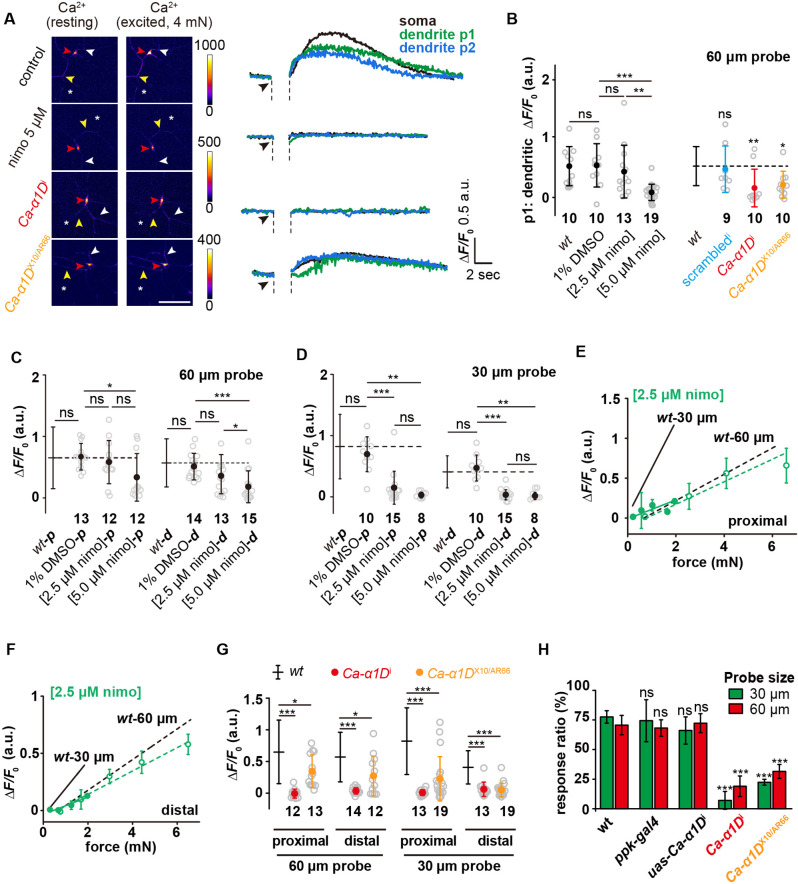
Figure 6. Active signal propagation in the dendrites of c4da.
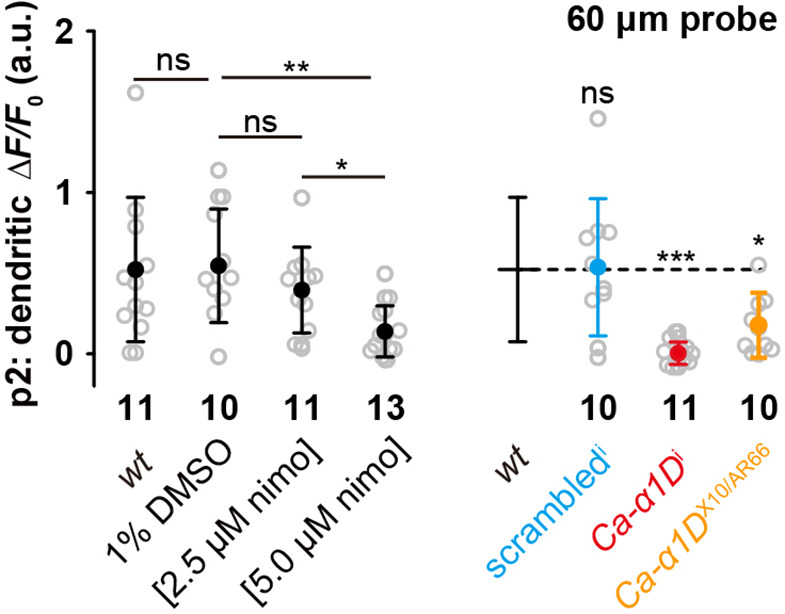
(A) Representative images and curves showing the responses (ΔF/F0) of the soma (red arrowhead), the homolateral dendrite (position p1: yellow arrowhead) and contralateral dendrite (position p2: white arrowhead) of c4da-wt, nimodipine-treated c4da-wt, c4da-Ca-α1Di and Ca-α1DX10/AR66 to the stimuli (4 mN, 60 μm probe). The asterisk indicated the force application point. Scale bar: 50 μm. C4da-wt: ppk-gal4/+; ppk-cd4-tdtom/20×uas-ivs-gcamp6s. Ca-α1Di: ppk-gal4/20×uas-ivs-gcamp6s; ppk-cd4-tdtom/ uas-Ca-α1Di. Ca-α1DX10/AR66: Ca-α1DX10/ Ca-α1DAR66; ppk-gal4/20×uas-ivs-gcamp6s. (B) Statistical quantification of calcium increases in the homolateral dendrites in response to the stimuli (4 mN, 60 μm probe). The genotypes were the same as those shown in panel (A). Mann Whitney U test was used for the comparison between c4da-wt and the mutants. Kruskal Wallis test was used for the comparison among the groups with drug treatment. ***: p<0.001. **: p<0.01. ns: no significance. (C) and (D) The responses (ΔF/F0) of nimodipine treated c4da-wt to the proximal and distal stimuli from the probes of different sizes. (C): 60 μm probe stimuli. (D) : 30 μm probe stimuli. Kruskal Wallis test was used. ***: p<0.001. **: p<0.01. *: p<0.05. ns: no significance. (E) and (F) The responses (ΔF/F0) of c4da-wt (n=10 cells) treated with 2.5 μM nimodipine to the proximal (E) and distal (F) stimuli delivered using the probes of two different sizes. Solid lines: 30 μm probe. Dashed lines: 60 μm probe. (G) The responses (ΔF/F0) of c4da-Ca-α1Di and c4da-Ca-α1DX10/AR66 to the stimuli applied at different dendritic regions (proximal and distal) and delivered using the probes of two different sizes (60 μm: 4 mN, 30 μm: 1.5 mN). (H) The behavioral responses of the c4da-Ca-α1Di larvae to mechanical poking using the probes of two different sizes. wt: w1118. ppk-gal4: ppk-gal4; +/+. uas-Ca-α1Di: +/+; uas-Ca-α1Di. Ca-α1Di: ppk-gal4/+; uas-Ca-α1Di /+. Ca-α1DX10/AR66: Ca-α1DX10/ Ca-α1DAR66; +/+. uas-Ca-α1Di larvae: 30 μm probe (n=61 larvae from three experiments), 60 μm probe (n=61 larvae from three experiments). Ca-α1Di larvae: 30 μm probe (n=56 larvae from three experiments), 60 μm probe (n=60 larvae from three experiments). Ca-α1DX10/AR66 larvae: 30 μm probe (n=43 larvae from three experiments), 60 μm probe (n=47 larvae from three experiments). One-way ANOVA test was used for the comparison among different genotypes. ***: p<0.001. ns: no significance. In panels (B), (C), (D), (G) and (H), data were presented as mean ± std. In panels (B), (C), (D) and (G), the numbers of cells were indicated below the scattered data points. In panels (E) and (F), data were presented as mean ± sem. In panels (C), (D), (E), (F), (G) and (H), the corresponding data from c4da-wt were provided for comparison.
Figure 6—figure supplement 1. Statistics quantification of the calcium increase in the contralateral dendrites in response to the force stimuli.