Abstract
Canine distemper virus (CDV) and canine circovirus (CanineCV) have been described worldwide in multi-systemic disease in dogs. Both agents may be occasionally associated with other viral pathogens, but reports of coinfection by CDV and CanineCV associated with disease are rare. In this article, we report a coinfection between CDV and CanineCV detected during an investigation of viral agents involved in multisystemic disease in dogs in Southern Brazil. Molecular testing by PCR in lungs and intestines of 77 dogs necropsied in pathology services (2015–2020) revealed several single and mixed viral infections, including a CDV/CanineCV coinfection. In the case reported here, gross and histological findings were compatible with CDV pathology (bronchointerstitial pneumonia and viral intracytoplasmatic inclusions in pneumocytes and transitional epithelial cells of urinary bladder). CanineCV DNA and CDV antigens were detected in lung and intestine fragments by PCR and immunohistochemistry, respectively. CanineCV PCR amplicons subjected to nucleotide sequencing showed > 98.6% nucleotide identity with CanineCV sequences from GenBank. Although the role of CanineCV in the pathogenesis of the reported case could not be determined, our results show that CanineCV may be associated with other viral infections in cases of multisystemic disease in dogs. These results reinforce the circulation of CanineCV in dogs in Brazil and highlight the importance of including this virus in the list of differential diagnoses of respiratory and gastroenteric infectious diseases in dogs.
Keywords: CDV, CanineCV, Canine, Viral infection, Diagnosis
Introduction
Canine distemper virus (CDV) is a non-enveloped, single-stranded negative-sense, RNA virus classified in Paramyxoviridae family, responsible for several clinical manifestations in dogs, varying from subclinical to fatal multi-systemic disease, including respiratory, gastroenteric, and neurological disorders [1]. Canine circovirus (CanineCV) is a non-enveloped, single-stranded circular DNA virus belonging to the family Circoviridae. CanineCV has been identified in healthy dogs and wild carnivores and also in animals presenting severe hemorrhagic gastroenteritis, lymphadenitis, vasculitis, and respiratory signs [1, 2]. Although CanineCV virulence and pathogenesis remains unclear, some reports suggested its potential role in worsening the severity of the disease caused by other agents [1, 3–5].
The pathogenic potential of CDV is widely known, and the virus is considered one of the main pathogens of dogs [6]. CanineCV has been associated to disease in dogs, and, in some cases, it has been detected in association with other agents [2, 5], but its role in mixed infections is still poorly known [3, 7]. Thus, the involvement of CanineCV in the pathogenesis of single or multiple infections in dogs is undetermined. However, coinfection may be important for the expression of some circovirus-associated syndromes, as in porcine circovirus associated disease [8].
Thus, a possible role of CanineCV in single and mixed infection remains to be investigated. CDV or CanineCV have been described in dogs [9, 10] in single and mixed infections [3, 5, 11–13]. However, to the best of our knowledge, no reports of CDV and CanineCV coinfections in domestic dogs from South America have been published to date. Thus, we herein report a CDV/CanineCV coinfection in a case of fatal disease in dog in Southern Brazil and describe the pathologic features of such coinfection.
Materials and methods
Seventy-six small intestinal fragments, two lung, one liver, and one spleen samples of 77 dogs that presented pathological lesions compatible with viral infections were submitted to molecular investigation searching for canine parvovirus (CPV-2), canine adenovirus (CAdV-1/2), and CanineCV. CDV was investigated when histopathological lesions were suggestive of distemper. The samples were collected between 2015 and 2020, during necropsy in pathology services from Southern Brazil and kept at − 20 °C until processing and molecular analysis.
The tissues were macerated and homogenized with 1 × TE buffer solution (10 mM Tris–HCl, 0.1 mM EDTA) and submitted to DNA extraction following a phenol protocol/chloroform. When CDV infection were suspected, samples were submitted to RNA extraction using TRIzol™ reagent (Thermo Fisher Scientific). Complementary strand of DNA (cDNA) was made using 8 ug of total RNA mixed to the GoScript™ Reverse Transcriptase Kit (Promega, Madison, Wisconsin, EUA), according to the manufacturer’s protocol. Total DNA was submitted to PCR to detect CanineCV, CPV-2, and CAdV-2, and cDNA was used in RT-PCR to CDV, using primers and protocols described by Kotsias et al. (2019) [11], Buonavoglia et al. (2001) [14], Hu et al. (2001) [15], and Wang et al. (2011) [16], respectively. All reactions were performed using a total volume of 25 μL, containing 2 μL of total DNA/cDNA (100–200 ng), 1 μL of each primer, 1.25 mM of MgCl2, 2.5 mM 10 × Taq buffer, 1 U of Taq DNA polymerase (0.2 μL) (Thermo Fisher Scientific®), 20 μM deoxyribonucleotides (dNTPs), and ultrapure water (q.s.p.). PCR positive samples were purified and submitted to nucleotide sequencing. Amplicons were sequenced in duplicates on an automatized sequencer. The Staden Package was used to obtaining the consensus sequence [17]. Nucleotide alignments were obtained in BioEdit 7.2.5 software [18], and phylogenetic tree were built in MEGA X [19].
Histopathology analysis was performed in tissue fragments fixed in 10% buffered formalin for 24 to 48 h. After processing, paraffin-embedded samples were cut in 3-μm-thick sections and stained with hematoxylin and eosin (H&E). Considering the possibility of RNA degradation due to − 20 °C storage, tissues fragments were submitted to immunohistochemistry (IHC) using a monoclonal antibody specific to nucleoprotein of CDV and a polymer-HRP system developed with 3,3′-diaminobenzidine (DAB), according to Areco et al. (2021) [20].
Results
Viral molecular investigation in samples of 77 dogs necropsied in pathology services (2015–2020) revealed several single and mixed infections (not showed), including a CDV/CanineCV co-infection. CanineCV DNA was detected in fragments of lungs and intestine from one dog (LDP 104/20, lung; and LDP 106/20, intestine). These samples were obtained from a 3-month-old male French Bulldog that had pathological lesions compatible with CDV-induced disease, as a multifocal neutrophilic and histiocytic bronchointerstitial pneumonia (Fig. 1a, b) with intracytoplasmatic viral inclusions in pneumocytes and transitional epithelial cells of urinary bladder.
Fig. 1.

Lung and intestine of a dog coinfected with canine distemper virus (CDV) and canine circovirus (CanineCV). a Multifocal red areas within the lungs are observed. The lungs did not collapse when the thoracic cavity was opened. b Histologically, there is a multifocal neutrophilic and histiocytic bronchointerstitial pneumonia (stained by hematoxylin and eosin, H&E). c The intestines show crypt dilation and necrosis (H&E)
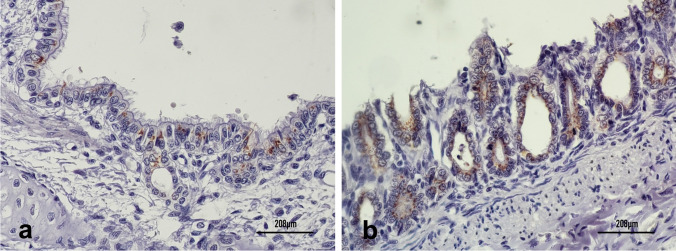
Interestingly, these tissues were CDV-negative in a RT-PCR. Considering that the clinical and pathological features were suggestive of CDV infection, lung and intestine fragments were submitted to IHC. CDV antigen expression was observed in the cytoplasm of ciliated bronchial cells and enterocytes, clearly demonstrating virus replication in these cells (Fig. 2a, b). It is possible that the lack of CDV detection by RT-PCR was due to RNA degradation, since the samples were stored at − 20 °C for a long time, or possible low sensitivity/restricted coverage of the used primers.
Fig. 2.
Immunohistochemistry (IHC) of lung and intestine samples of a dog coinfected with canine distemper virus (CDV) and canine circovirus (CanineCV) using a CDV-specific monoclonal antibody and a polymer-HRP system developed with 3,3′-diaminobenzidine (DAB). Bronchial ciliated epithelial cells (a) and enterocytes (b) show CDV antigen expression (brown labeling)
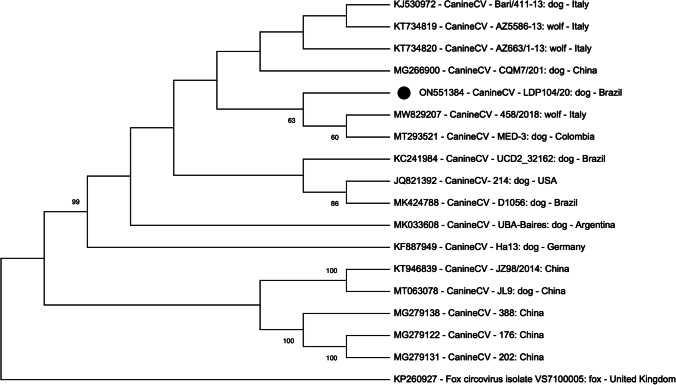
CanineCV PCR amplicons were submitted to nucleotide sequencing, and the consensus was analyzed by Basic Local Alignment Search Tool (BLAST). LDP 104/20 (lung) and LDP 106/20 (intestine) sequences (GenBank accession numbers ON551384 and ON551385) showed 100% of nucleotide identity, and presented > 98.6% of identity with CanineCV sequences deposited in GenBank (accession numbers MW829207.1; MW829206.1; MW829205.1; MW829204.1; MW829202.1, and MW829201.1). At phylogenetic tree, CanineCV sequence of this study clustered with CanineCV sequences obtained from wild and domestic animals from Italy, Colombia and China (Fig. 3).
Fig. 3.
Phylogenetic tree based on the partial nucleotide sequence of the CanineCV LDP 104/20 (positions 1 to 530). The tree was constructed with the Maximum Likelihood method (MEGA X) using the GTR + G as substitution model. The bootstrap of 1,000 replicates was used in the tree (bootstrap showed > 60%). The CanineCV sequence obtained in this study (ON551384) is highlighted as a black circle
Discussion
The present study reported a case of coinfection by CDV and CanineCV in a dog from Rio Grande do Sul, Brazil, confirmed by virus detection in the lungs and gut fragments using PCR, histological examination and/or IHC. CDV is a well-known agent associated with viral infections in puppies worldwide [21], but the potential pathogenic role of CanineCV in enteric and respiratory disease of dogs is still a matter of debate. Some reports have associated CanineCV with severe disease in dogs in single or mixed infections [4, 5, 22], whereas others showed no correlation [2].
CanineCV coinfections with other viral agents in domestic and wild carnivores presenting diarrhea and fatal gastroenteritis have been described before [1, 3, 5]. However, although this virus was already associated with systemic disease [1], investigations showed its involvement in respiratory disease [23]. In our study, CanineCV was detected in intestinal and lung tissues, both positive for CDV. Circovirus DNA detection does not suffice to confirm CanineCV involvement in pulmonary and intestinal lesions, yet it shows its systemic dissemination, since it was detected in both systems. Similar to what has been described to porcine circovirus 2, CanineCV might be potentialized upon replication of other agents in the same tissue/organ [5]. Then, although histological lesions observed in the dog of this study were compatible with CDV infection, it cannot be ruled out that they are the result of the replication of both viruses. IHC using CanineCV-specific monoclonal antibodies could help elucidate this hypothesis, but unfortunately, it was not performed due to the lack of specific reagents.
CanineCV and CDV coinfection has been reported in feces of domestic and wild carnivores in Italy, associated or not with diarrhea [2], but this association has not yet been described in dogs from South America. It is possible that CanineCV detected in samples of the dog in this study potentialized the CDV systemic infection, resulting in fatal lung and intestinal lesions. However, the CanineCV pathogenic role in CDV associated disease needs be more investigated, aiming to discard a circumstantial DNA detection without pathogenic implications and/or to confirm its involvement in the pathogenesis of this mixed infection.
In Brazil, there are few reports of CanineCV infection in dogs. The virus has been detected in a dog presenting intermittent hemorrhagic gastroenteritis [9] and in the serum of asymptomatic dog submitted to virome examination [24]. Mixed infections with other viruses have not been reported. In this study, we analyzed around 80 samples from 77 dogs, collected between 2015 and 2020, and only one animal was positive to CanineCV. Although CanineCV circulation in Brazil is largely unknown, the continuous search for this virus in canine samples is important to determine the real virus prevalence and its role in infectious diseases of dogs.
Thus, our report showed the occurrence of CanineCV and CDV coinfection in a dog; confirms the circulation of CanineCV in dogs from Brazil and shows that CanineCV may be present in lung samples—in addition to gut tissues—highlighting the need of its inclusion in differential diagnosis of respiratory disease of dogs.
Author contribution
CRediT taxonomy: Conceptualization: Ana Paula Gnocato Mortari, Fernanda Silveira Flôres Vogel, Juliana Felipetto Cargnelutti. Data curation: Ana Paula Gnocato Mortari, Eduardo Furtado Flores, Eduardo Kenji Masuda, Fernanda Silveira Flôres Vogel, Juliana Felipetto Cargnelutti. Formal analysis and investigation: Ana Paula Gnocato Mortari, Fernanda Silveira Flôres Vogel, Juliana Felipetto Cargnelutti. Methodology: Ana Paula Gnocato Mortari, Eduardo Kenji Masuda, Mariana Martins Flores. Writing—original draft preparation: Ana Paula Gnocato Mortari, Fernanda Silveira Flôres Vogel. Writing—review and editing: Ana Paula Gnocato Mortari, Eduardo Furtado Flores, Eduardo Kenji Masuda, Fernanda Silveira Flôres Vogel, Juliana Felipetto Cargnelutti, Mariana Martins Flores. Resources: Eduardo Furtado Flores, Eduardo Kenji Masuda, Mariana Martins Flores. Supervision: Fernanda Silveira Flôres Vogel.
Funding
This study was founded in part by the Coordenação de Aperfeico̧amento de Pessoal de Nível Superior (CAPES) – Finance Code 001, and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq). APGM, EFF, and FSFV are recipients of CNPq fellowships.
Availability of data and material
Not applicable.
Declarations
Ethics approval
No approval of research ethics committees was required to accomplish the goals of this study because.
experimental work was conducted with samples received for diagnosis.
Consent to participate
Not applicable.
Consent of publication
Not applicable.
Conflict of interest
The authors declare no competing interests.
Footnotes
Responsible Editor: Fernando R. Spilki
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Ana Paula G. Mortari, Email: ana-paula.gnocato@acad.ufsm.br
Eduardo K. Masuda, Email: masuda@axysanalises.com.br
Mariana M. Flores, Email: mariana.flores@ufsm.br
Eduardo F. Flores, Email: eduardofurtadoflores@gmail.com
Juliana F. Cargnelutti, Email: juliana.cargnelutti@ufsm.br
Fernanda S. F. Vogel, Email: fernanda.vogel@ufsm.br
References
- 1.Li L, McGraw S, Zhu K, et al. Circovirus in tissues of dogs with vasculitis and hemorrhage. Emerg Infect Dis. 2013;19(4):534–541. doi: 10.3201/eid1904.121390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zaccaria G, Malatesta D, Scipioni G, et al. Circovirus in domestic and wild carnivores: an important opportunistic agent? Virology. 2016;490:69–74. doi: 10.1016/j.virol.2016.01.007. [DOI] [PubMed] [Google Scholar]
- 3.Dowgier G, Lorusso E, Decaro N, et al. A molecular survey for selected viral enteropathogens revealed a limited role of Canine circovirus in the development of canine acute gastroenteritis. Vet Microbiol. 2017;204:54–58. doi: 10.1016/j.vetmic.2017.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson A, Hartmann K, Leutenegger CM, Proksch AL, Mueller RS, Unterer S. Role of canine circovirus in dogs with acute haemorrhagic diarrhoea. Vet Rec. 2017;180(22):542–542. doi: 10.1136/vr.103926. [DOI] [PubMed] [Google Scholar]
- 5.Thaiwong T, Wise AG, Maes RK, Mullaney T, Kiupel M. Canine circovirus 1 (CaCV-1) and canine parvovirus 2 (CPV-2): recurrent dual infections in a papillon breeding colony. Vet Pathol. 2016;53(6):1204–1209. doi: 10.1177/0300985816646430. [DOI] [PubMed] [Google Scholar]
- 6.Martinez-Gutierrez M, Ruiz-Saenz J. Diversity of susceptible hosts in canine distemper virus infection: a systematic review and data synthesis. BMC Vet Res. 2016;12(1):1–11. doi: 10.1186/S12917-016-0702-Z/FIGURES/2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Balboni A, Terrusi A, Urbani L, et al. Canine circovirus and canine adenovirus type 1 and 2 in dogs with parvoviral enteritis. Vet Res Commun. 2022;46(1):223–232. doi: 10.1007/S11259-021-09850-Y/FIGURES/2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Opriessnig T, Halbur PG. Concurrent infections are important for expression of porcine circovirus associated disease. Virus Res. 2012;164(1):20. doi: 10.1016/J.VIRUSRES.2011.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cruz TF, Batista TN, Vieira EM et al (2020) Genomic characterization of canine circovirus detected in a dog with intermittent hemorrhagic gastroenteritis in Brazil. Ciênc Rural 50(5). 10.1590/0103-8478cr20190909
- 10.Caddy SL. New viruses associated with canine gastroenteritis. Vet J. 2018;232:57–64. doi: 10.1016/j.tvjl.2017.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kotsias F, Bucafusco D, Nuñez DA, Lago Borisovsky LA, Rodriguez M, Bratanich AC. Genomic characterization of canine circovirus associated with fatal disease in dogs in South America. Schang LM, ed. PLoS One. 2019;14(6):e0218735. doi: 10.1371/journal.pone.0218735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Monteiro FL, Cargnelutti JF, Martins M, et al. Detection of respiratory viruses in shelter dogs maintained under varying environmental conditions. Braz J Microbiol. 2016;47(4):876–881. doi: 10.1016/j.bjm.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Headley SA, Alfieri AA, Fritzen JTT, et al. Concomitant canine distemper, infectious canine hepatitis, canine parvoviral enteritis, canine infectious tracheobronchitis, and toxoplasmosis in a puppy. J Vet Diagn Investig. 2013;25(1):129–135. doi: 10.1177/1040638712471344. [DOI] [PubMed] [Google Scholar]
- 14.Buonavoglia C, Martella V, Pratella A, et al. Evidence for evolution of canine parvovirus type 2 in Italy. J Gen Virol. 2001;82(12):3021–3025. doi: 10.1099/0022-1317-82-12-3021. [DOI] [PubMed] [Google Scholar]
- 15.Hu RL, Huang G, Qiu W, Zhong ZH, Xia XZ, Yin Z. Detection and differentiation of CAV-1 and CAV-2 by polymerase chain reaction. Vet Res Commun. 2001;25(1):77–84. doi: 10.1023/a:1006417203856. [DOI] [PubMed] [Google Scholar]
- 16.Wang F, Yan X, Chai X, et al. Differentiation of canine distemper virus isolates in fur animals from various vaccine strains by reverse transcription-polymerase chain reaction-restriction fragment length polymorphism according to phylogenetic relations in china. Virol J. 2011;8(1):85. doi: 10.1186/1743-422X-8-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Staden R. The Staden sequence analysis package. Mol Biotechnol. 1996;5(3):233–241. doi: 10.1007/BF02900361. [DOI] [PubMed] [Google Scholar]
- 18.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 19.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35(6):1547. doi: 10.1093/MOLBEV/MSY096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Areco WVC, Tondo LAS, de Avila NC, et al. Histopathological features of spinal cord lesions in dogs with distemper-associated demyelinating leucoencephalomyelitis. J Comp Pathol. 2021;189:110–119. doi: 10.1016/J.JCPA.2021.10.006. [DOI] [PubMed] [Google Scholar]
- 21.Beineke A, Puff C, Seehusen F, Baumgärtner W. Pathogenesis and immunopathology of systemic and nervous canine distemper. Vet Immunol Immunopathol. 2009;127(1–2):1–18. doi: 10.1016/j.vetimm.2008.09.023. [DOI] [PubMed] [Google Scholar]
- 22.Hsu H-S, Lin T-H, Wu H-Y, et al. High detection rate of dog circovirus in diarrheal dogs. BMC Vet Res. 2016;12(1):116. doi: 10.1186/s12917-016-0722-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Altan E, Seguin MA, Leutenegger CM, Phan TG, Deng X, Delwart E. Nasal virome of dogs with respiratory infection signs include novel taupapillomaviruses. Virus Genes. 2019;55(2):191. doi: 10.1007/S11262-019-01634-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weber MN, Cibulski SP, Olegário JC, et al. Characterization of dog serum virome from Northeastern Brazil. Virology. 2018;525:192–199. doi: 10.1016/j.virol.2018.09.023. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.