Abstract
Background
Helicobacter pylori (H. pylori) eradication has been reported to cause short-term disruption of gut microbiota. It is acknowledged that probiotics supplementation mitigates side effects induced by H. pylori eradication, yet its role on alleviating dysbiosis of microbiota is obscure.
Objectives
To evaluate the impact of probiotics on gastrointestinal microbiota after eradication therapy.
Methods
This was a multicenter, double-blinded, randomized trial done at seven centers in China. A total of 276 treatment-naïve H. pylori-positive patients were randomly assigned to receive 14-day bismuth-containing quadruple therapy (esomeprazole, bismuth, amoxicillin, furazolidone) combined with probiotics (Bifidobacterium Tetragenous viable Bacteria Tablets) (n=140) or placebo (n=136) for 28 days. Saliva, gastric mucosa and fecal samples were collected before and after therapy for 16S rRNA gene sequencing.
Results
The incidence of gastrointestinal adverse events was lower in probiotics group compared to placebo group (23.6% vs 37.7%, p=0.016), while there was no significant difference in eradication rate. We found dramatic perturbations of gut microbiota immediately following eradication, with the predominance of Proteobacteria in replacement of commensal Firmicutes and Bacteroidetes, and gradually restored after two weeks. The reduction of gut Bacteroidetes caused by eradication drugs was neutralized with probiotics supplementation. The gastric microbiota was completely reconstituted with H. pylori depleted and other taxa flourished. Of note, patients treated with probiotics showed smaller fluctuations of gastric microbiota compared to those with placebo. We also observed changes of saliva microbiota after H. pylori eradication, illustrated by the overgrowth of Neisseria and depletion of Streptococcus. The expansion of some pathogenic genera, including Porphyromonas, Leptotrichia, in the mouth was suppressed by probiotics.
Conclusion
This study not only demonstrated the beneficial effect of probiotics implementation on side events during H. pylori eradication but also provided a comprehensive profile of microbiome alterations along gastrointestinal tract that modulated by probiotics.
Keywords: helicobacter pylori eradication, probiotics, gut microbiota, gastric microbiota, saliva microbiota, 16S rRNA gene sequence
Introduction
Helicobacter pylori (H. pylori), the pathogenic bacterium in the stomach, infects approximately 50% of the population worldwide and causes a wide range of gastric diseases, including chronic gastritis, peptic ulcer and gastric cancer. Thus, it is recommended to screen and eradicate H. pylori to prevent those dieseases (1, 2). With the decreased eradication rate of standard triple regimen nowadays, the first-line regimen for H. pylori treatment is bismuth-containing quadruple therapy, which includes bismuth, proton pump inhibitor, and two antibiotics, in a series of global consensus (1, 3, 4). Accumulating evidence suggest that eradication drugs, especially antibiotics, lead to the disorder of gut microbiota, whose homeostasis play an important role in human health (5). The composition of gut microbiota changed immediately after H. pylori treatment with reduced alpha diversity and increased abundance of Enterobacteriaceae and Enterococcus (6, 7). Although most reported full recovery of bacterial diversity six months after eradication, some studies observed notable changes in abundance at genus level (8). Therefore, it is critical to alleviate the profound impact of drugs involved in H. pylori therapy on gut microbiota.
Probiotics, as live micro-organisms, are commonly used to modulate gut microbiota and ameliorate human diseases like metabolic syndrome, colitis, and antibiotic-associated diarrhea (9). A large body of studies has shown that probiotics supplementation mitigates the side effects during H. pylori eradication, although with controversial effect on improving eradication rate (10, 11). Moreover, several randomized controlled trials have observed that probiotics supplementation help to construct a beneficial profile of gut microbiota after eradication therapy (12). While the alpha diversity was increased with the combination of non-viable Lactobacillus reuteri, no significant changes of diversity was found with Medilac-S supplementation (13). The inconsistency between those studies is probably due to different probiotic strains and load. In addition to gut microbiota, probiotics have been reported to restore the dysbiosis of gastric microbiota induced by H. pylori eradication (14). Thus, it is of interest to explore whether probiotics administration could influence microbiota along gastrointestinal tract in patients receiving eradication therapy.
The live probiotic drug Bifidobacterium Tetragenous viable Bacteria Tablets is comprised of four strains, including Bifidobacterium infantis CGMCC0460.1, Lactobacillus acidophilus CGMCC0460.2, Enterococcus faecalis CGMCC0460.3, and Bacillus cereus CGMCC0460.4. It has been widely applied to modulate intestinal dysbacteriosis for patients with diarrhea, constipation and dyspepsia. The numbers of probiotic bacteria Bacteroides, Faecalibacterium and Akkermansia were significantly enhanced after probiotic treatment, while the richness of pathogenic Streptococcus was lowered (15, 16). Herein, a randomized, double-blinded, placebo-controlled trial was conducted to evaluate the efficacy and safety of probiotics combined with 14-day bismuth-containing quadruple therapy on H. pylori eradication and the changes of gastrointestinal microbiota.
Methods and materials
Study design
This multicenter, randomized, placebo-controlled study was performed at seven tertiary hospitals in China from March 2019 to November 2021. This study was approved by the Ethics Committee of the First Affiliated Hospital of Nanchang University (2019-072) and registered at ClinicalTrials.gov (NCT04034641).
Eligibility criteria
In brief, adult patients (18-65 years) diagnosed with chronic gastritis and tested positive for H. pylori were prospectively recruited. H. pylori infections were examined by the [13C] urea breath test. Patients with any one of the following criteria were excluded from the study: diagnosed with gastric cancer, peptic ulcer diseases, esophagitis, history of gastrectomy, previous eradication therapy for H. pylori, allergy to any of the medications, severe concurrent diseases, pregnant or lactating women, the use of antibiotics or probiotics in the preceding 4 weeks, alcohol abuse or drug addiction, and patients who could not give informed consent.
Interventions
Eligible patients were randomly allocated to probiotics group (Group A) or the placebo group (Group B) in a 1:1 ratio. Randomization was conducted by a computer-generated random number sequence, and the assignments were sealed in opaque envelops. Patients, clinical researchers and statistician were blinded to the randomization and study products. Both groups of patients received 14-day bismuth-containing quadruple therapy (esomeprazole 20mg, amoxicillin 1000mg, furazolidone 100mg, bismuth potassium citrate 220mg, dosed morning and evening), supplemented with probiotics (Bifidobacterium Tetravaccine Tablets, SiLianKang, Hangzhou Grand Biologic Pharmaceutical Inc., Hangzhou, China) or placebo (starch) (nine tablets once a day, dosed noon) for 4 weeks.
Procedures
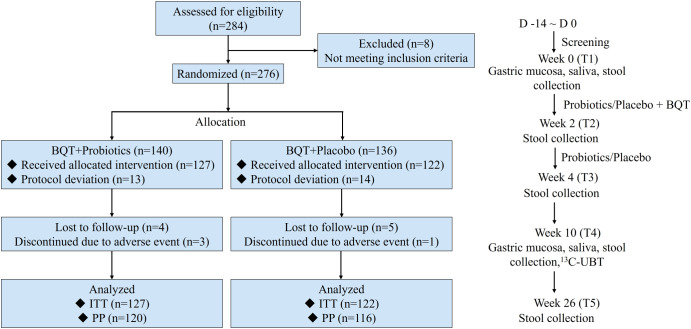
Patients were evaluated at 5 visits: screening at baseline (T1), end of 14-day eradication therapy (T2), end of probiotics/placebo treatment (T3), 8 weeks after eradication (T4), and 24 weeks after eradication (T5). Fecal samples were collected into a DNA stabilizer kit (Fecal DNA Storage Buffer, CoWin Biosciences Co. Ltd, China) from T1 to T5. The liquid DNA stabilization buffers protect the microorganisms from degradation. The stabilized samples were then stored in a refrigerator at -80°C immediately. The gastric antrum mucosa and saliva samples were collected at T1 and T4 and immediately frozen at -80°C. A flow diagram of this trial was shown in Figure 1 . The OMEGA Soil DNA Kit (M5635-02) (Omega Bio-Tek, USA) was used to extract the genomic DNA. High throughput sequencing of 16S rRNA was done by means of the Illumina NovaSeq platform (Shanghai Personal Biotechnology Co., Ltd, China). Detailed methods are shown in the Supplement Materials .
Figure 1.
Flow diagram showing the study design. BQT, bismuth quadruple therapy; ITT, intention-to-treat; PP, per protocol; D, day; 13C-UBT, 13C-Urea Breath Test.
Outcomes
The primary outcome of this study was the incidence of gastrointestinal adverse events, including vomiting, bloating, diarrhea, etc. The second outcomes included eradication rate of H. pylori, compliance with therapy, alterations of gastrointestinal microbiota.
Statistical analyses
Based on a literature review of H. pylori eradication-induced adverse events, we expected a difference between quadruple therapy combined with probiotics and placebo on the incidence of adverse events of 14.4% vs 30.1% (17). The calculation yielded 119 for probiotics group and 119 for placebo group, with a power of 80% and a two-sided significance level of 0.05 with an assumed 10% dropout rate.
Statistical analyses were performed using the software SAS (version 9.4). The data are presented as means ± standard deviations (SDs) or as percentages. The eradication rate was determined by both intention-to-treat (ITT)- and per-protocol (PP)-based analyses. All enrolled patients were included in the ITT analysis, but the PP analysis excluded those discontinued or loss to follow-up. We used the χ2 test or Fisher’s exact test for analysis of categorical data, and Student’s t test or Wilcoxon rank sum test for analysis of continuous data. All P-values were 2-tailed and P < 0.05 was considered as significant.
Microbiome bioinformatics were performed with QIIME2 and R packages (v3.2.0) (18). Alpha diversity of microbiota was determined by calculating indices, including Shannon and Chao1. Significant differences of beta diversity were evaluated by PERMANOVAR and visualized by Principal coordinate analysis on the basis of Bray-Curtis distance. Wilcoxon signed rank-sum test was used to analyze the differential taxa for paired samples. Co-occurrence network analysis was performed by SparCC and visualized using R package. Genera with significantly changed abundance in T4 were combined into Microbial Dysbiosis Index (MDI) for using follow formula: MDI=log(total abundance of genera decreased after H. pylori eradication/total abundance of genera increased after H. pylori eradication) (19). The difference between the MDI of the timepoint of T4 and the baseline T1 were defined as delta-MDI, which was calculated using the difference value of MDI between paired samples. Detailed methods are shown in the Supplement Materials .
Results
Trial profile
A total of 276 patients fulfilling the inclusion criteria were assigned randomly to either Group A (probiotics) or Group B (placebo). Of these 276 patients, 27 patients were excluded as they refused to take study drugs, and the remaining 249 patients were enrolled for intention-to-treat (ITT), with 127 patients in Group A and 122 patients in Group B. During the study, thirteen patients were excluded from the per-protocol (PP) analysis: 7 in Group A (5.5%) and 6 in Group B (4.9%). Three patients discontinued treatment and ten patients lost to follow-up ( Figure 1 ). Demographic and baseline characteristics of the patients in the two groups are shown in Supplement Table 1 .
H. pylori eradication rate and adverse events
As shown in Table 1 , ITT analysis demonstrated that the eradication rates were 86.6% for Group A and 87.7% for Group B (p=0.797). PP analysis indicated that the eradication results were 91.7% for Group A and 92.2% for Group B (p=0.871). Both ITT and PP analyses showed no statistical difference in the eradication rate between Group A and Group B.
Table 1.
Eradication rate of H. pylori in patients treated with probiotics (A) or placebo (B).
| Group | Eradication rate, %(n) (95% confidence interval) ITT analysis | PP analysis |
|---|---|---|
| Group A | 86.6 (11./127) (80.6-92.6) |
91.7 (110/120) (86.6-96.7) |
| Group B | 87.8 (107/122) (81.8-93.6) |
92.2 (107/116) (87.3-97.2) |
| P value | 0.797 | 0.871 |
The frequencies of gastrointestinal adverse events (GAE) were summarized in Table 2 . All of the GAE were graded as mild. The overall incidences of GAEs were significantly reduced in Group A compared to Group B (23.62% vs 37.7%, p=0.016). The nausea rate of Group B was higher than that of Group A (24.59% vs 13.39%, p=0.024). Meanwhile, the combination of probiotics tended to decrease the incidence of vomiting (2.36% vs 7.38%, p=0.065). There was no statistical difference between Group A and Group B in terms of diarrhea, bloating, abdominal pain, belching, dysgeusia. The compliance rate of Group A and Group B was 93.7% and 95.9%, respectively (p=0.435).
Table 2.
Gastrointestinal side effects of patients treated with probiotics (A) or placebo (B) during eradication therapy.
| Group A | Group B | P value | |
|---|---|---|---|
| Total | 30 (23.62%) | 46 (37.70%) | 0.016 |
| Vomitting | 3 (2.36%) | 9 (7.38%) | 0.065 |
| Abdominal pain | 21 (16.54%) | 17 (13.93%) | 0.568 |
| Bloating | 15 (11.81%) | 21 (27.21%) | 0.226 |
| Nausea | 17 (13.39%) | 30 (24.59%) | 0.024 |
| Diarrhea | 1 (0.79%) | 1 (0.82%) | 0.977 |
| Belching | 8 (6.30%) | 12 (9.84%) | 0.305 |
| Dysgeusia | 4 (3.15%) | 4 (3.28%) | 0.954 |
Disturbance and restoration of gut microbiota after H. pylori eradication
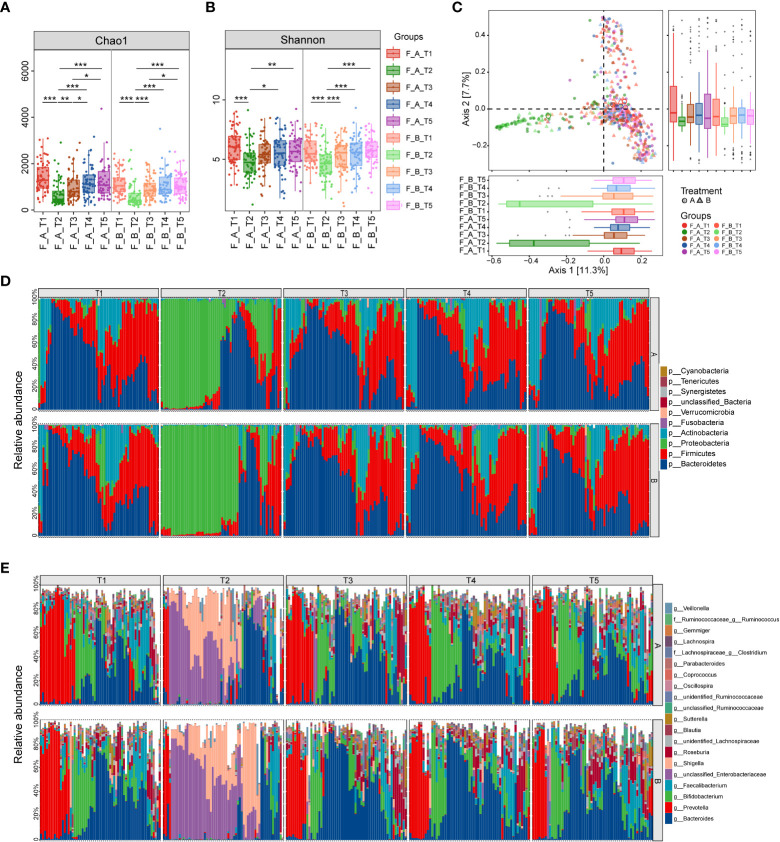
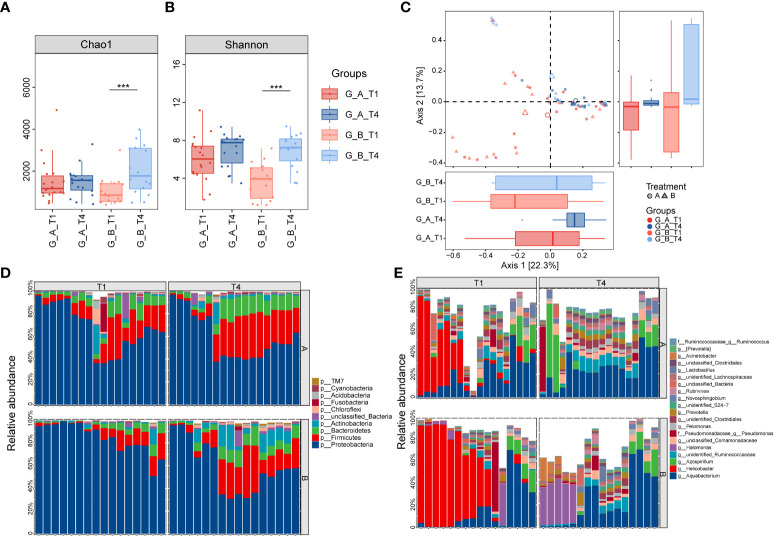
A total of 597 fecal samples were collected from 65 patients in Group A and 61 patients from Group B at different time points ( Figure 1 ). The indices of alpha diversity, including Chao1 and Shannon, were remarkably reduced at week 2 (T2) compared to baseline (T1). Those indexes were quickly increased to the level of baseline at week 4 (T3), and maintained at the steady level until 1 year (T5) ( Figures 2A, B ). However, no significant difference of those indices was observed between Group A and Group B.
Figure 2.
Comparison of diversity and composition of gut microbiota after eradication therapy. (A, B) Alpha diversity, representing the richness of microbiota, was significantly reduced immediately following eradication treatment (T2) and then gradually returned to baseline from T3 to T5. (C) Principal coordinate analysis (PCoA) based on Bray-Curtis distances revealed the overall microbiota composition differed between T2 and other time points. (D) Relative proportions of the top 10 bacterial phyla. (E) Relative proportions of the top 20 bacterial genera. A, probiotics group; B, placebo group; T1, pretreatment; T2, cessation of quadruple therapy; T3, 2 weeks after quadruple therapy; T4, 8 weeks after quadruple therapy; T5, 24 weeks after quadruple therapy; *, p<0.05; **, p<0.01, ***, p<0.001.
Principal coordinated analysis (PCoA) based on Bray-Curtis distance revealed that the overall microbial composition of T2 deviated from other time points ( Figure 2C , PERMANOVA, p=0.001), although there was no statistical difference between Group A and Group B. Pairwise permanova analysis showed the difference of beta diversity between T3 and T1 was greater in Group B compared to Group A (PERMANOVA, A_T3 vs A_T1: p=0.049, B_T3 vs B_T1: p=0.037), and the distinction relative to baseline was disappeared at T4 in both groups.
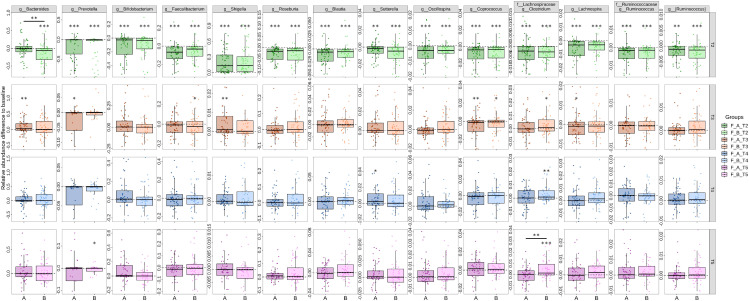
To investigate the specific microbiota that contribute to the fluctuations, we assessed the relative abundance of taxa at different periods. At the phylum level, the gut microbiota is dominated by Bacteroidetes and Firmicutes before treatment, which account for up to 60% in most samples. Of note, the relative abundance of Proteobacteria was dramatically elevated as the dominant phylum following the cessation of eradication drugs, while Bacteroidetes, Firmicutes and Actinobacteria were significantly reduced ( Figure 2D ). The dominance of Proteobacteria was replaced with Bacteroidetes and Firmicutes from T3 to T5, indicating the compositional restoration to baseline. The proportion of patients dominated with Proteobacteria was higher in Group B compared to Group A. The most abundant genera were Bacteroides, Prevotella, Bifidobacterium and Faecalibacterium at baseline, which were substituted by Shigella and an unclassified genus in the family of Enterobacteriaceae at T2 ( Figure 2E ). The percentage of patients with prevailing Shigella and the unclassified genus of Enterobacteriaceae was larger in Group B compared to Group A. The relative abundance of Bacteroides was distinctly decreased in Group B relative to Group A, which indicate that probiotics could partially resist the destruction of eradication drugs on Bacteroides ( Figure 3 ). The compositional disproportionation at T2, including the reduction of Bifidobacterium, Faecalibacterium, Roseburia, Blautia, etc, and over-representation of Shigella was gradually restored from T3 to T5 compared to baseline.
Figure 3.
Dynamic alterations of genus-level gut microbiota after different periods of H. pylori eradication. 14 most abundant genera that differed between T1 and T2 were identified, and their alterations from T2 to T5 were calculated based on their abundance relative to T1. Value that lower than zero represents reduced relative abundance compared to baseline, whereas higher than zero represents increased relative abundance. *, p<0.05; **, p<0.01; ***, p<0.001; star without a line, group compared to T1; star with a line, comparison between (A, B). (A) probiotics group; (B) placebo group; T1, pretreatment; T2, cessation of quadruple therapy; T3, 2 weeks after quadruple therapy; T4, 8 weeks after quadruple therapy; T5, 24 weeks after quadruple therapy.
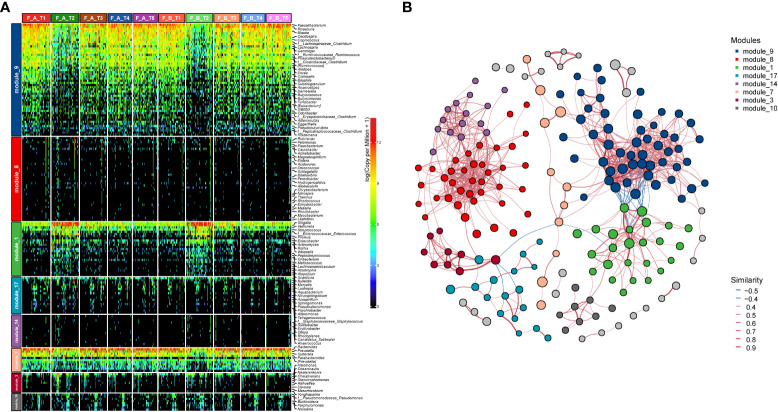
We further performed co-occurrence network analysis to assess the impact of eradication treatment on gut microbiota interactions at different time points ( Figure 4 ). The communities could be classified into eight modules with the prevalence of module_9, module_1 and module_7, while other modules distributed with individual variation. The community of module_9 was transformed to module_1 at T2 compared to T1, and subsequently restored from T3 to T5. The inter-correlations of taxa in module_9 was stronger than those in module_1. We also observed the co-occurring interactions between module_9 and module_7, while both of them, especially module_9, were co-excluded with module_1. Compared to baseline, the alterations of module_1 and module_7 at T2 were more evident in Group B compared to Group A.
Figure 4.
Co-occurrence network reveals the transformation of gut microbial pattern and their interconnection. (A) Heatmap shows the relative abundance of genera in each module. An obvious transformation of module_9 to module_1 was observed at T2, which was reverted from T3 to T5. (B) Significant co-occurrence and co-exclusion relationships among the modules. Each node represents a genus colored by module and its size is proportional to the relative abundance. Edge color indicates the positive (red) and negative (blue) association. The width of each line exhibits the correlation strength among genus.
Probiotics mitigated alterations of gastric microbiota induced by H. pylori eradication
To further explore the changes of gastric microbiota, a total of 68 paired gastric mucosa samples were collected from 34 patients, including 18 in Group A and 16 in Group B, before and 8 weeks after H. pylori treatment. An increased alpha diversity was observed after H. pylori eradication in Group B, measured by Chao1 and Shannon, while no significant change was observed in Group A ( Figures 5A, B ). PCoA based on Bray-Curtis distance revealed that the samples of T4 deviated in global microbiome structure from those of T1 ( Figure 5C ). PERMANOVA analysis demonstrated significant differences in gastric taxonomic composition between Group A and Group B at T4, while there was no obvious difference between these two groups at baseline.
Figure 5.
Longitudinal changes of gastric microbial diversity and composition after H. pylori eradication. (A, B) Box and whisker plots of the alpha diversity indices for richness and diversity of the bacterial communities on the ASV level in patients treated with either probiotics or placebo at T1 and T4, respectively. (C) Beta diversity analysis is represented by principal coordinate analysis based on Bray-Curtis distance. The red samples (T1) are deviated from the blue samples (T4). Relative abundance of the top 10 phyla (D) and the top 20 genera (E) in gastric mucosa. ***, p<0.001. A, probiotics group; B, placebo group; T1, pretreatment; T4, 8 weeks after quadruple therapy; T1, pretreatment; T4, 8 weeks after quadruple therapy.
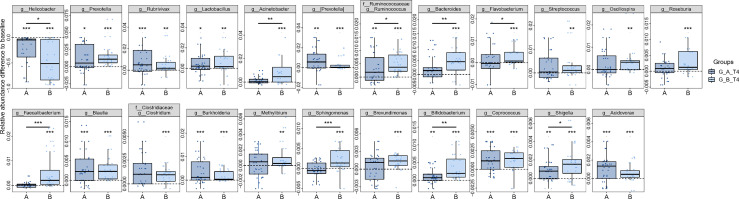
Different from gut microbiota, the gastric microbiota was predominated with Proteobacteria followed by Firmicutes as well as Bacteroidetes in patients with H. pylori infection ( Figure 5D ). With the reduction of Proteobacteria after eradication, the relative abundances of Firmicutes and Bacteroidetes were enhanced. At the genus level, Helicobacter was abundant at baseline albeit with inter-individual variation, and was approximately depleted after eradication, accompanied with the enrichment of Lactobacillus, Prevotella, Bifidobacterium, etc ( Figure 5E ). Notably, some taxa including Faecalibacterium, Bacteroides, Streptococcus, displayed differential abundance at T4 relative to T1 in Group B, while their abundances were not changed in Group A ( Figure 6 ). Additionally, the amplitude of fluctuation was smaller in Group A compared to Group B, such as Shigella, Ruminococcus, Bifidobacterium. In order to investigate the changes of microbial dysbiosis after H. pylori eradication, Microbial Dysbiosis Index (MDI) were calculated using differential genera between T4 and T1. The shift of MDI from T1 to T4 was greater in Group B compared to Group A ( Supplement Figure 1A ). The delta MDI of Group B was farther to zero than Group A, indicating that probiotics play a role in attenuating the turbulence of gastric microbiota after H. pylori eradication ( Supplement Figure 1B ). The co-occurrence network analysis showed that module_2, module_5 and module_3 were the predominant communities in the stomach, albeit with distinct intraindividual variation ( Supplement Figure 2 ). Negative correlation was observed between module_2 and module_5, while module_2 was co-occurred with module_4 and module_3, although relatively weak. The changes of module_2 and module_5 between T4 and T1 were remarkable in Group B compared to Group A.
Figure 6.
Alterations of distinct gastric genera after H. pylori eradication. The most abundant genera that differed between T1 and T4 were identified. The difference was illustrated based on the relative abundance of T4 compared to T1. Values above zero represent that the genera increased after therapy, whereas those below zero represent the decreased genera. *p<0.05; **p<0.01; ***p<0.001; star without a line, comparison between T1 and T4; star with a line, comparison between (A, B); (A) probiotics group; (B) placebo group.
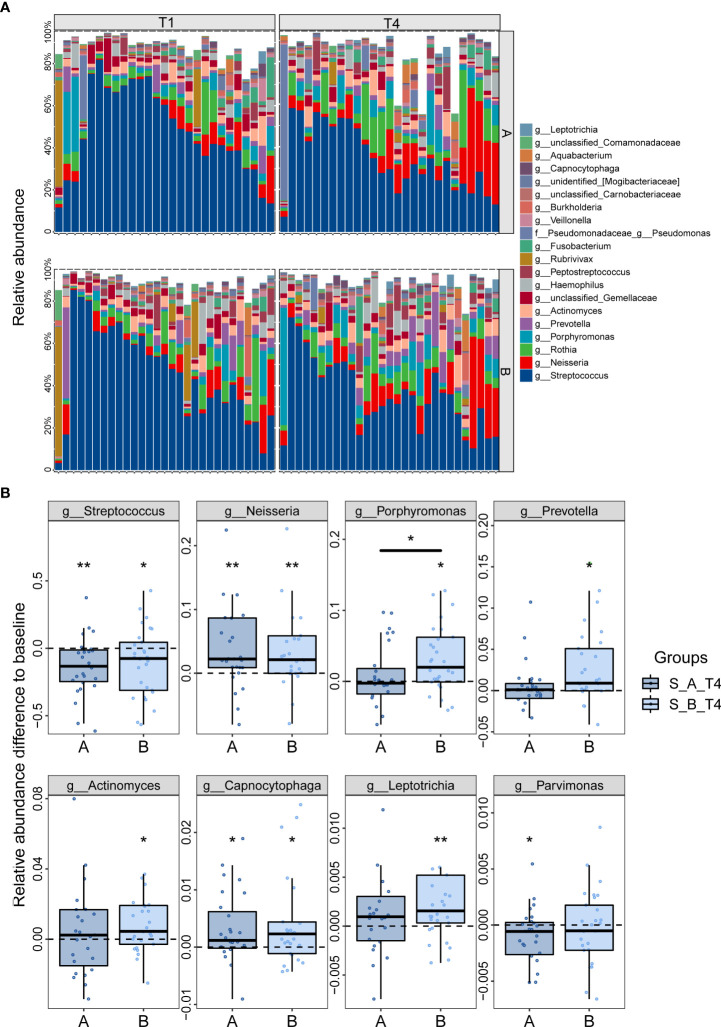
Changes of saliva microbiota after H. pylori eradication
Since the oral cavity is the entry port and the first component of the gastrointestinal system, the colonization of H. pylori in the stomach has been supposed to derive from a potential oral-oral transmission route. Thus, we explored the oral prevalence of H. pylori and the alterations of oral microbiota after eradication. 112 matched-saliva samples were collected from 56 patients (n=27 for Group A, n=29 for Group B) before and after 8 weeks of eradication therapy. The alpha diversity indexes showed no significant difference among the four groups ( Supplement Figure 3A ). Significant shift of the overall microbial structure, illustrated by PCoA analysis, was observed in Group B rather than Group A ( Supplement Figure 3B ). The oral microbiota is mainly composed of Streptococcus, Neisseria, Rothia, Porphyromonas, which account for more than 50% of the taxa detected ( Figure 7A ). The relative abundance of H. pylori in the saliva was extremely low and showed no correlation with the gastric H. pylori ( Supplement Figure 4 ). The eradication of gastric H. pylori did not influence the amount of H. pylori in the saliva. While Streptococcus was significantly decreased at T4 compared to baseline, the amount of Neisseria was enriched ( Figure 7B ). Moreover, some pathogenic taxa including Porphyromonas, Leptotrichia, Actinomyces, Prevotella elevated significantly in Group B after eradication, yet their abundance remained at pretreatment level in Group A.
Figure 7.
Effect of probiotics on saliva microbiota in patients following H. pylori eradication. (A) Compositional changes of the top 20 most abundant saliva genera. (B) Alterations of differential genera after H. pylori eradication. Values above zero represented the increased relative abundance at T4 relative to baseline, while those below zero represented decreased abundance. *p<0.05; **p<0.01; star without a line, comparison between T1 and T4; star with a line, comparison between (A, B); (A) probiotics group; (B) placebo group.
Discussion
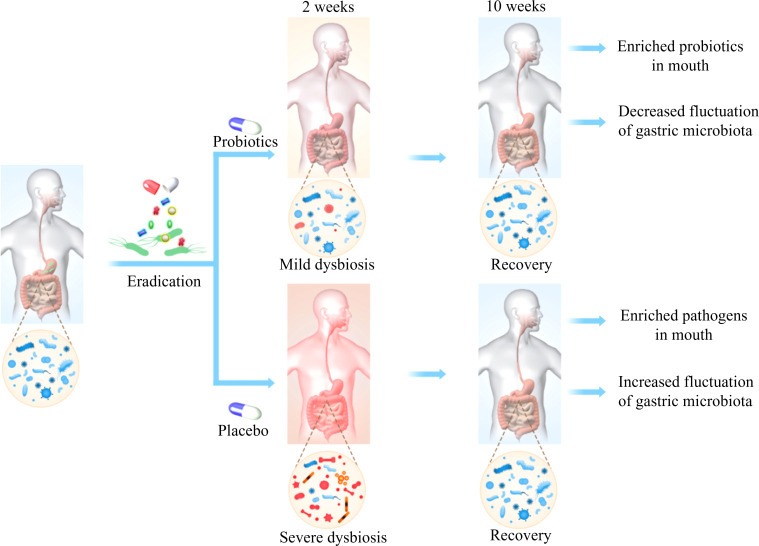
To our knowledge, this is the first multi-center, randomized, double-blinded, placebo-controlled trial to show the effect of probiotics (Bifidobacterium Tetravaccine Tablets) combined with quadruple Bismuth-containing regimen on H. pylori eradication rate, gastrointestinal adverse events (GAE), as well as microbiota along gastrointestinal tract. The probiotics adjuvant therapy reduced the incidence of GAE, while showed no effect on eradication rate. The composition of microbial community across saliva, stomach and gut changed significantly after eradication, with the gut microbiota swiftly restored to baseline as soon as two weeks after cessation of antibiotics. Interestingly, the disturbance of microbiota induced by eradication drugs could be partially alleviated by probiotics treatment ( Figure 8 ).
Figure 8.
There were dramatical changes in gut microbiota immediately after H. pylori eradication, which were swiftly restored after 8 weeks. These perturbations were partially counteracted by probiotics. Meanwhile, alterations of oral and gastric microbiota were also observed. Probiotics mitigated the fluctuations of gastric microbiota as well as promoted the enrichment of beneficial bacteria in the mouth.
The infection of H. pylori is associated with a variety of diseases, including gastritis, peptic ulcers and gastric cancer. Therefore, a subsequent of global consensus has recommended eradication therapy, unless there are competing considerations (1, 20, 21). With the prevalence of antibiotic resistance worldwide, the first-line regimen transformed from triple to quadruple therapy and the duration was prolonged from 7 days to 14 days, causing increased side effects and poor compliance. Recently, a large number of studies explored the efficacy of adjuvant probiotics therapy for patients with H. pylori infection, yet the clinical outcomes were controversial (3). While some studies reported improved eradication rate in patients given probiotics, others showed negative results (10, 22). We observed no difference between probiotics and placebo group in eradication rate, both of which achieved up to 90% as revealed by PP analysis. Additionally, we found that probiotics combination mitigated gastrointestinal adverse events induced by eradication therapy, which is in agreement with other studies (10, 12).
Accumulating evidence suggest that H. pylori therapy can lead to short-term impairment of gut microbiota, including decreased diversity and increased abundance of Proteobacteria (7). In line with previous reports, we observed a significant shift of gut microbiota immediately following treatment, and then began to recover after 2 weeks, indicating the resilience of the gut ecosystem. To further counteract the perturbations of gut microbiota caused by antibiotics involved in the eradication regimen, several studies investigated the effects of probiotics, such as Enterococcus faecium, Bacillus subtilis, Lactobacillus reuteri ( 12, 23). The probiotic compound consumed in this study contains four live strains, which was different from previous studies. Despite the overall structure of gut microbiota was not different between probiotic and placebo group, the genus Bacteroides was more abundant in patients treated with probiotics compared to placebo. As commensals and mutualists, Bacteroides establish stable, long-term associations with human hosts and confer numerous health benefits, including anti-inflammation, metabolic regulation as well as cancer prevention (24). Nevertheless, the expansion of pathogenic Proteobacteria after cessation of antibiotics was not counteracted by probiotics. The co-occurrence network analysis also showed the wane of commensal bacteria and the wax of pathogens such as Shigella after eradication no matter with or without probiotics. We speculate that the activity of probiotics was impaired by antibiotics as the probiotic strains were not detected in fecal samples. Thus, it is necessary to improve the type and dose of probiotic strains in the future in addition to the staggered dosing. The commensal bacteria including Faecalibacterium, Roseburia, Blautia, which were prevalent at baseline, are potential candidates for the development of novel probiotic.
Recent advances in sequencing technology enabled to identify a complex microbiota within the human stomach in addition to H. pylori that may be potentially associated with various disease states (25). Most reports show that H. pylori-infected patients exhibited unbalanced gastric microbiota compared to the negative individuals, which was reverted after eradication therapy (19). Our observation agreed with previous reports that show the depletion of one genus Helicobacter and the over-representation of other taxa such as Streptococcus, Prevotella, Bacteroides, et al. after treatment. Of interest, we found that the fluctuation of gastric microbiome was milder in probiotics group compared to those received placebo, indicating the ability of probiotics on keeping equilibrium of the ecosystem during the exposure of antibiotics. Consistently, several studies have demonstrated the modulation of probiotics on gastric microbiota (14, 26). We speculate that the reduced incidence of upper gastrointestinal side effects such as nausea and vomiting might be associated with the homeostasis of gastric microbiota in patients with probiotics consumption.
A potential oral-oral transmission route of H. pylori raises the question concerning whether it colonizes in the oral cavity. Our study showed very low levels of H. pylori in the saliva from patients with confirmed gastric H. pylori infection, and there was no significant change after eradication therapy. This observation has been supported by a previous study (27). Additionally, we found that H. pylori eradication brought about reduction of Streptococcus together with noteworthy elevation of Neisseria. Furthermore, probiotics influenced the oral microbiota by suppressing the expansion of some taxa, including Porphyromonas, Leptotrichia, and Actinomyces. The nitrate-reducing bacteria Neisseria are consistently found at higher levels in individuals with healthy oral status and linked with better cardiometabolic health (28). On the other hand, Streptococcus, Porphyromonas, Actinomyces were reported to be associated with dental caries and gingivitis (29).
Although our investigations attempt to provide a comprehensive insight into the gastrointestinal microbiome alterations after H. pylori eradication as well as the impact of probiotics on microbial configuration, there are several limitations to be addressed. First, we did not collect samples from H. pylori negative individuals. It is difficult to determine whether the eradication therapy restored the dysbiosis of gastrointestinal microbiota induced by H. pylori infection towards healthy status. Second, we were not able to elucidate the long-term effect of H. pylori eradication on gastric and oral microbiota, as the samples were collected after merely 2 months. Third, the samples collected at different sites were not identical to the number of patients enrolled due to the low compliance.
Conclusion
Our data reported the reconstitution of gastric and oral microbiota after H. pylori depletion, accompanied with transient perturbations of gut microbiota that generally returned to baseline by 2 weeks. The alterations of microbiota were not only observed in the gut but also saliva and stomach after probiotics treatment with different variations. These results highlight a need of developing high efficacy probiotics that target multiple parts of microbiota to improve resilience of the microbial ecosystem during disruptions and to restore its equilibrium afterwards.
Data availability statement
The raw Illumina sequencing data have been deposited in the NCBI Sequence Read Archive (SRA, http://www.ncbi.nlm.nih.gov/sra) under accession number PRJNA839582 for saliva samples, PRJNA801428 for gastric mucosa samples, PRJNA801350 for fecal samples.
Ethics statement
This study was approved by the Ethics Committee of the First Affiliated Hospital of Nanchang University (2019-072) and registered at ClinicalTrials.gov (NCT04034641). The patients/participants provided their written informed consent to participate in this study. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
Author contributions
All authors have read and approved the final manuscript NL, BL, ZZ designed and supervised the project CH performed bioinformatics analysis and drafted the manuscript YX and YZ collected samples, recorded clinical data and analyzed the clinical parameters KZ, LH, YY, QG, XS, ZX performed clinical diagnosis and enrolled eligible patients. All authors contributed to the article and approved the submitted version.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.1033063/full#supplementary-material
References
- 1. Liou JM, Malfertheiner P, Lee YC, Sheu BS, Sugano K, Cheng HC, et al. Screening and eradication of helicobacter pylori for gastric cancer prevention: The Taipei global consensus. Gut (2020) 69(12):2093–112. doi: 10.1136/gutjnl-2020-322368 [DOI] [PubMed] [Google Scholar]
- 2. Du Y, Zhu H, Liu J, Li J, Chang X, Zhou L, et al. Consensus on eradication of helicobacter pylori and prevention and control of gastric cancer in China (2019, shanghai). J Gastroenterol Hepatol (2020) 35(4):624–9. doi: 10.1111/jgh.14947 [DOI] [PubMed] [Google Scholar]
- 3. Malfertheiner P, Megraud F, O'Morain CA, Gisbert JP, Kuipers EJ, Axon AT, et al. Management of helicobacter pylori infection-the maastricht V/Florence consensus report. Gut (2017) 66(1):6–30. doi: 10.1136/gutjnl-2016-312288 [DOI] [PubMed] [Google Scholar]
- 4. Fallone CA, Chiba N, van Zanten SV, Fischbach L, Gisbert JP, Hunt RH, et al. The Toronto consensus for the treatment of helicobacter pylori infection in adults. Gastroenterology (2016) 151(1):51–69.e14. doi: 10.1053/j.gastro.2016.04.006 [DOI] [PubMed] [Google Scholar]
- 5. Ye Q, Shao X, Shen R, Chen D, Shen J. Changes in the human gut microbiota composition caused by helicobacter pylori eradication therapy: A systematic review and meta-analysis. Helicobacter (2020) 25(4):e12713. doi: 10.1111/hel.12713 [DOI] [PubMed] [Google Scholar]
- 6. Zhou Y, Ye Z, Wang Y, Huang Z, Zheng C, Shi J, et al. Long-term changes in the gut microbiota after triple therapy, sequential therapy, bismuth quadruple therapy and concomitant therapy for helicobacter pylori eradication in Chinese children. Helicobacter (2021) 26(4):e12809. doi: 10.1111/hel.12809 [DOI] [PubMed] [Google Scholar]
- 7. Liou JM, Chen CC, Chang CM, Fang YJ, Bair MJ, Chen PY, et al. Long-term changes of gut microbiota, antibiotic resistance, and metabolic parameters after helicobacter pylori eradication: a multicentre, open-label, randomised trial. Lancet Infect Dis (2019) 19(10):1109–20. doi: 10.1016/S1473-3099(19)30272-5 [DOI] [PubMed] [Google Scholar]
- 8. Tao ZH, Han JX, Fang JY. Helicobacter pylori infection and eradication: Exploring their impacts on the gastrointestinal microbiota. Helicobacter (2020) 25(6):e12754. doi: 10.1111/hel.12754 [DOI] [PubMed] [Google Scholar]
- 9. Chang CJ, Lin TL, Tsai YL, Wu TR, Lai WF, Lu CC, et al. Next generation probiotics in disease amelioration. J Food Drug Anal (2019) 27(3):615–22. doi: 10.1016/j.jfda.2018.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhao Y, Yang Y, Aruna, Xiao J, Song J, Huang T, et al. Saccharomyces boulardii combined with quadruple therapy for helicobacter pylori eradication decreased the duration and severity of diarrhea: A multi-center prospective randomized controlled trial. Front Med (Lausanne) (2021) 8:776955. doi: 10.3389/fmed.2021.776955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Plomer M, Iii Perez M, Greifenberg DM. Effect of bacillus clausii capsules in reducing adverse effects associated with helicobacter pylori eradication therapy: A randomized, double-blind, controlled trial. Infect Dis Ther (2020) 9(4):867–78. doi: 10.1007/s40121-020-00333-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yang C, Liang L, Lv P, Liu L, Wang S, Wang Z, et al. Effects of non-viable lactobacillus reuteri combining with 14-day standard triple therapy on helicobacter pylori eradication: A randomized double-blind placebo-controlled trial. Helicobacter (2021) 26(6):e12856. doi: 10.1111/hel.12856 [DOI] [PubMed] [Google Scholar]
- 13. Tang B, Tang L, Huang C, Tian C, Chen L, He Z, et al. The effect of probiotics supplementation on gut microbiota after helicobacter pylori eradication: A multicenter randomized controlled trial. Infect Dis Ther (2021) 10(1):317–33. doi: 10.1007/s40121-020-00372-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yuan Z, Xiao S, Li S, Suo B, Wang Y, Meng L, et al. The impact of helicobacter pylori infection, eradication therapy, and probiotics intervention on gastric microbiota in young adults. Helicobacter (2021) 26(6):e12848. doi: 10.1111/hel.12848 [DOI] [PubMed] [Google Scholar]
- 15. Zheng C, Chen T, Wang Y, Gao Y, Kong Y, Liu Z, et al. A randomised trial of probiotics to reduce severity of physiological and microbial disorders induced by partial gastrectomy for patients with gastric cancer. J Cancer (2019) 10(3):568–76. doi: 10.7150/jca.29072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Deng X, Tian H, Yang R, Han Y, Wei K, Zheng C, et al. Oral probiotics alleviate intestinal dysbacteriosis for people receiving bowel preparation. Front Med (Lausanne) (2020) 7:73. doi: 10.3389/fmed.2020.00073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang F, Feng J, Chen P, Liu X, Ma M, Zhou R, et al. Probiotics in helicobacter pylori eradication therapy: Systematic review and network meta-analysis. Clin Res Hepatol Gastroenterol (2017) 41(4):466–75. doi: 10.1016/j.clinre.2017.04.004 [DOI] [PubMed] [Google Scholar]
- 18. Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol (2019) 37(8):852–7. doi: 10.1038/s41587-019-0209-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Guo Y, Zhang Y, Gerhard M, Gao JJ, Mejias-Luque R, Zhang L, et al. Effect of helicobacter pylori on gastrointestinal microbiota: a population-based study in linqu, a high-risk area of gastric cancer. Gut (2020) 69(9):1598–607. doi: 10.1136/gutjnl-2019-319696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sugano K, Tack J, Kuipers EJ, Graham DY, El-Omar EM, Miura S, et al. Kyoto Global consensus report on helicobacter pylori gastritis. Gut (2015) 64(9):1353–67. doi: 10.1136/gutjnl-2015-309252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ding SZ, Du YQ, Lu H, Wang WH, Cheng H, Chen SY, et al. Chinese Consensus report on family-based helicobacter pylori infection control and management (2021 edition). Gut (2022) 71(2):238–53. doi: 10.1136/gutjnl-2021-325630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Viazis N, Argyriou K, Kotzampassi K, Christodoulou DK, Apostolopoulos P, Georgopoulos SD, et al. A four-probiotics regimen combined with a standard helicobacter pylori-eradication treatment reduces side effects and increases eradication rates. Nutrients (2022) 14(3):632. doi: 10.3390/nu14030632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tang B, Tang L, Huang C, Tian C, Chen L, He Z, et al. The effect of probiotics supplementation on gut microbiota after helicobacter pylori eradication: A multicenter randomized controlled trial. Infect Dis Ther (2020) 10(1):317–33. doi: 10.1007/s40121-020-00372-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tan H, Zhai Q, Chen W. Investigations of bacteroides spp. towards next-generation probiotics. Food Res Int (2019) 116:637–44. doi: 10.1016/j.foodres.2018.08.088 [DOI] [PubMed] [Google Scholar]
- 25. Rajilic-Stojanovic M, Figueiredo C, Smet A, Hansen R, Kupcinskas J, Rokkas T, et al. Systematic review: Gastric microbiota in health and disease. Aliment Pharmacol Ther (2020) 51(6):582–602. doi: 10.1111/apt.15650 [DOI] [PubMed] [Google Scholar]
- 26. Igarashi M, Nakae H, Matsuoka T, Takahashi S, Hisada T, Tomita J, et al. Alteration in the gastric microbiota and its restoration by probiotics in patients with functional dyspepsia. BMJ Open Gastroenterol (2017) 4(1):e000144. doi: 10.1136/bmjgast-2017-000144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ji Y, Liang X, Lu H. Analysis of by high-throughput sequencing: Helicobacter pylori infection and salivary microbiome. BMC Oral Health (2020) 20(1):84. doi: 10.1186/s12903-020-01070-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Morou-Bermudez E, Torres-Colon JE, Bermudez NS, Patel RP, Joshipura KJ. Pathways linking oral bacteria, nitric oxide metabolism, and health. J Dent Res (2022) 101(6):623–31. doi: 10.1177/00220345211064571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang Y, Wang X, Li H, Ni C, Du Z, Yan F. Human oral microbiota and its modulation for oral health. BioMed Pharmacother (2018) 99:883–93. doi: 10.1016/j.biopha.2018.01.146 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw Illumina sequencing data have been deposited in the NCBI Sequence Read Archive (SRA, http://www.ncbi.nlm.nih.gov/sra) under accession number PRJNA839582 for saliva samples, PRJNA801428 for gastric mucosa samples, PRJNA801350 for fecal samples.