistance and the predictor variables were 16 indicators of the presence of the following bacterial species or groups that tested positive in at least 30 samples; these were A. schaalii, A. urinae, A. omnicolens, CoNS, C. riegelii, E. faecalis, E. coli, K. oxytoca, K. pneumoniae, M. morganii, P. mirabilis, P. aeruginosa, S. aureus, S. agalactiae, U. urealyticum, and VGS. A backward stepwise model selection was performed on the model with all main effects and all pairwise interactions using an enter significance level of α=0.10 and an exit significance level of α=0.05 to obtain the best fitting model. This model was used to predict resistance rates when a specific bacterial species was present or when a specific pair of species was present.
Using the logistic regression model described above, the resistance rate for a pair of species was compared to the resistance rates for each species alone. Two different principles were applied to calculate the expected resistance rate to a pair of species that do not interact: (a) Highest Single Agent Principle (HSAP) (a commonly used model for drug interactions) and (b) Union Principle (UP) (also used to model drug interactions).
Using the HSAP, a pair of species was considered to have an interaction if the resistance rate of the pair of species was statistically different from the highest resistance rate of each of the two species alone. This model is based on the idea that a pooled specimen containing two species will survive application of a specific antibiotic only if the more resistant species survives. When an antibiotic is applied to the pool, it may kill off species A, but if species B survives, the pool is called resistant.
The UP assumes a pair of bacteria (species A and B) is made up of one genetic variant of species A and one genetic variant of species B, and that the pool is resistant if either species A is resistant or if species B is resistant. If species A is resistant with probability P(A), and species B is resistant with probability P(B), then the probability of resistance of the pool is:
This assumes the two species do not interact, and therefore act independently.
Interactions were statistically tested using bootstrapped samples of the 1,352 patients with antibiotic resistance results; each patient was randomly selected with replacement. A logistic regression with terms in the best fitting model selected as above was fit to each bootstrapped sample. The predicted resistance when the pair of species was present was compared to the predicted resistance to each species alone using the model fit to the bootstrap sample (assuming either the HSAP or UP). 5000 bootstrapped samples were generated and analyzed. If 97.5% or greater of the bootstrapped samples demonstrated a pool resistance higher than expected, the interaction was deemed to show a statistically significant interaction with increased resistance. If 97.5% or greater of the bootstrapped samples demonstrated a pool resistance lower than expected, the interaction was deemed to show a statistically significant interaction with decreased resistance.
Since resistance testing is not generally performed for monomicrobial infections of the following bacterial species, interactions with these species or groups were not tested: P. agglomerans, U. urealyticum, A. schaalii, A. urinae, A. omnicolens, C. riegelii, M. hominis, and M. genitalium, M. tuberculosis, S. anginosus, U. urealyticum, and VGS.
Results
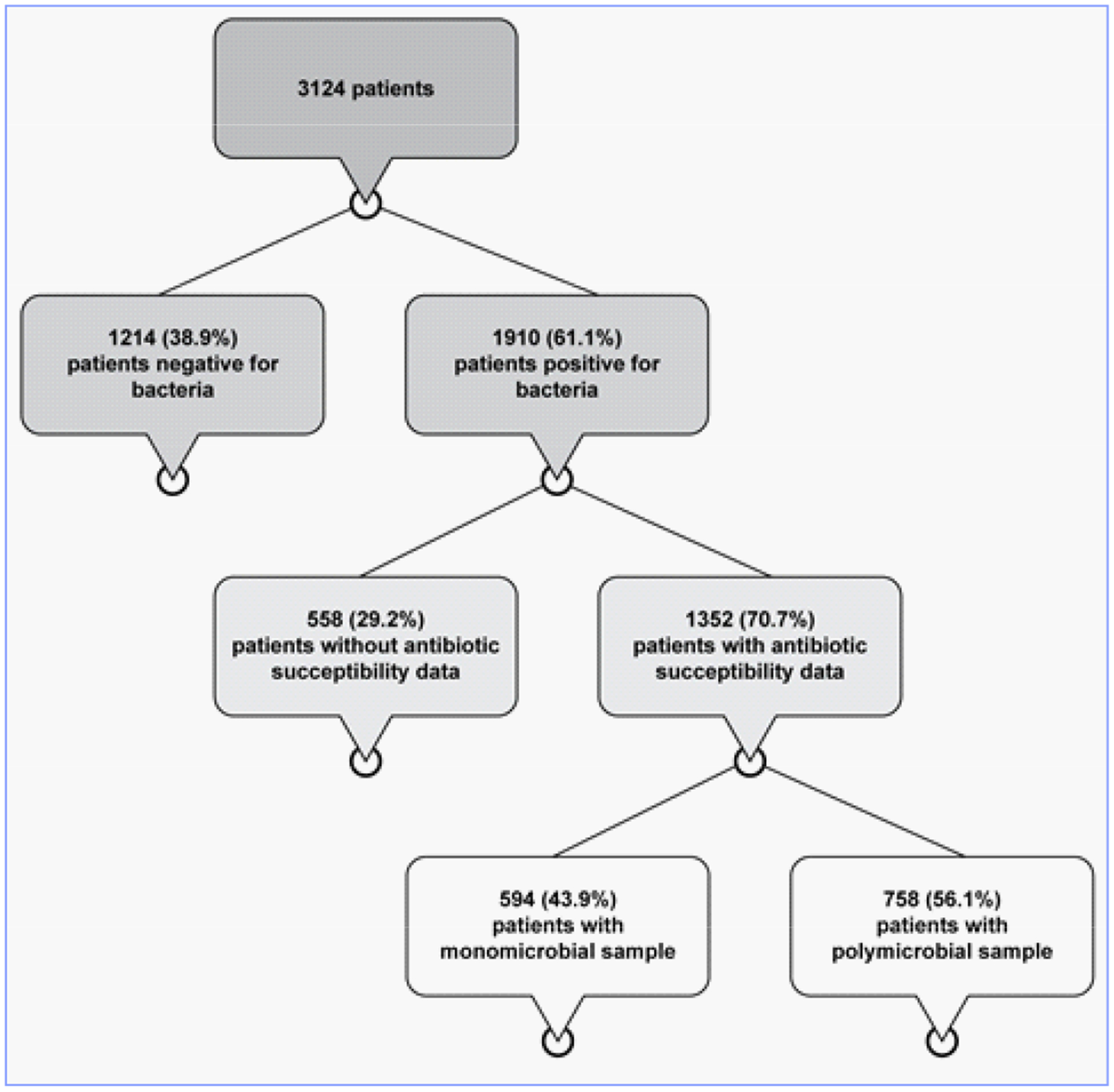
A total of 3,124 patients, from two studies and from 37 geographically disparate urology clinics in the United States, presenting with symptoms consistent with a UTI, were initially included in the study. P-AST data were available for 43.3% (1352) of these patients (Figure 1). The demographics and presenting symptoms for these 1352 patients are presented in Table 1. Their mean age was 75 years. Sixty-six percent (887/1,352) were female, whereas 34% (465/1,352) male.
Figure 1:
Flow chart of patient samples.
Table 1:
Demographics and symptoms.
| Parameter | All Subjects (N=1352) |
|---|---|
| Age (years) | |
| Mean (SD) | 74.9 (8.5) |
| Median | 75 |
| Min, Max | 31.0,100.0 |
| Sex – n (%) | |
| Male | 465 (34.4%) |
| Female | 887 (65.6%) |
| Baseline Symptoms – n (%) | |
| Burning sensation when urinating | 474 (35.1%) |
| Urinating incontinence | 459 (34.0%) |
| Cloudy or strong-smelting urine | 267 (19.8%) |
| Pain | 260 (19.2%) |
| - Abdominal | 119 (8.8%) |
| - Flank | 47 (3.5%) |
| - Lower back | 67 (5.0%) |
| - Base of penis or Behind scrotum | 7 (0.5%) |
| Pelvic discomfort | 161 (11.9%) |
| Lower grade fever | 23 (1.7%) |
| Frequency | 475 (35.1%) |
| Urgency | 515 (38.1%) |
| Nocturia | 456 (33.7%) |
| Baseline Atypical Symptoms – n (%) | |
| Acute change in mentation | 24 (1.8%) |
By M-PCR, 38.9% (1,214/3,124) of the specimens were negative for bacteria, whereas 61.1% (1,910/3,124) were positive. P-AST data were available for 1,352 (70.7%) of these 1,910 positive specimens,. Of these 1,352 specimens, 43.9% (594/1,352) were monomicrobial, whereas 56.1% (758/1,352) were polymicrobial.
Five hundred and fifty eight positive samples lacked antimicrobial susceptibility data for the following reasons:
no species were detected at ≥ 10,000 cells/mL (correlating to colony forming units/mL) and thus no species were tested against antibiotics;
the species detected by PCR were fastidious (i.e., they required specific growth conditions, extremely restrictive growth conditions, or extreme length of time in order to perform susceptibility testing);
prior antimicrobial use caused bacteria to fail to thrive in the P-AST assay; or
species were not identified because the M-PCR reaction was inhibited. M-PCR inhibition can occur when an interfering substance prevents the amplification and subsequent detection of the PCR product associated with targeted DNA.
Odds ratios of antibiotic resistance in polymicrobial versus monomicrobial specimens are shown in Table 2, along with the odds ratio of resistance for each increase in the number of bacterial species in polymicrobial specimens. The resistance rates of polymicrobial samples were generally higher than the rates of monomicrobial samples; 10 of 18 antibiotics or antibiotic combinations had statistically higher resistance rates for polymicrobial samples. The odds of resistance for each additional species identified in a polymicrobial specimen increased for ampicillin, amoxicillin/clavulanate, five of the six of the cephalosporins tested, vancomycin, and tetracycline. The opposite was true for piperacillin/tazobactam, where each additional species in a polymicrobial specimen resulted in a 75% decrease in the odds of resistance (95% CI 0.61, 0.94, p = 0.01).
Table 2:
Odds ratios of antibiotic resistance.
| Antibiotic | Odds Ratio of Resistance Polymicrobial v. Monomicrobial | p-value | Odds Ratio of Resistance for Each Additional Species | p-value |
|---|---|---|---|---|
| Penicillins | ||||
| Ampicillin | 1.37 (1.10, 1.70) | 0.005* | 1.14 (1.05, 1.24) | 0.001* |
| Combinations | ||||
| Amoxicilin/Clavulanate | 1.38 (1.09, 1.74) | 0.008* | 1.16 (1.07, 1.26) | 0.0005* |
| Ampicillin/Sulbactam | 1.13 (0.89, 1.42) | 0.32 | 1.05 (0.96, 1.14) | 0.30 |
| Trimethoprim/Sulfamethoxazole | 1.09 (0.88, l.37) | 0.43 | 0.98 (0.91, 1.07) | 0.69 |
| Piperacillin/TazobactBm | 0.69 (0.43, 1,11} | 0.12 | 0.75 (0.51, 0.94) | 0.010* |
| Cephalosporins | ||||
| Cefaclor | 1.42 (1.15, 1.77) | 0.001* | 1.15 (1.06, 1.25) | 0.0006* |
| Cefazolin | 1.38 (1.11, 1.72) | 0.004* | 1.15 (1.07, 1 25) | 0.0004* |
| Cefeprime | 1.45 (1.16, 1.80 | 0.001* | 1.12 (1.03, 1.21) | 0.006* |
| Cefoxitin | 1.41 (1.14, 1.76) | 0.002* | 1.10 (1.01, 1.19) | 0.020* |
| Ceftazidime | 1.31 (1.05, 1.62) | 0.02* | 1.07 (0.99, 1.15) | 0.10 |
| Caftriaxone | 1.25 (1.01, 1.56) | 0.04* | 1.09 (1.01, 1.18) | 0.03* |
| Carbapenams | ||||
| Meropenem | 1.28 (0.99, 1.65) | 0.06 | 1.08 (0.99, 1.18) | 0.10 |
| Aminoglycosides | ||||
| Gentamicin | 1.17 (0.90, 1.51) | 0.23 | 1.01 (0.92, 1.11) | 0.78 |
| Fluroroqinolones | ||||
| Ciprofloxacin | 1.20 (0.96, 1.51) | 0.11 | 1.03 (0.95, 1.12) | 0.45 |
| Levofloxacin | 1.20 (0.95, 1.53) | 0.13 | 1.05 (0.97, 1.14) | 0.25 |
| Nilrofurans | ||||
| Nitmiurantoin | 1.03 (0.64, 1.65) | 0.9 | 0.90 (0.74, 1.08) | 0.25 |
| Tetracyclines | ||||
| Tetracyclines | 1.26 (1.02, 1.57) | 0.04* | 1.11 (1.02, 1.20) | 0.010* |
| Glycopeptides | ||||
| Vancomycin | 2.15 (1.63, 2.84) | <00001 | 1.38 (1.21, 1.56) | <0.0001 |
Significant p-value
The numbers in parentheses are 95% confidence interval bounds. Column 2 represents odds ratios of resistance for polymicrobial versus monomicrobial specimens. Column 4 presents odds ratios of resistance for each additional bacterial species in a specimen.
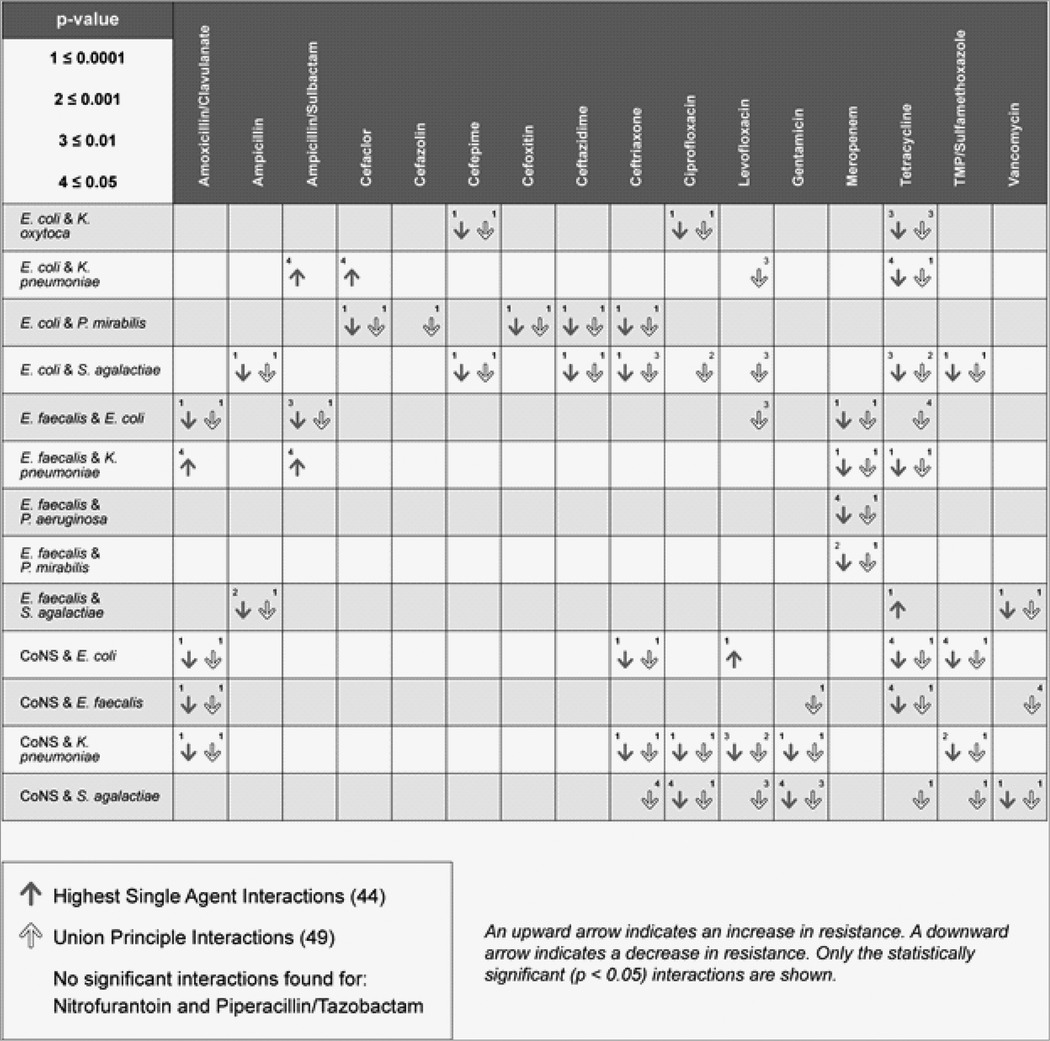
Table 3 shows the effect of specific species interactions on the probability of increased or decreased resistance to each antibiotic tested. No interactions were detected for nitrofurantoin and piperacillin/tazobactam. Whereas the odds of resistance to ampicillin, amoxicillin/clavulanate, 6 different cephalosporins, vancomycin, and tetracycline increased with increasing number of detected species, There were 19 instances for which 11 of the 13 bacterial pairs resulted in reduced susceptibility to the same antibiotics.
Table 3:
Effects of organism interactions on antibiotic resistance.
|
Using HSAP, there were 44 instances for which 13 pairs of bacteria showed statistically significant interactions that either increased or decreased the probability of resistance to the antibiotics tested. According to the HSAP principle, most interactions resulted in a decreased probability of resistance. Only 6/44 (13.6%) pairings resulted in increased odds of antibiotic resistance, whereas a decreased probability occurred in 38/44 (86.4%) of pairings.
The bacterial combinations that increased the probability of antibiotic resistance according to the HSAP model were E. faecalis and K. pneumoniae (amoxicillin/clavulanate, p = 0.02 and ampicillin/sulbactam, p = 0.03), E. coli and K. pneumoniae (ampicillin/sulbactam, p = 0.04 and cefaclor, p = 0.05), CoNS and E. coli and (levofloxacin, p < 0.001), E. faecalis and S. agalactiae (tetracycline, p < 0.001).
The UP model identified 49 statistically significant interactions, all of which showed decreased probability of resistance to the antibiotics tested.
To illustrate the model, one specific pair is presented in graphical form. Figure 2 shows the predicted probabilities of resistance to ampicillin/sulbactam, cefaclor, and tetracycline by monomicrobial positive cultures for E. coli and K. pneumoniae and a polymicrobial culture positive for both E. coli and K. pneumoniae. When the HSAP model was used, the pairing of E. coli and K. pneumoniae resulted in either a significant increase or significant decrease in the probability of resistance depending on the antibiotic tested. For example,when ampicillin/sulbactam or cefaclor was applied to the the combination of E. coli and K. pneumoniae, the resistance rate was higher than either E. coli or K. pneumoniae alone. In contrast, the resistance rate to tetracycline of the same combination of species, E. coli and K. pneumoniae, was intermediate between the resistance rates to each species alone.
Figure 2:
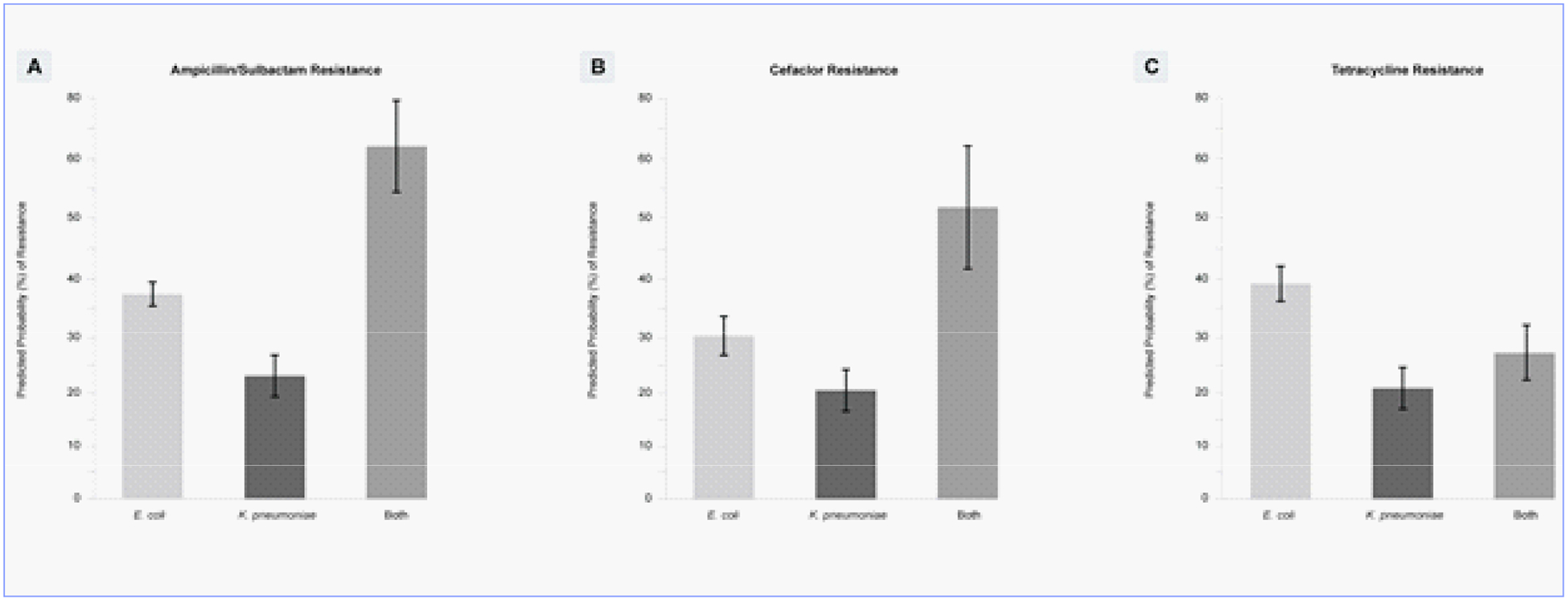
The effects of E. coli and K. pneumoniae interactions on resistance to ampicillin/sulbactam, cefaclor, and tetracycline (p = 0.05).
Discussion
These results demonstrate that polymicrobial infections, which constituted 56.1% (758/1,352) of positive samples with susceptibility results, can alter response to antibiotics. This alteration in susceptibility is associated with specific bacterial combinations, the antibiotic tested and the number of bacteria in the polymicrobial sample.
There is a growing concern that the number of multiresistant strains of bacteria is increasing despite the addition of antibiotic stewardship programs. In the polymicrobial sample, we see the effect of the increased resistance due to the presence of multiple resistance genes in that the odds of resistance increased per each organism added for 2 penicillin antibiotics, 5 cephalosporine, vancomycin, and tetracycline. Previous studies show that clinical isolates can protect other species from antibiotics with various interactions demonstrating a higher than a 3.5-fold increase in antimicrobial tolerance [8]. In the case of penicillin’s and cephalosporins, the whole population may be protected by the ability of a single resistant bacteria species, by breaking down the penicillin or cephalosporine with β-lactamase inhibitor [9]. The likelihood of the polymicrobial sample containing such a protective species increases with each additional bacterial species added to the polymicrobial mix. Additionally, antibiotic resistance can be conferred by one bacterium on another bacterium through Horizontal Gene Transfer (HGT) of antibiotic resistance genes [10].
Increasing the complexity of the interactions, thirteen specific bacterial pairs had one or more significant interactions when tested in the presence of 16 of the 18 antibiotics or antibiotic combinations using HSAP and UP. Of these interactions, 38 resulted in a decreased probability of resistance, while 6 resulted in an increased probability of resistance. The combination of E. coli and K. pneumoniae resulted in an increased probability of resistance to ampicillin/sulbactam and cefaclor, but decreased probability of resistance to tetracycline. E. faecalis together with K. pneumoniae resulted in increased resistance to amoxicillin/clavulanate and ampicillin/sulbactam, but decreased resistance to levofloxacin, meropenem, and tetracycline. E. faecalis combined with S. agalactiae produced an increase in resistance to tetracycline, but decreased resistance to ampicillin and vancomycin. Similarly, the combination of CoNS and E. coli produced an increased probability in resistance to levofloxacin, but the same combination produced a decreased probability in resistance to amoxicillin/clavulanate, ceftriaxone, tetracycline, and trimethoprim/sulfamethoxazole. These differences may be attributed to the unique mechanisms of action of the specific antibiotics.
A similar set of contrasts is observed from the perspective of individual antibiotics. Different pairs of bacteria caused both increased and decreased resistance to amoxicillin/clavulanate, ampicillin/sulbactam, cefaclor, levofloxacin, and tetracycline. For instance, E. coli combined with K. pneumoniae produced an increase in resistance to cefaclor, while E. coli combined with P. mirabilis produced a decrease in resistance to cefaclor. These results highlight the importance of accurate identification of bacteria in polymicrobial infections: a difference in identification of one species can influence antibiotic resistance.
The observed effects on antibiotic resistance in polymicrobial infections may be due to cooperative and/or competitive interactions between bacteria. Resistant bacteria can cooperatively protect susceptible bacteria by degrading antibiotics, as occurs when secreted beta-lactamase degrades beta-lactam antibiotics [11]. Antibiotic resistance can be conferred by one bacterium on another bacterium by means of horizontal gene transfer (HGT) of antibiotic resistance genes [10]. Bacterial interactions with host macrophages can promote HGT. For instance, P. aeruginosa, when present in biofilms, produces extracellular DNA that induces neutrophils to produce pro-inflammatory cytokines (IL-8 and IL-1 beta). The ensuing inflammation can promote HGT involving E. coli [12]. Interestingly, some antibiotics can also promote HGT: antibiotics that cause bacterial lysis release DNA and proteins that can be taken up by other bacteria [12]. In addition, one bacterium can stimulate gene expression in another bacterium, resulting in upregulation of efflux pumps leading to increased antibiotic resistance [7]. Bacterial community spatial structuring within a polymicrobial biofilm may also affect the efficacy of antibiotics [13,14].
Decreased resistance to antibiotics in polymicrobial specimens may also be due to competitive mechanisms between bacteria. P. aeruginosa has been documented to produce antibiotics, whereas Enterococcus species produce and secrete bacteriocins [15,16]. Gram-negative bacteria have developed a number of specialized secretion systems that can perform protective functions. Type V secretion systems secrete proteases that digest IgA, surface receptors that bind the constant region of IgG, and virulence factor/adhesin proteins that promote colonization [17–20]. Type VI secretion systems allow Gram-negative bacteria to secrete antibacterial toxins directly into other bacteria [21,22]. At the same time, Type VI systems mediate DNA acquisition via HGT; an example is the capacity for A. baumannii to rapidly acquire resistance genes from E. coli by means of Type VI transfer systems [23,24].
One type of bacterial interaction can cause a paradoxical result: cross-feeding between bacteria can produce decreased antibiotic resistance. This may explain our observed decreased probability of antibiotic resistance seen in most specific organism combinations. Cross-feeding is a process by which one organism produces metabolites that promote the survival of another organism [25,26]. However, this interaction can produce a chain of dependencies, leaving the entire chain only as resistant as the most susceptible bacterium. Adamowicz et al. [7] showed that bacterial species were inhibited at significantly lower antibiotic concentrations in cross-feeding communities than in monoculture, coined as the “weakest link” model.
None of these potentially important changes of drug responses would be captured through routine ATS testings done on bacterium in isolation. There is growing evidence that testing the bacterium in association with other organisms may improve clinical outcomes. In a retrospective research study involving 66,383 subjects cared for by house-call health care physicians, subjects with possible UTI were split into 2 cohorts; one treated based upon the results from SUC and an assay incorporating P-AST. The complete number of emergency department and hospitalization visits were compared. The P-AST cohort was associated with a significant decrease in hospital admissions and/or emergency department as compard to the SUC cohort [27].
Strengths of the study include the large number of samples coming from multiple urology clinics across the United States. Additionally, interactions were analyzed using specimens collected from the clinical setting.
Conclusions
Bacteria are social organisms that interact within and between species. Key interactions play critical roles in the growth, pathogenesis, and virulence of bacterial species. Because of these key and specific interactions, correct identification of bacterial species increases in significance. In order to accurately assess these interactions and provide clinical context, we have developed P-AST, a pooled antibiotic susceptibility test. Using this methodology, we observed both increased and decreased antibiotic susceptibility based on the type of species observed, as well as the class of antibiotic administered. Elucidation of the molecular mechanisms by which alterations in antibiotic response occur in polymicrobial infections will require additional research. Based on these findings, P-AST testing might more closely approximate the polymicrobial environment in the patient and possibly provide more clinically important information regarding antibiotic susceptibility. Clinical studies will need to be conducted and are underway to analyze clinical outcome benefits and/or cost effectiveness if results from the P-AST testing are to impact clinical antibiotic selections.
Acknowledgements
Funding for this project was provided by Pathnostics, Inc. (Irvine, CA) and ThermoFisher (Carlsbad, CA). David Baunoch, Natalie Luke, Michael Opel, Meghan Campbell, Miguel Penaranda, and David E. Smith are employed by Pathnostics, Inc. Alan Wolfe receives funding from NIH through Grant R01DK104718.
References
- 1.Price TK, Hilt EE, Dune TJ,Mueller ER,Wolfe AJ, et al. (2018) Urine trouble: should we think differently about UTI?. Int Urogynecol J 29: 205–210. [DOI] [PubMed] [Google Scholar]
- 2.Laudisio A, Marinosci F, Fontana D, Gemma A, Zizzo A, Coppola A, et al. (2015) The burden of comorbidity is associated with symptomatic polymicrobial urinary tract infection among institutionalized elderly. Aging Clinical and Experimental Research 27: 805–812. [DOI] [PubMed] [Google Scholar]
- 3.Wojno KJ, Baunoch D, Luke N, Opel M, Kormana H, et al. (2019) Multiplex PCR Based Urinary Tract Infection (UTI) Analysis Compared to Traditional Urine Culture in Identifying Significant Pathogens in Symptomatic Patients. Urology 136: 119–126. [DOI] [PubMed] [Google Scholar]
- 4.Price TK, Dune T, Hilt EE, Thomas-White KJ, Kliethermes S, et al. (2016) The Clinical Urine Culture: Enhanced Techniques Improve Detection of Clinically Relevant Microorganisms. Journal of Clinical Microbiology 54: 1216–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rogers GB, Hoffman LR, Whiteley M, Daniels TWV, Carroll MP, et al. (2010) Revealing the dynamics of polymicrobial infections: implications for antibiotic therapy. Trends in Microbiology 18: 357–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peters BM, Jabra-Rizk MA, O’May GA, Costerton JW, Shirtliff ME (2012) Polymicrobial Interactions: Impact on Pathogenesis and Human Disease. Clin Microbiol Rev 25: 193–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adamowicz EM, Flynn J, Hunter RC, Harcombe WR (2018) Cross-feeding modulates antibiotic tolerance in bacterial communities. The ISME Journal 12: 2723–2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vos MG de, Zagorski M, McNally A, Bollenbach T (2017) Interaction networks, ecological stability, and collective antibiotic tolerance in polymicrobial infections. Proceedings of the National Academy of Sciences 114: 10666–10671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yurtsev EA, Chao H, Datta MS, Artemova T, Gore J (2013) Bacterial cheating drives the population dynamics of cooperative antibiotic resistance plasmids. Molecular Systems Biology 9: 683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bolotin E, Hershberg R (2017) Horizontally Acquired Genes Are Often Shared between Closely Related Bacterial Species. Front Microbiol 8: 1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shaikh S, Fatima J, Shakil S, Rizvi SMD, Kamal MA (2014) Antibiotic resistance and extended spectrum beta-lactamases: Types, epidemiology and treatment. Saudi J Biol Sci 22: 90–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sitaraman R (2018) Prokaryotic horizontal gene transfer within the human holobiont: ecological-evolutionary inferences, implications and possibilities. Microbiome 6: 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Estrela S, Brown SP (2018) Community interactions and spatial structure shape selection on antibiotic resistant lineages. Plos Comput Biol0z 14: e1006179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee K, Periasamy S, Mukherjee, Xie C, Kjelleberg S, et al. (2014) Biofilm development and enhanced stress resistance of a model, mixed-species community biofilm. [DOI] [PMC free article] [PubMed]
- 15.Hibbing ME, Fuqua C, Parsek MR, Peterson SB (2010) Bacterial competition: surviving and thriving in the microbial jungle. Nat Rev Microbiol 8: 15–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kommineni S, Bretl DJ, Lam Vy, Chakraborty R, Hayward M, et al. (2015) Bacteriocin production augments niche competition by enterococci in the mammalian gastrointestinal tract. Nature 526: 719–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leo JC, Goldman A (2009) The immunoglobulin-binding Eib proteins from Escherichia coli are receptors for IgG Fc. Mol Immunol 46: 1860–1866. [DOI] [PubMed] [Google Scholar]
- 18.Roy K, Bartels S, Qadri F,Fleckenstein JM (2010) Enterotoxigenic Escherichia coli elicits immune responses to multiple surface proteins. Infect Immun 78: 3027–3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roy K, Hamilton D, Allen KP, Randolph MP, Fleckenstein JM (2008) The EtpA exoprotein of enterotoxigenic Escherichia coli promotes intestinal colonization and is a protective antigen in an experimental model of murine infection. Infect Immun 76: 2106–2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meuskens I, Saragliadis A, Leo JC, Linke D (2019) Type V Secretion Systems: An Overview of Passenger Domain Functions. Front Microbiol 10: 1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dörr NCD, Blokesch M (2018) Bacterial type VI secretion system facilitates niche domination. P Natl Acad Sci Usa 115: 8855–8857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hood RD, Singh P, Hsu FS, Güvener T, Carl MA, et al. (2010) A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe 7: 25–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Russell AB, Wexler AG, Harding BN, Whitney JC, Bohn AJ, et al. (2014) A type VI secretion-related pathway in Bacteroidetes mediates interbacterial antagonism. Cell Host Microbe 16: 227–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coulthurst SJ (2013) The Type VI secretion system - a widespread and versatile cell targeting system. Res Microbiol 164: 640–654. [DOI] [PubMed] [Google Scholar]
- 25.Rosenzweig RF, Sharp RR, Treves DS, Adams J (1994) Microbial evolution in a simple unstructured environment: genetic differentiation in Escherichia coli. Genetics 137: 903–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schink B (2002) Synergistic interactions in the microbial world. Antonie Van Leeuwenhoek 81: 257–261. [DOI] [PubMed] [Google Scholar]
- 27.Daly A, Baunoch D, Rehling K, Luke N, Campbell M, et al. (2020) Utilization of M-PCR and P-AST for Diagnosis and Management of Urinary Tract Infections in Home-Based Primary Care. JOJ Urology & Nephrology 7: 555707. [Google Scholar]


