Abstract
Staphylococcus pseudintermedius is the main coagulase-positive staphylococci associated with canine skin/soft tissue infections (SSTI), otitis externa, and surgical site infections. The international spread of an epidemic and multiresistant lineage of methicillin-resistant Staphylococcus pseudintermedius (MRSP), the so-called European clone—displaying sequence type (ST) 71—requires attention. The first isolation of an MRSP ST71 isolate in South America was reported in Rio de Janeiro city, in 2010; however, a limited number of canine isolates were analyzed. Thus, to have a better panel of the MRSP spread in this city, we were stimulated to continue this study and search for the presence of MRSP in 282 colonized or infected dogs in the city of Rio de Janeiro. Among the MRSP isolates collected (N = 17; 6.1%), the pulsed-field gel electrophoresis (PFGE) patterns were similar to those of European clone. All 17 isolates were classified as ST71 by multilocus sequence typing (MLST). In order to assess whether isolates of MRSP ST71 may have also spread to the Rio de Janeiro state countryside, we collected samples from 124 infected dogs in the city of Campos dos Goytacazes (232 km away from Rio de Janeiro city). Our data showed the presence of ST71 lineage in one isolate among three MRSP detected. S. pseudintermedius was isolated from 40.6% of the clinical samples (N = 165/406). A relatively high incidence of methicillin resistance, detected by a PCR-based method, was found in 12.1% of the S. pseudintermedius recovered from animals (N = 20/165). The resistance profile of these isolates was similar to that described for the international ST71 strains whose genomes are publicly available in the GenBank. The prospect of ST71 isolates being resistant to virtually all antimicrobials used in veterinary medicine is alarming and should be considered a central issue considering that MRSP ST71 spreads over large geographic distances and its transmission from animals to humans.
Supplementary Information
The online version contains supplementary material available at 10.1007/s42770-022-00852-9.
Keywords: Staphylococcus aureus, Staphylococcus pseudintermedius, MRSP, MRSA, Canine skin infections
Introduction
Staphylococcus pseudintermedius is a commensal bacterium that colonizes the skin and mucosa membranes of dogs. Furthermore, it is the most prevalent cause of canine bacterial otitis and pyoderma [1]. Methicillin-resistant S. pseudintermedius (MRSP) is of great concern in veterinary medicine due to its high-level resistance to several antimicrobials. High-level antimicrobial resistance makes infections difficult to treat with veterinary licensed systemic antimicrobial agents [2].
Since its first isolation, MRSP has quickly spread across the world [3–5]. Prolonged antimicrobial therapy, surgical interventions, and chronic infections are known risk factors for MRSP infections [6]. Previous studies have demonstrated a remarkable genetic diversity among S. pseudintermedius, with more than 1400 sequence types (STs) reported. ST71, ST68, and ST45 have been described as the most successful MRSP lineages [7]. The ST71 lineage initially identified in Europe is the most widespread [8]. It is concerning that some studies have reported zoonotic transmission of S. pseudintermedius from dog to human, including MRSP [9]. Studies on the dynamic and molecular epidemiology of infections caused by multiresistant staphylococci can generate critical data to guide public health programs. These data may untimely result in effective policies for preventing and controlling the spread of hypervirulent and multidrug-resistant microorganisms. Few studies have tracked MRSP lineages in Latin American countries, including Brazil. Thus, we performed the molecular characterization of the MRSP isolates from dogs attending different veterinary clinics in two cities in Rio de Janeiro State to better understand the spread of ST71 in this region. In addition, we compared their resistance gene profile with that of international ST71 isolates using genomic sequences deposited in the GenBank.
Material and methods
Bacterial isolates
Two hundred eighty-two dogs from Rio de Janeiro city were initially screened for the presence of MRSP ST71 (European clone). Then, to test whether ST71 isolates could also be found infecting dogs in a countryside town in the state of Rio de Janeiro, we also included infected dogs from Campos dos Goytacazes city (N = 124). Thus, a total of 406 were examined during the period of 2010 to 2013. The dogs were adults (1–8 years), healthy (n = 88 nose swabs), or infected (ear secretion and skin exudate swabs; N = 318), either male or female. The clinical material from Rio de Janeiro city was collected from dogs attending several private veterinary clinics and sent to the Laboratory of Animal Bacteriology at Universidade Federal Fluminense (UFF). Isolates from the city of Campos dos Goytacazes were collected from infected dogs assisted at the Department of Small Animal Practice of the Veterinary Hospital at Universidade Estadual do Norte Fluminense (UENF). Veterinarians collected all samples with the aid of sterile cotton swabs. The swab was streaked on mannitol salt agar (MSA; Merck, Darmstadt, Germany) and incubated at 37 °C/18 h. Cell morphology was examined by the Gram-staining procedure. The Staphylococcus intermedius group (SIG) was identified using MALFI-TOF MS (Biotyper 3.1, Bruker, Atibaia, São Paulo, Brazil) [10]. S. pseudintermedius was confirmed by PCR-based on the amplification of the nuc gene [11].
Antimicrobial susceptibility
The disk diffusion test was performed and interpreted according to the recommendations of the Clinical Laboratory Standard Institute (CLSI) for animals and humans (CLSIvet, 2018; CLSI, 2020). Antimicrobial disks evaluated were oxacillin (1 μg; OXA), ciprofloxacin (5 μg; CIP), clindamycin (2 μg; CLI), chloramphenicol (30 μg; CHL), erythromycin (15 μg; ERY), gentamicin (10 μg; GEN), penicillin G (10 UI; PEN), rifampicin (5 μg; RIF), sulfamethoxazole-trimethoprim (25 μg; SXT), and tetracycline (30 μg; TET) (Cecon, São Paulo, SP, Brazil). The Staphylococcus aureus ATCC 25,923 was used to control the test. Methicillin resistance was confirmed by the detection of the mecA gene by PCR [12]. Multiresistance was defined when the isolate displays resistance to at least three antimicrobial classes other than beta-lactams.
Molecular characterization
These studies were only performed for S. pseudintermedius isolates carrying the mecA gene. SmaI-fragmented DNA from MRSP isolates was analyzed by PFGE using a CHEF DR III System (Bio-Rad Laboratories, Richmond, CA, USA) under the following conditions: switch time, 2.0 to 20 s, and run time, 20 h; temperature, 11.3 °C; angle, 120°; and voltage, 6 V/cm. DNA fragments were stained with ethidium bromide and visualized with a UV transilluminator. Band patterns were assessed by visual inspection and interpreted according to Tenover criteria [13]. The BMBP02 isolate was used as a representative of the ST71 MRSP [5]. The multilocus sequence typing (MLST) was carried out for the 17 MRSP isolates from Rio de Janeiro and for the three MRSP isolates from Campos dos Goytacazes as described [14]. Sequence types were assigned by comparison with allele sequences present in the PubMLST database (http://pubmlst.org/spseudintermedius).
Genomic analysis for assessing antimicrobial resistance
A total of 206 genomes available in the GenBank were used in this study to compare the antimicrobial resistance pattern of Brazilian MRSP with that of international ST71 isolates. Supplementary Table 1 provides information about the genomes included in this analysis. Initially, the ST was determined using the Center for Genomic Epidemiology tool MLST 2.0 [15]. Then, genome sequences related to ST71 were selected and submitted to ResFinder 4.1 [16, 17] to assess patterns of antimicrobial resistance.
Statistical analyses
A chi-square test was used to analyze differences in antimicrobial susceptibility patterns and frequency of MRSP isolation.
Results
A total of 165 isolates were identified as S. pseudintermedius. Of these, 20 (12.1%) were MRSP according to the oxacillin disk diffusion test and the PCR for the mecA gene. Most MRSP originated from canine clinical cases (15/20; 75%) and five (25%) from asymptomatic dogs. The isolates were recovered from otitis (9/20; 45%), pyoderma (6/20; 30%), and nasal cavity (5/20; 25%).
The PFGE patterns of all 17 MRSP isolates from Rio de Janeiro were similar to that of the European clone, represented by isolate BMBSP02—an isolate MRSP ST71 previously detected in Rio de Janeiro, Brazil [5] (Table 1). Two isolates had precisely the same pattern of BMBSP02 (SA116 and SD12). Most isolates showed three-band differences when compared with BMBSP02 (Table 1). The MLST of these 17 MRSP isolates confirmed the allocation of all of them to the ST71 lineage (allelic profile: 2–2-2–2-6–3-2).
Table 1.
Phenotypic and genotypic characteristics of the MRSP isolates from Rio de Janeiro and Campo dos Goytacazes, RJ, Brazil (2010–2012)
| Isolate | Year of isolation | Source | Antimicrobial resistance** | PFGE | ST (MLST) |
|---|---|---|---|---|---|
| PA36 | 2011 | NC | ery, cli, sxt, tet | A4 | ST71 |
| PA41 | 2011 | NC | ery, cli, sxt, tet, cip | A2 | ST71 |
| PA76 | 2011 | NC | ery, cli, sxt, cip | A2 | ST71 |
| PA85 | 2011 | NC | ery, cli, sxt, cip | A2 | ST71 |
| PA31 | 2011 | NC | ery, cli, sxt, cip | A2 | ST71 |
| SA02 | 2011 | Ear | ery, cli, gen, sxt, cip | A2 | ST71 |
| SA41 | 2011 | Ear | ery, cli, gen, tet, cip | A2 | ST71 |
| SA62 | 2011 | Ear | ery, cli, gen, sxt, tet, cip | A2 | ST71 |
| SA63 | 2011 | Ear | ery, cli, gen, sxt, tet, cip | A4 | ST71 |
| SA71 | 2012 | Ear | ery, cli, gen, sxt, tet, cip | A4 | ST71 |
| SA89 | 2012 | Ear | ery, cli, sxt, cip | A2 | ST71 |
| SA93 | 2012 | Ear | ery, cli, gen, sxt, tet, cip | A3 | ST71 |
| SA103 | 2012 | Ear | ery, cli, sxt, cip | A3 | ST71 |
| SA116 | 2012 | Ear | ery, cli, sxt, cip | A1 | ST71 |
| SD12 | 2010 | PD | ery, cli, gen, sxt, tet, rif, cip | A1 | ST71 |
| SD71 | 2011 | PD | sxt, tet, rif | A2 | ST71 |
| SD87 | 2011 | PD | ery, cli, gen, sxt, cip | A4 | ST71 |
| Pa1.2* | 2012 | PD | ery, cli, tet | ND | ST330 |
| Pa1.3* | 2012 | PD | ery, cli, tet | ND | ST330 |
| Pa124* | 2012 | PD | ery, cli, sxt, cip | ND | ST71 |
NC, nasal cavity; PD, pyoderma; cip, ciprofloxacin (5 μg); cli, clindamycin (2 μg); ery, erythromycin (15 μg); gen, gentamicin (10 μg); rif, rifampicin (5 μg); sxt, sulfamethoxazole-trimethoprim (25 μg); tet, tetracycline (30 μg). The PFGE classification was based on the PFGE pattern of the BMBSP02 strain (a MRSP ST71 related to the European clone), which was taken as the A1 pattern; A2, three-band differences in relation to BMBSP02; A3, five-band differences; A4, six-band differences; ND, not done. ST, sequence type; and MLST, multilocus sequence typing. *Pal 2, Pal 3, and Pal 24 are MRSP isolates from Campos dos Goytacazes city. **All samples were resistant to oxacillin in the disk diffusion test
To investigate the possibility that ST71 has also spread to a rural area of Rio de Janeiro state, we analyzed the three isolates of MRSP obtained from Campos dos Goytacazes. One isolate was classified as ST71 (Pal 24), confirming the presence of this international lineage in a city located 232 km from the city of Rio de Janeiro. The other two MRSP isolates (Pal.2 and Pal.3) were classified as ST330.
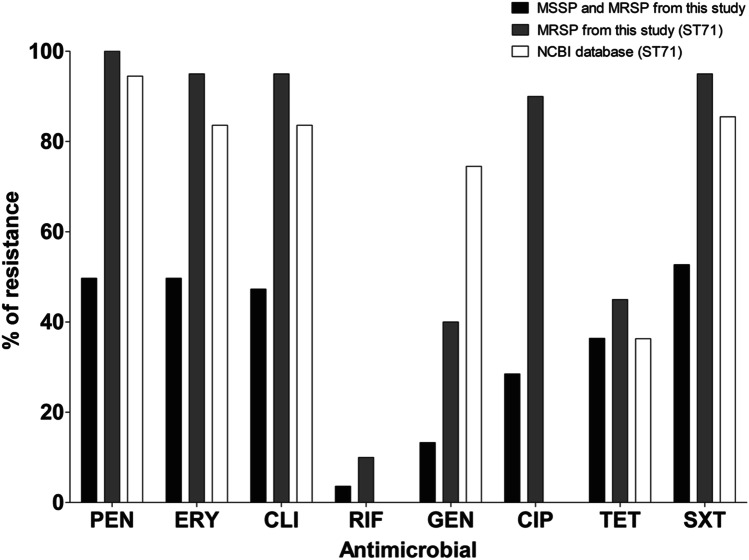
Among all S. pseudintermedius isolates detected in this study, sulfamethoxazole/trimethoprim was the antimicrobial agent that showed the higher resistance rate (N = 87/165; 52.7%). Penicillin (N = 82/165; 49.7%), erythromycin (N = 82/165, 49.7%), and clindamycin (N = 78/165, 47.3%) also showed high resistance rates. Rifampicin had the lowest rate (N = 6/165; 3.6%), followed by gentamycin (N = 22/165; 13.3%), ciprofloxacin (47/165; 28.5%), and tetracycline (60/165; 36.4%). Multidrug resistance was also a common finding (81/165; 49%), with multiresistant isolates among S. pseudintermedius (Fig. 1).
Fig. 1.
Resistance rates observed in the collection of isolates from this study and from international ST71 MRSP isolates whose genomes were publicly available in the NCBI database. MRSP, methicillin-resistant Staphylococcus pseudintermedius; MSSP, methicillin-susceptible Staphylococcus pseudintermedius. pen, penicillin; ery, erythromycin; cli, clindamycin; rif, rifampicin; gen, gentamicin; cip, ciprofloxacin; tet, tetracycline; sxt, sulfamethoxazole-trimethoprim. NCBI, the National Center for Biotechnology Information
Considering MRSP isolates, resistance to erythromycin, clindamycin, and sulfamethoxazole/trimethoprim was almost universally found (19/20; 95%). Resistance to ciprofloxacin was also observed frequently (18/20; 90%) and more present in MRSP isolates compared with methicillin-susceptible S. pseudintermedius (MSSP) (N = 27/141; 19.1%) (p < 0.0001). We also found resistance to gentamicin (8/20; 40%) and tetracycline (N = 9/20; 45%) at considerable rates. The most efficient drug in the present study was rifampicin, with two (N = 2/20; 10%) resistant MRSP isolates (Table 1 and Fig. 1). All twenty MRSPs were multiresistant. Despite being frequently reported in MSSP (N = 61/145, 42.1%), we found an important increase in multidrug resistance among methicillin-resistant isolates (p < 0.0001).
When we searched the genomic database for resistance markers, the resistance profiles of international ST71 MRSP isolates were comparable to the MRSP isolates ST71 from our study (Fig. 1). A total of 55 genomes were typed as belonging to ST71 (Supplementary Table S1). Of these 55 genomes, 52 (94.5%) showed resistance markers to beta-lactams, 50 (90.9%) to gentamycin, 41 (74.5%) to sulfamethoxazole/trimethoprim, 46 (83.6%) to erythromycin and clindamycin, 20 (36.3%) to tetracycline, and none of the genomes presented resistance traits to ciprofloxacin and rifampicin. When we compared the antimicrobial susceptibility pattern between our isolates and international ST71 genomes, a difference was observed in the resistance rates for gentamycin and ciprofloxacin (p < 0.01).
Discussion
Some studies have reported the occurrence and distribution of MRSP worldwide. However, comprehensive data on the prevalence and characterization of MRSP isolates in the clinical routine of veterinary settings are limited. This study provides information on clinical isolates of MRSP from canine origin collected for four years (2010–2013). The rates of MRSP collected in the present study did not differ from studies elsewhere, such as the one published in the Netherlands, in which an 8% frequency of MRSP was reported [18].
A high incidence of resistance to erythromycin and clindamycin in S. pseudintermedius collected from otitis and pyoderma has already been reported in two studies performed by our group in Rio de Janeiro [19, 20]. Faires et al. [21] reported resistance rates of 73.9% for clindamycin and erythromycin among 46 MRSP analyzed. Similar to this study, Van Damme et al. [18] found considerable rates of macrolide resistance and observed that MRSP was significantly more resistant (> 80%) than MSSP (32.4%). In addition to macrolides and lincosamides, high resistance rates to trimethoprim-sulfamethoxazole were also common. While not surprising, this could lead to unsuccessful empiric treatment. According to international guidelines, clindamycin, lincomycin, trimethoprim-potentiated sulfonamides, first-generation cephalosporins, and amoxicillin-clavulanate are the indicated empirical choice for systemic treatment [22].
Tetracycline resistance is one of the most prevalent among S. pseudintermedius isolates [23]. A high rate of tetracycline resistance (62.6%) has also been reported among Staphylococcus sp. in Korea [24]. Although we did not investigate this, the high level of resistance to tetracycline found in our study is likely to be associated with the prescription of its derivatives, such as doxycycline, to treat babesiosis and ehrlichiosis, both endemic diseases in dogs in the region studied.
A study carried out in European countries classified MRSP as a hospital pathogen in veterinary settings similar to HA-MRSA in human medicine [25]. Furthermore, the potential of S. pseudintermedius to cause infections in humans has been established [26]. Indeed, some cases of human infection by S. pseudintermedius have been reported in Spain [9], Canada [27], the USA [28], and Argentina [29].
The resistance rates among ST71 isolates circulating between 2010 and 2013 in our state were similar to those found in genomes from S. pseudintermedius strains isolated elsewhere from dogs and cats. The exception was gentamycin and ciprofloxacin, where the resistance rates were significantly different between the two groups (p < 0.01). This could be a bias for ciprofloxacin due to the typing method used for genomic analysis. ResFinder is an online tool developed to search for genes related antimicrobial resistance and search for point mutations that lead to a resistant phenotype [16, 17], which is the case of ciprofloxacin resistance. However, the database available for this tool is restricted to some specific pathogens, such as Campylobacter spp., Escherichia coli, and Staphylococcus aureus. Our analysis was performed against the S. aureus database. Based on this, the results obtained from the genomic analysis may not represent the actual scenario for this resistance among the international strains.
Another limitation of this study is that the S. pseudintermedius isolates were collected from 2010 to 2013. Despite that, in the last 10 years, there have been no significant advances worldwide to prevent the spread of multidrug-resistant strains. Therefore, it is more likely that this situation may be even more aggravating. However, additional studies are needed to have the most up-to-date picture of the incidence of MRSP, including MRSP ST71, in domestic animals.
Regarding resistance to gentamicin, our study revealed that ST71 isolates from our state have lower resistance rates (40%) when compared to the international ST1 genomes (74.5%; p < 0.01). This might be due to differences in the preferred use of antibiotics in veterinarian clinical settings. Data available in the scientific literature also demonstrate this wide range of gentamicin resistance rates among clinical isolates of S. pseudintermedius. For example, Kalhoro et al. [30] found a frequency of 70.8%, and Bourély et al. [31] showed a rate of 13.5% of resistance to this drug.
The potential for zoonotic infections caused by ST71 MRSP isolates and their geographic spread in the European and US regions has been documented [32, 33]. In 2013, an ST71 MRSP isolate was collected from a dog in Rio de Janeiro city [5]. In the present study, all 17 MRSP isolates from the city of Rio de Janeiro were isolated from the same period and identified as ST71. The presence of the ST71 in a canine isolate in Campos dos Goytacazes city also indicates the introduction of this MRSP lineage in a rural zone of Rio de Janeiro state. Unfortunately, only few epidemiological studies on MRSP detection and molecular epidemiology are available, especially in our country. A study from Argentina with infected dogs did not detect ST71 MRSP among 10 MRSP detected [34].
The prospective of ST71 isolates exhibiting resistance to virtually all antimicrobials used in veterinary medicine is alarming, especially when considering the potential for spread over large geographic distances of a single lineage, such as ST71 MRSP [5, 8, 22]. These data suggest that regular monitoring by public institutions of well-fit MRSP lineages is highly recommended to risk assessment and the development of rational intervention strategies to limit the spread of these hypervirulent, highly multiresistant bacteria.
Supplementary Information
Below is the link to the electronic supplementary material.
Author contribution
BP collected animal samples, carried out strain isolation and molecular characterization, and wrote the manuscript. MBS collected animal samples, carried out strain isolation and identification, and wrote the draft of the manuscript. AMNB performed the genomic database construction and genomics analysis and contributed to the final version of the manuscript. FAF and MSR performed molecular typing by MLST. MCSC and RFR performed molecular characterization by PFGE. AMSF, BF, and OVM were responsible for the study design and wrote the final version of the manuscript. All authors approved the final version of the manuscript.
Funding
This work was supported by the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), grant # 307672/2019–0, and by Fundação de Amparo à Pesquisa do Rio de Janeiro (FAPERJ), grants E-26/010.101098/2018, E-26/010.001280/2016, E-26/010.002435/2019, and E-26/200.952/2021. This study was also financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior—Brasil (CAPES) Finance Code 001.
Declarations
Ethics approval
This study was approved by the animal ethics committees from Universidade Federal Fluminense (#218/2010) and Universidade Estadual do Norte Fluminense (#145/2011).
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Bruno Penna and Marcella B. Silva contributed equally to this work.
References
- 1.Richards AC, O’Shea M, Beard PM et al (2018) Staphylococcus pseudintermedius surface protein L (SpsL) is required for abscess formation in a murine model of cutaneous infection. Infect Immun 86(11). 10.1128/IAI.00631-18 [DOI] [PMC free article] [PubMed]
- 2.Pires Dos Santos T, Damborg P, Moodley A, Guardabassi L. Systematic review on global epidemiology of methicillin-resistant Staphylococcus pseudintermedius: inference of population structure from multilocus sequence typing data. Front Microbiol. 2016;7:1599. doi: 10.3389/fmicb.2016.01599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kjellman EE, Slettemeås JS, Small H, Sunde M. Methicillin-resistant Staphylococcus pseudintermedius (MRSP) from healthy dogs in Norway - occurrence, genotypes and comparison to clinical MRSP. Microbiologyopen. 2015;4(6):857–866. doi: 10.1002/mbo3.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bergot M, Martins-Simoes P, Kilian H, et al. Evolution of the population structure of Staphylococcus pseudintermedius in France. Front Microbiol. 2018;9:3055. doi: 10.3389/fmicb.2018.03055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Quitoco IMZ, Ramundo MS, Silva-Carvalho MC, et al. First report in South America of companion animal colonization by the USA1100 clone of community-acquired methicillin-resistant Staphylococcus aureus (ST30) and by the European clone of methicillin-resistant Staphylococcus pseudintermedius (ST71) BMC Res Notes. 2013;6:336. doi: 10.1186/1756-0500-6-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lehner G, Linek M, Bond R, et al. Case-control risk factor study of methicillin-resistant Staphylococcus pseudintermedius (MRSP) infection in dogs and cats in Germany. Vet Microbiol. 2014;168(1):154–160. doi: 10.1016/j.vetmic.2013.10.023. [DOI] [PubMed] [Google Scholar]
- 7.Brooks MR, Padilla-Vélez L, Khan TA et al (2020) Prophage-mediated disruption of genetic competence in Staphylococcus pseudintermedius. mSystems 5(1). 10.1128/mSystems.00684-19 [DOI] [PMC free article] [PubMed]
- 8.Menandro ML, Dotto G, Mondin A, Martini M, Ceglie L, Pasotto D. Prevalence and characterization of methicillin-resistant Staphylococcus pseudintermedius from symptomatic companion animals in Northern Italy: Clonal diversity and novel sequence types. Comp Immunol Microbiol Infect Dis. 2019;66:101331. doi: 10.1016/j.cimid.2019.101331. [DOI] [PubMed] [Google Scholar]
- 9.Lozano C, Rezusta A, Ferrer I, et al. Staphylococcus pseudintermedius human infection cases in Spain: dog-to-human transmission. Vector Borne Zoonotic Dis. 2017;17(4):268–270. doi: 10.1089/vbz.2016.2048. [DOI] [PubMed] [Google Scholar]
- 10.Silva MB, Ferreira FA, Garcia LNN, et al. An evaluation of matrix-assisted laser desorption ionization time-of-flight mass spectrometry for the identification of Staphylococcus pseudintermedius isolates from canine infections. J Vet Diagn Invest. 2015;27(2):231–235. doi: 10.1177/1040638715573297. [DOI] [PubMed] [Google Scholar]
- 11.Sasaki T, Tsubakishita S, Tanaka Y, et al. Multiplex-PCR method for species identification of coagulase-positive staphylococci. J Clin Microbiol. 2010;48(3):765–769. doi: 10.1128/JCM.01232-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oliveira DC, de Lencastre H. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2002;46(7):2155–2161. doi: 10.1128/AAC.46.7.2155-2161.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tenover FC, Arbeit RD, Goering RV, et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33(9):2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Solyman SM, Black CC, Duim B, et al. Multilocus sequence typing for characterization of Staphylococcus pseudintermedius. J Clin Microbiol. 2013;51(1):306–310. doi: 10.1128/JCM.02421-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Larsen MV, Cosentino S, Rasmussen S, et al. Multilocus sequence typing of total-genome-sequenced bacteria. J Clin Microbiol. 2012;50(4):1355–1361. doi: 10.1128/JCM.06094-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bortolaia V, Kaas RS, Ruppe E, et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J Antimicrob Chemother. 2020;75(12):3491–3500. doi: 10.1093/jac/dkaa345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zankari E, Allesøe R, Joensen KG, Cavaco LM, Lund O, Aarestrup FM. PointFinder: a novel web tool for WGS-based detection of antimicrobial resistance associated with chromosomal point mutations in bacterial pathogens. J Antimicrob Chemother. 2017;72(10):2764–2768. doi: 10.1093/jac/dkx217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Damme CMM, Broens EM, Auxilia ST, Schlotter YM. Clindamycin resistance of skin derived Staphylococcus pseudintermedius is higher in dogs with a history of antimicrobial therapy. Vet Dermatol. 2020;31(4):305–e75. doi: 10.1111/vde.12854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Penna B, Varges R, Medeiros L, Martins GM, Martins RR, Lilenbaum W. In vitro antimicrobial susceptibility of staphylococci isolated from canine pyoderma in Rio de Janeiro. Brazil Braz J Microbiol. 2009;40(3):490–494. doi: 10.1590/S1517-83822009000300011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Penna B, Varges R, Medeiros L, Martins GM, Martins RR, Lilenbaum W. Species distribution and antimicrobial susceptibility of staphylococci isolated from canine otitis externa. Vet Dermatol. 2010;21(3):292–296. doi: 10.1111/j.1365-3164.2009.00842.x. [DOI] [PubMed] [Google Scholar]
- 21.Faires MC, Tater KC, Weese JS. An investigation of methicillin-resistant Staphylococcus aureus colonization in people and pets in the same household with an infected person or infected pet. J Am Vet Med Assoc. 2009;235(5):540–543. doi: 10.2460/javma.235.5.540. [DOI] [PubMed] [Google Scholar]
- 22.Hillier A, Lloyd DH, Weese JS, et al. Guidelines for the diagnosis and antimicrobial therapy of canine superficial bacterial folliculitis (Antimicrobial Guidelines Working Group of the International Society for Companion Animal Infectious Diseases) Vet Dermatol. 2014;25(3):163–e43. doi: 10.1111/vde.12118. [DOI] [PubMed] [Google Scholar]
- 23.Moodley A, Damborg P, Nielsen SS. Antimicrobial resistance in methicillin susceptible and methicillin resistant Staphylococcus pseudintermedius of canine origin: literature review from 1980 to 2013. Vet Microbiol. 2014;171(3–4):337–341. doi: 10.1016/j.vetmic.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 24.Youn J-H, Yoon JW, Koo HC, Lim S-K, Park YH. Prevalence and antimicrogram of Staphylococcus intermedius group isolates from veterinary staff, companion animals, and the environment in veterinary hospitals in Korea. J Vet Diagn Invest. 2011;23(2):268–274. doi: 10.1177/104063871102300211. [DOI] [PubMed] [Google Scholar]
- 25.Perreten V, Kadlec K, Schwarz S, et al. Clonal spread of methicillin-resistant Staphylococcus pseudintermedius in Europe and North America: an international multicentre study. J Antimicrob Chemother. 2010;65(6):1145–1154. doi: 10.1093/jac/dkq078. [DOI] [PubMed] [Google Scholar]
- 26.Kmieciak W, Szewczyk EM. Are zoonotic Staphylococcus pseudintermedius strains a growing threat for humans? Folia Microbiol (Praha) 2018;63(6):743–747. doi: 10.1007/s12223-018-0615-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blondeau LD, Rubin JE, Deneer H, et al. Bacteremia with Staphylococcus pseudintermedius in a 4 month old pediatric oncology patient. J Chemother. 2020;32(5):260–262. doi: 10.1080/1120009X.2020.1773627. [DOI] [PubMed] [Google Scholar]
- 28.Diaz MA, Gardner LB, Libertin CR (2019) Staphylococcus pseudintermedius catheter-related bloodstream infection after exposure to domestic dogs and a cat. BMJ Case Rep 12(12). 10.1136/bcr-2019-231489 [DOI] [PMC free article] [PubMed]
- 29.Gagetti P, Errecalde L, Wattam AR, et al. Characterization of the first mecA-positive multidrug-resistant Staphylococcus pseudintermedius Isolated from an Argentinian patient. Microb Drug Resist. 2020;26(7):717–721. doi: 10.1089/mdr.2019.0308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kalhoro DH, Kalhoro MS, Mangi MH, et al. Antimicrobial resistance of staphylococci and streptococci isolated from dogs. Trop Biomed. 2019;36(2):468–474. [PubMed] [Google Scholar]
- 31.Bourély C, Cazeau G, Jarrige N, et al. Antimicrobial resistance patterns of bacteria isolated from dogs with otitis. Epidemiol Infect. 2019;147:e121. doi: 10.1017/S0950268818003278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grönthal T, Ollilainen M, Eklund M, et al. Epidemiology of methicillin resistant Staphylococcus pseudintermedius in guide dogs in Finland. Acta Vet Scand. 2015;57:37. doi: 10.1186/s13028-015-0129-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paul NC, Moodley A, Ghibaudo G, Guardabassi L. Carriage of methicillin-resistant Staphylococcus pseudintermedius in small animal veterinarians: indirect evidence of zoonotic transmission. Zoonoses Public Health. 2011;58(8):533–539. doi: 10.1111/j.1863-2378.2011.01398.x. [DOI] [PubMed] [Google Scholar]
- 34.Gagetti P, Wattam AR, Giacoboni G, et al. Identification and molecular epidemiology of methicillin resistant Staphylococcus pseudintermedius strains isolated from canine clinical samples in Argentina. BMC Vet Res. 2019;15(1):264. doi: 10.1186/s12917-019-1990-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.