Abstract
Congenital diarrheal disorders (CDDs) with genetic etiology are uncommon hereditary intestinal diseases characterized by chronic, life-threatening, intractable watery diarrhea that starts in infancy. CDDs can be mechanistically divided into osmotic and secretory diarrhea. Congenital tufting enteropathy (CTE), also known as intestinal epithelial dysplasia, is a type of secretory CDD. CTE is a rare autosomal recessive enteropathy that presents with intractable neonatal-onset diarrhea, intestinal failure, severe malnutrition, and parenteral nutrition dependence. Villous atrophy of the intestinal epithelium, crypt hyperplasia, and irregularity of surface enterocytes are the specific pathological findings of CTE. The small intestine and occasionally the colonic mucosa include focal epithelial tufts. In 2008, Sivagnanam et al. discovered that mutations in the epithelial cell adhesion molecule (EpCAM, MIM# 185535) were the genetic cause of CTE (MIM# 613217). More than a hundred mutations have been reported to date. Furthermore, mutations in the serine peptidase inhibitor Kunitz type 2 (SPINT2, MIM# 605124) have been linked to syndromic CTE. In this study, we report the case of a 17-month-old male infant with congenital diarrhea. Despite extensive etiological workup, no etiology could be established before admission to our center. The patient died 15 hours after being admitted to our center in a metabolically decompensated state, probably due to a delay in admission and diagnosis. Molecular autopsy with exome sequencing revealed a previously reported homozygous missense variant, c.757G>A, in EpCAM, which was confirmed by histopathological examination.
Keywords: Infantile diarrhea, Epithelial cell adhesion molecule, Whole exome sequencing, Diagnostic molecular pathology
INTRODUCTION
Congenital diarrheal disorders (CDDs) are rare intestinal diseases characterized by chronic, life-threatening, intractable watery diarrhea that starts in infancy, and most cases are caused by genetic defects [1]. CDDs can be mechanistically divided into osmotic and secretory diarrhea. Congenital tufting enteropathy (CTE), also known as intestinal epithelial dysplasia [2], is a secretory CDD. CTE is a rare autosomal recessive enteropathy first defined by Reifen et al. [3] in 1994. The prevalence of CTE is 1 in 50,000-100,000 live births in western Europe. CTE is characterized by intractable watery diarrhea and severe malnutrition. Intractable diarrhea is independent of breast or formula feeding, and most patients with CTE depend on parenteral nutrition (PN) to gain weight [4]. Patients with CTE may present with extraintestinal findings, such as choanal atresia, ophthalmologic findings, and dysmorphic facial features representing syndromic forms of CTE [5,6]. Villous atrophy of the intestinal epithelium, crypt hyperplasia, and irregularity of surface enterocytes are specific pathological findings in CTE [7]. The small intestine and occasionally the colonic mucosa exhibit focal epithelial tufts [8]. Intestinal epithelial cells form tufts by rounding the plasma membrane and forming a teardrop-shaped structure [9]. In 2008, Sivagnanam et al. [10] discovered that mutations in the epithelial cell adhesion molecule (EpCAM, MIM#185535) cause CTE. Furthermore, mutations in the serine peptidase inhibitor Kunitz type 2 (SPINT2, MIM# 605124) have been linked to syndromic CTE [6].
A molecular autopsy involves the analysis of the DNA of deceased patients to determine whether they had a genetic mutation that contributed to their death. Recently, next-generation sequencing or exome sequencing (ES) has been used to identify new mutations that could underlie and perhaps explain the unknown cause in deceased patients [11,12]. In this study, we aimed to report the clinical, histopathological, and molecular features of a patient with CTE in whom molecular autopsy revealed a homozygous missense variant in EpCAM, along with a review of the literature.
Clinical report
A 17-month-old infant was admitted to our hospital with intractable watery diarrhea that began after birth and occurred 7-8 times per day. The patient was born to a G2P2PPex1 mother by cesarean section at 35 weeks of gestation with a birth weight of 2,700 g. The prenatal history was complicated by gestational hypertension. The parents were consanguineous (first cousins) and healthy. The family history was remarkable for a similarly affected sibling who was born at term and died on postnatal day 15 of intractable watery diarrhea. An autopsy was not performed, and no diagnosis was established.
Before his admission to our center, the patient was frequently hospitalized because of diarrhea and recurrent infections with malnutrition, dehydration, and electrolyte imbalance. Despite extensive laboratory workup, no definitive diagnosis could be established. Endoscopy and colonoscopy were performed, and histopathological examination of the duodenal biopsy specimen revealed villous atrophy, crypt hyperplasia, and intraepithelial lymphocyte accumulation. Stool microscopy revealed Giardia lamblia, which required treatment with metronidazole. Blood and stool cultures were negative. Although he gained weight within the first three months of life, his diarrhea worsened after the third month, and he started to lose weight. Despite lactose-free or amino-acid-based formulas, diarrhea continued, and he gained only 300 g within 17 months.
On admission, the patient had fever, tachycardia, and severe malnutrition. His body weight was 3,600 g (−9.7 standart deviation score [SDS]), body length was 61 cm (−7.7 SDS), and occipitofrontal circumference was 40 cm (−5.49 SDS). He looked dehydrated, and the skin turgor decreased. He was hypoactive. Laboratory evaluation revealed metabolic acidosis with a blood pH of 7.19 (normal range, 7.26–7.42) and bicarbonate of 9.9 mmoL/L (normal range, 22.5–26.9 mmoL/L). He had hyponatremia and hyperuricemia with serum sodium and uric acid levels of 127 mEq/L (normal range 136–146 mEq/L) and 8.9 mg/dL (normal range 3.5–7.2 mg/dL), respectively. The C-reactive protein test was negative. The patient received parenteral hydration with bicarbonate, enteral nutrition with lactose-free formula, and antibiotics. However, his clinical condition deteriorated during follow-up, and he died 15 hours after hospitalization. The family provided consent for the autopsy and molecular genetic tests.
MATERIALS AND METHODS
Genomic DNA was extracted using a salting-out procedure from the peripheral blood of the affected individual and unaffected parents after obtaining written informed consent. This study was approved by the Hacettepe University Ethics Committee (GO 15-530/25). To determine genetic etiology, ES was performed on the affected individual. Sanger sequencing was performed to validate this variant and analyze the segregation. We reviewed all published cases, including 22 articles and 116 patients. Articles were included if they reported clinical data of patients with CTE linked to EpCAM mutations. Articles without clinical data, such as reviews, animal models, or functional studies, were not included. A systematic search was conducted from 2008, when EpCAM was identified as a gene for CTE, to 2022 in PubMed using the following terms: “EpCAM tufting enteropathy,” “EpCAM diarrhea,” and “EpCAM exome”. For all patients, data on coding DNA, type of mutation, zygosity, ethnicity, symptoms, consanguinity, treatment, and outcomes were recorded (Table 1).
Table 1. Variants of EpCAM in CTE patients.
| Study | Patient | Year | Coding DNA | Type of mutation | Zygosity | Ethnicity | Sypmtoms | Consanguinity in familiy | Treatment | Outcome |
|---|---|---|---|---|---|---|---|---|---|---|
| Sivagnanam et al. [10] | P1.1, P1.2 (cousins) | 2008 | c.491+1G>A (intron 4) | Noncoding-splicing mutation | Homozygous | Mexican American | Diarrhea | None | PN | Unknown |
| P2 | c.427-1G>A (intron 4) | Noncoding-splicing mutation | Homozygous | Native Canadian | Diarrhea | None | PN | Unknown | ||
| P3 | c.200G>A (exon 3) | Missense mutation | Heterozygous | English/Italian | Diarrhea | Yes | PN | Unknown | ||
| Al-Mayouf et al. [15] | P1 | 2009 | c.498insC (exon 5) | Frameshift mutation | Homozygous | Arabic | Diarrhea, chronic arthritis† | Yes | PN | Unknown |
| Sivagnanam et al. [27] | P1 | 2010 | c.412C>T (exon 3) | Nonsense mutation | Homozygous | Pakistanian | Diarrhea | Yes | PN | Unknown |
| Ko et al. [22] | P1.1, P1.2 (siblings) | 2010 | c.491+1G>A (intron 4)/c.316A>T (exon 3) | Noncoding-splicing mutation/nonsense mutation | Compound heterozygous | Korean | Diarrhea, oligoarticular juvenile rheumatoid arthritis (P1.1)† | None | PN | Alive |
| Salomon et al. [24] | P1 | 2010 | c.492-2A>G (intron 5) | Noncoding-splicing mutation | Homozygous | Arabic | Diarrhea | Yes | PN | Unknown |
| P2.1, P2.2,(siblings) | c.498insC (exon 5) | Frameshift mutation | Homozygous | Arabic | Diarrhea | Yes | PN | Unknown | ||
| P3 | c.498insC (exon 5) | Frameshift mutation | Homozygous | Arabic | Diarrhea | Yes | PN | Unknown | ||
| P4 | c.492-2A>G (intron 5)/c.498insC (exon 5) | Noncoding-splicing mutation/frameshift mutation | Compound heterozygous | Arabic | Diarrhea | Yes | PN | Unknown | ||
| P5.1, P5.2 (siblings) | c.498insC (exon 5) | Frameshift mutation | Homozygous | Arabic | Diarrhea | Yes | PN | Unknown | ||
| P6.1, ,P6.2 (second cousins) | c.498insC (exon 5) (P6.1), c.492-2A>G (intron 5)/c.498insC (exon 5) (P6.2) | Frameshift mutation (P6.1), noncoding-splicing mutation/frameshift mutation (P6.2) | Homozygous (P6.1), compound heterozygous (P6.2) | Arabic | Diarrhea | Yes | PN | Unknown | ||
| P7.1, P7.2 (siblings) | c.498insC (exon 5) | Frameshift mutation | Homozygous | Arabic | Diarrhea | Yes | PN | Unknown | ||
| Thoeni et al. [29] | P1 | 2013 | c.227C>G (exon 3) | Nonsense mutation | Homozygous | East African | Diarrhea | Yes | PN | Alive |
| Pégas et al. [32] | P1 | 2014 | c.556-14A>G (intron 6) | Noncoding-splicing mutation | Homozygous | Brazilian | Diarrhea | Unknown | PN | Alive |
| Salomon et al. [25] (39 patients) | P1 | 2014 | c.139C>T/? (exon 2) | Noncoding-splicing mutation | Heterozygous | Italian | Diarrhea | 45% consanguineous in familiy | PN | Alive |
| P2 | c.227C>G (exon 3) | Nonsense mutation | Homozygous | Turkish | Diarrhea | PN | Alive | |||
| P3 | c.227C>G (exon 3)/c.555+1G>C (intron 5) | Nonsense mutation/noncoding-splicing mutation | Compound heterozygous | Italian | Diarrhea | PN | Alive | |||
| P4 | c.307G>A (exon 3) | Missense mutation | Homozygous | Italian | Diarrhea | PN | Alive | |||
| P5 | c.314T>G (exon 3) | Missense mutation | Homozygous | Sri Lankan | Diarrhea | PN | Alive | |||
| P6 | c.321delC (exon 3) | Noncoding-splicing mutation | Homozygous | Mali | Diarrhea | PN | Deceased | |||
| P7.1, P7.2 | c.352_368del (exon 3) | Frameshift mutation | Homozygous | Morroccan | Diarrhea | ITx (P7.1), PN (P7.2) | Alive | |||
| P8 | c.359A>T (exon3) | Missense mutation | Homozygous | Algerian | Diarrhea | PN | Alive | |||
| P9 | c.380C>T/? (exon3) | Missense mutation/? | Heterozygous | Algerian | Diarrhea | PN | Alive | |||
| P10 | c.394G>T (exon 3) | Nonsense mutation | Homozygous | Egyptian | Diarrhea | ITx (at 5 years) | Alive | |||
| P11 | c.467delC (exon 4)/c.492-1G>A (intron 5) | Frameshift mutation/noncoding-splicing mutation | Compound heterozygous | French | Diarrhea | ITx (at 7 years) | Alive | |||
| P12 | c.492-2A>G (intron 5) | Noncoding-splicing mutation | Homozygous | Kuwaiti | Diarrhea | Unknown | Unknown | |||
| P13 | c.492-2A>G (intron 5)/c.498insC (exon 5) | Noncoding-splicing mutation/frameshift mutation | Compound heterozygous | Kuwaiti | Diarrhea | PN | Alive | |||
| P14.1, P14.2 | c.492-2A>G (intron 5)/c.498insC (exon 5) | Noncoding-splicing mutation/frameshift mutation | Compound heterozygous | Kuwaiti | Diarrhea | 45% consanguineous in familiy | PN | Deceased (P14.1), alive (P14.2) | ||
| P15 | c.492_555del (exon 5) | Deletion | Homozygous | French | Diarrhea | ITx (at 4 years) | Alive | |||
| P16 | c.498insC (exon 5) | Frameshift mutation | Homozygous | Kuwaiti | Diarrhea | PN | Alive | |||
| P17.1, P17.2 | c.498insC (exon 5) | Frameshift mutation | Homozygous | Kuwaiti | Diarrhea | Unknown | Unknown | |||
| P18 | c.498insC (exon 5) | Frameshift mutation | Homozygous | Kuwaiti | Diarrhea | PN | Deceased | |||
| P19.1, P19.2 | c.498insC (exon 5) | Frameshift mutation | Homozygous | Kuwaiti | Diarrhea | PN | Deceased | |||
| P20.1, P20.2 | c.498insC (exon 5) | Frameshift mutation | Homozygous | Qatari | Diarrhea | Unknown | Unknown | |||
| P21 | c.498insC (exon 5) | Frameshift mutation | Homozygous | Kuwaiti | Diarrhea | Unknown | Unknown | |||
| P22 | c.555+1G>C (intron 5) | Noncoding-splicing mutation | Homozygous | Turkish | Diarrhea, keratodermy† | ITx (at 5 years) | Alive | |||
| P23 | c.555+1G>C (intron 5) | Noncoding-splicing mutation | Homozygous | Turkish | Diarrhea | ITx (at 11 years) | Alive | |||
| P24 | c.556-14A>G (intron 6) | Noncoding-splicing mutation | Homozygous | Morroccan | Diarhhea, arthritis† | PN | Alive | |||
| P25 | c.556-14A>G (intron 6) | Noncoding-splicing mutation | Homozygous | Morroccan | Diarhhea | PN | Alive | |||
| P26 | c.556-14A>G (intron 6) | Noncoding-splicing mutation | Homozygous | Algerian | Diarhhea | PN | Alive | |||
| P27 | c.556-14A>G (intron 6) | Noncoding-splicing mutation | Homozygous | Algerian | Diarhhea | PN | Alive | |||
| P28 | c.556-14A>G (intron 6) | Noncoding-splicing mutation | Homozygous | Portugal | Diarhhea | PN | Deceased | |||
| P29 | c.556-14A>G/? (intron 6) | Noncoding-splicing mutation | Heterozygous | Morroccan | Diarhhea, arthritis, oesophageal atresia† | PN | Alive | |||
| P30.1, P30.2 | c.556-14A>G (intron 6)/c.654delA (exon 6) | Noncoding-splicing mutation/frameshift mutation | Compound heterozygous | Italian | Diarhhea | ITx (at 9 years) | Alive | |||
| P31 | Del Ex 1–4 | Deletion | Homozygous | Turkish | Diarrhea | PN | Alive | |||
| P32 | Del Ex 1–7 | Deletion | Homozygous | Turkish | Diarrhea | PN | Alive | |||
| P33 | Del EpCAM | Deletion | Homozygous | Algerian | Diarrhea | ITx (at 12 years) | Alive | |||
| Haas et al. [20] | P1.1, P1.2 (siblings) | 2016 | NA | NA | Homozygous | Unknown | Diarrhea | Unknown | PN | Alive |
| d’Apolito et al. [18] | P1.1, P1.2 (siblings) | 2016 | c.654delA (exon 6)/c.556-14A>G (intron 6) (P1.1, P1.2) | Frameshift mutation/noncoding-splicing mutation (P1.1, P1.2) | Compound heterozygous (P1.1, P1.2) | Italian | Diarrhea | None | ITx (P1.1 at 10 years, P1.2 at 11 years) | Deceased (P1.1, P1.2) |
| P2 | c.437T>A (exon 4) | Missense mutation | Heterozygous | Italian | Diarrhea | None | PN | Alive | ||
| Tang et al. [7] | P1 | 2016 | c.307G>A (exon 3)/deletion (exon 2–5) | Missense mutation | Compound heterozygous | Chinese | Diarrhea | None | PN | Alive |
| AlMahamed and Hammo et al. [14] | P1 | 2017 | c.498insC (exon 5) | Frameshift mutation | Homozygous | Arabic | Diarrhea | Yes | PN, ITx at 4 years | Alive |
| P2 | c.498insC (exon 5) | Frameshift mutation | Homozygous | Arabic | Diarrhea | Yes | PN | Deceased | ||
| P3 | Homozygous deletion of whole gene | Deletion | Homozygous | Arabic | Diarrhea | None | PN | Deceased | ||
| P4 | c.412C>T (exon 3) | Nonsense mutation | Homozygous | Arabic | Diarrhea | Yes | PN, ITx at 2.5 years | Alive | ||
| Bodian et al. [17] | P1.1, P1.2 (second cousins) | 2017 | c.556-14A>G (intron 6) | Noncoding-splicing mutation | Homozygous | Hispanic | Diarrhea, dilate cardiomyopathy (unexplained) (P1)† | None (?) | PN | Deceased (P1.1), alive (P1.2) |
| Tan et al. [28] | P1 | 2017 | c.38_62dup25 (exon 1)+c.1A>C (exon1) | Frameshift mutation+ Missense mutation | Homozygous (for two different EpCAM variants) | Unknown | Diarrhea | Yes | PN | Alive |
| Shakhnovich et al. [26] | P1 | 2018 | c.426-1G>A (intron 4)/c.265C>T (exon 3) | Noncoding-splicing mutation/nonsense mutation | Compound heterozygous | Caucasian | Diarrhea | None | PN | Alive |
| Chuanjie et al. [33] | P1 | 2018 | c.412C>T (exon 3) | Missense mutation | Homozygous | Chinese | Diarrhea | Unknown | PN | Unknown |
| Pathak et al. [23] (17 patients; many of whom were from families without consanguinity) | P1 | 2019 | c.113G>A (exon 2)/c.48_68del21 (exon 1) | Missense mutation/inframe deletion | Compound heterozygous | Caucasian | Diarrhea | Unknown | PN | Alive |
| P2 | c.1A>C (exon 1)/c.38_62dup25 (exon 1) | Missense mutation/frameshift mutation | Compound heterozygous | Palestinian | Diarrhea | Unknown | PN | Alive | ||
| P3 | c.267G>C (exon 3)/c.118T>C (exon 2) | Missense mutation/unknown | Compound heterozygous | English | Diarrhea | Unknown | PN | Alive | ||
| P4 | c.307G>A (exon 3)/c.492-5T>C (intron 5) | Missense mutation/noncoding-splicing mutation | Compound heterozygous | English/Italian | Diarrhea | Unknown | PN | Alive | ||
| P5.1, P5.2 | c.491+1G>A (intron 4)/c.556-14A>G (intron 6) | Noncoding-splicing mutation | Compound heterozygous | Bangladeshi | Diarrhea | Unknown | PN | Alive | ||
| P6 | c.492-1G>A (intron 5)/c.491+1G>A (intron 4) | Noncoding-splicing mutation | Compound heterozygous | Hispanic | Diarrhea | Unknown | PN | Alive | ||
| P7 | c.509_511delTCA (exon 5) | In-frame deletion | Homozygous | Pakistani | Diarrhea | Unknown | PN | Alive | ||
| P8 | c.555+1G>C (intron 5) | Noncoding-splicing mutation | Homozygous | Turkish | Diarrhea | Unknown | PN | Alive | ||
| P9.1, P9.2 | c.579delT (exon 6) | Frameshift mutation | Homozygous | Bedouin | Diarrhea | Unknown | PN | Alive | ||
| P10 | c.589C>T (exon 6) | Nonsense mutation | Homozygous | Iraqi | Diarrhea | Unknown | PN | Alive | ||
| P11.1, P11.2 | c.540delT (exon 5)/c.491+1G>A (intron 4) | Frameshift mutation/noncoding-splicing mutation | Compound heterozygous | Unknown | Diarrhea | Unknown | PN | Alive | ||
| P12 | c.757G>A (exon 7) | Missense mutation | Homozygous | Turkish | Diarrhea | Unknown | PN | Alive | ||
| P13 | Deletion (end of exon 1 and intron 3–4) | Deletion | Homozygous | Hispanic | Diarrhea | Unknown | PN | Alive | ||
| P14 | c.227C>G (exon 3) | Nonsense mutation | Homozygous | Israeli | Diarrhea | Unknown | PN | Alive | ||
| Hassan et al. [21] (18 patients) | P1–P18* | 2019 | c.498insC (exon 5) | Frameshift mutation | Homozygous | Qataris | Diarrhea | Yes (86%) | PN, ITx (17%) | Deceased (50%), alive (50%) |
| Fang et al. [19] | P1 | 2019 | c.491+1G>A (intron 4) | Noncoding-splicing mutation | Homozygous | Chinese | Diarrhea | None | PN | Deceased |
| Zhou et al. [31] | P1 | 2020 | c.657+1G>A (intron 6) | Noncoding-splicing mutation | Homozygous | Chinese | Diarrhea | None | PN | Alive |
| Yan et al. [30] | P1 | 2021 | c.184+6T>G (intron 2)/large deletion (exon 1–9) | Noncoding-splicing mutation/large deletion | Compound heterozygous | Chinese | Diarrhea | None | PN | Deceased |
| P2 | c.96C>A (exon 2)/c.823delG (exon 7) | Nonsense mutation/frameshift mutation | Compound heterozygous | Chinese | Diarrhea | None | PN | Alive | ||
| Ayyıldız Cİvan et al. [16] | P1 | 2021 | c.1_46del (exon 1) | Frameshift mutation | Homozygous | Unknown | Diarrhea | Yes | PN | Alive |
| P2.1, P2.2 (siblings) | c.326A>G (exon 3) (P2.1, P2.2.) | Missense mutation (P2.1, P2.2) | Homozygous | Unknown | Diarrhea | Yes | PN | Deceased (P2.1), alive (P2.2) | ||
| P3 | c.429G>A (exon 4)/c.556-14A>G (intron 6) | Nonsense mutation/noncoding-splicing mutation | Compound heterozygous | Unknown | Diarrhea | None | PN | Alive |
EpCAM: epithelial cell adhesion molecule, CTE: congenital tufting enteropathy, NA: not available, PN: parenteral nutrition, ITx: intestinal transplantation.
*Two siblings as part of a previously published study (Salomon et al. [24]).
†Extraintestinal symptoms.
Exome sequencing
The Ion AmpliSeq Exome RDY Kit (Thermo Fisher Scientific, Waltham, MA, USA) was used for exome library preparation. Emulsion polymerase chain reaction was performed on an Ion OneTouch 2 instrument using the Ion PI Hi-Q OT2 200 Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. The enrichment of the Ion Sphere particles (ISPs) was performed using the OneTouch ES module. ISPs were loaded on Ion PI chips and sequenced with the Ion Proton platform (Thermo Fisher Scientific).
Bioinformatic analysis
ES was performed for the index case. The variant filtering steps were performed as previously described [13]. All variants that passed the quality score filters were annotated using Ion Reporter 5.10 software. Exonic and non-synonymous rare variants (minimum allele frequency<0.01) were selected for in-house filtering. Subsequently, alterations in the boundaries of runs of homozygosity (ROH) regions ≥1 Mb within all chromosomes were extracted. We used HomSI, a software that maps disease loci by single nucleotide polymorphism genotyping using a variant call format file obtained from the ES data, to identify the ROH in index cases. Finally, variants that were observed in the ExAC Browser (in the homozygous state) were ruled out.
Sanger sequencing
The EpCAM variant identified by ES was verified by DNA sequencing. For conventional sequencing, the BigDye Terminator v.3.1 Cycle Sequencing Kit and sequencing products were applied to an ABI 3500 genetic analyzer (Thermo Fisher Scientific). Sanger sequencing was also performed to evaluate cosegregation among family members.
RESULTS
The clinical findings of the present patient are shown in Table 1, along with the findings of patients reported to date in the literature. The initial histopathological findings of the patient’s autopsy were villous atrophy and crypt hyperplasia without inflammation in the duodenum and destruction of the intestinal surface epithelium. Electron microscopy revealed villous atrophy in the duodenum, jejunum, ileum, and colon and decreased size and number of microvilli. The epithelium was regular and did not contain microvillus inclusions. These findings were not compatible with CTE, microvillus inclusion disease, and immune dysregulation polyendocrinopathy enteropathy X-linked. Villous atrophy was remarkable in celiac disease, but an increase in the number of intraepithelial lymphocytes was absent.
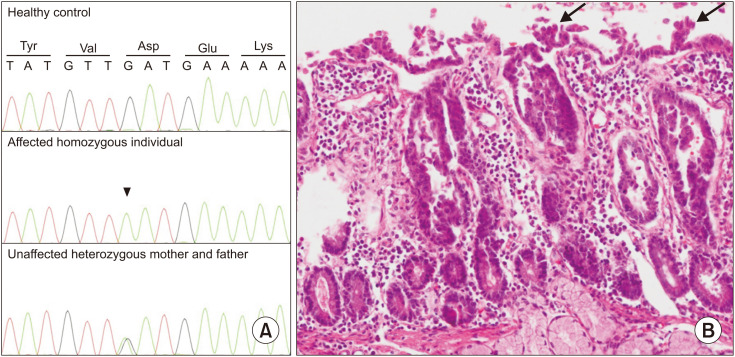
ES was performed for the index cases. After selection and filtering, we identified 35 candidate variants in 17 homozygous regions. Among these, we identified a homozygous c.757G>A missense variant in EpCAM (RefSeq Number: NM_002354.3; p.Asp253Asn) compatible with the disease phenotype in the affected individual (Fig. 1A). Genetic counseling for autosomal recessive disorders was provided to the family. The family was informed about the carrier testing of at-risk relatives, prenatal testing, and preimplantation genetic testing for future pregnancies.
Fig. 1. (A) Sanger Sequencing of healthy control, affected individual (homozygous c.757G>A variant), and unaffected mother and father (heterozygous c.757G>A variant). (B) Staining with hematoxylin and eosin revealed villus abnormalities, including epithelial crowding, disorganization, and focal tufting (arrows) with no epithelial lymphocytosis shown at 400× magnification.
After the detection of the EpCAM variant, the histopathological findings of the patient were reviewed. A thorough histological examination of the slides and deeper sections revealed villus abnormalities, including epithelial crowding, disorganization, and focal tufting with no epithelial lymphocytosis (Fig. 1B).
EpCAM variants causing the CTE
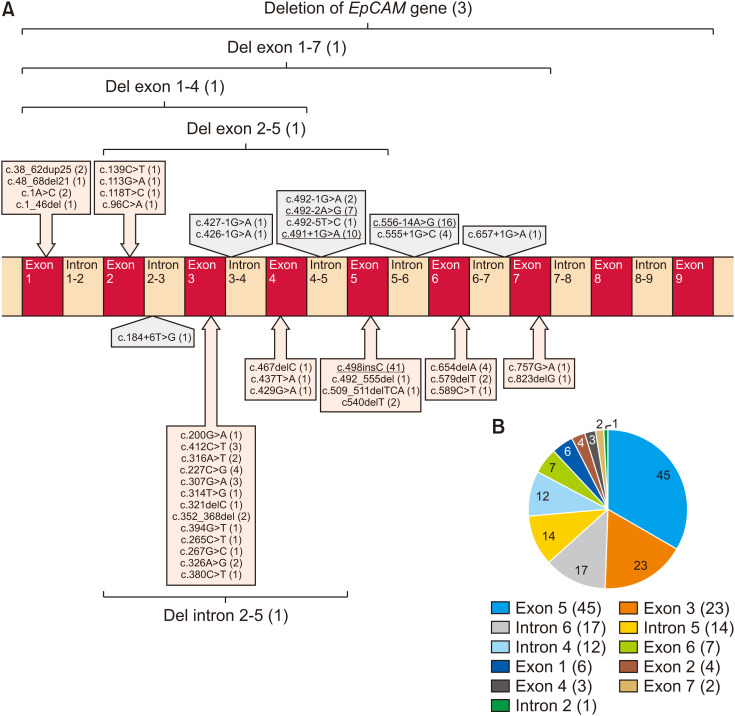
After a comprehensive analysis of previous reports on CTE and EpCAM, we found 140 mutations (48 different EpCAM mutations in 116 patients) in EpCAM from 22 studies, including missense, nonsense, frameshift, noncoding/splicing mutations, and chromosomal deletions (Table 1, Fig. 2) [7,10,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33]. Among them, pathogenic variants c.491+1G>A (intron 4), c.556-14A>G (intron 6), c.492-2A>G (intron 5), and c.498insC (exon 5) were reported in five or more patients (Fig. 2B). The frameshift mutation (c.498insC) was the most common mutation in all patients with CTE. Many patients (84 of 116, 72.4%) were homozygous for EpCAM mutations, and most of the remaining patients were compound heterozygotes (27 of 116 patients, 23.2%) (Table 1). The disease is more common in consanguineous families, such as those seen in Middle Eastern populations.
Fig. 2. (A) Pathogenic EpCAM variants identified in patients with CTE. Schematic diagram of the EpCAM gene illustrating 48 variants to date. The number of patients carrying variants in parentheses (the most common variants were reported in five or more patients are underlined). (B) The number of patients carrying mutations in different exons/introns (the number of patients carrying variants in parentheses).
EpCAM: epithelial cell adhesion molecule, CTE: congenital tufting enteropathy.
Clinically, patients with EpCAM variants mainly present with isolated congenital diarrhea. Only a few patients had extraintestinal symptoms, such as arthritis (2 patients) [15,25], oligoarticular juvenile rheumatoid arthritis (1 patient) [22], unexplained dilated cardiomyopathy (1 patient) [17], keratoderma (1 patient) [25], and esophageal atresia (1 patient) [25]. CTE is a life-threatening disease characterized by persistent, severe diarrhea. Of the 90 patients for whom data were available, 23 (25.5%) died. In most patients, CTE causes irreversible intestinal failure; therefore, intestinal transplantation (ITx) is required in severe cases. Of the 110 patients for whom data were available, 13 (11.8%) required ITx (Table 1).
DISCUSSION
In this study, we identified a homozygous missense variant of EpCAM in a patient presenting with congenital diarrhea. The initial histopathological examination during the patient’s autopsy was inconclusive. However, molecular autopsy with the aid of ES revealed a homozygous missense variant in EpCAM, which further contributed to the detection of tufts in the histopathological specimen after the reevaluation of the autopsy material.
CTE is a rare autosomal recessive enteropathy that presents with intractable neonatal-onset diarrhea, intestinal failure, severe malnutrition, and PN dependence [3]. Most patients with CTE have significant intestinal malabsorption and diarrhea, requiring long-term full PN; however, positive outcomes with definitive weaning from PN have also been reported [34]. In general, laboratory workup and imaging methods do not contribute to the diagnosis of CTE in most patients [2]. Patients suspected of having CTE usually undergo endoscopy and colonoscopy, during which mucosal biopsies are performed in multiple regions. Histological abnormalities in the intestines of CTE patients include total or partial villous atrophy, crypt hyperplasia without inflammation, and basement membrane abnormalities. Focal epithelial tufts were observed from the duodenum to the large intestine. In most patients, typical abnormalities are localized primarily to the surface epithelium, forming focal epithelial “tufts” that consist of tightly packed enterocytes with rounding of the plasma membrane, shaping a tear-like structure by the cells [3,8]. Sometimes, these characteristic tufts could be absent in the biopsy of patients with early CTE with typical clinical symptoms, as was the case in the present patient. Despite extensive laboratory workup, imaging methods, and histopathologic findings, no specific diagnosis could be established. However, molecular autopsy with the aid of ES identified a homozygous c.757G>A missense variant in EpCAM.
The EpCAM gene maps to chromosome 2p21 and encodes a 40 kDa transmembrane glycoprotein consisting of an extracellular domain (encoded by exon 1–6), a single transmembrane domain (encoded by exon 7), and an intracellular domain (encoded by exon 8–9). In addition, the thyroglobulin homology domain, N-domain, and C-domain are three compactly folded extracellular domains. EpCAM is involved in cell signaling, proliferation, differentiation, and organ morphology development and maintenance [35]. EpCAM (MIM #185535) variants were identified in the genetic etiology of CTE by Sivagnanam et al. [10] in 2008. Since then, more than 100 EpCAM variants have been identified (Table 1, Fig. 2) [36]. The frameshift pathogenic variant (c.498insC) in exon 5 is the most common form in all patients with CTE [15]. Although the c.498insC homozygous pathogenic variant in exon 5 is common in the Middle East, exon 3 pathogenic variants are more common in East Asia [7,24]. The exon regions in which the most frequent pathogenic variants are defined vary across different populations.
We identified a homozygous c.757G>A missense variant in exon 7 of our patient. The variant was classified as “uncertain significance” according to ACMG 2015 guidelines and predicted to be “disease causing” by MutationTaster. The combined annotation-dependent depletion score was 28.2 (deleterious). EpCAM missense mutations encoding single amino acid substitutions primarily affected the extracellular thyroglobulin homology domain and internal portions of the extracellular C-terminal domain of EpCAM, consistent with the hypothesis that defects in cell-cell interactions are causal in CTE. The missense variant c.757G>A, p.(Asp253Asn) affects residues at one of the two contact regions between the thyroglobulin homology domain and the C-terminal domain [23]. Although this variant has not been previously submitted to ClinVar, a Turkish patient with the same variant presenting with neonatal-onset intractable diarrhea who is still alive with full PN support has previously been reported in the literature [23].
Pathak et al. [23] hypothesized that some differential clinical outcomes, including nutritional data/PN status, need for intestinal transplantation, and mortality, could be used as markers of disease severity. The patients were classified into genotypic groups in which EpCAM alleles were mapped to categories (frameshift mutations, nonsense mutations, missense mutations, and splicing defects) to investigate these data for evidence of genotype-phenotype correlation. Patients with frameshift mutations were more likely to require total PN and to have died, suggesting that frameshift mutations are consistent with more severe disease. In contrast, splice site mutations are more common in patients who require PN only partially. Neither nonsense nor missense mutations were significantly associated with any clinical outcome [23]. Our findings verified that frameshift mutations were more common in patients who died (13 of 23 patients). In contrast, noncoding/splicing mutations (four patients), mixed mutations (four patients), missense mutations (one patient), and deletions (one patient) were less common in deceased patients (Table 1). These data suggest that frameshift mutations correlate with more aggressive treatment and poorer outcomes. Despite having a missense mutation, our patient died during infancy, probably due to a delay in the admission and diagnosis. Despite progress in nutritional management, the patient died of complications from severe malnutrition. Therefore, patients with early-onset severe diarrhea should be referred to a specialized center to avoid malnutrition and plan the diagnostic workup. Although there is evidence that frameshift mutations are associated with a more severe phenotype in general, establishing specific genotype/phenotype correlations in CTE is complicated by several factors, including the rarity of CTE, presence of compound heterozygotes, and lack of standard quantitative assessments for disease severity. The development of quantitative CTE severity assessments is required to investigate genotype-phenotype correlations and to guide patient treatment and prognosis.
CONCLUSION
Since the genetic etiology of CTE was discovered, there has been considerable progress in molecular diagnosis, particularly with the wide use of ES in clinical practice over the past ten years. Molecular diagnosis with ES provides early diagnosis and treatment and prevents invasive procedures in the diagnostic approach for these patients.
ACKNOWLEDGEMENTS
The authors thank the patient’s parents for their participation in the study. This study was supported by Hacettepe University Scientific Research Unit under the project entitled Hacettepe Exome Project (Grant ID: TAY 2015-7335).
Footnotes
Conflict of Interest: The authors have no financial conflicts of interest.
References
- 1.Terrin G, Tomaiuolo R, Passariello A, Elce A, Amato F, Di Costanzo M, et al. Congenital diarrheal disorders: an updated diagnostic approach. Int J Mol Sci. 2012;13:4168–4185. doi: 10.3390/ijms13044168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sherman PM, Mitchell DJ, Cutz E. Neonatal enteropathies: defining the causes of protracted diarrhea of infancy. J Pediatr Gastroenterol Nutr. 2004;38:16–26. doi: 10.1097/00005176-200401000-00007. [DOI] [PubMed] [Google Scholar]
- 3.Reifen RM, Cutz E, Griffiths AM, Ngan BY, Sherman PM. Tufting enteropathy: a newly recognized clinicopathological entity associated with refractory diarrhea in infants. J Pediatr Gastroenterol Nutr. 1994;18:379–385. [PubMed] [Google Scholar]
- 4.Gambarara M, Diamanti A, Ferretti F, Papadatou B, Knafelz D, Pietrobattista A, et al. Intractable diarrhea of infancy with congenital intestinal mucosa abnormalities: outcome of four cases. Transplant Proc. 2003;35:3052–3053. doi: 10.1016/j.transproceed.2003.10.053. [DOI] [PubMed] [Google Scholar]
- 5.Roche O, Putterman M, Salomon J, Lacaille F, Brousse N, Goulet O, et al. Superficial punctate keratitis and conjunctival erosions associated with congenital tufting enteropathy. Am J Ophthalmol. 2010;150:116–21.e1. doi: 10.1016/j.ajo.2010.01.034. [DOI] [PubMed] [Google Scholar]
- 6.Heinz-Erian P, Müller T, Krabichler B, Schranz M, Becker C, Rüschendorf F, et al. Mutations in SPINT2 cause a syndromic form of congenital sodium diarrhea. Am J Hum Genet. 2009;84:188–196. doi: 10.1016/j.ajhg.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tang W, Huang T, Xu Z, Huang Y. Novel mutations in EPCAM cause congenital tufting enteropathy. J Clin Gastroenterol. 2018;52:e1–e6. doi: 10.1097/MCG.0000000000000739. [DOI] [PubMed] [Google Scholar]
- 8.Goulet O, Kedinger M, Brousse N, Cuenod B, Colomb V, Patey N, et al. Intractable diarrhea of infancy with epithelial and basement membrane abnormalities. J Pediatr. 1995;127:212–219. doi: 10.1016/s0022-3476(95)70297-0. [DOI] [PubMed] [Google Scholar]
- 9.Goulet O, Salomon J, Ruemmele F, de Serres NP, Brousse N. Intestinal epithelial dysplasia (tufting enteropathy) Orphanet J Rare Dis. 2007;2:20. doi: 10.1186/1750-1172-2-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sivagnanam M, Mueller JL, Lee H, Chen Z, Nelson SF, Turner D, et al. Identification of EpCAM as the gene for congenital tufting enteropathy. Gastroenterology. 2008;135:429–437. doi: 10.1053/j.gastro.2008.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Opdal SH, Rognum TO. The sudden infant death syndrome gene: does it exist? Pediatrics. 2004;114:e506–e512. doi: 10.1542/peds.2004-0683. [DOI] [PubMed] [Google Scholar]
- 12.Heathfield LJ, Martin LJ, Ramesar R. A systematic review of molecular autopsy studies in sudden infant death cases. J Pediatr Genet. 2018;7:143–149. doi: 10.1055/s-0038-1668079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Utine GE, Taşkıran EZ, Koşukcu C, Karaosmanoğlu B, Güleray N, Doğan ÖA, et al. HERC1 mutations in idiopathic intellectual disability. Eur J Med Genet. 2017;60:279–283. doi: 10.1016/j.ejmg.2017.03.007. [DOI] [PubMed] [Google Scholar]
- 14.AlMahamed S, Hammo A. New mutations of EpCAM gene for tufting enteropathy in Saudi Arabia. Saudi J Gastroenterol. 2017;23:123–126. doi: 10.4103/1319-3767.203359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Al-Mayouf SM, Alswaied N, Alkuraya FS, Almehaidib A, Faqih M. Tufting enteropathy and chronic arthritis: a newly recognized association with a novel EpCAM gene mutation. J Pediatr Gastroenterol Nutr. 2009;49:642–644. doi: 10.1097/MPG.0b013e3181acaeae. [DOI] [PubMed] [Google Scholar]
- 16.Ayyıldız Civan H, Leitner C, Östreicher I, Schneider AM, Cremer M, Mayr JA, et al. Three novel EPCAM variants causing tufting enteropathy in three families. Children (Basel) 2021;8:503. doi: 10.3390/children8060503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bodian DL, Vilboux T, Hourigan SK, Jenevein CL, Mani H, Kent KC, et al. Genomic analysis of an infant with intractable diarrhea and dilated cardiomyopathy. Cold Spring Harb Mol Case Stud. 2017;3:a002055. doi: 10.1101/mcs.a002055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.d’Apolito M, Pisanelli D, Faletra F, Giardino I, Gigante M, Pettoello-Mantovani M, et al. Genetic analysis of Italian patients with congenital tufting enteropathy. World J Pediatr. 2016;12:219–224. doi: 10.1007/s12519-015-0070-y. [DOI] [PubMed] [Google Scholar]
- 19.Fang Y, Luo Y, Yu J, Chen J. A case of severe malnutrition infant with neonatal onset intractable diarrhea. BMC Pediatr. 2020;20:133. doi: 10.1186/s12887-020-1999-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haas K, Martin B, Martín M, Kerner J. Intractable diarrhea in two brothers: late diagnosis of tufting enteropathy in adolescence. Dig Dis Sci. 2016;61:381–383. doi: 10.1007/s10620-015-3766-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hassan K, Sher G, Hamid E, Hazima KA, Abdelrahman H, Al Mudahka F, et al. Outcome associated with EPCAM founder mutation c.499dup in Qatar. Eur J Med Genet. 2020;63:104023. doi: 10.1016/j.ejmg.2020.104023. [DOI] [PubMed] [Google Scholar]
- 22.Ko JS, Seo JK, Shim JO, Hwang SH, Park HS, Kang GH. Tufting enteropathy with EpCAM mutations in two siblings. Gut Liver. 2010;4:407–410. doi: 10.5009/gnl.2010.4.3.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pathak SJ, Mueller JL, Okamoto K, Das B, Hertecant J, Greenhalgh L, et al. EPCAM mutation update: variants associated with congenital tufting enteropathy and Lynch syndrome. Hum Mutat. 2019;40:142–161. doi: 10.1002/humu.23688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salomon J, Espinosa-Parrilla Y, Goulet O, Al-Qabandi W, Guigue P, Canioni D, et al. A founder effect at the EPCAM locus in Congenital Tufting Enteropathy in the Arabic Gulf. Eur J Med Genet. 2011;54:319–322. doi: 10.1016/j.ejmg.2011.01.009. [DOI] [PubMed] [Google Scholar]
- 25.Salomon J, Goulet O, Canioni D, Brousse N, Lemale J, Tounian P, et al. Genetic characterization of congenital tufting enteropathy: epcam associated phenotype and involvement of SPINT2 in the syndromic form. Hum Genet. 2014;133:299–310. doi: 10.1007/s00439-013-1380-6. [DOI] [PubMed] [Google Scholar]
- 26.Shakhnovich V, Dinwiddie D, Hildreth A, Attard T, Kingsmore S. A novel compound-heterozygous epithelial cell adhesion molecule mutation in tufting enteropathy. J Pediatr Gastroenterol Nutr. 2017;64:e14–e16. doi: 10.1097/MPG.0000000000000629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sivagnanam M, Janecke AR, Müller T, Heinz-Erian P, Taylor S, Bird LM. Case of syndromic tufting enteropathy harbors SPINT2 mutation seen in congenital sodium diarrhea. Clin Dysmorphol. 2010;19:48. doi: 10.1097/MCD.0b013e328331de38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tan QK, Cardona DM, Rehder CW, McDonald MT. Identification of EPCAM mutation: clinical use of microarray. Clin Case Rep. 2017;5:980–985. doi: 10.1002/ccr3.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thoeni C, Amir A, Guo C, Zhang S, Avitzur Y, Heng YM, et al. A novel nonsense mutation in the EpCAM gene in a patient with congenital tufting enteropathy. J Pediatr Gastroenterol Nutr. 2014;58:18–21. doi: 10.1097/MPG.0000000000000106. [DOI] [PubMed] [Google Scholar]
- 30.Yan W, Xiao Y, Zhang Y, Tao Y, Cao Y, Liu K, et al. Monogenic mutations in four cases of neonatal-onset watery diarrhea and a mutation review in East Asia. Orphanet J Rare Dis. 2021;16:383. doi: 10.1186/s13023-021-01995-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou YQ, Wu GS, Kong YM, Zhang XY, Wang CL. New mutation in EPCAM for congenital tufting enteropathy: a case report. World J Clin Cases. 2020;8:4975–4980. doi: 10.12998/wjcc.v8.i20.4975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pêgas KL, Cambruzzi E, Ferrelli RS, da Silva CS, Guedes RR, Adami M, et al. Tufting enteropathy with EpCAM mutation: case report. J Bras Patol Med Lab. 2014;50:234–237. [Google Scholar]
- 33.Yuan C, Wu J. Congenital tufting enteropathy caused by mutation of EPCAM gene: a case report and review of literature; Paper presented at: 57th Annual ESPE; 2018 Sep 27-29; Athens, Greece. [Google Scholar]
- 34.Lemale J, Coulomb A, Dubern B, Boudjemaa S, Viola S, Josset P, et al. Intractable diarrhea with tufting enteropathy: a favorable outcome is possible. J Pediatr Gastroenterol Nutr. 2011;52:734–739. doi: 10.1097/MPG.0b013e31820731db. [DOI] [PubMed] [Google Scholar]
- 35.Schnell U, Cirulli V, Giepmans BN. EpCAM: structure and function in health and disease. Biochim Biophys Acta. 2013;1828:1989–2001. doi: 10.1016/j.bbamem.2013.04.018. [DOI] [PubMed] [Google Scholar]
- 36.Cai C, Chen Y, Chen X, Ji F. Tufting enteropathy: a review of clinical and histological presentation, etiology, management, and outcome. Gastroenterol Res Pract. 2020;2020:5608069. doi: 10.1155/2020/5608069. [DOI] [PMC free article] [PubMed] [Google Scholar]