Abstract
The continued evolution of SARS-CoV-2 has led to the emergence of several new Omicron subvariants, including BQ.1, BQ.1.1, BA.4.6, BF.7, and BA.2.75.2. Here, we examine the neutralization resistance of these subvariants against sera from 3-dose vaccinated healthcare workers, hospitalized BA.1-wave patients, and BA.4/5-wave patients. We found enhanced neutralization resistance in all new subvariants, especially in the BQ.1 and BQ.1.1 subvariants driven by N460K and K444T mutations, as well as the BA.2.75.2 subvariant driven largely by its F486S mutation. All Omicron subvariants maintained their weakened infectivity in Calu-3 cells, with the F486S mutation driving further diminished titer for the BA.2.75.2 subvariant. Molecular modeling revealed the mechanisms of antibody-mediated immune evasion by R346T, K444T, F486S, and D1199N mutations. Altogether, these findings shed light on the evolution of newly emerging SARS-CoV-2 Omicron subvariants.
Keywords: Omicron subvariants, neutralization, immune evasion, mutations, molecular modeling
Graphical abstract
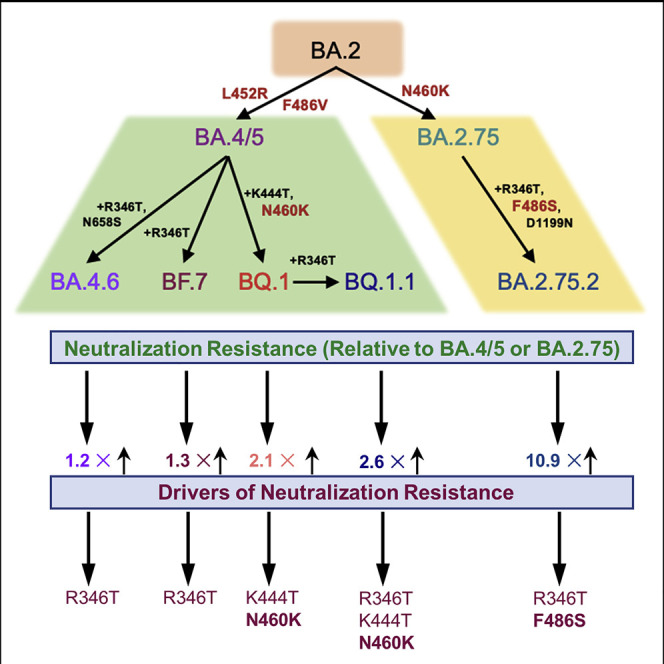
Numerous Omicron subvariants have emerged following BA.4/5 and BA.2.75 subvariants. Qu and colleagues investigate the neutralizing antibody resistance of these subvariants and their ancestral variants. BQ.1, BQ.1.1, BA.4.6, BF.7, and BA.2.75.2 exhibit enhanced neutralizing antibody escape, with BQ.1/BQ.1.1 and BA.2.75.2 driven by N460K/K444T and F486S mutations, respectively.
Introduction
Since its emergence in late 2021, the Omicron variant of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has led to the emergence of numerous subvariants that continue to evade vaccine- and infection-induced immunity and alter the virus biology.1 , 2 , 3 , 4 , 5 , 6 , 7 The initial BA.1 Omicron subvariant drove a large wave of coronavirus disease 2019 (COVID-19) cases and exhibited strong immune escape from 2-mRNA vaccine dose-induced immunity that was recovered by a booster mRNA vaccine administration.1 , 8 , 9 , 10 , 11 , 12 , 13 , 14 In addition, the BA.1 subvariant exhibited reduced cell-cell fusogenicity, impaired replication in lower airway epithelial cells and lung-derived CaLu-3 cells, as well as reduced plasma membrane entry through transmembrane protease serine 2 (TMPRSS2).15 , 16 , 17 , 18 , 19 , 20 These features correlated with reduced replication capacity of Omicron in lung tissues, enhanced nasopharyngeal tropism, and decreased pathogenicity in vivo.15 , 18 , 20 , 21 , 22 Importantly, these characteristics were largely maintained by subsequent Omicron subvariants.
The BA.2 subvariant overtook BA.1 due to its slightly enhanced transmissibility and immune evasion, with an ability to reinfect individuals who were previously infected with BA.1.23 , 24 , 25 From BA.2, several subvariants emerged in quick succession, often with concurrent circulations; these included the BA.4 and BA.5 subvariants (bearing identical S proteins, referred to as BA.4/5 hereafter) that next rose to dominance and exhibited further immune escape.4 , 26 , 27 , 28 , 29 , 30 , 31 In addition, BA.2 gave rise to the BA.2.75 subvariant, which is currently increasing the proportions of COVID-19 cases32 but does not exhibit as substantial immune escape compared with BA.4/5.3 , 33 , 34 , 35 Notably, the BA.4/5 and BA.2.75 subvariants have driven further diversification of the circulating SARS-CoV-2, with the emergence of several additional subvariants including the BA.4.6, BF.7, BQ.1, and BQ.1.1 (derived from BA.4/5), as well as BA.2.75.2 (derived from BA.2.75). Additionally, these new subvariants are currently increasing in frequency32 , 36 and may be the next major dominant Omicron subvariant.
The extent of immune evasion and functional alterations to the spike protein (S) in these emerging Omicron subvariants currently remains unclear. To address this, we examine the sensitivity of the BA.4.6, BF.7, BQ.1, BQ.1.1, and BA.2.75.2 subvariants to neutralization by serum from recipients of 3 mRNA vaccine doses, as well as COVID-19 patients infected with the BA.1 or BA.4/5 variants. Additionally, we examined the contribution of subvariant lineage-defining mutations to neutralization escape. We observe strong neutralization resistance in the BQ.1 and BQ.1.1 subvariants driven largely by their N460K mutation, as well as in the BA.2.75.2 subvariant driven largely by its F486S mutation. Notably, the F486S diminishes BA.2.75.2 infectivity in Calu-3 cells. Finally, structural modeling showed that the F486S mutation reduces binding affinity for both the angiotensin-converting enzyme 2 (ACE2) receptor and class I and II antibodies, whereas the R346T and K444T mutations are likely responsible for evasion of class III antibody recognition.
Results
New Omicron subvariants exhibit similarly increased infectivity in HEK293T-ACE2 cells but comparably decreased infectivity in Calu-3 cells
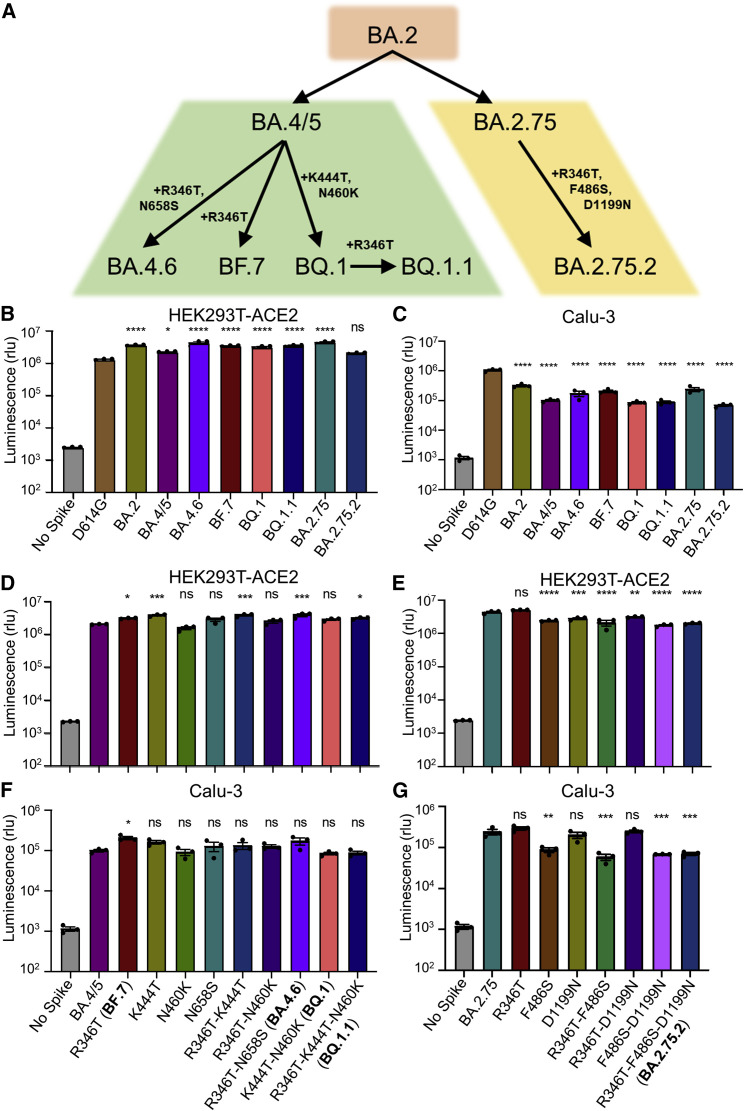
Derived from the BA.4/5 subvariant, BF.7 acquires an additional R346T mutation, whereas BA.4.6 and BQ.1 contain R346T/N658S mutations and K444T/N460K mutations, respectively; BQ.1.1 also bears an additional R346T mutation compared with BQ.1 (Figure 1 A). The BA.2.75.2 subvariant acquires R346T, F486S, and D1199N mutations in contrast to BA.2.75 (Figure 1A). These mutations, particularly those on the receptor-binding domain (RBD), have caused increasing concern over further immune escape.37
Figure 1.
Omicron subvariant-defining mutations and their impacts on pseudotyped viral infectivity in HEK293T-ACE2 and Calu-3 cells
(A) Displayed is a schematic of SARS-CoV-2 Omicron subvariant evolution indicating the mutations acquired by the BA.4.6, BF.7, BQ.1, BQ.1.1, and BA.2.75.2 subvariants.
(B and C) Infectivity of lentivirus pseudotyped with the indicated Omicron subvariant S constructs in HEK293T-ACE2 cells (B) (n = 3) or in Calu-3 cells (C) (n = 3). Bars represent means ± standard error. Significance relative to D614G was determined by one-way ANOVA with Bonferroni’s multiple testing correction. p values are represented as ns for p ≥ 0.05, ∗p < 0.05, and ∗∗∗∗p < 0.0001.
(D–G) Infectivity of lentivirus pseudotyped with the indicated BA.4/5-derived mutant S constructs (D and F) (n = 3) or BA.2.75-derived mutant S constructs (E and G) (n = 3) in HEK293T-ACE2 or Calu-3 cells. Bars represent means ± standard error. Significance relative to BA.4/5 or BA.2.75 was determined by one-way ANOVA with Bonferroni’s multiple testing correction. p values are represented as ns for p ≥ 0.05, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001.
We first examined the infectivity of these subvariants, along with the individual mutants and D614G by producing lentivirus pseudotyped with S from each of the critical S constructs (Figures 1B–1G). All Omicron subvariant-pseudotyped viruses exhibited modestly enhanced infectivity in HEK293T-ACE2 cells over the D614G variant with the exception of BA.2.75.2 (Figure 1B). Additionally, all Omicron subvariants exhibited comparably poor infectivity in lung-derived Calu-3 cells (Figure 1C) compared with D614G, consistent with prior Omicron subvariants and the weak lung tropism observed for Omicron.15 , 16 , 17 , 18 We found that these panels of mutants also exhibited similar infectivity compared with their parental BA.4/5 or BA.2.75 variants in HEK293T-ACE2 cells (Figures 1D and 1E) and Calu-3 cells (Figures 1F and 1G), although F486S-containing/BA.2.75-derived mutants, including the triple mutant R346T-F486S-D1199N (i.e., BA.2.75.2), had reduced titers in Calu-3 cells relative to the parental BA.2.75 (Figure 1G).
BQ.1, BQ.1.1, and BA.2.75.2 exhibit potent neutralization resistance to vaccine- and infection-induced sera
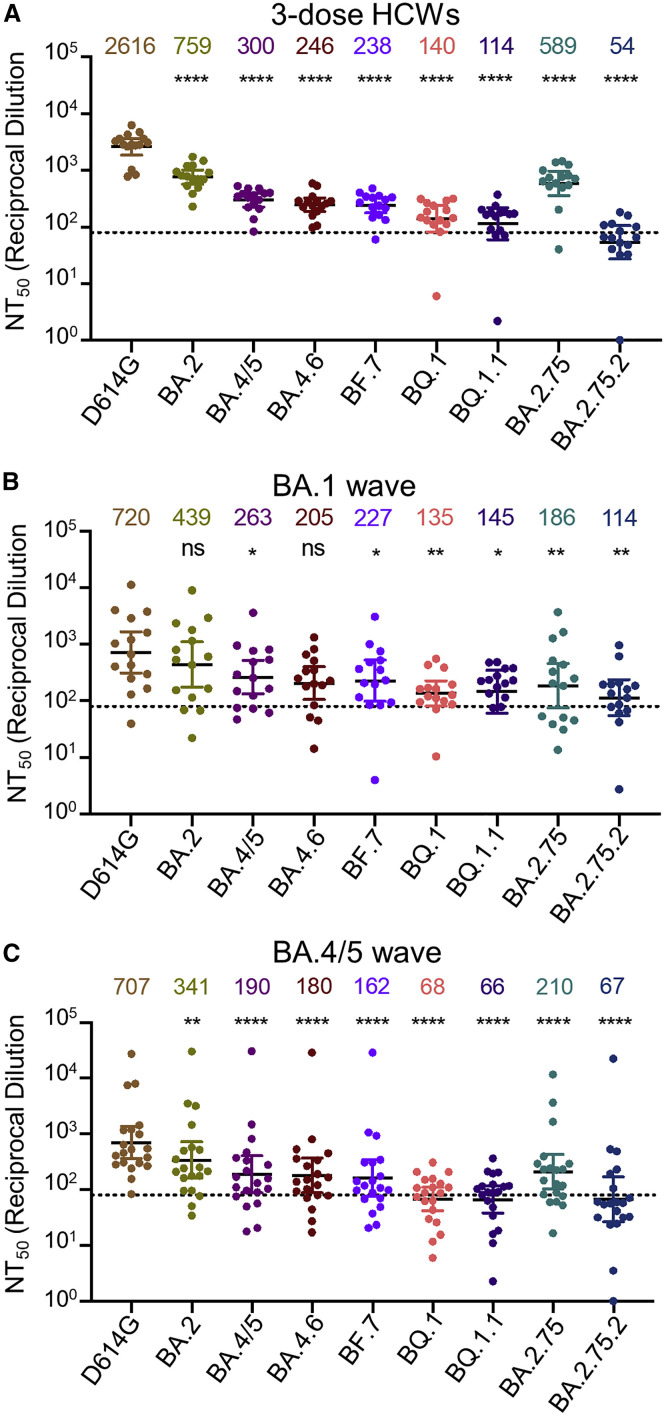
Using our previously reported pseudotyped lentivirus neutralization assay,38 we first examined the resistance of these emerging Omicron subvariants to sera from healthcare workers (HCWs) collected 2–13 weeks after vaccination with a homologous booster dose of monovalent Moderna mRNA-1273 (n = 3) or Pfizer/BioNTech BNT162b2 vaccine (n = 12). Compared with D614G, the BA.4.6, BF.7, BQ.1, and BQ.1.1 subvariants exhibited a 10.6-fold (p < 0.0001), 11.0-fold (p < 0.0001), 18.7-fold (p < 0.0001), and 22.9-fold (p < 0.0001) higher neutralization resistance, respectively, whereas BA.4/5 exhibited an 8.7-fold (p < 0.0001) higher neutralization resistance (Figures 2 A and S1A). Similarly, compared with D614G, the BA.2.75.2 subvariant exhibited a 48.4-fold (p < 0.0001) higher neutralization resistance, whereas BA.2.75 exhibited a 4.4-fold (p < 0.0001) higher neutralization resistance (Figure 2A). These data indicate further neutralization escape in emerging Omicron subvariants, with BA.Q.1, BA.Q.1.1, and BA.2.75.2, especially the latter, showing the most substantial neutralization resistance.
Figure 2.
Omicron subvariants, especially BQ.1, BQ.1.1, and BA.2.75.2, exhibit strong neutralization resistance
Neutralizing antibody titers were determined against lentivirus pseudotyped with S from the Omicron subvariants and ancestral D614G variant for sera from health care workers (HCWs) (n = 15) who received a single homologous monovalent Moderna mRNA-1273 (n = 3) or Pfizer/BioNTech BNT162b2 (n = 12) mRNA booster vaccination (A); for sera from BA.1-wave hospitalized COVID-19 patients (n = 15) (B); or for sera from BA.4/5-wave SARS-CoV-2 infected Columbus, Ohio first responders and household contacts (n = 20) (C). Bars represent geometric means with 95% confidence intervals. Geometric means are displayed at the top of the plots. Significance relative to D614G was determined by one-way repeated measures ANOVA with Bonferroni’s multiple testing correction. p values are displayed as ns for p ≥ 0.05, ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗∗p < 0.0001.
We also examined the resistance of these Omicron subvariants to neutralization by sera from hospitalized COVID-19 patients (n = 15) infected during the BA.1 wave of the pandemic. These include 6 unvaccinated patients, 5 patients vaccinated with 2 doses of Pfizer/BioNTech BNT162b2 or Moderna mRNA-1273, and 4 patients with 3 doses of Pfizer/BioNTech BNT162b2. BA.1-wave patient sera exhibited a similar pattern with the BA.4.6, BF.7, BQ.1, BQ.1.1, and BA.2.75.2 subvariants exhibiting the strongest immune evasion, with neutralization resistance 3.5-fold (p > 0.05), 3.2-fold (p < 0.05), 5.3-fold (p < 0.01), 5.0-fold (p < 0.05), and 6.3-fold (p < 0.01) higher than D614G, respectively (Figures 2B and S1B). As would be expected, BA.1 patient sera neutralized BA.2 with a higher efficiency compared with these BA.2-derived subvariants (Figures 2B and S1B).
To determine the breadth of immunity from individuals infected with more recent Omicron subvariants, we next examined sera from Columbus, Ohio first responders, and household contacts testing positive for COVID-19 during the BA.4/5 wave of the pandemic, with 11 of 20 subjects having the infecting variant confirmed by sequencing. Compared with D614G, the BA.4.6, BF.7, BQ.1, and BQ.1.1 subvariants exhibited a 3.9-fold (p < 0.0001), 4.4-fold (p < 0.0001), 10.4-fold (p < 0.0001), and 10.7-fold (p < 0.0001) higher neutralization resistance, respectively, whereas BA.4/5 exhibited a 3.7-fold (p < 0.0001) higher neutralization resistance than D614G (Figures 2C and S1C). Additionally, compared with D614G, the BA.2.75.2 subvariant exhibited a 10.6-fold (p < 0.0001) higher neutralization resistance, whereas BA.2.75 showed a 3.4-fold (p < 0.0001) higher neutralization resistance (Figure 2C). Interestingly, BA.2 exhibited less resistance to sera from BA.4/5-wave infected patients than BA.4/5, with 2.1-fold reduced neutralization sensitivity compared with D614G (p < 0.01) (Figures 2C and S1C). Similar trends were observed for both BA.1-wave and BA.4/5-wave patients, regardless of the vaccination status (Figure S2). Together, these results showed that BQ.1, BQ.1.1, and BA.2.75.2 are strongly resistant to neutralization by sera from subjects infected with the recently dominant BA.4/5 variant.
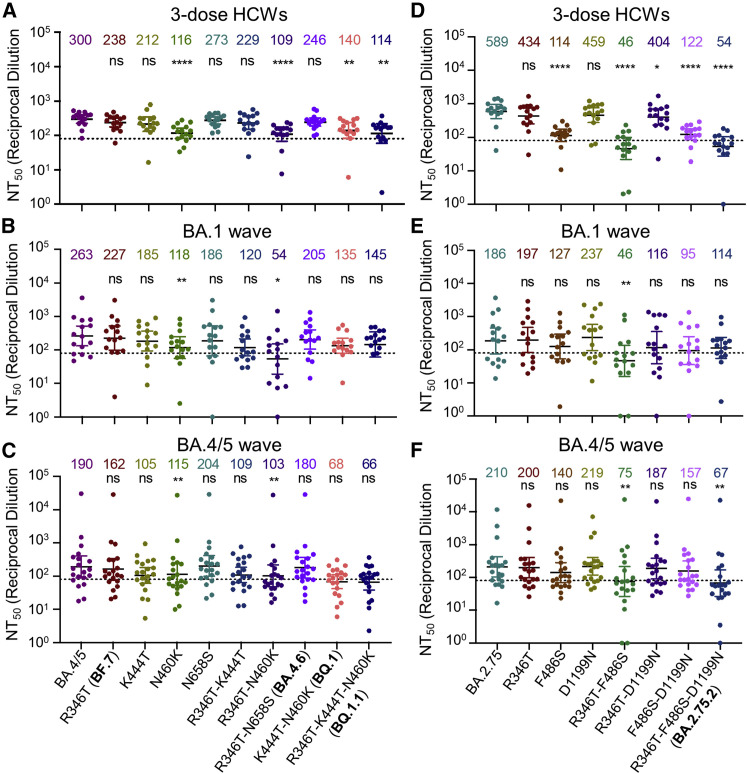
The N460K mutation is most critical for the enhanced neutralization resistance of BA.4.6, BF.7, BQ.1, and BQ.1.1
We further examined the amino acid residues critical for the neutralization resistance to sera from HCWs, BA.1 infection, and BA.4/5-infected individuals. Compared with the parental BA.4/5 subvariant, introduction of the N460K mutation reduced neutralization sensitivity by an additional 2.6-fold (p < 0.0001), with a similar 2.8-fold (p < 0.0001) reduction for the R346T/N460K double mutant (Figure 3 A). Consistent with this finding, the N460K-bearing subvariants BQ.1 and BQ.1.1 showed the strongest neutralization resistance with a 2.1-fold (p < 0.01) and 2.6-fold (p < 0.01) reduction in neutralization sensitivity compared with BA.4/5 (Figures 3A and S3A). Although not significant, two other individual mutations, R346T and K444T, were associated with a milder 1.3-fold (p = 0.0782) and 1.4-fold (p = 0.1886) reduction in neutralization sensitivity, respectively (Figure 3A). Of note, the N658S mutation did not appear to be strongly associated with neutralization resistance (Figure S3A), with a 1.1-fold (p > 0.05) reduction compared with the parental BA.4/5 (Figure 3A).
Figure 3.
Mutations N460K and F486S, and to a lesser extent, R346T and K444T, drive Omicron subvariant neutralization resistance
Neutralizing antibody titers were determined against lentivirus pseudotyped with S from the BA.4/5-derived (A–C) or BA.2.75 (D–F) mutants for sera from healthcare workers (HCWs) (A and D) (n = 15) who received a single homologous monovalent Moderna mRNA-1273 (n = 3) or Pfizer/BioNTech BNT162b2 (n = 12) mRNA booster vaccination; for sera from BA.1-wave hospitalized COVID-19 patients (B and E) (n = 15); or for sera from BA.4/5-wave SARS-CoV-2 infected Columbus, Ohio first responders, and household contacts (C and F) (n = 20). Bars represent geometric means with 95% confidence intervals. Geometric means are displayed at the top of the plots. Significance relative to parental variants was determined by one-way repeated measures ANOVA with Bonferroni’s multiple testing correction. p values are displayed as ns for p ≥ 0.05, ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗∗p < 0.0001.
We next examined the determinants of resistance of these Omicron subvariant mutants to neutralization by sera from hospitalized COVID-19 patients infected during the BA.1 wave of the pandemic. In addition, we tested sera from Columbus, Ohio first responders, and household contacts testing positive for COVID-19 during the BA.4/5 wave of the pandemic. We found that N460K and R346T/N460K mutations in the context of BA.4/5 reduced neutralization sensitivity to BA.1 sera 2.2-fold (p < 0.01) and 4.9-fold (p < 0.05), respectively (Figures 3B and S3B) and to BA.4/5 sera 1.7-fold (p < 0.01) and 1.8-fold (p < 0.01), respectively (Figures 3C and S3C). Notably, two BA.4/5-wave patients (BA.4/5_P5 and BA.4/5_P17) and one BA.1-wave patient (BA.1_P11) exhibited starkly reduced neutralization of K444T containing mutants, indicating the K444T mutation may be critical for evading the neutralizing antibody response for a subset of Omicron-infected patients (Figures S3B and S3C). Together with the results from HCWs, we concluded that the N460K mutation, and to a lesser extent, R346T, K444T, and N658S are critical for the enhanced resistance of the BQ.1 and BQ.1.1 subvariants.
The F486S mutation drives the enhanced neutralization resistance of BA.2.75.2
We also interrogated the critical amino acid residues responsible for the substantially enhanced nAb resistance of the BA.2.75.2 subvariant to sera from the same cohorts of HCWs, BA.1-wave patients, as well as BA.5-infected individuals mentioned above. We found that introduction of the F486S, R346T/F486S, or F486S/D1199N mutations to the parental BA.2.75 reduced neutralization sensitivity to HCW sera 5.2-fold (p < 0.0001), 12.8-fold (p < 0.0001), 4.8-fold (p < 0.0001), respectively; these were in comparison with the 10.9-fold (p < 0.0001) reduction seen for the BA.2.75.2 subvariant which contains all three mutations, i.e., R346T, F486S, and D1199N (Figure 3D). Notably, introduction of the R346T, D1199N, or R346T/D1199N resulted in only a 1.4-fold (p = 0.1074), 1.3-fold (p > 0.05), and 1.5-fold (p < 0.05) reduction in neutralization sensitivity, respectively (Figures 3D and S3D), indicating that R346T and D1199N mutations play less important roles compared with F486S in conferring the neutralization resistance. We noted that R346T/F486S double mutations reduced neutralization sensitivity to BA.1 wave patient sera 4.0-fold (p < 0.01) (Figures 3E and 3E), although the F486S mutation alone only conferred a 1.5-fold (p = 0.3698) reduction in neutralization sensitivity for this cohort (Figure 3E). Importantly, introduction of the R346T/F486S mutations into BA.2.75 reduced neutralization sensitivity to BA.4/5-infected patient sera 2.8-fold (p < 0.01) (Figures 3F and S3F). Overall, these results indicated that the F486S mutation in BA.2.75.2 is the major driver of enhanced nAb resistance, for both sera from HCWs and COVID-positive patients.
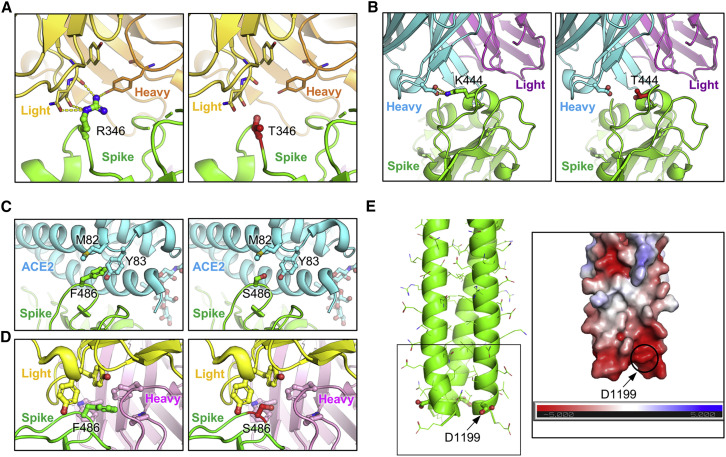
Structural modeling reveals mechanism of mutation-mediated antibody evasion
To further understand the functional impact of the mutations in these Omicron subvariants, we performed homology modeling-based structural analyses. Neutralizing antibodies targeting the SARS-CoV-2 S protein can largely be divided into four classes based on the region of the receptor-binding domain (RBD) targeted by the antibodies.39 , 40 Specifically, class I antibodies target RBDs in the “up” conformation, class II and class III antibodies target RBDs in either their up or “down” conformation, and class IV antibodies generally fail to block interactions with ACE2.40 Notably, R346 and K444 are located outside of SARS-CoV-2 receptor-binding motif (RBM) and are within the epitope of class III neutralizing antibodies. Structural analysis indicated an interference of antibody recognition introduced by R346T (Figure 4 A) and K444T (Figure 4B), where hydrogen bonds and salt-bridge can be abolished. In contrast, F486 is located within the RBM and is a key residue for binding to both the ACE2 receptor and to neutralizing antibodies; F486 interacts hydrophobically with M82 and Y83 on ACE2 and residues on antibody complementarity-determining regions region, whereas the F486S mutation negatively impacts the interaction with ACE2 (Figure 4C), as well as the recognition by some monoclonal antibodies in the class I and II categories (Figure 4D). Further structural analysis showed that residue D1199 is located in the heptad repeat 2 (HR2) region on a solvent-accessible surface close to the transmembrane domain or membrane (Figure 4E). Electrostatic surface potential (Figure 4E inset) of this region reveals a strong negative surface charge, which repulses the negatively charged membrane and could help keep the spike in an up-right position. However, the D1199N mutation in the S2 subunit would reduce the electrostatic repulsion, resulting in a more tilted spike and may impact receptor utilization. These analyses provided molecular mechanisms of mutation-mediated antibody evasion and alteration in receptor utilization by the BQ.1, BQ.1.1, BA.4.6, BF.7, and BA.2.75.2 subvariants.
Figure 4.
Homology modeling-based structural analyses of mutations essential for nAb resistance and fusogenicity
(A and B) Structures of spike-antibody binding interface shown as ribbons. Spike recognition of class III neutralizing antibodies C1365 (A) and SW186 (B) are interfered by R346T and K444T mutations, where multiple hydrogen bonds and salt-bridge (shown as yellow dot lines) are abolished.
(C) Structure of spike-ACE2 binding interface shown as ribbon. F486 interacts hydrophobically with M82 and Y83 on ACE2, whereas F486S impedes this interaction.
(D) Structure of class I antibody (AZD8895) recognition focusing on residue F486, with multiple antibody residues forming a surrounding hydrophobic cage. The F486S mutation abolishes this interaction.
(E) Structural model of HR2 region of SARS-CoV-2 S. Inset: electrostatic surface potential of HR2 membrane proximal region. D1199 contributes to the overall negative charge of this region.
Discussion
We examined the neutralization resistance of the emerging BA.4.6, BF.7, BQ.1, BQ.1.1, and BA.2.75.2 Omicron subvariants, as well as the impact on neutralization resistance of their lineage-defining mutations. All of these subvariants exhibit some degree of enhanced neutralization resistance over their parental BA.4/5 or BA.2.75.2 subvariants, with BQ.1, BQ.1.1, and BA.2.75.2 exhibiting the strongest resistance. Notably, this pattern is consistent for sera collected from HCWs following a homologous mRNA booster vaccination, from BA.1-wave hospitalized COVID-19 patients, as well as from BA.4/5-wave SARS-CoV-2 COVID-19-positive first responders and household contacts. Critically, we find that the neutralization resistance of BQ.1 and BQ.1.1 is driven largely by their N460K mutation, whereas the neutralization resistance of the BA.2.75.2 subvariant is determined by the F486S mutation. Our previous structural modeling analyses have shown that the N460K mutation present in BA.2.75 may potentiate ACE2 interactions,3 thus introduction of the N460K mutation into the BA.4/5-derived subvariants may contribute to enhanced cell-cell fusion and neutralization resistance. Our new structural modeling and additional analyses in this work revealed crucial roles of residues R346, K444, and F486 in antibody recognition, as well as the potential mechanisms of immune evasion through R346T, K444T, and F486S mutations present in Omicron subvariants. Notably, these mutations mediate resistance not only to 3-dose mRNA vaccine-induced sera but also to BA.1- and BA.4/5-infection-induced sera. We also found that BA.4/5-wave patient sera exhibited weaker neutralization of BA.4/5 than of BA.2, which could be related to prior exposure to SARS-CoV-2 variant antigen biasing patient neutralizing antibody response to BA.4/5 infection. In addition, conformational alterations to the S protein have been demonstrated to alter neutralization sensitivity.41 , 42 , 43
It is well established that the Omicron subvariants exhibit diminished TMPRSS2-mediated plasma membrane entry resulting in reduced infectivity in Calu-3 cells, although retaining cathepsin-mediated endosomal entry capacity allowing robust entry in HEK293T-ACE2 cells.15 , 17 , 20 This altered preference of entry route has been shown to be associated with the reduced lung tropism and reduced pathogenicity of the Omicron subvariants.15 , 17 , 18 , 20 Here, we show that the emerging Omicron subvariants BA.4.6, BF.7, BQ.1, BQ.1.1, and BA.2.75.2 retain this diminished infectivity in Calu-3 cells, but noticeably, the F486S mutation resulted in further diminished infectivity in Calu-3 cells. If this reduced infectivity in Calu-3 cell remains correlated with reduced lung tropism, the F486S mutation would induce further reduced pathogenicity of SARS-CoV-2. However, we have previously found increased syncytia formation capacity in several Omicron subvariants including BA.4/5, BA.2.75, BQ.1, BQ.1.1, and BA.2.75.2 compared with their ancestral Omicron strains.3 , 44 This may indicate a continuing shift toward more efficient TMPRSS2 utilization to allow for plasma membrane-mediated fusion. A thorough investigation of the entry route preferences and in vivo pathogenicity of emerging Omicron subvariants is warranted.
The perpetual emergence of SARS-CoV-2 variants with enhanced immune escape continues to threaten public health. Monitoring the immune escape of emerging variants will be critical to improving mRNA vaccine reformulation, assessing new broader coronavirus vaccine candidates, as well as directing ongoing public health measures. Further, emerging variants, especially those containing the N460K, F486S, R346T, and K444T mutations, must be monitored closely for enhanced neutralization resistance.
Limitations of this work include the lack of experiments using authentic viruses for the neutralization assay; however, prior reports from ours and other groups have confirmed that results of pseudotyped viruses can faithfully reflect the neutralization of infectious SARS-CoV-2 and variants.38 , 45 Additionally, whether these subvariants, as well as associated mutants, would use entry pathways distinct from the prototype Omicron and their parental lineages,37 is unknown, which will be determined in future studies. Finally, the impact of Omicron-defining mutations on ACE2 binding, as well as spike conformational changes warrants further biochemical and structural characterization.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological samples | ||
| 3-dose HCWs Sera | Evans et al.,1 Qu et al.4 | N/A |
| Omicron BA.1-wave Infected Patient Sera | Evans et al.,1 Qu et al.4 | N/A |
| Omicron BA.4/5-wave Infected First Responders and Household Contacts Sera | This paper | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Polyethylenimine (PEI) | Thermo Fisher Scientific | Cat# BMS1003 |
| Dulbecco’s Modified Eagles Medium (DMEM) | Sigma-Aldrich | Cat#: 11965-092 |
| Fetal Bovine Serum (FBS) | Thermo Fisher Scientific | Cat#: F1051 |
| 0.05% Trypsin + 0.53 mM EDTA | Corning | Cat# 25-052-CI |
| Penicillin-Streptomycin | HyClone | Cat#: SV30010 |
| Immobilon Crescendo Western HRP substrate | Millipore | Cat# WBLUR0500 |
| QIAprep Spin Miniprep Kit | QIAGEN | Cat# 27106 |
| Coelenterazine | GoldBio | Cat#: CZ2.5, CAS: 55779-48-1 |
| Deposited data | ||
| NT50 Values and De-identified patient data | SeroNet Coordinating Center, NCI, NIH | N/A |
| Experimental models: Cell lines | ||
| HEK293T | ATCC | Cat#: CRL-11268, RRID: CVCL_1926 |
| HEK293T-ACE2 | BEI Resources | Cat#: NR-52511, RRID: CVCL_A7UK |
| Calu-3 | ATCC | RRID: CVCL_0609 |
| Recombinant DNA | ||
| pNL4-3-inGluc | David Derse, NIH Maruzov et al.46 |
N/A |
| pcDNA3.1-SARS-CoV-2-Flag-S-Flag_D614G | GenScript Biotech Zeng et al.47 |
N/A |
| pcDNA3.1-SARS-CoV-2-Flag-S-Flag_BA.2 | GenScript Biotech Evans et al.1 |
N/A |
| pcDNA3.1-SARS-CoV-2-Flag-S-Flag_BA.4/5 | GenScript Biotech Qu et al.4 |
N/A |
| pcDNA3.1-SARS-CoV-2-Flag-S-Flag_BA.4.6 | This paper | N/A |
| pcDNA3.1-SARS-CoV-2-Flag-S-Flag_BF.7 | This paper | N/A |
| pcDNA3.1-SARS-CoV-2-Flag-S-Flag_BQ.1 | This paper | N/A |
| pcDNA3.1-SARS-CoV-2-Flag-S-Flag_BQ.1.1 | This paper | N/A |
| pcDNA3.1-SARS-CoV-2-Flag-S-Flag_BA.2.75 | GenScript Biotech Qu et al.3 |
N/A |
| pcDNA3.1-SARS-CoV-2-Flag-S-Flag_BA.2.75.2 | This paper | N/A |
| pcDNA3.1-SARS-CoV-2-Flag-S-Flag_BA.4/5_K444T | This paper | N/A |
| pcDNA3.1-SARS-CoV-2-Flag-S-Flag_BA.4/5_N460K | This paper | N/A |
| pcDNA3.1-SARS-CoV-2-Flag-S-Flag_BA.4/5_N658S | This paper | N/A |
| pcDNA3.1-SARS-CoV-2-Flag-S-Flag_BA.4/5_R346T_K444T | This paper | N/A |
| pcDNA3.1-SARS-CoV-2-Flag-S-Flag_BA.4/5_R346T_N460K | This paper | N/A |
| pcDNA3.1-SARS-CoV-2-Flag-S-Flag_BA.2.75_R346T | This paper | N/A |
| pcDNA3.1-SARS-CoV-2-Flag-S-Flag_BA.2.75_F486S | This paper | N/A |
| pcDNA3.1-SARS-CoV-2-Flag-S-Flag_BA.2.75_D1199N | This paper | N/A |
| pcDNA3.1-SARS-CoV-2-Flag-S-Flag_BA.2.75_R346T_F486S | This paper | N/A |
| pcDNA3.1-SARS-CoV-2-Flag-S-Flag_BA.2.75_R346T_D1199N | This paper | N/A |
| pcDNA3.1-SARS-CoV-2-Flag-S-Flag_BA.2.75_F486S_D1199N | This paper | N/A |
| Software and algorithms | ||
| GraphPad Prism | GraphPad Version 9.0.0 | www.graphpad.com |
| SWISS-MODEL | Guex et al.48 | https://swissmodel.expasy.org/ |
| PyMOL | Warren DeLano and Sarin Bromberg | https://pymol.org/ |
| Other | ||
| Cytation 5 Imaging Reader | BioTek | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to the lead contact, Dr. Shan-Lu Liu (liu.6244@osu.edu).
Materials availability
Plasmids generated in this study are available upon request made to the lead contact.
Experimental model and subject details
Samples and patient information
Sera samples were collected from HCWs at the Ohio State University Wexner Medical Center in Columbus, Ohio with approval from an institutional review board49 (IRB 2020H0228, IRB 2020H0527, and IRB 2017H0292). These HCWs samples were collected 2-13 weeks after vaccination with a third homologous dose of the monovalent Moderna mRNA-1273 (n = 3) or Pfizer BioNTech BNT162b2 (n = 12) vaccines. HCWs included 10 male and 5 female subjects with ages ranging from 26 to 61 (median 33).
Sera from BA.1-wave COVID-19 patients hospitalized in Columbus, Ohio were collected with approval from an IRB (IRB 2020H0527). The patient samples were collected 1-7 days after hospitalization with COVID-19. Hospitalizations occurred between the end of January and the end of February of 2022, a BA.1 dominant period in Columbus, Ohio. Patients included 12 male and 3 female patients with ages ranging from 29 to 78 (median 57). Patients included 6 unvaccinated patients, 5 patients vaccinated with 2 doses of Pfizer/BioNTech BNT162b2 (n = 2) or Moderna mRNA-1273 (n = 3), and 4 patients vaccinated and boosted with Pfizer/BioNTech BNT162b2.
Sera from BA.4/5-wave Columbus, Ohio first responders and household contacts who tested positive for SARS-CoV-2 infection were collected with IRB approval (IRB 2020H0527, 2020H0531, and 2020H0240). 11 patient nasal swab samples were sequenced to confirm infection with BA.4, BA.5, or derivative variants, with 4 patients infected with BA.4, 7 with BA.5, and 9 patients could not have their variant determined. Of those who could not have the specific variant identified, their samples were collected between late July and late September of 2022, a BA.4/5 dominant period based on CDC Ohio database. Except for one patient whose gender and age are unknown, patients included 4 male and 15 female with ages ranging from 27 to 58 (median 44). Patients included 17 unvaccinated, and 3 vaccinated and boosted with Pfizer/BioNTech BNT162b2 (n = 1) or Moderna mRNA-1273 (n = 2).
Cell lines and maintenance
Human female embryonic kidney cell lines HEK293T (ATCC CRL-11268, RRID: CVCL_1926) and HEK293T cells stably expressing human ACE2 (BEI NR-52511, RRID: CVCL_A7UK) were maintained in DMEM (Gibco, 11965-092) with 10% FBS (Sigma, F1051) and 1% penicillin-streptomycin (HyClone, SV30010) added. Human male adenocarcinoma lung epithelial cell line Calu-3 (RRID:CVCL_0609) were maintained in EMEM (ATCC, 30-2003) with 10% FBS and 1% penicillin-streptomycin added. All cells were passaged first by washing with Dulbecco’s phosphate buffered saline (Sigma, D5652-10X1L) then incubating in 0.05% Trypsin + 0.53 mM EDTA (Corning, 25-052-CI) until cells were completely detached. Cells were maintained at 37°C and 5.0% CO2 in 10 cm cell culture dishes (Greiner Bio-one, 664160).
Method details
Plasmids
The pNL4-3 inGluc lentiviral vector has been reported on in our previous publications.38 Briefly, the vector is in the HIV-1 pNL4-3 backbone with a deletion of Env and an addition of a Gaussia luciferase reported gene that is expressed in target cells without premature expression in producer cells. The S variant constructs were cloned into the pcDNA3.1 vector by GenScript Biotech (Piscataway, NJ) using restriction enzyme cloning by Kpn I and BamH I; alternatively, they were produced by PCR mutagenesis. The constructs bear FLAG tags on the N- and C-terminal ends. All constructs were confirmed by sequencing.
Pseudotyped lentivirus production and infectivity
Pseudotyped lentiviral vectors were produced as previously described.38 HEK293T cells were transfected with the pNL4-3-inGluc and S constructs in a 2:1 ratio using polyethyleneimine transfection (Transporter 5 Transfection Reagent, Polysciences) in order to generate viral particles. Virus was harvested 24, 48, and 72 hours post-transfection. Relative infectivity was determined by infection of HEK293T-ACE2 or Calu-3 cells and measurement of Gaussia luciferase activity 48 and 72 hours post-infection. Gaussia luciferase activity was measured by combining equal volumes of cell culture media and Gaussia luciferase substrate (0.1 M Tris pH 7.4, 0.3 M sodium ascorbate, 10 μM coelenterazine) with luminescence measured immediately by a BioTek Cytation5 plate reader.
Lentivirus neutralization assay
Neutralization assays with pseudotyped lentiviral vectors were performed as described previously.38 HCW and COVID-19 patient samples were 4-fold serially diluted and equal amounts of SARS-CoV-2 pseudotyped virus were added to the diluted sera. Final dilutions were 1:80, 1:320, 1:1280, 1:5120, 1:20480, and no serum control. The virus and sera mixture was incubated for 1 hour at 37°C then added to HEK293T-ACE2 cells to allow for infection. Gaussia luciferase activity was measured at 48 and 72 hours post-infection by combining equal volumes of cell culture media and Gaussia luciferase substrate with luminescence measured immediately by a BioTek Cytation5 plate reader. The 50% neutralization titers (NT50) were determined by least-squares-fit, non-linear regression in GraphPad Prism 9 (San Diego, CA).
Structural modeling and analysis
Homology modeling of Omicron spike protein complexes with either ACE2 receptor or neutralizing antibodies was performed on SWISS-MODEL server with published X-ray crystallography and cryo-EM structures as templates (PDB: 7K8Z, 8DT3, 7XB0, 7L7D, 2FXP). Molecular contacts of Omicron mutants were examined and illustrated with the programs PyMOL.
Quantification and statistical analysis
NT50 values were determined by least-squares-fit, non-linear-regression in GraphPad Prism 9 (San Diego, CA). NT50 values were log10 transformed for hypothesis testing to better approximate normality, and multiplicity was addressed by the use of Bonferroni corrections. The statistical analysis was performed using GraphPad Prism 9 and are referenced in the figure legends, including one-way ANOVA (Figures 1B–1G), one-way repeated measures ANOVA (Figures 2A–2C, 3A–3F, S2A, and S2B), and two-way repeated measures ANOVA (Figures S2A and S2B) for significance analysis between vaccination status.
Acknowledgments
We thank the NIH AIDS Reagent Program and BEI Resources for providing important reagents for this work. We also thank the Clinical Research Center/Center for Clinical Research Management of The Ohio State University Wexner Medical Center and The Ohio State University College of Medicine in Columbus, Ohio, specifically Francesca Madiai, Dina McGowan, Breona Edwards, Evan Long, and Trina Wemlinger, for logistics, collection, and processing of samples. In addition, we thank Sarah Karow, Madison So, Preston So, Daniela Farkas, and Finny Johns in the clinical trials team of The Ohio State University for sample collection and other supports. This work was supported by a fund provided by an anonymous private donor to OSU, United States. S.-L.L., S.F., D.J., G.L., A.P., R.J.G., L.J.S., and E.M.O. were supported by the National Cancer Institute of the NIH, United States, under award no. U54CA260582. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. J.P.E. was supported by Glenn Barber Fellowship from the Ohio State University College of Veterinary Medicine, United States. R.J.G. was additionally supported by the Robert J. Anthony Fund for Cardiovascular Research and the JB Cardiovascular Research Fund, and L.J.S. was partially supported by NIH R01 HD095881. K.X. was supported by the Ohio State University James Cancer Center and a Path to K award from the Ohio State University Office of Health Sciences and the Center for Clinical & Translational Science, United States. The content is solely the responsibility of the authors and does not necessarily represent the official views of the university, or the Center for Clinical & Translational Science.
Author contributions
S.-L.L. conceived and directed the project. P.Q. and J.P.E. performed the experiments. C.C., M.A., P.S., S.F., D.J., G.L., A.P., and R.J.G. provided clinical samples. K.X. performed homology modeling. P.Q., J.P.E., and S.-L.L. wrote the paper. J.N.F., Y.-M.Z., L.J.S., E.M.O., and K.X. provided insightful discussion and revision of the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: November 22, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.chom.2022.11.012.
Supplemental information
Data and code availability
-
•
NT50 values and de-identified patient information will be deposited to the National Cancer Institute SeroNet Coordinating Center. Additionally, NT50 values and de-identified patient information reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Evans J.P., Zeng C., Qu P., Faraone J., Zheng Y.M., Carlin C., Bednash J.S., Zhou T., Lozanski G., Mallampalli R., et al. Neutralization of SARS-CoV-2 Omicron sub-lineages BA.1, BA.1.1, and BA.2. Cell Host Microbe. 2022;30 doi: 10.1016/j.chom.2022.04.014. 1093.e3–1102.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kurhade C., Zou J., Xia H., Cai H., Yang Q., Cutler M., Cooper D., Muik A., Jansen K.U., Xie X., et al. Neutralization of omicron BA.1, BA.2, and BA.3 SARS-CoV-2 by 3 doses of BNT162b2 vaccine. Nat. Commun. 2022;13:3602. doi: 10.1038/s41467-022-30681-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qu P., Evans J.P., Zheng Y.M., Carlin C., Saif L.J., Oltz E.M., Xu K., Gumina R.J., Liu S.L. Evasion of neutralizing antibody responses by the SARS-CoV-2 BA.2.75 variant. Cell Host Microbe. 2022;30 doi: 10.1016/j.chom.2022.09.015. 1518.e4–1526.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qu P., Faraone J., Evans J.P., Zou X., Zheng Y.M., Carlin C., Bednash J.S., Lozanski G., Mallampalli R.K., Saif L.J., et al. Neutralization of the SARS-CoV-2 omicron BA.4/5 and BA.2.12.1 subvariants. N. Engl. J. Med. 2022;386:2526–2528. doi: 10.1056/NEJMc2206725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xia S., Wang L., Zhu Y., Lu L., Jiang S. Origin, virological features, immune evasion and intervention of SARS-CoV-2 Omicron sublineages. Signal Transduct. Target. Ther. 2022;7:241. doi: 10.1038/s41392-022-01105-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yamasoba D., Kimura I., Nasser H., Morioka Y., Nao N., Ito J., Uriu K., Tsuda M., Zahradnik J., Shirakawa K., et al. Virological characteristics of the SARS-CoV-2 Omicron BA.2 spike. Cell. 2022;185 doi: 10.1016/j.cell.2022.04.035. 2103.e19–2115.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu J., Collier A.Y., Rowe M., Mardas F., Ventura J.D., Wan H., Miller J., Powers O., Chung B., Siamatu M., et al. Neutralization of the SARS-CoV-2 omicron BA.1 and BA.2 variants. N. Engl. J. Med. 2022;386:1579–1580. doi: 10.1056/NEJMc2201849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abu-Raddad L.J., Chemaitelly H., Ayoub H.H., AlMukdad S., Yassine H.M., Al-Khatib H.A., Smatti M.K., Tang P., Hasan M.R., Coyle P., et al. Effect of mRNA vaccine boosters against SARS-CoV-2 omicron infection in Qatar. N. Engl. J. Med. 2022;386:1804–1816. doi: 10.1056/NEJMoa2200797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gruell H., Vanshylla K., Tober-Lau P., Hillus D., Schommers P., Lehmann C., Kurth F., Sander L.E., Klein F. mRNA booster immunization elicits potent neutralizing serum activity against the SARS-CoV-2 Omicron variant. Nat. Med. 2022;28:477–480. doi: 10.1038/s41591-021-01676-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cao Y., Wang J., Jian F., Xiao T., Song W., Yisimayi A., Huang W., Li Q., Wang P., An R., et al. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature. 2022;602:657–663. doi: 10.1038/s41586-021-04385-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xia H., Zou J., Kurhade C., Cai H., Yang Q., Cutler M., Cooper D., Muik A., Jansen K.U., Xie X., et al. Neutralization and durability of 2 or 3 doses of the BNT162b2 vaccine against Omicron SARS-CoV-2. Cell Host Microbe. 2022;30 doi: 10.1016/j.chom.2022.02.015. 485–488.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zou J., Xia H., Xie X., Kurhade C., Machado R.R.G., Weaver S.C., Ren P., Shi P.Y. Neutralization against Omicron SARS-CoV-2 from previous non-Omicron infection. Nat. Commun. 2022;13:852. doi: 10.1038/s41467-022-28544-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu L., Iketani S., Guo Y., Chan J.F., Wang M., Liu L., Luo Y., Chu H., Huang Y., Nair M.S., et al. Striking antibody evasion manifested by the Omicron variant of SARS-CoV-2. Nature. 2022;602:676–681. doi: 10.1038/s41586-021-04388-0. [DOI] [PubMed] [Google Scholar]
- 14.Cerutti G., Guo Y., Liu L., Liu L., Zhang Z., Luo Y., Huang Y., Wang H.H., Ho D.D., Sheng Z., Shapiro L. Cryo-EM structure of the SARS-CoV-2 Omicron spike. Cell Rep. 2022;38:110428. doi: 10.1016/j.celrep.2022.110428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shuai H., Chan J.F., Hu B., Chai Y., Yuen T.T., Yin F., Huang X., Yoon C., Hu J.C., Liu H., et al. Attenuated replication and pathogenicity of SARS-CoV-2 B.1.1.529 Omicron. Nature. 2022;603:693–699. doi: 10.1038/s41586-022-04442-5. [DOI] [PubMed] [Google Scholar]
- 16.Wang Q., Anang S., Iketani S., Guo Y., Liu L., Katsamba P.S., Shapiro L., Ho D.D., Sodroski J.G. Functional properties of the spike glycoprotein of the emerging SARS-CoV-2 variant B.1.1.529. Cell Rep. 2022;39:110924. doi: 10.1016/j.celrep.2022.110924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meng B., Abdullahi A., Ferreira I.A.T.M., Goonawardane N., Saito A., Kimura I., Yamasoba D., Gerber P.P., Fatihi S., Rathore S., et al. Altered TMPRSS2 usage by SARS-CoV-2 Omicron impacts infectivity and fusogenicity. Nature. 2022;603:706–714. doi: 10.1038/s41586-022-04474-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barut G.T., Halwe N.J., Taddeo A., Kelly J.N., Schön J., Ebert N., Ulrich L., Devisme C., Steiner S., Trüeb B.S., et al. The spike gene is a major determinant for the SARS-CoV-2 Omicron-BA.1 phenotype. Nat. Commun. 2022;13:5929. doi: 10.1038/s41467-022-33632-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cui Z., Liu P., Wang N., Wang L., Fan K., Zhu Q., Wang K., Chen R., Feng R., Jia Z., et al. Structural and functional characterizations of infectivity and immune evasion of SARS-CoV-2 Omicron. Cell. 2022;185 doi: 10.1016/j.cell.2022.01.019. 860–871.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suzuki R., Yamasoba D., Kimura I., Wang L., Kishimoto M., Ito J., Morioka Y., Nao N., Nasser H., Uriu K., et al. Attenuated fusogenicity and pathogenicity of SARS-CoV-2 Omicron variant. Nature. 2022;603:700–705. doi: 10.1038/s41586-022-04462-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Su W., Choy K.T., Gu H., Sia S.F., Cheng K.M., Nizami S.I.N., Krishnan P., Ng Y.M., Chang L.D.J., Liu Y., et al. Omicron BA.1 and BA.2 sub-lineages show reduced pathogenicity and transmission potential than the early SARS-CoV-2 D614G variant in Syrian hamsters. J. Infect. Dis. 2022 doi: 10.1093/infdis/jiac276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McMahan K., Giffin V., Tostanoski L.H., Chung B., Siamatu M., Suthar M.S., Halfmann P., Kawaoka Y., Piedra-Mora C., Jain N., et al. Reduced pathogenicity of the SARS-CoV-2 omicron variant in hamsters. Med. (NY) 2022;3 doi: 10.1016/j.medj.2022.03.004. 262–268.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lyngse F.P., Kirkeby C.T., Denwood M., Christiansen L.E., Mølbak K., Møller C.H., Skov R.L., Krause T.G., Rasmussen M., Sieber R.N., et al. Household transmission of SARS-CoV-2 Omicron variant of concern subvariants BA.1 and BA.2 in Denmark. Nat. Commun. 2022;13:5760. doi: 10.1038/s41467-022-33498-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stegger M., Edslev S.M., Sieber R.N., Ingham A.C., Ng K.L., Tang M.-H.E., Alexandersen S., Fonager J., Legarth R., Utko M., et al. Occurrence and significance of Omicron BA.1 infection followed by BA.2 reinfection. Preprint at medRxiv. 2022 doi: 10.1101/2022.02.19.22271112. [DOI] [Google Scholar]
- 25.Iketani S., Liu L., Guo Y., Liu L., Chan J.F., Huang Y., Wang M., Luo Y., Yu J., Chu H., et al. Antibody evasion properties of SARS-CoV-2 Omicron sublineages. Nature. 2022;604:553–556. doi: 10.1038/s41586-022-04594-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khan K., Karim F., Ganga Y., Bernstein M., Jule Z., Reedoy K., Cele S., Lustig G., Amoako D., Wolter N., et al. Omicron BA.4/BA.5 escape neutralizing immunity elicited by BA.1 infection. Nat. Commun. 2022;13:4686. doi: 10.1038/s41467-022-32396-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tuekprakhon A., Nutalai R., Dijokaite-Guraliuc A., Zhou D., Ginn H.M., Selvaraj M., Liu C., Mentzer A.J., Supasa P., Duyvesteyn H.M.E., et al. Antibody escape of SARS-CoV-2 Omicron BA.4 and BA.5 from vaccine and BA.1 serum. Cell. 2022;185 doi: 10.1016/j.cell.2022.06.005. 2422–2433.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hachmann N.P., Miller J., Collier A.Y., Ventura J.D., Yu J., Rowe M., Bondzie E.A., Powers O., Surve N., Hall K., Barouch D.H. Neutralization escape by SARS-CoV-2 Omicron subvariants BA.2.12.1, BA.4, and BA.5. N. Engl. J. Med. 2022;387:86–88. doi: 10.1056/NEJMc2206576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cao Y., Yisimayi A., Jian F., Song W., Xiao T., Wang L., Du S., Wang J., Li Q., Chen X., et al. BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection. Nature. 2022;608:593–602. doi: 10.1038/s41586-022-04980-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kimura I., Yamasoba D., Tamura T., Nao N., Suzuki T., Oda Y., Mitoma S., Ito J., Nasser H., Zahradnik J., et al. Virological characteristics of the SARS-CoV-2 Omicron BA.2 subvariants, including BA.4 and BA.5. Cell. 2022;185 doi: 10.1016/j.cell.2022.09.018. 3992–4007.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Q., Guo Y., Iketani S., Nair M.S., Li Z., Mohri H., Wang M., Yu J., Bowen A.D., Chang J.Y., et al. Antibody evasion by SARS-CoV-2 Omicron subvariants BA.2.12.1, BA.4 and BA.5. Nature. 2022;608:603–608. doi: 10.1038/s41586-022-05053-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Centers for Disease Control and Prevention . US Department of Health and Human Services, CDC; 2022. COVID data tracker.https://covid.cdc.gov/covid-data-tracker [Google Scholar]
- 33.Cao Y., Song W., Wang L., Liu P., Yue C., Jian F., Yu Y., Yisimayi A., Wang P., Wang Y., et al. Characterization of the enhanced infectivity and antibody evasion of Omicron BA.2.75. Cell Host Microbe. 2022;30 doi: 10.1016/j.chom.2022.09.018. 1527–1539.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Q., Iketani S., Li Z., Guo Y., Yeh A.Y., Liu M., Yu J., Sheng Z., Huang Y., Liu L., Ho D.D. Antigenic characterization of the SARS-CoV-2 Omicron subvariant BA.2.75. Cell Host Microbe. 2022;30 doi: 10.1016/j.chom.2022.09.002. 1512–1517.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saito A., Tamura T., Zahradnik J., Deguchi S., Tabata K., Anraku Y., Kimura I., Ito J., Yamasoba D., Nasser H., et al. Virological characteristics of the SARS-CoV-2 Omicron BA. 2.75 variant. Cell Host Microbe. 2022;30 doi: 10.1016/j.chom.2022.10.003. 1540–1555.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Iacobucci G. Covid-19: hospital admissions rise in England as some trusts reinstate mask requirements. BMJ. 2022;379:o2440. doi: 10.1136/bmj.o2440. [DOI] [PubMed] [Google Scholar]
- 37.Cao Y., Jian F., Wang J., Yu Y., Song W., Yisimayi A., An R., Zhang N., Wang Y., Wang P., et al. Imprinted SARS-CoV-2 humoral immunity induces converging Omicron RBD evolution. Preprint at bioRxiv. 2022 doi: 10.1101/2022.09.15.507787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zeng C., Evans J.P., Pearson R., Qu P., Zheng Y.M., Robinson R.T., Hall-Stoodley L., Yount J., Pannu S., Mallampalli R.K., et al. Neutralizing antibody against SARS-CoV-2 spike in COVID-19 patients, health care workers, and convalescent plasma donors. JCI Insight. 2020;5:e143213. doi: 10.1172/jci.insight.143213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Greaney A.J., Starr T.N., Barnes C.O., Weisblum Y., Schmidt F., Caskey M., Gaebler C., Cho A., Agudelo M., Finkin S., et al. Mapping mutations to the SARS-CoV-2 RBD that escape binding by different classes of antibodies. Nat. Commun. 2021;12:4196. doi: 10.1038/s41467-021-24435-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barnes C.O., Jette C.A., Abernathy M.E., Dam K.A., Esswein S.R., Gristick H.B., Malyutin A.G., Sharaf N.G., Huey-Tubman K.E., Lee Y.E., et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020;588:682–687. doi: 10.1038/s41586-020-2852-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weissman D., Alameh M.G., de Silva T., Collini P., Hornsby H., Brown R., LaBranche C.C., Edwards R.J., Sutherland L., Santra S., et al. D614G spike mutation increases SARS CoV-2 susceptibility to neutralization. Cell Host Microbe. 2021;29 doi: 10.1016/j.chom.2020.11.012. 23–31.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Plante J.A., Liu Y., Liu J., Xia H., Johnson B.A., Lokugamage K.G., Zhang X., Muruato A.E., Zou J., Fontes-Garfias C.R., et al. Spike mutation D614G alters SARS-CoV-2 fitness. Nature. 2021;592:116–121. doi: 10.1038/s41586-020-2895-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yurkovetskiy L., Wang X., Pascal K.E., Tomkins-Tinch C., Nyalile T.P., Wang Y., Baum A., Diehl W.E., Dauphin A., Carbone C., et al. Structural and functional analysis of the D614G SARS-CoV-2 spike protein variant. Cell. 2020;183 doi: 10.1016/j.cell.2020.09.032. 739–751.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qu P., Evans J.P., Faraone J., Zheng Y.-M., Carlin C., Anghelina M., Stevens P., Fernandez S., Jones D., Lozanski G., et al. Distinct neutralizing antibody escape of SARS-CoV-2 omicron subvariants BQ.1, BQ.1.1, BA.4.6, BF.7 and BA.2.75.2. Preprint at bioRxiv. 2022 doi: 10.1101/2022.10.19.512891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xiong H.L., Wu Y.T., Cao J.L., Yang R., Liu Y.X., Ma J., Qiao X.Y., Yao X.Y., Zhang B.H., Zhang Y.L., et al. Robust neutralization assay based on SARS-CoV-2 S-protein-bearing vesicular stomatitis virus (VSV) Pseudovirus and ACE2-overexpressing BHK21 cells. Emerg. Microbes Infect. 2020;9:2105–2113. doi: 10.1080/22221751.2020.1815589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mazurov D., Ilinskaya A., Heidecker G., Lloyd P., Derse D. Quantitative comparison of HTLV-1 and HIV-1 cell-to-cell infection with new replication dependent vectors. PLoS Pathog. 2010;6:e1000788. doi: 10.1371/journal.ppat.1000788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zeng C., Evans J.P., Faraone J.N., Qu P., Zheng Y.M., Saif L., Oltz E.M., Lozanski G., Gumina R.J., Liu S.L. Neutralization of SARS-CoV-2 variants of concern harboring Q677H. mBio. 2021;12:e0251021. doi: 10.1128/mBio.02510-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guex N., Peitsch M.C., Schwede T. Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: a historical perspective. Electrophoresis. 2009;30(Suppl 1):S162–S173. doi: 10.1002/elps.200900140. [DOI] [PubMed] [Google Scholar]
- 49.Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O’Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L., et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
NT50 values and de-identified patient information will be deposited to the National Cancer Institute SeroNet Coordinating Center. Additionally, NT50 values and de-identified patient information reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.