Abstract
The WRKY transcription factor (TF) belongs to one of the major plant protein superfamilies. The WRKY TF gene family plays an important role in the regulation of transcriptional reprogramming associated with plant stress responses. Change in the expression patterns of WRKY genes or the modifications in their action; participate in the elaboration of numerous signaling pathways and regulatory networks. WRKY proteins contribute to plant growth, for example, gamete formation, seed germination, post-germination growth, stem elongation, root hair growth, leaf senescence, flowering time, and plant height. Moreover, they play a key role in many types of environmental signals, including drought, temperature, salinity, cold, and biotic stresses. This review summarizes the current progress made in unraveling the functions of numerous WRKY TFs under drought, salinity, temperature, and cold stresses as well as their role in plant growth and development.
Keywords: WRKY TFs, drought-stress, salinity-stress, temperature-stress, cold-stress, plant development and growth, plants/crops
Introduction
The WRKY family is a group of transcription factors (TFs) that are widely distributed in plants and play important roles in plant growth and development, and biotic and abiotic stress management. The increased exposure in plants to various stresses, such as extreme temperatures, drought, and salinity is a global threat to key crops which significantly affect plant/crop growth and productivity. Many TF genes help plants withstand to adverse conditions and remain potential genomic candidates for widespread use in crop breeding. WRKY TFs represent important molecular switches that evaluate plant development processes and are involved in regulating responses to various stresses. Under stress conditions, plants can initiate a variety of changes at the molecular, cellular, and physiological levels, including stomatal closure, reduced photosynthesis, higher osmolality accumulation, and induction of many stress response genes (Shinozaki and Yamaguchi-Shinozaki, 2007; Masclaux-Daubresse et al., 2010; Kapoor et al., 2020). Genetic engineering is considered an alternative to increasing stress tolerance and has made significant contributions to changing the agronomic properties of crops. Many genes encoding functional proteins, TFs, and proteins involved in signal transduction pathways have been identified as genes responding to abiotic stresses (Turan et al., 2012; Rashid et al., 2020; Cohen et al., 2021). Many TF families, such as WRKY, AP2 (APETLA2)/ERF (ethylene responsive factor), and NAC (NAM, ATAF1/3, and CUC1/2), are unique to plants and have important and specific functions (Jiang et al., 2017).
Structural features and homology of the WRKY TFs
WRKY protein have the unaltered sequence WRKYGQK (hence called WRKY) and a 60 amino acid DNA binding domain comprising a zinc finger-like domain (CX7CX23HXC or CX4-5CX22-23HXH) (Rushton et al., 1996; Finatto et al., 2018). WRKY TFs are classified into different groups; several WRKY proteins are placed in group I, containing two WRKY domains. WRKY proteins comprising one WRKY domain and a Cys2-His2 zinc finger motif are placed in group II. Furthermore, based on additional structural motifs maintained outside the WRKY domain, group II is subdivided into five subgroups (group IIa, group IIb, group IIc, group IId, and group IIe). Group III proteins represent WRKY domains with different zinc finger motifs (Cys2-His/Cys Cys-His2) (Eulgem et al., 2000; Finatto et al., 2018). The genomes of various plants have sequenced—presenting important knowledge about WRKY TFs and revealed that the WRKY TF family consists of a large number of genes (Zhang et al., 2011b; Xiong et al., 2013; Ayadi et al., 2016; Li et al., 2016a; Mohanta et al., 2016; Liu et al., 2017; Finatto et al., 2018) ( Table 1 ). Plant-specific WRKY TFs, a major family of TFs, are a class of DNA-binding proteins found primarily in plants that have a variety of roles in plant processes, including growth, development, and stress signaling through autonomic and cross-regulation with TF and various other genes (Bakshi and Oelmüller, 2014). The first member of WRKY SPF1 superfamily was isolated from the sweet potato (Ipomoea batatas) (Ishiguro and Nakamura, 1994). In general, WRKY TF is expected to function as a key regulatory protein through precise binding to the W-box (TTGAC (C/T)) that regulates gene expression (Chi et al., 2013).
Table 1.
Number of WRKY TFs genes in plants.
| S. No | Name of plant | Number of WRKY TF gene |
|---|---|---|
| 1 | A. thaliana | 74 |
| 2 | B. distachyon | 81 |
| 3 | C. sinensis | 51 |
| 4 | C. clementina | 48 |
| 5 | D. carota | 38 |
| 6 | G. max | 179 |
| 7 | J. curcas | 58 |
| 8 | M. esculenta | 117 |
| 9 | M. domestica | 123 |
| 10 | M. notabilis | 54 |
| 11 | O. sativa Indica | 116 |
| 12 | O. sativa japonica | 137 |
| 13 | P. vulgaris | 88 |
| 14 | P. trichocarpa | 119 |
| 15 | S. lycopersicum | 79 |
| 16 | S. tuberosum | 82 |
| 17 | V. vinifera | 98 |
| 18 | Z. mays | 180 |
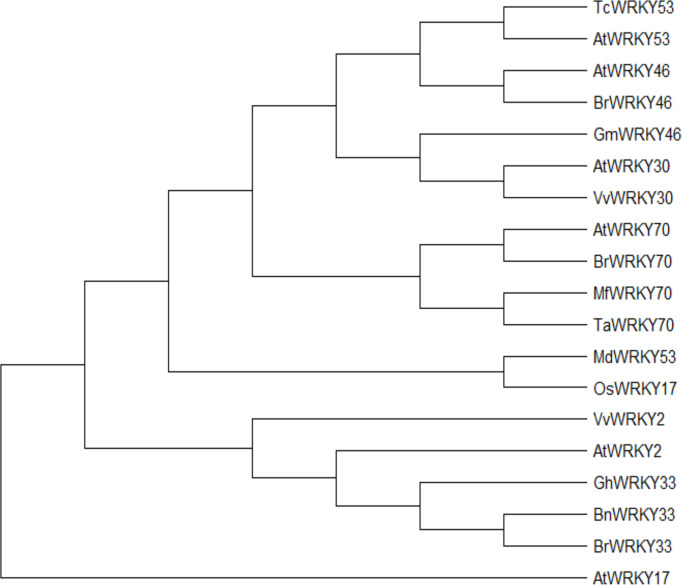
The coding sequence (CDS) of each WRKY gene was obtained from the National Center for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov/) to build the phylogenetic tree using MEGA X and 1000 BS. It was shown in ( Figure 1 ) that each homolog of WRKY genes showed the closest similarity, such as AtWRKY53 with TcWRKY53, AtWRKY46 with BrWRKY46, and AtWRKY70, BrWRKY70, MfWRKY70 with TaWRKY70. As it was mentioned before that, AtWRKY53 expression was induced by drought stress (Jiang et al., 2012). In contrast, TcWRKY53 was induced by cold stress (Wei et al., 2008), illustrating that these two WRKY genes have a different role under different abiotic stress and species as well. It was also assumed that each WRKY gene might also contribute to multiple abiotic stresses.
Figure 1.
Neighbor-joining phylogeny of WRKY-related protein. MEGA X reconstructed the neighbor-joining phylogeny with 1000 bootstrap replicates and used maximum composite likelihood.
Drought stress-related WRKY TFs
The expression of WRKY TF is induced when plants are exposed to various stresses or defense signals, including salicylic acid (SA) or other molecules. In addition to the fact that WRKY TF expression is rapid, transient, and tissue-specific, WRKY proteins also play diverse functions in plant defenses against different stresses including drought, plant growth, development, metabolism, trichome and embryonic morphogenesis, senescence, biosynthesis and regulation of hormonal signals (Wei et al., 2017) ( Figure 2 ). The WRKY TFs present important roles in response and adaptation to drought stress ( Table 2 ). Overexpression of AtWRKY57 increased drought tolerance in A. thaliana. It has been studied that the Arabidopsis WRKY57 transcription factor may confer drought tolerance to transgenic rice O. sativa plants. The overexpression of AtWRKY57 in rice improved drought, salinity, and polyethylene gylcol (PEG) tolerance, indicating a possible role of AtWRKY57 in crop development (Jiang et al., 2016). The MaWRKY80 was up-regulated under drought stress conditions and was identified as a TF capable of binding to the W-box in A. thaliana. MaWRKY80 overexpression exhibits improved phenotypic morphology, improved survival, lower water loss rate, and lower malondialdehyde (MDA) levels than WT (wild-type) under drought stress. Under drought stress, the transgenic MaWRKY80-leaves of A. thaliana showed lower reactive oxygen species (ROS) than WT. The MaWRKY80 also promoted leaf stomata motility and water retention by regulating 9-cis-epoxycarotenoid dioxygenase (NCED) transcript and abscisic acid (ABA) biosynthesis in A. thaliana (Liu et al., 2020).
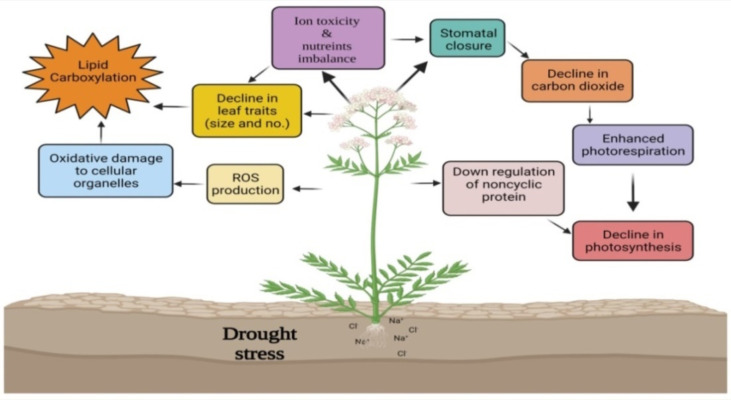
Figure 2.
Effect of drought stress and the role of WRKY TFs in mitigating drought stress. Drought stress causes ROS production, oxidative damage, ion toxicity, and nutrient imbalance impairing plant growth and development. WRKY TFs regulate the expression of stress response genes and ROS scavenging enzymes. Overexpression of various WRKY TFs reduces ion loss and ROS accumulation, induces leaf stomatal mobility, decreases water loss rate thereby promote water retention, which overall improves phenotypic morphology and plant survival.
Table 2.
Drought Stress-related WRKY TFs in plants.
| S. No. | Gene | Species | Tolerance to stress | Reference |
|---|---|---|---|---|
| 1 | AtWRKY53 | A. thaliana | drought | (Jiang et al., 2012) |
| 2 | SlWRKY81 | S. lycopersicum | drought | (Ahammed et al., 2020) |
| 3 | GhWRKY33 | G. hirsutum L. | drought | (Wang et al., 2019) |
| 4 | SlWRKY72 | S. lycopersicum | drought | (Karkute et al., 2018) |
| 5 | MuWRKY3 | M. uniflorum lam.verdc. | drought | (Kiranmai et al., 2018) |
| 6 | TaWRKY2 | T. aestivum L. | drought | (Gao et al., 2018) |
| 7 | TaWRKY1/33 | T. aestivum L. | drought | (He et al., 2016) |
| 8 | ZmWRKY40 | Z. mays | drought | (Wang et al., 2018b) |
| 9 | SbWRKY30 | S. bicolor | drought | (Yang et al., 2020) |
| 10 | AtWRKY30 | A. thaliana | drought | (El-Esawi et al., 2019) |
| 11 | VlWRKY48 | CV.kyoho | drought | (Zhao et al., 2018a) |
| 12 | XsWRKY20 | X. sorbifolium | drought | (Xiong et al., 2020) |
| 13 | GhWRKY41 | G. hirsutum L. | drought | (Chu et al., 2015) |
| 14 | ZmWRKY106 | Z. mays | drought | (Wang et al., 2018a) |
| 15 | VfWRKY1/2 | V. faba L. | drought | (Abid et al., 2017) |
| 16 | AhWRKY | A. hypogaea L. | drought | (Zhao et al., 2020b) |
| 17 | VvWRKY13 | V. vinifera L. | drought | (Hou et al., 2020) |
| 18 | BdWRKY36 | B. distachyon | drought | (Sun et al., 2015) |
| 19 | GhWRKY27a | G. hirsutum | drought | (Yan et al., 2015) |
| 20 | MbWRKY1 | M. baccata L. | drought | (Han et al., 2018a) |
| 21 | SpWRKY1 | Phy. infestans | drought | (Li et al., 2015) |
| 22 | EjWRKY17 | E. japonica | drought | (Wang et al., 2021a) |
| 23 | PheWRKY86 | Phy. edulis | drought | (Wu et al., 2022a) |
| 24 | BoWRKY1o | B. oleracea var. A. DC | drought | (Guo et al., 2021) |
| 25 | TaWRKY46 | T. aestivum L. | drought | (Yu and Zhang, 2021) |
| 26 | OsWRKY5 | O. sativa | drought | (Lim et al., 2021) |
| 27 | CsWRKY26 | C. sinensis | drought | (Chen et al., 2021) |
The sorghum WRKY TF, SbWRKY30 primarily expressed in leaves and roots was induced via drought stress. In A. thaliana and rice, heterologous expression of SbWRKY30 confers drought tolerance via disturbing root architecture. In addition, SbWRKY30 induced SbRD19 (a homologous gene of the drought stress response gene RD19 in A. thaliana) expression in sorghum and the overexpression of SbRD19 increased drought tolerance in Arabidopsis compared to WT plants. This suggests that SbWRKY30 functions as a positive regulator in response to drought stress (Yang et al., 2020). Suppression of GhWRKY21 has been shown to improve drought tolerance in cotton, although GhWRK21 exhibits a negative role in drought response in cotton (Wang et al., 2021b). Overexpression of the MuWRKY3 TF gene in peanuts (A. hypogaea L.) showed increased tolerance to drought stress and exhibited reduced and delayed wilting symptoms in transgenic plants than WT under drought stress imposition. This indicated that MuWRKY3 (nuclear-localized) TFs controlled the expression of stress response genes and the actions of ROS scavenging enzymes, thereby led to increased drought tolerance in peanuts (Kiranmai et al., 2018). The expression analysis of GhWRK25 revealed that GhWRK25 gene is induced by biotic stress and several defense-related signaling molecules (Liu et al., 2016). Overexpression of GhWRKY25 in N. benthamiana reduced plant tolerance to drought stress and increased tolerance to salt stress (Liu et al., 2016). The GmWRKY12, clustered in WRKYII, is 714 bp in length and encodes 237 amino acids. The GmWRKY12 is expressed in various tissues, not only under normal conditions in soybean, but also strongly expressed under drought and salt treatments (Shi et al., 2018).
The GhWRKY68 overexpression in N. benthamiana, a novel group of WRKY group IIC genes, responds to drought and salt stresses by regulating ABA signaling and modulating cellular ROS (Chi et al., 2013; Jia et al., 2019). The gene BdWRKY36 belongs to the WRKY IIe group, designated from B. distachyon, the BdWRKY36 localization in the nucleus is identified via the transient expression in onion epidermal cell. The C-terminal region of BdWRKY36 was found to be transcriptionally active by transactivation assays in transgenic tobacco lines under drought stress. Overexpression of BdWRKY36 resulted in less ion loss (IL) and ROS accumulation in tobacco lines. Whereas, under drought stress in BdWRKY36-overexpressing tobacco lines, the expression levels of ROS scavenging and stress response genes were up-regulated. Overall, BdWRKY36 was found to act as a positive regulator of drought stress response through regulation of ROS homeostasis and regulation of transcription of stress-related genes (Sun et al., 2015; Li et al., 2020b). The OsWRKY11 activates the drought-responsive gene transcription, namely RAB21, via binding directly to the promoter site, and the protein levels of OsWRKY11 controlled by the system known as ubiquitin-proteasome (Lee et al., 2018; Liu et al., 2020). It was studied that GmWRKY54 improved stomatal closure to reduce water loss, thus confirming drought tolerance in soybean through improved gene ontology (GO), co-expression network analysis, and physiological parameters. In transgenic soybean plants, expression of GmWRKY54 confers drought tolerance by the constitutive promoter (pCm) and drought-induced promoter (RD29a). In soybean, the GmWRKY54 activates genes (PYL8, SRK2A, CIPK11, and CPK3) by directly binding to the promoter region, and it has revealed that GmWRKY54 played its function via ABA and Ca2+ signaling pathways. In transgenic Arabidopsis, GmWRKY54 could also improve drought stress tolerance (He et al., 2016; Wei et al., 2019). The GhWRKY59 plays an important role in regulating cotton’s response to drought. Studies have identified that key WRKY TFs are activated and phosphorylated by the MAP kinase cascade, which exhibited GhMAP3K15, GhMKK4, GhMPK6, GhWRKY59, and GhDREB2, as regulatory modules involved in regulating the response of cotton to drought (Li et al., 2017a).
Wheat (Triticum aestivum) is the main crop worldwide; its production in various areas is affected by drought. Therefore, improving the drought tolerance of wheat via breeding cultivars is an essential step for food security. It has been examined that TaWRKY2 isolated from T. aestivum enhanced drought tolerance and increased grain productivity in wheat (Niu et al., 2012; Gao et al., 2018; El-Esawi et al., 2019). The WRKY30 TF, AtWRKY30, cloned from A. thaliana, overexpressed in wheat, which exhibited lower levels of hydrogen peroxide, electrolyte leakage, and malondialdehyde in transgenic plants compared to WT. Moreover, in transgenic wheat plants, some enzyme encoding stress-responsive genes (WRKY19, TIP2, ERF5a, DREB1, DREB3, and AQP7), were induced, which indicates AtWRKY30 to be a possible candidate gene to improve stress-tolerance in wheat (El-Esawi et al., 2019). A WRKY TF, GhWRKY33, established in cotton, localizes to the cell nucleus and can bind to (W-box) cis-acting elements of target promoters. Moreover, GhWRKY33 overexpression in Arabidopsis acts as negative regulator that mediates drought stress responses and contributes to ABA signaling (Wang et al., 2019; Khuman et al., 2020; Shaheen et al., 2020). It has been reported that the grape gene WRKY48 is upregulated due to drought stress, fungal infection, and response to exogenous addition of plant hormones. In A. thaliana, over-expressed V1WRKY48 form (cv. Kyoho), regulates a variety of drought stress responses and exhibits resistance to powdery mildew infection (Han et al., 2018c; Zhao et al., 2018a). The maize WRKY gene promoter region contains C-repeats, dehydration response element (DRE), cold response element (LTR), microbial biomass-C (MBC), and TCA elements that act on drought stress, flocculation, and SA. In transgenic Arabidopsis, the overexpression of ZmWRKY106 (from the maize member WRKY group II) acted as a positive factor, which improved the drought and heat tolerance (Wang et al., 2018a; Hou et al., 2020). It has been recognized that the WRKY TF gene ZmWRKY40, is located in the core of mesophyll protoplasts and the promoter region of ZmWRKY40 and has numerous transcriptional regulatory elements. A candidate gene, ZmWRKY40, improved drought tolerance in transgenic A. thaliana through regulation of stress-related genes under drought stress in transgenic lines where ROS levels decreased by enhancing the activity of two enzymes, peroxide dismutase (POD) and catalase (CAT) (Wang et al., 2018b; Leng and Zhao, 2020). The WRKY genes, TaWRKY1 and TaWRKY33 (group III and II) have reported to be localized in nucleus in wheat mesophyll protoplasts. In the promoter regions of these genes, several abiotic cis-acting elements were detected. Due to high temperature and ABA, TaWRKY1 gene was up-regulated and down-regulated via low temperature. In addition, the TaWRKY33 gene shows the higher response to ABA, jasmonic acid methyl ester, and to high and low temperatures. In Arabidopsis transgenic lines, TaWRKY33 exhibited less water loss than the TaWRKY1 gene, and the overexpressed TaWRKY1 and TaWRKY33 genes were associated in activation of various downstream stress-related genes, and higher germination rates under various stress conditions (He et al., 2016).
Temperature stress-related WRKY TFs
Most plants grow in specific environments and repeatedly experience changes in external conditions. As a result, plants have evolved many complex mechanisms to resist various stresses. WRKY TFs are key proteins that respond to environmental stimuli by regulating gene expression (Xu et al., 2018; He et al., 2019). WRKY TFs are major plant-specific TFs that regulate numerous downstream stress response genes and play important roles in plant biotic and abiotic stress responses. Abiotic stressors, such as drought, heat, salinity, and cold are the main reasons why plants are undermining productivity around the world (Surendran et al., 2017). At the molecular level, WRKY-TFs are one of the most important families of plant-specific regulatory proteins in the plant kingdom, and are known to contribute to biotic and abiotic stress responses (Sarris et al., 2015; Joshi et al., 2016).
The high and low temperatures cause widespread agricultural damage, reducing crop yields and plant quality. To Protect plant cells from damage caused by extreme temperature changes essential for increasing agricultural production (Ohama et al., 2017). Due to global change, extremely high temperatures are getting a lot of attention and there is evidence that heat stress is responsible for biochemical changes in plants (Li et al., 2020b). Extremely high temperatures have become a major factor affecting plant growth, crop yield, fruit quality, flowering, plant biochemistry, morphology, and physiology (Goraya et al., 2017; Li et al., 2018). WRKY TF plays an important role in plant responses to heat stress. Most studies have shown that WRKY TF responds positively to plant tolerance to high temperatures. For example, in A. thaliana high-temperature treatment induces the expression of AtWRKY25 and AtWRKY26, and inhibits AtWRKY33, whereas overexpression of AtWRKY25/26 increases tolerance to heat stress in A. thaliana (Li et al., 2011). In peppers, CaWRKY40 promotes stress resistance at high temperatures and the overexpression of CaWRKY40 in tobacco reduces susceptibility to heat treatment, whereas loss of CaWRKY40 reduces this tolerance (Liu et al., 2021). Inhibition of AtWRKY41 expression in A. thaliana leads to reduced seed dormancy and suppression of high temperature (Chen et al., 2012; Ding et al., 2014). The overexpression of TaWRKY33 in wheat enhances the high-temperature tolerance (El-Esawi et al., 2019). It has been studied that WRKY-TFs increase ROS production in the cell because of high-temperature stress in plants results in an excessive accumulation of ROS produced oxidative stress. Recent studies have shown that WRKY-TF is induced through ROS and contributes to the ROS elimination transformation pathway.
Oxidative stress is a severe stress caused by a variety of stresses, and ROS-mediated signaling is regulated by a delicate balance between production and clearance (Salvucci et al., 2001; Alvarez-Venegas et al., 2007). There are four types of reactive oxygen species in plants: oxygen, hydrogen peroxide, hydroxyl radicals, and superoxide anions. Several WRKY TFs (WRKY6, WRKY30, WRKY22, WRKY8, WRKY53, WRKY48, WRKY39, and WRKY75) are activated in A. thaliana in response to hydrogen peroxide treatment (Davletova et al., 2005; Jiang et al., 2017). It has been investigated that treatment of H2O2 activated higher expression of (WRKY6, WRKY8, WRKY22, WRKY30, WRKY39, WRKY48, WRKY53, and WRKY75) that could respond to a higher temperature in A. thaliana (Chen et al., 2010). OsWRKY42 has been shown to play an important role as a negative regulator of oxidative stress, and overexpression of OsWRKY42 in rice results in higher ROS accumulation (Han et al., 2014). Overexpression of TaWRKY10 in wheat showed reduced malonaldehyde (MDA) accumulation, and low MDA was associated with a low rate of lipid peroxidation. This showed that the transgenic seedlings exhibited high resistance to oxidative stress due to increased expression of TaWRKY10, which resists reduced heat damage. The AtWRKY28 was found to regulate the expression of downstream-associated genes through ROS in A. thaliana when exposed to oxidative stress (Niu et al., 2012; Babitha et al., 2013). The ClWRKY20 belongs to group III of the WRKY family, and intracellular localization of ClWRKY20 was found in the nucleus. The expression level of ClWRKY20 was increased due to salinity, drought, and phytohormones (ABA, ET, and SA) treatment. ClWRKY20 overexpression in transgenic Arabidopsis increased sensitivity to ABA at low temperatures, salinity, and during seed germination (Zhu et al., 2022). This study showed that WRKY-TF enhances plant tolerance to high temperature through transcriptional regulation ( Table 3 ).
Table 3.
Temperature stress-related WRKY TFs.
| S. No. | Gene | Species | Tolerance to stress | References |
|---|---|---|---|---|
| 1 | AtWRKY30 | A. thaliana | temperature | (El-Esawi et al., 2019) |
| 2 | AtWRKY46 | A. thaliana | temperature | (Suzuki et al., 2005) |
| 3 | OsWRKY77 | O. Sativa | temperature | (Lan et al., 2013) |
| 4 | CaWRKY27 | C. annuum | temperature | (Dang et al., 2018) |
| 5 | CaWRKY40 | C. annuum | temperature | (Dang et al., 2013) |
| 6 | AtWRKY41 | A. thaliana | temperature | (Ding et al., 2014) |
| 7 | TaWRKY70 | T. aestivum | temperature | (Wang et al., 2017) |
| 8 | AtWRKY54 | A. thaliana | temperature | (Li et al., 2020b) |
| 9 | PtWRKY13 | P. tomentosa | temperature | (Ren et al., 2019) |
| 10 | PtWRKY50 | P. tomentosa | temperature | (Ren et al., 2019) |
| 11 | ZmWRKY106 | Z. mays | temperature | (Wang et al., 2018a) |
| 12 | AtWRKY39 | A. thaliana | temperature | (Li et al., 2010b) |
| 13 | AtWRKY72 | A. thaliana | temperature | (Cheng et al., 2021) |
| 14 | AtWRKY7 | A. thaliana | temperature | (Park et al., 2005) |
| 15 | AtWRKY8 | A. thaliana | temperature | (Han et al., 2015) |
| 16 | AtWRKY15 | A. thaliana | temperature | (Han et al., 2015) |
| 17 | AtWRKY26 | A. thaliana | temperature | (Fu and Yu, 2010) |
| 18 | AtWRKY33 | A. thaliana | temperature | (Fu and Yu, 2010) |
| 19 | TaWRKY1 | T. aestivum | temperature | (Ren et al., 2019) |
| 20 | NtWRKY6 | N. tabacum | temperature | (Macková et al., 2013) |
| 21 | HaWRKY6 | H. annuus | temperature | (Giacomelli et al., 2012) |
| 22 | ClWRKY20 | C. lanatus | temperature | (Zhu et al., 2022) |
Cold stress-related WRKY TFs
Cold stress (cold below 20°C and freezing below 0°C) adversely affects plant growth and development and greatly limits agricultural productivity. Plants adapt tolerance to cold stress, chilling and freezing by various physiological, protective, and molecular response systems. It has been studied via analyzing regulatory mechanism in plants, many genes have been identified that respond to cold stress at the transcriptional level (Ahmadizadeh and Heidari, 2014; Ritonga et al., 2021). Many WRKY TFs known to have important role in cold stress tolerance in various species ( Table 4 ). Recent studies have shown that transgenic lines of Arabidopsis overexpressing CsWRKY46 and cucumber WRKY show higher seedling viability when frozen at 4°C. In addition, the study identified transgenic A. thaliana in which overexpression of GmWRKY21 (soybean WRKY) showed increased resistance to cold stress (Zhou et al., 2008; Zhang et al., 2016). Another study showed that CsWRKY46 (belonging to the group II WRKY family) was localized in the nucleus, as determined by transient expression analysis. After freezing treatment, Arabidopsis lines, overexpressing CsWRKY46, WRK46-OE1, and WRK46-OE5 had a higher survival rate than the WT. CsWRKY46 confers cold tolerance to transgenic plants and modulates cold signaling pathways in an ABA-dependent manner. Whereas, overexpression of OsWRKY76 was found to enhance cold stress tolerance at 4°C (Zhang et al., 2016). Overexpression lines compared to WT exhibited better surveillance under -20°C after 80 minutes and until 72 hours. The over-expressing plant lines had lower ion content leakage related to WT plants. From that, it could be assumed that overexpression lines could possess higher membrane stability (Yokotani et al., 2013).
Table 4.
Cold stress-related WRKY TFs.
| S. No. | Gene | Plant species | Factors | Responses | Reference |
|---|---|---|---|---|---|
| 1 | AtWRKY34 | A. thaliana | Cold | Play a role as a negative regulator in cold stress | (Zou et al., 2010) |
| 2 | VvWRKY24 | V. vinifera | Cold | Up-regulate regulation of hypothermia | (Wang et al., 2014b) |
| 3 | OsWRKY76 | O. sativa | Cold | Tolerance to cold | (Yokotani et al., 2013) |
| 4 | BcWRKY46 | B. campestris | Cold and Salt | Drought and salt tolerance | (Wang et al., 2012) |
| 5 | VbWRKY32 | V. bonariensis | Cold | Tolerance to cold stress | (Wang et al., 2020) |
| 6 | GmWRKY21 | G. max | Cold, Drought, | Tolerance to cold stress | (Zhou et al., 2008) |
| 7 | VpWRKY2 | V. pseudoreticulata | Cold, ABA, and Salt | Tolerance to cold and salt stress | (Li et al., 2010a) |
| 8 | TcWRKY53 | T. caerulescens | Cold, NaCl, and PEG | Play a role as a negative regulator in osmatic stress | (Wei et al., 2008) |
| 9 | JrWRKY2 | J. regia | Cold and Drought | Cold and drought tolerance | (Yang et al., 2017) |
| 10 | JrWRKY7 | J. regia | Cold and Drought | Tolerance to cold and drought stress | (Yang et al., 2017) |
| 11 | LchiWRKY33 | L. chinense (Lchi) | Cold | Tolerance to cold stress | (Wu et al., 2022b) |
Salinity stress-related WRKY TFs
Soil salinity is one of the major abiotic stresses that affect the productivity of crops. Because the ionic and osmotic stresses of high salt concentrations in the soil affect the growth and development of plants. Salt stress is highly common in arid regions because of excessive evaporation leading to the accumulation of inorganic salts, which affects plant metabolism. With the success of traditional breeding approaches to improve stress-tolerant traits, transformation methods appear to be particularly beneficial for breeding stress-tolerant crops. In this regard, TFs play an important role as mediators in genetic engineering due to their unique roles in the regulation and modification of various stress-sensitive genes (Chaudhry et al., 2021; Hussain et al., 2021).
WRKY TFs also present a key role in salt stress response and tolerance ( Table 5 ). Recent studies have shown that overexpression of AtWRKY46 enhances root development during salt stress in Arabidopsis through modulation of ABA signaling. In addition, overexpression of GhWRKY34 (G. hirsutum) enhances the plant’s ability to selectively absorb Na+ as well as K+ and maintain low Na+/K+ levels, thereby increasing resistance to salt stress in the leaves and roots of transgenic Arabidopsis plants (Dai et al., 2016). Overexpression of GmWRKY54 (WRKY soybean) in transgenic Arabidopsis plants shows salt tolerance, it has indicated that WT plants showed 25% survival while over-expressing lines showed 70% survival under 180 mM NaCl treatment (Zhou et al., 2008). Another study found that N. benthamiana GmWRKY17 (cotton WRKY) improved salinity stress tolerance as measured by physiological analyzes of germination rate, root growth, survival, and leaf water loss (Yan et al., 2014). A new WRKY gene was isolated from M. xiaojinensis, namely MxWRKY55, and it is localized in the nucleus. The expression level of MxWRKY55 in M. xiaojinensis seedlings was affected by salinity, low Fe, and high Fe stresses, and MxWRKY55 also increased salinity and iron tolerance when introduced into A. thaliana. Overexpression of MxWRKY55 in A. thaliana showed high levels of chlorophyll and proline, as well as increased activity of superoxide dismutase (SOD), peroxidase (POD), and catalase (CAT). Similarly, MxWRKY55 in A. thaliana resulted in lower levels of malondialdehyde (MAD), particularly in response to salt stress. In addition, overexpression of MxWRKY55 in transgenic A. thaliana showed greater root length, mass, chlorophyll, and iron content compared to WT (Han et al., 2020). Based on these properties, it has been demonstrated that MxWRKY55 can play a positive role in the process of salt resistance, resistance to high Fe, and low Fe content. Another study showed that the growth and development of M. xiaojinesis (semi-dwarf apple in China) was affected by the salinity and Fe. The novel WRKY MxWRKY53/64 gene isolated from M. xiaojinesis is a nuclear-localized protein and its expression level is strongly influenced by salt as well as Fe, when MxWRKY53/64 was introduced into transgenic A. thaliana, resistance to salinity and iron stress was significantly increased (Han et al., 2021a; Han et al., 2021b). Moreover, the over-expression of wheat WRKY TF, the TaWRKY93 in A. thaliana showed high salt tolerance, low temperature, and osmotic stress tolerance (Qin et al., 2015).
Table 5.
Salinity stress-related WRKY TFs.
| S. No. | Gene | Plant | Tolerance to stress | Reference |
|---|---|---|---|---|
| 1 | FcWRKY70 | F. crassifolia | Salt | (Wang et al., 2007) |
| 2 | GmWRKY17 | G. max | Salt | (Yan et al., 2014) |
| 3 | ZmWRKY17 | Z. mays | Salt | (Cai et al., 2017) |
| 4 | SbWRKY30 | S. bicolor | Salt | (Yang et al., 2020) |
| 5 | GbWRKY1 | G. barbadense | Salt | (Luo et al., 2020) |
| 6 | IbWRKY47 | I. batatas | Salt | (Qin et al., 2020) |
| 7 | PgWRKY33/62 | P. glaucum | Salt | (Chanwala et al., 2020) |
| 8 | SbWRKY50 | S. bicolor | Salt | (Song et al., 2020b) |
| 9 | VpWRKY1 | V. pseudoreticulata | Salt | (Li et al., 2010a) |
| 10 | VpWRKY2 | V. pseudoreticulata | Salt | (Li et al., 2010a) |
| 11 | MbWRKY5 | M. baccata | Salt | (Han et al., 2018b) |
| 12 | CmWRKY | C. pepo | Salt | (Bankaji et al., 2019) |
| 13 | PbWRKY40 | P. betulaefolia | Salt | (Lin et al., 2022) |
| 14 | ClWRKY20 | C. Lanatus | Salt | (Zhu et al., 2022) |
| 15 | MxWRKY53 | M. xiaojinensis | Salt | (Han et al., 2021b) |
| 16 | MxWRKY64 | M. xiaojinensis | Salt | (Han et al., 2021a) |
| 17 | AhWRKY75 | A. hypogaea L. | Salt | (Zhu et al., 2021) |
| 18 | MfWRKY70 | M. Flabellifolia | Salt | (Xiang et al., 2021) |
WRKY TFs as key regulators in plant growth and development
The WRKY TF is one of the largest TF families in plants, which in addition to stress response and defense regulation significantly contributes to plant growth and development. Various WRKY genes have been reported in different plant species that promote growth and development (Zhang et al., 2017) ( Table 6 ). The AtWRKY28 gene, AtWRKY2, and AtWRKY34 which are involved in macrospore fate, pollen tube extension, pollen production, seed germination, and early growth after germination. AtWRKY2 (a knockout mutant exhibiting high sensitivity to ABA) plays an important role in seed germination (Jiang and Yu, 2009). The overexpression of VvWRKY30 in Arabidopsis increased resistance to salt stress at various growth stages by regulating ROS clearance and osmotic accumulation (Zhu et al., 2019). In soybean, GmWRKY12 induced a positive role in ABA, salt, and drought stresses (Shi et al., 2018).
Table 6.
Role of WRKY TFs in plant growth and development.
| S. No. | Name | Plant | Function | References |
|---|---|---|---|---|
| 1 | VvWRKY30 | V. vinifera | Increasing salt stress resistance by ROS and accumulation of osmoticum. | (Zhu et al., 2019) |
| 2 | GmWRKY12 | G. max | Drought and salinity tolerance | (Shi et al., 2018; Zhang et al., 2020) |
| 3 | MdWRKY40 | M. domestica | Important regulators of wound-induced anthocyanin biosynthesis | (An et al., 2019) |
| 4 | TaWRKY51 | T. aestivum L. | Promotes lateral root formation due to negative regulation of ethylene biosynthesis | (Hu et al., 2018) |
| 5 | GhWRKY59 | G. hirsutum | Drought responses | (Li et al., 2017a) |
| 6 | HbWRKY82 | H. brasiliensis | Abiotic resistance and leaf aging | (Kang et al., 2021) |
| 7 | MfWRKY70 | M. Flabellifolia | Drought and salinity tolerance | (Xiang et al., 2021) |
| 8 | HmoWRKY40 | H. monacanthus | Betalain biosynthesis | (Zhang et al., 2021b) |
| 9 | MxWRKY64 | M. xiaojinensis | It plays an important role in response to Fe and salt stress | (Han et al., 2021a) |
| 10 | AhWRKY75 | A. hypogaea L | Conferred salt tolerance in transgenic peanut lines | (Zhu et al., 2021) |
| 11 | BoWRKY10 | B. oleracea var.acephala DC | Regulation of drought stress tolerance | (Guo et al., 2021) |
| 12 | AtWRKY28 | A. thaliana | Oocyte development | (Zhao et al., 2018b) |
| 13 | AtWRKY2 | A. thaliana | Seed germination, growth after germination | (Jiang and Yu, 2009) |
| 14 | AtWRKY10 | A. thaliana | The size of the seed | (Luo et al., 2005) |
| 15 | AtWRKY34 | A. thaliana | Seed germination, growth after germination | (Guan et al., 2014) |
| 16 | AtWRKY41 | A. thaliana | The dormancy of seed | (Ding et al., 2014) |
| 17 | AtWRKY44 | A. thaliana | In the proanthocy seed coat of tannins | (Gonzalez et al., 2016) |
| 18 | OsWRKY78 | O. sativa | The development of seed and stem elongation | (Zhang et al., 2011a) |
| 19 | OsWRKY24 | O. sativa | Increased lamina inclination and grain size through cell elongation. | (Jang and Li, 2018) |
| 20 | GhWRKY42 | G. hirsutum | Premature leaf senescence and stem development | (Gu et al., 2018) |
| 21 | AtWRKY23 | A. thaliana | Root growth and biosynthesis of flavanols | (Grunewald et al., 2012) |
| 22 | GhWRKY91 | G. hirsutum | Leaf senescence and stress response | (Gu et al., 2019b) |
| 23 | OsWRKY93 | O. sativa | Leaf senescence and in response to fungi attack | (Li et al., 2021) |
| 24 | BrWRKY6 | B. rapa ssp.pekinensis | Leaf senescence | (Fan et al., 2018) |
| 25 | GhWRKY27 | G. hirsutum | Leaf senescence | (Gu et al., 2019a) |
| 26 | PyMYB114 | Red-Skinned pears | Regulate anthocyanin biosynthesis and transport | (Li et al., 2020a) |
| 27 | WRKY6 | A. thaliana | Improve FA accumulation and seed yield | (Song et al., 2020a) |
| 28 | TaWRKY40-D | T. aestivum L. | Association to the promotion of leaf senescence with jasmonic acid and abscisic acid | (Zhao et al., 2020a) |
| 29 | WRKY46/6 | A. thaliana | PBZ/SA-mediated leaf senescence | (Zhang et al., 2021a) |
| 30 | WRKY45 | A. thaliana | Positive regulator of age-triggered leaf senescence | (Chen et al., 2017) |
| 31 | VvWRKY2 | V. venifera | Vigor, yield, and tuber quality | (Chiab, 2021) |
| 32 | AtWRKY26 | A. thaliana | Leaf senescence | (Li et al., 2017b) |
| 33 | WRKY12/13 | A. thaliana | Regulate flowering time | (Li et al., 2016b) |
| 34 | WRKY42 | A. thaliana | Root hair growth and development | (Moison et al., 2021) |
| 35 | OsWRKY11 | O. sativa | Flowering time and plant height | (Cai et al., 2014) |
| 36 | AtWRKY45 | A. thaliana | Play a key role in Phosphate uptake | (Wang et al., 2014a) |
| 37 | AtWRKY42 | A. thaliana | Play a great role in phosphate uptake | (Su et al., 2015) |
| 38 | AtWRKY71 | A. thaliana | Flowering time | (Yu et al., 2016) |
| 39 | MxWRKY55 | M. xiaojinensis | Tolerance to salt, low-iron and high-iron stress | (Han et al., 2020) |
There are several WRKY genes involved in plant root development. The TaWRKY51, an important WRKY TF that increases lateral root formation through the regulation of ethylene biosynthesis in wheat (Hu et al., 2018). The study also reported that TaWRKY51 regulates lateral root formation via the ethylene and auxin signaling pathways (Hu et al., 2018). AtWRKY23 expression induced by the auxin response factor7 (ARF7) and auxin response factor 19 (ARF19) (serve as part of the auxin feedback loop), help to regulate the growth of plant roots and the synthesis of flavonoids (Grunewald et al., 2012). Both AtWRKY75 and AtWRKY44 are involved in root hair development. AtWRKY44 is also a downstream gene (TTG1 and GLABROUS1) expressed in root hairs that act jiontly with GLABRA2 to regulate root hair growth in plants (Johnson et al., 2002). Studies have shown that the number and length of root hairs are increased in AtWRKY75 (Knockout mutant) compared to the WT, suggesting that AtWRKY75 is a negative regulator of root hair development (Devaiah et al., 2007).
A novel WRKY TF, designated HbWRKY82, was identified based on stress-related WRKY in rubber trees, encoded by nuclear proteins and present an important function as a transcriptional activator. Exogenous ethrel and ABA stimulation induce HbWRK82 transcriptional activity, which play important roles as transcriptional regulators in ethrel and in response to ABA-mediated leaf senescence and abiotic stress (Kang et al., 2021). The WRKY70 is involved in biological stress as a positive regulator and has a negative role in abiotic stress signaling in Arabidopsis and several other plant species. The localization of MfWRKY70 in the nucleus was confirmed by examining MfWRK70 from M. flabellifolia in Arabidopsis model plants. The MfWRKY70 is reported to have an essential role in drought, osmotic pressure, and salinity tolerance by promoting root growth and water retention. Under stress conditions, MfWRKY70 enhanced the antioxidant enzyme system, maintaining ROS homeostasis and stability of membrane lipids (Xiang et al., 2021).
A novel WRKY TF, the HmoWRKY40 was identified from the transcriptomic data of pitaya (H. monacanthus), and the HmoWRKY40 transcriptionally activates HmoCYP76AD, which regulates ptiaya betalain biosynthesis (Zhang et al., 2021b). Fe and high salinity affect the growth and development of M. xiaojiensis, a semi-dwarf apple in China. The newly isolated WRKY gene from M. xiaojinesis, namely MxWRKY64 (localization in the nucleus)was introduced into A. thaliana, which showed increased resistance to Fe and salts, and overexpression of MxWRKY64 in transgenic A. thaliana under Fe stress resulted in higher levels of mass, root length, chlorophyll, and Fe content compared to WT (Han et al., 2021a). A novel WRKY-TF gene AhWRKY75 (WRKYIIC) identified from M34 (salt-tolerant mutant) confers salt tolerance to transgenic peanut strains by increasing the efficiency of ROS removal system and photosynthesis during stress treatment (Zhu et al., 2021). In flowering plants, female gonadal megasporoblasts (MMCs) start as single cells in each ovule, and Arabidopsis cytochrome P450 (KLU) functions through the SWR1 chromatin remodeling complex to promote WRKY28 expression in oocyte primordial (Zhao et al., 2018b). The studies have suggested that WRKY genes play a key role in seed germination and post-germination growth. The Arabidopsis WRKY2 TF is involved in seed germination and post-emergence stunting (Jiang and Yu, 2009), plant (male) gamete formation with complex and dynamic changes in gene expression. Studies have shown that WRKY2 and its close homolog WRKY34 (pollen-specific) TFs participated in male gametogenesis in A. thaliana (Guan et al., 2014).
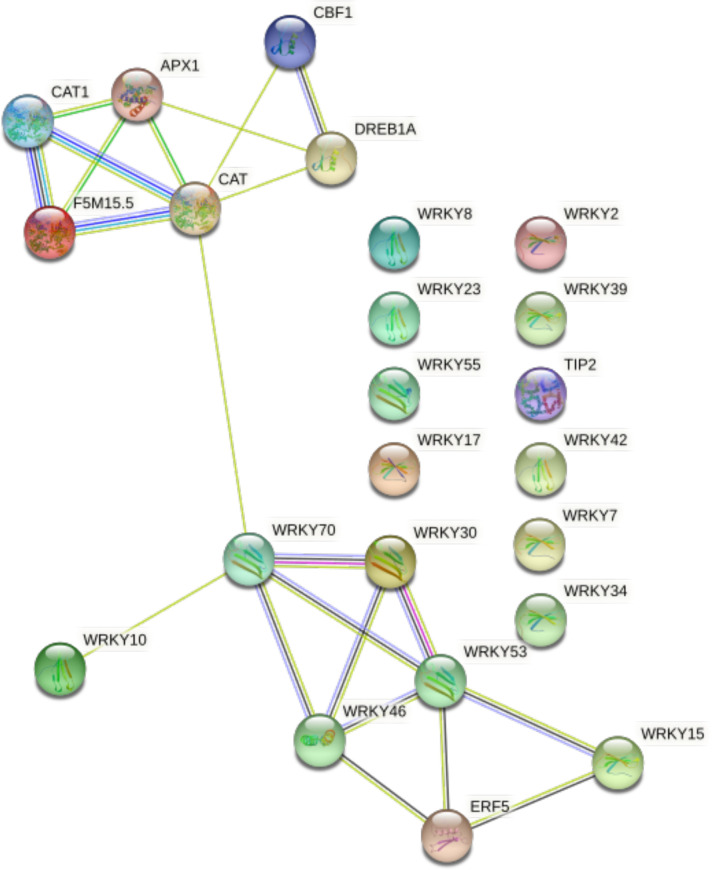
Interaction of WRKY genes with some stress-related genes to improve plant abiotic stress tolerance in plants was shown in Figure 3 . The interaction network with STRING (https://string-db.org/cgi/) was recognized. The result showed that several WRKY genes correlate with abiotic stress-related genes; for instance, the above mentioned AtWRKY30 cloned TFs from Arabidopsis; its over-expression in wheat showed improved stress tolerance. Moreover, in transgenic wheat, antioxidant genes such as APX1, CAT, CAT1, F5M15.5, ERF5, CBF1, and DREBIA play key roles as stress-responsive genes (El-Esawi et al., 2019). It was speculated that correlated genes might have a positive or negative correlation in response to abiotic stress.
Figure 3.
The interaction network analysis of some WRKY genes identified in Arabidopsis thaliana using STRING: Line colors are associated with interaction types. Around the green line gene, pink lines are experimentally determined, black lines mean co-expression, dark blue lines mean gene co-occurrence, and blue lines mean protein homology.
Conclusion and future prospects
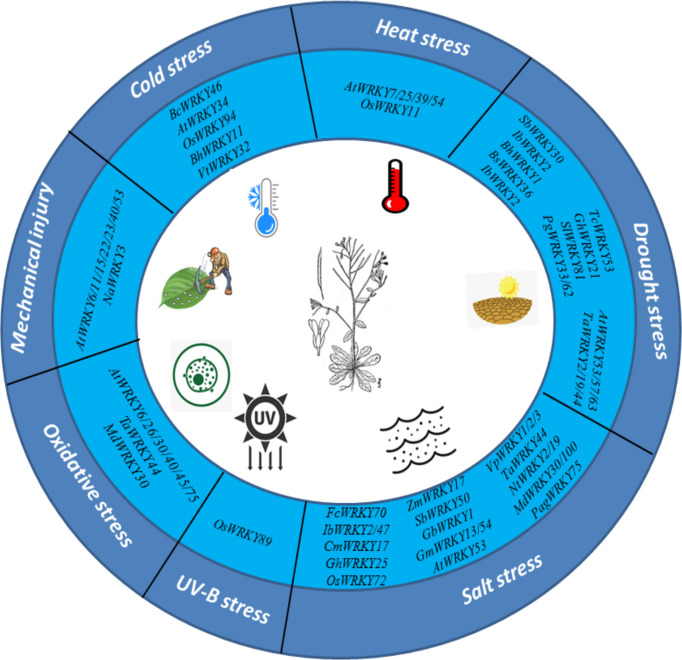
Plants are considered as sessile organisms that cannot avoid adverse abiotic stresses as well as other major environmental stresses and have developed complex signaling networks composed of multiple pathways. One of the largest TF families, WRKY-TFs act as molecular switches that regulates the expression of stress-sensitive genes. Stress-induced WRKY-TF expression is regulated by a complex transcriptional regulatory network that allows plants to maintain the proper balance between growth and stress response. This review discusses the recent studies of WRKY-TF. Many studies have shown that WRKY-TFs play important roles in abiotic stress tolerance ( Figure 4 ). Nowadays the sequencing of plant genomes has increased largely; especially in economically important crops and whole-genome identification of the WRKY gene (with respect to functional plant genes) facilitate screening. Previous studies have demonstrated that the WRKY gene primarily depends on its functional assumptions and transcriptome. In addition, genetic confirmations joined to the latest technologies are increasing to confirm the novel role of the WRKY genes, expression of WRKY-TF or downstream genes regulated by self-regulation of WRKY-TF, which helps to simplify the regulatory network of responses to abiotic stresses. Future studies should explore noncoding RNAs and epigenetic modifications involved in the regulation of WRKY-TFs. Based on current studies the role of WRKY-TFs in regulating plant responses related to abiotic stresses, particularly drought, salinity, and temperature stress, are not sufficiently detailed, particularly at the transcriptional level. Finally, the use of WRKY-TF screening for plant stress tolerance in context to increase climate change significantly improves crop yield and crop quality.
Figure 4.
WRKY-TFs response to various abiotic stresses.
Author contributions
MK, and AH planned and designed this review manuscript. MK, AH, and HM wrote this review paper. FR, QA, MC, QM, MA, WZ, RMA, and RB helped to improve the manuscript writing. FL and HM contributed to the critically revising of the manuscript. All the authors have reviewed, edited, and approved the manuscript before submission.
Funding
This work was supported by grant from the Lushan Botanical Garden, Chinese Academy of Sciences (No. 2021ZWZX28 to HM), and by grant from the National Natural Science Foundation of China (32100297 to FL).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Abid G., Muhovski Y., Mingeot D., Saidi M., Aouida M., Aroua I., et al. (2017). Identification and characterization of two faba bean (Vicia faba l.) WRKY transcription factors and their expression analysis during salt and drought stress. J. Agric. Sci. 155, 791–803. doi: 10.1017/S0021859616000885 [DOI] [Google Scholar]
- Ahammed G. J., Li X., Yang Y., Liu C., Zhou G., Wan H., et al. (2020). Tomato WRKY81 acts as a negative regulator for drought tolerance by modulating guard cell H2O2–mediated stomatal closure. Environ. Exp. Bot. 171, 103960. doi: 10.1016/j.envexpbot.2019.103960 [DOI] [Google Scholar]
- Ahmadizadeh M., Heidari P. (2014). Bioinformatics study of transcription factors involved in cold stress. Biharean Biol. 8, 83–86. [Google Scholar]
- Alvarez-Venegas R., Abdallat A. A., Guo M., Alfano J. R., Avramova Z. (2007). Epigenetic control of a transcription factor at the cross section of two antagonistic pathways. Epigenetics 2, 106–113. doi: 10.4161/epi.2.2.4404 [DOI] [PubMed] [Google Scholar]
- An J. P., Zhang X. W., You C. X., Bi S. Q., Wang X. F., Hao Y. J. (2019). Md WRKY 40 promotes wounding-induced anthocyanin biosynthesis in association with md MYB 1 and undergoes md BT 2-mediated degradation. New Phytol. 224, 380–395. doi: 10.1111/nph.16008 [DOI] [PubMed] [Google Scholar]
- Ayadi M., Hanana M., Kharrat N., Merchaoui H., Marzoug R. B., Lauvergeat V., et al. (2016). The WRKY transcription factor family in citrus: Valuable and useful candidate genes for citrus breeding. Appl. Biochem. Biotechnol. 180, 516–543. doi: 10.1007/s12010-016-2114-8 [DOI] [PubMed] [Google Scholar]
- Babitha K., Ramu S., Pruthvi V., Mahesh P., Nataraja K. N., Udayakumar M. (2013). Co-Expression of AtbHLH17 and AtWRKY28 confers resistance to abiotic stress in arabidopsis. Transgenic Res. 22, 327–341. doi: 10.1007/s11248-012-9645-8 [DOI] [PubMed] [Google Scholar]
- Bakshi M., Oelmüller R. (2014). WRKY transcription factors: Jack of many trades in plants. Plant Signaling Behav. 9, e27700. doi: 10.4161/psb.27700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankaji I., Sleimi N., Vives-Peris V., Gómez-Cadenas A., Pérez-Clemente R. M. (2019). Identification and expression of the cucurbita WRKY transcription factors in response to water deficit and salt stress. Scientia Hortic. 256, 108562. doi: 10.1016/j.scienta.2019.108562 [DOI] [Google Scholar]
- Cai Y., Chen X., Xie K., Xing Q., Wu Y., Li J., et al. (2014). Dlf1, a WRKY transcription factor, is involved in the control of flowering time and plant height in rice. PLos One 9, e102529. doi: 10.1371/journal.pone.0102529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai R., Dai W., Zhang C., Wang Y., Wu M., Zhao Y., et al. (2017). The maize WRKY transcription factor ZmWRKY17 negatively regulates salt stress tolerance in transgenic arabidopsis plants. Planta 246, 1215–1231. doi: 10.1007/s00425-017-2766-9 [DOI] [PubMed] [Google Scholar]
- Chanwala J., Satpati S., Dixit A., Parida A., Giri M. K., Dey N. (2020). Genome-wide identification and expression analysis of WRKY transcription factors in pearl millet (Pennisetum glaucum) under dehydration and salinity stress. BMC Genomics 21, 1–16. doi: 10.1186/s12864-020-6622-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhry U. K., Gökçe Z. N. Ö., Gökçe A. F. (2021). “The Influence of Salinity Stress on Plants and Their Molecular Mechanisms.” in Biology and Life Sciences Forum (Basel, Switzerland: MDPI; ) 11 (1), 31. [Google Scholar]
- Cheng Z., Luan Y., Meng J., Sun J., Tao J., Zhao D. (2021). WRKY transcription factor response to high-temperature stress. Plants 10, 2211. doi: 10.3390/plants10102211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Song Y., Li S., Zhang L., Zou C., Yu D. (2012). The role of WRKY transcription factors in plant abiotic stresses. Biochim. Biophys. Acta (BBA)-Gene Regul. Mech. 1819, 120–128. doi: 10.1016/j.bbagrm.2011.09.002 [DOI] [PubMed] [Google Scholar]
- Chen L., Xiang S., Chen Y., Li D., Yu D. (2017). Arabidopsis WRKY45 interacts with the DELLA protein RGL1 to positively regulate age-triggered leaf senescence. Mol. Plant 10, 1174–1189. doi: 10.1016/j.molp.2017.07.008 [DOI] [PubMed] [Google Scholar]
- Chen L., Zhang L., Yu D. (2010). Wounding-induced WRKY8 is involved in basal defense in arabidopsis. Mol. Plant-Microbe Interact. 23, 558–565. doi: 10.1094/MPMI-23-5-0558 [DOI] [PubMed] [Google Scholar]
- Chen W., Zheng C., Yao M., Chen L. (2021). The tea plant CsWRKY26 promotes drought tolerance in transgenic arabidopsis plants. Beverage Plant Res. 1, 1–11. doi: 10.48130/BPR-2021-0003 [DOI] [Google Scholar]
- Chiab N. (2021). The overexpression of the VvWRKY2 transcription factor in potato improved the agricultural performance and tubers’ physio-chemical and industrial properties even under non-stress conditions Research Square 1, 1–19. doi: 10.21203/rs.3.rs-361740/v1 [DOI] [Google Scholar]
- Chi Y., Yang Y., Zhou Y., Zhou J., Fan B., Yu J.-Q., et al. (2013). Protein–protein interactions in the regulation of WRKY transcription factors. Mol. Plant 6, 287–300. doi: 10.1093/mp/sst026 [DOI] [PubMed] [Google Scholar]
- Chu X., Wang C., Chen X., Lu W., Li H., Wang X., et al. (2015). The cotton WRKY gene GhWRKY41 positively regulates salt and drought stress tolerance in transgenic nicotiana benthamiana. PLos One 10, e0143022. doi: 10.1371/journal.pone.0143022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen I., Zandalinas S. I., Huck C., Fritschi F. B., Mittler R. (2021). Meta-analysis of drought and heat stress combination impact on crop yield and yield components. Physiologia Plantarum 171, 66–76. doi: 10.1111/ppl.13203 [DOI] [PubMed] [Google Scholar]
- Dai X., Wang Y., Zhang W.-H. (2016). OsWRKY74, a WRKY transcription factor, modulates tolerance to phosphate starvation in rice. J. Exp. Bot. 67, 947–960. doi: 10.1093/jxb/erv515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang F., Lin J., Xue B., Chen Y., Guan D., Wang Y., et al. (2018). CaWRKY27 negatively regulates H2O2-mediated thermotolerance in pepper (Capsicum annuum). Front. Plant Sci., 9, 1633. doi: 10.3389/fpls.2018.01633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang F. F., Wang Y. N., Yu L., Eulgem T., Lai Y., Liu Z. Q., et al. (2013). CaWRKY40, a WRKY protein of pepper, plays an important role in the regulation of tolerance to heat stress and resistance to ralstonia solanacearum infection. Plant. Cell Environ. 36, 757–774. doi: 10.1111/pce.12011 [DOI] [PubMed] [Google Scholar]
- Davletova S., Rizhsky L., Liang H., Shengqiang Z., Oliver D. J., Coutu J., et al. (2005). Cytosolic ascorbate peroxidase 1 is a central component of the reactive oxygen gene network of arabidopsis. Plant Cell 17, 268–281. doi: 10.1105/tpc.104.026971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaiah B. N., Karthikeyan A. S., Raghothama K. G. (2007). WRKY75 transcription factor is a modulator of phosphate acquisition and root development in arabidopsis. Plant Physiol. 143, 1789–1801. doi: 10.1104/pp.106.093971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z. J., Yan J. Y., Li G. X., Wu Z. C., Zhang S. Q., Zheng S. J. (2014). WRKY 41 controls arabidopsis seed dormancy via direct regulation of ABI 3 transcript levels not downstream of ABA. Plant J. 79, 810–823. doi: 10.1111/tpj.12597 [DOI] [PubMed] [Google Scholar]
- El-Esawi M. A., Al-Ghamdi A. A., Ali H. M., Ahmad M. (2019). Overexpression of AtWRKY30 transcription factor enhances heat and drought stress tolerance in wheat (Triticum aestivum l.). Genes 10, 163. doi: 10.3390/genes10020163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulgem T., Rushton P. J., Robatzek S., Somssich I. E. (2000). The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5, 199–206. doi: 10.1016/S1360-1385(00)01600-9 [DOI] [PubMed] [Google Scholar]
- Fan Z.-Q., Tan X.-L., Shan W., Kuang J.-F., Lu W.-J., Chen J.-Y. (2018). Characterization of a transcriptional regulator, BrWRKY6, associated with gibberellin-suppressed leaf senescence of Chinese flowering cabbage. J. Agric. Food Chem. 66, 1791–1799. doi: 10.1021/acs.jafc.7b06085 [DOI] [PubMed] [Google Scholar]
- Finatto T., Viana V. E., Woyann L. G., Busanello C., Maia L. C. D., Oliveira A. C. D. (2018). Can WRKY transcription factors help plants to overcome environmental challenges? Genet. Mol. Biol. 41, 533–544. doi: 10.1590/1678-4685-GMB-2017-0232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Q.-T., Yu D.-Q. (2010). Expression profiles of AtWRKY25, AtWRKY26 and AtWRKY33 under abiotic stresses. Yi chuan= Hereditas 32, 848–856. doi: 10.3724/SP.J.1005.2010.00848 [DOI] [PubMed] [Google Scholar]
- Gao H., Wang Y., Xu P., Zhang Z. (2018). Overexpression of a WRKY transcription factor TaWRKY2 enhances drought stress tolerance in transgenic wheat. Front. Plant Sci. 9, 997. doi: 10.3389/fpls.2018.00997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacomelli J. I., Weigel D., Chan R. L., Manavella P. A. (2012). Role of recently evolved miRNA regulation of sunflower HaWRKY6 in response to temperature damage. New Phytol. 195, 766–773. doi: 10.1111/j.1469-8137.2012.04259.x [DOI] [PubMed] [Google Scholar]
- Gonzalez A., Brown M., Hatlestad G., Akhavan N., Smith T., Hembd A., et al. (2016). TTG2 controls the developmental regulation of seed coat tannins in arabidopsis by regulating vacuolar transport steps in the proanthocyanidin pathway. Dev. Biol. 419, 54–63. doi: 10.1016/j.ydbio.2016.03.031 [DOI] [PubMed] [Google Scholar]
- Goraya G. K., Kaur B., Asthir B., Bala S., Kaur G., Farooq M. (2017). Rapid injuries of high temperature in plants. J. Plant Biol. 60, 298–305. doi: 10.1007/s12374-016-0365-0 [DOI] [Google Scholar]
- Grunewald W., De Smet I., Lewis D. R., Löfke C., Jansen L., Goeminne G., et al. (2012). Transcription factor WRKY23 assists auxin distribution patterns during arabidopsis root development through local control on flavonol biosynthesis. Proc. Natl. Acad. Sci. 109, 1554–1559. doi: 10.1073/pnas.1121134109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y., Meng X., Khanna R., Lamontagne E., Liu Y., Zhang S. (2014). Phosphorylation of a WRKY transcription factor by MAPKs is required for pollen development and function in arabidopsis. PLos Genet. 10, e1004384. doi: 10.1371/journal.pgen.1004384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu L., Dou L., Guo Y., Wang H., Li L., Wang C., et al. (2019. a). The WRKY transcription factor GhWRKY27 coordinates the senescence regulatory pathway in upland cotton (Gossypium hirsutum l.). BMC Plant Biol. 19, 1–14. doi: 10.1186/s12870-019-1688-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu L., Ma Q., Zhang C., Wang C., Wei H., Wang H., et al. (2019. b). The cotton GhWRKY91 transcription factor mediates leaf senescence and responses to drought stress in transgenic arabidopsis thaliana. Front. Plant Sci. 10, 1352. doi: 10.3389/fpls.2019.01352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J.-J., Li S., Li H.-Y., Li W., Li D.-H. (2021). Expression of the kale WRKY gene BoWRKY10 in transgenic tobacco confers drought stress tolerance. Russian J. Plant Physiol. 68, 147–157. doi: 10.1134/S1021443721010076 [DOI] [Google Scholar]
- Gu L., Wei H., Wang H., Su J., Yu S. (2018). Characterization and functional analysis of GhWRKY42, a group IId WRKY gene, in upland cotton (Gossypium hirsutum l.). BMC Genet. 19, 1–14. doi: 10.1186/s12863-018-0653-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han D., Ding H., Chai L., Liu W., Zhang Z., Hou Y., et al. (2018. a). Isolation and characterization of MbWRKY1, a WRKY transcription factor gene from malus baccata (L.) borkh involved in drought tolerance. Can. J. Plant Sci. 98, 1023–1034. doi: 10.1139/cjps-2017-0355 [DOI] [Google Scholar]
- Han D., Han J., Xu T., Li T., Yao C., Wang Y., et al. (2021. a). Isolation and preliminary functional characterization of MxWRKY64, a new WRKY transcription factor gene from malus xiaojinensis Cheng et jiang. In Vitro Cell. Dev. Biology-Plant 57, 202–213. doi: 10.1007/s11627-021-10171-7 [DOI] [Google Scholar]
- Han D., Hou Y., Wang Y., Ni B., Li Z., Yang G. (2018. b). Overexpression of a malus baccata WRKY transcription factor gene (MbWRKY5) increases drought and salt tolerance in transgenic tobacco. Can. J. Plant Sci. 99, 173–183. doi: 10.1139/cjps-2018-0053 [DOI] [Google Scholar]
- Han M., Kim C.-Y., Lee J., Lee S.-K., Jeon J.-S. (2014). OsWRKY42 represses OsMT1d and induces reactive oxygen species and leaf senescence in rice. Molecules Cells 37, 532. doi: 10.14348/molcells.2014.0128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X., Wang H., Pam J., Hu Y., Chen X., Yu D. (2015). Arabidopsis WRKY8 transcription factor-associated genes vq10 and vq11 are responsive to multiple abiotic stresses. Plant Diversity Resour. 37, 760–766. [Google Scholar]
- Han D., Xu T., Han J., Liu W., Wang Y., Li X., et al. (2021. b). Overexpression of MxWRKY53 increased iron and high salinity stress tolerance in arabidopsis thaliana. In Vitro Cell. Dev. Biology-Plant 58 (2), 266–278. doi: 10.1007/s11627-021-10241-w [DOI] [Google Scholar]
- Han D., Zhang Z., Ding H., Chai L., Liu W., Li H., et al. (2018. c). Isolation and characterization of MbWRKY2 gene involved in enhanced drought tolerance in transgenic tobacco. J. Plant Interact. 13, 163–172. doi: 10.1080/17429145.2018.1447698 [DOI] [Google Scholar]
- Han D., Zhou Z., Du M., Li T., Wu X., Yu J., et al. (2020). Overexpression of a malus xiaojinensis WRKY transcription factor gene (MxWRKY55) increased iron and high salinity stress tolerance in arabidopsis thaliana. In Vitro Cell. Dev. Biology-Plant 56, 600–609. doi: 10.1007/s11627-020-10129-1 [DOI] [Google Scholar]
- He X., Li J.-J., Chen Y., Yang J.-Q., Chen X.-Y. (2019). Genome-wide analysis of the WRKY gene family and its response to abiotic stress in buckwheat (Fagopyrum tataricum). Open Life Sci. 14, 80–96. doi: 10.1515/biol-2019-0010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He G.-H., Xu J.-Y., Wang Y.-X., Liu J.-M., Li P.-S., Chen M., et al. (2016). Drought-responsive WRKY transcription factor genes TaWRKY1 and TaWRKY33 from wheat confer drought and/or heat resistance in arabidopsis. BMC Plant Biol. 16, 1–16. doi: 10.1186/s12870-016-0806-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou L., Fan X., Hao J., Liu G., Zhang Z., Liu X. (2020). Negative regulation by transcription factor VvWRKY13 in drought stress of vitis vinifera l. Plant Physiol. Biochem. 148, 114–121. doi: 10.1016/j.plaphy.2020.01.008 [DOI] [PubMed] [Google Scholar]
- Hussain Q., Asim M., Zhang R., Khan R., Farooq S., Wu J. (2021). Transcription factors interact with ABA through gene expression and signaling pathways to mitigate drought and salinity stress. Biomolecules 11, 1159. doi: 10.3390/biom11081159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z., Wang R., Zheng M., Liu X., Meng F., Wu H., et al. (2018). Ta WRKY 51 promotes lateral root formation through negative regulation of ethylene biosynthesis in wheat (Triticum aestivum l.). Plant J. 96, 372–388. doi: 10.1111/tpj.14038 [DOI] [PubMed] [Google Scholar]
- Ishiguro S., Nakamura K. (1994). Characterization of a cDNA encoding a novel DNA-binding protein, SPF1, that recognizes SP8 sequences in the 5′ upstream regions of genes coding for sporamin and β-amylase from sweet potato. Mol. Gen. Genet. MGG 244, 563–571. doi: 10.1007/BF00282746 [DOI] [PubMed] [Google Scholar]
- Jang S., Li H.-Y. (2018). Overexpression of OsAP2 and OsWRKY24 in arabidopsis results in reduction of plant size. Plant Biotechnol., 35 (3), 273–279. doi: 10.5511/plantbiotechnology.18.0508a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y., Liang G., Yu D. (2012). Activated expression of WRKY57 confers drought tolerance in arabidopsis. Mol. Plant 5, 1375–1388. doi: 10.1093/mp/sss080 [DOI] [PubMed] [Google Scholar]
- Jiang J., Ma S., Ye N., Jiang M., Cao J., Zhang J. (2017). WRKY transcription factors in plant responses to stresses. J. Integr. Plant Biol. 59, 86–101. doi: 10.1111/jipb.12513 [DOI] [PubMed] [Google Scholar]
- Jiang Y., Qiu Y., Hu Y., Yu D. (2016). Heterologous expression of AtWRKY57 confers drought tolerance in oryza sativa. Front. Plant Sci. 7, 145. doi: 10.3389/fpls.2016.00145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W., Yu D. (2009). Arabidopsis WRKY2 transcription factor mediates seed germination and postgermination arrest of development by abscisic acid. BMC Plant Biol. 9, 1–14. doi: 10.1186/1471-2229-9-96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia H., Wang C., Wang F., Liu S., Li G., Guo X. (2019). Correction: GhWRKY68 reduces resistance to salt and drought in transgenic nicotiana benthamiana. PLos One 14, e0213540. doi: 10.1371/journal.pone.0213540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C. S., Kolevski B., Smyth D. R. (2002). TRANSPARENT TESTA GLABRA2, a trichome and seed coat development gene of arabidopsis, encodes a WRKY transcription factor. Plant Cell 14, 1359–1375. doi: 10.1105/tpc.001404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi R., Wani S. H., Singh B., Bohra A., Dar Z. A., Lone A. A., et al. (2016). Transcription factors and plants response to drought stress: current understanding and future directions. Front. Plant Sci. 7, 1029. doi: 10.3389/fpls.2016.01029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang G., Yan D., Chen X., Yang L., Zeng R. (2021). HbWRKY82, a novel IIc WRKY transcription factor from hevea brasiliensis associated with abiotic stress tolerance and leaf senescence in arabidopsis. Physiologia Plantarum 171, 151–160. doi: 10.1111/ppl.13238 [DOI] [PubMed] [Google Scholar]
- Kapoor D., Bhardwaj S., Landi M., Sharma A., Ramakrishnan M., Sharma A. (2020). The impact of drought in plant metabolism: How to exploit tolerance mechanisms to increase crop production. Appl. Sci. 10, 5692. doi: 10.3390/app10165692 [DOI] [Google Scholar]
- Karkute S. G., Gujjar R. S., Rai A., Akhtar M., Singh M., Singh B. (2018). Genome wide expression analysis of WRKY genes in tomato (Solanum lycopersicum) under drought stress. Plant Gene 13, 8–17. doi: 10.1016/j.plgene.2017.11.002 [DOI] [Google Scholar]
- Khuman A., Arora S., Makkar H., Patel A., Chaudhary B. (2020). Extensive intragenic divergences amongst ancient WRKY transcription factor gene family is largely associated with their functional diversity in plants. Plant Gene 22, 100222. doi: 10.1016/j.plgene.2020.100222 [DOI] [Google Scholar]
- Kiranmai K., Lokanadha Rao G., Pandurangaiah M., Nareshkumar A., Amaranatha Reddy V., Lokesh U., et al. (2018). A novel WRKY transcription factor, MuWRKY3 (Macrotyloma uniflorum lam. verdc.) enhances drought stress tolerance in transgenic groundnut (Arachis hypogaea l.) plants. Front. Plant Sci. 16 (9), 346. doi: 10.3389/fpls.2018.00346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan A., Huang J., Zhao W., Peng Y., Chen Z., Kang D. (2013). A salicylic acid-induced rice (Oryza sativa l.) transcription factor OsWRKY77 is involved in disease resistance of arabidopsis thaliana. Plant Biol. 15, 452–461. doi: 10.1111/j.1438-8677.2012.00664.x [DOI] [PubMed] [Google Scholar]
- Lee H., Cha J., Choi C., Choi N., Ji H.-S., Park S. R., et al. (2018). Rice WRKY11 plays a role in pathogen defense and drought tolerance. Rice 11, 1–12. doi: 10.1186/s12284-018-0199-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leng P., Zhao J. (2020). Transcription factors as molecular switches to regulate drought adaptation in maize. Theor. Appl. Genet. 133, 1455–1465. doi: 10.1007/s00122-019-03494-y [DOI] [PubMed] [Google Scholar]
- Li S., Fu Q., Chen L., Huang W., Yu D. (2011). Arabidopsis thaliana WRKY25, WRKY26, and WRKY33 coordinate induction of plant thermotolerance. Planta 233, 1237–1252. doi: 10.1007/s00425-011-1375-2 [DOI] [PubMed] [Google Scholar]
- Li B., Gao K., Ren H., Tang W. (2018). Molecular mechanisms governing plant responses to high temperatures. J. Integr. Plant Biol. 60, 757–779. doi: 10.1111/jipb.12701 [DOI] [PubMed] [Google Scholar]
- Li Y., Liao S., Mei P., Pan Y., Zhang Y., Zheng X., et al. (2021). OsWRKY93 dually functions between leaf senescence and in response to biotic stress in rice. Front. Plant Sci. 12, 327. doi: 10.3389/fpls.2021.643011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Li M., Wang P., Cox K. L., Jr., Duan L., Dever J. K., et al. (2017. a). Regulation of cotton (Gossypium hirsutum) drought responses by mitogen-activated protein (MAP) kinase cascade-mediated phosphorylation of gh WRKY 59. New Phytol. 215, 1462–1475. doi: 10.1111/nph.14680 [DOI] [PubMed] [Google Scholar]
- Li J. B., Luan Y. S., Liu Z. (2015). Overexpression of SpWRKY1 promotes resistance to phytophthora nicotianae and tolerance to salt and drought stress in transgenic tobacco. Physiologia plantarum 155, 248–266. doi: 10.1111/ppl.12315 [DOI] [PubMed] [Google Scholar]
- Lim C., Kang K., Shim Y., Yoo S.-C., Paek N.-C. (2021). Inactivating transcription factor OsWRKY5 enhances drought tolerance through abscisic acid signaling pathways. Plant Physiol. 188 (4), 1900–1916. doi: 10.1093/plphys/kiab492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L., Yuan K., Huang Y., Dong H., Qiao Q., Xing C., et al. (2022). A WRKY transcription factor PbWRKY40 from pyrus betulaefolia functions positively in salt tolerance and modulating organic acid accumulation by regulating PbVHA-B1 expression. Environ. Exp. Bot. 196, 104782. doi: 10.1016/j.envexpbot.2022.104782 [DOI] [Google Scholar]
- Li W., Pang S., Lu Z., Jin B. (2020. b). Function and mechanism of WRKY transcription factors in abiotic stress responses of plants. Plants 9, 1515. doi: 10.3390/plants9111515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G., Li B., Li X., Wei Y., He C., Shi H. (2020). MaWRKY80 positively regulates plant drought stress resistance through modulation of abscisic acid and redox metabolism. Plant Physiol. Biochem. 156, 155–166. doi: 10.1016/j.plaphy.2020.09.015 [DOI] [PubMed] [Google Scholar]
- Liu Q.-N., Liu Y., Xin Z.-Z., Zhang D.-Z., Ge B.-M., Yang R.-P., et al. (2017). Genome-wide identification and characterization of the WRKY gene family in potato (Solanum tuberosum). Biochem. Systematics Ecol. 71, 212–218. doi: 10.1016/j.bse.2017.02.010 [DOI] [Google Scholar]
- Liu Z. Q., Shi L. P., Yang S., Qiu S. S., Ma X. L., Cai J. S., et al. (2021). A conserved double-W box in the promoter of CaWRKY40 mediates autoregulation during response to pathogen attack and heat stress in pepper. Mol. Plant Pathol. 22, 3–18. doi: 10.1111/mpp.13004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Song Y., Xing F., Wang N., Wen F., Zhu C. (2016). GhWRKY25, a group I WRKY gene from cotton, confers differential tolerance to abiotic and biotic stresses in transgenic nicotiana benthamiana. Protoplasma 253, 1265–1281. doi: 10.1007/s00709-015-0885-3 [DOI] [PubMed] [Google Scholar]
- Li W., Wang H., Yu D. (2016. b). Arabidopsis WRKY transcription factors WRKY12 and WRKY13 oppositely regulate flowering under short-day conditions. Mol. Plant 9, 1492–1503. doi: 10.1016/j.molp.2016.08.003 [DOI] [PubMed] [Google Scholar]
- Li C., Wu J., Hu K.-D., Wei S.-W., Sun H.-Y., Hu L.-Y., et al. (2020. a). PyWRKY26 and PybHLH3 cotargeted the PyMYB114 promoter to regulate anthocyanin biosynthesis and transport in red-skinned pears. Hortic. Res. 7, 37. doi: 10.1038/s41438-020-0254-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M.-Y., Xu Z.-S., Tian C., Huang Y., Wang F., Xiong A.-S. (2016. a). Genomic identification of WRKY transcription factors in carrot (Daucus carota) and analysis of evolution and homologous groups for plants. Sci. Rep. 6, 1–17. doi: 10.1038/srep23101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Xu Y., Xiao Y., Zhu Z., Xie X., Zhao H., et al. (2010. a). Expression and functional analysis of two genes encoding transcription factors, VpWRKY1 and VpWRKY2, isolated from Chinese wild vitis pseudoreticulata. Planta 232, 1325–1337. doi: 10.1007/s00425-010-1258-y [DOI] [PubMed] [Google Scholar]
- Li Z., Zhao Y., Liu X., Jiang Z., Peng J., Jin J., et al. (2017. b). “Construction of the leaf senescence database and functional assessment of senescence-associated genes,” in Plant genomics databases (Springer; ) 1533, 315–333. doi: 10.1007/978-1-4939-6658-5_19 [DOI] [PubMed] [Google Scholar]
- Li S., Zhou X., Chen L., Huang W., Yu D. (2010. b). Functional characterization of arabidopsis thaliana WRKY39 in heat stress. Molecules Cells 29, 475–483. doi: 10.1007/s10059-010-0059-2 [DOI] [PubMed] [Google Scholar]
- Luo M., Dennis E. S., Berger F., Peacock W. J., Chaudhury A. (2005). MINISEED3 (MINI3), a WRKY family gene, and HAIKU2 (IKU2), a leucine-rich repeat (LRR) KINASE gene, are regulators of seed size in arabidopsis. Proc. Natl. Acad. Sci. 102, 17531–17536. doi: 10.1073/pnas.0508418102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X., Li C., He X., Zhang X., Zhu L. (2020). ABA signaling is negatively regulated by GbWRKY1 through JAZ1 and ABI1 to affect salt and drought tolerance. Plant Cell Rep. 39, 181–194. doi: 10.1007/s00299-019-02480-4 [DOI] [PubMed] [Google Scholar]
- Macková H., Hronková M., Dobrá J., Turečková V., Novák O., Lubovská Z., et al. (2013). Enhanced drought and heat stress tolerance of tobacco plants with ectopically enhanced cytokinin oxidase/dehydrogenase gene expression. J. Exp. Bot. 64, 2805–2815. doi: 10.1093/jxb/ert131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masclaux-Daubresse C., Daniel-Vedele F., Dechorgnat J., Chardon F., Gaufichon L., Suzuki A. (2010). Nitrogen uptake, assimilation and remobilization in plants: challenges for sustainable and productive agriculture. Ann. Bot. 105, 1141–1157. doi: 10.1093/aob/mcq028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanta T. K., Park Y.-H., Bae H. (2016). Novel genomic and evolutionary insight of WRKY transcription factors in plant lineage. Sci. Rep. 6, 1–22. doi: 10.1038/srep37309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moison M., Pacheco J. M., Lucero L., Fonouni-Farde C., Rodríguez-Melo J., Mansilla N., et al. (2021). The lncRNA APOLO interacts with the transcription factor WRKY42 to trigger root hair cell expansion in response to cold. Mol. Plant 14, 937–948. doi: 10.1016/j.molp.2021.03.008 [DOI] [PubMed] [Google Scholar]
- Niu C. F., Wei W., Zhou Q. Y., Tian A. G., Hao Y. J., Zhang W. K., et al. (2012). Wheat WRKY genes TaWRKY2 and TaWRKY19 regulate abiotic stress tolerance in transgenic arabidopsis plants. Plant. Cell Environ. 35, 1156–1170. doi: 10.1111/j.1365-3040.2012.02480.x [DOI] [PubMed] [Google Scholar]
- Ohama N., Sato H., Shinozaki K., Yamaguchi-Shinozaki K. (2017). Transcriptional regulatory network of plant heat stress response. Trends Plant Sci. 22, 53–65. doi: 10.1016/j.tplants.2016.08.015 [DOI] [PubMed] [Google Scholar]
- Park C. Y., Lee J. H., Yoo J. H., Moon B. C., Choi M. S., Kang Y. H., et al. (2005). WRKY group IId transcription factors interact with calmodulin. FEBS Lett. 579, 1545–1550. doi: 10.1016/j.febslet.2005.01.057 [DOI] [PubMed] [Google Scholar]
- Qin Z., Hou F., Li A., Dong S., Wang Q., Zhang L. (2020). Transcriptome-wide identification of WRKY transcription factor and their expression profiles under salt stress in sweetpotato (Ipomoea batatas l.). Plant Biotechnol. Rep. 14, 599–611. doi: 10.1007/s11816-020-00635-4 [DOI] [Google Scholar]
- Qin Y., Tian Y., Liu X. (2015). A wheat salinity-induced WRKY transcription factor TaWRKY93 confers multiple abiotic stress tolerance in arabidopsis thaliana. Biochem. Biophys. Res. Commun. 464, 428–433. doi: 10.1016/j.bbrc.2015.06.128 [DOI] [PubMed] [Google Scholar]
- Rashid M., Ejaz S., Shah K. H. (2020). “Regulatory role of transcription factors in abiotic stress responses in plants,” in Plant ecophysiology and adaptation under climate change: Mechanisms and perspectives II (Singapore: Springer; ) 543–565. doi: 10.1007/978-981-15-2172-0_19 [DOI] [Google Scholar]
- Ren S., Ma K., Lu Z., Chen G., Cui J., Tong P., et al. (2019). Transcriptomic and metabolomic analysis of the heat-stress response of populus tomentosa Carr. Forests 10, 383. doi: 10.3390/f10050383 [DOI] [Google Scholar]
- Ritonga F. N., Ngatia J. N., Wang Y., Khoso M. A., Farooq U., Chen S. (2021). AP2/ERF, an important cold stress-related transcription factor family in plants: A review. Physiol. Mol. Biol. Plants 27, 1953–1968. doi: 10.1007/s12298-021-01061-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton P. J., Torres J. T., Parniske M., Wernert P., Hahlbrock K., Somssich I. (1996). Interaction of elicitor-induced DNA-binding proteins with elicitor response elements in the promoters of parsley PR1 genes. EMBO J. 15, 5690–5700. doi: 10.1002/j.1460-2075.1996.tb00953.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvucci M. E., Osteryoung K. W., Crafts-Brandner S. J., Vierling E. (2001). Exceptional sensitivity of rubisco activase to thermal denaturation in vitro and in vivo . Plant Physiol. 127, 1053–1064. doi: 10.1104/pp.010357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarris P. F., Duxbury Z., Huh S. U., Ma Y., Segonzac C., Sklenar J., et al. (2015). A plant immune receptor detects pathogen effectors that target WRKY transcription factors. Cell 161, 1089–1100. doi: 10.1016/j.cell.2015.04.024 [DOI] [PubMed] [Google Scholar]
- Shaheen T., Bilal M. J., Ijaz U., Nahid N. (2020). “Improving cotton crop tolerance to drought stress through molecular approaches,” in Plant ecophysiology and adaptation under climate change: Mechanisms and perspectives II (Singapore: Springer; ) 17–37. doi: 10.1007/978-981-15-2172-0_2 [DOI] [Google Scholar]
- Shi W.-Y., Du Y.-T., Ma J., Min D.-H., Jin L.-G., Chen J., et al. (2018). The WRKY transcription factor GmWRKY12 confers drought and salt tolerance in soybean. Int. J. Mol. Sci. 19, 4087. doi: 10.3390/ijms19124087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K., Yamaguchi-Shinozaki K. (2007). Gene networks involved in drought stress response and tolerance. J. Exp. Bot. 58, 221–227. doi: 10.1093/jxb/erl164 [DOI] [PubMed] [Google Scholar]
- Song G., Li X., Munir R., Khan A. R., Azhar W., Yasin M. U., et al. (2020. a). The WRKY6 transcription factor affects seed oil accumulation and alters fatty acid compositions in arabidopsis thaliana. Physiologia plantarum 169, 612–624. doi: 10.1111/ppl.13082 [DOI] [PubMed] [Google Scholar]
- Song Y., Li J., Sui Y., Han G., Zhang Y., Guo S., et al. (2020. b). The sweet sorghum SbWRKY50 is negatively involved in salt response by regulating ion homeostasis. Plant Mol. Biol. 102, 603–614. doi: 10.1007/s11103-020-00966-4 [DOI] [PubMed] [Google Scholar]
- Sun J., Hu W., Zhou R., Wang L., Wang X., Wang Q., et al. (2015). The brachypodium distachyon BdWRKY36 gene confers tolerance to drought stress in transgenic tobacco plants. Plant Cell Rep. 34, 23–35. doi: 10.1007/s00299-014-1684-6 [DOI] [PubMed] [Google Scholar]
- Surendran U., Kumar V., Ramasubramoniam S., Raja P. (2017). Development of drought indices for semi-arid region using drought indices calculator (DrinC)–a case study from madurai district, a semi-arid region in India. Water Resour. Manage. 31, 3593–3605. doi: 10.1007/s11269-017-1687-5 [DOI] [Google Scholar]
- Su T., Xu Q., Zhang F.-C., Chen Y., Li L.-Q., Wu W.-H., et al. (2015). WRKY42 modulates phosphate homeostasis through regulating phosphate translocation and acquisition in arabidopsis. Plant Physiol. 167, 1579–1591. doi: 10.1104/pp.114.253799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N., Rizhsky L., Liang H., Shuman J., Shulaev V., Mittler R. (2005). Enhanced tolerance to environmental stress in transgenic plants expressing the transcriptional coactivator multiprotein bridging factor 1c. Plant Physiol. 139, 1313–1322. doi: 10.1104/pp.105.070110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turan S., Cornish K., Kumar S. (2012). Salinity tolerance in plants: breeding and genetic engineering. Aust. J. Crop Sci. 6 (9), 1337–1348. [Google Scholar]
- Wang D., Chen Q., Chen W., Liu X., Xia Y., Guo Q., et al. (2021. a). A WRKY transcription factor, EjWRKY17, from eriobotrya japonica enhances drought tolerance in transgenic arabidopsis. Int. J. Mol. Sci. 22, 5593. doi: 10.3390/ijms22115593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Hao J., Chen X., Hao Z., Wang X., Lou Y., et al. (2007). Overexpression of rice WRKY89 enhances ultraviolet b tolerance and disease resistance in rice plants. Plant Mol. Biol. 65, 799–815. doi: 10.1007/s11103-007-9244-x [DOI] [PubMed] [Google Scholar]
- Wang F., Hou X., Tang J., Wang Z., Wang S., Jiang F., et al. (2012). A novel cold-inducible gene from pak-choi (Brassica campestris ssp. chinensis), BcWRKY46, enhances the cold, salt and dehydration stress tolerance in transgenic tobacco. Mol. Biol. Rep. 39, 4553–4564. doi: 10.1007/s11033-011-1245-9 [DOI] [PubMed] [Google Scholar]
- Wang M.-Q., Huang Q.-X., Lin P., Zeng Q.-H., Li Y., Liu Q.-L., et al. (2020). The overexpression of a transcription factor gene VbWRKY32 enhances the cold tolerance in verbena bonariensis. Front. Plant Sci. 10, 1746. doi: 10.3389/fpls.2019.01746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C.-T., Ru J.-N., Liu Y.-W., Li M., Zhao D., Yang J.-F., et al. (2018. a). Maize WRKY transcription factor ZmWRKY106 confers drought and heat tolerance in transgenic plants. Int. J. Mol. Sci. 19, 3046. doi: 10.3390/ijms19103046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C.-T., Ru J.-N., Liu Y.-W., Yang J.-F., Li M., Xu Z.-S., et al. (2018. b). ). the maize WRKY transcription factor ZmWRKY40 confers drought resistance in transgenic arabidopsis. Int. J. Mol. Sci. 19, 2580. doi: 10.3390/ijms19092580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Tao F., An F., Zou Y., Tian W., Chen X., et al. (2017). Wheat transcription factor TaWRKY70 is positively involved in high-temperature seedling plant resistance to puccinia striiformis f. sp. tritici. Mol. Plant Pathol. 18, 649–661. doi: 10.1111/mpp.12425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Vannozzi A., Wang G., Liang Y.-H., Tornielli G. B., Zenoni S., et al. (2014. b). Genome and transcriptome analysis of the grapevine (Vitis vinifera l.) WRKY gene family. Horticult. Res. 1, 14016. doi: 10.1038/hortres.2014.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Wang L., Yan Y., Zhang S., Li H., Gao Z., et al. (2021. b). GhWRKY21 regulates ABA-mediated drought tolerance by fine-tuning the expression of GhHAB in cotton. Plant Cell Rep. 40, 2135–2150. doi: 10.1007/s00299-020-02590-4 [DOI] [PubMed] [Google Scholar]
- Wang H., Xu Q., Kong Y.-H., Chen Y., Duan J.-Y., Wu W.-H., et al. (2014. a). Arabidopsis WRKY45 transcription factor activates PHOSPHATE TRANSPORTER1; 1 expression in response to phosphate starvation. Plant Physiol. 164, 2020–2029. doi: 10.1104/pp.113.235077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N.-N., Xu S.-W., Sun Y.-L., Liu D., Zhou L., Li Y., et al. (2019). The cotton WRKY transcription factor (GhWRKY33) reduces transgenic arabidopsis resistance to drought stress. Sci. Rep. 9, 1–13. doi: 10.1038/s41598-018-37035-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W., Liang D. W., Bian X. H., Shen M., Xiao J. H., Zhang W. K., et al. (2019). GmWRKY54 improves drought tolerance through activating genes in abscisic acid and Ca2+ signaling pathways in transgenic soybean. Plant J. 100, 384–398. doi: 10.1111/tpj.14449 [DOI] [PubMed] [Google Scholar]
- Wei S., Ma X., Pan L., Miao J., Fu J., Bai L., et al. (2017). Transcriptome analysis of taxillusi chinensis (DC.) danser seeds in response to water loss. PLos One 12 (1), e0169177. doi: 10.1371/journal.pone.0169177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W., Zhang Y., Han L., Guan Z., Chai T. (2008). A novel WRKY transcriptional factor from thlaspi caerulescens negatively regulates the osmotic stress tolerance of transgenic tobacco. Plant Cell Rep. 27, 795–803. doi: 10.1007/s00299-007-0499-0 [DOI] [PubMed] [Google Scholar]
- Wu M., Zhang K., Xu Y., Wang L., Liu H., Qin Z., et al. (2022. a). The moso bamboo WRKY transcription factor, PheWRKY86, regulates drought tolerance in transgenic plants. Plant Physiol. Biochem. 170, 180–191. doi: 10.1016/j.plaphy.2021.10.024 [DOI] [PubMed] [Google Scholar]
- Wu W., Zhu S., Xu L., Zhu L., Wang D., Liu Y., et al. (2022. b). Genome-wide identification of the liriodendron chinense WRKY gene family and its diverse roles in response to multiple abiotic stress. BMC Plant Biol. 22, 1–27. doi: 10.1186/s12870-021-03371-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang X.-Y., Chen J., Xu W.-X., Qiu J.-R., Song L., Wang J.-T., et al. (2021). Dehydration-induced WRKY transcriptional factor MfWRKY70 of myrothamnus flabellifolia enhanced drought and salinity tolerance in arabidopsis. Biomolecules 11, 327. doi: 10.3390/biom11020327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong W., Xu X., Zhang L., Wu P., Chen Y., Li M., et al. (2013). Genome-wide analysis of the WRKY gene family in physic nut (Jatropha curcas l.). Gene 524, 124–132. doi: 10.1016/j.gene.2013.04.047 [DOI] [PubMed] [Google Scholar]
- Xiong C., Zhao S., Yu X., Sun Y., Li H., Ruan C., et al. (2020). Yellowhorn drought-induced transcription factor XsWRKY20 acts as a positive regulator in drought stress through ROS homeostasis and ABA signaling pathway. Plant Physiol. Biochem. 155, 187–195. doi: 10.1016/j.plaphy.2020.06.037 [DOI] [PubMed] [Google Scholar]
- Xu Y., Liu F., Han G., Cheng B. (2018). Genome-wide identification and comparative analysis of phosphate starvation-responsive transcription factors in maize and three other gramineous plants. Plant Cell Rep. 37, 711–726. doi: 10.1007/s00299-018-2262-0 [DOI] [PubMed] [Google Scholar]
- Yang Z., Chi X., Guo F., Jin X., Luo H., Hawar A., et al. (2020). SbWRKY30 enhances the drought tolerance of plants and regulates a drought stress-responsive gene, SbRD19, in sorghum. J. Plant Physiol. 246, 153142. doi: 10.1016/j.jplph.2020.153142 [DOI] [PubMed] [Google Scholar]
- Yang G., Zhang W., Liu Z., Yi-Maer A. Y., Zhai M., Xu Z. (2017). Both jr WRKY 2 and jr WRKY 7 of juglans regia mediate responses to abiotic stresses and abscisic acid through formation of homodimers and interaction. Plant Biol. 19, 268–278. doi: 10.1111/plb.12524 [DOI] [PubMed] [Google Scholar]
- Yan H., Jia H., Chen X., Hao L., An H., Guo X. (2014). The cotton WRKY transcription factor GhWRKY17 functions in drought and salt stress in transgenic nicotiana benthamiana through ABA signaling and the modulation of reactive oxygen species production. Plant Cell Physiol. 55, 2060–2076. doi: 10.1093/pcp/pcu133 [DOI] [PubMed] [Google Scholar]
- Yan Y., Jia H., Wang F., Wang C., Liu S., Guo X. (2015). Overexpression of GhWRKY27a reduces tolerance to drought stress and resistance to rhizoctonia solani infection in transgenic nicotiana benthamiana. Front. Physiol. 6, 265. doi: 10.3389/fphys.2015.00265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokotani N., Sato Y., Tanabe S., Chujo T., Shimizu T., Okada K., et al. (2013). WRKY76 is a rice transcriptional repressor playing opposite roles in blast disease resistance and cold stress tolerance. J. Exp. Bot. 64, 5085–5097. doi: 10.1093/jxb/ert298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y., Liu Z., Wang L., Kim S. G., Seo P. J., Qiao M., et al. (2016). WRKY 71 accelerates flowering via the direct activation of FLOWERING LOCUS T and LEAFY in arabidopsis thaliana. Plant J. 85, 96–106. doi: 10.1111/tpj.13092 [DOI] [PubMed] [Google Scholar]
- Yu Y., Zhang L. (2021). Overexpression of TaWRKY46 enhances drought tolerance in transgenic wheat. Cereal Res. Commun. 148, 605–614. doi: 10.1007/s42976-021-00215-4 [DOI] [Google Scholar]
- Zhang L., Chen C., Xie F., Hua Q., Zhang Z., Zhang R., et al. (2021. b). A novel WRKY transcription factor HmoWRKY40 associated with betalain biosynthesis in pitaya (Hylocereus monacanthus) through regulating HmoCYP76AD1. Int. J. Mol. Sci. 22, 2171. doi: 10.3390/ijms22042171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Jin J., Tang L., Zhao Y., Gu X., Gao G., et al. (2011. b). PlantTFDB 2.0: update and improvement of the comprehensive plant transcription factor database. Nucleic Acids Res. 39, D1114–D1117. doi: 10.1093/nar/gkq1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Liu Y., Cai H., Guo M., Chai M., She Z., et al. (2020). The bZIP transcription factor GmbZIP15 negatively regulates salt-and drought-stress responses in soybean. Int. J. Mol. Sci. 21, 7778. doi: 10.3390/ijms21207778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C.-Q., Xu Y., Lu Y., Yu H.-X., Gu M.-H., Liu Q.-Q. (2011. a). The WRKY transcription factor OsWRKY78 regulates stem elongation and seed development in rice. Planta 234, 541–554. doi: 10.1007/s00425-011-1423-y [DOI] [PubMed] [Google Scholar]
- Zhang Y., Yu H., Yang X., Li Q., Ling J., Wang H., et al. (2016). CsWRKY46, a WRKY transcription factor from cucumber, confers cold resistance in transgenic-plant by regulating a set of cold-stress responsive genes in an ABA-dependent manner. Plant Physiol. Biochem. 108, 478–487. doi: 10.1016/j.plaphy.2016.08.013 [DOI] [PubMed] [Google Scholar]
- Zhang H., Zhang J., Lang Z., Botella J. R., Zhu J.-K. (2017). Genome editing–principles and applications for functional genomics research and crop improvement. Crit. Rev. Plant Sci. 36, 291–309. doi: 10.1080/07352689.2017.1402989 [DOI] [Google Scholar]
- Zhang D., Zhu Z., Gao J., Zhou X., Zhu S., Wang X., et al. (2021. a). The NPR1-WRKY46-WRKY6 signaling cascade mediates probenazole/salicylic acid-elicited leaf senescence in arabidopsis thaliana. J. Integr. Plant Biol. 63, 924–936. doi: 10.1111/jipb.13044 [DOI] [PubMed] [Google Scholar]
- Zhao L., Cai H., Su Z., Wang L., Huang X., Zhang M., et al. (2018. b). KLU suppresses megasporocyte cell fate through SWR1-mediated activation of WRKY28 expression in arabidopsis. Proc. Natl. Acad. Sci. 115, E526–E535. doi: 10.1073/pnas.1716054115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao N., He M., Li L., Cui S., Hou M., Wang L., et al. (2020. b). Identification and expression analysis of WRKY gene family under drought stress in peanut (Arachis hypogaea l.). PLos One 15, e0231396. doi: 10.1371/journal.pone.0231396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Zhang X., Guo R., Wang Y., Guo C., Li Z., et al. (2018. a). Over-expression of a grape WRKY transcription factor gene, VlWRKY48, in arabidopsis thaliana increases disease resistance and drought stress tolerance. Plant Cell Tissue Organ Cult. (PCTOC) 132, 359–370. doi: 10.1007/s11240-017-1335-z [DOI] [Google Scholar]
- Zhao L., Zhang W., Song Q., Xuan Y., Li K., Cheng L., et al. (2020. a). A WRKY transcription factor, TaWRKY40-d, promotes leaf senescence associated with jasmonic acid and abscisic acid pathways in wheat. Plant Biol. 22, 1072–1085. doi: 10.1111/plb.13155 [DOI] [PubMed] [Google Scholar]
- Zhou Q. Y., Tian A. G., Zou H. F., Xie Z. M., Lei G., Huang J., et al. (2008). Soybean WRKY-type transcription factor genes, GmWRKY13, GmWRKY21, and GmWRKY54, confer differential tolerance to abiotic stresses in transgenic arabidopsis plants. Plant Biotechnol. J. 6, 486–503. doi: 10.1111/j.1467-7652.2008.00336.x [DOI] [PubMed] [Google Scholar]
- Zhu D., Hou L., Xiao P., Guo Y., Deyholos M. K., Liu X. (2019). VvWRKY30, a grape WRKY transcription factor, plays a positive regulatory role under salinity stress. Plant Sci. 280, 132–142. doi: 10.1016/j.plantsci.2018.03.018 [DOI] [PubMed] [Google Scholar]
- Zhu H., Jiang Y., Guo Y., Huang J., Zhou M., Tang Y., et al. (2021). A novel salt inducible WRKY transcription factor gene, AhWRKY75, confers salt tolerance in transgenic peanut. Plant Physiol. Biochem. 160, 175–183. doi: 10.1016/j.plaphy.2021.01.014 [DOI] [PubMed] [Google Scholar]
- Zhu L., Li S., Ouyang M., Yang L., Sun S., Wang Y., et al. (2022). Overexpression of watermelon ClWRKY20 in transgenic arabidopsis improves salt and low-temperature tolerance. Scientia Hortic. 295, 110848. doi: 10.1016/j.scienta.2021.110848 [DOI] [Google Scholar]
- Zou C., Jiang W., Yu D. (2010). Male Gametophyte-specific WRKY34 transcription factor mediates cold sensitivity of mature pollen in arabidopsis. J. Exp. Bot. 61, 3901–3914. doi: 10.1093/jxb/erq204 [DOI] [PMC free article] [PubMed] [Google Scholar]