Abstract
The gene family encoding a trypomastigote-specific protein restricted to the part of the flagellum in contact with the cell body of the trypomastigote form of Trypanosoma cruzi has been isolated, characterized, and expressed in a baculovirus expression system. The gene family contains three tandemly repeated members that have 97 to 100% sequence identity. The predicted protein encoded by the gene family has both significant amino acid sequence identity and other physical and biological features in common with the TolA proteins of Escherichia coli and Pseudomonas aeruginosa. Based on these similarities, we have designated this gene family tolT. Immunization of mice with recombinant TolT generates a population of CD4+ T lymphocytes that recognize T. cruzi-infected macrophages, resulting in the production of gamma interferon (IFN-γ), which leads to NO production and a 50 to 60% reduction in parasite numbers compared to that seen with infected macrophages incubated with naive T cells. This population of T cells also produces both IFN-γ and interleukin 2 (IL-2) but not IL-4 or IL-5 when incubated with spleen cells stimulated with TolT antigen, indicating that they are of the T-helper 1 type. T cells from mice chronically infected with T. cruzi also produce significant levels of IFN-γ when cocultured with macrophages and either TolT protein or paraflagellar rod protein, indicating that both of these flagellar proteins produce positive T-cell responses in mice chronically infected with T. cruzi.
Trypanosoma cruzi, a hemoflagellate protozoan, is the causative agent of American trypanosomiasis or Chagas’ disease. The disease is prevalent throughout most of Central America and South America, and it is estimated that 16 to 18 million people are infected with T. cruzi. At present, no adequate chemotherapeutic agents for treating the disease or efficacious immunoprophylactic vaccines against the parasite are available. Parasite antigens that have been shown to elicit a protective immune response in vaccination trials are scarce, and immunization regimens with these antigens generally provide only partial protection against a lethal challenge with the parasite in mice. In part, the lack of success in identifying protective parasite antigens may reside in the choice of immunological responses that are believed to be desirable in a vaccine candidate. The observations that (i) inbred mouse strains with an impaired ability to produce antibodies and rats treated with immunoglobulin M (IgM) antiserum both show a high degree of susceptibility to infection (12, 28) and (ii) passive transfer of T. cruzi immune serum to naive animals results in partial protection against challenge infection (12, 37) have resulted in much attention being focused on the identification of parasite antigens that produce strong antibody responses against T. cruzi. However, no parasite antigen that elicits a protective antibody response against T. cruzi infection has been identified.
More recent studies have demonstrated the importance of CD4+ and CD8+ T-cell responses in immunity to T. cruzi (29, 30, 39), and attention is now being directed toward antigens that elicit cell-mediated immune responses that result in immune recognition of parasite-infected host cells (18, 20, 21, 42). We have reported that parasite antigens that generate such an immune response are the paraflagellar rod (PFR) proteins present in the flagellum of T. cruzi (45). Immunization of mice with the PFR proteins induced an immune response capable of providing 100% survival against an otherwise lethal inoculum of the parasite. By use of genetic knockout mice lacking either CD4, CD8, β2-microglobulin, or the μ heavy chain of IgM, the immunological responses that play a critical role in PFR protein-mediated protection have been shown to require T-cell, but not B-cell, function (21, 44a). Furthermore, CD4+ T cells from PFR protein-immunized mice were found to release gamma interferon (INF-γ) when incubated with T. cruzi-infected macrophages, leading to the release of nitric oxide and a >90% reduction in parasite numbers compared to that seen with control macrophages. Since the PFR proteins are present in the flagellum of the invasive bloodstream trypomastigote form of the parasite but are almost absent in the intracellular amastigote form of the parasite, the conundrum arose as to why CD4+ T cells from PFR protein-immunized mice should recognize T. cruzi-infected macrophages in an antigen-specific fashion. One plausible explanation involved the obligatory transformation of the trypomastigote, which is in G0, into the amastigote. It was proposed that during transformation within the acidic phagolysosome, when the flagellum is reduced in length by >90% by catabolic mechanisms, the degradative products of the PFR proteins might be available for entry into the major histocompatibility complex (MHC) class II pathway (21). It was further suggested that if this mechanism is the means by which the PFR proteins are presented on the surface of infected macrophages, other trypomastigote-specific proteins that are degraded during the transformation event might also be available for association with MHC class II molecules.
In the present study, we have explored this possibility by examining the ability of T cells from mice immunized with a protein present on trypomastigotes, but not amastigotes, to stimulate T. cruzi-infected macrophages. Using a monoclonal antibody (MAb), designated 20H1, we have previously identified an antigen that is present on the part of the flagellum in contact with the cell body of trypomastigotes but that is absent from amastigotes (33). Western blot analysis showed that the antigen recognized by MAb 20H1 consists of four different molecules with sizes of 34 to 41 kDa and that these molecules are glycoproteins with an affinity for concanavalin A. We now report the cloning and characterization of the genes that encode this antigen. We also show that immunization of mice with a recombinant form of this antigen generates a CD4+ T-cell population that releases IFN-γ and stimulates T. cruzi-infected macrophages to release nitric oxide, leading to parasite reduction.
MATERIALS AND METHODS
Parasites.
The Peru clone 3, Y, and Esmeraldo clone 3 strains of T. cruzi were used. Epimastigotes were grown in modified L-15 media (FlowLabs, McLean, Va.) (13). Tissue culture-derived trypomastigotes used for the preparation of RNA and macrophage infection were obtained from infected monolayers as described elsewhere (1).
DNA library construction and screening.
A Peru strain trypomastigote poly(A)+ cDNA library was constructed with the phage λgt11 vector as previously described (27). A Peru strain genomic DNA library was constructed with the λ ZAP Express vector by use of epimastigote nuclear DNA as previously described (8). The cDNA library was screened with MAb 20H1 as described elsewhere (1), and positive phage clones were identified by use of the picoBlue immunoscreening system (Stratagene, La Jolla, Calif.). The λ genomic DNA library was screened with a radiolabeled oligonucleotide consisting of nucleotides 1145 to 1165 of the tolT1 gene (5′-AAAAAGGCTGCCATTGCTGAG-3′). The inserts present in phages showing positive hybridization were characterized by restriction enzyme mapping and direct nucleotide sequence analysis.
DNA sequencing.
DNA sequence information was obtained by use of the dideoxy chain termination method (35). Fragments to be sequenced were sequenced directly from the excised pBK-CMV phagemid or from DNA fragments subcloned into plasmid pBluescript KS(+). Oligonucleotide sequencing primers were obtained from Integrated DNA Technologies, Inc., Coralville, Iowa.
Nucleic acid isolation, radiolabeling, Southern and Northern transfer, and restriction enzymes.
Parasites were harvested, and DNA, RNA, and poly(A)+ RNA were isolated as described previously (2, 7). Plasmid DNA was isolated by use of the Strataclean boiling miniprep protocol according to the manufacturer’s instructions (Stratagene). Synthetic oligonucleotides were radiolabeled with [γ-32P]ATP by use of T4 polynucleotide kinase (Pharmacia, Piscataway, N.J.) as described previously (34). Agarose gel electrophoresis of DNA, Southern transfer, prehybridization, hybridization, and filter washing were performed as described previously (7), with the following exceptions. Gels were 1% agarose, and the washing temperature following hybridization with the end-labeled oligonucleotide was 42°C. RNA was electrophoresed in a formaldehyde gel, blotted to nylon, cross-linked by UV irradiation, prehybridized, hybridized, and washed as described previously (31). Northern and Southern blots were imaged with a Molecular Dynamics 435SI PhosphorImager. Analysis of the digital images was conducted with the program ImageQuant (Molecular Dynamics). All restriction enzymes were purchased from GIBCO BRL and used as recommended by the manufacturer.
Digestion with N-glycosidase F.
Tissue culture-derived trypomastigotes (5 × 108) were resuspended in 200 μl of 0.5% Triton X-100–1.0 M NaCl–20 mM Tris (pH 8.0), incubated for 10 min at 25°C, and clarified by centrifugation. Aliquots (25 μl) of the supernatant were removed, added to 75 μl of 50 mM sodium phosphate (pH 7.5)–20 mM EDTA–0.5% Triton X-100–1.0% β-mercaptoethanol containing 0, 1, 3, or 10 U of N-glycosidase F (Boehringer Mannheim Biochemicals, Indianapolis, Ind.), and incubated at 37°C for 2 h. Aliquots (20 μl) were removed for Western blot analysis.
PAGE and immunoblot analysis.
For analysis of whole-cell lysates, parasites were harvested from culture media by centrifugation, washed twice with phosphate-buffered saline (PBS), and solubilized by the direct addition of boiling 2% sodium dodecyl sulfate (SDS); boiling was continued for 5 min. These and all other samples were separated by one-dimensional polyacrylamide gel electrophoresis (PAGE) and transferred to nitrocellulose filters by previously described methods (33) with a Transblot Cell (Bio-Rad, Richmond, Calif.) overnight at 150 mA. Filters probed with MAb 20H1 were reacted with peroxidase-linked anti-mouse IgG and detected with an enhanced chemiluminescence system (DuPont, Wilmington, Del.).
Construction and expression of recombinant TolT1 in a baculovirus expression system.
For the production of recombinant TolT1, the entire tolT1 coding region (Fig. 1) was expressed in a baculovirus expression system by cloning nucleotides 377 to 1364 into the shuttle vector pVL1393. The fragment was inserted into the BamHI/XhoI sites in the polylinker of the shuttle vector by generating a BamHI site at nucleotides 375 to 380 and an XhoI site at nucleotides 1356 to 1361 by use of the following oligonucleotides as primers for PCR amplification of the tolT1 gene: 5′-ATGCCGTCAAAGGATCCCTTTAC-3′, representing nucleotides 364 to 386, and 5′-TCATCGCCTCGAGTACGC-3′, representing nucleotides 1349 to 1366. The production of recombinant baculovirus, the production of recombinant protein, and protein purification were done as described elsewhere (44).
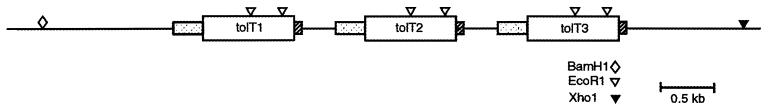
FIG. 1.
Schematic of the organization of the tolT genes in genomic clone pTcg20H1a. The open reading frames for the tolT1, tolT2, and tolT3 genes are indicated by open boxes. The stippled boxes denote the regions 5′ of the three open reading frames that have high sequence identity. The hatched boxes denote the regions 3′ of the open reading frames that have high sequence identity. EcoRI, XhoI, and BamHI restriction enzyme sites are indicated.
Immunization.
Six- to 8-week-old female C57BL/6J mice were immunized by subcutaneous injection with 40 μg of recombinant TolT1 protein emulsified with Freund’s complete adjuvant. Mice were boosted twice at 2-week intervals with 20 μg of protein emulsified with Freund’s incomplete adjuvant. Purification of PFR proteins from T. cruzi and immunization of mice were done as described previously (32, 45). Purification of baculovirus-produced recombinant trypomastigote surface antigen 1 (TSA-1) and immunization of mice were done as described previously (44). Mice immunized with heat-killed trypomastigotes (HKTC) were injected intraperitoneally with 5 × 107 HKTC prepared by incubation of tissue culture-derived strain Y trypomastigotes at 56°C for 1 h.
Preparation of shed parasite antigen and 35S-methionine-labeled shed parasite antigen.
Tissue-culture derived strain Y trypomastigotes were washed, resuspended in C-DMEM (25 mM HEPES buffer [pH 7.2], 1 mM sodium pyruvate, nonessential amino acids, l-glutamine, 5 × 10−5 M 2-mercaptoethanol, 50 U of penicillin per ml, 50 μg of streptomycin sulfate per ml, 10% fetal bovine serum) at 5 × 107 parasites/ml, and incubated at 37°C. At 2 and 24 h, aliquots were removed and parasites were removed by two rounds of centrifugation at 13,000 × g for 10 min each time. Supernatants were filtered through a 0.22-μm-pore-size filter, divided into aliquots, and stored at −80°C until use. Similar preparations were labeled with 35S-methionine by incubation of trypomastigotes in methionine-deficient C-DMEM supplemented with 100 μCi of 35S-methionine. At periodic intervals, aliquots were removed for assessment of culture viability. Cultures were >99% viable throughout the culture period.
Measurement of cytokines.
Culture supernatants were taken from triplicate cultures of T cells containing 3 μg of recombinant TolT1 protein and assayed for the presence of interleukin 2 (IL-2), IL-4, IL-5, and IFN-γ by a capture enzyme-linked immunosorbent assay (ELISA) (Pharmingen, San Diego, Calif.). Briefly, 1 to 2 μg of cytokine-specific capture antibody per ml was bound to 96-well microtiter plates containing 0.1 M NaHCO3 (pH 8.2) at 4°C overnight, washed with PBS–0.05% Tween 20 (PBST), and then blocked with 10% fetal calf serum in PBS for 2 h at room temperature. Wells were washed with PBST, 100 μl of either standards or samples was added, and the reaction mixture was incubated at 4°C overnight. Wells were again washed with PBST, and the appropriate concentration of biotinylated anticytokine detecting antibody was added in a 100-μl volume and incubated for 45 min at room temperature. The wells were thoroughly washed, 100 μl of streptavidin-peroxidase (2.5 μl/ml) was added, and the reaction mixture was incubated for 30 min at room temperature. After extensive washes with PBST, 100 μl of 2,2′-azino-di-3-ethylbenzthiazoline sulfonate (Boehringer) substrate was added, and plates were read at 405 nm by use of an automated ELISA plate reader. Concentrations were calculated from linear regions of a titration curve for cytokine standards, values for control wells were subtracted, and final concentrations were expressed in picograms per milliliter.
Macrophage and T-cell cultures.
Mice were immunized as described above. Seven to 10 days after the last injection, spleens were removed and single-cell suspensions were prepared with C-DMEM. Spleen cell suspensions were enriched for T cells by passage over nylon wool columns (9). IC-21 macrophages were plated in C-DMEM at 105 macrophages/well in 96-well plates. Macrophages were allowed to adhere overnight (37°C at 8% CO2). Cells were either infected overnight with trypomastigotes at a 10:1 parasite-to-macrophage ratio or incubated with recombinant TolT1 protein (3 μg/ml). Control wells were incubated overnight with C-DMEM containing no parasites. Infected monolayers were washed three times with C-DMEM to remove extracellular parasites. Cells were cultured for 7 days at 37°C in an atmosphere of 8% CO2, and supernatants were harvested and assayed for nitrite (NO2). All experimental groups were tested in triplicate. CD4+ and CD8+ T cells were positively selected with MACS MultiSort MicroBeads by following procedures recommended by the manufacturer (Miltenyi Biotec, Bergish-Gladbach, Germany).
Nitrite assays.
Nitrite levels in 4- and 7-day culture supernatants were measured with the Greiss reagent as previously described (21). Briefly, 50 μl of culture supernatant was combined in a 96-well plate with a 1:1 mixture of 1% sulfanilamide in 2.5% H3PO4 and 0.1% naphthylethlenediamide in 2.5% H3PO4. Plates were incubated for 10 min at room temperature, and the absorbance at 550 nm was determined with an automated microplate reader. Nitrite concentrations were determined in triplicate with a standard curve of sodium nitrite from 125 to 1 μM in culture media.
Inhibition of T. cruzi growth in vitro.
Parasite titers in infected IC-21 macrophage–T-cell culture supernatants were determined by pipetting media up and down vigorously several times to resuspend trypomastigotes. The parasite number was determined by counting with a Neubauer hemocytometer.
Nucleotide sequence accession number.
The GenBank accession no. for the nucleotide sequences discussed in this manuscript is AF099099.
RESULTS
Isolation of the gene encoding the protein recognized by MAb 20H1.
In order to isolate a cDNA fragment that contains at least a portion of the gene that encodes the protein recognized by MAb 20H1, a trypomastigote cDNA expression library was constructed with λgt11 and screened with MAb 20H1. Approximately 120,000 recombinant phage were screened, and eight positive plaques were identified; one of the latter, designated λTcc20H1, rescreened positive. To detect the cDNA insert in λTcc20H1, two oligonucleotides that flank the EcoRI cloning site in λgt11 were synthesized and used as primers in a PCR to amplify the cDNA insert. Agarose gel electrophoresis of the PCR product revealed a single DNA fragment of approximately 150 nucleotides. The amplified fragment was cloned into the EcoRI site of the plasmid vector pBluescript KS(+), and its length was determined by direct DNA sequence analysis to be 154 nucleotides. Since this cDNA insert contains only a portion of the gene that encodes the protein recognized by MAb 20H1 and T. cruzi genes lack intron sequences, it seemed reasonable that the complete nucleotide sequence of the gene could be obtained by analysis of genomic DNA.
To obtain genomic DNA fragments that putatively contained the gene of interest, an oligonucleotide whose sequence is present in the cDNA insert of λTcc20H1 was synthesized, radiolabeled, and used as a probe to screen a λ ZAP recombinant genomic DNA library. Ten recombinant phage rescreened positive following plaque purification. The phagemid in each λ phage was excised, and a partial restriction enzyme map of each T. cruzi DNA insert was generated (data not shown). The complete nucleotide sequence of the DNA insert in one phage, designated pTcg20H1a, was determined.
Analysis of the DNA sequence of pTcg20H1a showed three separate open reading frames, each of 930 bp. A schematic diagram depicting the arrangement of these three open reading frames within the DNA insert is shown in Fig. 1. The three putative genes are arranged in a head-to-tail tandem array and are separated by two intergenic regions, each of 727 bp. The first two genes in the array are identical in sequence and have 98.8% nucleotide sequence identity with the last gene in the array, while the predicted proteins encoded by these genes have 97.4% amino acid sequence identity. The predicted Mrs of the two conceptual proteins encoded by the first and last genes in the array are 33,019 and 32,952, respectively, values consistent with the observed Mrs of 34,000 to 41,000 for the four protein bands recognized by MAb 20H1.
The intergenic regions have 100% sequence identity. The intergenic regions also have significant nucleotide sequence identity with sequences immediately 5′ upstream of the first gene in the array and sequences 3′ downstream of the last gene in the array. The 396 nucleotides immediately 5′ of the three putative ATG translation start sites have 97% sequence identity, while the 86 nucleotides immediately following the termination stop site of the three genes have 97% sequence identity. Since gene families that are arranged in tandem arrays in trypanosomes are frequently transcribed as polycistronic mRNAs that are subsequently processed prior to translation, the high conservation of these nontranslated nucleotide sequences suggests that they may play an important role in RNA processing (14).
Analysis of the restriction enzyme maps and partial nucleotide sequences determined for the other nine phagemids revealed that all of these phagemids contain T. cruzi DNA inserts that show no variations from the gene arrangement and nucleotide sequence found in pTcg20H1a.
The closest matches for the protein recognized by MAb 20H1 in databases were proteins associated with membrane structure or function. The highest amino acid sequence identity was found with rat plectin (27% in a 197-amino-acid [aa] overlap), Drosophila axoneme-associated MST1 (27% identity in a 286-aa overlap) and MST2 (30% identity in a 204-aa overlap), and the TolA proteins of Escherichia coli (32% identity in a 229-aa overlap) and Pseudomonas aeruginosa (34% identity in a 195-aa overlap). As discussed below (see Discussion), analysis of the similarities between the protein recognized by MAb 20H1 and the bacterial TolA proteins suggests that the T. cruzi protein may be a homologue of the TolA proteins. Therefore, we have tentatively defined this T. cruzi protein as TolT.
Number and expression of the tolT genes.
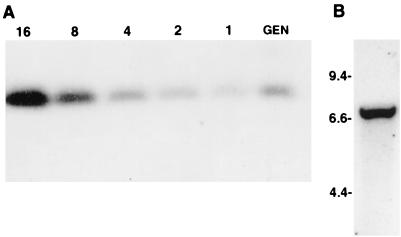
The copy number of the tolT gene sequences in the genome of T. cruzi was determined by methods previously described (2). The T. cruzi DNA insert in pTcg20H1a was digested with restriction enzyme EcoRI, and the 330-bp EcoRI fragment present within the tolT genes was subcloned into the pBluescript plasmid and designated pTcgTolR1. An oligonucleotide present in this EcoRI fragment (nucleotides 1145 to 1165 of tolT1) was synthesized, radiolabeled with 32P, and hybridized to a Southern blot containing epimastigote nuclear DNA digested with EcoRI (Fig. 2). Included on the Southern blot was pTcgTolR1 DNA restricted with EcoRI in amounts equivalent to 1, 2, 4, 8, and 16 copies per haploid genome. Strong hybridization of the probe was observed with a single genomic fragment of 330 bp. Quantification of the hybridization in the genomic plasmid DNAs with a PhosphorImager indicated that the EcoRI fragment occurs about three or four times per haploid genome. While this result is consistent with the possibility that the three genes in pTcg20H1a represent all of the tolT genes within the genome, it is possible that a fourth, outlying gene was not identified in the above cloning studies. To determine if this was the case, genomic DNA was digested with restriction enzymes BamHI and XhoI, enzymes that have predicted recognition sites immediately upstream and downstream, respectively, of the tandem array. A Southern blot of this DNA was hybridized with the 32P-labeled oligonucleotide described above, and hybridization only to a single fragment of 6.7 kb was observed (Fig. 2). This is the size of the BamHI/XhoI fragment predicted by sequence analysis of pTcg20H1a, thus confirming that this fragment contains all of the tolT genes within the T. cruzi genome.
FIG. 2.
Determination of the tolT gene copy number in trypomastigote DNA. (A) Determination of the tolT gene copy number in genomic DNA. Nuclear DNA (10 μg) was digested with EcoRI and electrophoresed on a 1% agarose gel (lane GEN). Included in the gel was EcoRI-restricted DNA from clone pTcgTolR1 in amounts equivalent to 1, 2, 4, 8, and 16 copies per haploid genome (lanes 1, 2, 4, 8, and 16, respectively). A Southern blot of the gel was hybridized with a 32P-labeled oligonucleotide containing nucleotides 1145 to 1165 of pTcg20H1a, which are contained in the 330-bp EcoRI fragments in the tolT1, tolT2, and tolT3 gene sequences. (B) Southern blot analysis of T. cruzi genomic DNA digested with BamHI and XhoI. Five micrograms of epimastigote genomic DNA was digested with BamHI and XhoI, electrophoresed on a 1% agarose gel, Southern blotted, and probed with the same 32P-labeled oligonucleotide as that used above. The numbers (in kilobases) indicate the migration of DNA molecular size markers.
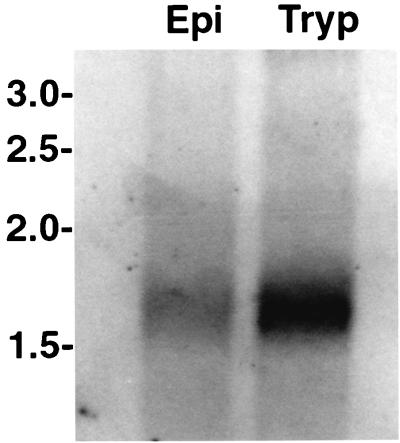
To determine the developmental expression pattern of the tolT genes, a Northern blot containing trypomastigote and epimastigote poly(A)+ RNAs was hybridized with the 32P-labeled oligonucleotide described above. As shown in Fig. 3, hybridization to a single RNA band of 1.6 kb in both RNAs was observed. However, the intensity of hybridization signals observed with the trypomastigote and epimastigote RNAs showed some variation. Quantification of the hybridization signals showed that the tolT mRNA is about four times more abundant in trypomastigotes than in epimastigotes.
FIG. 3.
Identification of poly(A)+ RNA from trypomastigotes and epimastigotes complementary to a tolT gene-specific probe. A Northern blot containing 5 μg of trypomastigote (Tryp) or epimastigote (Epi) poly(A)+ RNA was first stained with methylene blue to identify molecular size markers and ensure that both RNA samples were transferred in equivalent amounts. The blot was then hybridized with a 32P-labeled oligonucleotide containing nucleotides 1145 to 1165 of pTcg20H1a. Numbers (in kilobases) indicate the migration of RNA molecular size markers.
Posttranslational modification of the TolT protein.
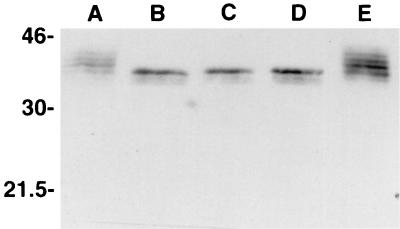
The predicted difference in the Mrs of the two conceptual TolT proteins cannot account for the observation that MAb 20H1 recognizes four separate protein bands with apparent Mrs of 34,000 to 41,000 in Western blots of trypomastigote lysates (33). Since previous studies have shown that these four proteins bind the lectin concanavalin A (33), it seemed possible that the four protein bands detected by MAb 20H1 might represent different glycosylated forms of TolT. To investigate this possibility, trypomastigote lysates were digested with N-glycosidase F, Western blotted, and reacted with MAb 20H1. As shown in Fig. 4, following digestion with N-glycosidase F, only a single protein band with an apparent Mr of 34,000 was recognized by MAb 20H1, consistent with the view that the multiple protein bands in the original lysates represented different glycosylated forms of proteins encoded by the tolT genes present in pTcg20H1a.
FIG. 4.
Treatment of trypomastigote lysates with N-glycosidase F. A trypomastigote lysate was prepared, divided into aliquots in the recommended buffer, incubated with 0, 1, 3, or 10 U of N-glycosidase F for 2 h at 37°C (A to D, respectively) or 4°C (E), fractionated by SDS-PAGE, blotted to nitrocellulose, and reacted with MAb 20H1. The numbers (in kilodaltons) are protein molecular size markers.
CD4+ T cells from TolT-immunized mice can activate T. cruzi-infected macrophages.
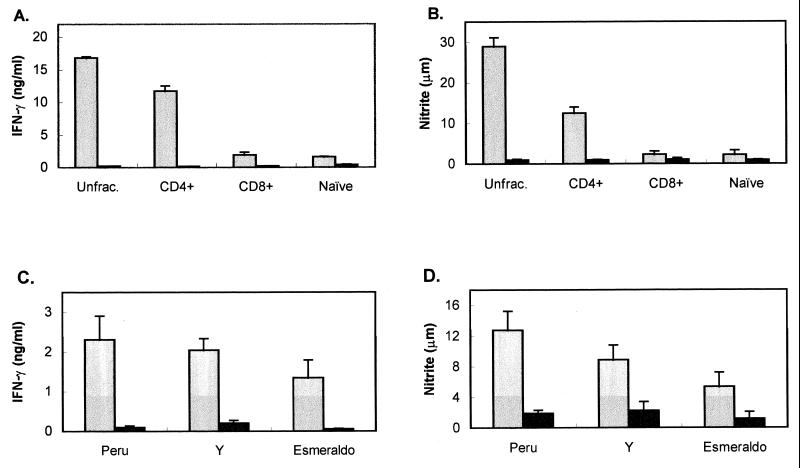
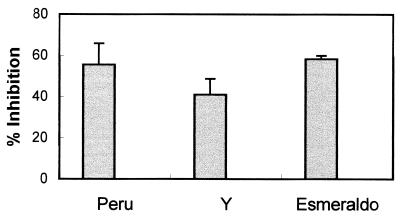
Previous results obtained with PFR-immunized mice suggested that proteins present in the invasive trypomastigote form and metabolized during transformation to the intracellular amastigote form of the parasite may be presented by MHC molecules on the surface of infected macrophages (21). These studies also indicated that in PFR-immunized mice, much of the parasite reduction observed following a T. cruzi challenge is a result of CD4+ T cells functioning as effector cells to activate T. cruzi-infected macrophages, leading to NO production and a concomitant reduction of parasite numbers (21). To determine whether other trypomastigote-specific proteins also may be presented by MHC molecules on T. cruzi-infected macrophages, mice were immunized with recombinant TolT protein, and the ability of T cells from TolT-immunized mice to recognize and activate T. cruzi-infected macrophages was determined. Nylon wool-purified T cells or MACS MultiSort MicroBeads-selected CD4+ or CD8+ T cells from both TolT-immunized and naive C57BL/6 mice were cultured with IC-21 macrophages that had been infected with T. cruzi, left untreated, or cultured in the presence of recombinant TolT antigen. T-cell–macrophage culture supernatants were collected on days 3, 5, and 7, and IFN-γ levels, nitrite concentrations, and parasite numbers were determined. Since the population structure of T. cruzi shows that different strains of the parasite are separated by great genetic distances (40), thus indicating high genetic diversity within the population, three strains of the parasite that differ in a number of biological traits were used in the macrophage activation assay. As shown in Fig. 5, levels of IFN-γ and NO2 in the culture supernatants of macrophages treated with TolT antigen in the presence of purified T cells or CD4+ T cells from TolT-immunized mice were significantly higher than those in cultures containing naive T cells. In contrast, no IFN-γ or NO2 above background levels could be detected in cultures containing CD8+ T cells. Significant levels of IFN-γ and NO2 also were observed in the culture supernatants of purified T cells obtained from TolT-immunized mice and incubated with macrophages infected with the different strains of T. cruzi. Consistent with previous results which showed that NO production in infected macrophages is accompanied by a reduction in parasite replication, culture supernatants in which significant levels of NO2 were observed also contained about 50 to 60% fewer parasites than culture supernatants that showed background levels of NO2 (Fig. 6).
FIG. 5.
Measurement of NO and IFN-γ induced by T cells from Tol-immunized mice. Non-nylon wool-adherent spleen cells were cultured with IC-21 macrophages in the presence of antigen or parasites. Supernatants were collected on days 4 and 7 and assayed for nitrite and IFN-γ as described in Materials and Methods. (A and B) Unfractionated T cells (Unfrac.), CD4+-selected T cells, CD8+-selected T cells, or naive T cells were cultured with TolT protein (3 μg/ml; gray bars) or no antigen (black bars), and culture supernatants were assayed for IFN-γ (A) and nitrite (B). μm, micromolar. (C and D) T cells were cultured in the presence of T. cruzi-infected IC-21 macrophages, and culture supernatants were assayed for IFN-γ (C) and nitrite (D). The gray bars represent cultures from TolT-immunized mice, and the black bars represent cultures from nonimmunized mice. Peru, Y, and Esmeraldo denote three genetically diverse clonal lines of T. cruzi. Each bar represents the average values ± standard deviations obtained from triplicate cultures. Each experiment was performed three times.
FIG. 6.
Parasiticidal activity of macrophages activated by T cells from TolT-immunized mice. T cells isolated by nylon wool nonadherence were cultured with T. cruzi-infected IC-21 macrophages. On days 4 and 7, the number of motile trypomastigotes in the culture supernatants was counted with a hemocytometer. The percentage of inhibition was calculated as 100 × [(1 − number of parasites in immunized cultures)/number of parasites in nonimmunized cultures]. Each bar represents the average values ± standard deviations obtained from triplicate cultures. Each experiment was performed three times.
T cells from TolT-immunized mice produce a T-helper 1 cytokine response.
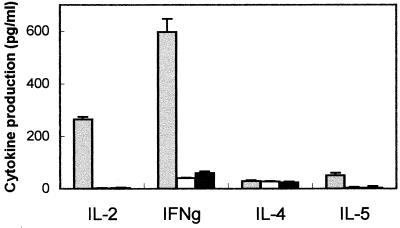
It has been shown that the parasiticidal activity in mice immunized with PFR proteins is associated with a T-helper 1 response (20). To determine whether this is also the case in mice immunized with TolT protein, we measured the in vitro levels of the cytokines IFN-γ, IL-2, IL-4, and IL-5 in the culture supernatants of TolT-primed T cells stimulated in vitro with recombinant TolT protein by using a capture ELISA. As shown in Fig. 7, the only cytokines whose concentrations were above background levels were IFN-γ and IL-2, the signature cytokines for T-helper 1 cells.
FIG. 7.
Cytokine profile induced by immunization with TolT protein. T cells were cultured with TolT protein (3 μg/ml) and irradiated syngeneic feeder cells. Supernatants were collected on days 2, 3, 5, and 7 of culturing and assayed for IL-2, IL-4, IL-5, and IFN-γ (IFNg) levels by a capture ELISA as described in Materials and Methods. Values for each cytokine are presented for the peak days of production, which were as follows: IL-2, day 2; IFN-γ, day 5; IL-4, day 3; and IL-5, day 5. The gray bars represent cultures of T cells from TolT-immunized mice cultured with antigen. The white bars represent cultures of T cells from TolT-immunized mice cultured without antigen. The black bars represent cultures of T cells from nonimmunized mice. Each bar represents the average values ± standard deviations obtained from triplicate cultures.
T cells from T. cruzi-infected mice recognize TolT.
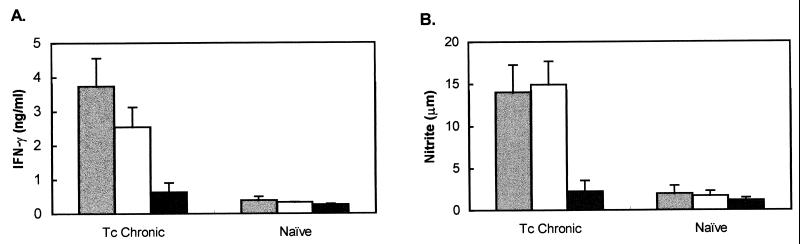
To determine whether mice that are chronically infected with T. cruzi contain T lymphocytes that recognize TolT and/or PFR antigen, 10 C57BL/6 mice were infected with five parasites/mouse. Of these, three survived the acute infection. At approximately 3 months postinfection, these mice were boosted with 103 bloodstream trypomastigotes. Two weeks after the boost, T cells were obtained from these mice, nylon wool purified, added to IC-21 macrophages, and cultured in the presence of either TolT protein, PAR protein, or no antigen. Identical control cultures were also established, except that purified T cells were obtained from naive mice. T-cell recognition and activation were monitored by measurement of IFN-γ and IL-4 levels in culture supernatants. IL-4 was not detected in any of the culture supernatants (data not shown). In contrast, levels of IFN-γ in the supernatants of cultures containing T cells from infected mice and macrophages incubated with either TolT or PFR protein were significantly higher than IFN-γ levels in control cultures containing T cells from naive mice (Fig. 8), thus indicating that both of these flagellar proteins produce positive T-cell responses in mice chronically infected with T. cruzi.
FIG. 8.
Measurement of NO and IFN-γ induced by T cells from mice chronically infected with T. cruzi. To establish chronic infections, mice were injected with five Peru strain trypomastigotes. At 3 months postinfection, mice were given a challenge infection of 103 trypomastigotes. Two weeks after the boost, T cells were isolated by nylon wool nonadherence. T cells from chronic mice and naive mice were cultured with PFR protein (gray bars), TolT protein (white bars), or no antigen (black bars) in the presence of IC-21 macrophages. Supernatants were removed from cultures at days 4 and 7 and assayed for IFN-γ (A) and nitrite (B). μm, micromolar. Bars represent average values ± standard deviations obtained from triplicate cultures.
TSA-1-specific CD4+ T lymphocytes do not recognize T. cruzi-infected macrophages.
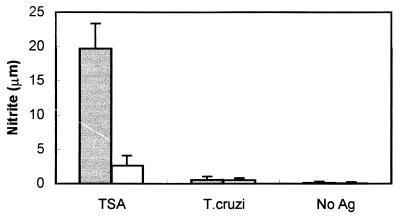
Since both PFR (21) and TolT protein generate CD4+ T lymphocytes that recognize and activate T. cruzi-infected macrophages, we wished to determine whether this immune response is common to other T. cruzi proteins, particularly those not associated with the flagellum. To address this question, mice were immunized with recombinant TSA-1 protein, which is not present on the flagellum (27). CD4+ T cells from these mice were purified and assayed for their ability to recognize and activate T. cruzi-infected macrophages. We determined the levels of NO2 in the supernatants of cultures containing nylon wool-purified CD4+ T cells from TSA-1-immunized and naive C57BL/6 mice and IC-21 macrophages that were either infected with T. cruzi, cultured in the presence of recombinant TSA-1, or left untreated. As shown in Fig. 9, the level of NO2 observed in the culture supernatants of macrophages treated with TSA-1 in the presence of purified immune CD4+ T cells was significantly higher than that observed in similar cultures containing T cells from naive mice. In contrast, the level of NO2 in cultures containing immune CD4+ T cells and T. cruzi-infected macrophages was the same as that in cultures containing naive T cells incubated with infected macrophages. These results clearly demonstrate that TSA-1 does not enter the class II MHC presentation pathway in T. cruzi-infected macrophages in amounts sufficient for activation by TSA-1-specific CD4+ T lymphocytes.
FIG. 9.
Measurement of NO induced by T cells from TSA-1-immunized mice. CD4+ T cells isolated by use of MACS MultiSort MicroBeads from naive mice (white bars) and mice immunized with TSA-1 (gray bars) were cultured with macrophages infected with T. cruzi or macrophages incubated in the presence of TSA-1. On days 4 and 7 of culturing, supernatants were removed and assayed for nitrite. Bars represent average values ± standard deviations obtained from triplicate cultures. μm, micromolar; Ag, antigen.
T cells from mice immunized with TolT fail to respond to shed-antigen preparations or HKTC.
Previous work has shown that trypomastigote-specific antigens are shed or secreted by parasites during in vitro culturing and are capable of activating CD4+ T cells (26). To investigate the possibility that the responses of T cells from TolT-immunized mice to parasite-infected macrophages were due to the secretion or shedding of TolT antigen into the cultures, preparations of antigen shed by live trypomastigotes were tested for their ability to activate T cells from TolT-immunized mice. Antigens derived from 2- and 24-h trypomastigote cultures were added to IC-21 macrophages and TolT-immunized T cells. On days 4 and 7 of culturing, supernatants were removed and assayed for nitrite and IFN-γ levels. All culture supernatants were negative for nitrite and IFN-γ production (data not shown), indicating that the shed-antigen preparation failed to activate T cells from TolT-immunized mice.
To monitor the antigenic composition of the shed or secreted antigens, trypomastigotes were labeled with 35S-methionine, and the supernatants were analyzed by SDS-PAGE and fluorography. The fluorograph of supernatants obtained from 35S-methionine-labeled trypomastigotes revealed diffuse bands of protein in the 150- to 200-kDa range and the 80- to 100-kDa range, as well as minor bands at 50 to 60 kDa (data not shown). Although no radioactivity was observed in the 31- to 40-kDa region, corresponding to the Mr of TolT, Western blots of the shed-antigen preparations were probed with MAb 20H1 to determine whether TolT protein was present. Control blots containing recombinant TolT protein ranging in amounts from 0.5 ng to 5 μg were also analyzed. Following reaction of the blots with antibody and detection by chemiluminescence, TolT protein was detected. A direct comparison with the control blots containing recombinant protein indicated that the concentration of TolT present in the shed-antigen preparation was <40 ng/ml. Therefore, it is not surprising that we observed no activation of T cells cultured with the shed-antigen preparation, since the amount of recombinant TolT protein required for induction of detectable levels of IFN-γ by T cells from TolT-immunized mice is >400 ng/ml (data not shown).
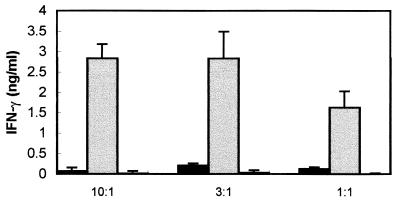
We next considered the possibility that the activation of the TolT-immunized T cells was due to the presence of dead parasites in the culture. To directly test this possibility, T cells from TolT-immunized mice were cultured with HKTC added at various ratios of trypomastigotes to macrophages. As a positive control, the responses of T cells isolated from mice immunized with HKTC were also analyzed. On days 4 and 7 of culturing, supernatants were removed and assayed for nitrite and IFN-γ levels. As shown in Fig. 10, T cells from mice immunized with HKTC at ratios of 10:1, 3:1, and 1:1 HKTC per macrophage produced IFN-γ. In contrast, T cells from mice immunized with TolT produced barely detectable levels of IFN-γ, even when cultured with 10:1 HKTC per macrophage, and no nitrite was detected (data not shown).
FIG. 10.
Measurement of IFN-γ from T cells cultured in the presence of HKTC. Non-nylon wool-adherent T cells from mice immunized with TolT protein (black bars) or HKTC (gray bars) or from naive mice (white bars) were cultured with IC-21 macrophages pulsed with HKTC at the ratios shown. On days 4 and 7 of culturing, supernatants were removed and assayed for IFN-γ levels by an ELISA. Bars represent average values ± standard deviations obtained from triplicate cultures.
DISCUSSION
The major purpose of this study was to test the hypothesis that flagellar proteins released during the transformation of the trypomastigote form to the amastigote form of T. cruzi are presented on the surface of infected cells in a configuration recognizable by antigen-primed T cells. To examine this possibility, we selected an antigen that previously was shown by immunofluorescence and immunoelectron microscopic analysis to be present in the trypomastigote stage but not the epimastigote or amastigote stage of the parasite (33). The gene that encodes this protein was cloned, characterized, and expressed in baculovirus and bacterial expression systems. Mice were immunized with the baculovirus-produced recombinant TolT antigen, and T cells from these mice were examined for their ability to recognize T. cruzi-infected macrophages or macrophages incubated with TolT antigen produced in bacteria. Our principle findings are as follows.
(i) The genes that encode the antigen recognized by MAb 20H1 belong to a small gene family containing three members arranged in a tandem array within the genome. (ii) The three members of the family are almost identical in their nucleotide sequence, predicting that very little amino acid sequence polymorphism would be present in predicted proteins encoded by these genes. (iii) The protein encoded by these genes is differentially glycosylated. (iv) The protein encoded by this gene family shares numerous physical and biological features with the TolA proteins of E. coli and P. aeruginosa. (v) CD4+ T cells from TolT-immunized mice are capable of stimulating T. cruzi-infected macrophages to produce NO, which results in strong inhibition of parasite growth. (vi) The fact that macrophages infected with the Esmeraldo and Y strains of T. cruzi were recognized by T cells from mice immunized with TolT protein produced by the tolT gene of the Peru strain suggests that immunological recognition is not parasite strain specific. (vii) T cells from mice chronically infected with T. cruzi recognize both TolT and PFR antigens presented by IC-21 macrophages.
Five different proteins were identified by a search of databases with the TolT protein. One of these proteins, plectin, is a large (4,687-aa) protein prominently located at the plasma membrane attachment sites of intermediate filaments and microfilaments (41). It contains a long alpha-helical central domain flanked by two large globular domains. Sequence identity between plectin and TolT occurs in the predicted alpha-helical regions of the central portions of both proteins. A similar situation exists with MST1 and MST2 of Drosophila. Both of these proteins are members of the sperm tail-specific axoneme-associated protein family Dhmst101, which forms extended alpha-helical rods within the spermatozoa of Drosophila hydei (23, 24). Regions of identity with TolT are localized to the alpha-helical regions of both proteins. Since the functional role of the alpha-helical regions of the plectin protein and the MST proteins is to provide structural integrity to the cell and to the sperm tail, respectively, it seems likely that the central alpha-helical region of the TolT protein performs a similar function in the membrane of T. cruzi. Consistent with this view is the observation that one of several structural features which TolT shares with the TolA proteins of E. coli and P. aeruginosa is significant sequence identity (i.e., 33 to 34%) within the predicted alpha-helical regions of the proteins.
In E. coli, TolA is an inner membrane protein with three distinct domains (15). The amino-terminal 40 residues contain a region of 21 hydrophobic aas capable of forming a transmembrane region that anchors the TolA protein in the inner membrane. The central domain contains a long periplasmic region of 260 aas with a predicted high alpha-helical content. Both the central domain and the C-terminal domain (ca. 120 residues) function to maintain the integrity of the outer membrane and facilitate the uptake of the group A colicins (16, 36).
Analysis of the predicted amino acid sequence of the TolT protein reveals that it has several chemical and physical characteristics in common with the TolA protein (3). (i) The predicted numbers of amino acid residues in the proteins are similar (i.e., for TolT, 310 aas; for E. coli TolA, 421 aas; and for P. aeruginosa TolA, 335 aas). (ii) All are integral membrane proteins. (iii) A transmembrane segment is predicted within the N-terminal 30 residues of each protein. (iv) The central and C-terminal domains are separated by polyglycine sequences which are believed to serve as a flexible hinge (15). (v) The amino acid compositions are very similar (i.e., 48 to 46% hydrophobic aas, 21 to 22% polar aas, 16 to 18% basic aas, and 14 to 16% acidic aas). Possibly of equal importance, analysis of the predicted secondary structures of these proteins with five separate algorithms (4, 5, 11, 17, 19) indicates coil or helix motifs at almost identical regions within the three proteins.
Since the bacterial TolA proteins and the T. cruzi protein share many biological, chemical, and physical features, it seems likely that they represent genetic homologues. Consistent with this suggestion is the observation that a dendrogram of the relative sequence distances among these three proteins shows that the sequence distance between the TolA protein of E. coli and the T. cruzi protein is approximately the same as the sequence distance between the TolA proteins of E. coli and P. aeruginosa. Based on these analyses, we have tentatively designated the protein recognized by MAb 20H1 as TolT.
One of the primary functions of the TolA protein of E. coli is to provide stability to the cell envelope. Deletion mutations in the central domain are sufficient to destabilize the interaction of the inner and outer membranes, leading to enhanced susceptibility to lysogenic agents (36). The importance of the TolA protein for cell viability is evidenced by the fact that attempts to inactivate the chromosomal copy of the tolA gene have been unsuccessful, suggesting that inactivation of the tolA gene results in a lethal phenotype (6). The many similarities observed between the TolA and TolT proteins suggest that TolT may also play an essential role in the trypomastigote surface membrane. However, since T. cruzi does not have a double-membrane structure similar to that of E. coli, it seems unlikely that TolT would have the same membrane-stabilizing function in T. cruzi as TolA has in E. coli.
T-cell responses directed toward the mammalian stages of T. cruzi have long been recognized as important to protection against this parasite (38). However, few defined antigens that generate protective T-cell responses have been identified. Recent studies have identified one trypomastigote surface antigen, TSA-1 (42), and two amastigote surface proteins, ASP-1 and ASP-2 (18), that are targets of T. cruzi-specific CD8+ T-cell responses. TSA-1-, ASP-1-, and ASP-2-specific cytotoxic T lymphocytes lyse cells infected with T. cruzi, thus demonstrating that these antigens enter the class I MHC presentation pathway in infected cells. Furthermore, immunization of mice with either recombinant TSA-1 (44) or DNA vaccines that encode TSA-1 (43) provides partial protection against a T. cruzi challenge.
Antigens that are known to enter the class II MHC presentation pathway in T. cruzi-infected cells and generate protective CD4+ T-cell-dependent responses in immunized mice are even fewer in number. In an effort to identify such antigens, Nickell et al. (25) isolated nine murine clonal CD4+ T-cell lines that specifically recognized antigens expressed by the trypomastigote stage of the parasite. Of these, three lines recognized and activated T. cruzi-infected macrophages, resulting in intracellular killing of the parasite. Adoptive transfer of two of these three lines also protected mice against a subsequent T. cruzi challenge. Unfortunately, subsequent studies were not successful in definitively identifying the antigen(s) recognized by these two protective T-cell lines (26). In a somewhat similar study (22), two CD4+ T-cell lines which specifically proliferated against parasite antigens were established from infected mice. However, only one of these cell lines was capable of inducing trypanocidal activity in T. cruzi-infected macrophages, and the T. cruzi antigen(s) recognized by this clonal line was not reported. While these studies did not identify the specific antigens recognized by the clonal T-cell lines, they clearly showed that some but not all CD4+ T. cruzi-specific T-cell lines activate macrophages to kill intracellular T. cruzi amastigotes. These results suggest that some T. cruzi antigens that generate CD4+ T-cell responses might not enter the class II MHC presentation pathway in infected macrophages.
To our knowledge, only one defined T. cruzi antigen, the PFR protein, has been shown to protect immunized mice against an otherwise lethal challenge with T. cruzi and to generate CD4+ T cells that recognize and activate trypanocidal activity in T. cruzi-infected macrophages (21). The results presented here now identify a second flagellar protein, TolT, which shows immunological properties similar to those of the PFR protein. First, immunization of mice with TolT generates CD4+ T cells that recognize and activate macrophages infected with T. cruzi, resulting in intracellular killing of the parasite; these findings definitively demonstrate that in the infected host cell, TolT protein enters the class II MHC presentation pathway. Second, T-cell responses to both the TolT and the PFR proteins occur in T. cruzi-infected mice (Fig. 8).
A feasible explanation of the observation that both the PFR and the TolT proteins enter the class II MHC presentation pathway in T. cruzi-infected cells may reside in the complex life cycle of the parasite. Prior to replication in the mammalian host, the invasive trypomastigote undergoes an obligatory transformation to the amastigote in the host cell. This event results in the catabolism of >90% of the flagellum, potentially providing degradation products of flagellar proteins for entry into the vacuolar class II MHC presentation pathway. However, an alternative explanation of the TolT and PFR protein results may be that one or a few parasites invading macrophages are killed during this process, allowing released antigens to be degraded in the phagolysosome and to enter the class II MHC presentation pathway. If the latter were the case, it might be anticipated that many, and possibly most, of the proteins present in the mammalian stage of the parasite might enter the class II MHC presentation pathway in infected macrophages. Although this possibility is not in accord with the finding that only some parasite-derived CD4+ T-cell lines recognize T. cruzi-infected macrophages (22, 25), we chose to further explore this possibility.
Previous studies showed that an antibody response against TSA-1 was generated in mice immunized with recombinant TSA-1 (44), suggesting that TSA-1-specific CD4+ T cells may also have been generated in these mice. As shown in Fig. 9, this is the case. Immunization of mice with baculovirus-produced TSA-1 generated CD4+ T cells that can recognize and activate macrophages cultured in the presence of bacterially produced TSA-1. In contrast, these same CD4+ T cells did not recognize or activate macrophages infected with T. cruzi, indicating that in T. cruzi-infected cells, TSA-1 does not enter the class II MHC presentation pathway in sufficient amounts to result in the activation of CD4+ T cells.
To directly investigate the possibility that the activation of T cells from TolT-immunized mice was due to the phagocytosis of dead trypomastigotes by IC-21 macrophages, we measured the responses of T cells from TolT-immunized mice to macrophages pulsed with HKTC. As shown in Fig. 10, the levels of IFN-γ produced by T cells from TolT-immunized mice cultured with HKTC and IC-21 macrophages were similar to those observed for naive mice, whereas T cells from mice immunized with HKTC produced substantial IFN-γ levels under identical culture conditions. We therefore believe that it is extremely unlikely that the TolT antigen presented by infected macrophages and recognized by T cells from TolT-immunized mice is derived from dead parasites in the culture.
Since several investigators have reported the ability of live trypomastigotes to release protein via shedding or secretion processes, we considered the possibility that antigen shed or secreted in cultures might be responsible for the activation of the T cells from TolT-immunized mice. To test this possibility, T cells from TolT-immunized mice were cultured with macrophages in the presence of a shed-antigen preparation from trypomastigotes. No IFN-γ or nitrite was detected in the culture supernatants, indicating that the TolT protein was not present in sufficient concentrations for T-cell activation. Therefore, the activation of T cells from TolT-immunized mice cannot be explained by the presentation of the TolT protein present in the culture media as the result of shedding or secretion by trypomastigotes. Rather, we believe that these results further support the hypothesis that the antigen(s) responsible for the activation of T cells from TolT-immunized mice is derived from live parasites during the invasion and transformation process.
In conclusion, these studies report the isolation and characterization of a small, highly conserved gene family, tolT, which encodes a trypomastigote-specific flagellar protein that induces a CD4+ immune response in immunized mice and mice chronically infected with T. cruzi. The TolT protein has several features that make it an attractive vaccine candidate. These are (i) its ability to induce a T-helper 1-type T-lymphocyte response that results in parasiticidal activity in T. cruzi-infected macrophages following immunization with recombinant TolT antigen, (ii) the production of TolT-specific T cells in mice chronically infected with T. cruzi, and (iii) its immunologic cross-reactivity among parasite strains. Also, the fact that both PFR and TolT are flagellar proteins that are degraded during the transformation of trypomastigotes to amastigotes in infected cells supports the proposition that flagellum-associated proteins may be a rich source of prospective vaccine candidates.
ACKNOWLEDGMENTS
We thank Barbara Granger for assistance with the maintenance and growth of parasite cultures.
This work was supported by Public Health Service grant AI18873 from the National Institutes of Health (to J.E.M.).
REFERENCES
- 1.Beard C A, Saborio J L, Tewari D, Krieglstein K G, Henschen A H, Manning J E. Evidence for two distinct major protein components, PAR 1 and PAR 2, in the paraflagellar rod of Trypanosoma cruzi. J Biol Chem. 1992;267:21656–21662. [PubMed] [Google Scholar]
- 2.Beard C A, Wrightsman R A, Manning J E. Stage and strain specific expression of the tandemly repeated 90 kDa surface antigen gene family in Trypanosoma cruzi. Mol Biochem Parasitol. 1988;28:227–234. doi: 10.1016/0166-6851(88)90007-2. [DOI] [PubMed] [Google Scholar]
- 3.Brendel V, Bucher P, Nourbakhsh I R, Blaisdell B E, Karlin S. Methods and algorithms for statistical analysis of protein sequences. Proc Natl Acad Sci USA. 1992;89:2002–2006. doi: 10.1073/pnas.89.6.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chou K C. A novel approach to predicting protein structural classes in a (20-1)-D amino acid composition space. Proteins. 1995;21:319–344. doi: 10.1002/prot.340210406. [DOI] [PubMed] [Google Scholar]
- 5.Deleage G, Roux B. Use of class prediction to improve protein secondary structure prediction: joint prediction with methods based on sequence homology. In: Fasman G D, editor. Prediction of protein structure and the principles of protein conformation. New York, N.Y: Plenum Press; 1989. pp. 147–196. [Google Scholar]
- 6.Dennis J J, Lafontaine E R, Sokol P A. Identification and characterization of the tolQRA genes of Pseudomonas aeruginosa. J Bacteriol. 1996;178:7059–7068. doi: 10.1128/jb.178.24.7059-7068.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fouts D L, Manning J E, Fox G M, Schmid C W. A complex repeated DNA sequence within the Drosophila transposable element copia. Nucleic Acids Res. 1981;9:7053–7064. doi: 10.1093/nar/9.24.7053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fouts D L, Ruef B J, Ridley P T, Wrightsman R A, Peterson D S, Manning J E. Nucleotide sequence and transcription of a trypomastigote surface antigen gene of Trypanosoma cruzi. Mol Biochem Parasitol. 1991;46:189–200. doi: 10.1016/0166-6851(91)90043-6. [DOI] [PubMed] [Google Scholar]
- 9.Julius M H, Simpson E, Herzenberg L A. A rapid method for the isolation of functional thymus-derived lymphocytes. Eur J Immunol. 1973;3:645–649. doi: 10.1002/eji.1830031011. [DOI] [PubMed] [Google Scholar]
- 10.Kierszenbaum F, Howard J G. Mechanisms of resistance against experimental Trypanosoma cruzi infection: the importance of antibodies and antibody-forming capacity in the Biozzi high and low responder mice. J Immunol. 1976;116:1208–1211. [PubMed] [Google Scholar]
- 11.King R D, Sternberg J E. Machine learning approach for the prediction of protein secondary structure. J Mol Biol. 1990;216:441–457. doi: 10.1016/S0022-2836(05)80333-X. [DOI] [PubMed] [Google Scholar]
- 12.Krettli A U, Brener Z. Protective effects of specific antibodies in Trypanosoma cruzi infections. J Immunol. 1976;116:755–761. [PubMed] [Google Scholar]
- 13.Lanar D. Growth and isolation of Trypanosoma cruzi cultivated with Triatoma infestans embryo cell line. J Protozool. 1979;26:457–462. doi: 10.1111/j.1550-7408.1979.tb04653.x. [DOI] [PubMed] [Google Scholar]
- 14.Lebowitz J H, Smith H Q, Rusche L, Beverley S M. Coupling of poly(A) site selection and transsplicing in Leishmania. Genes Dev. 1993;7:996–1007. doi: 10.1101/gad.7.6.996. [DOI] [PubMed] [Google Scholar]
- 15.Levengood S K, Beyer W F J, Webster R E. TolA: a membrane protein involved in colicin uptake contains an extended helical region. Proc Natl Acad Sci USA. 1991;88:5939–5943. doi: 10.1073/pnas.88.14.5939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levengood-Freyermuth S K, Click E M, Webster R E. Role of the carboxyl-terminal domain of TolA in protein import and integrity of the outer membrane. J Bacteriol. 1993;175:222–228. doi: 10.1128/jb.175.1.222-228.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levin J M, Garnier J. Improvements in a secondary structure prediction method based on a search for local sequence homologies and its use as a model building tool. Biochim Biophys Acta. 1988;95:283–295. doi: 10.1016/0167-4838(88)90206-3. [DOI] [PubMed] [Google Scholar]
- 18.Low H P, Santos M A M, Wizel B, Tarleton R L. Amastigote surface proteins of Trypanosoma cruzi are targets for CD8+ CTL. J Immunol. 1998;160:1817–1823. [PubMed] [Google Scholar]
- 19.Mao B, Chou K C, Zhang C T. Protein folding classes: a geometric interpretation of the amino acid composition of globular proteins. Protein Eng. 1994;7:319–330. doi: 10.1093/protein/7.3.319. [DOI] [PubMed] [Google Scholar]
- 20.Miller M J, Wrightsman R A, Manning J E. Trypanosoma cruzi: protective immunity in mice immunized with paraflagellar rod proteins is associated with a T-helper type 1 response. Exp Parasitol. 1996;84:156–167. doi: 10.1006/expr.1996.0101. [DOI] [PubMed] [Google Scholar]
- 21.Miller M J, Wrightsman R A, Stryker G A, Manning J E. Protection of mice against Trypanosoma cruzi by immunization with paraflagellar rod proteins requires T cell, but not B cell, function. J Immunol. 1997;158:5330–5337. [PubMed] [Google Scholar]
- 22.Munoz-Fernandez M A, Fernandez M A, Fresno M. Synergism between tumor necrosis factor-α and interferon-γ on macrophage activation for the killing of intracellular Trypanosoma cruzi through a nitric oxide-dependent mechanism. Eur J Immunol. 1992;22:301–307. doi: 10.1002/eji.1830220203. [DOI] [PubMed] [Google Scholar]
- 23.Neesen J, Padmanabhan S, Bunemann H. Tandemly arranged repeats of a novel highly charged 16-amino-acid motif representing the major component of the sperm-tail-specific axoneme-associated protein family Dhmst101 form extended alpha-helical rods within the extremely elongated spermatozoa of Drosophila hydei. Eur J Biochem. 1994;225:1089–1095. doi: 10.1111/j.1432-1033.1994.1089b.x. [DOI] [PubMed] [Google Scholar]
- 24.Neesen J, Bunemann H, Heilein U A. The Drosophila hydei gene Dhmst101(1) encodes a testis-specific, repetitive, axoneme-associated protein with differential abundance in Y chromosomal deletion mutant flies. Dev Biol. 1994;162:414–425. doi: 10.1006/dbio.1994.1098. [DOI] [PubMed] [Google Scholar]
- 25.Nickell S P, Gebremichael A, Hoff R, Boyer M H. Isolation and functional characterization of murine T cell lines and clones specific for the protozoan parasite Trypanosoma cruzi. J Immunol. 1987;138:914–921. [PubMed] [Google Scholar]
- 26.Nickell S P, Keane M, So M. Further characterization of protective Trypanosoma cruzi-specific CD4+ T-cell clones: T helper type 1-like phenotype and reactivity with shed trypomastigote antigens. Infect Immun. 1993;61:3250–3258. doi: 10.1128/iai.61.8.3250-3258.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peterson D S, Wrightsman R A, Manning J E. Cloning of a major surface-antigen gene of Trypanosoma cruzi and identification of a nonapeptide repeat. Nature. 1985;322:566–568. doi: 10.1038/322566a0. [DOI] [PubMed] [Google Scholar]
- 28.Rodriguez A M, Santor F, Afchain D, Bazin H, Capron A. Trypanosoma cruzi infection in B-cell-deficient rats. Infect Immun. 1981;31:524–529. doi: 10.1128/iai.31.2.524-529.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rottenberg M E, Bakhiet M, Olsson T, Kristensson K, Mak T, Wigzell H, Orn A. Differential susceptibilities of mice genomically deleted of CD4 and CD8 to infections with Trypanosoma cruzi or Trypanosoma brucei. Infect Immun. 1993;61:5129–5133. doi: 10.1128/iai.61.12.5129-5133.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rottenberg M E, Riarte A, Sporrong L, Altcheh J, Petray P, Ruiz A M, Wigzell H, Orn A. Outcome of infection with different strains of Trypanosoma cruzi in mice lacking CD4 and/or CD8. Immunol Lett. 1994;45:53–60. doi: 10.1016/0165-2478(94)00221-c. [DOI] [PubMed] [Google Scholar]
- 31.Rozed C E, Davidson N. Drosophila has one myosin heavy-chain gene with three developmentally regulated transcripts. Cell. 1983;32:23–34. doi: 10.1016/0092-8674(83)90493-2. [DOI] [PubMed] [Google Scholar]
- 32.Saborio J L, Hernandez J M, Narayanswami S, Wrightsman R A, Palmer E, Manning J E. Isolation and characterization of paraflagellar proteins from Trypanosoma cruzi. J Biol Chem. 1989;264:4071–4075. [PubMed] [Google Scholar]
- 33.Saborio J L, Wrightsman R A, Kazuko S G, Granger B S, Manning J E. Trypanosoma cruzi: identification of a surface antigen restricted to the flagellar region of the infective form of the parasite. Exp Parasitol. 1990;70:411–418. doi: 10.1016/0014-4894(90)90125-v. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 35.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schendel S L, Click E M, Webster R E, Cramer W A. The TolA protein interacts with colicin E1 differently than with other group A colicins. J Bacteriol. 1997;179:3683–3690. doi: 10.1128/jb.179.11.3683-3690.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takehara H A, Perini A, Da Silva M H, Mota I. Trypanosoma cruzi: role of different antibody classes in protection against infection in the mouse. Exp Parasitol. 1981;52:137–146. doi: 10.1016/0014-4894(81)90069-2. [DOI] [PubMed] [Google Scholar]
- 38.Tarleton R L. Immunity to Trypanosoma cruzi. In: Kaufman S H E, editor. Host response to intracellular pathogens. R. G. Austin, Tex: Landes Co.; 1997. pp. 227–247. [Google Scholar]
- 39.Tarleton R L, Koller B H, Latour A, Postan M. Susceptibility of β2-microglobulin-deficient mice to Trypanosoma cruzi infection. Nature. 1992;356:338–340. doi: 10.1038/356338a0. [DOI] [PubMed] [Google Scholar]
- 40.Tibayrenc M. Population genetics of parasitic protozoa and other microorganisms. Adv Parasitol. 1995;36:47–115. doi: 10.1016/s0065-308x(08)60490-x. [DOI] [PubMed] [Google Scholar]
- 41.Wiche G, Becker B, Luber K, Weitzer G, Castanon M J, Hauptmann R, Stratowa C, Stewart M. Cloning and sequencing of rat plectin indicates a 466-kD polypeptide chain with a three-domain structure based on a central alpha-helical coiled coil. J Cell Biol. 1991;114:83–99. doi: 10.1083/jcb.114.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wizel B, Nunes M, Tarleton R L. Identification of Trypanosoma cruzi trans-sialidase family members as targets of protective CD8+ Tc1 responses. J Immunol. 1997;159:6120–6130. [PubMed] [Google Scholar]
- 43.Wizel B, Garg N, Tarleton R L. Vaccination with trypomastigote surface antigen 1-encoding plasmid DNA confers protection against lethal Trypanosoma cruzi infection. Infect Immun. 1998;66:5073–5081. doi: 10.1128/iai.66.11.5073-5081.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wrightsman R A, Dawson B D, Fouts D L, Manning J E. Identification of immunodominant epitopes in Trypanosoma cruzi trypomastigote surface antigen-1 protein that mask protective epitopes. J Immunol. 1994;153:3148–3154. [PubMed] [Google Scholar]
- 44a.Wrightsman, R. A., and J. E. Manning. Unpublished data.
- 45.Wrightsman R A, Miller M J, Saborio J L, Manning J E. Pure paraflagellar rod protein protects mice against Trypanosoma cruzi infection. Infect Immun. 1995;63:122–125. doi: 10.1128/iai.63.1.122-125.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]