Abstract
Bat flies are obligate ectoparasitic dipterans that are highly specialised to bats and have apomorphic characteristics, such as absent or reduced wings, and specialised legs and claws, which contribute to their survival. They are often associated with fungi and harbour a fungal diversity that is still poorly understood. Fungi were found in association with the bat flies in a cave of the Caatinga dry forest in Brazil. In total, 43% of the captured bat flies were associated with fungi. Seventy-six flies were collected. DNA sequence analyses of 39 isolates showed that the isolates belonged to 13 species within nine genera, with 38 isolates belonging to Ascomycota and one isolate to Basidiomycota, and Aspergillus was the most frequently isolated genus. Most of the genera found have also been isolated from bat bodies and other substrates/hosts in caves in different regions of the world. Based on morphological and multi-locus phylogenetic analyses, two new species of Ascomycota were described: Allophoma brasiliensis sp. nov. and Pyrenochaetopsis cecavii sp. nov.
Supplementary Information
The online version contains supplementary material available at 10.1007/s42770-022-00841-y.
Keywords: Ascomycota, Fungal diversity, Fungal taxonomy, Trichobius
Introduction
Bats are the second most abundant order among mammals, with approximately 1450 known species (batnames.org). Similar to other mammals, and due to their ecology and biology, bats are parasitised by other organisms such as fleas, true bugs, earwigs, mites, and ticks [1–5]. Approximately 687 species of bat ectoparasitic insects which are distributed in six families are recognised, of which bat flies (Diptera: Hippoboscoidea) are the most frequently recorded and relatively well studied [5–8]. Bat flies are organised into the families Nycteribiidae and Streblidae, and are obligatory haematophagous parasites exclusively associated with bats [6]. Most species are found in tropical and subtropical regions, with few species found in temperate regions [2]. In Brazil, approximately 68 species belonging to 20 genera of Streblidae and 25 species of two genera of Nycteribiidae have been recorded [9–11].
Bat flies reproduce by adenotrophic viviparity [12] and part of their life cycle involves depositing larvae on roost substrates used by chiropteran hosts (e.g. twigs, leaves, mines, and cave walls); bats are the habitat for the adult stage of these dipterans [13]. The bat fly larvae have a pupal stage and emerge actively to seek their hosts [14, 15]. Once deposited on their hosts, the bat flies may also act as vectors of other organisms, as is the case with fungi. Studies on fungal species associated with bat flies have been conducted for over 100 years [16]. However, knowledge of the mycota associated with bat flies has been largely concentrated on the inventory and description of ectosymbiont fungi of the order Laboulbeniales (Ascomycota: Laboulbeniomycetes) [5, 17–20]. In addition to fungal species, studies have also reported that bat flies can serve as vectors for bacteria (e.g. Bartonella), blood parasites (e.g. Polychromophilus), protozoa (e.g. Nycteria), and viruses, and transmit these to and among bats [8].
Understanding the fungal community associated with bat flies is important because arthropods that parasitise vertebrates are often involved in the transmission of a large number of pathogens, including agents of many emerging diseases [21–24]. For example, in the Czech Republic, Lučan et al. [25] detected Pseudogymnoascus destructans (Ascomycota: Pseudeurotiaceae), which is the fungus that causes white-nose syndrome (WNS) in bats, in ectoparasitic haematophagous mites of the genus Spinturnix (Arachnida: Spinturnicidae) collected from the wing membranes of bats. WNS is a virulent fungal disease that has devastated North American bat populations during their hibernation [26–28]. Some studies have reported the presence of their DNA in substrates/hosts in caves in European countries and the USA, highlighting the importance of investigating the presence and distribution of this fungal pathogen [29–32].
Although Laboulbeniales are the most commonly studied fungi in association with bat flies [5, 18, 20], other fungal species of the genera Aspergillus, Penicillium, and Cladosporium have also been reported in bats and from bat caves in temperate and tropical countries [33, 34]. In Brazil, despite the scarcity of studies on the association of fungal species with bat flies, an interesting fungal richness has been reported. Some species of Laboulbeniales have been recorded in association with Streblidae in Brazilian regions (Distrito Federal by Graciolli and Aguiar [17] and São Paulo by Bertola et al. [35]). In addition, Pereira et al. [36] reported Cladosporium subuliforme associating with bat flies from bats in a cave in the Caatinga dry forest. This was the first record of this fungus in a tropical cave and in association with bat flies.
Although there are recent advances in terms of the interaction between bat flies and fungi, most studies to date have focused on the search for ectosymbiotic fungi [5] and there is a knowledge gap regarding other fungal species associated with bat flies. Therefore, we focused on the culturable fungi associated with bat flies from bats in a cave in the Caatinga dry forest of Northeastern Brazil as our sampling unit. We described two new Ascomycota species isolated from these flies.
Material and methods
Sampling site
The Catimbau National Park (IUCN Category II) (8°24ʺ00″ and 8°36′35ʺ S; 37°0′30ʺ and 37°1′40ʺ W) is located in the Caatinga domain, which is the largest and richest tropical dry forest in South America [37, 38]. The park consists of 62,294 ha and comprises parts of the municipalities of Buíque, Tupanatinga, and Ibimirim in the state of Pernambuco, Northeastern Brazil [39]. The flora of this region consists of a mosaic of herbaceous (height < 2 m), shrub (2–5 m), and arboreal (8–12 m) strata, mainly of the families Fabaceae, Euphorbiaceae, and Myrtaceae, as well as seasonal dry forests, humid forest enclaves, and rocky outcrops [37, 40, 41]. The climate of the region is classified as hot semi-arid (Bsh, according to the Köppen-Geiger classification), with an average annual temperature of 23 °C and seasonal rainfall, with average annual precipitation ranging from 486 to 975 mm, which is concentrated between March and July [42, 43].
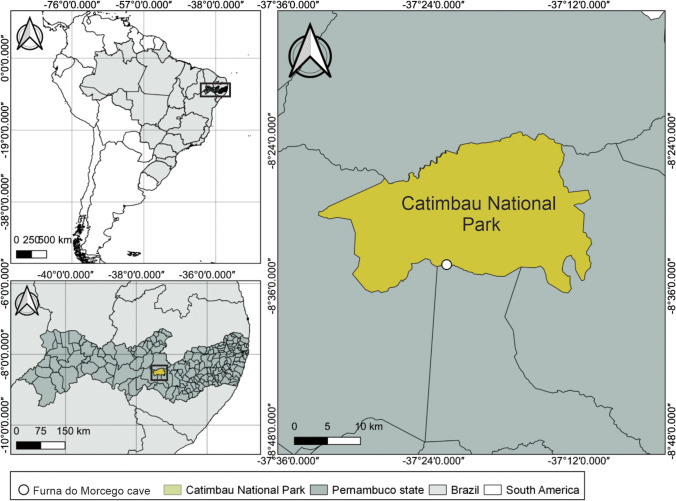
The Furna do Morcego cave (37°22′55ʺ S and 8°34′14ʺ W; 557 m above sea level) has a sandstone lithology and is located at the southern border of the Catimbau National Park (municipality of Ibimirim) near the Kapinawá Indigenous Land (Figs. 1 and 2). This cave is approximately 44 m in length, with a single entrance, an area of 200 m2, and an annual average temperature of 30 °C. This cave is occupied by large colonies of Pteronotus gymnonotus and P. personatus, which are both exclusively insectivorous species, and can harbour up to 71,000 bats in certain times of the year [39].
Fig. 1.
The geographical location of the Furna do Morcego cave in the Parque Nacional do Catimbau [Catimbau National Park], Brazil
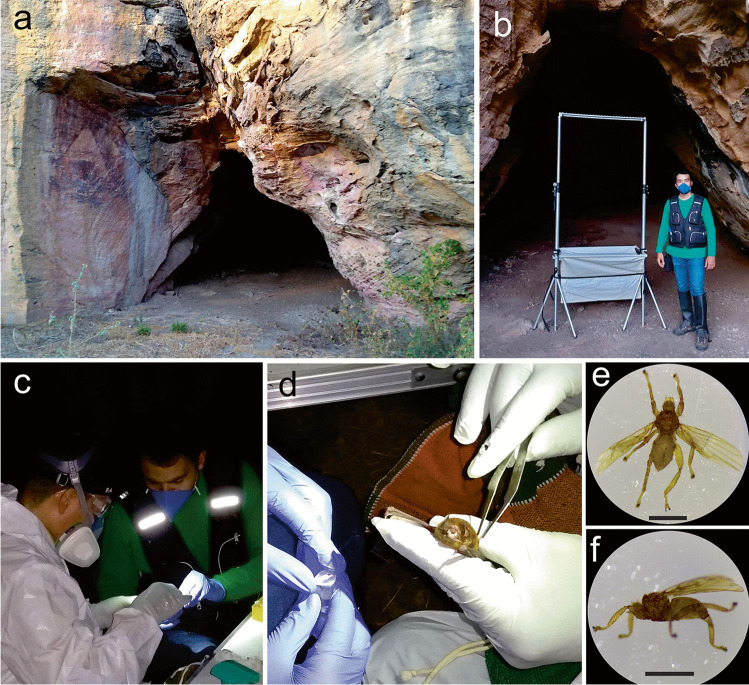
Fig. 2.
Collection of ectoparasites (Streblidae: Trichobius sp.) of bats (Mormoopidae: Pteronotus gymnonotus) captured at the Furna do Morcego cave located in the Parque Nacional do Catimbau [Catimbau National Park], Brazil. a Cave entrance. b Harp trap installed at the entrance of the cave. c–d Collection of bat flies. e–f Top and side view of the ectoparasitic bat fly. Scale bars: 1 mm. Photos were taken by E. Barbier, J. Carvalho and N. Pimentel
Capturing bats and bat flies
Field collections were conducted in October 2020, during the dry season. For the collection of ectoparasites (Trichobius sp.) (Fig. 2), Pteronotus gymnonotus bats were captured and identified, under the licenses of the Ministério do Meio Ambiente (MMA)/Instituto Chico Mendes de Conservação da Biodiversidade (ICMBio) (MMA/ICMBio/SISBIO 68,992–3) and the Ethics Committee on Animal Care–UFPE (CEUA/UFPE) 114/2019. Bat capture was performed using a harp trap (1.50 × 1.50 m) installed at the entrance of the cave before the emergence of bats. The harp trap was constantly inspected, and captured bats were placed in tissue bags for the identification of species, sex, age, and reproductive status, and measurements of forearm length and body mass. Bat identification followed the methods of Gardner [44] and Díaz et al. [45].
For bat fly collection, a visual analysis was conducted on each bat [46]. Using sterilised metal forceps, bat flies were removed from their hosts and stored individually in 2 mL microcentrifuge tubes containing sterilised distilled water. A total of 76 microcentrifuge tubes were used. The metal forceps used during the collection of bat flies were regularly cleaned with 70% alcohol.
Isolation of fungi associated with bat flies
Bat flies were incubated on the surface of the culture media Dichloran Rose Bengal Chloramphenicol (DRBC) agar, Brain–Heart Infusion (BHI) agar plus chloramphenicol (100 mg/L), and Sabouraud (SAB) agar plus chloramphenicol (100 mg/L). Six flies selected from each bat were placed on the surface of the culture media. All plates were incubated at 27 °C in the dark until fungal colonies developed (up to 30 days of incubation). Fungal colonies were purified in Petri dishes containing the same medium of origin as the isolates.
Representative isolates, holotypes (preserved as metabolically inactive culture), and ex-type cultures are deposited in the University Recife Mycology (URM) culture collection (Micoteca URM Profa. Maria Auxiliadora Cavalcanti, WDCM 604) at the Universidade Federal de Pernambuco (UFPE), Recife, Brazil. In addition, the isolates were deposited in the working collection of the Laboratório de Taxonomia e Biotecnologia Utilizando Fungos/UFPE, Recife, Brazil.
Morphologic and phylogenetic analyses
Morphological characterisation of fungal isolates was performed based on the observation of micro- and macro-features of the colonies (e.g. [47, 48].). The hyphal tip method was used to obtain pure cultures [49]. Isolates were sub-cultured to observe somatic and reproductive structures, and cultures on SAB culture medium were used for the preliminary identification of genera and morphotypes. Reproductive structures, when present, were observed and compared with specialised literature. Isolates identified as putative new species were sub-cultured on malt extract agar (MEA), oatmeal agar (OA), and potato dextrose agar (PDA) and incubated at 27 °C for 7 days in the dark. The colour chart of Rayner [50] was used to evaluate colony colours.
DNA from fungal isolates was extracted using cultures within 7 days of growth on the PDA or MEA incubated at 27 °C in the dark, following the instructions of the Wizard Genomic DNA Purification Kit. Primers ITS5/ITS4 [51], ACT-512F/ACT-783R [52], Bt2a/Bt2b [53], CMD5/CMD6 [54], EF-728F/EF-986R [52], LR0R/LR5 [55, 56], and RPB2-5f2/fRPB2-7cR [57, 58] were used to amplify the internal transcribed spacer (ITS) region, partial actin gene (ACT), partial β-tubulin gene (TUB2), calmodulin (CAM), partial translation elongation factor 1-α (TEF1), the large subunit (LSU) of the ribosomal RNA gene, and a fragment of the RNA polymerase second largest subunit gene (RPB2), respectively. PCR amplifications were conducted under the conditions described by Bezerra et al. [59, 60] or Samson et al. [61]. The obtained PCR products were purified using alkaline phosphatase/exonuclease I (EXO + SAP) and sent to the Plataforma Multiusuários de Sequenciamento at the Centro de Biociências of the Universidade Federal de Pernambuco (Recife, Brazil) for sequencing. The same primer pairs were used for amplicon sequencing using the BigDye Terminator Cycle Sequencing Kit v.3.1, following the manufacturer’s instructions.
Phylogenetic analyses were performed by recovering reference sequences from species related to those obtained in this study using the BLASTn tool in the NCBI GenBank database and following previously published studies (e.g. [62–65]). Sequences were aligned using the online tool MAFFT v.7 [66] and manually edited with the MEGA v.7 software [67]. For fungal sequence analyses, we built 10 alignments according to each fungal genus and performed an analysis based on the maximum likelihood (ML) (data not shown), following the instructions of Bezerra et al. [59]. For the phylogenetic inference of genera with new species, Bayesian inference (BI) and ML analyses were conducted using MrBayes v. 3.2.7a [68] with XSEDE and RAxML-HPC BlackBox v. 8.2.12 [69], respectively, at the CIPRES Science Gateway [70]. BI analysis was conducted with 1 × 106 generations and a burning value of 25%, with chains sampled every 1000 generations, and ML analysis with 1000 bootstrap replicates. The best nucleotide model for BI analysis was estimated using MrModelTest v.2.3 software [71]. The resulting phylogenetic trees were visualised using the FigTree software [72]. Values equal to or greater than 0.95 BI posterior probability (BPP) and 70% ML bootstrap support (ML-BS) are shown near nodes. New sequences were deposited in GenBank (Online Resource 1) and alignments of genera having new species were deposited in TreeBASE (study ID 29773).
Data analyses
Abundance (ab) was defined as the total number of isolates obtained for each species, and relative abundance (ra) was determined by the ab of each species divided by the total number of isolates and multiplied by 100 (ra = ab/total of isolates × 100). To evaluate the sufficiency of the sampling effort, the total richness (observed) was compared with the estimated richness using the first- and second-order Chao (Chao 1 and 2), first- and second-order Jackknife (Jackknife 1 and 2), and bootstrap estimators using the EstimateS software version 9.1.0 [73]. We tested whether the abundance of the most frequent fungal species was statistically significant compared with those of the other species using the Kruskal–Wallis test (H) followed by a post hoc Dunn’s test for multiple paired comparisons using SigmaPlot software (version 14.0; Systat Software, San Jose, CA, USA) with a significance level of P ≤ 0.05.
Results
Thirty-nine isolates were obtained by incubation of 76 bat flies on agar with DRBC, BHI, and SAB. These isolates were obtained from 33 (43%) flies belonging to the genus Trichobius (Streblidae), collected from 10 P. gymnonotus bats. The fungal isolates were identified as belonging to 10 genera based on their morphological characteristics and phylogenetic analyses of their rDNA sequences. Fungi from the flies belonged to five orders of Ascomycota (Pleosporales, Eurotiales, Capnodiales, Hypocreales, and Microascales) and one order of Basidiomycota (Tremellales). The total richness of fungi associated with bat flies was 13 species, 12 Ascomycota and one Basidiomycota (Hannaella cf. siamensis) (Table 1). Thirty-eight isolates were Ascomycota of the genera Allophoma, Alternaria, Aspergillus, Cladosporium, Fusarium, Penicillium, Pyrenochaetopsis, Stagonosporopsis, and Yunnania.
Table 1.
Absolute (ab) and relative (ra) abundance, number of isolates, and richness (S) of culturable fungal species isolated from ectoparasites (Streblidae: Trichobius sp.) of bats (Mormoopidae: Pteronotus gymnonotus) captured at the Furna do Morcego cave located in the Parque Nacional do Catimbau [Catimbau National Park], Brazil
| Bat | Bat 1 | Bat 2 | Bat 3 | Bat 4 | Bat 5 | Bat 6 | Bat 7 | Bat 8 | Bat 9 | Bat 10 | ab | ra (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Number of bat flies | 6 | 9 | 9 | 6 | 6 | 8 | 9 | 9 | 6 | 8 | 76 | |
| Allophoma brasiliensis sp. nov | 2 | 2 | 5.1 | |||||||||
| Alternaria alternata | 1 | 1 | 2.6 | |||||||||
| Aspergillus austroafricanus | 1 | 1 | 2.6 | |||||||||
| Aspergillus penicillioides | 2 | 2 | 1 | 4 | 2 | 2 | 2 | 15 | 38.5 | |||
| Aspergillus sydowii | 1 | 1 | 2 | 5.1 | ||||||||
| Cladosporium halotolerans | 1 | 1 | 2.6 | |||||||||
| Cladosporium subuliforme | 1 | 3 | 1 | 5 | 12.8 | |||||||
| Fusarium equiseti | 3 | 3 | 7.7 | |||||||||
| Hannaela cf. siamensis | 1 | 1 | 2.6 | |||||||||
| Penicillium citrinum | 1 | 2 | 3 | 7.7 | ||||||||
| Pyrenochaetopsis cecavii sp. nov | 2 | 2 | 5.1 | |||||||||
| Stagonosporopsis citruli | 1 | 1 | 2.6 | |||||||||
| Yunnania carbonaria | 2 | 2 | 5.1 | |||||||||
| Number of isolates | 3 | 5 | 6 | 11 | 3 | 8 | 1 | 2 | 39 | |||
| Richness (S) | 2 | 2 | 5 | 5 | 2 | 5 | 1 | 1 | 13 |
The most abundantly isolated species were from the Aspergillus (ra = 42.2%) and Cladosporium (ra = 15.4%) genera, while Fusarium (ra = 7.7%), Penicillium (ra = 7.7%), Allophoma (ra = 5.1%), Pyrenochaetopsis (ra = 5.1%), Yunnania (ra = 5.1%), Stagonosporopsis (ra = 2.6%), Alternaria (ra = 2.6%), and Hannaela (ra = 2.6%) had low absolute frequency (< 4 isolates). The genus Aspergillus had the highest number of taxa (three), followed by Cladosporium (two), with A. penicillioides being the species most frequently isolated from bat flies, and was found on the majority of bats sampled.
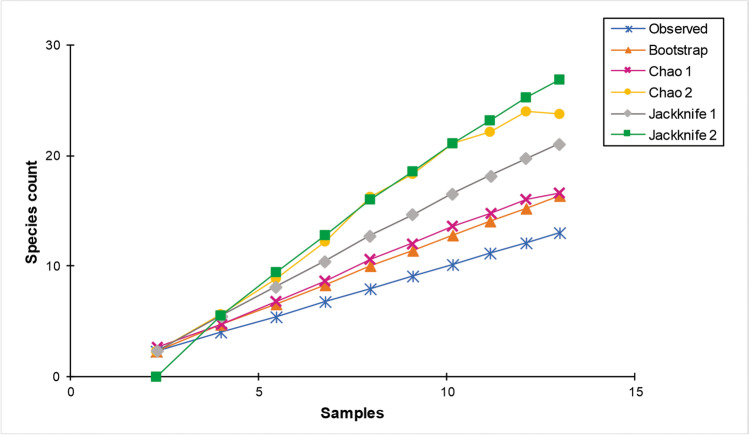
The species accumulation curves of fungi from bat flies did not reach an asymptote (Fig. 3); however, for example, using the Bootstrap estimator, the sampling effort was sufficient to recover 81% of species estimated, followed by the Chao 1 (76%) and Jackknife 1 (62%). However, Chao 2 and Jackknife 2 returned 54% and 48%, respectively, of the estimated species. Analysis of the fungal taxa obtained from bat flies showed significant differences between the most frequent species, A. penicillioides, and the others in the fungal community (H = 26.51, P = 0.009) (Online Resource 2).
Fig. 3.
Species accumulation curves for culturable fungal species observed and estimated (Bootstrap, Chao 1, Chao 2, Jackknife 1 and Jackknife 2) in association with bat flies collected from bats captured at the Furna do Morcego cave located in the Parque Nacional do Catimbau [Catimbau National Park], Brazil
New species
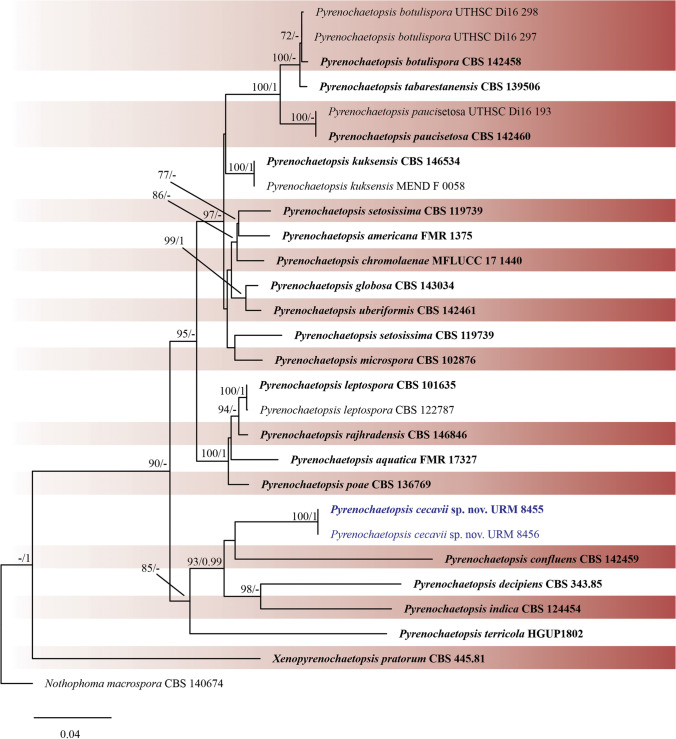
BLASTn searches in the NCBI GenBank database showed that two sequences of the isolates URM 8453 and URM 8454 were related to Allophoma spp., and two sequences (URM 8455 and URM 8456) were related to Pyrenochaetopsis spp. The matrix of Allophoma sequences was composed of 16 species, and the combined alignment contained 2,630 characters (ITS = 494, LSU = 960, RPB2 = 622, and TUB2 = 551), including gaps. For Pyrenochaetopsis, sequences from 22 species were used, and the combined alignment contained 2,727 characters (ITS = 552, LSU = 860, RPB2 = 947, and TUB2 = 365), including gaps.
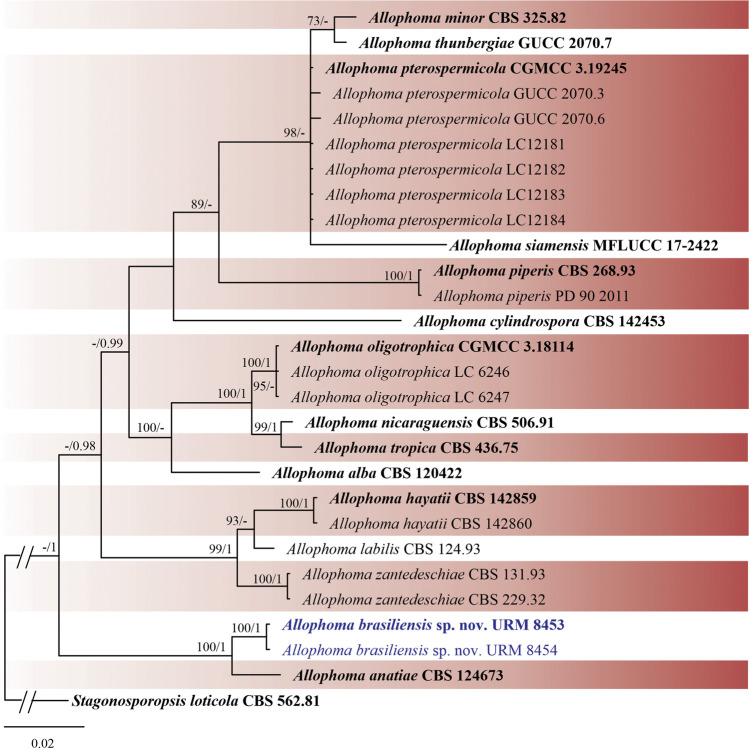
Phylogenetic trees were constructed for the combined matrices (Figs. 4 and 5) and individual datasets (data not shown). The best nucleotide models for BI analysis were SYM + G for ITS, GTR + G for TUB2 and SYM + G for RPB2. The GTR + I + G model was used for all ML analyses. Isolates URM 8453 and URM 8454 were placed as an independent lineage with Allophoma anatiae as sister species (ML-BS = 100 and BPP = 1) (Fig. 4), and isolates URM 8455 and URM 8456 were placed as unique well-supported clades (ML-BS = 100 and BPP = 1) (Fig. 5), with Pyrenochaetopsis confluens as a sister species, and both shared a clade with P. indica and P. decipiens. Four isolates from the two phylogenetically unknown species are described below as new species.
Fig. 4.
Bayesian phylogenetic tree using sequences of ITS, LSU, RPB2 and TUB2 of the genus Allophoma. The new species described in this study is highlighted in blue. Ex-type strains are in bold. Values for ML-BS ≥ 70% and BPP ≥ 0.95 are included near nodes. The tree was rooted to Stagonosporopsis loticola CBS 562.81
Fig. 5.
Bayesian phylogenetic tree using sequences of ITS, LSU, RPB2 and TUB2 of the genus Pyrenochaetopsis. The new species described in this study is highlighted in blue. Ex-type strains are in bold. Values for ML-BS ≥ 70% and BPP ≥ 0.95 are included near nodes. The tree was rooted to Notophoma macrospora CBS 140674
Taxonomy
Pleosporales, Didymellaceae.
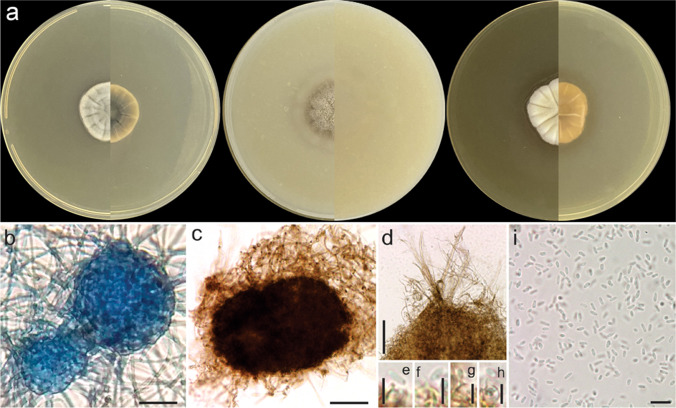
Allophoma brasiliensis J.L.V.R. Carvalho, J.D.P. Bezerra, & Souza-Motta, sp. nov. Figures 6 and 8
Fig. 6.
Allophoma brasiliensis URM 8453, ex-type. a Colonies on PDA, OA, and MEA (verse and reverse). b Aggregated pycnidia. c Details of the pycnidial wall. d–e Coniogenous cells and conidia. f–g Conidia. Scale bars: b = 100 μm and c–g = 10 μm
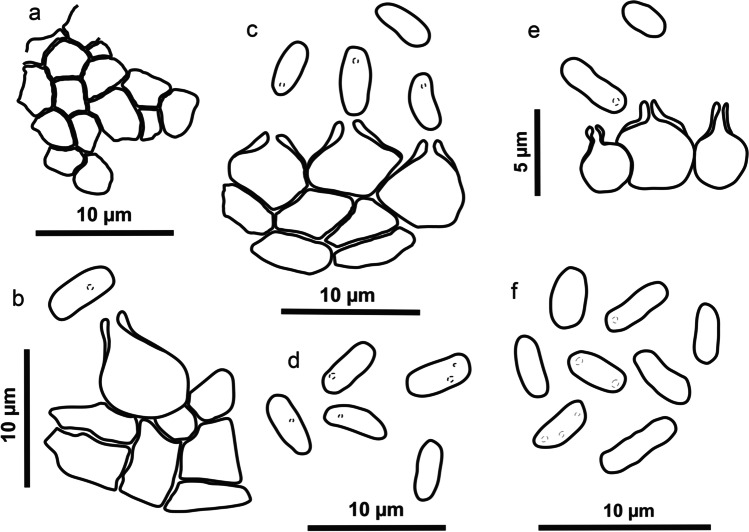
Fig. 8.
Schematic line drawing of the two new species Allophoma brasiliensis (a–d) and Pyrenochaetopsis cecavii (e–f). a Details of the pycnidial wall. b–c, e Conidiogenous cells and conidia. d, f Conidia
MycoBank: MB845564.
Type: Brazil, Pernambuco state, Ibimirim municipality, Catimbau National Park, Furna do Morcego bat cave, 37°22′55ʺ S and 8°34′14ʺ W; 557 m above sea level, isolated from bat flies (Trichobius sp.) on 6 Oct. 2020 J.L.V.R. Carvalho (holotype URM 8453, preserved as metabolically inactive culture; culture ex-type URM 8453).
Etymology: Named after Brazil, the country where the fungus was isolated.
Description: Coelomycetous. Conidiomata pycnidial, solitary or aggregated, globose to subglobose, with a single ostiole, without evident papillae or neck, brown, glabrous, superficial, or semi-immersed in culture media, 65–75(120) × (49)61–65(82) μm; conidiomatal wall pseudoparenchymatous, textura angularis. Conidiogenous cells phialidic, hyaline, smooth-walled, ampulliform, 10.5–15 × 10–11 μm. Conidia oblong, apices rounded, smooth- and thin-walled, hyaline, aseptate, guttulate, (1.8)3–3.5(4.5) × 1–2 μm. Teleomorph not observed.
Culture characteristics (27 °C, 7 days, in the dark): Colonies on OA: 58.8 mm diam., flat, wavy margin, buff colour in the centre, olive buff on the margin; reverse hazel at centre and grey at margin. Colonies on MEA: 60.8 mm diam., flat, wavy margin, buff colour, white near margins; reverse dark brick colour, pure yellow margin. Colonies on PDA: 78.6 mm diam., flat, entire margin, buff colour, white near margin; reverse purplish grey, primrose near margin. No growth at 36 °C.
Other materials analysed: Brazil, Pernambuco state, Ibimirim municipality, Catimbau National Park, Furna do Morcego bat cave, 37°22′55ʺ S and 8°34′14ʺ W; 557 m above sea level, isolated from bat flies (Trichobius sp.) on 6 Oct. 2020, J.L.V.R. Carvalho (URM 8454).
Notes: Allophoma brasiliensis was phylogenetically related to A. anatiae (Fig. 4). Allophoma anatiae [as 'anatii'] was described by Hou et al. [63] from Acropora formosa (Acroporidae, a coral) in Australia. The new species, A. brasiliensis, differs from A. anatiae by having pycnidia that are solitary, slightly ellipsoidal, and larger (90)130–400(460) × (75)120–370 μm in A. anatiae vs. 65–75(120) × (49)61–65(82) μm in A. brasiliensis). Conidia in A. anatiae are larger than those found in A. brasiliensis (3.5–5.5 × 2–3 μm in A. anatiae vs. (1.8)3–3.5(4.5) × 1–2 μm in A. brasiliensis). Only few or no pycnidia were produced during the morphological analysis of A. brasiliensis. Colonies of A. brasiliensis on OA have a buff colour in the centre and olive buff at the margin, wavy margin, and poor production of pycnidia, while A. anatiae has pale cinnamon colonies with a regular margin and abundant production of pycnidia. On MEA and PDA, colonies of A. anatiae have regular margins and sparse aerial mycelia [63]. Allophoma brasiliensis is also morphologically similar but phylogenetically distant from other Allophoma species such as A. alba (conidia size, 3 − 4.5 × 1.5 − 2.3 μm) [62], A. cylindrospora (conidia size, 3 − 4 × 2 μm) [74], A. nicaraguensis (conidia size, 2.5 − 4 × 1.5 − 2.5 μm) [63], A. siamensis (pycnidia size, 70 − 90 × 68 − 85 μm; conidia size, 3 − 4 × 2 − 3 μm) [75], A. thunbergiae (conidia size, 3 − 5 × 1.5 − 2.5 μm) [76], and A. tropica (conidia size, 3 − 4 × 1 − 2 μm) [64]; A. brasiliensis has the largest conidiogenous cell among Allophoma species.
Pleosporales, Cucurbitariaceae.
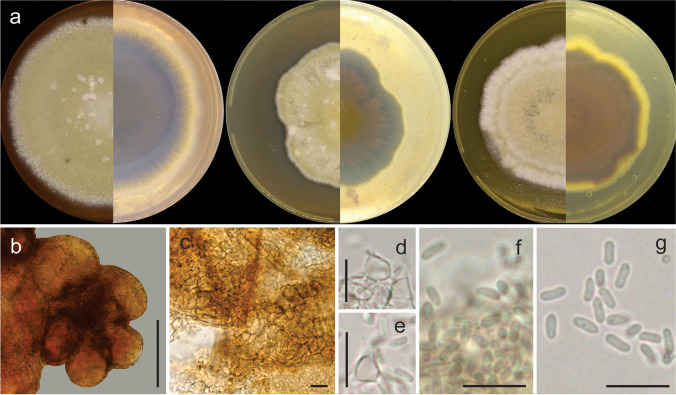
Pyrenochaetopsis cecavii J.L.V.R. Carvalho, J.D.P. Bezerra, & Souza-Motta, sp. nov. Figure 7 and 8
Fig. 7.
Pyrenochaetopsis cecavii URM 8455, ex-type. a Colonies on PDA, OA, and MEA (verse and reverse). b–c Pycnidia. d Details of setose pycnidium. e–h Coniogenous cells and conidia. i Conidia. Scale bars: = b–c = 10 μm, d–i = 10 μm and e–h = 5 μm
MycoBank: MB845565.
Type: Brazil, Pernambuco state, Ibimirim municipality, Catimbau National Park, Furna do Morcego bat cave, 37°22′55ʺ S and 8°34′14ʺ W; 557 m above sea level, isolated from bat flies (Trichobius sp.) on 6 Oct. 2020, J.L.V.R. Carvalho (holotype URM 8455, preserved as metabolically inactive culture; culture ex-type URM 8455).
Etymology: Named after the Brazilian federal institution devoted to the conservation and study of caves, the Centro Nacional de Pesquisa e Conservação de Cavernas (CECAV).
Description: Coelomycetous. Conidiomata pycnidial, brown, solitary or aggregated, superficial or semi-immersed in culture media, globose to sub-globose, setose, glabrous to slightly hairy, ostiolate, 325–365 × 236–327 μm; conidiomatal wall of textura angularis. Conidiogenous cells phialidic, hyaline, smooth-walled, ampulliform, 3.5–4.5(6) × 2–4 μm. Conidia hyaline, aseptate, cylindrical to allantoid, guttulate, 2.6–3.7(4) × 1–1.5(2) μm. Teleomorph not observed.
Culture characteristics (27 °C, 10 days, in the dark): Colonies on PDA reaching 24 mm diameter, umbonate elevation, entire margin, white colour; reverse pale olivaceous grey in centre and white on margin. Colonies on MEA reaching 28 mm diameter, raised, wavy margin, white colour, reverse honey colour. Colonies in OA reaching 25 mm diameter, raised, aerial mycelia, surface smoke grey in centre, and olivaceous grey on margin. No growth at 36 °C.
Other materials analysed: Brazil, Pernambuco state, Ibimirim municipality, Catimbau National Park, Furna do Morcego bat cave, 37°22ʺ55ʺ S and 8°34′14ʺ W; 557 m above sea level, isolated from bat fly (Trichobius sp.), 6 Oct. 2020, J.L.V.R. Carvalho (URM 8456).
Notes: Pyrenochaetopsis cecavii was phylogenetically related to P. indica (Fig. 5). Pyrenochaetopsis indica was introduced by de Gruyter et al. [62] based on Pyrenochaeta indica isolated from Saccharum officinarum leaves in India [77]. The new species, P. cecavii, has larger pycnidia than P. indica (325–365 × 236–327 μm in P. cecavii vs. 55–240 μm in P. indica) and smaller conidia (2.6–3.7(4) × 1–1.5(2) μm in P. cecavii vs. 4–5.5 × 1.5–2.5 μm in P. indica). During the morphological analysis of P. cecavii, only few or no pycnidia were produced. Chlamydospores were observed in P. indica but not in P. cecavii. In addition, colonies of P. cecavii growing on OA are raised, smoke grey in the centre, and olivaceous grey on the margin, while P. indica has a regular colony and olivaceous surface. On MEA, P. cecavii has a colony raised with a wavy margin, surface white, and reverse honey, differing from P. indica which has a regular colony with greyish colour to grey olivaceous near the margin [78]. Pyrenochaetopsis cecavii is morphologically similar but phylogenetically distant from P. botulispora (conidiogenous cell size, 4 × 5 μm) [74], P. uberiformis (pycnidia size, 200–440 × 130–410 μm; conidiogenous cell size, 3–4 × 4–5 μm) [74], P. kuksensis (conidia size, 2.5–4 × 1–2 μm) [79], and P. aquatica (conidia size, 3.5–5 µm × 1–1.8 µm) [80].
Discussion
To the best of our knowledge, our study is the first to isolate cultivable fungi that are associated with bat ectoparasitic flies in Brazil. Approximately 80% of the bats captured in Furna do Morcego, a cave in the dry portion of Northeastern Brazil, had fungi associated with their ectoparasites, and a total of 13 species were recorded. Among the isolates, Ascomycota was the most representative phylum (eight genera), and Aspergillus had the highest number of taxa (three species). Over the past several decades, mycological surveys have been conducted to identify ectosymbiont fungi of the order Laboulbeniales in association with bat flies [19, 20]. However, there is a gap in our knowledge regarding the richness of cultivable fungi associated with bat flies. In our study, we found fungal species commonly isolated from air samples, substrates found in caves, and bat bodies [33, 34, 79, 81, 82].
Our findings show that bat flies are associated with cultivable fungal species and may act as dispersers of fungal propagules because their lifestyle is closely related with the body of bats [83]. This type of dispersion of fungal propagules mediated by bat ectoparasites was observed in spinturnicid mites, which proved to be a potential vector showing a positive relationship between the amount of fungal infection in bats and the fungal load present in the mites [25]. Considering that bat flies of the genus Trichobius have a much larger body surface than spinturnicid mites, often occur in greater abundance, and have a marked capacity for locomotion within the shelter of their hosts, their role as potential mechanical carriers of fungal propagules among bats could be more relevant than that of mites. However, this issue remains to be resolved. Future studies should investigate whether bats that are more parasitised by flies have greater fungal diversity and/or abundance than those with less fly associations.
We observed a similar culturable fungal community in both bats and bat flies. Cunha et al. [34] isolated fungi from bats in a cave in the Caatinga dry forest in Brazil, and showed that the genus Aspergillus was the most representative among the isolates obtained. Johnson et al. [81] studied fungi from bat wings in the USA and observed that most fungal isolates were species of Cladosporium, Fusarium, Geomyces, Mortierella, Penicillium, and Trichosporon. Other studies isolating fungi from bats showed that the fur and skin of these animals can harbour a greater fungal diversity than that of the cave floor, which can be explained by the movement of bats (and their attached flies) between epigean and hypogean environments [33, 34, 84, 85]. In our study, Aspergillus penicillioides was the most frequently isolated species, and we also found two species of Cladosporium (C. halotolerans and C. subuliforme) (see [36]) and one species of Penicillium (P. citrinum) on the bat flies, showing that flies have fungi that are also present on bats and may contribute to the dispersion of fungal species because of their mode of life, reproduction, and changes of host.
The most abundantly found species, A. penicillioides, which was first found on the leaves of Saccharum officinarum in Argentina [86], has been reported as one of the most xerophilic species of the Aspergillus section Restricti [64, 87]. The section Restricti is composed of xerophilic and halophilic species that mainly grow on substrates with low water activity in extreme environments [87]. Aspergillus penicillioides has been reported on several substrates/hosts (e.g. human skin, green fabric covered binders, seeds of cereal, indoor air, dried corn) globally, including Brazil [87]. In a worldwide survey of fungi in house dust, A. penicillioides was a common OTU identified [88], and it was also found in caves [33]. Strains of this species are able to grow at 37 °C and they produce several exometabolites (e.g. asperglaucide and mycophenolic acid) [87]. They are described as true halophiles [89], and have the potential to be used in heavy metal bioremediation, for antibacterial activity [90, 91], and as enzyme producers [92]. However, A. penicillioides is also recorded as an opportunistic etiologic agent that causes human infections [93–95].
Among the 13 fungal species identified in our study, two (15%) were described as new species of the genera Allophoma and Pyrenochaetopsis. Recently, new species of fungi have been described from different substrates/hosts of caves in China (53 species, Zhang et al. [96, 97], and Cunha et al. [34] indicated that in Brazil approximately 15 (25%) of the 59 taxa reported in their study could be described as new species. In addition, other new species of the genera Malassezia and Geosmithia have been introduced as fungal isolates obtained from the body of bats [98, 99]. Species of the genera Allophoma and Pyrenochaetopsis are mainly associated with plants [100, 101]. The discovery of new species of Allophoma and Pyrenochaetopsis associated with bat flies expand the possibilities to find species of these genera associated with different hosts; in our case, the bat lifestyle could be an important mechanism for these fungi to have their conidia associated with flies and bats. In a tropical bat cave in Brazil, Cunha et al. [34] reported fungi from bats that were also generally associated with plants (also as pathogens) or acting as opportunistic pathogens of animals (e.g. Deniquelata, Fusarium, and Nothophoma). Other surveys have observed these fungal genera in caves in China (Zhang et al. [96, 97]. Therefore, research such as ours is important to provide evidence of a broader amplitude of adaptation and habitat use of known and unknown fungal species. Moreover, our results shed light on cryptic and poorly known parts of biodiversity and may contribute to a better estimation of national and global fungal diversity.
In this study of culturable fungal species associated with flies collected from bats in a tropical cave in Brazil, we discovered a previously unknown fungal diversity associated with these ectoparasites. Bat flies could be a method for fungi to ‘catch a ride’ along with their hosts (bats), and may contribute to fungal dispersion in the cave environment. This novelty of culturable fungal association with bat flies could indicate the ecological importance of these insects in the mode of life of several fungal species, and highlight the importance of protecting the cave environment, along with its inhabitants (e.g. microorganisms, plants, animals).
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We thank Narjara Pimentel (UFPE) for her invaluable support during the fieldwork. We also thank Fundação Nacional do Índio (FUNAI), and Jailton Fernandes (ICMBio/PARNA do Catimbau) for their logistical support.
Author contribution
João L. V. R. Carvalho, formal analysis, investigation, data curation, writing (original draft, review, and editing); Joenny M. S. Lima, formal analysis, investigation, data curation, writing (review and editing); Eder Barbier, Enrico Bernard, Jadson D. P. Bezerra, and Cristina M. Souza-Motta: conceptualization, supervision, project administration, funding acquisition, writing (original draft, review, and editing).
Funding
This study was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior – Brasil (CAPES, Finance Code 001; CAPES-PRInt process number 88887.311891/2018–00), the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq, processes 310298/2018–0 and 408788/2021-6), and the Fundação de Amparo à Ciência e Tecnologia do Estado de Pernambuco (FACEPE, process number APQ-0350-2.12/19). Funding for fieldwork was also partially provided by the Anglo American. E. Barbier is supported by a postdoctoral grant from CAPES and FACEPE (process #88887.353052/2019–00). C.M. Souza-Motta and E. Bernard have a fellowship from CNPq.
Data availability
DNA sequences are available in GenBank and alignments were deposited in TreeBASE.
Declarations
Ethics approval
The collection was authorised by the Ministério do Meio Ambiente (MMA)/Instituto Chico Mendes de Conservação da Biodiversidade (ICMBio) (SISBIO number 68992–3) and by the Ethics Committee on Animal Care–UFPE (number 114/2019).
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jadson D. P. Bezerra, Email: jadsonbezerra@ufg.br
Cristina M. Souza-Motta, Email: cristina.motta@ufpe.br
References
- 1.Allen GM. The parasites of bats. Cambridge: Museum of Comparative Zoology; 1967. [Google Scholar]
- 2.Whitaker JO., Jr . Collecting and preserving ectoparasites for ecological study. In: Kunz T, editor. Ecological and behavioral methods for the study of bats. Washington: Smithsonian Inst Press; 1988. pp. 459–474. [Google Scholar]
- 3.Naegle MA, Mugleston JD, Bybee SM, Whiting MF. Reassessing the phylogenetic position of the epizoic earwigs (Insecta: Dermaptera) Mol Phylogenet Evol. 2016;100:382–390. doi: 10.1016/j.ympev.2016.03.012. [DOI] [PubMed] [Google Scholar]
- 4.Taylor S, Durden LA, Foley EH, Reeves WK. The bat tick Carios azteci (Acari: Argasidae) from Belize with an endosymbiotic Coxiellaceae. Speleobiol Notes. 2016;8:16–21. doi: 10.5563/spbn.v8i0.81. [DOI] [Google Scholar]
- 5.Haelewaters D, Hiller T, Dick CW. Bats, bat flies, and fungi: a case of hyperparasitism. Trends Parasitol. 2018;34(9):784–799. doi: 10.1016/j.pt.2018.06.006. [DOI] [PubMed] [Google Scholar]
- 6.Marshall AG (1982) Ecology of insects ectoparasitic on bats. In: Kunz TH (ed) Ecology of bats. Springer, Boston, pp 369–401. 10.1007/978-1-4613-3421-7_10
- 7.Frank R, Münster J, Schulze J, Liston A, Klimpel S (2014) Macroparasites of Microchiroptera: bat ectoparasites of Central and South America. In: Klimpel S, Mehlhorn H (eds) Bats (Chiroptera) as Vectors of Diseases and Parasites. Parasitology Research Monographs, vol 5. Springer, Berlin, pp 87–130
- 8.Szentiványi T, Christe P, Glaizot O. Bat flies and their microparasites: current knowledge and distribution. Front Vet Sci. 2019;6:115. doi: 10.3389/fvets.2019.00115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Graciolli G, Autino AG, Claps GL. Catalogue of American Nycteribiidae (Diptera, Hippoboscoidea) Rev Bras Entomol. 2007;51:142–159. doi: 10.1590/S0085-56262007000200004. [DOI] [Google Scholar]
- 10.Graciolli G, Azevedo AA, Árzua M, Barros-Battesti DM, Linardi PM (2008) Artrópodos ectoparasitos de morcegos no Brasil. In: Pacheco SM, Marques RV, Esbérard CEL (orgs) Morcegos do Brasil: biologia sistemática ecologia e conservação. Armazém Digital, Porto Alegre, 123 –138
- 11.Graciolli G, Zortéa M, Carvalho LFAC. Bat flies (Diptera, Streblidae and Nycteribiidae) in a Cerrado area of Goiás state, Brazil. Rev Bras Entomol. 2010;54:511–514. doi: 10.1590/S008556262010000300025. [DOI] [Google Scholar]
- 12.Hagan HR. Embryology of the viviparus insects. New York: Ronald Press; 1951. [Google Scholar]
- 13.Kunz TH, Lumsden LF, Fenton MB (2003) Ecology of cavity and foliage roosting bats. In: Kunz TH, Fenton MB (eds) Bat Ecology. The University of Chicago Press, Chicago, pp 3–89
- 14.Bequart J. Moscas parasitas pupiparas de Colombia y Panamá. Rev Acad Colomb Cienc Exact Fis Nat. 1940;3:414–418. [Google Scholar]
- 15.Patterson BD, Dick CW, Dittmar K. Roosting habits of bats affect their parasitism by bat flies (Diptera: Streblidae) J Trop Ecol. 2007;23:177–189. doi: 10.1017/S0266467406003816. [DOI] [Google Scholar]
- 16.Thaxter R. Preliminary diagnoses of new species of Laboulbeniaceae. III Proc Am Acad Arts Sci. 1901;36:397–414. doi: 10.2307/20021044. [DOI] [Google Scholar]
- 17.Graciolli G, Aguiar LS. Ocorrência de moscas ectoparasitas (Diptera, Streblidae e Nycteribiidae) de morcegos (Mammalia, Chiroptera) no Cerrado de Brasília, Distrito Federal, Brasil. Rev Bras Zoo. 2002;19:177–181. doi: 10.1590/S0101-81752002000500012. [DOI] [Google Scholar]
- 18.Walker MJ, Dorrestein A, Camacho JJ, Meckler LA, Silas KA, Hiller T, Haelewaters D. A tripartite survey of hyperparasitic fungi associated with ectoparasitic flies on bats (Mammalia: Chiroptera) in a neotropical cloud forest in Panama. Parasite. 2018;25:19. doi: 10.1051/parasite/2018017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haelewaters D, Verhaeghen SJ, Rios Gonzalez TA, Bernal Vega JÁ, Villarreal Saucedo RV. New and interesting Laboulbeniales from Panama and neighboring areas. Nova Hedwigia. 2017;105:267–299. doi: 10.1127/nova_hedwigia/2017/0410. [DOI] [Google Scholar]
- 20.de Groot MD, Dumolein I, Hiller T, Sándor AD, Szentiványi T, Schilthuizen M, Catherine MA, Verbeken A, Haelewaters D. On the fly: tritrophic associations of bats bat flies and fungi. J Fungus. 2020;6:361. doi: 10.3390/jof6040361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tan JS. Human zoonotic infections transmitted by dogs and cats. Archives Intern Med. 1997;157:1933–1943. doi: 10.1001/archinte.1997.00440380035003. [DOI] [PubMed] [Google Scholar]
- 22.Parola P, Davoust B, Raoult D. Tick- and flea-borne rickettsial emerging zoonoses. Vet Res. 2005;36:469–492. doi: 10.1051/vetres:2005004. [DOI] [PubMed] [Google Scholar]
- 23.Förster M, Klimpel S, Mehlhorn H, Sievert K, Messler S, Pfeffer K. Pilot study on synanthropic flies (e.g. Musca, Sarcophaga, Calliphora, Fannia, Lucilia, Stomoxys) as vectors of pathogenic microorganisms. Parasitol Res. 2007;101:243–246. doi: 10.1007/s00436-007-0522-y. [DOI] [PubMed] [Google Scholar]
- 24.Beugnet F, Marie JL. Emerging arthropod–borne diseases of companion animals in Europe. Vet Parasitol. 2009;163:298–305. doi: 10.1016/j.vetpar.2009.03.028. [DOI] [PubMed] [Google Scholar]
- 25.Lučan RK, Bandouchova H, Bartonička T, Pikula J, Zahradníková A, Zukal J, Martínková N. Ectoparasites may serve as vectors for the white-nose syndrome fungus. Parasites Vectors. 2016;9:16. doi: 10.1186/s13071-016-1302-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Foley J, Clifford D, Castle K, Cryan P, Ostfeld RS. Investigating and managing the rapid emergence of white nose syndrome a novel fatal infectious disease of hibernating bats. Conserv Biol. 2011;25:223–231. doi: 10.1111/j.1523-1739.2010.01638.x. [DOI] [PubMed] [Google Scholar]
- 27.Turner JM, Warnecke L, Wilcox A, Baloun D, Bollinger TK, Misra V, Willis CK. Conspecific disturbance contributes to altered hibernation patterns in bats with white-nose syndrome. Physiol Behavior. 2015;140:71–78. doi: 10.1016/j.physbeh.2014.12.013. [DOI] [PubMed] [Google Scholar]
- 28.Hoyt JR, Kilpatrick AM, Langwig KE. Ecology and impacts of white-nose syndrome on bats. Nat Rev Microbiol. 2021;19:196–210. doi: 10.1038/s41579-020-00493-5. [DOI] [PubMed] [Google Scholar]
- 29.Bandouchova H, Bartonicka T, Berkova H, Brichta J, Cerny J, Kovacova V, Kolarik M, Köllner B, Kulich P, Martínková N, Rehak Z, Turner GG, Zukal J, Pikula J. Pseudogymnoascus destructans: evidence of virulent skin invasion for bats under natural conditions, Europe. Transbound Emerg Dis. 2015;62:1–5. doi: 10.1111/tbed.12282. [DOI] [PubMed] [Google Scholar]
- 30.Barlow AM, Worledge L, Miller H, Drees KP, Wright P, Foster JT, Sobek C, Borman AM, Fraser M. First confirmation of Pseudogymnoascus destructans in British bats and hibernacula. Vet Rec. 2015;177:73–73. doi: 10.1136/vr.102923. [DOI] [PubMed] [Google Scholar]
- 31.Garzoli L, Riccucci M, Patriarca E, Debernardi P, Boggero A, Pecoraro L, Picco AM. First isolation of Pseudogymnoascus destructans the fungal causative agent of white-nose disease in bats from Italy. Mycopathologia. 2019;184:637–644. doi: 10.1007/s11046-019-00371-6. [DOI] [PubMed] [Google Scholar]
- 32.Urbina J, Chestnut T, Allen JM, Levi T. Pseudogymnoascus destructans growth in wood soil and guano substrates. Sci Rep. 2021;11:763. doi: 10.1038/s41598-020-80707-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vanderwolf KJ, McAlpine DF, Malloch D, Forbes GJ. Ectomycota associated with hibernating bats in eastern Canadian caves prior to the emergence of white-nose syndrome. Northeastern Nat. 2013;20:115–130. doi: 10.1656/045.020.0109. [DOI] [Google Scholar]
- 34.Cunha AOB, Bezerra JDP, Oliveira TGL, Barbier E, Bernard E, Machado AR, Souza-Motta CM. Living in the dark: bat caves as hotspots of fungal diversity. PLoS ONE. 2020;15:e0243494. doi: 10.1371/journal.pone.0243494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bertola PB, Aires CC, Favorito SE, Graciolli G, Amaku M, Pinto-da-Rocha R. Bat flies (Diptera: Streblidae Nycteribiidae) parasitic on bats (Mammalia: Chiroptera) at Parque Estadual da Cantareira São Paulo Brazil: parasitism rates and host–parasite associations. Mem Inst Oswaldo Cruz. 2005;100:25–32. doi: 10.1590/S0074-02762005000100005. [DOI] [PubMed] [Google Scholar]
- 36.Pereira MLS, Carvalho JLVR, Lima JMS, Barbier E, Bernard E, Bezerra JDP, Souza-Motta CM. Richness of Cladosporium in a tropical bat cave with the description of two new species. Mycol Prog: In press; 2022. [Google Scholar]
- 37.Rito KF, Arroyo Rodríguez V, Queiroz RT, Leal IR, Tabarelli M. Precipitation mediates the effect of human disturbance on the Brazilian Caatinga vegetation. J Ecol. 2017;105:828–838. doi: 10.1111/13652745.12712. [DOI] [Google Scholar]
- 38.Silva JMC, Barbosa LCF, Leal IR, Tabarelli M (2017) The Caatinga: Understanding the challenges. In: Silva JMC, Leal IR, Tabarelli M (eds) Caatinga. Springer, Cham, pp 3–19
- 39.Leal ESB, Bernard E. Mobility of bats between caves: ecological aspects and implications for conservation and environmental licensing activities in Brazil. Stud Neotrop Fauna Environ. 2021 doi: 10.1080/01650521.2021.1964910. [DOI] [Google Scholar]
- 40.MMA Ministério do MeioAmbienteSecretaria de Biodiversidade e Florestas . Biodiversidade brasileira: avaliação e identificação de áreas e ações prioritárias para conservação utilização sustentável e repartição dos benefícios da biodiversidade nos biomas brasileiros. Brasília: MMA/SBF; 2002. [Google Scholar]
- 41.Gomes APDS, Rodal MJN, Melo ALD. Florística e fitogeografia da vegetação arbustiva subcaducifólia da Chapada de São José, Buíque, PE, Brasil. Acta Bot Bras. 2006;20:37–48. doi: 10.1590/S010233062006000100005. [DOI] [Google Scholar]
- 42.SNE – SociedadeNordestina de Ecologia . Projeto Técnico para a Criação do Parque Nacional do Catimbau/PE – versão final em cumprimento ao Contrato no 086–00/02 Subprojeto “Proposta para Criação do Parque Nacional do Catimbau/PE”. Recife: Sociedade Nordestina de Ecologia; 2002. [Google Scholar]
- 43.Specht MJ, Santos BA, Marshall N, Melo FPL, Leal IR, Tabarelli M, Baldauf C. Socioeconomic differences among resident users and neighbour populations of a protected area in the Brazilian dry forest. J Environ Manage. 2019;232:607–614. doi: 10.1016/j.jenvman.2018.11.101. [DOI] [PubMed] [Google Scholar]
- 44.Gardner AL. Mammals of South America: marsupials xenarthrans shrews and bats. Chicago: The University of Chicago Press; 2007. [Google Scholar]
- 45.Díaz MM, Solari S, Aguirre LF, Aguiar LMS, Barquez RM (2016) Clave de identificación de los murciélagos de Sudamérica/Chave de identificação dos morcegos da América do Sul. Programa de Conservación de los Murciélagos de Argentina, Tucumán
- 46.Barbier E, Bernard E. From the Atlantic Forest to the borders of Amazonia: species richness distribution and host association of ectoparasitic flies (Diptera: Nycteribiidae and Streblidae) in northeastern Brazil. Parasitol Res. 2017;116:3043–3055. doi: 10.1007/s00436-017-5615-7. [DOI] [PubMed] [Google Scholar]
- 47.Samson AR, Houbraken J, Thrane U, Frisvald JC, Andersen B. Food and indoor fungi. Utrecht: CBS-KNAW Fungal Biodiversity Centre; 2010. [Google Scholar]
- 48.Seifert K, Morgan-Jones G, Gams W, Kendrick B (2011) The genera of Hyphomycetes. CBS-KNAW Fungal Biodiversity Centre, Utrecht
- 49.Tuite J. Plant pathological methods. Fungi and bacteria: Fungi and bacteria. Plant pathological methods; 1969. [Google Scholar]
- 50.Rayner RW. A mycological colour chart. Kew: CMI and British Mycological Society; 1970. [Google Scholar]
- 51.White TJ, Bruns T, Lee S, Taylor L. Amplification and direct sequencing of fungal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols. A guide to methods and applications: Academic Press, Cambridge; 1990. pp. 315–322. [Google Scholar]
- 52.Carbone I, Kohn LM. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia. 1999;91:553–556. doi: 10.1080/00275514.1999.12061051. [DOI] [Google Scholar]
- 53.Glass NL, Donaldson GC. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl Environ Microbiol. 1995;61:1323–1330. doi: 10.1007/s11046-019-00371-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hong SB, Go SJ, Shin HD, Frisvad JC, Samson RA. Polyphasic taxonomy of Aspergillus fumigatus and related species. Mycologia. 2005;97:1316–1329. doi: 10.1080/15572536.2006.11832738. [DOI] [PubMed] [Google Scholar]
- 55.Vilgalys R, Hester M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol. 1990;172(8):4238–4246. doi: 10.1128/jb.172.8.4238-4246.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vilgalys R, Sun BL. Ancient and recent patterns of geographic speciation in the oyster mushroom Pleurotus revealed by phylogenetic analysis of ribosomal DNA sequences. Proc Natl Acad Sci USA. 1994;91(10):4599–4603. doi: 10.1073/pnas.91.10.4599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu YJ, Whelen S, Hall BD. Phylogenetic relationships among ascomycetes: evidence from an RNA polymerse II subunit. Mol Biol Evol. 1999;16:1799–1808. doi: 10.1093/oxfordjournals.molbev.a026092. [DOI] [PubMed] [Google Scholar]
- 58.Sung GH, Sung JM, Hywel Jones NL, Spatafora JW. A multi-gene phylogeny of Clavicipitaceae (Ascomycota, Fungi): identification of localized incongruence using a combinational bootstrap approach. Mol Phylogenet Evol. 2007;44:1204–1223. doi: 10.1016/j.ympev.2007.03.011. [DOI] [PubMed] [Google Scholar]
- 59.Bezerra JDP, Oliveira RJV, Paiva LM, Silva GA, Groenewald JZ, Crous PW, Souza-Motta CM. Bezerromycetales and Wiesneriomycetales ord. nov. (class Dothideomycetes) with two novel genera to accommodate endophytic fungi from Brazilian cactus. Mycol Prog. 2017;16:297–309. doi: 10.1007/s11557-016-1254-0. [DOI] [Google Scholar]
- 60.Bezerra JDP, Sandoval-Denis M, Paiva LM, Silva GA, Groenewald JZ, Souza-Motta CM, Crous PW. New endophytic Toxicocladosporium species from cacti in Brazil and description of Neocladosporium gen. nov. IMA Fungus. 2017;8:77–97. doi: 10.5598/imafungus.2017.08.01.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Samson RA, Visagie CM, Houbraken J, Hong SB, Hubka V, Klaassen CH, Perrone G, Seifert KA, Susca A, Tanney JB, Varga J, Kocsubé S, Szigeti G, Yaguchi T, Frisvad JC. Phylogeny identification and nomenclature of the genus Aspergillus. Stud Mycol. 2014;78:141–173. doi: 10.1016/j.simyco.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.de Gruyter J, Woudenberg JH, Aveskamp MM, Verkley GJ, Groenewald JZ, Crous PW. Systematic reappraisal of species in Phoma section Paraphoma, Pyrenochaeta and Pleurophoma. Mycologia. 2010;102:1066–1081. doi: 10.3852/09-240. [DOI] [PubMed] [Google Scholar]
- 63.Hou LW, Groenewald JZ, Pfenning LH, Yarden O, Crous PW, Cai L. The phoma-like dilemma. Stud Mycol. 2020;96:309–396. doi: 10.1016/j.simyco.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen Q, Jiang JR, Zhang GZ, Cai L, Crous PW. Resolving the Phoma enigma. Stud Mycol. 2015;82:137–217. doi: 10.1016/j.simyco.2015.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Houbraken J, Kocsubé S, Visagie CM, Yilmaz N, Wang XC, Meijer M, Kraak B, Hubka V, Bensch K, Samson RA, Frisvad JC. Classification of Aspergillus, Penicillium, Talaromyces and related genera (Eurotiales): an overview of families genera subgenera sections series and species. Stud Mycol. 2020;95:5–169. doi: 10.1016/j.simyco.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ronquist F, Teslenko M, Van Der Mark P, Kumar S, Atecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ronquist F, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic biology. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed]
- 70.Miller MA, Pfeiffer W, Schwartz T (2010) Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In: 2010 Gateway Computing Environments Workshop (GCE), pp 1–8
- 71.Nylander JAA (2004) MrModeltest 2.2. Computer program and documentation distributed by the author. Evolutionary Biology Centre, Uppsala University, Uppsala
- 72.Rambaut A (2010) FigTree v1.3.1. Institute of Evolutionary Biology, University of Edinburgh, Edinburgh. http://tree.bio.ed.ac.uk/software/figtree/. Accessed 17 Feb 2022
- 73.Cowell R K, Elsensohn J E. Estimates turns 20: statistical estimation of species richness and shared species from samples, with non‐parametric extrapolation. Ecography. 2014;37:609–613. doi: 10.1111/ecog.00814. [DOI] [Google Scholar]
- 74.De León JG, Méndez RG, Cadilla CL, Rivera-Mariani FE, Bolaños-Rosero B (2018) Identification of immunoglobulin E-Binding proteins of the xerophilic fungus Aspergillus penicillioides crude mycelial mat extract and serological reactivity assessment in subjects with different allergen reactivity profiles. International Archives of Allergy and Immunology 175:147–159. 10.1159/000484898 [DOI] [PMC free article] [PubMed]
- 75.Jayasiri S C. Diversity, morphology and molecular phylogeny of Dothideomycetes on decaying wild seed pods and fruits. 2019. p. 1. [Google Scholar]
- 76.Yuan J, Zeng XY, Geng K et al (2021) Allophoma species (Pleosporales: Didymellaceae) associated with Thunbergia grandiflora in Guangxi Province, China. Biodiversity Data Journal 9:e63643. 10.3897/BDJ.9.e63643 [DOI] [PMC free article] [PubMed]
- 77.Viswanathan T S. A new species of Pyrenochaeta from sugarcane in India. Curr Sci. 1957;26(4):117–118. [Google Scholar]
- 78.De Gruyter J, Boerema GH (2002) Contributions towards a monograph of Phoma (Coelomycetes) VIII. Section Paraphoma: Taxa with setose pycnidia. Persoonia 17:541–561
- 79.Spetik M, et al. Phytotaxa. 2021;498(3):177. doi: 10.11646/phytotaxa.498.3.3. [DOI] [Google Scholar]
- 80.Magaña-Dueñas V, Stchigel AM, Cano-Lira JF (2021) New Coelomycetous fungi from freshwater in Spain. J Fungi 7:368. 10.3390/jof7050368 [DOI] [PMC free article] [PubMed]
- 81.Johnson LJ, Miller AN, McCleery RA, McClanahan R, Kath JA, Lueschow S, Porras-Alfaro A (2013) Psychrophilic and psychrotolerant fungi on bats and the presence of Geomyces spp. on bat wings prior to the arrival of white nose syndrome. Appl Environ Microbiol 79:5465–5471. 10.1128/AEM.0142913 [DOI] [PMC free article] [PubMed]
- 82.Visagie CM, Yilmaz N, Vanderwolf K, Renaud JB, Sumarah MW, Houbraken J, Assebgui R, Seifert KA, Malloch D (2020) Penicillium diversity in Canadian bat caves including a new species P. speluncae. FUSE 5:1–16. 10.3114/fuse.2020.05.01 [DOI] [PMC free article] [PubMed]
- 83.Dick CW, Patterson BD (2006) Bat flies: obligate ectoparasites of bats. In: Morand S, Krasnov BR, Poulin R (eds) Micromammals and macroparasites. Springer, Tokyo, pp 179–194
- 84.Vanderwolf KJ, Malloch D, McAlpine DF, Forbes GJ (2013) A world review of fungi yeasts and slime molds in caves. Int J Speleol 42:77–96. 10.5038/1827-806X.42.1.9
- 85.Ogórek R, Kurczaba K, Cal M, Apoznański G, Kokurewicz T (2020) A culture-based ID of Micromycetes on the wing membranes of greater Mouse-Eared bats (Myotis myotis) from the “Nietoperek” site (Poland). Animals 10:1337. 10.3390/ani10081337 [DOI] [PMC free article] [PubMed]
- 86.Spegazzini C (1896) Hongos de la caña de azúcar. Revista Fac Agron Univ Nac La Plata 2:227–258
- 87.Sklenář F, Jurjević Ž, Zalar P, Frisvad JC, Visagie CM, Kolařík M, Hubka V (2017) Phylogeny of xerophilic aspergilli (subgenus Aspergillus) and taxonomic revision of section Restricti. Stud Mycol 88:161–236. 10.1016/j.simyco.2017.09.002 [DOI] [PMC free article] [PubMed]
- 88.Amend AS, Seifert KA, Bruns TD (2010) Quantifying microbial communities with 454 pyrosequencing: does read abundance count? Mol Ecol 19:5555–5565. 10.1111/j.1365-294X.2010.04898.x [DOI] [PubMed]
- 89.Nazareth SW, Gonsalves V (2014) Halophilic Aspergillus penicillioides from at halassohaline thalassohaline and polyhaline environments. Front Microbiol 5:412. 10.3389/fmicb.2014.00412 [DOI] [PMC free article] [PubMed]
- 90.Chi LP, Yang SQ, Li XM, Li XD, Wang BG, Li X (2020) A new steroid with 7β,8β–epoxidation from the deep sea–derived fungus Aspergillus penicillioides SD-311. J Asian Nat Prod Res 23:884–891. 10.1080/10286020.2020.1791096 [DOI] [PubMed]
- 91.Paria K, Chakraborty SK (2019) Eco-potential of Aspergillus penicillioides (F12): bioremediation and antibacterial activity. SN Appl Sci 1:1515. 10.1007/s42452-019-1545-6
- 92.Ali I, Akbar A, Anwar M, Prasongsuk S, Lotrakul P, Punnapayak H (2015) Purification and characterization of a polyextremophilic α–amylase from an obligate halophilic Aspergillus penicillioides isolate and its potential for souse with detergents. Biomed Res Int 2015:245649. 10.1155/2015/245649 [DOI] [PMC free article] [PubMed]
- 93.de Hoog G, Guarro J, Figueiras MJ, Gené J (2009) Atlas of clinical fungi. CBS–KNAW Fungal Biodiversity Centre, Utrecht
- 94.Gupta K, Gupta P, Mathew JL, Bansal A, Singh G, Singh M, Chakrabarti A (2016) Fatal disseminated Aspergillus penicillioides infection in a 3-month-old infant with suspected cystic fibrosis: Autopsy case report with review of literature. Pediatr Dev Pathol 19:506–511. 10.2350/15-10-1729-CR.1 [DOI] [PubMed]
- 95.Machowicz-Matejko E, Furmańczyk A, Zalewska ED (2018) Aspergillus penicillioides Speg. implicated in keratomycosis. Pol J Microbiol 67:407–416. 10.21307/pjm-2018-049 [DOI] [PMC free article] [PubMed]
- 96.Zhang ZF, Liu F, Zhou X, Liu XZ, Liu SJ, Cai L (2017) Culturable mycobiota from Karst caves in China with descriptions of 20 new species. Persoonia 39:1–31. 10.3767/persoonia.2017.39.01 [DOI] [PMC free article] [PubMed]
- 97.Zhang ZF, Zhou SY, Eurwilaichitr L, Ingsriswang S, Raza M, Chen Q, Zhao P, Liu F, Cai L (2021) Culturable mycobiota from Karst caves in China II with descriptions of 33 new species. Fungal Divers 106:29–136. 10.1007/s13225-020-00453-7
- 98.Crous PW, Luangsa-Ard JJ, Wingfield MJ et al (2018) Fungal Planet description sheets: 785–867. Persoonia 41:238–417. 10.3767/persoonia.2018.41.12 [DOI] [PMC free article] [PubMed]
- 99.Lorch JM, Palmer JM, Vanderwolf KJ, Schmidt KZ, Verant ML, Weller TJ, Blehert DS (2018) Malassezia vespertilionis sp. nov.: a new cold-tolerant species of yeast isolated from bats. Persoonia 41:56–70. 10.3767/persoonia.2018.41.04 [DOI] [PMC free article] [PubMed]
- 100.Valenzuela-Lopez N, Cano-Lira JF, Guarro J, Sutton DA, Wiederhold N, Crous PW, Stchigel AM (2018) Coelomycetous Dothideomycetes with emphasis on the families Cucurbitariaceae and Didymellaceae. Stud Mycol 90:1–69. 10.1016/j.simyco.2017.11.003 [DOI] [PMC free article] [PubMed]
- 101.Marin-Felix Y, Hernández-Restrepo M, Iturrieta-González I, García D, Gené J, Groenewald JZ, Crous PW (2019) Genera of phytopathogenic fungi: GOPHY 3. Stud Mycol 94:1–124. 10.1016/j.simyco.2019.05.001 [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
DNA sequences are available in GenBank and alignments were deposited in TreeBASE.