Abstract
Lactoferrin (LF) is a glycoprotein that exerts both bacteriostatic and bactericidal activities. The interaction of LF with lipopolysaccharide (LPS) of gram-negative bacteria seems to play a crucial role in the bactericidal effect. In this study, we evaluated, by means of an enzyme-linked immunosorbent assay, the binding of biotinylated LF to the S (smooth) and R (rough) (Ra, Rb, Rc, Rd1, Rd2, and Re) forms of LPS and different lipid A preparations. In addition, the effects of two monoclonal antibodies (AGM 10.14, an immunoglobulin G1 [IgG1] antibody, and AGM 2.29, an IgG2b antibody), directed against spatially distant epitopes of human LF, on the LF-lipid A or LF-LPS interaction were evaluated. The results showed that biotinylated LF specifically binds to solid-phase lipid A, as this interaction was prevented in a dose-dependent fashion by either soluble uncoupled LF or lipid A. The binding of LF to S-form LPS was markedly weaker than that to lipid A. Moreover, the rate of LF binding to R-form LPS was inversely related to core length. The results suggest that the polysaccharide O chain as well as oligosaccharide core structures may interfere with the LF-lipid A interaction. In addition, we found that soluble lipid A also inhibited LF binding to immobilized LPS, demonstrating that, in the whole LPS structure, the lipid A region contains the major determinant recognized by LF. AGM 10.14 inhibited LF binding to lipid A and LPS in a dose-dependent fashion, indicating that this monoclonal antibody recognizes an epitope involved in the binding of LF to lipid A or some epitope in its close vicinity. In contrast, AGM 2.29, even in a molar excess, did not prevent the binding of LF to lipid A or LPS. Therefore, AGM 10.14 may represent a useful tool for neutralizing selectively the binding of LF to lipid A. In addition, the use of such a monoclonal antibody could allow better elucidation of the consequences of the LF-lipid A interaction.
Lactoferrin (LF) is an iron-binding glycoprotein of ∼77 kDa and present in high levels in milk, tears, saliva, and other secretions (28, 32). It is also a constituent of specific granules of neutrophil granulocytes (PMN), from which it is released following PMN activation (6, 21). Several biological functions of LF have been demonstrated for host defense, mostly at mucosal surfaces (for a review, see reference 28). In addition, LF modulates inflammatory and immune responses and may act as a multifunctional immunoregulatory protein (8). Thus, LF decreases the release of interleukin (IL)-1, IL-2, and tumor necrosis factor alpha by endotoxin-stimulated mononuclear cells and enhances monocyte cytotoxicity and natural killer cell activity (10, 19, 20, 22, 29, 36).
LF exerts both a bacteriostatic effect, through its ability to sequester iron, and direct bactericidal activity, which is independent of the nutritional deprivation of iron. An N-terminal domain, the so-called lactoferricin, distinct from the iron-binding sites and isolated following pepsin cleavage of human LF (hLF) and bovine LF, is responsible for the bactericidal activity (3–5, 7, 30). In particular, it has been documented that the sequences showing antibacterial activity are located in a loop region corresponding to residues 20 to 37 of hLF and 19 to 36 of bovine LF (7).
LF causes the release of lipopolysaccharide (LPS) molecules from bacterial cells, thus damaging the outer membrane of gram-negative bacteria (13). Therefore, the binding of LF to LPS of gram-negative bacteria seems to play a crucial role in its bactericidal activity. In this respect, Appelmelk et al. (2) demonstrated that hLF specifically reacted with various types of lipid A isolated from clinically relevant serotypes of the species which most frequently cause bacteremia; they concluded that lipid A likely represents the major determinant of the whole LPS molecule recognized by LF. More recently, the involvement of a loop region (residues 28 to 34 of the N-terminal domain) of hLF in high-affinity binding to LPS was reported (11). Furthermore, synthetic peptides homologous to a loop region in hLF have been shown to possess antibacterial activity (25). It is noteworthy that Wang et al. have shown that PMN can inactivate LPS, the inactivation being primarily due to LF secreted by these cells (34).
We recently produced and characterized two murine monoclonal antibodies (MAbs) (AGM 10.14, an immunoglobulin G1 [IgG1] antibody, and AGM 2.29, an IgG2b antibody), directed against two spatially distant epitopes of hLF (1, 9). The objectives of this study were to analyze in vitro the binding of hLF to lipid A and to different smooth (S)- and rough (R)-form LPSs with different degrees of core depletion and to evaluate the potential neutralizing effect of anti-hLF MAb AGM 10.14 or AGM 2.29 on the hLF-lipid A or hLF-LPS interaction.
MATERIALS AND METHODS
Reagents.
RPMI 1640 was purchased from HyClone Europe Ltd., Cramlington, United Kingdom. Fetal calf serum was supplied by GIBCO, Eggenstein, Germany. hLF (purified from human milk; cod. L 0520), l-glutamine, streptomycin, penicillin, ammonium sulfate, caprylic acid, biotin-N-hydroxysuccinimide ester, dimethyl sulfoxide, horseradish peroxidase-coupled avidin, bovine serum albumin (BSA), casein from bovine milk, 2,6,10,14-tetramethylpentadecane (Pristane), o-phenylenediamine, merthiolate, triethylamine, and Tween 20 were obtained from Sigma Chemical Co., St. Louis, Mo.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (17) showed the purity of hLF, which migrated as an approximately 80-kDa band. LPS contamination of reagents and buffer solutions was less than 50 pg/ml, as estimated by a Limulus amoebocyte lysate assay (Chromogenix AB, Mölndal, Sweden).
LPS and lipid A preparations.
S-form LPSs were purified from Salmonella typhi, Bacteroides fragilis, Salmonella typhimurium, Salmonella abortus-equi, and Escherichia coli by a phenol-water extraction method (35). R-form LPSs, isolated from E. coli EH 100 (Ra chemotype), E. coli F 515 (Re chemotype), and R mutants of Salmonella minnesota with increasing core lengths, i.e., R 60 (Ra), R 345 (Rb), R 5 (Rc), R 7 (Rd1), R 3 (Rd2), and R 595 (Re), were prepared by the phenol-chloroform-petroleum ether procedure (14). The LPS preparations contained less than 0.2% protein, as determined by the Lowry procedure, and no detectable nucleid acid (absorbance at 260 nm). Lipid A from E. coli F 515 was prepared by hydrolysis of E. coli F 515 LPS in 1% acetic acid at 100°C for 2 h (15). The resulting lipid A precipitate was obtained by centrifugation at 3,000 × g (4°C for 30 min), washed three times with distilled water, and lyophilized. LPS and lipid A were solubilized by sonication and the addition of triethylamine to pH 7.5. Lipid A from S. typhimurium SH 9013 (R form) as well as lipid A from Helicobacter pylori NCTC 11637 (R form) were kind gifts from A. P. Moran, Department of Microbiology, National University of Ireland, Galway, Galway, Ireland.
Cell culture.
Hybridoma cells secreting murine anti-hLF MAbs AGM 10.14 (IgG1) and AGM 2.29 (IgG2b) were maintained in RPMI 1640 supplemented with 10% fetal calf serum, 2 mM l-glutamine, streptomycin (50 μg/ml), and penicillin (50 U/ml) (complete medium).
Production and purification of MAbs.
Ascitic fluid was produced by injecting 2 × 106 MAb-producing cells into Pristane-primed BALB/c mice. MAbs were purified from ascitic fluid by sequential precipitation with caprylic acid and 45% ammonium sulfate as previously described (31). The human CD4 internal antigen anti-idiotypic MAb 16 D7 (IgG1) (26), used as a negative control, was kindly provided by Federico Perosa, Department of Biomedical Sciences and Human Oncology, University of Bari, Bari, Italy. The purity of the MAbs was assessed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
Coupling of hLF to biotin.
One milliliter of hLF (1 mg/ml) was dialyzed against 0.1 M NaHCO3. Biotin-N-hydroxysuccinimide ester was dissolved in dimethyl sulfoxide (1 mg/ml), and 200 μl was added to 1 ml of dialyzed hLF. After 4 h of incubation at 25°C with occasional shaking, the mixture was extensively dialyzed at 4°C against phosphate-buffered saline (PBS) (pH 7.4).
Binding of hLF to lipid A and LPS.
The binding of hLF to lipid A and LPS was evaluated by means of an enzyme-linked immunosorbent assay (ELISA) with biotinylated hLF (bhLF). Lipid A or LPS suspensions were sonicated, diluted to 5 μg/ml in pyrogen-free PBS, and transferred (100 μl/well) to 96-well flat-bottom polyvinyl chloride plates (Falcon Micro Test III; Becton Dickinson, Oxnard, Calif.). After overnight incubation at 4°C, the plates were washed three times with PBS containing 0.05% (vol/vol) Tween 20 and 0.01% (wt/vol) merthiolate (PBST) and saturated with 200 μl of 1% (wt/vol) casein in PBS per well for 2 h at 25°C. The saturating solution was discarded, and the plates were washed three times with PBST. One hundred microliters of different concentrations of bhLF (depending on the experiment) was added to each well and incubated for 1 h at 25°C. After the plates were washed, 100 μl of 25 ng of horseradish peroxidase-coupled avidin was added to each well and incubated for 45 min. The plates were thoroughly washed and incubated with 100 μl of a freshly prepared solution of o-phenylenediamine (0.5 mg/ml) and hydrogen peroxide (0.015%) in citrate-phosphate buffer (pH 5) per well. After 30 min of incubation in the dark, the colorimetric reaction was stopped by adding 50 μl of 1 M sulfuric acid per well, and the absorbance at 492 nm was read with a Multiskan plate reader (Labsystem, Helsinki, Finland). The magnitude of binding was expressed as optical density (OD) units. All determinations were done in duplicate. Negative controls (nonspecific binding) for each plate included wells from which the coating antigen was omitted as well as lipid A- or LPS-coated wells incubated with PBST instead of bhLF. As a positive control, four wells on each plate were coated with 100 μl of a 5-μg/ml solution of anti-hLF MAb AGM 10.14 or AGM 2.29 per well. This positive control also ensured that the biotinylation of hLF did not affect its interaction with MAbs.
All ELISA determinations were done in duplicate with the same batch of reagents throughout the study.
Inhibition experiments.
To evaluate either the specificity of the interaction between hLF and lipid A or LPS or the effect of anti-hLF MAbs on hLF binding to lipid A or LPS, inhibition experiments were performed.
Cross-blocking of bhLF binding to solid-phase lipid A by uncoupled hLF.
E. coli F 515 lipid A-coated wells (5 μg/ml; 100 μl/well) were incubated with 100 μl of increasing concentrations of uncoupled hLF (twofold increments, final concentrations ranging from 0.156 to 20 μg/ml) or negative control antigen (BSA) for 1 h at 25°C. Thirty nanograms of bhLF in 50 μl of PBST was added without removal of the competitor. The experiment was then performed as for the binding assay. Wells incubated with diluent buffer instead of uncoupled hLF served as positive controls (100% binding).
Inhibition of bhLF binding to solid-phase lipid A by soluble lipid A or anti-hLF MAbs.
A fixed amount (final concentration, 300 ng/ml) of bhLF was mixed with increasing concentrations of soluble E. coli F 515 lipid A (fourfold dilutions, final concentrations ranging from 0.0012 to 20 μg/ml) or anti-hLF MAbs (twofold dilutions, final concentrations ranging from 0.02 to 2.5 μg/ml) and incubated for 1 h at 25°C. One hundred microliters of this mixture was transferred to E. coli F 515 lipid A-coated wells and incubated for 1 h. bhLF preincubated with diluent buffer served as a positive control (100% binding). The experiment was then carried out as for the binding assay. BSA or MAb 16 D7 was used as the antigen control, respectively.
RESULTS
Preliminary experiments were carried out to evaluate whether the biotinylation of hLF affected the hLF–anti-hLF MAb interaction. To this end, wells of ELISA plates were coated with MAb AGM 10.14 or AGM 2.29 and reacted with bhLF. Strong reactivity was demonstrated for both MAbs (OD, between 2.7 and 2.9), ensuring that the biotinylation procedure did not affect the binding of the MAbs to bhLF.
Binding of bhLF to LPS and lipid A.
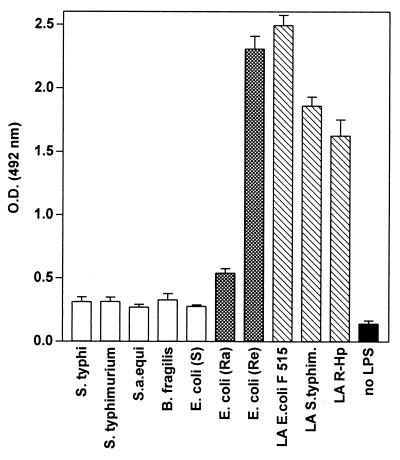
The binding of bhLF to various LPSs and lipid A purified from three different R-form LPSs is illustrated in Fig. 1. The degree of binding to all S-form LPSs and LPS from E. coli EH 100 (Ra mutant) was markedly lower than that observed with lipid A preparations as well as with LPS from E. coli F 515 (Re mutant).
FIG. 1.
Binding of bhLF to different forms of LPS or lipid A (LA). Each bar is representative of the mean OD ± standard deviation for three separate experiments. bhLF was used at a concentration of 300 ng/ml (100 μl/well). Nonspecific binding was assessed by adding bhLF to wells from which the coating antigen was omitted (black bar). S.a. equi, S. abortus-equi; S. typhim., S. typhimurium; R-Hp, H. pylori R-form.
Prolongation of the length of the bhLF incubation did not result in a significant increase in binding to lipid A but rather enhanced the degree of nonspecific binding (data not shown). Indeed, after 3 h of incubation, the magnitude of binding to lipid A increased no more than 10%, while the OD detected in wells lacking coating antigens increased 50%. Therefore, a 1-h incubation time was used throughout the study.
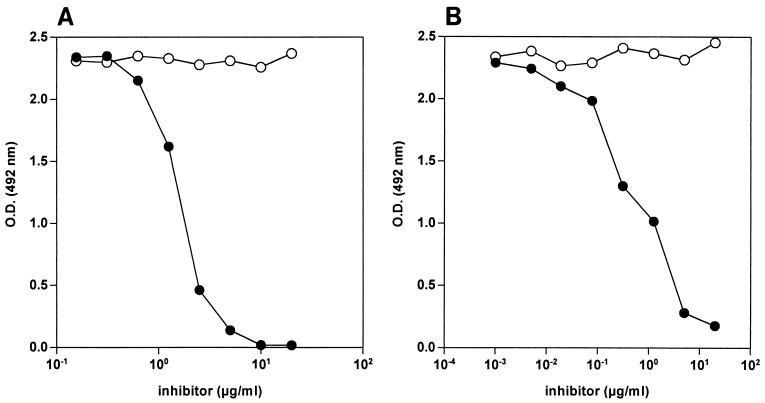
The specificity of the bhLF-lipid A interaction was checked in cross-blocking and inhibition experiments. As shown in Fig. 2A, the binding of bhLF to solid-phase lipid A was inhibited in a dose-dependent fashion by uncoupled hLF. Moreover, preincubation of bhLF with soluble lipid A inhibited in a dose-dependent fashion the bhLF–solid-phase lipid A interaction (Fig. 2B). These results demonstrate the specificity of the bhLF-lipid A interaction.
FIG. 2.
(A) Cross-blocking of bhLF binding to solid-phase lipid A by uncoupled hLF. E. coli F 515 lipid A-coated wells were preincubated for 1 h with 100 μl of increasing concentrations (twofold increments) of uncoupled hLF (solid circles) or BSA (negative control; open circles) per well. Without removal of the competitor, bhLF (30 ng/well) was added, and binding was evaluated as reported in Materials and Methods. Nonspecific binding was subtracted. Data are representative results obtained in duplicate in three independent experiments. (B) Inhibition of bhLF binding to solid-phase lipid A by soluble lipid A. A constant amount of bhLF (final concentration, 300 ng/ml) was mixed with increasing concentrations (fourfold increments) of soluble E. coli F 515 lipid A (solid circles) or BSA (negative control; open circles) and incubated for 1 h. Then, 100 μl of this mixture was added to E. coli F 515 lipid A-coated wells, and binding was evaluated as reported in Materials and Methods. Nonspecific binding was subtracted. Data are representative results obtained in duplicate in three independent experiments.
Preincubation of bhLF with soluble lipid A also inhibited the binding of bhLF to different forms of solid-phase LPS (Table 1), thus suggesting that lipid A represents the main structure recognized by hLF in the intact LPS molecule.
TABLE 1.
Inhibition of bhLF binding to solid-phase lipid A and LPS after preincubation with soluble lipid A, BSA, and MAbs AGM 10.14, AGM 2.29, and 16 D7
| Molecule | OD at 492 nm after preincubation with the following inhibitor at 2.5 μg/ml:
|
|||||
|---|---|---|---|---|---|---|
| Diluent buffera | Lipid Ab | BSA | AGM 10.14 | AGM 2.29 | 16 D7 | |
| Lipid A | 2.512 | 0.427 | 2.499 | 0.007 | 2.581 | 2.470 |
| E. coli Ra LPS | 0.560 | 0.055 | 0.547 | 0.012 | 0.613 | 0.591 |
| E. coli Re LPS | 2.260 | 0.475 | 2.222 | 0.022 | 2.315 | 2.198 |
| S. minnesota Ra LPS | 0.374 | 0.087 | 0.407 | 0.002 | 0.364 | 0.399 |
| S. minnesota Rb LPS | 0.382 | 0.053 | 0.354 | 0.018 | 0.421 | 0.402 |
| S. minnesota Rc LPS | 0.437 | 0.089 | 0.415 | 0.005 | 0.470 | 0.451 |
| S. minnesota Rd1 LPS | 0.712 | 0.178 | 0.687 | 0.037 | 0.743 | 0.698 |
| S. minnesota Rd2 LPS | 1.970 | 0.256 | 2.001 | 0.027 | 2.064 | 1.952 |
| S. minnesota Re LPS | 2.198 | 0.328 | 2.144 | 0.034 | 2.215 | 2.067 |
| S. typhi S LPS | 0.311 | 0.047 | 0.322 | 0.012 | 0.341 | 0.299 |
Preincubation of bhLF (final concentration, 300 ng/ml) with diluent buffer served as a positive control (100% binding).
E. coli F 515 lipid A was used.
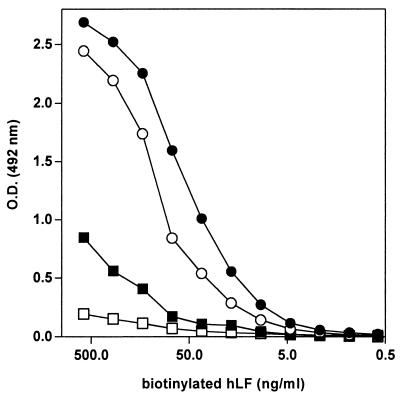
The dose-dependent binding of bhLF to E. coli F 515 lipid A, F 515 LPS (Re), and E. coli EH 100 LPS (Ra) was also evaluated. As shown in Fig. 3, bhLF specifically reacted in a dose-dependent fashion with all these structures, even though maximum binding was found when lipid A was used as a coating antigen. In particular, the magnitude of the bhLF interaction with lipid A was more than threefold higher than that achieved with Ra LPS, while the reactivity against Re LPS was slightly lower than that achieved with lipid A.
FIG. 3.
Comparative dose-dependent binding of bhLF to lipid A, Re LPS, and Ra LPS. One hundred microliters of bhLF (twofold dilutions) was added to wells coated with E. coli F 515 lipid A (solid circles), E. coli F 515 Re LPS (open circles), or E. coli EH 100 Ra LPS (solid squares). Binding was evaluated as reported in Materials and Methods. Nonspecific binding was assessed by adding bhLF to wells from which the coating antigen was omitted (open squares).
The results described above suggested that the polysaccharide O chain, as well as the oligosaccharide structure of the core, could hamper bhLF binding to lipid A. Therefore, the reactivity of bhLF against LPSs from six R mutants of S. minnesota with increasing core lengths was evaluated. The results demonstrated an inverse relationship between the degree of bhLF binding and core length (Table 1). While the binding intensity slowly increased from Ra LPS to Rd1 LPS, a dramatic enhancement was observed when Rd2 LPS was used as the coating antigen (Table 1).
Effect of anti-hLF MAbs on the hLF-lipid A or hLF-LPS interaction.
In preliminary experiments, we demonstrated that neither AGM 10.14 nor AGM 2.29 reacted with lipid A or LPS preparations (data not shown).
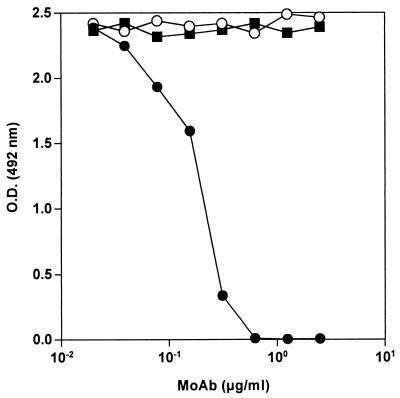
The binding of bhLF to lipid A was inhibited in a dose-dependent fashion by preincubation of bhLF with MAb AGM 10.14 (Fig. 4). At an MAb concentration of 0.625 μg/ml, the reactivity against lipid A was completely prevented. In contrast, no inhibition was found when bhLF was preincubated with MAb AGM 2.29 or control MAb 16 D7, even in a molar excess. Also, bhLF binding to different forms of LPS was completely abrogated by preincubation of bhLF with MAb AGM 10.14, whereas no inhibition was documented when MAb AGM 2.29 or 16 D7 was used as the inhibitor (Table 1).
FIG. 4.
Inhibition of bhLF binding to solid-phase lipid A by anti-hLF MAbs (MoAb). A constant amount of bhLF (final concentration, 300 ng/ml) was mixed with increasing concentrations (twofold increments) of anti-hLF MAb AGM 10.14 (solid circles), anti-hLF MAb AGM 2.29 (open circles), or control MAb 16 D7 (solid squares) and incubated for 1 h. Then, 100 μl of this mixture was added to E. coli F 515 lipid A-coated wells, and binding was evaluated as reported in Materials and Methods. Nonspecific binding was subtracted. Data are representative results obtained in duplicate in three independent experiments.
DISCUSSION
In this study, we clearly demonstrate a strong interaction between hLF and lipid A of various origins. Moreover, preincubation of bhLF with soluble lipid A inhibited bhLF binding to different forms of LPS, indicating that in the LPS molecular structure, lipid A likely represents the main determinant recognized by hLF.
Recently, Appelmelk and coworkers found high-affinity binding of hLF to the lipid A moiety of LPS (2). Moreover, a reduction in the intensity of binding to different LPSs, likely due to sterical hindrance exerted by the polysaccharide O chain and the oligosaccharide core structure, was also reported (2). However, when hLF was reacted with different LPSs from R mutants of S. minnesota, the degree of hLF binding did not follow the exact order of chain lengths (2). In the present study, we found a more evident inverse relationship between the magnitude of the hLF interaction with LPS and core lengths. In this respect, it is possible that the shorter incubation time that we used (1 h in our experiments versus 16 h in those of Appelmelk et al. [2]) allowed for better discrimination of differences in binding intensity.
Our data are consistent with those reported by Naidu et al., who demonstrated low or negligible LF binding to whole cells of S. typhimurium (S form) compared to that obtained with their isogenic R mutants (24). In particular, using a panel of R forms of S. typhimurium, these authors found a magnitude of LF binding to bacteria inversely related to the oligosaccharide core length. Interestingly, a higher level of susceptibility to the antibacterial effects of LF was demonstrated for bacteria with the shortest core, thus indicating that the polysaccharide O chain as well as the core oligosaccharide may protect gram-negative microorganisms from the antibacterial effects of LF (24). Even in some studies on the reactivity of anti-lipid A MAbs with different LPS preparations, an inverse correlation between the degree of anti-lipid A MAb binding to LPS and the stage of completion of the core was reported (23, 27).
Quite interestingly, even though in our investigation bhLF bound very well to the three lipid A preparations used, the degrees of reactivity were different. In a previous study on the epitope specificity of murine MAbs directed against lipid A, it was reported that the acylation pattern of lipid A strongly influenced the intensity of binding of such MAbs, probably by modulating the exposure of lipid A epitopes and/or by affecting the coating efficiency of compounds (16). It is therefore conceivable that in our study, the molecular structure of the lipid A preparations used might account for the differences observed in bhLF binding intensity.
It should be emphasized that our experiments were performed with lipid A and LPS preparations immobilized on polyvinyl chloride surfaces. It is possible that the interaction between LF and soluble LPS is not affected by the O chain or by core structures. In this respect, in a recent study the protective effects of LF feeding against lethal shock in germfree piglets challenged with E. coli O55:B5 LPS were reported (18).
The interaction between LPS and monocytes or macrophages results in the production and release of tumor necrosis factor alpha, IL-1, and IL-6, which play a crucial role in inducing septic shock (33). Thus, besides a bactericidal effect, LF may act by interfering with the access of endotoxin to its cell surface receptor. Indeed, evidence has recently been provided that hLF inhibits the interaction of LPS with CD14 on monocytes or macrophages by competition with the LPS-binding protein, a 60-kDa serum protein which binds to the lipid A portion of LPS, thus mediating the transfer of LPS to CD14 (12).
Therefore, since it is well established that lipid A represents the toxic moiety of endotoxin, it is conceivable that in the interaction between LF and circulating LPS, LF binding to lipid A is crucial for preventing the noxious effect of endotoxins.
Another aim of our study was to evaluate the effect of two anti-hLF MAbs on the LF-lipid A or LF-LPS interaction. We demonstrated that MAb AGM 10.14 was able to inhibit hLF binding to lipid A and LPS in a dose-dependent fashion, indicating that this MAb recognizes the epitope for the hLF-binding site for lipid A or LPS or an epitope closely related to it. To the best of our knowledge, this is the first anti-hLF MAb showing such peculiar activity. The finding that MAb AGM 2.29, even in a molar excess, did not affect the hLF-lipid A interaction is consistent with our previous results showing that this MAb reacts with an hLF epitope spatially distant from that recognized by MAb AGM 10.14 (9).
In most studies of the physiological activities of LF, polyclonal anti-LF antibodies have been used to inhibit LF functions. Polyclonal antisera, however, contain antibodies directed against several epitopes and thus are unable to neutralize selectively the domain involved in the mechanism(s) under study. In this respect, MAb AGM 10.14 may represent a useful tool for inhibiting specifically the binding of hLF to lipid A without affecting other sites putatively involved in different activities. In addition, this MAb may allow evaluation of whether the same LF epitope or epitopes in the vicinity of the lipid A-binding site are involved in interactions with other molecular structures, including LF-binding gram-negative outer membrane proteins or LF surface receptors on different cell types. Such information could contribute to a better understanding of the complex and intriguing patterns of LF functions.
ACKNOWLEDGMENTS
We are grateful to A. P. Moran for providing lipid A from S. typhimurium and from H. pylori and to F. Perosa for providing control MAb 16 D7.
This work was partially supported by grants from AIRC 1997, MURST, and BMBF (grant 01KI94747).
REFERENCES
- 1.Afeltra A, Caccavo D, Ferri G M, Addessi M A, De Rosa F G, Amoroso A, Bonomo L. Expression of lactoferrin on human granulocytes: analysis with polyclonal and monoclonal antibodies. Clin Exp Immunol. 1997;109:279–285. doi: 10.1046/j.1365-2249.1997.4351333.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Appelmelk B J, An Y-Q, Geerts M, Thijs B G, De Boer H A, MacLaren D M, de Graaff J, Nuijens J H. Lactoferrin is a lipid A-binding protein. Infect Immun. 1994;62:2628–2632. doi: 10.1128/iai.62.6.2628-2632.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arnold R R, Cole M F, McGhee J R. A bactericidal role for human lactoferrin. Science. 1977;197:263–265. doi: 10.1126/science.327545. [DOI] [PubMed] [Google Scholar]
- 4.Arnold R R, Russel J E, Champion W J, Gauthier J J. Bactericidal activity of human lactoferrin: influence of physical conditions and metabolic state of the target microorganism. Infect Immun. 1981;32:655–660. doi: 10.1128/iai.32.2.655-660.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arnold R R, Russel J E, Champion W J, Brewer M, Gauthier J J. Bactericidal activity of human lactoferrin: differentiation from the stasis of iron deprivation. Infect Immun. 1982;35:792–797. doi: 10.1128/iai.35.3.792-799.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baggiolini M, De Duve C, Masson P L, Heremans J F. Association of lactoferrin with specific granules in rabbit heterophil leukocytes. J Exp Med. 1970;131:559–570. doi: 10.1084/jem.131.3.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bellamy W, Takase M, Yamauchi K, Wakabayashi H, Kawase K, Tomita M. Identification of the bactericidal domain of lactoferrin. Biochim Biophys Acta. 1992;1121:130–136. doi: 10.1016/0167-4838(92)90346-f. [DOI] [PubMed] [Google Scholar]
- 8.Brock J. Lactoferrin: a multifunctional immunoregulatory protein? Immunol Today. 1995;16:417–419. doi: 10.1016/0167-5699(95)80016-6. [DOI] [PubMed] [Google Scholar]
- 9.Caccavo D, Afeltra A, Guido F, Di Monaco C, Ferri G M, Amoroso A, Vaccaro F, Bonomo L. Two spatially distant epitopes of human lactoferrin. Hybridoma. 1996;15:263–269. doi: 10.1089/hyb.1996.15.263. [DOI] [PubMed] [Google Scholar]
- 10.Crouch S P M, Slater K J, Fletcher J. Regulation of cytokine release from mononuclear cells by the iron-binding protein lactoferrin. Blood. 1992;80:235–240. [PubMed] [Google Scholar]
- 11.Elass-Rochard E, Roseanu A, Legrand D, Trif M, Salmon V, Motas C, Montreuil J, Spik G. Lactoferrin-lipopolysaccharide interaction: involvement of the 28-34 loop region of human lactoferrin in the high-affinity binding to Escherichia coli O55B5 lipopolysaccharide. Biochem J. 1995;312:839–845. doi: 10.1042/bj3120839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elass-Rochard E, Legrand D, Salmon V, Roseanu A, Trif M, Tobias P S, Mazurier J, Spik G. Lactoferrin inhibits the endotoxin interaction with CD14 by competition with the lipopolysaccharide-binding protein. Infect Immun. 1998;66:486–491. doi: 10.1128/iai.66.2.486-491.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ellison R T, III, Giehl T J. Killing of Gram-negative bacteria by lactoferrin and lysozyme. J Clin Investig. 1991;88:1080–1091. doi: 10.1172/JCI115407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galanos C, Lüderitz O, Westphal O. A new method for the extraction of R lipopolysaccharides. Eur J Biochem. 1969;9:245–249. doi: 10.1111/j.1432-1033.1969.tb00601.x. [DOI] [PubMed] [Google Scholar]
- 15.Galanos C, Rietschel E T, Lüderitz O, Westphal O. Interaction of lipopolysaccharide and lipid A with complement. Eur J Biochem. 1971;19:143–152. doi: 10.1111/j.1432-1033.1971.tb01298.x. [DOI] [PubMed] [Google Scholar]
- 16.Kuhn H-M, Brade L, Appelmelk B J, Kusumoto S, Rietschel E T, Brade H. Characterization of the epitope specificity of murine monoclonal antibodies directed against lipid A. Infect Immun. 1992;60:2201–2210. doi: 10.1128/iai.60.6.2201-2210.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 18.Lee W J, Farmer J L, Hilty M, Kim Y B. The protective effects of lactoferrin feeding against endotoxin lethal shock in germfree piglets. Infect Immun. 1998;66:1421–1426. doi: 10.1128/iai.66.4.1421-1426.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lima M F, Kierszenbaum F. Lactoferrin effects on phagocytic cell function. Increased uptake and killing of an intracellular parasite by murine macrophages and human monocytes. J Immunol. 1985;134:4176–4183. [PubMed] [Google Scholar]
- 20.Machnicki M, Zimecki M, Zagulski T. Lactoferrin regulates the release of tumor necrosis factor alpha and interleukin 6 in vivo. Int J Exp Pathol. 1993;74:433–439. [PMC free article] [PubMed] [Google Scholar]
- 21.Masson P L, Heremans J F, Schonne E. Lactoferrin, an iron-binding protein in neutrophilic leukocytes. J Exp Med. 1969;130:643–658. doi: 10.1084/jem.130.3.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCormick J A, Markey G M, Morris T C M. Lactoferrin-inducible monocyte cytotoxicity for K562 cells and decay of natural killer lymphocyte cytotoxicity. Clin Exp Immunol. 1991;83:154–156. doi: 10.1111/j.1365-2249.1991.tb05606.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mitov I, Freudenberg M A, Bamberger U, Galanos C. Cross-binding activity and protective capacity of monoclonal antibodies to lipid A. Immunobiology. 1993;188:1–12. doi: 10.1016/S0171-2985(11)80482-1. [DOI] [PubMed] [Google Scholar]
- 24.Naidu S S, Svensson U, Kishore A R, Satynarayan Naidu A. Relationship between antibacterial activity and porin binding of lactoferrin in Escherichia coli and Salmonella typhimurium. Antimicrob Agents Chemother. 1993;37:240–245. doi: 10.1128/aac.37.2.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Odell E W, Sarra R, Foxworthy M, Chapple D S, Evans R W. Antibacterial activity of peptides to a loop region in human lactoferrin. FEBS Lett. 1996;382:175–178. doi: 10.1016/0014-5793(96)00168-8. [DOI] [PubMed] [Google Scholar]
- 26.Perosa F, Scudelletti M, Imro M A, Dammacco F, Indiveri F. Human CD4-internal antigen anti-idiotypic monoclonal antibody: induction of a CD4-specific response in humans. J Immunol. 1996;156:3563–3569. [PubMed] [Google Scholar]
- 27.Salomao R, Rigato O, Pignatari A C C, Freudenberg M A, Galanos C. Blood stream infections: epidemiology, pathophysiology and therapeutic perspectives. Infection. 1999;27:1–11. doi: 10.1007/BF02565163. [DOI] [PubMed] [Google Scholar]
- 28.Sànchez L, Calvo M, Brock J H. Biological role of lactoferrin. Arch Dis Child. 1992;67:657–661. doi: 10.1136/adc.67.5.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shau H, Kim A, Golub S H. Modulation of natural killer and lymphokine-activated killer cell cytotoxicity by lactoferrin. J Leukocyte Biol. 1992;51:343–349. [PubMed] [Google Scholar]
- 30.Stuart J, Norrel S, Harrington J P. Kinetic effect of human lactoferrin on the growth of Escherichia coli O111. Int J Biochem. 1984;16:1043–1047. doi: 10.1016/0020-711x(84)90085-5. [DOI] [PubMed] [Google Scholar]
- 31.Temponi M, Kageshita T, Perosa F, Ono R, Okada H, Ferrone S. Purification of murine IgG monoclonal antibodies by precipitation with caprylic acid: comparison with other methods of purification. Hybridoma. 1989;8:85–92. doi: 10.1089/hyb.1989.8.85. [DOI] [PubMed] [Google Scholar]
- 32.Tenovuo J, Lehtonen O P J, Aaltonen A S, Vilja P, Tuohimaa P. Antimicrobial factors in whole saliva of human infants. Infect Immun. 1986;51:49–53. doi: 10.1128/iai.51.1.49-53.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Waage C S, Brandzaeg P, Halstensen A, Kierulf P, Espevik T. The complex pattern of cytokines in serum from patients with meningococcal septic shock. J Exp Med. 1989;169:333–338. doi: 10.1084/jem.169.1.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang D, Pabst K M, Aida Y, Pabst M J. Lipopolysaccharide-inactivating activity of neutrophils is due to lactoferrin. J Leukocyte Biol. 1995;57:865–874. doi: 10.1002/jlb.57.6.865. [DOI] [PubMed] [Google Scholar]
- 35.Westphal O, Lüderitz O, Bister F. Uber die Extraktion von Bakterien mit Phenol/Wasser. Z Naturforsch Teil B. 1952;7:148–155. [Google Scholar]
- 36.Zucali J R, Broxmeyer H E, Levy D, Morse C. Lactoferrin decreases monocyte-induced fibroblast production of myeloid colony-stimulating activity by suppressing monocyte release of interleukin-1. Blood. 1989;74:1531–1536. [PubMed] [Google Scholar]