Abstract
The reference material (RM) is a technical requirement for the quality assurance of analytical results and proficiency tests or interlaboratory comparisons. Microbiological RMs are most available in the dehydrated form, mainly by freeze-drying, and maintaining bacterial survival after preparation is a challenge. Thus, obtaining the most resistant cells is essential. Considering that bacteria present cross-response to dehydration after being submitted to an array of stress conditions, this study aimed to evaluate the influence of growth conditions on enterobacteria for the production of mixed microbiological RMs by freeze-drying in skim milk powder. Salmonella enterica serovar Enteritidis, Cronobacter sakazakii, Escherichia coli, and Citrobacter freundii were grown in a minimal medium with 0.5 M of NaCl and 0 to 5.0 mM of manganese sulfate (MnSO4) until stationary phase. Salmonella Enteritidis presented an increased resistance to dehydration in the presence of Mn, while C. sakazakii was the most resistant to freeze-drying and further storage for 90 days. Mixed microbiological RMs were produced by freeze-drying containing Salmonella Enteritidis and coliforms in skim milk powder with 100 mM of trehalose and the Salmonella survival rate was 91.2 to 93.6%. The mixed RM was stable after 30 days at -20 °C, and Salmonella and coliforms were detected by different methods being, the Rambach Agar the best for the bacterial differentiation. The results showed that the culture conditions applied in this study resulted in bacterial cells being more resistant to dehydration, freeze-drying, and stabilization for the production of mixed microbiological RMs more stable and homogeneous.
Supplementary Information
The online version contains supplementary material available at 10.1007/s42770-022-00808-z.
Keywords: Citrobacter freundii, Cronobacter sakazakii, Enterobacteria, Escherichia coli, Homogeneous, Resistance
Introduction
Reference material (RM) is an indispensable, reliable, and widely accepted tool for comparing the results of the analyses, representing one of the main bases for methodological quality control [1–5]. In the quality assurance context, the use of RM is mentioned in ISO/IEC 17025 [6] as a technical requirement for the quality assurance of analytical results, through regular use as the internal control of analysis and in the calibration, as well as for proficiency tests or interlaboratory comparisons [1, 5–9].
Microbiological reference materials (microbiological RMs) can be used in food microbiology laboratories as part of the quality assurance program, internal control of trials, validation of methods, training of workers, evaluation of laboratory accuracy, and scientific research [1, 5, 8, 10, 11]. Thus, microbiological RMs have to meet minimum requirements, such as showing homogeneity and stability within limits established over a period of time. Also, microbiological RMs should be representative of the intended use, that is, they should resemble routine samples [12–14] to guarantee to assess of participants performance and characterize measurement accuracy [15]. Therefore, mixed microbiological RMs containing the target microorganism plus interfering and, or competitors microorganisms may aid in the performance evaluation of laboratory analysis methods. The target microorganism's detection and, or quantification should be possible even in the presence of interfering microorganisms [1, 16, 17].
Most microbiological RMs are dehydrated by freeze-drying or spray drying of reconstituted skim milk powder containing bacteria and posteriorly packaged in vials or presented in capsule or lenticular forms [1, 4, 5]. After production, these materials must present desirable homogeneity and stability characteristics, and microbial cells must remain viable throughout storage and analysis [1, 4, 18]. For this, the cells need to preserve their biological integrity under dehydration [19]. However, the freeze-drying method combines freezing and drying stresses harmful to bacterial cells [20]. The use of the most resistant cells is recommended to reduce these cellular damages.
Bacteria present cross-response to dehydration after being submitted to nutritional, osmotic, oxidative, or thermal stresses [21–23]. Nutritional stress, that is, growth in a medium with deprivation of carbon, nitrogen, phosphate, or other nutrients, independent of the growth phase of the bacteria, also increases the resistance to dehydration and other stresses by cross-response [24–26]. Also, bacteria in the stationary growth phase are more adapted to multiple stresses when compared to bacteria in the logarithmic phase [27–31]. Thus, bacteria at the end of the logarithmic phase and the beginning of the stationary phase of growth should be used in freeze-drying since they have greater resistance to adverse conditions [19, 29, 32–35].
The response to osmotic stress induces intracellular accumulation of ions such as potassium (K+) and compatible solutes such as proline, glycine betaine, and trehalose, as well as activation of other osmoregulatory responses that also increase the survival of bacterial cells to dehydration [26, 36–43]. Trehalose is an osmoprotectant and cryoprotectant that can replace water molecules around macromolecules, preventing cell damage [44]. This disaccharide plays a central role in the protection of bacteria against dehydration through stabilizing membrane proteins and phospholipids [19, 35, 45–49]. Besides, trehalose may also exert a protective effect against cold stress in Salmonella and Escherichia coli [44, 50].
Adding manganese (Mn) to the culture medium may reduce the effects of oxidative stress from dehydration [51, 52]. This element can act as a cofactor of Mn-dependent superoxide dismutase (SOD) enzymes and replace iron (Fe), exerting a possible protective effect against oxidative damage in certain proteins [26, 53, 54]. In E. coli, when iron from the ribulose-5-phosphate-3-epimerase enzyme was replaced by Mn, the protein became less sensitive to hydrogen peroxide [55]. Fredrickson et al. [51] reported that bacteria with a high Mn/Fe ratio were less susceptible to oxidation of protein induced by dehydration. Salmonella enterica serovar Typhimurium cells showed higher activity of the enzyme SOD when grown in the presence of 5 mM of Mn [52].
Considering the interest in enhancing the resistance of cells subjected to stresses during the preparation of microbiological RMs, this study aimed to evaluate the influence of Mn on Salmonella and other enterobacteria under nutritional and osmotic stress. Besides, it was produced and analyzed mixed microbiological RMs with cells in the stationary phase of growth by freeze-drying in skim milk powder containing trehalose.
Materials and methods
Bacterial species
The enterobacteria species Salmonella enterica serovar Enteritidis phage type 4 (PT4) 578 (GenBank: 16S ribosomal RNA gene - MF066708.1), Cronobacter sakazakii ATCC 29004, Escherichia coli ATCC 29214, and Citrobacter freundii ATCC 8090 were used in this study. Bacterial cultures were stored at -20 °C in Brain Heart Infusion broth, pH 7.4 (BHI broth; Himedia, India) supplemented with 20% (v/v) of sterilized glycerol.
Evaluation of the growth of enterobacteria in a minimal medium containing NaCl
The bacterial species were cultivated in 3 mL of BHI broth for 12 h at 37 °C. Then, an aliquot of 200 µL of each specie was transferred to 20 mL of minimal medium (MM; 0.7% of K2HPO4, 0.2% of KH2PO4, 0.1% of (NH4)2SO4, 0.02% of MgSO4.7H2O, 0.4% of glycerol, 1 mM of CaCl2), pH 7.1 and incubated at 37 °C. After 24 h of incubation, 2 mL of each species was transferred to 200 mL of MM containing 0.5 M of NaCl (MMS), pH 7.1 and incubated at 37 °C. The optical density at 600 nm (OD 600 nm) was determined by spectrophotometry (Thermo Scientific, USA). Besides, the water activity (aw) of MM and MMS was measured in an automatic analyzer (Decagon Aqualab CX-2, USA).
Evaluation of the resistance of enterobacteria to dehydration in the presence of Mn
The resistance to dehydration was performed according to Breeuwer et al. [27], with modifications. The bacteria were cultivated at 37 °C in 200 mL of MMS containing 0.0, 0.1, 2.5, or 5.0 mM of manganese sulfate (MnSO4), pH 6.6, separately. After 38 h of incubation, an aliquot of 20 µL of each species in different concentrations of MnSO4 was transferred to 96-well microplates, incubated, and kept at 25 °C. Every seven days up to 49 days of dehydration, 20 μL of 0.85% (w/v) of saline solution was added to each well to resuspend the cells, diluted in 0.1% (w/v) of peptone saline. Then, an aliquot was plated by drop plate method in Plate Count Agar (PCA) for enumeration of viable and culturable cells [56].
Evaluation of enterobacteria survival after freeze-drying and stabilization
The treatment described in item "Evaluation of the resistance of enterobacteria to dehydration in the presence of Mn" that provided the cells most resistant to dehydration was selected and used to produce the inoculum. An aliquot of 20 mL of each species was harvested by centrifugation at 2,500 × g at 4 °C for 30 min (Sorvall, USA), washed with 0.1% of peptone saline, and the pellet resuspended in 12 mL of 10% (w/v) of reconstituted skim milk powder containing 100 mM of trehalose. Then, volumes of 1.5 mL were distributed in glass vials with a capacity of 10 mL, frozen immediately at -196 °C in liquid nitrogen (N2), stored at -80 °C for 18 h in an ultra-freezer (Thermo Scientific, USA) and freeze-dried for 24 h (Liotop, Brazil). These vials were kept at 4 °C for 90 days for stabilizing cells. Before and after freeze-drying, as well as after stabilization, the contents of a vial were resuspended in 1.5 mL of 0.1% of peptone saline, diluted in 0.1% of peptone saline and an aliquot was plated by drop plate method in PCA [56].
Selection of differential medium for enterobacteria
This analysis was performed to select at least one medium to differentiate colonies from Salmonella Enteritidis, C. sakazakii, E. coli, and C. freundii. These four enterobacteria species were cultivated in BHI broth for 12 h at 37 °C. Then, an aliquot of 0.1 and 1.0 mL of each one and their mixture were plated by spread plate and pour plate methods, respectively, in the differential medium, such as Brilliant Green Agar (BGA; Oxoid, England), Xylose-Lysine Deoxycholate Agar (XLD; Oxoid, England), Hektoen Enteric Agar (HE; Becton Dickinson, Germany), Violet Red Bile Glucose Agar (VRBG; Himedia, India), Eosin Methylene Blue Agar (EMB; Himedia, India), MacConkey Agar (Merck, Germany) and Rambach Agar (Merck, Germany). The inoculation was also carried in Petrifilm E. coli/Coliform Count Plates (Petrifilm EC; 3 M, USA) according to fabricant instructions.
Production of mixed microbiological RMs
Mixed microbiological RMs were produced according to Schulten et al. [57], with modifications. Mixed microbiological RMs were produced in three independent repetitions (RM1, RM2, and RM3) containing 3.0 to 4.0 log CFU.g−1 of Salmonella Enteritidis and 4.0 to 5.0 log CFU.g−1 of a mix of C. sakazakii, E. coli, and C. freundii. Initially, 1.5 mL of each species was freeze-dried and stabilized at 4 °C for 90 days as described in item "Evaluation of enterobacteria survival after freeze-drying and stabilization." Posteriorly, an aliquot of 0.6 g of Salmonella Enteritidis freeze-dried in 10% (w/v) of reconstituted skim milk powder containing 100 mM of trehalose was mixed aseptically with a mortar and pestle in a laminar flow cabinet with 59.4 g of skim milk powder sterilized by radiation. The same procedure was performed for 0.6 g of each previously mentioned coliform culture. Then, 0.6 g of skim milk powder contaminated with Salmonella Enteritidis was mixed with 59.4 g of skim milk powder contaminated with coliforms to obtain 60 g of mixed microbiological RMs. The mixed microbiological RMs were stored at -20 °C for 30 days. After storage, 0.3 g of each RM was weighed, and 2.7 mL of 0.1% of peptone saline was added. Then, dilutions were performed in 0.1% of peptone saline, and 1 mL of dilution was plated on PCA and incubated for 4 h at 37 °C. An overlay of Rambach Agar was added and incubated for another 24 h at 37 °C. It is noteworthy that Rambach Agar was used based on the results of the experiment described in item "Selection of differential medium for enterobacteria."
Assessment of homogeneity of mixed microbiological RMs
The homogeneity of mixed microbiological RMs was assessed according to Schulten et al. [57], with modifications. After 30 days of storage of the microbiological RMs at -20 °C, 10 replicates of 0.3 g of each microbiological RM were weighed into disposable Petri dishes, added 2.7 mL of 0.1% of peptone saline, performed dilutions in 0.1% of peptone saline and 1 mL of dilution plated on PCA with an overlay of Rambach Agar as described in item "Production of mixed microbiological RMs."
The statistical methods and models used to assess homogeneity depending on the microbiological contamination level of microbiological RM [58]. Homogeneity evaluation of microbiological RMs was performed in two stages [58, 59]. First, homogeneity was determined by calculating the dispersion index T1 to check for a variation between the duplicates of the analytical portions followed by a Poisson distribution. T1 was calculated as:
J represents the subsamples number. In this work, J was equal to 2;
I represents the replica number. In this case, I was equal to 10;
Zij is a colony-forming units (CFU) count of the subsample j of fraction i;
Zi + is a CFU sum of the number in all subsamples of a fraction i.
T1 is the Chi-square distribution with I (J-1) degrees of freedom. When 10 replicates of the microbiological RMs were used for duplicate enumeration, the value found should not exceed 18.3 (T1 ≤ 18.3, significance level α = 0.05) for the dispersion of the cells in the microbiological RM to follow the Poisson distribution. The homogeneous distribution of microorganisms in a liquid or powder is reported in the Poisson distribution. A value of T1 greater than the critical value of χ2 indicates low repeatability of the counts in the subsamples or analytical portions of the microbiological RMs [56].
The second assessment consisted of the application of test T2, which determines the variation between the counts of different replicates. T2 was calculated as:
Zi + represents the sum of counts in each replica;
Z + + is the total counts of all subsamples of all replicates;
I is the total number of replicas.
T2 test provides a valid result only when the T1 test result is not significant. The homogeneity of microbiological RM is determined by calculating the value of T2/(I-1) [58–60]. Microbiological RM showing T2/(I-1) value of at most 3.0 was accepted as homogeneous (T2/(I-1) ≤ 3.0) [61]. However, a more rigorous criterion of homogeneity determines a value of T2/(I-1) ≤ 2 for homogeneous microbiological RM [60].
Detection of Salmonella in mixed microbiological RMs
The detection of Salmonella in mixed microbiological RMs was performed by a conventional method, immunomagnetic separation and polymerase chain reaction (PCR).
Conventional method
The conventional method for research of Salmonella in mixed microbiological RMs was performed according to ISO 6579 [62], which involved first, the pre-enrichment of 25 g in buffered peptone water (Himedia, India) with incubation at 37 °C for 18 h. Subsequently, the selective enrichment step was performed in Rappaport Vassiliadis broth (Himedia, India) and tetrathionate broth (Himedia, India) with incubation at 41 and 37 ºC for 24 h, respectively. The selective isolation was carried out in XLD, HE, and Rambach Agar and incubated at 37 °C for 24 h. The typical colonies of Salmonella in each differential medium were grown in BHI broth at 37 °C for 12 h and subsequently striated in PCA. Then, selected colonies were characterized biochemically by the API 20E kit (Biomeriéux, USA) and by the oxidase test (Probac, Brazil), as well as performed the serological test using the Salmonella O Antiserum Poly A-I & VI (BD Difco, USA).
Immunomagnetic separation
The immunomagnetic separation for research of Salmonella in mixed microbiological RMs using the Dynabeads anti-Salmonella kit (Invitrogen, USA) was performed according to the manufacturer's recommendations. Initially, an aliquot of 1 mL of the pre-enrichment sample of the conventional method described in item "Conventional method" was added in a microtube containing 20 µL of Dynabeads anti-Salmonella and mixed for 30 min in Dynabeads MX4 Mixer (Invitrogen, USA). Then, the microtube was kept in the magnetic separator for 10 min; the liquid was removed and was added 1 mL of 1% of peptone saline solution buffered with 0.02% (v/v) of Tween 20. The mixture was homogenized by inversions without the magnetic separator, and then this separator was replaced and mixed for 5 min. This step was repeated two more times. Subsequently, the microtube was kept in the magnetic separator for 10 min; the liquid was removed and was added 1 mL of phosphate-buffered saline. This buffer was removed, and an additional 20 µL was added. Posteriorly, this volume plus Dynabeads anti-Salmonella was transferred to 10 mL of Rappaport Vassiliadis broth and followed the analysis of the conventional method described in item "Conventional method."
Polymerase chain reaction (PCR)
The PCR for research of Salmonella in mixed microbiological RMs was performed for the invA gene. Initially, an aliquot of 1 mL of the Rappaport Vassiliadis broth of the conventional method described in item "Conventional method" was used to extract the genomic DNA using the Wizard Genomic DNA Purification kit (Promega; USA), according to the recommendations of the manufacturer. The DNA was amplified using the oligonucleotide specific for the invA gene of Salmonella, 5′-GCATGAAATGGCAGAACAGC-3′ (forward) and 5′-ATGAGTGAAGGATCGCAACC-3′ (reverse). The PCR program was as follows: an initial denaturing step at 95 °C for one min, followed by 40 cycles of 95 °C for one min, 51 °C for one min and 72 °C for 2.5 min and, posteriorly, a final extension at 72 °C for 5 min. The amplified samples and molecular weight marker (1 kb DNA ladder; Promega, USA) with GelRed (Biotium, USA) were subjected to an electrophoretic run on 0.8% (w/v) of agarose gel at 80 V, and then the gel was exposed to ultra-violet light. The band with approximately 2 kb corresponds to the invA gene of Salmonella.
Enumeration of coliforms and E. coli in mixed microbiological RMs
The enumeration of coliforms and E. coli in mixed microbiological RMs was performed on Petrifilm EC and on PCA with an overlay of Rambach Agar and Violet Red Bile Agar (VRBA), separately. Initially, an aliquot of 1.0 g of each mixed microbiological RM was weighed into disposable Petri dishes and added 9 mL of 0.1% of peptone saline. This mixture was kept at room temperature for 45 min, further diluted with 0.1% of peptone saline and plated to enumerate coliforms as described below.
The enumeration of coliforms and E. coli in mixed microbiological RMs using the Petrifilm EC was performed according to AOAC [63]. An aliquot of 100 µL of dilution was plated on Petrifilm EC and incubated at 37 °C for 24 h. The red and blue colonies with and without gas were counted as coliforms, and the blue colonies with gas as E. coli.
The enumeration of coliforms in mixed microbiological RMs was performed using PCA with an overlay of differential media [64]. Aliquots of 1 mL of dilution were inoculated in duplicate in PCA by the pour-plate method. After incubation of 2 h at room temperature or 4 h at 37 °C, an overlay of VRBA (Merck, Germany) or Rambach Agar was added, respectively. The colony counts were performed after 24 h incubation at 37 °C.
Statistical analysis
The data were subjected to analysis of variance (ANOVA) followed by Tukey's test using the Statistical Analysis System and Genetics Software [65]. A p-value of < 0.05 (p < 0.05) was considered to be statistically significant.
Results
Enterobacteria survive and grow in minimal medium containing NaCl
Salmonella Enteritidis, C. sakazakii, E. coli, and C. freundii showed similar growth in MMS (p > 0.05) and entered in stationary phase after 18 h of cultivation (Fig. S1). These enterobacteria showed to be halotolerant due to their ability to survive and grow in the MM containing 0.5 M of NaCl (MMS), which corresponds to approximately 3% (w/v). It is noteworthy that the addition of this concentration of NaCl in the MM to produce MMS changes the aw from 0.993 to 0.977 at 25 °C.
Mn increases the resistance to dehydration of Salmonella Enteritidis
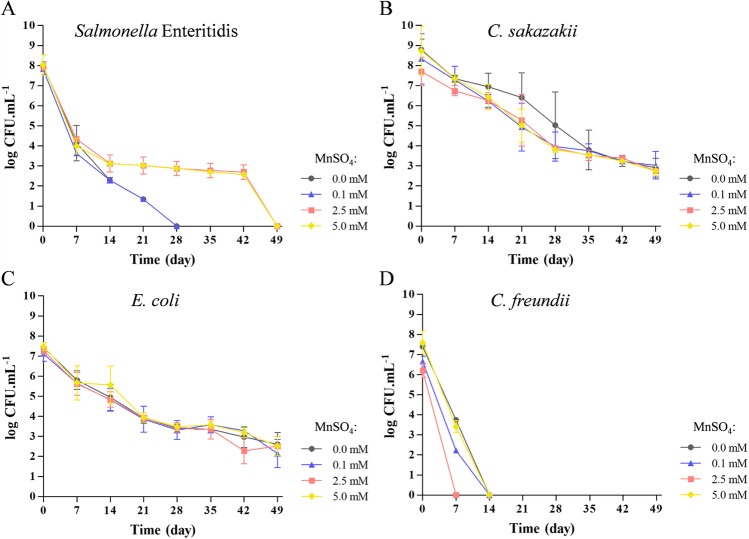
Growth in MMS containing 2.5 and 5.0 mM of MnSO4 promoted an increase in survival of Salmonella Enteritidis to dehydration for up to 42 days when compared to the control, i.e., cells growing in the absence of Mn (Fig. 1A). The high survival of C. sakazakii and E. coli to dehydration occurs independently of the presence of Mn (Fig. 1B and C, respectively). Among the coliforms evaluated, C. freundii was more sensitive to dehydration, and Mn did not increase its resistance (Fig. 1D).
Fig. 1.
Evaluation of resistance to dehydration at 25 °C up to 49 days of Salmonella Enteritidis (A), C. sakazakii (B), E. coli (C), and C. freundii (D), previously cultivated for 38 h in MMS containing 0.0, 0.1, 2.5, or 5.0 mM of MnSO4, separately
C. sakazakii is the most resistant to freeze-drying and stabilization
Considering that MMS containing 2.5 and 5.0 mM of MnSO4 increased resistance to dehydration of Salmonella Enteritidis, the lowest concentration was chosen for the preparation of cells for dehydration before freeze-dried. On the other hand, as Mn did not alter the survival of C. sakazakii, E. coli, and C. freundii for 49 days of dehydration, the cells were grown in the absence of MnSO4 before freeze-dried. After freeze-drying in reconstituted skim milk powder containing 100 mM of trehalose and stabilization at 4 °C for 90 days, the cultivable cells were quantified and the log CFU.mL−1 is presented in Table 1.
Table 1.
The means of the logarithm of colony-forming units per milliliter (means log CFU.mL−1) of Salmonella Enteritidis, C. sakazakii, E. coli, and C. freundii before and after freeze-drying in reconstituted skim milk powder containing 100 mM of trehalose and after stabilizing at 4 °C for 90 days
| Bacterial species | Means log CFU.mL−1 | Means log CFU.mL−1 reduced (% reduced) | ||||
|---|---|---|---|---|---|---|
| Before freeze-drying |
After freeze-drying |
After Stabilization |
After freeze-drying |
After stabilization |
After freeze-drying and stabilization |
|
| Salmonella Enteritidis | 8.48a | 7.59b | 6.24c | 0.89A (10.5) | 1.35AB (17.8) | 2.24A (26.4) |
| C. sakazakii | 8.64a | 8.51a | 7.92a | 0.13B (1.5) | 0.59B (6.9) | 0.72B (8.3) |
| E. coli | 8.06a | 7.83a | 5.27b | 0.23AB (2.7) | 2.56A (32.7) | 2.79A (34.6) |
| C. freundii | 7.31a | 6.70a | 4.66b | 0.61AB (8.3) | 2.04A (30.4) | 2.65A (36.3) |
log CFU.mL−1 = logarithm of colony-forming units per milliliter;
The comparisons can be drawn among processes or bacteria. Mean followed by different superscript lowercase letters in the same line (among processes for the same bacteria) and followed by different superscript uppercase letters in the same column for log CFU.mL−1 reduced (among bacteria for each process, separately) differs at 5% probability (p < 0.05) by Tukey's test
C. sakazakii was the most resistant to freeze-drying in reconstituted skim milk powder containing 100 mM of trehalose and after stabilization at 4 °C for 90 days (p < 0.05) (Table 1). This result can be better evidenced by the 0.72 cycle log CFU.mL−1 reduced after the whole process, including freeze-drying and stabilization (p < 0.05) (Table 1). The other evaluated enterobacteria presented up to 2 log CFU.mL−1 reduced after freeze-drying and stabilization (Table 1).
Rambach Agar allows differentiating Salmonella and coliforms colonies
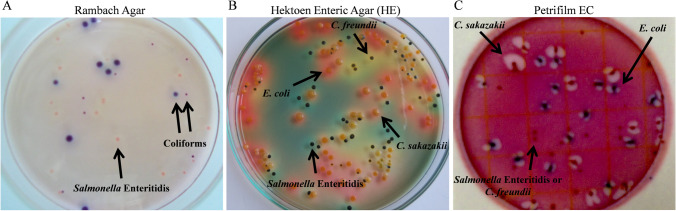
The definition of a differential medium for counting the four enterobacteria used in the mixed microbiological RM was necessary to evaluate the survival of each bacterial species. The plating by the spread plate method in the Rambach Agar allowed differentiating the colonies of Salmonella Enteritidis from the coliforms (Fig. 2A). In this medium, Salmonella Enteritidis showed pink colonies, while coliforms showed blue or purple colonies (Fig. 2A). On the other hand, in HE the colonies of four species were different (Fig. 2B). Salmonella Enteritidis and C. freundii showed black-bluish colonies, whereas C. freundii had a yellow halo around the colonies (Fig. 2B). E. coli and C. sakazakii showed colonies with yellow to orange coloration, but C. sakazakii may be differentiated by the mucoid appearance on the surface (Fig. 2B). Petrifilm EC allowed differentiation of E. coli and C. sakazakii from other species. In this medium, E. coli showed blue colonies with gas production, while C. sakazakii showed red coloration with gas production (Fig. 2C). Salmonella Enteritidis and C. freundii colonies were small with red coloration without gas production, which did not distinguish them (Fig. 2C).
Fig. 2.
Characteristics of colonies of Salmonella Enteritidis, C. sakazakii, E. coli, and C. freundii plated by spread plate method in Rambach Agar (A), Hektoen Enteric Agar (HE) (B) and Petrifilm EC (C)
The plating by pour plate method in Rambach Agar allowed differentiation of Salmonella colonies from coliforms, while in HE it was impossible to differentiate them with the same precision as the other plating method. It was impossible to differentiate the enterobacteria colonies when the BGA, XLD, VRBG, EMB, and MacConkey Agar were used.
Enterobacteria survive in mixed microbiological RMs
Salmonella counts in the mixed microbiological RMs ranged from 3.0 to 4.0 log CFU.g−1, and coliforms from 4.0 to 5.0 log CFU.g−1 after production and these values were statistically different between microbiological RMs repetitions (p < 0.05) (Table 2). Similar reductions of the cycles log CFU.g−1 of Salmonella were detected in all microbiological RMs after 30 days of storage at -20 °C with a Salmonella survival rate of 91.2 to 93.6% (p > 0.05). However, the same variation was not observed for coliforms (p < 0.05) (Table 2).
Table 2.
Microbiological evaluation of the homogeneity of the microbiological RMs after production and after storage for 30 days at -20 °C
| Bacteria | Microbiological RMs | Means log CFU.g−1 | Homogeneity | ||||
|---|---|---|---|---|---|---|---|
| After production | After storage | Reduced (% reduced) |
Mean 1 (log CFU.g−1) |
T1 2 | T2/(I-1) 3 | ||
| Salmonella | RM1 | 3.06aC | 2.79bC | 0.27 (8.8) | 2.70 | 10.3 | 3.0 |
| RM2 | 3.93B | 3.66B | 0.27 (6.9) | 3.43 | 9.3 | 5.1* | |
| RM3 | 4.56aA | 4.27bA | 0.29 (6.4) | 4.18 | 6.9 | 4.4* | |
| Coliforms | RM1 | 4.69aB | 4.35bB | 0.33B (7.0) | 4.26 | 13.9 | 1.9 |
| RM2 | 5.39aA | 4.81bA | 0.58A (10.8) | 4.72 | 4.0 | 5.4* | |
| RM3 | 4.39C | 4.07C | 0.32B (7.3) | 3.96 | 20.9* | 3.5** | |
log CFU.g−1 = logarithm of colony-forming units per gram of microbiological RM;
The comparisons can be drawn among time or microbiological RM. Mean followed by different superscript lowercase letters in the same line (among time of production and storage for the same microbiological RM) and followed by different superscript uppercase letters in the same column (among microbiological RMs for the same microorganism and time or among log CFU.g−1 reduced of microbiological RMs for the same microorganism, separately) differs at 5% probability (p < 0.05) by Tukey's test. Where a letter is not shown, no statistical difference among samples was observed;
1 Mean of log CFU.g−1 of 10 analytical portions or fraction of 0.3 g of each microbiological RM;
2 The T1 value of homogeneous microbiological RM is less than or equal to 18.3 (T1 ≤ 18.3);
3 The T2/(I-1) value of homogeneous microbiological RM is less than or equal to 3.0 (T2/(I-1) ≤ 3.0);
* The T1 or T2/(I-1) values exceed the limits for a mixed microbiological RM;
** The T2/(I-1) value is invalid;
Homogeneous mixed microbiological RM is shown in bold
The homogeneity of the mixed microbiological RMs was evaluated by two parameters: T1 and T2/(I-1) for Salmonella and coliforms, separately (Table 2). The numbers of viable Salmonella showed T1 ≤ 18.3 in mixed microbiological RMs (Table 2), indicating repeatability of the counts in the analytical portions or fractions of the mixed microbiological RMs. The counts of coliforms showed values of T1 within the limit for microbiological RM1 and RM2 (Table 2). On the other hand, only microbiological RM1 presented values of T2/(I-1) ≤ 3.0 for counts of Salmonella and coliforms (Table 2). However, considering a more rigorous value of T2/(I-1) ≤ 2.0, only the counts of coliforms of microbiological RM1 met this criterion (Table 2). Thus, only microbiological RM1 was homogeneous considering the two stages of homogeneity evaluated (Table 2).
Different methods detect Salmonella in mixed microbiological RMs
The three mixed microbiological RMs (RM1, RM2, and RM3) were analyzed by three methods for research of Salmonella after storage at -20 °C for 30 days. This pathogen was detected in these mixed microbiological RMs through the conventional method, immunomagnetic separation, and PCR (Fig. S2). In addition, all colonies suspected isolated in the conventional method and immunomagnetic separation were characterized biochemically by the API 20E kit and oxidase test and confirmed serologically by Salmonella O Antiserum Poly A-I & VI as Salmonella.
PCA with an overlay of Rambach Agar recovers more coliforms from mixed microbiological RMs
The enumeration of coliforms in mixed microbiological RMs after storage at -20 °C for 30 days showed a similarity to results obtained in Petrifilm EC and VRBA (p < 0.05) and evidenced the superiority of the Rambach Agar in recovering coliforms from RMs (Table 3). In addition, it was possible to enumerate E. coli in these RMs by Petrifilm EC (Table 3).
Table 3.
Enumeration of coliforms and E. coli by different methods in the mixed microbiological RMs after 30 days at -20 °C
| Microorganisms | Methods | Means log CFU.g−1 | ||
|---|---|---|---|---|
| Microbiological RMs | ||||
| RM1 | RM2 | RM3 | ||
| Coliforms | PCA + Rambach Agar overlay | 4.35bA | 4.81aA | 4.07cA |
| Petrifilm EC for coliform count plates | 3.65cB | 4.45aB | 3.78bB | |
| PCA + VRBA overlay | 3.56bB | 4.24aB | 3.78bB | |
| E. coli | Petrifilm EC for E. coli | 2.65b | 3.72a | 2.39c |
Microbiological RM = Microbiological reference material;
PCA = Plate Count Agar;
Petrifilm EC = Petrifilm E. coli/Coliform Count Plates;
VRBA = Violet Red Bile Agar;
log CFU.g−1 = logarithm of colony-forming units per gram of microbiological RM;
Mean followed by different superscript lowercase letters in the same line (among mixed microbiological RMs for the same method) and followed by different superscript uppercase letters in the same columns (among methods as Rambach Agar, Petrifilm EC for coliform count plates and VRBA for the same mixed microbiological RM) for log CFU.g−1 differs at 5% probability (p < 0.05) by Tukey's test
Discussion
The ability of Salmonella Enteritidis, C. sakazakii, E. coli, and C. freundii to survive and grow in the MMS that contains a high concentration of NaCl, that is, to be halotolerant, can be explained, at least in part, by some mechanisms, such as regulation and accumulation of osmoprotectants, filamentation, capsule production, and biofilm formation including the production of various extracellular polysaccharides [37, 66–69].
The varied resistance to dehydration, freeze-drying, and stabilization found in this study for the evaluated enterobacteria is recognized. According to Barron and Forsythe [70], enterobacteria may be divided into three groups considering the survival over time of storage of the dehydrated cells. The first group consists of C. freundii, Citrobacter koseri, and Enterobacter cloacae, which were not recovered after six months of storage under dehydration. The second group is composed of Salmonella Enteritidis, E. coli, and Klebsiella pneumoniae that were not recovered after 15 months of storage under dehydration. The third group is composed of Pantoea sp., Klebsiella oxytoca, and Escherichia vulneris that persisted for more than two years of storage under dehydration, and some strains of C. sakazakii that were recovered after 2.5 years.
C. sakazakii was the most resistant to dehydration, freeze-drying, and stabilization compared to Salmonella Enteritidis, E. coli, and C. freundii in this study. Breeuwer et al. [27] also showed that C. sakazakii was more resistant to osmotic stress and dehydration conditions than Salmonella, E. coli, and C. freundii and suggested that the resistance may be related to the accumulation of trehalose in the cells. The desiccation tolerance of C. sakazakii was also significantly higher than E. coli O157:H7, Salmonella enterica, and Listeria monocytogenes in powdered infant formula [71]. Besides, the freeze-drying of C. sakazakii ATCC 29544 in whole milk had the highest protective effect, followed by skim milk and tryptic soy broth [72]. Milk contains proteins that provide an additional protective layer for cells during drying by stabilizing membrane [45, 48, 73–77] and, when added with trehalose or sucrose, provide a higher survival rate during subsequent storage [73, 74, 78].
The high resistance of Cronobacter to desiccation may also be related to several genes as those of the Cpx system that encode an envelope stress response regulator, dnaK and dnaJ genes that encode two molecular chaperones, and the sigma factors RpoN and RpoS seem to be the main signals regulating the bacterial response to hyperosmotic conditions [79]. Furthermore, all Cronobacter contain genes for the production of β-carotene, which is believed to protect bacteria from harmful oxygen radicals and add tolerance to physical desiccation [80, 81].
The high survival of E. coli to dehydration shown in our study can be attributed to the intracellular accumulation of trehalose. E. coli MC4100 grown in an M9 medium containing 1% (w/v) of glucose, trace elements (0.015 mM of FeSO4, 0.015 mM of ZnSO4, and 0.015 mM of MnSO4) and 0.6 M of NaCl until the stationary phase presented high levels of intracellular trehalose and was more tolerant to desiccation [82]. Although the accumulation of trehalose increases the tolerance of E. coli to desiccation, this is not the only factor related to the resistance of this bacterium [36, 82]. The rpoS also was required by E. coli O157:H7 to tolerate osmotic and desiccation stress [83]. Furthermore, Chen and Goulian [32] showed that the general stress response regulator RpoS and the transcriptional regulators DksA, LexA, RecA, and ArcA play a critical role in the survival of E. coli to dehydration. These authors also identified two additional regulators, Crl and ArcZ that promote dehydration tolerance through modulation of RpoS.
Considering the effect of Mn, only Salmonella Enteritidis increased resistance to dehydration in the presence of this element, and this may be related to the observation by Maserati et al. [84]. These authors showed that the Mn transport transcriptional regulator (mntR) of Salmonella Typhimurium ATCC 14028 was induced by low aw, and consequently, the increase in intracellular Mn can reduce the effects of oxidative stress from dehydration, promoting cell survival. Interestingly, the RNA polymerase sigma factor RpoS (σS), which is required for stress resistance, such as starvation, hyper-osmolarity, and oxidative stress, induces transcription of the Mn transporter genes mntH and sitABCD and prevents their repression by the Mn-responsive regulator MntR and the ferric uptake regulator Fur in Salmonella Typhimurium ATCC 14028 [85].
Thus, we have applied the results found in this study to the development of mixed microbiological MRs of Salmonella Enteritidis and the three coliforms. The conditions adopted, such as growth in MMS containing 2.5 mM of MnSO4 until the stationary phase followed by freeze-drying in skim milk powder containing trehalose, may have contributed to the cell stability of Salmonella Enteritidis in the mixed microbiological RMs. It is noteworthy that minor variations of cell viability result from changes that may occur during dehydration and storage [35].
Of the mixed microbiological RMs produced, only one was homogeneous, and the lack of homogeneity of the others may be due to the difficulty in dispersing the bacterial cells in the milk powder during the various mixing steps. In microbiological RMs containing approximately 2.0 and 3.0 log CFU of Salmonella Enteritidis per capsule in the presence of competing microorganisms from chicken feces, the values of T2/(I-1) found were 1.7 and 3.1, respectively [86]. These authors also produced microbiological RMs containing, on average, 2.44 log CFU of Salmonella Typhimurium per capsule in the presence of a competing microbiota and observed a T2/(I-1) value of 5.57. In a second study, microbiological RMs containing about 2.0 and 2.7 log CFU of Salmonella Enteritidis per capsule and the same competing microorganisms presented T2/(I-1) values of 1.91 and 1.58, respectively [87]. Microbiological RMs produced with 2.0 log CFU.mL−1 of Salmonella Enteritidis ATCC 13076 by freeze-drying in 14% of fish protein hydrolysate (FPH) from catfish (Clarias batrachus) [3] and tuna (Euthynnus affinis) [7] were homogeneous and stable at a storage temperature of -20 ºC for 28 days, and survival rates were between 1.2 and 1.4 log CFU.mL−1, respectively. On the other hand, the microbiological RMs prepared with 10 to 14% of these matrices were not homogeneous [3, 7]. However, these authors assessed the homogeneity of the microbiological RMs by different analyzes from those used in this study.
Although not all mixed microbiological RMs developed in the present study meet the criteria for homogeneity, it was possible to detect Salmonella in all of them by the conventional method, immunomagnetic separation, and PCR. Thus, the competing microbiota in our mixed microbiological RMs did not interfere in the detection of Salmonella by the methods used, meeting an interesting criterion for microbiological RMs [1]. The invA gene was also used by Múrtula et al. [9] to quantify by quantitative PCR (qPCR) Salmonella spp. present in RM developed in tablet format.
Finally, Rambach Agar was allowed to differentiate Salmonella and coliforms colonies, as well as greater recovery of coliforms from mixed microbiological MRs when performed on PCA with an overlay of Rambach Agar. The highest count on Rambach Agar may be due to the recovery of the injured cells in a non-selective medium such as PCA for 4 h at 37 °C before adding an overlay of Rambach Agar, a selective and differential medium. In the VRBA, the previous recovery in PCA was also performed, but the count was statistically lower than in Rambach Agar. This difference can be attributed to the short time and inadequate temperature of recovery, which were room temperature and 2 h, respectively. In Petrifilm EC, the injured cells are directly plated in the selective medium without previous recovery, which may justify the lower enumeration of coliforms. Smith et al. [88] also showed that the number of cells recovered from E. coli O157:H7 using a non-selective medium with an overlay of the selective medium was significantly higher relative to direct inoculation in Petrifilm EC. However, most microbiological analyses of foods may involve direct inoculation of the samples into a specific selective medium for the subsequent enumeration of microorganisms. Most injured bacteria do not grow in a selective medium, taking a count to underestimate and threatening consumers' health [88, 89].
Therefore, the production of stable, homogeneous, and recoverable mixed microbiological RMs, containing a higher number of culturable cells and a lower number of injured cells, remains an important issue that needs to be further investigated as it is the critical point [1]. These challenges must be overcome until scientific knowledge can be transformed into innovative, affordable and low-cost products [1, 90, 91], as there is a shortage of microbiological RMs available [1].
Conclusion
The cell cultivation conditions to increase bacterial resistance to the stresses inherent in the production process of the microbiological RMs is necessary due to the importance of the cell viability in microbiological RMs for food microbiology. Therefore, Salmonella survival to freeze-drying was successfully obtained when cells were previously cultivated in MMS until the stationary phase, added with Mn and submitted to osmotic stress. This pathogen was also successfully recovered from the mixed microbiological MRs containing coliforms using different methods, and the Rambach Agar allowed the differentiation of colonies of Salmonella and coliforms. Thus, this knowledge generated will contribute to the development of other more stable and homogeneous mixed microbiological RMs in the dehydrated form.
Supplementary Information
Below is the link to the electronic supplementary material.
Growth of Salmonella Enteritidis, C. sakazakii, E. coli, and C. freundii in MMS at 37 °C for 48 h (28.1 KB)
Detection of Salmonella in the mixed microbiological RMs by PCR of invA gene. Lane 1: Molecular weight marker (1 kb DNA ladder); lanes 2, 3, and 4: amplification of a sample of microbiological RM1, RM2, and RM3 enriched in Rappaport Vassiliadis broth after 30 days storage at -20 °C, respectively; lane 5: positive control; lane 6: negative control (112 KB)
Acknowledgements
The authors thank the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), and Fundação de Amparo à Pesquisa do Estado de Minas Gerais (FAPEMIG) for financial support and scholarships.
Author contribution
Maria Roméria da Silva: conceptualization, methodology, investigation, data curation, writing-original draft, writing-review, and editing. Felipe Alves de Almeida: investigation, data curation, writing-original draft, writing-review, and editing. Ana Íris Mendes Coelho: methodology and investigation. Fernanda Lopes da Silva: investigation. Maria Cristina Dantas Vanetti: conceptualization, supervision, project administration, funding acquisition, resources, writing-original draft, writing-review, and editing.
Funding
This work was financially supported by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), and Fundação de Amparo à Pesquisa do Estado de Minas Gerais (FAPEMIG).
Declarations
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Consent to participate
Not applicable.
Consent for publication
The authors Maria Roméria da Silva, Felipe Alves de Almeida, Ana Íris Mendes Coelho, Fernanda Lopes da Silva, and Maria Cristina Dantas Vanetti are in accordance with the submission of this manuscript to the Brazilian Journal of Microbiology.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Singh KA, Rai R, Nair SS. Review on development of assigned value microbiological reference materials used in food testing. Food Microbiol. 2022;102:103904. doi: 10.1016/j.fm.2021.103904. [DOI] [PubMed] [Google Scholar]
- 2.Gouveia GJ, Shaver AO, Garcia BM, Morse AM, Andersen EC, Edison AS, McIntyre LM. Long-term metabolomics reference material. Anal Chem. 2021;93:9193–9199. doi: 10.1021/acs.analchem.1c01294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kurniawati E, Ibrahim B, Desniar D. Potency of catfish (Clarias sp.) protein hydrolysates as candidates matrices for microbiology reference material. Squalen Bull Mar Fish Postharvest Biotechnol. 2019;14:121–130. doi: 10.15578/squalen.v14i3.404. [DOI] [Google Scholar]
- 4.Philipp WJ, van Iwaarden P, Schimmel H, Meeus N, Kollmorgen N. Development of reference materials for microbiological analysis. Accredit Qual Assur. 2007;12:134–138. doi: 10.1007/s00769-006-0244-3. [DOI] [Google Scholar]
- 5.Rosas CO, Rodrigues JM, de la Cruz MHC, Lopes SMR, Souto ASS, Brandão MLL, Capasso IRVF. Microbiological reference material (bacterial and fungal domains): Definition, production rules, use and need for establishment in Brazil. Braz J Food Technol. 2019;22:e2017208. doi: 10.1590/1981-6723.20817. [DOI] [Google Scholar]
- 6.ISO/IEC 17025 (2017) ISO/IEC 17025 - General requirements for the competence of testing and calibration laboratories, 3rd ed. International Organization for Standardization (ISO). https://www.iso.org/publication/PUB100424.html. Accessed 01 Feb 2022
- 7.Kurniawati E, Ibrahim B, Desniar Homogeneity and stability of a secondary microbiological reference material candidate for Salmonella in fish matrix. IOP Conf Ser Earth Environ Sci. 2020;404:012036. doi: 10.1088/1755-1315/404/1/012036. [DOI] [Google Scholar]
- 8.ISO 17034:2016 (2016) ISO 17034 - General requirements for then competence of reference material producers, 1rd ed. International Organization for Standardization (ISO). https://www.iso.org/standard/29357.html. Accessed 07 Jun 2022
- 9.Múrtula R, Yáñez MA, Soria E, Catalán V. Development of a new reference material for the validation of molecular detection methods for microbiological analysis. Accredit Qual Assur. 2010;15:217–221. doi: 10.1007/s00769-009-0629-1. [DOI] [Google Scholar]
- 10.Abdelmassih M, Planchon V, Anceau C, Mahillon J. Development and validation of stable reference materials for food microbiology using Bacillus cereus and Clostridium perfringens spores. J Appl Microbiol. 2011;110:1524–1530. doi: 10.1111/j.1365-2672.2011.05007.x. [DOI] [PubMed] [Google Scholar]
- 11.ISO Guide 35 (2017) ISO Guide 35 - Reference materials - Guidance for characterization and assessment of homogeneity and stability, 4th ed. International Organization for Standardization (ISO)
- 12.Griepink B. Aiming at good accuracy with the Community Bureau of Reference (BCR) Química Analítica. 1989;10:1–21. [Google Scholar]
- 13.In’t Veld P. The use of reference materials in quality assurance programmes in food microbiology laboratories. Int J Food Microbiol. 1998;45:35–41. doi: 10.1016/S0168-1605(98)00145-7. [DOI] [PubMed] [Google Scholar]
- 14.Jarvis B. Microbiological reference materials. In: Batt CA, Tortorello ML, editors. Encyclopedia of food microbiology. 2. Elsevier; 2014. pp. 614–620. [Google Scholar]
- 15.Molinier O, Guarini P. Model of uncertainty for the variability of water microbiological enumeration in a proficiency testing scheme. Accredit Qual Assur. 2020;25:139–146. doi: 10.1007/s00769-019-01420-9. [DOI] [Google Scholar]
- 16.Bort M, Messineo E, Le Neve R, Boubetra A, Tirard A. Production of external reference materials in food microbiology. Int J Microbiol Infect Dis. 2018;2:1–3. doi: 10.33425/2639-9458.1025. [DOI] [Google Scholar]
- 17.Glaeser H. Reference materials for checking the performance of qualitative tests (presence/absence of pathogenic micro-organisms): properties, future needs, use in method validation. Accredit Qual Assur. 2004;9:205–208. doi: 10.1007/s00769-004-0763-8. [DOI] [Google Scholar]
- 18.Miyamoto-Shinohara Y, Sukenobe J, Imaizumi T, Nakahara T. Survival curves for microbial species stored by freeze-drying. Cryobiology. 2006;52:27–32. doi: 10.1016/j.cryobiol.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 19.Potts M. Desiccation tolerance of prokaryotes. Microbiol Rev. 1994;58:755–805. doi: 10.1128/MMBR.58.4.755-805.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heylen K, Hoefman S, Vekeman B, Peiren J, De Vos P. Safeguarding bacterial resources promotes biotechnological innovation. Appl Microbiol Biotechnol. 2012;94:565–574. doi: 10.1007/s00253-011-3797-y. [DOI] [PubMed] [Google Scholar]
- 21.Gruzdev N, Pinto R, Sela S. Effect of desiccation on tolerance of Salmonella enterica to multiple stresses. Appl Environ Microbiol. 2011;77:1667–1673. doi: 10.1128/AEM.02156-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maserati A, Lourenco A, Diez-Gonzalez F, Fink RC. iTRAQ-based global proteomic analysis of Salmonella enterica serovar Typhimurium in response to desiccation, low water activity, and thermal treatment. Appl Environ Microbiol. 2018;84:e00393–e418. doi: 10.1128/AEM.00393-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ricke SC, Dawoud TM, Kim SA, Park SH, Kwon YM (2018) Salmonella cold stress response: mechanisms and occurrence in foods. In: Advances in applied microbiology. pp 1–38 [DOI] [PubMed]
- 24.Kenyon WJ, Sayers DG, Humphreys S, Roberts M, Spector MP. The starvation-stress response of Salmonella enterica serovar Typhimurium requires σE-, but not CpxR-regulated extracytoplasmic functions. Microbiology. 2002;148:113–122. doi: 10.1099/00221287-148-1-113. [DOI] [PubMed] [Google Scholar]
- 25.Kenyon WJ, Humphreys S, Roberts M, Spector MP. Periplasmic peptidyl-prolyl isomerases SurA and FkpA play an important role in the starvation-stress response (SSR) of Salmonella enterica serovar Typhimurium. Antonie Van Leeuwenhoek. 2010;98:51–63. doi: 10.1007/s10482-010-9428-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spector MP, Kenyon WJ. Resistance and survival strategies of Salmonella enterica to environmental stresses. Food Res Int. 2012;45:455–481. doi: 10.1016/j.foodres.2011.06.056. [DOI] [Google Scholar]
- 27.Breeuwer P, Lardeau A, Peterz M, Joosten HM. Desiccation and heat tolerance of Enterobacter sakazakii. J Appl Microbiol. 2003;95:967–973. doi: 10.1046/j.1365-2672.2003.02067.x. [DOI] [PubMed] [Google Scholar]
- 28.Kosaka T, Murata M, Yamada M (2017) Survival strategy of Escherichia coli in stationary phase: involvement of σE-dependent programmed cell death. In: Samie A (ed) Escherichia coli - Recent Advances on Physiology, Pathogenesis and Biotechnological Applications. InTech, pp 383–404
- 29.Llorens JMN, Tormo A, Martínez-García E. Stationary phase in gram-negative bacteria. FEMS Microbiol Rev. 2010;34:476–495. doi: 10.1111/j.1574-6976.2010.00213.x. [DOI] [PubMed] [Google Scholar]
- 30.Meng XC, Stanton C, Fitzgerald GF, Daly C, Ross RP. Anhydrobiotics: the challenges of drying probiotic cultures. Food Chem. 2008;106:1406–1416. doi: 10.1016/j.foodchem.2007.04.076. [DOI] [Google Scholar]
- 31.Rychlik I, Barrow PA. Salmonella stress management and its relevance to behaviour during intestinal colonisation and infection. FEMS Microbiol Rev. 2005;29:1021–1040. doi: 10.1016/j.femsre.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 32.Chen AI, Goulian M. A network of regulators promotes dehydration tolerance in Escherichia coli. Environ Microbiol. 2018;20:1283–1295. doi: 10.1111/1462-2920.14074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hengge-Aronis R. Regulation of gene expression during entry into stationary phase. Cell Mol Biol. 1996;5:1497–1512. [Google Scholar]
- 34.Jørgensen F, Leach S, Wilde SJ, Davies A, Stewart GSAB, Humphrey T. Invasiveness in chickens, stress resistance and RpoS status of wild-type Salmonella enterica subsp. enterica serovar Typhimurium definitive type 104 and serovar Enteritidis phage type 4 strains. Microbiology. 2000;146:3227–3235. doi: 10.1099/00221287-146-12-3227. [DOI] [PubMed] [Google Scholar]
- 35.Morgan CA, Herman N, White PA, Vesey G. Preservation of micro-organisms by drying: a review. J Microbiol Methods. 2006;66:183–193. doi: 10.1016/j.mimet.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 36.Billi D, Potts M. Life and death of dried prokaryotes. Res Microbiol. 2002;153:7–12. doi: 10.1016/S0923-2508(01)01279-7. [DOI] [PubMed] [Google Scholar]
- 37.Burgess CM, Gianotti A, Gruzdev N, Holah J, Knøchel S, Lehner A, Margas E, Esser SS, Sela S, Tresse O. The response of foodborne pathogens to osmotic and desiccation stresses in the food chain. Int J Food Microbiol. 2016;221:37–53. doi: 10.1016/j.ijfoodmicro.2015.12.014. [DOI] [PubMed] [Google Scholar]
- 38.Castro AG, Bredholt H, Strom AR, Tunnacliffe A. Anhydrobiotic engineering of gram-negative bacteria. Appl Environ Microbiol. 2000;66:4142–4144. doi: 10.1128/AEM.66.9.4142-4144.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jin DJ, Cagliero C, Zhou YN. Growth rate regulation in Escherichia coli. FEMS Microbiol Rev. 2012;36:269–287. doi: 10.1111/j.1574-6976.2011.00279.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kempf B, Bremer E. Uptake and synthesis of compatible solutes as microbial stress responses to high-osmolality environments. Arch Microbiol. 1998;170:319–330. doi: 10.1007/s002030050649. [DOI] [PubMed] [Google Scholar]
- 41.Li H, Bhaskara A, Megalis C, Lou TM. Transcriptomic analysis of Salmonella desiccation resistance. Foodborne Pathog Dis. 2012;9:1143–1151. doi: 10.1089/fpd.2012.1254. [DOI] [PubMed] [Google Scholar]
- 42.Rojas E, Theriot JA, Huang KC. Response of Escherichia coli growth rate to osmotic shock. Proc Natl Acad Sci. 2014;111:7807–7812. doi: 10.1073/pnas.1402591111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou K, George SM, Métris A, Li PL, Baranyi J. Lag phase of Salmonella enterica under osmotic stress conditions. Appl Environ Microbiol. 2011;77:1758–1762. doi: 10.1128/AEM.02629-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leslie SB, Israeli E, Lighthart B, Crowe JH, Crowe LM. Trehalose and sucrose protect both membranes and proteins in intact bacteria during drying. Appl Environ Microbiol. 1995;61:3592–3597. doi: 10.1128/AEM.61.10.3592-3597.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carvalho AS, Silva J, Ho P, Teixeira P, Malcata FX, Gibbs P. Relevant factors for the preparation of freeze-dried lactic acid bacteria. Int Dairy J. 2004;14:835–847. doi: 10.1016/j.idairyj.2004.02.001. [DOI] [Google Scholar]
- 46.Crowe JH, Hoekstra FA, Crowe LM. Anhydrobiosis. Annu Rev Physiol. 1992;54:579–599. doi: 10.1146/annurev.ph.54.030192.003051. [DOI] [PubMed] [Google Scholar]
- 47.Elliott GD, Wang S, Fuller BJ. Cryoprotectants: a review of the actions and applications of cryoprotective solutes that modulate cell recovery from ultra-low temperatures. Cryobiology. 2017;76:74–91. doi: 10.1016/j.cryobiol.2017.04.004. [DOI] [PubMed] [Google Scholar]
- 48.Hubálek Z. Protectants used in the cryopreservation of microorganisms. Cryobiology. 2003;46:205–229. doi: 10.1016/S0011-2240(03)00046-4. [DOI] [PubMed] [Google Scholar]
- 49.Strom AR, Kaasen I. Trehalose metabolism in Escherichia coli: stress protection and stress regulation of gene expression. Mol Microbiol. 1993;8:205–210. doi: 10.1111/j.1365-2958.1993.tb01564.x. [DOI] [PubMed] [Google Scholar]
- 50.Gruzdev N, Pinto R, Sela Saldinger S. Persistence of Salmonella enterica during dehydration and subsequent cold storage. Food Microbiol. 2012;32:415–422. doi: 10.1016/j.fm.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 51.Fredrickson JK, Li SMW, Gaidamakova EK, Matrosova VY, Zhai M, Sulloway HM, Scholten JC, Brown MG, Balkwill DL, Daly MJ. Protein oxidation: key to bacterial desiccation resistance? ISME J. 2008;2:393–403. doi: 10.1038/ismej.2007.116. [DOI] [PubMed] [Google Scholar]
- 52.Rishi P, Jindal N, Bharrhan S, Tiwari RP. Salmonella-macrophage interactions upon manganese supplementation. Biol Trace Elem Res. 2010;133:110–119. doi: 10.1007/s12011-009-8406-x. [DOI] [PubMed] [Google Scholar]
- 53.Anjem A, Varghese S, Imlay JA. Manganese import is a key element of the OxyR response to hydrogen peroxide in Escherichia coli. Mol Microbiol. 2009;72:844–858. doi: 10.1111/j.1365-2958.2009.06699.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McEwan AG. New insights into the protective effect of manganese against oxidative stress. Mol Microbiol. 2009;72:812–814. doi: 10.1111/j.1365-2958.2009.06700.x. [DOI] [PubMed] [Google Scholar]
- 55.Sobota JM, Imlay JA. Iron enzyme ribulose-5-phosphate 3-epimerase in Escherichia coli is rapidly damaged by hydrogen peroxide but can be protected by manganese. Proc Natl Acad Sci U S A. 2011;108:5402–5407. doi: 10.1073/pnas.1100410108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ryser ET, Schuman JD (2015) Mesophilic aerobic plate count. In: Salfinger Y, Tortorello M Lou (eds) Compendium of Methods for the Microbiological Examination of Foods, 5th ed. American Public Health Association (APHA). 10.2105/MBEF.0222.013
- 57.Schulten SM, In’t Veld PH, Ghameshlou Ζ, Schimmel H, Linsinger T (2000) The certification of the number of colony forming particles of Salmonella Typhimurium and number fraction of negative capsules from artificially contaminated milk powder, CRM 507R, EUR 19622 EN, In: BCR Information Reference Materials, European Commission, Belgium, 2000. https://publications.jrc.ec.europa.eu/repository/handle/JRC21325. Accessed 15 January 2022
- 58.Heisterkamp SH, Hoekstra JA, Van Strijp-Lockefeer NGWM, Havelaar AH, Mooijman KA, In’t Veld PH, Notermans SHW, Maier EA, Griepink B (1993) Statistical analysis of certification trials for microbiological reference materials, EUR 15008 EN, In: BCR Information Chemical Analysis, Commission of the European Communities, Belgium, 1993. https://op.europa.eu/en/publication-detail/-/publication/c16db1a4-7cec-417c-a289-519be1f374db. Accessed 15 January 2022
- 59.In’t Veld PH, Soentoro PSS, Notermans SHW, Properties of Bacillus cereus spores in reference materials prepared from artificially contaminated spray dried milk. Int J Food Microbiol. 1993;20:23–26. doi: 10.1016/0168-1605(93)90057-N. [DOI] [PubMed] [Google Scholar]
- 60.Mooijman KA, During M, Nagelkerke NJD (2003) MICROCRM: preparation and control of batches of microbiological reference materials consisting of capsules. RIVM REPORT 250935001, Netherlands
- 61.Mooijman KA, Nagelkerke NJD, Demarquilly M, Lemdani M, Stewardson D, Fouweather T, Lightfoot N, Simonart T (2004) MICROCRM: feasibility certification studies of microbiological reference materials consisting of capsules. RIVM REPORT 250935002, Netherlands
- 62.ISO 6579 (2017) ISO 6579–1:2017 - Microbiology of the food chain - Horizontal method for the detection, enumeration and serotyping of Salmonella - Part 1: Detection of Salmonella spp., 1st ed. International Organization for Standardization (ISO) [DOI] [PubMed]
- 63.AOAC (2006) AOAC Official Method 991.14 Coliform and Escherichia coli Counts in Food. Petrifilm Coliform Count Plate and Petrifilm Coliform Count Plate Methods. In: Official Methods of Analysis of AOAC International, 18th ed. Association of Official Analytical Chemists (AOAC), p 32
- 64.Kornacki JL, Gurtler JB, Stawick BA (2015) Enterobacteriaceae, coliforms, and Escherichia coli as quality and safety indicators. In: Salfinger Y, Tortorello M Lou (eds) Compendium of Methods for the Microbiological Examination of Foods, 5th ed. American Public Health Association (APHA). 10.2105/MBEF.0222.014
- 65.Ferreira DF. Sisvar: a computer statistical analysis system. Ciência e Agrotecnologia. 2011;35:1039–1042. doi: 10.1590/S1413-70542011000600001. [DOI] [Google Scholar]
- 66.Culham DE, Dalgado C, Gyles CL, Mamelak D, MacLellan S, Wood JM. Osmoregulatory transporter ProP influences colonization of the urinary tract by Escherichia coli. Microbiology. 1998;144:91–102. doi: 10.1099/00221287-144-1-91. [DOI] [PubMed] [Google Scholar]
- 67.Sleator R. Bacterial osmoadaptation: the role of osmolytes in bacterial stress and virulence. FEMS Microbiol Rev. 2001;25:49–71. doi: 10.1016/S0168-6445(01)00071-7. [DOI] [PubMed] [Google Scholar]
- 68.Wood JM (2007) Bacterial Osmosensing Transporters. In: Haussinger D, Sies H (eds) Methods in Enzymology. Elsevier, pp 77–107. 10.1016/S0076-6879(07)28005-X [DOI] [PubMed]
- 69.Wood JM. Bacterial responses to osmotic challenges. J Gen Physiol. 2015;145:381–388. doi: 10.1085/jgp.201411296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Barron JC, Forsythe SJ. Dry stress and survival time of Enterobacter sakazakii and other Enterobacteriaceae in dehydrated powdered infant formula. J Food Prot. 2007;70:2111–2117. doi: 10.4315/0362-028X-70.9.2111. [DOI] [PubMed] [Google Scholar]
- 71.Koseki S, Nakamura N, Shiina T. Comparison of desiccation tolerance among Listeria monocytogenes, Escherichia coli O157:H7, Salmonella enterica, and Cronobacter sakazakii in powdered infant formula. J Food Prot. 2015;78:104–110. doi: 10.4315/0362-028X.JFP-14-249. [DOI] [PubMed] [Google Scholar]
- 72.Jiao R, Gao J, Zhang X, Zhang M, Chen J, Wu Q, Zhang J, Ye Y. Short communication: Effects of vacuum freeze-drying on inactivation of Cronobacter sakazakii ATCC 29544 in liquid media with different initial inoculum levels. J Dairy Sci. 2017;100:1674–1678. doi: 10.3168/jds.2016-11937. [DOI] [PubMed] [Google Scholar]
- 73.Abadias M, Benabarre A, Teixidó N, Usall J, Viñas I. Effect of freeze drying and protectants on viability of the biocontrol yeast Candida sake. Int J Food Microbiol. 2001;65:173–182. doi: 10.1016/S0168-1605(00)00513-4. [DOI] [PubMed] [Google Scholar]
- 74.Abadias M, Teixidó N, Usall J, Benabarre A, Viñas I. Viability, efficacy, and storage stability of freeze-dried biocontrol agent Candida sake using different protective and rehydration media. J Food Prot. 2001;64:856–861. doi: 10.4315/0362-028X-64.6.856. [DOI] [PubMed] [Google Scholar]
- 75.Bevilacqua A, Cagnazzo MT, Caldarola C, Ciuffreda E, Dragano AR, Franchino S, Lauriola R, Pacifico A, Corbo MR, Sinigaglia M. Bifidobacteria as potential functional starter cultures: a case study by MSc students in Food Science and Technology (University of Foggia, Southern Italy) Food Nutr Sci. 2012;03:55–63. doi: 10.4236/fns.2012.31010. [DOI] [Google Scholar]
- 76.Castro HP, Teixeira PM, Kirby R. Changes in the cell membrane of Lactobacillus bulgaricus during storage following freeze-drying. Biotechnol Lett. 1996;18:99–104. doi: 10.1007/BF00137819. [DOI] [Google Scholar]
- 77.Selmer-Olsen E, Birkeland SE, Sorhaug T. Effect of protective solutes on leakage from and survival of immobilized Lactobacillus subjected to drying, storage and rehydration. J Appl Microbiol. 1999;87:429–437. doi: 10.1046/j.1365-2672.1999.00839.x. [DOI] [PubMed] [Google Scholar]
- 78.Zayed G, Roos YH. Influence of trehalose and moisture content on survival of Lactobacillus salivarius subjected to freeze-drying and storage. Process Biochem. 2004;39:1081–1086. doi: 10.1016/S0032-9592(03)00222-X. [DOI] [Google Scholar]
- 79.Álvarez-Ordóñez A, Cummins C, Deasy T, Clifford T, Begley M, Hill C. Acid stress management by Cronobacter sakazakii. Int J Food Microbiol. 2014;178:21–28. doi: 10.1016/j.ijfoodmicro.2014.03.001. [DOI] [PubMed] [Google Scholar]
- 80.Johler S, Stephan R, Hartmann I, Kuehner KA, Lehner A. Genes involved in yellow pigmentation of Cronobacter sakazakii ES5 and influence of pigmentation on persistence and growth under environmental stress. Appl Environ Microbiol. 2010;76:1053–1061. doi: 10.1128/AEM.01420-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Joseph S, Cetinkaya E, Drahovska H, Levican A, Figueras MJ, Forsythe SJ. Cronobacter condimenti sp. nov., isolated from spiced meat, and Cronobacter universalis sp. nov., a species designation for Cronobacter sp. genomospecies 1, recovered from a leg infection, water and food ingredients. Int J Syst Evol Microbiol. 2012;62:1277–1283. doi: 10.1099/ijs.0.032292-0. [DOI] [PubMed] [Google Scholar]
- 82.Welsh D. Osmotically induced intracellular trehalose, but not glycine betaine accumulation promotes desiccation tolerance in Escherichia coli. FEMS Microbiol Lett. 1999;174:57–63. doi: 10.1016/S0378-1097(99)00122-6. [DOI] [PubMed] [Google Scholar]
- 83.Pratt Z, Shiroda M, Stasic AJ, Lensmire J, Kaspar CW. Osmotic and desiccation tolerance in Escherichia coli O157:H7 and Salmonella enterica requires rpoS (σ38) In: Bruijin FJ, editor. Stress and environmental regulation of gene expression and adaptation in bacteria. John Wiley & Sons; 2016. pp. 716–724. [Google Scholar]
- 84.Maserati A, Fink RC, Lourenco A, Julius ML, Diez-Gonzalez F. General response of Salmonella enterica serovar Typhimurium to desiccation: a new role for the virulence factors sopD and sseD in survival. PLoS ONE. 2017;12:e0187692. doi: 10.1371/journal.pone.0187692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Metaane S, Monteil V, Ayrault S, Bordier L, Levi-Meyreuis C, Norel F. The stress sigma factor σS/RpoS counteracts Fur repression of genes involved in iron and manganese metabolism and modulates the ionome of Salmonella enterica serovar Typhimurium. PLoS ONE. 2022;17(3):e0265511. doi: 10.1371/journal.pone.0265511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Raes M, Nagelkerke N, Henken AM (2000) Bacteriological detection of Salmonella in the presence of competitive micro-organisms. Bacteriological collaborative study IV amongst the National References Laboratories for Salmonella, the use of MSRV as selective enrichment. RIVM report 284500014
- 87.Raes M, NagelkerkE N, Henken AM (2001) Bacteriological detection of Salmonella in the presence of competitive micro-organisms. Bacteriological collaborative study V amongst the National References Laboratories for Salmonella. RIVM report 284500018
- 88.Smith AR, Ellison AL, Robinson AL, Drake M, McDowell SA, Mitchell JK, Gerard PD, Heckler RA, McKillip JL. Enumeration of sublethally injured Escherichia coli O157:H7 ATCC 43895 and Escherichia coli atrain B-41560 using selective agar overlays versus commercial methods. J Food Prot. 2013;76:674–679. doi: 10.4315/0362-028X.JFP-12-363. [DOI] [PubMed] [Google Scholar]
- 89.McKillip JL. Recovery of sublethally injured bacteria using selective agar overlays. Am Biol Teach. 2001;63:184–189. doi: 10.1662/0002-7685(2001)063[0184:rosibu]2.0.co;2. [DOI] [Google Scholar]
- 90.Emons H, Majoros L. Conference report “The Future of Reference Materials – Science and Innovation”. Accredit Qual Assur. 2011;16:327–328. doi: 10.1007/s00769-011-0773-2. [DOI] [Google Scholar]
- 91.Wertheim HFL, Huong VTL, Kuijper EJ. Clinical microbiology laboratories in low-resource settings, it is not only about equipment and reagents, but also good governance for sustainability. Clin Microbiol Infect. 2021;27:1389–1390. doi: 10.1016/j.cmi.2021.07.027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Growth of Salmonella Enteritidis, C. sakazakii, E. coli, and C. freundii in MMS at 37 °C for 48 h (28.1 KB)
Detection of Salmonella in the mixed microbiological RMs by PCR of invA gene. Lane 1: Molecular weight marker (1 kb DNA ladder); lanes 2, 3, and 4: amplification of a sample of microbiological RM1, RM2, and RM3 enriched in Rappaport Vassiliadis broth after 30 days storage at -20 °C, respectively; lane 5: positive control; lane 6: negative control (112 KB)